Abstract
The HIV-1 envelope (Env) glycoproteins play an essential role in the virus replication cycle by mediating the fusion between viral and cellular membranes during the entry process. The Env glycoproteins are synthesized as a polyprotein precursor, gp160, that is cleaved by cellular proteases to the mature surface glycoprotein gp120 and the transmembrane glycoprotein gp41. During virus assembly the gp120/gp41 complex is incorporated as heterotrimeric spikes into the lipid bilayer of nascent virions. These gp120/gp41 complexes then initiate the infection process by binding receptor and co-receptor on the surface of target cells. Much is currently known about the HIV-1 Env glycoprotein trafficking pathway and the structure of gp120 and the extracellular domain of gp41. However, the mechanism by which the Env glycoprotein complex is incorporated into virus particles remains incompletely understood. Genetic data support a major role for the cytoplasmic tail of gp41 and the matrix domain of Gag in Env glycoprotein incorporation. Still to be defined are the identities of host cell factors that may promote Env incorporation, and the role of specific membrane microdomains in this process. Here we review our current understanding of HIV-1 Env glycoprotein trafficking and incorporation into virions.
A. Overview of HIV-1 replication
In vivo, human immunodeficiency virus type 1 (HIV-1) infects predominantly CD4+ T cells and cells of the monocyte/macrophage lineage. This cell tropism, which is intimately linked to the pathogenesis induced by HIV-1, is determined by the viral envelope (Env) glycoproteins. The Env glycoprotein complex projects from the virion surface as highly glycosylated spikes, composed of a heterotrimer of the surface (SU) glycoprotein gp120 and the transmembrane (TM) glycoprotein gp41. The entry process is initiated by the binding of gp120 to CD4, the primary attachment receptor for primate lentiviruses. CD4 binding triggers a conformational change in gp120 that enhances its affinity for a secondary receptor, or “coreceptor”. The major physiologically relevant coreceptors for HIV-1 are CCR5 and CXCR4. Upon binding of gp120 to the coreceptor, further conformational changes in both gp120 and gp41 trigger a membrane fusion reaction that delivers the viral core into the cytoplasm. HIV-1 can enter host cells through two different mechanisms, either direct, pH-independent fusion with the plasma membrane or clathrin-mediated endocytosis followed by low-pH fusion with the endosomal membrane.1,2,3
The binding affinity between gp120 and CCR5 or CXCR4 to a large extent determines the cell tropism of a particular viral isolate. Macrophage-tropic HIV-1 isolates utilize CCR5 for entry and are thus designated as R5 viruses; these isolates can also infect primary CD4+ T cells but not T-cell lines. Viruses that primarily use CXCR4 are denoted X4-tropic; these strains can infect T-cell lines and primary T cells. R5X4 or “dual-tropic” strains can enter cells via either CCR5 and CXCR4.4 The CCR5 coreceptor is used during the primary or early, asymptomatic stage of HIV-1 infection and R5 viruses are largely responsible for person-to-person transmission.5 During the late stage of infection, viral isolates may arise that use CXCR4 for entry. The emergence in vivo of X4 or dual-tropic viruses is often associated with a sharp decline in the number of CD4+ T cells in infected individuals and the onset of AIDS-defining symptoms.
Following entry into the cytosol, the viral RNA genome is converted (reverse transcribed) into double-stranded DNA by the viral enzyme reverse transcriptase (RT). The newly synthesized viral DNA is then translocated across the nuclear pore as part of a high-molecular-weight structure known as the preintegration complex.6,7,8,9,10 In the nucleus, the viral DNA is integrated into the host cell chromosomal DNA; this integration process is catalyzed by a second viral enzyme, integrase (IN). The integrated viral DNA directs the transcription of viral RNAs, which are transported into the cytoplasm where translation of the viral proteins takes place. The newly synthesized viral proteins, together with two single-stranded copies of full-length (unspliced) viral RNA, assemble into a new generation of viral particles. Concomitant with release of virus particles from the infected cell, a third viral enzyme, protease (PR), cleaves the Gag and GagPol polyprotein precursors, triggering the conversion of the immature particle to the mature virion. The virus replication cycle is now complete, and the mature virus particle can initiate a new cycle of infection. 11,12
B. Introduction to HIV-1 assembly
The process of HIV-1 assembly is regulated by both viral and cellular factors. The Gag polyprotein precursor, Pr55Gag, is the major viral structural protein responsible for assembly; its expression is sufficient for the assembly, budding, and release of immature particles. Pr55Gag, usually simply referred to as Gag, is synthesized on cytosolic ribosomes and is composed of matrix (MA), capsid (CA), nucleocapsid (NC), and p6 domains, along with two spacer peptides (SP1 and SP2). Assembly of viral genomic RNA, the Env glycoprotein complex, and GagPol precursor protein (Pr160GagPol) into virus particles occurs in most cells at the plasma membrane (PM).12,13,14,15
Each of the major domains in Gag (MA, CA, NC, and p6) serves distinct functions during the viral assembly process. Intracellular trafficking of Pr55Gag and binding of Gag to the PM is regulated by MA. Membrane association is mediated by a bipartite membrane-binding domain, comprised of a covalently attached myristic acid linked to the N-terminus of MA and a highly basic patch of residues that forms a positively charged surface at the “top”, or membrane-proximal surface, of the folded MA domain.16,17,18 One of the primary functions of the basic patch is to bind the PM phosphoinositide phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2], which plays a major role in directing Gag to the PM.19,20 The MA domain also directs the incorporation of the Env glycoprotein complex into virions, as will be described in more detail below. CA contributes to Gag-Gag interactions during assembly and ultimately forms the outer shell of the viral core after virion release and maturation. The NC domain mediates the packaging of viral genomic RNA and also promotes Gag multimerization. The p6 domain stimulates the release of viral particles from the PM by recruiting the endosomal sorting complexes required for transport (ESCRT) machinery, a cellular apparatus that normally functions in cellular budding and membrane scission events.21,22,23,24,25
The question of where in the cell HIV-1 assembles has been the focus of debate in recent years. It is now clear that the major site of HIV-1 assembly is the PM.26,27,28,29 However, under certain circumstances, late endosomes can function as the site of productive virus assembly.30 In primary macrophages, it has long been appreciated that internal compartments that bear late endosomal markers (e.g., the tetraspanins) are the primary sites of virus assembly. Several studies have demonstrated that these internal, virus-containing compartments are not true late endosomes but rather are specialized PM invaginations into which virus assembles and buds.28,31,32 Virus particles contained in these internal, but PM-connected, compartments are rapidly released to the surface at sites of cell-to-cell contact known as virological synapses.33
C. Env biogenesis
Env synthesis and trafficking
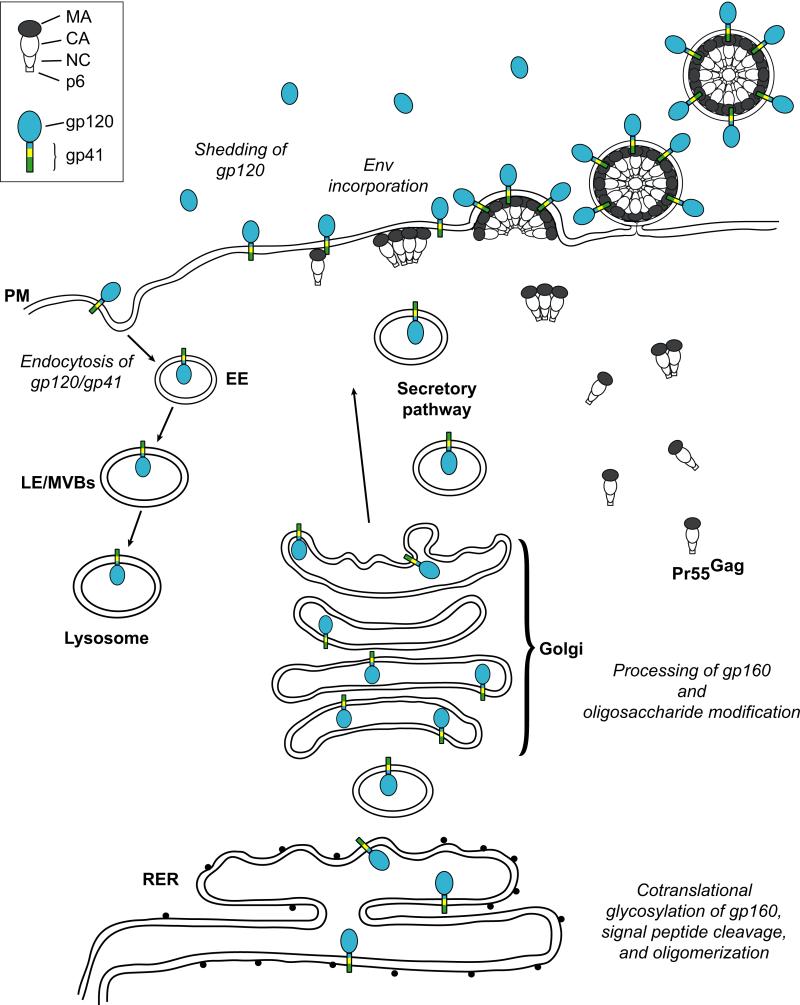
HIV-1 Env glycoproteins are synthesized as a polyprotein precursor from a singly spliced, bicistronic vpu/env mRNA on the rough endoplasmic reticulum (RER) (Fig. 1).34,35 The unprocessed Env glycoprotein precursor (gp160) contains an ER signal sequence at its N-terminus, which targets Env to the RER membrane. This signal peptide is cotranslationally removed by cellular signal peptidases within the ER. The transmembrane domain (TMD) of gp41 contains a hydrophobic stop-transfer signal (membrane anchor) that prevents gp160 from being fully released into the lumen of the ER.36,37 The extracellular portions of Env thus extend into the lumen of the ER, while the gp41 cytoplasmic tail (CT) extends into the cytoplasm and later the interior of the virion. Concomitant with translation, gp160 is glycosylated with N-linked and some O-linked oligosaccharide side chains.38,39,40 Monomers of gp160 oligomerize in the ER into predominantly trimers, although dimers and tetramers can also be observed.41,42,43,44 This oligomerization process is thought to facilitate the trafficking of gp160 to the Golgi complex. As gp160 traffics through the secretory pathway, high-mannose oligosaccharide side chains acquire complex modifications in the trans-Golgi network (TGN). In part to prevent premature interaction of the oligomerized Env with CD4 in the secretory pathway, the HIV-1 accessory protein Vpu binds to CD4 and down-regulates its expression via ubiquitin-mediated proteasomal degradation.45,46,47
Fig. 1.
HIV-1 Env trafficking. Env is synthesized and glycosylated in the rough endoplasmic reticulum (RER) as a 160 kDa precursor protein (gp160), which oligomerizes predominantly into trimers. Precursor Gag (Pr55Gag) is synthesized on cytosolic ribosomes and is directed to the plasma membrane (PM) where it multimerizes in lipid rafts (not shown) to form virus particles. Oligomerized gp160 is transported to the Golgi and trans-Golgi network where it is processed to yield mature SU gp120 and TM gp41. Complexes of gp120-gp41 traffic through the secretory pathway to the PM and are incorporated as trimeric spikes into virus particles. At the PM, endocytosis of Env into early endosomes (EE) occurs via interaction with clathrin adaptor complexes. The internalized Env can either traffic through the late endosomes/multivesicular bodies (LE/MVBs) for degradation in the lysosome, or be returned to the PM through recycling endosomes (not shown). Gag and Env domains are defined in the inset at the top left. Illustration adapted from Ono and Freed229 with permission from Elsevier.
In the Golgi, gp160 is proteolytically cleaved by cellular furin or furin-like proteases48 at a highly conserved K/R-X-K/R-R motif,49,50 to yield the mature surface (SU) glycoprotein gp120 and the transmembrane (TM) glycoprotein gp41. Proteolytic processing of gp160 is absolutely required for activation of the fusogenic activity of Env and is thus essential for viral infectivity. Following cleavage, gp120 and gp41 remain associated by noncovalent interactions. Three molecules each of gp120 and gp41 form a heterotrimeric HIV-1 glycoprotein spike. After its arrival at the PM, Env is rapidly recycled via endocytosis,51,52 driven by interactions with the cellular clathrin adaptor complexes (see below). The internalization of Env and the shedding of gp120 from the cell surface due to the relatively weak and non-covalent nature of the gp120-gp41 interaction are factors that contribute to relatively low levels of Env incorporation into virus particles (~10 spikes/virion).53 The maintenance of low cell-surface and virion-associated levels of gp120 and gp41 likely help HIV-1 evade the host immune response and decrease virus-induced cytopathicity.
HIV-1 spread from one cell to another occurs much more efficiently at points of cell-cell contact known as infectious or virological synapses (VS)54,55,56,57,58 than spread as cell-free virus.59 Env and Gag both accumulate at the VS, along with actin, CD4, coreceptors, adhesion molecules, and tetraspanins.56,60,61,62,63 Env–receptor contacts have been reported to be required for VS formation induced by HIV-1 in T cells60 and by murine leukemia virus (MLV) in fibroblasts.64,65 In the MLV system, the Env glycoprotein appears to direct Gag assembly to the VS in a manner dependent on the CT of Env.66 In macrophages, as in T cells, both Gag and Env accumulate at the VS but Gag localization to the site of cell-cell contact is fairly Env-independent.33,67 How Env is directed to the synapse remains a question of central importance, given the vital role that HIV-1 transfer across the VS is likely to play in vivo.
Env Structure
gp120
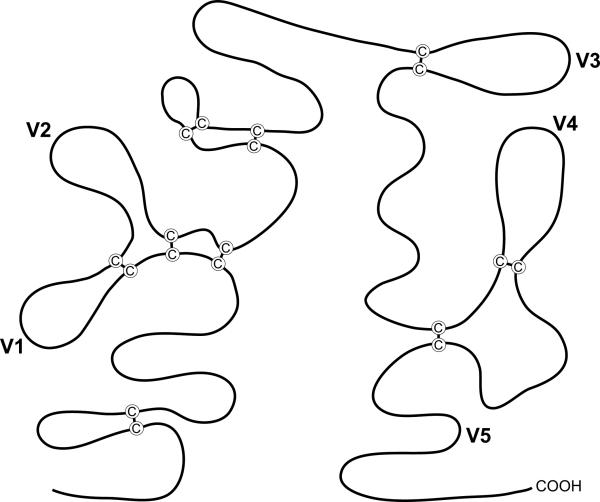
Comparison of env gene sequences from diverse HIV-1 isolates identified a high degree of variability in discontinuous segments within the gp120 subunit. gp120 has five variable domains (V1-V5) interspersed with five relatively constant domains (C1-C5) (Fig. 2).68,69 Also, a number of highly conserved Cys residues are located throughout gp120 and gp41. These Cys residues form intramolecular disulfide bonds that are crucial to the formation of Env tertiary structure (Fig. 3).39 Typically, gp120 contains 18 Cys residues that are covalently bound to form nine disulfide bridges. V1 is separated from V2 by two disulfide bonds but the two loops are contained in a larger loop by yet another disulfide bond; V3 and V4 loops are also delimited by a disulfide bond. Several studies have reported changes in the number and distribution of Cys residues that could cause changes in Env structure and antigenicity.70,71
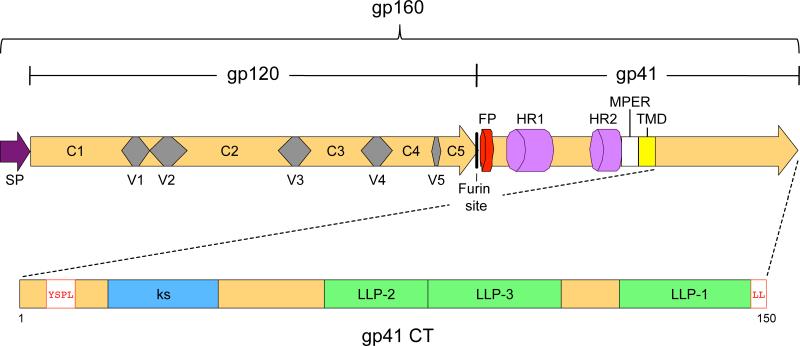
Fig. 2.
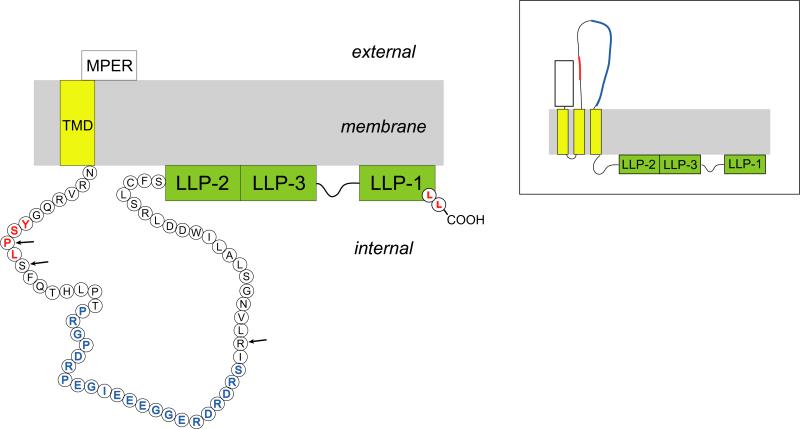
Domains of HIV-1 Env. Precursor gp160 contains the signal peptide (SP), which is cleaved during translation. The remaining precursor is cleaved into the surface subunit (gp120) and transmembrane subunit (gp41) in the Golgi complex at the furin site indicated. gp120 contains five variable domains (V1-V5) and five constant domains (C1-C5). gp41 consists of an extracellular domain, containing the fusion peptide (FP), heptad-repeats (HR1 and HR2), and the membrane-proximal external region (MPER), a transmembrane domain (TMD), and a cytoplasmic tail (CT). An enlarged representation of the gp41 CT is shown to highlight several motifs: the internalization signal YSPL, the Kennedy sequence (ks), the amphipathic α-helices LLP-1, -2, -3, and a C-terminal dileucine motif (LL) involved in endocytosis and intracellular distribution of Env.
Fig. 3.
Schematic representation of gp120 disulfide bonds. Highly conserved cysteine (C) residues in gp120 form disulfide bridges that are important for Env tertiary structure. Variable domains V1-V5 are indicated. Illustration adapted from Leonard et al.39, originally published in The Journal of Biological Chemistry. © The American Society for Biochemistry and Molecular Biology.
Approximately half of the molecular mass of gp120 is composed of N-linked glycans38 with a small additional contribution from O-linked sugars.40 There are 20-35 N-linked glycosylation sites in gp120 and 3-5 in gp41 that mask Env from host immune recognition,72 contribute to Env folding,73 and help virions bind to the host cell surface.74
Sequence variability in the V domains arose through recombination, point mutations, insertions and deletions. The V1V2 domain is the most variable in loop length and number of glycosylation sites.75,76,77,78,79,80 The V1V2 loop can range in length from 50-90 amino acids (aa), while the length variation of V4 and V5 loops ranges from 19-44 aa and 14-36 aa, respectively. The V3 loop and C2, C3, and C4 domains show relatively little length variation. Furthermore, a correlation exists between an increase in length and number of glycosylation sites in V1V2 and disease progression, suggesting that modification of V1V2 length may promote escape from the host humoral immune response.81
The CD4 receptor binding site is formed from conserved residues in discontinuous segments that are folded into proximity in the Env tertiary structure. The conserved domains within gp120, in particular C1, C3 and C4, are the principle Env determinants that bind CD4.82,83,84 The variable domains, which are exposed on the gp120 surface, do not have a direct role in CD4 binding, as a gp120 variant from which V1, V2, and V3 domains are deleted still binds to CD4 with high affinity.85,86 A number of studies have shown that the V3 loop is important for membrane fusion,87 coreceptor specificity,88,89,90,91,92 and contains dominant epitopes recognized by neutralizing antibodies.93,94,95,96 The switch from R5 to X4 tropism driven by V3 mutation has been linked to an increase in the net positive charge of V3, allowing an interaction with the negatively charged surface of CXCR4.97,98,99 Mutations in the tip and base of V3 as well as changes in the so-called “bridging sheet” of gp120 (see below) can influence coreceptor binding.100,101,102,103 The V3-CCR5 interaction is the target of the FDA-approved drug maraviroc, which binds directly to CCR5 to prevent gp120 association.104 As one would expect for a CCR5-based drug, maraviroc is not active against viral isolates that enter via CXCR4. Interestingly, resistance to maraviroc arises not by switching coreceptor use to CXCR4 but primarily by acquiring mutations that allow gp120 to effectively utilize drug-bound CCR5.105,106
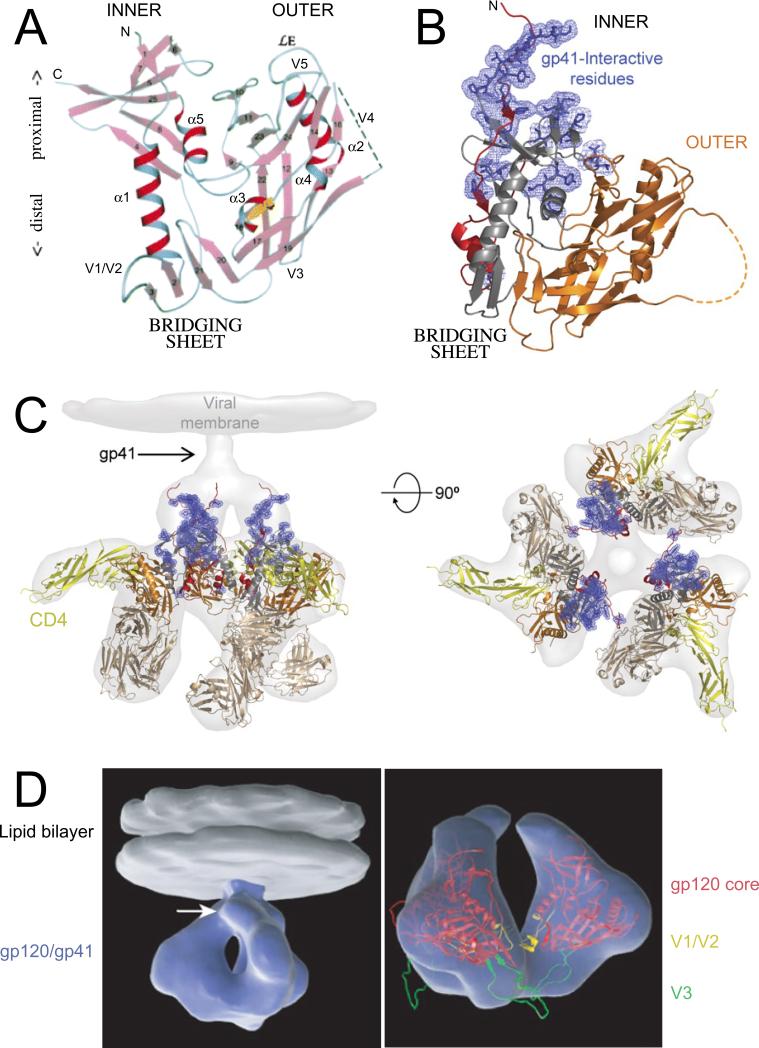
A number of gp120 structures have been solved by X-ray crystallography. These include the HIV-1 gp120 “core” (from which N- and C-termini and V1/V2 and V3 variable loops were deleted) in complex with fragments of CD4 and a neutralizing antibody;107 the SIV “unliganded” gp120 (not bound to CD4);108 HIV-1 gp120 unliganded and CD4-bound109; the gp120 core with V3 loop, bound to CD4 and neutralizing antibody;110 the gp120 core locked in the CD4-bound conformation associated with a CD4 binding-site-specific neutralizing antibody;111 the gp120 core in complex with poorly neutralizing CD4 binding-site-specific antibodies;112 and the gp120 core bound to the gp120-interacting portion of gp41.113 These studies have provided a wealth of information on the mechanism of gp120 interactions with CD4, coreceptor, gp41, neutralizing antibodies, and entry inhibitors. The overall fold of CD4-bound gp120 consists of an “inner” domain and an “outer” domain (so named for their putative orientation in the trimeric complex) linked by a four-stranded “bridging sheet” (Fig. 4A, B).107 CD4 is observed to directly contact 26 residues in gp120;107 these are widely dispersed in gp120, consistent with previous mutagenesis studies.83,84,114 Comparison of CD4-bound and unliganded gp120 structures suggests that gp120 undergoes a remarkable degree of refolding following CD4 binding. This large CD4-induced conformational change results in the formation of the coreceptor binding surface from residues that were spatially well separated prior to CD4 binding.108 This strategy allows the virus to delay the formation of the vulnerable coreceptor binding surface until the Env complex is in close contact with the target cell, at which time steric occlusion prevents effective access by neutralizing antibodies. Fitting the X-ray data into density maps obtained by electron tomography allows for models to be constructed for the gp120 trimer in unliganded and CD4-bound states (Fig. 4C, D).115 The picture that emerges from these structural studies is that the HIV-1 Env glycoprotein complex is exquisitely evolved to keep key functional surfaces hidden from antibody recognition and sufficiently plastic and tolerant of sequence variation to escape a robust host immune response.
Fig 4.
HIV-1 Env gp120 and gp41 structures. (A) Ribbon diagram of gp120 core containing α-helices (α1-5), β-strands (1-25), with relative positions of variable loops (V1-V5) and N and C-termini shown. The orientation of gp120 in this diagram places the viral membrane toward the top and the cell membrane toward the bottom. When gp120 is bound to CD4 it forms a “bridging sheet” consisting of four β-strands, which separates the inner and outer domains of gp120 relative to their orientation in the trimeric complex. Image reprinted by permission from Macmillan Publishers Ltd: Nature107, copyright 1998. (B) Ribbon diagram of gp120 core (as in panel A) with N- terminus (red) and gp41 interaction site (blue) shown. The inner domain is shown in red and grey and the outer domain is shown in orange. The bridging sheet, which shares elements from both inner and outer domains, is in grey and orange. (C) Trimeric gp120 (same colors as in panel B) bound to three molecules of CD4 (yellow) and Fab from neutralizing antibody 17b (brown), used to stabilize the gp120 structure, superimposed onto the electron density observed by cryoelectron tomography (light grey). Orientation of this structure rotated 90°, which places the viral membrane in the plane of the page, is also shown on the right. Images (panels B, C) reproduced from Pancera et al.113 with permission. (D) Three-dimensional representation of HIV-1 Env in its CD4-bound conformation. (Left) A trimeric Env spike (blue) anchored in the lipid bilayer of the viral membrane (grey) is shown. The white arrow indicates the predicted location of gp41. (Right) Ribbon diagram of gp120 core (red) superimposed on the density map (blue) with V1/V2 loop (yellow) and V3 loop (green) shown. Reprinted by permission from Macmillan Publishers Ltd: Nature115, copyright 2008.
gp41
The gp41 TM glycoprotein mediates fusion of the viral envelope with host cell membrane. The gp41 subunit is comprised of ~345 aa, organized into three major domains: an extracellular domain (or ectodomain), a transmembrane domain (TMD), and a C-terminal cytoplasmic tail (CT) (Fig. 2). The extracellular domain contains the major fusion determinants: an N-terminal hydrophobic region known as the fusion peptide,116,117,118 a polar region, two hydrophobic regions that form α-helical coiled-coil structures referred to as the heptad-repeat regions HR1 and HR2 (also known as N-helix and C-helix, respectively),119,120,121,122,123 and a Trp-rich domain referred to as the membrane-proximal external region (MPER).124,125 HR1 and HR2 are connected by a disulfide-bridge within a hydrophilic loop and their interaction drives the fusion process. The fusion peptide is normally buried in the gp120/gp41 quaternary complex. Binding of gp120 to CD4 and coreceptor changes the structure of gp41 leading to exposure of the fusion peptide and its penetration into target cell membrane, thus causing membrane destabilization126 and formation of the fusion pore. Three HR1 motifs form a core bundle in parallel and fold over a hydrophobic groove antiparallel to three HR2 domains within each trimer, thus forming a stable six-helix bundle that brings the viral and cell membranes into close enough proximity for fusion to occur.119,123 General structural features of the gp41-induced fusion mechanism are shared with those of other viral fusion proteins (e.g., hemagglutinin of influenza)127 and some cellular fusion proteins (e.g., the SNAREs).128 Formation of the gp41 six-helix bundle can be blocked by peptides derived from either HR1 or HR2.129,130,131 One such HR2-derived peptide, known as enfuvirtide, T-20, or fuzeon, is approved for use in HIV-1-infected patients. Its high cost and lack of oral bioavailability, however, limit its clinical application.132
The MPER consists of the last 24 amino acids of the gp41 extracellular domain. This region is required for fusogenicity and virus infectivity;124,125,133 however, the mechanism by which it functions in promoting membrane fusion is not entirely clear. The MPER is highly conserved and is recognized by several well-studied neutralizing antibodies (2F5, 4E10, and Z13).134,135
The gp41 TMD consists of ~25 highly conserved amino acids that anchor Env in the lipid bilayer. The highly conserved nature of the gp41 TMD suggests that it plays specific roles in Env function, and, indeed, mutations in the “core” region of the TMD influence Env-mediated fusion.136,137,138 The traditional model for gp41 topology proposes that the TMD is an α-helix that spans the membrane once and the CT is inside the virus particle (Fig. 5A). An alternative topology has been proposed in which gp41 spans the membrane three times, initially based on recognition of an epitope within the gp41 CT by neutralizing monoclonal Abs (Fig. 5B, “Kennedy” sequence). 139,140,141,142 Similarly, Lu et al. recently demonstrated that the LLP-2 region in gp41 CT may be transiently exposed on the cell surface during cell-cell fusion.143 However, more recent studies suggest that the traditional “single-pass” model most accurately reflects the topology of gp41 in virions. For example, antibodies that bind the Kennedy sequence on Env-expressing cells do not bind Env on intact virions.144 Furthermore, viral PR cleavage sites flanking the Kennedy sequence were acquired during selection for HIV-1 resistance to the cholesterol-binding compound amphotericin B methyl ester.145,146,147 The ability of PR to cleave this sequence in virions strongly implies that this region of gp41 must be inside the virus particle.
Fig 5.
Models of HIV-1 gp41 CT topology. (Left) Traditional model of the gp41 CT with a single membrane-spanning domain, most likely found in virions. This single-pass model contains the entire CT inside the virion (internal). gp41 domains are the same as indicated in Fig 2. Sites of mutation that confer resistance to AME, which were later shown to become new HIV-1 PR cleavage sites, are indicated (black arrows).145,146,147 (Right) Alternative three-membrane-spanning topology of gp41 CT, which exposes portions of the gp41 CT to the extracellular space, may also exist with a detectable frequency in HIV-1 Env-expressing cells in addition to the more traditional model. Illustration adapted from Steckbeck et al.144
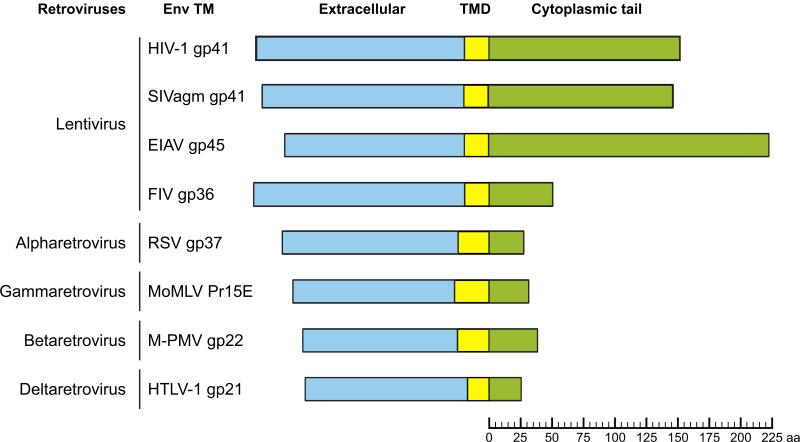
The TM subunit of most lentiviral Env glycoproteins has a very long CT compared to those found in other retroviruses (Fig. 6).34 For example, the gp41 CTs from HIV-1, HIV-2, and SIVs are approximately 150 aa in length; the equine infectious anemia virus (EIAV) CT is even longer (~200 aa).148,149 In contrast, the TM CTs of other prototypic retroviruses, such as Rous sarcoma virus (RSV, an alpharetrovirus), Mason-Pfizer monkey virus (M-PMV, a betaretrovirus), and murine leukemia virus (MLV, a gammaretrovirus), are only ~20-40 aa long (Fig. 6). The presence of a very long CT in the vast majority of lentiviral TM Env glycoproteins suggests that they evolved to serve specific functions in lentiviral replication, a hypothesis supported by the observation that truncation of the lentiviral CT leads to suppression of virus replication in animal models.150 The CT of HIV-1 gp41 influences multiple properties of the gp120/gp41 glycoprotein complex, such as Env incorporation into virus particles, virus infectivity, cell-surface Env expression, gp120 shedding, and Env-induced fusion151,152,153,154,155,156,157,158,159,160,161; discussed in more detail below). The CTs of HIV-1 and SIV gp41 also influence the conformation of gp120 and the ectodomain of gp41 such that CT mutations affect antibody recognition and neutralization.162,163,164,165 It is interesting to note that some HIV-1 isolates that have acquired CD4 independence bear truncations in their gp41 CTs,163,166 again consistent with a role for this domain of Env in regulating gp120 conformation.
Fig. 6.
Schematic representation of retroviral TM subunits, indicating the lengths [in amino acids (aa)] of their respective cytoplasmic tails. As illustrated, lentiviral TM CTs tend to be significantly longer than those of other retroviruses, with the exception of feline immunodeficiency virus (FIV). SIVagm, simian immunodeficiency virus from African green monkey; MoMLV, Moloney-MLV; HTLV-1, human T-cell lymphotropic virus type 1. Other viral acronyms are defined in the text.
The activities of the gp41 CT are modulated by several functional and structural domains (Fig. 2, 5). A membrane-proximal tyrosine-based sorting signal with the consensus sequence YxxL interacts with the clathrin adaptor protein complex 2 (AP-2) to mediate clathrin-dependent endocytosis of Env from the PM.167,168,169,170 This motif has also been implicated in the targeted release of virions from the basolateral surface of polarized epithelial cells, 171 cell-cell transmission in T cells,172 and particle infectivity.173,174 Mutation of the analogous signal in SIV resulted in attenuation of virus replication in vivo.175 The intracellular distribution of Env is also regulated by a dileucine motif in the gp41 CT; this signal interacts with the clathrin adaptor protein complex 1 (AP-1).155,170,176 Endocytosis of Env via this motif is independent of the function exerted by the YxxL motif. Mutation of both the C-terminal dileucine motif and the YxxL motif is sufficient to completely inhibit cell surface downregulation of Env, suggesting that both motifs account for most Env internalization.176
Three conserved amphipathic α-helical segments that are referred to as lentiviral lytic peptides (LLP-1, LLP-2, and LLP-3) are present in the central and C-terminal regions of the gp41 CT (Fig. 2, 5).177,178,179,180 LLP-1 and LLP-2 segments are highly positively charged due to the presence of Arg residues on one face of the α-helix. LLP-3 is located between the other two segments. The structure of the LLPs is highly conserved among lentiviruses.177 LLP domains have been implicated in Env fusogenicity,160 protein stability,181 multimerization,182 cell surface expression,183 and incorporation.159,184,185 Also, LLP fragments have been shown to bind and perturb membranes,186,187,188,189 causing cytolysis, hence the name “lytic peptides”.178 Mutation of the Arg residues in LLP-1 to Glu, which allows the preservation of the secondary structure and hydrophilicity of this segment, results in loss of cell lysis activity, implying that these residues are important for LLP function.190 Although the results are somewhat controversial, the CT contains two Cys residues that are palmitylated, a post-translational modification that has been proposed to target Env to lipid rafts.191 The LLP-1 segment has also been proposed to contain the determinants for lipid raft association, as truncation of LLP-1 led to a decrease in localization of Env to lipid rafts192 (see below for further discussion of this topic). It is clear from the discussion above that the biological functions of the gp41 CT are diverse and somewhat enigmatic; a more complete understanding of this region of Env will require careful analysis of its biological properties in relevant primary cell types, evaluation of its role in cell-cell transfer, and the identification of cellular interaction partners.
D. General models for HIV-1 Env glycoprotein incorporation
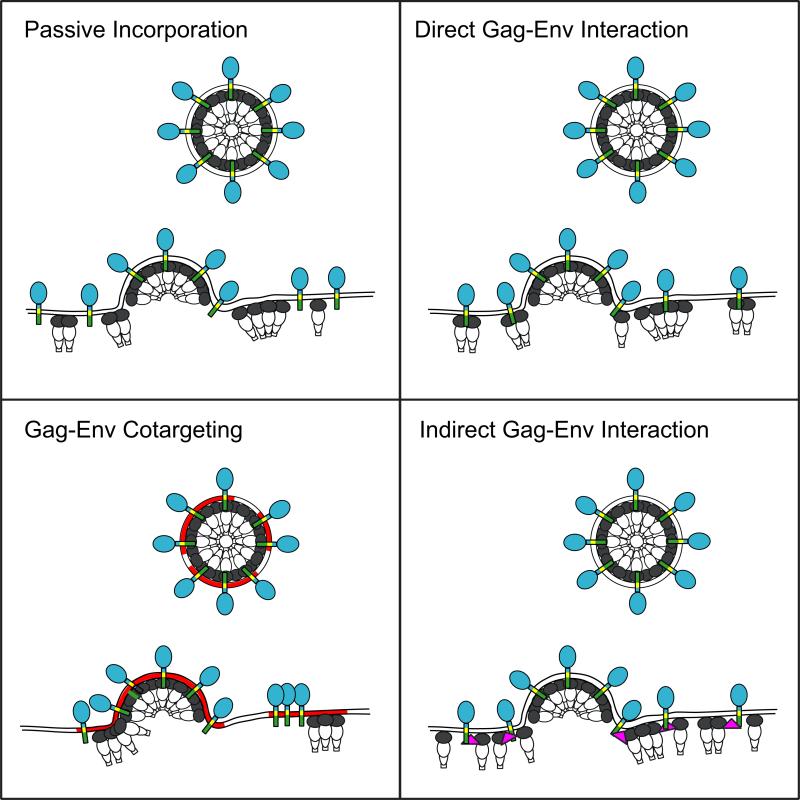
Several general models can be invoked to explain the incorporation of HIV-1 Env glycoproteins into virus particles: 1) The “passive” or “random” incorporation model (Fig. 7A) postulates that the Env glycoprotein complex is incorporated into virions simply as a result of its expression on the host cell PM. This model requires no mechanism for concentrating Env at sites of virus assembly. 2) The “direct Gag-Env interaction” model (Fig. 7B) proposes that direct binding between the MA domain of Gag and the CT of gp41 recruits Env glycoprotein spikes into virus particles. 3) The “Gag-Env cotargeting” model (Fig. 7C) postulates the existence of a cellular structure (e.g., a PM microdomain) that acts as an assembly platform to which both Gag and Env are recruited, thereby enhancing the efficiency of Env incorporation. 4) The “indirect Gag-Env interaction” model (Fig. 7D) invokes the existence of a host cell protein that serves as a bridge between Gag and Env to promote Env incorporation.
Fig. 7.
Models for HIV-1 Env incorporation. The passive incorporation model (A) postulates no interaction between Gag and Env. The direct Gag-Env interaction model (B) takes into account evidence that the MA domain of Gag interacts with the CT of gp41. The Gag–Env cotargeting model (C) suggests that Env is enriched in virus assembly domains through lipid raft (brown) association. The indirect Gag–Env interaction model (D) provides an alternative explanation for Env incorporation that involves an adaptor protein that binds both Env and Gag. The association of Gag with lipid rafts in all models is assumed but not shown. The color scheme for Gag and Env is the same as in Fig. 1.
Passive incorporation model
The passive model draws support from the observation that host cell membrane proteins are abundantly incorporated into virus particles. In contrast to some enveloped viruses that tightly restrict the packaging of host proteins (e.g., the alphaviruses),193,194 retroviruses are fairly promiscuous in their virion incorporation of host membrane proteins.195,196,197 Furthermore, non-HIV viral glycoproteins can be readily incorporated into HIV-1 virions to generate pseudotyped particles that display high levels of particle infectivity. For example, amphotropic murine leukemia virus (ampho-MLV) Env and vesicular stomatitis virus G glycoprotein (VSV-G) are frequently used to pseudotype HIV-1 particles.198,199 Finally, a number of studies have shown that removal of most or virtually all of the gp41 CT has only a modest effect on Env glycoprotein incorporation into HIV-1 particles produced in some commonly used laboratory cell lines (e.g., HeLa, 293T, CV-1, and COS).152,156,157,158,200 Based on these findings, it would seem plausible that passive incorporation could be sufficient to account for the ~10 copies53 of gp120/gp41 heterotrimeric spikes present on HIV-1 virions.
Direct Gag-Env interaction model
Despite the apparent lack of a requirement for the long gp41 CT in Env incorporation in some laboratory cell lines, a number of studies demonstrated that the MA domain of Gag is required for Env incorporation.156,157,201,202 Deletion of the gp41 CT reversed the Env incorporation defect imposed by mutations in MA,156,157,203 or MA deletion,204 suggesting that incorporation of full-length Env requires an interaction with MA, whereas incorporation of truncated Env does not require Env-MA binding. Although deletion of the entire gp41 CT has little effect on Env incorporation in HeLa and 293T (COS) cells, somewhat paradoxically, smaller deletions or truncations within the tail lead to lower levels of Env in virus particles.153,154,159,185,205,206 Although in some cases the effects of these gp41 CT mutations can be attributed to reduced cell-surface Env expression, in other cases the CT mutations appear to specifically impair Env incorporation. Interestingly, the effects of a gp41 CT deletion that reduced Env incorporation could be reversed by a point mutation in MA that was acquired during propagation of the gp41 mutant virus in culture.159
X-ray crystallography studies revealed that HIV-1 and SIV MA form a trimeric lattice containing holes that could potentially accommodate a trimer of gp41 CTs.18,207 This led to the model that the gp41 CTs might project into these holes, thereby permitting direct contacts to take place between residues in MA and gp41. Indeed, several key residues shown to be required for Env incorporation (MA aa 12, 30, and 34)156,157,159,208 appear to face the inside of the largest hole in the trimeric lattice. While this is still a viable model, there is currently little evidence supporting the formation of MA trimers in virions. Also, uncertainty remains about the orientation of the gp41 CT with respect to Gag; i.e., do the CTs “swim” in the plane of the membrane or do they project toward the center of the virion parallel to the radial orientation of Gag? It is also possible that MA-gp41 interactions, and perhaps the orientation of gp41 CTs relative to Gag, change during the transition from immature to mature particles that accompanies PR-mediated Gag processing.
Evidence of cross-talk between Gag and gp41 during assembly also derives from studies of Gag and Env localization in polarized epithelial cells. HIV-1 Env was shown to localize to the basolateral surface of Madin-Darby canine kidney (MDCK) polarized epithelial cells.209 In the absence of Env expression, Gag budded in a non-polarized manner in this cell type. However, when Env and Gag were coexpressed, budding was restricted to the basolateral surface.210 Mutations in MA that prevented Env incorporation, as well as truncations in the gp41 CT, reversed the ability of Env to direct basolateral budding of Gag.211 Polarized budding reportedly requires the Tyr-based (YxxL) sorting signal located in the gp41 CT close to the transmembrane domain.171 Together, these findings are consistent with the concept that Env and MA associate in some manner during virus assembly.
Additional findings provide support for the concept that MA and Env (specifically the gp41 CT) interact in virions. Mature virions, which contain fully processed Gag proteins, can fuse with the target cell membrane. This virion-cell membrane fusion activity can be detected in an assay in which virions are engineered to package the enzyme β-lactamase; upon fusion of the virion with target cells, β-lactamase is delivered into the target cell cytoplasm. This delivery of β-lactamase can be quantitatively detected by preloading the target cells with a fluorogenic β-lactamase substrate. In contrast to the robust virion-cell fusion activity observed with mature virions, immature virions, in which Gag remains unprocessed, are fusion defective.212 A similar defect in Env-mediated membrane fusion activity in immature virions can be measured in a “fusion-from-without” assay in which high concentrations of virions induce cell-cell fusion.213 In both of these studies, the fusion defect observed with immature particles could be reversed by full gp41 CT truncation,212,213 or, in a subsequent study, by removal of the C-terminal 28 amino acids of gp41.214 These results suggest that an interaction between the gp41 CT and uncleaved Gag locks Env in a non-fusogenic state; cleavage of Gag by the viral PR during virion maturation then activates the fusogenic activity of the Env glycoprotein complex. The suppression of Env-mediated fusion by uncleaved Gag could provide a mechanism for keeping fusion activity in check until virion maturation occurs, when the particle is ready to infect a new target cell. Intriguingly, virion maturation is reportedly accompanied by a significant reduction in particle “stiffness”, measured by nanoindentation atomic force microscopy methods.215 The degree of “stiffness” of the virus particle in this study was regulated by the gp41 CT, raising the possibility that this physical feature of the virion is influenced by Gag-gp41 interactions.215
Whereas HIV-1 modulates Env function via PR-mediated Gag processing (i.e., virion maturation), several other retroviruses have evolved an alternative strategy for regulating the fusogenicity of the Env glycoprotein complex until PR-mediated virion maturation takes place. In MLV, EIAV, and M-PMV particles, the viral PR cleaves the CT of the transmembrane Env protein. This PR-mediated cleavage of the Env CT is required for activating the fusogenicity of the Env glycoprotein complex. PR-mediated cleavage of the Env protein can occur in trans; MLV Env used to pseudotype HIV-1 virions undergoes HIV-1 PR-mediated processing of its Env CT. Interestingly, this PR-mediated Env cleavage can be blocked by specific mutations in MA.216 The simplest explanation for these results is that the MA mutations change the conformation of the MA shell underlying the lipid bilayer, thereby restricting the ability of PR to access and cleave the MLV Env CT. Although WT HIV-1 Env has not been observed to undergo PR-mediated gp41 CT processing, HIV-1 variants that have escaped the fusion-inhibitory activity of the cholesterol-binding compound AME acquire gp41 CT mutations that create novel cleavage sites for PR.146,147 PR-mediated cleavage of the gp41 CT allows virion-cell membrane fusion to proceed even in the presence of AME.
Evidence for modulation of Env function by MA also derives from a study demonstrating that a mutation in MA influences the association between gp120 and gp41, leading to increased shedding of gp120 from the virus particle.217 Again, the ability of MA to influence gp120 shedding requires the presence of an intact gp41 CT.217
A common theme in many of the studies discussed above is the apparent cross-talk between the MA domain of Gag and the gp41 CT. Despite the abundant circumstantial evidence in support of a MA-gp41 CT interaction, only a limited amount of supportive biochemical data have been obtained. Cosson reported a direct in vitro binding between the gp41 CT fused to glutathione-S-transferase (GST) and MA bound to β-Gal.184 Although the binding data were reinforced by the observation that mutations in both Env and MA that disrupt Env incorporation reduced the interaction, these results have not been independently confirmed. Similarly, Hourioux et al.218 used peptides representing the central region of HIV-1 gp41 CT (flanking LLP-2) to inhibit the immunocapture of membrane-free Pr55Gag particles by immobilized anti-p24CA antibody in vitro, again implying a direct Env-Gag interaction. Wyma and colleagues219 demonstrated that gp41 is retained in immature virus particles that were treated briefly with detergent to solubilize the viral lipid bilayer. The retention of gp41 and some gp120 in these detergent-stripped immature particles required the gp41 CT, consistent with the existence of an interaction (direct or indirect) between Gag and gp41. While successful co-immunoprecipitation of Gag and Env in HIV-1-expressing cells has not been reproducibly achieved, Vincent et al. reported such data for SIV.220
Gag-Env co-targeting model
Although, as discussed above, a considerable number of reports support the concept that the gp41 CT and MA interact during assembly to promote the recruitment of Env complexes into nascent virions, most of these findings do not distinguish between a direct gp41 CT-MA interaction and an indirect interaction mediated by a host cell protein or membrane structure. A compelling piece of evidence to support the role of host cell factor(s) in Env incorporation is the observation that the requirement for the gp41 CT in Env incorporation and virus replication is strongly cell-type-dependent. As mentioned above, in several popular laboratory cells lines (e.g., HeLa, COS, 293T), and in a small number of T-cell lines (notably MT-4 and M8166), the gp41 CT can be entirely deleted with little effect on Env levels in virions.152,158,221 These cell lines are referred to as being “permissive” for gp41 truncation. In contrast, the majority of T-cell lines and the more physiologically relevant primary cell types (T cells and monocyte-derived macrophages) are “non-permissive” for gp41 truncation; in these cells, removal of the gp41 CT induces a ~10-fold defect in Env incorporation and a complete block in virus replication.158 The basis for this cell-type-dependent requirement for the gp41 CT remains to be defined. A number of cellular factors (discussed in more detail below) have been reported that bind the gp41 CT; differences in the expression or localization of one or multiple host factors could account for the cell type dependence. SIVmac also displays interesting cell-type-dependent requirements for the gp41 CT in virus replication. When grown in human cells, SIVmac typically acquires truncations in the gp41 CT that facilitate efficient virus replication.222,223 When SIVmac variants with short CTs are passaged in monkey cells in culture or in vivo, strains encoding full-length gp41 predominate.222,223,224 As is the case for the HIV-1 gp41 truncations, the basis for this cell-type dependence remains to be defined; Vzorov et al. have suggested that gp41 truncation affects multiple stages of the virus replication cycle, including early, post-entry events.225
The host cell mediator of indirect Env incorporation could be a protein factor (discussed in more detail below) or a membrane microdomain or other cellular structure to which both Gag and Env are targeted. Membrane microdomains known as “lipid rafts”,226 which are enriched in cholesterol and highly saturated lipids (e.g., sphingolipids), have been studied extensively over the past two decades.227 Although somewhat controversial,228 there is now widespread agreement that cholesterol- and saturated lipid-enriched raft microdomains do exist and serve important functions in a variety of cellular processes.227 Lipid rafts have also been reported to promote the replication of a number of enveloped viruses, including retroviruses, by serving as organizing platforms for both virus entry and particle assembly (for reviews, see Refs.229,230,231). The highly saturated nature of lipid rafts causes them to be less soluble in cold non-ionic detergent (typically Triton X-100) than non-raft membrane.232 Although detergent-resistant membranes (DRM) isolated from lysed cells are unlikely to represent native membrane structures found in living cells,228 the presence of proteins in DRM is often used as evidence of their raft localization.233,234 Studies performed in the early 2000's showed that both HIV-1 Gag and, to a lesser extent, HIV-1 Env, are DRM-associated.191,235,236,237,238 Evidence that HIV-1 Gag and Env are raft associated also derives from microscopy analyses demonstrating copatching of Gag, Env, and cellular raft components.229,237,239 Furthermore, treating virus-producing cells with cholesterol-depleting, binding, or sequestering agents impairs HIV-1 particle production.145,236,237,240,241 In the case of cholesterol depletion, the defect in virus assembly and release is associated with reduced Gag–membrane binding and higher-order Gag multimerization.240 Lipid rafts often function as platforms for promoting protein–protein interactions; protein multimerization in turn stabilizes lipid rafts and induces their aggregation.227 Newly synthesized HIV-1 Gag appears to bind membrane rapidly, then more slowly acquires raft-associating properties, concomitant with Gag assembly and raft aggregation.236,242 Also consistent with the hypothesis that HIV-1 assembly takes place in cholesterol-enriched membrane microdomains is the observation that the viral envelope displays a rather striking raft-like composition (elevated levels of cholesterol and saturated lipids) relative to the host cell PM.243,244
The mechanism by which the HIV-1 Env glycoproteins associate with rafts and the domains required for this association have been somewhat controversial topics. An early report suggested that palmitylation of the gp41 CT is critical for both DRM association and Env incorporation. These findings were surprising given that the study was performed in 293T cells, and that as mentioned above, deletion of the entire gp41 CT has little effect on Env incorporation in this cell line.158 Subsequent studies found that mutating the gp41 CT Cys residues that serve as palmitate attachment sites either had no effect on Env–DRM association, Env incorporation, or virus replication245 or reduced DRM association with only a small effect on virus infectivity.246 While these two studies differ somewhat in their findings, probably due to the use of different viral isolates and producer cell types, both concluded that palmitylation of the gp41 CT is unlikely to play a major role in virus replication efficiency, at least in culture. So what is the mechanism by which Env is recruited to lipid rafts? Bhattacharya and colleagues247 reported that Gag directs the recruitment of Env glycoproteins to lipid rafts; Env co-expressed with Gag was found in DRM, whereas Env expressed in the absence of Gag was not DRM-associated. Likewise, mutations in either Env or MA that disrupt Env incorporation abrogated the putative raft association of Env. However, the role of Gag in targeting Env to rafts does not exclude a role for Env in polarized targeting of Gag. Many reports suggest that the membrane-proximal Tyr-based sorting signal in gp41 CT may polarize HIV release in some cell types.171,172,211
Two recent studies that examined the mechanism by which viral pseudotypes are generated have bearing on the Gag-Env co-targeting hypothesis. Leung et al.248 investigated whether HIV-1 Env glycoproteins and the Ebola virus glycoprotein (GP), when expressed in the same cell, are copackaged into HIV-1 virions or segregated into two distinct populations of virus particles. HIV-1 virions produced from cells expressing both HIV-1 and Ebola GP were treated with neutralizing antibodies against each glycoprotein, immunodepletion analyses were performed, and colocalization between HIV-1 and Ebola GP was analyzed. The surprising and interesting result was that virions appeared to contain either HIV-1 Env or Ebola GP, but not both. This result suggests that HIV-1 Env and GP segregate into distinct microdomains on the cell surface, each of which associates with individual virions. The microdomains to which HIV-1 Env and GP segregate could be lipid rafts or some other type of microdomain or subcellular structure. Jorgenson and colleagues249 examined the coclustering of three different retroviral Env glycoproteins – those of Rous sarcoma virus (RSV), MLV or HIV-1 – with RSV or HIV-1 Gag proteins. Scanning electron microscopy and immuno-electron microscopy showed that none of the viral glycoproteins demonstrated appreciable clustering in the absence of Gag expression. RSV Env clustered at budding sites in the presence of RSV but not HIV-1 Gag; the coclustering of RSV Env and Gag required the CT of RSV Env. MLV and HIV-1 Env both clustered with HIV-1 Gag at virus budding sites. These data support a role for Gag in recruiting Env to PM microdomains (lipid rafts or potentially other distinct structures) and highlight interesting differences in the segregation behavior of different retroviral Env glycoproteins. Direct interactions between Gag and Env from distantly related retroviruses seem unlikely, arguing for co-recruitment of Gag and Env to specific PM structures or microdomains. These data are also inconsistent with the passive incorporation model, according to which no mechanism for co-clustering of Gag and Env is required.
Indirect Gag-Env interaction model: gp41 CT-binding host factors
The Gag-Env co-targeting model discussed above posits that Gag and Env are recruited, by unknown mechanisms, to a common site on the PM (for example, a lipid raft). It also remains possible that, at least under some circumstances, a cellular protein could act as a bridge between MA and the gp41 CT to recruit Env complexes into virions. A number of host proteins have been reported to interact with MA and/or the gp41 CT.
The best understood binding partners of the gp41 CT are the clathrin adaptor protein complexes AP-1 and AP-2. These complexes direct the sorting and trafficking of proteins in the secretory and endocytic pathway. As mentioned earlier, AP-2 drives the clathrin-mediated endocytosis of Env from the PM by binding to the YXXL motif in the gp41 CT.167,168,169,170 AP-1 mediates the trafficking of vesicles between the TGN and endosomes and regulates the subcellular localization of Env via binding to a dileucine motif in the gp41 CT (Fig. 2).155,170,176 AP-1 and AP-2 have also been shown to bind HIV-1 Gag in the MA domain and at the MA-CA junction, respectively.250,251
Tail-interacting protein of 47 kDa (TIP47) was reported to play a direct role in Env incorporation. TIP47, also known as mannose-6-phosphate receptor (MPR) binding protein 1, was first described as having a role in the retrograde traffic of MPRs from late endosomes to the TGN252 via binding to Rab9.253 However, subsequent reports showed that TIP47 does not colocalize with MPR or Rab9, does not bind Rab9 in vitro, or affect the trafficking or function of MPRs. Instead, TIP47 appears to be involved in the biogenesis of lipid droplets,254,255,256 although early reports were not entirely consistent.257 Indeed, TIP47 is a member of the PAT [for perilipin/adipose differentiation-related protein (APRP)/TIP47] family of lipid droplet-associated proteins.258 Berlioz-Torrent and colleagues identified an interaction between TIP47 and the gp41 CT by yeast two-hybrid and GST pull-down assays. 259 As further evidence of a TIP47-gp41 CT interaction, mutation of a YW diaromatic motif in LLP-3 of the gp41 CT resulted in decreased binding of TIP47 by GST pull-down and yeast two-hybrid assays and decreased targeting of Env to the TGN. YW motif mutation disrupted virus replication in the Jurkat T-cell line and induced a defect in Env incorporation. Deletion of the YW motif had previously been shown to delay virus replication and impair Env incorporation, defects that were corrected upon acquisition by the virus of a compensatory mutation in MA.159 TIP47 was later proposed to act as a linker between the gp41 CT and MA, thus allowing efficient Env incorporation in both Jurkat and HeLa cells.260 In this study, yeast two-hybrid and coimmunoprecipitation (coIP) assays demonstrated that TIP47 interacts with a WE motif near the N-terminus of MA. Lopez-Verges et al. also showed that coIP of gp41 by Gag was reduced when TIP47 was depleted. Subsequently, it was shown that HIV-1 with mutations in the Env YW or MA WE motifs displayed defects in replication and infectivity in monocyte-derived macrophages, providing indirect evidence that TIP47 is involved in the production of infectious HIV-1 particles in this primary cell type.261 However, a role for TIP47 in HIV-1 biology has not been independently confirmed, and our own studies have failed to support a role for TIP47 in Env incorporation, particle production, or virus replication (Checkley and Freed in preparation). The role of TIP47 in HIV-1 replication thus remains uncertain.
Human discs large protein (Dlg1/hDlg1/Dlgh/synapse-associated protein 97) is a scaffolding protein implicated in the organization of multiprotein complexes at the immunological synapse.262 Blot and colleagues263 demonstrated that Dlg1 interacts with the CT of HTLV-1 Env and colocalizes with HTLV-1 Env and Gag in infected cells. In follow-up work, Perugi et al. investigated whether this protein functions in HIV-1 replication. They observed that Dlg1 binds HIV-1 Gag; Dlg1 depletion was found to upregulate HIV-1 Env expression and increase Env levels in virions. These effects correlated with an accumulation of both Gag and Env in putative late endosomal (CD63+) intracellular compartments and PM-associated tetraspanin-enriched microdomains.264 The mechanism responsible for the altered Gag and Env localization and the increased Env expression and incorporation induced by Dlg1 depletion remains to be elucidated.
Calmodulin, p155-RhoGEF, α-catenin, prenylated Rab acceptor (PRA1/PRAF1), luman, and the prohibitin 1/prohibitin 2 heterodimer have also been observed to interact with HIV-1 gp41, however a clear role for these proteins in Env function has not been established. HIV-1 gp41 and peptide sequences derived from LLP-1 and LLP-2 in the gp41 CT, which structurally resemble calmodulin-binding proteins, 179,265 have been shown to interact in vitro with calmodulin.190,266,267,268 Calmodulin senses and binds calcium to function as a signal transducer. Increased Env levels are associated with upregulated calmodulin expression269,270 and sensitivity to Fas-mediated apoptosis in certain cell types.269,271,272 Based on these findings, the interaction of the gp41 CT with calmodulin has been proposed to sequester calmodulin and consequently lead to T cell anergy and apoptosis, which are linked to AIDS progression.267,272,273 Recent studies have provided evidence that MA also binds calmodulin in a calcium-dependent manner in vitro.274,275 Nuclear magnetic resonance (NMR) data revealed that upon binding to calmodulin MA undergoes a marked conformational change,274,275 which triggers exposure of the myristic acid moiety covalently attached to the N-terminus of MA.275 Myristate exposure is crucial for binding of Gag to membrane.276
Kim et al.277 identified an interaction between gp41 from HIV-1 or SIV with the C-terminal domain of α-catenin in a yeast two-hybrid screen. α-catenin has been implicated in the regulation of actin filament assembly and dynamics278 and proposed to play a role in cell-cell contact formation.279 An interaction between the gp41 CT and α-catenin was later confirmed by in vitro binding assays and the binding domain was mapped to the LLP-3 domain of the gp41 CT.280 HIV-1 Env was shown to colocalize with α-catenin at the PM of HeLa cells microinjected with Env and α-catenin expression vectors.280
LLP-3 has been reported to interact with the C-terminal regulatory domain of the guanine nucleotide exchange factor p115-RhoGEF.281 This interaction may be physiologically important, as mutation of residues that abolish binding to p115-RhoGEF impairs HIV-1 replication. p115-RhoGEF activates RhoA GTPase, which stimulates actin stress fiber formation 282,283 and activates serum response factor (SRF), which in turn induces cell proliferation.284,285,286 These p115 activities were impaired when the gp41 CT was expressed in murine fibroblasts.281 A RhoA-derived peptide has been shown to inhibit CXCR4-dependent HIV-1 entry in culture;287 however, this anti-HIV activity is linked to the polyanionic properties of the RhoA peptide, suggesting that the peptide may disrupt the infection process simply by interfering with gp120/CXCR4 binding, and not as a result of a requirement for RhoA binding during entry.
PRA1 is a Rab protein regulator that binds prenylated Rab GTPases to mediate the trafficking of Rabs in vesicles.288 PRA1 localizes primarily to the Golgi289,290 and interacts with Rab3A and vesicle-associated membrane protein 2 (VAMP2) to regulate vesicle docking and fusion in the Golgi complex and the PM.291 PRA1 was first identified as a cellular binding partner of the SIV gp41 CT in a yeast two-hybrid screen.292 In this same study, PRA1 also interacted with the Env CT from various lentiviruses, including HIV-1, EIAV, and FIV, in a mammalian two-hybrid system. However, the significance of this interaction is not clear. Despite the observation that PRA1 colocalizes with SIV gp41 in 293T cells, modulation of PRA1 protein levels by RNAi or overexpression does not affect SIV or HIV-1 release, infectivity, or Env incorporation.293 Nonetheless, studies have also shown an interaction of PRA1 with proteins from other viruses, including a lipid raft-associated rotavirus spike protein (VP4)294,295 and the LMP1 oncoprotein of Epstein-Barr virus296 (reviewed in Ref.297).
Luman is an ER-membrane-bound transcription factor of the CREB3 family that is involved in ER stress responses, such as the unfolded protein response (UPR) and ER-associated degradation (ERAD). During ERAD, ER stress caused by an accumulation of misfolded proteins leads to cleavage of the cytoplasmic domain of luman, which translocates to the nucleus and activates factors that enhance proteasomal degradation.298,299 In 2006, Blot et al.300 identified an interaction between luman and the HIV-1 gp41 CT. Expression of Env bearing the gp41 CT led to a decrease in luman levels in cells. Luman was also observed to interact with Tat, and expression of a transcriptionally active form of luman led to a decrease in Tat-mediated HIV-1 gene expression.300 Env expression could thus potentially counteract luman's ability to suppress HIV-1 gene expression. Deciphering the biological implications of these observations awaits further analysis.
Using tandem affinity purification methods, the endogenous prohibitin 1 and 2 (Phb1 and Phb2) heterodimer was shown to bind a tagged HIV-1 gp41 CT.301 Phb1 and Phb2 are members of the prohibitin family of proteins, which are ubiquitously expressed and localize to diverse cellular compartments including the mitochondria, nucleus, lipid droplets, and the PM.302,303 They are involved in mitochondrial function and morphogenesis, cell proliferation, and transcriptional regulation.302,303 Prohibitin proteins contain the prohibitin homology domain, which is found in proteins known to associate with lipid rafts.304 The interaction between gp41 and Phb1 and Phb2 was validated by several in vitro binding assays and mapped to LLP-3. HIV-1 mutants with reduced Phb1/Phb2-binding were replication defective in a cell line non-permissive for gp41 truncation mutants (H9) but were replication competent in the highly permissive MT-4 T-cell line.301 However, the defective replication phenotype conferred by mutation of the putative Phb1/Phb2 binding site in gp41 was not associated with impaired Env incorporation.301
Conclusions
A key step in the production of infectious HIV-1 particles is the incorporation of the Env glycoprotein complex. Much is known about events leading to Env incorporation; however, the actual mechanism of incorporation still remains unclear. Four general models can be postulated (Fig. 7). The passive incorporation model, in which Env and Gag arrive at virus assembly sites independently and incorporation occurs simply as a result of gp120/gp41 expression on the cell surface. The direct Gag-Env interaction model, according to which binding of the gp41 CT to the MA domain of Gag mediates Env incorporation. The Gag-Env co-targeting model, in which Gag and Env are targeted to lipid rafts or other PM microdomains or structures. The indirect Gag-Env interaction model, which posits that Gag and Env associate with host proteins that serve as linkers or adapters to facilitate Env incorporation. Since these models are not mutually exclusive, it is possible that each contributes to Env incorporation to varying degrees in different cellular contexts.
Clearly distinguishing between these models will require progress in answering a number of key questions. For example, what is the structure and topology of the gp41 CT? While we now know quite a lot about the structures of gp120 and the ectodomain of gp41 at the atomic level, the structure of the gp41 CT, which plays a key role in Env incorporation, is still a matter of speculation. Although there is evidence for two, or several, distinct topologies of the gp41 CT in Env-expressing cells, the traditional model of a single membrane-spanning domain most likely reflects the conformation of the active Env complex in virions. But how are the LLP domains oriented with respect to the lipid bilayer? Do they associate with each other to form coiled-coil domains, do they “swim” in the plane of the membrane or are they oriented away from the membrane, do they bind directly to Gag and/or host factors, and does Gag processing and virion maturation alter their orientation? By analogy with gp120, which undergoes a number of major structural rearrangements during receptor/coreceptor binding and membrane fusion, one can imagine that the gp41 CT is also capable of adopting a variety of different conformations. Important advances have been made recently in our ability to solve the structures of membrane proteins by using techniques such as reverse micelle encapsulation305 and electron crystallography. 306,307 Application of these tools to solving the structure of the CT of membrane-associated Env appears warranted.
A related question is what domains of the gp41 CT are required for Env incorporation? To date, essential domains in the gp41 CT have been identified by amino acid substitutions or deletions followed by biochemical analyses of Env incorporation efficiency. However, robust assays for detecting and quantifying Gag-Env interactions are still lacking, and many of these mutations affect Env trafficking and/or protein stability, suggesting that they may alter global folding. Developing a genetic screen that identifies essential domains in the gp41 CT may provide further insight into regions required for interaction with viral or host proteins.
It is clear that the role of the gp41 CT in Env incorporation is cell-type-dependent. What is the basis for this cell-type specificity? If it is based on the pattern of cellular factors expressed, then which of these promote Env incorporation? Alternatively, the cell type-dependent differences elicited by gp41 CT deletion may reflect variations in PM lipid composition and microdomain formation in different cell types. Both Gag and Env display properties of being associated with lipid rafts at the PM. What are the nature, composition, and properties of these Gag- and Env-recruiting microdomains, and how are Env glycoproteins from different viruses segregated into different microdomains?
Also of great interest is the question of how Gag-Env interactions affect their co-recruitment to the VS and subsequent early events in the next round of viral replication. Is there a role for gp41 at a post-entry step, after fusion is completed, perhaps as a trigger for uncoating? The many potential contributions of Gag to Env function (e.g., fusogenicity, membrane localization in polarized cells, recruitment to the VS, gp120 shedding, and virus particle stiffness) need to be defined more completely.
Answers to these questions will not only enhance our understanding of Env incorporation but may open the way to the development of novel antiretrovirals that target Env incorporation. Given that diverse viruses likely utilize conserved mechanisms for directing Env incorporation, understanding this key step for HIV-1 would likely further our understanding of Env incorporation in other viral systems as well.
Acknowledgments
We thank P.D. Kwong, S. Subramaniam, A. Ono, J.D. Steckbeck, R. Montelaro, C.K. Leonard, and T.J. Gregory for permission to reproduce or adapt published figures. We are grateful to M. Johnson and members of Freed laboratory for helpful discussions. Research in our laboratory is supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and by the Intramural AIDS Targeted Antiviral Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 2.Daecke J, Fackler OT, Dittmar MT, Kräusslich H-G. Involvement of clathrin-mediated endocytosis in human immunodeficiency virus type 1 entry. J Virol. 2005;79:1581–94. doi: 10.1128/JVI.79.3.1581-1594.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miyauchi K, Kim Y, Latinovic O, Morozov V, Melikyan GB. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell. 2009;137:433–44. doi: 10.1016/j.cell.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger EA, Doms RW, Fenyö EM, Korber BT, Littman DR, Moore JP, Sattentau QJ, Schuitemaker H, Sodroski J, Weiss RA. A new classification for HIV-1. Nature. 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 5.Gorry PR, Ancuta P. Coreceptors and HIV-1 Pathogenesis. Curr HIV/AIDS Rep. 2011;8:45–53. doi: 10.1007/s11904-010-0069-x. [DOI] [PubMed] [Google Scholar]
- 6.Goff SP. Intracellular trafficking of retroviral genomes during the early phase of infection: viral exploitation of cellular pathways. J Gene Med. 2001;3:517–28. doi: 10.1002/1521-2254(200111)3:6<517::AID-JGM234>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki Y, Craigie R. The road to chromatin - nuclear entry of retroviruses. Nat Rev Microbiol. 2007;5:187–96. doi: 10.1038/nrmicro1579. [DOI] [PubMed] [Google Scholar]
- 8.Fassati A, Goff SP. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J Virol. 2001;75:3626–35. doi: 10.1128/JVI.75.8.3626-3635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bukrinsky M, Sharova N, Dempsey MP, Stanwick TL, Bukrinskaya AG, Haggerty S, Stevenson M. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc Natl Acad Sci USA. 1992;89:6580–4. doi: 10.1073/pnas.89.14.6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whittaker GR, Kann M, Helenius A. Viral entry into the nucleus. Annu Rev Cell Dev Biol. 2000;16:627–51. doi: 10.1146/annurev.cellbio.16.1.627. [DOI] [PubMed] [Google Scholar]
- 11.Freed EO, Martin MA. HIVs and their replication. In: Knipe David M., H. PM, Griffin Diane E, Lamb Robert A, Martin Malcolm A, Roizman Bernard, Straus Stephen E., editors. Fields Virology. 5th Edition Lippincott Williams & Wilkins; 2006. [Google Scholar]
- 12.Swanstrom R, Wills JW. Synthesis, assembly, and processing of viral proteins. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. [PubMed] [Google Scholar]
- 13.Freed EO. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 14.Adamson CS, Freed EO. Human immunodeficiency virus type 1 assembly, release, and maturation. Adv Pharmacol. 2007;55:347–87. doi: 10.1016/S1054-3589(07)55010-6. [DOI] [PubMed] [Google Scholar]
- 15.Bieniasz PD. The cell biology of HIV-1 virion genesis. Cell Host & Microbe. 2009;5:550–8. doi: 10.1016/j.chom.2009.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou W, Parent LJ, Wills JW, Resh MD. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–69. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Massiah MA, Starich MR, Paschall C, Summers MF, Christensen AM, Sundquist WI. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J Mol Biol. 1994;244:198–223. doi: 10.1006/jmbi.1994.1719. [DOI] [PubMed] [Google Scholar]
- 18.Hill CP, Worthylake D, Bancroft DP, Christensen AM, Sundquist WI. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc Natl Acad Sci USA. 1996;93:3099–104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc Natl Acad Sci USA. 2004;101:14889–94. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc Natl Acad Sci USA. 2006;103:11364–9. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demirov DG, Freed EO. Retrovirus budding. Virus Res. 2004;106:87–102. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 22.Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- 23.Bieniasz PD. Late budding domains and host proteins in enveloped virus release. Virology. 2006;344:55–63. doi: 10.1016/j.virol.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 24.Fujii K, Hurley JH, Freed EO. Beyond Tsg101: the role of Alix in ‘ESCRTing’ HIV-1. Nat Rev Microbiol. 2007;5:912–6. doi: 10.1038/nrmicro1790. [DOI] [PubMed] [Google Scholar]
- 25.Hurley JH, Boura E, Carlson L-A, Różycki B. Membrane budding. Cell. 2010;143:875–87. doi: 10.1016/j.cell.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jouvenet N, Neil SJD, Bess C, Johnson MC, Virgen CA, Simon SM, Bieniasz PD. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 2006;4:e435. doi: 10.1371/journal.pbio.0040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balasubramaniam M, Freed EO. New insights into HIV assembly and trafficking. Physiology. 2011 doi: 10.1152/physiol.00051.2010. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Welsch S, Keppler OT, Habermann A, Allespach I, Krijnse-Locker J, Kräusslich H-G. HIV-1 buds predominantly at the plasma membrane of primary human macrophages. PLoS Pathog. 2007;3:e36. doi: 10.1371/journal.ppat.0030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finzi A, Orthwein A, Mercier J, Cohen EA. Productive human immunodeficiency virus type 1 assembly takes place at the plasma membrane. J Virol. 2007;81:7476–90. doi: 10.1128/JVI.00308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joshi A, Ablan SD, Soheilian F, Nagashima K, Freed EO. Evidence that productive human immunodeficiency virus type 1 assembly can occur in an intracellular compartment. J Virol. 2009;83:5375–87. doi: 10.1128/JVI.00109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deneka M, Pelchen-Matthews A, Byland R, Ruiz-Mateos E, Marsh M. In macrophages, HIV-1 assembles into an intracellular plasma membrane domain containing the tetraspanins CD81, CD9, and CD53. J Cell Biol. 2007;177:329–41. doi: 10.1083/jcb.200609050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bennett AE, Narayan K, Shi D, Hartnell LM, Gousset K, He H, Lowekamp BC, Yoo TS, Bliss D, Freed EO, Subramaniam S. Ion-abrasion scanning electron microscopy reveals surface-connected tubular conduits in HIV-infected macrophages. PLoS Pathog. 2009;5:e1000591. doi: 10.1371/journal.ppat.1000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gousset K, Ablan SD, Coren LV, Ono A, Soheilian F, Nagashima K, Ott DE, Freed EO. Real-time visualization of HIV-1 GAG trafficking in infected macrophages. PLoS Pathog. 2008;4:e1000015. doi: 10.1371/journal.ppat.1000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunter E, Swanstrom R. Retrovirus envelope glycoproteins. Curr Top Microbiol Immunol. 1990;157:187–253. doi: 10.1007/978-3-642-75218-6_7. [DOI] [PubMed] [Google Scholar]
- 35.Freed EO, Martin MA. The role of human immunodeficiency virus type 1 envelope glycoproteins in virus infection. J Biol Chem. 1995;270:23883–6. doi: 10.1074/jbc.270.41.23883. [DOI] [PubMed] [Google Scholar]
- 36.Haffar O, Dowbenko DJ, Berman PW. Topogenic analysis of the human immunodeficiency virus type 1 envelope glycoprotein, gp160, in microsomal membranes. J Cell Biol. 1988;107:1677–87. doi: 10.1083/jcb.107.5.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berman PW, Nunes WM, Haffar O. Expression of membrane-associated and secreted variants of gp160 of human immunodeficiency virus type 1 in vitro and in continuous cell lines. J Virol. 1988;62:3135–42. doi: 10.1128/jvi.62.9.3135-3142.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allan JS, Coligan JE, Barin F, McLane MF, Sodroski JG, Rosen CA, Haseltine WA, Lee TH, Essex M. Major glycoprotein antigens that induce antibodies in AIDS patients are encoded by HTLV-III. Science. 1985;228:1091–4. doi: 10.1126/science.2986290. [DOI] [PubMed] [Google Scholar]
- 39.Leonard CK, Spellman MW, Riddle L, Harris RJ, Thomas JN, Gregory TJ. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J Biol Chem. 1990;265:10373–82. [PubMed] [Google Scholar]
- 40.Bernstein HB, Tucker SP, Hunter E, Schutzbach JS, Compans RW. Human immunodeficiency virus type 1 envelope glycoprotein is modified by O-linked oligosaccharides. J Virol. 1994;68:463–8. doi: 10.1128/jvi.68.1.463-468.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pinter A, Honnen WJ, Tilley SA, Bona C, Zaghouani H, Gorny MK, Zolla-Pazner S. Oligomeric structure of gp41, the transmembrane protein of human immunodeficiency virus type 1. J Virol. 1989;63:2674–9. doi: 10.1128/jvi.63.6.2674-2679.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schawaller M, Smith GE, Skehel JJ, Wiley DC. Studies with crosslinking reagents on the oligomeric structure of the env glycoprotein of HIV. Virology. 1989;172:367–9. doi: 10.1016/0042-6822(89)90142-6. [DOI] [PubMed] [Google Scholar]
- 43.Earl PL, Doms RW, Moss B. Oligomeric structure of the human immunodeficiency virus type 1 envelope glycoprotein. Proc Natl Acad Sci USA. 1990;87:648–52. doi: 10.1073/pnas.87.2.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Earl PL, Koenig S, Moss B. Biological and immunological properties of human immunodeficiency virus type 1 envelope glycoprotein: analysis of proteins with truncations and deletions expressed by recombinant vaccinia viruses. J Virol. 1991;65:31–41. doi: 10.1128/jvi.65.1.31-41.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujita K, Omura S, Silver J. Rapid degradation of CD4 in cells expressing human immunodeficiency virus type 1 Env and Vpu is blocked by proteasome inhibitors. J Gen Virol. 1997;78(Pt 3):619–25. doi: 10.1099/0022-1317-78-3-619. [DOI] [PubMed] [Google Scholar]
- 46.Schubert U, Antón LC, Bacík I, Cox JH, Bour S, Bennink JR, Orlowski M, Strebel K, Yewdell JW. CD4 glycoprotein degradation induced by human immunodeficiency virus type 1 Vpu protein requires the function of proteasomes and the ubiquitin-conjugating pathway. J Virol. 1998;72:2280–8. doi: 10.1128/jvi.72.3.2280-2288.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Margottin F, Bour SP, Durand H, Selig L, Benichou S, Richard V, Thomas D, Strebel K, Benarous R. A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol Cell. 1998;1:565–74. doi: 10.1016/s1097-2765(00)80056-8. [DOI] [PubMed] [Google Scholar]
- 48.Hallenberger S, Bosch V, Angliker H, Shaw E, Klenk HD, Garten W. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature. 1992;360:358–61. doi: 10.1038/360358a0. [DOI] [PubMed] [Google Scholar]
- 49.McCune JM, Rabin LB, Feinberg MB, Lieberman M, Kosek JC, Reyes GR, Weissman IL. Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell. 1988;53:55–67. doi: 10.1016/0092-8674(88)90487-4. [DOI] [PubMed] [Google Scholar]
- 50.Freed EO, Myers DJ, Risser R. Mutational analysis of the cleavage sequence of the human immunodeficiency virus type 1 envelope glycoprotein precursor gp160. J Virol. 1989;63:4670–5. doi: 10.1128/jvi.63.11.4670-4675.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rowell JF, Ruff AL, Guarnieri FG, Staveley-O'Carroll K, Lin X, Tang J, August JT, Siliciano RF. Lysosome-associated membrane protein-1-mediated targeting of the HIV-1 envelope protein to an endosomal/lysosomal compartment enhances its presentation to MHC class II-restricted T cells. J Immunol. 1995;155:1818–28. [PubMed] [Google Scholar]
- 52.Egan MA, Carruth LM, Rowell JF, Yu X, Siliciano RF. Human immunodeficiency virus type 1 envelope protein endocytosis mediated by a highly conserved intrinsic internalization signal in the cytoplasmic domain of gp41 is suppressed in the presence of the Pr55gag precursor protein. J Virol. 1996;70:6547–56. doi: 10.1128/jvi.70.10.6547-6556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu P, Chertova E, Bess J, Lifson JD, Arthur LO, Liu J, Taylor KA, Roux KH. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc Natl Acad Sci USA. 2003;100:15812–7. doi: 10.1073/pnas.2634931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jolly C, Sattentau QJ. Retroviral spread by induction of virological synapses. Traffic. 2004;5:643–50. doi: 10.1111/j.1600-0854.2004.00209.x. [DOI] [PubMed] [Google Scholar]
- 55.McDonald D, Wu L, Bohks SM, KewalRamani VN, Unutmaz D, Hope TJ. Recruitment of HIV and its receptors to dendritic cell-T cell junctions. Science. 2003;300:1295–7. doi: 10.1126/science.1084238. [DOI] [PubMed] [Google Scholar]
- 56.Sattentau QJ. Cell-to-Cell Spread of Retroviruses. Viruses. 2010;2:1306–1321. doi: 10.3390/v2061306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jolly C. T Cell Polarization at the Virological Synapse. Viruses. 2010;2:1261–1278. doi: 10.3390/v2061261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mothes W, Sherer NM, Jin J, Zhong P. Virus cell-to-cell transmission. J Virol. 2010;84:8360–8. doi: 10.1128/JVI.00443-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dimitrov DS, Willey RL, Sato H, Chang LJ, Blumenthal R, Martin MA. Quantitation of human immunodeficiency virus type 1 infection kinetics. J Virol. 1993;67:2182–90. doi: 10.1128/jvi.67.4.2182-2190.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jolly C, Kashefi K, Hollinshead M, Sattentau QJ. HIV-1 cell to cell transfer across an Env-induced, actin-dependent synapse. J Exp Med. 2004;199:283–93. doi: 10.1084/jem.20030648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jolly C, Mitar I, Sattentau QJ. Adhesion molecule interactions facilitate human immunodeficiency virus type 1-induced virological synapse formation between T cells. J Virol. 2007;81:13916–21. doi: 10.1128/JVI.01585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jolly C, Mitar I, Sattentau QJ. Requirement for an intact T-cell actin and tubulin cytoskeleton for efficient assembly and spread of human immunodeficiency virus type 1. J Virol. 2007;81:5547–60. doi: 10.1128/JVI.01469-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jolly C, Sattentau QJ. Human immunodeficiency virus type 1 assembly, budding, and cell-cell spread in T cells take place in tetraspanin-enriched plasma membrane domains. J Virol. 2007;81:7873–84. doi: 10.1128/JVI.01845-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sherer NM, Lehmann MJ, Jimenez-Soto LF, Horensavitz C, Pypaert M, Mothes W. Retroviruses can establish filopodial bridges for efficient cell-to-cell transmission. Nat Cell Biol. 2007;9:310–5. doi: 10.1038/ncb1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sherer NM, Jin J, Mothes W. Directional spread of surface-associated retroviruses regulated by differential virus-cell interactions. J Virol. 2010;84:3248–58. doi: 10.1128/JVI.02155-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jin J, Sherer NM, Heidecker G, Derse D, Mothes W. Assembly of the murine leukemia virus is directed towards sites of cell-cell contact. PLoS Biol. 2009;7:e1000163. doi: 10.1371/journal.pbio.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Waki K, Freed EO. Macrophages and Cell-Cell Spread of HIV-1. Viruses. 2010;2:1603–1620. doi: 10.3390/v2081603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Starcich BR, Hahn BH, Shaw GM, McNeely PD, Modrow S, Wolf H, Parks ES, Parks WP, Josephs SF, Gallo RC. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell. 1986;45:637–48. doi: 10.1016/0092-8674(86)90778-6. [DOI] [PubMed] [Google Scholar]
- 69.Willey RL, Rutledge RA, Dias S, Folks T, Theodore T, Buckler CE, Martin MA. Identification of conserved and divergent domains within the envelope gene of the acquired immunodeficiency syndrome retrovirus. Proc Natl Acad Sci USA. 1986;83:5038–42. doi: 10.1073/pnas.83.14.5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gao F, Morrison SG, Robertson DL, Thornton CL, Craig S, Karlsson G, Sodroski J, Morgado M, Galvao-Castro B, von Briesen H, Beddows S, Weber J, Sharp PM, Shaw GM, Hahn BH. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. The WHO and NIAID Networks for HIV Isolation and Characterization. J Virol. 1996;70:1651–67. doi: 10.1128/jvi.70.3.1651-1667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jobes DV, Daoust M, Nguyen V, Padua A, Michele S, Lock MD, Chen A, Sinangil F, Berman PW. High incidence of unusual cysteine variants in gp120 envelope proteins from early HIV type 1 infections from a Phase 3 vaccine efficacy trial. AIDS Res Hum Retroviruses. 2006;22:1014–21. doi: 10.1089/aid.2006.22.1014. [DOI] [PubMed] [Google Scholar]
- 72.Montefiori DC, Robinson WE, Mitchell WM. Role of protein N-glycosylation in pathogenesis of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1988;85:9248–52. doi: 10.1073/pnas.85.23.9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li H, Chien PC, Tuen M, Visciano ML, Cohen S, Blais S, Xu C-F, Zhang H-T, Hioe CE. Identification of an N-linked glycosylation in the C4 region of HIV-1 envelope gp120 that is critical for recognition of neighboring CD4 T cell epitopes. J Immunol. 2008;180:4011–21. doi: 10.4049/jimmunol.180.6.4011. [DOI] [PubMed] [Google Scholar]
- 74.Raska M, Novak J. Involvement of envelope-glycoprotein glycans in HIV-1 biology and infection. Arch Immunol Ther Exp (Warsz) 2010;58:191–208. doi: 10.1007/s00005-010-0072-3. [DOI] [PubMed] [Google Scholar]
- 75.Palmer C, Balfe P, Fox D, May JC, Frederiksson R, Fenyö EM, McKeating JA. Functional characterization of the V1V2 region of human immunodeficiency virus type 1. Virology. 1996;220:436–49. doi: 10.1006/viro.1996.0331. [DOI] [PubMed] [Google Scholar]
- 76.Masciotra S, Owen SM, Rudolph D, Yang C, Wang B, Saksena N, Spira T, Dhawan S, Lal RB. Temporal relationship between V1V2 variation, macrophage replication, and coreceptor adaptation during HIV-1 disease progression. AIDS. 2002;16:1887–98. doi: 10.1097/00002030-200209270-00005. [DOI] [PubMed] [Google Scholar]
- 77.Shioda T, Oka S, Xin X, Liu H, Harukuni R, Kurotani A, Fukushima M, Hasan MK, Shiino T, Takebe Y, Iwamoto A, Nagai Y. In vivo sequence variability of human immunodeficiency virus type 1 envelope gp120: association of V2 extension with slow disease progression. J Virol. 1997;71:4871–81. doi: 10.1128/jvi.71.7.4871-4881.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chohan B, Lang D, Sagar M, Korber B, Lavreys L, Richardson B, Overbaugh J. Selection for human immunodeficiency virus type 1 envelope glycosylation variants with shorter V1-V2 loop sequences occurs during transmission of certain genetic subtypes and may impact viral RNA levels. J Virol. 2005;79:6528–31. doi: 10.1128/JVI.79.10.6528-6531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sagar M, Wu X, Lee S, Overbaugh J. Human immunodeficiency virus type 1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity. J Virol. 2006;80:9586–98. doi: 10.1128/JVI.00141-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kitrinos KM, Hoffman NG, Nelson JAE, Swanstrom R. Turnover of env variable region 1 and 2 genotypes in subjects with late-stage human immunodeficiency virus type 1 infection. J Virol. 2003;77:6811–22. doi: 10.1128/JVI.77.12.6811-6822.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Curlin ME, Zioni R, Hawes SE, Liu Y, Deng W, Gottlieb GS, Zhu T, Mullins JI. HIV-1 envelope subregion length variation during disease progression. PLoS Pathog. 2010;6:e1001228. doi: 10.1371/journal.ppat.1001228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lasky LA, Nakamura G, Smith DH, Fennie C, Shimasaki C, Patzer E, Berman P, Gregory T, Capon DJ. Delineation of a region of the human immunodeficiency virus type 1 gp120 glycoprotein critical for interaction with the CD4 receptor. Cell. 1987;50:975–85. doi: 10.1016/0092-8674(87)90524-1. [DOI] [PubMed] [Google Scholar]
- 83.Kowalski M, Potz J, Basiripour L, Dorfman T, Goh WC, Terwilliger E, Dayton A, Rosen C, Haseltine W, Sodroski J. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science. 1987;237:1351–5. doi: 10.1126/science.3629244. [DOI] [PubMed] [Google Scholar]
- 84.Olshevsky U, Helseth E, Furman C, Li J, Haseltine W, Sodroski J. Identification of individual human immunodeficiency virus type 1 gp120 amino acids important for CD4 receptor binding. J Virol. 1990;64:5701–7. doi: 10.1128/jvi.64.12.5701-5707.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pollard SR, Rosa MD, Rosa JJ, Wiley DC. Truncated variants of gp120 bind CD4 with high affinity and suggest a minimum CD4 binding region. EMBO J. 1992;11:585–91. doi: 10.1002/j.1460-2075.1992.tb05090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wyatt R, Sullivan N, Thali M, Repke H, Ho D, Robinson J, Posner M, Sodroski J. Functional and immunologic characterization of human immunodeficiency virus type 1 envelope glycoproteins containing deletions of the major variable regions. J Virol. 1993;67:4557–65. doi: 10.1128/jvi.67.8.4557-4565.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Freed EO, Myers DJ, Risser R. Identification of the principal neutralizing determinant of human immunodeficiency virus type 1 as a fusion domain. J Virol. 1991;65:190–4. doi: 10.1128/jvi.65.1.190-194.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.O'Brien WA, Koyanagi Y, Namazie A, Zhao JQ, Diagne A, Idler K, Zack JA, Chen IS. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature. 1990;348:69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- 89.Hwang SS, Boyle TJ, Lyerly HK, Cullen BR. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science. 1991;253:71–4. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- 90.Shioda T, Levy JA, Cheng-Mayer C. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature. 1991;349:167–9. doi: 10.1038/349167a0. [DOI] [PubMed] [Google Scholar]
- 91.Chesebro B, Nishio J, Perryman S, Cann A, O'Brien W, Chen IS, Wehrly K. Identification of human immunodeficiency virus envelope gene sequences influencing viral entry into CD4-positive HeLa cells, T-leukemia cells, and macrophages. J Virol. 1991;65:5782–9. doi: 10.1128/jvi.65.11.5782-5789.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cann AJ, Churcher MJ, Boyd M, O'Brien W, Zhao JQ, Zack J, Chen IS. The region of the envelope gene of human immunodeficiency virus type 1 responsible for determination of cell tropism. J Virol. 1992;66:305–9. doi: 10.1128/jvi.66.1.305-309.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rusche JR, Javaherian K, McDanal C, Petro J, Lynn DL, Grimaila R, Langlois A, Gallo RC, Arthur LO, Fischinger PJ. Antibodies that inhibit fusion of human immunodeficiency virus-infected cells bind a 24-amino acid sequence of the viral envelope, gp120. Proc Natl Acad Sci USA. 1988;85:3198–202. doi: 10.1073/pnas.85.9.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Palker TJ, Clark ME, Langlois AJ, Matthews TJ, Weinhold KJ, Randall RR, Bolognesi DP, Haynes BF. Type-specific neutralization of the human immunodeficiency virus with antibodies to env-encoded synthetic peptides. Proc Natl Acad Sci USA. 1988;85:1932–6. doi: 10.1073/pnas.85.6.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Goudsmit J, Debouck C, Meloen RH, Smit L, Bakker M, Asher DM, Wolff AV, Gibbs CJ, Gajdusek DC. Human immunodeficiency virus type 1 neutralization epitope with conserved architecture elicits early type-specific antibodies in experimentally infected chimpanzees. Proc Natl Acad Sci USA. 1988;85:4478–82. doi: 10.1073/pnas.85.12.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Matsushita S, Robert-Guroff M, Rusche J, Koito A, Hattori T, Hoshino H, Javaherian K, Takatsuki K, Putney S. Characterization of a human immunodeficiency virus neutralizing monoclonal antibody and mapping of the neutralizing epitope. J Virol. 1988;62:2107–14. doi: 10.1128/jvi.62.6.2107-2114.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shioda T, Oka S, Ida S, Nokihara K, Toriyoshi H, Mori S, Takebe Y, Kimura S, Shimada K, Nagai Y. A naturally occurring single basic amino acid substitution in the V3 region of the human immunodeficiency virus type 1 env protein alters the cellular host range and antigenic structure of the virus. J Virol. 1994;68:7689–96. doi: 10.1128/jvi.68.12.7689-7696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fouchier RA, Groenink M, Kootstra NA, Tersmette M, Huisman HG, Miedema F, Schuitemaker H. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–7. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pollakis G, Abebe A, Kliphuis A, Chalaby MIM, Bakker M, Mengistu Y, Brouwer M, Goudsmit J, Schuitemaker H, Paxton WA. Phenotypic and genotypic comparisons of CCR5- and CXCR4-tropic human immunodeficiency virus type 1 biological clones isolated from subtype C-infected individuals. J Virol. 2004;78:2841–52. doi: 10.1128/JVI.78.6.2841-2852.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rizzuto CD, Wyatt R, Hernández-Ramos N, Sun Y, Kwong PD, Hendrickson WA, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–53. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 101.Rizzuto C, Sodroski J. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res Hum Retroviruses. 2000;16:741–9. doi: 10.1089/088922200308747. [DOI] [PubMed] [Google Scholar]
- 102.Cormier EG, Tran DN, Yukhayeva L, Olson WC, Dragic T. Mapping the determinants of the CCR5 amino-terminal sulfopeptide interaction with soluble human immunodeficiency virus type 1 gp120-CD4 complexes. J Virol. 2001;75:5541–9. doi: 10.1128/JVI.75.12.5541-5549.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cormier EG, Dragic T. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J Virol. 2002;76:8953–7. doi: 10.1128/JVI.76.17.8953-8957.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dragic T, Trkola A, Thompson DA, Cormier EG, Kajumo FA, Maxwell E, Lin SW, Ying W, Smith SO, Sakmar TP, Moore JP. A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5. Proc Natl Acad Sci USA. 2000;97:5639–44. doi: 10.1073/pnas.090576697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Westby M, Smith-Burchnell C, Mori J, Lewis M, Mosley M, Stockdale M, Dorr P, Ciaramella G, Perros M. Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. J Virol. 2007;81:2359–71. doi: 10.1128/JVI.02006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pugach P, Marozsan AJ, Ketas TJ, Landes EL, Moore JP, Kuhmann SE. HIV-1 clones resistant to a small molecule CCR5 inhibitor use the inhibitor-bound form of CCR5 for entry. Virology. 2007;361:212–28. doi: 10.1016/j.virol.2006.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–59. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 2005;433:834–41. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- 109.Wu S-R, Löving R, Lindqvist B, Hebert H, Koeck PJB, Sjöberg M, Garoff H. Single-particle cryoelectron microscopy analysis reveals the HIV-1 spike as a tripod structure. Proc Natl Acad Sci USA. 2010;107:18844–9. doi: 10.1073/pnas.1007227107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Huang C.-c., Tang M, Zhang M-Y, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310:1025–8. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang S-H, Yang X, Zhang M-Y, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–7. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen L, Kwon YD, Zhou T, Wu X, O'Dell S, Cavacini L, Hessell AJ, Pancera M, Tang M, Xu L, Yang Z-Y, Zhang M-Y, Arthos J, Burton DR, Dimitrov DS, Nabel GJ, Posner MR, Sodroski J, Wyatt R, Mascola JR, Kwong PD. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science. 2009;326:1123–7. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pancera M, Majeed S, Ban Y-EA, Chen L, Huang C.-c., Kong L, Kwon YD, Stuckey J, Zhou T, Robinson JE, Schief WR, Sodroski J, Wyatt R, Kwong PD. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci USA. 2010;107:1166–71. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cordonnier A, Montagnier L, Emerman M. Single amino-acid changes in HIV envelope affect viral tropism and receptor binding. Nature. 1989;340:571–4. doi: 10.1038/340571a0. [DOI] [PubMed] [Google Scholar]
- 115.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–13. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bosch ML, Earl PL, Fargnoli K, Picciafuoco S, Giombini F, Wong-Staal F, Franchini G. Identification of the fusion peptide of primate immunodeficiency viruses. Science. 1989;244:694–7. doi: 10.1126/science.2541505. [DOI] [PubMed] [Google Scholar]
- 117.Freed EO, Myers DJ, Risser R. Characterization of the fusion domain of the human immunodeficiency virus type 1 envelope glycoprotein gp41. Proc Natl Acad Sci USA. 1990;87:4650–4. doi: 10.1073/pnas.87.12.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Freed EO, Delwart EL, Buchschacher GL, Panganiban AT. A mutation in the human immunodeficiency virus type 1 transmembrane glycoprotein gp41 dominantly interferes with fusion and infectivity. Proc Natl Acad Sci USA. 1992;89:70–4. doi: 10.1073/pnas.89.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–73. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 120.Dubay JW, Roberts SJ, Brody B, Hunter E. Mutations in the leucine zipper of the human immunodeficiency virus type 1 transmembrane glycoprotein affect fusion and infectivity. J Virol. 1992;66:4748–56. doi: 10.1128/jvi.66.8.4748-4756.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lu M, Blacklow SC, Kim PS. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat Struct Biol. 1995;2:1075–82. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 122.Tan K, Liu J, Wang J, Shen S, Lu M. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc Natl Acad Sci USA. 1997;94:12303–8. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–30. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 124.Muñoz-Barroso I, Salzwedel K, Hunter E, Blumenthal R. Role of the membrane-proximal domain in the initial stages of human immunodeficiency virus type 1 envelope glycoprotein-mediated membrane fusion. J Virol. 1999;73:6089–92. doi: 10.1128/jvi.73.7.6089-6092.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Salzwedel K, West JT, Hunter E. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J Virol. 1999;73:2469–80. doi: 10.1128/jvi.73.3.2469-2480.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Brasseur R, Vandenbranden M, Cornet B, Burny A, Ruysschaert JM. Orientation into the lipid bilayer of an asymmetric amphipathic helical peptide located at the N-terminus of viral fusion proteins. Biochim Biophys Acta. 1990;1029:267–73. doi: 10.1016/0005-2736(90)90163-i. [DOI] [PubMed] [Google Scholar]
- 127.White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Südhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–7. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wild CT, Shugars DC, Greenwell TK, McDanal CB, Matthews TJ. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc Natl Acad Sci USA. 1994;91:9770–4. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Furuta RA, Wild CT, Weng Y, Weiss CD. Capture of an early fusion-active conformation of HIV-1 gp41. Nat Struct Biol. 1998;5:276–9. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- 131.Kilby JM, Hopkins S, Venetta TM, DiMassimo B, Cloud GA, Lee JY, Alldredge L, Hunter E, Lambert D, Bolognesi D, Matthews T, Johnson MR, Nowak MA, Shaw GM, Saag MS. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat Med. 1998;4:1302–7. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 132.LaBonte J, Lebbos J, Kirkpatrick P. Enfuvirtide. Nat Rev Drug Discov. 2003;2:345–6. doi: 10.1038/nrd1091. [DOI] [PubMed] [Google Scholar]
- 133.Poumbourios P, el Ahmar W, McPhee DA, Kemp BE. Determinants of human immunodeficiency virus type 1 envelope glycoprotein oligomeric structure. J Virol. 1995;69:1209–18. doi: 10.1128/jvi.69.2.1209-1218.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cao J, Bergeron L, Helseth E, Thali M, Repke H, Sodroski J. Effects of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 gp41 envelope glycoprotein. J Virol. 1993;67:2747–55. doi: 10.1128/jvi.67.5.2747-2755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, Moore JP, Stiegler G, Katinger H, Burton DR, Parren PW. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol. 2001;75:10892–905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shang L, Yue L, Hunter E. Role of the membrane-spanning domain of human immunodeficiency virus type 1 envelope glycoprotein in cell-cell fusion and virus infection. J Virol. 2008;82:5417–28. doi: 10.1128/JVI.02666-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shang L, Hunter E. Residues in the membrane-spanning domain core modulate conformation and fusogenicity of the HIV-1 envelope glycoprotein. Virology. 2010;404:158–67. doi: 10.1016/j.virol.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kondo N, Miyauchi K, Meng F, Iwamoto A, Matsuda Z. Conformational changes of the HIV-1 envelope protein during membrane fusion are inhibited by the replacement of its membrane-spanning domain. J Biol Chem. 2010;285:14681–8. doi: 10.1074/jbc.M109.067090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kennedy RC, Henkel RD, Pauletti D, Allan JS, Lee TH, Essex M, Dreesman GR. Antiserum to a synthetic peptide recognizes the HTLV-III envelope glycoprotein. Science. 1986;231:1556–9. doi: 10.1126/science.3006246. [DOI] [PubMed] [Google Scholar]
- 140.Chanh TC, Dreesman GR, Kanda P, Linette GP, Sparrow JT, Ho DD, Kennedy RC. Induction of anti-HIV neutralizing antibodies by synthetic peptides. EMBO J. 1986;5:3065–71. doi: 10.1002/j.1460-2075.1986.tb04607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cleveland SM, McLain L, Cheung L, Jones TD, Hollier M, Dimmock NJ. A region of the C-terminal tail of the gp41 envelope glycoprotein of human immunodeficiency virus type 1 contains a neutralizing epitope: evidence for its exposure on the surface of the virion. J Gen Virol. 2003;84:591–602. doi: 10.1099/vir.0.18630-0. [DOI] [PubMed] [Google Scholar]
- 142.Hollier MJ, Dimmock NJ. The C-terminal tail of the gp41 transmembrane envelope glycoprotein of HIV-1 clades A, B, C, and D may exist in two conformations: an analysis of sequence, structure, and function. Virology. 2005;337:284–96. doi: 10.1016/j.virol.2005.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lu L, Zhu Y, Huang J, Chen X, Yang H, Jiang S, Chen Y-H. Surface exposure of the HIV-1 env cytoplasmic tail LLP2 domain during the membrane fusion process: interaction with gp41 fusion core. J Biol Chem. 2008;283:16723–31. doi: 10.1074/jbc.M801083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Steckbeck JD, Sun C, Sturgeon TJ, Montelaro RC. Topology of the C-terminal tail of HIV-1 gp41: differential exposure of the Kennedy epitope on cell and viral membranes. PLoS ONE. 2010;5:e15261. doi: 10.1371/journal.pone.0015261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Waheed AA, Ablan SD, Mankowski MK, Cummins JE, Ptak RG, Schaffner CP, Freed EO. Inhibition of HIV-1 replication by amphotericin B methyl ester: selection for resistant variants. J Biol Chem. 2006;281:28699–711. doi: 10.1074/jbc.M603609200. [DOI] [PubMed] [Google Scholar]
- 146.Waheed AA, Ablan SD, Roser JD, Sowder RC, Schaffner CP, Chertova E, Freed EO. HIV-1 escape from the entry-inhibiting effects of a cholesterol-binding compound via cleavage of gp41 by the viral protease. Proc Natl Acad Sci USA. 2007;104:8467–71. doi: 10.1073/pnas.0701443104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Waheed AA, Ablan SD, Sowder RC, Roser JD, Schaffner CP, Chertova E, Freed EO. Effect of mutations in the human immunodeficiency virus type 1 protease on cleavage of the gp41 cytoplasmic tail. J Virol. 2010;84:3121–6. doi: 10.1128/JVI.02002-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rushlow K, Olsen K, Stiegler G, Payne SL, Montelaro RC, Issel CJ. Lentivirus genomic organization: the complete nucleotide sequence of the env gene region of equine infectious anemia virus. Virology. 1986;155:309–21. doi: 10.1016/0042-6822(86)90195-9. [DOI] [PubMed] [Google Scholar]
- 149.Rice NR, Henderson LE, Sowder RC, Copeland TD, Oroszlan S, Edwards JF. Synthesis and processing of the transmembrane envelope protein of equine infectious anemia virus. J Virol. 1990;64:3770–8. doi: 10.1128/jvi.64.8.3770-3778.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Shacklett BL, Weber CJ, Shaw KE, Keddie EM, Gardner MB, Sonigo P, Luciw PA. The intracytoplasmic domain of the Env transmembrane protein is a locus for attenuation of simian immunodeficiency virus SIVmac in rhesus macaques. J Virol. 2000;74:5836–44. doi: 10.1128/jvi.74.13.5836-5844.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lee SJ, Hu W, Fisher AG, Looney DJ, Kao VF, Mitsuya H, Ratner L, Wong-Staal F. Role of the carboxy-terminal portion of the HIV-1 transmembrane protein in viral transmission and cytopathogenicity. AIDS Res Hum Retroviruses. 1989;5:441–9. doi: 10.1089/aid.1989.5.441. [DOI] [PubMed] [Google Scholar]
- 152.Wilk T, Pfeiffer T, Bosch V. Retained in vitro infectivity and cytopathogenicity of HIV-1 despite truncation of the C-terminal tail of the env gene product. Virology. 1992;189:167–77. doi: 10.1016/0042-6822(92)90692-i. [DOI] [PubMed] [Google Scholar]
- 153.Gabuzda DH, Lever A, Terwilliger E, Sodroski J. Effects of deletions in the cytoplasmic domain on biological functions of human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 1992;66:3306–15. doi: 10.1128/jvi.66.6.3306-3315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yu X, Yuan X, McLane MF, Lee TH, Essex M. Mutations in the cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein impair the incorporation of Env proteins into mature virions. J Virol. 1993;67:213–21. doi: 10.1128/jvi.67.1.213-221.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wyss S, Berlioz-Torrent C, Boge M, Blot G, Höning S, Benarous R, Thali M. The highly conserved C-terminal dileucine motif in the cytosolic domain of the human immunodeficiency virus type 1 envelope glycoprotein is critical for its association with the AP-1 clathrin adaptor [correction of adapter]. J Virol. 2001;75:2982–92. doi: 10.1128/JVI.75.6.2982-2992.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Freed EO, Martin MA. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J Virol. 1995;69:1984–9. doi: 10.1128/jvi.69.3.1984-1989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Freed EO, Martin MA. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J Virol. 1996;70:341–51. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Murakami T, Freed EO. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc Natl Acad Sci USA. 2000;97:343–8. doi: 10.1073/pnas.97.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Murakami T, Freed EO. Genetic evidence for an interaction between human immunodeficiency virus type 1 matrix and alpha-helix 2 of the gp41 cytoplasmic tail. J Virol. 2000;74:3548–54. doi: 10.1128/jvi.74.8.3548-3554.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kalia V, Sarkar S, Gupta P, Montelaro RC. Rational site-directed mutations of the LLP-1 and LLP-2 lentivirus lytic peptide domains in the intracytoplasmic tail of human immunodeficiency virus type 1 gp41 indicate common functions in cell-cell fusion but distinct roles in virion envelope incorporation. J Virol. 2003;77:3634–46. doi: 10.1128/JVI.77.6.3634-3646.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Affranchino JL, González SA. Mutations at the C-terminus of the simian immunodeficiency virus envelope glycoprotein affect gp120-gp41 stability on virions. Virology. 2006;347:217–25. doi: 10.1016/j.virol.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 162.Spies CP, Ritter GD, Mulligan MJ, Compans RW. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein alters the conformation of the external domain. J Virol. 1994;68:585–91. doi: 10.1128/jvi.68.2.585-591.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Edwards TG, Wyss S, Reeves JD, Zolla-Pazner S, Hoxie JA, Doms RW, Baribaud F. Truncation of the cytoplasmic domain induces exposure of conserved regions in the ectodomain of human immunodeficiency virus type 1 envelope protein. J Virol. 2002;76:2683–91. doi: 10.1128/JVI.76.6.2683-2691.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kalia V, Sarkar S, Gupta P, Montelaro RC. Antibody neutralization escape mediated by point mutations in the intracytoplasmic tail of human immunodeficiency virus type 1 gp41. J Virol. 2005;79:2097–107. doi: 10.1128/JVI.79.4.2097-2107.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Vzorov AN, Gernert KM, Compans RW. Multiple domains of the SIV Env protein determine virus replication efficiency and neutralization sensitivity. Virology. 2005;332:89–101. doi: 10.1016/j.virol.2004.10.044. [DOI] [PubMed] [Google Scholar]
- 166.Edwards TG, Hoffman TL, Baribaud F, Wyss S, LaBranche CC, Romano J, Adkinson J, Sharron M, Hoxie JA, Doms RW. Relationships between CD4 independence, neutralization sensitivity, and exposure of a CD4-induced epitope in a human immunodeficiency virus type 1 envelope protein. J Virol. 2001;75:5230–9. doi: 10.1128/JVI.75.11.5230-5239.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Rowell JF, Stanhope PE, Siliciano RF. Endocytosis of endogenously synthesized HIV-1 envelope protein. Mechanism and role in processing for association with class II MHC. J Immunol. 1995;155:473–88. [PubMed] [Google Scholar]
- 168.Ohno H, Aguilar RC, Fournier MC, Hennecke S, Cosson P, Bonifacino JS. Interaction of endocytic signals from the HIV-1 envelope glycoprotein complex with members of the adaptor medium chain family. Virology. 1997;238:305–15. doi: 10.1006/viro.1997.8839. [DOI] [PubMed] [Google Scholar]
- 169.Boge M, Wyss S, Bonifacino JS, Thali M. A membrane-proximal tyrosine-based signal mediates internalization of the HIV-1 envelope glycoprotein via interaction with the AP-2 clathrin adaptor. J Biol Chem. 1998;273:15773–8. doi: 10.1074/jbc.273.25.15773. [DOI] [PubMed] [Google Scholar]
- 170.Berlioz-Torrent C, Shacklett BL, Erdtmann L, Delamarre L, Bouchaert I, Sonigo P, Dokhelar MC, Benarous R. Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adaptor complexes modulate intracellular and cell surface expression of envelope glycoproteins. J Virol. 1999;73:1350–61. doi: 10.1128/jvi.73.2.1350-1361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lodge R, Lalonde JP, Lemay G, Cohen EA. The membrane-proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cells. EMBO J. 1997;16:695–705. doi: 10.1093/emboj/16.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Deschambeault J, Lalonde JP, Cervantes-Acosta G, Lodge R, Cohen EA, Lemay G. Polarized human immunodeficiency virus budding in lymphocytes involves a tyrosine-based signal and favors cell-to-cell viral transmission. J Virol. 1999;73:5010–7. doi: 10.1128/jvi.73.6.5010-5017.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Day JR, Münk C, Guatelli JC. The membrane-proximal tyrosine-based sorting signal of human immunodeficiency virus type 1 gp41 is required for optimal viral infectivity. J Virol. 2004;78:1069–79. doi: 10.1128/JVI.78.3.1069-1079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.West JT, Weldon SK, Wyss S, Lin X, Yu Q, Thali M, Hunter E. Mutation of the dominant endocytosis motif in human immunodeficiency virus type 1 gp41 can complement matrix mutations without increasing Env incorporation. J Virol. 2002;76:3338–49. doi: 10.1128/JVI.76.7.3338-3349.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Fultz PN, Vance PJ, Endres MJ, Tao B, Dvorin JD, Davis IC, Lifson JD, Montefiori DC, Marsh M, Malim MH, Hoxie JA. In vivo attenuation of simian immunodeficiency virus by disruption of a tyrosine-dependent sorting signal in the envelope glycoprotein cytoplasmic tail. J Virol. 2001;75:278–91. doi: 10.1128/JVI.75.1.278-291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Byland R, Vance PJ, Hoxie JA, Marsh M. A conserved dileucine motif mediates clathrin and AP-2-dependent endocytosis of the HIV-1 envelope protein. Mol Biol Cell. 2007;18:414–25. doi: 10.1091/mbc.E06-06-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Miller MA, Garry RF, Jaynes JM, Montelaro RC. A structural correlation between lentivirus transmembrane proteins and natural cytolytic peptides. AIDS Res Hum Retroviruses. 1991;7:511–9. doi: 10.1089/aid.1991.7.511. [DOI] [PubMed] [Google Scholar]
- 178.Miller MA, Cloyd MW, Liebmann J, Rinaldo CR, Islam KR, Wang SZ, Mietzner TA, Montelaro RC. Alterations in cell membrane permeability by the lentivirus lytic peptide (LLP-1) of HIV-1 transmembrane protein. Virology. 1993;196:89–100. doi: 10.1006/viro.1993.1457. [DOI] [PubMed] [Google Scholar]
- 179.Eisenberg D, Wesson M. The most highly amphiphilic alpha-helices include two amino acid segments in human immunodeficiency virus glycoprotein 41. Biopolymers. 1990;29:171–7. doi: 10.1002/bip.360290122. [DOI] [PubMed] [Google Scholar]
- 180.Venable RM, Pastor RW, Brooks BR, Carson FW. Theoretically determined three-dimensional structures for amphipathic segments of the HIV-1 gp41 envelope protein. AIDS Res Hum Retroviruses. 1989;5:7–22. doi: 10.1089/aid.1989.5.7. [DOI] [PubMed] [Google Scholar]
- 181.Lee S-F, Ko C-Y, Wang C-T, Chen SS-L. Effect of point mutations in the N terminus of the lentivirus lytic peptide-1 sequence of human immunodeficiency virus type 1 transmembrane protein gp41 on Env stability. J Biol Chem. 2002;277:15363–75. doi: 10.1074/jbc.M201479200. [DOI] [PubMed] [Google Scholar]
- 182.Lee SF, Wang CT, Liang JY, Hong SL, Huang CC, Chen SS. Multimerization potential of the cytoplasmic domain of the human immunodeficiency virus type 1 transmembrane glycoprotein gp41. J Biol Chem. 2000;275:15809–19. doi: 10.1074/jbc.M000601200. [DOI] [PubMed] [Google Scholar]
- 183.Bültmann A, Muranyi W, Seed B, Haas J. Identification of two sequences in the cytoplasmic tail of the human immunodeficiency virus type 1 envelope glycoprotein that inhibit cell surface expression. J Virol. 2001;75:5263–76. doi: 10.1128/JVI.75.11.5263-5276.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Cosson P. Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J. 1996;15:5783–8. [PMC free article] [PubMed] [Google Scholar]
- 185.Piller SC, Dubay JW, Derdeyn CA, Hunter E. Mutational analysis of conserved domains within the cytoplasmic tail of gp41 from human immunodeficiency virus type 1: effects on glycoprotein incorporation and infectivity. J Virol. 2000;74:11717–23. doi: 10.1128/jvi.74.24.11717-11723.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chen SS, Lee SF, Wang CT. Cellular membrane-binding ability of the C-terminal cytoplasmic domain of human immunodeficiency virus type 1 envelope transmembrane protein gp41. J Virol. 2001;75:9925–38. doi: 10.1128/JVI.75.20.9925-9938.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Chernomordik L, Chanturiya AN, Suss-Toby E, Nora E, Zimmerberg J. An amphipathic peptide from the C-terminal region of the human immunodeficiency virus envelope glycoprotein causes pore formation in membranes. J Virol. 1994;68:7115–23. doi: 10.1128/jvi.68.11.7115-7123.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Gawrisch K, Han KH, Yang JS, Bergelson LD, Ferretti JA. Interaction of peptide fragment 828-848 of the envelope glycoprotein of human immunodeficiency virus type I with lipid bilayers. Biochemistry. 1993;32:3112–8. doi: 10.1021/bi00063a024. [DOI] [PubMed] [Google Scholar]
- 189.Comardelle AM, Norris CH, Plymale DR, Gatti PJ, Choi B, Fermin CD, Haislip AM, Tencza SB, Mietzner TA, Montelaro RC, Garry RF. A synthetic peptide corresponding to the carboxy terminus of human immunodeficiency virus type 1 transmembrane glycoprotein induces alterations in the ionic permeability of Xenopus laevis oocytes. AIDS Res Hum Retroviruses. 1997;13:1525–32. doi: 10.1089/aid.1997.13.1525. [DOI] [PubMed] [Google Scholar]
- 190.Tencza SB, Miller MA, Islam K, Mietzner TA, Montelaro RC. Effect of amino acid substitutions on calmodulin binding and cytolytic properties of the LLP-1 peptide segment of human immunodeficiency virus type 1 transmembrane protein. J Virol. 1995;69:5199–202. doi: 10.1128/jvi.69.8.5199-5202.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Rousso I, Mixon MB, Chen BK, Kim PS. Palmitoylation of the HIV-1 envelope glycoprotein is critical for viral infectivity. Proc Natl Acad Sci USA. 2000;97:13523–5. doi: 10.1073/pnas.240459697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Yang P, Ai L-S, Huang S-C, Li H-F, Chan W-E, Chang C-W, Ko C-Y, Chen SS-L. The cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein gp41 harbors lipid raft association determinants. J Virol. 2010;84:59–75. doi: 10.1128/JVI.00899-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Fuller SD, Berriman JA, Butcher SJ, Gowen BE. Low pH induces swiveling of the glycoprotein heterodimers in the Semliki Forest virus spike complex. Cell. 1995;81:715–25. doi: 10.1016/0092-8674(95)90533-2. [DOI] [PubMed] [Google Scholar]
- 194.Garoff H, Sjöberg M, Cheng RH. Budding of alphaviruses. Virus Res. 2004;106:103–16. doi: 10.1016/j.virusres.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 195.Lusso P, di Marzo Veronese F, Ensoli B, Franchini G, Jemma C, DeRocco SE, Kalyanaraman VS, Gallo RC. Expanded HIV-1 cellular tropism by phenotypic mixing with murine endogenous retroviruses. Science. 1990;247:848–52. doi: 10.1126/science.2305256. [DOI] [PubMed] [Google Scholar]
- 196.Arthur LO, Bess JW, Sowder RC, Benveniste RE, Mann DL, Chermann JC, Henderson LE. Cellular proteins bound to immunodeficiency viruses: implications for pathogenesis and vaccines. Science. 1992;258:1935–8. doi: 10.1126/science.1470916. [DOI] [PubMed] [Google Scholar]
- 197.Ott DE. Cellular proteins detected in HIV-1. Rev Med Virol. 2008;18:159–75. doi: 10.1002/rmv.570. [DOI] [PubMed] [Google Scholar]
- 198.Page KA, Landau NR, Littman DR. Construction and use of a human immunodeficiency virus vector for analysis of virus infectivity. J Virol. 1990;64:5270–6. doi: 10.1128/jvi.64.11.5270-5276.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Cronin J, Zhang X-Y, Reiser J. Altering the tropism of lentiviral vectors through pseudotyping. Curr Gene Ther. 2005;5:387–98. doi: 10.2174/1566523054546224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Chen SS, Ferrante AA, Terwilliger EF. Characterization of an envelope mutant of HIV-1 that interferes with viral infectivity. Virology. 1996;226:260–8. doi: 10.1006/viro.1996.0654. [DOI] [PubMed] [Google Scholar]
- 201.Yu X, Yuan X, Matsuda Z, Lee TH, Essex M. The matrix protein of human immunodeficiency virus type 1 is required for incorporation of viral envelope protein into mature virions. J Virol. 1992;66:4966–71. doi: 10.1128/jvi.66.8.4966-4971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Dorfman T, Mammano F, Haseltine WA, Gottlinger HG. Role of the matrix protein in the virion association of the human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1994;68:1689–96. doi: 10.1128/jvi.68.3.1689-1696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Mammano F, Kondo E, Sodroski J, Bukovsky A, Gottlinger HG. Rescue of human immunodeficiency virus type 1 matrix protein mutants by envelope glycoproteins with short cytoplasmic domains. J Virol. 1995;69:3824–30. doi: 10.1128/jvi.69.6.3824-3830.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Reil H, Bukovsky AA, Gelderblom HR, Gottlinger HG. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 1998;17:2699–708. doi: 10.1093/emboj/17.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Dubay JW, Roberts SJ, Hahn BH, Hunter E. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J Virol. 1992;66:6616–25. doi: 10.1128/jvi.66.11.6616-6625.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Iwatani Y, Ueno T, Nishimura A, Zhang X, Hattori T, Ishimoto A, Ito M, Sakai H. Modification of virus infectivity by cytoplasmic tail of HIV-1 TM protein. Virus Res. 2001;74:75–87. doi: 10.1016/s0168-1702(00)00249-5. [DOI] [PubMed] [Google Scholar]
- 207.Rao Z, Belyaev AS, Fry E, Roy P, Jones IM, Stuart DI. Crystal structure of SIV matrix antigen and implications for virus assembly. Nature. 1995;378:743–7. doi: 10.1038/378743a0. [DOI] [PubMed] [Google Scholar]
- 208.Freed EO, Englund G, Martin MA. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J Virol. 1995;69:3949–54. doi: 10.1128/jvi.69.6.3949-3954.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Owens RJ, Compans RW. Expression of the human immunodeficiency virus envelope glycoprotein is restricted to basolateral surfaces of polarized epithelial cells. J Virol. 1989;63:978–82. doi: 10.1128/jvi.63.2.978-982.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Owens RJ, Dubay JW, Hunter E, Compans RW. Human immunodeficiency virus envelope protein determines the site of virus release in polarized epithelial cells. Proc Natl Acad Sci USA. 1991;88:3987–91. doi: 10.1073/pnas.88.9.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Lodge R, Gottlinger HG, Gabuzda D, Cohen EA, Lemay G. The intracytoplasmic domain of gp41 mediates polarized budding of human immunodeficiency virus type 1 in MDCK cells. J Virol. 1994;68:4857–61. doi: 10.1128/jvi.68.8.4857-4861.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Wyma DJ, Jiang J, Shi J, Zhou J, Lineberger JE, Miller MD, Aiken C. Coupling of human immunodeficiency virus type 1 fusion to virion maturation: a novel role of the gp41 cytoplasmic tail. J Virol. 2004;78:3429–35. doi: 10.1128/JVI.78.7.3429-3435.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Murakami T, Ablan SD, Freed EO, Tanaka Y. Regulation of human immunodeficiency virus type 1 Env-mediated membrane fusion by viral protease activity. J Virol. 2004;78:1026–31. doi: 10.1128/JVI.78.2.1026-1031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Jiang J, Aiken C. Maturation-dependent human immunodeficiency virus type 1 particle fusion requires a carboxyl-terminal region of the gp41 cytoplasmic tail. J Virol. 2007;81:9999–10008. doi: 10.1128/JVI.00592-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kol N, Shi Y, Tsvitov M, Barlam D, Shneck RZ, Kay MS, Rousso I. A stiffness switch in human immunodeficiency virus. Biophys J. 2007;92:1777–83. doi: 10.1529/biophysj.106.093914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Kiernan RE, Ono A, Freed EO. Reversion of a human immunodeficiency virus type 1 matrix mutation affecting Gag membrane binding, endogenous reverse transcriptase activity, and virus infectivity. J Virol. 1999;73:4728–37. doi: 10.1128/jvi.73.6.4728-4737.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Davis MR, Jiang J, Zhou J, Freed EO, Aiken C. A mutation in the human immunodeficiency virus type 1 Gag protein destabilizes the interaction of the envelope protein subunits gp120 and gp41. J Virol. 2006;80:2405–17. doi: 10.1128/JVI.80.5.2405-2417.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Hourioux C, Brand D, Sizaret PY, Lemiale F, Lebigot S, Barin F, Roingeard P. Identification of the glycoprotein 41(TM) cytoplasmic tail domains of human immunodeficiency virus type 1 that interact with Pr55Gag particles. AIDS Res Hum Retroviruses. 2000;16:1141–7. doi: 10.1089/088922200414983. [DOI] [PubMed] [Google Scholar]
- 219.Wyma DJ, Kotov A, Aiken C. Evidence for a stable interaction of gp41 with Pr55(Gag) in immature human immunodeficiency virus type 1 particles. J Virol. 2000;74:9381–7. doi: 10.1128/jvi.74.20.9381-9387.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Vincent MJ, Melsen LR, Martin AS, Compans RW. Intracellular interaction of simian immunodeficiency virus Gag and Env proteins. J Virol. 1999;73:8138–44. doi: 10.1128/jvi.73.10.8138-8144.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Akari H, Fukumori T, Adachi A. Cell-dependent requirement of human immunodeficiency virus type 1 gp41 cytoplasmic tail for Env incorporation into virions. J Virol. 2000;74:4891–3. doi: 10.1128/jvi.74.10.4891-4893.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Hirsch VM, Edmondson P, Murphey-Corb M, Arbeille B, Johnson PR, Mullins JI. SIV adaptation to human cells. Nature. 1989;341:573–4. doi: 10.1038/341573a0. [DOI] [PubMed] [Google Scholar]
- 223.Kodama T, Wooley DP, Naidu YM, Kestler HW, Daniel MD, Li Y, Desrosiers RC. Significance of premature stop codons in env of simian immunodeficiency virus. J Virol. 1989;63:4709–14. doi: 10.1128/jvi.63.11.4709-4714.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Luciw PA, Shaw KE, Shacklett BL, Marthas ML. Importance of the intracytoplasmic domain of the simian immunodeficiency virus (SIV) envelope glycoprotein for pathogenesis. Virology. 1998;252:9–16. doi: 10.1006/viro.1998.9467. [DOI] [PubMed] [Google Scholar]
- 225.Vzorov AN, Weidmann A, Kozyr NL, Khaoustov V, Yoffe B, Compans RW. Role of the long cytoplasmic domain of the SIV Env glycoprotein in early and late stages of infection. Retrovirology. 2007;4:94. doi: 10.1186/1742-4690-4-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Pike LJ. Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J Lipid Res. 2006;47:1597–8. doi: 10.1194/jlr.E600002-JLR200. [DOI] [PubMed] [Google Scholar]
- 227.Simons K, Gerl MJ. Revitalizing membrane rafts: new tools and insights. Nat Rev Mol Cell Biol. 2010;11:688–99. doi: 10.1038/nrm2977. [DOI] [PubMed] [Google Scholar]
- 228.Munro S. Lipid rafts: elusive or illusive? Cell. 2003;115:377–88. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- 229.Ono A, Freed EO. Role of lipid rafts in virus replication. Adv Virus Res. 2005;64:311–58. doi: 10.1016/S0065-3527(05)64010-9. [DOI] [PubMed] [Google Scholar]
- 230.Waheed AA, Freed EO. Lipids and membrane microdomains in HIV-1 replication. Virus Res. 2009 doi: 10.1016/j.virusres.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Waheed AA, Freed EO. The Role of Lipids in Retrovirus Replication. Viruses. 2010 doi: 10.3390/v2051146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Brown DA, Rose JK. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 1992;68:533–44. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- 233.Brown DA, London E. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J Biol Chem. 2000;275:17221–4. doi: 10.1074/jbc.R000005200. [DOI] [PubMed] [Google Scholar]
- 234.Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1:31–9. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 235.Nguyen DH, Hildreth JE. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J Virol. 2000;74:3264–72. doi: 10.1128/jvi.74.7.3264-3272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ono A, Freed EO. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc Natl Acad Sci USA. 2001;98:13925–30. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Pickl WF, Pimentel-Muiños FX, Seed B. Lipid rafts and pseudotyping. J Virol. 2001;75:7175–83. doi: 10.1128/JVI.75.15.7175-7183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Lindwasser OW, Resh MD. Multimerization of human immunodeficiency virus type 1 Gag promotes its localization to barges, raft-like membrane microdomains. J Virol. 2001;75:7913–24. doi: 10.1128/JVI.75.17.7913-7924.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Holm K, Weclewicz K, Hewson R, Suomalainen M. Human immunodeficiency virus type 1 assembly and lipid rafts: Pr55(gag) associates with membrane domains that are largely resistant to Brij98 but sensitive to Triton X-100. J Virol. 2003;77:4805–17. doi: 10.1128/JVI.77.8.4805-4817.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Ono A, Waheed AA, Freed EO. Depletion of cellular cholesterol inhibits membrane binding and higher-order multimerization of human immunodeficiency virus type 1 Gag. Virology. 2007;360:27–35. doi: 10.1016/j.virol.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Waheed AA, Ablan SD, Soheilian F, Nagashima K, Ono A, Schaffner CP, Freed EO. Inhibition of human immunodeficiency virus type 1 assembly and release by the cholesterol-binding compound amphotericin B methyl ester: evidence for Vpu dependence. J Virol. 2008;82:9776–81. doi: 10.1128/JVI.00917-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Halwani R, Khorchid A, Cen S, Kleiman L. Rapid localization of Gag/GagPol complexes to detergent-resistant membrane during the assembly of human immunodeficiency virus type 1. J Virol. 2003;77:3973–84. doi: 10.1128/JVI.77.7.3973-3984.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Aloia RC, Tian H, Jensen FC. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc Natl Acad Sci USA. 1993;90:5181–5. doi: 10.1073/pnas.90.11.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Chan R, Uchil PD, Jin J, Shui G, Ott DE, Mothes W, Wenk MR. Retroviruses human immunodeficiency virus and murine leukemia virus are enriched in phosphoinositides. J Virol. 2008;82:11228–38. doi: 10.1128/JVI.00981-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Chan W-E, Lin H-H, Chen SS-L. Wild-type-like viral replication potential of human immunodeficiency virus type 1 envelope mutants lacking palmitoylation signals. J Virol. 2005;79:8374–87. doi: 10.1128/JVI.79.13.8374-8387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Bhattacharya J, Peters PJ, Clapham PR. Human immunodeficiency virus type 1 envelope glycoproteins that lack cytoplasmic domain cysteines: impact on association with membrane lipid rafts and incorporation onto budding virus particles. J Virol. 2004;78:5500–6. doi: 10.1128/JVI.78.10.5500-5506.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Bhattacharya J, Repik A, Clapham PR. Gag regulates association of human immunodeficiency virus type 1 envelope with detergent-resistant membranes. J Virol. 2006;80:5292–300. doi: 10.1128/JVI.01469-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Leung K, Kim J-O, Ganesh L, Kabat J, Schwartz O, Nabel GJ. HIV-1 assembly: viral glycoproteins segregate quantally to lipid rafts that associate individually with HIV-1 capsids and virions. Cell Host Microbe. 2008;3:285–92. doi: 10.1016/j.chom.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Jorgenson RL, Vogt VM, Johnson MC. Foreign glycoproteins can be actively recruited to virus assembly sites during pseudotyping. J Virol. 2009;83:4060–7. doi: 10.1128/JVI.02425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Camus G, Segura-Morales C, Molle D, Lopez-Vergès S, Begon-Pescia C, Cazevieille C, Schu P, Bertrand E, Berlioz-Torrent C, Basyuk E. The clathrin adaptor complex AP-1 binds HIV-1 and MLV Gag and facilitates their budding. Mol Biol Cell. 2007;18:3193–203. doi: 10.1091/mbc.E06-12-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Batonick M, Favre M, Boge M, Spearman P, Höning S, Thali M. Interaction of HIV-1 Gag with the clathrin-associated adaptor AP-2. Virology. 2005;342:190–200. doi: 10.1016/j.virol.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 252.Díaz E, Pfeffer SR. TIP47: a cargo selection device for mannose 6-phosphate receptor trafficking. Cell. 1998;93:433–43. doi: 10.1016/s0092-8674(00)81171-x. [DOI] [PubMed] [Google Scholar]
- 253.Carroll KS, Hanna J, Simon I, Krise J, Barbero P, Pfeffer SR. Role of Rab9 GTPase in facilitating receptor recruitment by TIP47. Science. 2001;292:1373–6. doi: 10.1126/science.1056791. [DOI] [PubMed] [Google Scholar]
- 254.Wolins NE, Rubin B, Brasaemle DL. TIP47 associates with lipid droplets. J Biol Chem. 2001;276:5101–8. doi: 10.1074/jbc.M006775200. [DOI] [PubMed] [Google Scholar]
- 255.Miura S, Gan J-W, Brzostowski J, Parisi MJ, Schultz CJ, Londos C, Oliver B, Kimmel AR. Functional conservation for lipid storage droplet association among Perilipin, ADRP, and TIP47 (PAT)-related proteins in mammals, Drosophila, and Dictyostelium. J Biol Chem. 2002;277:32253–7. doi: 10.1074/jbc.M204410200. [DOI] [PubMed] [Google Scholar]
- 256.Bulankina AV, Deggerich A, Wenzel D, Mutenda K, Wittmann JG, Rudolph MG, Burger KNJ, Höning S. TIP47 functions in the biogenesis of lipid droplets. J Cell Biol. 2009;185:641–55. doi: 10.1083/jcb.200812042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Barbero P, Buell E, Zulley S, Pfeffer SR. TIP47 is not a component of lipid droplets. J Biol Chem. 2001;276:24348–51. doi: 10.1074/jbc.M102468200. [DOI] [PubMed] [Google Scholar]
- 258.Bickel PE, Tansey JT, Welte MA. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim Biophys Acta. 2009;1791:419–40. doi: 10.1016/j.bbalip.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Blot G, Janvier K, Le Panse S, Benarous R, Berlioz-Torrent C. Targeting of the human immunodeficiency virus type 1 envelope to the trans-Golgi network through binding to TIP47 is required for env incorporation into virions and infectivity. J Virol. 2003;77:6931–45. doi: 10.1128/JVI.77.12.6931-6945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Lopez-Vergès S, Camus G, Blot G, Beauvoir R, Benarous R, Berlioz-Torrent C. Tail-interacting protein TIP47 is a connector between Gag and Env and is required for Env incorporation into HIV-1 virions. Proc Natl Acad Sci USA. 2006;103:14947–52. doi: 10.1073/pnas.0602941103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Bauby H, Lopez-Vergès S, Hoeffel G, Delcroix-Genête D, Janvier K, Mammano F, Hosmalin A, Berlioz-Torrent C. TIP47 is required for the production of infectious HIV-1 particles from primary macrophages. Traffic. 2010;11:455–67. doi: 10.1111/j.1600-0854.2010.01036.x. [DOI] [PubMed] [Google Scholar]
- 262.Round JL, Tomassian T, Zhang M, Patel V, Schoenberger SP, Miceli MC. Dlgh1 coordinates actin polymerization, synaptic T cell receptor and lipid raft aggregation, and effector function in T cells. J Exp Med. 2005;201:419–30. doi: 10.1084/jem.20041428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Blot V, Delamarre L, Perugi F, Pham D, Bénichou S, Benarous R, Hanada T, Chishti AH, Dokhélar M-C, Pique C. Human Dlg protein binds to the envelope glycoproteins of human T-cell leukemia virus type 1 and regulates envelope mediated cell-cell fusion in T lymphocytes. J Cell Sci. 2004;117:3983–93. doi: 10.1242/jcs.01266. [DOI] [PubMed] [Google Scholar]
- 264.Perugi F, Muriaux D, Ramirez BC, Chabani S, Decroly E, Darlix J-L, Blot V, Pique C. Human Discs Large is a new negative regulator of human immunodeficiency virus-1 infectivity. Mol Biol Cell. 2009;20:498–508. doi: 10.1091/mbc.E08-02-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Segrest J, De Loof H, Dohlman... J. Amphipathic helix motif: classes and properties. Proteins: Structure. 1990 doi: 10.1002/prot.340080202. [DOI] [PubMed] [Google Scholar]
- 266.Srinivas SK, Srinivas RV, Anantharamaiah GM, Compans RW, Segrest JP. Cytosolic domain of the human immunodeficiency virus envelope glycoproteins binds to calmodulin and inhibits calmodulin-regulated proteins. J Biol Chem. 1993;268:22895–9. [PubMed] [Google Scholar]
- 267.Miller MA, Mietzner TA, Cloyd MW, Robey WG, Montelaro RC. Identification of a calmodulin-binding and inhibitory peptide domain in the HIV-1 transmembrane glycoprotein. AIDS Res Hum Retroviruses. 1993;9:1057–66. doi: 10.1089/aid.1993.9.1057. [DOI] [PubMed] [Google Scholar]
- 268.Tencza SB, Mietzner TA, Montelaro RC. Calmodulin-binding function of LLP segments from the HIV type 1 transmembrane protein is conserved among natural sequence variants. AIDS Res Hum Retroviruses. 1997;13:263–9. doi: 10.1089/aid.1997.13.263. [DOI] [PubMed] [Google Scholar]
- 269.Micoli KJ, Pan G, Wu Y, Williams JP, Cook WJ, McDonald JM. Requirement of calmodulin binding by HIV-1 gp160 for enhanced FAS-mediated apoptosis. J Biol Chem. 2000;275:1233–40. doi: 10.1074/jbc.275.2.1233. [DOI] [PubMed] [Google Scholar]
- 270.Radding W, Pan ZQ, Hunter E, Johnston P, Williams JP, McDonald JM. Expression of HIV-1 envelope glycoprotein alters cellular calmodulin. Biochem Biophys Res Commun. 1996;218:192–7. doi: 10.1006/bbrc.1996.0034. [DOI] [PubMed] [Google Scholar]
- 271.Ishikawa H, Sasaki M, Noda S, Koga Y. Apoptosis induction by the binding of the carboxyl terminus of human immunodeficiency virus type 1 gp160 to calmodulin. J Virol. 1998;72:6574–80. doi: 10.1128/jvi.72.8.6574-6580.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Micoli KJ, Mamaeva O, Piller SC, Barker JL, Pan G, Hunter E, McDonald JM. Point mutations in the C-terminus of HIV-1 gp160 reduce apoptosis and calmodulin binding without affecting viral replication. Virology. 2006;344:468–79. doi: 10.1016/j.virol.2005.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Beary TP, Tencza SB, Mietzner TA, Montelaro RC. Interruption of T-cell signal transduction by lentivirus lytic peptides from HIV-1 transmembrane protein. J Pept Res. 1998;51:75–9. doi: 10.1111/j.1399-3011.1998.tb00419.x. [DOI] [PubMed] [Google Scholar]
- 274.Chow JYH, Jeffries CM, Kwan AH, Guss JM, Trewhella J. Calmodulin disrupts the structure of the HIV-1 MA protein. J Mol Biol. 2010;400:702–14. doi: 10.1016/j.jmb.2010.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Ghanam RH, Fernandez TF, Fledderman EL, Saad JS. Binding of calmodulin to the HIV-1 matrix protein triggers myristate exposure. J Biol Chem. 2010;285:41911–20. doi: 10.1074/jbc.M110.179093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Tang C, Loeliger E, Luncsford P, Kinde I, Beckett D, Summers MF. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc Natl Acad Sci USA. 2004;101:517–22. doi: 10.1073/pnas.0305665101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Kim EM, Lee KH, Kim JW. The cytoplasmic domain of HIV-1 gp41 interacts with the carboxyl-terminal region of alpha-catenin. Mol Cells. 1999;9:281–5. [PubMed] [Google Scholar]
- 278.Drees F, Pokutta S, Yamada S, Nelson WJ, Weis WI. Alpha-catenin is a molecular switch that binds E-cadherin-beta-catenin and regulates actin-filament assembly. Cell. 2005;123:903–15. doi: 10.1016/j.cell.2005.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Pokutta S, Drees F, Yamada S, Nelson WJ, Weis WI. Biochemical and structural analysis of alpha-catenin in cell-cell contacts. Biochem Soc Trans. 2008;36:141–7. doi: 10.1042/BST0360141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Kim JT, Kim EM, Lee KH, Choi J-E, Jhun BH, Kim JW. Leucine zipper domain of HIV-1 gp41 interacted specifically with alpha-catenin. Biochem Biophys Res Commun. 2002;291:1239–44. doi: 10.1006/bbrc.2002.6583. [DOI] [PubMed] [Google Scholar]
- 281.Zhang H, Wang L, Kao S, Whitehead IP, Hart MJ, Liu B, Duus K, Burridge K, Der CJ, Su L. Functional interaction between the cytoplasmic leucine-zipper domain of HIV-1 gp41 and p115-RhoGEF. Curr Biol. 1999;9:1271–4. doi: 10.1016/s0960-9822(99)80511-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–99. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 283.Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 284.Hill CS, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–70. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 285.Perona R, Montaner S, Saniger L, Sánchez-Pérez I, Bravo R, Lacal JC. Activation of the nuclear factor-kappaB by Rho, CDC42, and Rac-1 proteins. Genes Dev. 1997;11:463–75. doi: 10.1101/gad.11.4.463. [DOI] [PubMed] [Google Scholar]
- 286.Olson MF, Ashworth A, Hall A. An essential role for Rho, Rac, and Cdc42 GTPases in cell cycle progression through G1. Science. 1995;269:1270–2. doi: 10.1126/science.7652575. [DOI] [PubMed] [Google Scholar]
- 287.Maselko M, Ward C, Pastey M. A RhoA-Derived Peptide Inhibits Human Immunodeficiency Virus-1 Entry in vitro. Curr HIV Res. 2011;9:1–5. doi: 10.2174/157016211794582605. [DOI] [PubMed] [Google Scholar]
- 288.Figueroa C, Taylor J, Vojtek AB. Prenylated Rab acceptor protein is a receptor for prenylated small GTPases. J Biol Chem. 2001;276:28219–25. doi: 10.1074/jbc.M101763200. [DOI] [PubMed] [Google Scholar]
- 289.Hutt DM, Da-Silva LF, Chang LH, Prosser DC, Ngsee JK. PRA1 inhibits the extraction of membrane-bound rab GTPase by GDI1. J Biol Chem. 2000;275:18511–9. doi: 10.1074/jbc.M909309199. [DOI] [PubMed] [Google Scholar]
- 290.Abdul-Ghani M, Gougeon PY, Prosser DC, Da-Silva LF, Ngsee JK. PRA isoforms are targeted to distinct membrane compartments. J Biol Chem. 2001;276:6225–33. doi: 10.1074/jbc.M009073200. [DOI] [PubMed] [Google Scholar]
- 291.Martincic I, Peralta ME, Ngsee JK. Isolation and characterization of a dual prenylated Rab and VAMP2 receptor. J Biol Chem. 1997;272:26991–8. doi: 10.1074/jbc.272.43.26991. [DOI] [PubMed] [Google Scholar]
- 292.Evans DT, Tillman KC, Desrosiers RC. Envelope glycoprotein cytoplasmic domains from diverse lentiviruses interact with the prenylated Rab acceptor. J Virol. 2002;76:327–37. doi: 10.1128/JVI.76.1.327-337.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Blancou P, Evans DT, Desrosiers RC. PRA1 co-localizes with envelope but does not influence primate lentivirus production, infectivity or envelope incorporation. J Gen Virol. 2005;86:1785–90. doi: 10.1099/vir.0.80873-0. [DOI] [PubMed] [Google Scholar]
- 294.Enouf V, Chwetzoff S, Trugnan G, Cohen J. Interactions of rotavirus VP4 spike protein with the endosomal protein Rab5 and the prenylated Rab acceptor PRA1. J Virol. 2003;77:7041–7. doi: 10.1128/JVI.77.12.7041-7047.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Cuadras MA, Bordier BB, Zambrano JL, Ludert JE, Greenberg HB. Dissecting rotavirus particle-raft interaction with small interfering RNAs: insights into rotavirus transit through the secretory pathway. J Virol. 2006;80:3935–46. doi: 10.1128/JVI.80.8.3935-3946.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Liu H-P, Wu C-C, Chang Y-S. PRA1 promotes the intracellular trafficking and NF-kappaB signaling of EBV latent membrane protein 1. EMBO J. 2006;25:4120–30. doi: 10.1038/sj.emboj.7601282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Compton SL, Behrend EN. PRAF1: a Golgi complex transmembrane protein that interacts with viruses. Biochem Cell Biol. 2006;84:940–8. doi: 10.1139/o06-176. [DOI] [PubMed] [Google Scholar]
- 298.Liang G, Audas TE, Li Y, Cockram GP, Dean JD, Martyn AC, Kokame K, Lu R. Luman/CREB3 induces transcription of the endoplasmic reticulum (ER) stress response protein Herp through an ER stress response element. Mol Cell Biol. 2006;26:7999–8010. doi: 10.1128/MCB.01046-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.DenBoer LM, Hardy-Smith PW, Hogan MR, Cockram GP, Audas TE, Lu R. Luman is capable of binding and activating transcription from the unfolded protein response element. Biochem Biophys Res Commun. 2005;331:113–9. doi: 10.1016/j.bbrc.2005.03.141. [DOI] [PubMed] [Google Scholar]
- 300.Blot G, Lopez-Vergès S, Treand C, Kubat NJ, Delcroix-Genête D, Emiliani S, Benarous R, Berlioz-Torrent C. Luman, a new partner of HIV-1 TMgp41, interferes with Tat-mediated transcription of the HIV-1 LTR. J Mol Biol. 2006;364:1034–47. doi: 10.1016/j.jmb.2006.09.080. [DOI] [PubMed] [Google Scholar]
- 301.Emerson V, Holtkotte D, Pfeiffer T, Wang I-H, Schnölzer M, Kempf T, Bosch V. Identification of the cellular prohibitin 1/prohibitin 2 heterodimer as an interaction partner of the C-terminal cytoplasmic domain of the HIV-1 glycoprotein. J Virol. 2010;84:1355–65. doi: 10.1128/JVI.01641-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Mishra S, Ande SR, Nyomba BLG. The role of prohibitin in cell signaling. FEBS J. 2010;277:3937–46. doi: 10.1111/j.1742-4658.2010.07809.x. [DOI] [PubMed] [Google Scholar]
- 303.Merkwirth C, Langer T. Prohibitin function within mitochondria: essential roles for cell proliferation and cristae morphogenesis. Biochim Biophys Acta. 2009;1793:27–32. doi: 10.1016/j.bbamcr.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 304.Morrow IC, Parton RG. Flotillins and the PHB domain protein family: rafts, worms and anaesthetics. Traffic. 2005;6:725–40. doi: 10.1111/j.1600-0854.2005.00318.x. [DOI] [PubMed] [Google Scholar]
- 305.Kielec JM, Valentine KG, Wand AJ. A method for solution NMR structural studies of large integral membrane proteins: reverse micelle encapsulation. Biochim Biophys Acta. 2010;1798:150–60. doi: 10.1016/j.bbamem.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Ubarretxena-Belandia I, Stokes DL. Present and future of membrane protein structure determination by electron crystallography. Adv Protein Chem Struct Biol. 2010;81:33–60. doi: 10.1016/B978-0-12-381357-2.00002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Fujiyoshi Y. Structural physiology based on electron crystallography. Protein science : a publication of the Protein Society. 2011 doi: 10.1002/pro.621. [DOI] [PMC free article] [PubMed] [Google Scholar]