Abstract
Documenting and exploring the patterns of diversity of life on Earth has always been a central theme in biology. Species richness despite being the most commonly used measure of diversity in macroecological studies suffers from not considering the evolutionary and ecological differences among species. Phylogenetic diversity (PD) and functional diversity (FD) have been proposed as alternative measures to overcome this limitation. Although species richness, PD and FD are closely related, their relationships have never been investigated on a global scale. Comparing PD and FD with species richness corroborated the general assumptions of surrogacy of the different diversity measures. However, the analysis of the residual variance suggested that the mismatches between the diversity measures are influenced by environmental conditions. PD increased relative to species richness with increasing mean annual temperature, whereas FD decreased with decreasing seasonality relative to PD. We also show that the tropical areas are characterized by a FD deficit, a phenomenon, that suggests that in tropical areas more species can be packed into the ecological space. We discuss potential mechanisms that could have resulted in the gradient of spatial mismatch observed in the different biodiversity measures and draw parallels to local scale studies. We conclude that the use of multiple diversity measures on a global scale can help to elucidate the relative importance of historical and ecological processes shaping the present gradients in mammalian diversity.
Keywords: environmental heterogeneity, trait diversity, evolutionary history, niche conservatism
1. Introduction
Biodiversity is a concept that embraces many aspects of biological variation, ranging from genetic and taxonomic differences to phenetic diversity among species [1]. These different measures are thought to be related and, more importantly, to determine the resultant complexity of biological interactions which ultimately produce the patterns of species coexistence, productivity, nutrient cycling, decomposition and energy flow in ecosystems [2]. Most commonly, species richness (the number of species per unit area) has been used as a surrogate for all these different aspects. However, there is a growing consensus that species richness alone cannot appropriately describe the mechanisms involved in species coexistence and ecosystem processes and does not describe the differences in community structure well. Thus, using species richness as the sole measure of biodiversity may compromise our ability to understand the mechanistic basis linked to the spatial and temporal dynamics of biodiversity [3].
The inadequacy of species richness in representing the differences in evolutionary history, how communities function, and how the network of interactions within communities are organized has led to the development of alternative measures, mainly phylogenetic diversity (PD) and functional diversity (FD) [4–7]. The rationale behind the use of these alternative diversity measures is to better identify the underlying processes determining species richness and ecosystem functioning [8,9].
(a). Phylogenetic diversity
PD is a biodiversity measure that accounts for the phylogenetic relationship (hence evolutionary history) among taxa [4,10–12]. While species richness assigns equal value to all species in a community, PD provides additional value to theoretical and applied ecology [13]. This metric was initially proposed as a way of prioritising species and areas for conservation [4]. Because extinction risk is not phylogenetically random [14,15], there is, for instance, a clumping of threat towards species which are large, long-lived, slowly reproducing and with specialized habitats and high levels of endemism (e.g. [16]). Consequently, the concern over the preservation of evolutionary history highlights the importance of determining whether priority sites for species conservation are also important in respect to evolutionary history [14,17–21]. Also, there has been a recent increasing effort to bring information about evolutionary relationships of species to elucidate questions of community assembly and diversity patterns [7,10,22]. For example, large-scale assemblages of African carnivores are a non-random phylogenetic set of species from the biogeographic pool, an indication of early biome-filling radiations followed by competitive sorting within ecoregions [23]). This scenario corroborates the idea that density-dependent and ecologically limited models of clade growth are a more suitable explanation for patterns of diversity than models in which species richness is simply a function of clade age or divesification rate (see [23,24] for references). Thus, from an ecological perspective, understanding the phylogenetic structure of communities can shed light on present-day ecological interactions, as well as link community ecology with biogeography and the study of character evolution [10,22].
(b). Functional diversity
FD can be understood as a representation of how species are distributed in a multidimensional niche space defined by functional traits [6,25]. Based on the niche theory, it is easier to understand the processes related to community functioning and assembly rules using functional traits [3,26–28]. Therefore, the FD of a community will often be the most ecologically relevant biodiversity measure [29], predicting the functional consequences of changes caused by humans [30–34] and providing a mechanistic link between the composition of species assemblages and ecosystem functioning [5,6].
Although the concept of FD has received wide coverage in community ecology [25], mammalian communities, in contrast to plant communities for example, have rarely been subject to studies of FD. This lag is probably attributable to the difficulties involved in gathering quantitative measures of functional traits in mammals.
(c). Comparing species richness, phylogenetic and functional diversity
Few studies have investigated the potential large-scale gradients in FD in relationship with species richness [3,35,36]. Hence, we still have a poor understanding of the broad-scale variation in many measures of biodiversity other than species richness [37]. For example, the extent to which PD and FD exhibit latitudinal gradients similar to species richness remains to be fully investigated [3,23,36,37].
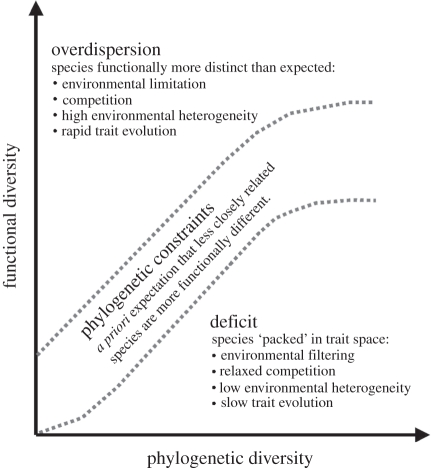
Although some relationship between PD and FD and species richness is expected, the precise shape of these relationships and their environmental determinants are still unclear [36]. Communities with the same richness level may differ both in the phylogenetic relatedness of the species as a consequence of differences in their evolutionary histories [7,38], as well as in their functional traits due to contrasting environmental conditions [3,28,35,37]. In addition, by comparing PD with FD we can investigate how evolutionary time relates to trait evolution. Following the rationale that PD represents the accumulated amount of evolutionary history and is essentially a measure of time we should be able to relate time to divergence of functional traits and thus understand how functionally and phylogenetically distinct members of communities must be to coexist (figure 1).
Figure 1.
How phylogenetic diversity (PD) and functional diversity (FD) together could inform about spatial and biophylogeographic patterns. The relationship between PD and FD is, although yet unknown, drawn as a nonlinear relationship, since studies of trait evolution suggest that for many life-history traits the tempo of trait evolution slows down after an initial boost in divergence. Therefore, we assumed that the differences in functional divergence will not follow phylogenetic divergence linearly. Overdispersion of FD characterizes communities in which species are distributed farther apart in functional niche space than expected by their PD. FD deficit in contrast describes the situation where species in a community share more functional traits than expected. And finally, evolutionary constraint describes the assumed general condition for communities where the FD of communities follows the phylogenetic divergence of the species composing the community.
Here, we explore the spatial distribution of mammal PD and FD in relation to species richness on a global scale. We investigate the relationship among these diversity measures and search for general rules for how PD, FD and species richness represent surrogates of diversity and seek to identify areas deviating from such a general relationship [38,39]. FD can be higher or lower than expected due to several non-mutually exclusive processes. Variation in the rate of trait evolution as shown, for example, for body size evolution in mammals which is faster in temperate areas [40] can cause FD to deviate locally from a global relationship. At the same time, if resources are limited, resources species need to occupy wider ecological niches in order to secure their energy demands and therefore communities would show signs of over-dispersion in functional traits (figure 1). In addition, high degrees of environmental heterogeneity could result in an over-dispersion in FD because coexisting species could adapt and specialize to the different environmental conditions. Ultimately, we want to investigate whether communities deviate from an expected relationship, and in cases where they do so, try to relate these deviances to environmental variables to understand the underlying processes determining accumulation and maintenance of biodiversity.
2. Methods
Using range distribution information for 4536 mammalian species, we derived the number and identity of mammal species for a global raster of 200 × 200 km grid size [41]. As a higher resolution of species richness based on mammal distribution range information would only increase spatial autocorrelation without a real gain in analytical performance we decided to use this particular spatial resolution. We used equal-area Mollweide projection given that different diversity measures should represent similar areas independent of their latitudinal position. Grid cells were considered occupied by those species where the grid cell centre intersected with the species' ranges. In cases where a range (or isolated sections of the entire range distribution) was smaller than a grid cell we regarded the next closest grid cell as occupied. Species richness (S) was in this case simply the number of occurrences of species per grid cell.
Using species composition on each cell, we calculated both PD and FD. We calculated the commonly used PD [4,12] using a recent mammalian phylogenetic tree [42]. PD is a continuous measure that uses the sum of branch lengths of the phylogenetic tree connecting all species within a community to assess species relatedness [4] and, thus, PD. We followed Rodrigues and Gaston's [12] advice to include in PD the length of the branch leading to the most basal node of the minimum spanning tree of each community (i.e. each grid cell) as this represents the phylogenetic history intimately connected to the community.
To calculate FD, we used body mass, diet (vertebrates, invertebrates, foliage, stems and bark, grass, fruits, seeds, flowers, nectar and pollen, roots and tubers), habit (aquatic, fossorial, ground dwelling, above ground dwelling) and activity period (diurnal, nocturnal, cathemeral, crepuscular). The traits represent many aspects of resource use, for example, the quantity and type of resources used by each species and what they do to acquire them. Thus, the available traits used here should relate to resource acquisition and define important niche dimensions of the studied species reasonably well. They also encompass the type of trait data previously used in other investigations of the FD of mammals (see, for example [3,30]). Trait information was collated from the PanTheria database [43] updated for 1900 species using several additional sources (data can be obtained from the authors upon request). Yet, where trait values were not available (for 853 species) we either used genus/family values or our own expertise. Overall, such extrapolations were more common for Rodentia and Soricomorpha.
One way to estimate species variation or dispersion in functional space is through the FD measure proposed by Petchey & Gaston [6,25]. FD is a continuous dendrogram-based measure based on the distribution of species in trait space: high FD means that species are distant in trait space (high complementarity), whereas low FD means that species are more similar and, thus, more clumped in trait space (low complementarity). Analytically, FD resembles Faith's PD since the FD of a given community will be the sum of branch lengths of the functional dendrogram necessary to connect all the species belonging to the community. FD measures diversity at all hierarchical scales simultaneously, including the small functional differences among species ignored by functional groups and the large functional differences that might delineate these groups [6]. We used Gower distance and the unweighted pair group method with arithmetic averages to produce the distance matrix and the functional dendrogram [25]. Subsequently, FD was calculated for each grid cell, using the sum of the dendrogram branches necessary to connect all species that occurred in the grid cell.
The environmental variables we used included actual evapotranspiration (AET), mean and standard deviation of monthly temperature, and elevation [44]. The data were obtained from GlobalGIS [44] originally published at a resolution of 0.5° (geographical degrees coordinate system). We decreased the resolution of the environmental variables to 2° averaging the 0.5° values using the aggregate function in the package ‘raster’ [45], available in the R statistical package 2.10 [46]. We subsequently changed the projection of the raster by applying a change of projection followed by an interpolation step to match the diversity raster with the environmental raster [45].
(a). Statistical analysis
To investigate the relationships among the diversity measures it is important to account for potential bias due to spatial autocorrelation since regression analysis assumes independence of the residuals which in a spatial framework is not necessarily given. In order to obtain unbiased estimates for the slopes of the independent variables in cases where the relationships were not linear we used generalized additive models (GAM) and included the coordinates of the grid cells as a smooth factor to account for spatial autocorrelation [47]. All GAMs used generalized cross validation to adjust the degrees of freedom for the smooth spline (the geographical coordinates) and we checked visually for normal distribution of the model residuals and lack of heteroscedascity in the correlations of the residuals against predictor values. These analyses were done using the R statistical package 2.10 [46] in conjunction with the ‘mgcv’ library [47].
For the environmental analysis, we regressed each of the multiple diversity metrics against the four environmental predictors using a standard ordinary least-squares (OLS) regression with the software package SAM [48,49]. Spatial correlograms were built using Moran's I coefficients at 15 geographical distance classes and used in the OLS [50,51]. Because residuals of all models showed very high spatial autocorrelation levels, mainly at the first distance class, we added to the OLS a set of eigenvectors extracted from the geographical distance matrix among cells, a procedure called spatial eigenvector mapping (SEVM) [52–54]. After that all Moran's I in the model residuals become smaller than (or close to) 0.1 indicating the successful removal of spatial inertia. Truncation distance used for SEVM was equal to 1000 km and eigenvectors to be added to the OLS model were selected according to their level of spatial autocorrelation. We used the first 40 eigenvectors which presented Moran's I larger than 0.1 in the first distance class (i.e. connecting adjacent cells).
The relative importance of the environmental predictors was evaluated using the software SAM [48,49] where we evaluated the standardized slopes of the full SEVM, after taking into account geographical effects expressed by the set of eigenvectors. We also used a partial regression approach to partition the variance among the ‘pure’ environmental components, the ‘pure’ geographical patterns and their overlap, for each diversity metric. Finally, PD and FD are known to show a monotonic relationship with species richness [13,55] since the addition of each species will invariably lead to an increase in PD/FD by adding branches to the minimum spanning tree/dendrogram. Therefore, to evaluate the environmental determinants of PD and FD independent of the inherent relationship with species richness we ran the same analysis using a three-way partial regression [51] with squared richness as a third set, and reported the partial R2 of SEVM and environmental variables independent of the effect of richness. Following the same reasoning, a three-way partial regression was also used to investigate the patterns of FD taking PD into account.
3. Results
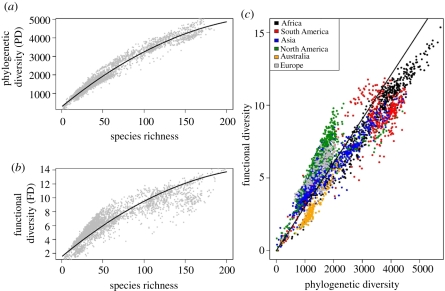
FD and PD showed a very similar spatial pattern with respect to latitude due to their monotonic relationship with species richness (compare [56]). The relationship between FD and richness as well as between PD and richness was nonlinear (table 1). In fact, a quadratic model had a better fit than either cubic or linear alternatives to model the relationships between S and PD, as well as between S and FD (figure 2a,b and table 1).
Table 1.
The parameter estimates for the quadratic relationship between species richness (S) and PD and FD was estimated using a trend surface analysis (in a generalized additive model with latitude and longitude of each grid cell used as smooth factor to account for large-scale spatial autocorrelation structures).
| PD:aR2 = 0.98 |
FD:bR2 = 0.96 |
|||||
|---|---|---|---|---|---|---|
| estimate ± s.e. | t | p | estimate ± s.e. | t | p | |
| intercept | 276.87 ± 8.43 | 32.84 | <0.0001 | 1.5 ± 0.036 | 41.49 | <0.0001 |
| S | 36.35 ± 0.25 | 142.90 | <0.0001 | 0.1 ± 0.001 | 89.96 | <0.0001 |
| S2 | −0.07 ± 0.002 | −45.32 | <0.0001 | −1.9E−04 ± 6.4E−06 | −29.16 | <0.0001 |
aSmooth term PD: estimated degrees of freedom = 28.17, F = 171.7, p < 0.0001.
bSmooth term FD: estimated degrees of freedom = 28.72, F = 278.2, p < 0.0001.
Figure 2.
Relationship between (a) PD and richness and (b) FD and richness as well as (c) the correlation between PD and FD colour coded for different continents. Generalized additive models were used (with latitude and longitude of each grid cell used as smooth factors) to account for biases in model estimates due to spatial autocorrelation. Smooth term: estimated degrees of freedom in (a) = 28.17, F = 171.7, p < 0.0001; in (b) = 28.72, F = 278.2, p < 0.0001; in (c) = 28.8, F = 344.3, p < 0.0001.)
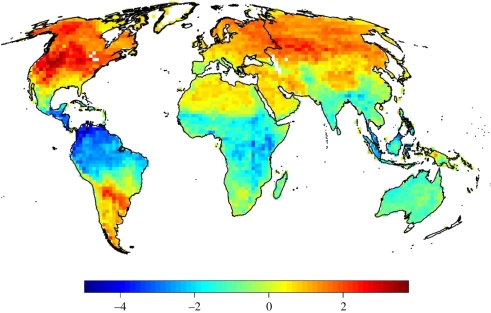
The relationship between PD and FD was linear over the entire global dataset (figure 2c). However, it was evident that different continents had different dispersion around the global relationship between PD and FD. The differences suggested that the amount of functional similarity at comparable levels of phylogenetic distance is limited in some areas more than in others. Visual inspection of the residuals between observed FD and predicted FD (according to a theoretical linear relationship with PD; see figure 2c) suggests that temperate communities have accumulated more FD than expected compared to tropical communities (figure 3).
Figure 3.
Deviance (residuals) of observed FD compared to the global linear relationship between PD and FD (FD = 0.003·PD; see figure 2c). Blue areas depict areas of lower than expected FD (according to the PD present in the area) whereas red areas are areas with more FD than expected from this linear relationship.
About 50 per cent of the variation in the three different diversity metrics were explained by environmental factors, according to the OLS (table 2), although significant spatial patterns remained in model residuals, according to the Moran's I coefficients (which were always larger than 0.5). Removing the residual autocorrelation increased R2 to about 70–95% of the original deviance when eigenvectors were added (table 2). Partial regression analysis showed a relatively small role of the unique, local effects of environment (which tended to be geographically structured and was expressed by the shared component with spatial eigenvectors). Given that species richness is intrinsically correlated with the FD and PD metrics we used, naturally the partial R2 for space, environment and their superposition were low; and species richness captured most of the variation of FD and PD.
Table 2.
Three-way partial regression analysis of species richness, and phylogenetic and functional diversity showing the amount of deviance explained by environmental variables (env) versus spatial autocorrelation partitioning between the ‘pure’ environmental components, the ‘pure’ geographical patterns and their overlap, for each diversity metric (S, species richness; PD, phylogenetic diversity; FD, functional diversity). SEVM, spatial eigenvectors; full, full model containing both environmental and spatial eigenvectors. a, b and c denote the contributions of environment (a), spatial structures (c) and their joint effects (b). ‘Covariate’ is squared richness or PD, and in these analyses the partial R2 and slopes are from a triple partial regression.
| partial regression model R2 |
|||||||
|---|---|---|---|---|---|---|---|
| env | SEVM | full | a (env) | b | c (geo) | covariate | |
| S | 0.536 | 0.650 | 0.798 | 0.148 | 0.388 | 0.262 | |
| PD | 0.552 | 0.684 | 0.817 | 0.133 | 0.419 | 0.265 | |
| FD | 0.569 | 0.548 | 0.738 | 0.190 | 0.379 | 0.169 | |
| PD × S2 | 0.646 | 0.688 | 0.861 | 0.056 | 0.000 | 0.161 | 0.668 |
| FD × S2 | 0.457 | 0.553 | 0.941 | 0.021 | 0.018 | 0.051 | 0.850 |
| FD · PD | 0.457 | 0.553 | 0.951 | 0.057 | 0.039 | 0.009 | 0.845 |
The environmental models using SEVM showed that S, PD and FD were well explained by AET (table 3). However, when taking species richness into account the most important predictors changed to temperature and altitude, although the overall effect of environment in these metrics was much smaller when compared to original PD and FD (most of the residual variation was explained by geographical structures expressed by eigenvectors; tables 2 and 3). The residuals of FD against PD, indicating the regions where FD deviates from the expectation by PD, were better explained by temperature as well, but with a more ‘balanced’ signal from AET and seasonality (table 3).
Table 3.
The slopes of the environmental predictors of the different diversity measures. S refers to species richness, PD to phylogenetic diversity, FD to functional diversity. FD·PD are the residuals of the three-way partial regression of functional diversity against phylogenetic diversity. Significant effects (p < 0.05) are in bold.
| slopes (SEVM + env) |
||||
|---|---|---|---|---|
| elevation | AET | annual temp | temp. (s.d.) | |
| S | −0.019 | 0.555 | −0.244 | −0.153 |
| PD | 0.008 | 0.550 | −0.077 | −0.086 |
| FD | 0.133 | 0.740 | 0.285 | 0.265 |
| PD × S2 | 0.061 | 0.224 | 0.341 | 0.073 |
| FD × S2 | 0.186 | 0.414 | 0.634 | 0.425 |
| FD · PD | 0.124 | 0.147 | 0.292 | 0.359 |
4. Discussion
Species richness in mammals follows the typical global trend for higher diversity in the tropics, showing a strong latitudinal gradient similar to those found for birds and amphibians [56–58]. We found a positive association between species richness and both PD and FD. This results from the property of these measures whereby increases in species richness can only increase PD and FD (or rarely cause no change) while decreases in species richness can only decrease them (or rarely cause no change). This makes some form of positive association largely inevitable [13]. However, the scatter of the positive association indicates that for a given level of species richness assemblages may have high or low PD and FD. The scatter represents the degree of importance of species identity (i.e. species composition), where increased scatter implies stronger effects of species identity as opposed to species richness. Also, both PD and FD showed the tendency to level-off with increasing species richness. This indicates that with increasing species richness the probability that additional species entering a community are distant relatives or functionally very dissimilar decreases [6,59].
The mammalian historical biogeography and colonization pattern is complex suggesting that the pattern of PD and FD could be complex as well. According to Davies et al. [57] ‘these patterns reflect a complex history of speciation, extinction, anagenesis, and dispersal, which each factor probably shaped by biological traits and changed through time’. Whereas mammal assemblages are much older in Africa, Middle East, India and Himalayan regions they are relatively younger in South America and Europe [56,57,60]. The correlation between PD and FD showed that FD generally accumulates with phylogenetic age in communities on a global scale. This relationship was linear, with a very informative pattern of deviances from the general relationship.
Whereas communities in the temperate regions showed signs of limiting functional similarity compared to the amount of PD, the tropical mammal communities proved to contain many more functionally similar species. There are several non-mutually exclusive mechanisms that could be made responsible for such a pattern (figure 1). Temperate mammal assemblages were composed of more complementary species (i.e. species which are less similar in their functional traits), which can be a result of competition pressure and low energy availability in high latitudes. Even though it is always problematic to invoke the ‘ghost of competition past’ [61] there is evidence of high rates of mammal extinction in the temperate regions in comparison to the tropics [62]. The temperate areas, with their higher seasonality and lower structural complexity, might prevent species from being packed as densely in niche space as they can be in the tropical areas. The fact that species tend to have larger range sizes in temperate regions [57] could also be interpreted as indicative of high levels of resource competition. Limited resource availability according to this rationale forces species to be functionally more distinct and to occupy wider ecological niches. Alternatively, or in addition to high competition, recent colonization events in combination with fast adaptive radiation (and high interspecific competition) could also result in high levels of FD and low levels of PD. In fact, species richness of young mammal clades is higher in temperate areas and the ages of these clades coincide with the expansion of temperate climate zones in the late Eocene [63]. Thus, the over-dispersion in FD we found in temperate mammals may, in addition to the competition hypothesis, be a result of recent adaptations to rather novel environments or recently recolonized areas.
Although the increased species richness at lower latitudes has been usually understood as a consequence of the great degree of species complementarity due to a high degree of niche specialization [64,65], we found a clear opposite pattern. Coexisting species according to our analysis are more similar in functional traits in the tropics for any given level of PD. This is in accordance with the phylogenetic conservatism hypothesis [8,9,36] which predicts that there is a tendency for species to retain most of their ancestral characters. Such ‘phylogenetic inertia’ has been corroborated by many studies at macroecological scales usually defining the niche by environmental variables [9,66]. Our study adds additional evidence that the high levels of speciation in the tropics seem to produce many species with ‘conserved niches’ that not only stayed within or near their ancestral geographical range but also maintained great functional similarities. Consequently, the lower observed abundance and small range sizes of tropical mammals, instead of being simply a consequence of narrow tolerances of species [57], may reflect an ‘ecological packing’ in geographical space [67]. This would allow for a higher degree of functional redundancy without necessarily causing high local species extinction rates through resource competition due to the higher levels of primary productivity. Thus, functional turnover (functional beta diversity) should be much lower than species turnover in the tropics, a point still remaining to be verified (see [3]).
The deficits in FD in the tropical areas are in line with the finding that high levels of species diversity tend to promote stabilizing selection on ecological traits, inhibiting evolutionary responses within species and ultimately leading to high levels of niche conservatism [68]. The rationale behind this is that as richness increases, species become more and more restricted to only those patches whose final optima are close to the species' initial optimal phenotype [68]. Therefore, in the species-rich tropical environment species are expected to disperse to occupy new patches with conditions closely matching their initial needs, rather than adapting to the changes in their original habitat [68,69]. This mechanism would consequently lead to a deficit in FD in geographical space in species-rich communities. Also, a direct consequence of this process is that in richer assemblages even subtle changes in the environmental conditions would potentially lead to dramatic changes in species abundances and have less impact on phenotypic changes or environmental adaptation [68]. This second prediction also fits well the pattern in tropical diversity since tropical species tend, on average, to exist at lower densities than species in temperate regions (e.g. [70,71]).
The rates of molecular evolution and substitution rates are known to vary across taxonomic units and in space; however, the factors influencing the rates of molecular evolution and the precise spatial pattern of the rate variation are still under debate [72–74]. Our results indicate that trait evolution in tropical areas is slower than in temperate areas under the assumption that the species evolved in the environments in which they are encountered. According to our results at similar average phylogenetic distances the differences between species in functional traits were found to be smaller in tropical areas. As a consequence niche conservatism is more pronounced in tropical areas. In other words, high levels of speciation in the tropics seem to result in many species with ‘conserved niches’ that not only stay within or near their ancestral geographical range (tropics), but also retain a lot of functional similarities (resulting in the FD deficit). On the other hand, the opposite pattern, niche evolution, seems to occur in temperate regions. Niche evolution can be understood as the expansion of niche breadth or specialization to new conditions that should enable lineages to invade new habitats and climatic regions which would normally limit the distribution of the ancestors [75]. Given that we observed higher levels of FD for mammal assemblages in temperate regions, and that temperate mammalian assemblages are composed of species that are the outcome of more recent speciation events [57,62], there is evidence for niche evolution as an important evolutionary force driving patterns of diversity in temperate mammal assemblages.
Current knowledge suggests that biodiversity is of paramount importance for environmental and biological processes and ultimately pivotal to human well-being [26,76–78]. However, despite the fact that many theories have been suggested to explain the global patterns of biodiversity [37,79–81] unravelling the mechanisms behind the global patterns and determinants of biodiversity still is one of the major intellectual challenges [82]. Here, we showed how combining the different diversity measures can provide interesting global-scale results. Discrepancies between FD and species richness as well as PD have also been identified recently in local-scale studies and mismatches between congruency of different diversity indices have challenged the claim of interchangeability of different diversity measures [35,39,57]. Clearly, studying such areas where discrepancies in different diversity measures occur can provide us with the means to understand what the central mechanisms in creating and maintaining biological diversity are.
Acknowledgements
We are grateful to Kate Jones and Bob Ricklefs for lively discussions and suggestions on the topic of comparing diversity measures. Natalie Cooper and an anonymous reviewer have provided insightful suggestions on earlier versions of the manuscript. The research of R.D.L. was supported by CNPq (project #475 886/2009-7) and CAPES (project #012/09) M.V.C. by CAPES (project #012/09) and J.A.F.D.F. by CNPq (projects #300 762/94-1, 300 367/96-1, 400 381-97.4) and CAPES (project #012/09).
References
- 1.Magurran A. 2004. Measuring biological diversity. Oxford, UK: Blackwell Science [Google Scholar]
- 2.Tilman D. 1997. Biodiversity and ecosystem function. In Nature's services: societal dependence on natural ecosystems, pp. 93–112 Wahington, DC: Island Press [Google Scholar]
- 3.Stevens R. D., Cox S. B., Strauss R. E., Willig M. R. 2003. Patterns of functional diversity across an extensive environmental gradient: vertebrate consumers, hidden treatments and latitudinal trends. Ecol. Lett. 6, 1099–1108 10.1046/j.1461-0248.2003.00541.x (doi:10.1046/j.1461-0248.2003.00541.x) [DOI] [Google Scholar]
- 4.Faith D. P. 1992. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10 10.1016/0006-3207(92)91201-3 (doi:10.1016/0006-3207(92)91201-3) [DOI] [Google Scholar]
- 5.Hooper D. U., et al. 2002. Species diversity, functional diversity, and ecosystem functioning. In Biodiversity and ecosystem functioning: synthesis and perspectives (eds Inchausti P., Loreau M., Naeem S.), pp. 195–208 New York, NY: Oxford University Press [Google Scholar]
- 6.Petchey O. L., Gaston K. J. 2002. Extinction and the loss of functional diversity. Proc. R. Soc. Lond. B 269, 1721–1727 10.1098/rspb.2002.2073 (doi:10.1098/rspb.2002.2073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Webb C. O., Ackerly D. D., McPeek M. A., Donoghue M. J. 2002. Phylogenies and community ecology. Annu. Rev. Ecol. Syst. 33, 475–505 10.1146/annurev.ecolsys.33.010802.150448 (doi:10.1146/annurev.ecolsys.33.010802.150448) [DOI] [Google Scholar]
- 8.Losos J. B., Glor R. E. 2003. Phylogenetic comparative methods and the geography of speciation. Trends Ecol. Evol. 18, 220–227 10.1016/S0169-5347(03)00037-5 (doi:10.1016/S0169-5347(03)00037-5) [DOI] [Google Scholar]
- 9.Wiens J. J., Graham C. H. 2005. Niche conservatism: integrating evolution, ecology, and conservation biology. Annu. Rev. Ecol. Evol. Syst. 36, 519–539 10.1146/annurev.ecolsys.36.102803.095431 (doi:10.1146/annurev.ecolsys.36.102803.095431) [DOI] [Google Scholar]
- 10.Cavender-Bares J., Kozak K. H., Fine P. V. A., Kembel S. W. 2009. The merging of community ecology and phylogenetic biology. Ecol. Lett. 12, 693–715 10.1111/j.1461-0248.2009.01314.x (doi:10.1111/j.1461-0248.2009.01314.x) [DOI] [PubMed] [Google Scholar]
- 11.Emerson B. C., Gillespie R. G. 2008. Phylogenetic analysis of community assembly and structure over space and time. Trends Ecol. Evol. 23, 619–630 10.1016/j.tree.2008.07.005 (doi:10.1016/j.tree.2008.07.005) [DOI] [PubMed] [Google Scholar]
- 12.Rodrigues A. S. L., Gaston K. J. 2002. Maximising phylogenetic diversity in the selection of networks of conservation areas. Biol. Conserv. 105, 103–111 10.1016/S0006-3207(01)00208-7 (doi:10.1016/S0006-3207(01)00208-7) [DOI] [Google Scholar]
- 13.Schweiger O., Klotz S., Durka W., Kuhn I. 2008. A comparative test of phylogenetic diversity indices. Oecologia 157, 485–495 10.1007/s00442-008-1082-2 (doi:10.1007/s00442-008-1082-2) [DOI] [PubMed] [Google Scholar]
- 14.Mace G. M., Gittleman J. L., Purvis A. 2003. Preserving the tree of life. Science 300, 1707–1709 10.1126/science.1085510 (doi:10.1126/science.1085510) [DOI] [PubMed] [Google Scholar]
- 15.Purvis A., Agapow P.-M., Gittleman J. L., Mace G. M. 2000. Nonrandom extinction and the loss of evolutionary history. Science 288, 328–330 10.1126/science.288.5464.328 (doi:10.1126/science.288.5464.328) [DOI] [PubMed] [Google Scholar]
- 16.Fritz S. A., Purvis A. 2010. Phylogenetic diversity does not capture body size variation at risk in the world's mammals. Proc. R. Soc. B 277, 2435–2441 10.1098/rspb.2010.0030 (doi:10.1098/rspb.2010.0030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diniz-Filho J. A. 2004. Phylogenetic diversity and conservation priorities under distinct models of phenotypic evolution. Conserv. Biol. 18, 698–704 10.1111/j.1523-1739.2004.00260.x (doi:10.1111/j.1523-1739.2004.00260.x) [DOI] [Google Scholar]
- 18.Loyola R., Oliveira-Santos L., Almeida-Neto M., Nogueira D., Kubota U., Diniz-Filho J., Lewinsohn T. 2009. Integrating economic costs and biological traits into global conservation priorities for carnivores. PLoS ONE 4, e6807. 10.1371/journal.pone.0006807 (doi:10.1371/journal.pone.0006807) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loyola R. D., Oliveira G., Diniz-Filho J. A. F., Lewinsohn T. M. 2008. Conservation of neotropical carnivores under different prioritization scenarios: mapping species traits to minimize conservation conflicts. Divers. Distrib. 14, 949–960 10.1111/j.1472-4642.2008.00508.x (doi:10.1111/j.1472-4642.2008.00508.x) [DOI] [Google Scholar]
- 20.Rodrigues A. S. L., Brooks T. M., Gaston K. J. 2005. Integrating phylogenetic diversity in the selection of priority areas for conservation: does it make a difference? In Phylogeny and conservation (eds Purvis A., Gittleman J. L., Brooks T.), pp. 101–119 Cambridge, UK: Cambridge University Press [Google Scholar]
- 21.Sechrest W., Brooks T. M., da Fonseca G. A. B., Konstant W. R., Mittermeier R. A., Purvis A., Rylands A. B., Gittleman J. L. 2002. Hotspots and the conservation of evolutionary history. Proc. Natl Acad. Sci. USA 99, 2067–2071 10.1073/pnas.251680798 (doi:10.1073/pnas.251680798) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vamosi S. M., Heard S. B., Vamosi J. C., Webb C. O. 2009. Emerging patterns in the comparative analysis of phylogenetic community structure. Mol. Ecol. 18, 572–592 10.1111/j.1365-294X.2008.04001.x (doi:10.1111/j.1365-294X.2008.04001.x) [DOI] [PubMed] [Google Scholar]
- 23.Cardillo M. 2011. Phylogenetic structure of mammal assemblages at large geographic scales: linking phylogenetic community ecology with macroecology. Phil. Trans. R. Soc. B 366, 2545–2553 10.1098/rstb.2011.0021 (doi:10.1098/rstb.2011.0021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kisel Y., McInnes L., Toomey N. H., Orme C. D. L. 2011. How diversification rates and diversity limits combine to create large-scale species–area relationships. Phil. Trans. R. Soc. B 366, 2514–2525 10.1098/rstb.2011.0022 (doi:10.1098/rstb.2011.0022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petchey O. L., Gaston K. J. 2006. Functional diversity: back to basics and looking forward. Ecol. Lett. 9, 741–758 10.1111/j.1461-0248.2006.00924.x (doi:10.1111/j.1461-0248.2006.00924.x) [DOI] [PubMed] [Google Scholar]
- 26.Díaz S., Fargione J., Chapin F. S., Tilman D. 2006. Biodiversity loss threatens human well-being. PLoS Biol. 4, e277. 10.1371/journal.pbio.0040277 (doi:10.1371/journal.pbio.0040277) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mouchet M. A., Villéger S., Mason N. W. H., Mouillot D. 2010. Functional diversity measures: an overview of their redundancy and their ability to discriminate community assembly rules. Funct. Ecol. 24, 867–876 10.1111/j.1365-2435.2010.01695.x (doi:10.1111/j.1365-2435.2010.01695.x) [DOI] [Google Scholar]
- 28.Petchey O. L., Evans K. L., Fishburn I. S., Gaston K. J. 2007. Low functional diversity and no redundancy in British avian assemblages. J. Anim. Ecol. 76, 977–985 10.1111/j.1365-2656.2007.01271.x (doi:10.1111/j.1365-2656.2007.01271.x) [DOI] [PubMed] [Google Scholar]
- 29.Diaz S., Cabido M. 2001. Vive la difference: plant functional diversity matters to ecosystem processes. Trends Ecol. Evol. 16, 646–655 10.1016/S0169-5347(01)02283-2 (doi:10.1016/S0169-5347(01)02283-2) [DOI] [Google Scholar]
- 30.Blackburn T. M., Petchey O. L., Cassey P., Gaston K. J. 2005. Functional diversity of mammalian predators and extinction in island birds. Ecology 86, 2916–2923 10.1890/04-1847 (doi:10.1890/04-1847) [DOI] [Google Scholar]
- 31.Burel F., et al. 1998. Comparative biodiversity along a gradient of agricultural landscapes. Acta Oecologica-Int. J. Ecol. 19, 47–60 10.1016/S1146-609X(98)80007-6 (doi:10.1016/S1146-609X(98)80007-6) [DOI] [Google Scholar]
- 32.Flynn D. F. B., Gogol-Prokurat M., Nogeire T., Molinari N., Richers B. T., Lin B. B., Simpson N., Mayfield M. M., DeClerck F. 2009. Loss of functional diversity under land use intensification across multiple taxa. Ecol. Lett. 12, 22–33 10.1111/j.1461-0248.2008.01255.x (doi:10.1111/j.1461-0248.2008.01255.x) [DOI] [PubMed] [Google Scholar]
- 33.Fox B. J., Fox M. D. 2000. Factors determining mammal species richness on habitat islands and isolates: habitat diversity, disturbance, species interactions and guild assembly rules. Global Ecol. Biogeogr. 9, 19–37 10.1046/j.1365-2699.2000.00184.x (doi:10.1046/j.1365-2699.2000.00184.x) [DOI] [Google Scholar]
- 34.Lidicker W. Z. 1999. Responses of mammals to habitat edges: an overview. Landscape Ecol. 14, 333–343 10.1023/A:1008056817939 (doi:10.1023/A:1008056817939) [DOI] [Google Scholar]
- 35.Devictor V., Mouillot D., Meynard C., Jiguet F., Thuiller W., Mouquet N. 2010. Spatial mismatch and congruence between taxonomic, phylogenetic and functional diversity: the need for integrative conservation strategies in a changing world. Ecol. Lett. 13, 1030–1040 10.1111/j.1461-0248.2010.01493.x (doi: 10.1111/j.1461-0248.2010.01493.x) [DOI] [PubMed] [Google Scholar]
- 36.Losos J. B. 2008. Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecol. Lett. 11, 995–1003 10.1111/j.1461-0248.2008.01229.x (doi:10.1111/j.1461-0248.2008.01229.x) [DOI] [PubMed] [Google Scholar]
- 37.Willig M. R., Kaufman D. M., Stevens R. D. 2003. Latitudinal gradients of biodiversity: pattern, process, scale, and synthesis. Annu. Rev. Ecol. Evol. Syst. 34, 273–309 10.1146/annurev.ecolsys.34.012103.144032 (doi:10.1146/annurev.ecolsys.34.012103.144032) [DOI] [Google Scholar]
- 38.Winter M., et al. 2009. Plant extinctions and introductions lead to phylogenetic and taxonomic homogenization of the European flora. Proc. Natl Acad. Sci. USA 106, 21 721–21 725 10.1073/pnas.0907088106 (doi:10.1073/pnas.0907088106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forest F., et al. 2007. Preserving the evolutionary potential of floras in biodiversity hotspots. Nature 445, 757–760 10.1038/nature05587 (doi:10.1038/nature05587) [DOI] [PubMed] [Google Scholar]
- 40.Cooper N., Purvis A. 2010. Body size evolution in mammals: complexity in tempo and mode. Am. Nat. 175, 727–738 10.1086/652466 (doi:10.1086/652466) [DOI] [PubMed] [Google Scholar]
- 41.IUCN 2009. IUCN red list of threatened species version 2009. 1. See http://www.iucnredlist.org
- 42.Bininda-Emonds O. R. P., et al. 2008. The delayed rise of present-day mammals (vol 446, pg 507, 2007). Nature 456, 271–274 10.1038/nature07347 (doi:10.1038/nature07347) [DOI] [PubMed] [Google Scholar]
- 43.Jones K. E., et al. 2009. PanTHERIA: a species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology 90, 2648. 10.1890/08-1494.1 (doi:10.1890/08-1494.1) [DOI] [Google Scholar]
- 44.Hearn P., Hare T., Schruben P., Sherrill D., LaMar C., Tsushima P. 2003. Global GIS Global Coverage DVD. American Geological Institute U.S. Geological Survey; See http://www.agiweb.org/pubs/globalgis [Google Scholar]
- 45.Hijmans R. J., van Etten J. 2009. Raster. Geographic analysis and modeling with raster data: R package version 0.9.8-40/r681. See http://www.R-Forge.R-project.org/projects/raster/
- 46.R Development Core Team 2010. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- 47.Woods S. N. 2006. Generalized additive models: an introduction with R. Texts in statistical science. Boca Raton, FL: Chapman & Hall/CRC [Google Scholar]
- 48.Rangel T., Diniz-Filho J. A. F., Bini L. M. 2006. Towards an integrated computational tool for spatial analysis in macroecology and biogeography. Global Ecol. Biogeogr. 15, 321–327 10.1111/j.1466-822X.2006.00237.x (doi:10.1111/j.1466-822X.2006.00237.x) [DOI] [Google Scholar]
- 49.Rangel T. F. L. V. B., Diniz-Filho J. A. F., Bini L. M. 2010. SAM: a comprehensive application for spatial analysis in macroecology. Ecography 33, 46–50 10.1111/j.1600-0587.2009.06299.x (doi:10.1111/j.1600-0587.2009.06299.x) [DOI] [Google Scholar]
- 50.Diniz-Filho J. A. F., Bini L. M., Hawkins B. A. 2003. Spatial autocorrelation and red herrings in geographical ecology. Global Ecol. Biogeogr. 12, 53–64 10.1046/j.1466-822X.2003.00322.x (doi:10.1046/j.1466-822X.2003.00322.x) [DOI] [Google Scholar]
- 51.Legendre P., Legendre L. 1998. Numerical ecology. Amsterdam, The Netherlands: Elsevier [Google Scholar]
- 52.Diniz-Filho J. A. F., Bini L. M. 2005. Modelling geographical patterns in species richness using eigenvector-based spatial filters. Global Ecol. Biogeogr. 14, 177–185 10.1111/j.1466-822X.2005.00147.x (doi:10.1111/j.1466-822X.2005.00147.x) [DOI] [Google Scholar]
- 53.Dormann C. F., et al. 2007. Methods to account for spatial autocorrelation in the analysis of species distributional data: a review. Ecography 30, 609–628 10.1111/j.2007.0906-7590.05171.x (doi:10.1111/j.2007.0906-7590.05171.x) [DOI] [Google Scholar]
- 54.Griffith D. A., Peres-Neto P. R. 2006. Spatial modeling in ecology: the flexibility of eigenfunction spatial analyses. Ecology 87, 2603–2613 10.1890/0012-9658(2006)87[2603:SMIETF]2.0.CO;2 (doi:10.1890/0012-9658(2006)87[2603:SMIETF]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 55.Poos M. S., Walker S. C., Jackson D. A. 2009. Functional-diversity indices can be driven by methodological choices and species richness. Ecology 90, 341–347 10.1890/08-1638.1 (doi:10.1890/08-1638.1) [DOI] [PubMed] [Google Scholar]
- 56.Davies T. J., Buckley L. B., Greyner R., Gittleman J. L. 2011. The influence of past and present climate on the biogeography of modern mammal diversity. Phil. Trans. R. Soc. B 366, 2526–2535 10.1098/rstb.2011.0018 (doi:10.1098/rstb.2011.0018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davies T. J., et al. 2008. Phylogenetic trees and the future of mammalian biodiversity. Proc. Natl Acad. Sci. USA 105, 11 556–11 563 10.1073/pnas.0801917105 (doi:10.1073/pnas.0801917105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jetz W., Rahbek C. 2002. Geographic range size and determinants of avian species richness. Science 297, 1548–1551 10.1126/science.1072779 (doi:10.1126/science.1072779) [DOI] [PubMed] [Google Scholar]
- 59.Cianciaruso M. V., Batalha M. A., Gaston K. J., Petchey O. L. 2009. Including intraspecific variability in functional diversity. Ecology 90, 81–89 10.1890/07-1864.1 (doi:10.1890/07-1864.1) [DOI] [PubMed] [Google Scholar]
- 60.Springer M. S., Meredith R. W., Janecka J. E., Murphy W. J. 2011. The historical biogeography of mammalia. Phil. Trans. R. Soc. B 366, 2478–2502 10.1098/rstb.2011.0023 (doi:10.1098/rstb.2011.0023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Connell J. H. 1980. Diversity and the coevolution of competitors, or the ghost of competition past. Oikos 35, 131–138 10.2307/3544421 (doi:10.2307/3544421) [DOI] [Google Scholar]
- 62.Weir J. T., Schluter D. 2007. The latitudinal gradient in recent speciation and extinction rates of birds and mammals. Science 315, 1574–1576 10.1126/science.1135590 (doi:10.1126/science.1135590) [DOI] [PubMed] [Google Scholar]
- 63.Buckley L. B., et al. 2010. Phylogeny, niche conservatism and the latitudinal diversity gradient in mammals. Proc. R. Soc. B 277, 2131–2138 10.1098/rspb.2010.0179 (doi:10.1098/rspb.2010.0179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hutchinson G. 1959. Homage to Santa Rosalia or why are there so many kinds of animals? Am. Nat. 93, 145. 10.1086/282070 (doi:10.1086/282070) [DOI] [Google Scholar]
- 65.Hutchinson G., MacArthur R. 1959. A theoretical ecological model of size distribution among species of mammals. Am. Nat. 93, 117–125 10.1086/282063 (doi:10.1086/282063) [DOI] [Google Scholar]
- 66.Harvey P. H., Pagel M. 1991. The comparative method in evolutionary biology. Oxford, UK: Oxford University Press [Google Scholar]
- 67.Palmer M. W. 1992. The coexistence of species in fractal landscapes. Am. Nat. 139, 375–397 10.1086/285332 (doi:10.1086/285332) [DOI] [Google Scholar]
- 68.de Mazancourt C., Johnson E., Barraclough T. G. 2008. Biodiversity inhibits species' evolutionary responses to changing environments. Ecol. Lett. 11, 380–388 10.1111/j.1461-0248.2008.01152.x (doi:10.1111/j.1461-0248.2008.01152.x) [DOI] [PubMed] [Google Scholar]
- 69.Ackerly D. D. 2003. Community assembly, niche conservatism, and adaptive evolution in changing environments. Int. J. Plant Sci. 164, S165–S184 10.1086/368401 (doi:10.1086/368401) [DOI] [Google Scholar]
- 70.Currie D. J., Fritz T. 1993. Global patterns of animal abundance and species energy use. Oikos 67, 56–68 10.2307/3545095 (doi:10.2307/3545095) [DOI] [Google Scholar]
- 71.Johnson C. 1998. Rarity in the tropics: latitudinal gradients in distribution and abundance in Australian mammals. J. Anim. Ecol. 67, 689–698 10.1046/j.1365-2656.1998.00232.x (doi:10.1046/j.1365-2656.1998.00232.x) [DOI] [Google Scholar]
- 72.Bromham L. 2011. The genome as a life history character: why rate of molecular evolution varies between mammal species. Phil. Trans. R. Soc. B 366, 2503–2513 10.1098/rstb.2011.0014 (doi:10.1098/rstb.2011.0014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gillman L. N., McBride P., Keeling D. J., Ross H. A., Wright S. D. 2011. Are rates of molecular evolution in mammals substantially accelerated in warmer environments? Reply. Proc. R. Soc. B 278, 1298–1305 10.1098/rspb.2010.2618 (doi:10.1098/rspb.2010.2618) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weir J. T., Schluter D. 2011. Are rates of molecular evolution in mammals substantially accelerated in warmer environments? Proc. R. Soc. B 278, 1291–1293 10.1098/rspb.2010.0388 (doi:10.1098/rspb.2010.0388) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wiens J. J., Donoghue M. J. 2004. Historical biogeography, ecology and species richness. Trends Ecol. Evol. 19, 639–644 10.1016/j.tree.2004.09.011 (doi:10.1016/j.tree.2004.09.011) [DOI] [PubMed] [Google Scholar]
- 76.European Communities 2008. The economics of ecosystems and biodiversity: an interim report. See www.teebweb.org
- 77.Sachs J. D., et al. 2009. Biodiversity conservation and the Millennium development goals. Science 325, 1502–1503 10.1126/science.1175035 (doi:10.1126/science.1175035) [DOI] [PubMed] [Google Scholar]
- 78.Wilkins D. 1999. Assessing ecosystem health. Trends Ecol. Evol. 14, 69. 10.1016/S0169-5347(98)01526-2 (doi:10.1016/S0169-5347(98)01526-2) [DOI] [PubMed] [Google Scholar]
- 79.Isaac N. J. B., Jones K. E., Gittleman J. L., Purvis A. 2005. Correlates of species richness in mammals: body size, life history, and ecology. Am. Nat. 165, 600–607 10.1086/429148 (doi:10.1086/429148) [DOI] [PubMed] [Google Scholar]
- 80.Ricklefs R. E. 2004. A comprehensive framework for global patterns in biodiversity. Ecol. Lett. 7, 1–15 10.1046/j.1461-0248.2003.00554.x (doi:10.1046/j.1461-0248.2003.00554.x) [DOI] [Google Scholar]
- 81.Stehli F. G., Douglas R. G., Newell N. D. 1969. Generation and maintenance of gradients in taxonomic diversity. Science 164, 947–949 10.1126/science.164.3882.947 (doi:10.1126/science.164.3882.947) [DOI] [PubMed] [Google Scholar]
- 82.Gaston K. J. 2000. Global patterns in biodiversity. Nature 405, 220–227 10.1038/35012228 (doi:10.1038/35012228) [DOI] [PubMed] [Google Scholar]