Abstract
The pressing need to develop antivirals active against resistant strains of HIV-1 has led to efforts to target steps in the virus life cycle other than reverse transcription and Gag proteolysis. Among those steps are entry, integration, and assembly and/or maturation. Advances in understanding the structural biology of both the immature and mature forms of the HIV capsid have made it possible to design or discover small molecules and peptides which interfere with both assembly and maturation. In this article we review the current state of the art in assembly and maturation inhibitors.
Roughly twenty years ago, Salunke et al observed that under a particular set of solution conditions the subunits of polyoma virus capsid protein assembled into octahedra 1. They suggested that “If, in fact, the variant octahedral structures occur naturally, they could be stabilized by a tetrameric reagent designed to bind selectively to the four unoccupied VP, bonding sites facing each fourfold axis.Such a reagent might interfere with virion assembly without impeding normal cell functions. Thus, further studies on conditions for stabilizing octahedral assemblies of papovavirus capsomeres could lead to applications for blocking assembly of infectious papillomaviruses.” Two years later Teschke et al, demonstrated that the small hydrophobic molecule bis-ANS could block in vitro assembly of bacteriophage P22 through binding to the capsid protein with a micromolar Kd 2. The compound did not appear to significantly alter the protein conformation and it was therefore suggested that binding at inter-subunit interfaces directly inhibited assembly. Subsequent studies suggested that the mechanism was actually the promotion of subunit association into assembly inactive dimers 3. The recognition that small molecule inhibition of capsid assembly was possible led to mathematical modeling of the process 4. A key insight was the recognition that it was not necessary to completely prevent subunit/subunit association. Modeling suggested that it was sufficient and perhaps preferable to misdirect the assembly pathway resulting in the accumulation of non-viable aberrant forms of capsids that would act as “sinks” for large numbers of subunits. Viewed in this way, viral capsids represent a single target containing hundreds to thousands of identical potentially inhibitory binding sites. Substantial progress has been made over the past 10 years in applying these concepts to targeting hepatitis B assembly 5; 6; 7.
Despite the success of HAART in treating HIV-1 infections, the emergence of resistance drives a pressing need to develop new antivirals. Cross-resistance, a phenomenon in which development of resistance to one particular therapeutic, concurrently results in development of resistance to other agents in that class suggests the need for not only novel compounds but compounds active against novel targets. While the capsids of polyoma, bacteriophage P22, and hepatitis are icosahedral, the fullerene core of the mature HIV particle, and the striking rearrangement of subunit interactions during the transition from immature to mature lattice suggests the possibility of identifying compounds that target either or both immature assembly and maturation. Central to this strategy is the fact that mutational studies indicate that not only Gag cleavage but also proper core formation is required for infectivity 8. Malformed cores appear to be defective at the stage of reverse transcription 9; 10, and in fact, mutations as subtle as those that increase or decrease core stability result in a loss of infectivity 11. Thus, it seems reasonable that compounds that alter the assembly pathway or the stability of the viral core would exhibit antiviral effects.
Targeting the N-terminal Domain of HIV Capsid Protein
In proof of concept experiments the entire C-terminal domain (CTD) of CA or even simply a peptide derived from helix nine, the helix driving CTD dimerization, were shown to inhibit polymerization of CA into helical tubes 12 (M.G. Mateau, personal communication). However, the first small molecule inhibitors of HIV assembly were the compounds CAP-1 and CAP-2 reported by Tang et al 13. Their discovery was based on a computational screen of public domain compounds for molecules that could bind to clefts on the N-terminal domain (NTD) of CA (CAN). NMR titration experiments determined that CAP-1 bound to CAN with a Kd of 1 mM and CAP-2 with a Kd of 52 uM. Mapping of the binding site by NMR perturbation experiments indicated that both compounds bound at the same site, the apex of a helical bundle composed of helices 1, 2, 3, 4, and 7 (Figure 1C). This region has been demonstrated to be involved in an inter-subunit CA NTD-CTD interaction unique to the mature lattice (Figure 1B) suggesting that CAP compound binding might act to block the formation of this interaction 14; 15; 16. The structure of CAN crystallized in the presence of CAP-1 indicated that CAP-1 binding induces a conformational rearrangement in CAN th resulted in the formation of a hydrophobic docking pocket 17. Despite the fact that the CAP-1 compound itself was not seen in the crystal structure computational docking experiments generated a docking model consistent with all X-ray and NMR data in which CAP-1 docking forces the exposure of a buried phenyalanine group (Phe32). The energetic penalty of exposing a hydrophobic Phe residue might be offset by relief of a strained main chain conformation seen in the unliganded CAN.
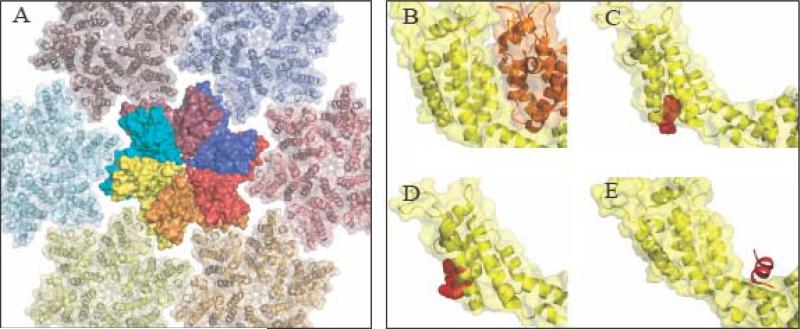
Figure 1.
Intersubunit Interactions and Inhibitor Binding Sites in HIV-1 Capsid Protein. (A) The structure of the mature CA hexamer. One full hexamer is shown in space filling form, and six surrounding hexamers are shown as ribbon diagrams. The individual subunits of the full hexamer are displayed with different colors. Note how the C-terminal domain of one subunit sits underneath the N-terminal domain of the adjacent (counter-clockwise) subunit. (B) Structure of two interacting CA subunits extracted from the mature HIV-1 hexamer. The NTD of both subunits is pictured. The CTD of the yellow subunit contacts the NTD of the adjacent (orange) subunit. This NTD/CTD interaction is unique to the mature form. (C) CAP-1 (red) is a small organic compound that binds to the NTD of HIV-1 CA and prevents assembly of the mature lattice, presumably by interfering with the NTD/CTD interaction. (D). (D) PF-74 (red) is a small organic compound that binds to the NTD of HIV-1 CA and destabilizes cores. (E) CAP-1 (red) is a small α-helical peptide that binds to the CA CTD and prevents assembly of both the immature and mature forms. The compounds were docked into subunits extracted from the hexamer crystal structure (PDB code 3MGE) by alignment of overlapping regions using domain/compound co-crystal structures (PDB Code 2JPR for CAP-1, 2XDE for PF-74, and 2BU0 for CAI) and hence do not reflect conformational changes induced by compound docking. The viewpoint varies slightly from panel to panel to best illustrate the relevant interaction.
Both CAP-1 and CAP-2 block in vitro assembly of CA into helical tubes which have been demonstrated to recapitulate the intersubunit interactions seen in the mature virus 14; 15. CAP-2 proved to be cytotoxic but 100 μM concentrations of CAP-1 were non-toxic and resulted in greater than 95% inhibition of virus replication in cell culture 13. Mechanism of action studies demonstrated that CAP-1 treatment did not reduce the amount of virus produced or the incorporation of envelope glycoprotein (Env). CAP-1 itself was not virucidal. Morphological analysis indicated that virions produced in the presence of CAP-1 were more heterogeneous in size than those produced in its absence and displayed aberrant core morphology. Taken together these results indicate that CAP-1 functions to block the subunit/subunit interactions formed during maturation. While no direct measurements of the number of CAP-1 molecules bound per virion have been made, based on the measured CAP-1/CAN affinity, it is estimated that as few as ~25 molecules of CAP-1 per particle are sufficient to inhibit the formation of a functional core particle, an observation consistent with the concept of considering the capsid as a single target with multiple binding sites.
These findings paved the way for subsequent efforts to screen for small molecule inhibitors of HIV-1 assembly and/or maturation. A screen of 10,000 compound using an in vitro mature lattice assembly assay led to the identification of approximately100 inhibitors of assembly with an average IC50 of10 μM 18. Of these 100 compounds six displayed favorable selectivity indices in cell culture and mechanism of action studies suggested a subset selectively blocked maturation (Prevelige, unpublished).
Larger scale screens have been carried out in the pharmaceutical industry. Boehringer-Ingelheim recently reported screens using an in vitro assembly system that resulted in the identification of several classes of small molecules with antiviral activity. Optimization of two of these compounds led to compounds with an EC50 < 100 nM and low cytotoxicity. These compounds bound the CAN and induced the formation of a binding pocket that overlapped the binding pocket for CAP-1. EM studies of virus produced under treatment conditions showed dramatically altered core morphology. Although these observations are consistent with their acting to block subunit/subunit interactions required for maturation mechanistic studies indicated that some of the compounds had a dual mode of action, acting as both capsid assembly inhibitors and non-nucleoside inhibitors of reverse transcriptase. However, it was possible to drive optimization of these compounds towards assembly inhibition as the single mode of action 19. Passage of virus in the presence of these inhibitors led to the appearance of escape mutants, the mutations were primarily localized to the NTD near the binding pocket although some CTD escape mutations were also seen.
In a search for novel antiviral inhibitors of HIV replication Pfizer developed and employed a cell based screen that required completion of the entire viral replication cycle to screen 106 compounds 20. Among the compounds with antiviral activity discovered was a series of compounds that appear to interfere with both assembly and uncoating. The preferred compound, known as PF-74, bound CA with a Kd of 3 μM, and had an EC50 of 0.6 μM and a therapeutic index of 121 when tested against HIV NL4-3 in MT-2 cells 21. The compound was co-crystallized with CAN and unlike CAP-1 did not induce a conformational change in the protein. Rather, it bound in a preformed pocket defined by helices 3, 4, 5, and 7 with the indole group protruding into NTD-CTD inter-subunit interface, a location in which it might act to prevent mature core formation (Fig 1D). Morphological analysis of particles produced in the presence of PF-74 bear this out as they appear highly heterogeneous and lack the central conical core 21. Selection of escape mutants resulted in strains carrying mutations in or near the compound binding pocket. Multiple mutations were required to reach a high level of resistance to the compound. Interestingly there was no obvious decrease in fitness associated with the escape mutations.
In addition to acting late in the replication cycle to block assembly, PF-74 was able to act early by altering post-entry uncoating. It is well documented that the stability of the core is a key determinant of infectivity. Cores which are either too stable, or too unstable, show reduced infectivity. Single cycle infectivity experiments suggested that PF-74 was capable of acting early during infection, and this was born out when virus that was incubated with PF-74 was shown to be capable of entry but incapable of undergoing reverse transcription. Experiments to address the mechanism of inhibition took advantage of both in vitro and in vivo assays of core stability 22. PF-74 bound to preformed mature HIV particles but as expected was incapable of binding to the escape mutant. Cores purified from PF-74 treated particles displayed a dose dependent decrease in stability when analyzed by sucrose gradient sedimentation whereas the cores derived from the escape mutant were unaffected by compound treatment. Similar results were found when naked cores (rather than viral particles) were treated with PF-74. To ascertain whether PF-74 destablized cores in vivo, VSV-G pseudotyped particles were used to infect cells in the presence of absence of PF-74. Four hours post infection, the cell were lysed and the fraction of soluble and pelletable CA was determined. Once again treatment with PF-74 increased the extent of dissociation of wild type cores but had no effect on the escape mutants. Finally, the effect of PF-74 on CA mutants known to result in hyperstable or destabilized cores was tested. Hyperstabilized cores were insensitive to PF-74 while intrinsically destabilized cores proved more sensitive. Taken together these studies provide compelling evidence that PF-74 interferes with protein/protein interactions in the mature capsid and that this interference can result in failed maturation or premature post-entry core dissociation.
Targeting the C-terminal Domain of HIV Capsid Protein
While the compounds described above bind to the NTD and inhibit the assembly or the stability of the mature capsid lattice, a peptide that inhibits the assembly of both the immature and mature form has been identified using phage display techniques. Sticht et al, panned random 12-mer phage display libraries against the entire CA molecule or a protein comprising the CTD, SP1 and NC regions of Gag (C-CANC) 23. They were able to identify sixteen different peptides capable of binding to both CA and C-CANC. These sixteen peptides could be classified into four groups based on sequence similarity with one group being dominant. Several synthetic peptides based on the discovered sequence were tested for binding and one peptide, called CAI, was selected for further study based on its favorable solubility profile. The authors demonstrated the ability of a five-fold molar excess of CAI to inhibit the in vitro assembly of a Gag-derived protein lacking p6 and residues 16-99 of MA (ΔMACANCSP2) which assembles into immature like particles. In addition to inhibiting immature assembly the peptide was also capable of effectively inhibiting the assembly of CANC or CA only into tubes that reflect the mature capsid lattice thereby displaying dual function. The interaction of CAI with CAC was mapped by deletion analysis, NMR experiments and ultimately a co-crystal structure 23; 24. The CAI peptide adopts an α-helical conformation and inserts in a groove between helices 1, 2, and 4 of the CTD (Figure 1E). Binding is mediated primarily by hydrophobic interactions and binding of the peptide induces a conformational change in the N-terminus of helix 2 that alters the CTD/CTD dimer interface in a manner that roughly halves the buried surface area. However, NMR rotational correlation time measurements indicated that CAI binding does not disrupt CAC dimerization leading to the suggestion that altered flexibility in the CTD might be the underlying cause of the assembly deficit. In a series of experiments whose outcome was surprising, using the co-crystal structure as a guide, a series of alanine substitutions were introduced into the CAI binding site on CA25. As expected, the mutations increased the Kd from ~1.6 μM as seen for the wild type to values as high as ~65 μM. While the mutations had no effect on the in vitro assembly of immature particles, most of them prevented the in vitro assembly of mature particles. When the mutations introduced into an infectious clone, similar results were seen. By and large the mutant viruses budded similar amounts of properly processed CA. However, mutations at two residues (Y169A and L211S) rendered the viruses non-infectious and thin section EM analysis revealed that the cores had failed to mature properly. The surprising finding was that the structure of the mutant CA in the absence of CAI was similar to the structure of the wild type protein with CAI bound. The implication of this finding for their mechanism of inhibition is that CAI does not inhibit maturation through exclusively through steric occlusion but may also function to lock the CTD in a conformation incompatible with maturation. In the case of inhibition of assembly of the immature form the mechanism is apparently more subtle as the mutations did not prevent immature core formation.
The CAI peptide itself was not capable of cell entry and therefore its antiviral efficacy could not be directly tested. However, using the structure of the co-crystal as a guide it was possible to take a structure based design approach to stabilize the α-helical peptide and convert it to a form that could penetrate cells using an approach known as hydrocarbon stapling. 26; 27. The resulting molecule was named NYAD-1. Fluorescently labeled NYAD-1 was capable of entering cells as determined by both FACS analysis and confocal microscopy. Confocal microscopy demonstrated colocalization of NYAD-1 with Gag in transit to the plasma membrane.
When cells were treated with NYAD-1 and transiently transfected with HIV-1 the yield of virus was reduced and an intracellular buildup of Gag was detected. EM analysis indicated that the production of both immature and mature virus was inhibited in NYAD-1 treated cells, results consistent with the in vitro findings. In experiments in which virus was incubated with NYAD-1 and then used to infect cells entry was not affected but infectivity was reduced and the virus morphology was perturbed. These results suggest that NYAD-1 might also act as an early post-entry inhibitor 27. An important consideration for any HIV antiviral is whether it is capable of broad spectrum anti-viral activity. NYAD-1 showed anti-viral activity against multiple laboratory adapted strains, a diverse collection of clinical isolates, and two RT- resistant HIV strains.
While the studies with CAI and NYAD-1 clearly demonstrated the therapeutic potential of targeting the hydrophobic pocket formed by helices 1, 2, and 4, small molecule inhibitors are likely to represent more promising lead compounds than stapled peptides. Towards this end, Curreli et al peformed a high throughput virtual docking screen for compounds binding in the pocket and selected the 200 top scoring compounds for further analysis from 100,000 compounds analyzed 28. Based on visual analysis they selected and purchased eight compounds for testing, two of which showed anti-viral activity and were deemed lead compounds . These compounds prevented mature but not immature assembly in vitro. When tested in cell culture low micromolar concentrations of the compounds effectively suppressed the release of a broad range of laboratory adapted and primary isolates of HIV and the released particles were less infectious. These studies validate both the N- and C-terminal domains of HIV-1 capsid protein as potential antiviral targets.
Targeting the CA-SP1 Cleavage Site
A natural products screen identified betulinic acid as a weak inhibitor of viral replication 29 and subsequent activity based modification led to a more effective molecule called alternatively, Beviramat, DSB, or PA-457. Beviramat inhibits the in vitro replication of a variety of HIV-1 strains including those resistant to protease or reverse transcriptase inhibitors and does so with IC50 in the nM range. It is specific for HIV-1 and displays no activity against HIV-2 or SIV.
Beviramat inhibition of viral replication occurs by a unique mechanism. In contrast to the protease inhibitors which inhibit all Gag processing, Beviramat inhibits only the final cleavage event in Gag processing, cleavage at the CA-SP1 junction. Mutational studies have demonstrated that this cleavage event is necessary for maturation, and therefore for viral replication. Tomographic reconstructions of the immature virus suggest that the CA-SP1 of Gag forms a helical bundle that serves to anchor the CTD below the plane of the NTD. Maturation requires the upward translation of this region to form the NTD/CTD interactions that stabilize the mature hexameric lattice (see article by Yeager in this issue). CEM studies indicate that in the absence of this cleavage event, immature cores appear to have been disassembled but mature cores cannot be formed 30; 31. Hydrogen/deuterium exchange studies on VLPs that do not undergo CA-SP1 cleavage suggest that extended order is lost but most of the local intersubunit interactions remain intact 32. Bevirimat treated particles, in contrast to the CA-SP1 cleavage mutants, appear to be incapable of disassembly and immature appearance. This observation suggests that Bevirimat not only prevents CA-SP1 cleavage but also may stabilize the immature lattice 33.
Passage of HIV-1 in the presence of increasing amounts of Beviramat led to the selection of escape mutations that mapped to the CA-SP1 junction 34; 35; 36; 37. Three of the mutations mapped at the C-terminus of CA, and three resistance mutations occurred in SP1, one at the first residue and two at the third residue suggesting this region might correspond to the Bevirimat binding site. Further support for this model is the fact that binding studies using radiolabeled Beviramant showed that it bound to the immature core but was incapable of binding to either monomeric protein or the mature core suggesting that the binding site was required subunit multimerization and was destroyed by CA-SP1 cleavage. Biochemical studies using labeled Beviramat indicated that it bound less well or not at all to strains carrying the CA-SP1 junction Beviramat escape mutations. Finally, moving the HIV-1 CA-SP1 region into HIV-2 rendered it susceptible to Beviramat inhibition 38. While this evidence strongly supports the hypothesis that Beviramat binds to the CA-SP1 region, the inability to obtain a high resolution structure of the immature virus with or without Beviramat bound limits the depth of mechanistic understanding or our ability to rationally design small molecules that target this presumptive binding site.
Beviramat is the only assembly or maturation inhibitor to undergo clinical testing and development to date. The compound showed promise in a mouse model, and promising phase I clinical trial data led to testing in HIV infected individuals 39;40; 41; 42; 43. These phase II trials produced mixed results. Some patients responded well, displaying an average of 1.26 log decrease in viral load. Others displayed no significant decrease in viral load and were classified as non-responders . Genotypic analysis suggested that poor response was associated with baseline polymorphisms in residues 6-8 of SP1 , and these observations were born out during in vitro testing of Beviramat susceptibility using genotyped patient isolates 44. It is noteworthy that this is not the region where escape mutations occurred during in vitro selection experiments.
Future Prospects
Despite the success of current anti-retroviral therapies, selection towards resistance remains a significant problem and drives the need for new antivirals which are ideally directed towards new targets. The complexity and delicate balance of HIV-1 viral protein interactions with both viral and host proteins suggests that these interactions could make attractive antiviral targets and agents that stabilize or destabilize them might prove effective. While protein/protein interactions have long been considered unattractive targets, the recognition of intersubunit “hotspots” responsible for the bulk of the binding energy and some recent successes suggest that this approach is tractable 45; 46; 47. Recent advances in understanding the structural biology of HIV coupled with the promising results presented in this review suggest that it will be possible to identify therapeutically useful compounds that interfere with HIV-1 assembly and maturation.
Acknowledgements
This work was supported by NIH grant AI44626. The author would like to thank H.G. Krausslich for critical comments on the manuscript and Eric Monroe for assistance with Figure 1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Salunke DM, Caspar DL, Garcea RL. Polymorphism in the assembly of polyomavirus capsid protein VP1. Biophys J. 1989;56:887–900. doi: 10.1016/S0006-3495(89)82735-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teschke CM, King J, Prevelige PE., Jr. Inhibition of viral capsid assembly by 1,1'-bi(4-anilinonaphthalene-5-sulfonic acid). Biochemistry. 1993;32:10658–65. doi: 10.1021/bi00091a016. [DOI] [PubMed] [Google Scholar]
- 3.Stafford WF, Liu S, Prevelige PE. New High Sensitivity Sedimentation Methods: Application to the Analysis of the Assembly of Bateriophage P22. In: Crabb JW, editor. Techniques in Protein Chemistry VI. Academic Press; San Diego: 1995. pp. 427–432. [Google Scholar]
- 4.Prevelige PE., Jr. Inhibiting virus-capsid assembly by altering the polymerisation pathway. Trends Biotechnol. 1998;16:61–5. doi: 10.1016/s0167-7799(97)01154-2. [DOI] [PubMed] [Google Scholar]
- 5.Zlotnick A, Ceres P, Singh S, Johnson JM. A small molecule inhibits and misdirects assembly of hepatitis B virus capsids. J Virol. 2002;76:4848–54. doi: 10.1128/JVI.76.10.4848-4854.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stray SJ, Bourne CR, Punna S, Lewis WG, Finn MG, Zlotnick A. A heteroaryldihydropyrimidine activates and can misdirect hepatitis B virus capsid assembly. Proc Natl Acad Sci U S A. 2005;102:8138–43. doi: 10.1073/pnas.0409732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stray SJ, Zlotnick A. BAY 41-4109 has multiple effects on Hepatitis B virus capsid assembly. J Mol Recognit. 2006;19:542–8. doi: 10.1002/jmr.801. [DOI] [PubMed] [Google Scholar]
- 8.von Schwedler UK, Stray KM, Garrus JE, Sundquist WI. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J Virol. 2003;77:5439–50. doi: 10.1128/JVI.77.9.5439-5450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang S, Murakami T, Agresta BE, Campbell S, Freed EO, Levin JG. Human immunodeficiency virus type 1 N-terminal capsid mutants that exhibit aberrant core morphology and are blocked in initiation of reverse transcription in infected cells. J Virol. 2001;75:9357–66. doi: 10.1128/JVI.75.19.9357-9366.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scholz I, Arvidson B, Huseby D, Barklis E. Virus particle core defects caused by mutations in the human immunodeficiency virus capsid N-terminal domain. J Virol. 2005;79:1470–9. doi: 10.1128/JVI.79.3.1470-1479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forshey BM, von Schwedler U, Sundquist WI, Aiken C. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J Virol. 2002;76:5667–77. doi: 10.1128/JVI.76.11.5667-5677.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lanman J, Sexton J, Sakalian M, Prevelige PE., Jr. Kinetic analysis of the role of intersubunit interactions in human immunodeficiency virus type 1 capsid protein assembly in vitro. J Virol. 2002;76:6900–8. doi: 10.1128/JVI.76.14.6900-6908.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang C, Loeliger E, Kinde I, Kyere S, Mayo K, Barklis E, Sun Y, Huang M, Summers MF. Antiviral inhibition of the HIV-1 capsid protein. J Mol Biol. 2003;327:1013–20. doi: 10.1016/s0022-2836(03)00289-4. [DOI] [PubMed] [Google Scholar]
- 14.Lanman J, Lam TT, Barnes S, Sakalian M, Emmett MR, Marshall AG, Prevelige PE., Jr. Identification of novel interactions in HIV-1 capsid protein assembly by high-resolution mass spectrometry. J Mol Biol. 2003;325:759–72. doi: 10.1016/s0022-2836(02)01245-7. [DOI] [PubMed] [Google Scholar]
- 15.Lanman J, Lam TT, Emmett MR, Marshall AG, Sakalian M, Prevelige PE., Jr. Key interactions in HIV-1 maturation identified by hydrogen-deuterium exchange. Nat Struct Mol Biol. 2004;11:676–7. doi: 10.1038/nsmb790. [DOI] [PubMed] [Google Scholar]
- 16.Pornillos O, Ganser-Pornillos BK, Kelly BN, Hua Y, Whitby FG, Stout CD, Sundquist WI, Hill CP, Yeager M. X-Ray Structures of the Hexameric Building Block of the HIV Capsid. Cell. 2009 doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelly BN, Kyere S, Kinde I, Tang C, Howard BR, Robinson H, Sundquist WI, Summers MF, Hill CP. Structure of the Antiviral Assembly Inhibitor CAP-1 Complex with the HIV-1 CA Protein. J Mol Biol. 2007;373:355–66. doi: 10.1016/j.jmb.2007.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prevelige P. Small Molecule Inhibitors of HIV-1 Capsid Assembly United States patent application number 20090176776. 2006. inventor.
- 19.Fader LD, Bethell R, Bonneau P, Bos M, Bousquet Y, Cordingley MG, Coulombe R, Deroy P, Faucher AM, Gagnon A, Goudreau N, Grand-Maitre C, Guse I, Hucke O, Kawai SH, Lacoste JE, Landry S, Lemke CT, Malenfant E, Mason S, Morin S, O'Meara J, Simoneau B, Titolo S, Yoakim C. Discovery of a 1,5-dihydrobenzo[b][1,4]diazepine-2,4-dione series of inhibitors of HIV-1 capsid assembly. Bioorg Med Chem Lett. 2011;21:398–404. doi: 10.1016/j.bmcl.2010.10.131. [DOI] [PubMed] [Google Scholar]
- 20.Cao J, Isaacson J, Patick AK, Blair WS. High-throughput human immunodeficiency virus type 1 (HIV-1) full replication assay that includes HIV-1 Vif as an antiviral target. Antimicrob Agents Chemother. 2005;49:3833–41. doi: 10.1128/AAC.49.9.3833-3841.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blair WS, Pickford C, Irving SL, Brown DG, Anderson M, Bazin R, Cao J, Ciaramella G, Isaacson J, Jackson L, Hunt R, Kjerrstrom A, Nieman JA, Patick AK, Perros M, Scott AD, Whitby K, Wu H, Butler SL. HIV capsid is a tractable target for small molecule therapeutic intervention. PLoS Pathog. 2010;6:e1001220. doi: 10.1371/journal.ppat.1001220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi J, Zhou J, Shah VB, Aiken C, Whitby K. Small-molecule inhibition of human immunodeficiency virus type 1 infection by virus capsid destabilization. J Virol. 2011;85:542–9. doi: 10.1128/JVI.01406-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sticht J, Humbert M, Findlow S, Bodem J, Muller B, Dietrich U, Werner J, Krausslich HG. A peptide inhibitor of HIV-1 assembly in vitro. Nat Struct Mol Biol. 2005;12:671–7. doi: 10.1038/nsmb964. [DOI] [PubMed] [Google Scholar]
- 24.Ternois F, Sticht J, Duquerroy S, Krausslich HG, Rey FA. The HIV-1 capsid protein C-terminal domain in complex with a virus assembly inhibitor. Nat Struct Mol Biol. 2005;12:678–82. doi: 10.1038/nsmb967. [DOI] [PubMed] [Google Scholar]
- 25.Bartonova V, Igonet S, Sticht J, Glass B, Habermann A, Vaney MC, Sehr P, Lewis J, Rey FA, Krausslich HG. Residues in the HIV-1 capsid assembly inhibitor binding site are essential for maintaining the assembly-competent quaternary structure of the capsid protein. J Biol Chem. 2008;283:32024–33. doi: 10.1074/jbc.M804230200. [DOI] [PubMed] [Google Scholar]
- 26.Bhattacharya S, Zhang H, Debnath AK, Cowburn D. Solution Structure of a Hydrocarbon Stapled Peptide Inhibitor in Complex with Monomeric C-terminal Domain of HIV-1 Capsid. J Biol Chem. 2008;283:16274–8. doi: 10.1074/jbc.C800048200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, Zhao Q, Bhattacharya S, Waheed AA, Tong X, Hong A, Heck S, Curreli F, Goger M, Cowburn D, Freed EO, Debnath AK. A cell-penetrating helical peptide as a potential HIV-1 inhibitor. J Mol Biol. 2008;378:565–80. doi: 10.1016/j.jmb.2008.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Curreli F, Zhang H, Zhang X, Pyatkin I, Victor Z, Altieri A, Debnath AK. Virtual screening based identification of novel small-molecule inhibitors targeted to the HIV-1 capsid. Bioorg Med Chem. 2011;19:77–90. doi: 10.1016/j.bmc.2010.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujioka T, Kashiwada Y, Kilkuskie RE, Cosentino LM, Ballas LM, Jiang JB, Janzen WP, Chen IS, Lee KH. Anti-AIDS agents, 11. Betulinic acid and platanic acid as anti-HIV principles from Syzigium claviflorum, and the anti-HIV activity of structurally related triterpenoids. J Nat Prod. 1994;57:243–7. doi: 10.1021/np50104a008. [DOI] [PubMed] [Google Scholar]
- 30.Briggs JA, Riches JD, Glass B, Bartonova V, Zanetti G, Krausslich HG. Structure and assembly of immature HIV. Proc Natl Acad Sci U S A. 2009;106:11090–5. doi: 10.1073/pnas.0903535106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Marco A, Muller B, Glass B, Riches JD, Krausslich HG, Briggs JA. Structural analysis of HIV-1 maturation using cryo-electron tomography. PLoS Pathog. 6:e1001215. doi: 10.1371/journal.ppat.1001215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monroe EB, Kang S, Kyere SK, Li R, Prevelige PE., Jr. Hydrogen/Deuterium Exchange Analysis of HIV-1 Capsid Assembly and Maturation. Structure. 2010;18:1483–91. doi: 10.1016/j.str.2010.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keller PW, Adamson CS, Heymann JB, Freed EO, Steven AC. HIV-1 maturation inhibitor bevirimat stabilizes the immature Gag lattice. J Virol. 2011;85:1420–8. doi: 10.1128/JVI.01926-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adamson CS, Ablan SD, Boeras I, Goila-Gaur R, Soheilian F, Nagashima K, Li F, Salzwedel K, Sakalian M, Wild CT, Freed EO. In vitro resistance to the human immunodeficiency virus type 1 maturation inhibitor PA-457 (Bevirimat). J Virol. 2006;80:10957–71. doi: 10.1128/JVI.01369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li F, Zoumplis D, Matallana C, Kilgore NR, Reddick M, Yunus AS, Adamson CS, Salzwedel K, Martin DE, Allaway GP, Freed EO, Wild CT. Determinants of activity of the HIV-1 maturation inhibitor PA-457. Virology. 2006;356:217–24. doi: 10.1016/j.virol.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 36.Li F, Goila-Gaur R, Salzwedel K, Kilgore NR, Reddick M, Matallana C, Castillo A, Zoumplis D, Martin DE, Orenstein JM, Allaway GP, Freed EO, Wild CT. PA-457: a potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proc Natl Acad Sci U S A. 2003;100:13555–60. doi: 10.1073/pnas.2234683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou J, Chen CH, Aiken C. Human immunodeficiency virus type 1 resistance to the small molecule maturation inhibitor 3-O-(3',3'-dimethylsuccinyl)-betulinic acid is conferred by a variety of single amino acid substitutions at the CA-SP1 cleavage site in Gag. J Virol. 2006;80:12095–101. doi: 10.1128/JVI.01626-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou J, Huang L, Hachey DL, Chen CH, Aiken C. Inhibition of HIV-1 maturation via drug association with the viral Gag protein in immature HIV-1 particles. J Biol Chem. 2005;280:42149–55. doi: 10.1074/jbc.M508951200. [DOI] [PubMed] [Google Scholar]
- 39.Bullock P, Larsen D, Press R, Wehrman T, Martin DE. The absorption, distribution, metabolism and elimination of bevirimat in rats. Biopharm Drug Dispos. 2008;29:396–405. doi: 10.1002/bdd.625. [DOI] [PubMed] [Google Scholar]
- 40.Wen Z, Martin DE, Bullock P, Lee KH, Smith PC. Glucuronidation of anti-HIV drug candidate bevirimat: identification of human UDP-glucuronosyltransferases and species differences. Drug Metab Dispos. 2007;35:440–8. doi: 10.1124/dmd.106.012815. [DOI] [PubMed] [Google Scholar]
- 41.Wen Z, Stern ST, Martin DE, Lee KH, Smith PC. Structural characterization of anti-HIV drug candidate PA-457 [3-O-(3',3'-dimethylsuccinyl)-betulinic acid] and its acyl glucuronides in rat bile and evaluation of in vitro stability in human and animal liver microsomes and plasma. Drug Metab Dispos. 2006;34:1436–42. doi: 10.1124/dmd.106.009233. [DOI] [PubMed] [Google Scholar]
- 42.Martin DE, Blum R, Doto J, Galbraith H, Ballow C. Multiple-dose pharmacokinetics and safety of bevirimat, a novel inhibitor of HIV maturation, in healthy volunteers. Clin Pharmacokinet. 2007;46:589–98. doi: 10.2165/00003088-200746070-00004. [DOI] [PubMed] [Google Scholar]
- 43.Martin DE, Blum R, Wilton J, Doto J, Galbraith H, Burgess GL, Smith PC, Ballow C. Safety and pharmacokinetics of Bevirimat (PA-457), a novel inhibitor of human immunodeficiency virus maturation, in healthy volunteers. Antimicrob Agents Chemother. 2007;51:3063–6. doi: 10.1128/AAC.01391-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Baelen K, Salzwedel K, Rondelez E, Van Eygen V, De Vos S, Verheyen A, Steegen K, Verlinden Y, Allaway GP, Stuyver LJ. Susceptibility of human immunodeficiency virus type 1 to the maturation inhibitor bevirimat is modulated by baseline polymorphisms in Gag spacer peptide 1. Antimicrob Agents Chemother. 2009;53:2185–8. doi: 10.1128/AAC.01650-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–6. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 46.Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov. 2004;3:301–17. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 47.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–9. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]