Abstract
Local delivery of DNA through a hydrogel scaffold would increase the applicability of gene therapy in tissue regeneration and cancer therapy. However, the delivery of DNA/cationic polymer nanoparticles (polyplexes) using hydrogels is challenging due to the aggregation and inactivation of polyplexes during their incorporation into hydrogel scaffolds. We developed a novel process (termed Caged Nanoparticle Encapsulation or CnE) to load concentrated and unaggregated non-viral gene delivery nanoparticles into various hydrogels. Previously, we showed that PEG hydrogels loaded with DNA/PEI polyplexes through this process were able to deliver genes both in vitro and in vivo. In this study, we found that hyaluronic acid and fibrin hydrogels with concentrated and unaggregated polyplexes loaded through CnE were able to deliver genes in vivo as well, demonstrating the universality of the process.
Keywords: Gene transfer, Gene delivery, Hyaluronic acid hydrogel, Fibrin hydrogel, Scaffold-mediated gene delivery
Introduction
Gene therapy is the genetic modification of cells through the delivery of DNA or RNA (e.g. small interfering RNA, siRNA), which can up- or down-regulate genes respectively. Gene therapy can be achieved with the use of modified viruses (viral delivery) or non-viral gene delivery vectors (non-viral delivery) that encapsulate or condense plasmid DNA or siRNA into nanoparticles that transport the genetic material into the cell [1–4]. Although non-viral delivery is generally less efficient than viral delivery in vivo, it has the following advantages: (1) the large-scale production of non-viral gene delivery nanoparticles can be more readily accomplished than that for viruses, (2) repeated injection non-viral nanoparticles is possible since they can be engineered not to elicit a severe immune response, (3) the risk of insertion mutagenesis is low since the non-viral gene delivery vectors do not integrate the delivered DNA into the genomic DNA and (4) larger genes can be delivered since the non-viral vectors can complex with large DNA molecules.
Local delivery of DNA or siRNA through a hydrogel scaffold would increase the applicability of gene therapy in tissue regeneration and cancer therapy [5, 6]. Genes encoding for tissue inductive factors can be delivered to the diseased site with the use of a hydrogel scaffold. DNA is transferred to the infiltrating cells to produce protein factors in situ to promote tissue regeneration [6, 7]. For cancer therapy, lethal genes or siRNAs can be delivered with a hydrogel scaffold to induce the apoptosis of tumor cells [8]. Naked DNA was first incorporated into hydrogels, such as collagen [9–11], pluronic-hyaluronic acid [12], PEG-poly(lactic acid)-PEG [13], alginate [14], oligo(polyethylene glycol) fumarate [15] and engineered silk elastin [16] for local gene therapy. Although naked DNA encapsulated in collagen hydrogels was able to promote bone regeneration in vivo [9, 17], the gene transfer efficiency was very low and most of the encapsulated DNA diffused out from the hydrogel scaffold rapidly. This motivated researchers to use non-viral gene delivery vectors to condense DNA into nanoparticles. These nanoparticles can both retain the DNA in the scaffold and significantly enhance the gene transfer efficiency. Cationic peptides, lipids, or polymers have been used to prepare nanoparticles that were encapsulated into collagen [18–20], fibrin [21–26], enzymatically degradable PEG hydrogels [27] and PEG-hyaluronic acid [22] hydrogels. Although gene delivery has been observed from these hydrogels, limitations with low DNA loading and low transgene expression motivated us to find an effective approach for introducing DNA/cationic polymer polyplexes into hydrogel scaffolds.
Previously, we reported on a strategy to incorporate concentrated, un-aggregated and highly active DNA/PEI polyplexes inside polyethylene glycol (PEG) hydrogel scaffolds, termed Caged Nanoparticle Encapsulation or CnE [28]. The approach utilizes neutral saccharides (sucrose) and polysaccharides (agarose) to protect the polyplexes from aggregation and inactivation during lyophilization and hydrogel formation, respectively. Results showed that matrix metalloproteinase (MMP) degradable polyethylene glycol (PEG) hydrogels with concentrated DNA/PEI polyplexes loaded through CnE were able to deliver genes in vivo and induced extensive angiogenesis when genes encoding for VEGF were delivered. In this report, we investigated whether CnE could be used to introduce active DNA/PEI polyplexes into charged hyaluronic acid and protein fibrin hydrogels and achieve efficient gene transfer in vivo. Both HA and fibrin pose extreme challenges in incorporating PEI/DNA polyplexes. HA is highly negatively charged and would compete with DNA for the positively charged PEI, causing polyplex decomplexation. Fibrin is a protein that can absorb the polyplexes and cause aggregation.
Materials and methods
Materials
Human fibrinogen, Bovine plasma thrombin and Linear poly(ethylene imine) (25 kDa, PEI) were bought from Enzyme Research Laboratories, Sigma and Polysciences, respectively. Vectors expressing vascular endothelial growth factor (pVEGF) [29] and beta-galactosidase (pβgal) were gifts from Prof. Lonnie Shea’s laboratory (Northwestern University). pVEGF, has the VEGF gene is inserted in a pcD3.1 vector with a CMV promoter. pβgal has the βgal gene inserted in a pNGVL1 promoter with a CMV promoter. A Giga Prep kit from Qiagen was used to expand the plasmid. Sodium hyaluronan (HA) was a kind gift from Genzyme Corporation (60 KDa MW, Cambridge, MA). All other chemicals were purchased from Fisher Scientific unless otherwise noted.
Hyaluronic acid modification
Acrylated hyaluronic acid (HA-AC) was prepared as previously described [28]. Briefly, 1.0 g hyaluronic acid was reacted with 18.0 g adipic dihydrazide (ADH) at pH 4.75 in the presence of 2.0 g 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC) overnight and purified through dialysis (8000 MWCO) in DI water for 1 week. 38.8% of the carboxyl groups were modified with ADH based on the TNBSA assay. One gram of HA-ADH was reacted with 0.75 g N-Acryloxysuccinimide in HEPES buffer (pH 7.2) overnight and purified through dialysis in DI water for 1 week. All the primary amines were acrylated based on the 2,4,6-Trinitrobenzene sulfonic acid (TNBSA, Thermo Scientific) assay performed following the product manual.
Polyplex lyophilization
Plasmid DNA (0–300 μg) and PEI (0–587 μg) were mixed in 3.5 mL water in the presence of 35 mg (0.10 mmole) of sucrose (Ultra pure, MP Biomedicals) and incubated at room temperature for 15 min. Agarose (1.0 mg, UltraPure™ Agarose, Tm = 34.5~37.5 °C, Invitrogen) in 1.5 mL water was added before lyophilization.
Hydrogels synthesis and characterization
To load polyplexes into HA hydrogels through CnE, lyophilized polyplexes were reconstituted with 100 μL HA-AC solution (in 0.3 M triethanolamine or TEOA, pH = 8.0) containing crosslinker, dithiothreitol (DTT) and gelled through incubation at 37 °C for 30 min. To load polyplexes through the regular encapsulation process, DNA and PEI were mixed in the HA-AC solution, vortexed for 15 s and incubated for 15 min at room temperature before adding crosslinker to form hydrogels at 37 °C for 30 min. To encapsulate polyplexes into fibrin hydrogels through CnE, 100μL fibrinogen solution in PBS (5 mg/mL) was mixed with lyophilized polyplexes and initiated to form gels at 37°C for 20 min with thrombin (2 U/mL). For the regular encapsulation, DNA and PEI were mixed and incubated in fibrin solution for 15 min at room temperature before adding thrombin to form hydrogels at 37 °C for 20 min. All hydrogels were swelled in PBS for 2-hrs. The swelled gels were stained with ethidium bromide (12 μM) for 2-hrs before imaging with a fluorescent microscope (Observer Z1, Zeiss) or a confocal microscope (Leica TCS SP MP) to visualize the distribution of polyplexes inside hydrogels. To test the DNA release from the hydrogel, a 30 μL hydrogel with polyplexes loaded through CnE was incubated in 200 μL PBS (pH 7.4) at 37 °C. Medium was collected daily and released DNA was quantified with HOECHST dye (H33258) [27].
Choriallantoic Membrane (CAM) assay
CAM assay was performed as previously described [28]. Briefly, fertilized chicken eggs were released into tissue culture dishes after 3-days of incubation in a 38°C ventilated, humid egg incubator and were incubated in a sterile humid incubator at 37°C for another 10 days. Hydrogels with polyplexes were placed on the CAM away from major veins and incubated for 3 additional days. Gross pictures were recorded (Stemi 2000-C, Zeiss) before the embryo was infused with 1 mL of FITC-dextran (0.5 mg/mL in PBS, Sigma). The CAM with the hydrogel was cut and fixed in 4% paraformaldehyde for 2-hrs before imaging with a fluorescent microscope (Observer Z1 Zeiss). Fixed CAM was immersed in 37°C X-gal solution (5 mM Potassium ferricyanide, 5 mM Potassium ferrocyanide, 1 mM MgCl2, 1 mg/mL X-Gal (Gold Biotechnology) in PBS) for 48-hrs to visualize the β-Gal expression, which was recorded with a color camera attached to a dissecting microscope (Stemi 2000-C, Zeiss)
Results
HA and fibrin induced the aggregation of DNA/PEI polyplexes
Directly mixing DNA/PEI polyplexes with a hydrogel precursor solution before gelation (regular encapsulation) resulted in severe polyplex aggregations (Fig. 1). Due to the soft, loose and charged structures, DNA/PEI polyplexes tend to aggregate at high concentrations or in salt solution during their incorporation into hydrogels. We tested if HA or fibrin induce polyplex aggregation by measuring the polyplex size using dynamic light scattering (DLS) in the absence or presence of HA and fibrin molecules. The average diameter of DNA/PEI polyplexes in water was about 50–100 nm when the DNA concentration was low (5 μg DNA in 100 μL H2O, N/P = 15). Adding 1% HA shifted the average diameter of the polyplexes to ~1 μm. Severe aggregations occurred in the presence of 1% HA when the DNA concentration was increased to 50 μg in 100 μL as shown by appearance of precipitate and the disappearance of the particles within the solution (Fig. 1A). Polyplexes containing 100 μg DNA (N/P = 15) were observed to aggregate into one large clot inside the 100 μL HA hydrogel (Fig. 1B). Fibrin proteins also induced polyplex aggregation. The average diameters of DNA/PEI polyplexes containing 5 or 50 μg DNA (N/P = 15) in 100 μL 5 mg/mL fibrin solution were about 4.1 and 5.5 μm respectively (Fig. 1C). Large aggregates were found when 100 μg DNA (N/P = 15) were incorporated into 100 μL fibrin hydrogels (Fig. 1D).
Figure 1.
HA or fibrin induced aggregation of polyplexes. (A, C) DNA/PEI polyplexes containing 5 or 50 μg DNA at N/P = 15 were prepared in 100 μL water or 1% HA or 5 mg/mL fibrinogen solution. The size distributions of the formed polyplexes were measured with DLS. (B, D) DNA/L-PEI polyplexes containing 100 μg DNA at N/P = 15 were encapsulated in 100 μL HA (B) or fibrin (D) hydrogels. The DNA was stained with ethidium bromide and visualized with a fluorecence microscope. Scale bar: 100 μm.
Encapsulating DNA/PEI polyplexes into HA and fibrin hydrogels through CnE
Caged nanoparticle encapsulation (CnE) was used to introduce DNA/PEI polyplexes into HA and fibrin hydrogels. Dilute DNA/PEI polyplexes were prepared in the presence of sucrose and agarose and lyophilized. The lyophilized powder containing polyplexes was reconstituted with the hydrogel precursor solution before being initiated to form the hydrogels. Using CnE, up to 300 μg DNA (N/P = 15) was encapsulated into 100 μL HA hydrogel crosslinked through Michael addition. The polyplexes were stained with ethidium bromide and visualized through fluorescence microscopy (Fig. 2A–C and G–I) or confocal microscopy (Fig. 2D–F and J–L). In contrast to the regular encapsulation (Fig. 1), no polyplex aggregations were found inside the HA hydrogels through CnE. Without DNA, the hydrogel scaffold had limited auto-fluorescence (Fig. 2A, B). Polyplexes were distributed throughout the whole gel (Fig. 2B, C). At high magnification, polyplexes were observed to be suspended as unaggregated nanoparticles (Fig. 2E, F). The distribution of the polyplexes inside the hydrogels was not completely homogeneous and there were some micro-domains with dense concentration of polyplexes. However, both inside and outside these micro-domains, the polyplexes were unaggregated. Similar distributions were seen when polyplexes with 50 μg DNA at N/P = 15 were encapsulated in 100 μL 5 or 15 mg/mL fibrin hydrogels through CnE (Fig. 2G–L). Without DNA, the fibrin hydrogel scaffold had no fluorescence (Fig. 2G, L). Polyplexes were distributed throughout the whole gel (Fig. 2H, I). At high magnification, polyplexes were observed to be suspended as unaggregated nanoparticles (Fig. 2K, L). The fibrin concentrations had no significant influence on the polyplex distribution.
Figure 2.
Polyplex distribution in HA and fibrin hydrogels. DNA/PEI polyplexes containing 0 to 300 μg DNA at N/P = 15 were encapsulated in 100 μL HA (A–F) or fibrin (5 or 15mg/mL) (G–L) hydrogels through CnE. The DNA was stained with ethidium bromide and visualized with a fluorescence (A–C, G–I) or confocal microscope (D–F, J–L). Scale bar: 50 μm for (A–C, G–I) and 1 μm for (D–F, J–L).
For DNA/PEI polyplexes encapsulated in HA hydrogels through CnE, less than 1% DNA in the hydrogel was released in PBS over 3 days (Fig. 3A). Polyplexes were retained in the 5 or 15 mg/mL fibrin hydrogel as well as shown that no DNA was released in PBS during the 3 days incubation (Fig. 3B).
Figure 3.
DNA release from HA and fibrin hydrogels. (A) polyplexes containing 100 μg DNA at N/P = 15 were encapsulated into 100 μL HA hydrogel. The hydrogel was incubated in PBS (pH 7.4) at 37 °C. (B) polyplexes containing 50 μg DNA at N/P = 15 were encapsulated into 100 μL 5 or 15 mg/mL fibrin hydrogel. The hydrogels were incubated in PBS (pH 7.4) at 37 °C. The released DNA was quantified with HOECHST.
HA hydrogels with DNA/PEI polyplexes loaded through CnE were able to deliver genes in vivo
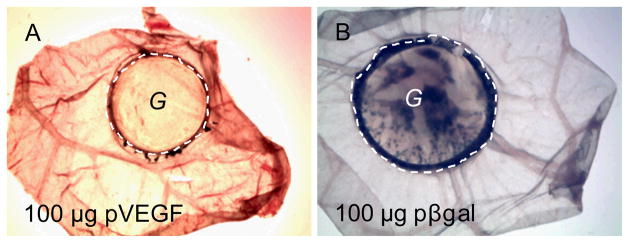
The ability of HA hydrogels loaded with DNA/PEI polyplexes through CnE to mediate gene transfer in vivo was assessed using a chorionic chick embryo (CAM) assay. Hydrogels loaded with DNA encoding for β-galactosidase (pβgal) or VEGF (pVEGF) were placed on top of the CAM. pβgal was transferred from HA gels to cells and resulted in β-galactosidase gene expression determined from the extensive blue X-gal staining (Fig. 4B), which was not found when hydrogels containing pVEGF were used (Fig. 4A).
Figure 4.
DNA loaded HA hydrogels resulted in gene transfer in CAM model. A HA hydrogel with polyplexes was placed on top of CAM for 3 days. The gel with CAM was cut, fixed and stained with x-gal solution for 48 hrs. Positive β-galactosidase expression resulted in blue color. 100 μg pVEGF or pβgal at N/P = 15 was loaded into 100 μL gel. The dashed line highlights the edge of the hydrogel and “G” indicates the gel area
The delivery of DNA encoding for VEGF was able to induce blood vessel formation in the CAM assay. HA hydrogels with polyplexes containing pVEGF were placed on top of the CAM for 3 days. pVEGF was transferred to cells and produced a high concentration of VEGF at the gel area, which resulted in hyperbranced neovessels (Fig. 5, arrows). VEGFs diffusing out the gel created a decreasing VEGF gradient around the gel and led to radial neovessels around the gel (Fig. 5, arrowheads). Gross evaluation of the blood vessels around the implanted hydrogels showed that hydrogel with no DNA (Fig. 5A) and hydrogel loaded with DNA polyplexes encoding for β-galactosidase (Fig. 5B) did not result in enhanced blood vessel formation. However, hydrogel containing DNA encoding for VEGF resulted in enhanced blood vessel formation around the implant site (Fig. 5C, D, arrow heads). Increasing the dose of DNA did not increase the level of angiogenesis around the implant. Evaluation of the blood vessels underneath/inside the hydrogel scaffold after perfusion showed enhanced micro-vessel formation for all hydrogels that contained DNA/PEI polyplexes encoding for VEGF (Fig. 5G, H, arrows). Hydrogels with no DNA, or pβgal resulted in no micro-vessel formation (Fig. 5E, F).
Figure 5.
pVEGF loaded HA hydrogels resulted in enhanced angiogenesis in a CAM model. A HA hydrogel with polyplexes containing pVEGF was placed on top of the CAM for 3 days. pVEGF was transferred to cells and produced a high concentration of VEGF at the gel area, which resulted in hyperbranced neovessels (arrow). VEGF diffusing out the gel created a decreasing VEGF gradient around the gel and led to radial neovessels around the gel (arrowheads). (A–D) Gross pictures on the gel edge were recorded before the CAM was infused with FITC-dextran for fluorescent imaging (E–H) at the gel area. Induced neovessels were found both around the gel (C–D, arrow heads) and at the gel area (G–H, arrows) with pVEGF, which were not found in the negative control (No DNA). Polyplexes at N/P = 15 were used. The dashed line highlights the edge of the hydrogel and “G” indicates the gel area. Scale bar: E–H: 200 μm
Fibrin hydrogels with DNA/PEI polyplexes loaded through CnE were able to deliver genes in vivo
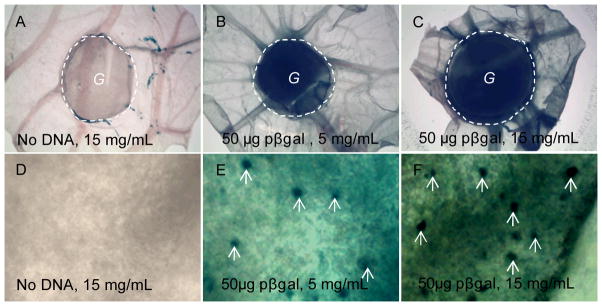
We tested the ability of DNA loaded fibrin hydrogels to deliver genes in vivo using CAM assay. Extensive X-gal staining was only observed for the 5 and 15 mg/mL hydrogels loaded with polyplexes containing pβgal indicating successful gene transfer occurred from the scaffold to the cells (Fig. 6B, C). We examined the density of the X-gal stained cell nucleus at high magnification (Fig. 6E, F, white arrows). Within the view, the number of stained nuclei was 0, 9 and 11 for the gel with no DNA, 5 and 15 mg/mL gels with DNA (Fig. 6D–F), respectively. However, we failed to see significant differences between the 5 and 15 mg/mL gels after examining a larger area (data not shown).
Figure 6.
DNA loaded fibrin hydrogels resulted in gene transfer in the CAM model. A fibrin hydrogel with polyplexes was placed on top of CAM for 3 days. The gel with CAM was cut, fixed and stained with x-gal solution for 48 hrs. Positive β-galactosidase expression resulted in blue color. (A–C) and (D–F) showed the low and high magnifications, respectively. 50 μg pβgal at N/P = 15 was loaded to 100μL 5 or 15 mg/mL fibrin gels. The dashed line highlights the edge of the hydrogel and “G” indicates the gel area. The white arrows in (E and F) point out some of the stained nucleus.
The delivery of DNA encoding for VEGF was able to induce angiogenesis in the CAM assay. Gross evaluation of the blood vessels around the implanted hydrogels showed that hydrogel with no DNA (Fig. 7A) did not result in enhanced blood vessel formation. However, all hydrogels containing DNA encoding VEGF resulted in enhanced blood vessel formation around the implant site (Fig. 7B, C, arrow heads). Evaluation of the blood vessels underneath/inside the hydrogel scaffold after perfusion showed enhanced micro-vessel formation for all hydrogels that contained polyplexes encoding for VEGF (Fig. 7E, F, arrows). Hydrogels with no DNA resulted in no micro-vessel formation (Fig. 7D).
Figure 7.
pVEGF loaded fibrin hydrogels resulted in enhanced angiogenesis in a CAM model. A fibrin hydrogel with polyplexes containing pVEGF was placed on top of the CAM for 3 days. pVEGF was transferred to cells and produced a high concentration of VEGF at the gel area, which resulted in hyperbranced neovessels (arrow). VEGF diffusing out the gel created a decreasing VEGF gradient around the gel and led to radial neovessels around the gel (arrowheads). (A–C) Gross pictures on the gel edge were recorded before the CAM was infused with FITC-dextran for fluorescent imaging (D–F) at the gel area. Induced neovessels were found both around the gel (B–C, arrow heads) and at the gel area (E–F, arrows) with pVEGF, which were not found in the negative control (No DNA). 50 μg pVEGF at N/P = 15 were used for both the 5 and 15 mg/mL gels. The dashed line outlines the edge of the hydrogel and “G” indicates the gel area. Scale bar: D–F: 200μm.
Discussion
Local delivery of DNA through a hydrogel scaffold can extend the application of gene therapy to tissue regeneration and cancer therapy [6]. However, the delivery of DNA/cationic polymer nanoparticles (polyplexes) using hydrogels has not been successful partially due to the aggregation and inactivation of polyplexes inside hydrogel scaffolds [28]. To overcome this challenge, we developed a strategy to introduce unaggregated and highly active DNA/PEI polyplexes to PEG hydrogel scaffolds [28]. Specifically, we have found that sucrose was able to retain the activity of polyplexes during lyophilization, which agrees with previous findings [30–32]. We also found that agarose could prevent polyplex aggregation during the gelation. We hypothesized this was due to an increase in the viscosity of the gel precursor solution and a caging effect of the agarose polymers to the polyplexes [28]. MMP degradable PEG hydrogels with concentrated DNA/PEI polyplexes loaded through CnE were able to deliver genes in vivo. In this study, we used the CnE to load DNA/PEI polyplexes into HA and fibrin hydrogels and tested their abilities to mediate gene transfer in vivo. HA and fibrin are extensively used as scaffolds for tissue engineering, tissue regeneration and wound healing [33, 34].
Due to the large numbers of carboxylic acid groups on the HA backbone, HA polymers exhibit a high density of negative charge that can interact with the DNA/PEI polyplexes through two potential mechanisms. HA can neutralize the positive charges at the polyplex surface and eliminate the repulsive force between the polyplexes, leading to severe aggregation. Alternatively, HA can interact with the PEI polymers and replace DNA from the DNA/PEI polyplexes similarly to what is found with heparin, causing decomplexation [35]. In dilute solution, HAs induced polyplex aggregation as showed by the fact that 1% HA increased the average diameter of the polyplexe from 50~100 nm to 1.0 μm (Fig. 1A). In a solution with high concentration of polyplexes, adding 1% HA completely precipitated the polyplexes (Fig. 1B). Fibrin proteins induced polyplex aggregation as well (Fig. 1C, D). Through CnE, polyplex aggregations were prevented in both HA and fibrin hydrogels (Fig. 2). The aggregation of nanoparticles occurs through three basic steps [36]. Nanoparticles are under random motion in solution that leads to frequent collisions. Part of the collisions results in aggregations when the collided particles fail to separate. We hypothesize that agarose can slow down the particle motion and reduce particle/particle interactions through a combination of increasing the viscosity of the hydrogel precursor solution and caging the polyplexes with the long agarose polymers, which minimizes nanoparticle aggregation. Although no aggregation was observed (Fig. 2), micro-scale domains displaying higher concentration of polyplexes were found inside the hydrogel scaffolds. Inside the microdomains unaggregated polyplexes were found. We hypothesize that these microdomains are a result of incomplete dissolution of the lyophilized powder during the formation of hydrogels, which keep the polyplexes trapped inside the microspheres [28].
Polyplexes were not free to diffuse inside the hydrogels as shown by the fact that no DNA was released from the HA or fibrin hydrogels in PBS (Fig. 3). Thus hydrogel degradation was required for the gene transfer from hydrogels to cells, which allowed highly localized gene delivery in vivo. We speculate that DNA/PEI polyplexes could be released from the scaffold during the hydrogel degradation uptake by cells surrounding the hydrogel or they could be uptake by cells infiltrating the scaffold. From our results using a reporter plasmid encoding for β-galactosidase (Fig. 4 and 6), for HA hydrogels it appears that gene transfer only occurs in the area of the hydyrogel, indicating that the polyplexes are transfecting either infiltrating or very near by cells and not diffusion far from the site of implantation. In fibrin hydrogels transfection is observed at regions further from the implantation site. However, most of the transfection was still observed in the area of the hydrogel. In contrast, for MMP degradable PEG hydrogels gene transfer occur mostly at sites adjacent to the implantation site [28], indicating that for HA and fibrin hydrogels the scaffold retains the polyplexes at the implantation site and that cells can more easily infiltrate these scaffolds. We speculate that the trafficking of DNA/PEI polyplexes once internalized by cells occurs through similar pathways as have been observed for cells cultured in tissue culture plates [37].
Polyplexes encapsulated inside the hydrogels through the CnE process were found highly active in vivo as shown by the β-galactosidase expression (Fig. 4, 6). To ensure that the expressed transgene was present at sufficient concentration to induce an angiogenic response in and around the hydrogel, plasmid DNA encoding for VEGF was entrapped within the hydrogel using the CnE. An angiogenic response was observed in all CAMs that contained a hydrogel with pVEGF/PEI polyplexes entrapped through CnE. The angiogenic response extended out in a radial orientation toward the CAM (Fig. 5 and 7, arrow heads), suggesting that the expressed VEGF diffused out of the hydrogel area and created a VEGF gradient with the highest concentration near the hydrogel. In contrast, the vessels observed at the hydrogel area are highly branched neovessels without preferred orientations (Fig. 5 and 7, arrows), suggesting that the VEGF concentration at the gel area was high and relatively constant. We previously demonstrated that the encapsulation of polyplexes through direct encapsulation and in the absence of CnE led to extensively aggregated polyplexes that were not active in vitro or in vivo [28].
Conclusion
In summary, polyplexes are known to aggregate in HA or fibrin solutions. The CnE process prevents polyplex aggregation during their incorporation into HA or fibrin hydrogels. HA or fibrin hydrogels with concentrated polyplexes loaded through CnE were able to deliver genes in a CAM model.
Acknowledgments
The authors would like to thank the National Institute of Health (R21EB007730) and the National Science Foundation (CAREER grant 0747539) for funding the research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kay MA, Glorioso JC, Naldini L. Viral vectors for gene therapy: the art of turning infectious agents into vehicles of therapeutics. Nat Med. 2001;7:33–40. doi: 10.1038/83324. [DOI] [PubMed] [Google Scholar]
- 2.Leong KW, Mao HQ, Truong-Le VL, Roy K, Walsh SM, August JT. DNA-polycation nanospheres as non-viral gene delivery vehicles. J Control Release. 1998;53:183–193. doi: 10.1016/s0168-3659(97)00252-6. [DOI] [PubMed] [Google Scholar]
- 3.Mulligan RC. The basic science of gene therapy. Science. 1993;260:926–932. doi: 10.1126/science.8493530. [DOI] [PubMed] [Google Scholar]
- 4.Pouton CW, Seymour LW. Key issues in non-viral gene delivery. Adv Drug Deliv Rev. 2001;46:187–203. doi: 10.1016/s0169-409x(00)00133-2. [DOI] [PubMed] [Google Scholar]
- 5.Bonadio J. Tissue engineering via local gene delivery: update and future prospects for enhancing the technology. Adv Drug Deliv Rev. 2000;44:185–194. doi: 10.1016/s0169-409x(00)00094-6. [DOI] [PubMed] [Google Scholar]
- 6.De Laporte L, Shea LD. Matrices and scaffolds for DNA delivery in tissue engineering. Adv Drug Deliv Rev. 2007;59:292–307. doi: 10.1016/j.addr.2007.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheema SK, Chen E, Shea LD, Mathur AB. Regulation and guidance of cell behavior for tissue regeneration via the siRNA mechanism. Wound Repair Regen. 2007;15:286–295. doi: 10.1111/j.1524-475X.2007.00228.x. [DOI] [PubMed] [Google Scholar]
- 8.Krebs MD, Jeon O, Alsberg E. Localized and Sustained Delivery of Silencing RNA from Macroscopic Biopolymer Hydrogels. J Am Chem Soc. 2009;131:9204–7. doi: 10.1021/ja9037615. [DOI] [PubMed] [Google Scholar]
- 9.Bonadio J, Smiley E, Patil P, Goldstein S. Localized, direct plasmid gene delivery in vivo: prolonged therapy results in reproducible tissue regeneration. Nat Med. 1999;5:753–759. doi: 10.1038/10473. [DOI] [PubMed] [Google Scholar]
- 10.Tyrone JW, Mogford JE, Chandler LA, Ma C, Xia Y, Pierce GF, Mustoe TA. Collagen-embedded platelet-derived growth factor DNA plasmid promotes wound healing in a dermal ulcer model. J Surg Res. 2000;93:230–236. doi: 10.1006/jsre.2000.5912. [DOI] [PubMed] [Google Scholar]
- 11.Ochiya T, Takahama Y, Nagahara S, Sumita Y, Hisada A, Itoh H, Nagai Y, Terada M. New delivery system for plasmid DNA in vivo using atelocollagen as a carrier material: the Minipellet. Nat Med. 1999;5:707–710. doi: 10.1038/9560. [DOI] [PubMed] [Google Scholar]
- 12.Chun KW, Lee JB, Kim SH, Park TG. Controlled release of plasmid DNA from photo-cross-linked pluronic hydrogels. Biomaterials. 2005;26:3319–3326. doi: 10.1016/j.biomaterials.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 13.Quick DJ, Anseth KS. DNA delivery from photocrosslinked PEG hydrogels: encapsulation efficiency, release profiles, and DNA quality. J Control Release. 2004;96:341–351. doi: 10.1016/j.jconrel.2004.01.021. [DOI] [PubMed] [Google Scholar]
- 14.Kong HJ, Kim ES, Huang YC, Mooney DJ. Design of biodegradable hydrogel for the local and sustained delivery of angiogenic plasmid DNA. Pharm Res. 2008;25:1230–1238. doi: 10.1007/s11095-007-9526-7. [DOI] [PubMed] [Google Scholar]
- 15.Kasper FK, Jerkins E, Tanahashi K, Barry MA, Tabata Y, Mikos AG. Characterization of DNA release from composites of oligo(poly(ethylene glycol) fumarate) and cationized gelatin microspheres in vitro. J Biomed Mater Res A. 2006;78A:823–835. doi: 10.1002/jbm.a.30736. [DOI] [PubMed] [Google Scholar]
- 16.Megeed Z, Haider M, Li D, O’Malley BW, Jr, Cappello J, Ghandehari H. In vitro and in vivo evaluation of recombinant silk-elastinlike hydrogels for cancer gene therapy. J Control Release. 2004;94:433–445. doi: 10.1016/j.jconrel.2003.10.027. [DOI] [PubMed] [Google Scholar]
- 17.Jang JH, Rives CB, Shea LD. Plasmid delivery in vivo from porous tissue-engineering scaffolds: transgene expression and cellular transfection. Mol Ther. 2005;12:475–483. doi: 10.1016/j.ymthe.2005.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scherer F, Schillinger U, Putz U, Stemberger A, Plank C. Nonviral vector loaded collagen sponges for sustained gene delivery in vitro and in vivo. J Gene Med. 2002;4:634–643. doi: 10.1002/jgm.298. [DOI] [PubMed] [Google Scholar]
- 19.Cohen-Sacks H, Elazar V, Gao J, Golomb A, Adwan H, Korchov N, Levy RJ, Berger MR, Golomb G. Delivery and expression of pDNA embedded in collagen matrices. J Control Release. 2004;95:309–320. doi: 10.1016/j.jconrel.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Holladay C, Keeney M, Greiser U, Murphy M, O’Brien T, Pandit A. A matrix reservoir for improved control of non-viral gene delivery. J Control Release. 2009;136:220–225. doi: 10.1016/j.jconrel.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 21.Lei P, Padmashali RM, Andreadis ST. Cell-controlled and spatially arrayed gene delivery from fibrin hydrogels. Biomaterials. 2009;30:3790–3799. doi: 10.1016/j.biomaterials.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wieland JA, Houchin-Ray TL, Shea LD. Non-viral vector delivery from PEG-hyaluronic acid hydrogels. J Control Release. 2007;120:233–241. doi: 10.1016/j.jconrel.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saul JM, Linnes MP, Ratner BD, Giachelli CM, Pun SH. Delivery of non-viral gene carriers from sphere-templated fibrin scaffolds for sustained transgene expression. Biomaterials. 2007;28:4705–4716. doi: 10.1016/j.biomaterials.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 24.Trentin D, Hall H, Wechsler S, Hubbell JA. Peptide-matrix-mediated gene transfer of an oxygen-insensitive hypoxia-inducible factor-1 alpha variant for local induction of angiogenesis. Proc Natl Acad Sci USA. 2006;103:2506–2511. doi: 10.1073/pnas.0505964102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trentin D, Hubbell J, Hall H. Non-viral gene delivery for local and controlled DNA release. J Control Release. 2005;102:263–275. doi: 10.1016/j.jconrel.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 26.Kulkarni M, Breen A, Greiser U, O’Brien T, Pandit A. Fibrin-lipoplex system for controlled topical delivery of multiple genes. Biomacromolecules. 2009;10:1650–1654. doi: 10.1021/bm900248n. [DOI] [PubMed] [Google Scholar]
- 27.Lei YG, Segura T. DNA delivery from matrixmetalloproteinase degradable poly(ethylene glycol) hydrogels to mouse cloned mesenchymal stem cells. Biomaterials. 2009;30:254–265. doi: 10.1016/j.biomaterials.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lei Y, Huang S, Sharif-Kashani P, Chen Y, Kavehpour P, Segura T. Incorporation of active DNA/cationic polymer polyplexes into hydrogel scaffolds. Biomaterials. 31:9106–9116. doi: 10.1016/j.biomaterials.2010.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jang JH, Rives CB, Shea LD. Plasmid delivery in vivo from porous tissue-engineering scaffolds: Transgene expression and cellular Transfection. Mol Ther. 2005;12:475–483. doi: 10.1016/j.ymthe.2005.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anchordoquy TJ, Carpenter JF, Kroll DJ. Maintenance of transfection rates and physical characterization of lipid/DNA complexes after freeze-drying and rehydration. Arch Biochem Biophys. 1997;348:199–206. doi: 10.1006/abbi.1997.0385. [DOI] [PubMed] [Google Scholar]
- 31.Brus C, Kleemann E, Aigner A, Czubayko F, Kissel T. Stabilization of oligonucleotide-polyethylenimine complexes by freeze-drying: physicochemical and biological characterization. J Control Release. 2004;95:119–131. doi: 10.1016/j.jconrel.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 32.Allison SD, Anchordoquy TJ. Mechanisms of protection of cationic lipid-DNA complexes during lyophilization. J Pharm Sci. 2000;89:682–691. doi: 10.1002/(SICI)1520-6017(200005)89:5<682::AID-JPS14>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 33.Ahmed TA, Dare EV, Hincke M. Fibrin: A Versatile Scaffold for Tissue Engineering Applications. Tissue Eng Part B Rev. 2008;14:199–215. doi: 10.1089/ten.teb.2007.0435. [DOI] [PubMed] [Google Scholar]
- 34.des Rieux A, Shikanov A, Shea LD. Fibrin hydrogels for non-viral vector delivery in vitro. J Control Release. 2009;136:148–154. doi: 10.1016/j.jconrel.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moret I, Esteban Peris J, Guillem VM, Benet M, Revert F, Dasi F, Crespo A, Alino SF. Stability of PEI-DNA and DOTAP-DNA complexes: effect of alkaline pH, heparin and serum. J Control Release. 2001;76:169–181. doi: 10.1016/s0168-3659(01)00415-1. [DOI] [PubMed] [Google Scholar]
- 36.Friedlander SK. Smoke, dust, and haze: fundamentals of aerosol dynamics. Oxford University Press; New York: 2000. [Google Scholar]
- 37.Lechardeur D, Verkman AS, Lukacs GL. Intracellular routing of plasmid DNA during non-viral gene transfer. Adv Drug Deliv Rev. 2005;57:755–767. doi: 10.1016/j.addr.2004.12.008. [DOI] [PubMed] [Google Scholar]