Abstract
Hyaluronan exerts a variety of biological effects on cells including changes in cell migration, proliferation, and matrix metabolism. However, the signaling pathways associated with the action of hyaluronan on cells have not been clearly defined. In some cells, signaling is induced by the loss of cell-hyaluronan interactions. The goal of this study was to use hyaluronan oligosaccharides as a molecular tool to explore the effects of changes in cell-hyaluronan interactions and determine the underlying molecular events that become activated. In this study, hyaluronan oligosaccharides induced the loss of extracellular matrix proteoglycan and collagen from cultured slices of normal adult human articular cartilage. This loss was coincident with an increased expression of matrix metalloproteinase (MMP)-13. MMP-13 expression was also induced in articular chondrocytes by hyaluronan (HA) hexasaccharides but not by HA tetrasaccharides nor high molecular weight hyaluronan. MMP-13 promoter-reporter constructs in CD44-null COS-7 cells revealed that both CD44-dependent and CD44-independent events mediate the induction of MMP-13 by hyaluronan oligosaccharides. Electromobility gel shift assays demonstrated the activation of chondrocyte NFκB by hyaluronan oligosaccharides. NFκB activation was also documented in C-28/I2 immortalized human chondrocytes by luciferase promoter assays and phosphorylation of IKK-α/β. The link between activation of NFκB and MMP-13 induction by HA oligosaccharides was further confirmed through the use of the NFκB inhibitor helenalin. Inhibition of MAP kinases also demonstrated the involvement of p38 MAP kinase in the hyaluronan oligosaccharide induction of MMP-13. Our findings suggest that hyaluronan-CD44 interactions affect matrix metabolism via activation of NFκB and p38 MAP kinase.
In many tissues cell-matrix interactions serve to regulate cellular homeostasis (1, 2). Interference with these associations typically signal cells to initiate matrix repair, cell proliferation, or even cell migration. Such interactions are especially important in tissues such as articular cartilage, a tissue rich in extracellular matrix but with limited vascular access for systemic control of homeostasis. Chondrocytes are thus highly dependent on cell-matrix interactions as a primary means to “sense” changes in the extracellular environment (3). Most investigations in this area focus on cell signaling induced through the interaction of integrin receptors with collagens, fibronectin, laminin, matrilins, etc. (3–7). However, cell-matrix interactions involving hyaluronan (HA),2 once thought to be predominately structural in nature, are now beginning to receive increasing attention as initiators of cell signaling.
HA is a high molecular weight polysaccharide consisting of repeating disaccharide units glucuronic acid and N-acetylglucosamine (8). In cartilage and many other connective tissues, HA serves as a central filamentous scaffold to which proteoglycans such as aggrecan and link protein become bound (9). This complex matrix network of HA and proteoglycan in turn is anchored to the cell surface via a continued association of HA with a HA synthase and/or the binding of HA to a HA receptor such as CD44 (10, 11). HA-rich extracellular matrices tethered to cells can be visualized on living cells in vitro using a particle exclusion assay. These matrices are seen as a gel-like, transparent zone extending by up to one cell diameter away from the plasma membrane (2, 10). In cells such as chondrocytes that exhibit large cell-associated matrices, the presence of high molecular mass HA appears to promote quiescence, dampens matrix degradation induced by inflammatory cytokines (12–14) or fragmented fibronectin (15, 16), and inhibits FAS ligand-induced apoptosis (17). We have also recently demonstrated that high molecular mass HA promotes the interaction between CD44 and Smad1, an interaction that appears necessary for optimal presentation of Smad1 to the bone morphologic protein-7 receptor complex (18).
The mechanism for HA-CD44 signaling events has not been clearly defined. In some cells CD44 functions as a co-receptor, for example, CD44 coupled to Smad1 (18), CD44 coupled to members of the epidermal growth factor receptor family of receptor tyrosine kinases (19, 20) or c-Src kinase (21), CD44 coupled to transforming growth factor β receptor (22, 23), or CD44 coupled to phosphatidylinositol 3-kinase (24, 25). One property that appears common to all of these interactions is that CD44-mediated signaling likely involves multivalent interactions of HA with numerous CD44 receptors. The rationale for this model is that: 1) HA is a high molecular weight polymer with a highly repetitive structure that can support multivalent interactions, and 2) cells respond differentially to HA of different molecule size. Thus, signaling is believed to be initiated by the clustering of CD44 receptors, an event that gives rise to subsequent downstream effects on the underlying cytoskeleton or other partnered co-receptors (24, 25). We have used a variety of methods to selectively interfere with the clustering potential of high molecular mass HA. These include the use of testicular or Streptomyces hyaluronidase (11, 26), pCD44Δ67 (a dominant negative recombinant CD44 that cannot bind HA) (18, 27, 28), and small HA oligosaccharides (HAoligo) such as HA hexasaccharides (HA6) (29–31). Streptomyces hyaluronidase and pCD44Δ67 block the ability of CD44 to interact with Smad1 (18). We have also shown previously that interfering with HA-cell interactions in the developing limb bud by the use of HAoligo results in a delay in the formation of precartilage condensations as well as a delay in chondrogenic differentiation of mesenchymal cells (32). In adult cartilage HAoligo activate a “matrix repair” response that includes both a massive loss of extracellular matrix components because of the activation of gelatinolytic enzymes and aggrecanase as well as the induction of new matrix biosynthesis. Similar results are obtained when cartilage is treated with antisense oligonucleotides directed against CD44 (33) or chondrocytes are treated with hyaluronidase (26). Thus interfering with CD44-HA interactions by perturbing either the HA or the CD44, both induce metabolic changes away from matrix homeostasis. However, the cell signaling events responsible for these changes have not been well clarified.
Recently, we performed protein-DNA array analyses to detect changes in the activation profile of transcription factors in chondrocytes following treatment with HAoligo (31). Our results demonstrated that the transcription factors retinoic acid receptor, retinoid X receptor, Sp1, and NFκB were activated by the stimulation of HAoligo. Thus, HAoligo do, in fact, affect cell signaling pathways resulting in changes in transcription factor activation. Defining the linkage between these altered transcription factor activities and downstream events that affect changes in cartilage matrix metabolism was one of the goals of this study.
During cartilage repair as well as progressive cartilage degeneration associated with osteoarthritis, the synthesis and activation of matrix metalloproteinases (MMPs) are considered to be of critical importance (34–36). The initial degradation of collagen fibrils within the triple-helical region depends on cleavage at the “collagenase site.” Two major candidate enzymes, namely collagenase-1 (MMP-1) and collagenase-3 (MMP-13), are considered physiologically significant collagenases. MMP-13 has the potential to cleave type II collagen into ¾ and ¼ fragments at a rate 10 times faster than MMP-1 (37–39) and, in addition, can cleave aggrecan (40). Because the expression of MMP-13 is elevated in the arthritic joints (35, 39), this proteinase is thought to be a key enzyme associated with osteoarthritis. Recent studies have shown that retinoid-related signals strongly enhance the induction of MMP-13 in chondrocytic cells through the action of retinoic acid receptor-retinoid X receptor heterodimers (41). In addition, it has been demonstrated that MMP-13 induction by inflammatory cytokines requires the activation of NFκB (42). Furthermore, Fieber et al. (43) reported that HAoligo induced the expression of MMP-13 in murine embryonic fibroblasts and tumor cells coincident with an activation of NFκB. These findings together with the results from our protein-DNA array analyses led us to explore the effects of HAoligo on the expression of MMP-13 in chondrocytes.
In this study we demonstrate that HAoligo enhance the expression of MMP-13 in chondrocytes at mRNA and protein levels, mediated at least in part through CD44. We also demonstrate that the induction mechanism is associated with the activation of the transcription factor NFκB as well as p38 MAP kinase.
EXPERIMENTAL PROCEDURES
Materials
Antibodies for mouse anti-human type II collagen and mouse anti-human MMP-13 were purchased from R & D Systems, Inc. (Minneapolis, MN). Mouse anti-human phospho-IKK-α (Ser180) and phospho-IKK-β (Ser181) antibodies were purchased from Cell Signaling Technology (Beverly, MA), and β-actin antibody was from Sigma. Streptavidin peroxidase (component of Vectastain ABC kit) was purchased from Vector Laboratories (Burlingame, CA). Dulbecco’s modified Eagle’s medium (DMEM) for cell and tissue cultures was obtained from Invitrogen. Fetal bovine serum (FBS) was purchased from Summit Biotechnology (Fort Collins, CO). Pronase and collagenase P used in dissociation of the tissue were purchased from Calbiochem and Roche Applied Science, respectively. Trizol® reagent for RNA extraction was purchased from Invitrogen. SYBR® green nucleic acid gel stain was purchased from Molecular Probes, Inc. (Eugene, OR).
Hyaluronan Oligosaccharides
Hyaluronan from human umbilical cord (type I; Sigma) was used to generate HAoligo by testicular hyaluronidase (type I-S; Sigma) cleavage at a ratio of 320 units/mg hyaluronan in 0.1 m sodium acetate buffer with 0.5 m NaCl (pH 5) (44). Digestion condition was selected at 37 °C for 16 h so as to yield preparations with a predominant proportion of HA6 (29). After precipitation of heat-in-activated hyaluronidase in 80% ethanol, the oligosaccharides were blown dry, dissolved in sterile phosphate-buffered saline, and used. Purified HA4 and HA6 obtained from Seikagaku Corporation (Japan) were also used (45).
Cells
Bovine articular chondrocytes were isolated from metacarpophalangeal joints of 18-month-old steers obtained from a local slaughterhouse. Human articular chondrocytes were isolated from the talocrural ankle cartilage obtained from tissue donors through the Gift of Hope Organ and Tissue Donor Network (formerly the Regional Organ Bank of Illinois). These donor tissues were obtained, with appropriate institutional approval, less than 24 h from the time of death and were from donors with no history of arthritic disease. Full thickness slices of articular cartilages were dissected, and chondrocytes were isolated by use of a sequential Pronase/collagenase digestion as described previously (28). The primary chondrocytes were cultured as a high density monolayer (5 × 106 cells/35-mm diameter dish) in DMEM supplemented with 50 units/ml penicillin and 50 µg/ml streptomycin containing 10% FBS. The immortalized human chondrocytes C-28/I2 cells were kindly provided by Dr. Mary Goldring (Harvard Institutes of Medicine). The mammalian COS-7 cell line (SV40-transformed African green monkey kidney cells) was purchased from American Type Culture Collection (Manassas, VA). Immortalized chondrocytes and COS-7 cells were seeded in 12-well dishes at a density of 5 × 104/well and 2 × 105/well, respectively, and maintained in DMEM containing 10% FBS until transfection. The cultures were maintained in an atmosphere of 5% CO2 at 37 °C in a humidified incubator.
Treatment of Cells
The cells were brought to serum-free conditions by a gradual reduction to 1% FBS for 12 h and to 0% for 12 h prior to the start of the experiment. For gene analysis of MMP-13 in human and bovine articular chondrocytes, HAoligo at 0–500 µg/ml were applied to cells in serum-free DMEM for 0–48 h. For investigation of transcription factor activation, bovine chondrocytes were treated with HAoligo at 250 µg/ml for 1–12 h. For the Western blot analysis of phospho-IKK-α/β, human immortalized chondrocytes were treated with 250 µg/ml HAoligo for 5–60 min. For signaling inhibition assays, bovine chondrocytes were pretreated with inhibitors of NFκB (1 nm helenalin; Calbiochem), p38 MAP kinase (5 nm SB203580; Calbiochem) or MAP kinase kinase (MEK) (10 nm PD98059; Calbiochem) for 1 h and then treated with 250 µg/ml HAoligo for 24 h in the presence or absence of each inhibitor. In some recovery experiments, cultures of COS-7 cells were washed excessively with DMEM after a 12-h treatment with 250 µg/ml HA6 and then incubated in fresh media with or without 500 µg/ml high molecular mass HA (HMW-HA) (~5.0 × 105-7.3 × 105 Da; HYAL-GAN®, Sanofi-Synthelabo Inc., New York, NY).
Staining Cartilage Explants
Full thickness slices (~1 × 10 × 10 mm) of human articular cartilage dissected from talocrural ankle joints were cultured directly in 1.0 ml of DMEM containing 10% FBS. Following a 2-day recovery period, the tissue slices were treated with or without 250 µg/ml HAoligo under serum-free conditions. Following 7 days of incubation, the slices were fixed with 4% paraformaldehyde for 2 h and embedded in paraffin, and 7-µm sections were prepared. Sections were stained with safranin O and counterstained with fast green (46). Other paraffin sections were incubated with type II collagen or MMP-13 antibodies overnight after blocking endogenous peroxidase with 0.3% H2O2 in methanol and blocking nonspecific IgG binding with 10% FBS. These antibodies were detected with biotinylated anti-mouse IgG (diluted 1:1500; Vector Laboratories) and the avidin-biotin-peroxidase complex visualized with 3,3′-diaminobenzidine (FAST Tablet set; Sigma).
Real Time Reverse Transcription (RT)-PCR
Total RNA was isolated from the bovine and human chondrocytes cultures according to the manufacturer’s instructions for the use of Trizol® reagent. The RNA was reverse transcribed with GENE Amp RNA PCR kit reagents (PerkinElmer Life Sciences) and amplified using the PTC-100™ programmable thermal controller (MJ Research, Inc., Watertown, MA). For real time RT-PCR, the PCR products were detected by SYBR® green. Primer-specific amplification was at 57 °C for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and at 60 °C for MMP-13. However, fluorescence quantification was performed at a higher temperature, 76 °C for GAPDH and 82 °C for MMP-13. These quantification temperatures were set below the individual melting peak of each PCR product. The primers sequences are follows: GAPDH, forward, 5′-GTC AAC GGA TTT GGT CGT ATT GGG-3′, and reverse, 5′-TGC CAT GGG TGG AAT CAT ATT GG-3′; and MMP-13, forward, 5′-CGC CAG AAG AAT CTG TCT TTA AA-3′, and reverse, 5′-CCA AAT TAT GGA GGA GAT GC-3′. Thermal cycling and fluorescence detection was performed using a Smart Cycler system (Cepheid, Sunnyvale, CA). Real time PCR efficiency (E) was calculated according to the equation provided by Rasmussen et al. (47) as E = 10[−1/slope] for GAPDH and MMP-13. The slope was determined from the graph of ng of cDNA substrate (x axis) versus the cycle number at the crossing point (CP) (y axis). The CP is the PCR cycle number that represents the peak of the second derivative of change in SYBR® green fluorescence intensity. The fold increase in copy numbers of mRNA is calculated as a relative ratio of MMP-13 to GAPDH, following the mathematical model (Equation 1) introduced by Pfaffl (48).
| (Eq. 1) |
Assays for MMP-13 Activity
Ten times concentrated conditioned media from serum-free bovine articular chondrocyte cultures or full thickness slices of bovine cartilage tissue (~1 × 10 × 10 mm) were assayed for protease activity using casein zymography. In some experiments, the conditioned media samples were preactivated by treatment with 2.5 mm aminophenylmercuric acetate (APMA) for 1 h at 37 °C. The samples were next separated in 10% SDS-PAGE containing casein (0.5 mg/ml; Sigma). After electrophoresis, SDS was removed by washing the gel with 50 mm Tris-HCl (pH 7.5) and 2.5% Triton X-100 twice for 30 min and twice more for 10 min with 50 mm Tris-HCl (pH 7.5), 0.15 m NaCl, 10 mm CaCl2, 0.1% Triton X-100, and 0.02% NaN3. The gels were then incubated overnight at 37 °C in 50 mm Tris-HCl, 5 mm CaCl2, 1 µm ZnCl2, 1% Triton X-100, 0.02% NaN3 containing 1 mm APMA for MMP activation. The staining was performed for 1 h at room temperature with 0.5% Coomassie Brilliant Blue R-250 in 10% acetic acid until clear bands over a dark background were observed. In other experiments, MMP-13 specific enzymatic activity was assessed using a MMP-13 fluorogenic substrate (1 µm, Mca-Pro-Cha-Gly-Nva-His-Ala-Dpa-NH2; Calbiochem) as previously described (49). MMP activation was performed by treating conditioned media with 2.5 mm APMA for 1 h at 37 °C. The assays were performed in 10 mm Tris (pH 7.4), 2 mm CaCl2, 0.01% Triton X-100, 0.01% NaN3 substrate buffer. Fluorescence reading were made as specified by the manufacture (excitation, 325 nm; emission, 393 nm) using a fluorescence microreader (Wollec 1450; PerkinElmer Life Sciences).
Electrophoretic Mobility Shift Assay
25-µg nuclear extracts from control or experimental cultures were mixed with biotin labeled transcription factor probes (EMSA kit; Panomics) and incubated at 15 °C for 30 min. Also included were control assays in which excess unlabeled double-stranded DNA was added for DNA-protein binding competition assay during the hybridization period. The mixture was then loaded onto a 6% polyacrylamide gel and electrophoresed at 120 V in 0.5% Tris-borate-EDTA for 1 h. The sample was transferred in 0.5% Tris-borate-EDTA onto a nylon membrane (Biodyne; Pall Co., East Hills, NY) at 300 mA for 30 min. After transfer, the sample was fixed on the membrane by UV cross-linking for 3 min (120,000 microjoules). The bands were visualized after exposure to chemiluminescence film based on the Streptavidin horseradish peroxidase reaction.
Western Blot for Phospho-IKK-α/β
Aliquots of total cellular lysates from C-28/I2 cells containing 30 µg of protein were loaded onto 10% SDS-polyacrylamide gels for electrophoresis. Following electroblot transfer onto polyvinylidine difluoride membrane and blocking in 5% nonfat dry milk, phospho-IKK-α, phospho-IKK-β, and β-actin were detected with primary antibodies followed by horseradish peroxidase-conjugated anti-mouse IgG antibodies. Detection was performed using chemiluminescence (ECL Western blotting analysis system; Amersham Biosciences).
Promoter Analysis in Human Immortalized Chondrocytes and COS-7 Cells
The human immortalized chondrocytes C-28/I2 were plated 24 h prior to transfection at a density of 2 × 105 cells/well in 12-well plates and transiently transfected with 2 µg of MMP-13 promoter-reporter construct (50) or NFκB promoter-reporter construct (Stratagene, La Jolla, CA) using FuGENE 6 transfection reagent (Roche Applied Science) following the manufacturer’s instructions. In some experiments, COS-7 cells were plated 24 h prior to transfection at a density of 2 × 105 cells/well in 12-well plate and transiently co-transfected with 0.5 µg of MMP-13 reporter construct and a full-length pCD44H plasmid subcloned into pTracer (Invitrogen) (27, 51). After 24 h of incubation, the cells were rinsed in DMEM and changed to serum-free conditions for 20–24 h followed by treatment with 250 µg/ml HAoligo. The cells were harvested after 24 h, and luciferase activity was assayed using a luciferase reporter assay system (Promega). The firefly luciferase promoterless vector, pGL2-Enhancer (Promega) was used as a control, and the data were normalized by the total protein concentration in each well.
Statistical Analysis
Analysis of variance and Student’s t test were performed as appropriate, using the statistical programs in Statview (Abacus Concepts, Inc., Berkeley, CA). Analysis of variance was used to evaluate the effects of HAoligo on the promoter activity of MMP-13 and NFκB in C-28/I2 immortalized chondrocytes and COS-7 cells.
RESULTS
HAoligo Enhance Cartilage Degradation
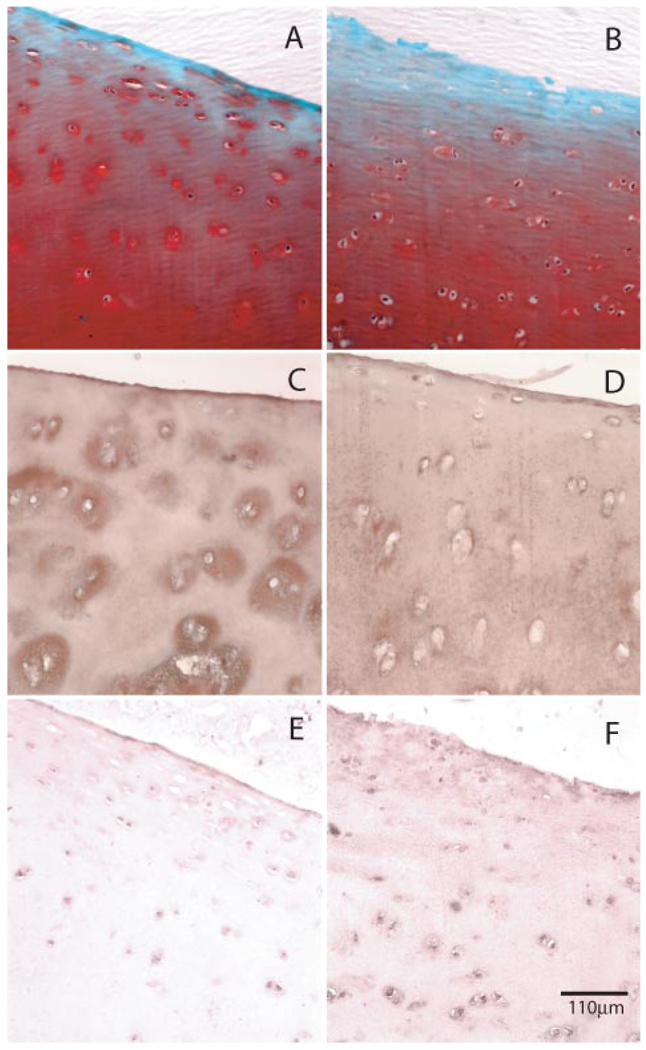
We have previously shown that HAoligo readily penetrate bovine articular cartilage and induce dramatic changes in matrix metabolism, most notably a loss of proteogly can from the tissue (29). This activating potential was confirmed in the present study using normal human ankle articular cartilage. As shown in Fig. 1A, a small deficit in safranin O staining was observed in the upper layer of control, untreated human articular cartilage slices after 7 days of culture. However, in the presence of HAoligo (Fig. 1B), the loss of safranin O was more pronounced, extending progressively into the deeper layers of the tissue. However, given that cartilage proteoglycan turnover is relatively rapid as compared with that of collagen (52–54), such losses could be due to effects on proteoglycan biosynthesis, degradation, or both. Therefore, of more interest was whether the HAoligo affected changes in type II collagen within the cartilage. Control cartilage slices cultured for 7 days exhibited prominent immunohistochemical staining for type II collagen within pericellular and interterritorial matrix regions of the tissues (Fig. 1C). After 7 days of treatment with HAoligo, the human articular cartilage exhibited reductions of type II collagen in the pericellular and interterritorial regions extending throughout the superficial, middle, and deep zones of the tissue (Fig. 1D).
FIGURE 1. Effects of HAoligo on cartilage matrix loss and expression of MMP-13.
Human articular cartilage explants were incubated in the absence (A, C, and E) or presence (B, D, and F) of HAoligo for 7 days under serum-free conditions. Sections were stained for proteoglycan by safranin O/fast green (A and B) or immunostained for type II collagen (C and D) or MMP-13 (E and F). The bar indicates 110 µm.
Induction of MMP-13 byHAoligo
Because matrix metalloproteinases such as MMP-13 are considered essential mediators of type II collagen degradation in cartilage (37–39), an immunohistochemical analysis for MMP-13 was undertaken. Whereas the staining for MMP-13 in control cartilage slices was modest (Fig. 1E), articular cartilage tissues treated with HAoligo for 7 days exhibited enhanced expression of MMP-13 protein (Fig. 1F). The enhanced MMP-13 expression was localized to the cells and pericellular region of chondrocytes in the middle zone of the cartilage tissue treated with HAoligo (Fig. 1F). Thus, HAoligo affect the loss of proteoglycan as well as type II collagen, and this loss is coincident with an increase in MMP-13 expression.
To better explore the mechanism of MMP-13 induction, cultured bovine articular chondrocytes were treated with HAoligo for varying times and concentrations. As shown in Fig. 2A, the induction of MMP-13 mRNA by HAoligo was first observed at a concentration of 100 µg/ml, with stimulations ≥2.5-fold above control observed using concentrations of 250 and 500 µg/ml HAoligo. The induction of MMP-13 mRNA was readily apparent following 12 h of incubation with HAoligo to a level that remained stable throughout the 48 h of treatment (Fig. 2B). Changes in MMP-13 enzymatic activity were also examined using a fluorogenic peptide substrate that displays selectivity for MMP-13. As shown in Fig. 2C, after chondrocytes were treated with HAoligo (solid circles), there was an enhanced cleavage of the MMP-13 peptide substrate as compared with control, untreated cultures (open circles).
FIGURE 2. Effect of HAoligo on the expression of MMP-13 in bovine articular chondrocytes.
Real time RT-PCR was performed on total RNA isolated from bovine articular chondrocytes treated without or with HAoligo to characterize changes in MMP-13 mRNA copy numbers. The ratio of copy numbers in treated to control cultures was calculated from the CP value using Equation 1. The data shown are representative of three experiments, analyzed in triplicate (means ± S.D.). The effect of varying concentrations of HAoligo (0–500 µg/ml) on MMP-13 mRNA copy numbers (24 h of treatment) are depicted in A. The effect of time of incubation of HAoligo (6–48 h) on MMP-13 mRNA copy numbers is shown in B. Open bars represent no HAoligo addition; black bars represent cells incubated with 250 µg/ml HAoligo. MMP-13-specific enzymatic activity, present in the conditioned medium of chondrocytes treated without (open circles) or with (closed circles) HAoligo for 48 h, was measured by cleavage of an MMP-13 selective fluorogenic substrate (C).
Next, the conditioned media of bovine articular chondrocyte cultures were analyzed for caseinolytic activity, indicative of MMP-13 enzymatic activity. As shown in Fig. 3A, caseinolytic activity was undetectable in control cultures at both 24 and 48 h. However, a distinct band of caseinolytic activity was visible in both the 24- and 48-h culture media of HAoligo-treated cultures. The expression of this activity is consistent with the changes in MMP-13 mRNA copy numbers (Fig. 2B). Nonetheless, without APMA preactivation (Fig. 3, A and B, lanes 1), all of the caseinolytic activity released by these chondrocyte cultures represented latent pro-enzyme. Following pretreatment with APMA, a 50-kDa band of caseinolytic activity began to appear (Fig. 3B, lane 2) in the media of chondrocytes treated with HAoligo for 24 h. This suggests that only pro-enzyme is present in the media of the cultured cells. Interestingly, however, in the medium of bovine cartilage slices treated with HAoligo for 48 h (Fig. 3C, lane 4), a 50-kDa band of caseinolytic activity can be seen, even in the absence of APMA activation.
FIGURE 3. Effect of HAoligo on the caseinolytic activity.
Caseinolytic activity, present in the conditioned medium of bovine articular chondrocytes treated without (−) or with (+) HAoligo for 24 or 48 h, is shown as labeled in A. The bands shown in A represent the caseinolytic activity present in the medium obtained without pretreatment with APMA prior to electrophoresis. B is another 24 h HAoligo-stimulated media sample pretreated without (lane 1) or with APMA (lane 2) prior to electrophoresis. C depicts caseinolytic activity present in the media of bovine articular cartilage slices treated without (lanes 1 and 3) or with (lanes 2 and 4) HAoligo for 24 or 48 h. As in A, these samples have not been pretreated with APMA prior to electrophoresis. The wet weights of cartilage slices that relate to media samples shown in C, lanes 1–4 were 45.7, 57.3, 54.9, and 49.3 mg, respectively. Shown in all three panels are reverse image zymograms wherein the original image has been digitally inverted from light bands on a dark background to dark bands on a light background.
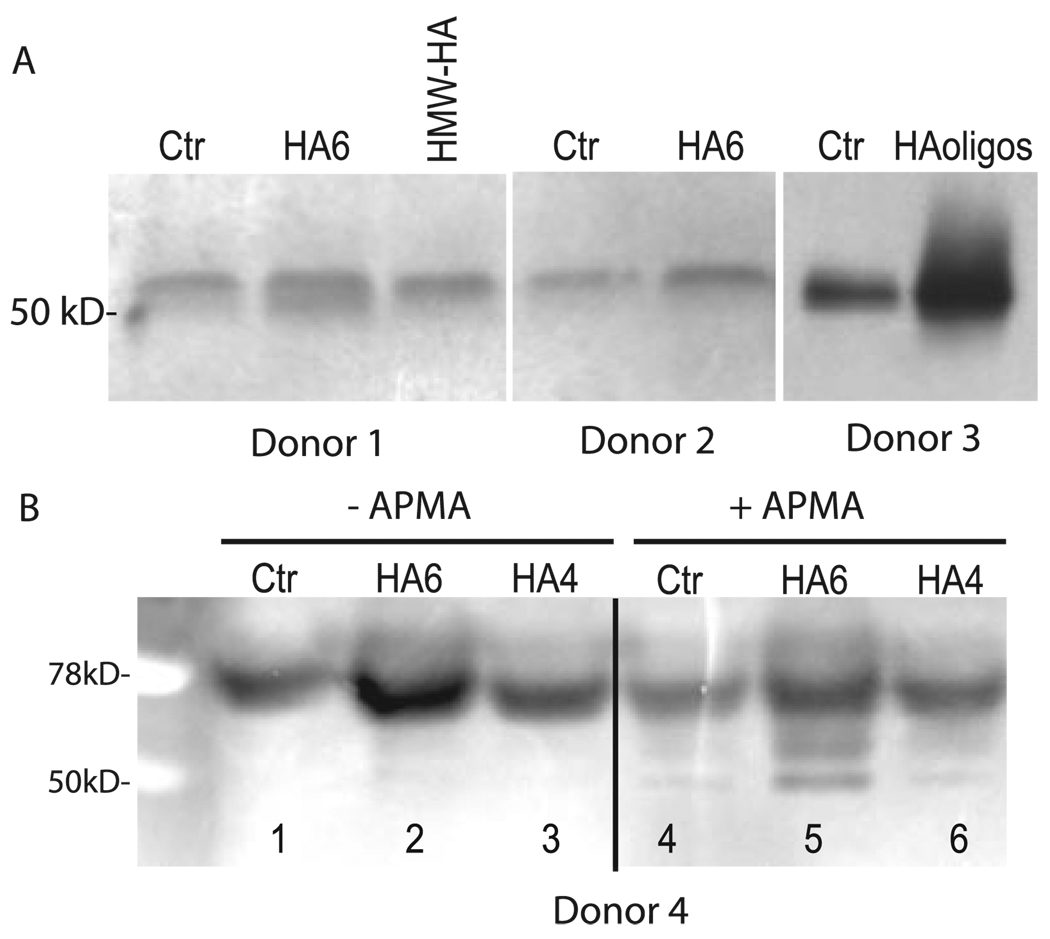
Normal human articular chondrocytes also respond to HAoligo however, the effect on MMP-13 mRNA copy number is not as pronounced (~1.5-fold increase; data not shown) as compared with the response observed in the bovine cells (Fig. 2, A and B). Nonetheless, through the use of anti-human MMP-13 antibodies, changes in the expression of MMP-13 protein can be determined. As shown in Fig. 4A,Western blot analysis showed an increase in the accumulated MMP-13 protein present in the conditioned medium of chondrocyte cultures treated with purified HA6 or HAoligo as compared with control, untreated cells. The data were obtained from chondrocytes isolated from three different human donors. As can be seen, the amount of MMP-13 protein in control cultures varies from donor to donor. However, stimulation in MMP-13 protein is observed in all three cultures in response to treatment with either HA6 or HAoligo. Interestingly, the addition of 250 µg/ml HMW-HAto chondrocytes from the first donor had no effect on MMP-13 protein expression. As with the bovine cells, all of the MMP-13 present was in the pro-enzyme form.
FIGURE 4. Effect of HAoligo on the expression of MMP-13 protein in human articular chondrocytes.
A, normal human chondrocytes derived from three different donors as labeled were exposed to HA6, HAoligo or HMW-HA for 24 h. Aliquots (equivalent protein) of the conditioned media from these cultures were then examined by Western blot analysis for MMP-13 protein expression as labeled. B, normal human articular chondrocytes were incubated in media alone as a control (Ctr) or media containing 250 µg/ml HA6 or HA4 for 24 h. Aliquots (equivalent protein) of the conditioned media from these cultures were then examined by Western blot analysis for MMP-13 protein expression, with or without pretreatment with APMA as labeled.
As shown in Fig. 4B, caseinolytic activity could also be detected in the conditioned media of cultures derived from a fourth human articular chondrocyte donor. Again, there is significant activity in the media of control, untreated cells (Fig. 4B, lanes 1 and 4) but substantial up-regulation upon treatment of the chondrocytes for 24 h with purified HA6. Interestingly, treatment of the human chondrocytes with HA4, oligosaccharides that do not interact efficiently with CD44, did not affect a stimulation of caseinolytic activity. Like the bovine chondrocytes, activation of the media samples with APMA resulted in the formation of mature MMP-13 at 50 kDa, again demonstrating that the MMP that is released in human chondrocyte cultures is also predominately the proenzyme form. In summary, HA6 or HAoligo induce the expression of MMP-13 mRNA, increase MMP-13 protein, and enhance MMP-13-related enzymatic activity.
Size-dependent Effects of HAoligo on the Induction of MMP-13 Transcription
To examine whether the induction effect of MMP-13 was due to transcriptional activation, the effects of various sized HA on MMP-13 promoter activity was evaluated in C-28/I2 human immortalized chondrocytes. As a positive control, the C-28/I2 cells were treated with IL-1β, conditions known to induce activation of MMP-13 (55). As shown in Fig. 5, IL-1β treatment affected approximately a 2-fold increase in MMP-13 promoter-directed luciferase activity. MMP-13 promoter activity was stimulated to approximately the same extent by treatment of the C-28/I2 cells with purified HA6. On the other hand, purified HA4 exhibited minimal activation of the MMP-13 promoter. Similar minimal activation was also observed with C-28/I2 cells treated with HMW-HA (~170–250 kDa). Thus, the level of transcriptional activation of a MMP-13 promoter luciferase construct (Fig. 5) is consistent with the level of stimulation of MMP-13 in chondrocytes (Figs. 1–4). These data also suggest that the minimum size of HA oligosaccharide necessary to activate chondrocyte expression of MMP-13 is a HA hexasaccharide.
FIGURE 5. Effect of HAoligo on the MMP-13 promoter activation.
C-28/I2 cells were transiently transfected with an MMP-13 luciferase construct (50). Twenty-four hours post-transfection the cells were incubated without or with 1 ng/ml IL-1β or 250 µg/ml HA6, HA4, or HMW-HA for an additional 24 h. The results are shown as fold increase in luciferase activity as compared with control, untreated cells (Ctr). The data shown are representative of two experiments, analyzed in triplicate (means ± S.D.). Analysis of variance was performed for the statistical analysis (*, p < 0.01).
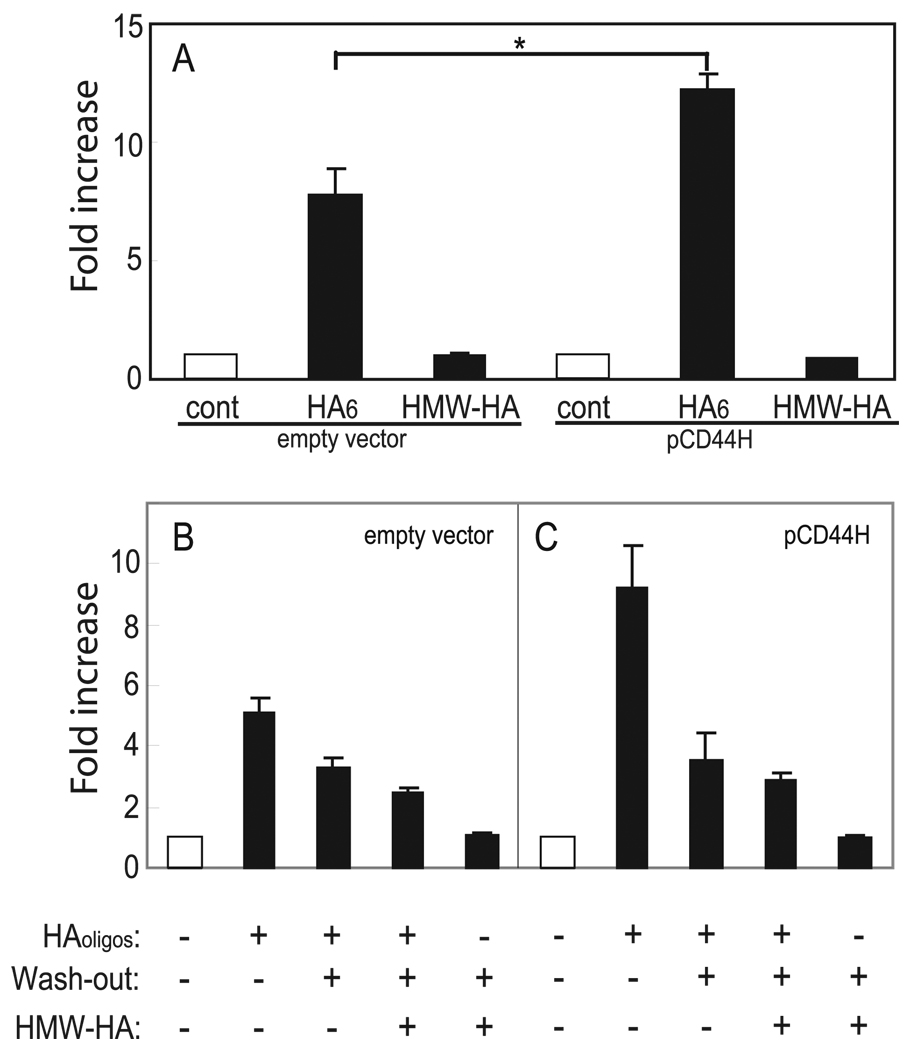
Involvement of CD44 in the Induction of MMP-13 by HAoligo
To clarify the participation of CD44 in the induction of MMP-13 by HAoligo, these oligosaccharides were added to CD44-negative COS-7 cells. To quantify activation of MMP-13, the COS-7 cells were transiently transfected with the MMP-13 promoter construct. As shown in Fig. 6A, CD44-negative, parental COS-7 cells exhibited a 7.5-fold enhancement in MMP-13 promoter luciferase activity following treatment with HAoligo. The addition of an equivalent concentration of HMW-HA to the CD44-negative cells had no effect and served as a negative control. These results would indicate CD44 is not responsible for the induction of MMP-13. To address this issue further, the COS-7 cells were transiently transfected with full-length pCD44H and then tested again for their sensitivity to HAoligo. As shown in Fig. 6A, CD44-positive COS-7 cells now exhibited a 12.5-fold enhancement in MMP-13 promoter luciferase activity following treatment with HAoligo. Again, HMW-HA served as a negative control. During subsequent replication of these experiments, the effect of wash-out of HAoligo in the presence or absence of exogenous HMW-HA was explored. As shown in Fig. 6B, a 12 h wash-out of HAoligo in the presence of HMW-HA reduced the level of MMP-13 promoter activation from 5.5- to 2.5-fold in CD44-negative COS-7 cells. However, once again, wash-out alone without HMW-HA reduced the level to 3.2-fold. In the CD44-positive cells (Fig. 6C), the addition of HMW-HA reduced the MMP-13 promoter activation from 9.2- to 2.8-fold. Wash-out alone reduced the level to 3.5-fold. Thus, it is clear that although other receptor mechanisms may contribute to HAoligo-induced signaling, CD44 is an important mediator. It also remains to be determined whether CD44-independent pathways participate in HAoligo-induced signaling in chondrocytes.
FIGURE 6. The participation of CD44 in HAoligo-mediated induction of MMP-13.
COS-7 cells were transiently co-transfected with an MMP-13 luciferase construct and either empty vector or full-length pCD44H. In A, 24 h post-transfection the cells were incubated without (cont) or with 250 µg/ml HA6 or HMW-HA for an additional 24 h as labeled. The data are shown as fold increases in luciferase activity as compared with control, untreated cells. Analysis of variance was performed for the statistical analysis (*, p < 0.05). In B, 24 h post-transfection the cells were incubated without or with 250 µg/ml HAoligo or HMW-HA for an additional 12 h, followed by a 12-h washout period in culture media without or with 500 µg/ml HMW-HA. The data are shown as fold increases in luciferase activity as compared with control, untreated cells and represent one experiment of three, analyzed in triplicate (means ± S.D.).
Enhancement of the NFκB Activity by HAoligo
Stimulation of MMP-13 transcription by fibronectin fragments or IL-1β has been shown to involve the activation of p38 MAP kinase and NFκB (50). To explore whether these signaling intermediates also participate in the activation of chondrocyte MMP-13 by HAoligo, NFκB EMSA analyses were performed. As shown in Fig. 7A, chondrocytes treated with HAoligo for 5 h exhibited an increase in nuclear NFκB by gel shift analysis as compared with control, untreated chondrocytes. This result was confirmed by competition with an excess of unlabeled NFκB probe (Fig. 7A, lanes 3 and 4). Time course experiments for EMSA analysis (Fig. 7B) showed that the highest level of activation of NFκB in nuclei occurred between 1 and 3 h, with little enhancement above control, untreated chondrocytes, 6 and 12 h following the addition of HAoligo. Activation of the NFκB pathway by HAoligo was also confirmed through the use of a validated NFκB promoter-reporter construct expressed in C-28/I2 cells. As shown in Fig. 7C, treatment of C-28/I2 cells with HA6 resulted in a 7-fold increase in the NFκB luciferase promoter activity. This increase was higher than the stimulation induced by IL-1β, used as a positive control. On the other hand, neither HMW-HA nor purified HA4 altered the basal NFκB promoter activity.
FIGURE 7. HAoligo activation of NFκB in chondrocytes.
A, EMSA was performed using biotin-labeled probes incubated with 25 µg of nuclear extracts derived from untreated bovine articular chondrocytes (lanes 1 and 3) or chondrocytes treated with HA6 (lanes 2 and 4). For competition analyses, an EMSA reaction was performed in the presence of excess unlabeled probe (lanes 3 and 4). Lane 5 shows the probes without addition of nuclear extract. B, EMSA analysis of the time dependent effects of HA6 on the activation of NFκB in bovine articular chondrocytes. C, changes in NFκB promoter activity in C-28/I2 cells are depicted following 24 h of treatment with 1 ng/ml IL-1β or 250 µg/ml HA6, HA4, or HMW-HA. The data are shown as fold increases in luciferase activity as compared with control, untreated cells (cont). The data represent the means ± S.D. of three experiments. D, changes in the phosphorylation of IKK-α and IKK-β in the C-28/I2 cells following treatment with HAoligo for 5–60 min as labeled were evaluated by Western blotting. β-Actin was used as a control for total protein loading.
To further validate the activation of NFκB by HAoligo, changes in phosphorylated IKK-α,β were examined by Western blot analysis. It has previously been shown that IKK-α,β phosphorylation is increased during the activation of the NFκB pathway (56). As shown in Fig. 7D, two bands corresponding to 87 (IKK-α) and 90 kDa (IKK-β) can be detected in extracts from C-28/I2 immortalized chondrocytes. Treatment of C-28/I2 cells with HA6 resulted in an increase in the levels of both phosphorylated IKK-α and phosphorylated IKK-β as early as 15 min after the treatment, with progressive increases through 60 min of incubation with the HA6.
To further establish the link between NFκB activation and HAoligo-mediated induction of MMP-13, the effects of an NFκB inhibitor, helenalin (57), was tested in bovine articular chondrocytes. As shown in Fig. 8A, 50-kDa caseinolytic activity in the conditioned medium of chondrocyte cultures (lane 1) was significantly enhanced following treatment with HAoligo for 24 h (lane 3). However, in the presence of 1 nm of the NFκB inhibitor, helenalin (lane 2, untreated cultures) stimulation by HAoligo was substantially diminished (lane 4). These results were further validated by examining the effect on MMP-13 enzymatic activity using the MMP-13 selective substrate assay. As shown in Fig. 8B (closed triangles, untreated control cultures), 24 h treatment of bovine articular chondrocytes with HAoligo resulted in enhanced capacity for cleavage of the MMP-13 selective fluorogenic substrate (closed squares). However, in the presence of 1 nm helenalin, there was an ~50% reduction in HAoligo-induced MMP-13 enzymatic activity (open squares), whereas little discernable affects on the basal activity present in untreated chondrocytes (open triangles). These results demonstrated that the activation of NFκB is essential to the stimulation of MMP-13 transcription by HAoligo.
FIGURE 8. Inhibition of HAoligo-induced MMP-13 enzymatic activity by NFκB inhibition.
A depicts caseinolytic activity present in the conditioned medium of bovine articular chondrocytes treated without (lanes 1 and 2) or with HAoligo (lanes 3 and 4) for 24 h and, in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of 1 nm NFκB inhibitor, helenalin. The concentrated media samples used in these zymograms was pretreated with APMA prior to electrophoresis, and thus the MMP-13 activity is shown as a dark band at 50 kDa in this digitally reversed image. B represents MMP-13 specific enzymatic activity as determined by the cleavage of an MMP-13 selective fluorogenic substrate. MMP-13 activity present in the conditioned medium of bovine articular chondrocytes cultured without (triangles) or with HAoligo (squares) for 24 h and, in the presence (open symbols) or absence (closed symbols) of the inhibitor of NFκB helenalin (1 nm) is shown. Units represent relative fluorescence.
Activation of p38 MAP Kinase by HAoligo
Previous studies have implicated the involvement of the MAP kinase signaling pathway in the activation of MMP-13 (42, 50). To explore whether these kinases participate in the activation of MMP-13 transcription byHAoligo, the effects of p38 MAP kinase and MEK inhibitors were examined. As shown in Fig. 9A, bovine articular chondrocytes exhibited a 3-fold increase in MMP-13 mRNA copy numbers following 24 h of exposure to HAoligo. In the presence of p38 MAP kinase or MEK inhibitors, there was some reduction in the basal expression level of MMP-13. However, whereas the MEK inhibitor affected no change in MMP-13 mRNA copy number induced by HAoligo (Fig. 9A, sixth bar), the induction of MMP-13 mRNA was completely abrogated by the inhibitor of p38 MAP kinase (Fig. 9A, fifth bar). This inhibition was also observed at the protein level in cultures of human articular chondrocytes. As shown in Fig. 9B, 24-h incubation of cells with purified HA6 resulted in enhanced MMP-13 protein expression in the culture medium. However, incubation in the presence of either the NF-κB inhibitor helenalin or the p38 MAP kinase inhibitor (SB203580) blocked the stimulation of MMP-13 protein expression.
FIGURE 9. Inhibition of HAoligo-induced stimulation of MMP-13 mRNA and MMP-13 protein by inhibition of MAP kinases.
A, real time RT-PCR was performed on total RNA isolated from bovine articular chondrocytes cultured without (open bars) or with 250 µg/ml HAoligo (black bars) for 24 h and in the absence or presence of inhibitors of p38 MAP kinase (SB203580) or MEK (PD98059) as labeled. MMP-13 mRNA was assessed using real time RT-PCR. The ratio of treated to control (cont) cultures (no HAoligo and no inhibitor of MAP kinase) was calculated from the CP value using Equation 1. The data represent the means ± S.D. of three experiments. B, normal human articular chondrocytes were incubated for 24 h in media alone (Ctr) or media containing 250 µg/ml HA6 in the absence (lanes 1 and 2) or presence of inhibitors of the NFκB inhibitor, helenalin (lanes 3 and 4) or the p38 MAP kinase inhibitor, SB203580 (lanes 5 and 6) as labeled. Aliquots (equivalent protein) of the conditioned media from these cultures were then examined by Western blot analysis for MMP-13 protein expression.
DISCUSSION
In the present study, disruption of HA-cell interactions was investigated by the use of low molecular mass HAoligo. These small oligosaccharides can readily penetrate into cartilage and affect not only a loss of proteoglycan but also depletion of type II collagen (Fig. 1D). The effect of HAoligo represents the activation of a catabolic cascade by the resident chondrocytes, as evidenced by the up-regulation of MMP-13 protein in the cartilage slices (Fig. 1F). Thus, HA-mediated cell signaling does occur and can be perturbed (through the use of HAoligo) within the environment of an intact tissue. Examining both human and bovine chondrocytes in vitro, we demonstrated that HAoligo activate MMP-13 gene transcription, resulting in an increase in MMP-13 mRNA copy number, an increase in MMP-13 protein, and an increase in MMP-13-related enzymatic activity.
The receptor that mediates these events remains somewhat ambiguous. Recent studies by Fieber et al. (43) demonstrated that a mixture of HA6 and HA4 induced the transcriptional activation of MMPs in wild-type mouse embryo fibroblasts but to a significantly lesser extent in fibroblasts isolated from the CD44-null mouse (C67BL6/J). These data led the authors to conclude that CD44 is not responsible for MMP activation by HAoligo in those embryonic mouse fibroblasts. However, another study using CD44 knock-out mice implicated a clear requirement for CD44 in the metabolic response to HAoligo (58). We also observed HAoligo-induced MMP-13 promoter activation in CD44-negative COS-7 cells (Fig. 5), indicative of CD44-independent cell signaling. However, in CD44-positive COS-7 cells, the induction of MMP-13 promoter activity by HAoligo was nearly double (Fig. 6, A and C). Thus, signaling through CD44 does have the capacity to contribute to cell signaling initiated by HAoligo. However, this still leaves open the possibility that different cell types can use other HA receptors such as RHAMM (receptor for hyaluronan-mediated motility), TLR-4 (Toll-like receptor 4), and LYVE-1 (lymphatic vessel endothelial hyaluronan receptor-1) or combinations of these receptors to initiate cell signaling. The receptor responsible for signaling in primary cultures of chondrocytes or cartilage slices thus remains to be defined. One approach to address this question is to use small interfering RNA or antisense oligonucleotides to selectively inhibit CD44 expression in adult, differentiated cells. Our previous studies demonstrated that CD44 antisense oligonucleotide treatment of cartilage tissue slices resulted in the induction of a catabolic cascade and a dramatic loss of extracellular matrix, nearly identical to the loss of matrix induced byHAoligo (33). We interpreted these results to be another example of what occurs when HA-cell interactions are perturbed. Again, this suggests that CD44 is a major mediator of HA and HAoligo-initiated cell signaling.
HAoligo represent an approximately equal mixture of HA octa-, hexa-, and tetrasaccharides (29). Both HAoligo and purified HA6 were effective at inducing MMP-13 transcription. HA4 on the other hand exhibited little signaling potential. The differential effects of HA6 versus HA4 provide a degree of specificity to the response. HA6 are too small to interfere with HA-aggrecan or HA-link protein interactions (59–61) but represent the minimum size for binding to HA receptors such as CD44 (62, 63). HA4 cannot interact with receptors such as CD44 but serve as a control for potential interactions of HAoligo with lectins, metabolic effects of sugar oligosaccharides, or alterations of pH or culture nutrients by the presence of HAoligo. Another important difference between HA4 and HA6 is that HA4 cannot affect the displacement of HMW-HA from the surface of chondrocytes or cells such as human bladder carcinoma cells (64), binding that is mediated via the interaction of HA with CD44. Thus, the observation that HA6 and not HA4 have the potential to activate MMP-13 transcription is consistent with our hypothesis that the cell signaling in chondrocytes is initiated by the displacement of HMW-HA, resulting in the declustering of CD44 in the cell membrane and destabilization of the cortical cytoskeletal conformation. Cytoskeletal rearrangements are known to be involved in the induction of MMPs (65, 66) and include activation of protein kinase C and NFκB (67).
In this study using EMSA analyses, we demonstrated that HAoligo increase the levels of active NFκB present in nuclear extracts of treated chondrocytes. In addition we also demonstrated an increase in the phosphorylation of IKK-α and IKK-β less than 15 min after stimulation, resulting in a peak level of nucleus-localized NFκB 1 h after stimulation. The induction of MMP-13 by HAoligo was also preferentially blocked by the NFκB inhibitor helenalin, an inhibitor that blocks NFκB-DNA binding activity by selectively alkylating the p65 subunit of NFκB (57). Together these results demonstrate that HAoligo induce MMP-13 expression through activation and nuclear translocation of NFκB. The pathway upstream of NFκB is less clear. In these studies, an inhibitor of p38 MAP kinase, but not MEK, completely blocked HAoligo-mediated induction of MMP-13. Activated p38 phosphorylates multiple kinases and transcription factors, including MAP kinase-activated protein kinase kinase, Elk-1, and ATF-2 (68). Phosphorylated Elk-1 and ATF-2 activate the transcription of AP-1 family members, c-Fos and c-Jun, respectively. Induction of MMP-13 by fibronectin fragments and IL-1β has been shown to involve the activation of p38 MAP kinase, NFκB, and AP-1 (50). However, HA6 activation in mouse embryo fibroblasts required the activation of NFκB but not the participation of p38 MAP kinase nor AP-1 (43). Our studies in chondrocytes suggest a third series of events occur, namely HA6 activation p38 MAP kinase and NFκB but no activation of AP-1 (31).
In conclusion, we demonstrated herein that HAoligo enhances the expression of MMP-13 in chondrocytes by activation of NFκB and p38 MAP kinase signaling pathways. CD44-mediated signaling supports this activation and is likely critical to certain cell types such as chondrocytes. These results suggest that articular chondrocytes have the capacity to sense changes in HA-cell interactions, resulting in the initiation of a chondrocytic chondrolysis response. Whether fragmentation of HA occurs in vivo remains to be determined. There are suggestions that HA oligosaccharides might be generated during periods of inflammation (69, 70) or otherwise generated through the action of chondrocyte-derived reactive oxygen species or other free radicals (71, 72). Nonetheless, several mechanisms can lead to a disruption of HA-cell interactions, all with similar results that include the activation of potent matrix metalloproteinases.
Acknowledgments
We thank the donor families and the Gift of Hope Organ and Tissue Donor Network. The generosity and beneficence of the donor families for access to the human tissues is greatly appreciated. The authors thank Dr. Mary Goldring (Harvard Institutes of Medicine) for the C-28/I2 cells, Dr. Richard Loeser (Rush Medical College) for the MMP-13 promoter construct, and the Seikagaku Corporation (Japan) for providing us with HA and HA oligosaccharides.
Footnotes
This work was supported in part by National Institutes of Health Grants RO1-AR43384, T32-AR07590, and P50-AR39239. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The abbreviations used are: HA, hyaluronan; CD, clusters of differentiation; HAoligo, hyaluronan oligosaccharide(s); HA4, hyaluronan tetrasaccharide; HA6, hyaluronan hexasaccharide; HMW-HA, high molecular weight hyaluronan; NFκB, nuclear factor κB; DMEM, Dulbecco’s modified Eagle’s medium; FBS, fetal bovine serum; RT, reverse transcriptase; MMP, matrix metalloproteinase; MAP, mitogen-activated protein; IKK, IκB kinase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; APMA, aminophenylmercuric acetate; EMSA, electrophoretic mobility shift assay; IL-1β, interleukin-1β; MEK, MAP kinase/extracellular signal-regulated kinase kinase.
REFERENCES
- 1.McDonald JA. Am. J. Physiol. 1989;257:331–337. [Google Scholar]
- 2.Knudson CB, Knudson W. FASEB J. 1993;7:1233–1241. [PubMed] [Google Scholar]
- 3.Knudson W, Loeser RF. Cell Mol. Life Sci. 2002;59:36–44. doi: 10.1007/s00018-002-8403-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Homandberg GA, Costa V, Wen C. Osteoarthritis Cartilage. 2002;10:938–949. doi: 10.1053/joca.2002.0854. [DOI] [PubMed] [Google Scholar]
- 5.Loeser RF, Forsyth CB, Samarel AM, Im HJ. J. Biol. Chem. 2003;278:24577–24585. doi: 10.1074/jbc.M304530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Homandberg GA, Costa V, Ummadi V, Pichika R. Osteoarthritis Cartilage. 2002;10:381–393. doi: 10.1053/joca.2002.0524. [DOI] [PubMed] [Google Scholar]
- 7.Makihira S, Yan W, Ohno S, Kawamoto T, Fujimoto K, Okimura A, Yoshida E, Noshiro M, Hamada T, Kato Y. J. Biol. Chem. 1999;274:11417–11423. doi: 10.1074/jbc.274.16.11417. [DOI] [PubMed] [Google Scholar]
- 8.Laurent TC, Fraser RE. FASEB J. 1992;6:2397–2404. [PubMed] [Google Scholar]
- 9.Knudson W, Knudson CB. Cur. Opin. Orthop. 2004;15:369–375. [Google Scholar]
- 10.Knudson CB. J. Cell Biol. 1993;120:825–834. doi: 10.1083/jcb.120.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knudson W, Aguiar DJ, Hua Q, Knudson CB. Exp. Cell Res. 1996;228:216–228. doi: 10.1006/excr.1996.0320. [DOI] [PubMed] [Google Scholar]
- 12.Shimazu A, Jikko A, Iwamoto M, Koike T, Yan W, Okada Y, Shinmei M, Nakamura S, Kato Y. Arthritis Rheum. 1993;36:247–253. doi: 10.1002/art.1780360217. [DOI] [PubMed] [Google Scholar]
- 13.Stove J, Gerlach C, Huch K, Gunther KP, Puhl W, Scharf HP. J. Orthop. Res. 2002;20:551–555. doi: 10.1016/S0736-0266(01)00141-3. [DOI] [PubMed] [Google Scholar]
- 14.Homandberg GA, Ummadi V, Kang H. Osteoarthritis Cartilage. 2003;11:177–186. doi: 10.1016/s1063-4584(02)00371-0. [DOI] [PubMed] [Google Scholar]
- 15.Kang Y, Eger W, Koepp H, Williams JM, Kuettner KE, Homandberg GA. J. Orthop. Res. 1999;17:858–869. doi: 10.1002/jor.1100170611. [DOI] [PubMed] [Google Scholar]
- 16.Williams JM, Zhang J, Kang H, Ummadi V, Homandberg GA. Osteoarthritis Cartilage. 2003;11:44–49. doi: 10.1053/joca.2002.0864. [DOI] [PubMed] [Google Scholar]
- 17.Lisignoli G, Grassi F, Zini N, Toneguzzi S, Piacentini A, Guidolin D, Bevilacqua C, Facchini A. Arthritis Rheum. 2001;44:1800–1807. doi: 10.1002/1529-0131(200108)44:8<1800::AID-ART317>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 18.Peterson RS, Andhare RA, Rousche KT, Knudson W, Wang W, Grossfield JB, Thomas RO, Hollingsworth RE, Knudson CB. J. Cell Biol. 2004;166:1081–1091. doi: 10.1083/jcb.200402138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wobus M, Rangwala R, Sheyn I, Hennigan R, Coila B, Lower EE, Yassin RS, Sherman LS. Appl. Immunohistochem. Mol. Morphol. 2002;10:34–39. doi: 10.1097/00129039-200203000-00006. [DOI] [PubMed] [Google Scholar]
- 20.Bourguignon LY, Zhu H, Zhou B, Diedrich F, Singleton PA, Hung MC. J. Biol. Chem. 2001;276:48679–48692. doi: 10.1074/jbc.M106759200. [DOI] [PubMed] [Google Scholar]
- 21.Bourguignon LYW, Zhu H, Shao L, Chen Y. J. Biol. Chem. 2001;276:7327–7336. doi: 10.1074/jbc.M006498200. [DOI] [PubMed] [Google Scholar]
- 22.Bourguignon LY, Singleton PA, Zhu H, Zhou B. J. Biol. Chem. 2002;277:39703–39712. doi: 10.1074/jbc.M204320200. [DOI] [PubMed] [Google Scholar]
- 23.Ito T, Williams JD, Fraser D, Phillips AO. Am. J. Pathol. 2004;164:1979–1988. doi: 10.1016/s0002-9440(10)63758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghatak S, Misra S, Toole BP. J. Biol. Chem. 2002;277:38013–38020. doi: 10.1074/jbc.M202404200. [DOI] [PubMed] [Google Scholar]
- 25.Bourguignon LY, Singleton PA, Zhu H, Diedrich F. J. Biol. Chem. 2003;278:29420–29434. doi: 10.1074/jbc.M301885200. [DOI] [PubMed] [Google Scholar]
- 26.Ohno-Nakahara M, Honda K, Tanimoto K, Tanaka N, Doi T, Suzuki A, Yoneno K, Nakatani Y, Ueki M, Ohno S, Knudson W, Knudson CB, Tanne K. J. Biochem. (Tokyo) 2004;135:567–575. doi: 10.1093/jb/mvh069. [DOI] [PubMed] [Google Scholar]
- 27.Jiang H, Peterson RS, Wang W, Bartnik E, Knudson CB, Knudson W. J. Biol. Chem. 2002;277:10531–10538. doi: 10.1074/jbc.M108654200. [DOI] [PubMed] [Google Scholar]
- 28.Embry J, Knudson W. Arthritis Rheum. 2003;48:3431–3441. doi: 10.1002/art.11323. [DOI] [PubMed] [Google Scholar]
- 29.Knudson W, Casey B, Nishida Y, Eger W, Kuettner KE, Knudson CB. Arthritis Rheum. 2000;43:1165–1174. doi: 10.1002/1529-0131(200005)43:5<1165::AID-ANR27>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 30.Nishida Y, Knudson CB, Knudson W. Osteoarthritis Cartilage. 2004;12:374–382. doi: 10.1016/j.joca.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 31.Ohno S, Im HJ, Knudson CB, Knudson W. Arthritis Rheum. 2005;52:800–809. doi: 10.1002/art.20937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maleski MP, Knudson CB. Exp. Cell Res. 1996;225:55–66. doi: 10.1006/excr.1996.0156. [DOI] [PubMed] [Google Scholar]
- 33.Chow G, Nietfeld J, Knudson CB, Knudson W. Arthritis Rheum. 1998;41:1411–1419. doi: 10.1002/1529-0131(199808)41:8<1411::AID-ART10>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 34.Woessner JF, Jr, Gunja-Smith Z. J. Rheumatol. 1991;27 suppl.:99–101. [PubMed] [Google Scholar]
- 35.Shlopov BV, Lie WR, Mainardi CL, Cole AA, Chubinskaya S, Hasty KA. Arthritis Rheum. 1997;40:2065–2074. doi: 10.1002/art.1780401120. [DOI] [PubMed] [Google Scholar]
- 36.Freemont AJ, Hampson V, Tilman R, Goupille P, Taiwo Y, Hoyland JA. Ann. Rheum. Dis. 1997;56:542–549. doi: 10.1136/ard.56.9.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freije JM, Diez-Itza I, Balbin M, Sanchez LM, Blasco R, Tolivia J, Lopez-Otin C. J. Biol. Chem. 1994;269:16766–16773. [PubMed] [Google Scholar]
- 38.Knauper V, Will H, Lopez-Otin C, Smith B, Atkinson SJ, Stanton H, Hembry RM, Murphy G. J. Biol. Chem. 1996;271:17124–17131. doi: 10.1074/jbc.271.29.17124. [DOI] [PubMed] [Google Scholar]
- 39.Mitchell PG, Magna HA, Reeves LM, Lopresti-Morrow LL, Yocum SA, Rosner PJ, Geoghegan KF, Hambor JE. J. Clin. Investig. 1996;97:761–768. doi: 10.1172/JCI118475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fosang AJ, Last K, Knauper V, Murphy G, Neame PJ. FEBS Lett. 1996;380:17–20. doi: 10.1016/0014-5793(95)01539-6. [DOI] [PubMed] [Google Scholar]
- 41.Jimenez MJ, Balbin M, Alvarez J, Komori T, Bianco P, Holmbeck K, Birkedal-Hansen H, Lopez JM, Lopez-Otin C. J. Cell Biol. 2001;155:1333–1344. doi: 10.1083/jcb.200106147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mengshol JA, Vincenti MP, Coon CI, Barchowsky A, Brinckerhoff CE. Arthritis Rheum. 2000;43:801–811. doi: 10.1002/1529-0131(200004)43:4<801::AID-ANR10>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 43.Fieber C, Baumann P, Vallon R, Termeer C, Simon JC, Hofmann M, Angel P, Herrlich P, Sleeman JP. J. Cell Sci. 2004;117:359–367. doi: 10.1242/jcs.00831. [DOI] [PubMed] [Google Scholar]
- 44.Knudson W, Gundlach MW, Schmid TM, Conrad HE. Biochemistry. 1984;23:368–375. doi: 10.1021/bi00297a028. [DOI] [PubMed] [Google Scholar]
- 45.Tawada A, Masa T, Oonuki Y, Watanabe A, Matsuzaki Y, Asari A. Glycobiology. 2002;12:421–426. doi: 10.1093/glycob/cwf048. [DOI] [PubMed] [Google Scholar]
- 46.Williams JM, Ongchi DR, Thonar EJ. J. Orthop. Res. 1993;11:705–716. doi: 10.1002/jor.1100110513. [DOI] [PubMed] [Google Scholar]
- 47.Rasmussen TB, Uttenthal A, de Stricker K, Belak S, Storgaard T. Arch. Virol. 2003;148:2005–2021. doi: 10.1007/s00705-003-0145-2. [DOI] [PubMed] [Google Scholar]
- 48.Pfaffl MW. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Knauper V, Lopez-Otin C, Smith B, Knight G, Murphy G. J. Biol. Chem. 1996;271:1544–1550. doi: 10.1074/jbc.271.3.1544. [DOI] [PubMed] [Google Scholar]
- 50.Im HJ, Pacione C, Chubinskaya S, Van Wijnen AJ, Sun Y, Loeser RF. J. Biol. Chem. 2003;278:25386–25394. doi: 10.1074/jbc.M302048200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Knudson W, Bartnik E, Knudson CB. Proc. Natl. Acad. Sci. U. S. A. 1993;90:4003–4007. doi: 10.1073/pnas.90.9.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morales TI, Hascall VC. J. Biol. Chem. 1988;263:3632–3638. [PubMed] [Google Scholar]
- 53.Ng CK, Handley CJ, Mason RM, Robinson HC. Biochem. J. 1989;263:761–767. doi: 10.1042/bj2630761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maroudas A, Bayliss MT, Uchitel-Kaushansky N, Schneiderman R, Gilav E. Arch. Biochem. Biophys. 1998;350:61–71. doi: 10.1006/abbi.1997.0492. [DOI] [PubMed] [Google Scholar]
- 55.Shlopov BV, Gumanovskaya ML, Hasty KA. Arthritis Rheum. 2000;43:195–205. doi: 10.1002/1529-0131(200001)43:1<195::AID-ANR24>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 56.Baldwin AS., Jr Annu. Rev. Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 57.Lyss G, Knorre A, Schmidt TJ, Pahl HL, Merfort I. J. Biol. Chem. 1998;273:33508–33516. doi: 10.1074/jbc.273.50.33508. [DOI] [PubMed] [Google Scholar]
- 58.Teder P, Vandivier RW, Jiang D, Liang J, Cohn L, Pure E, Henson PM, Noble PW. Science. 2002;296:155–158. doi: 10.1126/science.1069659. [DOI] [PubMed] [Google Scholar]
- 59.Hascall VC, Heinegard D. J. Biol. Chem. 1974;249:4242–4249. [PubMed] [Google Scholar]
- 60.Solursh M, Hardingham TE, Hascall VC, Kimura JH. Dev. Biol. 1980;75:121–129. doi: 10.1016/0012-1606(80)90148-7. [DOI] [PubMed] [Google Scholar]
- 61.Kimura JH, Hardingham TE, Hascall VC, Solursh M. J. Biol. Chem. 1979;254:2600–2609. [PubMed] [Google Scholar]
- 62.Kohda D, Morton CJ, Parkar AA, Hatanaka H, Inagaki FM, Campbell ID, Day AJ. Cell. 1996;86:767–775. doi: 10.1016/s0092-8674(00)80151-8. [DOI] [PubMed] [Google Scholar]
- 63.Teriete P, Banerji S, Noble M, Blundell CD, Wright AJ, Pickford AR, Lowe E, Mahoney DJ, Tammi MI, Kahmann JD, Campbell ID, Day AJ, Jackson DG. Mol. Cell. 2004;13:483–496. doi: 10.1016/s1097-2765(04)00080-2. [DOI] [PubMed] [Google Scholar]
- 64.Nemec RE, Toole BP, Knudson W. Biochem. Biophys. Res. Commun. 1987;149:249–257. doi: 10.1016/0006-291x(87)91632-9. [DOI] [PubMed] [Google Scholar]
- 65.Ogata Y, Pratta MA, Nagase H, Arner EC. Exp. Cell Res. 1992;201:245–249. doi: 10.1016/0014-4827(92)90271-9. [DOI] [PubMed] [Google Scholar]
- 66.Tanaka S, Hamanishi C, Kikuchi H, Fukuda K. Semin. Arthritis Rheum. 1998;27:392–399. doi: 10.1016/s0049-0172(98)80019-x. [DOI] [PubMed] [Google Scholar]
- 67.Fitzgerald KA, Bowie AG, Skeffington BS, O’Neill LA. J. Immunol. 2000;164:2053–2063. doi: 10.4049/jimmunol.164.4.2053. [DOI] [PubMed] [Google Scholar]
- 68.Garrington TP, Johnson GL. Curr. Opin. Cell Biol. 1999;11:211–218. doi: 10.1016/s0955-0674(99)80028-3. [DOI] [PubMed] [Google Scholar]
- 69.McKee CM, Penno MB, Cowman M, Burdick MD, Strieter RM, Bao C, Noble PW. J. Clin. Investig. 1996;98:2403–2413. doi: 10.1172/JCI119054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hodge-Dufour J, Noble PW, Horton MR, Bao C, Wysoka M, Burdick MD, Strieter RM, Trinchieri G, Pure E. J. Immunol. 1997;159:2492–2500. [PubMed] [Google Scholar]
- 71.Al-Assaf S, Navaratnam S, Parsons BJ, Phillips GO. Arch. Biochem. Biophys. 2003;411:73–82. doi: 10.1016/s0003-9861(02)00724-5. [DOI] [PubMed] [Google Scholar]
- 72.Baker MS, Green SP, Lowther DA. Arthritis Rheum. 1989;32:461–467. doi: 10.1002/anr.1780320416. [DOI] [PubMed] [Google Scholar]