Abstract
Alterations in dendrite branching and morphology are present in many neurodegenerative diseases. These variations disrupt postsynaptic transmission and affect neuronal communication. Thus, it is important to understand the molecular mechanisms that regulate dendritogenesis and how they go awry during disease states. Previously, our laboratory showed that cypin, a mammalian guanine deaminase, increases dendrite number when overexpressed and decreases dendrite number when knocked down in cultured hippocampal neurons. Here, we report that exposure to brain-derived neurotrophic factor (BDNF), an important mediator of dendrite arborization, for 72 h but not for 24 h or less increases cypin mRNA and protein levels in rat hippocampal neurons. BDNF signals through cypin to regulate dendrite number, since knocking down cypin blocks the effects of BDNF. Furthermore, BDNF increases cypin levels via mitogen-activated protein kinase and transcription-dependent signaling pathways. Moreover, the cypin promoter region contains putative conserved cAMP response element (CRE) regions, which we found can be recognized and activated by CRE-binding protein (CREB). In addition, exposure of the neurons to BDNF increased CREB binding to the cypin promoter and, in line with these data, expression of a dominant negative form of CREB blocked BDNF-promoted increases in cypin protein levels and proximal dendrite branches. Together, these studies suggest that BDNF increases neuronal cypin expression by the activation of CREB, increasing cypin transcription leading to increased protein expression, thus identifying a novel pathway by which BDNF shapes the dendrite network.
Introduction
Dendrite patterning and development are responsible for determining how input signals are processed in the CNS. The degree of dendrite branching can regulate the electrical properties of a neuron (Miller and Jacobs, 1984), and dendrite development is regulated by intrinsic and extrinsic factors (Landgraf and Evers, 2005; Libersat, 2005). Extrinsic factors can modulate specific patterns of dendrite growth and branching by activating intrinsic cues that directly affect the cytoskeleton or the transcriptional regulation of gene expression (Whitford et al., 2002). Neurotrophic factors play important roles in regulating neurite growth and branching in neurons. The major neurotrophic factors are the neurotrophins: nerve growth factor (NGF) (Levi-Montalcini, 1987; Shooter, 2001), brain-derived neurotrophic factor (BDNF) (Barde et al., 1982), neurotrophin-3 (NT-3) (Maisonpierre et al., 1990), and neurotrophin-4/5 (Hallböök et al., 1991). These factors are secreted from neurons and glial cells to exert their effects through the tyrosine receptor kinase (Trk) family in response to neuronal activity (Chao et al., 1992; Levine et al., 1995; Müller et al., 1995). BDNF is one of the most studied extrinsic factors regulating dendrite outgrowth and branching (Segal et al., 1995; McAllister et al., 1996, 1997; Schwartz et al., 1997; Baker et al., 1998; Jin et al., 2003) and increasing proximal dendrite growth and number in pyramidal neurons (McAllister et al., 1995; Baker et al., 1998; Horch et al., 1999).
Cypin (cytosolic PSD-95 interactor) was first identified as a protein that decreases the synaptic localization of PSD-95 (Firestein et al., 1999). Cypin is a guanine deaminase and contains a zinc-binding motif, CRMP (collapsin response mediator protein) homology domain and a PDZ-binding motif (Fernández et al., 2008). Our laboratory has reported that cypin plays an important role in regulating dendrite number in rat hippocampal neurons. Overexpression of cypin results in an increase in dendrite branching, which correlates with cypin's guanine deaminase activity (Akum et al., 2004). Cypin binds directly to tubulin heterodimers and promotes microtubule assembly (Akum et al., 2004), suggesting that cypin regulates dendrite branching via cytoskeletal rearrangement.
Here, we report a novel signaling pathway by which BDNF increases proximal dendrite branching. BDNF activates the mitogen-activated protein kinase (MAPK) signaling pathway, resulting in increased cypin mRNA and protein levels. Furthermore, this increase is only seen after 72 h of BDNF treatment, suggesting that cypin may mediate the effects of BDNF under chronic exposure. We also show that cAMP response element (CRE)-binding protein (CREB) binds to the cypin promoter to increase cypin transcription, and this binding is increased upon exposure to BDNF. Most importantly, we show that BDNF signals through CREB and cypin to increase proximal dendrite number. Our work links an important extrinsic regulator of dendrite branching to an important intrinsic factor that increases dendrite number, further uncovering molecular mechanisms that shape dendrites during development.
Materials and Methods
Antibodies and reagents.
Rabbit polyclonal antibody raised against cypin has been described previously (Firestein et al., 1999). Mouse monoclonal antibody recognizing β-actin was purchased from Sigma. Mouse anti-MAP2 was from BD-PharMingen, and chicken anti-GFP was purchased from Rockland Immunochemicals. Mouse monoclonal antibody to CREB was from Cell Signaling Technology. Rabbit monoclonal antibodies recognizing phospho-Akt, Akt, phospho-p70S6 kinase, p70S6 kinase, phospho-Erk1/2, and Erk1/2 were from Cell Signaling Technology. Cyanine (Cy)2-, Cy3-, and Cy5-conjugated secondary antibodies were from Jackson ImmunoResearch. Recombinant human BDNF (rhBDNF), mouse NGF, and U0126 were purchased from Promega, and myristoylated protein kinase A (PKA) inhibitory peptide 14–22 amide, wortmannin, and rapamycin were from Calbiochem/EMD Chemicals. Actinomycin D was purchased from Sigma. PKA kinase activity kit was from Enzo Life Sciences.
RNA interference and DNA constructs.
pSuper GFP vector (Oligoengine), containing shRNA against the cypin transcript and an unrelated sequence as a negative control, was used as described previously (Chen and Firestein, 2007). cDNA encoding rat CREB isoform A (NCBI accession no. NM_114443.1) was subcloned into pEGFP-C1 or dsRed2-C1 vector (Clontech Inc.). A dominant negative (DN) mutant of CREB and a constitutively active (CA) mutant of CREB were constructed using the QuikChange II Site-Directed Mutagenesis Kit (Stratagene) following the manufacturer's protocol. The serine 119 residue of CREB was replaced with alanine or aspartate using the primer 5′-CCT TTC AAG GAG GCC TGC CTA CAA GAA AAT CTT GAA TGA CTT ATC-3′ or the primer 5′- CTT TCA AGG AGG CCT GAC TAC AAG AAA ATC TTG AAT GAC TTA TC-3′, respectively.
Primary culture of hippocampal neurons.
Neuronal cultures were prepared from hippocampi of rat embryos of either sex at 18 d gestation as described previously (Firestein et al., 1999). The hippocampi were dissociated, and cells were plated on poly-d-lysine-coated glass coverslips (12 mm diameter) at a density of 1800 cells/mm2 or 1 × 106 cells on 35 mm culture dishes. Cultures were maintained in Neurobasal medium (Invitrogen) supplemented with B27 (Invitrogen), penicillin, streptomycin, and GlutaMAX (Invitrogen). Cells were grown for 7 days in vitro (DIV) and used for specific experiments as indicated below.
Western blotting.
Hippocampal neurons were plated with 1 × 106 cells per dish. Neurons were treated with neurotrophins, kinase inhibitors, or DMSO vehicle (0.01% final concentration) at the indicated concentrations for 72 h. Neurons were washed with ice-cold PBS and lysed in TEE (25 mm Tris-HCl, 1 mm EDTA, 1 mm EGTA, pH 7.4). Cells were further lysed by passing the extract through a 26 gauge needle 20 times, and solubilized using Triton X-100 at a final concentration of 1%. Insoluble material was pelleted at 12,000 × g at 4°C for 15 min. Proteins were resolved on a 10% SDS-polyacrylamide gel and transferred to PVDF membrane. The blot was probed with the indicated antibodies. Experiments were repeated three times. Blots were scanned and intensities of bands were quantitated using NIH ImageJ software as we have performed previously (Chen and Firestein, 2007; Carrel et al., 2009). An area close to the bands was used as a reference for background intensity. The difference between the intensity of the background and intensity of the band is the absolute intensity of the band. The number of pixels for the bands was normalized to the intensity of the internal control (β-actin) and then compared with that of the control condition.
Quantitative RT-PCR.
Neurons were plated as described above in the previous paragraph. At 7 DIV, neurons were treated with indicated concentrations of neurotrophins, kinase inhibitors, or DMSO vehicle. At 10 DIV, RNA was isolated using a Qiagen RNeasy Kit following the manufacturer's instructions. Total cDNA was then generated using an Applied Biosystems High-Capacity Reverse Transcription Kit with 1 μg of total RNA and following the manufacturer's protocol. We used a Stratagene Mx3000P qPCR system to perform multiplex assays using 50 ng of total cDNA for cypin/deaminase (GDA) and GAPDH as an internal control. The TaqMan Gene Expression assays (Applied Biosystems) containing primers and probes were used in our experiments. The cypin/GDA probe contained the FAM490 fluorophore, and the GAPDH probe contained the HEX fluorophore, both with the MGB (minor groove binder) quencher. Results were analyzed following the 2−ΔΔCt method using GAPDH as an internal control and nontreated or vehicle control.
PKA kinase activity assay.
Cultured hippocampal neurons (7 DIV) were treated with PKA inhibitory peptide for 72 h. Neurons were washed with ice-cold PBS and lysed in buffer containing 20 mm 3-(N-morpholino)propanesulfonic acid, 50 mm β-glycerolphosphate, 50 mm sodium fluoride, 1 mm sodium vanadate, 5 mm EGTA, 2 mm EDTA, 1% NP-40, 1 mm DTT, and 1 mm PMSF. Insoluble material was pelleted at 12,000 × g at 4°C for 15 min. Lysates were assayed following the manufacturer's instructions. Samples were added to plates precoated with PKA substrate, and ATP was added to the reactions. After 90 min incubation at 30°C, phospho-specific substrate antibody was added, and the plate was incubated for 1 h. The plate was washed four times, and a secondary antibody conjugated with HRP was applied. After 30 min incubation, the plate was washed and TMB substrates were added followed by the addition of Stop solution. The absorbance was measured at 450 nm using a SpectraMax 250 microplate reader (Molecular Devices).
Transfection of cultured cells.
Cultured hippocampal neurons were transfected 5 DIV for shRNAs and 6 DIV for cDNAs using Lipofectamine LTX with Plus reagent following the manufacturer's protocol (Invitrogen).
Immunocytochemistry.
Neurons were fixed in 4% paraformaldehyde in PBS for 15 min and then incubated in blocking solution (PBS containing 0.1% Triton X-100, 2% normal goat serum, and 0.02% sodium azide) for 1 h. All antibodies used were diluted in blocking solution, and dilutions of 1:500 for chicken anti-GFP, anti-MAP2, and anti-cypin were used. Neurons were incubated in primary antibody-containing solution at 4°C overnight. Neurons were then washed with PBS three times. The secondary antibody solution consisted of a 1:250 dilution of Cy2-conjugated anti-chicken IgY, Cy3-conjugated anti-mouse IgG, and Cy-5 conjugated anti-rabbit IgG. Coverslips were then mounted onto frosted glass microscope slides using Fluoromount G (Southern Biotechnology). Labeled cells were visualized by immunofluorescence on an Olympus Optical IX50 microscope with a Cooke SensiCam charge-coupled device (CCD) cooled camera fluorescence imaging system and Image Pro software (Media Cybernetics).
Assessment of dendrite number.
Dendrite morphology was processed as described previously (Kutzing et al., 2010; Langhammer et al., 2010) using custom scripts written in MATLAB (MathWorks. The axon was excluded based on the absence of MAP2 immunostaining. We performed Sholl analysis with a 6 μm ring interval starting at 9.3 μm from the soma. The experimenter was blinded to conditions during all data analysis. Dendrites less than 3 μm in length were not counted (Yu and Malenka, 2004; Charych et al., 2006).
Chromatin immunoprecipitation.
At 7 DIV, neurons were treated with 25 ng/ml rhBDNF and DMSO for 24 h. Cells were treated with 1% formaldehyde at 37°C for 10 min to cross-link histones to DNA. After washing the cells twice with ice-cold PBS containing protease inhibitors, cells were scraped and pelleted at 2000 rpm at 4°C for 4 min. The pellet was resuspended in SDS lysis buffer (1% SDS, 10 mm EDTA and 50 mm Tris, pH 8.1), and the lysate was sonicated. After centrifuging the lysates at 12,000 × g at 4°C for 10 min, the supernatant was diluted in chromatin immunoprecipitation (ChIP) dilution buffer (Millipore). Lysate was precleared with salmon sperm DNA/protein A agarose 50% slurry for 1 h and then immunoprecipitated with monoclonal CREB antibody or mouse IgG at 4°C overnight. Salmon sperm DNA/protein A agarose slurry was added for 1 h of incubation, and beads were washed one time each with MilliporeLow Salt Immune Complex Wash Buffer, zcomMilliporeHigh Salt Immune Complex Wash Buffer, MilliporeLiCl Immune Complex Wash Buffer, and then twice with TE buffer (10 mm Tris-HCl, 1 mm EDTA). Immunoprecipitated histone complex was eluted with elution buffer (1% SDS, 0.1 m NaHCO3). NaCl (5 m) was added to the eluates, followed by heating at 65°C for 4 h. After 1 h incubation with proteinase K, the DNA was recovered by phenol/chloroform extraction and ethanol precipitation. PCR was performed with following primers: forward, 5′-GAG GAC TTT AGA CTG GAA ACT TGC AAT TGG-3′; reverse, 5′-TTC CTG AGT GTG AGG GAT GCT GAC TAT G-3′.
Quantification of fluorescence intensity.
Hippocampal neurons were prepared, cultured, and transfected as described above. The neurons were immunostained using our polyclonal antibody against cypin. Fluorescence intensities of cypin were measured using ImageJ software as we have described previously (Chen and Firestein, 2007). Briefly, the cell bodies for each neuron were traced, and intensities were measured as integrated pixel intensity within the selected region corrected for background. Fluorescence was visualized using an Olympus LC Plan FL 20×/0.4 (air) objective. To quantitate the fluorescence levels of endogenous proteins, images of neurons were captured by a CCD camera as described above (see Immunohistochemistry) using a constant gain and exposure time for all samples. Images were corrected for coverslip fluorescence by subtracting a background image generated using an 11 × 11 erosion filter. The experimenter was blinded to the condition when taking images and assaying fluorescence intensities.
Results
BDNF increases cypin protein levels in hippocampal neurons
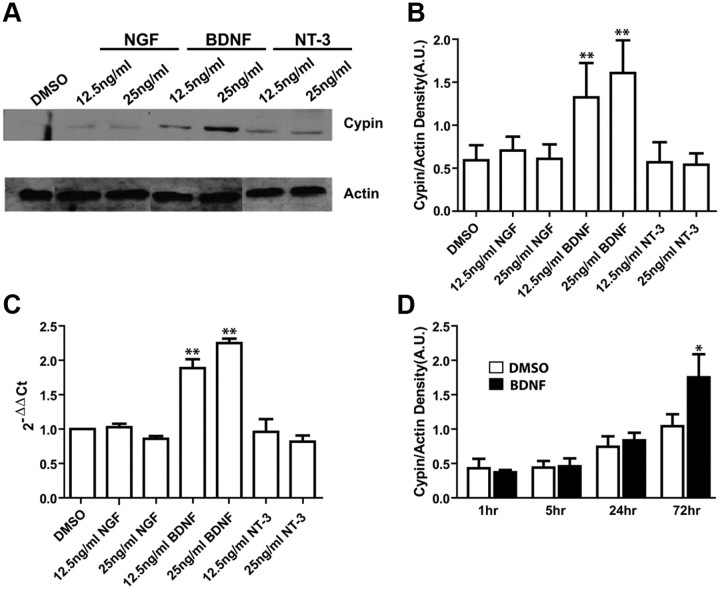
To investigate the possibility that neurotrophic factors may modulate cypin expression to promote increases in dendrite number, we treated cultures of primary hippocampal neurons with three neurotrophins: nerve growth factor, brain-derived neurotrophic factor, and neurotrophin-3. These neurotrophins have a degree of receptor specificity when applied at low levels, with NGF binding to TrkA, BDNF and NT-4/5 binding to TrkB, and NT-3 preferentially binding to TrkC (Reichardt, 2006). We investigated the role of these neurotrophins in promoting changes in cypin expression by examining mRNA levels with quantitative RT-PCR (qRT-PCR) and protein levels with Western blotting. Since different concentrations of neurotrophins can have different affinities for Trk receptors (Davies et al., 1993; Mahadeo et al., 1994), we used two different concentrations of each neurotrophin. Only exposure to BDNF significantly increases cypin mRNA and protein levels after 72 h of treatment (Fig. 1A–C). This increase is dose dependent and is due to TrkB activation, because at the concentrations tested no apparent cross-stimulation of TrkA, TrkC, or p75 should occur (Ji et al., 2005). Furthermore, since it has been reported that exposure of cortical neurons to BDNF for as little as 5 h is sufficient to increase the number of primary dendrites (Dijkhuizen and Ghosh, 2005), and TrkB is maximally phosphorylated in 5 min by 25 ng/ml BDNF (Ji et al., 2005), we tested whether a shorter exposure of hippocampal neurons to BDNF results in this increase in cypin expression. As seen in Figure 1D, 72 h is the minimum time of BDNF exposure needed to see the increase in cypin expression. It should be noted that cypin levels increase as neurons mature in culture (Kuwahara et al., 1999), and thus we observed this baseline increase in cultures treated with DMSO. Thus, we used a 72 h incubation time for the rest of our studies, acknowledging the fact that we are studying pathways involved in longer term exposure to BDNF rather than a short pulse of BDNF exposure.
Figure 1.
Endogenous cypin expression increases in response to BDNF but not NGF or NT-3. A, Cells were treated with the indicated concentrations of neurotrophins beginning at 7 DIV for 72 h. Extracts from untreated and treated cultures of hippocampal neurons were analyzed by SDS-PAGE and Western blotting using antibodies that recognize cypin or actin. Representative blot is shown. B, Densitometry analysis of cypin normalized to actin expression. C, Quantitative RT-PCR assay for the rat cypin gene (GDA) after indicated treatments of cultured hippocampal neurons. **p < 0.01, determined by ANOVA followed by Student-Newman–Keuls multiple-comparison test compared to DMSO as control. Cypin mRNA and protein levels are upregulated by increasing doses of BDNF. D, Exposure of hippocampal neurons to BDNF for 72 h, but not 24 h or less, results in increased cypin levels. Cells were treated with BDNF at 7 DIV for each indicated duration. Cypin protein levels were compared by Western blotting. *p < 0.05 by two-tailed t test compared to control. Error bars indicate SEM. A.U., Arbitrary units; n = 3 experiments for all panels.
BDNF signals through cypin to increase dendrite numbers
To determine whether BDNF signals via cypin to regulate dendrite patterning, we used an shRNA against cypin as we have described previously and shown to be specific for cypin by rescue experiments (Chen and Firestein, 2007). Consistent with results from other laboratories (Dijkhuizen and Ghosh, 2005), BDNF increases primary and secondary dendrite numbers in neurons treated during DIV 7–10 in the presence of the expression of a control shRNA. However, we did not see changes in dendrite number in our hippocampal neurons in response to shorter exposures of BDNF as has been seen in cortical neurons (Dijkhuizen and Ghosh, 2005), which is in line with a previous report showing that pathways that control dendrite outgrowth differ between cortical and hippocampal neurons (Ko et al., 2005). As seen in Figure 2A and B, knockdown of cypin blocks increases in primary and secondary dendrite numbers in response to 72 h treatment with BDNF. We also observed some pruning in primary dendrite number from DIV 7–10, consistent with previous results (Cline, 2001; Wong and Ghosh, 2002; Charych et al., 2006). In addition, we performed Sholl analysis by measuring the number of dendrites that cross ellipsoids at different radial distances from the cell body to analyze the roles of BDNF and cypin in higher-order dendritic branching (Fig. 2D). Sholl analysis shows that treatment with BDNF for 72 h results in an increase in dendrite numbers proximal to the soma, the 75 μm region closest to the cell body (Fig. 2E). In contrast, this treatment with BDNF does not increase dendrites when cypin is knocked down, strongly suggesting that BDNF promotes dendrite branching via cypin at DIV 7–10. We also noted that knocking down cypin at an earlier time window, DIV 5–7 (for the 5 h treatment) (Fig. 2D,E), has no effect on dendrite numbers, suggesting that cypin may not regulate neurite outgrowth in early stages of development.
Figure 2.
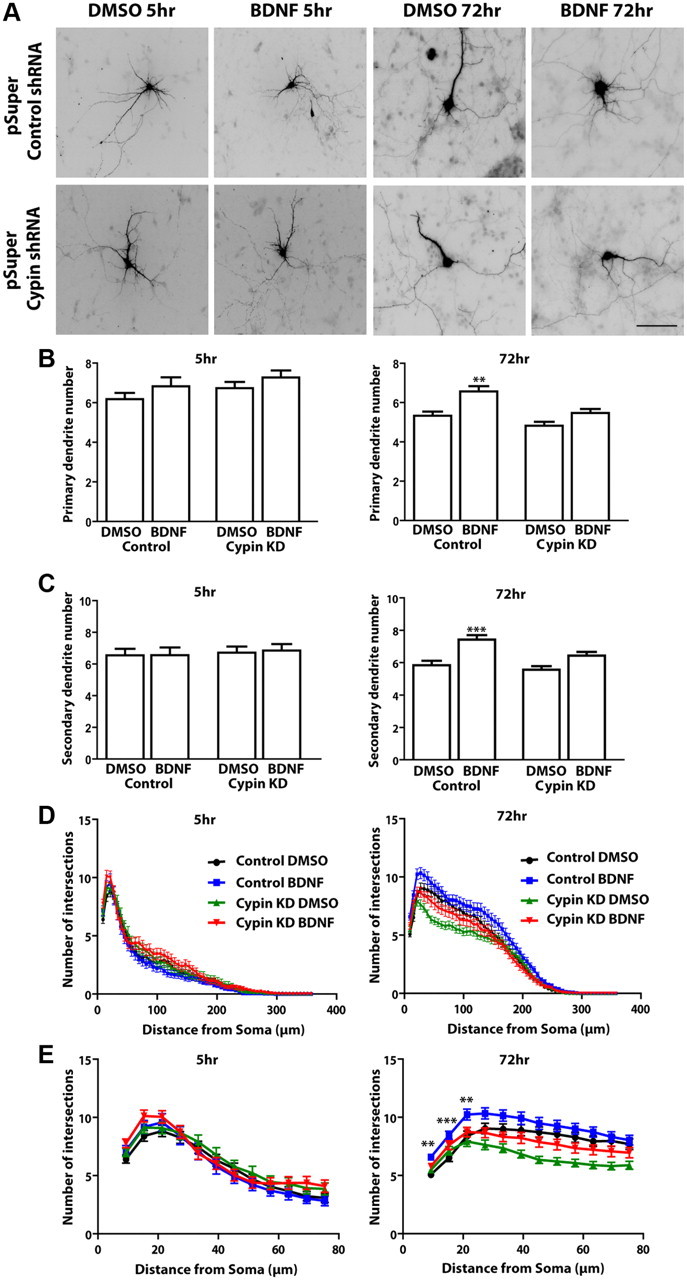
BDNF treatment results in increased proximal dendrites via a cypin-dependent pathway. A, Hippocampal neurons were transfected with shRNA against GST (control) or with shRNA against cypin at 5 DIV and treated with either DMSO (vehicle) or BDNF (25 ng/ml) at 7 DIV for either 5 or 72 h. Dendrite number was assessed at 7 or 10 DIV. Scale bar, 100 μm. B, C, Cypin knockdown (KD) blocks BDNF-promoted increases in primary dendrites (B) and secondary dendrites (C) with 72 h of treatment (right). BDNF treatment for 5 h does not increase dendrite numbers (left). ***p < 0.001, **p < 0.01, comparing control shRNA plus BDNF 72 h treatment to control shRNA with DMSO 72 h treatment and no significance comparing cypin knockdown plus DMSO 72 h treatment and cypin knockdown plus BDNF 72 h treatment. p values were determined by Kruskal–Wallis test followed by Dunn's multiple comparison test. D, Sholl analysis. BDNF treatment for 72 h increases the number of intersections close to the soma (proximal dendrites) E, Proximal Sholl analysis (from D) within the first 75 μm of the soma. (*p < 0.05 at 9 μm, ***p < 0.001 at 15 μm,**p < 0.01 at 33 μm when comparing control shRNA plus BDNF 72 h treatment and control shRNA plus DMSO 72 h treatment and no significance when comparing cypin knockdown plus DMSO 72 h treatment and cypin knockdown with BDNF 72 h treatment. Cypin knockdown results in a significant decrease (***p < 0.001) in intersections in comparison to control shRNA, consistent with our previous work. p values were determined by two-way ANOVA followed by Bonferroni multiple-comparisons test. Error bars indicate SEM. n = 51 neurons, control shRNA plus DMSO 5 h treatment; n = 40, control shRNA plus BDNF 5 h treatment; n = 48, cypin knockdown plus DMSO 5 h treatment; n = 52, cypin knockdown plus BDNF 5 h treatment; n = 76, control shRNA plus DMSO 72 h treatment; n = 79, control shRNA plus BDNF 72 h treatment; n = 90, cypin knockdown plus DMSO 72 h treatment; and n = 87, cypin knockdown plus BDNF 72 h treatment.
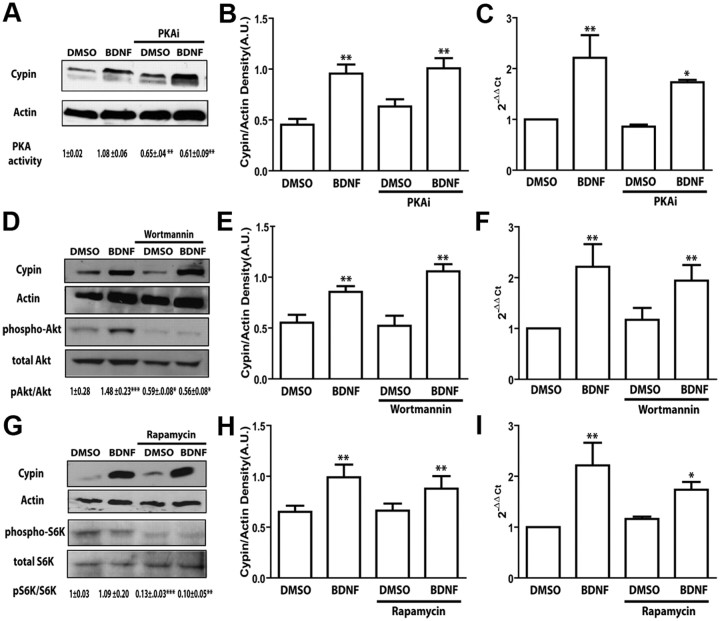
The cAMP/PKA pathway is not required for BDNF-promoted increases in cypin expression
We performed experiments to further identify the downstream pathways that mediate BDNF-promoted cypin gene expression. TrkB receptors dimerize in response to BDNF and phosphorylate one another. These phosphotyrosines generate binding sites for Shc and phospholipase C, which can recruit the signaling molecules to trigger activation of major pathways, such as cAMP/PKA, phosphoinositide 3-kinase (PI3K), mammalian target of rapamycin (mTOR), and MAPK signaling pathways (Chao, 2003; Segal, 2003; Kumar et al., 2005; Spencer et al., 2008). Therefore, we first tested the possibility that BDNF-mediated increases in cypin expression are mediated by the cAMP/PKA pathway. We treated cultured hippocampal neurons with a myristoylated membrane-permeable PKA inhibitor peptide and assessed BDNF action on cypin expression. Inhibition of PKA (Fig. 3A, bottom) does not block BDNF-promoted increases in cypin mRNA or protein (Fig. 3A–C). These results suggest that the activation of PKA by BDNF is not required for increased cypin expression. Interestingly, the appearance of a lower molecular weight band in our Western blot analysis suggests that the cAMP signaling pathway may alter expression of a lower molecular weight cypin variant (data not shown).
Figure 3.
The cAMP/PKA, PI3K/Akt/PKB, and mTOR pathways are not involved in BDNF-mediated increases in cypin expression. A–I, Hippocampal neurons were treated with membrane-permeable PKA inhibitory (PKAi) peptide (25 ng/ml; Myr-N-Gly-Arg-Thr-Gly-Arg-Arg-Asn-Ala-Ile-NH2) (A–C), wortmannin (100 nm) (D–F), or rapamycin (25 ng/ml) (G–I) at 7 DIV and treated with BDNF for 72 h. Cytoplasmic protein extracts were resolved by SDS-PAGE and Western blotting using antibodies that recognize indicative proteins. PKA activity was measured by absorbance at 450 nm with an ELISA-based PKA activity assay. PKA activity is shown as mean normalized absorbance ± SEM. Densitometric analysis of active phospho-proteins normalized to total proteins are indicated as mean ± SEM. Densitometric analysis of cypin is normalized to actin protein expression. Extracts from treated cultures were then analyzed by quantitative RT-PCR using specific rat cypin/GDA primers. *p < 0.05, **p < 0.01, ***p < 0.001 by ANOVA followed Student-Newman–Keuls multiple-comparison test compared to DMSO as vehicle control. Error bars indicate SEM. n = 3 experiments for all panels with representative blots shown in A, D, G. S6K, p70S6 kinase.
The PI3K-Akt-mTOR pathway is not required for BDNF-promoted increases in cypin expression
Given that the cAMP/PKA pathway appears not to be involved in BDNF-promoted expression of cypin in neurons, we further tested other signaling pathways reported to be activated by BDNF. PI3K/Akt pathways are triggered by BDNF to regulate primary dendrite formation in cortical neurons (Dijkhuizen and Ghosh, 2005). In addition, there are several studies that support the idea that BDNF can regulate dendrite branching through the PI3K pathway by activating mTOR (Schratt et al., 2004; Jaworski et al., 2005; Kumar et al., 2005). Therefore, we tested the possibility that BDNF uses these pathways to increase cypin expression. We treated cultured neurons with wortmannin and rapamycin, potent and specific PI3 kinase and mTOR inhibitors, respectively. We then measured cypin mRNA and protein expression. Inhibition of PI3K (Fig. 3D, bottom) or the mTOR (Fig. 3G, bottom) does not affect BDNF-mediated increases in cypin mRNA or protein expression (Fig. 3D–F, G–I), suggesting that the activation of PI3K/Akt and mTOR by BDNF is not required for increased cypin expression.
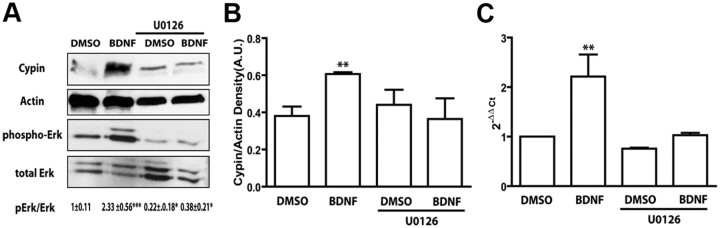
The MAPK signaling pathway is responsible for increased cypin expression in hippocampal neurons
The Ras/MAPK pathways have also been reported to mediate BDNF-mediated increases in primary dendrite number in cortical neurons (Kumar et al., 2005). To test the involvement of MAPK in BDNF-mediated increases in cypin expression, we used the highly selective MAP kinase kinase (MAPK/MEK) inhibitor U0126 (Favata et al., 1998). As seen in Figure 4, U0126 inhibits BDNF-promoted increases in cypin mRNA and protein overexpression. These results suggest that BDNF specifically increases cypin protein expression in hippocampal neurons via activation of the MAPK signaling pathway.
Figure 4.
BDNF increases cypin expression via the MAPK signaling pathway. A, Hippocampal neurons were treated with DMSO or 10 nm U0126, a MEK inhibitor, which consequently inhibits MAPK, in the absence or presence of 25 ng/ml BDNF on DIV 7 for 72 h. Representative Western blots using antibodies that recognize cypin, actin, phospho-Erk1/2, and total Erk1/2 are shown. Densitometric analysis of active MAPK normalized to total MAPK is indicated as mean ± SEM. B, Densitometric analysis of cypin is normalized to actin protein expression. C, Quantitative RT-PCR using specific rat cypin primers. *p < 0.05, **p < 0.01, ***p < 0.001 by ANOVA followed Student-Newman–Keuls multiple-comparison test compared to DMSO as vehicle control. n = 3 experiments for all panels.
BDNF treatment increases cypin expression via a transcription-dependent mechanism
Because BDNF not only increases cypin protein levels but also cypin mRNA levels, and MAPK pathways can trigger the activation of transcription factors to perform various mechanisms of cellular action, we hypothesized that BDNF promotes cypin expression in a transcription-dependent manner. We used a pharmacological drug, actinomycin D, to inhibit transcription by binding DNA at the transcription initiation complex and preventing elongation of transcription by the RNA polymerase (Sobell, 1985). As shown in Figure 5, actinomycin D treatment blocks BDNF-mediated increases in cypin mRNA and protein levels. We then tested the possibility that actinomycin D acts by decreasing MAPK activity, which would lead to decreased cypin levels, rather than by directly blocking BDNF-promoted transcription of cypin. This is not the case, as we found the opposite effect of actinomycin D. Actinomycin increases MAPK activity (Fig. 5A,D), suggesting that actinomycin D may inhibit the transcription of inhibitors of MAPK and that MAPK activity is upstream of BDNF-promoted increases in cypin transcription. Together with our other results, these data suggest that BDNF increases dendrite number by upregulating cypin transcription and, ultimately, cypin translation.
Figure 5.
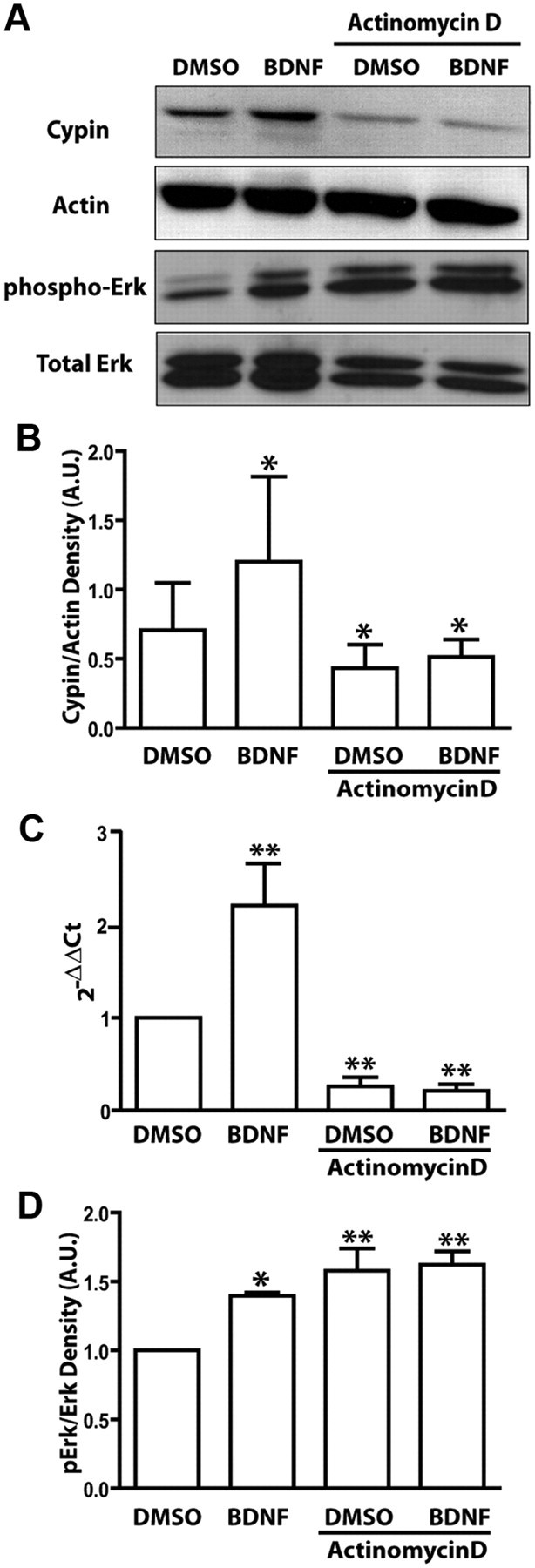
BDNF promotes cypin expression via a transcription-dependent mechanism. A, Cultured hippocampal neurons were treated with 25 ng/ml BDNF concurrent with or without 5 μm actinomycin D at 7 DIV for 72 h. Proteins were extracted at 10 DIV, and Western blotting was performed using a cypin antibody. A representative blot is shown. B, Densitometric analysis of cypin protein expression normalized to actin protein expression is shown. *p < 0.05 by Kruskal–Wallis test followed by Dunnett's multiple-comparison test compared to untreated control. C, Quantitative RT-PCR using specific rat cypin/GDA primers. D, Densitometric analysis of phospho-Erk1/2 protein levels normalized to total Erk1/2 protein levels is shown. **p < 0.01 by ANOVA followed Student-Newman–Keuls multiple-comparison test compared to DMSO as control. n = 3 experiments.
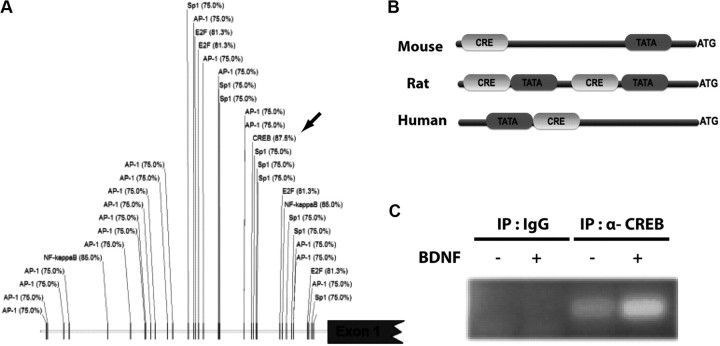
BDNF increases CREB binding to the cypin gene promoter
To identify possible cis-acting regulators of the cypin gene promoter, we performed a bioinformatics-based analysis to predict regulatory binding regions. Because the minimal promoter region of the rodent or human promoter region of cypin has not yet been characterized, we analyzed a vast region upstream of the first exon start region (10 kb). We compiled a list of predicted transcription factors consisting of AP-1, Sp-1, E2F, NF-κB, and CREB that may bind to a regulatory region of the cypin gene (Fig. 6A). Both NF-κB and CREB obtained higher binding probability than the other factors, and thus we analyzed whether there is conservation of these regions between rodent and human genes. CRE regions were conserved between the mouse, rat, and human cypin genes as shown in Fig. 6B, although the positions of these regions differed within the gene. In addition, distinct TATA box domains were also found in this region and in proximity to the CRE elements, suggesting that together these domains may represent an important regulatory region of the cypin gene. To assess whether CREB is involved in BDNF-promoted increases in cypin transcription, we performed a chromatin immunoprecipitation assay. We found that CREB binds to the cypin promoter under basal conditions and that this binding is enhanced by extracellular treatment of BDNF (Fig. 6C). Together, these data suggest that BDNF increases CREB binding to the cypin promoter to promote transcription of the cypin gene.
Figure 6.
BDNF increases CREB binding to the cypin promoter. A, Prediction of transcription factor binding sites in the promoter region of rat cypin. The 10 kb upstream sequence of the first exon of cypin was used to predict consensus binding sequences using the TF Search algorithm and TRANSFAC database (Heinemeyer et al., 1998). Each position is represented by the transcription factor, and the binding probability (expressed as percentage of identity) is indicated as predicted by TFSearch-specific matrices. B, Conserved CRE regions are present in rodent and human cypin gene promoters. Genomic sequences from mouse, rat, and human were retrieved from the NCBI database up to 2 kb upstream from the first exon of cypin. These sequences were analyzed for the occurrence of conserved CRE and TATA box regions. C, Hippocampal neurons were treated with 25 ng/ml BDNF on DIV 7 for 24 h. Chromatin was isolated on DIV 8, and ChIP was performed using specific primers to the cypin promoter. n = 3 experiments. A representative ChIP is shown.
BDNF activates CREB to increase cypin expression and dendrite branching
CREB is activated by its phosphorylation at Ser119 (equivalent position to Ser133 in CREB-341 isoform B, NCBI accession no. NP112279.1) to promote cellular gene expression, and this Ser119 phosphorylation of CREB is sufficient to recruit CBP-RNA polymerase II complexes and activate transcription of gene expression (Nakajima et al., 1997). We used a dominant negative mutant of CREB (CREB DN), in which serine119 is mutated to alanine and cannot be phosphorylated, to further investigate the role of CREB in BDNF-mediated increases in cypin and dendrite number. Wild-type CREB (CREB WT) was used as a control, and BDNF treatment increased cypin expression in neurons expressing CREB WT (Fig. 7A,B). Expression of CREB DN blocked BDNF-promoted increases in cypin expression (Fig. 7A,B). We then tested whether CREB activation is sufficient to increase cypin levels. We used a constitutively active mutant of CREB (CREB CA) in which serine119 is mutated to aspartate to mimic the phosphorylated state. Neurons that express CREB CA showed increased cypin protein levels. Furthermore, expression of CREB CA occludes BDNF-induced increases in cypin expression. Our data strongly suggest that BDNF activates CREB, which in turn increases cypin expression. We further investigated whether BDNF regulates dendritogenesis via CREB activation. Neurons were cotransfected with a DsRed-tagged CREB DN construct and enhanced GFP-containing vector to assess dendrite number. Sholl analysis shows that neurons that express CREB DN do not show increased proximal dendrite numbers when exposed to BDNF (Fig. 8). Our data support the idea that BDNF activates CREB to increase the expression of cypin and, as a result, proximal dendrite numbers.
Figure 7.
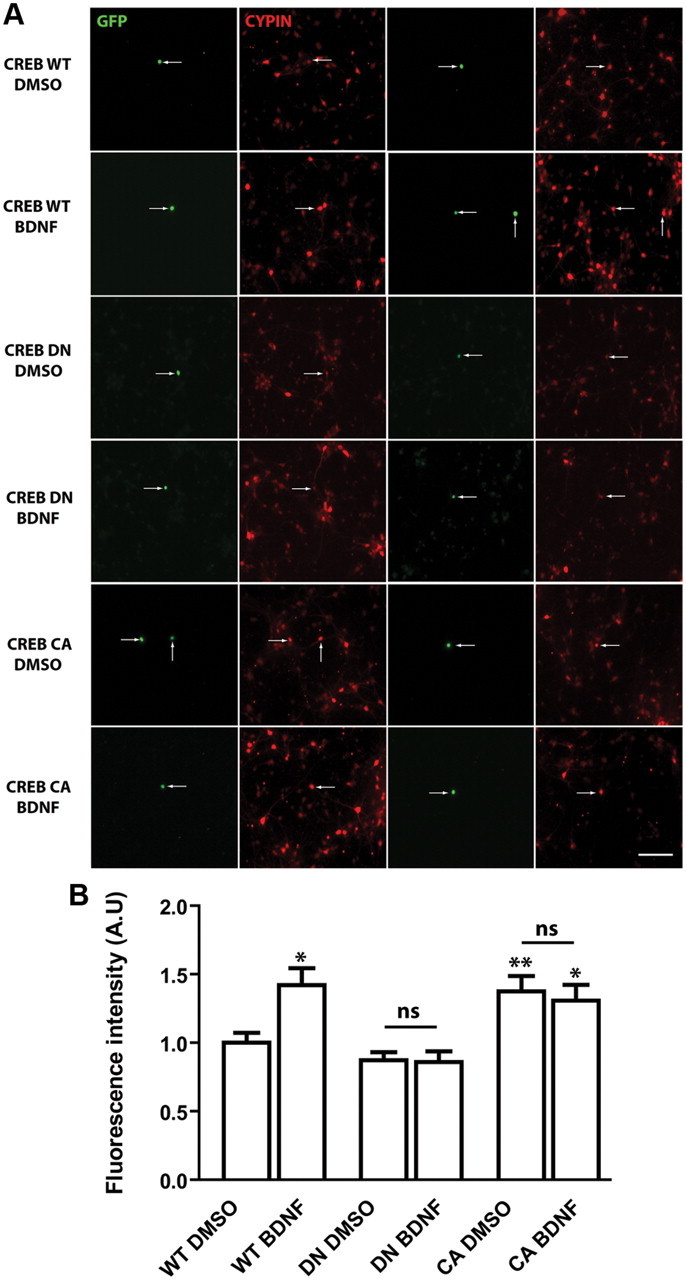
Expression of a dominant negative mutant or a constitutively active mutant of CREB attenuates BDNF-promoted increases in cypin expression. A, Hippocampal neurons were transfected with pEGFP-CREB WT (wild type, control) vector (control) with pEGFP-CREB DN (dominant negative form, Ser119Ala) or pEGFP-CREB CA (constitutively active form, Ser119Asp) at 6 DIV and treated with either DMSO (vehicle) or BDNF (25 ng/ml) at 7 DIV for 72 h. Neurons were immunostained using a polyclonal cypin antibody. Arrows point to transfected neurons. Scale bar, 100 μm. B, Average fluorescence intensity (A.U., Arbitrary units) of cypin immunostaining was assessed in individual cells. Expression of CREB DN blocks and expression of CREB CA occludes BDNF-promoted increases in cypin expression. n = 20 neurons for each condition, *p < 0.05, **p < 0.01 by ANOVA followed by Student-Newman–Keuls multiple-comparison test compared to DMSO-treated neurons expressing CREB WT. ns, No significance compared to DMSO-treated neurons.
Figure 8.
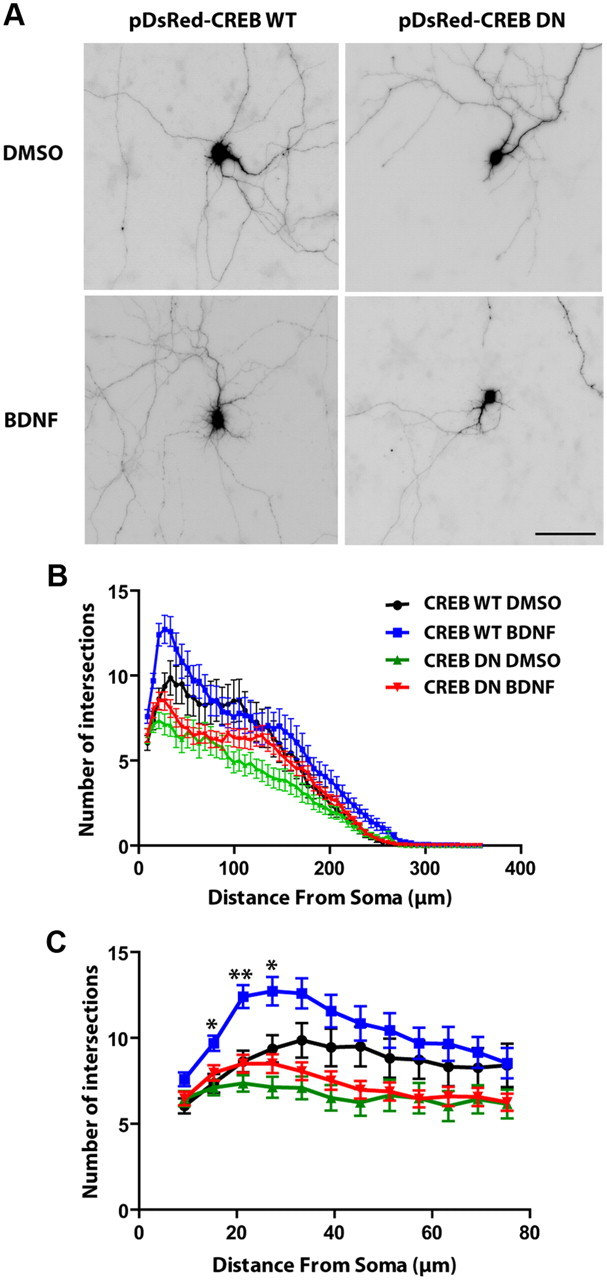
Expression of a dominant negative mutant of CREB attenuates BDNF-promoted increases in dendrite number. A, Hippocampal neurons were cotransfected with pEGFP and either DsRed-CREB WT(control) or ds-Red-CREB DN at 6 DIV and treated with either DMSO (vehicle) or BDNF (25 ng/ml) at 7 DIV for 72 h. Representative GFP images are shown. Dendrite number was assessed at 10 DIV. Scale bar, 100 μm. B, Sholl analysis. CREB DN overexpression blocks BDNF-promoted increases in the number of intersections close to the soma (proximal dendrites). C, Proximal Sholl analysis (from B) within the first 75 μm of the soma. *p < 0.05 at 15 and 27 μm, **p < 0.01 at 21 μm when comparing CREB WT plus BDNF to CREB WT plus DMSO and no significance when comparing CREB DN plus DMSO to CREB DN plus BDNF treatment. p values were determined by two-way ANOVA followed by Bonferroni multiple-comparisons test. Error bars indicate SEM. n = 22 neurons, CREB WT plus DMSO; n = 32, CREB WT plus BDNF; n = 33, CREB DN plus DMSO; n = 42, CREB DN plus BDNF.
Discussion
Dendrite number and branching are regulated by the interplay between extrinsic factors and intrinsic factors. Much progress has been made in identifying the key players and elucidating the signaling mechanisms that regulate dendrite morphology and patterning. There is a long list of external factors, including neurotrophins (for review, see McAllister et al., 1996; McAllister, 2000), estrogen (Audesirk et al., 2003; Sakamoto et al., 2003), and electrical activity (Vaillant et al., 2002). Some of the known intrinsic factors are the small GTPases RhoA, Rac1, and Cdc42 (Threadgill et al., 1997; Ruchhoeft et al., 1999; Li et al., 2000; Chen and Firestein, 2007), calcium/calmodulin-dependent protein kinase II (Fink et al., 2003), β-catenin (Yu and Malenka, 2004), CREST (Aizawa et al., 2004), Dishevelled (Rosso et al., 2005), some novel genes in Drosophila (Gao et al., 1999; Moore et al., 2002; Grueber et al., 2003; Emoto et al., 2004), cypin (Akum et al., 2004), and PSD-95 (Charych et al., 2006). Our laboratory has focused on cypin, a protein that regulates dendrite formation by promoting microtubule assembly and negatively regulates trafficking of PSD-95, which is associated with signaling networks at excitatory synapses and decreases dendrite branching. Our current studies are the first to demonstrate that extracellular factors, specifically BDNF, can regulate cypin gene expression.
Our results suggest that the MAPK pathway regulates BDNF-mediated cypin expression in hippocampal neurons. This signaling cascade is involved in promoting neuritogenesis and neurite outgrowth via CREB-mediated gene expression (Tojima et al., 2003). Therefore, we suggest that cypin is a candidate gene that is regulated by this signaling pathway via activation of CREB. Our results support this hypothesis, since pharmacological data confirm that a MEK-specific inhibitor and expression of a dominant negative form of CREB block BDNF-mediated increases in cypin expression.
Because BDNF is an important molecule for dendrite growth, much work has focused on elucidating BDNF signaling pathways. Ligand binding to TrkB triggers a cascade of phosphorylation events. Phosphorylated tyrosine residues on the receptor allow for Shc and phospholipase C (PLC) to attach. PLC becomes phosphorylated and activated (Segal, 2003). In addition to PLC, TrkB can signal via the PI3K pathway. Tyrosine phosphorylation allows SHC and Grb2 to recruit PI3K as well as SOS. PI3K activates the AKT pathway, and SOS activates the Ras/MAPK cascade (Segal, 2003). In contrast, there are also several studies that support the idea that BDNF can regulate dendrite branching through the PI3K pathway by activating the mTOR (Jaworski et al., 2005; Kumar et al., 2005), which can then activate the translation of a group of mRNAs (Schratt et al., 2004). In fact, local administration of BDNF to dendrites of cultured hippocampal neurons results in mTOR-dependent phosphorylation of S6, which is involved in local protein synthesis (Takei et al., 2004). While this increased phosphorylation was observed within 5 min of stimulation, our studies were performed using longer treatment times, suggesting the possibility that the duration of BDNF influences the activation of different signaling pathways. Recent studies support this idea that there would be temporal and spatial regulation of signaling by BDNF in neurons and have shown that acute and gradual increases in BDNF give rise to differential expression of Homer1 and Arc in cultured hippocampal neurons (Ji et al., 2010). Moreover, mRNAs with the same BDNF coding sequence but distinct 3′ UTRs have distinct cellular localizations and function (An et al., 2008), increasing the possibility that BDNF regulates neuronal morphology and function via several distinct cellular mechanisms. Our studies support the idea that increases in proximal dendrites resulting in response to BDNF do not depend on the activation of either PI3K or mTOR (Fig. 3), suggesting an alternative pathway of activation for BDNF-mediated increases in cypin expression and proximal dendrites in 72 h.
The MAPK signaling cascade, which can be activated by BDNF, is involved in promoting neuritogenesis and neurite outgrowth via CREB-mediated gene expression (Tojima et al., 2003). Similarly, in cortical neurons, primary dendrite number is regulated by BDNF via MAPK signaling pathways, and this effect is independent of nascent protein synthesis (Dijkhuizen and Ghosh, 2005). This is in contrast to our results in hippocampal neurons, whereby BDNF increases cypin transcription and translation to regulate proximal dendrite number. These differences may be due to the fact that different neuronal types were studied. This idea is supported by recent evidence that neurite outgrowth is regulated differently in cortical and hippocampal neurons (Ko et al., 2005).
Differences in signal transduction pathways activated by hippocampal versus cortical neurons may also be due to the BDNF receptor that is activated. Like other neurotrophins, BDNF exists in two states: proteolytically processed, which is the active form that can bind Trk receptors, or unprocessed, which allows BDNF to bind with high affinity to p75NTR. Thus, the complex actions of BDNF on the dendritic arbor may be mediated, in part, by multiple receptors resulting from alternative splicing of TrkB. Three alternative splice variants of TrkB have been identified. The full-length TrkB.FL contains a cytoplasmic tyrosine kinase domain, which is responsible for autophosphorylation and clustering of the receptor when activated by BDNF (Chao, 2003; Segal, 2003). Two truncated isoforms, termed TrkB.T1 and TrkB.T2, which lack the cytoplasmic tyrosine kinase domains, have also been identified (Barbacid, 1994). TrkB.FL and TrkB.T2 are expressed exclusively in neurons, and TrkB.T1 is expressed in both neurons and non-neuronal cells (Frisén et al., 1993; Rudge et al., 1994; Armanini et al., 1995; Biffo et al., 1995; Wetmore and Olson, 1995). Importantly, in rat, expression of the TrkB.FL in the hippocampus is barely detectable at embryonic day l5 but increases throughout development to reach adult levels by postnatal day (P)5 (Muragaki et al., 1995). Conversely, TrkB.FL reaches adult levels in cortex by P0. In addition, TrkB.T1 is expressed in the hippocampus at low levels until birth and reaches adult levels by P10–15. The expression of TrkB.T1 in cortex is similar, but adult levels are reached between P10 and P20 (Muragaki et al., 1995). This difference in developmental expression pattern, which encompasses the developmental time points of our cultures, coupled with the fact that TrkB.FL controls proximal branching while TrkB.T1 increases elongation of distal dendrites in cortical culture (Yacoubian and Lo, 2000), may explain the different pathways activated by BDNF in our study and other studies. In addition, TrkB.FL can inhibit the effects of TrkB.T1 and vice versa when both receptors are overexpressed, adding an additional layer of receptor crosstalk (Yacoubian and Lo, 2000; Hartmann et al., 2004).
MAPK phosphorylates extracellular signal-regulated kinases (ERKs), and these activated ERKs phosphorylate CREB (Bonni et al., 1999). CREB has been characterized as a critical molecule for the transcriptional regulation of dendritic complexity (Redmond and Ghosh, 2005; Wayman et al., 2006). Loss of CREB function has been shown to impair dendrite growth and arborization of newborn hippocampal neurons (Jagasia et al., 2009). In line with these reports, we have found that cypin is a target gene for CREB regulation and that BDNF enhances the binding of CREB to the cypin promoter. Thus, we have demonstrated, by pharmacological and cell biological methods, that BDNF, an important extrinsic factor needed for normal neuronal development, activates CREB-mediated increases in the expression of cypin, an essential intrinsic regulator of microtubule assembly and dendrite arborization. Since BDNF is thought to play a role in the development of cognitive disorders (Zuccato et al., 2001; Neves-Pereira et al., 2002; Sklar et al., 2002; He et al., 2004; Schumacher et al., 2005; Strauss et al., 2005; Lynch et al., 2007; Nagahara et al., 2009; Peng et al., 2009), our results will add to our understanding of how dendrite arborization is disrupted in these disorders.
Footnotes
This work was supported in part by a Busch Biomedical Grant, National Science Foundation (NSF) Grants IBN-0919747 and IBN-0548543, March of Dimes Foundation Grants 1-FY04-107 and 1-FY08-464, and a Grant-In-Aid from the American Heart Association (to B.L.F.). J.R.F. was supported in part by the IGERT (Integrative Graduate Education and Research Traineeship) Program on Integratively Engineered Biointerfaces at Rutgers: NSF Grant DGE-0333196. M.K. was awarded an Anna B. and James Leathem Fellowship, and G.F.Z. and S.B.L. received School of Arts and Sciences Honors Awards in support of this research. We acknowledge Maxine Chen for some of the pilot studies leading to this work.
References
- Aizawa H, Hu SC, Bobb K, Balakrishnan K, Ince G, Gurevich I, Cowan M, Ghosh A. Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science. 2004;303:197–202. doi: 10.1126/science.1089845. [DOI] [PubMed] [Google Scholar]
- Akum BF, Chen M, Gunderson SI, Riefler GM, Scerri-Hansen MM, Firestein BL. Cypin regulates dendrite patterning in hippocampal neurons by promoting microtubule assembly. Nat Neurosci. 2004;7:145–152. doi: 10.1038/nn1179. [DOI] [PubMed] [Google Scholar]
- An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, Torre ER, Jones KR, Feng Y, Lu B, Xu B. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanini MP, McMahon SB, Sutherland J, Shelton DL, Phillips HS. Truncated and catalytic isoforms of trkB are co-expressed in neurons of rat and mouse CNS. Eur J Neurosci. 1995;7:1403–1409. doi: 10.1111/j.1460-9568.1995.tb01132.x. [DOI] [PubMed] [Google Scholar]
- Audesirk T, Cabell L, Kern M, Audesirk G. Enhancement of dendritic branching in cultured hippocampal neurons by 17beta-estradiol is mediated by nitric oxide. Int J Dev Neurosci. 2003;21:225–233. doi: 10.1016/s0736-5748(03)00032-7. [DOI] [PubMed] [Google Scholar]
- Baker RE, Dijkhuizen PA, Van Pelt J, Verhaagen J. Growth of pyramidal, but not non-pyramidal, dendrites in long-term organotypic explants of neonatal rat neocortex chronically exposed to neurotrophin-3. Eur J Neurosci. 1998;10:1037–1044. doi: 10.1046/j.1460-9568.1998.00118.x. [DOI] [PubMed] [Google Scholar]
- Barbacid M. The Trk family of neurotrophin receptors. J Neurobiol. 1994;25:1386–1403. doi: 10.1002/neu.480251107. [DOI] [PubMed] [Google Scholar]
- Barde YA, Edgar D, Thoenen H. Purification of a new neurotrophic factor from mammalian brain. EMBO J. 1982;1:549–553. doi: 10.1002/j.1460-2075.1982.tb01207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffo S, Offenhäuser N, Carter BD, Barde YA. Selective binding and internalisation by truncated receptors restrict the availability of BDNF during development. Development. 1995;121:2461–2470. doi: 10.1242/dev.121.8.2461. [DOI] [PubMed] [Google Scholar]
- Bonni A, Brunet A, West AE, Datta SR, Takasu MA, Greenberg ME. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science. 1999;286:1358–1362. doi: 10.1126/science.286.5443.1358. [DOI] [PubMed] [Google Scholar]
- Carrel D, Du Y, Komlos D, Hadzimichalis NM, Kwon M, Wang B, Brzustowicz LM, Firestein BL. NOS1AP regulates dendrite patterning of hippocampal neurons through a carboxypeptidase E-mediated pathway. J Neurosci. 2009;29:8248–8258. doi: 10.1523/JNEUROSCI.5287-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao MV. Neurotrophins and their receptors: a convergence point for many signalling pathways. Nat Rev Neurosci. 2003;4:299–309. doi: 10.1038/nrn1078. [DOI] [PubMed] [Google Scholar]
- Chao W, Liu H, Hanahan DJ, Olson MS. Protein tyrosine phosphorylation and regulation of the receptor for platelet-activating factor in rat Kupffer cells. Effect of sodium vanadate. Biochem J. 1992;288:777–784. doi: 10.1042/bj2880777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charych EI, Akum BF, Goldberg JS, Jörnsten RJ, Rongo C, Zheng JQ, Firestein BL. Activity-independent regulation of dendrite patterning by postsynaptic density protein PSD-95. J Neurosci. 2006;26:10164–10176. doi: 10.1523/JNEUROSCI.2379-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Firestein BL. RhoA regulates dendrite branching in hippocampal neurons by decreasing cypin protein levels. J Neurosci. 2007;27:8378–8386. doi: 10.1523/JNEUROSCI.0872-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline HT. Dendritic arbor development and synaptogenesis. Curr Opin Neurobiol. 2001;11:118–126. doi: 10.1016/s0959-4388(00)00182-3. [DOI] [PubMed] [Google Scholar]
- Davies AM, Lee KF, Jaenisch R. p75-deficient trigeminal sensory neurons have an altered response to NGF but not to other neurotrophins. Neuron. 1993;11:565–574. doi: 10.1016/0896-6273(93)90069-4. [DOI] [PubMed] [Google Scholar]
- Dijkhuizen PA, Ghosh A. BDNF regulates primary dendrite formation in cortical neurons via the PI3-kinase and MAP kinase signaling pathways. J Neurobiol. 2005;62:278–288. doi: 10.1002/neu.20100. [DOI] [PubMed] [Google Scholar]
- Emoto K, He Y, Ye B, Grueber WB, Adler PN, Jan LY, Jan YN. Control of dendritic branching and tiling by the Tricornered-kinase/Furry signaling pathway in Drosophila sensory neurons. Cell. 2004;119:245–256. doi: 10.1016/j.cell.2004.09.036. [DOI] [PubMed] [Google Scholar]
- Favata MF, Horiuchi KY, Manos EJ, Daulerio AJ, Stradley DA, Feeser WS, Van Dyk DE, Pitts WJ, Earl RA, Hobbs F, Copeland RA, Magolda RL, Scherle PA, Trzaskos JM. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J Biol Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- Fernández JR, Welsh WJ, Firestein BL. Structural characterization of the zinc binding domain in cytosolic PSD-95 interactor (cypin): Role of zinc binding in guanine deamination and dendrite branching. Proteins. 2008;70:873–881. doi: 10.1002/prot.21683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink CC, Bayer KU, Myers JW, Ferrell JE, Jr, Schulman H, Meyer T. Selective regulation of neurite extension and synapse formation by the beta but not the alpha isoform of CaMKII. Neuron. 2003;39:283–297. doi: 10.1016/s0896-6273(03)00428-8. [DOI] [PubMed] [Google Scholar]
- Firestein BL, Brenman JE, Aoki C, Sanchez-Perez AM, El-Husseini AE, Bredt DS. Cypin: a cytosolic regulator of PSD-95 postsynaptic targeting. Neuron. 1999;24:659–672. doi: 10.1016/s0896-6273(00)81120-4. [DOI] [PubMed] [Google Scholar]
- Frisén J, Verge VM, Fried K, Risling M, Persson H, Trotter J, Hökfelt T, Lindholm D. Characterization of glial trkB receptors: differential response to injury in the central and peripheral nervous systems. Proc Natl Acad Sci U S A. 1993;90:4971–4975. doi: 10.1073/pnas.90.11.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FB, Brenman JE, Jan LY, Jan YN. Genes regulating dendritic outgrowth, branching, and routing in Drosophila. Genes Dev. 1999;13:2549–2561. doi: 10.1101/gad.13.19.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber WB, Jan LY, Jan YN. Different levels of the homeodomain protein cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell. 2003;112:805–818. doi: 10.1016/s0092-8674(03)00160-0. [DOI] [PubMed] [Google Scholar]
- Hallböök F, Ibañez CF, Persson H. Evolutionary studies of the nerve growth factor family reveal a novel member abundantly expressed in Xenopus ovary. Neuron. 1991;6:845–858. doi: 10.1016/0896-6273(91)90180-8. [DOI] [PubMed] [Google Scholar]
- Hartmann M, Brigadski T, Erdmann KS, Holtmann B, Sendtner M, Narz F, Lessmann V. Truncated TrkB receptor-induced outgrowth of dendritic filopodia involves the p75 neurotrophin receptor. J Cell Sci. 2004;117:5803–5814. doi: 10.1242/jcs.01511. [DOI] [PubMed] [Google Scholar]
- He XP, Kotloski R, Nef S, Luikart BW, Parada LF, McNamara JO. Conditional deletion of TrkB but not BDNF prevents epileptogenesis in the kindling model. Neuron. 2004;43:31–42. doi: 10.1016/j.neuron.2004.06.019. [DOI] [PubMed] [Google Scholar]
- Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, Podkolodny NL, Kolchanov NA. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horch HW, Krüttgen A, Portbury SD, Katz LC. Destabilization of cortical dendrites and spines by BDNF. Neuron. 1999;23:353–364. doi: 10.1016/s0896-6273(00)80785-0. [DOI] [PubMed] [Google Scholar]
- Jagasia R, Steib K, Englberger E, Herold S, Faus-Kessler T, Saxe M, Gage FH, Song H, Lie DC. GABA-cAMP response element-binding protein signaling regulates maturation and survival of newly generated neurons in the adult hippocampus. J Neurosci. 2009;29:7966–7977. doi: 10.1523/JNEUROSCI.1054-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski J, Spangler S, Seeburg DP, Hoogenraad CC, Sheng M. Control of dendritic arborization by the phosphoinositide-3′-kinase-Akt-mammalian target of rapamycin pathway. J Neurosci. 2005;25:11300–11312. doi: 10.1523/JNEUROSCI.2270-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Pang PT, Feng L, Lu B. Cyclic AMP controls BDNF-induced TrkB phosphorylation and dendritic spine formation in mature hippocampal neurons. Nat Neurosci. 2005;8:164–172. doi: 10.1038/nn1381. [DOI] [PubMed] [Google Scholar]
- Ji Y, Lu Y, Yang F, Shen W, Tang TT, Feng L, Duan S, Lu B. Acute and gradual increases in BDNF concentration elicit distinct signaling and functions in neurons. Nat Neurosci. 2010;13:302–309. doi: 10.1038/nn.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Hu H, Mathers PH, Agmon A. Brain-derived neurotrophic factor mediates activity-dependent dendritic growth in nonpyramidal neocortical interneurons in developing organotypic cultures. J Neurosci. 2003;23:5662–5673. doi: 10.1523/JNEUROSCI.23-13-05662.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Zou K, Minagawa H, Yu W, Gong JS, Yanagisawa K, Michikawa M. Cholesterol-mediated neurite outgrowth is differently regulated between cortical and hippocampal neurons. J Biol Chem. 2005;280:42759–42765. doi: 10.1074/jbc.M509164200. [DOI] [PubMed] [Google Scholar]
- Kumar V, Zhang MX, Swank MW, Kunz J, Wu GY. Regulation of dendritic morphogenesis by Ras-PI3K-Akt-mTOR and Ras-MAPK signaling pathways. J Neurosci. 2005;25:11288–11299. doi: 10.1523/JNEUROSCI.2284-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutzing MK, Langhammer CG, Luo V, Lakdawala H, Firestein BL. Automated Sholl analysis of digitized neuronal morphology at multiple scales. J Vis Exp. 2010;pii:2354. doi: 10.3791/2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara H, Araki N, Makino K, Masuko N, Honda S, Kaibuchi K, Fukunaga K, Miyamoto E, Ogawa M, Saya H. A novel NE-dlg/SAP102-associated protein, p51-nedasin, related to the amidohydrolase superfamily, interferes with the association between NE-dlg/SAP102 and N-methyl-d-aspartate receptor. J Biol Chem. 1999;274:32204–32214. doi: 10.1074/jbc.274.45.32204. [DOI] [PubMed] [Google Scholar]
- Landgraf M, Evers JF. Control of dendritic diversity. Curr Opin Cell Biol. 2005;17:690–696. doi: 10.1016/j.ceb.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Langhammer CG, Previtera ML, Sweet ES, Sran SS, Chen M, Firestein BL. Automated Sholl analysis of digitized neuronal morphology at multiple scales: Whole cell Sholl analysis versus Sholl analysis of arbor subregions. Cytometry A. 2010;77:1160–1168. doi: 10.1002/cyto.a.20954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi-Montalcini R. The nerve growth factor 35 years later. Science. 1987;237:1154–1162. doi: 10.1126/science.3306916. [DOI] [PubMed] [Google Scholar]
- Levine ES, Dreyfus CF, Black IB, Plummer MR. Brain-derived neurotrophic factor rapidly enhances synaptic transmission in hippocampal neurons via postsynaptic tyrosine kinase receptors. Proc Natl Acad Sci U S A. 1995;92:8074–8077. doi: 10.1073/pnas.92.17.8074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Van Aelst L, Cline HT. Rho GTPases regulate distinct aspects of dendritic arbor growth in Xenopus central neurons in vivo. Nat Neurosci. 2000;3:217–225. doi: 10.1038/72920. [DOI] [PubMed] [Google Scholar]
- Libersat F. Maturation of dendritic architecture: lessons from insect identified neurons. J Neurobiol. 2005;64:11–23. doi: 10.1002/neu.20142. [DOI] [PubMed] [Google Scholar]
- Lynch G, Kramar EA, Rex CS, Jia Y, Chappas D, Gall CM, Simmons DA. Brain-derived neurotrophic factor restores synaptic plasticity in a knock-in mouse model of Huntington's disease. J Neurosci. 2007;27:4424–4434. doi: 10.1523/JNEUROSCI.5113-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadeo D, Kaplan L, Chao MV, Hempstead BL. High affinity nerve growth factor binding displays a faster rate of association than p140trk binding. Implications for multi-subunit polypeptide receptors. J Biol Chem. 1994;269:6884–6891. [PubMed] [Google Scholar]
- Maisonpierre PC, Belluscio L, Squinto S, Ip NY, Furth ME, Lindsay RM, Yancopoulos GD. Neurotrophin-3: a neurotrophic factor related to NGF and BDNF. Science. 1990;247:1446–1451. doi: 10.1126/science.247.4949.1446. [DOI] [PubMed] [Google Scholar]
- McAllister AK. Cellular and molecular mechanisms of dendrite growth. Cereb Cortex. 2000;10:963–973. doi: 10.1093/cercor/10.10.963. [DOI] [PubMed] [Google Scholar]
- McAllister AK, Lo DC, Katz LC. Neurotrophins regulate dendritic growth in developing visual cortex. Neuron. 1995;15:791–803. doi: 10.1016/0896-6273(95)90171-x. [DOI] [PubMed] [Google Scholar]
- McAllister AK, Katz LC, Lo DC. Neurotrophin regulation of cortical dendritic growth requires activity. Neuron. 1996;17:1057–1064. doi: 10.1016/s0896-6273(00)80239-1. [DOI] [PubMed] [Google Scholar]
- McAllister AK, Katz LC, Lo DC. Opposing roles for endogenous BDNF and NT-3 in regulating cortical dendritic growth. Neuron. 1997;18:767–778. doi: 10.1016/s0896-6273(00)80316-5. [DOI] [PubMed] [Google Scholar]
- Miller JP, Jacobs GA. Relationships between neuronal structure and function. J Exp Biol. 1984;112:129–145. doi: 10.1242/jeb.112.1.129. [DOI] [PubMed] [Google Scholar]
- Moore AW, Jan LY, Jan YN. hamlet, a binary genetic switch between single- and multiple- dendrite neuron morphology. Science. 2002;297:1355–1358. doi: 10.1126/science.1072387. [DOI] [PubMed] [Google Scholar]
- Müller HW, Junghans U, Kappler J. Astroglial neurotrophic and neurite-promoting factors. Pharmacol Ther. 1995;65:1–18. doi: 10.1016/0163-7258(94)00047-7. [DOI] [PubMed] [Google Scholar]
- Muragaki Y, Timothy N, Leight S, Hempstead BL, Chao MV, Trojanowski JQ, Lee VM. Expression of trk receptors in the developing and adult human central and peripheral nervous system. J Comp Neurol. 1995;356:387–397. doi: 10.1002/cne.903560306. [DOI] [PubMed] [Google Scholar]
- Nagahara AH, Merrill DA, Coppola G, Tsukada S, Schroeder BE, Shaked GM, Wang L, Blesch A, Kim A, Conner JM, Rockenstein E, Chao MV, Koo EH, Geschwind D, Masliah E, Chiba AA, Tuszynski MH. Neuroprotective effects of brain-derived neurotrophic factor in rodent and primate models of Alzheimer's disease. Nat Med. 2009;15:331–337. doi: 10.1038/nm.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima T, Uchida C, Anderson SF, Parvin JD, Montminy M. Analysis of a cAMP-responsive activator reveals a two-component mechanism for transcriptional induction via signal-dependent factors. Genes Dev. 1997;11:738–747. doi: 10.1101/gad.11.6.738. [DOI] [PubMed] [Google Scholar]
- Neves-Pereira M, Mundo E, Muglia P, King N, Macciardi F, Kennedy JL. The brain-derived neurotrophic factor gene confers susceptibility to bipolar disorder: evidence from a family-based association study. Am J Hum Genet. 2002;71:651–655. doi: 10.1086/342288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S, Garzon DJ, Marchese M, Klein W, Ginsberg SD, Francis BM, Mount HT, Mufson EJ, Salehi A, Fahnestock M. Decreased brain-derived neurotrophic factor depends on amyloid aggregation state in transgenic mouse models of Alzheimer's disease. J Neurosci. 2009;29:9321–9329. doi: 10.1523/JNEUROSCI.4736-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond L, Ghosh A. Regulation of dendritic development by calcium signaling. Cell Calcium. 2005;37:411–416. doi: 10.1016/j.ceca.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Reichardt LF. Neurotrophin-regulated signalling pathways. Philos Trans R Soc Lond B Biol Sci. 2006;361:1545–1564. doi: 10.1098/rstb.2006.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso SB, Sussman D, Wynshaw-Boris A, Salinas PC. Wnt signaling through Dishevelled, Rac and JNK regulates dendritic development. Nat Neurosci. 2005;8:34–42. doi: 10.1038/nn1374. [DOI] [PubMed] [Google Scholar]
- Ruchhoeft ML, Ohnuma S, McNeill L, Holt CE, Harris WA. The neuronal architecture of Xenopus retinal ganglion cells is sculpted by rho-family GTPases in vivo. J Neurosci. 1999;19:8454–8463. doi: 10.1523/JNEUROSCI.19-19-08454.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudge JS, Li Y, Pasnikowski EM, Mattsson K, Pan L, Yancopoulos GD, Wiegand SJ, Lindsay RM, Ip NY. Neurotrophic factor receptors and their signal transduction capabilities in rat astrocytes. Eur J Neurosci. 1994;6:693–705. doi: 10.1111/j.1460-9568.1994.tb00981.x. [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Mezaki Y, Shikimi H, Ukena K, Tsutsui K. Dendritic growth and spine formation in response to estrogen in the developing Purkinje cell. Endocrinology. 2003;144:4466–4477. doi: 10.1210/en.2003-0307. [DOI] [PubMed] [Google Scholar]
- Schratt GM, Nigh EA, Chen WG, Hu L, Greenberg ME. BDNF regulates the translation of a select group of mRNAs by a mammalian target of rapamycin-phosphatidylinositol 3-kinase-dependent pathway during neuronal development. J Neurosci. 2004;24:7366–7377. doi: 10.1523/JNEUROSCI.1739-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher J, Jamra RA, Becker T, Ohlraun S, Klopp N, Binder EB, Schulze TG, Deschner M, Schmäl C, Höfels S, Zobel A, Illig T, Propping P, Holsboer F, Rietschel M, Nöthen MM, Cichon S. Evidence for a relationship between genetic variants at the brain-derived neurotrophic factor (BDNF) locus and major depression. Biol Psychiatry. 2005;58:307–314. doi: 10.1016/j.biopsych.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Schwartz PM, Borghesani PR, Levy RL, Pomeroy SL, Segal RA. Abnormal cerebellar development and foliation in BDNF−/− mice reveals a role for neurotrophins in CNS patterning. Neuron. 1997;19:269–281. doi: 10.1016/s0896-6273(00)80938-1. [DOI] [PubMed] [Google Scholar]
- Segal RA. Selectivity in neurotrophin signaling: theme and variations. Annu Rev Neurosci. 2003;26:299–330. doi: 10.1146/annurev.neuro.26.041002.131421. [DOI] [PubMed] [Google Scholar]
- Segal RA, Pomeroy SL, Stiles CD. Axonal growth and fasciculation linked to differential expression of BDNF and NT3 receptors in developing cerebellar granule cells. J Neurosci. 1995;15:4970–4981. doi: 10.1523/JNEUROSCI.15-07-04970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shooter EM. Early days of the nerve growth factor proteins. Annu Rev Neurosci. 2001;24:601–629. doi: 10.1146/annurev.neuro.24.1.601. [DOI] [PubMed] [Google Scholar]
- Sklar P, Gabriel SB, McInnis MG, Bennett P, Lim YM, Tsan G, Schaffner S, Kirov G, Jones I, Owen M, Craddock N, DePaulo JR, Lander ES. Family-based association study of 76 candidate genes in bipolar disorder: BDNF is a potential risk locus. Brain-derived neutrophic factor. Mol Psychiatry. 2002;7:579–593. doi: 10.1038/sj.mp.4001058. [DOI] [PubMed] [Google Scholar]
- Sobell HM. Actinomycin and DNA transcription. Proc Natl Acad Sci U S A. 1985;82:5328–5331. doi: 10.1073/pnas.82.16.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer TK, Mellado W, Filbin MT. BDNF activates CaMKIV and PKA in parallel to block MAG-mediated inhibition of neurite outgrowth. Mol Cell Neurosci. 2008;38:110–116. doi: 10.1016/j.mcn.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss J, Barr CL, George CJ, Devlin B, Vetró A, Kiss E, Baji I, King N, Shaikh S, Lanktree M, Kovacs M, Kennedy JL. Brain-derived neurotrophic factor variants are associated with childhood-onset mood disorder: confirmation in a Hungarian sample. Mol Psychiatry. 2005;10:861–867. doi: 10.1038/sj.mp.4001685. [DOI] [PubMed] [Google Scholar]
- Takei N, Inamura N, Kawamura M, Namba H, Hara K, Yonezawa K, Nawa H. Brain-derived neurotrophic factor induces mammalian target of rapamycin-dependent local activation of translation machinery and protein synthesis in neuronal dendrites. J Neurosci. 2004;24:9760–9769. doi: 10.1523/JNEUROSCI.1427-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill R, Bobb K, Ghosh A. Regulation of dendritic growth and remodeling by Rho, Rac, and Cdc42. Neuron. 1997;19:625–634. doi: 10.1016/s0896-6273(00)80376-1. [DOI] [PubMed] [Google Scholar]
- Tojima T, Kobayashi S, Ito E. Dual role of cyclic AMP-dependent protein kinase in neuritogenesis and synaptogenesis during neuronal differentiation. J Neurosci Res. 2003;74:829–837. doi: 10.1002/jnr.10754. [DOI] [PubMed] [Google Scholar]
- Vaillant AR, Zanassi P, Walsh GS, Aumont A, Alonso A, Miller FD. Signaling mechanisms underlying reversible, activity-dependent dendrite formation. Neuron. 2002;34:985–998. doi: 10.1016/s0896-6273(02)00717-1. [DOI] [PubMed] [Google Scholar]
- Wayman GA, Impey S, Marks D, Saneyoshi T, Grant WF, Derkach V, Soderling TR. Activity-dependent dendritic arborization mediated by CaM-kinase I activation and enhanced CREB-dependent transcription of Wnt-2. Neuron. 2006;50:897–909. doi: 10.1016/j.neuron.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Wetmore C, Olson L. Neuronal and nonneuronal expression of neurotrophins and their receptors in sensory and sympathetic ganglia suggest new intercellular trophic interactions. J Comp Neurol. 1995;353:143–159. doi: 10.1002/cne.903530113. [DOI] [PubMed] [Google Scholar]
- Whitford KL, Dijkhuizen P, Polleux F, Ghosh A. Molecular control of cortical dendrite development. Annu Rev Neurosci. 2002;25:127–149. doi: 10.1146/annurev.neuro.25.112701.142932. [DOI] [PubMed] [Google Scholar]
- Wong RO, Ghosh A. Activity-dependent regulation of dendritic growth and patterning. Nat Rev Neurosci. 2002;3:803–812. doi: 10.1038/nrn941. [DOI] [PubMed] [Google Scholar]
- Yacoubian TA, Lo DC. Truncated and full-length TrkB receptors regulate distinct modes of dendritic growth. Nat Neurosci. 2000;3:342–349. doi: 10.1038/73911. [DOI] [PubMed] [Google Scholar]
- Yu X, Malenka RC. Multiple functions for the cadherin/catenin complex during neuronal development. Neuropharmacology. 2004;47:779–786. doi: 10.1016/j.neuropharm.2004.07.031. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, Conti L, MacDonald ME, Friedlander RM, Silani V, Hayden MR, Timmusk T, Sipione S, Cattaneo E. Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science. 2001;293:493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]