Abstract
Segmentation of the left ventricle is important in the assessment of cardiac functional parameters. Manual segmentation of cardiac cine MR images for acquiring these parameters is time-consuming. Accuracy and automation are the two important criteria in improving cardiac image segmentation methods. In this paper, we present a comprehensive approach to segment the left ventricle from short axis cine cardiac MR images automatically. Our method incorporates a number of image processing and analysis techniques including thresholding, edge detection, mathematical morphology, and image filtering to build an efficient process flow. This process flow makes use of various features in cardiac MR images to achieve high accurate segmentation results. Our method was tested on 45 clinical short axis cine cardiac images and the results are compared with manual delineated ground truth (average perpendicular distance of contours near 2 mm and mean myocardium mass overlapping over 90%). This approach provides cardiac radiologists a practical method for an accurate segmentation of the left ventricle.
Key words: Image segmentation, cardiac imaging, image analysis, left ventricle
Background
Cardiac magnetic resonance images (CMR) have become significantly important in the last decade because of the rapid improvement of hardware techniques, computing power, coil design, and sequence development. Given the high quality of the CMR images, this has surpassed echocardiography which is now no longer the gold standard for cardiac parameters. It is believed that in the future, when the cost and time required for image acquisition become comparable to that of echocardiography, CMR will be the primary diagnostic modality in cardiovascular medicine because of its unique versatility and noninvasive nature.1
Quantitative analysis of clinical parameters such as ejection fraction, left ventricle myocardium mass and stroke volume can be obtained by delineation of the left ventricle from cine MR images. These parameters play important roles in cardiac functional assessment, heart failure, cardiomyopathies, and congenital heart disease.
The main challenges in left ventricle (LV) segmentation from cine MR images lie in the following aspects: the intensity distribution of cardiac images varies significantly, which makes automatic classification or clustering methods ineffective, and the shape of the ventricles varies from person to person; hence, edge information is inconsistent with the epicardial surface of the left ventricular wall appearing indistinct. Due to cardiac dynamic nature, motion artifacts are inevitable in cine images. Besides, at each 3D volume of time snap, slices are taken from different breath holds, which caused vicinity slices to not align correctly; this phenomenon is usually called slice shift. All these problems inherent in cardiac MR imaging are difficult to eliminate and have to be resolved by segmentation algorithms.
In recent years, quite a number of methods have been proposed for (semi-) automated LV segmentation. To deal with the aforementioned problems, different strategies have been used in these methods, including image-driven methods,2,3 probabilistic atlas,4 dynamic programming,5,6 fuzzy clustering,7 deformable model,8 active appearance model,9 level set and its variations,10–12 and graph cut approach.13,14 A comprehensive review of techniques commonly used in cardiac image segmentation can be found in Frangi et al..15 However, fully automatic accurate segmentation of the left and right ventricles is still attracting research interest as CMR is becoming more widely used and manual segmentation is still a time-consuming task for cardiac radiologists. The vast amount of images acquired from one cardiac study exhausted radiologist capacity and resources, besides inter- and intra-operator differences.
Our study found that characteristics of cine image from different scans can be quite variable, and hence, it is not easy to achieve high accuracy results by applying a single technique automatically. Therefore, we proposed a comprehensive approach by combining several techniques: radial region growing, image edge detection, and mathematical morphology. This approach demonstrates robustness in performing the endocardial and epicardial contours segmentation task from cine MR images with achievement of a highly accurate method of automated segmentation.
The paper is organized as follows: “Material and Methods” describes the details of the segmentation methods. “Results and Discussions” presents and analyzes the results, followed by “Conclusion and Discussion” with a conclusion and discussion on issues and future work.
Material and Methods
Image Data
The datasets used in this paper are cine steady-state free precession (SSFP) MR short axis (SAX) images published by the MICCAI 2009 Clinical Image Segmentation Grand Challenge Workshop organizer on the Internet, available to the public at http://sourceforge.net/projects/cardiac-mr/, thanks to the efforts by Dr. Rauda Perry from Sunnybrook Health Sciences Centre. Forty-five cardiac cine MR datasets are made available for the event in three batches: 15 datasets for training, 15 for qualification testing, and 15 for the online contest. Each batch contains four heart failures with ischemia (HF-I) cases, four heart failure without ischemia (HF-NI) cases, four hypertrophy (HYP) cases, and three normal cases. All the data are in anonymized DICOM format following HIPAA regulations. Permission for public use has been granted by the ethics review board of the same center.
All image data were randomly selected from the clinical database from Sunnybrook Health Sciences Centre. All the images were obtained during 10- to 15-s breath holds with a temporal resolution of 20 cardiac phases over the heart cycle. Six to 12 SAX images were obtained from the atrioventricular ring to the apex (thickness = 8–10 mm, FOV = 320 × 320 mm, matrix = 256 × 256).
The goals of the contest were to compare LV segmentation methods by providing an evaluation system and a database of images and expert contours. Comparing segmentation results across research studies can be difficult due to unspecified differences in the method or implementation of evaluation matrices. This contest provides an open-source code for contour evaluation. Furthermore, the database provides a set of images such that confounding segmentation differences due to image quality or pathology can be eliminated.
Approach Overview
CMR images include the lower chest and upper abdomen. The structures surrounding the heart include the lungs, chest wall/musculature, ribs, liver, gall bladder, spleen, bowel, and peritoneal/omental fat. Different tissues present different signal intensities regions in quite flexible locations, which make automatic segmentation algorithms difficult to recognize patterns. Meanwhile, the heartbeat motion combined with heart position changes caused by patients’ breath holding moments and physical position changes during scan instigate misalignment of structures in their shape. Another issue that needs to be noted is to shorten the scan time; the slice thickness usually is big, which makes the partial volume effect obvious. To overcome these problems, we carefully designed a workflow using a series of image analysis techniques to identify and extract image features step by step and finally reveal the whole left ventricle from the cine CMR data.
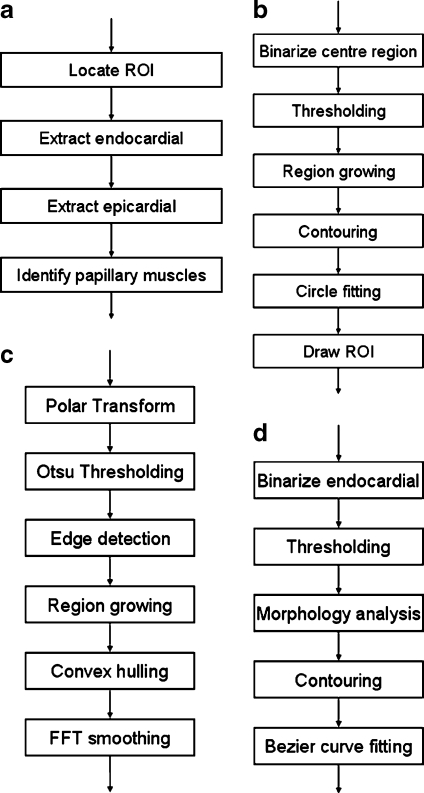
The complete procedure is shown in Figure 1. The algorithm starts from locating the region of interest (ROI) from the images. ROI is defined as a cylinder encapsulating the LV structures. To find the ROI, we look for the center point of the LV blood pool and then define a ROI using this center point. Subsequently, the image in ROI is transformed to a rectangular image by polar transformation. Using the transformed image, we apply canny edge detection and one-way region growing methods iteratively to compute the endocardial boundary and epicardial boundary. After the contours are extracted, we apply reverse polar transformation on the result back to the original image space. The endocardial contours and epicardial contours are transformed back to the enclosed curves. Next, we apply thresholding onto the image surrounded by endocardial to find candidate regions of papillary muscles, then use mathematical morphology to pick up the two biggest regions as the papillary muscles. More details of our segmentation method are described in the following sections.
Fig. 1.
Workflow of the whole segmentation process: a overall flow; b steps in locating LV seed point and ROI; c steps in segment endocardial and epicardial; d steps in indentifying papillary muscles.
Several image segmentation techniques and mathematical transformations are repeatedly used in our algorithm. They are: polar transformation, thresholding, edge detection, hull convex, fast Fourier transformation (FFT) smoothing, radial region growing, image morphology, etc.
LV Center and ROI Localization
In LV segmentation from SAX CMR images scanning, we are only interested in the LV structure. And because the whole heart is to be scanned generally in clinical practice, other structures are presented in images. We defined an ROI first to exclude the areas we are not interested in.
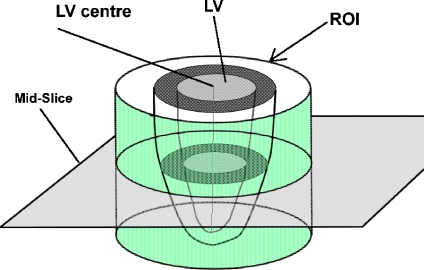
Several ROI allocation approaches have been used in previous studies to narrow down the particular area in the image. For example, in Lu et al.,2 a square area is plotted in the center of the image as ROI, and in Rezaee et al.,7 a Hough transform is applied to locate the ROI center. We used a slightly different approach here. We defined the ROI in a circle shape because the shape of the LV cross-section is similar to a circle shape; therefore, a cylinder-shaped ROI will be more effective for excluding regions we are not interested in and hence will improve the result of image-based algorithms. To define the ROI, we seek for a seed point in LV blood pool first and extract the LV blood pool by thresholding and region growing. Then, the mass center of the LV blood pool is calculated to be used as the circle center of our ROI. The illustration of ROI in SAX images is shown in Figure 2. More detailed explanation is described in the following.
Fig. 2.
Illustration of ROI and LV center point in SAX images.
Obtain Seed Point in LV Blood Pool
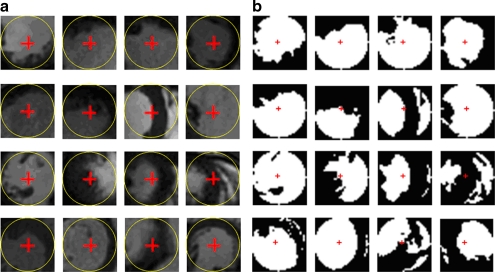
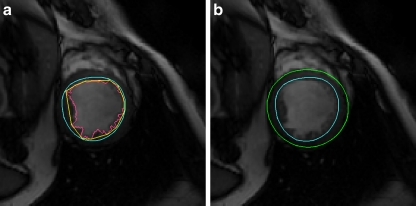
While conducting SAX image scans, radiologists usually try to place the LV in the center of the image because it is the most interesting structure in study. Therefore, the center of LV are usually near the center of the image, but not necessary exactly at the center. We started from seeking the seed point of LV blood pool on the middle slice (mid-slice) of the 3D volume because this slice typically is neither near the apex nor left ventricle outflow track. That means the cross-section of the LV area in the slice is sufficiently big and close to the circle shape. Blood appears brighter than the myocardium in SSFP SAX images. Based on these features, we can roughly estimate the center of the LV and hence define the ROI which closely fit the LV in the images. Figure 3a shows the center circle images extract from the test images. We can see that the center circle areas are mostly occupied by LV. These areas consist of two types of pixels, blood and myocardium. The popularly used Otsu thresholding was first proposed by Otsu16 for automatic selection of the optimal threshold value that minimizes weighted within-class variance of bimodal images. Therefore, we used Otsu thresholding to classify the images in the center circle into two classes, blood and myocardium. The results are shown in Figure 3b. Next, we examine the blood region in the result if the result contains only one foreground region; that region belongs to the LV blood pool region. Otherwise, we take the biggest sized region as the LV blood pool region. A pixel inside this area can be used as seed point.
Fig. 3.
Some examples of image center circle area and their thresholding results. We can find the image center either inside or very close to the LV blood pool. Typically, the LV is the biggest or most circle-like region in the center circle area. a Samples of the center region of original images. b Circle center region in a after binarizing using two-level Otsu thresholding.
Obtain LV Blood Pool and Its Center
Using the thresholding value and the seed point computed from the previous step, we apply a region growing operation. The resulting foreground region is the LV blood pool. Although the cross-section of LV is a circle-like convex shape anatomically, in most of the SAX image slices, the LV region we derived here may not be in a convex shape because of the existence of papillary muscle and trabeculae muscles. We should make up the concaves before we proceed to the next step. To achieve this, we calculate the convex hull based on the region growing result.
This convex hull is then used as the initial location of the LV to approximate the ROI circle. Performing a circle fitting the convex hull with linear least square method,17 we obtain the center of the blood pool of the LV. Using this center point as origin, and a diameter that is slightly bigger than the average diastolic LV (we used 11 cm after observing all CMR data we have had), a compact circular ROI is generated for subsequent segmentation processes.
Left Ventricle Segmentation
A cardiac cine image scan acquires 20–25 phases in a heartbeat cycle. Each phase constituting a stack of images containing 10–20 slices captured the complete 3D shape of the left ventricle at one moment. A stack of images in one phase is also called a volume, similar to other 3D image analysis studies. Our algorithm processes one volume at a time and can be repeated for all phases without user interference. In clinical practice, the end-diastole (ED) phase and the end-systole (ES) phase are the most interesting phases because they are directly used in cardiac functional assessment, for instance stroke volume and ejection fraction calculation. In the work of this paper, ES and ED phase numbers are provided by the data supplier. However, our algorithm is fully automated; therefore, it can simply segment all phases if necessary. ES and ED phases can be identified by computing the LV volumes from the segmentation results. Completely segmenting all the phases is not practical by manual segmentation but is easy for automatic segmentation. The outcome of this advantage may also be used for exploring more precise quantification of cardiac functions in the future.
Endocardial Contouring
Given a circular ROI generated in the aforementioned steps covering the LV compactly, tissues within the region are more predictable. The LV blood pool and myocardium occupied a major volume of the ROI region and other structures such as lung. Chest spaces are minimized; hence, the intensity distribution is relatively easy to classify. We observed that image intensities in the ROI include various tissues, i.e., blood, muscle, fat, lung, and chest space. Pixel intensities can be classified into three classes, namely, blood/fat, muscle, and lung/space. An extended Otsu method of multilevel thresholding18 is used to classify the intensity of image in ROI to obtain threshold values of the blood, muscle, and lung/chest space.
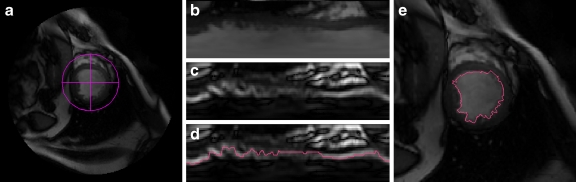
Using the computed blood threshold, a radial region growing is applied to extract the blood pool of the left ventricle. Radial region growing is similar to convention region growing; however, it starts from a center point of a circle and grows by increasing the radius of the circle pixel by pixel. To ease calculation and illustration, a polar transformation is applied on the region of interest to form a flat image (Fig. 4a, b) beforehand. Transforming the image into a polar coordinate system is an effective method in LV segmentation, and it has been used by a few other works.2,5 After transformation, radius region growing has become one-way directional region growing, which is easy to implement. However, our study discovered that due to intensity inhomogeneity caused by blood dynamic and low signal noise rate, there are possible leakages which appeared in the region growing. To deal with this problem, we use an additional measure to overcome this issue. We apply canny edge detector19 onto the transformed image to find the boundary of LV blood pool in the transform image (Fig. 4c, d). Combining the radial region growing results with the edge detection results, we obtained an edge contour of the endocardial in the transformed image. The contours are transformed back to the original coordinates, which become the candidates of the endocardial contours (Fig. 4e). Given the uneven nature of the inner surface of the ventricle due to papillary muscles, chordae tendineae and trabeculae, we compute the convex hull to warp the region growing result so that it includes the papillary muscle region (Fig. 5a), and then a low-pass filter of FFT (six coefficients are used) is applied on the convex hull to remove the irregular uneven ventricle surface disturbing and to form a smooth endocardial contour (Fig. 5b). The whole process flow is shown in Figure 1c.
Fig. 4.
Segmentation of endocardial contour: a Region of interest. b ROI image transformed to polar coordinates. c Canny filtering result of b. d Edge of LV detected by combined region growing in polar transformed image and edge detection. e LV transformed back to Cartesian coordinates system.
Fig. 5.
Segmentation of endocardial and epicardial: a Endocardial is generated by Fourier transformation from the convex hull of the blood pool edges. b Epicardial contour produced by radial region growing from endocardial.
Epicardial Contouring
The process of extracting epicardial is almost the same as extracting endocardial. However, radial region growing will be started from the endocardial contour instead of from the origin. The region growing will use the threshold value of muscle class instead of blood class. Similarly, the radial region growing detects both the intensity threshold and the canny filter results to determine the stop criteria. When the radial region growing is stopped, the initial edge of epicardial computed thresholding and canny edging is jaggy, and again, FFT low-pass filter is applied to produce a good smooth contour that fit the surface of epicardial (Fig. 5b). The process flow is similar to Figure 1c; however, because some operations like thresholding and edge detection have been carried out in the previous step, results can be used directly without repeating the operation.
Papillary Muscle Segmentation
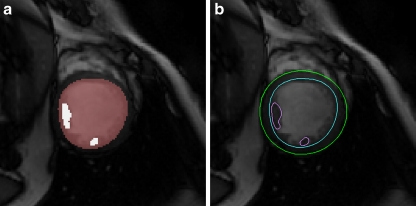
Papillary muscles occupy a relatively significant space in ventricles; therefore, these muscles, particularly the two biggest papillary muscles (indicated as P1 and P2), also need to be delineated and considered when calculating the ventricle eject factors. To outline the papillary muscles, we used the blood’s low threshold derived in “Endocardial Contouring” again onto the endocardial region. Generally, several dark regions similar to the papillary muscles can be identified as candidate regions. We apply a mathematical morphology opening operation, with 3 × 3 square structure element, on these regions to disconnect weak connections between these candidate regions. The two biggest regions are then selected automatically as papillary muscles P1 and P2 (Fig. 6a). Contours of the two regions are extracted by a marching square, a 2D version of the marching cube.20 The contours are finalized by smoothing using Bezier curve fitting (Fig. 6b).
Fig. 6.
Segmentation of papillary muscles: a Apply thresholding in the region of endocardial. b Extract contours of papillary muscles.
Results and Discussions
Our method has been tested on 45 datasets published by the MICCAI Clinical Image Segmentation Grand Challenge Workshop organizer (described in “Image Data”). We used the same evaluation methods as the ones used in the conference workshop to analyze our results. The publicity of the data and evaluation methods allow people to test and compare their methodology easier and objectively.
Evaluation Methods
Evaluation of left ventricle segmentation is conducted using the framework used in the segmentation competition. Both automatic and visual methods are used in benchmarking the segmentation results. Three automatic evaluation measures are used in this framework; these are contour good percentage, average perpendicular distances, overlapping dice metric (Eq. 1):
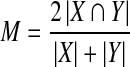 |
1 |
where X is the contour area segmented and Y is the ground truth manually segmented. X∩Y is the intersection of both areas and |A| stands for the number of pixels in area A.
The ground truth of LV is manually delineated by the clinical experts from the data from the same institute. Contours generated by automatic algorithms is considered as good if the maximum distance from ground truth is <5 mm. Contour good percentage is calculated by the number of good contours over the number of all contours of ground truth. Average perpendicular distance evaluation calculates the distance from the automatically segmented contour to the corresponding one manually drawn by an expert, averaged over all contour points. Overlapping dice metric calculates the contour areas overlapping proportion between the automatic segmentation results and the ground truth. Besides automatic evaluation, visual evaluation has also been conducted by experienced cardiologists based on their general impression on the resulting contours.
Evaluation Results and Discussions
The statistics of segmentation results compared to the ground truth contours are listed in Table 1. The average perpendicular distances of our resulting contours to ground truth is about 2 mm for both endocardial and epicardial contours. This is comparable to other methods developed recently2,12, although they are not directly comparable because of the difference in data and ground truth. Considering inter-operator and intra-operator segmentation errors, the automatic segmentation results should be acceptable clinically.
Table 1.
Contour Accuracy in Terms of Average Perpendicular Distance, Dice Metric, and Good Percentage (Mean Value ± standard deviation)
| Group | Studies | Average perpendicular distance | Overlapping dice metric | Good percent | |||
|---|---|---|---|---|---|---|---|
| Endocardial | Epicardial | Endocardial | Epicardial | Endocardial | Epicardial | ||
| HF-I | 12 | 1.84 ± 0.32 | 2.10 ± 0.57 | 0.93 ± 0.02 | 0.94 ± 0.02 | 86.5 ± 18.9 | 87.8 ± 21.2 |
| HF-NI | 12 | 2.17 ± 0.40 | 2.08 ± 0.36 | 0.91 ± 0.02 | 0.94 ± 0.01 | 83.4 ± 14.1 | 84.5 ± 14.9 |
| HYP | 12 | 2.29 ± 0.52 | 2.34 ± 0.34 | 0.87 ± 0.03 | 0.92 ± 0.02 | 73.0 ± 17.6 | 82.6 ± 13.7 |
| Normal | 9 | 2.40 ± 0.40 | 2.39 ± 0.41 | 0.87 ± 0.04 | 0.92 ± 0.01 | 72.0 ± 24.0 | 80.0 ± 18.0 |
| All | 45 | 2.16 ± 0.46 | 2.22 ± 0.43 | 0.89 ± 0.04 | 0.93 ± 0.02 | 79.2 ± 19.0 | 83.9 ± 16.8 |
| Overall | 45 | 2.19 ± 0.44 | 0.91 ± 0.03 | 81.5 ± 18.0 | |||
The test cases are categorized by the subjects’ syndromes: heart failure with ischemic (HF-I), heart failure without ischemic (HF-NI), hypotrophy (HYP), and normal subjects
Table 1 presented the evaluation results of both endocardial and epicardial delineated by automatic method compared with ground truth categorized by the pathology of subjects. The results of heart failure groups appear very slightly better, but are not significant. That means our approach is not sensitive to pathology. The average distance to ground truth is about 2 mm plus, which is close to the manual segmentation error.
Finally, the resulting contours of the validation batch (15 studies) have also been visually evaluated by experienced cardiologists at Sunnybrook Health Sciences Centre (Kim Connelly and Gideon Paul). Results are rated in a four-point scale (1: Excellent, little correction required; 2: Good, over 50% of contours are acceptable; 3: Poor, <50% of the contours are acceptable; and 4: Not usable). The visual evaluation results are shown in Table 2. In the 15 results being evaluated, 11 of them (73%) are rated 1 and four of them (27%) are rated 2. This evaluation result, on the other hand, further confirms that our segmentation approach generated acceptable results to clinicians.
Table 2.
Visual Evaluation Results for 15 Segmentation Outputs
| Group | Studies | Number of cases rated 1 | Number of cases rated 2 | Number of cases rated 3 | Number of cases rated 4 | Average rating |
|---|---|---|---|---|---|---|
| HF-I | 4 | 4 | 0 | 0 | 0 | 1 |
| HF-NI | 4 | 3 | 1 | 0 | 0 | 1.25 |
| HYP | 4 | 2 | 2 | 0 | 0 | 1.5 |
| Normal | 3 | 2 | 1 | 0 | 0 | 1.33 |
| Total | 15 | 11 | 4 | 0 | 0 | 1.27 |
(Results are rated with a four-point scale (1: Excellent, little correction required; 2: Good, over 50% of contours are acceptable; 3: Poor, <50% of the contours are acceptable; and 4: Not usable). For group HF-I, all the results are visually excellent. Other groups have some small problems, but all are in good category, which means that most of the contours segmented are acceptable to experienced cardiologists; only small corrections are needed
During the MICCAI 2009 clinical image segmentation grant challenge on-site testing, our segmentation result on the on-site datasets was ranked number 3. This shows that our algorithm is competitive with frontal competitors. Nevertheless, our result on the qualification testing datasets was ranked number 1. This positive outcome gives us confidence that our approach has the potential to achieve better results.
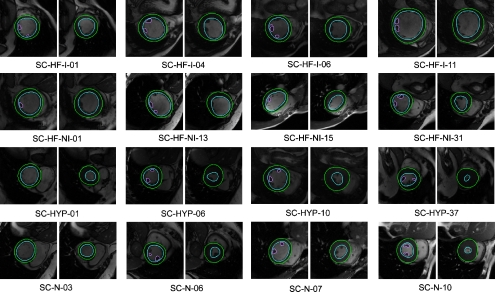
Figure 7 shows some segmentation outputs from our method. As we mentioned above, data of four categories of studies are used in this experiment. The images are arranged in four rows where each row presents one pathology group studies. The first row presents heart failure with ischemia studies, the second row shows heart failure with no ischemia cases, the third row contains hypertrophy cases, and the last row presents normal cases. Four datasets are presented in each group. For each dataset, two resulting images are shown. The left one is an image of the ED phase and the right one of the ES phase. Papillary muscles are segmented on the ED images whenever possible. The name below the image data is from the data source.
Fig. 7.
Some segmentation outputs by our method; each row presents one pathology group study. The first row presents heart failure with ischemia studies, the second row shows the heart failure with no ischemia cases, the third row contains hypertrophy cases, and the last row presents normal cases. Four datasets are present in each group. For each dataset, two resulting images are shown. The left one is the image of the ED phase and the right one of the ES phase. The names below the image data are from the data source. Papillary muscles are segmented on ED images whenever possible.
From the results, we found that the performance of our method is very promising, comparable to many other more complex methods, although the techniques used in our algorithm are relatively simple classic techniques in image analysis. We believe that classic methods, like Otsu thresholding, have very good performance in classifying tissues in CMR images given ROIs are well defined with our LV center allocating method. Simultaneously, skillful integration of radial region growing, canny edge detection, and FFT filtering are well suited to the target object shape, that is, a circle similar region of the LV. Nevertheless, they performed better than some sophisticated algorithms. Sophisticated methods are typically not easy to control in the context of CMR images because of the complex chest structures surrounding the LV region and blood flow disturbance; these artifacts often misguide the mathematical models to produce undesirable output. We trust that using appropriate methods, regardless of whether they are simple or sophisticated, suitable to the image nature and the problem target is the key factor in addressing image analysis challenges.
Conclusion and Discussion
We have developed a comprehensive segmentation approach to segment the left ventricle from cine MR images by using a processing pipeline of image transformation, thresholding, region growing, edge detection, and image filtering technologies. Endocardial and epicardial contours as well as two papillary muscles are delineated automatically. Testing results on the data randomly selected from clinical images demonstrated that our comprehensive approach of the left ventricle segmentation is a feasible and fast method for cardiac cine MR image segmentation. Its accuracy is comparable to other methods published recently.
Automatic segmentation of LV from cine CMR images is a challenging task. Given the fact of the rapid increase of CMR scans and overwhelming time required by manual segmentation, automatic methods are attractive solutions for clinicians. Although many different approaches have been attempted to resolve this problem, we see a bottleneck in increasing the accuracy of automatic algorithms. Given the dynamic nature of CMR images, we believe that image-based techniques are still attractive to be integrated into solutions aiming for the objective of CMR image automatic segmentation. Our method has achieved competitive results so far. We will still be working on to improve the accuracy.
Although we aim for a fully automatic segmentation procedure, in some extreme cases, manual enhancement to the results might still be required because unforeseeable vast variations in source images quality come from different scanners, operators, and scan conditions. Our design allows users to have the final touch to the automatic computed contours in order to output high-quality segmentation results. We believe that such a system would increase users’ confidence to accept the segmentation results. Users’ operation and time of finalizing modification to the contours can be minimized by using appropriate contour deforming technology. We have also implemented deformable model method21,22 to allow propagation of modification from one phase to other phases. Our test shows that this process is efficient and practical in refining segmentation outputs.
Remarks from visual evaluation reviewers underline that the most problematic issue is in the left ventricle outflow tract area, which is a generic problem to most LV segmentation methods. Resolving this problem would be able to significantly improve the accuracy, and we will spend much effort on this issue in our future work.
Several areas still can be improved and enhanced to increase the accuracy of segmentation. First, automatic allocation of the center and radius of circular ROI can be optimized to achieve better result in fully automatic segmentation. Meanwhile, end-diastolic phase and end-systolic phase automatically determination is a nice function to be integrated into the approach. We also found that in some situations where the image intensity of LV blood pool is not homogenously distributed due to blood flow motion effect, adaptive measures can be added into the region growing method to increase the accuracy of endocardial contours. More intelligent methods, for instance knowledge-driven and feature-driven techniques, can be implemented to trace LV contours from cine MR images with high accuracy.
Acknowledgments
We thank Sunnybrook Health Sciences Centre for making their clinical image data, ground truth contour data and evaluation software accessible to public.
We gratefully acknowledge funding for this research by the Biomedical Research Council, Agency for Science, Technology and Research, Singapore.
Contributor Information
Su Huang, Phone: +65-64788406, FAX: +65-64789049, Email: huangsu@sbic.a-star.edu.sg.
Jimin Liu, Email: jmliu@sbic.a-star.edu.sg.
Looi Chow Lee, Email: lee_looi_chow@sbic.a-star.edu.sg.
Sudhakar K Venkatesh, Email: dnrskv@nus.edu.sg.
Lynette Li San Teo, Email: lslteo@yahoo.com.
Christopher Au, Email: christopher_au@nuh.com.sg.
Wieslaw L. Nowinski, Email: wieslaw@sbic.a-star.edu.sg
References
- 1.Selvanayagam JB, Robson MD, Francis JM, Neubauer S. Cardiovascular Magnetic Resonance: Basic Principles, Methods and Techniques. In: Dilsizian V, Pohost GM, editors. Cardiac CT, PET and MRI. Oxford: Blackwell; 2007. pp. 28–68. [Google Scholar]
- 2.Lu Y, Radau P, Connelly K, Dick A, Wright GA: Segmentation of Left Ventricle in Cardiac Cine MRI: An Automatic Image-Driven Method: LNCS 5528:339–347, 2008
- 3.Cocosco CA, Niessen WJ, Netsch T, Vonken EPA, Lund G, Stork A, Viergever MA. Automatic image-driven segmentation of the ventricles in cardiac cine MRI. J Magn Reson Imaging. 2008;28:366–374. doi: 10.1002/jmri.21451. [DOI] [PubMed] [Google Scholar]
- 4.Lorenzo-Valdes M, Sanchez-Ortiz GI, Elkington AG, Mohiaddin RH, Rueckert D. Segmentation of 4D cardiac MR images using a probabilistic atlas and the EM algorithm. Med Image Anal. 2004;8:255–265. doi: 10.1016/j.media.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Pednekar A, Kurkure U, Muthupillai R, Flamm S, Kakadiaris IA. Automated left ventricular segmentation in cardiac MRI. IEEE Transactions on Biomedical Engineering. 2006;53:1425–1428. doi: 10.1109/TBME.2006.873684. [DOI] [PubMed] [Google Scholar]
- 6.Uzümcü M, Geest RJ, Swingen C, Reiber JH, Lelieveldt BP. Time continuous tracking and segmentation of cardiovascular magnetic resonance images using multidimensional dynamic programming. Invest Radiol. 2006;41:52–62. doi: 10.1097/01.rli.0000194070.88432.24. [DOI] [PubMed] [Google Scholar]
- 7.Rezaee MR, Zwet PMJ, Lelieveldt BPE, Geest RJ, Reiber JHC. A multiresolution image segmentation technique based on pyramidal segmentation and fuzzy clustering. IEEE Transactions on Image Processing. 2000;9:1238–1248. doi: 10.1109/83.847836. [DOI] [PubMed] [Google Scholar]
- 8.Kaus MR, Berg Jv, Weese J, Niessen W, Pekar V. Automated segmentation of the left ventricle in cardiac MRI. Med Image Anal. 2004;8:245–254. doi: 10.1016/j.media.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 9.Mitchell SC, Lelieveldt BPF, Geest RJ, Bosch HG, Reiver JHC, Sonka M. Multistage hybrid active appearance model matching: segmentation of left and right ventricles in cardiac MR images. IEEE Trans Med Imaging. 2001;20:415–423. doi: 10.1109/42.925294. [DOI] [PubMed] [Google Scholar]
- 10.Paragios N. A level set approach for shape-driven segmentation and tracking of the left ventricle. IEEE Trans Med Imaging. 2003;22:773–776. doi: 10.1109/TMI.2003.814785. [DOI] [PubMed] [Google Scholar]
- 11.Fradkin M, Ciofolo C, Mory B, Hautvast G, Breeuwer M. Comprehensive segmentation of cine cardiac MR images. Med Image Comput Comput Assist Interv. 2008;11:178–185. doi: 10.1007/978-3-540-85988-8_22. [DOI] [PubMed] [Google Scholar]
- 12.Lynch M, Ghita O, Whelan PF. Segmentation of the left ventricle of the heart in 3-D+t MRI data using an optimized nonrigid temporal model. IEEE Trans Med Imaging. 2008;27:195–203. doi: 10.1109/TMI.2007.904681. [DOI] [PubMed] [Google Scholar]
- 13.Boykov Y, Jolly M-P: Interactive Organ Segmentation Using Graph Cuts. Proceedings of MICCAI, 2000, pp 276–286
- 14.Lin X, Cowan B, Young A: Model-Based Graph Cut Method for Segmentation of the Left Ventricle. 27th Annual International Conference Proceedings of the Engineering in Medicine and Biology Society, IEEE-EMBS, 2005, pp 3059–3062 [DOI] [PubMed]
- 15.Frangi AF, Niessen WJ, Viergever MA. Three-dimensional modeling for functional analysis of cardiac images: a review. IEEE Trans Med Imaging. 2001;20:2–5. doi: 10.1109/42.906421. [DOI] [PubMed] [Google Scholar]
- 16.Otsu N. A threshold selection method from gray-level histograms. IEEE Trans Systems Man Cybernet. 1979;9:62–66. doi: 10.1109/TSMC.1979.4310076. [DOI] [Google Scholar]
- 17.Coope ID. Circle fitting by linear and nonlinear least squares. J Optim Theory Appl. 1993;76:381–388. doi: 10.1007/BF00939613. [DOI] [Google Scholar]
- 18.Liao PS, Chen TS, Chung PC. A fast algorithm for multilevel thresholding. J Inf Sci Eng. 2001;17:713–727. [Google Scholar]
- 19.Canny J. A computational approach to edge detection. Pattern analysis and machine intelligence. IEEE Transactions on PAMI. 1986;8:679–698. doi: 10.1109/TPAMI.1986.4767851. [DOI] [PubMed] [Google Scholar]
- 20.Lorensen WE, Cline HE. Marching cubes: a high resolution 3D surface construction algorithm. Comput Graph. 1987;21:163–169. doi: 10.1145/37402.37422. [DOI] [Google Scholar]
- 21.Kass M, Witkin A, Terzopoulos D. Snakes: active contour models. Int J Comput Vision. 1988;1:321–331. doi: 10.1007/BF00133570. [DOI] [Google Scholar]
- 22.Liu J, Huang S, Nowinski WL. A hybrid approach for segmentation of anatomic structures in medical images. International Journal of Computer Assisted Radiology and Surgery. 2008;3:213–219. doi: 10.1007/s11548-008-0229-2. [DOI] [Google Scholar]