SUMMARY
HIV-1 has evolved a cunning mechanism to circumvent the antiviral activity of the APOBEC3 family of host-cell enzymes. The HIV-1 virion infectivity factor, one of several HIV accessory proteins, targets APOBEC3 proteins for proteasomal degradation and down-regulates their expression at the mRNA level. Despite the importance of Vif for HIV-1 infection, there is little conformational data on Vif alone or in complex with other cellular factors due to incompatibilities with many structural techniques and difficulties in producing suitable quantities of protein for biophysical analysis. As an alternative, we have turned to hydrogen exchange mass spectrometry (HX MS), a conformational analysis method well suited for proteins that are difficult to study using X-ray crystallography and/or NMR. HX MS was used to probe the solution conformation of recombinant full-length HIV-1 Vif. Vif specifically interacted with the previously identified binding partner Hck and was able to cause kinase activation suggesting that the Vif studied by HX MS retained a biochemically competent conformation relevant to Hck interaction. HX MS analysis of Vif alone revealed low deuteration levels in the N-terminal portion indicating that this region contained structured or otherwise protected elements. In contrast, high deuteration levels in the C-terminal portion of Vif indicated that this region was likely unstructured in the absence of cellular interacting proteins. Several regions within Vif displayed conformational heterogeneity in solution including the APOBEC3G/F binding site and HCCH zinc finger. Taken together, these HX MS results provide new insights into the solution conformation of Vif.
Keywords: Accessory protein, APOBEC3F/G, E3 Ligase, hydrogen exchange, mass spectrometry, deuterium, Vif
INTRODUCTION
The Human Immunodeficiency Virus (HIV) belongs to the Retroviridae family of viruses and differs from primate retroviruses in that HIV requires the expression of additional proteins besides Gag, Pol, and Env for efficient and productive viral infection (see Refs 1; 2 for reviews). These additional proteins can be classified into regulatory and accessory proteins. The regulatory proteins consist of Tat and Rev and are responsible for viral gene regulation, while the accessory proteins consist of Vif, Vpr, Nef, and Vpu 3; 4. The accessory proteins play a pivotal role in viral pathogenesis by acting as versatile adaptors bridging viral and cellular pathways necessary for infection and immune evasion 5–8.
The HIV-1 Viral (also called Virion) Infectivity Factor (Vif) has been shown to be essential for viral pathogenesis 9–11. Vif is a 22.5 kDa highly basic protein that interacts with an array of both cellular proteins and DNA/RNA 12–16. In the absence of Vif, members of the APOBEC3 family of cytidine deaminases, including APOBEC3F/G, are packaged into HIV virions 14; 17; 18. Upon virus entry into subsequent cells, APOBEC3F/G inhibits viral replication through mechanisms both dependent and independent of its deaminase activity 19; 20. In the deaminase independent mechanism, APOBEC3’s inhibit viral mRNA reverse transcription 19. The deaminase dependent mechanism involves deamination of cytidines to uridines in the (−) strand of the viral DNA, causing crippling G to A hyper-mutation that renders subsequent viral infection non-productive 20. HIV-1 Vif, however, circumvents the antiviral activities of the APOBEC3 proteins by several mechanisms including: 1) inhibition of APOBEC3G mRNA translation 21; 22, 2) promoting the formation of high molecular weight APOBEC3 complexes 23, and 3) by targeted proteasomal degradation wherein Vif links the APOBEC3 enzymes with components of the Elongin BC-Cullin 5 ubiquitin ligase complex 24; 25.
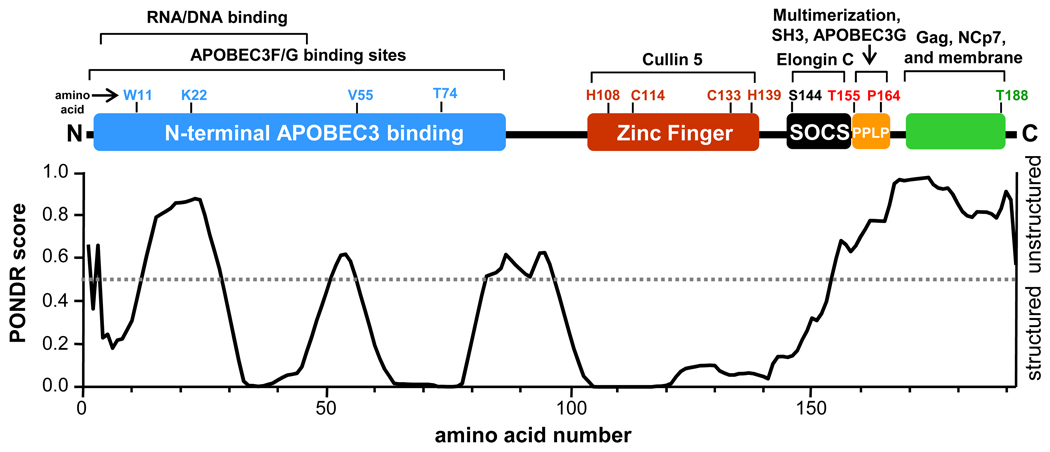
Vif lacks known intrinsic enzymatic activity, and functions instead by interacting directly with APOBEC3 proteins, the ubiquitylation machinery, as well as other cellular factors. Figure 1 summarizes the structural regions of Vif and the known functions ascribed to each region. The N-terminal portion of Vif (Fig. 1, blue) contains several APOBEC3F/G binding motifs and is also essential for interaction DNA/RNA 18; 26–28. There is a novel HCCH zinc finger in the central region of Vif (Fig. 1, red) that is responsible for interaction with the E3 ligase component Cullin5 27; 29; 30. The C-terminal portion of Vif contains multiple motifs including a novel viral Suppressor Of Cytokine Signaling (SOCS) box (Fig. 1, black) which recruits the E3 ligase scaffolding protein complex Elongin BC 31; 32. The PPLP domain (Fig. 1, orange) is just C-terminal to the viral SOCS box and has been implicated in several functions including Vif multerimization 33; 34, interaction with both the tyrosine kinase Hck 35; 36, and APOBEC3G 37. The extreme C-terminal domain of Vif is required for association with gag, NCp7, and the cellular membrane 38.
Figure 1.
Cartoon depicting known HIV-1 Vif functional regions aligned with the PONDR VL-XT prediction of Vif order/disorder 39–41. Interaction partners are described along the top of the figure. The grey dotted line in the PONDR plot denotes a PONDR score of 0.5: scores below 0.5 indicate regions that are likely structured while a score above 0.5 indicates regions that are likely unstructured. The indicated amino acids are provided to show the relative positions of each Vif functional region. This figure and the information therein have been adapted from 33;79.
Structural features of the Vif protein that enable interactions with such a diverse group of host cell factors are not well defined. No three-dimensional structure of full-length Vif has been reported to date, although lower resolution structural techniques and algorithms such as PONDR VL-XT 39–41 indicate that Vif contains some structure spread throughout the protein 42 (Figure 1, bottom). Segments in the N-terminus and the last forty residues in the C-terminus are predicted to be unstructured. These unstructured regions are likely to be conformationally flexible, thus enabling Vif to adopt the correct shape required for interaction with APOBEC3F/G and other host cell factors. Other regions are predicted to contain some structural elements. To characterize the various regions of Vif, both those predicted by algorithms to contain structure and those that are essential for interacting with other proteins, we turned to an alternative biophysical technique for conformational characterization: hydrogen exchange monitored by mass spectrometry (HX MS).
HX MS as a technique is advantageous over other biophysical techniques such as X-ray crystallography and NMR because it is compatible with proteins such as Vif that are aggregation prone and difficult to obtain in suitable quantities for analysis 43. Very little material is required (as little as 500–1000 pmol for an entire experiment) and the concentration of the material can be quite low (as low as 0.1 µM). Thus for proteins that are difficult to purify or otherwise hard to obtain in suitable quantities for characterization by other more classical tools, some information can still be obtained. HX MS cannot be used to solve the structure of a protein, but it can be used to glean conformational information, detect conformational changes, and differentiate protein subpopulations in solution 43; 44. We recently used HX MS to probe the binding between full-length recombinant Vif and the Elongin BC complex 45.
In the current study, HX MS was used to probe Vif conformation in detail. HX MS binding studies and tyrosine kinase assays revealed that recombinant full-length Vif specifically interacted with both the isolated Hck SH3 domain as well as near full-length Hck. Interaction with Vif induced activation of Hck, a member of the Src-kinase family prone to activation by SH3 binding proteins 46, consistent with the hypothesis that recombinant purified Vif adopts a biochemically competent conformation. HX MS analysis of Vif alone revealed low deuteration levels in the N-terminal part of the protein, indicating that this region contained structured and/or protected elements. Conversely, high deuteration levels were found in the C-terminal portion of the protein indicating that this region was likely unstructured in the absence of interacting proteins such as Hck, Elongin BC, and Cullin 5. Several regions within Vif displayed heterogeneous conformations in solution including the APOBEC3G/F binding site and HCCH zinc finger. Such results suggest that recombinant Vif is highly dynamic and likely populates an ensemble of conformational states in solution.
RESULTS AND DISCUSSION
Recombinant Vif retains a biochemically competent conformation amenable to biophysical analysis
Recombinant Vif expressed in E. coli without the aid of solubility enhancement tags (such as GST) localizes to insoluble inclusion bodies 47. Vif in inclusion bodies can be isolated under denaturing conditions, purified with Ni-NTA affinity chromatography and then refolded by subsequent dialysis with buffers containing decreasing concentrations of denaturant 33; 45; 47; 48. Vif produced in this manner is multimeric as shown by size exclusion chromatography and dynamic light scattering 33; 48 but retains essential biological functions such as APOBEC3G binding 42, Elongin BC binding 45, and oligonucleotide binding 15.
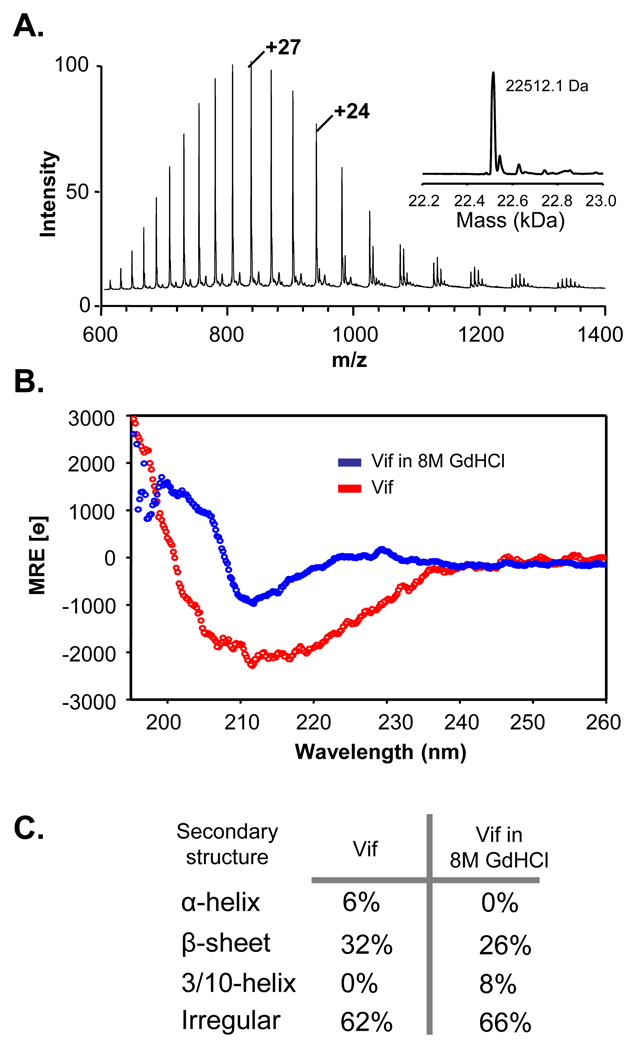
Recombinant full-length Vif was overexpressed in E. coli, isolated from inclusion bodies and refolded, according to the original protocol 47. The refolded recombinant Vif was highly amenable to analysis by ESI-MS, as shown in Figure 2A. To determine if the refolded material had any structure, refolded Vif was analyzed with circular dichroism, as shown in Figure 2B. Refolded Vif was also placed in 8M guanidine hydrochloride and a CD spectrum obtained. Subsequent analysis of the CD spectra with the software program CDPro 49 (Figure 2C) suggested that much of Vif was unstructured, with some secondary structure present as β-sheet. In guanidine hydrochloride, the secondary structure distribution relative to refolded Vif shifted to less alpha helical content, although beta sheet elements were largely retained. Our CD results are consistent with previous work in which the majority of secondary structural elements in recombinant refolded Vif were β-sheet 48.
Figure 2.
Characterization of recombinant, refolded, full-length HIV-1 Vif. (A) Electrospray mass spectrum of 200 pmol of recombinant HIV-1 Vif. The high quality of this spectrum shows that the protein was pure and was amenable to analysis by MS. The inset shows the result of transforming the raw m/z spectrum to the mass only scale. The observed mass (shown) is very close to the theoretical molecular weight of Vif (22512.9 Da). (B) Circular dichroism spectra of recombinant Vif in native buffer (red) and in 8M GdHCl (blue). (C) Secondary structure calculations from the Vif CD spectra in panel B, as determined with the CDPro algorithm 49.
To verify that recombinant full-length Vif expressed and purified according to the Yang protocol 47 had a biochemically competent conformation relevant to interaction with and modulation of the Src family tyrosine kinases, we monitored the interaction between Vif and the tyrosine kinase Hck. We have previously shown that Vif prepared in the same manner bound efficiently to the Elongin BC complex 45. Hck Src homology 3 (SH3) domain binding is a known biochemical function of Vif, and this binding alters Hck kinase activity 35; 36. The SH3 domain is a central regulatory component of Hck and is also targeted by another HIV-1 accessory protein, Nef 50; 51. The Nef:Hck SH3 interaction has been extensively studied using HX MS 46; 51–53. In those prior studies, it was shown that the Hck SH3 domain itself undergoes slow cooperative unfolding in solution which can be monitored with HX MS due to the appearance of two mass distributions corresponding to folded and unfolded species 52; 54. Protein/peptide binding slows the rate of SH3 domain unfolding; therefore, by monitoring the rate of unfolding, protein/peptide binding to SH3 can be detected and quantified 52; 55; 56. To interrogate the ability of recombinant Vif to interact with Hck SH3, the rate of SH3 unfolding was monitored by HX MS.
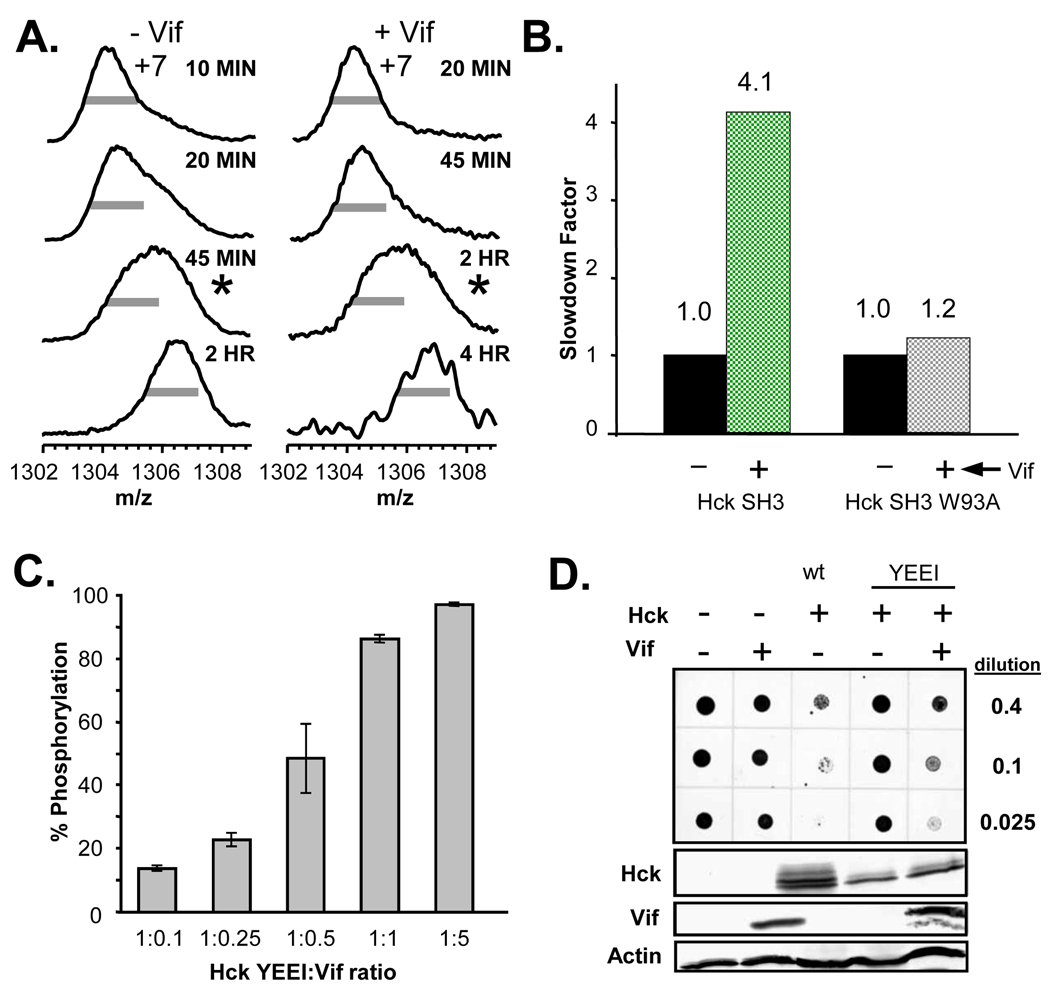
Figure 3 shows the results of the Vif:Hck SH3 binding assay. Hck SH3 was labeled with 2H2O in the absence or presence of Vif. A wide m/z distribution characteristic of protein unfolding 54; 55 in the mass spectra (Fig. 3A, indicated by the asterisks), occurred after approximately 30 minutes of deuterium labeling in the absence of Vif but at a much longer time (~2 hours) in the presence of Vif (note that when Nef binds to Hck SH3, almost all unfolding is abolished until very long incubation times, see Refs. 46; 51–53). This shift in the unfolding half-life indicates that Vif bound to and altered the solution dynamics of Hck SH3. Changes in Hck SH3 unfolding can be quantified by calculating a slow-down factor 46; 53; 56 as shown in Figure 3B. The presence of Vif slowed wild-type Hck SH3 unfolding by a factor of 4.1, a value that is indicative of relatively strong binding 53. To confirm that this Vif:SH3 association was specific, the wild-type Hck SH3:Vif interaction was compared to interaction with a non-binding Hck SH3 mutant. Mutation of W93 to alanine in Hck SH3 abolishes the Vif:SH3 interaction 36 as well as other Hck SH3 binding events with other proteins 57; 58. Slow-down factor analysis for Hck SH3 W93A alone and in the presence of Vif revealed that Vif had little effect on the unfolding of W93A and thus was no longer capable of binding to Hck SH3 W93A in vitro. These results confirm that suppression of Hck SH3 unfolding was a direct result of specific binding between recombinant, refolded Vif and wild-type Hck SH3.
Figure 3.
Recombinant Vif specifically interacted with Hck SH3 and caused kinase activation in vitro and in vivo. (A) Mass spectra showing the unfolding of the Hck SH3 domain during deuterium labeling, in the absence or presence of Vif. The grey bars are of equal length in all cases and are provided as a visual reference for peak width. The half-life for unfolding of Hck SH3 46 was approximately 30 minutes (calculated according to 80) in the absence of Vif and upon incubation with Vif the half-life of SH3 unfolding was shifted to >2 hours. The spectra closest to the calculated unfolding half-life are marked with *. The incubation times in 2H2O are indicated to the right of each mass spectrum. (B) Slowdown factor analysis of wild-type Hck SH3 and Hck SH3 W93A. The slow-down factor was calculated as described in 53. (C) Vif-mediated activation of Hck in vitro. Recombinant Vif was incubated with downregulated, near full length Hck (Hck-YEEI; 51) at the molar ratios indicated. Kinase activity was assessed using a FRET-peptide substrate and the Z’-Lyte assay as described in the Materials and Methods section. Each reaction condition was monitored in quadruplicate, and results are presented as mean percent of control peptide phosphorylation ± S.D. (D) Vif activates Hck in yeast cells. Yeast cultures were transformed with galactose-inducible expression plasmids for HIV Vif, Hck-YEEI, and wild-type Hck (wt) either alone or in the combinations shown. Liquid cultures were normalized for cell density, and equal volumes were spotted on galactose-agar plates at increasing dilutions and incubated at 30 °C for three days. Scanned images of the plates are shown at the top, in which the yeast patches appear as dark circles. Note that co-expression of Vif and Hck inhibits yeast cell growth, indicative of kinase activation. Expression of Vif and Hck proteins was verified by immunoblotting with actin as a loading control (lower panels).
The ability of Vif to associate with the Hck SH3 domain in vitro confirmed that the protein had refolded into a biochemically competent conformation relevant to Hck interaction. This observation raised the question of whether this recombinant Vif could also activate near full-length Hck through SH3 domain displacement as shown previously for Nef 46. As shown in Figure 3C, purified Vif was a remarkably efficient activator of Hck in vitro, with enhanced kinase activity observed at a sub-stoichiometric Vif to kinase molar ratio. Similar results were also obtained for several other Src family tyrosine kinases expressed in HIV target cells, including Lyn and Lck (see Supplemental Material, Figure S1).
To determine whether Vif could activate Hck in a cell-based system, we employed the yeast growth suppression assay previously used to probe Src-family kinase activation by HIV-1 Nef 51; 56. Yeast cells do not express orthologs of mammalian tyrosine kinases, and ectopic expression of Src family members perturbs normal cellular signaling resulting in growth suppression 59; 60. We reasoned that if Vif was co-expressed with a down-regulated form of Hck (Hck YEEI) in yeast, growth would only be suppressed if the kinase became activated by Vif. To test this idea, yeast cultures were transformed with Vif and downregulated Hck either alone or in combination, and spotted on agar plates in the presence of galactose to induce protein expression. As shown in Figure 3D, expression of either Vif or downregulated Hck alone did not alter yeast growth. In contrast, co-expression of Vif and Hck resulted in strong growth suppression, equivalent to that observed with wild-type, active Hck. Suppression of yeast growth correlated with enhanced phosphorylation of yeast cell proteins, and similar results were obtained for the Src family tyrosine kinases Lck and Lyn (see Supplemental Material, Figure S2). These data indicate that Vif associated with Hck in yeast cells and stimulated kinase activity, consistent with the in vitro kinase assays. Taken together, the in vitro and in vivo data support previous biochemical reports 35; 36 that Vif interacts with and alters the kinase activity of Hck.
Taken together, the in vitro HX MS Hck SH3 domain binding experiments and the Hck kinase activation assay results (in vitro and in vivo), indicate that recombinant Vif expressed and purified under denaturing conditions adopts a conformation upon refolding that is compatible with SH3 binding and Src-family kinase activation. This biochemically active conformation was next probed with HX MS to understand how the protein was folded in solution.
Vif displayed protection from exchange that was modulated by zinc
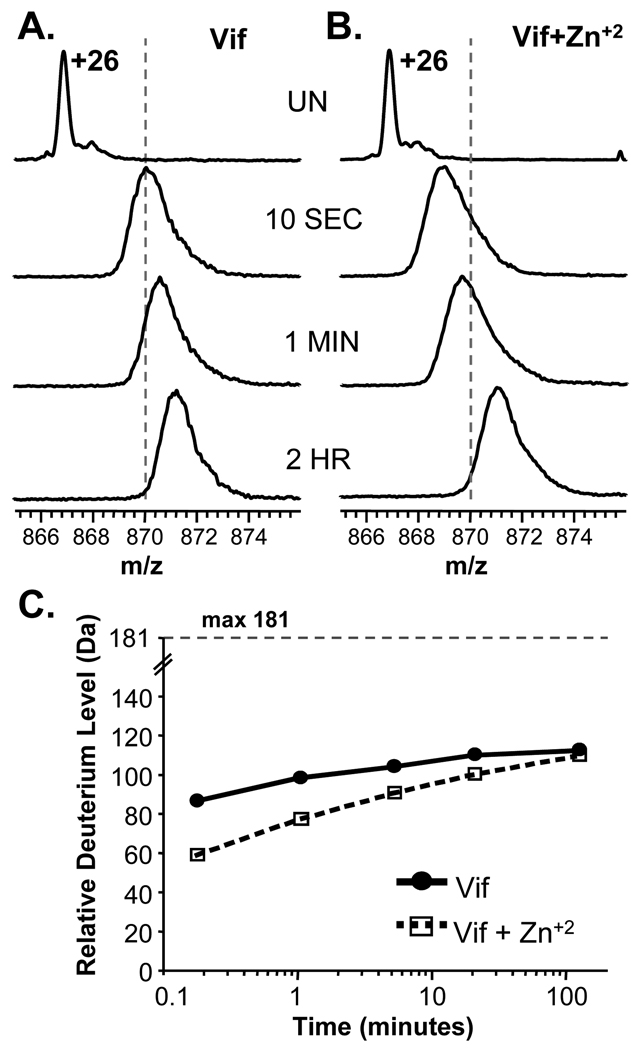
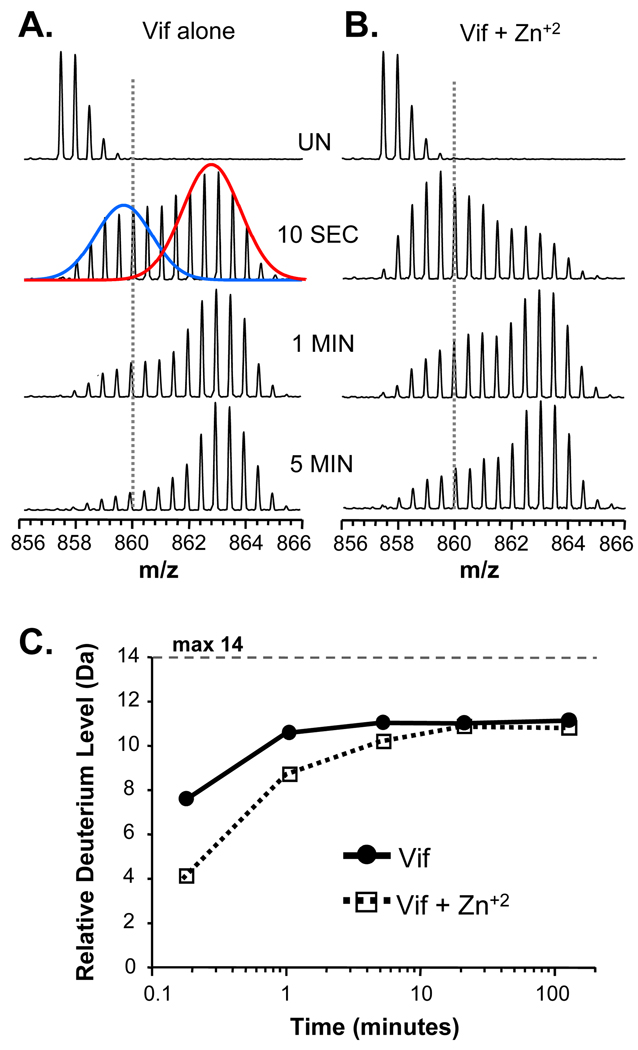
We next utilized HX MS to probe the solution conformation and dynamics of full length Vif alone or in the presence of Zn+2. The coordination of Zn+2 by the novel HCCH zinc finger is thought to result in a conformational change in Vif that exposes key residues that interact with Cullin 5 61. To our knowledge the dissociation constant for the full-length Vif:Zn +2 interaction has not been reported. To ensure Vif was maximally bound to zinc, an elevated concentration of Zn+2 (133 µM) was used during HX MS labeling. The results of the global conformational HX MS analysis of Vif are shown in Figure 4. Deuterium incorporation was faster in Vif alone (Fig. 4A) compared to the Zn+2-bound state (Fig. 4B). The isotope distribution for the +26 charge state of Vif alone shifted to the right of the dotted line (fixed at m/z 870) after only 10 seconds in 2H2O whereas it took more than 5 minutes for Vif incubated with Zn+2 to cross the line. The change in deuteration is also apparent in the deuterium uptake curves (Fig. 4C).
Figure 4.
HX MS of full-length Vif. Representative raw mass spectra during the time course of deuteration are shown for the +26 charge state of Vif when it was deuterated alone (A) or in the presence of 2 mM zinc chloride (B). The time of incubation in deuterium is shown between panels A and B. UN represents undeuterated Vif. The dotted lines are fixed at m/z=870 in both panels. (C) Deuterium incorporation graph derived from the data shown in panels A and B. The maximum amount of deuterium incorporation that is possible for Vif is 181. The data shown are the average of two replicate labeling experiments. The error of intact HX MS measurements was ± 3 Da as determined with replicate analysis of standard proteins of similar size.
The global HX MS analyses also gave a relative indication as to how much of Vif was highly exposed/not structured versus highly protected/structured. Vif contains 181 exchangeable backbone amide hydrogen positions. Only approximately 110 of these positions became deuterated in two hours. Note that these deuteration numbers are reported as relative deuterium incorporation, not absolute deuterium levels, because we have not corrected for back-exchange of deuterium during analysis. With approximately 20–25% deuterium loss due to back exchange, this value of ~110 may be as high as 138. Even without back-exchange correction, it is clear that a significant number of backbone amide hydrogens (between 43 and 70) did not exchange in the timescale of these measurements. Such results indicate that one or more regions of Vif are significantly protected from hydrogen exchange. Factors that could contribute to the observed amide protection are secondary structural elements, oligomeric complexes, and/or burying of amide positions in the core of the protein. In addition to the information about protection from exchange after 2 hours of labeling, incorporation of deuterium at the short labeling times also provides information. Backbone amide hydrogens that become deuterated quickly (within the 10 second exchange time-point) are primarily those on the surfaces of proteins and/or in highly solvent exposed regions 62; 63. For Vif alone, there were approximately 85–105 amide positions (higher number takes into account back-exchange) that were very unprotected from exchange and were presumably in totally unstructured and highly solvent exposed regions of Vif. The incubation of Vif with zinc resulted in a decrease in the amount of deuterium incorporation at shorter time points which became less apparent with time. This protection from exchange is consistent with the formation of structure such that a number of amide hydrogens exchanged more slowly. However, over time, the difference in deuterium level between free and zinc-bound Vif became smaller and smaller. Such results are typical of proteins that are protected initially from exchange due to decreases in solvent accessibility but that retain high protein dynamics and are able to breath/flex in solution over time to obtain the same amount of deuterium at longer time points (reviewed recently in 64). These results are in agreement with the previous reports in which a synthetic portion of the zinc binding region of Vif underwent a conformational change as a result of zinc coordination 65. Our results show that when full length recombinant Vif coordinated zinc, structural changes occurred; these changes might be important for Cullin 5 association 61 since that is a zinc-finger dependent interaction.
The Vif N-terminal region was protected while the C-terminal region was not
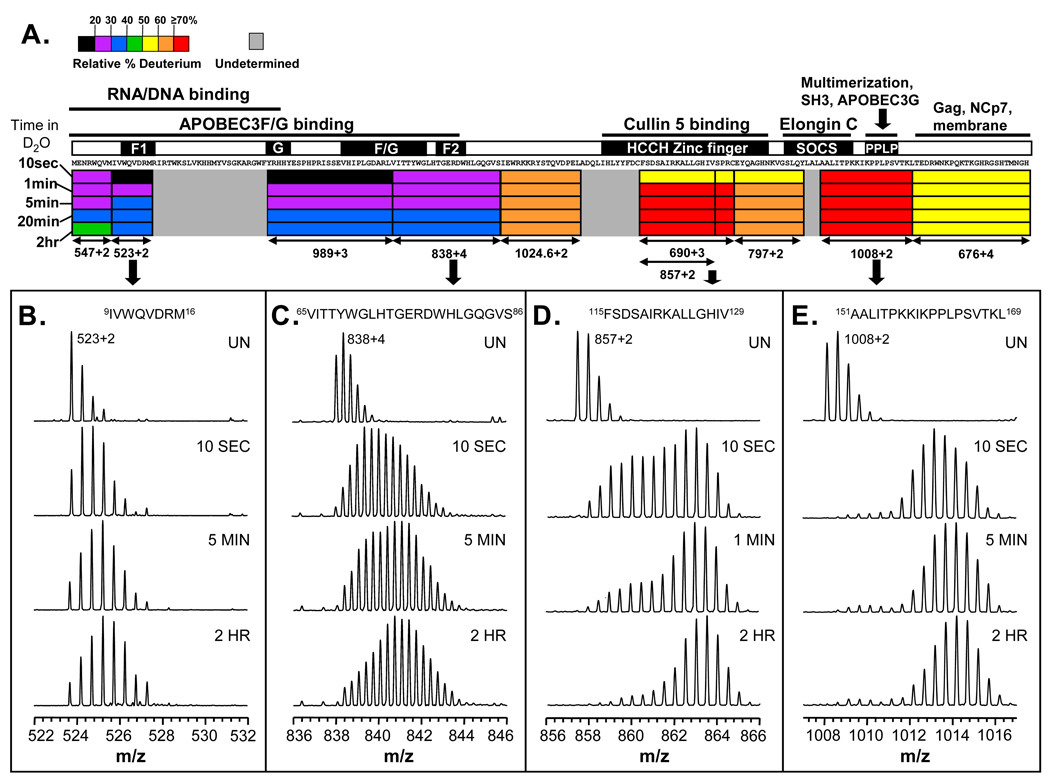
Although the intact HX MS experiments indicated that Vif was partially protected from exchange, they did not indicate where protection occurred in the protein. To localize deuterium incorporation, pepsin digestion was utilized after deuterium labeling 66 and the mass of each peptide followed. Note that the digestion occurs after the labeling is quenched when the conformational information about the protein at physiological pH has already been captured in the pattern of deuteration. The results of the peptide-level exchange experiments are summarized in Figure 5 (all deuterium incorporation graphs for the peptides are found in Supplemental Material, Figure S3). The N-terminal region of Vif displayed the least deuterium incorporation including residues 1–16 and 40–86 which only became ≤ 40% deuterated after 20 minutes in 2H2O (Figure 5A). Interestingly these highly protected regions consist of the APOBEC3G/F and RNA/DNA binding sites. In contrast, deuterium incorporation into the C-terminal half of Vif was markedly different in that most C-terminal peptides became ≥ 60% deuterated after only 1 minute in 2H2O. Recall that the C-terminal half of Vif consists of the Vif HCCH zinc finger, viral SOCS box, C-terminal APOBEC3G binding site, Hck binding site, and multimerization domain (Figure 1). The rapid deuteration of this C-terminal region is consistent with the idea that the C-terminal portion of Vif is unstructured in the unbound state 67.
Figure 5.
Deuterium incorporation at the peptic peptide level. (A) The deuteration map for recombinant Vif. The bars are colored according to the relative percent deuterium incorporation (color code at the top) at each time in deuterium (at the left-hand side). Also indicated are the different regions of Vif and their corresponding biological functions (see also Figure 1). (B–E) Mass spectra for several Vif peptic peptides. The amino acid sequence of each peptide is shown at the top. Deuteration times (not the same for all peptides: time points along the labeling time course were chosen that best illustrate the isotopic distributions and the change with time) are shown in each panel; UN represents undeuterated protein. The location of the peptides in panels B–E is indicated with arrows below panel A and with the m/z of each ion.
For most of Vif, the isotopic distributions for peptic peptides were characteristic of a traditional, single population of one protein conformation in solution (recently well described in 55). Two examples of this are shown in Figure 5B and 5E. The region in Figure 5B is protected and slow to become deuterated whereas the region in Figure 5E is not protected and rapidly becomes deuterated. Several regions of Vif, however, including a portion of the N-terminal APOBEC3G/F binding site and the HCCH zinc finger, displayed multiple isotopic distributions indicative of multiple conformational states in solution (Figure 5C and 5D). Rather than a sharp single isotopic distribution typical of deuterated proteins, there is a much wider distribution. These wide spectra indicate that in some parts of Vif, there is conformational heterogeneity such that at least two populations of molecules exist that are uniquely protected from exchange 55. These populations are in equilibrium with each other, interconvert and as there is a vast excess of deuterium relative to protium in this type of continuous labeling experiment, eventually all molecules become heavily deuterated. Capturing Vif populations existing simultaneously in solution and their interconversion is analogous to the dynamics and labeling for the Hck SH3 domain as described in the preceding section.
One region of particular interest is the 161PPLP164 motif, which has been suggested to be important in Vif multimerization 33; 68; 69. The deuteration of a peptide that encompassed this region (residues 151–169) is shown in Figure 5E. After only 10 seconds in 2H2O, the peptide was highly deuterated and only gained a small amount more deuterium after 2 hours of labeling (see also Supplemental Material, Figure S3). If this region were directly involved in oligomerization as a result of association of multiple Vif molecules, the amide exchange in this region and in the C-terminus would likely show some protection against deuterium incorporation. No such protection was seen and it is therefore likely that the role of the 161PPLP164 region in Vif oligomerization is not apparent in these in vitro solution measurements. The poly-proline type II (PPII) helix of the PPLP domain could possibly act as a conformational hinge bringing the necessary regions of Vif together required for multimer formation, such as the N- and C-termini. This bridging idea is consistent with previous studies 42 in which chemical cross linking was observed between the N- and C-termini of Vif and studies showing that when the prolines of the PPLP domain were substituted with alanines (PPLP to AALA), there was decreased, but not abolished, oligomerization 33.
Zinc coordination stabilizes the Vif HCCH motif
From whole protein HX MS analysis of Vif (Figure 4), we determined that zinc binding caused a significant conformational change. To localize the regions of Vif that were differentially deuterated in the presence of zinc, HX experiments were repeated in the presence and absence of zinc and the deuterium labeled protein digested into peptides. The mass spectra of all the peptides were obtained and deuterium incorporation determined. The main region that showed changes in deuterium incorporation in the presence of zinc included residues 115–129. As shown in Figure 6, a peptide covering this region displayed multiple conformational states in solution, apparent in the 10 second time point (see also Figure 5D). As has become standard practice for bimodal isotopic distributions 55, two Gaussian distributions were fit to the 10 second time point using the program Peak Fit™, Systat Software Inc (San Jose, CA). The lower mass distribution (blue) was more protected from amide exchange, likely corresponding to a more folded or structured state of the zinc finger while the higher mass distribution (red) was more easily deuterated, likely corresponding to an unfolded state(s). Upon addition of zinc, the Vif HCCH peptide, residues 115–129 (an overlapping peptide, residues 115–133, showed similar deuterium incorporation), was stabilized and more time was required to achieve deuteration. The average change in deuterium uptake of Vif alone or in the presence of zinc for residues 115–129 was approximately four deuterons which is less than that observed in global HX measurements (Figure 4C). These discrepancies could likely be attributed to several factors including the absence of peptic peptides in the remainder of the zinc finger (residues 108–114) along with loss of the deuterium label during pepsin digestion and HPLC separation (back exchange).
Figure 6.
Localization of conformational changes in Vif induced by zinc coordination. Hydrogen exchange mass spectra of the Vif peptide covering residues 115FSDSAIRKALLGHIV129 in the absence (A) or presence (B) of zinc. The dotted lines are fixed at m/z=860 in both panels. The multiple populations present in the 10 second time point (panel A) are indicated with the blue and red distributions. (C) deuterium incorporation curves for the peptide shown in panels A and B. The maximum amount of deuterium incorporation that is possible for this peptide is 14. The data shown here are the average of the two populations shown in panel (A). The error of peptide HX MS measurements was ± 0.50 Da as determined with replicate analysis of peptides standards and prior HX MS work with this experimental setup 77.
The zinc finger in Vif corresponds to residues H108C114C133H139 and is essential for its function 61; 70; 71. A synthetic peptide encompassing the Vif HCCH (residues 101–142) motif was unstructured in the absence of zinc 65 but upon zinc incubation, the motif underwent changes in secondary structure along with increased tertiary packing near the zinc site as determined with circular dichroism and fluorescence spectroscopy. Our HX MS results suggest that zinc coordination not only alters the secondary structure and tertiary packing of the HCCH motif 65, but also changes the structural dynamics of the HCCH region (Figure 6). We hypothesize that these changes in dynamics could play a role in efficient, productive interaction with Cullin 5 and other components of the E3 ligase machinery which are required for the proper degradation of APOBEC3 enzymes 24.
CONCLUSIONS
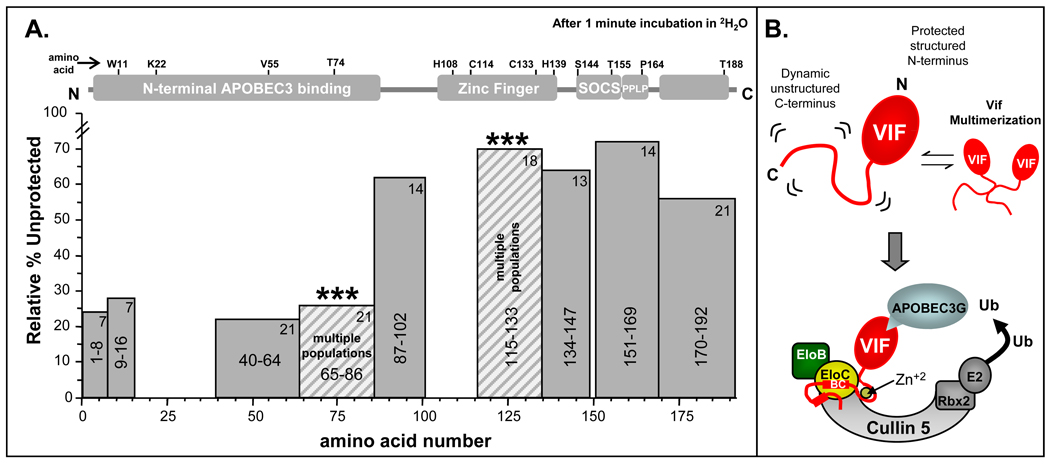
The difficulty in producing full-length soluble Vif in suitable quantities for structural and biophysical analysis has hampered advancement in the understanding of Vif biology. Our HX MS analysis of recombinant full-length Vif represents the most detailed conformational study to date. The results are summarized in Figure 7 where the relative %unprotection from amide exchange after one minute in 2H2O is shown for each peptic peptide of Vif. A low %unprotected (less easily deuterated) is indicative of features such as secondary structural elements, quaternary structure, oligomeric complexes, and/or burying of amide hydrogens such that exchange is slow. A high %unprotection (more easily deuterated) is indicative of features that are dynamic, solvent exposed, and most likely do not contain stable higher order structural elements.
Figure 7.
Summary of results and model of Vif. (A) Summary of deuterium incorporation into Vif after 1 minute in 2H2O. Each bar represents a Vif peptic peptide. Shown is the relative %unprotected for each region of Vif. The residues that each peptide encompasses are indicated in the center of each bar and the maximum number of exchangeable amide hydrogens is indicated by the number in the top right of each bar. Relative %unprotection was determined by dividing the relative deuterium incorporation at 1 minute by the maximum possible exchangeable hydrogens. The peptides that displayed multiple populations in HX MS are indicated by stripes and asterisk (***). The Relative %unprotection for these peptides was calculated using the average deuterium incorporation across the entire isotopic distribution of the multiple populations observed. The N-terminal portion showed the lowest amount of deuterium incorporation (most protection from amide exchange) and therefore likely contains the majority of structural elements while the C-terminal portion showed the highest (least protection from amide exchange) which is consistent with a dynamic and unstructured protein. (B) Model of HIV-1 Vif based on the HX MS results. The N-terminal portion of Vif has structure while the C-terminal portion does not. The C-terminal portion of Vif becomes partially organized upon binding zinc and components of the E3 ligase machinery. Parts of this figure were adapted from 27.
The N-terminal portion of Vif displayed the most protection from amide exchange. If there is any structure in Vif, this region likely contains the majority of it. This region of Vif is essential for interactions with APOBEC3 F/G and DNA/RNA. Interestingly a portion of the N-terminal part of Vif that binds APOBEC3F/G displays conformational exchange in solution (indicated by striped bars and asterisks, Figure 7A), i.e. simultaneously existing conformation one that is more protected from exchange than the other. The conformational heterogeneity in this region might be a result of Vif adopting different conformational states for binding to different APOBEC3 proteins, nucleic acids, or other cellular proteins. Alternatively, the more protected conformation of Vif in this heterogenous mixture of conformations may be the result of oligomerization in these particular regions.
Amide exchange in the C-terminal portion was markedly different than in the N-terminus in that a majority of the C-terminal portion became highly deuterated even after short 2H2O incubation times. This behavior suggests that the majority of the C-terminus is solvent exposed and likely unstructured in the absence of the E3 ligase machinery and zinc. Our results for full-length Vif are consistent with several reports in which synthetic peptides based on the C-terminus of Vif were found to be unstructured in solution 65; 67. The lack of structure in the absence of interacting partners in the C-terminus of Vif likely allows this region to adopt different conformations in solution enabling Vif to associate with multiple binding partners such as Cullin 5, Elongin BC, Hck, and the APOBEC3 proteins.
It was important in these structural studies of Vif to use recombinant protein that was as biochemically functional as possible. As Vif has no enzymatic activity of its own, and instead functions by binding to other proteins and protein complexes, binding to partner proteins was the assay that had to be used to test for biochemical functionality. We previously showed that recombinant Vif prepared exactly as described in the current work could effectively interact with Elongin B/C complex 45. We now show that the same recombinant, refolded Vif was capable of binding to the tyrosine kinase Hck, via its SH3 domain and that the interaction was specific for the known Hck SH3 PxxP binding motif (Figure 3). We showed that recombinant Vif potently activates Hck and other Src family members both in vitro and in a defined, cell-based system (yeast). There have only been a few other reports of Hck:Vif interaction in the literature 35; 36. The ability of Vif to alter kinase activity supports a meaningful role for Vif-induced regulation of Src-family kinases, perhaps through a connection between kinase activation and cell type restriction of APOBEC3 proteins. Activation of Src-family tyrosine kinases seems to be an inherent biochemical property of recombinant Vif, raising the possibility that compounds targeting the Vif:SFK interaction may exhibit antiretroviral activity. A similar approach has been successfully applied to Nef:Hck interaction, leading to the identification of small molecules that inhibit Nef-dependent HIV replication 72.
Our HX MS results for full-length Vif along with recent reports in the literature on synthetic Vif peptides 65; 67, provide molecular insight into the structure of this essential HIV-1 viral accessory protein. Figure 7B depicts a cartoon model of Vif which is consistent with our all of the data. Vif likely contains a folded core domain in the N-terminal portion of the protein which acts as the adaptor region for APOBEC3F/G. The remainder of Vif is highly dynamic and mobile, allowing for recruitment of the E3 ligase machinery to facilitate poly-ubiquitylation of APOBEC3F/G, and for interaction with Src-family kinases. These binding regions may or may not require structure to bind effectively to their targets. The ability of HX MS to probe and detect conformational properties in Vif provides another tool in which the molecular mechanism governing Vif folding and complex assembly can be ascertained. With this foundation of HXMS for Vif alone, future work can address the dynamic changes in Vif that accompany Vif-E3-APOBEC3 ligase complex assembly.
MATERIALS and METHODS
Protein expression and purification
Full length HIV-1 Vif (from HIV strain HXB2) was over-expressed from a codon optimized pET28b vector (a gift from Dana Gabuzda at the Dana Farber Cancer Institute). E. coli strain Rosetta 2 DE3 pLysS was transformed with the pET28b plasmid and the cells were grown to OD600 values of 0.6–1.0. Protein expression was induced by addition of 100 µM IPTG and allowed to proceed for 4 hours at 37 °C. Cells were harvested by centrifugation and lysed in 10 mL of lysis buffer (6 M guanidine hydrochloride, 0.1 M sodium phosphate, pH 8.0). The lysate was centrifuged at 20,000 g for 40 minutes at 4 C to remove the insoluble component; the supernatant was incubated with Ni+2-NTA agarose (Qiagen, Valencia, CA) and mixed end-over-end for 30 minutes at room temperature. The Ni+2-NTA agarose was washed 3 times with 10 mL each of lysis buffer. Vif was eluted from the Ni+2-NTA resin using a step gradient of lysis buffer with decreasing pH values (pH 6.5, pH 6.0, pH 5.5, pH 5.0). All fractions of pH values of ≤ 5.5 were checked for protein with mass spectrometry and SDS PAGE. Fractions containing Vif were pooled and diluted so the concentration was below 35 µM (as determined by Bradford assay 73 from Bio-Rad, Hercules, CA)). The diluted protein was dialyzed against Vif buffer (20 mM HEPES, 150 mM NaCl, 1 mM DTT, 10% glycerol pH 7.0) for 1.5 hours each with Vif buffer containing 3.0, 1.5, 0.75, 0.21, and 0 M guanidine HCl, respectively. A second 0 M guanidine HCl dialysis was conducted overnight to ensure the complete removal of denaturant. The final dialyzed material was rechecked with mass spectrometry and stored at −80 °C.
Human wild type Hck SH3 was over-expressed and purified as described previously 57. The coding sequence for the human Hck SH3 domain with the inactivating Trp93 to alanine (W93A) mutation was PCR-amplified from a full-length Hck template bearing this mutation 58. The resulting cDNA fragment was subcloned into the bacterial expression vector pET28b vector using the Xho I and Nde I restriction sites. Hck SH3 W93A was over-expressed in E. coli Rosetta 2 DE3 pLysS cells as described above, and the soluble protein was purified with Ni-NTA affinity chromatography. The affinity tags were removed from the SH3 domains (wild type and W93A) by overnight cleavage with thrombin, followed by dialysis with Vif buffer and storage at −80 °C. Protein concentrations were determined by Bradford assay and the purity verified with SDS-PAGE and electrospray mass spectrometry.
Circular dichroism measurements
Circular dichroism measurements were taken with a J-715 CD spectrometer (JASCO instruments, Easton, MD) with a path length of 1 mm at 20 °C. For analysis of refolded Vif, the buffer was 20 mM HEPES, 150 mM NaCl, 1mM DTT, 10 % glycerol pH 7.0. Denatured Vif was analyzed in the same buffer containing 8 M guanidine HCl, pH 7.0. Deconvolution of the CD spectra and assignment of the secondary structural percentages was conducted using the software program CDPro 49.
In vitro Src-family kinase activation assay
The Src family kinases Hck, Lck,and Lyn were expressed in their downregulated forms in Sf9 insect cells and purified as described elsewhere 51; 72. Briefly, recombinant purified Vif was incubated with each Src-family member at various molar ratios, and kinase assays were performed using the FRET-based Z’-Lyte method and Tyr-2 peptide substrate according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). Assays were performed in quadruplicate in 384-well plates in a final volume of 10 µL. Final ATP and Tyr-2 peptide substrate concentrations were held constant at 50 µM and 2 µM, respectively. Kinases were pre-incubated with Vif in kinase assay buffer for 30 min, followed by incubation with ATP and Tyr-2 peptide for 1 h and development buffer for an additional 1 h. Following reaction quench, substrate fluorescence was assessed on a Gemini XS microplate spectrofluorometer (Molecular Devices, Sunnyvale, CA).
Yeast growth suppression assay
The coding sequence for HIV-1 Vif (HXB2) was amplified and subcloned into the yeast expression vector pYC2/CT vector (Invitrogen, Carlsbad, CA) downstream of the Gal10 promoter. The Hck, Lck, and Lyn yeast expression vectors were prepared as described previously 51. The coding region of the negative regulatory tail of each SFK was modified by PCR to encode the sequence Tyr-Glu-Glu-Ile (“YEEI”) which enables auto-downregulation in the absence of Csk 56. For the growth suppression assay, the yeast strain YPH 499 (Stratagene) was transformed with the Vif and SFK expression plasmids, and colonies were selected on standard synthetic drop-out plates lacking uracil and tryptophan (SD/-Ura-Trp) with glucose as sole carbon source to repress protein expression. Transformed colonies were grown in SD/-Ura-Trp medium with glucose, normalized to OD600 = 0.4 in water, and then spotted as three serial dilutions onto SD/-Ura-Trp agar plates with galactose as sole carbon source to induce protein expression. Plates were incubated for 3 days at 30 °C and imaged using a flatbed scanner, which makes the yeast patches appear as dark patches against the agar background. Expression of SFK and Vif proteins and tyrosine phosphorylation of yeast proteins was confirmed by immunoblotting as described elsewhere 51; 56. All growth suppression assays were performed in triplicate and yielded comparable results; representative examples are shown.
Deuterium labeling
Vif (30 µM) was thawed from −80 °C and incubated at 4 °C for 5 minutes before labeling. The Vif solution was diluted 15-fold with 20 mM MOPS, 150 mM NaCl, 1 mM DTT, 99.99% 2H2O, pD 7.0 at 20 °C. For experiments that included zinc, Vif was equilibrated in the presence of 2 mM ZnCl2 before labeling; upon 15-fold dilution with labeling buffer, the zinc concentration was 133 µM. The labeling reaction was quenched at various times by adjusting the pH to 2.6 with a 1:1 ratio of a solution containing 0.8 M guanidine HCl, 0.8 % formic acid, 100% H2O. For SH3 binding experiments with recombinant Vif, wild-type Hck SH3 or W93A Hck SH3 were diluted to 6 µM and incubated with a 5-fold molar excess of Vif (30 µM) for 30 minutes at 4 °C. The mixture was labeled and quenched exactly the same as described for Vif alone.
Intact protein mass analysis
Protein samples were injected immediately after the exchange quench into a Shimadzu SCL-10A VP HPLC flowing water containing 0.05% formic acid, pH 2.6 at 50 µL/min coupled to a Waters LCT premier mass spectrometer with a standard electrospray interface. Proteins were eluted directly into the mass spectrometer with a gradient of 15–98 % acetonitrile (containing 0.05% formic acid, pH 2.6) in 3.5 minutes. The injector, column (Alltech analytical in-line guard column, packed with POROS 20-R2 reversed-phase media, PerSeptive Biosystems), and all associated tubing were kept at 0 °C to minimize back exchange 66. All experiments were conducted under identical conditions so the relative deuterium levels could be compared 74.Deuterium levels were not corrected for back exchange and are therefore reported as relative deuterium levels 74. Intact mass spectra were deconvoluted using the software MagTran 75. The intact protein relative deuterium uptake was calculated by subtracting the centroid mass of an undeuterated control from the centroid mass of deuterium labeled samples. All intact HX MS experiments were conducted at least twice and the results averaged.
Peptide mass analysis
Using a Shimadzu SCL-10A VP HPLC, protein samples were injected into an immobilized pepsin digestion column (2.1 mm × 50 mm stainless steel column packed with immobilized pepsin 76 on POROS-20AL beads from PerSeptive Biosystems) flowing 0.05% formic acid (pH 2.6) at 20 °C at a flow rate of 200 µL/min. Peptic peptides produced in the pepsin column were trapped on an Michrom Bioresources (Auburn, CA) peptide micropeptide trap at 0 °C and desalted for 3 minutes before separation using a POROS 20 R2 (PerSeptive Biosystems) column. The column was 0.20 × 100 mm and was operated at 0 °C with a flow rate of 50 µL/min. A 9 minute 8–40 % acetonitrile gradient (both mobile phases contained 0.05 % formic acid, pH 2.6) was used to elute the peptides directly into a Waters QToF API US mass spectrometer with standard electrospray interface. Mass accuracy of <5 ppm was maintained through continuous lock mass correction was carried out using Glu-fibrinogen peptide. The excel-based software program HX-Express 55 was used to process the data. The deuterium uptake for each peptide was determined by subtracting the centroid mass of each undeuterated control from the centroid mass of deuterium labeled samples. All peptide-level HX MS experiments were conducted at least twice and the results averaged. In this experimental setup, the error of measuring the deuterium incorporation was less than ±0.50 Da 77. All Peptic peptides were identified by a combination of exact mass analysis MS/MS, and using Waters IdentityE software 78. All peptide-level HX MS experiments were conducted at least twice.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Dana Gabuzda at HMS/DFCI for the HIV-1 Vif pET28b codon optimized construct. We also thank Prof. Thomas Wales and Prof. Penny Beuning for their technical support and critical insight. This work was supported in part by funding from the National Institutes of Health (GM070590 and GM086507 to JRE; CA081398 and AI077444 to TES) and a research collaboration with the Waters Corporation. L.E.-S. is the recipient of a Ruth L. Kirschstein NRSA Fellowship from the NIH. This is contribution 982 from the Barnett Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Frankel AD, Young JA. HIV-1: fifteen proteins and an RNA. Annu Rev Biochem. 1998;67:1–25. doi: 10.1146/annurev.biochem.67.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Seelamgari A, Maddukuri A, Berro R, de la Fuente C, Kehn K, Deng L, Dadgar S, Bottazzi ME, Ghedin E, Pumfery A, Kashanchi F. Role of viral regulatory and accessory proteins in HIV-1 replication. Front Biosci. 2004;9:2388–2413. doi: 10.2741/1403. [DOI] [PubMed] [Google Scholar]
- 3.Haseltine WA. Molecular biology of the human immunodeficiency virus type 1. FASEB J. 1991;5:2349–2360. doi: 10.1096/fasebj.5.10.1829694. [DOI] [PubMed] [Google Scholar]
- 4.Trono D. HIV accessory proteins: leading roles for the supporting cast. Cell. 1995;82:189–192. doi: 10.1016/0092-8674(95)90306-2. [DOI] [PubMed] [Google Scholar]
- 5.Miller RH, Sarver N. HIV accessory proteins as therapeutic targets. Nat Med. 1997;3:389–394. doi: 10.1038/nm0497-389. [DOI] [PubMed] [Google Scholar]
- 6.Bour S, Strebel K. HIV accessory proteins: multifunctional components of a complex system. Adv Pharmacol. 2000;48:75–120. doi: 10.1016/s1054-3589(00)48004-x. [DOI] [PubMed] [Google Scholar]
- 7.Malim MH, Emerman M. HIV-1 accessory proteins--ensuring viral survival in a hostile environment. Cell Host Microbe. 2008;3:388–398. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Kirchhoff F. Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe. 2009;8:55–67. doi: 10.1016/j.chom.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Fisher AG, Ensoli B, Ivanoff L, Chamberlain M, Petteway S, Ratner L, Gallo RC, Wong-Staal F. The sor gene of HIV-1 is required for efficient virus transmission in vitro. Science. 1987;237:888–893. doi: 10.1126/science.3497453. [DOI] [PubMed] [Google Scholar]
- 10.Strebel K, Daugherty D, Clouse K, Cohen D, Folks T, Martin MA. The HIV 'A' (sor) gene product is essential for virus infectivity. Nature. 1987;328:728–730. doi: 10.1038/328728a0. [DOI] [PubMed] [Google Scholar]
- 11.Gabuzda DH, Lawrence K, Langhoff E, Terwilliger E, Dorfman T, Haseltine WA, Sodroski J. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J Virol. 1992;66:6489–6495. doi: 10.1128/jvi.66.11.6489-6495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 13.Goncalves J, Santa-Marta M. HIV-1 Vif and APOBEC3G: multiple roads to one goal. Retrovirology. 2004;1:28. doi: 10.1186/1742-4690-1-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malim MH. Natural resistance to HIV infection: The Vif-APOBEC interaction. C R Biol. 2006;329:871–875. doi: 10.1016/j.crvi.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 15.Bernacchi S, Henriet S, Dumas P, Paillart JC, Marquet R. RNA and DNA binding properties of HIV-1 Vif protein: a fluorescence study. J Biol Chem. 2007;282:26361–26368. doi: 10.1074/jbc.M703122200. [DOI] [PubMed] [Google Scholar]
- 16.Goila-Gaur R, Strebel K. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology. 2008;5:51. doi: 10.1186/1742-4690-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheehy AM, Gaddis NC, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med. 2003;9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 18.Chiu YL, Greene WC. APOBEC3G: an intracellular centurion. Philos Trans R Soc Lond B Biol Sci. 2009;364:689–703. doi: 10.1098/rstb.2008.0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li XY, Guo F, Zhang L, Kleiman L, Cen S. APOBEC3G inhibits DNA strand transfer during HIV-1 reverse transcription. J Biol Chem. 2007;282:32065–32074. doi: 10.1074/jbc.M703423200. [DOI] [PubMed] [Google Scholar]
- 20.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 21.Mercenne G, Bernacchi S, Richer D, Bec G, Henriet S, Paillart JC, Marquet R. HIV-1 Vif binds to APOBEC3G mRNA and inhibits its translation. Nucleic Acids Res. 2010;38:633–646. doi: 10.1093/nar/gkp1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stopak K, de Noronha C, Yonemoto W, Greene WC. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol Cell. 2003;12:591–601. doi: 10.1016/s1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- 23.Goila-Gaur R, Khan MA, Miyagi E, Kao S, Opi S, Takeuchi H, Strebel K. HIV-1 Vif promotes the formation of high molecular mass APOBEC3G complexes. Virology. 2008;372:136–146. doi: 10.1016/j.virol.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mehle A, Strack B, Ancuta P, Zhang C, McPike M, Gabuzda D. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J Biol Chem. 2004;279:7792–7798. doi: 10.1074/jbc.M313093200. [DOI] [PubMed] [Google Scholar]
- 25.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 26.Kataropoulou A, Bovolenta C, Belfiore A, Trabatti S, Garbelli A, Porcellini S, Lupo R, Maga G. Mutational analysis of the HIV-1 auxiliary protein Vif identifies independent domains important for the physical and functional interaction with HIV-1 reverse transcriptase. Nucleic Acids Res. 2009;37:3660–3669. doi: 10.1093/nar/gkp226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mehle A, Thomas ER, Rajendran KS, Gabuzda D. A zinc-binding region in Vif binds Cul5 and determines cullin selection. J Biol Chem. 2006;281:17259–17265. doi: 10.1074/jbc.M602413200. [DOI] [PubMed] [Google Scholar]
- 28.Mehle A, Wilson H, Zhang C, Brazier AJ, McPike M, Pery E, Gabuzda D. Identification of an APOBEC3G binding site in human immunodeficiency virus type 1 Vif and inhibitors of Vif-APOBEC3G binding. J Virol. 2007;81:13235–13241. doi: 10.1128/JVI.00204-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu B, Sarkis PT, Luo K, Yu Y, Yu XF. Regulation of Apobec3F and human immunodeficiency virus type 1 Vif by Vif-Cul5-ElonB/C E3 ubiquitin ligase. J Virol. 2005;79:9579–9587. doi: 10.1128/JVI.79.15.9579-9587.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo K, Xiao Z, Ehrlich E, Yu Y, Liu B, Zheng S, Yu XF. Primate lentiviral virion infectivity factors are substrate receptors that assemble with cullin 5-E3 ligase through a HCCH motif to suppress APOBEC3G. Proc Natl Acad Sci U S A. 2005;102:11444–11449. doi: 10.1073/pnas.0502440102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mehle A, Goncalves J, Santa-Marta M, McPike M, Gabuzda D. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 2004;18:2861–2866. doi: 10.1101/gad.1249904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stanley BJ, Ehrlich ES, Short L, Yu Y, Xiao Z, Yu XF, Xiong Y. Structural insight into the human immunodeficiency virus Vif SOCS box and its role in human E3 ubiquitin ligase assembly. J Virol. 2008;82:8656–8663. doi: 10.1128/JVI.00767-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernacchi S, Mercenne G, Tournaire C, Marquet R, Paillart JC. Importance of the proline-rich multimerization domain on the oligomerization and nucleic acid binding properties of HIV-1 Vif. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq979. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, Pomerantz RJ, Dornadula G, Sun Y. Human immunodeficiency virus type 1 Vif protein is an integral component of an mRNP complex of viral RNA and could be involved in the viral RNA folding and packaging process. J Virol. 2000;74:8252–8261. doi: 10.1128/jvi.74.18.8252-8261.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Douaisi M, Dussart S, Courcoul M, Bessou G, Lerner EC, Decroly E, Vigne R. The tyrosine kinases Fyn and Hck favor the recruitment of tyrosine-phosphorylated APOBEC3G into vif-defective HIV-1 particles. Biochem Biophys Res Commun. 2005;329:917–924. doi: 10.1016/j.bbrc.2005.02.057. [DOI] [PubMed] [Google Scholar]
- 36.Hassaine G, Courcoul M, Bessou G, Barthalay Y, Picard C, Olive D, Collette Y, Vigne R, Decroly E. The tyrosine kinase Hck is an inhibitor of HIV-1 replication counteracted by the viral vif protein. J Biol Chem. 2001;276:16885–16893. doi: 10.1074/jbc.M009076200. [DOI] [PubMed] [Google Scholar]
- 37.Donahue JP, Vetter ML, Mukhtar NA, D'Aquila RT. The HIV-1 Vif PPLP motif is necessary for human APOBEC3G binding and degradation. Virology. 2008;377:49–53. doi: 10.1016/j.virol.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wissing S, Galloway NL, Greene WC. HIV-1 Vif versus the APOBEC3 cytidine deaminases: an intracellular duel between pathogen and host restriction factors. Mol Aspects Med. 2010;31:383–397. doi: 10.1016/j.mam.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X, Romero P, Rani M, Dunker AK, Obradovic Z. Predicting Protein Disorder for N-, C-, and Internal Regions. Genome Inform Ser Workshop Genome Inform. 1999;10:30–40. [PubMed] [Google Scholar]
- 40.Romero Obradovic, Dunker K. Sequence Data Analysis for Long Disordered Regions Prediction in the Calcineurin Family. Genome Inform Ser Workshop Genome Inform. 1997;8:110–124. [PubMed] [Google Scholar]
- 41.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 42.Auclair JR, Green KM, Shandilya S, Evans JE, Somasundaran M, Schiffer CA. Mass spectrometry analysis of HIV-1 Vif reveals an increase in ordered structure upon oligomerization in regions necessary for viral infectivity. Proteins. 2007;69:270–284. doi: 10.1002/prot.21471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engen JR. Analysis of protein conformation and dynamics by hydrogen/deuterium exchange MS. Anal Chem. 2009;81:7870–7875. doi: 10.1021/ac901154s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marcsisin SR, Engen JR. Hydrogen exchange mass spectrometry: what is it and what can it tell us? Anal Bioanal Chem. 2010;397:967–972. doi: 10.1007/s00216-010-3556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marcsisin SR, Engen JR. Molecular insight into the conformational dynamics of the Elongin BC complex and its interaction with HIV-1 Vif. J Mol Biol. 2010;402:892–904. doi: 10.1016/j.jmb.2010.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Engen JR, Wales TE, Hochrein JM, Meyn MA, Banu Ozkan S, 3rd, Bahar I, Smithgall TE. Structure and dynamic regulation of Src-family kinases. Cell Mol Life Sci. 2008;65:3058–3073. doi: 10.1007/s00018-008-8122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang X, Goncalves J, Gabuzda D. Phosphorylation of Vif and its role in HIV-1 replication. J Biol Chem. 1996;271:10121–10129. doi: 10.1074/jbc.271.17.10121. [DOI] [PubMed] [Google Scholar]
- 48.Gallerano D, Devanaboyina SC, Swoboda I, Linhart B, Mittermann I, Keller W, Valenta R. Biophysical characterization of recombinant HIV-1 subtype C virus infectivity factor. Amino Acids. 2010 doi: 10.1007/s00726-010-0725-x. in press. [DOI] [PubMed] [Google Scholar]
- 49.Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 50.Briggs SD, Sharkey M, Stevenson M, Smithgall TE. SH3-mediated Hck tyrosine kinase activation and fibroblast transformation by the Nef protein of HIV-1. J Biol Chem. 1997;272:17899–17902. doi: 10.1074/jbc.272.29.17899. [DOI] [PubMed] [Google Scholar]
- 51.Trible RP, Emert-Sedlak L, Smithgall TE. HIV-1 Nef selectively activates Src family kinases Hck, Lyn, and c-Src through direct SH3 domain interaction. J Biol Chem. 2006;281:27029–27038. doi: 10.1074/jbc.M601128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Engen JR, Smithgall TE, Gmeiner WH, Smith DL. Identification and localization of slow, natural, cooperative unfolding in the hematopoietic cell kinase SH3 domain by amide hydrogen exchange and mass spectrometry. Biochemistry. 1997;36:14384–14391. doi: 10.1021/bi971635m. [DOI] [PubMed] [Google Scholar]
- 53.Hochrein JM, Lerner EC, Schiavone AP, Smithgall TE, Engen JR. An examination of dynamics crosstalk between SH2 and SH3 domains by hydrogen/deuterium exchange and mass spectrometry. Protein Sci. 2006;15:65–73. doi: 10.1110/ps.051782206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wales TE, Engen JR. Partial unfolding of diverse SH3 domains on a wide timescale. J Mol Biol. 2006;357:1592–1604. doi: 10.1016/j.jmb.2006.01.075. [DOI] [PubMed] [Google Scholar]
- 55.Weis DD, Wales TE, Engen JR, Hotchko M, Ten Eyck LF. Identification and characterization of EX1 kinetics in H/D exchange mass spectrometry by peak width analysis. J Am Soc Mass Spectrom. 2006;17:1498–1509. doi: 10.1016/j.jasms.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 56.Trible RP, Emert-Sedlak L, Wales TE, Ayyavoo V, Engen JR, Smithgall TE. Allosteric loss-of-function mutations in HIV-1 Nef from a long-term non-progressor. J Mol Biol. 2007;374:121–129. doi: 10.1016/j.jmb.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Poe JA, Smithgall TE. HIV-1 Nef dimerization is required for Nef-mediated receptor downregulation and viral replication. J Mol Biol. 2009;394:329–342. doi: 10.1016/j.jmb.2009.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schreiner SJ, Schiavone AP, Smithgall TE. Activation of STAT3 by the Src family kinase Hck requires a functional SH3 domain. J Biol Chem. 2002;277:45680–45687. doi: 10.1074/jbc.M204255200. [DOI] [PubMed] [Google Scholar]
- 59.Superti-Furga G, Fumagalli S, Koegl M, Courtneidge SA, Draetta G. Csk inhibition of c-Src activity requires both the SH2 and SH3 domains of Src. EMBO J. 1993;12:2625–2634. doi: 10.1002/j.1460-2075.1993.tb05923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takashima Y, Delfino FJ, Engen JR, Superti-Furga G, Smithgall TE. Regulation of c-Fes tyrosine kinase activity by coiled-coil and SH2 domains: analysis with Saccharomyces cerevisiae. Biochemistry. 2003;42:3567–3574. doi: 10.1021/bi0272499. [DOI] [PubMed] [Google Scholar]
- 61.Xiao Z, Xiong Y, Zhang W, Tan L, Ehrlich E, Guo D, Yu XF. Characterization of a novel Cullin5 binding domain in HIV-1 Vif. J Mol Biol. 2007;373:541–550. doi: 10.1016/j.jmb.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 62.Dharmasiri K, Smith DL. Mass spectrometric determination of isotopic exchange rates of amide hydrogens located on the surfaces of proteins. Anal Chem. 1996;68:2340–2344. doi: 10.1021/ac9601526. [DOI] [PubMed] [Google Scholar]
- 63.Truhlar SM, Croy CH, Torpey JW, Koeppe JR, Komives EA. Solvent accessibility of protein surfaces by amide H/2H exchange MALDI-TOF mass spectrometry. J Am Soc Mass Spectrom. 2006;17:1490–1497. doi: 10.1016/j.jasms.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 64.Morgan CR, Engen JR. Investigating solution-phase protein structure and dynamics by hydrogen exchange mass spectrometry. Curr Protoc Protein Sci. 2009;Chapter 17:1–17. doi: 10.1002/0471140864.ps1706s58. Unit 17 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Giri K, Maynard EL. Conformational analysis of a peptide approximating the HCCH motif in HIV-1 Vif. Biopolymers. 2009;92:417–425. doi: 10.1002/bip.21209. [DOI] [PubMed] [Google Scholar]
- 66.Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reingewertz TH, Benyamini H, Lebendiker M, Shalev DE, Friedler A. The C-terminal domain of the HIV-1 Vif protein is natively unfolded in its unbound state. Protein Eng Des Sel. 2009;22:281–287. doi: 10.1093/protein/gzp004. [DOI] [PubMed] [Google Scholar]
- 68.Wolfe LS, Stanley BJ, Liu C, Eliason WK, Xiong Y. Dissection of the HIV Vif interaction with human E3 ubiquitin ligase. J Virol. 2010;84:7135–7139. doi: 10.1128/JVI.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang S, Sun Y, Zhang H. The multimerization of human immunodeficiency virus type I Vif protein: a requirement for Vif function in the viral life cycle. J Biol Chem. 2001;276:4889–4893. doi: 10.1074/jbc.M004895200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiao Z, Ehrlich E, Luo K, Xiong Y, Yu XF. Zinc chelation inhibits HIV Vif activity and liberates antiviral function of the cytidine deaminase APOBEC3G. FASEB J. 2007;21:217–222. doi: 10.1096/fj.06-6773com. [DOI] [PubMed] [Google Scholar]
- 71.Xiao Z, Ehrlich E, Yu Y, Luo K, Wang T, Tian C, Yu XF. Assembly of HIV-1 Vif-Cul5 E3 ubiquitin ligase through a novel zinc-binding domain-stabilized hydrophobic interface in Vif. Virology. 2006;349:290–299. doi: 10.1016/j.virol.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 72.Emert-Sedlak L, Kodama T, Lerner EC, Dai W, Foster C, Day BW, Lazo JS, Smithgall TE. Chemical library screens targeting an HIV-1 accessory factor/host cell kinase complex identify novel antiretroviral compounds. ACS Chem Biol. 2009;4:939–947. doi: 10.1021/cb900195c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 74.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 75.Zhang Z, Marshall AG. A universal algorithm for fast and automated charge state deconvolution of electrospray mass-to-charge ratio spectra. J Am Soc Mass Spectrom. 1998;9:225–233. doi: 10.1016/S1044-0305(97)00284-5. [DOI] [PubMed] [Google Scholar]
- 76.Wang L, Pan H, Smith DL. Hydrogen exchange-mass spectrometry: optimization of digestion conditions. Mol Cell Proteomics. 2002;1:132–138. doi: 10.1074/mcp.m100009-mcp200. [DOI] [PubMed] [Google Scholar]
- 77.Houde D, Berkowitz SA, Engen JR. The utility of hydrogen/deuterium exchange mass spectrometry in biopharmaceutical comparability studies. J Pharm Sci. 2010 doi: 10.1002/jps.22432. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics. 2006;5:144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- 79.Zhang W, Chen G, Niewiadomska AM, Xu R, Yu XF. Distinct determinants in HIV-1 Vif and human APOBEC3 proteins are required for the suppression of diverse host anti-viral proteins. PLoS One. 2008;3:e3963. doi: 10.1371/journal.pone.0003963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen S, Brier S, Smithgall TE, Engen JR. The Abl SH2-kinase linker naturally adopts a conformation competent for SH3 domain binding. Protein Sci. 2007;16:572–581. doi: 10.1110/ps.062631007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.