Abstract
HIV-1 becomes enveloped while budding through the plasma membrane, and the release of nascent virions requires a membrane fission event that separates the viral envelope from the cell surface. To facilitate this crucial step in its life cycle, HIV-1 exploits a complex cellular membrane remodeling and fission machinery known as the ESCRT pathway. HIV-1 Gag directly interacts with early-acting components of this pathway, which ultimately triggers the assembly of the ESCRT-III membrane fission complex at viral budding sites. Surprisingly, HIV-1 requires only a subset of ESCRT-III components, indicating that the membrane fission reaction that occurs during HIV-1 budding differs in crucial aspects from topologically related cellular abscission events.
Viruses are obligate intracellular parasites due to their dependence on host cell factors to complete their “life cycle”. Human Immunodeficiency Virus type 1 (HIV-1) is no exception, as it relies exclusively on cellular machinery to express the components required for the assembly of progeny virions. Furthermore, HIV-1 is exquisitely dependent on cellular factors for its ultimate release from virus-producing cells.
HIV-1 assembly and release are driven by the viral Gag protein, which associates with the inner leaflet of the plasma membrane and oligomerizes into a spherical protein shell that deforms the attached membrane. Ultimately, the growing bud pinches off from the cell surface, leading to the release of an immature virion. During or after budding, Gag is cleaved by the viral protease, which is necessary for the morphological maturation of the virion and for infectivity. The major cleavage products derived from Gag are the matrix (MA), capsid (CA), and nucleocapsid (NC) proteins. MA remains associated with the lipid envelope of the virion, CA rearranges to form the typically cone-shaped mature HIV-1 capsid, and NC covers the viral RNA genome within the capsid. The processing of HIV-1 Gag by the viral protease also yields a C-terminal peptide called p6, whose location in the mature virion has not yet been definitively determined.1
Because HIV-1 buds from the plasma membrane and thereby acquires a lipid envelope, the membrane covering the Gag shell ultimately must be severed from the plasma membrane to release the nascent virion into the extracellular medium. Although membrane fission could conceivably occur spontaneously once the assembling Gag shell approaches completion, retroviruses such as HIV-1 in fact exploit a cellular membrane fission machinery to facilitate their release. Specifically, the detachment of the virion depends on the host cell’s ESCRT pathway, which promotes membrane scission from the cytosolic side of bud necks, such as those formed during retroviral assembly. This review focuses on how HIV-1 engages the ESCRT pathway, and on how various components of the pathway contribute to HIV-1 release.
Retroviral late domains
The first indication that the release of retroviruses does not occur spontaneously came from a study showing that the unstructured p6 region of HIV-1 Gag is specifically required for the detachment of budded virions from the cell surface.2 In adherent cells, viral particles lacking p6 assembled at the plasma membrane and initiated the budding process, but remained trapped at the cell surface via a membranous tether.2 The phenotype of p6 mutants differs somewhat in T cell lines and primary lymphocytes, where the defect is primarily at the level of virion-virion detachment, resulting in the release of chains of virions.3 The major determinant required for HIV-1 virion detachment turned out to be a highly conserved PTAP motif within the otherwise rather variable C-terminal p6 domain of Gag.2; 4 All other known retroviruses harbor functionally equivalent short peptide motifs in Gag, which are commonly referred to as “late assembly” or L domains.5 In addition to the PTAP-type L domain found in HIV-1 and most other lentiviruses, two other types of L domains have been well characterized to date. These include the LYPxnL-type L domain originally identified in the Gag protein of equine infectious anemia virus (EIAV),6 and the PPxY-type L domain present in the Gag proteins of Rous sarcoma virus and of many other retroviruses.7 Different types of L domains often occur in combination; for instance, HIV-1 p6 harbors an auxiliary L domain of the LYPxnL-type, in addition to its primary PTAP-type L domain.8
Remarkably, different types of L domains can sometimes be exchanged between unrelated viruses, and the function of L domains does not strictly depend on their position within Gag.5 Together, these observations supported the concept that L domains are conserved docking sites that serve to recruit host factors to the site of virus assembly.5 This notion was confirmed by the discovery that the PTAP motif in HIV-1 p6 interacts with human TSG101, and that the interaction is required for HIV-1 budding.9; 10; 11; 12 Intriguingly, TSG101 is the mammalian homolog of a yeast protein required for the budding of cellular vesicles into the lumen of endosomal compartments called multivesicular bodies (MVB). This cellular budding process is mediated by a complex and highly conserved protein network, the ESCRT machinery, and serves to deliver transmembrane proteins in their entirety into the interior of endosomes for their eventual degradation.13; 14 Notably, the ESCRT-mediated budding of vesicles into MVB, like the budding of HIV-1 from the plasma membrane, occurs away from the cytosol, and thus has an identical topology.
In addition to TSG101, HIV-1 engages the ESCRT pathway component ALIX via its auxiliary LYPxnL-type L domain.8; 15; 16 ALIX is also the functionally relevant interaction partner for the LYPxnL-type L domain of EIAV, which is essential for EIAV budding.8; 15; 16 The third type of L domain (PPxY), which is not present in HIV-1, engages HECT ubiquitin ligases of the NEDD4 family.17; 18; 19 Interestingly, NEDD4 family ubiquitin ligases regulate the sorting of cargo into MVB.20 Thus, it appears that all types of L domains ultimately connect to the machinery involved in MVB sorting and biogenesis.
The ESCRT pathway
The ESCRT pathway exists in all eukaryotes, and consists of five heterooligomeric complexes (ESCRT-0, -I, -II, -III, and VPS4 complexes) as well as accessory proteins such as ALIX.21 ESCRT complexes are thought to be recruited sequentially to endosomal membranes, although there is evidence that upstream ESCRT complexes may act in parallel.22 ESCRT-0, -I, and –II are recruited as preformed complexes, whereas the ESCRT-III complex transiently assembles on endosomes from soluble cytosolic components. Mammalian ESCRT-III is formed by the CHMP proteins, a family of structurally related but highly divergent α-helical proteins that has twelve known members in humans. A recent report by Wollert and Hurley demonstrates that the individual ESCRT complexes perform discrete functions.23 Using purified ESCRT components and giant unilaminar vesicles (GUVs), they showed that: 1) the ESCRT-0 complex functions to concentrate ubiquitinated cargo on endosomal membranes, 2) the ESCRT-I and ESCRT-II complexes together induce bud formation without themselves entering the lumen of the budding vesicles, and 3) the ESCRT-III complex mediates membrane scission from the cytosolic side of the bud. Rather than being consumed, ESCRT-III is disassembled and recycled by the VPS4-VTA1 complex, enabling further rounds of sorting. Thus, in the MVB pathway, all of the ESCRT complexes are required.
Although the ESCRT machinery was first characterized based on its requirement for MVB biogenesis, it has recently emerged that the ESCRT system is also involved in membrane remodeling and fission events that lead to midbody abscission during the final stages of cytokinesis.24; 25; 26 Additionally, the ESCRT machinery controls earlier stages of cell division. At least a subset of ESCRT proteins is required for the proper function of centrosomes, and it has been noted that certain ESCRT-III components localize to kinetochores.27 Interestingly, ESCRT-III and Vps4 homologs are also key components of the cell divison system of certain archaea.28; 29 Since eukaryotes and archaea diverged at least a billion years ago, the promotion of cell division is likely to be the ancestral function of the ESCRT machinery.
ESCRT-I
The PTAP L domain of HIV-1 Gag binds directly to TSG101, the central component of ESCRT-I, a stable cytosolic heterotetramer formed by one copy each of TSG101, VPS28, VPS37, and MVB12.21; 30 Human cells express four versions of VPS37 (termed VPS37A-D) and two versions of MVB12 (termed MVB12A and B), which are encoded by different genes.30; 31; 32 Thus, eight combinations of TSG101, VPS28, VPS37, and MVB12 are theoretically possible, and all of these can indeed form stable ESCRT-I variants in cells.30 The complete ESCRT-I heterotetramer consists of a core formed by a headpiece and a long stalk, and of conformationally dynamic TSG101 UEV and VPS28 C-terminal domains (CTD) that are appended to opposite ends of the extended core.33
The flexible attachment of the UEV domain of TSG101 to the ESCRT-I core is thought to facilitate the binding of ligands, such as the PTAP L domain in HIV-1 p6. In the endocytic pathway, the TSG101 UEV domain binds to PSxP motifs in ESCRT-0 component HRS,34; 35 which has led to the proposal that HIV-1 Gag mimics the ability of HRS to recruit TSG101.34 In support of this notion, an HRS fragment capable of recruiting TSG101 rescues virus-like particle production when fused to an HIV-1 Gag construct that lacks its native PTAP L domain.34 Interestingly, P(T/S)AP binding-deficient alleles of TSG101 do not support HIV-1 budding as expected, but retain full function in EGF receptor degradation.32; 36 Therefore, compounds that target the HIV-1 Gag-TSG101 interaction to inhibit HIV-1 budding are not necessarily expected to have global inhibitory effects on the ESCRT pathway.
The role of the TSG101 UEV domain in HIV-1 budding is limited to the recognition of the PTAP L domain, because the N-terminal UEV domain is dispensable for the rescue of L domain-defective HIV-1 when TSG101 is artificially recruited to assembly sites.11 However, under these artificial conditions, a C-terminal region of TSG101 that binds to VPS28 remains necessary to confer budding activity.32; 37 Interestingly, the VPS28 CTD, which is attached to the ESRCT-I core via a flexible tether,33 is sufficient to rescue budding when appended to an L domain-defective Gag construct, apparently by acting as an adaptor for ESCRT-III component CHMP6.38 TSG101 also interacts with VPS37 proteins, which again can provide L domain function when fused directly to Gag.31; 32 Furthermore, binary TSG101-VPS37 complexes interact with MVB12,30 which is thought to stabilize the extended stalk of the ESCRT-I core.33 Nevertheless, MVB12 does not appear to play a central role in HIV-1 release.30 However, the depletion of MVB12 proteins induces aberrant virion morphologies and inhibits HIV-1 infectivity.30
ESCRT-II
Together with ESCRT-I, ESCRT-II mediates the deformation of endosomal membranes into buds during MVB biogenesis.23; 39 Yeast ESCRT-II binds to ESCRT-I component Vps28 and to ESCRT-III component Vps20, and thereby links ESCRT-I to ESCRT-III.40 Human ESCRT-I and ESCRT-II also interact, albeit in a different manner,41 and human ESCRT-II binds directly to ESCRT-III component CHMP6.41; 42 Together, these observations indicate that ESCRT-II also connects ESCRT-I to ESCRT-III in humans. Just as one might expect, human ESCRT-II transiently associates with endosomal membranes like its yeast counterpart, and functions in receptor downregulation.41 However, human ESCRT-II is dispensable for HIV-1 budding,41 indicating that HIV-1 p6-associated ESCRT-I recruits ESCRT-III membrane scission machinery in some other way.
ESCRT-III
The ESCRT-III complex is the main engine that carries out membrane scission in the ESCRT pathway.23; 43 The core ESCRT-III complex is formed by Vps20, SNF7, Vps24, and Vps2 in yeast, and by their homologs CHMP6, CHMP4A-C, CHMP3, and CHMP2A-B in humans. As its yeast ortholog Vps20, human CHMP6 is N-myristoylated and thought to nucleate the oligomerization of CHMP4 proteins on membranes.42; 44; 45 The remaining human ESCRT-III core components CHMP3 and CHMP2 appear to function as termination factors that cap CHMP4 filaments and recruit VPS4 for ESCRT-III disassembly.44
An essential role for the membrane fission activity of ESCRT-III in HIV-1 release is strongly implied by the fact that various dominant-negative versions of VPS4, the ATPase that disassembles ESCRT-III, arrest the budding of HIV-1 and other retroviruses.10; 46; 47 HIV-1 budding is also strongly inhibited if both human VPS4 proteins are depleted simultaneously via siRNA. 48 Moreover, individual ESCRT-III components, which are autoinhibited to prevent premature polymerization, can be activated to become extremely potent inhibitors of HIV-1 budding.8; 15; 16; 49; 50; 51; 52; 53 For instance, CHMP6 fused to RFP strongly inhibits HIV-1 release in a dominant-negative manner (Bettina Strack and Heinrich Göttlinger, unpublished observation). It therefore came as a surprise that CHMP6, the ESCRT-III subunit activated by ESCRT-II to trigger ESCRT-III assembly,45 is not required for HIV-1 budding.41 It is possible that activated CHMP proteins such as CHMP6-RFP sequester other ESCRT-III components that are directly involved in HIV-1 release. CHMP3 is also dispensable for HIV-1 budding (Alessia Zamborlini and Heinrich Göttlinger, unpublished observation), leaving only the core ESCRT-III components CHMP2 and CHMP4 as potential players in HIV-1 release. Indeed, a very recent study demonstrates that the co-depletion of CHMP2 or CHMP4 family members profoundly impairs HIV-1 release, whereas other CHMP proteins are not required.54
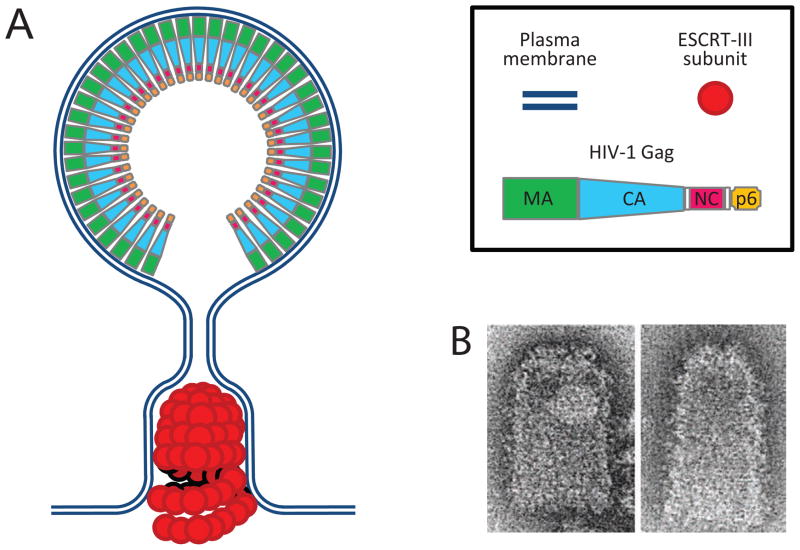
When overexpressed, human CHMP4A and CHMP4B polymerize into circular filaments that deform the plasma membrane into buds and tubules that emanate from the cell surface.55 Filament formation by SNF7, the yeast homolog of the three human CHMP4 proteins, is required to sequester endosomal cargo,44 and appears to be the most critical event during ESCRT-III-catalyzed membrane scission.43 Indeed, when recruited via ESCRT-II and the nucleation factor Vps20 (CHMP6), SNF7 (CHMP4A-C) was sufficient to catalyze the scission of bud necks in an in vitro reaction.23 Thus, during HIV-1 assembly, the formation of circular CHMP4 polymers within Gag-induced bud necks could in principle lead to membrane constriction and drive virus release.56 CHMP4-mediated membrane constriction in concert with Gag may also explain the observation that HIV-1 virions are often released before the Gag shell is completely assembled.57 In one model,58 which is based on the dome shape of certain CHMP2A/CHMP3 co-polymers,59 the assembly of dome-shaped CHMP4 polymers within HIV-1 buds would tighten the neck, owing to an affinity of CHMP4 for membranes, ultimately causing spontaneous fission (Fig. 1). However, the critical role of CHMP2 proteins, which recruit VPS4, also suggests an active participation of the VPS4-VTA1 complex in HIV-1 release.54 Recent live cell imaging studies reveal that ESCRT-III and VPS4 are rapidly and transiently recruited before HIV-1 particle release, which supports the notion that VPS4 has a direct role in HIV-1 budding.60; 61
Figure 1.
Model of ESCRT-III-mediated HIV-1 release. (A) HIV-1 Gag assembles into a plasma membrane-covered spherical protein shell that protrudes from the cell surface. The engagement of the ESCRT machinery by L domains in the C-terminal p6 domain of Gag leads to the assembly of an ESCRT-III tube with a dome-like membrane binding surface within the neck of the budding virion. The presence of a curved membrane-binding surface causes constriction of the bud neck until spontaneous fission occurs. (B) In vitro assembled CHMP2A-CHMP3 tubes exhibiting a dome-like cap. (Reprinted from Lata et al.53)
ALIX
Although HIV-1 relies predominantly on a PTAP-type L domain to recruit the CHMP4 fission factor via ERSCRT-I, it can also engage CHMP4 in alternative manner. As noted earlier, HIV-1 p6 harbors a secondary L domain of the LYPxnL-type, which functions as a docking site for ALIX, another early-acting factor in the ESCRT pathway.8; 15; 16 ALIX is composed of a banana-shaped Bro1 domain, a V-shaped middle domain, and a presumably unstructured proline-rich C-terminal domain (PRD) that harbors binding sites for various interaction partners.62 Through its Bro1 domain, ALIX interacts with C-terminal residues of the human CHMP4 proteins.8; 15; 16; 63; 64 ALIX thus directly links HIV-1 Gag to ESCRT-III.
The central V domain of ALIX harbors the binding site for the LYPxnL-type L domains in HIV-1 p6Gag and EIAV p9Gag.63; 65 Interestingly, the LYPxnL motif in HIV-1 p6Gag (n = 3) adopts a helical main chain conformation when bound to ALIX, whereas the EIAV L domain (n = 1) is bound in an extended conformation.66 Although the conserved residues in the LYPxnL-type L domains of HIV-1 and EIAV make equivalent interactions with ALIX, HIV-1 p6Gag has a significantly lower affinity for ALIX than EIAV p9Gag.66 Presumably, the difference in affinity is related to the fact that HIV-1 p6Gag also has a docking site for ESCRT-I, whereas EIAV relies solely on ALIX for budding.
HIV-1 budding can be potently inhibited by dominant-negative ALIX fragments, as long as a V domain capable of mediating ALIX dimerization is maintained.8; 67; 68 Dominant-negative ALIX fragments also induce Gag processing defects resembling those seen in the absence of a functional L domain.8; 67 However, mutations in the ALIX binding site of HIV-1 p6Gag generally have more modest effects on HIV-1 release and replication than mutations that disrupt the TSG101 binding site.63; 69 Also, HIV-1 p6Gag-mediated budding was significantly less affected by ALIX depletion than EIAV p9Gag-mediated budding.16 Overall, these findings are consistent with the notion that HIV-1 p6Gag contains a bipartite L domain in which the TSG101 binding site predominates, but does not account for all of the budding activity.70
Remarkably, the release and infectivity of HIV-1 mutants that lack the PTAP L domain can be dramatically stimulated by overexpressing ALIX.63; 71 This implies that ALIX, if expressed at sufficiently high levels, can essentially fully substitute for TSG101 in HIV-1 budding. As expected, the ALIX-mediated rescue is dependent on a V domain capable of binding to LYPxnL.63 Moreover, the presence of an intact LYPxnL motif in p6Gag is required, confirming that this motif constitutes a genuine L domain that functions through ALIX.63; 71 The activity of ALIX also depends on the ability of its Bro1 domain to interact with CHMP4, implying that the recruitment of this fission factor is crucial for ALIX-mediated HIV-1 budding. In contrast, interaction sites for TSG101, endophilin, CIN85, CMS/CD2AP, and CEP55 in the PRD of ALIX are dispensable for its function in HIV-1 budding.24; 63; 71 Nevertheless, the very C-terminus of the PRD is essential,24; 63; 71 apparently because PRD-dependent ALIX multimerization is required to promote HIV-1 budding.24
It has recently emerged that HIV-1 Gag harbors a second binding site for ALIX in the NC domain,72; 73 in addition to the LYPxnL ALIX binding site in p6. ALIX interacts with HIV-1 NC via its N-terminal Bro1 domain 72; 73 and the widely divergent Bro1 domains of other human proteins share an affinity for HIV-1 NC, indicating that NC recognizes a structural feature of the Bro1 domain rather than a specific sequence.74 The ability of NC to interact with Bro1 domains depends on two conserved CCHC-type zinc finger motifs in NC that are also required for the selective packaging of genomic viral RNA during assembly.72; 74 Nevertheless, nucleic acid does not appear to be involved in the ALIX-NC interaction, because the interaction was maintained in the presence of nuclease.72 Interestingly, mutations that disrupt the ALIX binding site in NC cause defects in Gag processing and HIV-1 release resembling those typically seen with HIV-1 L domain mutants.72 Furthermore, electron microscopy analyses of Rous sarcoma virus and HIV-1 NC mutants have revealed budding defects similar to those of L domain mutants.75; 76 Together, these observations suggest that NC may play a role in ESCRT engagement and virus release, in addition to its previously documented role in virus assembly.77
NEDD4L
PPxY-type L domains serve as docking sites for the WW domains of NEDD4 family ubiquitin ligases, and ultimately depend on the ESCRT pathway for function.17; 78 However, how NEDD4-type ubiquitin ligases connect to the ESCRT membrane fission machinery remains unknown. The nine human family members all possess an N-terminal C2 domain involved in membrane binding, multiple WW domains, and a C-terminal catalytic HECT domain. PPxY-mediated virus budding requires the engagement of a NEDD4 family member with a catalytically active HECT domain,17 but the functionally relevant target for ubiquitin attachment is uncertain. At least in principle, PPxY-mediated budding does not depend on the presence of ubiquitin acceptor sites in Gag,79; 80 which suggests the existence of an unknown cellular target.
Surprisingly, even though HIV-1 Gag lacks PPxY motifs, HIV-1 L domain mutants can be potently rescued by NEDD4L/NEDD4-2, particularly by a specific isoform with a naturally truncated C2 domain (termed NEDD4-2s).81; 82 Most regions of HIV-1 Gag, including all L domains, are known to be dispensable for the NEDD4L-mediated rescue, which in that regard differs clearly from the rescue mediated by ALIX.81; 82 The truncated C2 domain of NEDD4-2s functions as an autonomous HIV-1 Gag targeting module, and is sufficient to transfer the ability to stimulate HIV-1 budding to other NEDD4 family members, and even to their isolated catalytic HECT domains.83 The rescue of HIV-1 budding does not correlate with the overall levels of Gag ubiquitination, but rather with the synthesis of a particular type of chain in which ubiquitin molecules are linked through lysine 63.83 Ubiquitin chains of this type can serve as specific signals for the ESCRT-dependent sorting of cargo,84 and thus provide a possible link to the ESCRT membrane scission machinery. In a physiological context, NEDD4L may contribute to HIV-1 budding from cells with relatively low ESCRT-I levels, since the depletion of endogenous NEDD4L further reduced the residual release of HIV-1 L domain mutants.82
Concluding remarks
Although remarkable progress has been made in identifying the major cellular players involved in HIV-1 release, important questions remain to be addressed. For instance, it remains a mystery how ESCRT-I connects to ESCRT-III during HIV-1 budding, given that ESCRT-II is not required. Also, the mechanism by which ESCRT-III catalyzes membrane fission without being consumed in the reaction remains to be fully determined. Unlike topologically equivalent cellular budding events during yeast MVB biogenesis, HIV-1 budding depends only on a subset of core ESCRT-III components,54 but the reason for this difference is not known. It also is not entirely clear why HIV-1 maintains both a TSG101 binding site and a low-affinity ALIX binding site in p6, whereas some other primate lentiviruses appear to rely exclusively on a high-affinity interaction site for ALIX. Finally, it has recently emerged that certain HIV-1 Gag constructs do not depend on the ESCRT pathway for the efficient production of virus-like particles.85 Thus, a full understanding of why native HIV-1 Gag is so exquisitely dependent on the ESCRT machinery is still lacking.
Acknowledgments
Work in the authors’ laboratory is supported by grant number R37AI029873 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gottlinger HG, Weissenhorn W. Assembly and release. In: Kurth R, Bannert N, editors. Retroviruses: Molecular Microbiology and Genomics. Caister Academic Press; Norfolk, UK: 2010. pp. 187–215. [Google Scholar]
- 2.Gottlinger HG, Dorfman T, Sodroski JG, Haseltine WA. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci U S A. 1991;88:3195–9. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Demirov DG, Orenstein JM, Freed EO. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J Virol. 2002;76:105–17. doi: 10.1128/JVI.76.1.105-117.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang M, Orenstein JM, Martin MA, Freed EO. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–8. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parent LJ, Bennett RP, Craven RC, Nelle TD, Krishna NK, Bowzard JB, Wilson CB, Puffer BA, Montelaro RC, Wills JW. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1995;69:5455–60. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puffer BA, Parent LJ, Wills JW, Montelaro RC. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J Virol. 1997;71:6541–6. doi: 10.1128/jvi.71.9.6541-6546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wills JW, Cameron CE, Wilson CB, Xiang Y, Bennett RP, Leis J. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J Virol. 1994;68:6605–18. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–99. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 9.VerPlank L, Bouamr F, LaGrassa TJ, Agresta B, Kikonyogo A, Leis J, Carter CA. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag) Proc Natl Acad Sci U S A. 2001;98:7724–9. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, Myszka DG, Sundquist WI. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 11.Martin-Serrano J, Zang T, Bieniasz PD. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001;7:1313–9. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 12.Demirov DG, Ono A, Orenstein JM, Freed EO. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc Natl Acad Sci U S A. 2002;99:955–60. doi: 10.1073/pnas.032511899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saksena S, Sun J, Chu T, Emr SD. ESCRTing proteins in the endocytic pathway. Trends Biochem Sci. 2007;32:561–73. doi: 10.1016/j.tibs.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–52. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 15.von Schwedler UK, Stuchell M, Muller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, Scott A, Krausslich HG, Kaplan J, Morham SG, Sundquist WI. The protein network of HIV budding. Cell. 2003;114:701–13. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 16.Martin-Serrano J, Yarovoy A, Perez-Caballero D, Bieniasz PD. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc Natl Acad Sci U S A. 2003;100:12414–9. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin-Serrano J, Eastman SW, Chung W, Bieniasz PD. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J Cell Biol. 2005;168:89–101. doi: 10.1083/jcb.200408155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kikonyogo A, Bouamr F, Vana ML, Xiang Y, Aiyar A, Carter C, Leis J. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc Natl Acad Sci U S A. 2001;98:11199–204. doi: 10.1073/pnas.201268998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vana ML, Tang Y, Chen A, Medina G, Carter C, Leis J. Role of Nedd4 and ubiquitination of Rous sarcoma virus Gag in budding of virus-like particles from cells. J Virol. 2004;78:13943–53. doi: 10.1128/JVI.78.24.13943-13953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 21.Hurley JH. ESCRT complexes and the biogenesis of multivesicular bodies. Curr Opin Cell Biol. 2008;20:4–11. doi: 10.1016/j.ceb.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hurley JH, Ren X. The circuitry of cargo flux in the ESCRT pathway. J Cell Biol. 2009;185:185–7. doi: 10.1083/jcb.200903013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864–9. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlton JG, Agromayor M, Martin-Serrano J. Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc Natl Acad Sci U S A. 2008;105:10541–6. doi: 10.1073/pnas.0802008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–12. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 26.Morita E, Sandrin V, Chung HY, Morham SG, Gygi SP, Rodesch CK, Sundquist WI. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. Embo J. 2007;26:4215–27. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morita E, Colf LA, Karren MA, Sandrin V, Rodesch CK, Sundquist WI. Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc Natl Acad Sci U S A. 2010;107:12889–94. doi: 10.1073/pnas.1005938107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindas AC, Karlsson EA, Lindgren MT, Ettema TJ, Bernander R. A unique cell division machinery in the Archaea. Proc Natl Acad Sci U S A. 2008;105:18942–6. doi: 10.1073/pnas.0809467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in archaea. Science. 2008;322:1710–3. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morita E, Sandrin V, Alam SL, Eckert DM, Gygi SP, Sundquist WI. Identification of human MVB12 proteins as ESCRT-I subunits that function in HIV budding. Cell Host Microbe. 2007;2:41–53. doi: 10.1016/j.chom.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eastman SW, Martin-Serrano J, Chung W, Zang T, Bieniasz PD. Identification of human VPS37C, a component of endosomal sorting complex required for transport-I important for viral budding. J Biol Chem. 2005;280:628–36. doi: 10.1074/jbc.M410384200. [DOI] [PubMed] [Google Scholar]
- 32.Stuchell MD, Garrus JE, Muller B, Stray KM, Ghaffarian S, McKinnon R, Krausslich HG, Morham SG, Sundquist WI. The human endosomal sorting complex required for transport (ESCRT-I) and its role in HIV-1 budding. J Biol Chem. 2004;279:36059–71. doi: 10.1074/jbc.M405226200. [DOI] [PubMed] [Google Scholar]
- 33.Kostelansky MS, Schluter C, Tam YY, Lee S, Ghirlando R, Beach B, Conibear E, Hurley JH. Molecular architecture and functional model of the complete yeast ESCRT-I heterotetramer. Cell. 2007;129:485–98. doi: 10.1016/j.cell.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pornillos O, Higginson DS, Stray KM, Fisher RD, Garrus JE, Payne M, He GP, Wang HE, Morham SG, Sundquist WI. HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. J Cell Biol. 2003;162:425–34. doi: 10.1083/jcb.200302138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bouamr F, Houck-Loomis BR, De Los Santos M, Casaday RJ, Johnson MC, Goff SP. The C-terminal portion of the Hrs protein interacts with Tsg101 and interferes with human immunodeficiency virus type 1 Gag particle production. J Virol. 2007;81:2909–22. doi: 10.1128/JVI.01413-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Im YJ, Kuo L, Ren X, Burgos PV, Zhao XZ, Liu F, Burke TR, Jr, Bonifacino JS, Freed EO, Hurley JH. Crystallographic and functional analysis of the ESCRT-I/HIV-1 Gag PTAP interaction. Structure. 2010;18:1536–47. doi: 10.1016/j.str.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin-Serrano J, Zang T, Bieniasz PD. Role of ESCRT-I in Retroviral Budding. J Virol. 2003;77:4794–804. doi: 10.1128/JVI.77.8.4794-4804.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pineda-Molina E, Belrhali H, Piefer AJ, Akula I, Bates P, Weissenhorn W. The crystal structure of the C-terminal domain of Vps28 reveals a conserved surface required for Vps20 recruitment. Traffic. 2006;7:1007–16. doi: 10.1111/j.1600-0854.2006.00440.x. [DOI] [PubMed] [Google Scholar]
- 39.Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell. 2002;3:283–9. doi: 10.1016/s1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- 40.Hurley JH. The ESCRT complexes. Crit Rev Biochem Mol Biol. 2010;45:463–87. doi: 10.3109/10409238.2010.502516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langelier C, von Schwedler UK, Fisher RD, De Domenico I, White PL, Hill CP, Kaplan J, Ward D, Sundquist WI. Human ESCRT-II complex and its role in human immunodeficiency virus type 1 release. J Virol. 2006;80:9465–80. doi: 10.1128/JVI.01049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yorikawa C, Shibata H, Waguri S, Hatta K, Horii M, Katoh K, Kobayashi T, Uchiyama Y, Maki M. Human CHMP6, a myristoylated ESCRT-III protein, interacts directly with an ESCRT-II component EAP20 and regulates endosomal cargo sorting. Biochem J. 2005;387:17–26. doi: 10.1042/BJ20041227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–7. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teis D, Saksena S, Emr SD. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev Cell. 2008;15:578–89. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 45.Teis D, Saksena S, Judson BL, Emr SD. ESCRT-II coordinates the assembly of ESCRT-III filaments for cargo sorting and multivesicular body vesicle formation. Embo J. 2010;29:871–83. doi: 10.1038/emboj.2009.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scott A, Chung HY, Gonciarz-Swiatek M, Hill GC, Whitby FG, Gaspar J, Holton JM, Viswanathan R, Ghaffarian S, Hill CP, Sundquist WI. Structural and mechanistic studies of VPS4 proteins. Embo J. 2005;24:3658–69. doi: 10.1038/sj.emboj.7600818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gonciarz MD, Whitby FG, Eckert DM, Kieffer C, Heroux A, Sundquist WI, Hill CP. Biochemical and structural studies of yeast Vps4 oligomerization. J Mol Biol. 2008;384:878–95. doi: 10.1016/j.jmb.2008.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kieffer C, Skalicky JJ, Morita E, De Domenico I, Ward DM, Kaplan J, Sundquist WI. Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev Cell. 2008;15:62–73. doi: 10.1016/j.devcel.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 49.Zamborlini A, Usami Y, Radoshitzky SR, Popova E, Palu G, Gottlinger H. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc Natl Acad Sci U S A. 2006;103:19140–5. doi: 10.1073/pnas.0603788103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muziol T, Pineda-Molina E, Ravelli RB, Zamborlini A, Usami Y, Gottlinger H, Weissenhorn W. Structural basis for budding by the ESCRT-III factor CHMP3. Dev Cell. 2006;10:821–30. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 51.Shim S, Kimpler LA, Hanson PI. Structure/Function Analysis of Four Core ESCRT-III Proteins Reveals Common Regulatory Role for Extreme C-Terminal Domain. Traffic. 2007;8:1068–79. doi: 10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- 52.Bajorek M, Schubert HL, McCullough J, Langelier C, Eckert DM, Stubblefield WM, Uter NT, Myszka DG, Hill CP, Sundquist WI. Structural basis for ESCRT-III protein autoinhibition. Nat Struct Mol Biol. 2009;16:754–62. doi: 10.1038/nsmb.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lata S, Roessle M, Solomons J, Jamin M, Gottlinger HG, Svergun DI, Weissenhorn W. Structural basis for autoinhibition of ESCRT-III CHMP3. J Mol Biol. 2008;378:816–25. doi: 10.1016/j.jmb.2008.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morita E, Sandrin V, McCullough J, Katsuyama A, Baci Hamilton I, Sundquist WI. ESCRT-III Protein Requirements for HIV-1 Budding. Cell Host Microbe. 2011;9:235–42. doi: 10.1016/j.chom.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peel S, Macheboeuf P, Martinelli N, Weissenhorn W. Divergent pathways lead to ESCRT-III-catalyzed membrane fission. Trends Biochem Sci. 2010 Oct 26; doi: 10.1016/j.tibs.2010.09.004. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 57.Carlson LA, Briggs JA, Glass B, Riches JD, Simon MN, Johnson MC, Muller B, Grunewald K, Krausslich HG. Three-dimensional analysis of budding sites and released virus suggests a revised model for HIV-1 morphogenesis. Cell Host Microbe. 2008;4:592–9. doi: 10.1016/j.chom.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fabrikant G, Lata S, Riches JD, Briggs JA, Weissenhorn W, Kozlov MM. Computational model of membrane fission catalyzed by ESCRT-III. PLoS Comput Biol. 2009;5:e1000575. doi: 10.1371/journal.pcbi.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, Weissenhorn W. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321:1354–7. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baumgartel V, Ivanchenko S, Dupont A, Sergeev M, Wiseman PW, Krausslich HG, Brauchle C, Muller B, Lamb DC. Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat Cell Biol. 2011;13:469–74. doi: 10.1038/ncb2215. [DOI] [PubMed] [Google Scholar]
- 61.Jouvenet N, Zhadina M, Bieniasz PD, Simon SM. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat Cell Biol. 2011;13:394–401. doi: 10.1038/ncb2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fujii K, Hurley JH, Freed EO. Beyond Tsg101: the role of Alix in ‘ESCRTing’ HIV-1. Nat Rev Microbiol. 2007;5:912–6. doi: 10.1038/nrmicro1790. [DOI] [PubMed] [Google Scholar]
- 63.Fisher RD, Chung HY, Zhai Q, Robinson H, Sundquist WI, Hill CP. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 2007;128:841–52. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 64.McCullough J, Fisher RD, Whitby FG, Sundquist WI, Hill CP. ALIX-CHMP4 interactions in the human ESCRT pathway. Proc Natl Acad Sci U S A. 2008;105:7687–91. doi: 10.1073/pnas.0801567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee S, Joshi A, Nagashima K, Freed EO, Hurley JH. Structural basis for viral late-domain binding to Alix. Nat Struct Mol Biol. 2007;14:194–9. doi: 10.1038/nsmb1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhai Q, Fisher RD, Chung HY, Myszka DG, Sundquist WI, Hill CP. Structural and functional studies of ALIX interactions with YPX(n)L late domains of HIV-1 and EIAV. Nat Struct Mol Biol. 2008;15:43–9. doi: 10.1038/nsmb1319. [DOI] [PubMed] [Google Scholar]
- 67.Munshi UM, Kim J, Nagashima K, Hurley JH, Freed EO. An Alix fragment potently inhibits HIV-1 budding: Characterization of binding to retroviral YPXL late domains. J Biol Chem. 2007;282:3847–55. doi: 10.1074/jbc.M607489200. [DOI] [PubMed] [Google Scholar]
- 68.Pires R, Hartlieb B, Signor L, Schoehn G, Lata S, Roessle M, Moriscot C, Popov S, Hinz A, Jamin M, Boyer V, Sadoul R, Forest E, Svergun DI, Gottlinger HG, Weissenhorn W. A crescent-shaped ALIX dimer targets ESCRT-III CHMP4 filaments. Structure. 2009;17:843–56. doi: 10.1016/j.str.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fujii K, Munshi UM, Ablan SD, Demirov DG, Soheilian F, Nagashima K, Stephen AG, Fisher RJ, Freed EO. Functional role of Alix in HIV-1 replication. Virology. 2009;391:284–92. doi: 10.1016/j.virol.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martin-Serrano J, Bieniasz PD. A bipartite late-budding domain in human immunodeficiency virus type 1. J Virol. 2003;77:12373–7. doi: 10.1128/JVI.77.22.12373-12377.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Usami Y, Popov S, Gottlinger HG. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J Virol. 2007;81:6614–22. doi: 10.1128/JVI.00314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Popov S, Popova E, Inoue M, Gottlinger HG. Human immunodeficiency virus type 1 Gag engages the Bro1 domain of ALIX/AIP1 through the nucleocapsid. J Virol. 2008;82:1389–98. doi: 10.1128/JVI.01912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dussupt V, Javid MP, Abou-Jaoude G, Jadwin JA, de La Cruz J, Nagashima K, Bouamr F. The nucleocapsid region of HIV–1 Gag cooperates with the PTAP and LYPXnL late domains to recruit the cellular machinery necessary for viral budding. PLoS Pathog. 2009;5:e1000339. doi: 10.1371/journal.ppat.1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Popov S, Popova E, Inoue M, Gottlinger HG. Divergent Bro1 domains share the capacity to bind human immunodeficiency virus type 1 nucleocapsid and to enhance virus-like particle production. J Virol. 2009;83:7185–93. doi: 10.1128/JVI.00198-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee EG, Linial ML. Deletion of a Cys-His motif from the Alpharetrovirus nucleocapsid domain reveals late domain mutant-like budding defects. Virology. 2006;347:226–33. doi: 10.1016/j.virol.2005.11.048. [DOI] [PubMed] [Google Scholar]
- 76.Dussupt V, Sette P, Bello NF, Javid MP, Nagashima K, Bouamr F. Basic Residues in the Nucleocapsid Domain of Gag Are Critical for Late Events of HIV-1 Budding. J Virol. 2011;85:2304–15. doi: 10.1128/JVI.01562-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Y, Barklis E. Effects of nucleocapsid mutations on human immunodeficiency virus assembly and RNA encapsidation. J Virol. 1997;71:6765–76. doi: 10.1128/jvi.71.9.6765-6776.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pincetic A, Leis J. The Mechanism of Budding of Retroviruses From Cell Membranes. Adv Virol. 2009;2009:6239691–6239699. doi: 10.1155/2009/623969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhadina M, McClure MO, Johnson MC, Bieniasz PD. Ubiquitin-dependent virus particle budding without viral protein ubiquitination. Proc Natl Acad Sci U S A. 2007;104:20031–6. doi: 10.1073/pnas.0708002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhadina M, Bieniasz PD. Functional interchangeability of late domains, late domain cofactors and ubiquitin in viral budding. PLoS Pathog. 2010;6:e1001153. doi: 10.1371/journal.ppat.1001153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Usami Y, Popov S, Popova E, Gottlinger HG. Efficient and specific rescue of human immunodeficiency virus type 1 budding defects by a Nedd4-like ubiquitin ligase. J Virol. 2008;82:4898–907. doi: 10.1128/JVI.02675-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chung HY, Morita E, von Schwedler U, Muller B, Krausslich HG, Sundquist WI. NEDD4L overexpression rescues the release and infectivity of human immunodeficiency virus type 1 constructs lacking PTAP and YPXL late domains. J Virol. 2008;82:4884–97. doi: 10.1128/JVI.02667-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weiss ER, Popova E, Yamanaka H, Kim HC, Huibregtse JM, Gottlinger H. Rescue of HIV-1 release by targeting widely divergent NEDD4-type ubiquitin ligases and isolated catalytic HECT domains to Gag. PLoS Pathog. 2010;6:e1001107. doi: 10.1371/journal.ppat.1001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lauwers E, Erpapazoglou Z, Haguenauer-Tsapis R, Andre B. The ubiquitin code of yeast permease trafficking. Trends Cell Biol. 2010;20:196–204. doi: 10.1016/j.tcb.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 85.Popova E, Popov S, Gottlinger HG. Human immunodeficiency virus type 1 nucleocapsid p1 confers ESCRT pathway dependence. J Virol. 2010;84:6590–7. doi: 10.1128/JVI.00035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]