Abstract
NifEN plays an essential role in the biosynthesis of the nitrogenase FeMo cofactor (M-cluster). It is an α2β2 tetramer that is homologous to the catalytic MoFe protein (NifDK) component of nitrogenase. NifEN serves as a scaffold for the conversion of an iron-only precursor to a matured form of the M-cluster before delivering the latter to its target location within NifDK. Here, we present the structure of the precursor-bound NifEN of Azotobacter vinelandii at 2.6 Å resolution. From a structural comparison of NifEN with des-M-cluster NifDK and holo NifDK, we propose similar pathways of cluster insertion for the homologous NifEN and NifDK proteins.
Nitrogenase is a complex metalloenzyme that catalyzes a key step in the global nitrogen cycle: the reduction of atmospheric dinitrogen to bioavailable ammonia. The Mo-dependent nitrogenase is a two-component system, in which the Fe protein (NifH) mediates the ATP-dependent transfer of electrons to the catalytic MoFe protein (NifDK) during substrate turnover (1). The MoFe protein is an α2β2-tetramer that contains two unusual metalloclusters per αβ-dimer, the P-cluster and the M-cluster. The P-cluster is an [8Fe-7S] cluster at the α/β-subunit interface, coordinated by three Cys ligands from the α-subunit and three Cys ligands from the β-subunit. The M-cluster (or FeMo cofactor) is a [Mo-7Fe-9S-X-homocitrate] cluster (where X = C, N or O) located within the α-subunit, coordinated by a His ligand at the Mo end and a Cys ligand at the opposite Fe atom, with a Lys residue binding to the homocitrate entity (2, 3). During catalysis, the P-cluster is thought to mediate the electron flow from the Fe protein to the M-cluster, where substrate reduction occurs.
NifEN is an essential player in M-cluster biosynthesis (4–6). It presumably receives a precursor form of the M-cluster from NifB and hosts the conversion of this precursor to a mature M-cluster before delivering the latter to the MoFe protein (4, 5). A role for NifEN in FeMoco biosynthesis was initially hypothesized based on a considerable degree of similarity between the primary sequences of NifEN and MoFe protein, suggesting that NifEN may contain P- and M-like clusters (6). Subsequently, the NifEN-associated clusters were identified through the biochemical and spectroscopic analyses of three forms of NifEN. The first, apo NifEN, is free of any cofactor species and contains a [4Fe-4S] cluster in place of the [8Fe-7S] P-cluster (7). The second, NifEN, contains, in addition to the [4Fe-4S] cluster, an all-iron precursor that closely resembles the Fe/S core of the M-cluster (8). The third, holo NifEN, contains a mature M-cluster and the [4Fe-4S] cluster (9). NifEN could be readily converted to holo NifEN by Fe protein-mediated insertion of Mo and homocitrate into the precursor (10), and holo NifEN could directly serve as a cofactor donor for the apo MoFe protein (11). These observations not only establish the role of NifEN in cofactor biosynthesis, but also illustrate the dynamic nature of the cofactor site in NifEN during the assembly process. However, the mechanistic details of the biosynthetic events on NifEN have remained unclear without structural information on this protein. In this study, the structure of the precursor-bound NifEN of Azotobacter vinelandii has been solved to 2.6 Å resolution (12). From a structural comparison of NifEN with des-M-cluster MoFe protein (apo NifDK) (11) and holo MoFe protein (NifDK) (2, 3), we propose similar pathways of cluster insertion for the homologous NifEN and NifDK proteins.
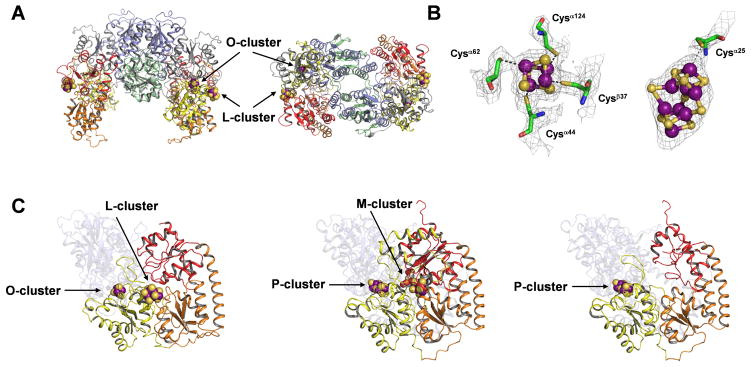
The crystal structure determination of NifEN is summarized in Tables S1 and S2. Like the α2β2-tetrameric NifDK, NifEN consists of a pair of αβ-dimers that are related by a molecular twofold rotation axis (Fig. 1A). The α- and β-subunits of NifEN, like those of NifDK and apo NifDK, are composed of three domains each—αI, αII, and αIII, and βI, βII, and βIII, respectively. All domains of NifEN, as for NifDK and apo NifDK, are organized around a common core of a four-stranded, parallel β-sheet flanked with α-helices and additional β-strands (13). Moreover, NifEN contains two types of clusters that correspond to the P- and M-clusters in NifDK: one, termed the O-cluster, is a [4Fe-4S] cluster that is coordinated by Cys α37, Cys α62, Cys α124 and Cys β44 at the same site in the α/β-subunit interface as the P-cluster; the other, termed the L-cluster, is an iron-only precursor form of the M-cluster that is at least ligated by Cys α25 at one end (Fig. 1B) (14). Although the electron density is not sufficiently well resolved to unambiguously establish the structure of the L-cluster, the shape and extent of the density is compatible with the core geometry of the M-cluster and, therefore, consistent with the previously proposed 8Fe-model (8) of this M-cluster precursor (Fig. 1B).
Fig. 1.
(A) The structure of the NifEN tetramer with the molecular twofold axis oriented vertically (left) and along the viewing direction (right). The domains of the α-subunits are colored yellow (αI), orange (αII), and red (αIII); and the domains of β-subunits are colored gray (βI), blue (βII), and green (βIII). The L-cluster (precursor of M-cluster) and O-cluster ([4Fe-4S] cluster) are illustrated as space-filling models, with atoms colored as follows: Fe, purple; S, yellow. (B) Structures and ligands of O-cluster (left) and L-cluster (right) overlaid with electron density maps. Both clusters are illustrated as ball-and-stick models, whereas the ligands are shown in stick presentation. Note that additional L-cluster ligands may be present (14). The atoms are colored as follows: Fe, purple; S, yellow; O, red; C, green; N, blue. The structure of L-cluster overlaid with anomalous difference electron density map is shown in Fig. S1. (C) Structures of the αβ-pairs of NifEN (left), NifDK (middle, PDB entry 1M1N) and apo NifDK (right, PDB entry 1L5H). The α-subunits are presented in the foreground, and the β-subunits are rendered transparent in the background. The domains of the subunits in all three proteins are colored as in A. All clusters are illustrated as space-filling models, with atoms colored as follows: Fe, purple; S, yellow; O, red; C, gray. The Mo atom and the interstitial ligand of the M-cluster are not visible.
Strikingly, although the L- and M-clusters are both positioned at the junction between the αI-, αII- and αIII-domains of their respective proteins, the L-cluster of NifEN is nearly surface exposed, possibly shielded from solvent by only a small stretch of disordered polypeptide between residues α14 and α24. In contrast, the M-cluster of NifDK occupies a buried site ~10 Å below the surface of the protein (2). Given the homology between NifEN and NifDK, it is possible that, following maturation, the M-cluster is inserted into a site in NifEN that is analogous to its binding site in NifDK. Significantly, the α-subunit of NifEN assumes a conformation that is more “open” than that of NifDK (presumably corresponding to holo NifEN) yet less “open” than that of apo NifDK (presumably corresponding to apo NifEN), suggesting that the α-subunit undergoes major structural rearrangements upon the maturation of the L-cluster on NifEN (Fig. 1C). Consistent with this proposal, a comparison of the molecular surfaces of these proteins reveals the presence of a positively charged funnel in apo NifDK, which is closed in NifDK containing a buried M-cluster (Fig. S2). There is a similar accumulation of positive charges on the surface of NifEN; however, an insertion funnel could not be clearly defined, perhaps due to a partial closure of the entrance by docking of the L-cluster (Fig. S2). In contrast, the β-subunits of the three proteins are similar in structure, except for the presence of an extended loop and an extra helix-turn-helix structure in the holo and apo NifDK (Fig. S3).
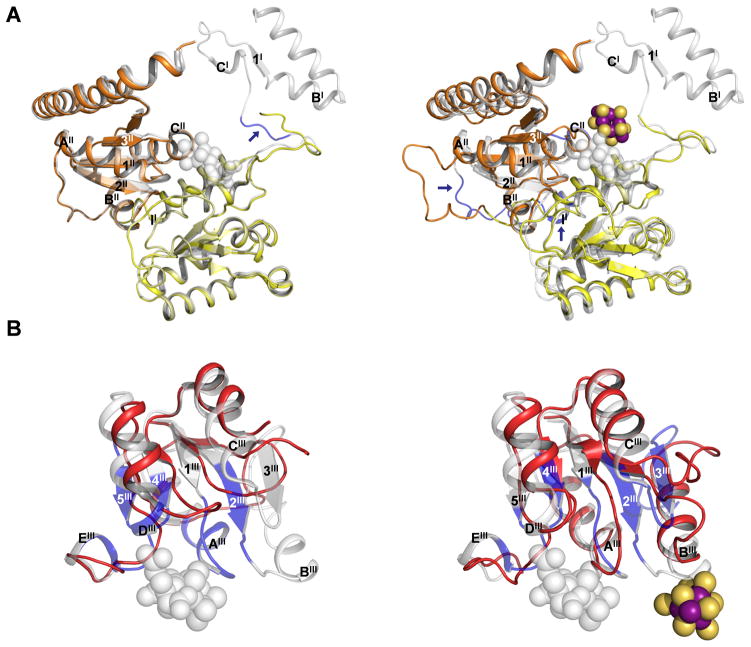
The αI- and αII-domains of apo NifDK do not show significant Cα-deviations from those of NifDK, except for a short loop (α49–52) that follows a disordered region (α1–48) at the N-terminus of the protein (Fig. S4A, upper; Fig. 2A, left). A similar loop (α24–27) at the N-terminus of NifEN aligns more closely with the corresponding region in NifDK (Fig. S4A, lower; Fig. 2A, right). Cys α25, which ligates one of the terminal Fe atoms of the L-cluster, is located near the end of this loop. Thus, this loop is likely oriented by docking of the L-cluster on Cys α25. Apart from this feature, NifEN contains an extra sequence (α187–192), which forms a larger external loop between the αI (α helix II)- and αII (β strand 1II)-domains (Fig. S4A and 2A). At the site that would contain a buried M-cluster in NifDK, the area surrounding the potential Cys α250 ligand (α248–252) features an α-helix (CII) that is shorter toward the N-terminus, with an extended loop in its place that could add flexibility to the dynamic M-cluster site in NifEN (Fig. S4A and 2A).
Fig. 2.
(A) Superposition of αI- and αII-domains of apo NifDK and NifDK (left) and those of NifEN and NifDK (right). The domains of apo NifDK and NifEN are color coded as in Fig. 1. NifDK is shown in gray (transparent). Regions with significant shifts in Cα-positions are colored blue and indicated by arrows, and the secondary structural elements of these regions are labeled according to those in Fig. S4A. The L-cluster is illustrated as in Fig. 1. The M-cluster is shown as a space-filling model in gray (transparent). PYMOL was used to prepare the figure (18). (B) Superposition of the αIII-domains of apo NifDK and NifDK (left) and those of NifEN and NifDK (right). Apo NifDK and NifEN are colored red; whereas NifDK is shown in gray (transparent). Regions with significant shifts in Cα-positions are colored blue. The secondary structural elements are labeled according to those shown in Fig. S4A. The L-cluster is illustrated as in Fig. 1. The M-cluster is shown as a space-filling model in gray (transparent). PYMOL was used to prepare the figure (19).
The αIII-domain of apo NifDK shows significant Cα-deviations from that of NifDK in four distinct areas: a “lid loop” (α353-α364), a disordered region (α381-α407), and regions surrounding the homocitrate anchor (α421-α428) and the histidine ligand (α436-α446) of the M-cluster, respectively (Fig. S4B, upper). By comparison, the αIII-domain of NifEN shows Cα-deviations from NifDK that are reminiscent of, yet much less significant than, those of apo NifDK from NifDK (Fig. S4B, lower). The β-strands (1III, 2III and 4III) and α-helices (AIII, CIII, and DIII) of apo NifDK are significantly shortened and displaced from those of NifDK (Fig. 2B, left). These changes result in a significant relocation of the lid loop and the adjacent (disordered) region in apo NifDK, which completely opens up the M-cluster site for cluster insertion (11). The same β-strands (1III, 2III and 4III) and α-helices (AIII, CIII, and DIII) in NifEN are shorter than those in NifDK; however, they are more extended and less displaced than those in apo NifDK, resulting in a conformation of NifEN intermediate between those of apo and holo NifDK (Fig. 2B, right). The changes of these structural elements, along with the structuring of α helix BIII, lead to the formation of an ordered, yet partially closed M-cluster site in NifEN (Fig. 2B, right). In particular, the downwardly bent β-strand 1III anchors the lid loop in a position close to that in NifDK; whereas β-strand 2III, together with α-helix BIII, extend toward the L-cluster at the surface of the protein (Fig. 2B, right). Two β-strands (3III and 5III) and one α-helix (EIII) are missing from the structures of both NifEN and apo NifDK (Fig. 2B). The presence of irregular loops in places of β-strand 3III (which “hinges” the α-helix BIII at the L-cluster site) and β-strand 5III/α-helix EIII (which flank the Asn ligand at the M-cluster site) may contribute to the conformational flexibility of the polypeptides surrounding the L- and M-cluster sites of NifEN, which could facilitate the transfer of the cluster from the L-cluster site to the M-cluster site upon maturation.
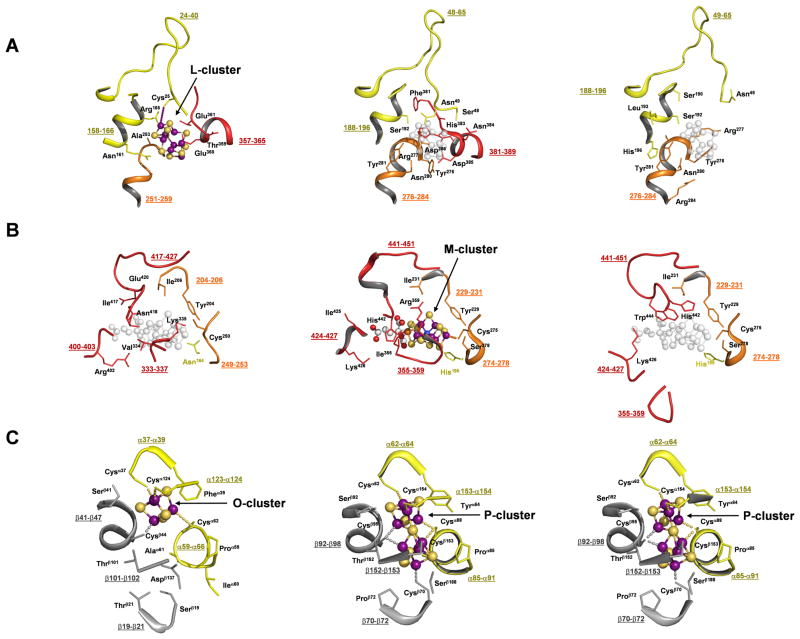
A closer examination of the L- and M-cluster sites in the three proteins provides additional insights into the pathway of M-cluster assembly. In all cases, an “L-cluster site” can be identified at the surface of the structure, located directly above the M-cluster site within the protein. The “L-cluster site” in apo NifDK is wide open, with a disordered region (α381–389) and a loop (α49–65) adjacent to Cys α45 shifted away from those in holo NifDK (Fig. 3A and S5A). The corresponding regions in NifEN (α357–365 and α24–40) are ordered, suggesting a concerted action between the two regions upon the ligation of the L-cluster to Cys α25 at the tip of the loop. The arrangement of the L-cluster site in NifEN is similar to that of the “L-cluster site” in NifDK; however, NifDK contains amino acids with bulky side chains (e.g., Phe α381, His α383, and Tyr α276) that close up the M-cluster site (located below the “L-cluster site”), whereas NifEN contains amino acids with small side chains (e.g., Ala α253 and Ser α251) that may provide the cluster with more freedom to move between the L- and M-sites during cluster assembly and delivery. A similar pattern is observed when the M-cluster sites of the three proteins are compared (Fig. 3B and S5B). The M-cluster site of apo NifDK is opened by a significant displacement of the lid loop (α355–359), while the corresponding loop in NifEN (α333–337) is positioned similarly to that in NifDK. Likewise, the likely ligands (Asn α418 and Cys α250) and homocitrate anchor (Arg α402) of the M-cluster in NifEN occupy positions that are similar to those of the M-cluster (His α442, Cys α275, and Lys α426) in NifDK. Despite these similarities, Tyr α444, which switches positions with His α442 and locks the M-cluster in place in NifDK through its bulky side chain (4), is not present in NifEN. This could facilitate the eventual release of the cluster from the M-cluster site of NifEN and its subsequent delivery to the homologous M-cluster site in NifDK. In contrast, the O-cluster site in NifEN assumes a nearly identical conformation to the P-cluster sites in apo and holo NifDK, except for the presence of an incomplete complement of Cys ligands that only accommodates a [4Fe-4S] cluster (Fig. 3C).
Fig. 3.
Close-up of the protein environment surrounding the L-cluster (A), M-cluster (B) and O/P-cluster (C) of NifEN (left), NifDK (middle) and apo NifDK (right). Shown are parts of the backbones (with numbers of residues underlined) and some side chain residues in the close vicinity of the clusters. The domains of the α- and β-subunits are colored as in Fig. 1A. All clusters are illustrated as ball-and-stick models, with atoms colored as follows: Fe, purple; S, yellow; O, red; C, gray; Mo, orange; X, blue. The superimposed positions of the L-cluster in NifDK and apo NifDK (A, middle and right), and of the M-cluster in NifEN and apo NifDK (B, left and right), are indicated in gray (transparent). PYMOL was used to prepare the figure (19).
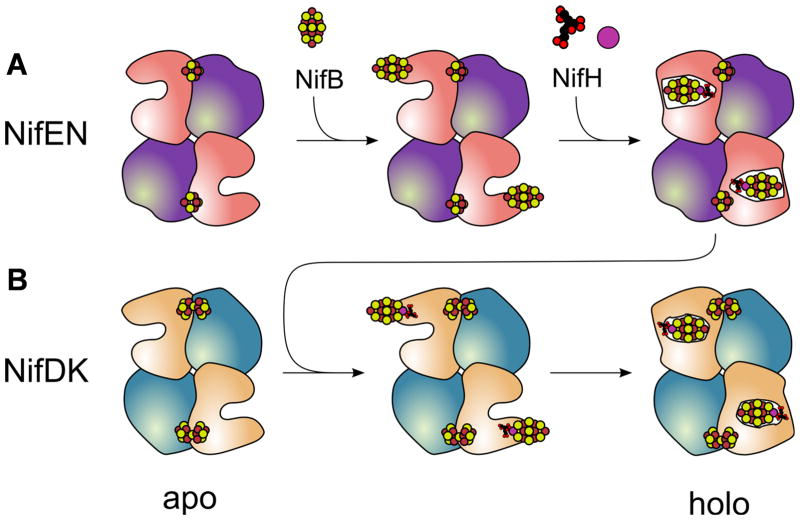
Similar pathways of cluster insertion can be proposed for NifEN and NifDK on the basis of these structural analyses (Fig. 4). It involves the formation of an apo protein that contains an open cluster insertion funnel, the docking of the cluster at (or near) the entrance of the funnel, and the subsequent insertion of the cluster into the funnel. In the case of NifEN, the L-cluster is delivered from NifB to the surface of apo NifEN, where it is poised to be transformed into a mature M-cluster upon the insertion of Mo and homocitrate by NifH (Fig. 4A). The M-cluster is then relocated from the surface to the binding site within NifEN. Interactions between NifEN and NifDK may cause conformational changes that release the M-cluster and facilitate its movement back to the surface of NifEN. Subsequently, the M-cluster is transferred to and incorporated into NifDK in a manner similar to the insertion of M-cluster into NifEN; only the M-cluster is securely locked at its binding site in this case (Fig. 4B). The coordination of the various cluster intermediates to cysteines located in loops (Cys α25 of NifEN and, potentially, the corresponding Cys α45 of NifDK) may serve to tether and facilitate efficient intermolecular transfer of these species without them escaping to the surrounding environment. Mechanistic parallels in this respect are evident between the functions of NifEN and metallochaperones for copper and other transition metals (15).
Fig. 4.
Schematic presentation of the proposed biosynthesis events that occur on NifEN (A) and NifDK (B). (A) Following the synthesis of an O-cluster containing, yet L-cluster deficient apo NifEN (left), the L-cluster is delivered from NifB to the entrance of the cluster insertion funnel in NifEN (middle). Subsequently, the Fe protein (NifH) inserts Mo and homocitrate into the L-cluster, resulting in a matured form of the M-cluster that is transferred to its binding site in NifEN (right). (B) NifDK first appears as a P-cluster containing, yet M-cluster deficient apo form (left). The mature M-cluster is then transferred from NifEN to the entrance of the cluster insertion funnel in NifDK upon direct protein-protein interactions (middle). Subsequently, the M-cluster is inserted into its binding site in NifDK, leading to the formation of holo NifDK (right). The proposed biosynthetic events on NifEN parallel those on NifDK, which is consistent with the close similarities between the overall structure and the cluster topology of the two proteins. The presence of O-cluster ([4Fe-4S]) and P-cluster ([8Fe-7S]) in apo NifEN and apo NifDK, respectively, may further imply an evolutionary connection between the two proteins. It can be hypothesized that the P-cluster site evolved from the O-cluster site upon the acquisition of missing ligands, thereby accommodating a second [4Fe-4S] cluster at this site for the subsequent coupling of two [4Fe-4S] modules into a mature P-cluster (20).
The presence of similar insertion funnels in these homologous proteins is further supported by the recently solved crystal structure of protochlorophyllide reductase (BchNB) (16, 17). Responsible for the reduction of a specific double bond of protochlorophyllide (Pchlide), BchNB is an α2β2 tetramer that assumes a conformation highly homologous to those of NifEN and NifDK (Fig. S6). Moreover, it contains one [4Fe-4S] cluster and one Pchlide at locations similar to those of the O/P-clusters and L/M-clusters in NifEN and NifDK. Interestingly, based on their locations in the respective homologous proteins, the L-cluster, M-cluster and Pchlide can be sequentially placed along the insertion funnel in apo NifDK (Fig. S7). This observation supports the hypothesis that a common ancestral protein—which appears to have adapted to accommodate different ligands—gave rise to all of these homologous proteins (18).
Supplementary Material
Acknowledgments
This work was supported by NIH grants GM-67626 (M.W.R.) and GM-45162 (D.C.R.) D.C.R. is an investigator of the Howard Hughes Medical Institute. We acknowledge the Gordon and Betty Moore Foundation for support of the Molecular Observatory at Caltech. Operations at SSRL are supported by the US DOE and NIH. The coordinates and structure factors have been deposited with the Protein Database, accession number 3PDI.
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
References and Notes
- 1.Burgess BK, Lowe DJ. Chem Rev. 1996;96:2983. doi: 10.1021/cr950055x. [DOI] [PubMed] [Google Scholar]
- 2.Kim J, Rees DC. Nature. 1992;360:553. doi: 10.1038/360553a0. [DOI] [PubMed] [Google Scholar]
- 3.Einsle O, et al. Science. 2002;297:1696. doi: 10.1126/science.1073877. [DOI] [PubMed] [Google Scholar]
- 4.Hu Y, Fay AW, Lee CC, Yoshizawa J, Ribbe MW. Biochemistry. 2008;47:3973. doi: 10.1021/bi7025003. [DOI] [PubMed] [Google Scholar]
- 5.Schwarz G, Mendel RR, Ribbe MW. Nature. 2009;460:839. doi: 10.1038/nature08302. [DOI] [PubMed] [Google Scholar]
- 6.Dos Santos PC, Dean DR, Hu Y, Ribbe MW. Chem Rev. 2004;104:1159. doi: 10.1021/cr020608l. [DOI] [PubMed] [Google Scholar]
- 7.Hu Y, Fay AW, Ribbe MW. Proc Natl Acad Sci USA. 2005;102:3236. doi: 10.1073/pnas.0409201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corbett MC, et al. Proc Natl Acad Sci USA. 2006;103:1238. doi: 10.1073/pnas.0507853103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Y, et al. Proc Natl Acad Sci USA. 2006;103:17119. doi: 10.1073/pnas.0602647103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Y, et al. Proc Natl Acad Sci USA. 2006;103:17125. doi: 10.1073/pnas.0602651103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmid B, et al. Science. 2002;296:352. doi: 10.1126/science.1070010. [DOI] [PubMed] [Google Scholar]
- 12.Materials and methods are detailed in supporting material on Science online.
- 13.Rees DC, Howard JB. Curr Opin Chem Biol. 2000;4:559. doi: 10.1016/s1367-5931(00)00132-0. [DOI] [PubMed] [Google Scholar]
- 14.The electron density around the L-cluster suggests that the cluster may be coordinated by more than one ligand to NifEN. However, since this region is largely disordered, it is difficult to accurately assign ligand(s) other than Cys α25.
- 15.Ma Z, et al. Chem Rev. 2009;109:4644. doi: 10.1021/cr900077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muraki N, et al. Nature. 2010;465:110. doi: 10.1038/nature08950. [DOI] [PubMed] [Google Scholar]
- 17.Bröcker MJ, et al. J Biol Chem. 2010;285:27336. doi: 10.1074/jbc.M110.126698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raymond J, Siefert JL, Staples CR, Blankenship RE. Mol Biol Evol. 2004;21:541. doi: 10.1093/molbev/msh047. [DOI] [PubMed] [Google Scholar]
- 19.www.pymol.com
- 20.Lee CC, et al. Proc Natl Acad Sci USA. 2009;106:18747. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.