Abstract
Palaeobiogeographic reconstructions are underpinned by phylogenies, divergence times and ancestral area reconstructions, which together yield ancestral area chronograms that provide a basis for proposing and testing hypotheses of dispersal and vicariance. Methods for area coding include multi-state coding with a single character, binary coding with multiple characters and string coding. Ancestral reconstruction methods are divided into parsimony versus Bayesian/likelihood approaches. We compared nine methods for reconstructing ancestral areas for placental mammals. Ambiguous reconstructions were a problem for all methods. Important differences resulted from coding areas based on the geographical ranges of extant species versus the geographical provenance of the oldest fossil for each lineage. Africa and South America were reconstructed as the ancestral areas for Afrotheria and Xenarthra, respectively. Most methods reconstructed Eurasia as the ancestral area for Boreoeutheria, Euarchontoglires and Laurasiatheria. The coincidence of molecular dates for the separation of Afrotheria and Xenarthra at approximately 100 Ma with the plate tectonic sundering of Africa and South America hints at the importance of vicariance in the early history of Placentalia. Dispersal has also been important including the origins of Madagascar's endemic mammal fauna. Further studies will benefit from increased taxon sampling and the application of new ancestral area reconstruction methods.
Keywords: ancestral areas, dispersal, historical biogeography, Mammalia, vicariance
1. Introduction
Class Mammalia is impressive for its taxonomic, ecological and morphological diversity [1]. A fundamental goal of mammalian palaeobiogeography is to reconstruct the underlying history of vicariant and dispersal events that have shaped this diversity. Here, we highlight the importance of phylogeny reconstruction, ancestral area reconstruction and molecular dating for producing ancestral area chronograms. We compare different approaches for reconstructing ancestral areas, and illustrate similarities and differences between these approaches using a dataset for placental mammals. We conclude with a review of selected topics in placental mammal palaeogeography that illustrates how phylogenies, ancestral area reconstructions, molecular dates and palaeographic histories have reshaped our views on mammalian historical biogeography. Finally, we identify important areas for future inquiry.
2. Phylogeny reconstruction
Phylogeny reconstruction begins with character data. Large molecular datasets have yielded robust phylogenies for many groups, thereby reducing the number of phylogenetic hypotheses that must be considered when formulating ancestral area chronograms. The inclusion of morphological data from fossils allows for taxonomically richer phylogenies, while also providing key data points that bear on the geographical provenance of a taxonomic group. In the words of Simpson [2], fossils ‘are the historical documents of animal distribution’. Fossils are also more difficult to place with confidence in a phylogenetic framework owing to missing (molecular) data, and the inability of current methods to separate homology and homoplasy with some morphological datasets [3,4].
Maximum parsimony (MP) and maximum likelihood (ML) yield a single best tree(s), whereas Bayesian methods yield a sampling of trees from posterior probability space. ML and Bayesian methods have the advantage of incorporating models of sequence evolution and yield trees with branch lengths. Some ancestral area reconstruction methods, such as those implemented in SIMMAP [5], can take advantage of trees with branch lengths, as well as multiple trees from posterior probability space.
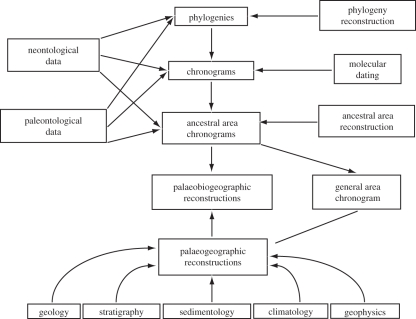
Phylogeny reconstruction is usually the first step in constructing an ancestral area chronogram, followed by the estimation of divergence times at each of the nodes. However, BEAST [6] allows for simultaneous estimation of branching relationships and divergence times. After reconstructing a phylogeny, molecular dating analyses and ancestral area reconstructions can be performed in parallel or in series, and then integrated to yield an ancestral area chronogram (figure 1).
Figure 1.
A flowchart of the approach used for incorporating different types of data, in conjunction with methods in phylogeny reconstruction, molecular dating and ancestral area reconstruction, for inferring ancestral area chronograms and palaeobiogeographic history.
3. Molecular dating analyses
Molecular clocks were introduced by Zuckerkandl & Pauling [7], but have fallen out of favour owing to the prevalence of lineage-specific rate variation. The emergence of relaxed molecular clock methods has promoted a resurgence of studies that have examined both interordinal and intraordinal divergence times in Mammalia [8–23]. Relaxed clock methods include penalized likelihood approaches [24,25], and Bayesian Markov chain Monte Carlo methods such as multidivtime [26], BEAST [6] and mcmctree [27]. It is useful to compare both the results of different programs and the results of the same program under different model and parameter settings [28–30].
An important difference between BEAST and multidivtime is that BEAST allows rates to vary randomly over lineages in a phylogeny, whereas multidivtime assumes autocorrelated rates. In simulation studies, BEAST performed poorly when rates were autocorrelated, whereas multidivtime performed poorly when there was uncorrelated rate variation [28]. Given these results, Battistuzzi et al. [28] recommended composite 95% credibility intervals.
Relaxed clock methods allow for multiple calibrations, including minimum and maximum constraints on individual nodes. Multidivtime only allows for ‘hard’ constraints, whereas BEAST and mcmctree provide other options including ‘soft’ constraints that permit specification of a given percentage (e.g. 95%) of the normal distribution between the minimum and maximum, with half of the remainder (e.g. 2.5%) allocated to each tail.
4. Ancestral area reconstruction
Methods for reconstructing historical biogeography include dispersalism, phylogenetic biogeography, panbiogeography, parsimony analysis of endemicity and cladistic biogeography [31]. Early reconstructions of mammalian historical biogeography were based on dispersalism and land bridges [2,32], and pre-date the general acceptance of plate tectonic theory. Subsequently, cladistic biogeography emphasized vicariance as the most important factor in diversification by discovering dichotomous area relationships (area cladograms) from taxon cladograms. In response to this paradigm, which paid little regard to dispersal and extinction, Ronquist [33] proposed dispersal–vicariance analysis (DIVA) for reconstructing patterns of historical biogeography [34]. DIVA infers ancestral areas by minimizing the number of dispersal and extinction events. Recent methods that build on Ronquist's work include Bayes-DIVA [35] and dispersal–extinction cladogenesis (DEC) [36,37].
A fundamental issue in ancestral area reconstruction is area coding. Areas are usually coded to include the entire geographical range of each species. Other options include coding the entire area of the monophyletic clade that is represented by the species, or the geographical area of the oldest fossil belonging to each lineage. An additional topic worthy of investigation is the problem of coding geographical areas for taxa from the geological past versus the present given that areas, as well as their boundaries and physical relationships to each other, can fluctuate over time. Parametric methods such as DEC, which allow for changing dispersal probabilities over time, provide a mechanism to accommodate the impact of continental fragmentation and suturing on historical biogeography.
Three general approaches are available for coding areas (box 1), whether for living species, fossils or larger monophyletic groups. The first method is single character, multi-state coding with non-overlapping character states. The second method is binary character coding with multiple characters, where each character represents the presence or absence of a taxon in a single area. In contrast to the first method, this approach allows ancestral nodes to encompass more than one area. Ancestral area reconstructions are simply the sum of the individual area reconstructions. A disadvantage of this approach is that ancestral areas may receive ‘no-state’ assignments, which imply empty ancestral areas. No-state assignments are an artefact of the character independence assumption [38]. Finally, string character coding [37] allows individual character states to include one or more geographical areas. Specifically, the geographical range of a species is coded as a string denoting its presence/absence in a set of individual areas. Ree & Smith's [37] string character coding is equivalent to Maddison & Maddison's [39] polymorphism coding.
Box 1. Methods for coding areas and analysing area-coded data matrices.
I. Area coding
1. single multi-state character coding. Individual character states are non-overlapping and correspond to a single area
disadvantages: ranges are limited to a single area (character state) unless they are coded as polymorphic
2. binary character coding with multiple characters. Each binary character corresponds to the presence/absence of a taxon in a single area
advantages: allows for the occupation of multiple areas
disadvantages: ancestral areas may receive no state assignments
3. string character coding (=polymorphism coding)
advantages: individual character states may include one or more areas
disadvantages: the number of character states becomes intractable when there are too many individual areas
II. Ancestral area reconstruction
1. monomorphic ancestral area reconstruction methods. These methods are used in conjunction with area data that have been coded as a single multi-state character
a. Fitch parsimony (e.g. MacClade)
b. stochastic mapping (e.g. SIMMAP)
advantages: stochastic mapping allows for branch lengths and multiple trees
disadvantages: methods in this category implicitly assume that different character states (areas) are homologous to each other, and attempt to find a single ancestral area (character state) at each node
2. polymorphic ancestral area reconstruction methods. These methods allow for ancestral areas that encompass more than one area, and employ either binary character data for multiple characters or string character data
a. Fitch parsimony (e.g. MacClade) with multiple, binary characters
b. stochastic mapping (e.g. SIMMAP) with multiple, binary characters
c. dispersal–vicariance (DIVA)
d. Bayes-DIVA
e. dispersal–extinction cladogenesis (DEC)
f. minimum area change (MAC) parsimony
advantages: all methods in this category allow for reconstructions that include multiple areas per node. Stochastic mapping and DEC incorporate branch lengths; stochastic mapping and Bayes-DIVA allow for multiple trees
disadvantages: methods that employ multiple binary characters can result in empty ancestral area reconstructions. Fitch parsimony, MAC parsimony and DIVA ignore branch length information. DIVA, Bayes-DIVA and DEC are biased towards ancestral reconstructions that include numerous individual areas
Ancestral reconstruction methods can be divided into parsimony versus Bayesian/likelihood approaches [40]. Only the latter takes advantage of branch lengths. Another useful distinction is between methods that reconstruct ancestral nodes as monomorphic character states versus those that allow for range expansion and contraction.
MP and ML methods employ discrete-state transition models, and reconstruct ancestral nodes as monomorphic. Monomorphic methods for character state reconstruction assume that different character states are homologous to each other, as is the case for characters that pass Patterson's [41] conjunction test, which states that two structures that are found in the same organism cannot be homologous. However, this test is nonsensical when applied to geographical areas because the presence of a species in one area does not rule out its presence in another area.
Other ancestral range reconstruction methods have the advantage of allowing for polymorphic ancestral states and thereby accommodating range expansion and contraction (box 1) [40]. Polymorphic reconstructions can be achieved using (i) monomorphic methods with multiple, binary characters, each of which codes for the presence/absence of a taxon in one area and (ii) polymorphic methods that allow ancestral nodes to include one or more areas.
Fitch parsimony and stochastic mapping can be used to reconstruct ancestral nodes for multiple binary characters, and then summed over all character reconstructions to obtain the complete set of areas for each ancestral node. One difficulty is ancestral nodes with no-state assignments. In these instances, multiple interpretations are possible, including vicariance of an ancestral area that was not included in the original analysis. If there is geological evidence for formerly contiguous areas, this information may be incorporated into ancillary characters to assist ancestral area reconstructions.
In contrast to methods that were co-opted from phylogenetics, DIVA [33] and DEC [36,37,42] were developed explicitly for historical biogeographic reconstruction. DIVA assigns no cost to widespread ancestral areas that are subdivided by vicariance, but assigns a cost to dispersal and local extinction events. DIVA ignores branch lengths. DEC uses a continuous time model for geographical range evolution, and employs string character coding to accommodate polymorphic areas. DEC permits range expansion through dispersal events and range contraction through local extinction events. DEC also allows areas of implausible distribution to be excluded, such as those that are geographically discontinuous [43]. DIVA and DEC are prone to reconstructing ancestral areas that include too many individual areas, especially towards the root of the tree. However, both programmes have options for limiting the number of ancestral areas.
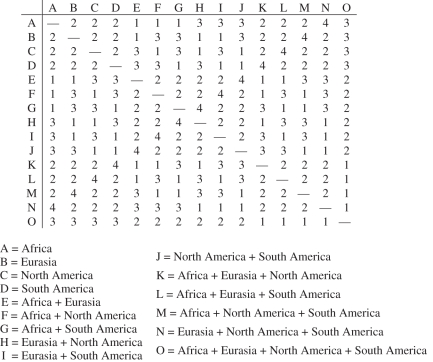
An additional approach that we introduce is minimum area change (MAC) parsimony, which uses polymorphic character coding [39] and Sankoff optimization, and can be implemented with Mesquite [44]. MAC parsimony requires a step matrix (figure 2). In contrast to DIVA, MAC parsimony assigns equal cost to all gains and losses of an area, whether through dispersal, local extinction or vicariance. An advantage of this approach is that it should be less prone than DIVA to reconstructing ancestral areas that are too broad relative to terminal taxa.
Figure 2.
Example of a step matrix for minimum area change (MAC) parsimony. MAC parsimony assigns equal cost to all gains and losses of an area. For example, a change in area from A (Africa) to G (Africa + South America) requires one step (gain South America), whereas a change from A to H (Eurasia + North America) requires three steps (Africa loss, Eurasia gain, North America gain). The step matrix is fully symmetrical.
Another recent approach that builds on earlier cladistic biogeography methods is phylogenetic analysis of comparing trees (PACT) [45–47]. Unlike earlier cladistic biogeography methods, PACT explicitly incorporates molecular dates into general area cladograms.
5. Ancestral area chronograms and palaeogeography
Ancestral area chronograms are similar to ancestral area cladograms, but additionally incorporate temporal information into their framework. Alternate approaches for reconstructing phylogeny, estimating divergence times and reconstructing ancestral areas may yield different ancestral area chronograms, each of which may be interpreted in the context of geology-based palaeogeographical hypotheses (figure 1). Ancestral area chronograms, in conjunction with geology-based palaeogeographical reconstructions, provide a framework for proposing, testing and refining palaeobiogeographic hypotheses. Ancestral area chronograms, when interpreted in the context of palaeogeographical hypothesis, yield insights into dispersal, vicariance and area extinctions, all of which are incorporated into palaeobiogeographic hypotheses (figure 1).
Ancestral area chronograms are taxon-specific, but ancestral area chronograms for multiple taxa that co-occur in the same region can yield general area chronograms. General area chronograms are similar to general area cladograms, but include temporal information that is absent from general area cladograms. The fundamental idea behind cladistic biogeography is that broad patterns, which are revealed through general area cladograms, demand comprehensive causal explanations. However, general area cladograms ignore temporal information and may result from pseudo-congruence when taxonomic groups with the same area relationships have different divergence times, and presumably different underlying causes [48]. Temporal information is critical for discriminating between groups that diversified during the same time period, and therefore may have experienced the same causal events, and groups that diversified during different time periods and require different causal explanations [48].
Just as there may be multiple ancestral area chronograms for a taxonomic group, there may also be multiple palaeogeographical hypotheses regarding the history of connections of formerly connected landmasses. For example, the ‘pan-Gondwanan’ and ‘Africa-first’ hypotheses represent alternate scenarios for the breakup of Gondwana [49]. Both hypotheses agree that the initial rift was between the African component of West Gondwana (Africa, South America) and the Indo-Madagascar component of East Gondwana, although connections between Africa and Indo-Madagascar were maintained via South America–Antarctica. Subsequent to this initial rift, the pan-Gondwanan hypothesis [50] postulates that three vicariant separations, South America from Africa, South America from Antarctica and Antarctica from Indo-Madagascar, all occurred during a narrow time window (100–90 Ma). The Africa-first hypothesis, in turn, suggests that Africa was the first Gondwanan continent to become completely separated from other Gondwanan landmasses when it separated from South America by approximately 100 Ma. Indo-Madagascar separated from Antarctica–Australia at approximately 130–110 Ma, but maintained subaerial connections with Antarctica via the Kerguelen Plateau, and possibly the Gunnerus Ridge to the west, well into the Late Cretaceous (approx. 80 Ma). The final separation was between the Antarctica Peninsula and the tip of South America in the Eocene.
Krause et al. [49] compared Cretaceous vertebrate faunas from different Gondwanan landmasses, and concluded that palaeontological data are most compatible with a modified version of the Africa-first hypothesis. Krause et al.'s [49] work also illustrates how biogeographic hypotheses based on fossils can be compared with geology-based palaeogeographical hypotheses in an arena that allows for reciprocal illumination. Thus, ancestral and general area chronograms provide a framework for evaluating competing geology-based palaeogeographical reconstructions just as geology-based palaeogeographical reconstructions provide a framework for evaluating alternate ancestral area chronograms (figure 1). Krause et al. [49] noted that there is no a priori reason to assume that geological data trump palaeontological data, or vice versa, insofar as each type of data can be used to reveal large-scale biogeographic patterns.
6. Placental phylogeny and a comparison of different ancestral area reconstruction methods
Most placental orders have first fossil occurrences and probable origins in Laurasia, but there are also orders with Gondwanan origins based on first fossil occurrences in South America (Xenarthra) or Africa (most afrotherian orders). Traditional morphological phylogenies [51,52] have suggested close relationships between Laurasian and Gondwanan orders, e.g. Edentata (Xenarthra (Gondwanan) + Pholidota (Laurasian)). By contrast, molecular phylogenies have recovered three superordinal groups, Afrotheria, Laurasiatheria and Euarchontoglires [3,53–63], that were not recovered on morphological trees. These three groups, plus Xenarthra, comprise the four major clades of placental mammals. There is also robust molecular support for Boreoeutheria (Euarchontoglires + Laurasiatheria) [60–62,64]. This overhaul of placental phylogeny, in conjunction with the results of molecular dating analyses, laid the foundation for new biogeographic hypotheses. We discuss these in §7, after first comparing the results of different ancestral area reconstruction methods in the remainder of this section.
Ancestral area chronograms were reconstructed for 43 fully terrestrial placental taxa from Springer et al. [3]. Chiropterans and fully aquatic forms were excluded because of their different modes of dispersal (i.e. flight, swimming), and also because most fully aquatic taxa inhabit areas (i.e. oceans) that are not contained in the four-area scheme used in our analyses (see below). Ancestral area chronograms were reconstructed using a ML phylogram obtained with RAxML [65], molecular divergence dates estimated with BEAST [6] and ancestral areas reconstructed with a variety of methods.
Four areas (Africa, Eurasia, North America and South America) were recognized, and two methods were used to code areas for terminal taxa. First, areas were coded based on the geographical ranges of extant species. Second, areas were coded based on the geographical provenance of the oldest fossil for each lineage. The step matrix that was used in MAC parsimony analysis is shown in figure 2. Given that the number of character states that are chosen for geographical range subdivision is arbitrary, it may be instructive to compare the results of analyses with coarser (e.g. Gondwana versus Laurasia) and finer (e.g. Europe and Asia instead of Eurasia) scales for area coding, although the analyses reported here are confined to the four areas listed above.
We reconstructed ancestral areas using nine methods: (i) MAC parsimony, (ii) Fitch parsimony with multiple binary characters (FP-MBC), (iii) Fitch parsimony with a single multi-state character (FP-SMC), (iv) DIVA with no constraints on the maximum number of areas per node, (v) DIVA with a maximum of two areas per node (DIVA-2), (vi) DEC with no constraints on the maximum number of areas per node, (vii) DEC with a maximum of two areas per node (DEC-2), (viii) stochastic mapping with multiple binary characters (SM-MBC), and (ix) stochastic mapping with a single multi-state character (SM-SMC). Ancestral area chronograms (MAC parsimony) based on the geographical ranges of extant species and fossil lineages are shown in figures 3 and 4, respectively. Tables 3 and 4 summarize the results of analyses with all nine methods.
Table 1.
Fossil constraints. Minimum ages are based on the age of the oldest unequivocal fossils belonging to the clade. Maximum ages are based on the maximum of stratigraphic bounding [66], phylogenetic bracketing [67,68] and phylogenetic uncertainty. Stratigraphic bounding encompassed two successive underlying fossil-bearing deposits that did not contain any fossils from the lineage of interest, phylogenetic bracketing encompassed the age of the oldest fossils that were up to two nodes below the divergence event, and phylogenetic bracketing allowed for the possibility that taxa of uncertain phylogenetic affinities belong to the crown clade, first outgroup or second outgroup. Dates used in stratigraphic bounding are from Gradstein et al. [69]. We recognized the following chronological units in succession from youngest to oldest: Pleistocene, Pliocene, Late Miocene, Middle Miocene, Early Miocene, Late Oligocene, Early Oligocene, Late Eocene, Middle Eocene, Early Eocene, Late Palaeocene, Middle Palaeocene, Early Palaeocene, Maastrichtian and Campanian.
| node numbera | fossil constraints (Ma) |
oldest fossil for minimum | reference(s) | |
|---|---|---|---|---|
| minimum | maximum | |||
| 3 | 55.6 | 71.2 | Eritherium | [70] |
| 8 | 58.5 | 71.2 | Riostegotherium | [66,71] |
| 10 | 33.8 | 65.5 | Antarctic specimenb | [72,73] |
| 16 | 61.1 | 84.2 | Adunator | [74] |
| 19 | 37.1 | 65.8 | Hesperocyon gregarious | [75–77] |
| 21 | 55.5 | 61.1 | Hyracotherium | [78] |
| 32 | 48.4 | 61.1 | leporid tarsals | [79] |
| 34 | 48.4 | 61.1 | Eogliravus | [80] |
| 36 | 33.8 | 56 | Gaudeamus | [81,82] |
| 38 | 11.8 | 34 | Prodolichotis | [83] |
| 41 | 52.4 | 61.1 | Mattimys | [84] |
bThe Eocene Antarctic specimen is an ungual phalanx that Carlini et al. [72] identified as a megatheroid sloth. Marenssi et al. [85] revised the identification of the phalanx to include either Tardigrada (sloths) or Vermilingua (anteaters). Subsequently, Vizcaíno & Scillato-Yané [73] described a fragmentary tooth from the Eocene of Antarctica and referred this tooth to Tardigrada, but MacPhee & Reguero [86] reinterpreted this tooth fragment as Mammalia incertae sedis based on histological evidence.
Table 2.
Geographical area of extant taxa and oldest fossils used in ancestral area reconstruction.
| taxona | area of extant species | area of oldest fossilb |
|---|---|---|
| Choloepus didactylus | SA | SA; Megalonychidae; Miocene [87] |
| Tamandua tetradactyla | SA | SA; Tamandua; Pleistocene [87] |
| Myrmecophaga tridactyla | SA | SA; Neotamandua; Miocene [87,88] |
| Euphractus sexcinctus | SA | SA; Zaedyus; Pliocene [87,89] |
| Chaetophractus villosus | SA | SA; Chaetophractus; Pliocene [90] |
| Erinaceus europaeus | Eurasia | NA; Adunator; Palaeocene [74] |
| Talpa altaica | Eurasia | Eurasia; Eotalpa; Eocene [91] |
| Sorex araneus | Eurasia | NA; Domnina; Eocene [92] |
| Echinops telfairi | Africa | Africa; Widanelfarasia; Eocene [93] |
| Amblysomus hottentotus | Africa | Africa; Eochrysochloris; Oligocene [93] |
| Procavia capensis | Africa | Africa; Seggeurius; Eocene [94] |
| Loxodonta africana | Africa | Africa; Eritherium; Palaeocene [70] |
| Macroscelides proboscideus | Africa | Africa; Macroscelides; Pliocene [95] |
| Elephantulus rufescens | Africa | Africa; Elephantulus; Pliocene [95] |
| Orycteropus afer | Africa | Africa; Orycteropus; Miocene [96] |
| Tamias striatus | NA | NA; Spurimus; Eocene [97] |
| Muscardinus avellanarius | Eurasia | Eurasia; Eogliravus; Eocene [80] |
| Mus musculus | Eurasia | Eurasia; Progonomys; Miocene [74] |
| Rattus norvegicus | Eurasia | Eurasia; Karnimata; Miocene [74] |
| Pedetes capensis | Africa | Africa; Pondaungimys; Eocene [98] |
| Hystrix brachyurus | Eurasia | Africa; Gaudeamus; Eocene [81] |
| Castor canadensis | NA | NA; Mattimys; Eocene [84] |
| Dipodomys merriami | NA | NA; Proheteromys; Oligocene [99] |
| Cavia porcellus | SA | SA; Prodolichotis; Miocene [83,100] |
| Hydrochaeris hydrochaeris | SA | SA; Cardiatherium; Miocene [101] |
| Erethizon dorsatum | NA | SA; Eopululo; Eocene? [102] |
| Sylvilagus floridanus | NA; SA | Eurasia; tarsal elements; Eocene [79] |
| Ochotona princeps | NA | Eurasia; Sinolagomys; Oligocene [103,104] |
| Cynocephalus variegatus | Eurasia | Eurasia; Dermotherium; Eocene [105] |
| Tupaia minor | Eurasia | Eurasia; Eodendrogale; Eocene [106] |
| Lemur catta | Africa | Africa; Pachylemur; Quaternary [107] |
| Homo sapiens | Eurasia, NA; SA, Africa | Eurasia; Anthrasimias; Palaeocene [108] |
| Tarsius syrichta | Eurasia | Eurasia; Tarsius; Eocene [109] |
| Hippopotamus amphibius | Africa | Africa; Morotochoerus; Miocene [110] |
| Lama glama | SA | NA; Poebrodon; Eocene [111] |
| Tragelaphus eurycerus | Africa | Eurasia; Archaeomeryx; Eocene [112] |
| Sus scrofa | Eurasia, Africa | Eurasia; Eocenchoerus; Eocene [113] |
| Equus caballus | Eurasia | Eurasia, NA; Hyracotherium; Eocene [78,114,115] |
| Ceratotherium simum | Africa | NA; Hyracodontidae; Eocene [116] |
| Tapirus indicus | Eurasia | NA; Helaletes; Eocene [117] |
| Felis catus | Africa | Eurasia; Stenoplesictis; Eocene [118,119] |
| Canis familiaris | Eurasia | NA; Hesperocyon; Eocene [120] |
| Manis pentadactyla | Eurasia | Eurasia; Eomanis; Eocene [121] |
aIn cases of chimeric taxa, we used the most common species from Springer et al.'s [3] concatenated supermatrix. NA, North America; SA, South America.
bArea of the oldest stem fossil belonging to the terminal branch represented by each living taxon.
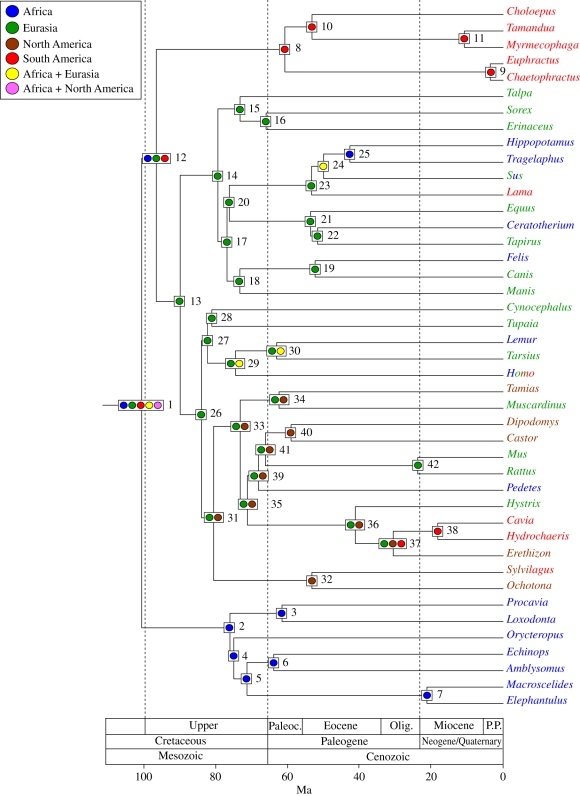
Figure 3.
Ancestral area chronogram for 43 placental taxa from Springer et al. [3] with area coding based on extant ranges for terminal taxa. RAxML was used to infer phylogenetic relationships, BEAST was used to infer divergence times, MAC parsimony was used to infer ancestral areas with the step matrix in figure 2. We employed soft constraints (nodes 3, 8, 10, 16, 19, 21, 32, 34, 36, 38, 41) that followed a normal distribution, with 95% of the normal distribution between the specified minimum and maximum constraints (table 1). Areas for extant taxa are enumerated in table 2 and are colour-coded as follows: Africa, blue; Eurasia, green; North America, brown; South America, red. Multi-coloured names denote taxa that occur in more than one area (table 2). Nodes with unambiguous ancestral area reconstructions are shown with a single coloured circle; nodes with ambiguous reconstructions are shown with two or more circles, and each coloured circle corresponds to a different reconstruction.
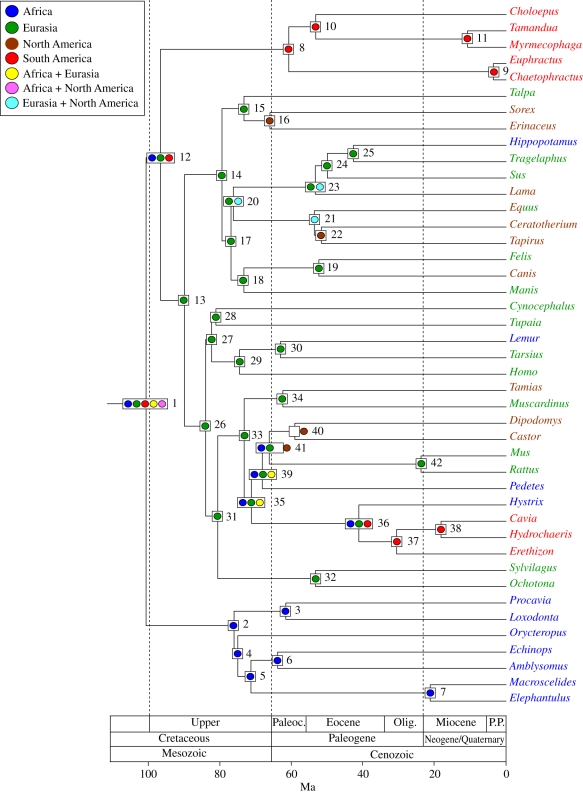
Figure 4.
Ancestral area chronogram for 43 placental taxa from Springer et al. [3] with area coding based on the oldest fossil for each lineage. RAxML was used to infer phylogenetic relationships, BEAST was used to infer divergence times, and MAC parsimony was used to infer ancestral areas with the step matrix in figure 2. Areas for the oldest fossil lineage are enumerated in table 2 and are colour-coded as follows: Africa, blue; Eurasia, green; North America, brown; South America, red. Nodes with unambiguous ancestral area reconstructions are shown with a single coloured circle; nodes with ambiguous reconstructions are shown with two or more circles, and each coloured circle corresponds to a different reconstruction.
Table 3.
Ancestral area reconstructions with areas coded for extant taxa. The order of coded areas in cells with character states is Africa, Eurasia, North America and South America. FP, Fitch parsimony; MBC, multiple binary characters; SMC, single multistate character; NC, no constraints on the maximum number of areas; MAC, minimum area change parsimony; DIVA, dispersal–vicariance with no constraints on the maximum number of areas; DIVA-2, dispersal–vicariance with the maximum number of areas per ancestral node set at 2; DEC, dispersal–extinction cladogenesis with no constraints on the maximum number of areas; DEC-2, dispersal–extinction cladogenesis with the maximum number of areas for each terminal taxon and ancestral range subdivision/inheritance scenario (‘split’) was set at 2, which required coding Homo as present in Africa and Eurasia only given that Homo sapiens inhabited these areas prior to inhabiting North America and South Americaa.
| clade | node no. (figure 3) | FP-MBC | FP-SMC | MAC | DIVA | DIVA-2 | DECb | DEC-2b | SM-MBC | SM-SMCc |
|---|---|---|---|---|---|---|---|---|---|---|
| Placentalia | 1 | (01)000 | 1000 | 1000 | 1101 | 1100 | 0.90, 0.90, 0.58, 0.90 | 0.74, 0.59, 0.00, 0.15 | 0.87, 0.01, 0.00, 0.02 | 0.90, 0.08, 0.00, 0.02 |
| 0100 | 0100 | 1111 | 1001 | |||||||
| 0001 | 0001 | |||||||||
| 1100 | ||||||||||
| 1010 | ||||||||||
| Afrotheria | 2 | 1000 | 1000 | 1000 | 1000 | 1000 | 1.00, 0.00, 0.00, 0.00 | 0.93, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 |
| Paenungulata | 3 | 1000 | 1000 | 1000 | 1000 | 1000 | 1.00, 0.00, 0.00, 0.00 | 0.96, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 |
| Afroinsectiphilia | 4 | 1000 | 1000 | 1000 | 1000 | 1000 | 1.00, 0.00, 0.00, 0.00 | 0.98, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 |
| Afroinsectivora | 5 | 1000 | 1000 | 1000 | 1000 | 1000 | 1.00, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 |
| Afrosoricida | 6 | 1000 | 1000 | 1000 | 1000 | 1000 | 1.00, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 |
| Macroscelidea | 7 | 1000 | 1000 | 1000 | 1000 | 1000 | 1.00, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 |
| Exafroplacentalia | 12 | 0000 | 1000 | 1000 | 0101 | 0101 | 0.12, 0.79, 0.57, 0.79 | 0.07, 0.93, 0.00, 0.23 | 0.01, 0.02, 0.00, 02 | 0.03, 0.71, 0.00, 0.26 |
| 0100 | 0100 | 1011 | ||||||||
| 0001 | 0001 | |||||||||
| Xenarthra | 8 | 0001 | 0001 | 0001 | 0001 | 0001 | 0.00, 0.00, 0.00, 0.98 | 0.00, 0.16, 0.00, 0.85 | 0.00, 0.00, 0.00, 1.00 | 0.00, 0.00, 0.00, 1.00 |
| Dasypodidae | 9 | 0001 | 0001 | 0001 | 0001 | 0001 | 0.00, 0.00, 0.00, 1.00 | 0.00, 0.00, 0.00, 0.90 | 0.00, 0.00, 0.00, 1.00 | 0.00, 0.00, 0.00, 1.00 |
| Pilosa | 10 | 0001 | 0001 | 0001 | 0001 | 0001 | 0.00, 0.00, 0.00, 0.99 | 0.00, 0.00, 0.00, 0.93 | 0.00, 0.00, 0.00, 1.00 | 0.00, 0.00, 0.00, 1.00 |
| Vermilingua | 11 | 0001 | 0001 | 0001 | 0001 | 0001 | 0.00, 0.00, 0.00, 1.00 | 0.00, 0.00, 0.00, 0.99 | 0.00, 0.00, 0.00, 1.00 | 0.00, 0.00, 0.00, 1.00 |
| Boreoeutheria | 13 | 0(01)00 | 0100 | 0100 | 0100 | 0100 | 0.12, 0.86, 0.60, 0.06 | 0.00, 0.84, 0.10, 0.00 | 0.01, 0.58, 0.00, 0.00 | 0.00, 1.00, 0.00, 0.00 |
| 0110 | ||||||||||
| Laurasiatheria | 14 | 0100 | 0100 | 0100 | 0100 | 0100 | 0.00, 0.94, 0.00, 0.00 | 0.00, 0.87, 0.00, 0.00 | 0.01, 0.99, 0.00, 0.00 | 0.00, 1.00, 0.00, 0.00 |
| Eulipotyphla | 15 | 0100 | 0100 | 0100 | 0100 | 0100 | 0.00, 1.00, 0.00, 0.00 | 0.00, 0.99, 0.00, 0.00 | 0.00, 1.00, 0.00, 0.00 | 0.00, 1.00, 0.00, 0.00 |
| Sorex + Erinaceus | 16 | 0100 | 0100 | 0100 | 0100 | 0100 | 0.00, 1.00, 0.00, 0.00 | 0.00, 1.00, 0.00, 0.00 | 0.00, 1.00, 0.00, 0.00 | 0.00, 1.00, 0.00, 0.00 |
| Fereuungulata | 17 | 0100 | 0100 | 0100 | 0100 | 0100 | 0.00, 0.91, 0.00, 0.00 | 0.00, 0.84, 0.00, 0.00 | 0.05, 0.85, 0.00, 0.00 | 0.00, 1.00, 0.00, 0.00 |
| Ostentoria | 18 | 0100 | 0100 | 0100 | 0100 | 0100 | 0.00, 0.95, 0.00, 0.00 | 0.00, 0.93, 0.00, 0.00 | 0.03, 0.95, 0.00, 0.00 | 0.00, 1.00, 0.00, 0.00 |
| Caniformia | 19 | 0100 | 0100 | 0100 | 1100 | 1100 | 0.56, 0.90, 0.00, 0.00 | 0.57, 0.98, 0.00, 0.00 | 0.24, 0.76, 0.00, 0.00 | 0.11, 0.89, 0.00, 0.00 |
| Euungulata | 20 | 0100 | 0100 | 0100 | 0100 | 0100 | 0.00, 0.89, 0.00, 0.00 | 0.00, 0.81, 0.00, 0.00 | 0.22, 0.51, 0.00, 0.00 | 0.01, 0.99, 0.00, 0.00 |
| 1100 | 1100 | |||||||||
| 0101 | 0101 | |||||||||
| 1101 | ||||||||||
| Perissodactyla | 21 | 0100 | 0100 | 0100 | 0100 | 0100 | 0.27, 0.99, 0.00, 0.00 | 0.26, 0.97, 0.00, 0.00 | 0.03, 0.95, 0.00, 0.00 | 0.01, 0.99, 0.00, 0.00 |
| Ceratomorpha | 22 | 0100 | 0100 | 0100 | 1100 | 1100 | 0.33, 0.94, 0.00, 0.00 | 0.26, 0.90, 0.00, 0.00 | 0.12, 0.88, 0.00, 0.00 | 0.04, 0.96, 0.00, 0.00 |
| Cetartiodactyla | 23 | 0(01)00 | 1000 | 0100 | 0101 | 0101 | 0.56, 0.64, 0.00, 0.00 | 0.63, 0.77, 0.00, 0.25 | 0.80, 0.02, 0.00, 0.01 | 0.56, 0.35, 0.00, 0.09 |
| 0100 | 1001 | 1001 | ||||||||
| 0001 | 1101 | |||||||||
| Sus + Bos + | 24 | 1(01)00 | 1000 | 1100 | 1000 | 1000 | 0.88, 0.73, 0.00, 0.00 | 0.91, 0.61, 0.00, 0.00 | 1.00, 0.02, 0.00, 0.00 | 0.97, 0.03, 0.00, 0.00 |
| Hippopotamus | 0100 | 1100 | 1100 | |||||||
| Bos + Hippopotamus | 25 | 1000 | 1000 | 1000 | 1000 | 1000 | 0.99, 0.00, 0.00, 0.00 | 0.97, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 |
| Euarchontoglires | 26 | 0(01)00 | 0100 | 0100 | 0100 | 0100 | 0.10, 0.81, 0.61, 0.00 | 0.00, 0.94, 0.37, 0.00 | 0.01, 0.53, 0.01, 0.00 | 0.00, 0.99, 0.01, 0.00 |
| 0110 | 0110 | |||||||||
| Euarchonta | 27 | 0100 | 0100 | 0100 | 0100 | 0100 | 0.00, 0.78, 0.00, 0.00 | 0.00, 0.98, 0.00, 0.00 | 0.03, 0.99, 0.00, 0.00 | 0.02, 0.98, 0.00, 0.00 |
| Paraprimates | 28 | 0100 | 0100 | 0100 | 0100 | 0100 | 0.00, 1.00, 0.00, 0.00 | 0.00, 1.00, 0.00, 0.00 | 0.00, 1.00, 0.00, 0.00 | 0.00, 1.00, 0.00, 0.00 |
| Primates | 29 | (01)100 | 0100 | 0100 | 0100 | 0100 | 0.00, 0.66, 0.00, 0.00 | 0.00, 0.87, 0.00, 0.00 | 0.92, 0.99, 0.02, 0.02 | 0.93, 0.07, 0.00, 0.00 |
| 1100 | ||||||||||
| Prosimii | 30 | (01)100 | 0100 | 0100 | 1100 | 1100 | 0.40, 0.86, 0.00, 0.00 | 0.27, 0.87, 0.00, 0.00 | 0.94, 0.70, 0.00, 0.00 | 0.98, 0.02, 0.00, 0.00 |
| 1100 | ||||||||||
| Glires | 31 | 0000 | 0100 | 0100 | 0010 | 0010 | 0.00, 0.40, 0.80, 0.00 | 0.00, 0.68, 0.74, 0.00 | 0.00, 0.01, 0.78, 0.00 | 0.00, 0.01, 0.99, 0.00 |
| 0010 | 0010 | 0110 | 0110 | |||||||
| Lagomorpha | 32 | 0010 | 0010 | 0010 | 0010 | 0010 | 0.00, 0.00, 0.86, 0.00 | 0.00, 0.00, 0.89, 0.11 | 0.00, 0.00, 1.00, 0.17 | 0.00, 0.00, 1.00, 0.00 |
| Rodentia | 33 | 0000 | 0100 | 0100 | 0100 | 0100 | 0.00, 0.48, 0.78, 0.00 | 0.00, 0.76, 0.76, 0.00 | 0.00, 0.00, 0.44, 0.00 | 0.00, 0.00, 1.00, 0.00 |
| 0010 | 0010 | 0010 | 0010 | |||||||
| squirrel-related clade | 34 | 0000 | 0100 | 0100 | 0110 | 0110 | 0.00, 0.59, 0.78, 0.00 | 0.00, 0.78, 0.77, 0.00 | 0.00, 0.06, 0.46, 0.00 | 0.00, 0.11, 0.89, 0.00 |
| 0010 | 0010 | |||||||||
| mouse-related clade + | 35 | 0000 | 0100 | 0100 | 0100 | 0100 | 0.00, 0.13, 0.52, 0.00 | 0.00, 0.44, 0.60, 0.00 | 0.00, 0.00, 0.09, 0.00 | 0.00, 0.03, 0.97, 0.00 |
| Hystricognathi | 0010 | 0010 | 0010 | 0010 | ||||||
| Hystricognathi | 36 | 0000 | 0100 | 0100 | 0110 | 0110 | 0.00, 0.75, 0.75, 0.54 | 0.00, 0.62, 0.56, 0.18 | 0.00, 0.06, 0.02, 0.01 | 0.01, 0.34, 0.55, 0.11 |
| 0010 | 0010 | 0101 | 0101 | |||||||
| 1011 | ||||||||||
| Caviomorpha | 37 | 0000 | 0100 | 0100 | 0011 | 0011 | 0.00, 0.00, 0.82, 0.82 | 0.00, 0.00, 0.76, 0.65 | 0.00, 0.00, 0.13, 0.21 | 0.00, 0.01, 0.41, 0.58 |
| 0010 | 0010 | |||||||||
| 0001 | 0001 | |||||||||
| Cavioidea | 38 | 0001 | 0001 | 0001 | 0001 | 0001 | 0.00, 0.00, 0.00, 0.97 | 0.00, 0.00, 0.00, 0.91 | 0.00, 0.00, 0.00, 1.00 | 0.00, 0.00, 0.00, 1.00 |
| mouse-related clade | 39 | 0000 | 0100 | 0100 | 1100 | 1100 | 0.24, 0.40, 0.84, 0.00 | 0.00, 0.45, 0.66, 0.00 | 0.01, 0.00, 0.13, 0.00 | 0.05, 0.02, 0.93, 0.00 |
| 0010 | 0010 | 1010 | 1010 | |||||||
| 1110 | ||||||||||
| Castorimorpha + | 41 | 0000 | 0100 | 0100 | 0110 | 0110 | 0.00, 0.41, 0.87, 0.00 | 0.00, 0.46, 0.76, 0.00 | 0.00, 0.01, 0.69, 0.00 | 0.00, 0.02, 0.98, 0.00 |
| Muridae | 0010 | 0010 | ||||||||
| Castorimorpha | 40 | 0010 | 0010 | 0010 | 0010 | 0010 | 0.00, 0.00, 0.94, 0.00 | 0.00, 0.00, 0.83, 0.00 | 0.00, 0.00, 0.99, 0.00 | 0.00, 0.00, 1.00, 0.00 |
| Muridae | 42 | 0100 | 0100 | 0100 | 0100 | 0100 | 0.00, 0.87, 0.00, 0.00 | 0.00, 0.85, 0.00, 0.00 | 0.00, 0.99, 0.01, 0.00 | 0.00, 1.00, 0.00, 0.00 |
aFP, MAC, DIVA and DEC used an ML topology that was recovered by RAxML [65], but without the associated branch lengths. The RAxML analysis was performed with GTRMIXI, individual parameter estimates for each gene partition and 500 bootstrap replicates with the rapid bootstrap algorithm. Bootstrap analyses started from randomized MP starting trees, employed the fast hill-climbing algorithm and estimated free model parameters. ML analyses were run with the ‘-f a’ command, which combines bootstrap analysis with the search for the best ML tree(s) by using every fifth bootstrap tree as a starting tree to search for the ML tree(s) with the original dataset. DEC analyses were performed with the ultrametric tree shown in figure 3. Stochastic mapping analyses were performed with SIMMAP [5] using default settings, and used 3750 post-burnin trees that were sampled from a Bayesian analysis with MrBayes v. 3.1.1 [122,123]. Parameters of the Bayesian analysis included five million generations with sampling once every 1000 generations, four chains (three heated, one cold) and the same gene partitions that were used in Springer et al. [3]. The average standard deviation of split frequencies was <0.007 after completion of the five million generation chain for two independent runs.
bDEC records the likelihood and relative probability of ancestral range subdivision/inheritance scenarios (‘splits’) at internal nodes, and only shows splits that are within two log-likelihood units of the maximum for each node. Some ancestral area reconstructions contain the same ancestral areas for a given node, but have different splits. For example, the DEC-2 reconstruction for the base of Placentalia included three different splits as follows: Africa|Eurasia = 0.54; Africa|South America = 0.14; and Africa|Eurasia + Africa = 0.10. Both the first and the third splits contain Africa and Eurasia. The difference is that the first split assigns an area of Africa to the left-descendant branch and an area of Eurasia to the right-descendant branch, whereas the third split assigns an area of Africa to the left branch and an area of Africa + Eurasia to the right-descendant branch. However, both splits imply the same ancestral area (Africa + Eurasia) for the base of Placentalia. The relative probability that an ancestral area for Placentalia included Africa is the sum of all inheritance scenarios that contain Africa, i.e. 0.54 + 0.14 + 0.10 = 0.78.
cPolymorphic characters are not accommodated by SIMMAP. So for SM-SMC analyses, we arbitrarily chose a single area for taxa with multiple areas as follows: Homo (Africa, extant coding); Sus (Eurasia, extant coding); Sylvilagus (North America, extant coding); Equus (North America, extinct coding).
Table 4.
Ancestral area reconstructions with areas coded for fossil lineages. The order of coded areas in cells with character states is Africa, Eurasia, North America and South Americaa. Abbreviations as in table 3. Also see table 3 for an explanation of methodological details.
| clade | node no. (figure 4) | FP-MBC | FP-SMC | MAC | DIVA | DIVA-2 | DEC | DEC-2 | SM-MBC | SM-SMC |
|---|---|---|---|---|---|---|---|---|---|---|
| Placentalia | 1 | (01)000 | 1000 | 1000 | 1101 | 1100 | 0.88, 0.88, 0.60, 0.88 | 0.78, 0.64, 0.00, 0.14 | 0.72, 0.01, 0.00, 0.00 | 0.86, 0.10, 0.01, 0.03 |
| 0100 | 0100 | 1111 | 1001 | |||||||
| 0001 | 0001 | |||||||||
| 1100 | ||||||||||
| 1010 | ||||||||||
| Afrotheria | 2 | 1000 | 1000 | 1000 | 1000 | 1000 | 1.00, 0.00, 0.00, 0.00 | 0.94, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 |
| Paenungulata | 3 | 1000 | 1000 | 1000 | 1000 | 1000 | 1.00, 0.00, 0.00, 0.00 | 0.97, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 |
| Afroinsectiphilia | 4 | 1000 | 1000 | 1000 | 1000 | 1000 | 1.00, 0.00, 0.00, 0.00 | 0.98, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 |
| Afroinsectivora | 5 | 1000 | 1000 | 1000 | 1000 | 1000 | 1.00, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 |
| Afrosoricida | 6 | 1000 | 1000 | 1000 | 1000 | 1000 | 1.00, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 |
| Macroscelidea | 7 | 1000 | 1000 | 1000 | 1000 | 1000 | 1.00, 0.00, 0.00, 0.00 | 0.99, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 | 1.00, 0.00, 0.00, 0.00 |
| Exafroplacentalia | 12 | 0000 | 1000 | 1000 | 0101 | 0101 | 0.11, 0.84, 0.58, 0.84 | 0.07, 0.82, 0.00, 0.28 | 0.00, 0.01, 0.00, 0.00 | 0.03, 0.78, 0.01, 0.18 |
| 0100 | 0100 | 0111 | ||||||||
| 0001 | 0001 | |||||||||
| Xenarthra | 8 | 0001 | 0001 | 0001 | 0001 | 0001 | 0.00, 0.00, 0.00, 0.98 | 0.00, 0.16, 0.00, 0.86 | 0.00, 0.00, 0.00, 1.00 | 0.00, 0.01, 0.00, 0.99 |
| Dasypodidae | 9 | 0001 | 0001 | 0001 | 0001 | 0001 | 0.00, 0.00, 0.00, 1.00 | 0.00, 0.00, 0.00, 0.99 | 0.00, 0.00, 0.00, 1.00 | 0.00, 0.00, 0.00, 1.00 |
| Pilosa | 10 | 0001 | 0001 | 0001 | 0001 | 0001 | 0.00, 0.00, 0.00, 1.00 | 0.00, 0.00, 0.00, 0.94 | 0.00, 0.00, 0.00, 1.00 | 0.00, 0.00, 0.00, 1.00 |
| Vermilingua | 11 | 0001 | 0001 | 0001 | 0001 | 0001 | 0.00, 0.00, 0.00, 1.00 | 0.00, 0.00, 0.00, 1.00 | 0.00, 0.00, 0.00, 1.00 | 0.00, 0.00, 0.00, 1.00 |
| Boreoeutheria | 13 | 0100 | 0100 | 0100 | 0100 | 0100 | 0.00, 0.74, 0.45, 0.00 | 0.00, 0.85, 0.00, 0.00 | 0.00, 0.67, 0.00, 0.00 | 0.00, 1.00, 0.00, 0.00 |
| 0110 | ||||||||||
| Laurasiatheria | 14 | 0100 | 0100 | 0100 | 0100 | 0100 | 0.00, 0.88, 0.69, 0.00 | 0.00, 0.97, 0.28, 0.00 | 0.00, 0.21, 0.25, 0.00 | 0.00, 0.95, 0.05, 0.00 |
| 0010 | ||||||||||
| Eulipotyphla | 15 | 0100 | 0100 | 0100 | 0110 | 0110 | 0.00, 0.67, 0.86, 0.00 | 0.00, 0.84, 0.76, 0.00 | 0.00, 0.26, 0.65, 0.00 | 0.01, 0.37, 0.62, 0.01 |
| Sorex + Erinaceus | 16 | 0010 | 0010 | 0010 | 0010 | 0010 | 0.00, 0.00, 0.94, 0.00 | 0.00, 0.00, 0.79, 0.00 | 0.00, 0.01, 0.99, 0.00 | 0.01, 0.02, 0.97, 0.01 |
| Fereuungulata | 17 | 0100 | 0100 | 0100 | 0100 | 0100 | 0.00, 0.81, 0.41, 0.00 | 0.00, 0.84, 0.11, 0.00 | 0.00, 0.44, 0.37, 0.00 | 0.00, 0.82, 0.18, 0.00 |
| 0010 | 0110 | |||||||||
| 0110 | ||||||||||
| Ostentoria | 18 | 0100 | 0100 | 0100 | 0100 | 0100 | 0.00, 0.97, 0.24, 0.00 | 0.00, 0.88, 0.00, 0.00 | 0.00, 0.89, 0.08, 0.00 | 0.00, 0.94, 0.05, 0.00 |
| 0110 | ||||||||||
| Caniformia | 19 | 0100 | 0100 | 0100 | 0110 | 0110 | 0.00, 0.91, 0.70, 0.00 | 0.00, 0.85, 0.49, 0.00 | 0.00, 0.87, 0.13, 0.00 | 0.01, 0.87, 0.12, 0.01 |
| Euungulata | 20 | 01(01)0 | 0100 | 0100 | 0100 | 0100 | 0.00, 0.67, 0.43, 0.00 | 0.00, 0.84, 0.22, 0.00 | 0.00, 0.07, 0.89, 0.00 | 0.00, 0.43, 0.56, 0.00 |
| 0010 | 0110 | 0010 | 0010 | |||||||
| 0110 | 0110 | |||||||||
| Perissodactyla | 21 | 0110 | 0100 | 0110 | 0010 | 0010 | 0.00, 0.53, 0.96, 0.00 | 0.00, 0.71, 0.93, 0.00 | 0.00, 0.74, 1.00, 0.00 | 0.00, 0.10, 0.90, 0.00 |
| 0010 | 0110 | 0110 | ||||||||
| Ceratomorpha | 22 | 0010 | 0010 | 0010 | 0010 | 0010 | 0.00, 0.00, 1.00, 0.00 | 0.00, 0.00, 0.99, 0.00 | 0.00, 0.00, 1.00, 0.00 | 0.00, 0.00, 1.00, 0.00 |
| Cetartiodactyla | 23 | 01(01)0 | 0100 | 0100 | 0110 | 0110 | 0.19, 0.90, 0.79, 0.00 | 0.00, 0.86, 0.55, 0.00 | 0.00, 0.09, 0.33, 0.00 | 0.04, 0.46, 0.48, 0.01 |
| 0010 | 0110 | |||||||||
| Sus + Bos + Hippopotamus | 24 | 0100 | 0100 | 0100 | 0100 | 0100 | 0.24, 0.96, 0.00, 0.00 | 0.00, 0.91, 0.00, 0.00 | 0.01, 0.82, 0.00, 0.00 | 0.06, 0.92, 0.01, 0.00 |
| Bos + Hippopotamus | 25 | 0100 | 0100 | 0100 | 1100 | 1100 | 0.60, 0.92, 0.00, 0.00 | 0.35, 0.91, 0.00, 0.00 | 0.29, 0.71, 0.00, 0.00 | 0.29, 0.69, 0.01, 0.01 |
| Euarchontoglires | 26 | 0100 | 0100 | 0100 | 0100 | 0100 | 0.00, 0.86, 0.00, 0.00 | 0.00, 0.92, 0.00, 0.00 | 0.00, 0.91, 0.00, 0.00 | 0.00, 1.00, 0.00, 0.00 |
| Euarchonta | 27 | 0100 | 0100 | 0100 | 0100 | 0100 | 0.00, 0.99, 0.00, 0.00 | 0.00, 0.99, 0.00, 0.00 | 0.00, 1.00, 0.00, 0.00 | 0.00, 1.00, 0.00, 0.00 |
| Paraprimates | 28 | 0100 | 0100 | 0100 | 0100 | 0100 | 0.00, 1.00, 0.00, 0.00 | 0.00, 1.00, 0.00, 0.00 | 0.00, 1.00, 0.00, 0.00 | 0.00, 1.00, 0.00, 0.00 |
| Primates | 29 | 0100 | 0100 | 0100 | 0100 | 0100 | 0.00, 0.94, 0.00, 0.00 | 0.00, 0.95, 0.00, 0.00 | 0.01, 0.99, 0.00, 0.00 | 0.00, 0.99, 0.00, 0.00 |
| Prosimii | 30 | 0100 | 0100 | 0100 | 1100 | 1100 | 0.46, 0.93, 0.00, 0.00 | 0.28, 0.90, 0.00, 0.00 | 0.30, 0.70, 0.00, 0.00 | 0.30, 0.68, 0.01, 0.01 |
| Glires | 31 | 0100 | 0100 | 0100 | 0100 | 0100 | 0.00, 0.74, 0.00, 0.00 | 0.00, 0.89, 0.00, 0.00 | 0.00, 0.81, 0.00, 0.00 | 0.00, 0.99, 0.01, 0.00 |
| Lagomorpha | 32 | 0100 | 0100 | 0100 | 0100 | 0100 | 0.00, 1.00, 0.00, 0.00 | 0.00, 0.99, 0.00, 0.00 | 0.00, 0.98, 0.00, 0.00 | 0.01, 0.98, 0.01, 0.00 |
| Rodentia | 33 | 0(01)00 | 0100 | 0100 | 0100 | 0100 | 0.41, 0.80, 0.25, 0.00 | 0.30, 0.90, 0.09, 0.00 | 0.00, 0.00, 0.03, 0.00 | 0.05, 0.23, 0.71, 0.02 |
| 1100 | 1100 | |||||||||
| 1110 | ||||||||||
| squirrel-related clade | 34 | 0(01)00 | 0100 | 0100 | 0110 | 0110 | 0.00, 0.84, 0.55, 0.00 | 0.00, 0.87, 0.30, 0.00 | 0.00, 0.03, 0.35, 0.00 | 0.02, 0.15, 0.81, 0.01 |
| mouse-related clade + | 35 | 0000 | 0100 | 1000 | 1000 | 1000 | 0.76, 0.76, 0.32, 0.00 | 0.59, 0.69, 0.00, 0.00 | 0.00, 0.00, 0.01, 0.00 | 0.19, 0.04, 0.71, 0.06 |
| Hystricognathi | 1000 | 0100 | 1100 | 1100 | ||||||
| 1100 | 1110 | 0101 | ||||||||
| 0101 | ||||||||||
| 1101 | ||||||||||
| 0111 | ||||||||||
| 1111 | ||||||||||
| Hystricognathi | 36 | 0000 | 1000 | 1000 | 1001 | 1001 | 0.85, 0.00, 0.00, 0.85 | 0.65, 0.10, 0.00, 0.75 | 0.08, 0.00, 0.00, 0.26 | 0.28, 0.02, 0.03, 0.67 |
| 0100 | 0100 | |||||||||
| 0001 | 0001 | |||||||||
| Caviomorpha | 37 | 0001 | 0001 | 0001 | 0001 | 0001 | 0.00, 0.00, 0.00, 0.97 | 0.00, 0.00, 0.00, 0.93 | 0.00, 0.00, 0.00, 1.00 | 0.00, 0.00, 0.00, 1.00 |
| Cavioidea | 38 | 0001 | 0001 | 0001 | 0001 | 0001 | 0.00, 0.00, 0.00, 1.00 | 0.00, 0.00, 0.00, 0.99 | 0.00, 0.00, 0.00, 1.00 | 0.00, 0.00, 0.00, 1.00 |
| mouse-related clade | 39 | 0000 | 1000 | 1000 | 1100 | 1100 | 0.78, 0.78, 0.58, 0.00 | 0.59, 0.69, 0.08, 0.00 | 0.03, 0.00, 0.06, 0.00 | 0.31, 0.04, 0.64, 0.02 |
| 0100 | 0100 | 1010 | 1010 | |||||||
| 1100 | 1110 | |||||||||
| Castorimorpha + | 41 | 0000 | 1000 | 1000 | 0110 | 0110 | 0.00, 0.73, 0.73, 0.00 | 0.00, 0.71, 0.44, 0.00 | 0.00, 0.01, 0.69, 0.00 | 0.02, 0.06, 0.92, 0.00 |
| Muridae | 0100 | 0100 | ||||||||
| 0010 | 0010 | |||||||||
| Castorimorpha | 40 | 0010 | 0010 | 0010 | 0010 | 0010 | 0.00, 0.00, 0.88, 0.00 | 0.00, 0.31, 0.89, 0.00 | 0.00, 0.00, 0.99, 0.00 | 0.00, 0.01, 0.99, 0.00 |
| Muridae | 42 | 0100 | 0100 | 0100 | 0100 | 0100 | 0.00, 0.97, 0.00, 0.00 | 0.00, 0.94, 0.00, 0.00 | 0.00, 0.99, 0.01, 0.00 | 0.00, 0.98, 0.01, 0.00 |
Ambiguous ancestral area reconstructions were a problem for all methods, and the number of nodes with equivocal reconstructions ranged from four (SM-SMC with extant coding) to 26 (DEC-2 with extant coding). For some methods, the number of ambiguous nodes was higher with extant coding than with fossil coding (FP-MBC, FP-SMC, MAC parsimony, DIVA, DIVA-2, DEC, DEC-2), but in other cases, this pattern was reversed (SM-MBC, SM-SMC). Ancestral areas for Placentalia, Exafroplacentalia (=Boreoeutheria + Xenarthra) and several nodes within Rodentia were reconstructed as ambiguous by nearly all methods. Other nodes were consistently reconstructed with unambiguous ancestral areas, including clades with ancestral areas in Africa (Afrotheria and its internal nodes), Eurasia (Euarchonta, Paraprimates [=Dermoptera + Scandentia], Muridae), North America (Erinaceidae + Soricidae) and South America (Xenarthra and its internal nodes, Cavioidea). Most analyses reconstructed Eurasia as the ancestral area for Boreoeutheria, Laurasiatheria and Euarchontoglires. This finding is discussed below.
The importance of fossils is illustrated by reconstructions for Lagomorpha (tables 3 and 4). All methods returned North America as the ancestral area when extant taxa were used for area coding, but identified Eurasia with fossil coding.
DIVA and DEC analyses reconstructed more nodes with multiple areas than did the other methods. Analyses with DEC reconstructed 17–20 nodes with two or more areas and four to six nodes with three or more areas. DIVA analyses resulted in 15–18 nodes with at least two areas, and five to six nodes with three or more areas. None of the other methods reconstructed ancestral nodes to include three or more areas in a single reconstruction, although three or four areas were sometimes represented by the full complement of alternate reconstructions for a given node.
FP-MBC returned nine empty nodes with extant coding, and five empty areas with fossil coding. SM-MBC with extant coding resulted in three or four empty nodes with extant coding, and four empty nodes with extinct coding (table 5).
Table 5.
Comparison of different methods for reconstructing ancestral areas. NA1, not applicable for monomorphic reconstruction methods; NA2, not applicable when the maximum number of areas is set at two; NA2, not applicable for methods that employ single multistate charactersa.
| FP-MBC | FP-SMC | MAC Parsimony | DIVA | DIVA-2 | DEC | DEC-2 | SM-MBC | SM-SMC | |
|---|---|---|---|---|---|---|---|---|---|
| nodes with ambiguous | 7/5 | 12/9 | 12/8 | 12/11 | 10/7 | 23/23 | 26/23 | 16/17 | 6/14 |
| reconstructionsb | 19/20 | 17/18 | 10/12 | 4/10 | |||||
| nodes with ≥ 2 areasc | 3/3 | NA1 | 4/6 | 16/18 | 15/16 | 18/20 | 20/19 | 7/7 | NA1 |
| 17/20 | 17/17 | 4/6 | |||||||
| nodes with ≥ 3 areasd | 0/0 | NA1 | 0/0 | 6/5 | NA2 | 6/6 | NA2 | 0/0 | NA1 |
| 4/5 | 0/0 | ||||||||
| empty nodese | 9/5 | NA3 | NA3 | NA3 | NA3 | NA3 | NA3 | 3/4 | NA3 |
| 4/4 |
aNumbers before slashes are based on analyses with area coding for extant taxa, and numbers after slashes are based on analyses with area coding for the oldest fossil. See table 3 for abbreviations.
bFor FP-MBC, nodes were considered ambiguous if at least one area was reconstructed as (01). For SM-MBC and SM-SMC, nodes were considered ambiguous if the posterior probability (PP) of at least one area was 0.1 < PP < 0.9 (top line) or 0.2 < PP < 0.8 (bottom line). For DEC and DEC-2, nodes were considered ambiguous if the frequency (f) of at least one area was 0.1 < f < 0.9 (top line) or 0.2 < p < 0.8 (bottom line).
cAt least two areas in at least one of the alternate resolutions for an ancestral node. For FP-MBC, each occurrence of 1 or (01) was taken to include an ancestral area. For SM-MBC, areas were counted as present at a node if posterior probabilities were ≥0.10 (top line) or ≥0.20 (bottom line). For DEC and DEC-2, areas were counted as present at a node if frequencies were ≥0.1 (top line) or ≥0.2 (bottom line).
dAt least three areas in more than one of the alternate resolutions for an ancestral node. For FP-MBC, each occurrence of 1 or (01) was taken to include an ancestral area. For SM-MBC, areas were counted as present at a node if posterior probabilities were ≥0.10 (top line) or ≥0.20 (bottom line). For DEC and DEC-2, areas were counted as present at a node if frequencies were ≥0.1 (top line) or ≥0.2 (bottom line).
eFor FP-MBC, nodes were considered empty if all areas were reconstructed as 0. For SM-MBC, nodes were considered empty if posterior probabilities were <0.10 (top line) or <0.20 (bottom line) for all four areas.
7. Placental biogeography
Afrotheria (Afrosoricida, Hyracoidea, Macroscelidea, Proboscidea, Sirenia, Tubulidentata) was the first of the new superordinal groups to receive robust molecular support [53,55,56]. With the exception of Sirenia, all afrotherian orders have first fossil occurrences in Africa, and two orders (Macroscelidea, Afrosoricida) have evolutionary histories that are restricted to the Afro-Malagasy region. Springer et al. [53] suggested that interordinal separation of afrotherian orders commenced during a window of isolation that began in the Cretaceous, after Africa separated from South America, and lasted until the early Cenozoic when Africa docked with Europe. Consistent with this scenario, Africa was unambiguously reconstructed as the ancestral area for Afrotheria (figures 3 and 4). This hypothesis contrasts with traditional views wherein the African mammal fauna arrived from the north, including a condylarth stock that arrived in Africa from Europe in the early Cenozoic, and insectivores that arrived in the Neogene [124].
Asher et al. [125], Zack et al. [126] and Tabuce et al. [127] suggested that the geographical distributions of living afrotherians are not representative of the historical geographical distribution of this clade, and that Afrotheria is Holarctic in origin based on the placement of extinct taxa from the Palaeocene of Laurasia within or at the base of Afrotheria. However, pseudoextinction tests call into question the reliability of the placement of fossil taxa in morphological cladistic analyses [3].
The oldest xenarthran fossils are scutes from the Palaeocene of South America [71]. Living members of Xenarthra (anteaters, sloths, armadillos) are restricted to South and Central America with the exception of the nine-banded armadillo, whose ancestors dispersed to North America during the Great American Interchange [128]. Simpson [129,130] supported the view that South American xenarthrans evolved in situ during South America's isolation from other continents in the early Tertiary. All of our analyses are consistent with the hypothesis that South America was the ancestral area for Xenarthra (figures 3 and 4).
The remaining placental orders are placed in Laurasiatheria (Eulipotyphla, Chiroptera, Perissodactyla, Cetartiodactyla, Carnivora, Pholidota) and Euarchontoglires (Primates, Dermoptera, Scandentia, Rodentia, Lagomorpha). With the exception of bats, these orders have first fossil occurrences that are exclusively Laurasian. Our reconstructions provide support for Eurasia, but not North America, as the ancestral area for these clades (figures 3 and 4). These results are consistent with previous suggestions that Cretaceous zhelestids and zamlambdalestids from Asia are members of crown Placentalia [131,132]. Further, the fossil record suggests that Eutheria were dominant in Eurasia throughout the Cretaceous, but were absent from North America through much of the Late Cretaceous and only attained appreciable diversity there during the last approximately 10 Myr of the period [133,134]. Boyer et al. [135] concluded that the Indian subcontinent, Eurasia and Africa are more likely places of origin for Euarchonta than is North America. This agrees with our ancestral area reconstructions (figures 3, 4 and tables 3, 4).
Although there is robust support for the monophyly of Xenarthra, Afrotheria and Boreoeutheria, relationships among these three groups and the root of the placental tree remain contentious [10,54,60–63,136]. Murphy et al. [62] and Springer et al. [10] suggested a causal relationship between the sundering of Africa and South America, and basal cladogenesis among crown-group placental mammals given the coincidence of molecular dates for the base of placentals and the vicariant separation of Africa and South America approximately 100–120 Ma.
Asher et al. [125] analysed a combined matrix and recovered Afrotheria in a nested position within Placentalia, which contradicts the hypothesis that the plate tectonic separation of Africa and South America played a causal role in the early cladogenesis of placental mammals. However, the nested position for Afrotheria resulted from the paraphyly of Euarchontoglires, Glires and Rodentia. Rare genomic changes confirm the monophyly of Xenarthra [137], Afrotheria [138–142], Euarchontoglires [139,141,142], Laurasiatheria [139,141,142] and Boreoeutheria [139,141,142], and preclude a nested position for Afrotheria in the placental tree.
Rare genomic changes have also been used to examine the position of the placental root. Kriegs et al. [139] reported LINE insertions that are shared by Epitheria, whereas Murphy et al. [16] discovered rare genomic changes that support Atlantogenata. Nishihara et al. [142] performed genome-wide retroposon analyses and found 22, 25 and 21 LINE insertions for Exafroplacentalia, Epitheria and Atlantogenata, respectively. Based on these results, Nishihara et al. [142] concluded that Xenarthra, Afrotheria and Boreoeutheria diverged from one another nearly simultaneously. They also suggested a new palaeogeographical model for the breakup of Pangaea and Gondwana in which Africa becomes isolated from both South America and Laurasia at approximately 120 Ma, and argued that these coeval plate tectonic events provide an explanation for the simultaneous divergence of Afrotheria, Xenarthra and Boreoeutheria. However, relaxed clock dates for the base of Placentalia are closer to 100 Ma than to 120 Ma (figures 3 and 4). A second difficulty concerns the opening of the South Atlantic. Nishihara et al. [142] suggested that the Brazilian Bridge, which represented the last connection between Africa and South America, was severed at approximately 120 Mya, but other recent reconstructions suggest that the connection between the South Atlantic and Central Atlantic was not established until late Aptian/mid-Albian times (approx. 110–100 Ma) [143,144].
8. The importance of dispersal
In the context of pre-plate tectonic views of the Earth, Simpson [2] proposed three types of migration routes to describe the movement of animals: corridors, filter bridges and sweepstakes dispersal. Corridors connect two areas and are permeable to all animals; filter bridges impose selective barriers that affect some, but not all animals; and sweepstakes dispersal is required when there are strong barriers to migration such as high mountain barriers or oceans.
Simpson [2] suggested that Madagascar's living mammals were the product of sweepstakes dispersal from Africa to Madagascar. Sweepstakes dispersal hypotheses fell out of favour with the validation of plate tectonic theory and were summarily dismissed as ‘miraculous’ hypotheses with no scientific basis [145]. However, it has become apparent that some distributional patterns can only be explained by sweepstakes dispersal [146]. Observational data also provide support for long-distance vertebrate dispersal [147]. Examples of low probability sweepstakes dispersal involving mammals include the origins of the endemic mammal fauna in Madagascar, and the occurrence of caviomorph rodents and platyrrhine primates in South America.
Madagascar's strictly terrestrial extant mammal fauna includes endemic lineages from four placental orders: tenrecs (Afrosoricida), euplerids (Carnivora), nesomyines (Rodentia) and lemurs (Primates). In each lineage, Madagascar endemics comprise monophyletic assemblages with closest living relatives in Africa [148,149]. Madagascar separated from Africa approximately 165 Ma, but maintained its connection with Antarctica via the Kerguelen Plateau until as late as 80 Ma, at which time it became permanently separated from other Gondwanan landmasses. This history suggests that Madagascar's terrestrial endemic mammals are either the ancient descendants of vicariant events that occurred prior to 80 Ma, or reached Madagascar via transoceanic sweepstakes dispersal at a later time. Another possibility is that a land bridge connected Africa and Madagascar between 45 and 26 Ma [150].
Molecular divergence dates suggest that all four endemic lineages last shared a common ancestor with their African sister group in the Cenozoic [148,149,151,152]. Poux et al. [148] concluded that dispersal by lemurs, rodents and carnivorans must have occurred by transoceanic dispersal rather than land bridge dispersal based on molecular dates for the colonization of Madagascar that were outside of the land bridge window, i.e. 60–50 Ma for lemurs, 26–19 Ma for carnivorans and 24–20 Ma for rodents. However, present ocean currents allow for dispersal from Madagascar to Africa, but oppose reciprocal dispersal from Africa to Madagascar across the Mozambique Channel. If ocean currents were the same for most of the Cenozoic as they are today, they would not have facilitated west to east transoceanic dispersal across the Mozambique Channel because of the strong south–southwest flow of the Mozambique Current [153].
Ali & Huber [154] addressed this problem by simulating surface ocean currents in the Indian Ocean during the Eocene. They concluded that large-scale ocean current systems in the Eocene were profoundly different from modern observed circulatory patterns, and that the flow along the African coast was eastward towards Madagascar instead of southward through the Mozambique Channel (figure 5). Ali & Huber [154] further suggested that dispersal probabilities were enhanced by tropical storms that (i) generated large, floating tree islands that would have allowed for a successful oceanic voyage and (ii) accelerated transportation rates from Africa to Madagascar that would have allowed for complete crossing of the Mozambique Channel in 25–30 days.
Figure 5.
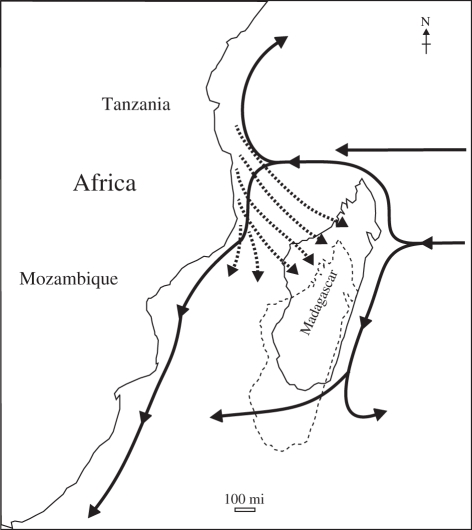
Present day surface ocean currents in the Mozambique Channel (solid arrows) are south–southwest and would not have facilitated west to east transoceanic dispersal from Africa to Madagascar [153]. By contrast, westerly surface ocean currents in the Eocene (dashed arrows) would have facilitated dispersal across the Mozambique Channel from Africa to Madagascar, especially during tropical storms [154]. The outline of Madagascar with dashed lines shows its approximate position relative to Africa during the Eocene.
The dispersal of four groups of fully terrestrial mammals from Africa to Madagascar, at a time when there was no land bridge, is a testament to the importance of rare sweepstakes events in the evolutionary history of Placentalia. Even more remarkable is the occurrence of two different groups of placental mammals, hystricognath rodents and anthropoid primates, in Africa and South America.
Hystricognathi includes Hystricidae (Old World porcupines) and Phiomorpha (e.g. cane rats, dassie rats) from the Old World, and Caviomorpha (e.g. porcupines, chinchillas) from the New World. The oldest hystricognaths are from the late Eocene Egypt, and have been dated at approximately 37 Ma [81]. Old World hystricognaths are paraphyletic, usually with phiomorphs having closer phylogenetic affinities to South American caviomorphs than to hystricids [14,155,156]. Relaxed clock dates suggest that South American caviomorphs last shared a common ancestor with phiomorphs between 45 and 36 Ma [81,155,157]. The most recent common ancestor of Caviomorpha has been dated at 45–31 Ma [81,155,157,158].
Among anthropoids, Old World catarrhines (e.g. macaques, apes) and South American platyrrhines (e.g. marmosets, capuchins, spider monkeys) are reciprocally monophyletic sister taxa. The oldest anthropoid fossils are from the Old World, although whether the most recent common ancestor of Anthropoidea is African or Asian is uncertain [108,159,160]. Poux et al. [155] dated the split between catarrhines and platyrrhines at approximately 37 Ma and the base of Platyrrhini at approximately 17 Ma.
The vicariant separation of Africa and South America (110–100 Ma) is too old to explain the separation of either Phiomorpha and Caviomorpha, or Catarrhini and Platyrrhini. Similarly, Arnason et al.'s [161] hypothesis of land bridge dispersal during the Late Cretaceous–Early Palaeocene is too old for relaxed clock dates, which instead rule out the colonization of South America by Caviomorpha and Platyrrhini prior to the Eocene. Other hypotheses for the colonization of South America by caviomorphs and/or platyrrhines include: (i) trans-Atlantic dispersal from Africa to South America [162], (ii) dispersal from Asia through North America to South America [163,164], and (iii) dispersal from Asia to South America via Australia and Antarctica after two ocean crossings (figure 6) [165].
Figure 6.
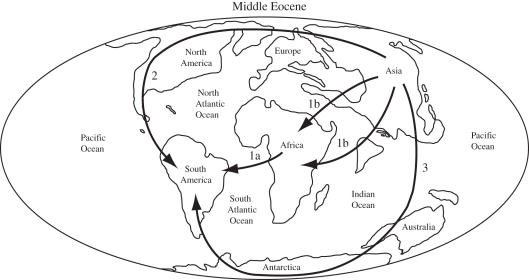
Alternate hypotheses for the dispersal of platyrrhine and caviomorph ancestors, respectively, from Africa/Asia to South America. Hypothesis 1: transoceanic dispersal (1a) from Africa to South America, possibly with an earlier dispersal from Asia to Africa (1b) if origination occurred in Asia. Hypothesis 2: dispersal from Asia through North America to South America. Hypothesis 3: dispersal from Asia to South America via Australia and Antarctica after two transoceanic crossings. Middle Eocene world map based on Palaeomap Project (http://www.scotse.com/newpage9.htm).
Most workers favour transoceanic dispersal from Africa to South America for both Caviomorpha and Platyrrhini. Dispersal through Asia and North America is an intriguing possibility, but palaeontological data provide no support for migrations through North America. Similarly, dispersal from Asia to South America through Australia and Antarctica lacks palaeontological support, requires multiple transoceanic dispersals and becomes even less likely after the Eocene because of the severed connection between Antarctica and South America, and climatic deterioration in Antarctica associated with the opening of the Drake Passage. In view of phylogenetic, geological, palaeontological and molecular data, trans-Atlantic dispersal is the most likely scenario for colonization of South America by caviomorphs and platyrrhines.
9. Bat biogeography
In contrast to other mammals, bats are capable of powered flight, which has profoundly enhanced their dispersal capabilities. The occurrence of seven different families of extant bats in Madagascar, including the endemic sucker-footed bats (Family Myzopodidae), and of another family in New Zealand, the short-tailed bats (Family Mystacinidae), provides abundant evidence of the dispersal capabilities of bats [166].
The oldest bat fossils are from the Early Eocene of North America [167,168]. Early Eocene bats are also known from Europe, Africa and Australia [167]. The prevalent view is that bats originated in Laurasia, but a minority view holds that bats originated in Gondwana [169,170]. Teeling et al. [13] reconstructed ancestral areas for bats with (i) multistate-coded data for the current global distribution of each lineage with nine different character states (Europe, Africa, Asia, Madagascar, Australia, New Zealand, North America, Central + South America and West Indies) and (ii) binary-coded data for the earliest fossil occurrence for each lineage (Laurasia versus Gondwana). Teeling et al.'s [13] results suggested North America or Laurasia as the ancestral area for bats, and Asia, Europe or Laurasia as the ancestral area for both Yinpterochiroptera and Yangochiroptera. Eick et al. [12] used DIVA [33] to estimate ancestral areas for Chiroptera and its subclades, and coded areas based on current distributions for each family. Seven areas (Africa, Asia, Australia, Europe, North America, South America and New Zealand) were recognized, and Africa was reconstructed as the ancestral area for the most recent common ancestors of Chiroptera, Yinpterochiroptera and Yangochiroptera. Lim [47] used parsimony to reconstruct ancestral areas, and also recovered Africa as the ancestral area for Yangochiroptera and its deepest nodes.
We recovered more inclusive ancestral areas for Chiroptera, Yinpterochiroptera and Yangochiroptera when we performed analyses with DIVA using the same data and settings that were reported by Eick et al. [12] (figure 7 and table 6). The reconstruction for the base of Chiroptera was equivocal and included seven different possibilities, all of which were equally parsimonious based on DIVA's criteria for minimizing dispersal and extinction (figure 7 and table 6). Each of these reconstructions included at least five areas, and four areas (Africa, Asia, North America and South America) were common to all seven reconstructions.
Figure 7.
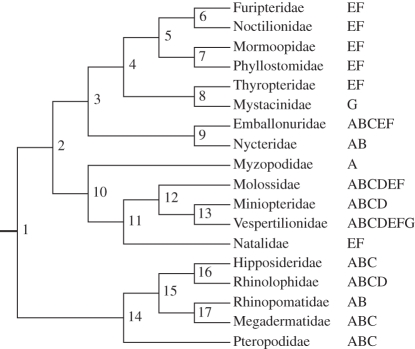
Eick et al.'s [12] phylogeny and area coding for extant bat families. Ancestral area reconstructions based on DIVA analyses are shown in table 6 for nodes 1–17. Africa, A; Asia, B; Australia, C; Europe, D; North America, E; South America, F; New Zealand, G.
Table 6.
A comparison of ancestral area reconstructions for bats based on DIVA analyses. Eick et al. [12] coded the presence or absence of extant bat families in seven different areas and performed DIVA analyses with no constraints on the maximum number of areas. We re-analysed Eick et al.'s [12] dataset with DIVA using the same settings reported by these authors. Africa, A; Asia, B; Australia, C; Europe, D; North America, E; South America, F; New Zealand, G.
| node number (figure 7) | Eick et al. [12] | re-analysis |
|---|---|---|
| 1 | A | ABCEF, ABDEF, ABCDEF, ABEFG, ABCEFG, ABDEFG, ABCDEFG |
| 2 | A | ACEF, BCEF, ABCEF, DEF, ADEF, BDEF, ABDEF, ACDEF, BCDEF, ABCDEF, AEFG, ABEFG, ACEFG, BCEFG, ABCEFG, DEFG, ADEFG, BDEFG, ABDEFG, ACDEFG, BCDEFG, ABCDEFG |
| 3 | AE, AF | E, AE, BE, CE, ACE, BCE, ABCE, F, AF BF, CF, ACF, BCF, ABCF, CEF, ACEF, BCEF, ABCEF, AG, BG, CG, ACG, BCG, ABCG AEG, BEG, CEG, ACEG, BCEG, ABCEG, AFG, BFG, CFG, ACFG, BCFG, ABCFG, AEFG, BEFG, CEFG, ACEFG, BCEFG, ABCEFG |
| 4 | E, F | E, F, EG, FG, EFG |
| 5 | E, F | E, F |
| 6 | E, F | E, F |
| 7 | E, F | E, F |
| 8 | EG, FG, EFG | EG, FG, EFG |
| 9 | A | A, B, AC, BC, ABC, AE, BE, ABE ACE, BCE, ABCE, AF, BF, ABF, ACF, BCF, ABCF, AEF, BEF, ABEF, ACEF, BCEF |
| 10 | A | A, AC, AD, ACD, ABCD, ACE, ADE ACDE, ABCDE, ACF, ADF, ACDF, ABCDF, ACEF, ADEF, ACDEF, ABCDEF, ACDEG ABCDEG, ACDFG, ABCDFG, ACDEFG, ABCDEFG |
| 11 | AE, AF, AEF | AE, CE, DE, CDE, ACDE, BCDE ABCDE, AF, CF, DF, CDF, ACDF, BCDF, ABCDF, AEF, CEF, DEF, CDEF, ACDEF BCDEF, ABCDEF, CDEG, ACDEG, BCDEG, ABCDEG, CDFG, ACDFG, BCDFG, ABCDFG, CDEFG, ACDEFG, BCDEFG, ABCDEFG |
| 12 | A | A, C, D, CD, ACD, BCD, ABCD, CDE ACDE, BCDE, ABCDE, CDF, ACDF, BCDF, ABCDF, CDEF, ACDEF, BCDEF, ABCDEF, CDG ACDG, BCDG, ABCDG, CDEG, ACDEG, BCDEG, ABCDEG, CDFG, ACDFG, BCDFG, ABCDFG, CDEFG, ACDEFG, BCDEFG, ABCDEFG |
| 13 | A | A B C, D, AG, BG, ABG, CG, ACG BCG, ABCG, DG, ADG, BDG, ABDG, CDG, ACDG, BCDG, ABCDG, AEG, BEG, ABEG, CEG ACEG, BCEG, ABCEG, DEG, ADEG, BDEG, ABDEG, CDEG, ACDEG, BCDEG, ABCDEG, AFG, BFG, ABFG, CFG, ACFG, BCFG, ABCFG, DFG, ADFG, BDFG, ABDFG, CDFG, ACDFG BCDFG, ABCDFG, AEFG, BEFG, ABEFG, CEFG, ACEFG, BCEFG, ABCEFG, DEFG, ADEFG, BDEFG, ABDEFG, CDEFG, ACDEFG, BCDEFG, ABCDEFG |
| 14 | A | A, B, C, AC, BC, ABC |
| 15 | A | A, B, C, AC, BC |
| 16 | A | A, B, C |
| 17 | A | A, B, AC, BC, ABC |
Among the most comprehensive studies in mammalian historical biogeography are Lim's [46,47] analyses of South American bats. Ancestral reconstructions provided evidence for multiple dispersals from Africa to South America. One dispersal occurred in Noctilionoidea (Eocene, approx. 42 Ma) and another occurred in Emballonuroidea (Oligocene, approx. 30 Ma). Vespertilionoidea have a more complex history that involves numerous independent dispersals from Africa (Eocene, earliest event approx. 50 Ma), as well as from North America. Lim [46] used PACT to examine evolutionary processes that have been important in the diversification of South American emballonurids. His general area cladogram revealed a complex history with multiple vicariant, within-area and dispersal events all playing a role. Within-area speciation during the Miocene, particularly in the northern Amazon area, was the most important diversification process in this group. Lim [47] correlated Miocene speciation with contemporaneous climatic and habitat changes that occurred in the Amazon Basin. Construction of an ancestral area cladogram for all bat species will provide an unprecedented opportunity to examine the importance of transoceanic dispersal in promoting taxonomic diversity in this highly successful group of mammals.
10. Marsupial biogeography
The oldest metatherian is Sinodelphys from China [171]. Cretaceous marsupial fossils are also known from Europe [172,173] and North America [174–178]. The consensus is that metatherians originated in Asia, and subsequently dispersed to North America and Europe [173].
In contrast to the Cretaceous record of Metatheria, almost all living metatherians have geographical distributions that are entirely Gondwanan. Case et al. [179] suggested that the ancestor of living marsupials dispersed to South America in the Late Cretaceous or early Palaeocene. The South American marsupial cohort Ameridelphia, which includes Paucituberculata (shrew opossums) and Didelphimorphia (opossums), is paraphyletic at the base of Australidelphia, which includes the South American order Microbiotheria (monito del monte), and the Australasian orders Diprotodontia (e.g. wombats, kangaroos), Dasyuromorphia (e.g. quolls, numbats), Peramelemorphia (e.g. bandicoots, bilbies) and Notoryctemorphia (marsupial moles) [17,21,180–182].
Subsequent to Kirsch et al.'s [183] single-copy DNA hybridization study of marsupials, which placed South American microbiotheres within Australidelphia, marsupial biogeographers have focused on the monophyly or paraphyly of Australasian taxa. Australasian monophyly is consistent with a single dispersal from South America to Australia via Antarctica, but Australasian paraphyly requires either multiple dispersals to Australia, or dispersal to Australia followed by back dispersal to South America [183–185]. Molecular phylogenies based on concatenated nuclear gene sequences [21,182] and retroposon insertions [186] support the monophyly of Australasian marsupials, and suggest that Australasian marsupials last shared a common ancestor with microbiotheres between 65 and 58 Ma. This phylogeny is compatible with a single dispersal event from South America to Australia via Antarctica [21]. This dispersal would have been overland if it occurred prior to the complete submergence of the South Tasman Rise approximately 64 Ma [187].
In contrast, Beck et al. [181] analysed a dataset comprising living and fossil taxa, including the early Eocene genus Djarthia from Australia, and recovered a sister-group relationship between Djarthia and living australidelphians. Beck et al.'s [181] topology suggest that South American microbiotheres back-dispersed from eastern Gondwana to South America even though living Australasian marsupials comprise a monophyletic taxon. However, the decay index that associates crown Australidelphia to the exclusion of Djarthia is only one step. This result highlights the potential importance of fossils for inferring biogeographic history, and the precarious nature of conclusions based on a fragmentary fossil record.
11. Monotreme biogeography
Living monotremes include the semi-aquatic platypus (Ornithorhynchus), which occurs in Australia and Tasmania, and echidnas, which occur in Australia (Tachyglossus) and New Guinea (Zaglossus). The oldest monotreme is Teinolophos (121–112.5 Ma) of Australia. Rowe et al. [188] suggested that Teinolophos is a crown monotreme based on cladistic analyses.
In contrast to this ancient fossil record, relaxed clock estimates for the platypus-echidna divergence range from 88.9 to 27.7 Ma [188–191] and are too young to accommodate Teinolophos in crown-group Monotremata. Rather, these dates suggest that Teinolophos lies on the monotreme stem branch. Younger monotreme fossils, whether stem or crown, are exclusively from the Southern Hemisphere. Luo et al. [192,193] and Kielan-Jaworowska et al. [194] suggested that Monotremata belongs to the more inclusive Gondwanan clade Australosphenida, although other studies place these Mesozoic taxa closer to Theria than to Monotremata [188,189].
With or without these Mesozoic taxa, it appears that the entire evolutionary history of Monotremata is restricted to Gondwana. Details of this history are difficult to reconstruct owing to Monotremata's depauperate taxonomic diversity and meagre fossil record. Future fossil discoveries and more robust phylogenetic analyses are essential for revealing the full biogeographic range of ancient monotremes in Gondwana. The occurrence of monotremes in Australia and South America suggests that their ancestral distribution may have included other fragments of Gondwana such as Africa, Antarctica, India and Madagascar.
12. Conclusions
Contemporary methods for deciphering palaeobiogeography are underpinned by phylogenies, divergence times and ancestral area reconstructions, which together yield ancestral area chronograms that provide a powerful framework for proposing and testing hypotheses of dispersal and vicariance when evaluated in the context of palaeographic hypotheses. The toolkit for unravelling historical patterns of vicariance and dispersal that have moulded the evolutionary history of Mammalia now includes molecular data, fossils, reconstructions of palaeogeography and palaeo-ocean currents, and a burgeoning array of methods in phylogeny reconstruction, molecular dating and ancestral area reconstruction. Larger and taxonomically more complete molecular datasets, new fossil discoveries and the application of new techniques will lead to significant advances in our understanding of the historical biogeography of Mammalia.
Acknowledgements
M.S.S. and W.J.M. acknowledge support from NSF. Three anonymous reviewers provided helpful comments on an earlier draft of this manuscript. We thank Kate Jones for inviting us to contribute to this volume.
References
- 1.Jones K. E., Safi K. 2011. Ecology and evolution of mammalian biodiversity. Phil. Trans. R. Soc. B 366, 2451–2461 10.1098/rstb.2011.0090 (doi:10.1098/rstb.2011.0090) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simpson G. G. 1940. Mammals and land bridges. J. Washington D.C. Acad. Sci. 30, 137–163 [Google Scholar]
- 3.Springer M. S., Burk-Herrick A., Meredith R., Eizirik E., Teeling E., O'Brien S. J., Murphy W. J. 2007. The adequacy of morphology for reconstructing the early history of placental mammals. Syst. Biol. 56, 673–684 10.1080/10635150701491149 (doi:10.1080/10635150701491149) [DOI] [PubMed] [Google Scholar]
- 4.Springer M. S., Meredith R. W., Eizirik E., Teeling E., Murphy W. J. 2008. Morphology and placental mammal phylogeny. Syst. Biol. 57, 499–503 10.1080/10635150802164504 (doi:10.1080/10635150802164504) [DOI] [PubMed] [Google Scholar]
- 5.Bollback J. P. 2006. SIMMAP: stochastic character mapping of discrete traits on phylogenies. BMC Bioinform. 7, 88. 10.1186/1471-2105-7-88 (doi:10.1186/1471-2105-7-88) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drummond A. J., Ho S. Y. W., Phillips M. J., Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4, e88. 10.1371/journal.pbio.0040088 (doi:10.1371/journal.pbio.0040088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuckerkandl E., Pauling L. 1962. Molecular disease, evolution, and genetic heterogeneity. In Horizons in biochemistry (eds Kasha M., Pullman B.), pp. 189–225 New York, NY: Academic Press [Google Scholar]
- 8.Douady C. J., Douzery E. J. P. 2003. Molecular estimation of eulipotyphlan divergence times and the evolution of ‘Insectivora’. Mol. Phylogenet. Evol. 28, 285–296 10.1016/S1055-7903(03)00119-2 (doi:10.1016/S1055-7903(03)00119-2) [DOI] [PubMed] [Google Scholar]
- 9.Douady C. J., Catzeflis F., Raman J., Springer M. S., Stanhope M. J. 2003. Molecular evidence for the Sahara as a vicariant agent, and the role of Miocene climatic events, in the diversification of the mammalian order Macroscelidea (elephant shrews). Proc. Natl Acad. Sci. USA 100, 8325–8330 10.1073/pnas.0832467100 (doi:10.1073/pnas.0832467100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Springer M. S., Murphy W. J., Eizirik E., O'Brien S. J. 2003. Placental mammal diversification and the Cretaceous–Tertiary boundary. Proc. Natl Acad. Sci. USA 100, 1056–1061 10.1073/pnas.0334222100 (doi:10.1073/pnas.0334222100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delsuc F., Vizcaíno S. F., Douzery E. J. P. 2004. Influence of Tertiary paleoenvironmental changes on the diversification of South American mammals: a relaxed molecular clock study within xenarthrans. BMC Evol. Biol. 4, 11. 10.1186/1471-2148-4-11 (doi:10.1186/1471-2148-4-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eick G. N., Jacobs D. S., Matthee C. A. 2005. A nuclear DNA phylogenetic perspective on the evolution of echolocation and historical biogeography of extant bats (Chiroptera). Mol. Biol. Evol. 22, 1869–1886 10.1093/molbev/msi180 (doi:10.1093/molbev/msi180) [DOI] [PubMed] [Google Scholar]
- 13.Teeling E. C., Springer M. S., Madsen O., Bates P., O'Brien S. J., Murphy W. J. 2005. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science 307, 580–584 10.1126/science.1105113 (doi:10.1126/science.1105113) [DOI] [PubMed] [Google Scholar]
- 14.Huchon D., Chevret P., Jordan U., Kilpatrick C. W., Ranwez V., Jenkins P. D., Brosius J., Schmitz J. 2007. Multiple molecular evidences for a living mammalian fossil. Proc. Natl Acad. Sci. USA 104, 7495–7499 10.1073/pnas.0701289104 (doi:10.1073/pnas.0701289104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janecka J. E., Miller W., Pringle T. H., Wiens F., Zitzmann A., Helgen K. M., Springer M. S., Murphy W. J. 2007. Molecular and genomic data identify the closest living relative of Primates. Science 318, 792–974 10.1126/science.1147555 (doi:10.1126/science.1147555) [DOI] [PubMed] [Google Scholar]
- 16.Murphy W. J., Pringle T. H., Crider T. A., Springer M. S., Miller W. 2007. Using genomic data to unravel the root of the placental mammal tree. Genome Res. 17, 413–421 10.1101/gr.5918807 (doi:10.1101/gr.5918807) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beck R. M. 2008. A dated phylogeny of marsupials using a molecular supermatrix and multiple fossil constraints. J. Mammal. 89, 175–189 10.1644/06-MAMM-A-437.1 (doi:10.1644/06-MAMM-A-437.1) [DOI] [Google Scholar]
- 18.Meredith R. W., Westerman M., Springer M. S. 2008. A timescale and phylogeny for ‘bandicoots’ (Peramelemorphia: Marsupialia) based on sequences for five nuclear genes. Mol. Phylogenet. Evol. 47, 1–20 10.1016/j.ympev.2008.01.002 (doi:10.1016/j.ympev.2008.01.002) [DOI] [PubMed] [Google Scholar]
- 19.Meredith R. W., Westerman M., Springer M. S. 2008. Phylogeny and timescale for the living genera of kangaroos and kin (Macropodiformes: Marsupialia) based on nuclear sequences. Aust. J. Zool. 56, 395–410 10.1071/ZO08044 (doi:10.1071/ZO08044) [DOI] [Google Scholar]
- 20.Meredith R. W., Westerman M., Springer M. S. 2009. A phylogeny of Diprotodontia (Marsupialia) based on sequences for five nuclear genes. Mol. Phylogenet. Evol. 51, 554–571 10.1016/j.ympev.2009.02.009 (doi:10.1016/j.ympev.2009.02.009) [DOI] [PubMed] [Google Scholar]
- 21.Meredith R. W., Krajewski C., Westerman W., Springer M. S. 2009. Relationships and divergence times among the orders and families of marsupials. Mus. N. Ariz. Bull. 65, 383–406 [Google Scholar]
- 22.Chatterjee H. J., Ho S. W. Y., Barnes I., Groves C. 2009. Estimating the phylogeny and divergence times of primates using a supermatrix approach. BMC Evol. Biol. 9, 259. 10.1186/1471-2148-9-259 (doi:10.1186/1471-2148-9-259) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eizirik E., Murphy W. J., Koepfli K. P., Johnson W. E., Dragoo J. W., Wayne R. K., O'Brien S. J. 2010. Pattern and timing of diversification of the mammalian order Carnivora inferred from multiple nuclear gene sequences. Mol. Phylogenet. Evol. 56, 49–63 10.1016/j.ympev.2010.01.033 (doi:10.1016/j.ympev.2010.01.033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanderson M. J. 1997. A nonparametric approach to estimating divergence times in the absence of rate constancy. Mol. Biol. Evol. 14, 1218–1231 [Google Scholar]
- 25.Sanderson M. J. 2002. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol. Biol. Evol. 19, 101–109 [DOI] [PubMed] [Google Scholar]
- 26.Thorne J. L., Kishino H. 2002. Divergence time and evolutionary rate estimation with multilocus data. Syst. Biol. 51, 689–702 10.1080/10635150290102456 (doi:10.1080/10635150290102456) [DOI] [PubMed] [Google Scholar]
- 27.Yang Z., Rannala B. 2006. Bayesian estimation of species divergence times under a molecular clock using multiple fossil calibrations with soft bounds. Mol. Biol. Evol. 23, 212–226 10.1093/molbev/msj024 (doi:10.1093/molbev/msj024) [DOI] [PubMed] [Google Scholar]
- 28.Battistuzzi F. U., Filipski A., Hedges S. B., Kumar S. 2010. Performance of relaxed-clock methods in estimating evolutionary divergence times and their credibility intervals. Mol. Biol. Evol. 27, 1289–1300 10.1093/molbev/msq014 (doi:10.1093/molbev/msq014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown R. P., Yang Z. 2010. Bayesian dating of shallow phylogenies with a relaxed molecular clock. Syst. Biol. 59, 119–131 10.1093/sysbio/syp082 (doi:10.1093/sysbio/syp082) [DOI] [PubMed] [Google Scholar]
- 30.Inoue J., Donoghue P. C. J., Yang Z. 2010. The impact of the representation of fossil calibrations on Bayesian estimation of species divergence times. Syst. Biol. 59, 74–89 10.1093/sysbio/syp078 (doi:10.1093/sysbio/syp078) [DOI] [PubMed] [Google Scholar]
- 31.Morrone J. J., Crisci J. V. 1995. Historical biogeography: introduction to methods. Annu. Rev. Ecol. Syst. 26, 373–401 10.1146/annurev.es.26.110195.002105 (doi:10.1146/annurev.es.26.110195.002105) [DOI] [Google Scholar]
- 32.Simpson G. G. 1965. The geography of evolution: collected essays. Philadelphia/New York, PA/NY: Chilton Books [Google Scholar]
- 33.Ronquist F. 1997. Dispersal–vicariance analysis: a new approach to the quantification of historical biogeography. Syst. Biol. 45, 195–203 10.1093/sysbio/46.1.195 (doi:10.1093/sysbio/46.1.195) [DOI] [Google Scholar]
- 34.Wen J., Xiang Q.-Y., Qian H., Li J., Want X.-W., Ickert-Bond S. M. Intercontinental and intracontinental biogeography—patterns and methods. J. Syst. Evol. 4, 327–329 [Google Scholar]
- 35.Nylander J. A. A., Olsson U., Alström P., Sanmartín I. 2008. Accounting for phylogenetic uncertainty in biogeography: a Bayesian approach to dispersal–vicariance analysis of the thrushes (Aves: Turdus). Syst. Biol. 57, 257–268 10.1080/10635150802044003 (doi:10.1080/10635150802044003) [DOI] [PubMed] [Google Scholar]
- 36.Ree R. H., Moore B. R., Webb C. O., Donoghue M. J. 2005. A likelihood framework for inferring the evolution of geographic range on phylogenetic trees. Evolution 59, 2299–2311 [PubMed] [Google Scholar]
- 37.Ree R. H., Smith S. A. 2008. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst. Biol. 57, 4–14 10.1080/10635150701883881 (doi:10.1080/10635150701883881) [DOI] [PubMed] [Google Scholar]
- 38.Hardy C. R., Linder H. P. 2005. Intraspecific variability and timing in ancestral ecology reconstruction: a test case from the Cape flora. Syst. Biol. 54, 299–316 10.1080/10635150590923317 (doi:10.1080/10635150590923317) [DOI] [PubMed] [Google Scholar]
- 39.Maddison W. P., Maddison D. R. 1992. MacClade version 3: analysis of phylogeny and character evolution Sunderland, MA: Sinauer Associates [Google Scholar]
- 40.Clark J. R., Ree R. H., Alfaro M. E., King M. G., Wagner W. L., Roalson E. H. 2008. A comparative study in ancestral range reconstruction methods: retracing the uncertain histories of insular lineages. Syst. Biol. 57, 693–707 10.1080/10635150802426473 (doi:10.1080/10635150802426473) [DOI] [PubMed] [Google Scholar]
- 41.Patterson C. 1982. Morphological characters and homology. In Problems of phylogenetic reconstruction (eds Joysey K. A., Friday A. E.), pp. 21–74 London, UK: Academic Press [Google Scholar]
- 42.Ree R. H., Sanmartín I. 2009. Prospects and challenges for parametric models in historical biogeographical inference. J. Biogeogr. 36, 1211–1220 10.1111/j.1365-2699.2008.02068.x (doi:10.1111/j.1365-2699.2008.02068.x) [DOI] [Google Scholar]
- 43.Lamm K. S., Redelings B. D. 2009. Reconstructing ancestral ranges in historical biogeography: properties and prospects. J. Syst. Evol. 47, 369–382 10.1111/j.1759-6831.2009.00042.x (doi:10.1111/j.1759-6831.2009.00042.x) [DOI] [Google Scholar]
- 44.Maddison W. P., Maddison D. R.Mesquite: a modular system for evolutionary analysis, version 2.72. 2009. See http://mesquiteproject.org .
- 45.Wojcicki M., Brooks D. R. 2005. PACT: an efficient and powerful algorithm for generating area cladograms. J. Biogeogr 32, 755–774 10.1111/j.1365-2699.2004.01148.x (doi:10.1111/j.1365-2699.2004.01148.x) [DOI] [Google Scholar]
- 46.Lim B. K. 2008. Historical biogeography of New World emballonurid bats (Tribe Diclidurini): taxon pulse diversification. J. Biogeogr. 35, 1385–1401 10.1111/j.1365-2699.2008.01888.x (doi:10.1111/j.1365-2699.2008.01888.x) [DOI] [Google Scholar]
- 47.Lim B. K. 2009. Review of the origins and biogeography of bats in South America. Chiroptera Neotropical 15, 391–410 [Google Scholar]
- 48.Donoghue M. J., Moore B. R. 2003. Toward an integrative historical biogeography. J. Int. Comp. Biol. 43, 261–270 10.1093/icb/43.2.261 (doi:10.1093/icb/43.2.261) [DOI] [PubMed] [Google Scholar]
- 49.Krause D. W., O'Connor P. M., Rogers K. C., Sampson S. D., Buckley G. A., Rogers R. R. 2006. Late Cretaceous terrestrial vertebrates from Madagascar: implications for Latin American biogeography. Ann. Mo. Bot. Gard. 93, 178–208 10.3417/0026-6493(2006)93[178:LCTVFM]2.0.CO;2 (doi:10.3417/0026-6493(2006)93[178:LCTVFM]2.0.CO;2) [DOI] [Google Scholar]
- 50.Sereno P. C., Wilson J. A., Conrad J. L. 2004. New dinosaurs link southern landmasses in the Mid-Cretaceous. Proc. R. Soc. Lond. B 271, 1325–1330 10.1098/rspb.2004.2692 (doi:10.1098/rspb.2004.2692) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Novacek M. J. 1992. Mammalian phylogeny: shaking the tree. Nature 356, 121–125 10.1038/356121a0 (doi:10.1038/356121a0) [DOI] [PubMed] [Google Scholar]
- 52.Novacek M. J. 1993. Reflections on higher mammalian phylogenetics. J. Mamm. Evol. 1, 1064–7554 [Google Scholar]
- 53.Springer M. S., Cleven G. C., Madsen O., de Jong W. W., Waddell V. G., Amrine H. M., Stanhope M. J. 1997. Endemic African mammals shake the phylogenetic tree. Nature 388, 61–64 10.1038/40386 (doi:10.1038/40386) [DOI] [PubMed] [Google Scholar]
- 54.Springer M. S., Murphy W. J., Eizirik E., O'Brien S. J. 2005. Molecular evidence for major placental clades. In The rise of placental mammals: origins and relationships of the major extant clades (eds Rose K. D., Archibald J. D.), pp. 37–49 Baltimore, MD: Johns Hopkins University Press [Google Scholar]
- 55.Stanhope M. J., Madsen O., Waddell V. G., Cleven G. C., de Jong W. W., Springer M. S. 1998. Highly congruent molecular support for a diverse superordinal clade of endemic African mammals. Mol. Phylogenet. Evol. 9, 501–508 10.1006/mpev.1998.0517 (doi:10.1006/mpev.1998.0517) [DOI] [PubMed] [Google Scholar]
- 56.Stanhope M. J., Waddell V. G., Madsen O., de Jong W. W., Hedges S. B., Cleven G. C., Kao D., Springer M. S. 1998. Molecular evidence for multiple origins of Insectivora and for a new order of endemic African insectivore mammals. Proc. Natl Acad. Sci. USA 95, 9967–9972 10.1073/pnas.95.17.9967 (doi:10.1073/pnas.95.17.9967) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waddell P., Okada N., Hasegawa M. 1999. Towards resolving the interordinal relationships of placental mammals. Syst. Biol. 48, 1–5 10.1093/sysbio/48.1.1 (doi:10.1093/sysbio/48.1.1) [DOI] [PubMed] [Google Scholar]
- 58.Waddell P. J., Kishino H., Ota R. 2001. A phylogenetic foundation for comparative mammalian genomics. Genome Inform. 12, 141–154 [PubMed] [Google Scholar]
- 59.Eizirik E., Murphy W. J., O'Brien S. J. 2001. Molecular dating and biogeography of the early placental mammal radiation. J. Hered. 92, 212–219 10.1093/jhered/92.2.212 (doi:10.1093/jhered/92.2.212) [DOI] [PubMed] [Google Scholar]
- 60.Madsen O., et al. 2001. Parallel adaptive radiations in two major clades of placental mammals. Nature 409, 610–614 10.1038/35054544 (doi:10.1038/35054544) [DOI] [PubMed] [Google Scholar]
- 61.Murphy W. J., Eizirik E., Johnson W. E., Zhang Y. P., Ryder O. A., O'Brien S. J. 2001. Molecular phylogenetics and the origins of placental mammals. Nature 409, 614–618 10.1038/35054550 (doi:10.1038/35054550) [DOI] [PubMed] [Google Scholar]
- 62.Murphy W. J., et al. 2001. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science 294, 2348–2351 10.1126/science.1067179 (doi:10.1126/science.1067179) [DOI] [PubMed] [Google Scholar]
- 63.Scally M., Madsen O., Douady C. J., de Jong W. W., Stanhope M. J., Springer M. S. 2001. Molecular evidence for the major clades of placental mammals. J. Mamm. Evol. 8, 239–277 10.1023/A:1014446915393 (doi:10.1023/A:1014446915393) [DOI] [PubMed] [Google Scholar]
- 64.Springer M. S., de Jong W. W. 2001. Which mammalian supertree to bark up? Science 291, 1709–1711 [DOI] [PubMed] [Google Scholar]
- 65.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analysis with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 10.1093/bioinformatics/btl446 (doi:10.1093/bioinformatics/btl446) [DOI] [PubMed] [Google Scholar]
- 66.Benton M. J., Donoghue P. C. J. 2007. Paleontological evidence to date the tree of life. Mol. Biol. Evol. 24, 26–53 10.1093/molbev/msl150 (doi:10.1093/molbev/msl150) [DOI] [PubMed] [Google Scholar]
- 67.Reisz R. R., Müller J. 2004. Molecular timescales and the fossil record: a paleontological perspective. Trends Genet. 20, 237–241 10.1016/j.tig.2004.03.007 (doi:10.1016/j.tig.2004.03.007) [DOI] [PubMed] [Google Scholar]
- 68.Müller J., Reisz R. R. 2005. Four well-constrained calibration points from the vertebrate fossil record for molecular clock estimates. BioEssays 27, 1069–1075 10.1002/bies.20286 (doi:10.1002/bies.20286) [DOI] [PubMed] [Google Scholar]
- 69.Gradstein F. M., Ogg J. G. 2009. The geologic time scale. In The timetree of life (eds Hedges S. B., Kumar S.), pp. 26–34 Oxford, UK: Oxford University Press [Google Scholar]
- 70.Gheerbrant E. 2009. Paleocene emergence of elephant relatives and the rapid radiation of African ungulates. Proc. Natl Acad. Sci. USA 106, 10717–10721 10.1073/pnas.0900251106 (doi:10.1073/pnas.0900251106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bergqvist L. P., Abrantes É. A. L., Avilla L. D. S. 2004. The Xenarthra (Mammalia) of São José de Itaboraí Basin (upper Paleocene, Itaboraian), Rio de Janeiro, Brazil. Geodiversitas 26, 323–337 [Google Scholar]
- 72.Carlini A. A., Pascual R., Reguero M. A., Scillato-Yané G. J., Tonni E. P., Vizcaíno S. F. 1990. The first Paleogene land placental mammal from Antarctica: its paleoclimatic and paleobiogeographical bearings. In Abstracts IV International Congress of Systematic and Evolutionary Biology (eds Cox B., Reveal J.), 325 p. Baltimore, MD: University of Maryland [Google Scholar]
- 73.Vizcaíno S. F., Scillato-Yané G. J. 1995. An Eocene Tardigrada (Mammalia, Xenarthra) from Seymour Island, Antarctica. Antarctic Sci. 7, 407–408 [Google Scholar]
- 74.Benton M. J., Donoghue P. C. J., Asher R. J. 2009. Calibrating and constraining molecular clocks. In The timetree of life (eds Hedges S. B., Kumar S.), pp. 35–86 Oxford, UK: Oxford University Press [Google Scholar]
- 75.Flynn J. J. 1996. Carnivoran phylogeny and rates of evolution: morphological, taxonomic, and molecular. In Carnivore behavior, ecology, and evolution, vol. 2 (ed. Gittleman J. L.), pp. 542–581 Ithaca, NY: Cornell University Press [Google Scholar]
- 76.Hunt R. M., Jr, Tedford R. H. 1993. Phylogenetic relationships within the aeluroid Carnivora and implications of their temporal and geographic distribution. In Mammal phylogeny: placentals, vol. 2 (eds Szalay F. S., Novacek M. J., McKenna M. C.), pp. 53–74 Berlin, Germany: Springer [Google Scholar]
- 77.Wesley-Hunt G. D., Flynn J. J. 2005. Phylogeny of the Carnivora: basal relationships among the carnivoramorphans, and assessment of the position of ‘Miacoidea’ relative to Carnivora. J. Syst. Palaeontol. 3, 1–28 10.1017/S1477201904001518 (doi:10.1017/S1477201904001518) [DOI] [Google Scholar]
- 78.Woodburne M. O., Gunnell G. F., Stucky R. K. 2009. Climate directly influences Eocene mammal faunal dynamics in North America. Proc. Natl Acad. Sci. USA 106, 13 399–13 403 10.1073/pnas.0906802106 (doi:10.1073/pnas.0906802106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rose K. D., DeLeon V. B., Missiaen P., Rana R. S., Sahni A., Singh L., Smith T. 2008. Early Eocene lagomorph (Mammalia) from Western India and the early diversification of Lagomorpha. Proc. R. Soc. B 275, 1203–1208 10.1098/rspb.2007.1661 (doi:10.1098/rspb.2007.1661) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Storch G., Seiffert C. 2007. Extraordinarily preserved specimen of the oldest known glirid from the middle Eocene of Messel (Rodentia). J. Vertebr. Paleontol. 27, 189–194 10.1671/0272-4634(2007)27[189:EPSOTO]2.0.CO;2 (doi:10.1671/0272-4634(2007)27[189:EPSOTO]2.0.CO;2) [DOI] [Google Scholar]
- 81.Sallam H. M., Seiffert E. R., Steiper M. E., Simons E. L. 2009. Fossil and molecular evidence constrain scenarios for the early evolutionary and biogeographic history of hystricognathous rodents. Proc. Natl Acad. Sci. USA 106, 16 722–16 727 10.1073/pnas.0908702106 (doi:10.1073/pnas.0908702106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hartenberger L. 1998. Description of the radiation of the Rodentia (Mammalia) from the Late Paleocene to the Miocene; phylogenetic consequences. C. R. Acad. Sci. II A 326, 439–444 [Google Scholar]
- 83.Vucetich M. G., Verzi D. H., Hartenberger L. 1999. Review and analysis of the radiation of the South American Hystricognathi (Mammalia, Rodentia). C. R. Acad. Sci. II A 329, 763–769 [Google Scholar]
- 84.Flynn L. J., Jacobs L. L. 2008. Castoridea. In Evolution of tertiary mammals of North America: small mammals, xenarthrans, and marine mammals (eds Janis C. M., Gunnell G. F., Uhen M. D.), vol. 2, pp. 391–405 Cambridge, UK: Cambridge University Press [Google Scholar]
- 85.Marenssi S. A., Reguero M. A., Santillana S. N., Vizcaíno S. F. 1994. Eocene land mammals from Seymour Island, Antarctica: paleobiogeographical implications. Antarctic Sci. 6, 3–15 10.1017/S0954102094000027 (doi:10.1017/S0954102094000027) [DOI] [Google Scholar]
- 86.MacPhee R. D. E., Reguero M. A. 2010. Reinterpretation of a middle Eocene record of Tardigrada (Pilosa, Xenarthra, Mammalia) from La Meseta Formation, Seymour Island, West Antarctica. Am. Mus. Novit. 3689, 1–21 10.1206/703.1 (doi:10.1206/703.1) [DOI] [Google Scholar]
- 87.McKenna M. C., Bell S. K. 1997. Classification of mammals above the species level. New York, NY: Columbia University Press [Google Scholar]
- 88.Gaudin T. J., Branham D. G. 1998. The phylogeny of the Myrmecophagidae (Mammalia, Xenarthra, Vermilingua) and relationship of Eurotamandua to the Vermilingua. J. Mamm. Evol. 5, 237–265 10.1023/A:1020512529767 (doi:10.1023/A:1020512529767) [DOI] [Google Scholar]
- 89.Galliari F. C., Carlini A. A., Sanchez-Villagra M. R. 2010. Evolution of the axial skeleton in armadillos (Mammalia, Dasypodidae). Mamm. Biol. 75, 326–333 10.1016/j.mambio.2009.03.014 (doi:10.1016/j.mambio.2009.03.014) [DOI] [Google Scholar]
- 90.Poljak S., Confalonieri V., Fasanella M., Gabrielli M., Lizarralde M. S. 2010. Phylogeography of the armadillo Chaetophractus villosus (Dasypodidae Xenarthra): post-glacial range expansion from Pampas to Patagonia (Argentina). Mol. Phylogenet. Evol. 55, 38–46 10.1016/j.ympev.2009.12.021 (doi:10.1016/j.ympev.2009.12.021) [DOI] [PubMed] [Google Scholar]
- 91.Sigé B., Crochet J.-Y., Insole A. 1977. Les plus vielles taupes. Geobios Mem. Spec. 1, 141–157 10.1016/S0016-6995(77)80014-4 (doi:10.1016/S0016-6995(77)80014-4) [DOI] [Google Scholar]
- 92.Gunnell G. F., Bown T. M., Hutchinson J. H., Bloch J. I. 2008. Lipotyphla. In Evolution of Tertiary mammals of North America: small mammals, xenarthrans, and marine mammals, vol. 2 (eds Janis C. M., Gunnell G. F., Uhen M. D.), pp. 89–125 Cambridge, UK: Cambridge University Press [Google Scholar]
- 93.Seiffert E. R., Simons E. L., Ryan T. M., Bown T. M., Attia Y. 2007. New remains of Eocene and Oligocene Afrosoricida (Afrotheria) from Egypt, with implications for the origin(s) of afrosoricid zalambdodonty. J. Vertebr. Paleontol. 27, 963–972 10.1671/0272-4634(2007)27[963:NROEAO]2.0.CO;2 (doi:10.1671/0272-4634(2007)27[963:NROEAO]2.0.CO;2) [DOI] [Google Scholar]
- 94.Tabuce R., Asher R. J., Lehmann T. 2008. Afrotherian mammals: a review of current data. Mammalia 72, 2–14 10.1515/MAMM.2008.004 (doi:10.1515/MAMM.2008.004) [DOI] [Google Scholar]
- 95.Butler P. M. 1995. Fossil Macroscelidea. Mammal Rev. 25, 3–14 10.1111/j.1365-2907.1995.tb00432.x (doi:10.1111/j.1365-2907.1995.tb00432.x) [DOI] [Google Scholar]
- 96.Milledge S. 2003. Fossil aardvarks from the Lothagam beds. In Lothagam: the dawn of humanity in Eastern Africa (eds Leakey J., Harris J.), pp. 363–368 New York, NY: Columbia University Press [Google Scholar]
- 97.Flynn L. J., Jacobs L. L. 2008. Aplodontia. In Evolution of tertiary mammals of North America: small mammals, xenarthrans, and marine mammals, vol. 2 (eds Janis C. M., Gunnell G. F., Uhen M. D.), pp. 377–390 Cambridge, UK: Cambridge University Press [Google Scholar]
- 98.Marivaux L., Ducrocq S., Jaeger J.-J., Marandat B., Sudre J., Chaimanee Y., Tun S. T., Htoon W., Soe A. N. 2005. New remains of Pondaungimys anomaluropsis (Rodentia, Anomaluroidea) from the latest middle Eocene Pondaung Formation of Central Myanmar. J. Vertebr. Paleontol. 25, 214–227 10.1671/0272-4634(2005)025[0214:NROPAR]2.0.CO;2 (doi:10.1671/0272-4634(2005)025[0214:NROPAR]2.0.CO;2) [DOI] [Google Scholar]
- 99.Flynn L. J., Lindsay E. H., Martin R. A. 2008. Geomorpha. In Evolution of Tertiary mammals of North America: small mammals, xenarthrans, and marine mammals, vol. 2 (eds Janis C. M., Gunnell G. F., Uhen M. D.), pp. 428–455 Cambridge, UK: Cambridge University Press [Google Scholar]
- 100.Antoine P.-O., et al. 2007. The middle Miocene (Laventan) Fitzgcarrald Fauna, Amazonian Peru. In Proc. 4th Eur. Meeting on the Palaeontology and Stratigraphy of Latin America (eds Díaz-Martínez E., Rábano I.), pp. 19–24 Madrid, Spain: Instituto Geológico y Minero de España [Google Scholar]
- 101.Deschamps C. M., Olivares A. I., Vieytes E. C., Vucetich M. G. 2007. Ontogeny and diversity of the oldest capybaras (Rodentia: Hydrochoeridae; late Miocene of Argentina). J. Vertebr. Paleontol. 27, 683–692 10.1671/0272-4634(2007)27[683:OADOTO]2.0.CO;2 (doi:10.1671/0272-4634(2007)27[683:OADOTO]2.0.CO;2) [DOI] [Google Scholar]
- 102.Frailey C. D., Campbell K. E. 2004. The rodents of the Santa Rosa Local Fauna. In The Paleogene mammalian fauna of Santa Rosa, Amazonian Peru (ed. Campbell K. E., Jr), pp. 1–130 Los Angeles, CA: Natural History Museum of Los Angeles County; Science Series 40 [Google Scholar]
- 103.Martin T. 2004. Evolution of incisor enamel microstructure in Lagomorpha. J. Vertebr. Paleontol. 24, 411–426 10.1671/2513 (doi:10.1671/2513) [DOI] [Google Scholar]
- 104.Lopez-Martinez N. 2008. The lagomorph fossil record and the origin of the European rabbit. In Lagomorph biology: evolution, ecology, and conservation (eds Alves P. C., Ferrand N., Hackländer K.), pp. 26–47 Amsterdam, The Netherlands: Springer [Google Scholar]
- 105.Marivaux L., Bocat L., Chaimanee Y., Jaeger J.-J., Marandat B., Srisuk P., Tafforeau P., Yamee C., Welcomme L. 2006. Cynocephalid dermopterans from the Palaeogene of South Asia (Thailand, Myanmar and Pakistan): systematic, evolutionary and palaeobiogeographic implications. Zool. Scripta 35, 395–420 10.1111/j.1463-6409.2006.00235.x (doi:10.1111/j.1463-6409.2006.00235.x) [DOI] [Google Scholar]
- 106.Tong Y. 1988. Fossil tree shrews from the Eocene Hetaoyuan Formation of Xichuan, Henan. Vertebrata Palasiatica 26, 214–220 [Google Scholar]
- 107.Godfrey L. R., Jungers W. L. 2002. Quaternary fossil lemurs. In The primate fossil record (ed. Hartwig W. C.), pp. 97–121 Cambridge, UK: Cambridge University Press [Google Scholar]
- 108.Bajpai S., Kay R. F., Williams B. A., Das D. P., Kapur V. V., Tiwari B. N. 2008. The oldest Asian record of Anthropoidea. Proc. Natl Acad. Sci. USA 105, 11 093–11 098 10.1073/pnas.0804159105 (doi:10.1073/pnas.0804159105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Beard K. C., Qi T., Dawson M. R., Wang B., Li C. 1994. A diverse new primate fauna from middle Eocene fissure-fillings in southeastern China. Nature 368, 604–609 10.1038/368604a0 (doi:10.1038/368604a0) [DOI] [PubMed] [Google Scholar]
- 110.Orliac M., Boisserie J.-R., MacLatchy L., Lihoreau F. 2010. Early Miocene hippopotamids (Cetartiodactyla) constrain the phylogenetic and spatiotemporal settings of hippopotamid origin. Proc. Natl Acad. Sci. USA 107, 11 871–11 876 10.1073/pnas.1001373107 (doi:10.1073/pnas.1001373107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Honey J. G., Harrison J. A., Prothero D. R., Stevens M. S. 1998. Camelidae. In Evolution of Tertiary mammals of North America: terrestrial carnivores, ungulates, and ungulatelike mammals, vol. 1 (eds Janis C. M., Scott K. M., Jacobs L. L.), pp. 439–462 Cambridge, UK: Cambridge University Press [Google Scholar]
- 112.Métais G., Vislobokova I. 2008. Basal ruminants. In The evolution of artiodactyls (eds Prothero D. R., Foss S. E.), pp. 189–212 Baltimore, MD: Johns Hopkins University Press [Google Scholar]
- 113.Harris J. M., Li-Ping L. 2008. Superfamily Suoidea. In The evolution of artiodactyls (eds Prothero D. R., Foss S. E.), pp. 130–150 Baltimore, MD: Johns Hopkins University Press [Google Scholar]
- 114.Bowen G. J., Clyde W. C., Koch P. L., Ting S., Alroy J., Tsubamoto T., Wang Y., Wang Y. 2002. Mammalian dispersal at the Paleocene/Eocene boundary. Science 295, 2062–2065 10.1126/science.1068700 (doi:10.1126/science.1068700) [DOI] [PubMed] [Google Scholar]
- 115.Smith T., Rose K. D., Gingerich P. D. 2006. Rapid Asia-Europe-North America geographic dispersal of earliest Eocene primate Teilhardina during the Paleocene–Eocene thermal maximum. Proc. Natl Acad. Sci. USA 103, 11223–11227 10.1073/pnas.0511296103 (doi:10.1073/pnas.0511296103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Prothero D. R. 1998. Hyracodontidae. In Evolution of Tertiary mammals of North America: terrestrial carnivores, ungulates, and ungulatelike mammals, vol. 1 (eds Janis C. M., Scott K. M., Jacobs L. L.), pp. 589–594 Cambridge, UK: Cambridge University Press [Google Scholar]
- 117.Colbert M. W., Schoch R. M. 1998. Tapiroidea and other moropomorphs. In Evolution of Tertiary mammals of North America: terrestrial carnivores, ungulates, and ungulatelike mammals, vol. 1 (eds Janis C. M., Scott K. M., Jacobs L. L.) pp. 569–582 Cambridge, UK: Cambridge University Press [Google Scholar]
- 118.Dashzeveg D. 1996. Some carnivorous mammals from the Paleogene of the Eastern Gobi Desert, Mongolia, and the application of Oligocene carnivores to stratigraphic correlation. Am. Mus. Novit. 3179, 1–14 [Google Scholar]
- 119.Hunt R. M., Jr 1998. Evolution of the aeluroid Carnivora: diversity of the earliest aeluroids from Eurasia (Quercy, Hsanda-Gol) and the origin of felids. Am. Mus. Novit. 3252, 1–65 [Google Scholar]
- 120.Spaulding M., Flynn J. J. 2009. Anatomy of the postcranial skeleton of ‘Miacis’ uintensis (Mammalia: Carnivoramorpha). J. Vertebr. Paleontol. 29, 1212–1223 10.1671/039.029.0408 (doi:10.1671/039.029.0408) [DOI] [Google Scholar]
- 121.Storch G. 2003. Fossil Old World ‘edentates’. In Morphological studies in fossil and extant Xenarthra (Mammalia), Senckenbergiana biologica 83 (eds Fariña R. A., Vizcaíno S. F., Storch G.), pp. 51–60 Germany: Schweizerbart Science Publishers [Google Scholar]
- 122.Huelsenbeck J. P., Ronquist F. 2001. MrBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 10.1093/bioinformatics/17.8.754 (doi:10.1093/bioinformatics/17.8.754) [DOI] [PubMed] [Google Scholar]
- 123.Ronquist F., Huelsenbeck J. P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 10.1093/bioinformatics/btg180 (doi:10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- 124.Carroll R. L. 1988. Vertebrate paleontology and evolution. New York, NY: W. H. Freeman and Company [Google Scholar]
- 125.Asher R. J., Novacek M. J., Geisler J. G. 2003. Relationships of endemic African mammals and their fossil relatives based on morphological and molecular evidence. J. Mamm. Evol. 10, 131–194 10.1023/A:1025504124129 (doi:10.1023/A:1025504124129) [DOI] [Google Scholar]
- 126.Zack S. P., Penkrot T. A., Bloch J. I., Rose K. D. 2005. Affinities of ‘hyopsodontids’ to elephant shrews and a Holarctic origin of Afrotheria. Nature 434, 497–501 10.1038/nature03351 (doi:10.1038/nature03351) [DOI] [PubMed] [Google Scholar]
- 127.Tabuce R., Marivaux L., Adaci M., Bensalah M., Hartenberger J.-L., Mahboudi M., Mebrouk F., Tafforeau P., Jaeger J. 2007. Early Tertiary mammals from North Africa reinforce the molecular Afrotheria clade. Proc. R. Soc. Lond. B 274, 1159–1166 10.1098/rspb.2006.0229 (doi:10.1098/rspb.2006.0229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Marshall L. G., Webb S. D., Sepkowski J. J., Raup D. M. 1982. Mammalian evolution and the Great American Interchange. Science 215, 1351–1357 10.1126/science.215.4538.1351 (doi:10.1126/science.215.4538.1351) [DOI] [PubMed] [Google Scholar]
- 129.Simpson G. G. 1951. History of the fauna of Latin America. In Science in progress, 7th series (ed. Baitsell G. A.), pp. 369–408 New Haven, CT: Yale University Press [Google Scholar]
- 130.Simpson G. G. 1978. Early mammals in South America: fact, controversy, and mystery. Proc. Am. Phil. Soc. 122, 318–328 [Google Scholar]
- 131.Archibald J. D. 1996. Fossil evidence for a Late Cretaceous origin of ‘hoofed’ mammals. Science 272, 1150–1153 10.1126/science.272.5265.1150 (doi:10.1126/science.272.5265.1150) [DOI] [PubMed] [Google Scholar]
- 132.Archibald J. D., Averianov A. O., Ekdale E. G. 2001. Oldest relatives of rabbits, rodents, and other extant eutherian mammals. Nature 414, 62–65 10.1038/35102048 (doi:10.1038/35102048) [DOI] [PubMed] [Google Scholar]
- 133.Cifelli R. L., Davis B. M. 2003. Marsupial origins. Science 302, 1899–1900 10.1126/science.1092272 (doi:10.1126/science.1092272) [DOI] [PubMed] [Google Scholar]
- 134.Davis B. M., Cifelli R. L., Kielan-Jaworowska Z. 2008. Earliest evidence of Deltatheroida (Mammalia: Metatheria) from the Early Cretaceous of North America. In Mammalian evolutionary morphology: a tribute to Frederick S. Szalay (eds Sargis E. J., Dagosto M.), pp. 3–24 Amsterdam, The Netherlands: Springer [Google Scholar]
- 135.Boyer D. M., Prasad G. V. R., Krause D. W., Godinot M., Goswami A., Verma O., Flynn J. J. 2010. New postcrania of Deccanolestes from the Late Cretaceous of India and their bearing on the evolutionary and biogeographic history of euarchontan mammals. Naturwissenschaften 97, 365–377 10.1007/s00114-010-0648-0 (doi:10.1007/s00114-010-0648-0) [DOI] [PubMed] [Google Scholar]
- 136.Nikolaev S., Montoya-Burgos J. I., Margulies E. H., Program N. C. S., Rougemont J., Nyffeler B., Antonarakis S. E. 2007. Early history of mammals is elucidated with the ENCODE multiple species sequencing data. PLoS Genet. 3, e2. 10.1371/journal.pgen.0030002 (doi:10.1371/journal.pgen.0030002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.van Dijk M. A. M., Paradis E., Catzeflis F., de Jong W. W. 1999. The virtues of gaps: xenarthran (edentate) monophyly supported by a unique deletion in αA-crystallin. Syst. Biol. 48, 94–106 10.1080/106351599260463 (doi:10.1080/106351599260463) [DOI] [PubMed] [Google Scholar]
- 138.Nikaido M., Nishihara H., Hukumoto Y., Okada N. 2003. Ancient SINEs from African endemic mammals. Mol. Biol. Evol. 20, 522–527 10.1093/molbev/msg052 (doi:10.1093/molbev/msg052) [DOI] [PubMed] [Google Scholar]
- 139.Kriegs J. O., Churakov G., Kiefmann M., Jordan U., Brosius J., Schmitz J. 2006. Retroposed elements as archives for the evolutionary history of placental mammals. PLoS Biol. 4, e91. 10.1371/journal.pbio.0040091 (doi:10.1371/journal.pbio.0040091) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Nishihara H., Satta Y., Nikaido M., Thewissen J. G. M., Stanhope M. J., Okada N. 2005. A retroposon analysis of afrotherian phylogeny. Mol. Biol. Evol. 22, 1823–1833 10.1093/molbev/msi179 (doi:10.1093/molbev/msi179) [DOI] [PubMed] [Google Scholar]
- 141.Nishihara H., Hasegawa M., Okada N. 2006. Pegasoferae, an unexpected mammalian clade revealed by tracking ancient retroposon insertions. Proc. Natl Acad. Sci. USA 103, 9929–9934 10.1073/pnas.0603797103 (doi:10.1073/pnas.0603797103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nishihara H., Maruyamab S., Okada N. 2009. Retroposon analysis and recent geological data suggest near-simultaneous divergence of the three superorders of mammals. Proc. Natl Acad. Sci. USA 106, 5235–5240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Eagles G. 2007. New angles on South Atlantic opening. Geophys. J. Int. 166, 353–361 10.1111/j.1365-246X.2006.03206.x (doi:10.1111/j.1365-246X.2006.03206.x) [DOI] [Google Scholar]
- 144.Torsvik T. H., Rousse S., Labails C., Smethurst M. A. 2009. A new scheme for the opening of the South Atlantic and the dissection of an Aptian salt basin. Geophys. J. Int. 177, 1315–1333 10.1111/j.1365-246X.2009.04137.x (doi:10.1111/j.1365-246X.2009.04137.x) [DOI] [Google Scholar]
- 145.Nelson G. 1978. From Candolle to Croizat: comments on the history of biogeography. J. Hist. Biol. 11, 269–305 10.1007/BF00389302 (doi:10.1007/BF00389302) [DOI] [PubMed] [Google Scholar]
- 146.de Queiroz A. 2005. The resurrection of oceanic dispersal in historical biogeography. Trends Ecol. Evol. 20, 68–73 10.1016/j.tree.2004.11.006 (doi:10.1016/j.tree.2004.11.006) [DOI] [PubMed] [Google Scholar]
- 147.Censky E. J., Hodge K., Dudley J. 1998. Over-water dispersal of lizards due to hurricanes. Nature 395, 556. 10.1038/26886 (doi:10.1038/26886) [DOI] [Google Scholar]
- 148.Poux C., Madsen O., Marquard E., Vieites D. R., de Jong W. W., Vences M. 2005. Asynchronous colonization of Madagascar by the four endemic clades of primates, tenrecs, carnivores, and rodents as inferred from nuclear genes. Syst. Biol. 54, 719–730 10.1080/10635150500234534 (doi:10.1080/10635150500234534) [DOI] [PubMed] [Google Scholar]
- 149.Yoder A. D., Nowak M. D. 2006. Has vicariance or dispersal been the predominant biogeographic force in Madagascar? Only time will tell. Annu. Rev. Ecol. Evol. Syst. 37, 405–431 10.1146/annurev.ecolsys.37.091305.110239 (doi:10.1146/annurev.ecolsys.37.091305.110239) [DOI] [Google Scholar]
- 150.McCall R. 1997. Implications of recent geological investigations of the Mozambique Channel for the mammalian colonization of Madagascar. Proc. R. Soc. Lond. B 264, 663–665 10.1098/rspb.1997.0094 (doi:10.1098/rspb.1997.0094) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yoder A. D., Burns M. M., Zehr S., Delefosse T., Veron G., Goodman S. M., Flynn J. J. 2003. Single origin of Malagasy Carnivora from an African ancestor. Nature 421, 734–737 10.1038/nature01303 (doi:10.1038/nature01303) [DOI] [PubMed] [Google Scholar]
- 152.Poux C., Madsen O., Glos J., de Jong W. W., Vences M. 2008. Molecular phylogeny and divergence times of Malagasy tenrecs: influence of data partitioning and taxon sampling on dating analyses. BMC Evol. Biol. 8, 102. 10.1186/1471-2148-8-102 (doi:10.1186/1471-2148-8-102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Stankiewicz J., Thiart C., Masters J. C., de Wit M. J. 2006. Did lemurs have sweepstake tickets? An exploration of Simpson's model for the colonization of Madagascar by mammals. J. Biogeogr. 33, 221–235 10.1111/j.1365-2699.2005.01381.x (doi:10.1111/j.1365-2699.2005.01381.x) [DOI] [Google Scholar]
- 154.Ali J. R., Huber M. 2010. Mammalian biodiversity on Madagascar controlled by ocean currents. Nature 463, 653–656 10.1038/nature08706 (doi:10.1038/nature08706) [DOI] [PubMed] [Google Scholar]
- 155.Poux C., Chevret P., Huchon D., de Jong W. W., Douzery E. J. P. 2006. Arrival and diversification of caviomorph rodents and platyrrhine primates in South America. Syst. Biol. 55, 228–244 10.1080/10635150500481390 (doi:10.1080/10635150500481390) [DOI] [PubMed] [Google Scholar]
- 156.Blanga-Kanfi S., Miranda H., Penn O., Pupko T., DeBry R. W., Huchon D. 2009. Rodent phylogeny revised: analysis of six nuclear genes from all major rodent clades. BMC Evol. Biol. 9, 71. 10.1186/1471-2148-9-71 (doi:10.1186/1471-2148-9-71) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hasegawa M., Thorne J. L., Kishino H. 2003. Time scale of eutherian evolution estimated without assuming a constant rate of molecular evolution. Genes Gen. Syst. 78, 267–283 10.1266/ggs.78.267 (doi:10.1266/ggs.78.267) [DOI] [PubMed] [Google Scholar]
- 158.Rowe D. L., Dunn K. A., Adkins R. M., Honeycutt R. L. 2010. Molecular clocks keep dispersal hypotheses afloat: evidence for trans-Atlantic rafting by rodents. J. Biogeogr. 7, 305–324 [Google Scholar]
- 159.Beard K. C., Wang B., Dawson M., Huang X., Tong Y. 1996. Earliest complete dentition of an anthropoid primate from the late middle Eocene of Shanxi Province, China. Science 272, 82–85 10.1126/science.272.5258.82 (doi:10.1126/science.272.5258.82) [DOI] [Google Scholar]
- 160.Beard K. C., Wang J. 2004. The eosimiid primates (Anthropoidea) of the Heti Formation, Yuanqu Basin, Shanxi and Henan Provinces, People's Republic of China. J. Hum. Evol. 46, 401–432 10.1016/j.jhevol.2004.01.002 (doi:10.1016/j.jhevol.2004.01.002) [DOI] [PubMed] [Google Scholar]
- 161.Arnason U., Gullberg A., Schweizer B. A., Janke A. 2000. Molecular estimates for primate dispersal and the origin of modern humans. Hereditas 133, 217–228 10.1111/j.1601-5223.2000.00217.x (doi:10.1111/j.1601-5223.2000.00217.x) [DOI] [PubMed] [Google Scholar]
- 162.Lavocat R. 1969. La systématique des rongeurs hystricomorphes et la dérive des continents. C. R. Acad. Sci. Ser. D 269, 1496–1497 [PubMed] [Google Scholar]
- 163.Hussain S. T., de Bruijn H., Leinders J. M. 1978. Middle Eocene rodents from the Kala Chitta Range (Punjab, Pakistan) (III). Proc. Kon. Ned. Akad. Wetensch. Ser. B 81, 101–112 [Google Scholar]
- 164.Wood A. E. 1985. The relationships, origin and dispersal of the hystricognathous rodents. In Evolutionary relationships among rodents: a multidisciplinary analysis (eds Luckett W. P., Hartenberger J.-L.), pp. 475–513 New York, NY: Plenum [Google Scholar]
- 165.Houle A. 1999. The origin of platyrrhines: an evaluation of the Antarctic scenario and the floating island model. Am. J. Phys. Anthropol. 109, 541–559 (doi:10.1002/(SICI)1096-8644(199908)109:4<541::AID-AJPA9>3.0.CO;2-N) [DOI] [PubMed] [Google Scholar]
- 166.Tattersall I. 2005. Mechanisms of faunal origin and diversity in island environments: the case of Madagascar's mammals. Hellenic J. Geosci. 41, 35–46 [Google Scholar]
- 167.Gunnell G. F., Simmons N. B. 2005. Fossil evidence and the origin of bats. J. Mamm. Evol. 12, 209–246 10.1007/s10914-005-6945-2 (doi:10.1007/s10914-005-6945-2) [DOI] [Google Scholar]
- 168.Simmons N. B., Seymour K. L., Habersetzer J., Gunnel G. F. 2008. Primitive early Eocene bat from Wyoming and the evolution of flight and echolocation. Nature 451, 818–822 [DOI] [PubMed] [Google Scholar]
- 169.Sigé B. 1991. Rhinolophoidae et Vespertilionoidea (Chiroptera) du Chambi (Eocene inferieur de Tunisie). Aspects biostratigraphique, biogeographique and paleoecologique de l'origine des chiropters modernes. Neues Jahrb. Geol. Palaontol. Abh. 182, 355–376 [Google Scholar]
- 170.Hand S. J., Kirsch J. A. W. 1998. A southern origin for the Hipposideridae (Microchiroptera)? Evidence from the Australian fossil record. In Bats: phylogeny, morphology, echolocation, and conservation biology (eds Kunz T. H., Racey P. A.), pp. 72–90 Washington, DC: Smithsonian Institution Press [Google Scholar]
- 171.Luo Z.-X., Ji Q., Wible J. R., Yuan X. 2003. An Early Cretaceous tribosphenic mammal and metatherian evolution. Science 302, 1934–1939 10.1126/science.1090718 (doi:10.1126/science.1090718) [DOI] [PubMed] [Google Scholar]
- 172.Martin J. E., Case J. A., Jagt J. W. M., Schulp A. S., Mulder E. W. A. 2005. A new European marsupial indicates a Late Cretaceous high-latitude transatlantic dispersal route. J. Mamm. Evol. 12, 495–511 10.1007/s10914-005-7330-x (doi:10.1007/s10914-005-7330-x) [DOI] [Google Scholar]
- 173.Vullo R., Gheerbrant E., de Muizon C., Néraudeau D. 2009. The oldest modern therian mammal from Europe and its bearing on stem marsupial paleobiogeography. Proc. Natl Acad. Sci. USA 106, 19 910–19 915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Cifelli R. L. 1990. Cretaceous mammals of Southern Utah, II: marsupials and marsupial–like mammals from the Wahweap Formation (Early Campanian). J. Vertebr. Paleontol. 10, 320–331 10.1080/02724634.1990.10011817 (doi:10.1080/02724634.1990.10011817) [DOI] [Google Scholar]
- 175.Cifelli R. L. 1993. Theria of metatherian–eutherian grade and the origin of marsupials. In Mammal phylogeny Mesozoic differentiation, multituberculates, monotremes, early therians, and marsupials, vol. 1 (eds Szalay F. S., Novacek M. J., McKenna M. C.), pp. 205–215 Berlin, Germany; Springer [Google Scholar]
- 176.Cifelli R. L. 2004. Marsupial mammals from the Albian-Cenomanian (Early-Late Cretaceous) boundary, Utah. Bull. Am. Mus Nat. Hist. 285, 62–79 (doi:10.1206/0003-0090(2004)285<0062:C>2.0.CO;2) [DOI] [Google Scholar]
- 177.Wible J. R. 1990. Late Cretaceous marsupial petrosal bones from North America and a cladistic analysis of the petrosal in therian mammals. J. Vertebr. Paleontol. 10, 183–205 10.1080/02724634.1990.10011807 (doi:10.1080/02724634.1990.10011807) [DOI] [Google Scholar]
- 178.Cifelli R. L., de Muizon C. 1997. Dentition and jaw of Kokopellia juddi, a primitive marsupial or near-marsupial from the medial Cretaceous of Utah. J. Mamm. Evol. 4, 241–258 10.1023/A:1027394430433 (doi:10.1023/A:1027394430433) [DOI] [Google Scholar]
- 179.Case J. A., Goin F. J., Woodburne M. O. 2005. ‘South American’ marsupials from the Late Cretaceous of North America and the origin of marsupial cohorts. J. Mamm. Evol. 11, 223–255 10.1023/B:JOMM.0000047339.39630.82 (doi:10.1023/B:JOMM.0000047339.39630.82) [DOI] [Google Scholar]
- 180.Amrine-Madsen H., Scally M., Westerman M., Stanhope M. J., Krajewski C., Springer M. S. 2003. Nuclear gene sequences provide evidence for the monophyly of australidelphian marsupials. Mol. Phylogenet. Evol. 28, 186–196 10.1016/S1055-7903(03)00122-2 (doi:10.1016/S1055-7903(03)00122-2) [DOI] [PubMed] [Google Scholar]
- 181.Beck R. M., Godthelp H., Weisbecker V., Archer M., Hand S. J. 2008. Australia's oldest marsupial fossils and their biogeographical implications. PLoS ONE 3, e1858. 10.1371/journal.pone.0001858 (doi:10.1371/journal.pone.0001858) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Springer M. S., Krajewski C., Meredith R. W. 2009. Marsupials (Metatheria). In The timetree of life (eds Hedges S. B., Kumar S.), pp. 466–470 Oxford, UK: Oxford University Press [Google Scholar]
- 183.Kirsch J. A. W., Dickerman A. W., Reig O. A., Springer M. S. 1991. DNA hybridization evidence for the Australasian affinity of the American marsupial Dromiciops australis. Proc. Natl Acad. Sci. USA 88, 10 465–10 469 10.1073/pnas.88.23.10465 (doi:10.1073/pnas.88.23.10465) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kirsch J. A. W., Lapointe F.-J., Springer M. S. 1997. DNA-hybridisation studies of marsupials and their implications for metatherian classification. Aust. J. Zool. 45, 211–280 10.1071/ZO96030 (doi:10.1071/ZO96030) [DOI] [Google Scholar]
- 185.Springer M. S., Westerman M., Kavanagh J. R., Burk A., Woodburne M. O., Kao D., Krajewski C. 1998. The origin of the Australasian marsupial fauna and the phylogenetic affinities of the enigmatic monito del monte and marsupial mole. Proc. R. Soc. Lond. B 265, 2381–2386 10.1098/rspb.1998.0587 (doi:10.1098/rspb.1998.0587) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Nilsson M. A., Churakov G., Sommer M., Tran N. V., Zemann A., Brosius J., Schmitz J. 2010. Tracking marsupial evolution using archaic genomic retroposon insertions. PLoS Biol. 8, e1000436. 10.1371/journal.pbio.1000436 (doi:10.1371/journal.pbio.1000436) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Woodburne M. O., Case J. A. 1996. Dispersal, vicariance, and the Late Cretaceous to early Tertiary land mammal biogeography from South America to Australia. J. Mamm. Evol. 3, 121–161 10.1007/BF01454359 (doi:10.1007/BF01454359) [DOI] [Google Scholar]
- 188.Rowe T., Rich T. H., Vickers–Rich P., Springer M. S., Woodburne M. O. 2008. The oldest platypus and its bearing on divergence timing of the platypus and echidna clades. Proc. Natl Acad. Sci. USA 105, 1238–1242 10.1073/pnas.0706385105 (doi:10.1073/pnas.0706385105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Woodburne M. O., Rich T. H., Springer M. S. 2003. The evolution of tribospheny and the antiquity of mammalian clades. Mol. Phylogenet. Evol. 28, 360–385 10.1016/S1055-7903(03)00113-1 (doi:10.1016/S1055-7903(03)00113-1) [DOI] [PubMed] [Google Scholar]
- 190.Phillips M. J., Bennetta T. H., Lee M. S. Y. 2009. Molecules, morphology, and ecology indicate a recent, amphibious ancestry for echidnas. Proc. Natl Acad. Sci. USA 106, 17 089–17 094 10.1073/pnas.0904649106 (doi:10.1073/pnas.0904649106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Springer M. S., Krajewski C. 2009. Monotremes (Prototheria). In The timetree of life (eds Hedges S. B., Kumar S.), pp. 462–465 Oxford, UK: Oxford University Press [Google Scholar]
- 192.Luo Z.-X., Cifelli R. L., Kielan-Jaworowska Z. 2001. Dual origin of tribosphenic mammals. Nature 409, 53–57 10.1038/35051023 (doi:10.1038/35051023) [DOI] [PubMed] [Google Scholar]
- 193.Luo Z.-X., Cifelli R. L., Kielan-Jaworowska Z. 2002. In quest for a phylogeny of Mesozoic mammals. Acta Palaeont. Polon. 47, 1–78 [Google Scholar]
- 194.Kielan-Jaworowska Z., Cifelli R. L., Luo X. 2004. Mammals from the age of dinosaurs—origins, evolution, and structure. New York, NY: Columbia University Press [Google Scholar]