Abstract
The comparison of the small molecule metabolism pathways in Escherichia coli and Saccharomyces cerevisiae (yeast) shows that 271 enzymes are common to both organisms. These common enzymes involve 384 gene products in E. coli and 390 in yeast, which are between one half and two thirds of the gene products of small molecule metabolism in E. coli and yeast, respectively. The arrangement and family membership of the domains that form all or part of 374 E. coli sequences and 343 yeast sequences was determined. Of these, 70% consist entirely of homologous domains, and 20% have homologous domains linked to other domains that are unique to E. coli, yeast, or both. Over two thirds of the enzymes common to the two organisms have sequence identities between 30% and 50%. The remaining groups include 13 clear cases of nonorthologous displacement. Our calculations show that at most one half to two thirds of the gene products involved in small molecule metabolism are common to E. coli and yeast. We have shown that the common core of 271 enzymes has been largely conserved since the separation of prokaryotes and eukaryotes, including modifications for regulatory purposes, such as gene fusion and changes in the number of isozymes in one of the two organisms. Only one fifth of the common enzymes have nonhomologous domains between the two organisms. Around the common core very different extensions have been made to small molecule metabolism in the two organisms.
[Online supplementary material available a http://www.genome.org.]
Here we compare the enzymes of small molecule metabolism found in the prokaryote Escherichia coli and the unicellular eukaryote Saccharomyces cerevisiae (yeast). There is evidence for the existence of prokaryotes 3.8 billion years ago (bya) and of eukaryotes 2.7 bya (Mojzsis et al. 1996; Brocks et al. 1999). Endosymbiosis of an α-proteobacterium is widely accepted as the origin of mitochondria, and mitochondrial genes, in the eukaryotes (Margulis 1970). This endosymbiosis event must have occured before the divergence of plants, 1.6 bya (Lang et al. 1999; Wang et al. 1999), and arguments have been made for it being much earlier (Martin and Müller 1998). Thus according to these estimates, most of the enzymes of small molecule metabolism in E. coli and yeast have had between 1.6 and 2.7 by of separate evolution, depending on whether the yeast enzymes originate from the eukaryotic ancestor or the protomitochondrial genome (Brown and Doolittle 1997).
Regardless of the origin of the enzymes, during this time there have been countless chances for orthologous genes in the two organisms to diverge by mutation, to undergo recombinations resulting in domain loss or accretion, and to change gene structure by gene fusion or fission. New genes for an existing function could be acquired by horizontal transfer or functional displacement of one gene by another within a genome. In addition, many new genes have arisen by duplication and divergence to produce new enzymatic functions and pathways.
Until now, investigations of these evolutionary processes have been limited to studying one aspect, such as gene fusion (Enright et al. 1999) or nonorthologous displacement (Koonin et al. 1996; Makarova et al. 1999), or have focused on differences in pathway topologies rather than the evolution of common enzymes (Huynen et al. 1999). Here we investigate, and to some extent quantify, the frequency of all these evolutionary processes in a large set of enzymes common to the two very distantly related organisms. The extensive information available on the enzymes and pathways of small molecule metabolism in E. coli and yeast allows us to determine the extent to which different evolutionary processes have taken place since they separated from their last common ancestor. At present such a comparison would be much less successful in any other pair of organisms due to the lack of knowledge of their enzymes and pathways. E. coli and yeast have long been model organisms and have been the subjects of very extensive experimental characterization of their genes and proteins, including the determination of their complete genome sequence.
We show that over half of the gene products involved in small molecule metabolism of E. coli and yeast carry out common reactions in the two organisms. Our approach is to use sequence and structural information to characterise the domain structure and the evolutionary relationships of these shared enzymes. The use of structural information together with powerful multiple sequence comparison methods, as well as assignment to sequence families, provides us with an almost complete picture of the protein families that the enzymes belong to, including very distant evolutionary relationships.
Knowledge of the domain architecture of common enzymes allows us to assess the extent of conservation between enzymes, but also provides insight into aspects of the regulation of enzymes, such as differing numbers of isozymes in E. coli and yeast and instances of gene fusion. As well as affecting regulation of otherwise separate genes, gene fusion serves to co-localize gene products. Protein–protein interactions have the same effect, and we survey and compare protein-protein interactions as well as gene fusions in yeast.
NOMENCLATURE
Genes, Gene Products, Domains, Enzymes, and Proteins
Before describing the pathways and their enzymes it is useful to provide a glossary of terms we use throughout the text.
Genes and Gene Products
These refer to the DNA entity and the polypeptide produced by its expression.
Proteins and Enzymes
These are the functional units. They can consist of one gene product with one or more domains, multiple copies of one gene product, or a combination of gene products.
Common or Equivalent Enzymes
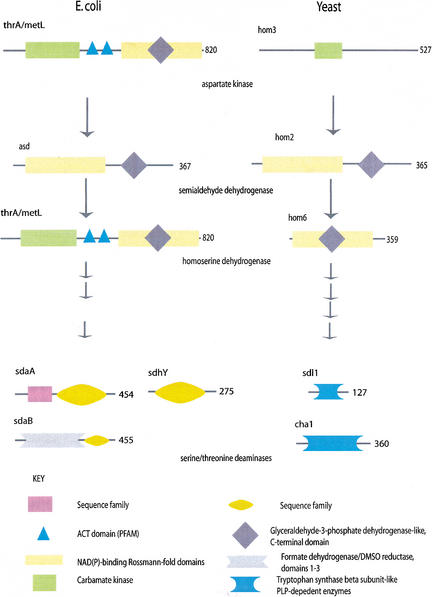
E. coli and yeast enzymes are described as common or equivalent when they play the same role, i.e., catalyze the same reaction step, in the pathways common to the two organisms. The E. coli and yeast enzymes can be, but don't have to be, homologous. For instance, in Figure 1, the E. coli and yeast aspartate semialdehyde dehydrogenases are homologous, whereas the serine/threonine deaminase enzymes are not. For one reaction step, there can be multiple E. coli and yeast gene products that constitute the common enzymes in the two organisms. For instance, in Figure 1 there are three serine/threonine deaminase isozymes in E. coli, and two serine/threonine dehydratases in yeast, so there are five gene products that constitute the common enzymes for this reaction step.
Figure 1.
A selection of enzymes from the KEGG Glycine, serine and threonine metabolism pathway in Escherichia coli and yeast. The domain architectures of selected enzymes from this pathway are shown as cartoons along polypeptide chains represented as black lines. Domains are assigned from structure or sequence domain databases, or identified by simple pairwise sequence similarity; these latter domains are described as belonging to ‘sequence families’. Domains can be inserted into other domains such as the Glyceraldehyde-3-phosphate dehydrogenase domain into the NAD(P)-binding Rossmann fold domains in thrA and metL. These two gene products contain the domains and catalyze the reactions of both the yeast hom3 and hom6, and are thus likely to have evolved by gene fusion. Other enzymes are identical in domain architecture, such as asd and hom2. The last enzymes on the diagram, for which there are three isozymes in E. coli and two in yeast, catalyze the same reaction, but do not have any shared domains. A nonorthologous displacement has occurred among these enzymes.
Domain
This is the evolutionary unit in proteins. Small- and most medium-sized proteins consist of a single domain. Large proteins usually consist of two or more domains that have been brought together by recombination. Domains may combine with more than one partner and may also occur in isolation as functional units. Throughout the figures accompanying this text, gene products are represented by black lines, and their domains are represented by colored shapes. For example, in Figure 1 the yeast serine/threonine dehydratases consist of a single domain represented by a light blue rectangular shape, which is a domain of the Tryptophan synthase beta subunit-like PLP-dependent enzyme family. Other gene products consist of multiple domains, as in the five domains of the E. coli aspartate kinase/homoserine dehydrogenases thrA and metL.
Family
Domains that are related, having evolved from a common ancestor by gene duplication, belong to the same family. Family membership can be detected by straightforward sequence comparison, but more distant relationships are only detectable through conservation of three-dimensional structure of proteins rather than amino acid sequence. Families that correspond to proteins of known three-dimensional structure are sometimes referred to as ‘structural families’, whereas families inferred on the basis of sequence alone are sometimes referred to as ‘sequence families’ in the text. In the figures, all domains belonging to one family are represented by the same colored shape.
SCOP
The Structural Classification of Proteins (SCOP) database (Murzin et al. 1995; LoConte et al. 2000), a hierarchical classification scheme of the proteins of known three-dimensional structure. This database organizes the protein structures according to their domains and evolutionary relationships in terms of protein families, as described in more detail below. By comparing the sequences of the proteins of known structure to the sequences of the yeast and E. coli enzymes, we can infer the domain architecture and evolutionary relationships of the enzymes.
SUPERFAMILY (Gough et al. 2001)
The database of Hidden Markov Models (HMMs) that represent the SCOP domains, as well as their assignments to the proteins of completely sequenced genomes.
PFAM (Bateman et al. 2000)
The database that currently contains 3360 multiple alignments of protein families and HMMs of these protein families. Some of these correspond to SCOP families with known structures, whereas others are purely sequence families.
HMM
Abbreviation for Hidden Markov Model. In our context, this means a probabilistic model of a set of aligned related protein sequences. This model can be used to match other protein sequences to themselves to see whether they are related to the family in the model (Eddy 1996). Because HMMs describe the average characteristics of a set of sequences, this is a more powerful way of detecting relationships between sequences than using simple pairwise sequence comparison methods such as FASTA.
FASTA (Pearson and Lipman 1988)
This is a pairwise sequence comparison method that allows one to find significant sequence similarities between pairs of sequences at a time. The enzymes were compared to each other, and relationships detected in this way are referred to as ‘sequence families.’
METHODS AND RESULTS
Pathways and Enzymes in E. Coli and Yeast
To compare the components of small molecule metabolism pathways in E. coli and yeast in a detailed and efficient manner it is necessary to have them in a form that allows the comparison to be made using computational procedures. To establish such a data set of pathways, we made use of four different databases. Though there is considerable overlap in the information they contain, each has features that made significant contributions to this work. We used the information from the databases to compare the sets of common and unique enzymes in E. coli and yeast rather than the pathways themselves. The enzymes are components of pathways of course, and we do mention the extent of shared and unique pathways according to the KEGG pathway definitions (see below), but our main focus is the common enzymes.
The Dataset of Pathways and Enzymes
The four databases used here are KEGG (Kanehisa and Goto 2000), EcoCyc (Karp et al. 2000), MetaCyc (Karp et al., 1999) and ERGO/WIT (Overbeek et al. 2000).
KEGG database
In this database, the pathways of individual organisms are all superimposed on reference or template pathways. This feature makes it easy to draw parallels between pathways in different organisms and KEGG provides the starting point for constructing the set of small molecule metabolic pathways and enzymes used here.
In KEGG, pathways are described purely in terms of Enzyme Commission (EC) numbers (NC-IUBMB 1992). The EC system uses four numbers that define in a hierarchical manner the function of an enzyme. However, there can be different enzymes that have the same EC number and which function in different pathways. This means that, in cases where EC numbers refer to more than one enzyme, we have to determine which enzyme is appropriate to particular KEGG pathways. To do this, we processed the KEGG pathways and enzymes using (1) information from the EcoCyc, MetaCyc, and WIT databases, and (2) a combination of automatic and manual analyses.
EcoCyc database
The E. coli proteins are well understood and well documented compared to other organisms, particularly through the work of Monica Riley and her colleagues (Riley 1998). Almost all of the enzymes in small molecule metabolism have been characterized, and their position in a pathway verified, experimentally. This information on pathways and enzymes are described in the EcoCyc database.
Differences between KEGG (January 2001) and EcoCyc (March 2000) mean that, of the 716 protein entries in KEGG and the 569 protein entries in EcoCyc (used by Teichmann et al. 2001), only 486 are common to both. The discrepancy in the two sets of protein entries has two main causes. First, enzymes that have not been assigned an EC number cannot be allocated to a pathway in KEGG. Second, sequences in KEGG that are not in the EcoCyc pathways generally do exist but as entries that have not been assigned to a particular pathway. This is because there is no good experimental evidence for the activity of the enzymes; the gene products are part of complexes or reactions that are not connected to a sequence of reactions, or the reactions are at the junction between small molecule and macromolecule metabolism and have been assigned to the latter.
MetaCyc database
This database is related to EcoCyc, but contains the pathways and enzymes of other organisms, including S. cerevisiae. The MetaCyc pathways are constructed by comparing the EC numbers and enzyme names of the organism of interest to pathways already established for E. coli or other organisms (Karp et al. 1999). Support for the existence of each pathway is quantified using a score based on the total number of reactions in the template pathway and the number of reactions identified in the organism whose pathways are being constructed. We were not able to use all information on yeast pathways now available from MetaCyc because only part of it was publicly available when we started work in January 2001.
ERGO or WIT metabolic pathway database
The public version of the database is a repository of detailed information for, at present, 39 organisms (Overbeek et al. 2000). It has an extensive set of E. coli and yeast pathways and gives detailed information on the localization of enzymes within each pathway. This information is particularly useful for the work described here.
Metabolic pathways used in this work
The automatic procedure we used to process the KEGG pathways is similar to that used in the construction of MetaCyc (see above), except that we increased the score if the reactions in a pathway in E. coli or yeast were actually connected to each other, as opposed to being separated by steps that were present in the template pathway but were not identified in the individual organism. In some cases this involved removing some pathways all together and modifying others. For example, we removed the KEGG photosynthesis and tetracycline biosynthesis pathways from E. coli as these are clearly not relevant to this organism and were assigned in KEGG as an artefact of the assignment system based on EC number alone.
The final number of KEGG pathways in our processed set is 55 in E. coli and 57 in yeast (Supplementary data are available at www.genome.org and URL's for all the public databases mentioned above). Of these pathways 48 are shared, meaning that at least a subset of the enzymes catalyzing reactions in these pathways are found in both E. coli and yeast. There are also seven pathways in E. coli not present in yeast, and nine pathways in yeast not present in E. coli. The set of common enzymes are members of the shared pathways, and these were used in our detailed comparison of the enzymes in the two organisms.
Identification of Common Enzymes
Overall, the comparisons of shared EC numbers and gene products show that at least half of the enzymes of small molecule metabolism in E. coli and one third in yeast are not shared between the two organisms, as shown in Table 1. We wanted to establish the extent and nature of the enzymes shared by E. coli and yeast, so we created groups of gene products that are the common enzymes.
Table 1.
Common Pathways and Enzymes in the Small Metabolic Pathways of E. coli and Yeast
| E. coli | Yeast | |
|---|---|---|
| Number of gene products in pathways | 716 | 599 |
| Number of enzymes in pathways | 504 | 368 |
| Number of common enzymes | 271 | 271 |
| Number of gene products that form common enzymes | 384 | 390 |
The common enzymes were identified by matching equivalent position in pathways in the two organisms and grouping together the enzymes that occur in two organisms at that position. A simple example is the fructose-1,6 bisphosphatase in E. coli (fbp) and yeast (FBP1) that occur at equivalent positions in the gluconeogenesis pathway.
Several cases are more complicated than this simple example, however, and these were treated according to the following rules: (1) If an enzyme occurred in more than one pathway, we assigned it to a single group. (2) Where reaction steps with the same EC number in different pathways are catalyzed by different sets of gene products, we made separate groups. (In E. coli, there are 11 EC numbers whose reactions are catalyzed by two nonidentical, but possibly homologous, combinations of gene products and two EC numbers [2.7.1.69, 1.1.1.-] catalyzed by three different combinations. In yeast, there are 10 EC numbers with two different combinations of gene products and two EC numbers [1.1.1.37, 2.5.1.-] with three different combinations. (3) Where enzymes catalyze two or more reaction steps corresponding to two or more EC numbers, there are two or more EC numbers that are associated with exactly the same gene products in both E. coli and yeast. The groups of gene products that are common enzymes were made nonredundant according to the gene product identifiers as well, so some of the groups correspond to multiple EC numbers.
After these filtering processes, we obtained a set of 271 groups that contain, in all, 384 E. coli and 390 yeast gene products. For a list of these see Supplementary data at www.genome.org. As described in Table 2, the contents of these groups vary. Two hundred and thirty four groups contain the same or similar small numbers of E. coli and yeast proteins. Five groups have large numbers of gene products from one organism and few from the other. Two of these groups are reactions that are only described by three EC numbers (acyltransferases, 2.3.1.- and galactosyl/mannosyltransferases, 2.4.1.-) and hence are not well defined. The other three groups involve large complexes: two representing NADH dehydrogenases and one for the ATP synthase complex. The remaining 32 groups have slightly different small numbers of gene products, such as one in one organism and three or four in the other and so forth. The cases of gene fusion have as many entries as there are component enzymes, so there are 37 groups of equivalent enzymes that correspond to the entries in Table 7 (see below).
Table 2.
Numbers of Chains in Equivalent Enzyme Groups
| No. of E. coli gene products | No. of yeast gene products | No. of groups |
|---|---|---|
| 1 | 1 | 152 |
| 1 | 2 | 39 |
| 2 | 1 | 27 |
| 2 | 2 | 13 |
| 3 | 3 | 2 |
| 4 | 4 | 1 |
| 1 | 12 | 1 |
| 8 | 1 | 1 |
| 9 | 26 | 1 |
| 13 | 1 | 2 |
| Other | Other | 32 |
| Total | 271 |
Table 7.
Gene Fusion or Fission Events Ordered by Gene Structure
| E. coli single gene, S. cerevisiae multiple genes | |||
| No. | E. coli | S. cerevisiae | Reactions |
|---|---|---|---|
| 1 | thrA | hom6, hom3 | 1 step apart |
| 2 | metL | hom6, hom3 | 1 step apart |
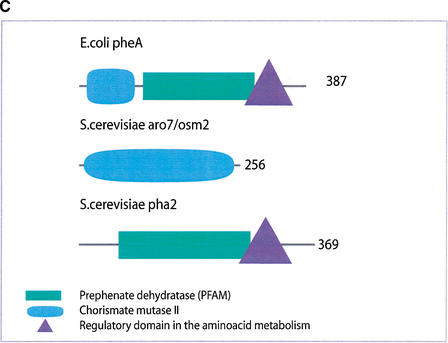
| 3 | pheA | pha2, osm2 | consecutive |
| 4 | hisB | his2, his3 | 1 step apart |
| 5 | aceE | pda1, pdb1 | single reaction |
| S. cerevisiae single gene, E. coli multiple genes adjacent on chromosome | |||
| No. | S. cerevisiae | E. coli | Reactions |
| 6 | arg56 | argC, argB | consecutive |
| 7 | fas3 | accB, accC, accD, (accA) | consecutive |
| 8 | fas1, fas2 | fabI, fabD, fabG, fabH/F/B | consecutive |
| 9 | ura2 | carA, carB, pyrB | consecutive |
| 10 | trp5 | trpA, trpB | single reaction |
| 11 | leu1 | leuD, leuC | single reaction |
| 12 | glt1 | gltB, gltD | single reaction |
| 13 | ade2 | purK, purE | single reaction |
| S. cerevisiae single gene, E. coli multiple genes close (within 10 genes) on chromosome | |||
| No. | S. cerevisiae | E. coli | Reactions |
| 14 | thi6 | thiE, thiM | consecutive |
| 15 | gal10 | galE, galM | consecutive (at junction of different pathways) |
| S. cerevisiae single gene, E. coli multiple genes further than 10 genes away on chromome | |||
| No. | S. cerevisiae | E. coli | Reactions |
| 16 | aro1 | aroA, aroL/K, aroD, aroB, (aroE) | consecutive |
| 17 | his4 | hisD, hisI | consecutive |
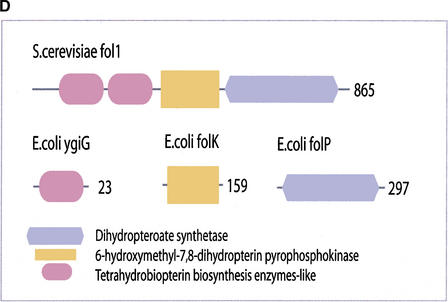
| 18 | fol1 | folK, ygiG, folP | consecutive |
| 19 | ade57 | purM, purD | 2 steps apart |
| 20 | abz1 | pabA, pabB | single reaction |
The accuracy of the assignments of enzymes to EC numbers and pathways could affect our comparison of common enzymes in the following way. If a yeast protein and an E. coli protein were erroneously assigned as being the same enzyme, our analysis would be affected. This is particularly likely to occur during assignment of putative enzymes, in other words enzymes that are assigned through homology rather than experimental characterization. There are at most 192 such enzymes in our data set, and 87 of these are part of the set of common enzymes we analyze in detail. Excluding these 87 enzymes would not affect our general conclusions, however, and so we retain them in our data set.
Enzymes Unique to E. coli or Yeast
In our dataset, there are 332 gene products constituting 233 enzymes unique to E. coli, and 209 gene products constituting 97 enzymes unique to yeast. These enzymes occur in the small number of pathways that are unique to each organism (seven and nine in E. coli and yeast, respectively), but also across the 48 KEGG pathways that contain common enzymes. The organism-specific extensions to pathways with common enzymes involve mostly one or two reactions that are connected to one or both ends of the common part of the pathway. However there are some cases where several separate organism-specific runs of reactions are added to different parts of the pathway, and not all the reactions in a KEGG pathway are necessarily connected. In the Aminosugars Metabolism pathway, there is a series of E. coli-specific reactions, followed by a few common reactions, which are followed by a series of yeast-specific reactions. This clear linear division between series of E. coli-specific and yeast-specific reactions is unique.
To ensure that the enzymes annotated as unique in KEGG do not have hitherto unidentified counterparts in the other genome, we compared the distribution of sequence identities for the 332 and 209 gene products in E. coli and yeast that represent enzymes unique to each of these organisms with that of the common enzymes, whose distribution of sequence identities is discussed below. Only 13% of the 541 unique enzymes have matches above 30%, as compared to 75% of the common enzymes. We inspected the 23 matches above 40% sequence identity because matches at these sequence identities are very likely to have identical EC numbers, according to Wilson et al. (2000) and Todd et al. (2001), and found only eight such cases, which are likely candidates for reclassification. Therefore, it is likely that most of the enzymes classified as present in one of the two organisms but absent in the other are classified correctly, as there is no reason why the pattern of sequence divergence should differ between this set of enzymes and the common enzymes. There remains the possibility that there are as yet unidentified enzymes that are unique to one of the two organisms. For E. coli, this is very unlikely though, as small molecule metabolism has been experimentally investigated for decades and even putative enzymes are included in our data set and the above calculations. Therefore, newly discovered enzymes in small molecule metabolism are most likely to be yeast enzymes that are not shared with E. coli. This would decrease the fraction of common enzymes out of all yeast enzymes, and therefore we view the fraction of common enzymes of one half and two thirds of all enzymes of small molecule metabolism as a lower bound.
The Domain Structure and Family Membership of the Common Enzymes
As described above, just over half of the gene products involved in small molecule metabolism in E. coli and two thirds of those in yeast carry out reactions that are common to both organisms. To compare these common enzymes in terms of their evolutionary relationships, we need to define the domain structure and the protein families to which these domains belong.
Identification of Domains in the E. coli and Yeast Enzymes
To identify the nature of domains in the E. coli and yeast gene products we used three sets of calculations. First, gene products were matched to hidden Markov models of the domains that occur in proteins of known structure (SUPERFAMILY HMMs); second, they were matched to the Pfam HMMs, and third they were matched to each other using FASTA. By these calculations, of the 384 E. coli gene products in shared enzymes, 374 (97%) were matched in nonoverlaping regions by 603 domains using the three methods. Of the 390 yeast gene products in shared enzymes, 343 (88%) were matched in nonoverlaping regions by 607 domains by all three methods (Table 3a).
SUPERFAMILY HMMs
The domains in proteins of known structure (and the superfamilies they belong to) are described in the SCOP database. Gough et al (2001) used the domains from SCOP version 1.53 as seed sequences to build HMMs (Eddy 1996). These were built using the iterative HMM method SAM-T99, described in Karplus et al. (1998), with parameters optimized using SCOP as the standard for detection of distant evolutionary relationships. The database of HMMs is available at: http://stash.mrc-lmb.cam.ac.uk/SUPERFAMILY/. These models were scanned against the E. coli and yeast gene products belonging to shared enzymes, and the resulting matches were processed to identify the domain(s) that made a match. Among the shared E. coli gene products, 481 domains were matched by HMMs from 171 SCOP superfamilies, and 522 yeast domains were matched by HMMs from 161 SCOP superfamilies.
Pfam HMMs
The gene products, or parts of them, that remained unassigned after the identification of SCOP domains were scanned against the Pfam database (Bateman et al. 2000). The Pfam database has a collection of HMMs based on alignments of families of related sequences. Some of the Pfam families have a homolog of known structure and therefore their HMMs may give results similar to those given by the SUPERFAMILY HMMs. But many Pfam families are not related to known structures and we identified 94 additional domains in 83 E. coli gene products and 61 domains in 53 yeast gene products. Of these gene products, 36 in E. coli and 22 in yeast had been matched in other regions by the SUPERFAMILY HMMs.
Pairwise Sequence Comparisons
Even after the Pfam search, some amino acid regions longer than 75 residues remained without a domain assignment. We compared these regions to each other with FASTA (Pearson and Lipman 1988) and clustered them into families in the manner described in Park and Teichmann (1998). This identified a further 29 domains in 25 E. coli sequences and 25 domains in 21 yeast sequences.
Taking together the matches made by SUPERFAMILY HMMs, the Pfam HMMs, and FASTA to the common enzymes, there are 336 (87%) completely assigned gene products in E. coli and 282 (72%) in yeast. In addition, 38 (10%) gene products in E. coli and 61 (16%) in yeast have at least one domain assigned, but contain an unassigned stretch of residues longer than 75 residues. These assignments are available from the Supplementary data at www.genome.org. An illustration of domain assignments for enzymes in glycine, serine and threonine metabolism is given in Figure 1.
Domain Structure of the E. coli and Yeast Enzymes
In total, 603 domains were identified in to the 384 E. coli gene products part of common enzymes and 607 domains in the 390 yeast gene products, as described in Table 3a. The single-domain gene products consist of one domain that matches the whole sequence. Among the E. coli gene products with assignments there are 202 (53%) such cases, and among the yeast gene products there are 148 (38%) such cases. The other gene products matched between two and six domains, except one sequence in yeast that has 11 domains. Overall, there is a slightly larger fraction of multi-domain gene products in yeast than in E. coli, and the multi-domain gene products tend to have somewhat more domains.
Previous work on the EcoCyc list of the gene products that form small molecule metabolism in E. coli showed that about half contain just one domain and half are the product of the recombination of two or more domains (Teichmann et al. 2001).
Protein Families of the E. coli and Yeast Enzymes
The sequences used to build the SUPERFAMILY HMMs are those of the domains in the proteins of known structure. In SCOP, on the basis of an examination of their structures, sequences, and functions, these domains have been clustered into superfamilies whose members can have distant or close evolutionary relationships. We can use this information to cluster into families the domains of the common enzymes matched by the SUPERFAMILY HMMs. Four hundred and eighty one E. coli domains belong to one of 171 different SCOP superfamilies and 522 yeast domains belong to one of 161 superfamilies, as shown in Table 3a. One hundred and forty of the SCOP superfamilies are common to both organisms.
Ninety four domains detected in E. coli by Pfam belong to 73 families and 61 domains in yeast belong to 51 families, 35 of which are present in both E. coli and yeast. The FASTA calculations identify another 22 families in E. coli and 20 in yeast, with 16 of the sequence families present in both organisms.
In all, 191 families are common to both organisms. These include all the large families and, in total, 488 of the 603 E. coli domains and 552 of the 607 yeast domains.
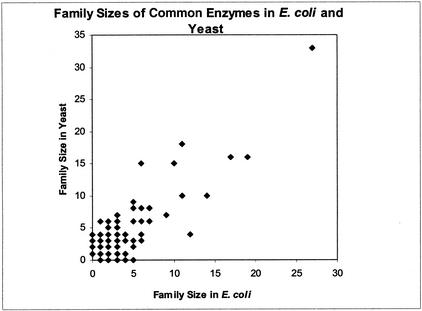
Among the common enzymes, the domains belong to families ranging in size from 1 to 33 (yeast) and 27 (E. coli) members within one organism. The distribution of the sizes of all three types of families is shown in Table 4a, and the 10 largest families are given in Table 4b. In both organisms, the largest family contains NAD(P)-binding Rossmann domains, and the second largest is the PLP-dependent transferase family in E. coli and the Class II aminoacyl tRNA and biotin synthetase family in yeast. Twenty-one of the 30 largest families are the same in the two organisms. As can be seen from Figure 2, most of the 191 common families have only roughly a similar number of members. For the small families the discrepancies tend to be larger: a number of families with a single domain in E. coli or yeast are several-fold larger in the other organism.
Figure 2.
Family sizes in Escherichia coli and Saccharomyces cerevisiae common enzymes. The family sizes in number of domains is shown for E. coli on the X axis and yeast on the Y axis. Families on these axes are unique to one of the organisms, whereas most families with more than one domain are within two- to three-fold size in the two organisms.
A Comparison of the Sequences and Domain Architectures of Common Enzymes
Above we discussed the general features of the domains and families of the common enzymes. Now we turn our attention to the similarity in sequence and domain architecture of the proteins within groups of common enzymes.
Sequence Identity Among Common Enzymes
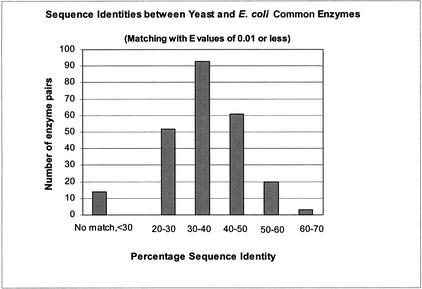
To find the distribution of sequence identities between the E. coli and yeast proteins in the 271 groups of common enzymes, a FASTA search was done between the proteins of the two organisms. The matches at an expectation value threshold of 0.01 or lower were accepted as significant, and the sequence identities for these matches were extracted. The resulting distribution of sequence identities is shown in Figure 3. The distribution of sequence identities is drawn from the best match between an E. coli and yeast sequence in 229 of the 271 common enzyme groups.
Figure 3.
Sequence identities between yeast and Escherichia coli common enzymes. The sequence identity for the best match with an expectation value of 0.01 or less among the gene products of yeast and E. coli common enzymes is shown. Two thirds of the matches are between 30% and 50% sequence identity.
From the domain assignments, we know that there are ⋝13 further common enzymes that share a homologous domain, and for which there is no match below the E-value threshold of 0.01. Their sequence identities are bound to be <30% as described in Brenner et al. (1998).
Inspection of Figure 3 shows that just under two thirds of the common enzyme pairs have sequence identities between 20% and 40%, and just over one third have identities of 40% to 60%. The average sequence identity is 38%. The most highly conserved are three enzymes with sequence identities of 60% to 70%: isopropylmalate isomerase (leuC and LEU1, 61%), the beta subunit of the ATP synthase (atpD and ATP2, 68%), and glyceraldehyde-3-phosphate dehydrogenase (gapA and GPD1, 70%).
Identity and Divergence of Domain Architectures
As mentioned above, significant sequence identity is detected in 229 of the 271 groups of common enzymes, but in 88 the matching region only covers part of the two most similar gene products out of the sets of gene products in these groups. Therefore, to obtain more information about the extent of homology between E. coli and yeast enzymes, we compared domain architectures i.e., the order and family identity of domains of enzymes within each group of gene products. The results are described in Table 5 and below.
Table 5.
Comparison of Domain Architectures in the 271 Groups of Common Enzymes
| Type of group | Number of groups | Totals |
|---|---|---|
| Identical domain architecture and number of chains | 80 | Groups that have same or very similar |
| Different domain architecture, same number of chains, potentially same | 16 | domain architecture: 179 |
| Same domains in a different order | 4 | |
| Same domain architecture, yeast has more chains with this domain architecture (likely larger number of isozymes) | 26 | |
| Same domain architecture, E. coli has more chains with this domain architecture (likely larger number of isozymes) | 14 | |
| Same domain, but either the yeast or E. coli chain has two copies of the domain (internal duplication) | 2 | |
| Same domains, cases of gene fusion (These correspond to the cases listed in Table 7; several common enzyme groups can correspond to one fusion case.) | 37 | |
| E coli has extra domains and/or chains | 26 | Groups that have shared domains and |
| Yeast has extra domains and/or chains | 21 | varied domains: 56 |
| Both organisms have extra domains and/or chains | 9 | |
| No domains shared (non-orthologous displacement) | 13 | Groups that do not share domains: 13 |
| Potential cases of non-orthologous displacement (incomplete assignments) | 6 | Groups that cannot be classified: 23 |
| Neither E. coli nor yeast chains have any assignment | 2 | |
| Either yeast or E. coli chains have no assignment | 15 |
Comparing the 271 groups of gene products of common enzymes. (1) We found that, 84 pairs that have identical domain architectures and identical numbers of gene products in E. coli and yeast, such as the aspartate semialdehyde dehydrogenases hom2 and asd in Figure 1, for instance.
(2) 16 pairs that share at least some domains and could potentially be identical (for example, if the yeast protein consists of domains A and B followed by a gap region, whereas the E. coli protein consists of domains A, B, and C, there may be a domain homologous to C in yeast that has diverged beyond the point where it can be recognized.).
(3) 40 sets of gene products have identical domain architectures, but either yeast or E. coli has more gene products with that domain architecture. These are likely to be additional isozymes in one of the two organisms. However, KEGG does not explicitly annotate isozymes and we have not checked these sequences, so they could also be additional subunits of an enzyme that happen to have the same domain architecture as the other gene products. There are a further three cases where yeast and E. coli have identical numbers of isozymes, and in all 43 of these isozyme cases inspection of sequence identities and phylogenetic analysis suggests that the isozymes within each organism have evolved after the last common ancestor of E. coli and yeast. This suggests that in almost one sixth of the common enzymes, the regulation of a metabolic step has been fine-tuned by adding additional copies of the enzyme in one or both organisms, like thrA/metL and sdl1/cha1 in Figure 1. E. coli isozymes are analyzed in Rison et al. (2002).
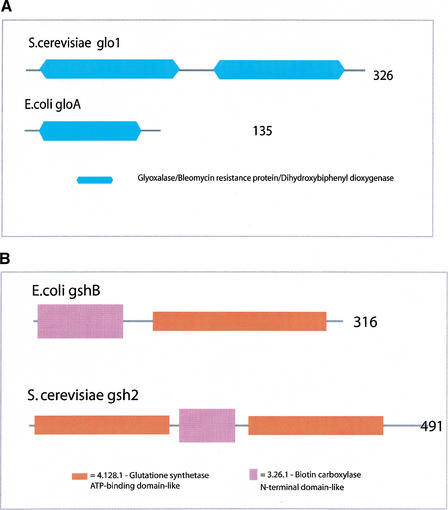
(4) Two cases of internal duplication, one in E. coli and one in yeast, where the enzyme in one organism consists of a gene product with one domain, whereas the enzyme in the other organism consists of a gene product with two copies of that domain. One of these cases is lactoylglutathione lyase, shown in Figure 4a, where the E. coli enzyme is a homodimer of two copies of gloA and the yeast enzyme has an internal duplication in glo1.
Figure 4.
Examples of internal duplication and domain shuffling. (a) The yeast lactoylglutathione lyase (glyoxalase I) consists of two domains of the Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase family, whereas the Escherichia coli enzyme consists of one domain and is active as a homodimer. (b) The yeast glutathione synthetase contains an additional N-terminal copy of the Glutathione ATP-binding domain-like family. It is known that the E. coli enzyme is a homotetramer.
(5) Four cases where E. coli and yeast have the same domains in their enzymes, but in a different order, or with additional copies of domains of the same type. An example of this is the glutathione synthetase enzymes shown in Figure 4b, where the yeast enzyme has an additional N-terminal domain of the Glutathione synthetase ATP-binding domain-like family.
(6) 37 cases of gene fusion or fission that correspond to the 20 gene fusions or fissions described below. An example of fusion is given in Figure 1: thrA/metL represent the domains and functions of both hom3 and hom6. Thus there are 183 of the 262 classifiable groups that have identical or roughly similar domain architecture.
(7) 60 groups where one or more domains are shared, but there are additional domains and/or gene products in one or both organisms that are not shared. These are enzymes and reaction steps that have gained or lost domains in one of the two organisms.
(8) 19 groups have no shared domains. These correspond to the 13 cases of nonorthologous displacement discussed below as well as six cases that we are not confident about; these could be attributable to KEGG misclassifications of proteins into reaction steps. An example of nonorthologous displacement is given in Figure 1: sdaA/sdaB/sdhY and sdl1/cha1.
(9) Nine pairs in which there is no domain assignment for either one or both organisms.
This means that overall some two thirds of the common enzymes have identical or very similar domain architecture. Just under one quarter of these groups have additional domains in one or both organisms beyond the shared set of domains. The remaining small fraction shares no domains at all, and these cases of nonorthologous displacement will be discussed in the next section.
Nonorthologous Displacement
There are 19 cases where pairs of common enzymes share no structural or sequence domains (Table 5). These 19 cases were investigated for evidence of nonorthologous displacement. This involved the retrieval of extra information from resources such as EcoCyc, MetaCyc, MIPS (Mewes et al. 2000), and WIT to establish that the genes were genuinely functionally identical.
From this investigation we identified the 13 possible cases of nonorthologous displacement (Table 6). They occur in 12 different pathways, representing a quarter of all the pathways shared by the two organisms. The structural details and biological explanations of two cases of nonorthologous displacement are given in Figure 5a,b, and another example is the last set of enzymes in Figure 1. Figures for all 13 cases are available as part of the Supplementary data located at www.genome.org.
Table 6.
Instances of Non-Orthologous Displacements
| No. | Pathway | EC number | Gene name(s) in E. coli | Gene name(s) in S. cerevisiae |
|---|---|---|---|---|
| 1 | Glycolysis/Gluconeogenesis | 2.7.1.2 | glk | glk1 |
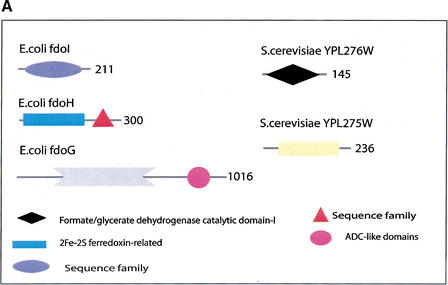
| 2 | Glyoxylate and dicarboxylate metabolism | 1.2.1.2 | fdoI, fdoH, fdoG, fdnI, fdnH, fdnG, fdhf | YPL275W, YPL276W |
| 3 | Sulfur metabolism | 2.7.7.4 | cysN, cysD | met3 |
| 4 | Purine metabolism | 4.6.1.1 | cyaA | cyr1/cdc35/hsr1/sra4 |
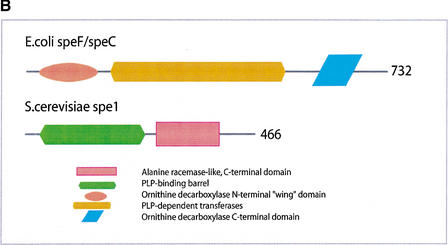
| 5 | Arginine and proline metabolism | 4.1.1.17 | speF/speC | spe1 |
| 6 | Glycine, serine and threonine metabolism | 4.2.1.13 | sdaA, sdaB, sdhY | sdl1/cha1 |
| 7 | Glycine, serine and threonine metabolism | 3.1.3.3 | serB | ser2 |
| 8 | Aminosugars metabolism | 2.7.7.23 | glmU | qril1 |
| 9 | Glycerolipid metabolism | 3.1.4.46 | glpQ, ugpQ | YPL206C |
| 10 | Phospholipid degradation | 3.1.1.5 | tesA | plb1, plb3, lag2 |
| 11 | Porphyrin and chlorophyll metabolism | 1.3.3.4 | hemG | hem14 |
| 12 | Riboflavin metabolism | 2.7.7.2 | ribF | fad1 |
| 13 | Galactose metabolism | 2.7.7.9 | galU, galF | YHL012W, ugp1 |
Figure 5.
(a) This figure corresponds to the second entry in Table 8, formate dehydrogenase in Glyoxylate and dicarboxylate metabolism. This enzyme is involved in the metabolism of formate under anaerobic conditions. The reaction catalyzed is: NAD + formate → NADH + H+ + CO2. The yeast chains YPL275W and YPL276W originate from genes that are adjacent on the same yeast chromosome and make up a putative enzyme complex. The E.coli chains are known to be subunits of the formate dehydrogenase complex. (b) This figure corresponds to the fifth entry in Table 6: Ornithine decarboxylase in Arginine and proline metabolism. The reaction for this enzyme is: L-ornithine CO2 + putrescine. The Escherichia coli genes speF and speC are isozymes and so share the same structure, but differ in their regulation. SpeF is the degradative form and speC is biosynthetic. (c) This figure corresponds to entry 3 in Table 7. The enzymes shown here are from the Phenylalanine, tyrosine and tryptophan biosynthesis pathway. The Escherichia coli chain, pheA, has the functions of chorismate mutase-P and prephenate dehydratase. These functions are matched by the yeast chains aro7 (chorismate mutase) and pha2 (prephenate dehydratase). The yeast chains are not known to physically interact although they are positioned consecutively in the pathway. The discrepancy in the size (a difference of 165 residues) of the chorismate mutase domain between pheA and aro7 is interesting, suggesting it either became truncated during the fusion of the yeast chains, or possibly was expanded after the fission of the E. coli protein. The other domains involved have remained very similar in size. (d) This figure corresponds to entry 18 in Table 7. The enzymes in this example are all from folate biosynthesis. The yeast chain fol1 has the functions of dihydroneopterin aldolase, dihydro-6-hydroxymethylpterin pyrophosphokinase and dihydropteroate synthetase. YgiG is a putative kinase, folK is known as 7,8-dihydro-6-hydroxymethylpterin-pyrophosphokinase, and folP is 7,8-dihydropteroate synthase. Given the structural similarity between ygiG and the first two domains of fol1 it seems likely that these two are functionally equivalent, making ygiG dihydroneopterin aldolase. The E. coli enzymes are consecutive in the pathway.
Of the 13 cases we identified (Table 6), nine seem to be very likely examples of nonorthologous displacement, but four are less sure because of structural assignments to the sequences are incomplete. It is clear from the absolute numbers involved (13 cases out of 254 classifiable groups, or 5%) that the process of displacement by a nonorthologous enzyme is not a frequent occurrence in metabolic pathways.
We tried to investigate the origin of these nonorthologous displacements by characterizing the common enzymes in other organisms. KEGG provides lists of orthologs for many of the reactions in the pathways, but in this set of 13 reactions, there were only ortholog tables for three of the cases, and for these cases there were few or no domain assignments in the other organisms. Therefore, we carried out simple sequence searches of the E. coli and yeast enzymes against 40 other completely sequenced genomes with FASTA (Pearson and Lipman 1988) and selected those matching sequences that had an expectation value of 0.01 or less and a sequence identity of 40% or more, thus ensuring that these gene products had the same function as the query sequence (Wilson et al. 2000; Todd et al. 2001). These searches gave the expected results: E. coli sequences matched proteobacterial or other bacterial proteins and in one of the 13 cases, archaeal proteins and the yeast sequences matched proteins from the four eukaryote genomes. These results show that the 13 cases of nonorthologous displacements in this set of enzymes are not recent events of horizontal transfer between bacteria and eukaryotes.
Gene Fusions and Protein–Protein Interactions
In the previous sections, we have seen that there is extensive conservation of domain architecture in the set of shared enzymes in small molecule metabolism of E. coli and yeast. In common enzymes that have identical domain architectures in E. coli and yeast, there can be a difference between the enzymes at the level of gene structure. Whether the domains belonging to enzymes come from one or several genes affects both the regulation and localization of enzymes, as the parts of a single gene will, by definition, be completely coregulated and colocalized. Colocalization, and potentially regulation, can also be achieved through protein–protein interactions, and we investigate the protein–protein interactions among yeast enzymes in the second part of this section.
Gene Fusions or Fissions
We identified 20 cases of gene fusion or fission with this system, listed in Table 7. The first five cases involve a single E. coli protein and pairs of S. cerevisiae proteins. In the other 15 cases, the yeast enzyme consists of a single gene product and the E. coli proteins are pairs (10 cases), triplets (two cases), quadruplets (two cases), or five different gene products (one case). In 10 of these cases, the E. coli enzymes are adjacent or close to each other on the bacterial chromosome, suggesting that they are coregulated in E. coli in some way as well. In the five cases where the E. coli enzymes are far apart on the chromosome, the fingerprint of gene fusion is lost and it is not clear to what extent the individual enzymes are coregulated.
Figure 5d,c and the first set of enzymes in Figure 1 are examples of the gene fusion events. The style of the diagrams is as for the cases of nonorthologous displacement above, and the numbering is as in Table 7. Figures for all cases of gene fusion are available at www.genome. org (Supplementary data). Taken as a whole, the cases of gene fusion identified in the data set indicate that, although the process is not common in metabolism, it does happen and usually involves a single enzyme or consecutive enzymes in the same pathway in yeast, whereas three of the five E. coli cases are in reactions one step apart. Furthermore, where the component genes are in E. coli, they are often in the same operon and therefore already adjacent on the chromosome. As to those cases where the component genes are not close on the E. coli chromosome, it may be that they have been relocated since the fusion event or were brought together in the eukaryote through transposition or another process.
Because of the absence of operons in eukaryotes, gene fusion appears to be a simple way of achieving coregulation of genes for these organisms, provided that the gene products can be permanently colocalized. However, the pattern of gene fusion of the yeast enzymes does not appear to be conserved across all eukaryotes. We compared the 15 instances of long yeast gene products and the equivalent E. coli component proteins to the gene predictions of four other completely sequenced eukaryote genomes (human, Drosophila melanogaster, Caenorhabditis elegans, Arabidopsis thaliana) as well as 34 completely sequenced prokaryote genomes with FASTA. There is only one case where all the eukaryotes have a better match to the yeast fused protein than to the E. coli proteins, and particularly Arabidopsis enzymes consistently appear to be more E. coli-like than yeast-like in their gene structure.
In five of the cases involving single gene products in E. coli and multiple gene products in yeast, historically either gene fission occurred to allow independent regulation of enzyme subunits in yeast (or a predecessor) or gene fusion occurred in E. coli (or a predecessor). When the five cases of fused or long E. coli genes were compared to the 38 other genomes mentioned above with FASTA, it became clear that not all the prokaryote genomes have fused versions of these genes. In fact, in two of the five cases, only the five gamma-subdivsion proteobacteria, which is the phylogenetic group that E. coli belongs to, have their best match to the E. coli long gene. In another of the five cases, only the beta and gamma-subdivision bacteria as well as Deinococcus radiodurans, Helicobacter pylori and Arabidopsis have their best match to the long E. coli gene. This evidence points towards the fused version of the gene in these prokaryotes being a later event and the separate version of the genes being the earlier, more common version. This begs the question as to what is the evolutionary pressure to obtain and conserve a fused version of genes in prokaryotes that possess operons to coordinate gene regulation. In the absence of an interface for protein–protein interaction, fusion may provide an evolutionarily fast way of forcing otherwise separate gene products to physically associate.
Protein–Protein Interactions in Yeast
Gene fusions provide a means of coregulation and colocalization of enzymes. We found that fusions occur in subunits of enzymes as well as separate enzymes that are at most two steps apart within a pathway. Protein–protein interactions can also serve to colocalize consecutive enzymes to improve flux by minimizing diffusion of the substrate. At the same time, protein–protein interactions between enzymes further apart in the metabolic network can occur for regulatory reasons: an example of this being the regulation of isoleucyl- and valyl-tRNA synthetase by threonine deaminase, two enzymes that are four steps apart in leucine and valine biosynthesis (Savageau and Jacknow 1979; Singer et al. 1984), though this particular interaction is not in the databases used here. Using the large set of protein–protein interactions experimentally determined for yeast proteins, we investigated to what extent enzyme protein–protein interactions occur between enzymes close in the metabolic network in the same way as gene fusions.
The raw data consisted of 7424 pairs of interacting gene products extracted from the MIPS database (Mewes et al. 2000). This database curates the published protein interactions in yeast from individual and large-scale experiments. The interactions are divided into genetic or physical interactions depending on the technique used to determine them. The genetic interactions relate to complementary mutations in two gene products. The physical interactions are typically inferred from methods such as yeast-two-hybrid assays, affinity chromatography, or coimmunoprecipitation.
We extracted the 148 interactions between pairs of gene products in our set of pathways, and these were subdivided into those where the participants were in the same pathway and those where they were in different pathways. For the interactions within pathways, there were seven cases where the gene products represented different enzymes in a pathway not part of the same multifunctional complex. These are shown in Table 8: It is clear that the interactions between enzymes at different steps within the same pathway are all very close to each other, at most two reaction steps apart.
Table 8.
Interactions between Different Enzymes within the Same Pathway
| Consecutive enzymes (3 pairs): |
| Glycoprotein biosynthesis: |
| RHK1 (putative Dol-P-Man dependent alpha(1-3) mannosyltransferase ⇔ oligosaccharyl transferase glycoprotein complex |
| Pyrimidine metabolism: |
| UPRTase ⇔ Uridine kinase |
| Oxidative phosphorylation: |
| ATP synthase subunit h ⇔ COX5B (Cytochrome-c oxidase chain Vb) |
| 1 enzymatic step apart (3 pairs): |
| Glycoprotein biosynthesis: |
| dolichol-P-glucose synthetase ⇔ ... ⇔ components of oligosaccharyl transferase glycoprotein complex |
| Pyruvate metabolism: |
| carbon-catabolite sensitive malate synthease ⇔ ... ⇔ pyruvate carboxylase isozymes |
| Oxidative phosphorylation: |
| ATP synthase subunit h ⇔ ... ⇔ Ubiquinol cytochrome-c reductase subunit 8 |
| 2 enzymatic steps apart (1 pair): |
| Starch and sucrose metabolism: |
| Hexokinase B ⇔ ... ⇔ ... ⇔ multifunctional trehalose-6-phosphate synthase/phosphatase complex |
There were 72 interactions across pathways, but many of these involved membrane proteins or proteins from different subcellular compartments, so we restricted our analysis to the 12 genetic interactions in this set. These 12 interactions follow the same pattern as interactions within pathways to some extent: five of the 12 genetic interactions across pathways are at junctions of pathways in the metabolic network, and hence are actually close to each other in terms of reaction steps. The remaining seven genetic interactions across pathways are cases where the pathway assignment is unclear, so some of these may also be interactions between proteins close to each other in terms of reaction steps.
A similar trend is observed on inspection of the set of complexes in intermediate and energy metabolism identified by mass spectrometry by Gavin et al. (2002). Thirty-three complexes have two or more enzymes in our set of KEGG pathways, and once enzymes that recur in several complexes are accounted for, there are 74 gene products in total with three enzymes that are double-counted because they recur in complexes. Of these, 36 occur in a complex with other gene products involved in the same reaction, 16 occur in a complex with at least one other protein within two metabolic steps, and 6 more occur with another enzyme of the same pathway that is further away. Thirty-five of these 56 interactions are confirmed by other experimental evidence, and generally, there are at most two reaction steps from one pathway in each complex. The exceptions to this are one complex that contains six enzymes from glycolysis and one that contains three enzymes from the citric acid cycle and pyruvate dehydrogenase. Only 18 enzymes occur in a complex without another enzyme from the same pathway, and there are two cases where multiple enzymes from a pair of different pathways occur in the same complex. (None of the 18 interactions of enzymes in different pathways are confirmed by other experimental evidence.) Therefore, the picture gained from these experimental results is that enzymes occasionally cluster with another enzyme close by in the pathway, and very rarely form larger complexes with many enzymes in a pathway, or interact with enzymes in different pathways.
Thus it appears that the main function of these interactions is to colocalize proteins that are in reaction steps close in a pathway. The most likely reason for this is to decrease efficiency lost in diffusion of intermediates between enzymes. Interactions between enzymes further apart in the metabolic network, for instance for regulatory reasons, do not appear to be common.
DISCUSSION
Our comparison of yeast and E. coli small molecule metabolic pathways and enzymes shows that over half of the proteins in this central set of pathways are present in both of these two distantly related organisms. This means that almost as many enzymes of small molecule metabolism are unique to each of the two organisms as are common. Of the sets of enzymes common to both organisms, over two thirds have very closely conserved domain architecture. Just under one quarter of the common enzymes have domain architectures that are partly shared and partly unique to one or both organisms. Among the enzymes that have some similarity in domain architecture, almost all have <50% sequence identity between the E. coli and yeast enzymes, and about a quarter have <30% sequence identity. There are only 13 cases of clear nonorthologous displacement where there is no homology whatsoever between the yeast and E. coli enzyme.
In one seventh of the sets of common enzymes, there are differing numbers of isozymes in E. coli and yeast. There are a few groups of common enzymes with identical numbers of isozymes in the two organisms, and analysis of all sets of isozymes suggests that they occurred after the last common ancestor of E. coli and yeast. The isozymes indicate that even if domain architecture is conserved, regulation of an enzymatic step may be different between the two organisms.
This is also evident in the cases of gene fusion. Fifteen of the 20 cases of gene fusion or fission involve a single yeast enzyme and several individual E. coli enzymes. The balance may be tilted towards the eukaryote due to the absence of operons, but the five cases of fusion in E. coli suggests that fusion may be more than just a means of coregulation, but rather a way of colocalizing otherwise separate gene products.
Colocalization through gene fusions is observed between enzymes at most two steps apart in pathways. A survey of the protein–protein interactions between yeast enzymes shows that a large fraction of these is also between enzymes that are either consecutive or very close to each other in a pathway in terms of reaction steps. Although there is this tendency for physical association of enzymes close to each other in the reaction network, this is by no means the general rule for all consecutive reactions. From our analysis, the frequency of both gene fusions and protein–protein interactions in metabolic pathways appears to be limited, with 15 cases of gene fusions and a small number of protein–protein interactions between separate enzymes among the 368 yeast enzymes considered here.
WEB SITE REFERENCES
http://stash.mrc-lmb.cam.ac.uk/SUPERFAMILY/; The database of HMMs.
Table 3a.
The Domains and Protein Families Identified by the Three Sequence Matching Procedures among the Common Enzymes
| Procedure | E. coli | Yeast | Families common to E. coli and yeast | ||||
|---|---|---|---|---|---|---|---|
| Domains | Families | Domains | Families | Families | E. coli domains | Yeast domains | |
| SUPERFAMILY | 481 | 171 | 522 | 161 | 140 | 429 | 492 |
| Pfam | 93 | 73 | 61 | 51 | 35 | 42 | 44 |
| FASTA | 29 | 22 | 24 | 20 | 16 | 17 | 16 |
| Total | 603 | 266 | 607 | 231 | 191 | 488 | 552 |
Table 3b.
Numbers of Domains in Common Enzymes
| No.domains | E. coli | Yeast | ||
|---|---|---|---|---|
| Complete match | Partial match | Complete match | Partial match | |
| 1 | 202 | 13 | 148 | 21 |
| 2 | 96 | 13 | 90 | 27 |
| 3 | 29 | 8 | 30 | 10 |
| 4 | 6 | 2 | 9 | 1 |
| 5 | 2 | 1 | 3 | |
| 6 | 1 | 1 | 1 | 2 |
| 11 | 1 | |||
| Total no. of proteins | 336 | 38 | 282 | 61 |
Table 4a.
Family Size Distributions among Common Enzymes
| Family size in no.domains | E. coli | S. cerevisiae | Family size in no.domains | E. coli | S. cerevisiae |
|---|---|---|---|---|---|
| 1 | 147 | 118 | 11 | 2 | |
| 2 | 62 | 49 | 12 | 1 | |
| 3 | 23 | 23 | 14 | 1 | |
| 4 | 6 | 13 | 15 | 2 | |
| 5 | 9 | 4 | 16 | 2 | |
| 6 | 7 | 8 | 17 | 1 | |
| 7 | 2 | 4 | 18 | 1 | |
| 8 | 4 | 19 | 1 | ||
| 9 | 2 | 1 | 27 | 1 | |
| 10 | 1 | 2 | 33 | 1 |
Table 4b.
Families with Ten or More Domains among E. coli and Yeast Common Enzymes
| Family | E. coli | Yeast |
|---|---|---|
| NAD(P)-binding Rossman domains | 27 | 33 |
| PLP-dependent transferases | 19 | 16 |
| P-loop containing nucleotide triphosphate hydrolases | 17 | 16 |
| Thiamin diphosphate-binding fold | 14 | 10 |
| Class II amino acyl tRNA synthetases and biotin synthetases | 11 | 18 |
| FAD/NAD(P)-binding domain | 11 | 10 |
| Nucleotidyl transferases | 10 | 15 |
Acknowledgments
We are grateful to Adrian Shepherd and Stuart Rison for help when we set up our relational database. S.A.T. has a Beit Memorial Fellowship for Medical Research and O.J. has a BBSRC studentship.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sat@mrc-lmb.cam.ac.uk; FAX +44-(0)1223-213556.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.228002.
REFERENCES
- Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner SE, Chothia C, Hubbard TJ. Assessing sequence comparison methods with reliably identified distant evolutionary relationships. Proc Natl Acad Sci. 1998;95:6073–6078. doi: 10.1073/pnas.95.11.6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocks JJ, Logan GA, Buick R, Summons RE. Archean molecular fossils and the early rise of eukaryotes. Science. 1999;285:1033–1036. doi: 10.1126/science.285.5430.1033. [DOI] [PubMed] [Google Scholar]
- Brown JR, Doolittle WF. Archaea and the prokaryote-to-eukaryote transition. Microbiol Mol Biol Rev. 1997;61:456–502. doi: 10.1128/mmbr.61.4.456-502.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Hidden markov models. Curr Op Struct Biol. 1996;6:361–365. doi: 10.1016/s0959-440x(96)80056-x. [DOI] [PubMed] [Google Scholar]
- Enright AJ, Ilioupoulos I, Kyrpides NC, Ouzounis CA. Protein interactions maps for complete genomes based on gene fusion events. Nature. 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- Gavin A-G, Bösche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden markov models that represent all proteins of known structure. J Mol Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- Huynen MA, Dandekar T, Bork P. Variation and evolution of the citric-acid cycle: A genomic perspective. Trends Microbiol. 1999;7:281–291. doi: 10.1016/s0966-842x(99)01539-5. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD, Krummenacker M, Paley S, Wagg J. Integrated pathway-genome databases and their role in drug discovery. Trends Biotechnol. 1999;17:275–281. doi: 10.1016/s0167-7799(99)01316-5. [DOI] [PubMed] [Google Scholar]
- Karp PD, Riley M, Saier M, Paulsen IT, Paley SM, Pellegrini-Toole A. The EcoCyc and MetaCyc databases. Nucleic Acids Res. 2001;28:56–59. doi: 10.1093/nar/28.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus K, Barrett C, Hughey R. Hidden markov models for detecting remote protein homologies. Bioinformatics. 1998;14:846–856. doi: 10.1093/bioinformatics/14.10.846. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Mushegian AR, Bork P. Non-orthologous gene displacement. Trends Genet. 1996;12:334–339. [PubMed] [Google Scholar]
- Lang BF, Gray MW, Burger G. Mitochondrial genome evolution and the origin of eurkaryotes. Annu Rev Genet. 1999;33:351–397. doi: 10.1146/annurev.genet.33.1.351. [DOI] [PubMed] [Google Scholar]
- LoConte L, Ailey B, Hubbard TJ, Brenner SE, Murzin AG, Chothia C. SCOP: A structural classification of proteins database. Nucleic Acids Res. 2000;28:257–259. doi: 10.1093/nar/28.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Galperin MG, Grishin NV, Tatusov RL, Wolf YI, Koonin EV. Comparative genomics of the archaea (euryarchaeota): Evolution of conserved protein families, the stable core, and the variable shell. Genome Res. 1999;9:608–628. [PubMed] [Google Scholar]
- Margulis L. Yale University Press, New Haven, CT. 1970. Origin of eukaryotic cells. [Google Scholar]
- Martin W, Müller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- Mewes HW, Frishman D, Gruber S, Geier B, Haase D, Kaps A, Lemcke K, Mannhaupt G, Preiffer K, Schuller C, et al. MIPS: A database for genomes and protein sequences. Nucleic Acids Res. 2000;28:37–40. doi: 10.1093/nar/28.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojzsis SJ, Arrhenius G, Mckeegan KD, Harrison TM, Nutman AP, Friend CRL. Evidence for life on earth before 3,800 million years ago. Nature. 1996;384:55–59. doi: 10.1038/384055a0. [DOI] [PubMed] [Google Scholar]
- Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB). 1992. Enzyme nomenclature, Academic Press, New York.
- Overbeek R, Larsen N, Pusch GD, D'Souza M, Selkov E, Jr, Kyrpides N, Fonstein M, Maltsev N, Selkov E. WIT: Integrated system for high-throughput genome sequence analysis and metabolic reconstruction. Nucleic Acids Res. 2000;28:123–125. doi: 10.1093/nar/28.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Teichmann SA. DIVCLUS: An automatic method in the GEANFAMMER package that finds homologous domains in single- and multi-domain proteins. BioInformatics. 1998;14:144–150. doi: 10.1093/bioinformatics/14.2.144. [DOI] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M. Genes and proteins of Escherichia coli K-12. Nucleic Acids Res. 1998;26:54. doi: 10.1093/nar/26.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rison SCG, Teichmann SA, Thornton JM. Homology, pathway distance and chromosomal localization of the small molecule metabolism enzymes in Escherichia coli J. Mol Biol. 2002;318:911–932. doi: 10.1016/S0022-2836(02)00140-7. [DOI] [PubMed] [Google Scholar]
- Savageau MA, Jacknow G. Feedforward inhibition in biosynthetic pathways: Inhibition of the aminoacyl-tRNA synthetase by intermediates of the pathway. J Theor Biol. 1979;77:405–425. doi: 10.1016/0022-5193(79)90017-1. [DOI] [PubMed] [Google Scholar]
- Singer PA, Levinthal M, Williams LS. Synthesis of the isoleucyl- and valyl-tRNA synthetases and the isoleucine biosynthetic enzymes in a threonine deaminase regulatory mutant of Escherichia coli K-12. J Mol Biol. 1984;175:39–55. doi: 10.1016/0022-2836(84)90444-3. [DOI] [PubMed] [Google Scholar]
- Teichmann SA, Rison SCG, Thornton JM, Riley M, Gough J, Chothia C. The evolution and structural anatomy of the small molecule metabolic pathways in Escherichia coli. J Mol Biol. 2001;311:693–708. doi: 10.1006/jmbi.2001.4912. [DOI] [PubMed] [Google Scholar]
- Todd AE, Orengo CA, Thornton JM. Evolution of function in protein superfamilies, from a structural perspective. J Mol Biol. 2001;307:1113–1143. doi: 10.1006/jmbi.2001.4513. [DOI] [PubMed] [Google Scholar]
- Wang Y-C, Kumar S, Hedges SB. Divergence time estimates for the early history of animal phyla and the origin of plants, animals and fungi. Proc R Soc Lond B. 1999;266:163–171. doi: 10.1098/rspb.1999.0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CA, Kreychman J, Gerstein M. Assessing annotation transfer for genomics: Quantifying the relations between protein sequence, structure and function through traditional and probabilistic scores. J Mol Biol. 2000;297:233–249. doi: 10.1006/jmbi.2000.3550. [DOI] [PubMed] [Google Scholar]