Abstract
With the increasing availability of high-resolution isotropic three- or four-dimensional medical datasets from sources such as magnetic resonance imaging, computed tomography, and ultrasound, volumetric image visualization techniques have increased in importance. Over the past two decades, a number of new algorithms and improvements have been developed for practical clinical image display. More recently, further efficiencies have been attained by designing and implementing volume-rendering algorithms on graphics processing units (GPUs). In this paper, we review volumetric image visualization pipelines, algorithms, and medical applications. We also illustrate our algorithm implementation and evaluation results, and address the advantages and drawbacks of each algorithm in terms of image quality and efficiency. Within the outlined literature review, we have integrated our research results relating to new visualization, classification, enhancement, and multimodal data dynamic rendering. Finally, we illustrate issues related to modern GPU working pipelines, and their applications in volume visualization domain.
Key words: Visualization, medical imaging, image processing, volume rendering, raycasting, splatting, shear-warp, shell rendering, texture mapping, Fourier transformation, shading, transfer function, classification, graphics processing unit (GPU)
Introduction
Volumetric medical image rendering is a method of extracting meaningful information from a three-dimensional (3D) dataset, allowing disease processes and anatomy to be better understood, both by radiologists as well as physicians and surgeons. Since the advent of computerized imaging modalities such as computed tomography (CT), magnetic resonance imaging (MRI), and positron emission tomography (PET) in the 1970s and 1980s, medical images have evolved from two-dimensional projection or slice images, to fully isotropic three- and even four-dimensional images, such as those used in dynamic cardiac studies. This evolution has made the traditional means of viewing such images (the view-box format) inappropriate for navigating the large data sets that today comprise the typical 3D image. While an early CT slice (320 × 320 pixels) contained about 100k pixels, today’s 5123 CT image contains ∼1,000 times as much data (or roughly 128 M pixels).1
This enormous increase in image size requires a more efficient means to manipulate and visualize these datasets.2,3 This is necessary on two fronts: The radiologist must navigate the data to make a diagnosis, while the surgeon needs an accurate 3D representation of the patient (and associated pathology) to plan a therapeutic procedure.4 To appreciate the full three-dimensional attributes of the data, the individual voxels in the dataset must be mapped onto the viewing screen in such a way that their 3D relationships are maintained, while making their screen appearance meaningful to the observer.5,6 The shift from traditional axial sections toward primary 3D volume visualization may provide improved visualization of the anatomical structures and thereby enhance radiologists’ performance in reviewing large medical data sets efficiently and comprehensively. The radiologists’ confidence in diagnosing diseases is therefore increased and the patient care is improved.7
Volumetric Image Visualization Methods
Three principle volume-rendering algorithms have been evolved for medical image visualization, i.e., multiplanar reformation (MPR), surface rendering (SR), and volume rendering (VR). As illustrated in the following sections, the rendering techniques can be classified as direct and indirect rendering. Direct rendering includes direct volume rendering (DVR) and direct surface rendering (DSR), while indirect rendering includes indirect surface rendering (ISR). Here, we refer to both DSR and ISR as SR.
Multiplanar Reformation
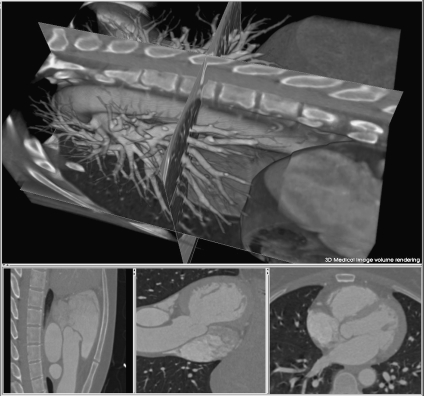
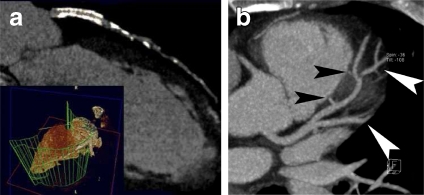
MPR is an image processing technique, which extracts two-dimensional (2D) slices from a 3D volume using arbitrarily positioned orthogonal or oblique planes.8 Although it is still a 2D method, it has the advantages of ease of use, high speed, and no information loss. The observer can display a structure of interest in any desired plane within the data set, and four-dimensional (4D) MPR can be performed in real time using graphics hardware.9 In addition, to accurately visualize tubular structures such as blood vessels, curved MPR may be employed to sample a given artery along a predefined curved anatomic plane.10 Curved MPR is an important visualization method for patients with bypass grafts and tortuous coronary arteries.11 Figure 1 illustrates a typical implementation of plane-based MPR showing three cross planes with arbitrary orientation, displayed within a volume-rendered heart. Figure 2a describes a curved MPR, defined by the green lines within a volume-rendered cardiac image.12 MPR can also serve as a useful adjunct to the axial images for detecting an inflamed appendix13 (Fig. 2b). In addition, curved MPR was combined with DVR to enhance the visualization of tubular structures such as trachea and colon.14
Fig 1.
MPR of 3D cardiac CT image. Top, three arbitrary cross planes. Bottom, synchronized 2D displays of cross planes.
Fig 2.
MPR illustration: a curved MPR12 (at 2003 RSNA); b coronal oblique MPR, displaying a tubular area (black arrows) surrounded by the abscess (white arrows)13 (at 2008 RSNA).
A major disadvantage of MPR is that it is a 2D display technique and orientation dependent. Therefore, when MPR is used to display coronary arteries, it may introduce false-positive or false-negative stenoses.15 Furthermore, although algorithms have been developed for automatically defining vessel centerlines, the curved MPR is still operator dependent, which limits the diagnostic accuracy.16
Surface Rendering
SR17 is a common method of displaying 3D images. ISR can be considered as surface modeling, while DSR is a special case of DVR. ISR requires that the surfaces of relevant structure boundaries within the volume be identified a priori by segmentation. In recently years, the performance of surface reconstruction and display was improved by employing graphics processing unit (GPU).18,19
For DSR, the surfaces are rendered directly from the volume without intermediate geometric representations, setting thresholds or using object labels to define a range of voxel intensities to be viewed. Recently, there have been many publications on isosurface extraction and rendering.
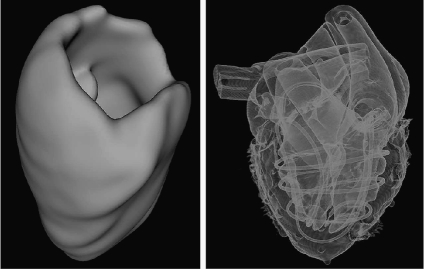
Hadwiger et al.20 proposed a real-time pipeline for implicit surface extraction and display, which was based on a two-level hierarchical data representation. Hirano et al.21 illustrated a semi-transparent multiple isosurface algorithm by employing the span space to rapidly search the intersections of cells with isosurfaces, while Petrik and Skala22 transformed the data into an alternative one-dimensional (1D) space to speed up isosurface extraction. A fully parallel isosurface extraction algorithm was presented by Zhang et al.23 In addition, a high degree of realism can be achieved with lighting models that simulate realistic viewing conditions. The first image of Figure 3 illustrates an implementation of ISR.
Fig 3.
Left, ISR of cardiac structures: myocardium; right, DSR of an MR heart phantom.
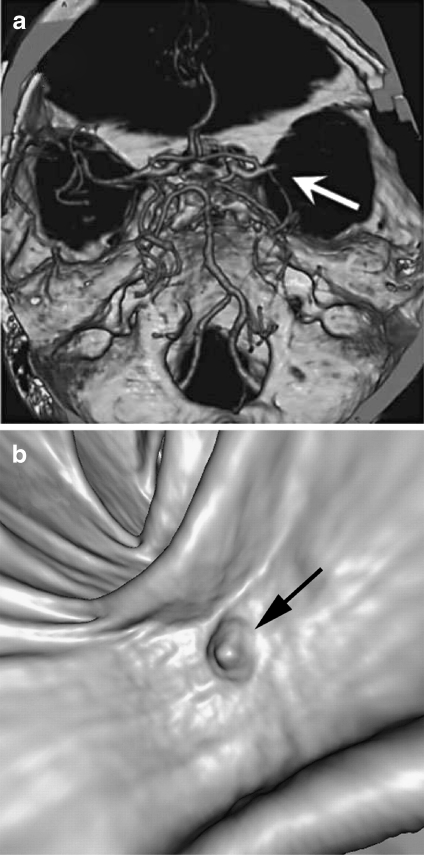
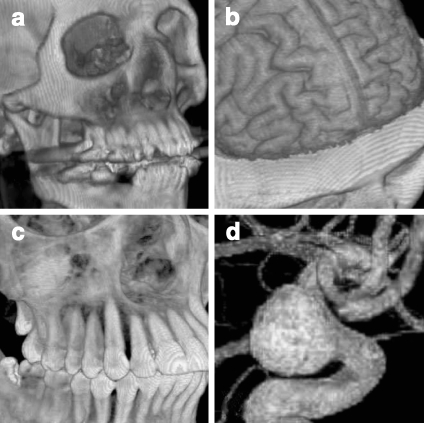
SR is often applied to contrast-enhanced CT data for displaying skeletal and vascular structures, and is also usually used in describing vascular disease and dislocations.24 In the process of detecting acute ischemic stroke, Schellinger and colleagues25 combined maximum intensity projection (MIP) with shaded SR to visualize proximal middle cerebral artery mainstem occlusion (Fig. 4a). SR has also been exploited to describe polyps within the 3D endoluminal CT colonographic images26 (Fig. 4b).
Fig 4.
SR applications: a image generated with MIP and shaded SR, demonstrating the proximal middle cerebral artery mainstem occlusion (arrow)25 (at 2007 S. Karger AG); b SR display of the endoluminal CT colonographic data, showing sessile morphology of the lesion (arrow)26 (at 2008 RSNA).
It is difficult to justify the accuracy and reliability of the images generated with shaded SR, i.e., the shiny surfaces might be misleading, causing the relation between image data and brightness in the resultant image to become more complex, a property which could affect the diagnosis. One example illustrating this phenomenon might be that vascular structures displayed with shaded SR may be easily interpreted as a severe stenosis just because the threshold slightly deviates from an optimal value.1,5
Volume Rendering
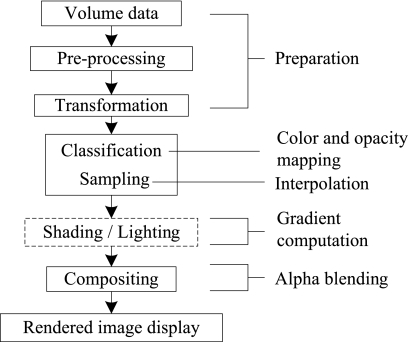
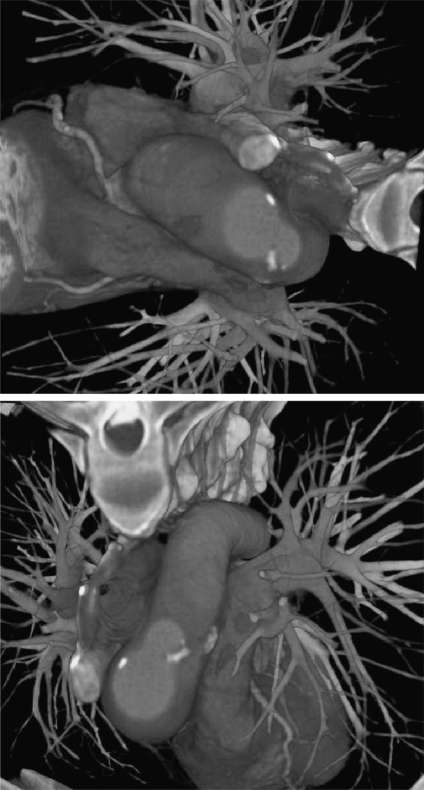
DVR displays the entire 3D dataset as a 2D image, without computing any intermediate geometry representations.27–29 This algorithm can be further divided into image-space DVR, such as software- 30,31 and GPU-based raycasting,32 and object-space DVR, such as splatting,33,34 shell rendering,35 texture mapping (TM),36 and cell projection.37 Shear-warp38 can be considered as a combination of these two categories. In addition, MIP,39 minimum intensity projection (MinIP), and X-ray projection40 are also widely used methods for displaying 3D medical images. This paper focuses its attentions on DVR, and the datasets discussed here are assumed to be represented on cubic and uniform rectilinear grids, such as are provided by standard 3D medical imaging modalities. Figure 5 describes the DVR pipeline with corresponding computations. Figure 6 shows an example of our DVR results applied to a CT cardiac volume.
Fig 5.
Volume-rendering pipeline and corresponding numerical operations.
Fig 6.
Volume-rendered images of volumetric human cardiac CT data set.
DVR is an efficient method for evaluating tissue structures, showing the vascular distribution and anatomic abnormality.41 This technique frees the user from making a decision as to whether or not surfaces are presented. When compared with SR, the main advantage of DVR is that interior information is retained, and so provides more information about the spatial relationships of different structures.27 In clinical applications, sensitivity and specificity are the major diagnostic criteria that must be considered. Even though DVR generally has high sensitivity and specificity for diagnosis,42 it is computationally intensive, so interactive performance is not always optimum. Another disadvantage is that it may be difficult to interpret the “cloudy” interiors. In addition, for analyzing large amounts of high-resolution medical data, DVR alone is probably not an optimal solution, since so many individual voxels are aggregated in a pixel on the screen during the global rendering process. The use of DVR alone may increase the chance that important information is hidden and overlooked. Therefore, a great deal of effect is currently being executed on merging DVR with newly developed computer-aided detection techniques,43 providing more intelligent visualization for early detection, diagnosis, and follow-up. Moreover, even though DVR is a powerful tool to show entire anatomic structures (for more detailed observation of specific lesions), slab imaging, where thick slices are rendered with DVR or MIP, has generally been used in clinical diagnosis.44 In addition, the combination of cross-sectional MPR and DVR can significantly increase the interpretation rate of anatomical structures.45
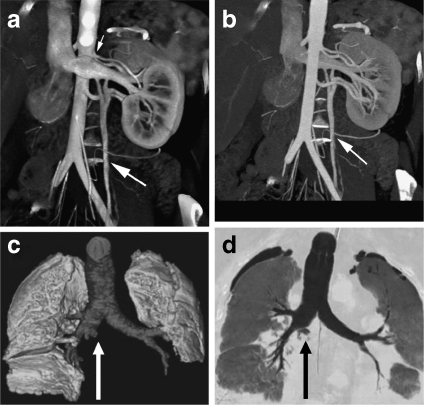
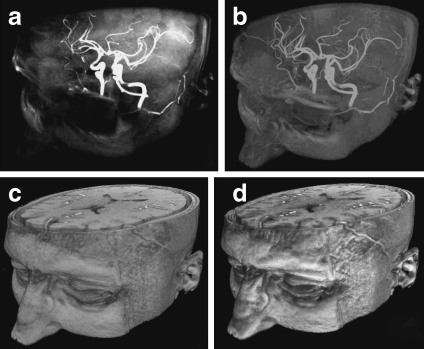
DVR has had a wide range of clinical applications. For example, to perform renal donor evaluation, Fishman et al.46 used DVR and MIP to display the renal vein of CT angiography data, as shown in Figure 7; for the DVR-generated image, the left gonadal vein (large arrow) is well defined (a), while the locations of the renal vein and gonadal vein (arrow) are inaccurately depicted in the MIP generated image (b). Gill et al.47 used DVR and MinIP to show the central airway and vascular structures of a 60-year-old male who underwent double lung transplantation for idiopathic pulmonary fibrosis, evaluating posttreatment in a noninvasive manner (Fig. 7c, d).
Fig 7.
DVR, MIP, and MinIP applications: a coronal oblique DVR image of kidney and veins; b MIP display of the same data as (a)46 (at 2006 RSNA); c DVR-generated image of the central airway and vascular structures; d coronal MinIP image of the same data as (c)47 (at 2008 AJR).
Volume-Rendering Principles
The core component of DVR is to solve an integral that describes the optical model. Different DVR techniques share similar components in the rendering pipeline, the main difference being the order in which they are applied.
Optical Models
The volume-rendering integral is based on an optical model developed by Blinn,48 which describes a statistical simulation of light passing through, and being reflected by, clouds of similar small particles. Such optical models may be based on emission or absorption individually, or both, depending on the applications.49 To reduce the computational cost, Blinn assumed that the volume is in a low albedo environment. In this case, a light emission–absorption model is an optimal balance between realism and computational complexity.29 Equation 1 demonstrates this procedure.
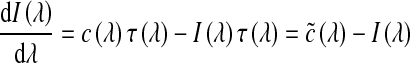 |
1 |
The solution to this equation is given below, showing the intensity of each pixel.
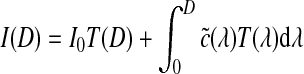 |
2 |
Here 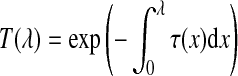 describes the volume transparency. The first term I0 illustrates light coming from the background, and D is the extent of the ray over which light is emitted. The last term demonstrates the behavior of the volume emitting and absorbing incoming light. The source term
describes the volume transparency. The first term I0 illustrates light coming from the background, and D is the extent of the ray over which light is emitted. The last term demonstrates the behavior of the volume emitting and absorbing incoming light. The source term 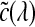 indicates the color change, and the extinction coefficient τ(λ) defines the occlusion of light per unit length due to light scattering or extinction.
indicates the color change, and the extinction coefficient τ(λ) defines the occlusion of light per unit length due to light scattering or extinction.
Color and Opacity Mapping
Classification
To display 3D medical images with DVR, the scalar values must first be mapped to optical properties such as color and opacity through a transfer function (TF), a process referred to as classification.
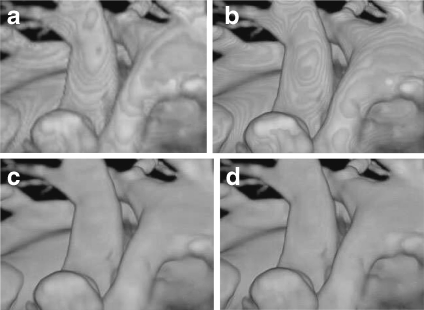
Pre- and post-classification approaches differ with respect to the order in which the TF and sampling interpolation are applied.29,50 Pre-classification first maps every scalar value at the grid into color and opacity, where the color and opacity are assigned at the resampling points. On the other hand, for post-classification, we first sample the scalar value by interpolation and then map the acquired values to colors and opacity through TFs. Both the pre- and post-classification operations introduce high-frequency components via the nonlinear TFs.51–53 Pre-classification suppresses this high-frequency information, so the rendered image appears blurry (Fig. 8a) while post-classification maintains all the high frequencies, but introduces “striping” artifacts in the final images (Fig. 8b).
Fig 8.
DVR of cardiac vessels via different classification algorithms: a pre-classification, b post-classification, c pre-integrated classification, d post color-attenuated classification.
To address the under-sampling problem, Röttger et al.54 proposed a pre-integrated classification algorithm for hardware-accelerated tetrahedra projection, which was first introduced by Max et al.55 Later, Engel et al.51 applied this classification algorithm for 3D texture-mapping-based volume visualization of regular-grid volume data. Pre-integrated classification separates the DVR integral into two parts, one for the continuous scalar value and the other for the TF parameters 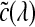 (colors) and τ(λ) (extinction). This algorithm renders the volume segment by segment instead of point by point. In this manner, the Nyquist frequency for reconstructing the continuous signal is not increased by the TF nonlinear properties.
(colors) and τ(λ) (extinction). This algorithm renders the volume segment by segment instead of point by point. In this manner, the Nyquist frequency for reconstructing the continuous signal is not increased by the TF nonlinear properties.
Compared with the pre- and post-classification algorithms, pre-integrated classification improves the image quality with decreased under-sampling artifacts (Fig. 8c). However, it is time consuming to construct the color lookup table. To address the problem, Engel et al.51 proposed a hardware acceleration technique, and Roettger et al.56 applied this hardware-accelerated approach, along with a classification algorithm, to GPU-based raycasting. We proposed a post color-attenuated classification algorithm57 to rapidly evaluate the optical properties of each segment, which eliminated the extinction coefficient from the opacity-associated color integral, and computed the color attenuation segment by segment instead of point by point (Fig. 8d). Furthermore, to improve the lookup table generation speed, Kye et al.58 presented an opacity approximation algorithm that used the arithmetic mean instead of the geometric mean in the integral computation. To improve the accuracy of approximating the pre-integrated TF integral, Hajjar and colleagues59 used a second-order function as a substitute for the piecewise linear segment. Recently, Kraus60 proposed a space-covering volume sampling technique to extend the pre-integrated volume rendering to multidimensional TFs.
Transfer Function
In DVR, the TF was typically used for tissue classification based on local intensities in the 3D dataset.61,62 Multidimensional TFs are efficient for multiple spatial feature detection, as demonstrated by Kniss et al.63 who designed such a TF and demonstrated its medical applications. Abellán et al.64 introduced a 2D fusion TF to facilitate the visualization of merged multimodal volumetric data, and a 3D TF was designed by Hadwiger et al.65 to interactively explore feature classes within the industrial CT volumes, where individual features and feature size curves can be colored, classified, and quantitatively measured with the help of TF specifications. Furthermore, Honigmann et al.66 designed an adaptive TF for 3D and 4D ultrasound display, and a default TF template was built to fit specific requirements. Later, to facilitate the TF specification, these authors67 also introduced semantic models for TF parameter assignment, while a similar idea was used by Rautek et al.68 to add a semantic layer in the TF design. Freiman et al.69 presented an automatic method for a patient-specific, feature-based multidimensional TF generation method, and used it to visualize liver blood vessels and tumors in CT datasets. Pinto and Freitas70 developed an interactive DVR platform for data exploration through adapting both image- and data-driven TFs.
Multiple factors can be exploited to improve TF specifications. A hardware-accelerated TF was designed in our own work71 to interactively manipulate medical image volumes during visualization, while Bruckner and Gröller72 proposed a style TF concept, which defines color at a sample point as a function of the data value and the eye-space normal. At the same time, to highlight structures of interest, they also designed an interactive halo TF to classify these structures based on data value, direction, and position.73 Moreover, Wu and Qu74 fused features captured by different TFs, and a size-based TF was proposed by Correa and Ma,75 mapping the relative size of local features to optical values for tissue detection within complex medical datasets. Caban and Rheingans76 introduced a texture-based TF, mapping the local textural properties of voxels to opacity and color, so that anatomical structures with the same intensity and gradients could be distinguished. As described by Hadwiger and Rezk-Salama, presenting image galleries with predefined TFs by setting visual parameters was one of the most frequently used operations in TF design.66,77
In the process of TF specification, to identify the distribution of different material types and reduce partial-volume effects, Laidlaw et al.78 used a probabilistic Bayesian algorithm to analyze the voxel histograms, and a similar idea was employed by Lundstrom et al.79 to include anatomical domain knowledge into the TF. Souza80 exploited the fraction measure and uncertainty principle to address this issue. Jani et al.81 used Powell’s method to optimize the opacity selection in the TF adjustment process, and demonstrated its applicability for prostate CT volume rendering.
Composition
During the DVR process, a number of composition schemes are commonly employed, including X-ray projection, MIP, MinIP, and alpha blending. In X-ray projection,40 the interpolated samples are simply summed, giving rise to an image typical of those obtained in projective diagnostic imaging. It has the advantage of clearly representing vessels or bones at the cost of losing the depth information (Fig. 9a). MIP39,82 and MinIP are widely used techniques in 3D CT and MR angiography. The image results from the voxels having the MIP or MinIP along the viewing rays traversing through the object. MIP and MinIP images can be generated rapidly and can clearly display vessels, tumor, or bones,24 and the image generation has been accelerated by graphics hardware.83 These two techniques still have some inherent disadvantages because only a small fraction of the data is used, and static MIPs or MinIPs are not particularly useful due to lack of depth information. However, MIP and MinIP can be interactively explored or used as animation sequences, where the limited-depth-perception drawback is less obvious. Local MIP, MinP, or closest vessel projection (CVP) are often used in slab imaging, i.e., thin-slab MIP or MinP, for vascular structure diagnosis.44,84 For vascular diagnosis, CVP is superior to MIP; however, the user needs to set an appropriate threshold for the local maximum, which is determined by a specific dataset, making the application of CVP more difficult than MIP. Figure 9b shows the result of MIP.
Fig 9.
DVR results of human head with angiographic contrast using different compositing techniques: a X-ray projection, b MIP, c alpha blending without shading, and d alpha blending with shading.
Alpha blending30,49 is a popular optical blending technique, often implemented by using the Riemann sum to discretize the continuous function Eq. 2, resulting front-to-back and back-to-front alpha blending Eqs. 3 and 4 respectively, depending on the compositing order.
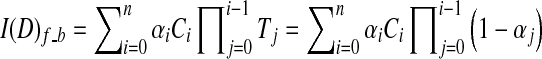 |
3 |
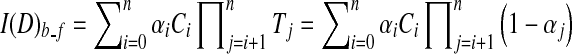 |
4 |
where now the pre-multiplied αiCi is the total light emitted from a point i, which is also called the opacity associated color.85Tj(0 ≤ Tj ≤ 1) is the transparency of a point, and αj is its opacity, which is defined as 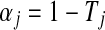 . Figure 9c describes the DVR result using alpha blending without shading, while (d) shows the result with shading.
. Figure 9c describes the DVR result using alpha blending without shading, while (d) shows the result with shading.
Volume Illumination and Illustration
In volume illumination, the normal at every sampling point is calculated by interpolation using the intensity changes across that voxel. These approximated voxel normals are then used in a Phong or Blinn–Phong model for shading computations, with the results being employed in the DVR composition. The shading computations may be accelerated by graphics hardware.86 In addition, the shading model can be used with volumetric shadows to capture chromatic attenuation for simulating translucent rendering,87 and can be combined with clipped volumes to increase the visual cues.88 Figure 10 illustrates a DVR of CT cardiac data sets with and without illumination, demonstrating that images with shading are visually more pleasing. However, clinicians sometimes deliberately disable illumination since it may complicate certain diagnostic tasks.84
Fig 10.
DVR images of CT pulmonary data with (right) and without illumination (left).
This technique is helpful to provide insight into medical data, aiding cognition during data exploration. For example, to illustratively define a mapping from volumetric attributes to visual styles, a fuzzy-logic rule-based semantic layer was introduced by Rautek et al.68 who extended their work89 by introducing an interaction-dependent semantic model. To achieve realistic anatomical volume rendering and illustration, Silverstein et al.90 presented a color-mapping algorithm, which combines natural colorization with the perceptual discriminating capacity of grayscale. For the purpose of enhancing anatomical structure explanation, Wang et al.91 introduced a knowledge-based system to encode a set of illustrative models into the volume-rendered images. Rezk-Salama et al.92 generated light field representations from volumetric data, so users could illustratively explore resultant images at run time. Jainek et al.93 presented a hybrid rendering method, combining an illustrative mesh of brain structures with DVR, resulting in a focus-and-context data representation. In addition, to effectively emphasize data in the focus-and-context style, Svakhine et al.94 illustrated an illustration-guided DVR algorithm, enhancing anatomical structures and their spatial relationships.
Illumination is an important aspect of volume illustration. Rheingans and Ebert95 proposed an illumination approach similar to volume shading, using non-photorealistic rendering to enhance physics-based DVR. Dong and Clapworthy96 extended the texture synthesis approach from surface to non-photorealistic DVR, and this volume illumination model was used in conjunction with a TF by Bruckner et al.97 to preserve contextual information during the rendering process of interior structures. To enhance material discrimination, Noordmans and colleagues98 extended the color- and opacity-based illumination model to wavelength-dependent spectral DVR, and the performance of this spectral DVR was optimized with a linear spectral factor model by Bergner et al.99 Recently, for the purpose of generating specified colors or spectral properties in surface or volume rendering, this group used constructed spectra to enhance a palette of given lights and material reflectances.100 In addition, Strengert et al.101 subsequently employed this spectral model to GPU-based raycasting.
To increase depth perception, Kersten et al.102 used a purely absorptive lighting model in DVR to enhance the display of translucent volume, while Bruckner and Gröller72 employed a style TF to add data-dependent image-based shading to volume-rendered images, and Joshi et al.103 exploited tone shading with distance color blending in vascular structure volume visualization. Furthermore, to facilitate tissue illumination, Ropinski and colleagues104 added a dynamic ambient occlusion simulation model to DVR, and Mora et al.105 introduced a rendering pipeline for isotropically emissive visualization to facilitate intuitive medical data exploration. To intuitively explore hidden structures and analyze their spatial relations in volumes, Chan et al.106 introduced a relation-aware visualization pipeline, in which inner structural relationships were defined by a region connection calculus and were represented by a relation graph interface. In addition, Rautek et al.107 gave a comprehensive review on illustrative visualization and envisioned their potential medical applications. Such illustrative techniques are sometimes integrated in commercial systems; however, there have been no systematic investigations regarding whether any of these techniques actually provides a clinical benefit.1
Software-Based Raycasting
Algorithm Overview
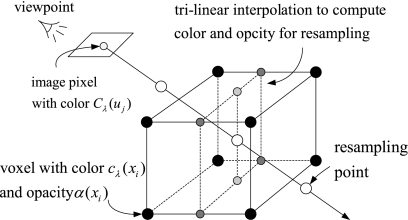
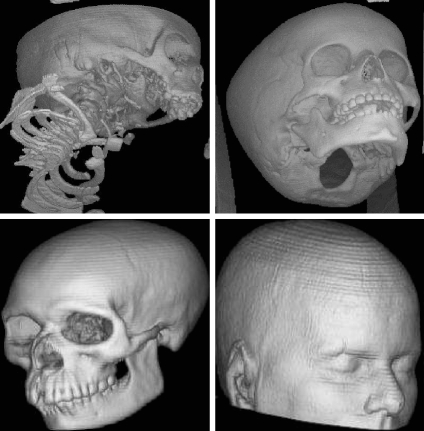
Raycasting27 is a popular technique used to display a 3D dataset in two dimensions, whose basic idea is to cast a ray from each pixel in the viewing plane into the volume, sampling the ray with a predetermined step in a front-to-back or back-to-front order by trilinear interpolation. A TF is used to map the scalar value to color and opacity. Finally, the acquired optical values at the sampling points along the casting ray are composited using Eqs. 3 or 4 to approximately compute the DVR integral Eq. 2, obtaining the corresponding final pixel color on the output image. Figure 11 illustrates the pipeline, and Figure 12 shows an implementation on four medical images.
Fig 11.
The raycasting pipeline: casting a ray, sampling, optical mapping, and compositing.
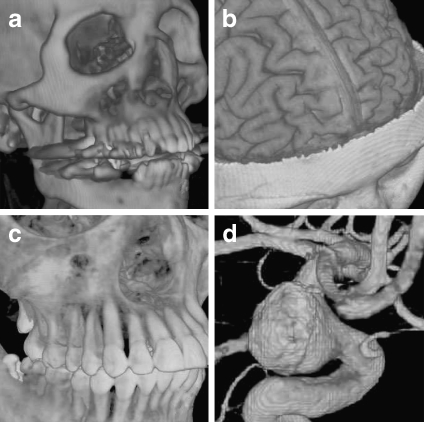
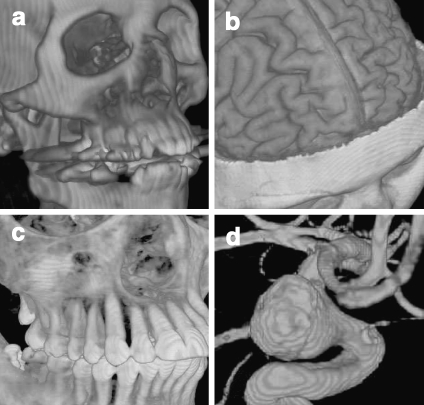
Fig 12.
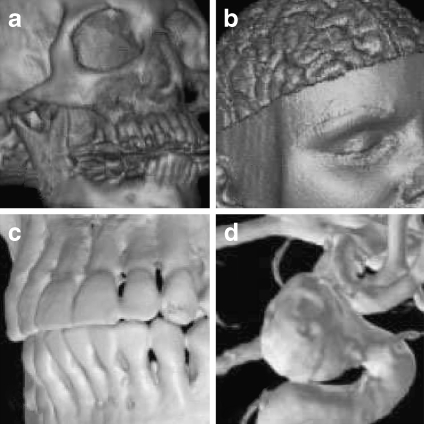
Medical images rendered with software-based raycasting: a CT skull, b MR brain, c CT jaw, and d MR cerebral blood vessel.
Applications
This technique was a significant 3D display method in some applications during the time when graphics hardware-accelerated DVR is not commonly available. For example, Sakas et al.108 used raycasting to display 3D ultrasound data of a fetus, and Höhne31 and Tiede et al.109 employed this algorithm to display and investigate the structures of human anatomy.
Improvements
Levoy110 presented an adaptive refinement approach for empty space skipping, and a similar idea was used in a hierarchical spatial enumeration technique,111 which first created a binary pyramid, which was used to build a min–max octree to encode the empty and non-empty space, and this min–max octree structure was exploited with a range tree by Lee and Ihm112 to skip coherent space in volume. A similar approach was used by Grimm et al.113 who proposed an algorithm based on a bricked volume layout.
Splatting Algorithms
Algorithm Overview
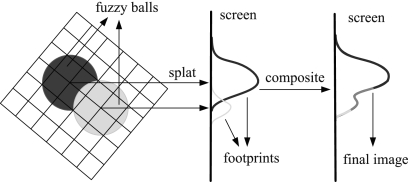
This technique was developed to improve the calculation speed of DVR at the expense of lower accuracy.33,34 Splatting calculates the influence of each voxel in the volume upon multiple pixels in the output image. This algorithm represents the volume as an array of overlapping basis functions called reconstruction kernels, which are commonly rotationally symmetric Gaussian functions with amplitudes scaled by the voxel values. These kernels are classified as colors and opacities by a TF, and are projected onto the output image plane in a depth-sorted order. Each projected kernel leaves a footprint (or splat) on the screen, and all of these splats are composed in back-to-front order to yield the final image. This process is described in Figure 13, and Figure 14 presents examples of images rendered with the splatting algorithm.
Fig 13.
Splatting pipeline: the optical model is evaluated for each voxel and projected onto the image plane, leaving a footprint (splat). Then, these footprints are composited to create the final image.
Fig 14.
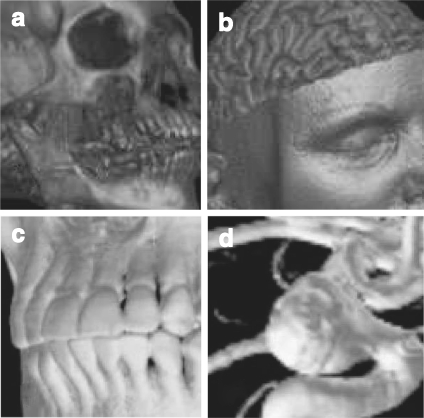
Medical images rendered with splatting: a CT skull with shading, b MR brain with shading114 (at 2000 Elsevier), c CT jaw, and d MR cerebral blood vessel115 (at 2000 IEEE).
Applications
Vega-Higuera et al.116 exploited texture-accelerated splatting to visualize the neurovascular structures surrounded by osseous tissue in CT angiography data. Splatting was also employed to rapidly generate digitally rendered radiographs in the iterative registration of medical images.117 Audigier and colleagues118 used splatting with raycasting to guide the interactive 3D medical image segmentation, providing the users with visual feedback at the end of each iterative segmentation step.
Improvements
Since basic splatting algorithms33 suffer from “color bleeding” artifacts, Westover34 employed an axis-aligned sheet buffer to solve this problem. However, this technique needs to maintain three stacks of sheets and introduces “popping” artifacts, which refers to the abrupt change in image appearance as the view switches from one of the three image stacks to another. To address this issue, Mueller et al.119 aligned the sheet buffers parallel to the image plane, and they later accelerated this image aligned splatting algorithm with GPUs.120 Recently, Subr et al.121 introduced a self-sufficient data structure called FreeVoxels to eliminate the color bleeding errors. In addition, the aliasing problem in splatting was addressed by Zwicker et al.122 with the use of an elliptical weighted average filter. To improve splatting performance, Ihm and Lee114 employed an octree to encode the object- and image-space coherence, where only voxels of interest were visited and splatted, while Xue et al.123 increased the speed of splatting through texture mapping.
Shear-Warp Algorithm
Algorithm Overview
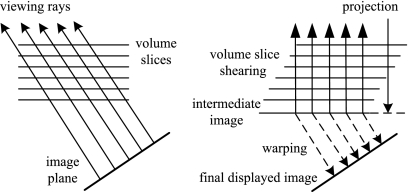
Shear-warp is a hybrid algorithm that attempts to combine the advantages of the image- or object-order based volume-rendering methods,38 and is based on factorizing the viewing transformation into 3D shear and 2D warp. The shear matrix transforms all the viewing rays so that they are parallel to the principal viewing axis in “sheared object” space, allowing the volume and image to be traversed simultaneously. Figure 15 illustrates the transformation from the original object space to sheared object space for parallel projection.
Fig 15.
The standard transformation (left) and shear-warp factorized transformation (right) for a parallel projection.
We can mathematically express the factorization of the viewing matrix Mview, which comprises a permutation matrix P and a view matrix M, such that 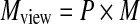 , and the view matrix M is factorized into a shear S and a warp component
, and the view matrix M is factorized into a shear S and a warp component 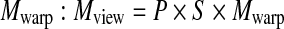 . Figure 16 shows the rendering examples.
. Figure 16 shows the rendering examples.
Fig 16.
Medical images rendered with the shear-warp DVR algorithm: a CT skull with shading, b MR brain with shading38 (at 1994 ACM), c CT jaw, and d MR cerebral blood vessel (distal common carotid artery)115 (at 2000 IEEE).
Applications
Perspective shear-warp was implemented by Schulze et al.124 in a virtual reality environment, while Anagnostou et al.125 used this algorithm to display time-varying data. Furthermore, Wan et al.126 used it to apply 3D warping in an interactive stereoscopic rendering for virtual navigation.
Improvements
This algorithm was thoroughly analyzed by Sweeney and Mueller,127 and they proposed several measures to alleviate the short-comings of this algorithm, such as blurry images on zoom and staircase artifacts on flat surfaces. The VolumePro graphics processing board is a hardware implementation of the raycasting using shear-warp factorization for real-time visualization.128 One limitation of VolumePro is that it can only provide orthographic projections, which renders it unsuitable for applications such as virtual endoscopy. Therefore, Lim and Shin129 employed the subvolume feature in VolumePro to approximate perspective projection, only rendering the subvolumes within the viewing frustum. To alleviate blur in the zoomed image, Schulze et al.130 applied a pre-integrated classification to shear-warp. Later, Kye and Oh131 used a super-sampling to improve the image quality, and a min–max block data structure was designed to maintain the rendering performance.
Shell Rendering
Algorithm Overview
The shell rendering algorithm is a software-based hybrid of surface and volume rendering, which is based on a compact data structure referred to as a shell, i.e., a set of nontransparent voxels near the extracted object boundary with a number of attributes associated with each related voxel for visualization.35
The shell data structure can store the entire 3D scene or only the hard (binary) boundary. For the hard boundary, the shell is crisp and only contains the voxels on the object surface, and shell rendering degenerates to SR. For a fuzzy boundary, the shell includes some voxels in the vicinity of the extracted surface, and shell rendering is identified as DVR. Figure 17 shows examples of shell SR and DVR, and the detailed descriptions of this algorithm can be found in Udupa and Odhner.35
Fig 17.
Shell rendering example. Top row, shell SR132 (at 2000 IEEE): skull CT data (left) and CT data of a “dry” child’s skull (right). Bottom row, shell DVR133 (at 2003 SPIE): CT skull (left), and MR head (right).
Applications
Lei et al.134 employed this algorithm to render the segmented structures of vessels and arteries of contrast-enhanced magnetic resonance angiography image. However, the explicit surface extraction creates errors. To address the problem, Bullitt et al.135 selectively dilated the segmented object boundaries along all axes, and visualized the extracted fuzzy shell with raycasting.
Improvements
To accelerate the shell rendering speed, Falcão and colleagues136 added the shear-warp factorization to the shell data structure, and Botha and Post137 used a splat-like elliptical Gaussian to compute the voxel contribution energy to the rendered image. Later, Grevera et al.138 extended the point-based shell element to a new T-shell element for isosurface rendering.
Texture Mapping
The pioneering work of exploiting texture hardware for DVR was performed by Cullip and Neumann139 and Cabral et al.140.
Algorithm Overview
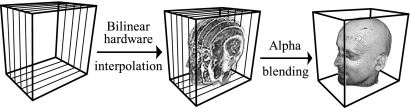
In 2D texture mapping (2DTM), the volume is decomposed into three stacks of perpendicularly object-aligned polygons. For the current viewing direction, the stack whose slicing direction (normal) is within ±45° of the current viewing direction is chosen for rendering. During rasterization, each of the polygonal slices is textured with the image information obtained from the volume via bilinear interpolation. Finally, the textured slices are alpha-blended in a back-to-front order to produce the final image. Figure 18 describes the working pipeline, while Figure 19 illustrates rendering results with this algorithm.
Fig 18.
The working pipeline of 2DTM for the 3D dataset (head) visualization.
Fig 19.
Medical image rendered with 2DTM: a CT skull, b MR brain, c CT jaw, and d MR cerebral blood vessel.
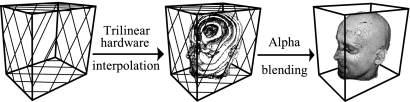
Three-dimensional texture mapping (3DTM) uploads the volume to the graphics memory as a single 3D texture, and a set of polygons perpendicular to the viewing direction is placed within the volume and textured with the image information by trilinear interpolation. Compared with 2DTM, there are no orientation limitations for the decomposed polygon slices, making the texture access more flexible. The compositing process is similar to 2DTM, in that the set of textured polygon planes are alpha-blended in a back-to-front order. Figure 20 illustrates the algorithm pipeline, and Figure 21 demonstrates an implementation of 3DTM.
Fig 20.
The working pipeline of 3DTM for 3D head rendering.
Fig 21.
Medical image rendered with 3DTM: a CT skull, b MR brain, c CT jaw, and d MR cerebral blood vessel.
Applications
Applications of TM-based medical volume rendering, as reported in the literature, can be divided into three main applications. The first is multimodal medical image rendering and tissue separation. Sato et al.141 designed a multidimensional TF to identify tissue structures in multimodal medical images generated with 3DTM. A two-level rendering technique was integrated with 3DTM by Hauser et al.,142 allowing different rendering techniques to be selected for different tissues. Later, Hadwiger et al.143 improved this two-level DVR algorithm with respect to image quality and performance, minimizing the computational cost of each pass, adding dynamic filtering, and using depth and stencil buffering to achieve correct compositing of objects created with different rendering techniques. Sereda et al.144 used the LH histogram in a multidimensional TF domain for volume boundary enhancement and tissue detection. In an attempt to improve arterial stenosis diagnosis, Ljung et al.145 applied a probabilistic TF model to 3DTM, replacing the optical lookup table with a tissue material selection table. Xie et al.146 used Boolean operations in a 3DTM-based DVR for interactive multi-object medical volume clipping and manipulation, and Kim and colleagues147 used a dual TF-based algorithm to render fused images of PET and CT in real time. Abellán and Tost64 exploited 3DTM for multimodal medical data rendering, in which various fusion techniques were integrated into a common hardware-accelerated DVR framework, which were employed to display registered dual-modal brain datasets.
The second application area is the display of specific tissues and organs for diagnosis and therapy. Holmes et al.148 used 3DTM to achieve a real-time display of trans-urethral ultrasound for prostate treatment. This technique was also used by Etlik et al.149 to find bone fractures, and also by Wenger et al.150 to visualize diffusion tensor MRI data in the brain. Wang et al.151 exploited 3DTM to display 3D ultrasound for cardiac ablation guidance. To provide visual information relating to structures located beneath the skin surface, Sharp et al.152 employed this technique to visualize the light diffusion in 3D inhomogeneous tissue. This visualization approach was also employed by Lopera et al.153 to view a stenosis in arbitrary image planes, and the authors used the results to demonstrate the subsequent arterial bypass. For the purpose of showing early opacification of hepatic veins, Siddiki and colleagues154 exploited 3DTM to display the arterial phase images. Aspin et al.155 combined this rendering method with data enhancement techniques to increase the users’ cognitive ability in diagnosing soft tissue pathologies in volume-rendered MR images.
The third area of application is in dynamic imaging and deformation. Levin et al.156 developed a TM-based platform that was used to interactively render multimodality 4D cardiac datasets. In addition, Yuan et al.157 designed a TF for nonlinear mapping density values in the dynamic range medical volumes, while Correa and colleagues158 presented a 3DTM algorithm that sampled the deformed space instead of the regular-grid points, simulating volume deformations caused by clinical manipulations, such as cuts and dissections.
Improvements
Shading Inclusion
The shading was first included into TM approaches by Van Gelder et al.,36 and both diffuse and specular shading models were later integrated by Rezk-Salama et al.159 using register combiners and paletted texture. Kniss et al.87 proposed a simple shading model that captured volumetric light attenuation to produce volumetric shadows and translucency. This shading model was also used with depth-based volume clipping by Weiskopf and colleagues88 in a 3DTM pipeline. Recently, Abellán and Tost64 defined three types of shadings for the TM-accelerated DVR of dual-modality dataset.
Empty Space Skipping
Li et al.160,161 computed texture hulls of all connected non-empty regions, and they later improved this technique with “growing boxes” and an orthogonal binary space partitioning tree. Bethune and Stewart162 proposed an adaptive slice DVR algorithm based on 3DTM. In this algorithm, the volume was partitioned into transparent and nontransparent regions, axis-aligned bounding boxes were used to enclose those that were opaque, and an octree was used to encode these boxes for efficient empty space skipping. In addition, Keleş et al.163 exploited slab silhouette maps and an early depth test to skip empty spaces along the direction normal to the texture slice. The volume partition technique was also employed by Xie et al.164 to subdivide the volume dataset into uniform-sized blocks, and several acceleration techniques, such as early ray termination, empty space skipping, and occlusion clipping, were applied to interactively display CT dataset. Recently, a kd-tree and dynamic programming were used by Frank and Kaufman165 to optimize the non-empty space tracking and the data distribution balance on a volume-rendering cluster.
Multiresolution
Multiresolution provides users with high-fidelity-rendering results in the region of interest while maintaining this region within the context of the whole volume, albeit at a lower resolution. LaMar et al.166 used a multiresolution data representation for large dataset rendering, in which an octree was employed to encode a hierarchical volume “bricking.” A similar octree-based multiresolution approach was presented by Boada and her colleagues.167 Kähler and Hege168 rendered the adaptive mesh refinement (AMR) data using 3DTM, in which volume was partitioned into axis-aligned bricks of the same level of resolution, and they also extended the AMR tree-based 3DTM to render large and sparse data sets.169 In addition, Gao et al. proposed a parallel multiresolution technique to accelerate large dataset visualization.170
Wavelet Compression
Wavelet decomposition is an efficient means of representing data in a multiscale form. A blockwise wavelet compression was used by Nguyen et al.171 to split 3D data into small blocks of equal size that could be compressed and uncompressed individually along the three axes. Guthe et al.172 employed a block based wavelet to compress datasets and extract the levels of detail necessary for the current viewpoint on the fly, and Xie et al.173 further extended this concept by compressing input data using hierarchical wavelet representation.
Multitexturing
Zhang and colleagues developed a hardware-accelerated DVR algorithm based on register combiners and multitexturing,71 in which a viewing-direction-based dynamic texture slice resampling scheme was proposed. The slicing steps are dynamically updated according to viewing directions, maintaining a constant step size during the DVR process, and therefore avoiding striping artifacts caused by nonuniform sampling step while maintaining high-rendering performance.
GPU-Based Raycasting
The GPU on today’s commodity video cards has become a powerful computation engine capable of a variety of applications, especially in the fields of computer graphics and visualization.174 By taking advantage of the stream computation and parallel architecture, GPU-accelerated raycasting can provide an interactive frame rate for medium-sized dataset visualization on commodity hardware.175
Introduction to GPU Architecture
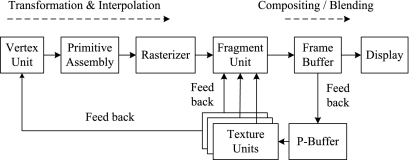
GPUs incorporate multi-pipelined stream processors and high bandwidth memory access, and include their own DRAM (device memory) optimized to process 2D and 3D geometries in parallel.176 Data are transferred between the CPU main memory and the GPU graphics memory via a dedicated Accelerated Graphics Port or a Peripheral Component Interconnect Express (PCI-Express) bus. Modern GPUs are programmable and employ a SWAR (SIMD, i.e., Single Instruction Stream Multiple Data Stream, Within A Register) execution model. Figure 22 illustrates the architecture and working pipeline of a typical GPU.
Fig 22.
GPU architecture and working pipeline: data flow, multi-pass rendering, and P-Buffer, taking the data from the frame buffers and feeding them back to the fragment and vertex units through the P-Buffer.
As illustrated in Figure 22, the geometry is specified as a set of vertices and a set of facets. The vertex stream is processed by the vertex units and then assembled into primitives, which are grouped together over triangles and are scan-converted by the rasterizer. The outcome of this operation is a large stream of elements (fragments), which are further processed by the fragment units. These fragments are written into a frame buffer and then copied into the video memory to produce an image on the screen. Recent GPUs include a non-visible rendering buffer, called the pixel buffer (P-Buffer), which can be employed if higher precision is required in the intermediate calculations. The nature of the data on which GPUs operate makes them suitable for a stream processing architecture.177
Recently, NVidia and AMD announced their new software development platforms, i.e., Compute Unified Device Architecture (CUDA)178 and Close To Metal (CTM),179 respectively, which are designed to directly access the native instruction set and memory in parallel on GeForce (G8X and later generations) and Radeon (R580 and later generations) graphics cards, respectively. CUDA is a C language development environment, while CTM is a virtual machine running assembler code. The goal of these two new architectures is to provide developers with a streaming-like programming model for compute-intensive parallel computations, allowing low-level, deterministic, and repeatable access to hardware. However, these new generalizations sacrifice some of the efficiencies and graphic specified features.180 In addition, the bus bandwidth and latency between CPU and GPU still remains a bottleneck for the further performance enhancement.
Algorithm Overview
A general strategy of exploiting the programmable GPU for raycasting was proposed by Krüger and Westermann32 and Purcell et al.,113 who described a stream model that exploits the intrinsic parallelism and efficient communication on modern GPUs. The main core of this algorithm is to generate a single ray per screen pixel and to trace this ray through the volume on a GPU fragment shader. Assuming that the origin point is O and the ray r(t) direction is v, then r(t) is defined as O + v × t. The algorithm is described by the following items:
Store the dataset as a 3D texture, whose bounding box is a colored unit cube. The texture coordinates at the eight corners are encoded in the RGB color channels.
Render the front face of the color cube and store the result into an intermediate texture (Fig. 23a).
Render the back face (Fig. 23b), subtract the color values of the back face from the front face, normalize the result, and store it in a 2D texture, which represents the direction vectors of the casting ray for each pixel. The origin of the ray is the pixel of the bounding cube front face.
Cast a ray for each pixel into the 3D texture volume, and uniformly sample the volume along the ray. This is all done in a single fragment program using the GPU Shader Model 3.0 loop capabilities.
Blend the TF-mapped color and opacity at each sampling point along the ray in a front-to-back order. When the ray leaves the volume or the accumulated opacity exceeds the preset threshold, terminate the ray.
Fig 23.
Rendering results of the front face (left) and back face (right) of a unit bounding box. The texture coordinates at the corners are encoded in the RGB color channels, which are used to calculate the casting ray direction.
Performance Comparisons
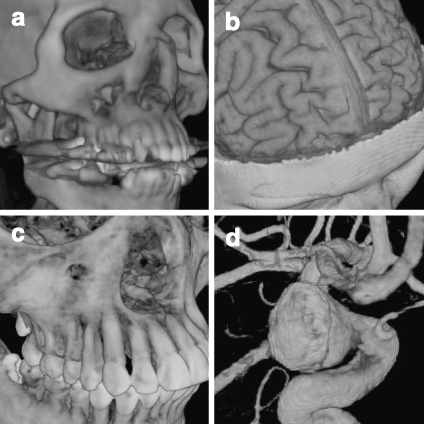
TM-based DVR and GPU-based raycasting are all graphics hardware-accelerated DVR algorithms. The TM uses fixed graphics pipelines, while the raycasting encodes the algorithm on the GPU programmable shaders. For DVR based on TM, the shading calculation is expensive since it needs to apply the computationally expensive shading model to every single fragment, even if some of them do not contribute to the final image. In addition, it is not easy to include some of the traditional acceleration techniques. Finally, for perspective projection, a uniform sampling step cannot be maintained, resulting in striping artifacts. For GPU-based raycasting, since it is a flexible rendering pipeline, the acceleration techniques such as early ray termination, empty space skipping can easily be included. Also, since the shading computation is fragment-based, the shading model need only be applied to the fragments that contribute to the final image. In addition, the sampling step is adaptive, so for perspective projection, uniform sampling steps can be maintained, resulting in improved image quality. Figure 24 illustrates our implementation of GPU-accelerated raycasting of four 3D medical datasets, from which we note that the image quality for raycasting is superior to the images generated with 2DTM (Fig. 19) or 3DTM (Fig. 21).
Fig 24.
Medical images rendered with the GPU-based raycasting: a CT skull, b MR brain, c CT jaw, and d MR cerebral blood vessel.
Applications
As exemplified by Beyer et al.,181 who used GPU-based raycasting to display fused multimodal brain datasets in real time for preoperative planning of neurosurgical procedures, there is a trend toward using TF-based methods for anatomical feature enhancement, tissue separation, and obtaining complementary diagnostic information from different image modalities. In a GPU-based raycasting framework, Weber et al.182 segmented a volume into regions in the form of branches of a hierarchical contour tree, and created a unique TF for each of these regions. These contour tree-based segmentations were used to optimize the TF design for highlighting the volume-rendered anatomical structures of thoracic CT data, and Termeer et al.183 used this rendering technique to build a visualization software platform for the diagnosis of coronary artery disease. The GPU-based raycasting algorithm was also used by Rieder et al.184 to display multimodal volume data for neurosurgical tumor treatment. They designed individual TFs for each imaging modality, and proposed a distance-based enhancement technique for functional data display and lesion detection, as well as for path to target exploration.
The second main application is volume illustration, applying semantic information to simplify the complexity in the transfer function design and intuitively revealing anatomical structures. Rezk-Salama and colleagues67 designed some high-level semantic models for intuitive TF parameter assignments. Rautek et al.68 introduced a fuzzy-logic rule based semantic layer to linguistically define a mapping from volumetric attributes to visual styles, and later extended their work89 by introducing an interaction-dependent semantic model, and employing graphics hardware to accelerate the fuzzy-logic arithmetic evaluation during the semantic parameter specification process.
The third main application trend is the real-time display of specific tissues or organs. For example, Ljung185 used this rendering approach to interactively visualize a multi-detector CT-scanned human cadaver to perform a “virtual autopsy.” To accelerate lung cancer screening, Wang and her colleagues186 developed a software platform based on GPU-accelerated raycasting for displaying volumetric CT images stereoscopically at interactive rates. Krüger et al.187 used GPU-based rendering technique with MPR to realistically represent volumetric images in a virtual endoscopy application, directly visualizing the individual patient data without time-consuming segmentation. This hardware-accelerated volume visualization technique was also exploited with a vessel detection algorithm by Joshi et al.103 to display vascular structures in an angiographic image, while Mayerich et al.188 designed an anatomical structure encoding algorithm that is used with a GPU-accelerated raycasting to detect and visualize the microvascular network and its anatomical relationships with the surrounding tissues in real time. Recently, Lee and Shin189 employed the GPU-based volume-rendering algorithm for fast perspective visualization in a virtual colonoscopy application, while Lim et al.190 used a GPU-based raycasting to visualize 3D ultrasound datasets, during which points within the native pyramidal ultrasound coordinate system were adaptively converted on the fly to a Cartesian grid.
Finally, a DVR system191,192 was developed in our group, which is based on a GPU-accelerated raycasting. In this system, a novel dynamic texture binding technique was employed to render 4D cardiac datasets with the heart rate being synchronized with the electrocardiogram signal. Subsequently, techniques were developed for dynamic volume rendering, manipulation, and enhancement.193,194 Furthermore, a new algorithm was presented for rendering segmented datasets with anatomical feature enhancement.195,196 When compared with previously published results, instead of using different TF for segmented object separation, the new algorithm “paints” various colors acquired from segmentation to the rendered volume, and delivers a higher image quality with smoother subvolume boundaries. Recently, to facilitate image manipulation and image-guided intervention, a new “depth texture indexing” algorithm was developed197 to allow virtual surgical tools to be included into the 4D medical image visualization environment.
Improvements
The increasing capabilities of modern GPUs provide great potential to enhance the performance of the GPU-based raycasting algorithms. For example, the intrinsically parallel nature of GPUs was exploited by Roettger et al.56 to build a real-time raycaster, in which adaptive pre-integrated voxel classification and volume clipping were included. The original GPU-based solutions were based on multiple rendering passes, casting rays through the volume and storing the intermediate results in a temporary framebuffer, which could be accessed in subsequent rendering passes. However, on current graphics hardware architecture, such a multi-pass-based rendering pipeline suffers from low efficiency. To address this issue, Stegmaier and colleagues built a single-pass raycasting framework and extended this framework to spectral DVR for refractive material selective absorption and dispersion using the Kubelka–Munk theory.101
For single-pass raycasting, Hadwiger et al.20 employed this visualization technique to display implicit isosurfaces from a volume defined on a rectilinear 3D grid. A two-level hierarchical grid representation was exploited to allow object- and image-order empty space skipping to be employed to improve the rendering performance, and to circumvent the memory limitations in visualizing datasets that are too large to fit into GPU texture memory. For the similar task of releasing on-board memory size restrictions, Wang and JaJa198 recently proposed a multicore processor-based raycasting algorithm for isosurface extraction and rendering. In addition, exploded views were introduced into the raycasting algorithm by Bruckner and Gröller199 to display inner occluded object of interest. Their approach employed a force-based model to partition the volume into several segments for displaying hidden structural details, and the partition manner was controlled by a number of forces and constraints. Similarly, to reveal the interior hidden spatial structures, opacity peeling was employed by Rezk-Salama and Kolb200 in the rendering pipeline. For the purpose of emphasizing objects of interest within the contextual volume, Viola et al.201 presented an importance-driven strategy, in which intelligent viewpoint selection and alternative visual representations of the volume were exploited. Furthermore, the importance-driven approach was also used by Wang et al.202 to visualize time-varying data for the purpose of unveiling and enhancing the dynamic features exhibited by these 4D volumetric data.
Compared with multi-pass rendering,56 a single-pass solution has the advantages of efficiency, accuracy, and flexibility. Klein et al.203 proposed techniques to improve the performance and quality of GPU-based volume raycasting. An empty space skipping algorithm that exploits the spatial coherence between consecutively rendered images was employed to estimate the optimal initial ray sampling point for each image pixel. A similar idea to exploit the empty space skipping and frame-to-frame coherence was later used by Lee and Shin189 to accelerate GPU-based raycasting; however, instead of using super-sampling, they designed a new algorithm to improve image quality by eliminating the artifacts caused by holes or illegal starting positions. Müller et al.204 presented a sort-last parallel architecture to accelerate single-pass volume raycasting on GPU, wherein they partitioned the texture volume into uniform texture bricks. A similar volumetric texture bricking and octree organization technique was later used by Gobbetti et al.205 to improve the performance of GPU raycasting on massive datasets. Furthermore, to render large out-of-core datasets, Moloney et al.206 presented a volume bricking-based sort-first algorithm to balance parallelized data loading and frame-to-frame coherence. The bricked volume technique was also used by Beyer et al.181 to display multimodal volume using GPU raycasting. Similarly, Ljung185 presented a multiresolution scheme to render large out-of-core CT human cadaver volumes using GPU raycasting, in which techniques such as load-on-demand data management and TF-based data reduction were employed. In addition, to address the difficulties of rendering large datasets on a GPU, a real-time volume compression and decompression technique based on block transformation coding was proposed by Fout and Ma.207 In the GPU-based raycasting pipeline, to render large volumetric data and improve image quality, Wang et al.208 developed a level-of-detail selection algorithm using an image-based quality metric. In addition, to provide novice audiences access to interactive volume renditions, Rezk-Salama et al.92 introduced a technique to generate high-quality light field representation and integrated it into the GPU-based raycasting pipeline.
Fourier Domain Volume Rendering
In Fourier transform (FT) based DVR, the rendering computation is shifted to the spatial frequency domain to create line integral projections of a 3D scalar field according to the projection-slice theorem.209,210 The goal is to compute 2D Fourier projections of 3D data, reducing the computational complexity that exists in spatial domain. Levoy209 and Malzbender210 independently applied this theorem to DVR, while Napel et al.211 proposed an application specific to MR angiography. Additionally, Jansen et al.212 performed Fourier domain volume rendering (FDVR) directly on the graphics hardware by using the split-stream fast Fourier transform (FFT) to map the recursive structure of the FFT to the GPU.
While GPU-accelerated FDVR is efficient, this algorithm has three drawbacks. First, it has limited ability for 3D texture handling; secondly, two copies of the data must be present in the graphics memory, one as the source and the other one as the destination array; finally, it is not possible to implement any local (image-space) operations in the Fourier domain, eliminating the possibility of incorporating lighting models.
Discussion and Outlook
Discussion
DVR is an efficient technique to explore complex anatomical structures within volumetric medical data, with requirement for real-time volumetric and dynamic image visualization; techniques to address these requirements must keep pace with the development of related technologies, and failure to do so will result in visualization being the rate-limiting step for the development of new treatment modalities. Real-time DVR of clinical datasets needs efficient data structures, algorithms, parallelization, and hardware acceleration. The progress of programmable GPUs has dramatically accelerated the performance and medical applications of volume visualization, and opened a new door for future developments of real-time 3D and 4D rendering techniques.
As the capabilities of programmable GPUs continue to increase, the distinction between algorithms that are appropriate for the GPU and those written for the CPU will disappear. The evolution from software-based raycasting to GPU-based raycasting is a good example. For DVR algorithms such as splatting, shell rendering, and shear-warp, hardware accelerations have been proposed and implemented; however, most of these improvements are based on fixed graphics pipelines and do not take full advantages of the programmable features of GPU. As mentioned previously, even if the TM-based DVR algorithms make use of graphics hardware in the volume-rendering pipeline, they differ considerably from the programmable GPU algorithms, such as raycasting, in that all of the volume-rendering computations and corresponding acceleration techniques are implemented on the GPU fragment shaders. The TM-based DVR algorithms only use fixed graphics pipelines that include relative simple texture operations, such as texture extraction, blending, and interpolation.
Since most DVR algorithms are application driven, and medicine is a major application area, in this paper, we described the general techniques of volumetric data visualization with a focus on medical applications. However, for a specific application, different DVR algorithms may generally be used interchangeably when sufficient image quality and rendering performance are available. For the evaluation and comparison of DVR algorithms, Meißner et al.115 quantitatively analyzed four algorithms: software-based raycasting, splatting, shear-warp, and 3DTM. Recently, Boucheny et al.213 presented three perceptual studies to examine the information transformation effectiveness of DVR algorithms, and unsurprisingly, they showed that a static representation is ambiguous. When tuning the rendering parameters, this ambiguity can be counterbalanced by using depth cues (i.e., obtained through motion parallax or stereoscopy). Furthermore, the quantitative evaluation of the four mainstream DVR algorithms was also reported in a publication of our group.197
Outlook
Modern GPUs support the Shader Model 4.0 features and floating point precision throughout the graphics pipeline. Currently, most of the advanced features of programmable GPUs are accessible through high-level programming languages, such as OpenGL, HLSL, GLSL, and Cg. A promising research direction is in the design of new image rendering and manipulation algorithms, specially for programmable GPU architectures, taking advantage of cache coherence and parallelism to achieve efficiency. With the newly released GPU programmable APIs CUDA178 and CTM,179 more complex algorithms can efficiently be implemented on GPUs, and we believe that these two APIs will more efficiently support graphics applications in the future.
In addition, to further enhance the visualization environment, DVR algorithms should be effectively combined with realistic haptic (i.e., force) feedback and crucial diagnostic and functional information extracted from medical data using new algorithms in the research fields such as machine learning and pattern recognition. Furthermore, innovative techniques of robotics, human–computer interaction, and computational vision should be developed to facilitate the medical image exploration, interpretation, processing, and analysis.
Finally, we note that the current volume visualization algorithms should be integrated into standard graphical and imaging processing frameworks such as the Insight Segmentation and Registration Toolkit 214 and the Visualization ToolKit,215 enabling them to be readily used in clinical medical imaging applications. We also note that even though there have been many efficient and advanced techniques developed for all parts of the volume-rendering pipeline, only a few simple approaches have been developed for clinical visualization. The translation of these advances into clinical practice nevertheless remains a problem. However, we must ensure that such challenges are addressed so as to not introduce roadblocks with respect to related technologies such as image-guided interventions.
Acknowledgments
The authors would like to thank J. Moore, C. Wedlake, and Dr. U. Aladl for technical support and assistance; K. Wang and Dr. M. Wachowiak for valuable discussions and assistance; and Ms. J. Williams for editorial assistance. They would also like to thank the anonymous editors and reviewers. This project was supported by the Canadian Institutes for Health Research (Grant MOP 74626), the National Science and Engineering Research Council of Canada (Grant R314GA01), the Ontario Research and Development Challenge Fund, the Canadian Foundation for Innovation and the Ontario Innovation Trust, as well as the Graduate Scholarships from the Ontario Ministry of Education and the University of Western Ontario.
References
- 1.Preim B, Bartz D. Visualization in Medicine: Theory, Algorithms, and Applications (The Morgan Kaufmann Series in Computer Graphics) 1. San Mateo: Morgan Kaufmann; 2007. [Google Scholar]
- 2.Robb RA, Hanson DP. Biomedical image visualization research using the visible human datasets. Clin Anat. 2006;19(3):240–253. doi: 10.1002/ca.20332. [DOI] [PubMed] [Google Scholar]
- 3.Linsen L, Hagen H, Hamann B. Visualization in Medicine and Life Sciences (Mathematics and Visualization) 1. Berlin: Springer; 2007. [Google Scholar]
- 4.Vidal F, et al. Principles and applications of computer graphics in medicine. Comput Graph Forum. 2006;25(1):113–137. [Google Scholar]
- 5.Udupa J, Herman G. 3D imaging in medicine. 2. Boca Raton: CRC; 2000. [Google Scholar]
- 6.Peters TM. Topical review: images for guidance for surgical procedures. Phys Med Biol. 2006;51(14):R505–R540. doi: 10.1088/0031-9155/51/14/R01. [DOI] [PubMed] [Google Scholar]
- 7.Sielhorst T, Feuerstein M, Navab N. Advanced medical displays: a literature review of augmented reality. J Display Technol. 2008;4(4):451–467. [Google Scholar]
- 8.Baek SY, Sheafor DH, Keogan MT, DeLong DM, Nelson RC. Two-dimensional multiplanar and three-dimensional volume-rendered vascular CT in pancreatic carcinoma: interobserver agreement and comparison with standard helical techniques. Am J Roentgenol. 2001;176(6):1467–1473. doi: 10.2214/ajr.176.6.1761467. [DOI] [PubMed] [Google Scholar]
- 9.Shekhar R, Zagrodsky V. Cine MPR: interactive multiplanar reformatting of four-dimensional cardiac data using hardware-accelerated texture mapping. IEEE Trans Inf Technol Biomed. 2003;7(4):384–393. doi: 10.1109/titb.2003.821320. [DOI] [PubMed] [Google Scholar]
- 10.Roos JE, Fleischmann D, et al. Multipath curved planar reformation of the peripheral arterial tree in CT angiography. Radiology. 2007;244:281–290. doi: 10.1148/radiol.2441060976. [DOI] [PubMed] [Google Scholar]
- 11.Lv X, Gao X, Zou H. Interactive curved planar reformation based on snake model. Comput Med Imaging Graph. 2008;32(8):662–669. doi: 10.1016/j.compmedimag.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 12.Ooijen PMA, Ho KY, Dorgelo J, Oudkerk M. Coronary artery imaging with multidetector CT: visualization issues. Radiographics. 2003;23:e16. doi: 10.1148/rg.e16. [DOI] [PubMed] [Google Scholar]
- 13.Kim HC, Yang DM, Jin W, Park SJ. Added diagnostic value of multiplanar reformation of multidetector CT data in patients with suspected appendicitis. Radiographics. 2008;28:393–405. doi: 10.1148/rg.282075039. [DOI] [PubMed] [Google Scholar]
- 14.Williams D, Grimm S, Coto E, Roudsari A, Hatzakis H. Volumetric curved planar reformation for virtual endoscopy. IEEE Trans Vis Comput Graph. 2008;14(1):109–119. doi: 10.1109/TVCG.2007.1068. [DOI] [PubMed] [Google Scholar]
- 15.Rodallec MH, Marteau V, Gerber S, Desmottes L, Zins M. Craniocervical arterial dissection: spectrum of imaging findings and differential diagnosis. Radiographics. 2008;28:1711–1728. doi: 10.1148/rg.286085512. [DOI] [PubMed] [Google Scholar]
- 16.Joemai RMS, Geleijns J, Veldkamp WJH, Roos A, Kroft LJM. Automated cardiac phase selection with 64-MDCT coronary angiography. AJR Am J Roentgenol. 2008;191:1690–1697. doi: 10.2214/AJR.08.1039. [DOI] [PubMed] [Google Scholar]
- 17.Tiede U, et al. Surface rendering. IEEE Comput Graph Appl. 1990;10(2):41–53. [Google Scholar]
- 18.Tatarchuk N, Shopf J, Coro C. Advanced interactive medical visualization on the GPU. J Parallel Distrib Comput. 2008;68(10):1319–1328. [Google Scholar]
- 19.Buchart C, Borro D, Amundarain A. GPU local triangulation: an interpolating surface reconstruction algorithm. Comput Graph Forum. 2008;27(3):807–814. [Google Scholar]
- 20.Hadwiger M, Sigg C, Scharsach H, Bühler K, Gross M. Realtime ray-casting and advanced shading of discrete isosurfaces. Comput Graph Forum. 2005;24(3):303–312. [Google Scholar]
- 21.Hirano M, Itoh T, Shirayama S. Numerical visualization by rapid isosurface extractions using 3D span spaces. J Vis. 2008;11(3):189–196. [Google Scholar]
- 22.Petrik S, Skala V. Technical section: space and time efficient isosurface extraction. Comput Graph. 2008;32(6):704–710. [Google Scholar]
- 23.Zhang X, Bajaj C. Scalable isosurface visualization of massive datasets on commodity off-the-shelf clusters. J Parallel Distrib Comput. 2009;69(1):39–53. doi: 10.1016/j.jpdc.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim S, Choi B, Kim S, Lee J. Three-dimensional imaging for hepatobiliary and pancreatic diseases: emphasis on clinical utility. Indian J Radiol Imaging. 2009;19:7–15. doi: 10.4103/0971-3026.45336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schellinger PD, Richter G, Köhrmann M, Dörfler A. Noninvasive angiography (magnetic resonance and computed tomography) in the diagnosis of ischemic cerebrovascular disease. Cerebrovasc Dis. 2007;24(1):16–23. doi: 10.1159/000107375. [DOI] [PubMed] [Google Scholar]
- 26.Park SH, Choi EK, et al. Linear polyp measurement at CT colonography: 3D endoluminal measurement with optimized surface-rendering threshold value and automated measurement. Radiology. 2008;246:157–167. doi: 10.1148/radiol.2453061930. [DOI] [PubMed] [Google Scholar]
- 27.Drebin R, Carpenter L, Hanrahan P: Volume rendering. In: Proceedings SIGGRAPH88, 1988, pp 65–74
- 28.Johnson C, Hansen C. Visualization Handbook. Orlando: Academic; 2004. [Google Scholar]
- 29.Hadwiger M, Kniss JM, Rezk-salama C, Weiskopf D, Engel K. Real-Time Volume Graphics. 1. Natick: A. K. Peters; 2006. [Google Scholar]
- 30.Levoy M. Display of surfaces from volume data. IEEE Comput Graph Appl. 1988;8(3):29–37. [Google Scholar]
- 31.Höhne KH, Bomans M, Pommert A, Riemer M, Schiers C, Tiede U, Wiebecke G. 3D visualization of tomographic volume data using the generalized voxel model. Vis Comput. 1990;6(1):28–36. [Google Scholar]
- 32.Krüger J, Westermann R: Acceleration techniques for GPU-based volume rendering. In: VIS’03: Proceedings of the 14th IEEE Visualization 2003 (VIS’03), IEEE Computer Society, Washington, DC, USA, 2003, pp. 287–292
- 33.Westover L: Interactive volume rendering. In: Proceedings of the 1989 Chapel Hill Workshop on Volume visualization, ACM, 1989, pp 9–16
- 34.Westover L: Footprint evaluation for volume rendering. In: SIGGRAPH ’90: Proceedings of the 17th annual conference on Computer graphics and interactive techniques, ACM Press, New York, NY, USA, 1990, pp. 367–376
- 35.Udupa JK, Odhner D. Shell rendering. IEEE Comput Graph Appl. 1993;13(6):58–67. [Google Scholar]
- 36.Gelder AV, Kim K: Direct volume rendering with shading via three-dimensional textures. In: Proc. of the 1996 Symposium on Volume Visualization. IEEE, 1996, pp 23–30
- 37.Marroquim R, Maximo A, Farias RC, Esperança C. Volume and isosurface rendering with GPU-accelerated cell projection. Comput Graph Forum. 2008;27(1):24–35. [Google Scholar]
- 38.Lacroute P, Levoy M. Fast volume rendering using a shear-warp factorization of the viewing transformation. Comput Graph. 1994;28(Annual Conference Series):451–458. [Google Scholar]
- 39.Mroz L, Hauser H, Gröller E. Interactive high-quality maximum intensity projection. Comput Graph Forum. 2000;19(3):341–350. [Google Scholar]
- 40.Robb R. X-ray computed tomography: from basic principles to applications. Annu Rev Biophys Bioeng. 1982;11:177–182. doi: 10.1146/annurev.bb.11.060182.001141. [DOI] [PubMed] [Google Scholar]
- 41.Mor-Avi V, Sugeng L, Lang RM. Real-time 3-dimensional echocardiography: an integral component of the routine echocardiographic examination in adult patients? Circulation. 2009;119:314–329. doi: 10.1161/CIRCULATIONAHA.107.751354. [DOI] [PubMed] [Google Scholar]
- 42.Ahmetoglu A, Kosucu P, Kul S, et al. MDCT cholangiography with volume rendering for the assessment of patients with biliary obstruction. Am J Roentgenol. 2004;183:1327–1332. doi: 10.2214/ajr.183.5.1831327. [DOI] [PubMed] [Google Scholar]
- 43.Yeh C, Wang J-F, Wu M-T, Yen C-W, Nagurka ML, Lin C-L. A comparative study for 2D and 3D computer-aided diagnosis methods for solitary pulmonary nodules. Comput Med Imaging Graph. 2008;32(4):270–276. doi: 10.1016/j.compmedimag.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 44.Fishman EK, Horton KM, Johnson PT. Multidetector CT and three-dimensional CT angiography for suspected vascular trauma of the extremities. Radiographics. 2008;28:653–665. doi: 10.1148/rg.283075050. [DOI] [PubMed] [Google Scholar]
- 45.Singh AK, Sahani DV, Blake MA, Joshi MC, Wargo JA, Castillo CF. Assessment of pancreatic tumor respectability with multidetector computed tomography: semiautomated console-generated images versus dedicated workstation-generated images. Acad Radiol. 2008;15(8):1058–1068. doi: 10.1016/j.acra.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 46.Fishman EK, Ney DR, et al. Volume rendering versus maximum intensity projection in CT angiography: what works best, when, and why. Radiographics. 2006;26:905–922. doi: 10.1148/rg.263055186. [DOI] [PubMed] [Google Scholar]
- 47.Gill RR, Poh AC, Camp PC, et al. MDCT evaluation of central airway and vascular complications of lung transplantation. Am J Roentgenol. 2008;191:1046–1056. doi: 10.2214/AJR.07.2691. [DOI] [PubMed] [Google Scholar]
- 48.Blinn JF. Light reflection functions for simulation of clouds and dusty surfaces. Comput Graph. 1982;16(3):21–29. [Google Scholar]
- 49.Max N. Optical models for direct volume rendering. IEEE Trans Vis Comput Graph. 1995;1(2):99–108. [Google Scholar]
- 50.Rezk-Salama C: Volume Rendering Techniques for General Purpose Graphics Hardware. Ph.D. thesis, University of Siegen, Germany, 2001
- 51.Engel K, Kraus M, Ertl T: High-quality pre-integrated volume rendering using hardware-accelerated pixel shading. In: Eurographics/SIGGRAPH Workshop on Graphics Hardware ’01, Annual Conference Series, Addison-Wesley Publishing Company, Inc., 2001, pp 9–16
- 52.Kraus M: Direct Volume Visualization of Geometrically Unpleasant Meshes. Ph.D. thesis, Universität Stuttgart, Germany, 2003
- 53.Kraus M, Ertl T. Pre-integrated volume rendering. In: Hansen C, Johnson C, editors. The Visualization Handbook. New York: Academic; 2004. pp. 211–228. [Google Scholar]
- 54.Röttger S, Kraus M, Ertl T: Hardware-accelerated volume and isosurface rendering based on cell-projection. In: VIS ’00: Proc. of the Conf. on Visualization ’00. IEEE Computer Society, Los Alamitos, 2000, pp 109–116
- 55.Max N, Hanrahan P, Crawfis R. Area and volume coherence for efficient visualization of 3D scalar functions. SIGGRAPH Comput Graph. 1990;24(5):27–33. [Google Scholar]
- 56.Roettger S, Guthe S, Weiskopf D, Ertl T, Strasser W: Smart hardware-accelerated volume rendering. In: Proc. of the Symposium on Data Visualisation 2003. Eurographics Association, 2003, pp 231–238
- 57.Zhang Q, Eagleson R, Peters TM: Rapid voxel classification methodology for interactive 3D medical image visualization. In: MICCAI (2), 2007, pp 86–93 [DOI] [PubMed]
- 58.Kye H, Shin B-S, Shin YG. Interactive classification for pre-integrated volume rendering of high-precision volume data. Graph Models. 2008;70(6):125–132. [Google Scholar]
- 59.Hajjar J-FE, Marchesin S, Dischler J-M, Mongenet C: Second order pre-integrated volume rendering. In: PacificVis, 2008, pp 9–16
- 60.Kraus M: Pre-integrated volume rendering for multidimensional transfer functions. In: IEEE/EG Symposium on Volume and Point-based Graphics, 2008, pp 97–104
- 61.Sato Y, Westin C-F, Bhalerao A, Nakajima S, Shiraga N, Tamura S, Kikinis R. Tissue classification based on 3D local intensity structures for volume rendering. IEEE Trans Vis Comput Graph. 2000;6:2000. [Google Scholar]
- 62.Pfister H, Lorensen B, Bajaj C, Kindlmann G, Schroeder W, Avila LS, Martin K, Machiraju R, Lee J. The transfer function bake-off. IEEE Comput Graph Appl. 2001;21(3):16–22. [Google Scholar]
- 63.Kniss J, Kindlmann GL, Hansen CD. Multidimensional transfer functions for interactive volume rendering. IEEE Trans Vis Comput Graph. 2002;8(3):270–285. [Google Scholar]
- 64.Abellán P, Tost D. Multimodal volume rendering with 3D textures. Comput Graph. 2008;32(4):412–419. [Google Scholar]
- 65.Hadwiger M, Laura F, Rezk-Salama C, Höllt T, Geier G, Pabel T. Interactive volume exploration for feature detection and quantification in industrial CT data. IEEE Trans Vis Comput Graph. 2008;14(6):1507–1514. doi: 10.1109/TVCG.2008.147. [DOI] [PubMed] [Google Scholar]
- 66.Petersch B, Hadwiger M, Hauser H, Hönigmann D. Real time computation and temporal coherence of opacity transfer functions for direct volume rendering of ultrasound data. Comput Med Imaging Graph. 2005;29(1):53–63. doi: 10.1016/j.compmedimag.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 67.Rezk-Salama C, Keller M, Kohlmann P. High-level user interfaces for transfer function design with semantics. IEEE Trans Vis Comput Graph. 2006;12(5):1021–1028. doi: 10.1109/TVCG.2006.148. [DOI] [PubMed] [Google Scholar]
- 68.Rautek P, Bruckner S, Gröller E. Semantic layers for illustrative volume rendering. IEEE Trans Vis Comput Graph. 2007;13(6):1336–1343. doi: 10.1109/TVCG.2007.70591. [DOI] [PubMed] [Google Scholar]
- 69.Freiman M, Joskowicz L, Lischinski D, Sosna J. A feature-based transfer function for liver visualization. Int J CARS. 2007;2(1):125–126. [Google Scholar]
- 70.Pinto FM, Freitas CMDS. Volume visualization and exploration through flexible transfer function design. Comput Graph. 2008;32(5):540–549. [Google Scholar]
- 71.Zhang Q, Eaglesona R, Peters TM: Graphics hardware based volumetric medical dataset visualization and classification. In: Proc. of SPIE Med. Imaging, Vol. 6141, San Diego, CA, 2006, pp 61412T-1–11
- 72.Bruckner S, Gröller ME. Style transfer functions for illustrative volume rendering. Comput Graph Forum. 2007;26(3):715–724. [Google Scholar]
- 73.Bruckner S, Gröller E. Enhancing depth-perception with flexible volumetric halos. IEEE Trans Vis Comput Graph. 2007;13(6):1344–1351. doi: 10.1109/TVCG.2007.70555. [DOI] [PubMed] [Google Scholar]
- 74.Wu Y, Qu H. Interactive transfer function design based on editing direct volume rendered images. IEEE Trans Vis Comput Graph. 2007;13(5):1027–1040. doi: 10.1109/TVCG.2007.1051. [DOI] [PubMed] [Google Scholar]
- 75.Correa C, Ma K-L. Size-based transfer functions: a new volume exploration technique. IEEE Trans Vis Comput Graph. 2008;14(6):1380–1387. doi: 10.1109/TVCG.2008.162. [DOI] [PubMed] [Google Scholar]
- 76.Caban J, Rheingans P. Texture-based transfer functions for direct volume rendering. IEEE Trans Vis Comput Graph. 2008;14(6):1364–1371. doi: 10.1109/TVCG.2008.169. [DOI] [PubMed] [Google Scholar]
- 77.Rezk-Salama C. Visual parameters and transfer functions. In: Zudilova-Seinstra E, Adriaansen T, Liere R, editors. Trends in Interactive Visualization: State-of-the-Art Survey. Berlin: Springer; 2008. pp. 99–116. [Google Scholar]
- 78.Laidlaw DH, Fleischer KW, Barr AH. Partial-volume bayesian classification of material mixtures in MR volume data using voxel histograms. IEEE Trans Med Imaging. 1998;17(1):74–86. doi: 10.1109/42.668696. [DOI] [PubMed] [Google Scholar]
- 79.Lundström C, Ljung P, Ynnerman A. Local histograms for design of transfer functions in direct volume rendering. IEEE Trans Vis Comput Graph. 2006;12(6):1570–1579. doi: 10.1109/TVCG.2006.100. [DOI] [PubMed] [Google Scholar]
- 80.Souza A, Udupa JK, Saha PK. Volume rendering in the presence of partial volume effects. IEEE Trans Med Imaging. 2005;24(2):223–235. doi: 10.1109/tmi.2004.840295. [DOI] [PubMed] [Google Scholar]
- 81.Jani AB, Johnstone PAS, Fox T, Pelizzari C. Optimization of opacity function for computed tomography volume rendered images of the prostate using magnetic resonance reference volumes. Int J Comput Assist Radiol Surg. 2007;1(5):285–293. [Google Scholar]
- 82.Mora B, Ebert DS. Low-complexity maximum intensity projection. ACM Trans Graph. 2005;24(4):1392–1416. [Google Scholar]
- 83.Kye H, Jeong D. Accelerated MIP based on GPU using block clipping and occlusion query. Comput Graph. 2008;32(3):283–292. [Google Scholar]
- 84.Rubin GD, Rofsky NM. CT and MR Angiography: Comprehensive Vascular Assessment. 1. Baltimore: Williams & Wilkins; 2008. [Google Scholar]
- 85.Blinn JF. Compositing. 1. Theory. IEEE Trans Vis Comput Graph. 1994;14(5):83–87. [Google Scholar]
- 86.Kautz J. Hardware lighting and shading: a survey. Comput Graph Forum. 2004;23(1):85–112. [Google Scholar]
- 87.Kniss J, Premoze S, Hansen CD, Shirley P, McPherson A. A model for volume lighting and modeling. IEEE Trans Vis Comput Graph. 2003;9(2):150–162. [Google Scholar]
- 88.Weiskopf D, Engel K, Ertl T. Interactive clipping techniques for texture-based volume visualization and volume shading. IEEE Trans Vis Comput Graph. 2003;9(3):298–312. [Google Scholar]
- 89.Rautek P, Bruckner S, Gröller ME. Interaction-dependent semantics for illustrative volume rendering. Comput Graphics Forum. 2008;27(3):847–854. [Google Scholar]
- 90.Silverstein JC, Parsad NM, Tsirline V. Automatic perceptual color map generation for realistic volume visualization. J Biomed Inform. 2008;41(6):927–935. doi: 10.1016/j.jbi.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang L, Giesen J, McDonnell KT, Zolliker P, Mueller K. Color design for illustrative visualization. IEEE Trans Vis Comput Graph. 2008;14(6):1739–1754. doi: 10.1109/TVCG.2008.118. [DOI] [PubMed] [Google Scholar]
- 92.Rezk-Salama C, Todt S, Kolb A. Raycasting of light field galleries from volumetric data. Comput Graph Forum. 2008;27(3):839–846. [Google Scholar]
- 93.Jainek WM, Born S, Bartz D, Straßer W, Fischer J. Illustrative hybrid visualization and exploration of anatomical and functional brain data. Comput Graph Forum. 2008;27(3):855–862. [Google Scholar]
- 94.Svakhine NA, Ebert DS, Andrews WM. Illustration-inspired depth enhanced volumetric medical visualization. IEEE Trans Vis Comput Graph. 2009;15(1):77–86. doi: 10.1109/TVCG.2008.56. [DOI] [PubMed] [Google Scholar]
- 95.Rheingans P, Ebert D. Volume illustration: nonphotorealistic rendering of volume models. IEEE Trans Vis Comput Graph. 2001;7(3):253–264. [Google Scholar]
- 96.Dong F, Clapworthy G. Volumetric texture synthesis for nonphotorealistic volume rendering of medical data. Vis Comput. 2005;21(7):463–473. [Google Scholar]
- 97.Bruckner S, Grimm S, Kanitsar A, Gröller ME. Illustrative context-preserving exploration of volume data. IEEE Trans Vis Comput Graph. 2006;12(6):1559–1569. doi: 10.1109/TVCG.2006.96. [DOI] [PubMed] [Google Scholar]
- 98.Noordmans HJ, Voort HTM, Smeulders AWM. Spectral volume rendering. IEEE Trans Vis Comput Graph. 2000;6(3):196–207. [Google Scholar]
- 99.Bergner S, Möller T, Tory M, Drew MS. A practical approach to spectral volume rendering. IEEE Trans Vis Comput Graph. 2005;11(2):207–216. doi: 10.1109/TVCG.2005.19. [DOI] [PubMed] [Google Scholar]
- 100.Bergner S, Drew MS, Möller T. A tool to create illuminant and reflectance spectra for light-driven graphics and visualization. ACM Trans Graph. 2009;28(1):1–11. [Google Scholar]
- 101.Strengert M, Klein T, Botchen RP, Stegmaier S, Chen M, Ertl T. Spectral volume rendering using GPU-based raycasting. Vis Comput. 2006;22(8):550–561. [Google Scholar]
- 102.Kersten M, Stewart J, Troje N, Ellis R. Enhancing depth perception in translucent volumes. IEEE Trans Vis Comput Graph. 2006;12:1117–1124. doi: 10.1109/TVCG.2006.139. [DOI] [PubMed] [Google Scholar]
- 103.Joshi A, Qian X, Dione DP, Bulsara KR, Breuer CK, Sinusas AJ, Papademetris X. Effective visualization of complex vascular structures using a non-parametric vessel detection method. IEEE Trans Vis Comput Graph. 2008;14(6):1603–1610. doi: 10.1109/TVCG.2008.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ropinski T, Meyer-Spradow J, Diepenbrock S, Mensmann J, Hinrichs K. Interactive volume rendering with dynamic ambient occlusion and color bleeding. Comput Graph Forum. 2008;27(2):567–576. [Google Scholar]
- 105.Mora B, Maciejewski R, Chen M, Ebert DS. Visualization and computer graphics on isotropically emissive volumetric displays. IEEE Trans Vis Comput Graph. 2009;15(2):221–234. doi: 10.1109/TVCG.2008.99. [DOI] [PubMed] [Google Scholar]
- 106.Chan M-Y, Qu H, Chung K-K, Mak W-H, Wu Y. Relation-aware volume exploration pipeline. IEEE Trans Vis Comput Graph. 2008;14(6):1683–1690. doi: 10.1109/TVCG.2008.159. [DOI] [PubMed] [Google Scholar]
- 107.Rautek P, Bruckner S, Gröller E, Viola I. Illustrative visualization: new technology or useless tautology? SIGGRAPH Comput Graph. 2008;42(3):1–8. [Google Scholar]
- 108.Sakas G, Schreyer L-A, Grimm M. Preprocessing and volume rendering of 3D ultrasonic data. IEEE Comput Graph Appl. 1995;15(4):47–54. [Google Scholar]
- 109.Tiede U, Schiemann T, Höhne KH. Visualizing the visible human. IEEE Comput Graph Appl. 1996;16(1):7–9. [Google Scholar]
- 110.Levoy M. Volume rendering by adaptive refinement. Vis Comput. 1990;6(1):2–7. [Google Scholar]
- 111.Levoy M. Efficient ray tracing of volume data. ACM Trans Graph. 1990;9(3):245–261. [Google Scholar]
- 112.Lee RK, Ihm I. On enhancing the speed of splatting using both object-and-image space coherence. Graph models image process. 2000;62(4):263–282. [Google Scholar]
- 113.Grimm S, Bruckner S, Kanitsar A, Gröller ME. A refined data addressing and processing scheme to accelerate volume raycasting. Comput Graph. 2004;28(5):719–729. [Google Scholar]
- 114.Lee RK, Ihm I. On enhancing the speed of splatting using both object- and image-space coherence. Graph Models. 2000;62(4):263–282. [Google Scholar]
- 115.Meißner M, Huang J, Bartz D, Mueller K, Crawfis R: A practical evaluation of popular volume rendering algorithms. In: VVS ’00: Proc. of the 2000 IEEE symposium on Volume Visualization, ACM Press, New York, NY, USA, 2000, pp 81–90
- 116.Higuera FV, Hastreiter P, Fahlbusch R, Greiner G: High performance volume splatting for visualization of neurovascular data. In: IEEE Visualization, 2005, p 35
- 117.Spoerk J, Bergmann H, Wanschitz F, Dong S, Birkfellner W. Fast DRR splat rendering using common consumer graphics hardware. Med Phys. 2007;34(11):4302–4308. doi: 10.1118/1.2789500. [DOI] [PubMed] [Google Scholar]
- 118.Audigier R, Lotufo R, Falcão A. 3D visualization to assist iterative object definition from medical images. Comput Med Imaging Graph. 2006;4(20):217–230. doi: 10.1016/j.compmedimag.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 119.Mueller K, Shareef N, Huang J, Crawfis R. High-quality splatting on rectilinear grids with efficient culling of occluded voxels. IEEE Trans Vis Comput Graph. 1999;5(2):116–134. [Google Scholar]
- 120.Neophytou N, Mueller K: Gpu accelerated image aligned splatting. In: Volume Graphics, 2005, pp 197–205
- 121.Subr K, Diaz-Gutierrez P, Pajarola R, Gopi M: Order Independent, Attenuation-Leakage Free Splatting Using Freevoxels, Tech. Rep. IFI-2007.01, Department of Informatics, University of Zürich, 2007
- 122.Zwicker M, Pfister H, Baar J, Gross MH. EWA splatting. IEEE Trans Vis Comput Graph. 2002;8(3):223–238. [Google Scholar]
- 123.Xue D, Crawfis R. Efficient splatting using modern graphics hardware. J Graphics Tools. 2004;8(3):1–21. [Google Scholar]
- 124.Schulze JP, Niemeier R, Lang U: The perspective shear-warp algorithm in a virtual environment. In: Proc. of the conference on Visualization ’01. IEEE Computer Society, 2001, pp 207–214
- 125.Anagnostou K, Atherton T, Waterfall A: 4D volume rendering with the shear warp factorisation: extensions and quantitative results. In: Proc. of the FifthInt. Conf. on Information Visualisation (IV’01). IEEE Computer Society, 2001, p 435
- 126.Wan M, Zhang N, Qu H, Kaufman AE. Interactive stereoscopic rendering of volumetric environments. IEEE Trans Vis Comput Graph. 2004;10(1):15–28. doi: 10.1109/TVCG.2004.1260755. [DOI] [PubMed] [Google Scholar]
- 127.Sweeney J, Mueller K: Shear-warp deluxe: the shear-warp algorithm revisited. In: VISSYM ’02: Proceedings of the symposium on Data Visualisation. Eurographics Association, Aire-la-Ville, 2002, pp 95–104
- 128.Pfister H, Hardenbergh J, Knittel J, Lauer H, Seiler L: The volumepro real-time ray-casting system. In: Proc. of the 26th Annual Conference on Computer Graphics and Interactive Techniques. ACM/Addison-Wesley, 1999, pp 251–260
- 129.Lim S, Shin B-S: An efficient perspective projection using volumepro. In: Int. Conf. on Computational Science, 2004, pp 396–403
- 130.Schulze JP, Kraus M, Lang U, Ertl T: Integrating pre-integration into the shear-warp algorithm. In: Proc. of the 2003 Eurographics/IEEE TVCG Workshop on Volume graphics. ACM, 2003, pp 109–118
- 131.Kye H, Oh K: High-quality shear-warp volume rendering using efficient supersampling and pre-integration technique. In: ICAT, 2006, pp 624–632
- 132.Grevera GJ, Udupa JK, Odhner D. An order of magnitude faster isosurface rendering in software on a pc than using dedicated, general purpose rendering hardware. IEEE Trans Vis Comput Graph. 2000;6(4):335–345. [Google Scholar]
- 133.Falcão AX, Rocha LM, Udupa JK: A combined approach of shell and shear-warp rendering for efficient volume visualization. In: Proc. of SPIE Med. Imaging, Vol. 5029, 2003, pp 569–580
- 134.Lei T, Udupa JK, Saha PK, Odhner D. Artery–vein separation via MRA—an image processing approach. IEEE Trans Med Imaging. 2001;20(8):689–703. doi: 10.1109/42.938238. [DOI] [PubMed] [Google Scholar]
- 135.Bullitt E, Aylward S. Volume rendering of segmented image objects. IEEE Trans Med Imag. 2002;21(8):998–1002. doi: 10.1109/TMI.2002.803088. [DOI] [PubMed] [Google Scholar]
- 136.Falcao AX, Rocha LM, Udupa JK: A comparative analysis of shell rendering and shear-warp rendering. In: Proc. of SPIE Med. Imaging, Vol. 4681, San Diego, CA, 2002, pp 472–482
- 137.Botha CP, Post FH: Shellsplatting: interactive rendering of anisotropic volumes. In: Proc. of the Symposium on Data Visualization 2003. Eurographics Association, 2003, pp 105–112
- 138.Grevera GJ, Udupa JK, Odhner D: T-shell rendering and manipulation. In: Proc. of SPIE Med. Imaging, Vol. 5744, San Diego, CA, 2005, pp 22–33
- 139.Cullip TJ, Neumann U. Accelerating volume reconstruction with 3D texture hardware, Tech. rep. Chapel Hill: University of North Carolina; 1994. [Google Scholar]
- 140.Cabral B, Cam N, Foran J: Accelerated volume rendering and tomographic reconstruction using texture mapping hardware. In: Proc. of the 1994 Symposium on Volume Visualization. ACM, 1994, pp 91–98
- 141.Sato Y, Westin C-F, Bhalerao A, Nakajima S, Shiraga N, Tamura S, Kikinis R. Tissue classification based on 3D local intensity structures for volume rendering. IEEE Trans Vis Comput Graph. 2000;6(2):160–180. [Google Scholar]
- 142.Hauser H, Mroz L, Bischi GI, Gröller E. Two-level volume rendering. IEEE Trans Vis Comput Graph. 2001;7(3):242–252. [Google Scholar]
- 143.Hadwiger M, Berger C, Hauser H: High-quality two-level volume rendering of segmented data sets on consumer graphics hardware, in: VIS ’03: Proceedings of the 14th IEEE Visualization 2003 (VIS’03), IEEE Computer Society, Washington, DC, USA, 2003, p 40
- 144.Sereda P, Bartrolí AV, Serlie I, Gerritsen FA. Visualization of boundaries in volumetric data sets using lh histograms. IEEE Trans Vis Comput Graph. 2006;12(2):208–218. doi: 10.1109/TVCG.2006.39. [DOI] [PubMed] [Google Scholar]
- 145.Ljung P, Persson A, Ynnerman A, Lundström C. Uncertainty visualization in medical volume rendering using probabilistic animation. IEEE Trans Vis Comput Graph. 2007;13(6):1648–1655. doi: 10.1109/TVCG.2007.70518. [DOI] [PubMed] [Google Scholar]
- 146.Xie K, Sun G, Yang J, Zhu YM. Interactive volume cutting of medical data. Comput Biol Med. 2007;37(8):1155–1159. doi: 10.1016/j.compbiomed.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 147.Kim J, Eberl S, Feng D. Visualizing dual-modality rendered volumes using a dual-lookup table transfer function. Comput Sci Eng. 2008;9(1):20–25. [Google Scholar]
- 148.Holmes D, Davis B, Bruce C, Robb R. 3D visualization, analysis, and treatment of the prostate using trans-urethral ultrasound. Comput Med Imaging Graph. 2003;27(5):339–349. doi: 10.1016/s0895-6111(02)00102-7. [DOI] [PubMed] [Google Scholar]
- 149.Etlik Ö, Temizöz O, Dogan A, Kayan M, Arslan H, Unal Ö. Three-dimensional volume rendering imaging in detection of bone fractures. Eur J Gen Med. 2004;1(4):48–52. [Google Scholar]
- 150.Wenger A, Keefe DF, Zhang S, Laidlaw DH. Interactive volume rendering of thin thread structures within multivalued scientific data sets. IEEE Trans Vis Comput Graph. 2004;10(6):664–672. doi: 10.1109/TVCG.2004.46. [DOI] [PubMed] [Google Scholar]
- 151.Wang A, Narayan G, Kao D, Liang D: An evaluation of using real-time volumetric display of 3D ultrasound data for intracardiac catheter manipulation tasks. Comput Med Imaging Graph 41–45, 2005
- 152.Sharp R, Adams J, Machiraju R, Lee R, Crane R. Physics-based subsurface visualization of human tissue. IEEE Trans Vis Comput Graph. 2007;13(3):620–629. doi: 10.1109/tvcg.2007.1003. [DOI] [PubMed] [Google Scholar]
- 153.Lopera JE, et al. Multidetector CT angiography of infrainguinal arterial bypass. Radiographics. 2008;28:529–548. doi: 10.1148/rg.282075032. [DOI] [PubMed] [Google Scholar]
- 154.Siddiki H, et al. Abdominal findings in hereditary hemorrhagic telangiectasia: pictorial essay on 2D and 3D findings with isotropic multiphase CT. Radiographics. 2008;28:171–183. doi: 10.1148/rg.281075037. [DOI] [PubMed] [Google Scholar]
- 155.Aspin R, Smith M, Hutchinson C, Funk L: Medivol: An initial study into real-time, interactive 3D visualisation of soft tissue pathologies. In: DS-RT ’08: Proceedings of the 2008 12th IEEE/ACM International Symposium on Distributed Simulation and Real-Time Applications. IEEE Computer Society, Washington, 2008, pp 103–110
- 156.Levin D, Aladl U, Germano G, Slomka P. Techniques for efficient, real-time, 3D visualization of multi-modality cardiac data using consumer graphics hardware. Comput Med Imaging Graph. 2005;29(6):463–475. doi: 10.1016/j.compmedimag.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 157.Yuan X, Nguyen MX, Chen B, Porter DH. Hdr volvis: high dynamic range volume visualization. IEEE Trans Vis Comput Graph. 2006;12(4):433–445. doi: 10.1109/TVCG.2006.72. [DOI] [PubMed] [Google Scholar]
- 158.Correa CD, Silver D, Chen M. Feature aligned volume manipulation for illustration and visualization. IEEE Trans Vis Comput Graph. 2006;12(5):1069–1076. doi: 10.1109/TVCG.2006.144. [DOI] [PubMed] [Google Scholar]
- 159.Rezk-Salama C, Engel K, Bauer M, Greiner G, Ertl T: Interactive volume rendering on standard PC graphics hardware using multi-textures and multi-stage rasterization. SIGGRAPH/Eurographics Workshop on Graphics Hardware (August) (2000) 109–118
- 160.Li W, Kaufman A: Accelerating volume rendering with texture hulls. IEEE/SIGGRAPH Symposium on Volume Visualization and Graphics, 2002, 115–122
- 161.Li W, Mueller K, Kaufman A: Empty space skipping and occlusion clipping for texture-based volume rendering. IEEE Vis 317–324, 2003
- 162.Bethune C, Stewart AJ. Adaptive slice geometry for hardware-assisted volume rendering. J Graphics Tools. 2005;10(1):55–70. [Google Scholar]
- 163.Keleş HY, Es A, İşler V. Acceleration of direct volume rendering with programmable graphics hardware. Vis Comput. 2007;23(1):15–24. [Google Scholar]
- 164.Xie K, Yang J, Zhu YM. Real-time visualization of large volume datasets on standard pc hardware. Comput Methods Programs Biomed. 2008;90(2):117–123. doi: 10.1016/j.cmpb.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 165.Frank S, Kaufmanl A. Out-of-core and dynamic programming for data distribution on a volume visualization cluster. Comput Graph Forum. 2009;28(1):141–153. [Google Scholar]
- 166.Lamar EC, Hamann B, Joy KI: Multiresolution techniques for interactive texture-based volume visualization. IEEE Vis 355–362, 1999
- 167.Boada I, Navazo I, Scopigno R. Multiresolution volume visualization with a texture-based octree. Vis Comput. 2001;17(3):185–197. [Google Scholar]
- 168.Käehler R, Hege H-C. Texure-based volume rendering of adaptive mesh refinement data. Vis Comput. 2002;18(8):481–492. [Google Scholar]
- 169.Kähler R, Simon M, Hege H-C. Interactive volume rendering of large sparse data sets using adaptive mesh refinement hierarchies. IEEE Trans Vis Comput Graph. 2003;9(3):341–351. [Google Scholar]
- 170.Gao J, Wang C, Li L, Shen H-W. A parallel multiresolution volume rendering algorithm for large data visualization. Parallel Comput. 2005;31(2):185–204. [Google Scholar]
- 171.Nguyen KG, Saupe D: Rapid high quality compression of volume data for visualization. Comput Graph Forum 20(3), 2001
- 172.Guthe S, Straßer W. Advanced techniques for high-quality multi-resolution volume rendering. Comput Graph. 2004;28(1):51–58. [Google Scholar]
- 173.Xie K, Yang J, Zhu YM. Real-time rendering of 3D medical data sets. Future Gener Comput Syst. 2005;21(4):573–581. [Google Scholar]
- 174.Goodnight N, Wang R, Humphreys G. Computation on programmable graphics hardware. IEEE Comput Graph Appl. 2005;25(5):12–15. doi: 10.1109/mcg.2005.101. [DOI] [PubMed] [Google Scholar]
- 175.Owens JD, Luebke D, Govindaraju N, Harris M, Krger J, Lefohn AE, Purcell TJ. A survey of general-purpose computation on graphics hardware. Comput Graph Forum. 2007;26(1):80–113. [Google Scholar]
- 176.Lefohn AE, Sengupta S, Kniss J, Strzodka R, Owens JD. Glift: generic, efficient, random-access GPU data structures. ACM Trans Graph. 2006;25(1):60–99. [Google Scholar]
- 177.Buck I, Foley T, Horn DR, Sugerman J, Fatahalian K, Houston M, Hanrahan P. Brook for GPUs: stream computing on graphics hardware. ACM Trans Graph. 2004;23(3):777–786. [Google Scholar]
- 178.NVidia: NVidia compute unified device architecture (CUDA) programming guide, version 1.0, 2007
- 179.AMD: “Close to Metal” technology unleashes the power of stream computing: AMD Press Release, 2006
- 180.Fatahalian K, Houston M. A closer look at gpus. Commun ACM. 2008;51(10):50–57. [Google Scholar]
- 181.Beyer J, Hadwiger M, Wolfsberger S, Bühler K. High-quality multimodal volume rendering for preoperative planning of neurosurgical interventions. IEEE Trans Vis Comput Graph. 2007;13(6):1696–1703. doi: 10.1109/TVCG.2007.70560. [DOI] [PubMed] [Google Scholar]
- 182.Dillard SE, Weber GH, Carr H, Pascucci V, Hamann B. Topology-controlled volume rendering. IEEE Trans Vis Comput Graph. 2007;13(2):330–341. doi: 10.1109/TVCG.2007.47. [DOI] [PubMed] [Google Scholar]
- 183.Termeer M, Bescós JO, Breeuwer M, Vilanova A, Gerritsen F, Gröller E. Covicad: comprehensive visualization of coronary artery disease. IEEE Trans Vis Comput Graph. 2007;13(6):1632–1639. doi: 10.1109/TVCG.2007.70550. [DOI] [PubMed] [Google Scholar]
- 184.Rieder C, Ritter F, Raspe M, Peitgen H-O. Interactive visualization of multimodal volume data for neurosurgical tumor treatment. Comput Graph Forum. 2008;27(3):1055–1062. [Google Scholar]
- 185.Ljung P, Winskog C, Persson A, Lundström C, Ynnerman A. Full body virtual autopsies using a state-of-the-art volume rendering pipeline. IEEE Trans Vis Comput Graph. 2006;12(5):869–876. doi: 10.1109/TVCG.2006.146. [DOI] [PubMed] [Google Scholar]
- 186.Wang X, Good W. Real-time stereographic rendering and display of medical images with programmable GPUs. Comput Med Imaging Graph. 2008;13(2):118–123. doi: 10.1016/j.compmedimag.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Krüger A, Kubisch C, Strauß G, Preim B. Sinus endoscopy—application of advanced GPU volume rendering for virtual endoscopy. IEEE Trans Vis Comput Graph. 2008;14(6):1491–1498. doi: 10.1109/TVCG.2008.161. [DOI] [PubMed] [Google Scholar]
- 188.Mayerich D, Abbott L, Keyser J. Visualization of cellular and microvascular relationships. IEEE Trans Vis Comput Graph. 2008;14(6):1611–1618. doi: 10.1109/TVCG.2008.179. [DOI] [PubMed] [Google Scholar]
- 189.Lee TH, Shin YG. Coherence aware GPU-based ray casting for virtual colonoscopy. J Vis Comput Animat. 2009;20(1):1–9. [Google Scholar]
- 190.Lim S, Kwon K, Shin B-S. GPU-based interactive visualization framework for ultrasound datasets. Comput Anim Virtual Worlds. 2009;20(1):11–23. [Google Scholar]
- 191.Zhang Q, Eagleson R, Peters TM: Real-time visualization of 4D cardiac MR images using graphics processing units. In: ISBI, 2006, pp 343–346
- 192.Zhang Q, Eagleson R, Peters T. Dynamic real-time 4D cardiac MDCT image display using GPU-accelerated volume rendering. Comput Med Imaging Graph. 2009;33(6):461–476. doi: 10.1016/j.compmedimag.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 193.Zhang Q, Eagleson R, Peters TM: GPU-based image manipulation and enhancement techniques for dynamic volumetric medical image visualization. In: ISBI, 2007, pp 1168–1171
- 194.Zhang Q, Huang X, Eagleson R, Guiraudon G, Peters TM: Real-time dynamic display of registered 4D cardiac MR and ultrasound images using a GPU. In: Proc. of SPIE Med. Imaging, Vol. 6509. San Diego, CA, USA, 2007, pp 65092D-1–11
- 195.Zhang Q, Eagleson R, Guiraudon G, Peters TM: High-quality anatomical structure enhancement for cardiac image dynamic volume rendering. In: Proc. of SPIE Med. Imaging, Vol. 6509, San Diego, CA, USA, 2008, pp 65092D-1–10
- 196.Zhang Q, Eagleson R, Peters TM. High-quality cardiac image dynamic visualization with feature enhancement and virtual surgical tool inclusion. Vis Comput. 2009;25(11):1019–1035. [Google Scholar]
- 197.Zhang Q, Eagleson R, Peters TM: A software platform for real-time visualization and manipulation of 4D cardiac images. In: FIMH, 2009, pp 396–406
- 198.Wang Q, JaJa J. Interactive high-resolution isosurface ray casting on multicore processors. IEEE Trans Vis Comput Graph. 2008;14(3):603–614. doi: 10.1109/TVCG.2007.70630. [DOI] [PubMed] [Google Scholar]
- 199.Bruckner S, Gröller ME. Exploded views for volume data. IEEE Trans Vis Comput Graph. 2006;12(5):1077–1084. doi: 10.1109/TVCG.2006.140. [DOI] [PubMed] [Google Scholar]
- 200.Rezk-Salama C, Kolb A. Opacity peeling for direct volume rendering. Comput Graph Forum. 2006;25(3):579–606. [Google Scholar]
- 201.Viola I, Feixas M, Sbert M, Gröller ME. Importance-driven focus of attention. IEEE Trans Vis Comput Graph. 2006;12(5):933–940. doi: 10.1109/TVCG.2006.152. [DOI] [PubMed] [Google Scholar]
- 202.Wang C, Yu H, Ma K-L. Importance-driven time-varying data visualization. IEEE Trans Vis Comput Graph. 2008;14(6):1547–1554. doi: 10.1109/TVCG.2008.140. [DOI] [PubMed] [Google Scholar]
- 203.Klein T, Strengert M, Stegmaier S, Ertl T: Exploiting frame-to-frame coherence for accelerating high-quality volume raycasting on graphics hardware. In: IEEE Visualization, 2005, pp 223–230
- 204.Müller C, Strengert M, Ertl T: Optimized volume raycasting for graphics-hardware-based cluster systems. In: Eurographics Symposium on Parallel Graphics and Visualization (EGPGV06), Eurographics Association, 2006, pp 59–66
- 205.Gobbetti E, Marton F, Guitián JAI. A single-pass GPU ray casting framework for interactive out-of-core rendering of massive volumetric datasets. Vis Comput. 2008;24(7–9):797–806. [Google Scholar]
- 206.Moloney B, Weiskopf D, Möller T, Strengert M: Scalable sort-first parallel direct volume rendering with dynamic load balancing. In: Eurographics Symposium on Parallel Graphics and Visualization (EGPGV07), Eurographics Association, 2007, pp 45–52
- 207.Fout N, Ma K-L. Transform coding for hardware-accelerated volume rendering. IEEE Trans Vis Comput Graph. 2007;13(6):1600–1607. doi: 10.1109/tvcg.2007.70516. [DOI] [PubMed] [Google Scholar]
- 208.Wang C, Garcia A, Shen H-W. Interactive level-of-detail selection using image-based quality metric for large volume visualization. IEEE Trans Vis Comput Graph. 2007;13(1):122–134. doi: 10.1109/TVCG.2007.15. [DOI] [PubMed] [Google Scholar]
- 209.Levoy M: Volume rendering using the Fourier projection-slice theorem. In: Proc. of the Conf. on Graphics Interface ’92. Morgan Kaufmann, 1992, pp 61–69
- 210.Malzbender T. Fourier volume rendering. ACM Trans Graph. 1993;12(3):233–250. [Google Scholar]
- 211.Napel S, Dunne S, Rutt BK. Fast Fourier projection for MR angiography. Magn Reson Med. 1991;19:393–405. doi: 10.1002/mrm.1910190230. [DOI] [PubMed] [Google Scholar]
- 212.Jansen T, von Rymon-Lipinski B, Hanssen N, Keeve E: Fourier volume rendering on the GPU using a Split-Stream-FFT. In: VMV, 2004, pp 395–403
- 213.Boucheny C, Bonneau G-P, Droulez J, Thibault G, Ploix S. A perceptive evaluation of volume rendering techniques. ACM Trans Appl Percept. 2009;5(4):1–24. [Google Scholar]
- 214.Ibánez L, Schroeder W, Ng L, Cates J: The Insight Software Consortium, The ITK Software Guide, 2nd edition. Updated for itk version 2.4 (November 2005)
- 215.Schroeder W, Martin K, Lorensen B: The Visualization Toolkit, 3rd edition. Kitware Inc., 2004