Abstract
The content of sulfur amino acid (SAA) in a meal affects postprandial plasma cysteine concentrations and the redox potential of cysteine/cystine. Because such changes can affect enzyme, transporter, and receptor activities, meal content of SAA could have unrecognized effects on metabolism during the postprandial period. This pilot study used proton NMR (1H-NMR) spectroscopy of human plasma to test the hypothesis that dietary SAA content changes macronutrient metabolism. Healthy participants (18–36 y, 5 males and 3 females) were equilibrated for 3 d to adequate SAA, fed chemically defined meals without SAA for 5 d (depletion), and then fed isoenergetic, isonitrogenous meals containing 56 mg·kg−1·d−1 SAA for 4.5 d (repletion). On the first and last day of consuming the chemically defined meals, a morning meal containing 60% of the daily food intake was given and plasma samples were collected over an 8-h postprandial time course for characterization of metabolic changes by 1H-NMR spectroscopy. SAA-free food increased peak intensity in the plasma 1H-NMR spectra in the postprandial period. Orthogonal signal correction/partial least squares-discriminant analysis showed changes in signals associated with lipids, some amino acids, and lactate, with notable increases in plasma lipid signals (TG, unsaturated lipid, cholesterol). Conventional lipid analyses confirmed higher plasma TG and showed an increase in plasma concentration of the lipoprotein lipase inhibitor, apoC-III. The results show that plasma 1H-NMR spectra can provide useful macronutrient profiling following a meal challenge protocol and that a single meal with imbalanced SAA content alters postprandial lipid metabolism.
Introduction
Variation in metabolism of sulfur amino acids (SAA)10 has been implicated in cardiovascular disease (CVD). For instance, individuals with homocysteinemia have accelerated CVD (1, 2) and high plasma total homocysteine has been associated with increased risk of CVD in several studies (1–4). High plasma cystine (CySS) and redox potentials for Cys/CySS and glutathione/glutathione disulfide have also been associated with CVD risk factors, including increased carotid intima media thickness (5), decreased flow mediated dilation (6), reversible myocardial perfusion defects (7), and persistent atrial fibrillation (8). Currently, however, there is no consensus concerning mechanisms relating SAA metabolites and CVD development.
One of the obstacles to elucidation of the underlying mechanisms is that clinical data are largely obtained under fasting conditions while individuals are in a postprandial state most of the day. Hence, an understanding of the metabolic responses to dietary SAA during the postprandial period could be useful to determine the roles of SAA in CVD development. Such information can be obtained from challenge studies where effects of variation in diet can be measured. Although such studies cannot directly establish causation in disease, they can reveal cause-effect relationships between diet and blood variables, which can then be used to design nutritional intervention trials.
In a diurnal variation study, we found that Cys, CySS, glutathione, and the redox potential for Cys/CySS varied in temporal patterns suggestive of meal-related changes (9), which was confirmed by meal challenges without and with SAA (10). In a subset of participants from the diurnal variation study, proton NMR (1H-NMR) spectroscopy of plasma showed the ability to discriminate metabolic patterns of macronutrients (blood lipids, amino acids, glucose) within participants (11). Fractal analysis of this data revealed that the metabolic patterns could predict plasma Cys concentration (12). Thus, the data show that, in principle, 1H-NMR spectroscopy of human plasma can provide an approach to gain understanding of metabolic changes associated with dietary SAA intake.
1H-NMR spectroscopy has been used to measure biologic responses to nutritional deficiency or excess in a number of studies (13–15). The spectra have some overlap of signals from different chemicals, but statistical pattern recognition techniques for data reduction and analysis (13, 16–18) show that useful metabolic information can be obtained for macronutrient metabolism (19). Such studies demonstrated that 1H-NMR spectroscopy of human plasma discriminates individuals with CVD based upon differences in lipid content (20, 21) and that the approach can be used for clinical measurement of blood lipids (22, 23).
The present research was designed as a pilot study to test the hypothesis that dietary SAA content altered postprandial macronutrient metabolism as measured by 1H-NMR spectroscopy of human plasma. The study was conducted concurrently with research to determine the effects of SAA content on postprandial redox potentials (10). In this research, we used a semisynthetic, chemically defined diet based upon the studies of Fukagawa et al. (24), Raguso et al. (25), and Lyons et al. (26), which allowed specific changes in Met and Cys content while controlling total energy and protein nitrogen (amino acid) content. Healthy participants were studied while consuming food without SAA and under comparable conditions with SAA (Met:Cys, 2:1) at an amount (56 mg·kg−1·d−1) approximating the mean American intake. The study design also provided postprandial measurements obtained after 3-d equilibration to a SAA-free or replete diet to determine whether recent history of SAA intake could affect postprandial responses.
Materials and Methods
Human participants.
This study was reviewed and approved by the Emory Investigational Review Board and was performed in accordance with the ethical standards of Emory University and the Emory IRB. The study was performed in the Emory University Hospital General Clinical Research Center (GCRC) as previously described (10). Eight participants (5 male, 3 female) aged 18–36 y were studied (Supplemental Table 1). Four were African American, 3 were white, and 1 was Asian. Age (mean ± SD) was 25 ± 6 y. BMI ranged from 20 to 26 (22.5 ± 2.1). Participants reported no acute or chronic illness and none were taking regular prescription medications.
A 3-d equilibration period with meals prepared by the GCRC Bionutrition Unit and providing the RDA for SAA (12.2 mg·kg−1 Met plus 6.6 mg·kg−1 Cys) and other amino acids was used to normalize participants with regard to diet. Participants were admitted as inpatients to the GCRC in the evening prior to initiation of the study. The study involved successive phases in which participants received 0 mg·kg−1 ·d−1 SAA for 5 d followed by 56 mg·kg−1·d−1 SAA for 5 d, with a distribution of Met:Cys of 2:1 (Supplemental Fig. 1). The protein equivalents were supplied in the form of l-amino acid mixtures (Ajinomoto USA) providing 1.0·g·kg−1·d−1 (25, 26). To compensate for the difference in Met + Cys between the equilibration, 0 and 56 mg·kg−1·d−1 SAA diets, the amount of all nonessential amino acids was proportionally changed to maintain a constant dietary nitrogen content while maintaining them as isoenergetic (10). Blood samples were taken from fasting participants daily at 0830. On days without kinetic analysis, meals were provided at 0830, 1230, and 1730 h, and a snack at 2230. On d 1 and d 5 of each study period, all participants were provided a single meal that combined the energy intake of the morning and midday meals and postprandial samples were collected at 0930, 1030, 1130, 1230, 1430, and 1630 h (Supplemental Fig. 1). The participants finished the study on d 5 after the 1630 time point, so the final 24-h time point on d5 of repletion was not available. Adequate hydration and vitamin, mineral, and electrolyte requirements were provided to all participants to meet or exceed recommended allowances (25) and body weights were determined daily and vital signs were obtained every 8 h.
Sampling and analyses.
Blood samples were collected, processed, and stored as previously described (11). A total of 34 samples were collected from each of 5 participants; 25 samples were available from a participant who discontinued after d 8, and 33 and 32 samples were available for 2 participants who left early on d 10. Two samples were excluded because of hemolysis. Consequently, the data analyzed were from a total of 258 samples.
For analysis, plasma samples (600 μL) were mixed with 66 μL of D2O containing 3-(trimethylsilyl)-1-propanesulfonic acid sodium salt (C6H15NaO3SSi, 1% wt:wt), and 1H-NMR spectra were measured at 600 MHz on a Varian INOVA 600 spectrometer under conditions where stability and reproducibility of the NMR analysis were previously established (11). Preprocessing of 1H-NMR spectra was performed to provide baseline-corrected, aligned, and normalized spectra containing 11,708 data points (11). Briefly, this included baseline correction with a polynomial regression (NUTS program, Acorn NMR), spectral alignment using a beam search algorithm (27) to enhance the computational efficiency of the genetic algorithm (28), elimination of uninformative spectral regions, and normalization relative to the internal standard. Chemicals contributing to plasma 1H-NMR spectra have been described in detail (29); we reconfirmed major signals (11) using addition of known standards (amino acids, sugars, organic acids, energy intermediates), comparisons to spectra in chemical databases, analysis with Chenomx software, and validation with 2-dimensional (2-D) NMR techniques. Additional analyses of lipid fractions were consistent with the results of Ala-Korpela et al. (22, 23).
Lipid analyses were done using a Beckman CX7 automatic chemistry analyzer. Plasma cholesterol and TG were measured by enzymatic methods using reagents from Beckman Diagnostics. HDL-cholesterol and LDL-cholesterol were measured by homogeneous enzymatic methods. Immunoturbinometric methods were used for high-sensitivity C-reactive protein (hsCRP; Equal Diagnostics), ApoC-III (Wako Chemicals) and ApoB (diaSorin). TG and phospholipids were quantified by liquid chromatography-MS in the positive ion mode. For analysis, 10 μL was injected onto an Ultrasphere 5 μm Spherical 80 Å pore C-18 (2.0 × 150 mm; Beckman Coulter) column with a Targa C18 precolumn (Higgins Analytical) for desalting and optimum chromatographic resolution. The flow rate was 0.5 mL·min−1 with 5% water/95% acetonitrile with 0.1% formic acid for 15 min. The eluate from the HPLC was connected to a Thermo LTQ-FT mass spectrometer (Thermo-Fisher Scientific). The LTQ-FT used an atmospheric pressure chemical ionization source that was operated with a source current of 5 μA, atmospheric pressure chemical ionization vaporization temperature of 450°C, sheath gas setting of 50 (arbitrary units), auxiliary gas setting of 50 (arbitrary units), capillary temperature of 275°C, capillary voltage of 35 V, and tube lens of 55 V. Analyses were done using the MS mode scanning from m/z 500 to 1000 in the FT detector at a resolution of 50,000 with the wide range scan mode and 400,000 ions/scan. Maximum ion injection time was 500 ms. Triolein (1,1,1-13C3, 99%; Cambridge Isotope Laboratories) was used as an internal standard.
Bioinformatics and statistics.
Data reduction by a principal component (PC) analysis (PCA) was performed using Pirouette software (Infometrix) after centering the mean of each variable. A previous analysis of diurnal variation in 1H-NMR spectra of human plasma showed that individual differences in rates of absorption and clearance of dietary components obscured common metabolic patterns, which were apparent when spectra for respective time points were averaged to minimize the contribution of inter-individual differences (11). Consequently, for analyses as indicated, inter-individual variation was minimized by averaging the intensity for the 8 participants at each of the 11,708 frequencies. Two-dimensional PCA score and loading plots were used to visualize patterns and discriminatory factors associated with SAA content. For supervised analyses, variation not correlated to classification was removed with orthogonal signal correction/partial least squares-discriminant analysis (OPLS-DA) using Pirouette software (Infometrix) after centering the mean value for each frequency (30–32). One of the limitations to the use of OPLS-DA is that data can give a false impression of the accuracy of classification, because class information is implicitly used to remove variability of input data orthogonal to the response. To validate the OPLS-DA classification model, a random permutation test was applied to obtain misclassification counts and Q2, a measure for classification prediction ability (33, 34). This permutation test evaluates whether the specific classification of the individuals in the 2 designed groups is significantly better than any other random classification in 2 arbitrary groups. In the OPLS-DA model, each time point (except baseline on d1) for d 1 and 5 was designated as SAA depletion, and each for d 6 and 10 was designated as SAA repletion. Baseline on d 1 was designated as SAA repletion, because this time point followed equilibration to adequate SAA intake.
To complement the use of loading plots in identification of spectral regions that differ due to SAA-content of meals, false discovery rate (FDR) (35, 36) was performed using MATLAB. For spectral regions identified by FDR, we integrated values associated with known plasma metabolites for each individual to generate plots of relative changes in metabolite concentration as a function of time. In the FDR model, we grouped the data according to day of measurement as was done with the OPLS-DA model. We performed a mixed model, repeated-measures ANOVA to determine the effect of SAA, time, and the interaction of SAA and time (SAA × time) and then used a paired t test for each frequency to find the significant regions that discriminated between SAA depletion and repletion at P < 0.05. For the lipid analyses, the same statistical procedure with a mixed model, repeated-measures ANOVA was performed to determine the effect of SAA, time, and interaction of SAA and time followed by a paired t test to find the significance for each time point that discriminated between SAA depletion and repletion at P < 0.05. To obtain detailed information on the effect of SAA in a single meal on postprandial plasma metabolites, we compared each time point between d 1 and 10 (equilibrated to SAA-containing food) and between d 5 and 6 (equilibrated to SAA-free food) using mixed measures ANOVA and then paired t test as described above.
Results
PCA of 1H-NMR spectra of plasma samples.
PCA of spectra for all 258 samples was performed to determine if differences between SAA deficiency and SAA repletion periods were apparent in macronutrient profiles. However, even though the first 2 PC accounted for 65% of the total variance, no separation was evident between the study periods in the 2-D PCA score plot (Supplemental Fig. 2A). Further examination of score plots for different combinations of the first 9 PC did not reveal any differences (not shown); however, some separation was apparent for individuals when they were separately color-coded within this analysis (Supplemental Fig. 3).
To determine whether response patterns to SAA deficiency could be identified within individuals, PCA was performed separately for the series of 34 spectra from each participant. Results showed separation in 2-D PCA plots according to SAA intake (Supplemental Fig. 2B–I). For some individuals, additional separation was visible in 3-D PCA plots in which the axes were rotated to enhance visualization (Supplemental Fig. 4). These data indicate that common response patterns are at least partially obscured by inter-individual variation.
Examination of metabolic patterns in averaged 1H-NMR spectra.
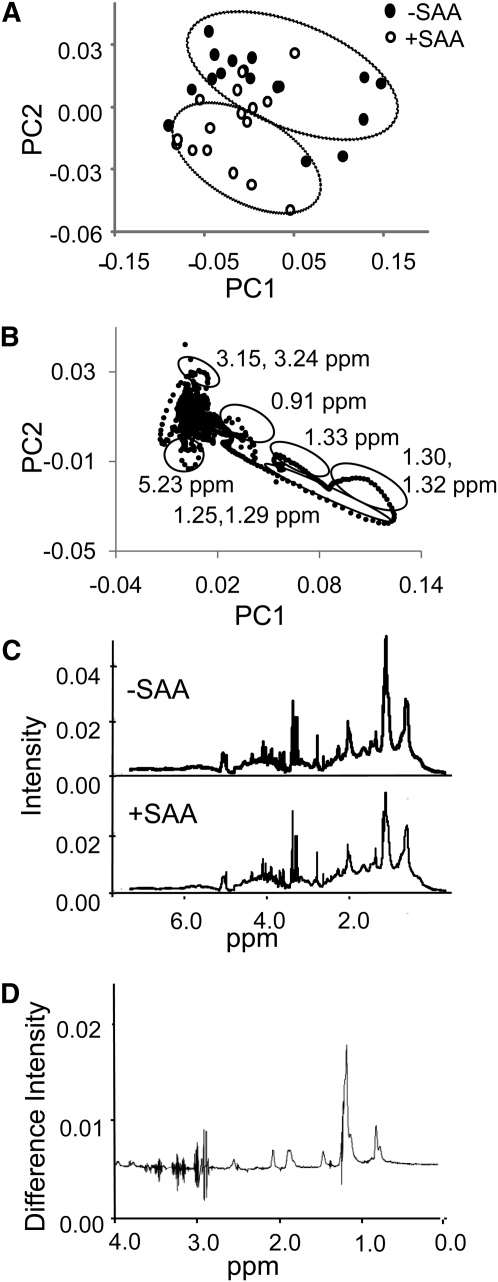
To decrease contribution from inter-individual variation, spectra from the 8 individuals were averaged for each time point to assess SAA-dependent metabolic changes. With this averaged dataset for 34 time points, PCA results showed that PC1 and PC2 accounted for 75 and 8% of variation, respectively, and trajectories for SAA-free and SAA intake were separated (Fig. 1A). The corresponding loading plot indicated that major discriminatory regions for the SAA-free period included those containing lipid (1.25, 1.29, 1.30, 1.32, and 5.23 ppm) (Fig. 1B). This plot also indicated that signals associated with amino acids (3.15, 3.24) contributed to the separation of the SAA-sufficient period. These conclusions were verified by visual inspection of the average spectra for the 1030 time point with and without SAA (Fig. 1C), which showed differences in signal intensities for these regions. The differences were also apparent in the difference spectrum obtained by subtracting the average spectrum with SAA from the average spectrum without SAA (Fig. 1D).
FIGURE 1.
Principal component analysis of 1H-NMR spectra of human plasma that were averaged for 8 participants at each time point during sulfur amino acid (SAA)-depletion and SAA-repletion study periods. (A) Two-dimensional score plot. (B) Loading plot corresponding to A. The principal components (PC1 and PC2) are unit free. Some of the regions (illustrated by circles in B) contributed to separation according to SAA content of food. These included spectral regions associated with LDL and VLDL (1.25, 1.29), cholesterol (0.91), lipid (1.30, 1.32), unsaturated lipid (5.23), lactate (1.33), and amino acids (3.15, 3.24). (C) Representative spectra illustrating change due to SAA-free meal 2 h after eating. Spectrum for 1030 during SAA-free period is shown in the upper panel and the corresponding spectrum for the SAA-replete period is shown in the lower panel. (D) Difference spectrum created by subtracting SAA-replete spectrum from SAA-free spectrum (1030 time points, C) illustrates spectral regions affected by dietary SAA. The predominant 2 peaks are associated with lipids.
OPLS-DA of 1H-NMR spectra according to SAA intake.
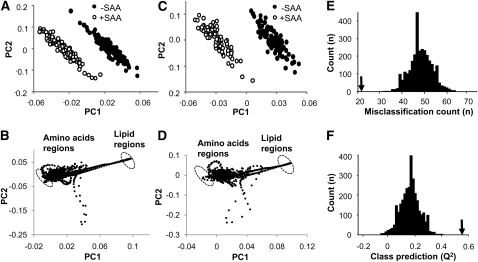
To more specifically examine spectral features that distinguished the SAA-free and SAA-sufficient conditions, an orthogonal signal correction method, OPLS-DA, was used to remove noncorrelated variation. The results showed separation according to SAA content, with better separation for the postprandial data alone (Fig. 2C) than for the entire sample set (Fig. 2A). The large distance between classes implies that the classification model is sufficient to separate classes from each other. The loading plots (Fig. 2B,D) confirmed that discriminatory regions included lipid and amino acid signals, which were represented by circles. These analyses further indicated that spectral regions associated with VLDL (1.29 ppm) and cholesterol (0.91 ppm) contributed to the discrimination due to SAA. These supervised analyses also showed contributions due to lactate (1.33 ppm) and amino acids (isoleucine, 1.00; valine, 1.02; glutamine, 2.09) (29). A histogram showing misclassification for 3000 randomly permuted data is shown in Figure 2E along with misclassification for the OPLS-DA model, shown by an arrow. Q2 for the random permutation distribution is shown by the histogram in Figure 2F and the original OPLS-DA classification is indicated by the arrow.
FIGURE 2.
Orthogonal signal correction partial least squares-discriminant analysis (OPLS-DA) of proton NMR (1H-NMR) spectra of human plasma to identify spectral regions that discriminate according to dietary sulfur amino acid (SAA) content. A shows the score plot and B the corresponding loading plot for all 258 spectra. (C,D) The score plot and corresponding loading plot of OPLS-DA for 181 spectra obtained during the postprandial periods at 0930, 1030, 1130, 1230, 1430, and 1630 h. The principal components (PC1 and PC2) in A–D are unit free. (B,D) The discriminatory regions between SAA deficiency and repletion on loading plots are shown by circles. These included signals associated with lipid and amino acid (B,D). (E,F) Class model validation for OPLS-DA. In E, the arrow corresponds to the misclassification in the OPLS-model; results for 3000 random permutations are shown in the histogram. In F, Q2 for the random permutation distribution is shown in the histogram and was between –0.1 and 0.4. The arrow corresponds to the value for the OPLS-DA model.
FDR analysis of spectral differences due to SAA content of food.
As an alternative means to test for spectral frequencies that contributed significantly to discriminate based upon SAA intake, we used a multiple testing procedure based on the concept of the FDR (35,36). Results of this analysis (Table 1) showed several frequencies associated with lipid regions, including those of cholesterol, SFA, unsaturated fatty acids, LDL cholesterol, VLDL cholesterol, and HDL cholesterol, were significantly different according to SAA intakes. This analysis also showed significant differences in spectral regions associated with lactate and amino acids (Table 1). Taken together with the unsupervised and supervised data reduction approaches, these statistical analyses show that consumption of SAA-free food has effects on postprandial plasma concentrations of lipids and amino acids as measured by 1H-NMR spectroscopy.
TABLE 1.
Regions of 1H-NMR spectra of human plasma with significant differences due to SAA intake according to false discovery rate (FDR) analysis1
| Region (ppm) | Possible feature | Assignment | P |
| Lipid signals | |||
| 0.66 | Cholesterol | C18 (in HDL) | 0.002 |
| 0.70 | Cholesterol | C18 (in VLDL) | <0.001 |
| 0.84 | Lipid (mainly LDL) | C26 and C27 | <0.001 |
| 0.91 | Cholesterol | C21 | 0.002 |
| 0.93 | Lipid | CH3CH2 | <0.001 |
| 1.22 | Lipid | CH3CH2CH2 | <0.001 |
| 1.25 | Lipid | CH3CH2(CH2)n | 0.006 |
| 1.29 | Lipid (mainly VLDL) | CH2CH2CH2CO | <0.001 |
| 1.30 | Lipid | CH2 | 0.004 |
| 1.32 | Lipid | CH2CH2CH2CO | 0.004 |
| 1.97 | Lipid | CH2C = C | 0.008 |
| 2.00 | Lipid | CH2C = C | <0.001 |
| 2.72 | Lipid | C = CCH2C = C | 0.009 |
| 3.66 | Choline (lipid) | NCH2 | <0.001 |
| 5.23 | Unsaturated lipid | CH = CHCH2CH = CH | <0.001 |
| 5.26 | Unsaturated lipid | CH = CHCH2CH = CH | <0.001 |
| 5.27 | Unsaturated lipid | =CHCH2CH2 | <0.001 |
| 5.29 | Unsaturated lipid | CH = CHCH2CH = CH | <0.001 |
| 5.31 | Unsaturated lipid | =CHCH2CH2 | <0.001 |
| 5.33 | Unsaturated lipid | =CHCH2CH2 | <0.001 |
| Amino acid signals | |||
| 0.93 | Isoleucine | δ-CH3 | 0.003 |
| 0.95 | Leucine | δ-CH3 | 0.002 |
| 0.97 | Leucine | δ-CH3 | 0.002 |
| 0.97 | Valine | CH3 | <0.001 |
| 1.00 | Isoleucine | β-CH3 | 0.004 |
| 1.02 | Valine | CH3 | <0.001 |
| 1.28 | Isoleucine | half γ-CH2 | 0.003 |
| 1.99 | Proline | γ-CH2 | <0.001 |
| 2.00 | Glutamate | half β-CH2 | <0.001 |
| 2.05 | Proline | half β-CH2 | <0.001 |
| 2.08 | Glutamine | half β-CH2 | <0.001 |
| 2.09 | Glutamine | half β-CH2 | <0.001 |
| 2.81 | Aspartate | half β-CH2 | 0.009 |
| 3.15 | Citrulline | γ-CH2 | 0.009 |
| 3.24 | Arginine | δ-CH2 | 0.002 |
| 3.94 | Tyrosine | α-CH | 0.009 |
| 4.12 | Proline | α-CH | <0.001 |
| Other signals | |||
| 1.13 | Isobutyrate | CH3 | <0.001 |
| 1.20 | 3-Hydroxybutyrate | γ-CH3 | <0.001 |
| 1.33 | Lactate | CH3 | 0.004 |
| 4.11 | Lactate | CH | <0.001 |
| 3.21 | Choline | N(CH3)3 | <0.001 |
| 4.13 | 3-Hydroxybutyrate | β-CH | 0.002 |
| 2.04 | Glycoprotein | NHCOCH3 | <0.001 |
Spectral regions with significant differences are associated with major plasma metabolites known to have H-NMR signal with the respective ppm. Regions are grouped according to common macronutrient groups.
Postprandial time course of signal changes in selective regions due to SAA-free meal.
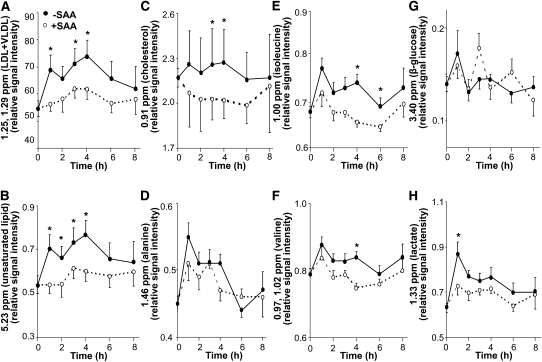
To visualize changes with time in spectral regions identified by FDR, we integrated signals within individual spectra corresponding to known metabolites and then constructed plots to test for changes due to SAA depletion in the postprandial period for the study periods equilibrated to adequate SAA intake (Fig. 3). Using a mixed model, repeated-measures ANOVA, effects of time and SAA intake were observed for signals corresponding to lipids ( ; 0.91 and 1.25, 1.29, 5.23 ppm; P < 0.0001). However, signals for amino acids differed in response characteristics, with alanine showing an effect of time but not SAA (Fig. 3D), whereas isoleucine (Fig. 3E; P < 0.001) and valine (Fig. 3F; P < 0.001) showed an effect of SAA and time. The values for the major ppm regions corresponding to glucose showed no effect of time or SAA (Fig. 3G; P < 0.35), but the region associated with lactate showed effects with both time and SAA (Fig. 3H; P < 0.005).
FIGURE 3.
Time-dependent changes in amplitude of selected spectral regions of human plasma proton NMR (1H-NMR) spectra in the postprandial period following a meal without or with sulfur amino acid (SAA). Regions were selected from those identified as significant in false discovery rate (FDR) analysis (Table 1) and are shown for the postprandial effect of dietary SAA content on d 1 and 10, which were equilibrated to adequate SAA intake. (A) Regions of LDL+VLDL centered at 1.25 and 1.29 ppm. (B) Region of unsaturated lipid centered at 5.23 ppm. (C) Region of cholesterol centered at 0.91 ppm. (D) Region of alanine centered at 1.46 ppm. (E) Region of isoleucine centered at 0.93 and 1.00 ppm. (F) Region of valine centered at 0.97 and 1.02 ppm. (G) Region of β-glucose centered at 3.24 ppm. (H) Region of lactate centered at 1.33 ppm. To determine the effect of SAA, time, and the interaction of SAA and time, a mixed model, repeated-measures ANOVA was performed. Significant main effects were SAA and time (A–C,E,F,H) or time (D). *Different from SAA-free at that time, P < 0.05.
Measurement of blood lipids in the postprandial period by conventional analysis.
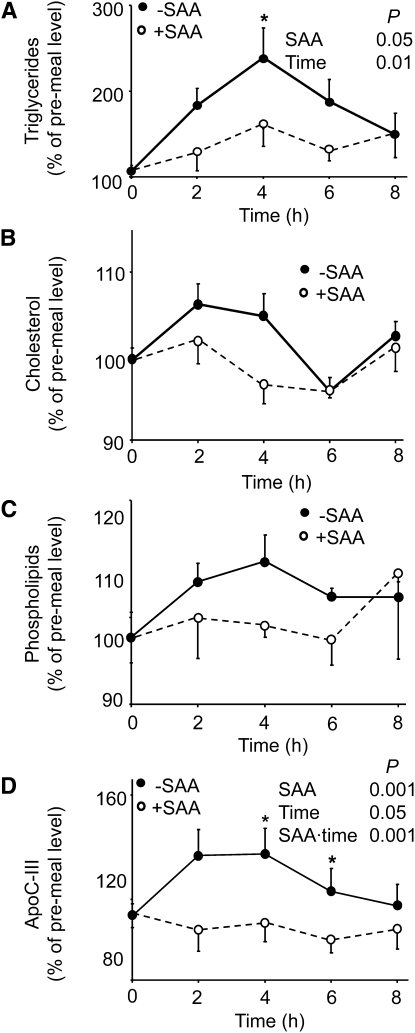
To confirm the results of 1H-NMR spectroscopy showing that consumption of SAA-free food resulted in higher plasma lipids, we used conventional chemical analysis for TG, total phospholipids, cholesterol, HDL, LDL, apoB, apoC-III, and hsCRP plasma concentrations. The results for the postprandial period equilibrated with adequate SAA diet were analyzed by repeated-measures ANOVA (Fig. 4). The plasma concentrations of TG and ApoC-III were different by dietary SAA contents and time after meal (Fig. 4; P < 0.05). Plasma cholesterol levels showed no difference by repeated-measures ANOVA, but showed a trend (P = 0.057) by paired t test at 4 h. Postprandial phospholipid concentrations did not significantly differ (Fig. 4C), and no significant differences according to SAA intake were observed for plasma HDL, LDL, apoB, or hsCRP levels (data not shown). Analysis following equilibration to SAA-free food also showed significant effects of SAA content on postprandial plasma TG and cholesterol (repeated measurement ANOVA; data not shown). Thus, direct lipid analyses confirmed that plasma TG were higher following consumption of SAA-free food.
FIGURE 4.
Effects of sulfur amino acid (SAA) content of meal on postprandial human plasma TG (A), cholesterol (B), phospholipids (C), and lipoprotein lipase inhibitor, apoC-III (D) following 3-d equilibration to a diet with adequate SAA content. Data are expressed as percentage of respective premeal concentration for each individual. Dietary SAA intake was 56 mg·kg−1·d−1. Values are given as mean ± SEM, n = 8. To determine the effect of SAA, time, and the interaction of SAA and time, a mixed model, repeated-measures ANOVA was performed. Significant main effects were SAA and time (A) or SAA, time, and SAA × time (D). *Different from SAA-free at that time, P < 0.05.
Additional information on changes in amounts of TG and phospholipid due to dietary SAA status were obtained using LC-MS. This method allowed detection of TG and phospholipid species in a small volume (10 μL), but most of these did not reveal significant differences in the small number of samples available for analysis. The TG 16:0/16:0/20:4 (Supplemental Fig. 5) showed the same pattern of intensity change as that observed by 1H-NMR spectroscopy (Fig. 3A) and conventional analysis (Fig. 4A). However, phosphatidylcholine, 14:1(9Z)/14:1(9Z) (Supplemental Fig. 6), showed a trend opposite to that for total phospholipid analysis (Fig. 4C).
Discussion
The present study used 1H-NMR spectroscopy of plasma to test for metabolic changes associated with intake of a SAA-free diet in humans. The spectroscopic method requires minimal sample processing and no chemical derivatization, allowing a rapid characterization of changes in high-abundance plasma metabolites. The method is attractive for development of personalized nutritional challenge protocols, because automated analysis with computer-based algorithms could provide near real-time evaluation of gastrointestinal dysfunction and macronutrient malabsorption. In previous studies, the method has been used for detection and quantification of metabolites related to fat, carbohydrate, and protein nutrition (37–39).
A key finding of the present study is that 1H-NMR spectroscopy can be used to study macronutrient metabolism following a meal challenge, specifically detecting lipid changes in the postprandial period upon consumption of a nutritionally imbalanced (SAA-free) meal. Although beyond the scope of this study, detailed studies of the SAA-dependent lipid changes are needed to understand this effect, because the 1H-NMR analysis provides global information, discriminating some lipid classes but not individual lipid species. This point is illustrated by the MS finding that change in a specific phosphatidylcholine did not match the total phospholipid analysis.
Plasma TG were increased by SAA-free food even though the content of fat was equivalent in the 2 diets. Because the total carbohydrate and amino acid-nitrogen were also constant, the data indicate that variables linked to Met or Cys, or some aspect of the amino acid imbalance created by SAA-free food, affects plasma concentrations of these lipids. Importantly, the changes in postprandial TG and apoC-III were immediate and seen after a single meal with SAA-free food. We have previously observed Cys/CySS redox effects on metalloproteinase activity and cell surface receptor (40), but in the present studies, we have no mechanistic evidence linking diet-dependent redox changes to changes in lipid metabolism.
The measured change in apoC-III, a known inhibitor of lipoprotein lipase responsible for TG hydrolysis (41), suggests that effects on TG concentrations could be mediated in part through interference with TG hydrolysis or increased TG production. Lipoprotein lipase is an enzyme produced in fat cells (adipocytes) and bound to the walls of capillaries. It breaks down triacylglycerols (TG) into FFA and monoglycerides, which can enter cells for storage. Changes in the N-methylation of phosphatidylethanolamine, which can be affected by SAA (1, 2), alter the composition of surface lipids on the TG-rich lipoproteins and thus affect the binding affinity of apoC-III. The rapid increase in apoC-III following the consumption of the first SAA-free meal would suggest that increased apoC-III production is unlikely to be responsible for this effect. These findings suggest a need for additional studies designed specifically to examine the role of dietary SAA on the regulation of TG metabolism.
As previously recognized, differences in rates of gastric emptying, intestinal absorption, and clearance of metabolites can affect plasma time courses on an individual basis (11). Individual differences are also apparent in the trajectories for individuals in the PCA due to SAA-free food. Individual trajectories in the PCA of all 256 samples show that individual metabolic responses differ even though all participants were studied under identical clinical research conditions and fed identical food. Research will be needed to characterize time courses and magnitudes of change for specific spectral regions to study dietary responses associated with risk of CVD or other disease process.
Metabolic effects of inadequate intake of an essential amino acid, Met, are expected due to an ensuing amino acid deficiency. However, deficiency does not occur with a single meal, so the altered amino acid signals probably reflect an amino acid imbalance of the meal rather than a nutritional deficiency. Surprisingly, the plasma 1H-NMR spectra showed that differences occurred in spectral regions associated with the essential amino acids isoleucine and valine, for which consumption was constant in the SAA depletion and SAA repletion periods. Although such changes in plasma amino acid concentrations in response to altered dietary SAA intake need independent validation, the complex patterns of response indicate that sophisticated bioinformatic analyses will be required for interpretation of metabolic profiles in dietary challenge studies.
The complexity of individual metabolic responses to dietary SAA content reinforces the need to improve metabolic profiling capabilities for individual health evaluation. Loscalzo et al. (42) addressed the inadequacies of contemporary approaches to address pathobiology of complex systems at the level of an individual. Information-rich metabolic profiling methods, such as 1H-NMR spectroscopy, can support new systems biology approaches in personalized medicine by providing a simple and reproducible approach to gain quantitative information on macronutrients in plasma. This measurement can be automated for routine use (38) and is complementary to MS and chemical approaches that measure individual metabolites (43).
In summary, the present study shows that 1H-NMR spectroscopy of human plasma can be used to evaluate macronutrient metabolism in a controlled meal challenge study. The results showed that lipid concentrations in humans are higher in the postprandial period following consumption of food without SAA. The results indicate a need to more specifically evaluate possible effects of SAA intake, as well as amino acid balance, on postprandial lipid concentrations in disease mechanisms.
Supplementary Material
Acknowledgments
Y.P. participated in NMR and mass spectral data collection, data analysis, interpretation, and manuscript preparation; N.A.L. participated in lipid analysis, data interpretation, and manuscript preparation; T.Y. participated in statistical analysis and interpretation; F.S. participated in mass spectral analysis and interpretation; N.G.M. participated in design detail, participant recruitment, and coordination of the clinical aspects of this study; C.J.A. participated in experimental planning and coordination of the clinical and analytic aspects of this study; K.S.L. participated in statistical analysis and interpretation; S.W. participated in NMR spectral data collection and interpretation; T.R.Z. participated in clinical design, supervision, data interpretation, and manuscript preparation; and D.P.J. participated in study design, analytic supervision, data interpretation, and manuscript preparation. All authors read and approved of the final manuscript.
Footnotes
Supported by NIH grants ES012929, ES011195, and DK066008 (D.P.J.), DK55850 and K24 RR023356 (T.R.Z.), and the Emory University General Clinical Research Center grant M01 RR00039/UL1 RR025008.
Supplemental Table 1 and Supplemental Figures 1–6 are available from the “Online Supporting Material” link in the online posting of the article and from the same link in the online table of contents at jn.nutrition.org.
Abbreviations used: CVD, cardiovascular disease; Cys, cysteine; CySS, cystine; D, dimensional; FDR, false discovery rate; GCRC, General Clinical Research Center; 1H-NMR, proton NMR; hsCRP, high-sensitivity C-reactive protein; PC, principal component; PCA, principal component analysis; OPLS-DA, orthogonal signal correction/partial least squares-discriminant analysis; SAA, sulfur amino acid.
Literature Cited
- 1.Splaver A, Lamas GA, Hennekens CH. Homocysteine and cardiovascular disease: biological mechanisms, observational epidemiology, and the need for randomized trials. Am Heart J. 2004;148:34–40 [DOI] [PubMed] [Google Scholar]
- 2.Wald DS, Law M, Morris JK. Homocysteine and cardiovascular disease: evidence on causality from a meta-analysis. BMJ. 2002;325:1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boushey CJ, Beresford SA, Omenn GS, Motulsky AG. A quantitative assessment of plasma homocysteine as a risk factor for vascular disease. Probable benefits of increasing folic acid intakes. JAMA. 1995;274:1049–57 [DOI] [PubMed] [Google Scholar]
- 4.Tofler GH, DAgostino RB, Jacques PF, Bostom AG, Wilson PW, Lipinska I, Mittleman MA, Selhub J. Association between increased homocysteine levels and impaired fibrinolytic potential: potential mechanism for cardiovascular risk. Thromb Haemost. 2002;88:799–804 [PubMed] [Google Scholar]
- 5.Ashfaq S, Abramson JL, Jones DP, Rhodes SD, Weintraub WS, Hooper WC, Vaccarino V, Harrison DG, Quyyumi AA. The relationship between plasma levels of oxidized and reduced thiols and early atherosclerosis in healthy adults. J Am Coll Cardiol. 2006;47:1005–11 [DOI] [PubMed] [Google Scholar]
- 6.Essex DW, Li M. Redox control of platelet aggregation. Biochemistry. 2003;42:129–36 [DOI] [PubMed] [Google Scholar]
- 7.Go YM, Jones DP. Intracellular proatherogenic events and cell adhesion modulated by extracellular thiol/disulfide redox state. Circulation. 2005;111:2973–80 [DOI] [PubMed] [Google Scholar]
- 8.Neuman RB, Bloom HL, Shukrullah I, Darrow LA, Kleinbaum D, Jones DP, Dudley SC., Jr Oxidative stress markers are associated with persistent atrial fibrillation. Clin Chem. 2007;53:1652–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanco RA, Ziegler TR, Carlson BA, Cheng PY, Park Y, Cotsonis GA, Accardi CJ, Jones DP. Diurnal variation in glutathione and cysteine redox states in human plasma. Am J Clin Nutr. 2007;86:1016–23 [DOI] [PubMed] [Google Scholar]
- 10.Park Y, Ziegler TR, Gletsu-Miller N, Liang Y, Yu T, Accardi CJ, Jones DP. Postprandial cysteine/cystine redox potential in human plasma varies with meal content of sulfur amino acids. J Nutr. 2010;140:760–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park Y, Kim SB, Wang B, Blanco RA, Le NA, Wu S, Accardi CJ, Alexander RW, Ziegler TR, et al. Individual variation in macronutrient regulation measured by proton magnetic resonance spectroscopy of human plasma. Am J Physiol Regul Integr Comp Physiol. 2009;297:R202–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoon YJ, Park Y, Jones DP, Ziegler TR, Vidakovic B. Self-similarity in NMR spectra: an application in assessing the level of cysteine. J Data Sci. 2010;8:1–19 [PMC free article] [PubMed] [Google Scholar]
- 13.Serkova NJ, Niemann CU. Pattern recognition and biomarker validation using quantitative 1H-NMR-based metabolomics. Expert Rev Mol Diagn. 2006;6:717–31 [DOI] [PubMed] [Google Scholar]
- 14.van Ommen B, Stierum R. Nutrigenomics: exploiting systems biology in the nutrition and health arena. Curr Opin Biotechnol. 2002;13:517–21 [DOI] [PubMed] [Google Scholar]
- 15.Watkins SM, German JB. Toward the implementation of metabolomic assessments of human health and nutrition. Curr Opin Biotechnol. 2002;13:512–6 [DOI] [PubMed] [Google Scholar]
- 16.Lindon JC, Holmes E, Nicholson JK. Pattern recognition methods and applications in biomedical magnetic resonance. Prog Nucl Magn Reson Spectrosc. 2001;39:1–40 [Google Scholar]
- 17.Steuer R. Review: on the analysis and interpretation of correlations in metabolomic data. Brief Bioinform. 2006;7:151–8 [DOI] [PubMed] [Google Scholar]
- 18.Trygg J, Holmes E, Lundstedt T. Chemometrics in metabonomics. J Proteome Res. 2007;6:469–79 [DOI] [PubMed] [Google Scholar]
- 19.Eriksson L, Antti H, Gottfries J, Holmes E, Johansson E, Lindgren F, Long I, Lundstedt T, Trygg J, et al. Using chemometrics for navigating in the large data sets of genomics, proteomics, and metabonomics (gpm). Anal Bioanal Chem. 2004;380:419–29 [DOI] [PubMed] [Google Scholar]
- 20.Bathen TF, Krane J, Engan T, Bjerve KS, Axelson D. Quantification of plasma lipids and apolipoproteins by use of proton NMR spectroscopy, multivariate and neural network analysis. NMR Biomed. 2000;13:271–88 [DOI] [PubMed] [Google Scholar]
- 21.Brindle JT, Antti H, Holmes E, Tranter G, Nicholson JK, Bethell HW, Clarke S, Schofield PM, McKilligin E, et al. Rapid and noninvasive diagnosis of the presence and severity of coronary heart disease using 1H-NMR-based metabonomics. Nat Med. 2002;8:1439–44 [DOI] [PubMed] [Google Scholar]
- 22.Ala-Korpela M, Hiltunen Y, Jokisaari J, Eskelinen S, Kiviniity K, Savolainen MJ, Kesaniemi YA. A comparative study of 1H NMR lineshape fitting analyses and biochemical lipid analyses of the lipoprotein fractions VLDL, LDL and HDL, and total human blood plasma. NMR Biomed. 1993;6:225–33 [DOI] [PubMed] [Google Scholar]
- 23.Ala-Korpela M, Korhonen A, Keisala J, Horkko S, Korpi P, Ingman LP, Jokisaari J, Savolainen MJ, Kesaniemi YA. 1H NMR-based absolute quantitation of human lipoproteins and their lipid contents directly from plasma. J Lipid Res. 1994;35:2292–304 [PubMed] [Google Scholar]
- 24.Fukagawa NK, Ajami AM, Young VR. Plasma methionine and cysteine kinetics in response to an intravenous glutathione infusion in adult humans. Am J Physiol. 1996;270:E209–14 [DOI] [PubMed] [Google Scholar]
- 25.Raguso CA, Regan MM, Young VR. Cysteine kinetics and oxidation at different intakes of methionine and cystine in young adults. Am J Clin Nutr. 2000;71:491–9 [DOI] [PubMed] [Google Scholar]
- 26.Lyons J, Rauh-Pfeiffer A, Yu YM, Lu XM, Zurakowski D, Tompkins RG, Ajami AM, Young VR, Castillo L. Blood glutathione synthesis rates in healthy adults receiving a sulfur amino acid-free diet. Proc Natl Acad Sci USA. 2000;97:5071–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee G-C, Woodruff DL. Beam search for peak alignment of NMR signals. Anal Chim Acta. 2004;513:413–6 [Google Scholar]
- 28.Forshed J, Schuppe-Koistinen I, Jacobbson SP. Peak alignment of NMR signals by means of a genetic algorithm. Anal Chim Acta. 2003;487:189–99 [Google Scholar]
- 29.Nicholson JK, Foxall PJ, Spraul M, Farrant RD, Lindon JC. 750 MHz 1H and 1H–13C NMR spectroscopy of human blood plasma. Anal Chem. 1995;67:793–811 [DOI] [PubMed] [Google Scholar]
- 30.Bylesjo M, Eriksson D, Sjodin A, Jansson S, Moritz T, Trygg J. Orthogonal discriminant analysis: combining the strengths of PLS-DA and SIMCA classification. J Chemometrics. 2006;20:341–51 [Google Scholar]
- 31.Trygg J, Wold S. Orthogonal projections to latent structures (O-PLS). J Chemometrics. 2002;16:119–28 [Google Scholar]
- 32.Wiklund S, Johansson E, Sjostrom L, Mellerowicz EJ, Edlund U, Shockcor JP, Gottfries J, Moritz T, Trygg J. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal Chem. 2008;80:115–22 [DOI] [PubMed] [Google Scholar]
- 33.Westerhuis JA, Hoefsloot HC, Smit S, Vis DJ, Smilde AK, van Velzen EJ, van Duijnhoven JM, van Dorsten FA. Assessment of PLSDA cross validation. Metabolomics. 2008;4:81–9 [Google Scholar]
- 34.Westerhuis JA, van Velzen EJ, Hoefsloot HC, Smilde AK. Multivariate paired data analysis: multilevel PLSDA versus OPLSDA. Metabolomics. 2010;6:119–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc, B. 1995;57:289–300 [Google Scholar]
- 36.Storey JD. A direct approach to false discovery rates. J R Stat Soc, B. 2002;64:479–98 [Google Scholar]
- 37.Frolkis A, Knox C, Lim E, Jewison T, Law V, Hau DD, Liu P, Gautam B, Ly S, et al. SMPDB: the small molecule pathway database. Nucleic Acids Res. 2010;38:D480–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inouye M, Kettunen J, Soininen P, Silander K, Ripatti S, Kumpula LS, Hamalainen E, Jousilahti P, Kangas AJ, et al. Metabonomic, transcriptomic, and genomic variation of a population cohort. Mol Syst Biol. 2010;6:441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Psychogios N, Hau DD, Peng J, Guo AC, Mandal R, Bouatra S, Sinelnikov I, Krishnamurthy R, Eisner R, et al. The human serum metabolome. PLoS ONE. 2011;6:e16957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Go YM, Jones DP. Cysteine/cystine redox signaling in cardiovascular disease. Free Radic Biol Med. 2011;50:495–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eurlings PMH, van der Kallen CJH, Geurts JMW, Flavell DM, de Bruin TWA. Identification of the PPARA locus on chromosome 22q 13.3 as a modifier gene in familial combined hyperlipidemia. Mol Genet Metab. 2002;77:274–81 [DOI] [PubMed] [Google Scholar]
- 42.Loscalzo J, Kohane I, Barabasi AL. Human disease classification in the postgenomic era: a complex systems approach to human pathobiology. Mol Syst Biol. 2007;3:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson JM, Yu T, Strobel FH, Jones DP. A practical approach to detect unique metabolic patterns for personalized medicine. Analyst. 2010;135:2864–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.