Abstract
A large body of structural work conducted over the past ten years has elucidated mechanistic details related to 3′ to 5′ processing and decay of RNA substrates by the RNA exosome. This chapter will focus on the structural organization of eukaryotic exosomes and their evolutionary cousins in bacteria and archaea with an emphasis on mechanistic details related to substrate recognition and to 3′ to 5′ phosphorolytic exoribonucleolytic activities of bacterial and archaeal exosomes as well as the hydrolytic exoribonucleolytic and endoribonucleolytic activities of eukaryotic exosomes. These points will be addressed in large part through presentation of crystal structures of phosphorolytic enzymes such as bacterial RNase PH, PNPase, and archaeal exosomes and crystal structures of the eukaryotic exosome and exosome sub-complexes in addition to standalone structures of proteins that catalyze activities associated with the eukaryotic RNA exosome, namely Rrp44, Rrp6 and their bacterial counterparts.
Keywords: 3′ to 5′ RNA decay, exoribonuclease, endoribonuclease, exosome, RNA degradation, RNA processing, multi-subunit, structure, hydrolytic, phosphorolytic, RNA binding, PH domain, KH domain, S1 domain, cold-shock domain
Introduction
Enzymes that catalyze 3′ to 5′ RNA decay share evolutionary relationships throughout prokaryotic, archaeal and eukaryotic phylogeny (Fig. 1). 3′ to 5′ RNA decay is promoted by three distinct classes of enzymes that catalyze exoribonuclease activity in bacteria. One includes two related enzymes, RNase II and RNase R, which catalyze processive hydrolytic RNA decay. Another class includes the enzyme RNase D which catalyzes distributive hydrolytic RNA decay. The third class includes PNPase, a processive phosphorolytic exoribonuclease that is associated with the degradosome, a RNA decay complex comprised of PNPase, the endoribonuclease RNase E, the RNA helicase RhlB, and enolase1–2. PNPase is a multi-domain protein that homooligomerizes to form a ring-like structure with a central channel that harbors the phosphorolytic active sites.
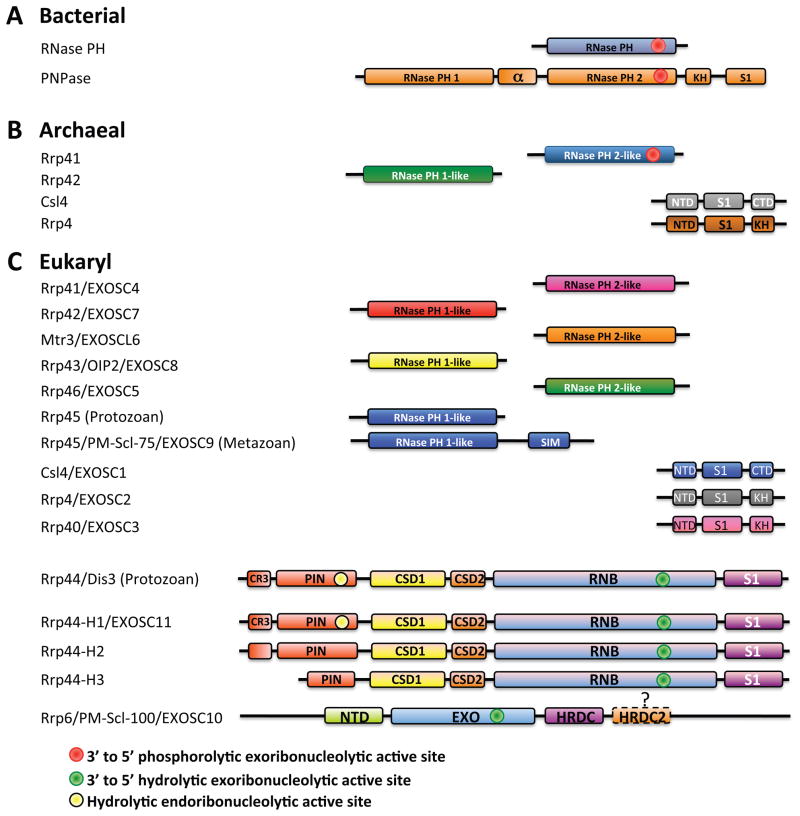
Figure 1. Schematics of domains in ‘exosomes’ from bacteria, archaea, and eukaryotes.
A) Bacterial RNase PH and PNPase subunits. Bacterial RNase PH contains a 3′ to 5′ phosphorolytic exoribonucleolytic active site (red circle). Bacterial PNPase contains five domains: RNase PH 1, the alpha domain, RNase PH 2, a KH domain, and an S1 domain. A 3′ to 5′ phosphorolytic exoribonucleolytic active site resides in the RNase PH 2 domain (red circle). B) Archaeal exosome subunits. Archaeal exosomes include four subunits: Rrp41, that contains a RNase PH 2-like domain with a 3′ to 5′ phosphorolytic exoribonucleolytic active site (red circle), Rrp42, that contains a RNase PH 1-like domain, and either Csl4 or Rrp4. Csl4 contains an N-Terminal Domain (NTD), a S1 domain, and a KH domain. Rrp4 contains an N-Terminal Domain (NTD), a S1 domain, and a Carboxy Terminal Domain (CTD). C) Eukaryotic exosome subunits. Protozoan and metazoan exosomes contain either ten or eleven components consisting of nine catalytically inert core components (Rrp41, Rrp42, Mtr3, Rrp43, Rrp46, Rrp45, Csl4, Rrp4, and Rrp40) and two active components Rrp44 and Rrp6. Alternative names for each of the eukaryotic subunits are included. Rrp44 contains five annotated domains: a PIN (PIlus N terminal) domain with a Cysteine-Rich sequence (CR3), two Cold Shock Domains (CSD1 and CSD2), a Ribo Nuclease Binding (RNB) domain, and an S1 domain. The “hydrolytic” endoribonucleolytic activity is located within the PIN domain (yellow circle), and the processive 3′ to 5′ hydrolytic exoribonucleolytic activity resides in the RNB domain (green circle). Metazoan exosomes may vary in their utilization of Rrp44 (e.g. three in human H1, H2, and H3). Rrp6 contains three known domains: NTD (N-Terminal Domain), EXO (EXOribonuclease domain), and HRDC (Homology to RNase D domain C-terminal). It is hypothesized that a second HRDC domain (HRDC2) may be located C-terminal to HRDC. 3′ to 5′ distributive hydrolytic endoribonucleolytic activity is located within the EXO domain (green circle).
Archaeal exosomes are processive phosphorolytic enzymes that share mechanistic and structural similarities to bacterial PNPase3–4 (Fig. 1B). Archaeal exosomes are composed of up to four individually encoded proteins that oligomerize to form an analogous structure to PNPase, although in this instance intact exosomes form by oligomerization of six subunits that make the ring and three additional subunits that cap the ring5–6. As with PNPase, archaeal exosomes have a central channel through which the RNA substrate must pass to gain access to the phosphorolytic active sites7–8.
The eukaryotic exosome core is architecturally similar to PNPase and archaeal exosomes, although it is more complex because it is composed of nine individually encoded subunits9. The eukaryotic exosome also differs fundamentally from PNPase and archaeal exosomes, because it is not a phosphorolytic enzyme and instead has developed the ability to directly associate with Rrp44 and Rrp6, hydrolytic exoribonucleases that share evolutionary relationships to bacterial RNase II/R and RNase D, respectively9–11.
In this chapter, we will describe the individual domains and overall architectures of enzymes and proteins that contribute to 3′ to 5′ decay through formation of exosomes or exosome-related complexes in bacterial, archaeal, and eukaryl organisms with an emphasis on what is currently known about their respective catalytic mechanisms and how the architecture of the intact exosome cores impacts their activities and function.
Global structure of the exosome
RNase PH, PNPase, archaeal and eukaryotic exosome cores are composed of evolutionarily related domains (Fig. 1) that oligomerize to form rings with central pores large enough to accommodate single stranded RNA (Fig. 2). RNase PH achieves this architecture through oligomerization of six RNase PH proteins (Fig. 1), resulting in a pseudo-hexameric ring with three-fold symmetry (Fig. 2A)12–14. The RNase PH ring includes six phosphorolytic active sites that are located in the interface between respective RNase PH proteins. The head to tail arrangement of RNase PH proteins around the ring generates a molecular two fold axis that situates three active sites on the bottom of the ring, another three active sites on the top of the ring, and RNA binding surfaces situated within the pore (Fig. 3A).
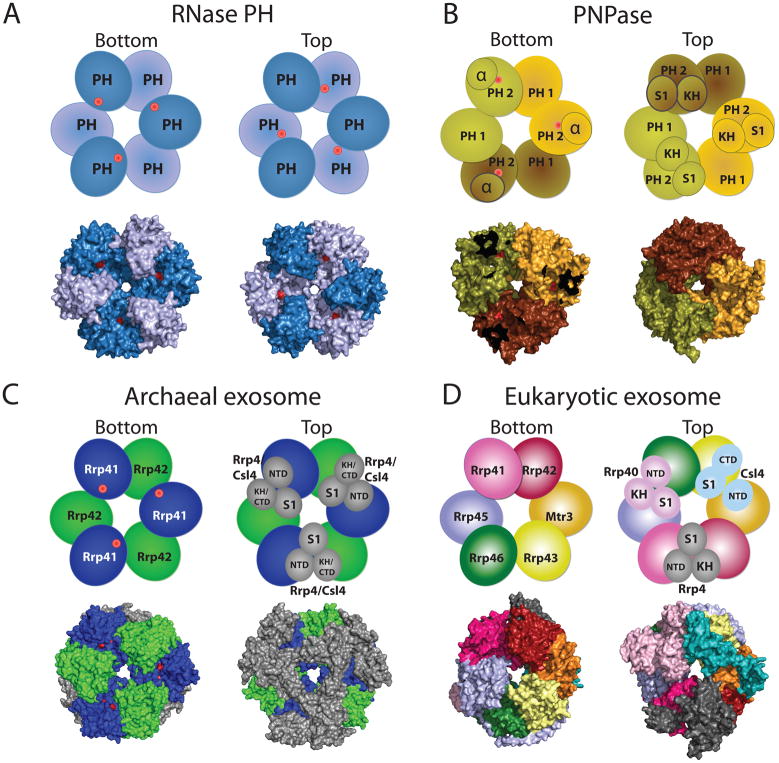
Figure 2. ‘Exosomes’ from bacteria, archaea, and eukaryotes have a similar architecture.
RNase PH, PNPase, archaeal exosome, and eukaryotic core exosome structures and schematics are depicted in two orientations which we denote bottom and top. Architectures emphasize a six-component ring with or without phosphorolytic active sites (shown as red dots in cartoon representation and red surfaces in the surface representations of the respective structures). A) RNase PH. The Aquifex aeolicus RNase PH structure (PDB ID = 1UDN) forms a homohexamer of PH subunits (colored dark blue and light blue, for clarity). B) PNPase. The S. antibioticus PNPase structure (PDB ID = 1E3P) forms a homotrimer. PNPase protomers are colored light yellow, dark yellow, and light brown to distinguish the homotrimer of RNase PH 1-like (PH 1) and RNase PH 2-like (PH 2) domains. The α domain was omitted to enable visualization of the phosphorolytic active sites in the bottom view (red dots). C) Archaeal exosome. The S. solfataricus archaeal exosome (PDB ID = 2JE6) is depicted with Rrp41 subunits (blue) and Rrp42 subunits (green) which form the six-component ring. Top view in the schematic shows the orientation of Csl4 (N-Terminal Domain, S1 domain, and C-Terminal Domain) and Rrp4 (N-Terminal Domain, S1 domain, and KH domain) labeled and shown in grey. The surface representation of the structure depicts the Rrp4-bound archaeal exosome. Residues in the active site are partially occluded from view such that the active site appears as two discontinuous red surfaces. D) Eukaryotic core exosome. The eukaryotic exosome is shown from H. sapiens (PDB ID = 2NN6). Subunits Rrp41 (magenta), Rrp42 (red), Mtr3 (orange), Rrp43 (yellow), Rrp46 (green), Rrp45 (blue) form the six-component ring. Subunits Rrp40 (light pink), Csl4 (cyan), and Rrp4 (grey) form the three-component cap. No phosphorolytic active site exists in the eukaryotic core exosome.
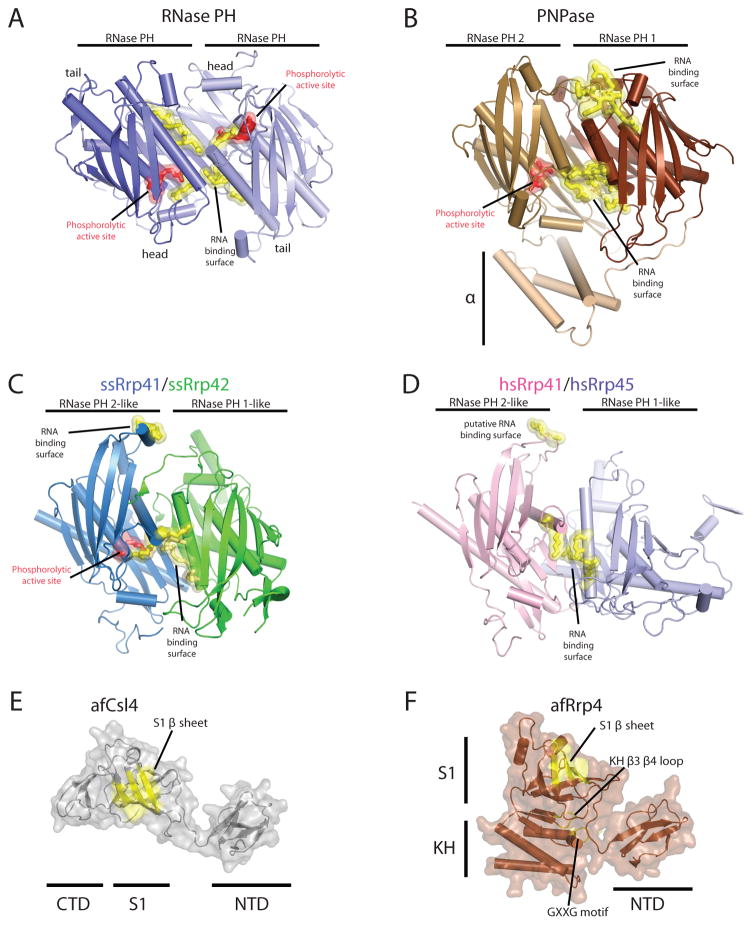
Figure 3. Structures of exosome domains.
Residues in red indicate phosphate binding regions and residues in yellow highlight RNA binding surfaces. Structures depicted in cartoons with helices as tubes and β-strands as arrows. A) RNase PH homodimerization interface (PDB ID = 1UDN). Phosphate binding residues include T125 and R126. RNA binding residues: R86, R92, R96, and R99. B) PNPase RNase PH 1/RNase PH 2 domain binding interface (PDB ID = 1E3P). Phosphate binding residues include: T462 and S463. RNA binding interface residues: R100, R104, R107, R422, and R423. A second RNA binding site includes residues F84, F85, R86, and R87. C) Archaeal S. solfataricus exosome Rrp41/Rrp42 heterodimer interface (PDB ID = 2JE6). Phosphate binding residues are from ssRrp41: S138 and R139. RNA binding interface residues from ssRrp41 are R98 and R99 and R112, R116, and R119 (ssRrp42). The second RNA binding region includes residues R67 and H68 (from ssRrp41) D) Eukaryotic H. sapiens Rrp41/Rrp45 hetero-dimerization interface. Putative RNA binding interface residues include: K94, S95, R104, R108, and R111, and the second putative RNA binding region includes residues R61 and A62. Structures for the archaeal three-component cap subunits (PDB ID = 2BAO and 2BA1): E) A. fulgidus Csl4 and F) A. fulgidus Rrp4. Putative RNA binding surfaces are highlighted in yellow for the S1 domain and KH domain on a transparent surface representation. Similar structures exist for the human three-component cap subunits Csl4, Rrp4 and Rrp40 as discussed in the text (PDB ID = 2NN6).
Similar to RNase PH, PNPase forms a related pseudo-hexameric ring through oligomerization of three PNPase molecules that contain an N-terminal RNase PH-like domain which we term RNase PH 1, an alpha domain, a second RNase PH-like domain which we term RNase PH 2 which is then followed by a KH domain and an S1 domain (Fig. 1A). The RNase PH 2 domain contains residues responsible for phosphorolytic activity, while the amino terminal RNase PH 1 domain is catalytically inactive2. The phosphorolytic active site and RNA binding surfaces are formed at the interface between the RNase PH 2 and RNase PH 1 domains (Fig. 3B). Because only one RNA PH-like domain in PNPase contains residues that form the phosphorolytic active site, only three active sites are formed in PNPase. In addition, the RNase PH 2 domain of PNPase is partially occluded from solvent by the alpha domain (bottom orientation) while additional putative RNA binding surfaces are formed by the KH and S1 domains (top orientation, Figs. 2B & 3B).
Crystal structures from the hyper-thermophiles Sulfolobus solfataricus, Archaeoglobus fulgidus, and Pyrococcus abyssi revealed that archaeal exosomes are composed of trimers of Rrp41-Rrp42 heterodimers which oligomerize to form pseudo-hexameric rings (Fig. 2C). Rrp41 contains residues that comprise the phosphorolytic active site that share sequence similarity with both the RNase PH 2 domain of PNPase and RNase PH (Fig. 1). Rrp42 shares sequence similarity with the RNase PH 1 domain and is catalytically inert (Figs. 2C & 3C). Analogous to bacterial PNPase, the RNA binding surfaces and active sites are located in a composite surface formed between the Rrp41 and Rrp42 heterodimer (Fig. 3C). The six-subunit rings are capped by three copies of Rrp4, Csl4, or combinations therein5–7, 15. Rrp4 and Csl4 both contain putative sites for RNA interaction via their S1 and KH domains or S1 domain, respectively. The phosphorolytic active sites are exposed to solvent and visible at the bottom of the ring while Rrp4 or Csl4 cap the top of the ring to presumably restrict access or guide substrates into the pore for degradation (Fig. 2C, top view).
The human exosome core features a pseudo-hexameric six-component ring, three-component cap, and a central pore, an architecture common to bacterial PNPase and archaeal exosomes (Figs. 1 & 2D)9. With that said, the human exosome architecture differs somewhat from archaeal exosomes because the nine-subunit core is formed through oligomerization of nine individually encoded subunits that form the ring (Rrp41, Rrp45, Rrp42, Rrp43, Mtr3 and Rrp46) or the three-component cap (Rrp4, Rrp40, and Csl4). While it is likely that the general architecture observed for the human exosome core is predictive of other eukaryotic exosomes, subtle distinctions between protozoa and metazoa are expected; for instance, metazoan Rrp45 subunits include a large (~150 amino acid) C-terminal extension that is absent in lower eukaryotes (Fig. 1C). Interestingly in human Rrp45, this extension contains a phosphorylation-dependent SUMO interaction motif suggesting that this region of Rrp45 may be important for regulation of exosome activities or assembly16.
Subunits that comprise the six-component ring of eukaryotic exosomes share higher sequence and structural similarities to either archaeal Rrp41 or PNPase RNase PH 2-like proteins (Rrp41, Mtr3, and Rrp46) or archaeal Rrp42 or PNPase RNase PH 1-like proteins (Rrp42, Rrp43, and Rrp45). The six-component ring is formed by oligomerization of three distinct RNase PH 2-like and RNase PH 1-like heterodimers: Rrp41-Rrp45, Rrp43-Rrp46, and Mtr3-Rrp42. However, unlike the archaeal exosome or PNPase, both classes of eukaryotic RNase PH-like domains are devoid of catalytic activity and do not contain key catalytic residues that are conserved in PNPase or archaeal exosome phosphorolytic active sites (Figs. 1C & 2D)5, 9–11, 17.
Csl4, Rrp4, and Rrp40 contain N-terminal Domains (NTD) and putative RNA binding S1 domains, but they differ with respect to inclusion of either a KH domain (as observed in Rrp4 and Rrp40) or a C-Terminal Domain (CTD), as observed for Csl4 (Fig. 1C). In addition, subunits of the cap are required to stabilize interactions between the different RNase PH-like heterodimers. Specifically in the human exosome, Rrp4 bridges interactions between Rrp41 and Rrp42, Rrp40 bridges the Rrp45 and Rrp46 interface, and Csl4 interacts with Mtr3 and to a lesser extent with Rrp43 (Fig. 2D). This phenomenon of the three-component cap stabilizing the hexameric core is unique to eukaryotes, insofar as the archaeal exosome forms stable six-component rings in the absence of the three-component cap5. A feature unique to eukaryotic and archaeal exosomes is that the S1 domains of the three-component cap face the central pore surface, while in bacterial PNPase the KH domains face the central pore. The significance of the orientations for the S1 and KH putative RNA binding domains with respect to the central pore has not been ascertained.
RNase PH-like domains in bacterial, archaeal and eukaryotic core exosomes
RNase PH domains are comprised of a βαβα fold and are conserved in RNase PH, PNPase, archaeal exosomes, and eukaryotic exosomes2,5–6,9. In RNase PH, two PH domains form a head to tail dimer generating a composite surface that includes residues that constitute RNA surfaces and the phosphorolytic active site (Fig. 3A). The location of the active site was determined by structures in which a sulfate ion or phosphate ion was observed in complex with RNase PH of B. subtillis or A. aeolicus, respectively13–14. Because RNase PH is a homodimer, two equivalent RNA binding surfaces are generated along the interdomain surface, one at the entrance of the central pore and one proximal to the active site (Fig. 3A). The functional significance of this symmetry is not understood.
In PNPase, two RNase PH-like domains are fused in a single polypeptide, but they come together in a pseudo-dimeric head to tail interaction to form a similar ‘dimerization’ interface to that observed in RNase PH between its respective RNase PH 1 and RNase PH 2 domains (Fig. 3B). The phosphorolytic active site is encompassed by residues from the RNase PH 2 domain and is positioned along the bottom of the inter-domain interface. Two distinct RNA binding surfaces are also present in this interface, one composed of residues from the RNase PH 1 domain at the entrance to the central pore and one proximal to the active site that is primarily composed by residues from the RNase PH 1 and RNase PH 2 domains (Fig. 3B).
The archaeal exosome is structurally analogous to PNPase with respect to the location of the two RNA binding surfaces and the phosphorolytic active site, however in this instance the interface is formed by two separately encoded subunits, archaeal Rrp41 and Rrp42 (Fig. 3C)12–14. Rrp41 contains key catalytic residues that constitute the active site, but it also contributes residues in combination with those from Rrp42 to form one of the two RNA binding surfaces7,17–18. In contrast to PNPase which uses a RNase PH 1 domain surface to interact with RNA at the top of the interface, a second distinct RNA binding surface is present at the top of the heterodimeric interface in archaeal exosomes and is comprised solely by residues from its RNase PH 2-like domain, archaeal Rrp41 (Fig. 3C).
The eukaryotic exosome contains three heterodimeric RNase PH-like pairs (Rrp41-Rrp45, Rrp43-Rrp46, and Mtr3-Rrp42) that are arranged in similar head to tail configurations as observed in RNase PH, PNPase, and archaeal exosomes9. While the key catalytic residues in RNase PH, PNPase, and the archaeal exosomes are not conserved in any of the human or budding yeast RNase PH-like proteins, a few of the subunits, namely Rrp41 and Rrp45, include several basic residues that are conserved across evolution that are believed to be important for RNA interactions, located near the top of the Rrp41-Rrp45 heterodimeric interface and proximal to the location where the phosphorolytic active site resides in archaeal exosomes and PNPase (Fig. 3D)9,19.
S1 and KH containing domains in bacterial, archaeal, and eukaryotic core exosomes
Bacterial PNPase, archaeal exosomes, and eukaryotic exosomes include putative RNA binding domains, KH type I and S1, in the three-component cap subunits in their respective core complexes. KH type I domains feature a β1-α1-α2-β2-β3-α3 secondary structure topology, and a tertiary structure that consists of three beta-strands that form a sheet which packs against three alpha helices20. Single stranded RNA typically binds a KH type I domain via surfaces formed by residues within helix α1, a conserved GXXG motif between helices α1 and α2, helix α2, the variable loop between strands β2 and β3, and residues within strand β2 (Fig. 3F). The S1 domain originally observed in the E. coli ribosomal protein S121–22 contains an OB (Oligonucleotide/oligosaccharide Binding) fold with a five-stranded β-sheet coiled to form a closed β-barrel (Figs. 3E & F). A typical OB domain binds nucleic acid through surfaces composed of positively charged and hydrophobic residues on the solvent exposed β-sheet (Figs. 3E and 3F). For instance, the RNase E S1 domain binds polymeric single-stranded nucleic acids via a positively charged surface that comprises strands β2 and β3 and the loops between strands β2 and β3 and strands β3 and β523. Both the KH and S1 domains of PNPase contribute to RNA binding, as simultaneous deletion of both domains impairs the apparent affinity of PNPase for RNA substrates24–25. Interestingly, the orientation of these domains situates the canonical RNA binding surfaces of the KH domain toward the central pore while the putative RNA binding surfaces of the S1 domain face outward near the exterior of the complex.
Archaeal Csl4 contains three domains: the NTD, S1 domain, and CTD. The NTD consists of 2 symmetrical three stranded β-sheets, and the CTD contains a 3-stranded β-sheet that coordinates a Zn2+ via four cysteine residues that is similar to the iron-binding portion of rubredoxins (Fig. 3E)26–27. Eukaryotic Csl4 shares structural similarity to archaeal Csl4 and contains an NTD, S1 domain, and CTD; however, despite having a similar rubredoxin-like fold, the four cysteine residues in the CTD that coordinate zinc in archaeal Csl4 are not conserved in eukaryotes9. Archaeal Rrp4 contains three domains: the NTD, KH type I domain, and a C-terminal S1 domain6. Eukaryotic Rrp4 and Rrp40 share structural similarity to archaeal Rrp4 and each contains an NTD, a central KH type I domain and a C-terminal S1 domain; however both subunits lack the canonical GXXG motif in their KH domains that is believed to be important for RNA interactions.
The arrangement of Csl4 and Rrp4 subunits on the six-subunit ring in the archaeal exosome positions the positively charged putative RNA binding S1 domain surfaces facing toward the central pore while the NTD and KH domains are position nearer to the periphery of the complex (Fig. 2C). It remains unclear how these domains interact with RNA. For instance, Rrp4 from S. solfataricus promotes interactions with a poly(A) RNA substrate in the context of the exosome, as evidenced by its ability to increase the affinity for RNA by ~30 fold compared to the archaeal Rrp41-Rrp42 six-component ring alone28. However, x-ray structures of archaeal exosomes bound to RNA have so far only elucidated interactions between RNA substrates and residues within the central pore of the six-component Rrp41-Rrp42 ring, despite the presence of the three-component cap7. Eukaryotic Rrp4, Rrp40, and Csl4 subunits are similarly positioned on the human exosome core9, 28, directing putative RNA binding surfaces of the respective S1 domains toward the central channel and the putative RNA binding surfaces of the KH domains toward the periphery of the complex. Additional experiments will be required to characterize the relevance of the putative RNA binding surfaces in the three-subunit exosome cap.
Mechanism of phosphorolytic activity in bacterial PNPase and archaeal exosomes
Bacterial PNPase and archaeal exosomes contain three identical active sites within the central pore that catalyze phosphorolytic 3′ to 5′ exoribonuclease activity (Figs. 2B & C)5,8,29. By analysis of the RNA-free and RNA-bound x-ray structures of PNPase, it was determined that the alpha domain, which partially obstructs the bottom entrance to the central pore, can transition from partially disordered to ordered upon RNA coordination2,8. In addition, two narrow constrictions in the pore are believed to regulate access to the phosphorolytic active sites. The first lies near the entrance to the central pore and features three phenylalanine side chains (Fig. 3B), one from each PNPase protomer, that each base stack with one nucleotide (presumably from three different RNA oligomers). The second constriction is located deep within the PNPase central channel near the active site (Fig. 3B).
Archaeal exosomes also recruit RNA to the active site via interactions with at least two RNA binding surfaces that reside in the Rrp41-Rrp42 heterodimer within the central pore (Fig. 3C). Interactions between the two RNA binding surfaces and RNA have been observed for x-ray crystal structures of archaeal exosomes using poly(A) or poly(U) RNA polymers of varying length7,17–18. The first interaction surface is located within a loop at the top of the central pore near the three-component cap interface, and it features a histidine residue from Rrp41 that stacks with a nitrogenous base near the 5′ end of the RNA. The second RNA interaction surface is proximal to the phosphorolytic active site and includes extensive contacts to the RNA substrate; this surface forms a 10 Å constriction of the central pore and thus it is believed to allow only one RNA molecule to pass through the pore at one time. Protein contacts to the RNA include ribose specific interactions at the 3′OH terminal nucleotide and contacts to the fourth to last nucleotide position through phosphate backbone interactions and nitrogenous base stacking interactions (Fig. 4C). No electron density has yet been observed for RNA nucleotides between these two RNA binding surfaces, thus it has been speculated that the intervening RNA nucleotides are not coordinated in any particular configuration7,15,17.
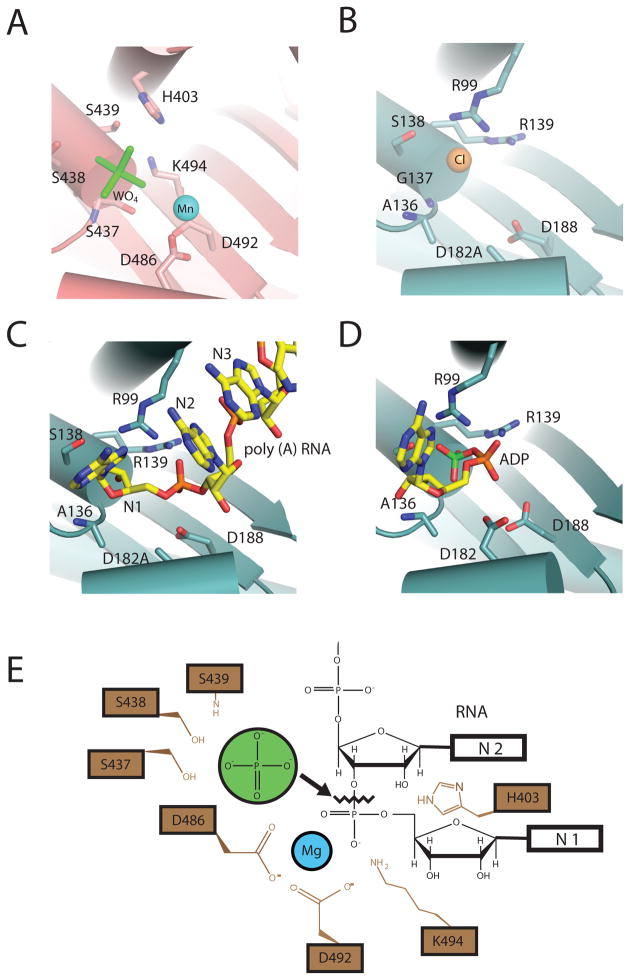
Figure 4. Phosphorolytic exoribonuclease catalytic mechanism for PNPase and the archaeal exosome.
A) PNPase active site. A composite structure is depicted that shows the phosphate-mimic tungstate (green) and magnesium ion-mimic manganese (blue sphere) (PDB ID = 1E3P and 3GME). Residues that bind “phosphate” include: S439, S438, and S437. Residues that coordinate “magnesium” are D486 and D492. B) S. solfataricus exosome active site. The phosphate-mimic chloride is shown as a yellow sphere, and residues that bind the proposed phosphate-mimic chloride are: A136, G137, and S138 (PDB ID = 2BR2). C) D182A mutant S. solfataricus in complex with a five-nucleotide poly(A) RNA substrate (PDB ID = 2C38). Nucleotides are colored yellow and numbered in such a manner that the first nucleotide (N1) is at the 3′OH end. D) S. solfataricus in complex with the product ADP (PDB ID = 2C39). ADP is colored in yellow with the α-phosphate colored orange and β-phosphate colored green. E) Proposed phosphorolytic exoribonuclease mechanism. Ser437 and Ser439 provide a binding pocket for phosphate (green). Asp486 and Asp492 coordinate a magnesium ion. The magnesium, with the aid of K494 and H403, positions the bridging phosphate between N1 and penultimate N2 nucleotides to facilitate in-line attack by the phosphate.
Structural insight into the catalytic mechanism during phosphorolysis can be gleaned by comparing active sites from a variety of x-ray crystal structures of bacterial PNPase and archaeal exosomes in complex with different ligands (Fig. 4). X-ray structures of PNPase with either manganese cations or tungstate revealed the identity of active site residues that coordinate magnesium or phosphate (Fig. 4A), respectively8. Residues that coordinate the phosphate include Ser437, Ser438, Ser439 and key residues that coordinate the magnesium include Asp486 and Asp492. A phosphate-binding site composed of similar amino acid side chains was also deduced in an analogous position for the S. solfataricus archaeal exosome through identification of a chloride ion7 and for the A. fulgidus exosome through identification of a tungstate ion6. Structures of the S. solfataricus archaeal exosome in complex with poly (A) RNA and the ADP product (Fig. 4C,D) further identified residues that coordinate the phosphate of the NDP product and phosphodiester backbone of the RNA substrate (Arg99 and Arg139)17.
A composite active site can be extrapolated from these structures onto the PNPase structure to provide a structural rationale for the phosphorolytic reaction mechanism (Fig. 4E). Two serine residues position a phosphate ion proximal to the phosphodiester linkage between the terminal and penultimate nucleotides. The magnesium ion, His403, and Lys494 position the terminal bridging phosphate in the substrate in an appropriate configuration to facilitate in-line attack by the phosphate nucleophile, which ultimately results in the formation of the NDP product. Although S. solfataricus Asp182 and Asp188 are predicted to be required for metal coordination based on sequence similarity, no metal ion has yet been observed in the active sites of S. solfataricus exosomes.
Rrp44, a eukaryotic exosome subunit with hydrolytic endoribonuclease and processive exoribonuclease activities
As discussed earlier, none of the human or budding yeast subunits that comprise the 9-component exosome core retain phosphorolytic exoribonuclease activity because most of the key active site residues required for RNA binding or for metal and phosphate coordination have not been conserved across evolution9–10,30. Studies with the budding yeast exosome have demonstrated that the tenth exosome subunit, Rrp44 (also known as Dis3), is solely responsible for the processive hydrolytic activity that is associated with the exosome in the cytoplasm9–10,31. It is important to note that while human encodes three apparent homologs of budding yeast Rrp44, human Rrp44 has not yet been shown to associate with the human exosome core9,32. Rrp44 exoribonuclease activity results in hydrolysis of RNA one nucleotide at a time in a 3′ to 5′ direction, releasing 5′ nucleotide monophosphates in a sequence independent manner30.
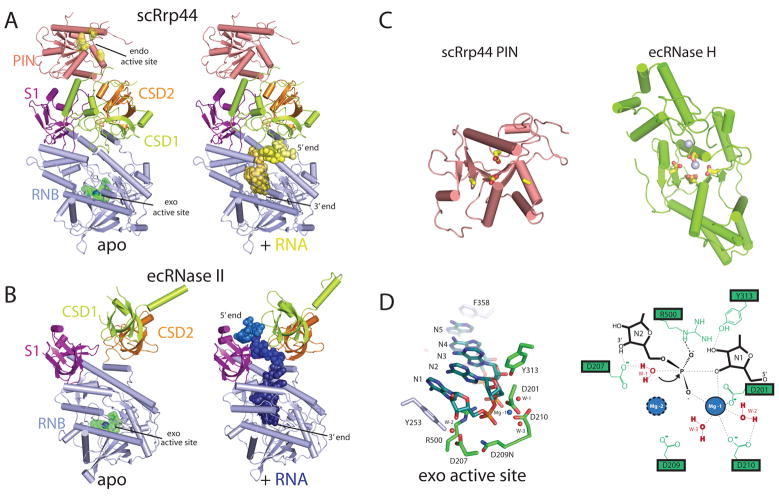
Rrp44 contains five domains: an endoribonucleolytic active site containing PIN (PIlus-forming N-terminus) domain, two cold shock domains (CSD1 and CSD2), a central hydrolytic exoribonucleolytic active site containing domain (RNB), and an S1 domain (Fig.1C)31. The overall architecture of Rrp44 has been determined based on two structures of Rrp44: one determined in complex with RNA in the absence of the PIN domain and one for full-length Rrp44 in complex with Rrp41 and Rrp45. These structures reveal the modular architecture of Rrp44 in which the PIN domain is located above the two CSDs and S1 domain with the RNB domain located below the two CSDs and S1 domain (Fig. 5A)19,30.
Figure 5. Eukaryotic Rrp44 structure and catalytic mechanism.
A) Structures of S. cerevisiae Rrp44 with and without poly(A) RNA. The Rrp44 domains are PIN (pink), CSD1 (lime), CSD2 (orange), RNB (blue), and S1 (purple) (PDB ID = 2WP8). Residues D91, E120, D171, and D198 are colored yellow and indicate the position of the endoribonucleolytic active site. Residues D543, D540, D551N, and D552 are colored green and indicate the position of the exoribonucleolytic active site. RNA (yellow spheres) was modeled into the full length Rrp44 (right panel) by alignment to the poly(A) RNA bound Rrp44ΔPIN structure (PDB ID = 2VNU). B) Structures of E. coli RNase II with and without poly(A) RNA. RNase II domains are CSD1 (lime), CSD2 (orange), RNB (blue), and S1 (purple). Residues D201, D207, D209, and D210 are colored green and indicate the position of the exoribonucleolytic active site. The magnesium ion is shown as a blue sphere (PDB ID = 2IXO). The poly(A) RNA (blue) bound structure of RNase II is shown in a similar orientation in the right panel (PDB ID = 2IX1). C) Structures shown for the S. cerevisiae PIN from Rrp44 (left, PDB ID = 2WP8), and E. coli bacteriophage T4 RNase H (right, PDB ID = 1TFR). Active site residues are highlighted in yellow, and magnesium ions shown as blue spheres. D) The hydrolytic exoribonucleolytic active site of RNase II. Left panel: Structural representation of relevant residues that coordinate magnesium ions, Mg-1 and Mg-2 (not detected), and residues that coordinate RNA are shown in green. Y253 and F358 are shown in blue and make base stacking interactions with the RNA substrate. Water molecules (W-1, W-2, and W-3), important for the reaction mechanism, are shown as red spheres. Right panel: Schematic representation of the active site. Representation depicts the binding of W-1 by magnesium ions and charged residues to facilitate nucleophilic attack of the bridging phosphate between nucleotides N1 and N2.
Rrp44 is structurally and mechanistically related to bacterial RNase II and RNase R, however comparison of Rrp44 to structures of RNase II in apo- and RNA-bound states reveals that RNase II has a slightly different arrangement of the cold shock domains and S1 domain (Fig. 5B)33. An Rrp44-RNA complex showed that CSD1 engages in interactions with the RNA substrate to guide it into the RNB domain exoribonucleolytic active site (Fig. 5A, right panel), and interactions between the single stranded RNA substrate and CSD1 facilitate recruitment to the exoribonucleolytic catalytic site by a specific orientation of the three OB-containing domains (CSD1, CSD2 and S1). In comparison, RNase II positions the three OB-containing domains in a different conformation that allows for single stranded RNA interactions with CSD2 and the S1 domains (Fig. 5B, right panel). As will be discussed, these two alternative modes of RNA interaction present fundamentally different paths that serve to guide the RNA to the exoribonucleolytic active site.
Rrp44 PIN domain
The PIN domain family, named after its apparent homology with the N-terminal domain of the pili biogenesis protein detected in some bacteria, includes over 300 members found in bacteria, archaea, and eukaryotes34. The PIN domain consists of a central twisted five-stranded β sheet flanked by α helices and an active site that is capable of cleaving nucleic acid (Fig. 5C, left panel). Biophysical and structural studies of PAE2754 from Pyrobaculum aerophilum and the OT3 protein from Pyrococcus horkoshii revealed that PIN domains can form dimers and dimers of dimers, respectively35–36. While the precise nature of the binding surfaces required for recruiting nucleic acids to the PIN domain active site remain unknown, it was postulated for PAE2754 that nucleotides thread through a central channel that is formed via tetramerization. Although many structures of PIN domains reveal dimers within the crystal lattice, it remains uncertain if dimerization is a functionally relevant feature for all catalytically active PIN domains.
The nucleolytic active site, detected within some but not all PIN domains37, consists of four conserved acidic residues that coordinate two divalent cations. For the PIN domain in budding yeast Rrp44, all four acidic residues are present. In higher eukaryotes such as humans, the three Rrp44 paralogs vary with respect to the conservation of the acidic residues or the presence of an intact PIN domain (Fig. 1C). The functional consequence of this variation within higher eukaryotes has yet to be investigated. Structures of the PIN domains also reveal a striking structural homology to T4 RNase H, despite a dearth in sequence identity (Fig. 5C), and therefore the endoribonucleolytic activity of PIN domains is predicted to utilize a similar two metal-dependent catalytic mechanism for hydrolysis of nucleotides38–39.
Rrp44 RNB domain
3′ to 5′ exoribonuclease activity is catalyzed in an active site within the RNB domain which is located at the end of a narrow channel and composed by acidic residues that coordinate two divalent metal ions, motifs conserved in bacterial RNase II and RNase R. Analysis of the RNase II apo- and RNA bound structures provide insights to the catalytic mechanism of exoribonucleolytic decay33. The active site of RNase II is composed by acidic residues Asp201, Asp207, Asp209, and Asp210 that coordinate two magnesium ions (Fig. 5D)33. The first (Mg-1) is coordinated by residues Asp201 and Asp210 and two waters, W-2 and W-3. The second (Mg-2) was not detected in x-ray structures of RNase II, but is speculated to be coordinated by Asp207 and Asp209. Interestingly, electron density for the Arg500 side chain was only detected when in the presence of RNA where it was observed coordinating the phosphate bridging the last two 3′ nucleotides (N1 and N2), conceivably to stabilize the transition state during cleavage of the phosphodiester bond. The exoribonucleolytic reaction is believed to proceed through a two-metal-ion bimolecular nucleophilic substitution mechanism. W-1, coordinated by Asp207 and Mg-2, is the nucleophile for in-line attack of the phosphate between the last and penultimate nucleotide, ultimately resulting in release of the NMP product.
A ‘clamp’ action is thought to promote translocation between successive cleavage events, thus leading to the processive degradation activities observed for this enzyme family. RNase II achieves this function by employing base stacking interactions between N5 and F358 as well as N1 and Y253 to stabilize the RNA substrate within the active site33. A similar mechanism for catalysis and translocation can be inferred for budding yeast Rrp44 by comparing the conserved active site residues of the RNA bound Rrp44ΔPIN x-ray crystal structure30. It should be noted that RNase II is unable to process structured RNA substrates, unlike the bacterial RNase R and eukaryotic Rrp449,19,30,40–41.
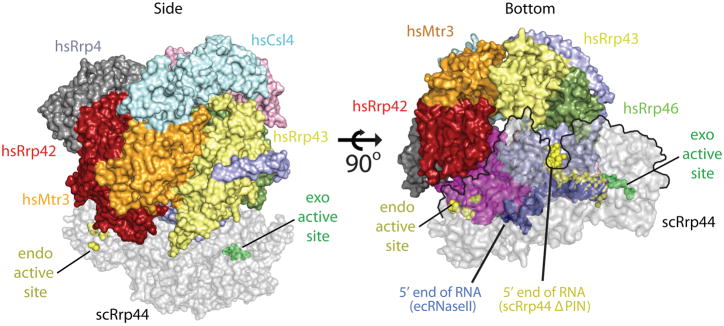
Rrp44 and the 10-component exosome
Models for the structure of the eukaryotic 10-component exosome have been proposed based on the x-ray structure of the human nine-component exosome core (Liu et al., 2006), the x-ray structure of the budding yeast Rrp41-Rrp45-Rrp44 trimer19, and a 20 Å resolution negative-stain EM structure of budding yeast Rrp44-bound to the core exosome (in the absence of Csl4)42. In the case of the Rrp41-Rrp45-Rrp44 trimer, the ten-component exosome was modeled by aligning the budding yeast Rrp41-Rrp45 proteins to the respective human counterparts (Fig. 6)19. In the case of the EM structure, the human exosome core structure was positioned in the EM density followed by docking the Rrp44 RNB domain into remaining density. In each of these models, the PIN domain of Rrp44 interacts with the ‘bottom’ of the exosome core principally through interactions with Rrp41 and Rrp45. Although these models are in general agreement, additional interactions have been reported that include surfaces identified in the EM structure between the CSD1 of Rrp44 and Rrp4342 and protomer interactions between Rrp44, Rrp41, Rrp45, Rrp42, and Rrp4, detected by the presence of sub-complexes of budding yeast exosomes by mass spectrometry experiments43. However, it should be noted that Rrp42 and Rrp4 may interact indirectly with Rrp44 through Rrp41 and Rrp45. Based on analysis of the architecture of the 10-component exosome and comparisons to the RNA bound Rrp44-ΔPIN and RNA bound RNase II structures, it was predicted that RNA threads into the exosome core through the three-component cap, progressing through the central pore of the exosome to direct the 3′ OH end of the RNA substrate into the hydrolytic exoribonucleolytic active site of the Rrp44 RNB domain (Fig. 6).
Figure 6. Model for RNA recruitment to the hydrolytic active site of Rrp44 within the eukaryotic exosome.
A 10-component exosome model was created by aligning the S. cerevisiae Rrp41-Rrp45-Rrp44 trimer (PDB ID = 2WP8) onto the Rrp41-Rrp45 subunits of the human exosome (PDB ID = 2NN6). Coloring for the 9-component exosome is described in Fig. 2, and the Rrp44 component is shaded grey. The left panel depicts a side view of the complex with the Rrp44 exoribonucleolytic active site indicated as green spheres, and the endoribonucleolytic active site as yellow spheres. The right panel depicts a bottom view of the complex. The RNA complexes determined for RNase II (blue spheres, PDB ID = 2IX1) and Rrp44ΔPIN (yellow spheres, PDB ID = 2VNU) were superimposed into the full-length Rrp44 structure to illustrate the paths of RNA in the complex. The Rrp44 molecule is outlined by a black line in the right panel where it overlaps with the exosome core.
As discussed in previous sections, the path of the RNA substrate into the exoribonucleolytic active site differs significantly in structures of budding yeast Rrp44 and bacterial RNase II despite conservation of the CSD and S1 domains. Extrapolating the path of RNA observed in these structures within the context of a 10-component exosome model, RNA would be required to exit the bottom of the 9-component exosome, become solvent exposed, and would then be required to make a ~45º turn around CSD1 of Rrp44 in order to enter into the Rrp44 channel that leads to the RNB active site (Fig. 6). While this route appears circuitous, it is consistent with both RNase protection and RNA exoribonuclease decay assays which indicated that RNA substrates required inclusion of at least 31–34 single stranded nucleotides at the 3′ end to be engaged by the 10-component exosome19. However, it is also conceivable that RNA binding could induce conformational changes in the complex to facilitate a more direct path for RNA substrates into the Rrp44 active site.
In contrast to the RNA path predicted for the 10-component exosome based on the RNA bound Rrp44-ΔPIN structure, the path predicted for RNA based on the structure of an RNase II-RNA complex, would place the RNA perpendicular to the central pore of the 9-component exosome (Fig. 6). This model is not consistent with the utilization of the central pore for RNA interactions but is consistent with a role for the PIN domain in RNA interactions, as this RNA path, if extended, points directly toward the PIN domain. Taken together, these structural models suggest that alternative binding modes may exist to engage the RNB domain of Rrp44 either via interactions with the PIN or the central pore of the exosome.
Rrp6, a eukaryotic exosome subunit with distributive hydrolytic activities
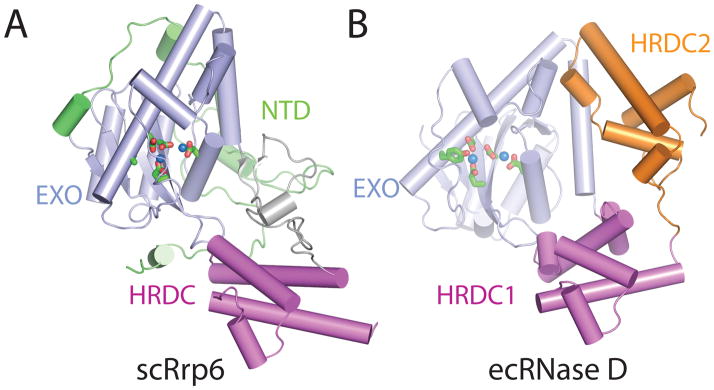
Rrp6 contains at least three domains: an N-terminal domain (NTD), an exoribonuclease domain (EXO) that contains the DEDD-Y active site amino acid motif detected in many DNA and RNA nucleases, and a Helicase and RNase D Carboxy terminal (HRDC) domain (Fig. 1C)44. The structure of a catalytically active domain of budding yeast Rrp6 shares structural homology to RNase D from E. coli (Fig. 7)45. RNase D contains the EXO domain with a DEDD-Y active site, but differs in that it contains two flanking HRDC domains that together form a funnel shaped ring. The two HRDC domains in RNase D were proposed to recruit RNA substrates, channeling them to the active site for processing. While a second HRDC domain has not been identified in the ~200 C-terminal residues in eukaryotic Rrp6, a similar hypothesis for RNA binding and recruitment to the active site has been suggested for the HRDC domain based on sequence similarity to the RecQ helicase protein family, and the fact that this domain is critical for processing RNAs such as 5.8 S rRNA and snR40 snoRNA, as determined using Rrp6 isolated from S. cerevisiae46.
Figure 7. Structure of eukaryotic Rrp6 and the bacterial homolog RNase D.
A) S. cerevisiae Rrp6 structure (PDB ID = 2HBL). Domains of Rrp6 are NTD (green), EXO (blue), and HRDC (pink). The active site residues (D238, E240, D296, D365, and Y361A) are shown coordinating two manganese ions (blue spheres). B) E. coli RNase D structure (PDB ID = 1YT3). Domains of RNase D are EXO (blue), HRDC1 (pink), and HRDC2 (orange). The active site residues (D28, E30, D85, D155, and Y151) colored green are coordinating two zinc ions (blue spheres).
The DEDD active site is observed in a variety of nucleolytic enzymes that catalyze degradation of DNA and RNA as exemplified by the Klenow fragment of DNA polymerase I47. A two-metal assisted catalytic mechanism has been proposed based on x-ray crystal structures and mutational analysis, in which the negatively charged DEDD residues coordinate two metal ions that are required for cleavage of the phosphodiester bond48. A similar model has been proposed for RNase D and Rrp6: a magnesium ion acts as a Lewis acid to deprotonate a water molecule, and then the phosphodiester backbone is attacked by the resulting nucleophilic water at the penultimate nucleotide of the RNA substrate. The DEDD-Y active site of RNase D, Rrp6, and related enzymes is unique compared to other enzymes containing DEDD active sites because they employ an additional tyrosine proximal to the DEDD active site to coordinate the nucleophilic water. The distributive 3′ to 5′ exoribonuclease activity observed for Rrp6 is consistent with the structure because unlike Rrp44, whose active site is sequestered at the end of a deep channel, the Rrp6 active site is exposed on the surface of the enzyme. Further mechanistic insight to Rrp6 interactions with RNA substrates will require additional structures and biochemical analysis of Rrp6 complexes with RNA substrates as none are yet resolved.
Rrp6 interactions with the exosome core
No detailed atomic resolution structures exist for Rrp6 in association with the exosome core, although a 35 Å resolution negative-stain EM structure of the L. tarentolae exosome core has been determined in complex with Rrp6 and Rrp47, an accessory protein that was reported to increase the exoribonuclease activity of the exosome49. From this work, the authors proposed a model whereby Rrp6 and Rrp47 interact with the 9-component exosome core near the ‘top’ and adjacent to the three-component cap. While this organization may apply to the L. tarentolae exosome, it remains unclear if this organization will apply to other eukaryotic exosomes because yeast two-hybrid data demonstrated that human Rrp6 interacts with Rrp41, Rrp43, Rrp46, and Mtr3 suggesting that Rrp6 may also interact with the six-subunit ring of the exosome50.
Unlike the exoribonucleolytic activities of Rrp44 which are clearly modulated or regulated via association with the exosome core9, 19, similar activities were observed in vitro for budding yeast Rrp6 prior to and after its association with the exosome core9. With that said, it is clear that Rrp6 association with the exosome core is important for targeting Rrp6 to its physiological substrates, as evidenced by the fact that a fragment of Rrp6 that loses its ability to interact with the core (but retains catalytic function) is not sufficient to complement many of the functions of Rrp6 in vivo51. However, it is also interesting that the activities of Rrp6 can be stimulated without the core exosome by association with members of the TRAMP complex, in a manner independent of the Trf4 poly(A) polymerase and Mtr4 RNA helicase activities52. Further investigations will be required to determine the structural basis for these seemingly disparate activities.
Conclusions
Structures and models derived for exosomes from bacteria, archaea, and eukaryotes demonstrate a striking architectural similarity with respect to 1) the six RNase PH-like domains that oligomerize to form a pseudo-hexameric ring and 2) the orientation of the S1 and KH RNA binding domains that form a trimeric cap on top of the exosome. This structural framework results in formation of a central channel. In bacterial PNPase and archaeal exosomes, this channel harbors RNA binding surfaces and phosphorolytic active sites, and because the central channel is narrow, only single stranded RNAs can thread through the central pore via interactions with two conserved RNA binding surfaces. Furthermore, the two RNA binding surfaces confer processivity during RNA decay, presumably by preventing RNA substrates from diffusing away from the complex between successive rounds of cleavage.
Eukaryotic exosomes have been reported to use the same strategy to engage RNA substrates by utilizing the inactive 9-component exosome core to bind and transport RNA substrates through the pore to ultimately engage the hydrolytic exoribonuclease activities of Rrp44, although this has not been demonstrated in any structural detail. It also remains unclear how Rrp6 engages the exosome core, and whether it too is influenced by the RNA binding properties of the exosome core channel. On a final note, it is known that the RNA exosome interacts with several other factors including the TRAMP and SKI complexes among others53–55, suggesting that additional surfaces of the exosome core may be required for recruitment of these effectors to alter or regulate exosome activity. Although much has been accomplished since the discovery of the eukaryotic exosome more than ten years ago56, it is clear that much work remains to fully understand how the molecular architecture of the eukaryotic exosome impacts on its biochemical and cellular functions.
Acknowledgments
K.J. and C.D.L. are supported in part by a grant from the National Institutes of Health (GM079196).
References
- 1.Carpousis AJ. The Escherichia coli RNA degradosome: structure, function and relationship in other ribonucleolytic multienzyme complexes. Biochem Soc Trans. 2002 Apr;30(2):150–155. [PubMed] [Google Scholar]
- 2.Symmons MF, Jones GH, Luisi BF. A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Structure. 2000 Nov 15;8(11):1215–1226. doi: 10.1016/s0969-2126(00)00521-9. [DOI] [PubMed] [Google Scholar]
- 3.Evguenieva-Hackenberg E, Walter P, Hochleitner E, Lottspeich F, Klug G. An exosome-like complex in Sulfolobus solfataricus. EMBO Rep. 2003 Sep;4(9):889–893. doi: 10.1038/sj.embor.embor929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koonin EV, Wolf YI, Aravind L. Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res. 2001 Feb;11(2):240–252. doi: 10.1101/gr.162001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lorentzen E, Walter P, Fribourg S, Evguenieva-Hackenberg E, Klug G, Conti E. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat Struct Mol Biol. 2005 Jul;12(7):575–581. doi: 10.1038/nsmb952. [DOI] [PubMed] [Google Scholar]
- 6.Buttner K, Wenig K, Hopfner KP. Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol Cell. 2005 Nov 11;20(3):461–471. doi: 10.1016/j.molcel.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 7.Lorentzen E, Dziembowski A, Lindner D, Seraphin B, Conti E. RNA channelling by the archaeal exosome. EMBO Rep. 2007 May;8(5):470–476. doi: 10.1038/sj.embor.7400945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nurmohamed S, Vaidialingam B, Callaghan AJ, Luisi BF. Crystal structure of Escherichia coli polynucleotide phosphorylase core bound to RNase E, RNA and manganese: implications for catalytic mechanism and RNA degradosome assembly. J Mol Biol. 2009 May 29;389(1):17–33. doi: 10.1016/j.jmb.2009.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006 Dec 15;127(6):1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 10.Dziembowski A, Lorentzen E, Conti E, Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol. 2007 Jan;14(1):15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 11.Liu QS, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome (vol 127, pg 1223, 2006) Cell. 2007 Oct 5;131(1):188–189. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 12.Shi Z, Yang WZ, Lin-Chao S, Chak KF, Yuan HS. Crystal structure of Escherichia coli PNPase: central channel residues are involved in processive RNA degradation. RNA. 2008 Nov;14(11):2361–2371. doi: 10.1261/rna.1244308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harlow LS, Kadziola A, Jensen KF, Larsen S. Crystal structure of the phosphorolytic exoribonuclease RNase PH from Bacillus subtilis and implications for its quaternary structure and tRNA binding. Protein Sci. 2004 Mar;13(3):668–677. doi: 10.1110/ps.03477004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishii R, Nureki O, Yokoyama S. Crystal structure of the tRNA processing enzyme RNase PH from Aquifex aeolicus. J Biol Chem. 2003 Aug 22;278(34):32397–32404. doi: 10.1074/jbc.M300639200. [DOI] [PubMed] [Google Scholar]
- 15.Navarro MV, Oliveira CC, Zanchin NI, Guimaraes BG. Insights into the mechanism of progressive RNA degradation by the archaeal exosome. J Biol Chem. 2008 May 16;283(20):14120–14131. doi: 10.1074/jbc.M801005200. [DOI] [PubMed] [Google Scholar]
- 16.Stehmeier P, Muller S. Phospho-regulated SUMO interaction modules connect the SUMO system to CK2 signaling. Mol Cell. 2009 Feb 13;33(3):400–409. doi: 10.1016/j.molcel.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 17.Lorentzen E, Conti E. Structural basis of 3′ end RNA recognition and exoribonucleolytic cleavage by an exosome RNase PH core. Mol Cell. 2005 Nov 11;20(3):473–481. doi: 10.1016/j.molcel.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 18.Ramos CR, Oliveira CL, Torriani IL, Oliveira CC. The Pyrococcus exosome complex: structural and functional characterization. J Biol Chem. 2006 Mar 10;281(10):6751–6759. doi: 10.1074/jbc.M512495200. [DOI] [PubMed] [Google Scholar]
- 19.Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell. 2009 Oct 30;139(3):547–559. doi: 10.1016/j.cell.2009.08.042. [DOI] [PubMed] [Google Scholar]
- 20.Valverde R, Edwards L, Regan L. Structure and function of KH domains. FEBS J. 2008 Jun;275(11):2712–2726. doi: 10.1111/j.1742-4658.2008.06411.x. [DOI] [PubMed] [Google Scholar]
- 21.Bycroft M, Hubbard TJ, Proctor M, Freund SM, Murzin AG. The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cell. 1997 Jan 24;88(2):235–242. doi: 10.1016/s0092-8674(00)81844-9. [DOI] [PubMed] [Google Scholar]
- 22.Subramanian AR. Structure and functions of ribosomal protein S1. Prog Nucleic Acid Res Mol Biol. 1983;28:101–142. doi: 10.1016/s0079-6603(08)60085-9. [DOI] [PubMed] [Google Scholar]
- 23.Schubert M, Edge RE, Lario P, et al. Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces. J Mol Biol. 2004 Jul 30;341(1):37–54. doi: 10.1016/j.jmb.2004.05.061. [DOI] [PubMed] [Google Scholar]
- 24.Briani F, Curti S, Rossi F, Carzaniga T, Mauri P, Deho G. Polynucleotide phosphorylase hinders mRNA degradation upon ribosomal protein S1 overexpression in Escherichia coli. RNA. 2008 Nov;14(11):2417–2429. doi: 10.1261/rna.1123908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stickney LM, Hankins JS, Miao X, Mackie GA. Function of the conserved S1 and KH domains in polynucleotide phosphorylase. J Bacteriol. 2005 Nov;187(21):7214–7221. doi: 10.1128/JB.187.21.7214-7221.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blake PR, Park JB, Zhou ZH, Hare DR, Adams MW, Summers MF. Solution-state structure by NMR of zinc-substituted rubredoxin from the marine hyperthermophilic archaebacterium Pyrococcus furiosus. Protein Sci. 1992 Nov;1(11):1508–1521. doi: 10.1002/pro.5560011112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Day MW, Hsu BT, Joshua-Tor L, et al. X-ray crystal structures of the oxidized and reduced forms of the rubredoxin from the marine hyperthermophilic archaebacterium Pyrococcus furiosus. Protein Sci. 1992 Nov;1(11):1494–1507. doi: 10.1002/pro.5560011111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oddone A, Lorentzen E, Basquin J, et al. Structural and biochemical characterization of the yeast exosome component Rrp40. EMBO Rep. 2007 Jan;8(1):63–69. doi: 10.1038/sj.embor.7400856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jarrige A, Brechemier-Baey D, Mathy N, Duche O, Portier C. Mutational analysis of polynucleotide phosphorylase from Escherichia coli. J Mol Biol. 2002 Aug 16;321(3):397–409. doi: 10.1016/s0022-2836(02)00645-9. [DOI] [PubMed] [Google Scholar]
- 30.Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E. Structure of the active subunit of the yeast exosome core, Rrp44: diverse modes of substrate recruitment in the RNase II nuclease family. Mol Cell. 2008 Mar 28;29(6):717–728. doi: 10.1016/j.molcel.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 31.Schneider C, Anderson JT, Tollervey D. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol Cell. 2007 Jul 20;27(2):324–331. doi: 10.1016/j.molcel.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen CY, Gherzi R, Ong SE, et al. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001 Nov 16;107(4):451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 33.Frazao C, McVey CE, Amblar M, et al. Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature. 2006 Sep 7;443(7107):110–114. doi: 10.1038/nature05080. [DOI] [PubMed] [Google Scholar]
- 34.Wall D, Kolenbrander PE, Kaiser D. The Myxococcus xanthus pilQ (sglA) gene encodes a secretin homolog required for type IV pilus biogenesis, social motility, and development. J Bacteriol. 1999 Jan;181(1):24–33. doi: 10.1128/jb.181.1.24-33.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arcus VL, Backbro K, Roos A, Daniel EL, Baker EN. Distant structural homology leads to the functional characterization of an archaeal PIN domain as an exonuclease. J Biol Chem. 2004 Apr 16;279(16):16471–16478. doi: 10.1074/jbc.M313833200. [DOI] [PubMed] [Google Scholar]
- 36.Jeyakanthan J, Inagaki E, Kuroishi C, Tahirov TH. Structure of PIN-domain protein PH0500 from Pyrococcus horikoshii. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005 May 1;61(Pt 5):463–468. doi: 10.1107/S1744309105012406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glavan F, Behm-Ansmant I, Izaurralde E, Conti E. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J. 2006 Nov 1;25(21):5117–5125. doi: 10.1038/sj.emboj.7601377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Vivo M, Dal Peraro M, Klein ML. Phosphodiester cleavage in ribonuclease H occurs via an associative two-metal-aided catalytic mechanism. J Am Chem Soc. 2008 Aug 20;130(33):10955–10962. doi: 10.1021/ja8005786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci U S A. 1993 Jul 15;90(14):6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng ZF, Deutscher MP. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J Biol Chem. 2002 Jun 14;277(24):21624–21629. doi: 10.1074/jbc.M202942200. [DOI] [PubMed] [Google Scholar]
- 41.Cheng ZF, Deutscher MP. An important role for RNase R in mRNA decay. Mol Cell. 2005 Jan 21;17(2):313–318. doi: 10.1016/j.molcel.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 42.Wang HW, Wang J, Ding F, et al. Architecture of the yeast Rrp44 exosome complex suggests routes of RNA recruitment for 3′ end processing. Proc Natl Acad Sci U S A. 2007 Oct 23;104(43):16844–16849. doi: 10.1073/pnas.0705526104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hernandez H, Dziembowski A, Taverner T, Seraphin B, Robinson CV. Subunit architecture of multimeric complexes isolated directly from cells. EMBO Rep. 2006 Jun;7(6):605–610. doi: 10.1038/sj.embor.7400702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Midtgaard SF, Assenholt J, Jonstrup AT, Van LB, Jensen TH, Brodersen DE. Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain. Proc Natl Acad Sci U S A. 2006 Aug 8;103(32):11898–11903. doi: 10.1073/pnas.0604731103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuo Y, Wang Y, Malhotra A. Crystal structure of Escherichia coli RNase D, an exoribonuclease involved in structured RNA processing. Structure. 2005 Jul;13(7):973–984. doi: 10.1016/j.str.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 46.Phillips S, Butler JS. Contribution of domain structure to the RNA 3′ end processing and degradation functions of the nuclear exosome subunit Rrp6p. RNA. 2003 Sep;9(9):1098–1107. doi: 10.1261/rna.5560903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ollis DL, Brick P, Hamlin R, Xuong NG, Steitz TA. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature. 1985 Feb 28 6;Mar 28 6;313(6005):762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- 48.Beese LS, Steitz TA. Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991 Jan;10(1):25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cristodero M, Bottcher B, Diepholz M, Scheffzek K, Clayton C. The Leishmania tarentolae exosome: purification and structural analysis by electron microscopy. Mol Biochem Parasitol. 2008 May;159(1):24–29. doi: 10.1016/j.molbiopara.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 50.Lehner B, Sanderson CM. A protein interaction framework for human mRNA degradation. Genome Res. 2004 Jul;14(7):1315–1323. doi: 10.1101/gr.2122004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Callahan KP, Butler JS. Evidence for core exosome independent function of the nuclear exoribonuclease Rrp6p. Nucleic Acids Res. 2008 Dec;36(21):6645–6655. doi: 10.1093/nar/gkn743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Callahan KP, Butler JS. TRAMP complex enhances RNA degradation by the nuclear exosome component Rrp6. J Biol Chem. 2010 Feb 5;285(6):3540–3547. doi: 10.1074/jbc.M109.058396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bayne EH, White SA, Allshire RC. DegrAAAded into silence. Cell. 2007 May 18;129(4):651–653. doi: 10.1016/j.cell.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 54.Houseley J, Tollervey D. The nuclear RNA surveillance machinery: the link between ncRNAs and genome structure in budding yeast? Biochim Biophys Acta. 2008 Apr;1779(4):239–246. doi: 10.1016/j.bbagrm.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 55.Lebreton A, Seraphin B. Exosome-mediated quality control: substrate recruitment and molecular activity. Biochim Biophys Acta. 2008 Sep;1779(9):558–565. doi: 10.1016/j.bbagrm.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 56.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′-->5′ exoribonucleases. Cell. 1997 Nov 14;91(4):457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]