Abstract
Coping with variations in network dosage is crucial for maintaining optimal function in gene networks. Here we explore how network architecture facilitates network-level dosage compensation. Using the yeast galactose network as a model, we combinatorially deleted one of the two copies of its four regulatory genes and found that network activity was robust to the change in network dosage. A mathematical analysis revealed that a 2-component genetic circuit with elements of opposite regulatory sign (activator and inhibitor) constitutes a minimal requirement for network-dosage invariance. Specific interaction topologies and a 1-to−1 interaction stoichiometry between the activating and inhibiting agents were additional essential elements facilitating dosage invariance. This mechanism of network-dosage invariance could represent a general design for gene network architecture in cells.
The number of copies of a gene network in a cell, or network dosage, has a direct effect on cellular phenotypes (1). Network dosage is altered in situations such as the switching of some organisms between haploid and diploid life forms (2), doubling of chromosomes during cell cycle (3), genome-wide duplication of genetic content (4, 5), and global variation (6) in gene expression. Different phenotypes have different levels of sensitivity to such variations and the need for effective compensation mechanisms arises when cells cannot tolerate these alterations.
It is believed that in the transition between haploid and diploid forms of life cells utilize a volume-mediated compensation mechanism to keep the concentrations of transcription factors constant as cell volume increases with ploidy (2). However, this mechanism cannot subdue the effects of global expression variation and genome duplication or loss events as they affect cellular phenotypes independently of cell volume. For example, variability in ribosome numbers can cause significant fluctuations in global expression levels. These observations raise the question of whether there are alternative layers of dosage compensation mechanisms independent of external factors such as cell volume. To what extent would network activity be robust to alterations in network dosage if we fixed cell volume and therefore excluded its compensatory effect? Could there be a molecular mechanism intrinsic to the network structure that helps cells diminish the effects of dosage variations? Despite the fundamental nature of these questions, what these mechanisms are and how they can be implemented has remained unclear.
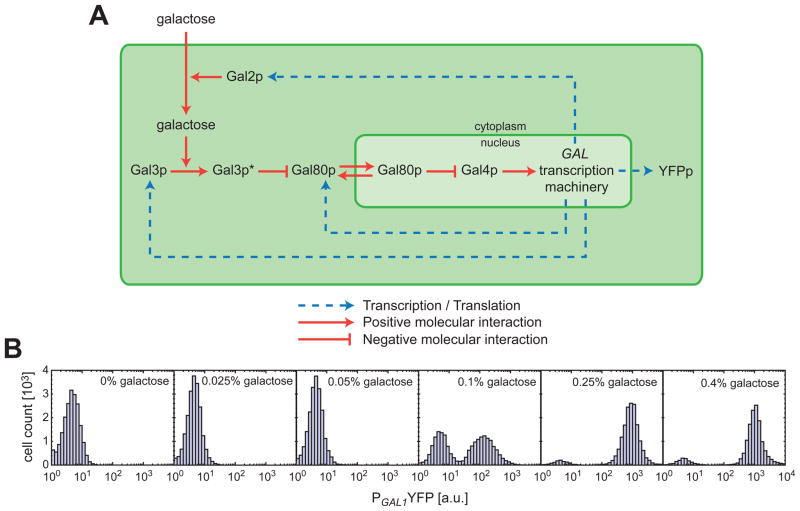
Using experimental and computational approaches, we investigate these questions by utilizing the galactose signalling pathway (GAL pathway) of the yeast Saccharomyces cerevisiae as a model system (Fig. 1A). The GAL network has a well-characterized (7) bistable expression profile. Bistability (7–9) is a dynamical system property giving rise to two distinct gene expression states (OFF and ON) for isogenic cells grown in the same environment. In a bistable gene network, the fraction of cells occupying the ON-state can be defined as the inducibility of the system and serves as a quantitative phenotypic trait. In the GAL network, four genes (GAL2, GAL3, GAL4, and GAL80) play key roles in regulating gene expression. The constitutively expressed Gal4p protein is a transcriptional activator that regulates expression of the other GAL pathway genes (10). Gal80p binds (11) to this protein and prevents Gal4p-mediated transcriptional activation. The protein Gal3p is activated (12) by galactose molecules that are imported into the cell by the galactose permease Gal2p. In its active form, Gal3p sequesters the Gal80p repressor to the cytoplasm, indirectly promoting transcription (13, 14). Except the constitutive GAL4 promoter, the activities of the different GAL pathway promoters are similar to each other (7). To quantify the activity of the GAL pathway at the single-cell level, we used the yellow fluorescent protein (YFP) driven by the GAL1 promoter as our reporter and measured expression profiles at different galactose concentrations using flow cytometry (Fig. 1A–B). We interpreted these experimental results in the context of an effective model (15).
Fig. 1. The galactose utilization pathway as a model gene network and bistability as a quantitative phenotype.
(A) Gal3p* represents the galactose-bound, active form of Gal3p. The shuttling of Gal80p between the cytoplasm and the nucleus is denoted by the bidirectional red arrows. The dotted blue arrows show how the transcriptional feedback loops are established through Gal2p, Gal3p, and Gal80p. (B) Histograms show the induction profile of the wild type galactose pathway for different galactose concentrations.
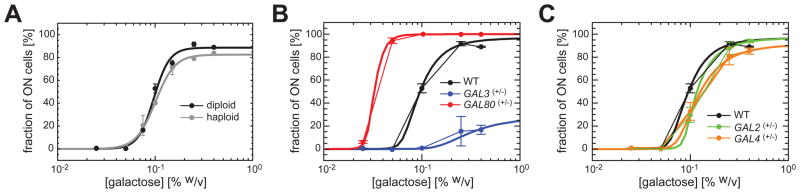
We observed similar inducibility profiles between haploid and diploid strains that contain the same reporter system (Fig. 2A), demonstrating that the system is invariant to ploidy changes. To dissect how network-dosage variations affect the inducibility of the network in the absence of volume effects, we systematically reduced the number of copies of the 4 regulatory genes in the GAL network from 2 to 1 in diploid backgrounds by using KanMX4 and NatMX4 cassettes (15), obtaining 16 different diploid yeast strains including the hemizygous and the wild type strains that have all 4 genes at one and two copies, respectively (15).
Fig. 2. Haploid-diploid comparison and measurement of the contribution of each regulatory gene to network inducibility.
(A) Fraction of ON cells as a function of galactose concentration for both diploid and haploid strains. The solid lines are guides to the eye constructed by fitting a sigmoidal function to the data. (B) The inducibility profile of the GAL network heterozygous in GAL3 (blue) or GAL80 (red) relative to the wild-type profile (black). (C) The inducibility profile of the GAL network heterozygous in GAL2 (green) or GAL4 (orange) relative to the wild-type profile (black). In both (B) and (C), the thick solid lines represent the model best fit to the 5 different inducibility profiles shown in Fig. 2B–C.
Halving the dosage of GAL3 dramatically reduced wild type inducibility levels whereas halving the dosage of GAL80 made the cells need less galactose for full induction (Fig. 2B). Varying GAL2 or GAL4 dosage levels did not have a large effect on network activity (Fig. 2C).
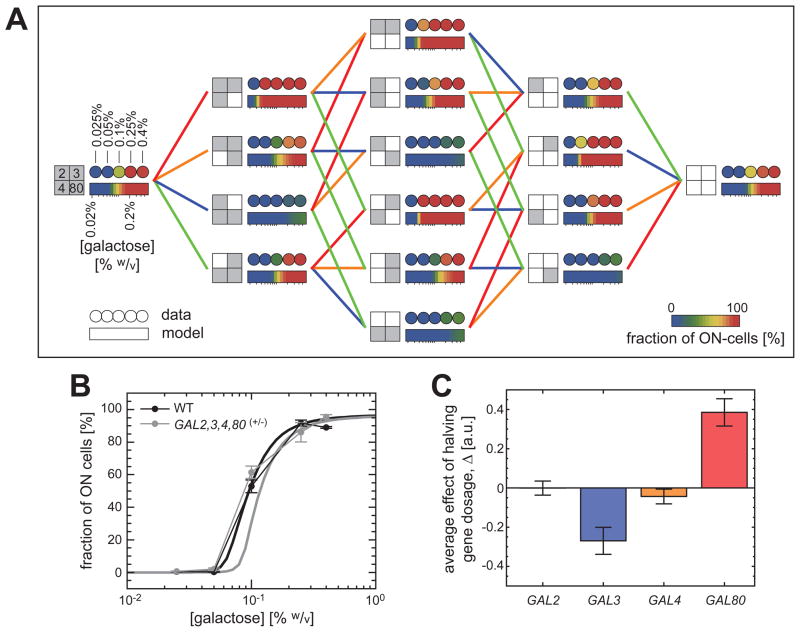
To comprehensively explore the degree of dosage compensation in the GAL network, we measured the inducibility profiles of all 16 strains, grouped the measurements in 4 dosage-perturbation orders, and compared the profiles to one another (Fig. 3A) (15). We observed similar inducibility profiles for the fourth-order hemizygous strain and the wild type strain, implying the presence of network-dosage invariance in the GAL network, even in the absence of volume-mediated compensation effects (Fig. 3A–B).
Fig. 3. Systematic dosage variations and network-dosage compensation.
(A) The color of each filled circle represents the network inducibility level. The rectangular, color-coded bars reflect the predictions of the model based on the best fit to the data presented in Fig. 2B–C. The genetic background of each strain is specified by a big square at its immediate left. The small squares represent the four regulatory genes of the GAL network. Grey (white) color marks the presence of two (one) copies of a specific gene. A line between two strains indicates that the two genetic backgrounds differ by a single copy of a specific gene and the color of the line codifies that gene (blue for GAL3, red for GAL80, green for GAL2, and orange for GAL4). (B) The similarity between the inducibility profiles of the wild-type strain (black) and the strain containing one copy of each regulatory gene (grey). The thick solid lines represent the model predictions. (C) Average contribution of the second copy of each regulatory gene to network inducibility (15).
To determine the relative significance of each regulatory gene in affecting the wild type inducibility levels, we quantified the average contribution of the second copy of each gene to inducibility (15). Fig. 3C depicts the greater importance of GAL3 as an activator and GAL80 as an inhibitor compared to the relatively smaller contributions of GAL2 and GAL4 to the inducibility profiles (15). These results suggest that it may be possible to build a dosage-invariant network using only 2 components, but they do not by themselves indicate how the wiring topology of the network components contributes to network-dosage invariance.
To pinpoint the minimal general conditions that can facilitate dosage invariance in the absence of volume effects, we moved away from the specific case of the GAL pathway and analyzed generic network structures consisting of a set of genes all regulated by the same factor (15). We first found that any network with only one component cannot be dosage invariant. For networks with two components, dosage invariance is possible only if the components have opposite regulatory signs (i.e., if one is an activator and the other is an inhibitor).
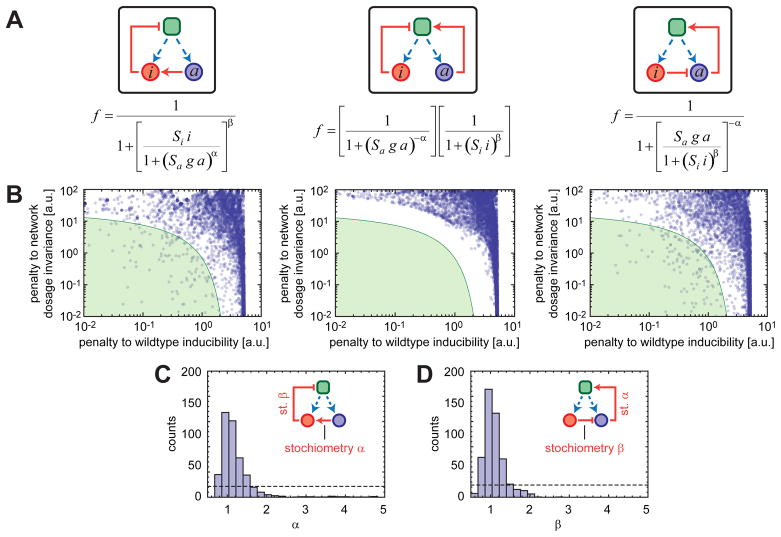
To further explore how certain wiring topologies of the 2-component generic networks would affect dosage invariance, we performed numerical investigations on the possible network topologies and analyzed their inducibility properties. Alternative network configurations are achieved based upon the following interaction topologies: the activator indirectly activates transcription; the activator directly activates transcription; the inhibitor gives up its direct-repressor role and the activator assumes a direct-activator role (Fig. 4A). Each interaction topology is represented by a 4-parameter functional form (Fig. 4A).
Fig. 4. Numerical analysis of general network features producing an inducible and network-dosage invariant system.
(A) Each functional form represents the relationship between the fraction of transcriptionally active cells and the total concentrations of the activating (a) and inhibiting (i) agents. Blue and red circles represent activating and inhibiting agents, respectively. Dashed blue arrows denote the transcriptional production of the network components. The green square represents a transcriptional center. Pointing red arrows show direct activation while blunt red arrows represent inhibition. Each configuration is described by 4 parameters: the scales of action of the activator and inhibitor (Sa and Si respectively) and coefficients (α and β) quantifying the typical nonlinearity of the interaction with downstream components. (B) For each configuration depicted in (A), the degree of inducibility and network-dosage invariance of systems are plotted on the x and y axes, respectively. The green region corresponds to systems that are both inducible and network-dosage invariant. (C) For the left configuration in (A), histogram of the parameter values corresponding to the green region shown in (B). (D) As in (C) but for the right configuration shown in (A). In (C–D), the dotted lines show what one would expect had the parameters had no effect in determining whether the system was in the green region or not.
We randomly sampled the parameters characterizing these forms over large ranges and fed them into the quantitative model to obtain numerical inducibility curves corresponding to the networks carrying one or two copies of the network genes (15). For each pair of these numerical curves, we calculated the level of dosage invariance by quantifying the area between the two curves, large areas corresponding to large penalties to network-dosage invariance, and vice versa (Fig. 4B). In principle, a high degree of dosage invariance can be observed at several different inducibility levels. For example, a biological network always staying in its OFF state is network-dosage invariant, but it lacks the ability to respond to signals of any kind. Thus, it is important to determine if a dosage-compensated system is also inducible or not. We quantified the relative inducibility levels of our numerical curves relative to a reference induction profile. Large differences from the reference curve corresponded to large penalties to inducibility (Fig. 4B). An examination of the dot-plots reveals that the topologies at left and right exhibit both dosage-invariance and inducibility for a wide range of parameter sets. The specific interaction configuration in the two networks is essential for the systems to display such behavior (Fig. 4A). However, the choice between activator and inhibitor in directly influencing transcription is not essential, so long as the other component regulates indirectly.
The green areas in Fig. 4B enclose the parameter sets corresponding to dosage-invariant and inducible networks (low penalties in both axes). For each point populating these areas, we extracted out the values of the 4 parameters (Fig. 4C–D) (15). The parameter quantifying the nonlinearity of the interaction between the inhibiting and activating agents (α in Fig. 4C and β in Fig. 4D) was the only one severely restricted in its values, which displayed a narrow distribution centered around 1. Thus, the effective stoichiometry of the interaction between the activating and inhibiting agents has to be close to 1-to−1 for a system that is both inducible and network-dosage invariant (15).
To understand why an inducible, network-dosage invariant system requires these specific interaction topologies and a 1-to−1 stoichiometry, consider how the system would respond to coordinated changes in the activator and inhibitor levels. For the system in the center of Fig. 4A, the output depends on independent contributions from the activator and inhibitor. For compensation, the increase in the activator concentration would have to be exactly compensated by the downregulation effect by the inhibitor. However, given the nonlinear effect of each component on output, compensation cannot be maintained over a large range of input levels. The system thus fails to be both inducible and network-dosage invariant. For the other systems analyzed, when the 1-to−1 stoichiometry condition is satisfied, an increase in the activator concentration is compensated by an increase in the inhibitor, as the regulation function is dependent on just the ratio of these levels (15).
The network-dosage invariant GAL system satisfies the dosage compensation requirements identified by the minimal model: the interaction topology between its activator (GAL3) and inhibitor (GAL80) is similar to the topology depicted in Fig. 4 (left panel). In addition, it has been experimentally shown (16) that GAL3 and GAL80 interact with 1-to−1 stoichiometry. These observations further validate our findings.
Using a constitutive promoter (CYC1) to eliminate the feedback regulation through the GAL3 and GAL80 genes, earlier work (17) measured the contribution of the GAL3 and GAL80 feedback loops to the noise in the network activity. It was found that without the feedback regulation the activity of the GAL network became noisier compared to the wild type network. Here, we have kept feedback regulation intact by maintaining at least one copy of the GAL3 and GAL80 genes, and probed the effect of gene and network dosage variations on the network activity, elucidating the contribution of network structure on dosage compensation.
These results provide a volume-independent mechanism that is sufficient for network-dosage invariance. The mechanism requires at least two network components: one positive and one negative regulator. These components have to interact with a 1-to−1 effective stoichiometry and specific topologies allowing only one of them to directly affect transcription. Interestingly, this type of interaction topology is frequently observed (18–21) in natural gene circuits that use sequestration-based signal transduction schemes. Robust network properties such as network-dosage invariance might be selected over evolutionary time scales, and network-dosage invariance could therefore represent a general design principle for gene network architecture in cells (22–29).
Supplementary Material
Acknowledgments
The authors would like to thank J. J. Collins, M. Thattai, and H. Youk for helpful discussions and/or comments on the manuscript. M.A. was supported by a fellowship grant from the Center for Biological Circuit Design at Caltech. B.F.P. and A.v.O were supported by grants from NIH (R01-GM-068957-07 and 1DP1OD003936-02) and NSF. Work in the Elowitz lab was supported by the Packard Foundation, NSF (MCB-0644463), and NIH (R01GM079771-02). Work in the Arnold lab was supported by the NIH (R01 GM068664-05A1 and R01 DA028299-02).
REFERENCES AND NOTES
- 1.Lee JA, Lupski JR. Genomic rearrangements and gene copy-number alterations as a cause of nervous system disorders. Neuron. 2006;52:103. doi: 10.1016/j.neuron.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 2.Galitski T, Saldanha AJ, Styles CA, Lander ES, Fink GR. Ploidy regulation of gene expression. Science. 1999;285:251. doi: 10.1126/science.285.5425.251. [DOI] [PubMed] [Google Scholar]
- 3.Di Talia S, et al. Daughter-specific transcription factors regulate cell size control in budding yeast. PLoS Biol. 2009;7:e1000221. doi: 10.1371/journal.pbio.1000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 5.Rancati G, et al. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell. 2008;135:879. doi: 10.1016/j.cell.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pedraza JM, van Oudenaarden A. Noise propagation in gene networks. Science. 2005;307:1965. doi: 10.1126/science.1109090. [DOI] [PubMed] [Google Scholar]
- 7.Acar M, Becskei A, van Oudenaarden A. Enhancement of cellular memory by reducing stochastic transitions. Nature. 2005;435:228. doi: 10.1038/nature03524. [DOI] [PubMed] [Google Scholar]
- 8.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 9.Xiong W, Ferrell JE., Jr A positive-feedback-based bistable ‘memory module’ that governs a cell fate decision. Nature. 2003;426:460. doi: 10.1038/nature02089. [DOI] [PubMed] [Google Scholar]
- 10.Mizutani A, Tanaka M. Regions of GAL4 critical for binding to a promoter in vivo revealed by a visual DNA-binding analysis. EMBO J. 2003;22:2178. doi: 10.1093/emboj/cdg220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melcher K, Xu HE. Gal80-Gal80 interaction on adjacent Gal4p binding sites is required for complete GAL gene repression. EMBO J. 2001;20:841. doi: 10.1093/emboj/20.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki-Fujimoto T, et al. Analysis of the galactose signal transduction pathway in Saccharomyces cerevisiae: interaction between Gal3p and Gal80p. Mol Cell Biol. 1996;16:2504. doi: 10.1128/mcb.16.5.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng G, Hopper JE. Evidence for Gal3p’s cytoplasmic location and Gal80p’s dual cytoplasmic-nuclear location implicates new mechanisms for controlling Gal4p activity in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:5140. doi: 10.1128/mcb.20.14.5140-5148.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng G, Hopper JE. Gene activation by interaction of an inhibitor with a cytoplasmic signaling protein. Proc Natl Acad Sci USA. 2002;99:8548. doi: 10.1073/pnas.142100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.See supporting material on Science Online.
- 16.Timson DJ, Ross HC, Reece RJ. Gal3p and Gal1p interact with the transcriptional repressor Gal80p to form a complex of 1:1 stoichiometry. Biochem J. 2002;363:515. doi: 10.1042/0264-6021:3630515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramsey SA, et al. Dual feedback loops in the GAL regulon suppress cellular heterogeneity in yeast. Nat Genet. 2006;38:1082. doi: 10.1038/ng1869. [DOI] [PubMed] [Google Scholar]
- 18.Buchler NE, Louis M. Molecular titration and ultrasensitivity in regulatory networks. J Mol Biol. 2008;384:1106. doi: 10.1016/j.jmb.2008.09.079. [DOI] [PubMed] [Google Scholar]
- 19.Bardwell L, et al. Repression of yeast Ste12 transcription factor by direct binding of unphosphorylated Kss1 MAPK and its regulation by the Ste7 MEK. Genes Dev. 1998;12:2887. doi: 10.1101/gad.12.18.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Belote JM. Protein-protein interactions among components of the Drosophila primary sex determination signal. Mol Gen Genet. 1995;248:182. doi: 10.1007/BF02190799. [DOI] [PubMed] [Google Scholar]
- 21.Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 22.Kacser H, Burns JA. The control of flux. Symp Soc Exp Biol. 1973;27:65. [PubMed] [Google Scholar]
- 23.Kollmann M, Løvdok L, Bartholomé K, Timmer J, Sourjik V. Design principles of a bacterial signalling network. Nature. 2005;438:504. doi: 10.1038/nature04228. [DOI] [PubMed] [Google Scholar]
- 24.Fell D. Understanding the Control of Metabolism. Portland Press; London: 1997. [Google Scholar]
- 25.Alon U, Surette MG, Barkai N, Leibler S. Robustness in bacterial chemotaxis. Nature. 1999;397:168. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- 26.Eldar A, et al. Robustness of the BMP morphogen gradient in Drosophila embryonic patterning. Nature. 2002;419:304. doi: 10.1038/nature01061. [DOI] [PubMed] [Google Scholar]
- 27.Shinar G, Milo R, Martinez MR, Alon U. Input output robustness in simple bacterial signaling systems. Proc Natl Acad Sci USA. 2007;104:19931. doi: 10.1073/pnas.0706792104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savageau MA. Comparison of classical and autogenous systems of regulation in inducible operons. Nature. 1974;252:546. doi: 10.1038/252546a0. [DOI] [PubMed] [Google Scholar]
- 29.Alon U. Design Principles of Biological Circuits. Chapman & Hall/CRC; Boca Raton, FL: 2007. An Introduction to Systems Biology. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.