Abstract
In mammalian cells, repair of DNA double-strand breaks (DSBs) occurs by both homologous and non-homologous mechanisms. By definition, homologous recombination requires a template with sufficient sequence identity to the damaged molecule in order to direct repair. We now show that the sister chromatid acts as a repair template in a substantial proportion of DSB repair events. The outcome of sister chromatid repair is primarily gene conversion unassociated with reciprocal exchange. This contrasts with expectations from the classical DSB repair model originally proposed for yeast meiotic recombination, but is consistent with models in which recombination is coupled intimately with replication. These results may explain why cytologically observable sister chromatid exchanges are induced only weakly by DNA-damaging agents that cause strand breaks, since most homologous repair events would not be observed. A preference for non-crossover events between sister chromatids suggests that crossovers, although genetically silent, may be disfavored for other reasons. Possibly, a general bias against crossing over in mitotic cells exists to reduce the potential for genome alterations when other homologous repair templates are utilized.
Keywords: double-strand break repair/genomic stability/homologous recombination/replication/sister chromatid
Introduction
Cells have evolved a number of repair pathways to contend with various types of DNA damage (Friedberg et al., 1995). One type of lesion, a DNA double-strand break (DSB), poses a particular threat to genomic integrity. In mammalian cells, the repair of DSBs occurs by both homologous recombination and non-homologous end joining (NHEJ) (Rouet et al., 1994b; Sargent et al., 1997; Liang et al., 1998; Lin et al., 1999). In order to repair a DSB by homologous recombination, a second DNA sequence with homology to the region to be repaired must be available to serve as a repair template. In Saccharomyces cerevisiae, the preferred substrate for homologous repair is the sister chromatid (Kadyk and Hartwell, 1992). Since sister chromatids are identical to each other, DNA damage can be repaired faithfully with no genetic consequence. Other potential repair templates are homologs and sequence repeats. With the exception of the X–Y pair, each chromosome is present as a pair of homologs that have a high degree of sequence identity. Sequence repeats comprise a large fraction of mammalian genomes (Schmid, 1996) and, although they can be quite divergent from each other, their enormous number and dispersal throughout the genome also makes them potential repair templates. Unlike sister chromatids, use of either of these repair templates has the potential to compromise genetic integrity. The biological significance of this is exemplified by the observation that homozygosis of a defective tumor suppressor gene by mitotic recombination of homologs promotes tumorigenesis (Lasko et al., 1991).
A widely accepted paradigm for DSB-promoted homologous recombination is the DSB repair model (Szostak et al., 1983). This model was proposed to account for the frequent association of gene conversion with crossing over during yeast meiotic recombination. Central to this model is a recombination intermediate containing two Holliday junctions, whose formation involves leading strand repair synthesis. Notably, resolution of the Holliday junctions results in either non-crossover or crossover gene conversion products. Direct physical examination of meiotic recombination intermediates in yeast strongly supports central aspects of this model (Schwacha and Kleckner, 1995).
Although the DSB repair model was also thought to be applicable to mitotic homologous recombination, a growing body of evidence is inconsistent with predictions of the model. In many instances, mitotic gene conversions, unlike meiotic events, are not associated with reciprocal exchange. Moreover, the unbroken donor allele is almost always unaltered in these events. For example, S.cerevisiae mating-type switching, which is a gene conversion event initiated by a DSB at the MAT locus, does not result in crossing over nor does it alter the donor sequence at the HML or HMR locus (Strathern et al., 1982). Similarly, DSB repair within tandem repeats in yeast frequently results in expansions and contractions of the repeats present on the broken recipient molecule, but the donor DNA molecule nearly always remains unchanged (Paques et al., 1998). Homologous recombination between heterologous chromosomes in Drosophila melanogaster and in mammalian cells also occurs without concomitant conversion of the template and is not associated with crossing over (Gloor et al., 1991; Nassif et al., 1994; Richardson et al., 1998). Thus, recombination mechanisms that suppress crossing over, either actively or passively, may be operational in mitotically dividing cells. To explain these results, a number of models have been proposed in which recombination does not require the resolution of Holliday junctions (Nassif et al., 1994; Richard et al., 1994; Ferguson and Holloman, 1996; Kogoma, 1997; Paques et al., 1998; Richardson et al., 1998; Holmes and Haber, 1999). In these models, variously called the SDSA (synthesis-dependent strand annealing; Nassif et al., 1994) and migrating D-loop models (Ferguson and Holloman, 1996), recombination is coupled intimately with replication, which unlike the DSB repair model, may involve both leading and lagging strand synthesis (Holmes and Haber, 1999).
In this report, we have investigated the role of sister chromatids in DSB repair in mammalian cells. For this, we used a recombination reporter substrate that allows us to detect unequal sister chromatid recombination events initiated by a DSB (Johnson et al., 1999). We examined both the frequency of DSB repair events involving the sister chromatid and the outcome of sister chromatid repair, using a type of random clone analysis in which all of the products of repair are recovered. Our analysis demonstrates that recombination between sister chromatids is much higher than reported cases of interchromosomal recombination in mammalian cells (Moynahan and Jasin, 1997; Richardson et al., 1998). We also find that gene conversion between sister chromatids is not usually associated with reciprocal exchange. These results are discussed in the context of a replication-based recombination model.
Results
Sister chromatid recombination is a major DSB repair pathway in mammalian cells
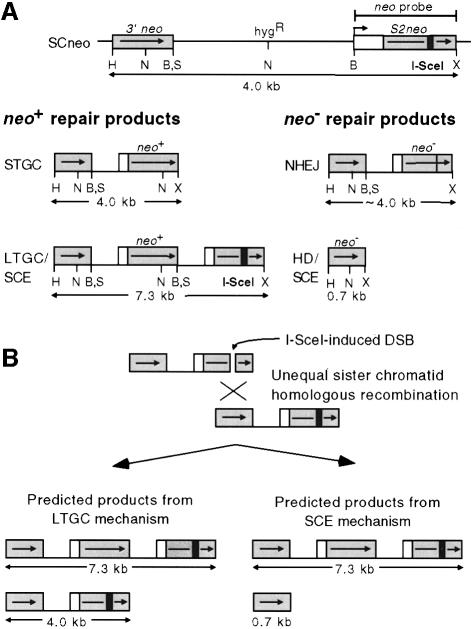
Homologous recombination between direct repeats, which are frequently used to examine DSB-promoted recombination, can occur between sequences on the same chromatid (intrachromatid) or on sister chromatids. We sought to determine specifically if recombination between sister chromatids is a major homologous repair pathway and by what mechanism sister chromatid recombination occurs. We previously constructed the recombination reporter substrate SCneo (Johnson et al., 1999), which can be used to address these questions. The SCneo substrate is composed of two defective neomycin phosphotransferase genes, 3′ neo, which is a 5′ truncation of the neo gene, and S2neo, which is mutated by a small internal deletion (Figure 1A). The S2neo deletion destroys an NcoI site and is accompanied by the insertion of the 18 bp I-SceI endonuclease site (Colleaux et al., 1988). Homologous recombination between the two defective neo genes can result in a neo+ gene, which is scored by resistance of cells to the drug G418. Expression of I-SceI allows DSB-promoted recombination events to be scored specifically.
Fig. 1. Recombination reporter substrate SCneo. (A) Schematic representation of SCneo and predicted neo+ and neo– DSB repair products after I-SceI expression. Shaded boxes represent the 0.7 kb neo gene repeats and the open box the promoter of the S2neo gene. The small black box is the 18 bp I-SceI site. During a short tract gene conversion (STGC), the I-SceI site is replaced by the NcoI site, but SCneo is not otherwise altered. Both long tract gene conversion and unequal sister chromatid exchange (LTGC/SCE) results in expansion of the SCneo locus. The expansion typically results in the middle neo repeat containing NcoI and SalI sites. Other LTGC products are also possible, depending on the length of the conversion tract. In a non-homologous end joining (NHEJ) event, the I-SceI site is disrupted, as these events usually involve small deletions or insertions. The 0.7 kb product can arise either from a homologous deletion (HD) or as the reciprocal contraction product of an unequal SCE. HD events arise from annealing of single strands at the two neo repeats, such that one repeat and the intervening hyg gene is deleted. The relevant restriction enzyme sites are shown: H, HindIII; N, NcoI; S, SalI; B, BamHI; X, XhoI. (B) Predicted products from two distinct mechanisms of unequal sister chromatid recombination. In an LTGC, the donor of information will remain unaltered during conversion of sequences downstream of the I-SceI site. In an SCE, expansion of one chromatid to a triplication will be associated with contraction on the other chromatid.
At least two distinct neo+ products result from homologous repair of the DSB in the SCneo substrate, both of which involve gene conversion. One product arises from gene conversion at the DSB without any gross alteration to the structure of the locus (Figure 1A). In this event, termed a short tract gene conversion (STGC), the 3′ neo gene used for repair can be on the same chromatid or, after DNA replication, on the sister chromatid. The products of these two STGC events are identical. Alternatively, repair mechanisms can result in expansion of the SCneo locus from two to three neo repeats through homologous repair from the 3′ neo gene on the sister chromatid (Figure 1A). An expansion product can arise from either a long tract gene conversion (LTGC) or an unequal sister chromatid exchange (SCE). An SCE event is expected to give rise to a unique neo+ product that contains three neo gene repeats (Figure 1). An LTGC event can also give rise to this product (Figure 1B), but in addition it can give rise to other products that vary in size due to the amount of sequence that is converted (see below).
The SCneo substrate was stably introduced into the genome of two recombination-proficient hamster cell lines, V79 and AA8, by selecting for hygromycin-resistant cells. Cell lines containing an intact SCneo substrate were verified by Southern blotting. One V79-derived cell line (4-18) (Johnson et al., 1999) and one AA8-derived cell line (10-4) (data not shown), each determined to have integrated a single copy of SCneo, were used in subsequent analyses. Consistent with previous results (Johnson et al., 1999), spontaneous recombination within the SCneo substrate is rare, between 10–5 and 10–6, but can be induced 100- to 1000-fold in both cell lines to >10–3 of electroporated cells by expression of I-SceI endonuclease. Although the DSB-induced recombination frequency is similar for both lines, it is possible that the absolute frequency may be found to be affected by the integration site or the distance between the neo repeats.
Previously we demonstrated that both STGC and LTGC/SCE products are readily detected in the V79 cell line after I-SceI expression (Johnson et al., 1999). In addition to events that restore an intact neo gene, other types of DSB repair are expected to occur within the SCneo substrate that do not give rise to a functional gene. These include NHEJ and homologous deletion (HD) events (Figure 1A). NHEJ involves joining of the two broken ends using little or no homology and, like homologous repair, is a major DSB repair pathway in mammalian cells. HD events, sometimes termed ‘popout’ events, result from deletion of one neo gene repeat, as well as the sequence between the two repeats, such that only a 3′ neo gene is retained at the SCneo locus. HD events are thought to arise primarily by non-conservative single-strand annealing (see, for example Liang et al., 1998). An identical product to that derived from an HD event can also arise as the reciprocal contraction product of an SCE event (Figure 1B).
To determine the relative proportion of each DSB repair product, we analyzed randomly generated clones following I-SceI expression. The V79 and AA8 cell lines were transiently transfected with the I-SceI expression vector and immediately thereafter plated in non-selective media in 96-well plates at a concentration of ∼1 cell per well. Each well was examined for the presence of a single cell and only those colonies that arose from a single cell were analyzed further. Colonies were examined by Southern analysis to identify the type of repair. Those that maintained the I-SceI site may never have been cleaved during transient I-SceI expression or they may have been cleaved but then repaired to restore the site (i.e. by precise ligation or equal sister chromatid recombination). These clones were excluded from further analysis because of the ambiguity arising from how the site was maintained.
A total of 300 clones were analyzed from the V79 and AA8 cell lines, in which 42 repair events were detected by loss of the I-SceI cleavage site (Table I). Of these, 18 (43%) were homologous repair events and 24 (57%) were NHEJ events. This is similar to results obtained previously with another hamster cell line containing a related DSB repair substrate, and is consistent with the notion that both homologous and non-homologous repair are important DSB repair pathways (Liang et al., 1998). In clones in which an NHEJ product was detected, the NHEJ product was generally the only product that was found (data not shown). However, homologous repair events were often found as part of mixed genotypes (see below).
Table I. Random clone analysis of DSB repair events.
| V79 cell line | AA8 cell line | Total | |
|---|---|---|---|
| Clones analyzed | 200 | 100 | 300 |
| Clones which lost an I-SceI site | 27 | 15 | 42 |
| Repair productsa | |||
| STGC | 5 | 3 | 8 (19%)b |
| LTGC/SCEc | 4 | 2 | 6 (14%) |
| HD/SCEc | 1 | 3 | 4 (10%) |
| NHEJ | 17 | 7 | 24 (57%) |
aRepair products are described in Figure 1.
bThe percentage is equal to the number of clones with products in the specified category divided by the total number of clones that had lost the I-SceI site.
cSince LTGC/SCE and HD/SCE products were never found within the same clone, these products arose from LTGC and HD events, respectively, rather than from SCE events (see text).
Of the total DSB repair events, 33% were neo+ gene conversions and 10% were HD/SCE events (Table I). Both STGC and LTGC/SCE events were represented in the gene conversions (19 and 14% of the total, respectively). The substantial contribution of expansion events to the gene conversion class demonstrates that sister chromatids are involved in DSB repair events. Considering that STGC events could also involve the sister chromatid, an even larger percentage of events are likely to be sister chromatid repair events.
Sister chromatid recombination within SCneo is not associated with reciprocal exchange
We next wanted to determine if sister chromatid recombination was associated with reciprocal exchange. In the case of unequal SCE, expansion of SCneo on one chromatid is associated with contraction of SCneo on the sister chromatid (Figure 1B). Conversely, if expansion of SCneo is the result of an LTGC event, the reciprocal crossover product is not produced (Figure 1B). The sister chromatid serves as a donor of information, but is itself unaltered by the event. Therefore, although LTGC and SCE events cannot always be distinguished by the structure of the neo+ product, they can be distinguished unequivocally by examining the structure of the SCneo locus on the participating sister chromatid.
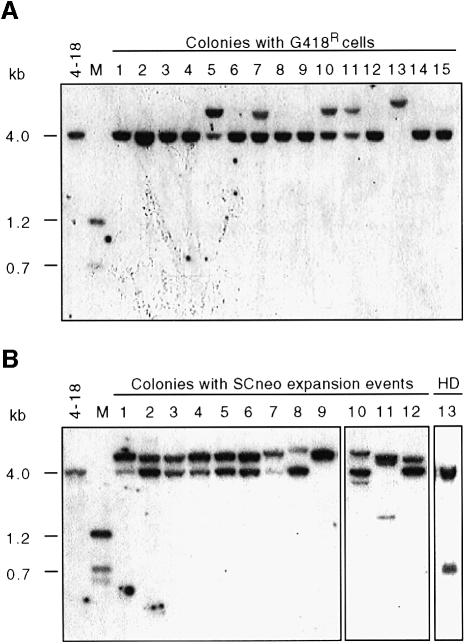
To examine the structure of both the neo+ expansion product and the product on the participating sister chromatid, random clones arising from unselected repair events were analyzed. Because sister chromatids segregate to different daughter cells upon cell division, sister chromatid recombination events that result in an expansion of SCneo would form a mixed colony. One portion of the cells would contain the neo+ expansion product, whereas the other portion would contain the product found on the participating chromatid. Because only a small number of randomly generated clones in the previous experiment contained the expansion product, we generated a larger number of clones. Rather than directly examining all clones by Southern blot analysis as in the previous experiment, in this experiment clones randomly generated after I-SceI expression were first screened by replica plating for the presence of G418R cells. This screen allowed us to specifically identify clones containing neo+ repair events for subsequent Southern analysis. For those clones in which neo+ cells were found, cells from the unselected replica plate were examined for the presence of the expansion product using XhoI–HindIII Southern analysis, since these enzymes cleave at sites flanking the neo gene repeats (Figure 1A).
Of the 2267 colonies tested from both the V79 and AA8 cell lines, 119 contained cells that were G418R (Table II), indicating that there had been a homologous recombination event in the colony. Of the 119 G418R colonies, 90 contained the recombination reporter on a 4.0 kb XhoI–HindIII fragment, indicating that these had arisen from an STGC event (Figure 2A and data not shown). The remaining 29 colonies contained the recombination reporter on a larger XhoI–HindIII fragment, indicating that these had undergone expansion events (Figure 2A and data not shown). Thus, 76% of the gene conversions were STGCs and 24% were LTGC/SCEs. We have noted some variation in the ratio of the two events, with LTGC/SCEs in some cases predominating over STGC events (Johnson et al., 1999). Nevertheless, this ratio is derived from the most comprehensive analysis of gene conversion within the SCneo substrate to date and is in good agreement with results obtained from analysis of spontaneous gene conversion events (Bollag and Liskay, 1991).
Table II. Repair mechanisms of SCneo homologous recombination events.
| V79 cell line | AA8 cell line | Total | |
|---|---|---|---|
| Clones analyzed | 1457 | 810 | 2267 |
| Clones with G418R cells | 75 | 44 | 119 |
| neo+ repair products | |||
| STGC | 55 (74%)a | 35 (80%) | 90 (76%) |
| LTGC | 19 (25%) | 7 (16%) | 26 (22%) |
| SCE | 0 | 0 | 0 |
| other | 1 (1%) | 2 (4%) | 3 (2%) |
aThe percentage is equal to the number of clones with products in that category divided by the total number of G418R colonies for the cell line.
Fig. 2. Sister chromatid recombination within the SCneo locus is not associated with reciprocal exchange. After I-SceI expression in cell lines containing the SCneo substrate, single cells were grown in non-selective medium and then replica plated to test for resistance to G418. Southern blot analysis was performed on clones containing G418R cells using XhoI and HindIII digestion of genomic DNA. The probe was a 1.2 kb BamHI–XhoI DNA fragment containing the complete neo sequence (Figure 1A). (A) Parental V79 (4-18) cells and clones in which G418R cells were identified. STGC events, lanes 1–4, 6, 8, 9, 12, 14 and 15; LTGC events, lanes 5, 7, 10, 11 and 13. (B) Parental V79 (4-18) cells and clones in which cells were identified to have undergone an LTGC event (lanes 1–12) or an HD event (lane 13). In both (A) and (B), each of the expansion products is presumed to be derived from an LTGC event, as none of the clones contains an associated 0.7 kb reciprocal SCE product. M, marker derived from V79 (4-18) genomic DNA digested with XhoI, BamHI and HindIII.
The expansion clones were subjected to further Southern analysis to determine the structure of the SCneo locus on both chromatids. Southern blot analysis of several of the expansion clones is presented in Figure 2B and the restriction fragment sizes for all of the 29 expansion clones are indicated in Table III. In most of these clones, two bands were detected, although none of the 29 clones contained the 0.7 kb band expected from an SCE event. Rather, most contained the 4.0 kb band expected from an LTGC event. Thus, in the SCneo substrate, sister chromatid recombination is consistent with non-crossover gene conversion, but not with an associated reciprocal exchange. Of the 29 expansion clones, the majority (17 clones; 57%) contained an ∼7.3 kb expansion product together with the 4.0 kb fragment (e.g. Figure 2B, lanes 1–7). The remaining clones contained a different set of bands or a single band. Three clones contained two bands, a 4.0 kb band and a second larger band that was not 7.3 kb (e.g. Figure 2B, lanes 8 and 12). The sizes of the second band ranged from 6.0 to 8.3 kb. Five clones contained a single band (e.g. Figure 2B, lane 9). Three clones contained three bands, in some cases of novel size (e.g. Figure 2B, lanes 10 and 11). The final colony contained 7.3 and 2.4 kb bands (data not shown). When two bands were found in a clone, they frequently had nearly equal intensity, although in some clones one was fainter.
Table III. Summary of genotypes of clones with SCneo expansion products.
| Clonea | XhoI–HindIII fragment(s) | Segregationb | Classc |
|---|---|---|---|
| A-35 | 4.0 kb, 7.3 kb | + | 1a |
| U1-50 | 4.0 kb, 7.3 kb | + | 1a |
| U3-352 | 4.0 kb, 7.3 kb | + | 1a |
| U3-399 | 4.0 kb, 7.3 kb | + | 1a |
| U3-537 | 7.3 kb | NA | 1a |
| U4-34 | 4.0 kb, 7.3 kb | + | 1a |
| U4-449 | 4.0 kb, 7.3 kb | + | 1a |
| A-26 | 4.0 kb, 7.3 kb | + | 1b |
| A-30 | 4.0 kb, 7.3 kb | + | 1b |
| A-9 | 4.0 kb, 7.3 kb | + | 1b |
| U1-55 | 4.0 kb, 7.3 kb | + | 1b |
| U2-136 | 4.0 kb, 7.3 kb | + | 1b |
| U2-98 | 4.0 kb, 7.3 kb | + | 1b |
| U4-422 | 4.0 kb, 7.3 kb | + | 1b |
| U4-44 | 7.2 kb | NA | 2 |
| U4-65 | 3.5 kb, 4.0 kb, 7.5 kb | + | 2 |
| U4-84 | 4.0 kb, 7.0 kb | – | 2 |
| A-24 | 3.5 kb, 4.0 kb, 5.0 kb | + | 3 |
| U1-69 | 4.0 kb, 7.3 kb | + | 3 |
| U4-490 | 4.0 kb, 7.3 kb | + | 4 |
| A-20 | 7.3 kb | NA | 5 |
| U3-450 | 4.0 kb, 7.3 kb | + | 5 |
| U4-466 | 4.0 kb, 7.3 kb | – | 5 |
| A-40 | 4.0 kb, 8.3 kb | + | 6 |
| U4-567 | 8.0 kb | NA | 6 |
| U4-68 | 2.0 kb, 7.0 kb, 7.3 kb | + | 6 |
| A-29 | 4.0 kb, 8.0 kb | + | ND |
| A-4 | 2.4 kb, 7.3 kb | – | ND |
| U1-63 | 7.3 kb | NA | ND |
a‘A’ clones are derived from the parental AA8 (10-4) cell line; ‘U’ clones are derived from the parental V79 (4-18) cell line.
bSegregation of the different sized XhoI–HindIII bands into subclones of the indicated clone. +, bands segregate; –, bands do not segregate; NA, not applicable since only a single band was present. All of the NA clones were subcloned, and each was found to contain the described band.
cAs defined in Figure 4 or not determined (ND).
We also examined the genotypes of the four unselected clones obtained in the first random clone analysis that contained the 0.7 kb HD/SCE product (Table I). None of the four clones had the 0.7 kb band associated with an expansion, indicating that this band was derived from an HD event involving single strand annealing rather than unequal SCE. In two of these clones, the HD product was the only one present (data not shown), suggesting that the DSB repair event occurred prior to DNA replication in the parental cell. In the other two clones, the 0.7 kb band was found with a 4.0 kb band, suggesting that the HD event occurred after DNA replication. In one case, the 4.0 kb band was of parental genotype, having maintained the I-SceI site (Figure 2B, lane 13 and data not shown), whereas in the other case it contained an STGC product (data not shown).
Segregation of repair products derived from sister chromatid recombination events
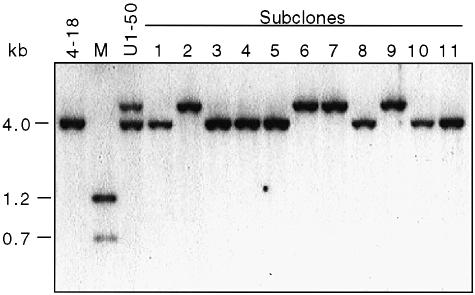
We expected that clones derived from sister chromatid recombination events would be mixed as a result of the segregation of the two sister chromatids to daughter cells. Thus, a clone found to contain 4.0 and 7.3 kb XhoI–HindIII fragments should be comprised of cells containing either of the two bands, but not both. To demonstrate this, we subcloned each of the clones containing an expansion and determined the genotype of the SCneo recombination reporter in the subclones by Southern blot analysis. Figure 3 shows the analysis of the subclones derived from a representative clone. As expected, each of the subclones from this clone contained only one SCneo genotype, on either a 4.0 or a 7.3 kb XhoI–HindIII fragment.
Fig. 3. Segregation of donor and recipient sequences after sister chromatid recombination. A clone containing 7.3 and 4.0 kb XhoI–HindIII SCneo fragments was subcloned. Each subclone contains either the 4.0 or the 7.3 kb XhoI–HindIII fragment as expected for segregation of sister chromatids to daughter cells. Presumably the 4.0 kb fragment served as the donor of information during DSB repair by LTGC to generate the 7.3 kb fragment on the recipient chromatid. Genomic DNA from each subclone was digested with XhoI and HindIII and subjected to Southern blot analysis using a neo gene probe.
Results from all of the clones are summarized in Table III. The majority of the clones with two XhoI–HindIII fragments segregated them to daughter cells, as expected if repair occurred after DNA replication in the parental cell. In each of the clones in which the fragments segregated, the subclone containing the expansion product was G418R. Those clones containing three fragments also segregated each of the fragments to subclones, and the subclone with the expansion product was G418R. The presence of three genotypes in these clones suggests that DSB repair occurred after more than one round of DNA replication. Clones in which a single XhoI–HindIII fragment was observed gave rise to subclones containing only that band. This raises the possibility that DSB repair occurred prior to DNA replication in the parental cell. However, in at least one case, the structure of the repair product clearly indicated that a sister chromatid participated in the repair event (see below).
Structure of sister chromatid recombination products
Clones with a 7.3 kb LTGC product could be expected to have conversion tracts encompassing both neo gene repeats on the sister chromatid, whereas those with different sized expansion product are predicted to have shorter gene conversion tracts. To verify this, we analyzed each LTGC product in detail to determine their structure. In many cases, the analysis was performed on subclones, rather than the original clone, so that the expansion product could be examined in the absence of the product from the other chromatid. Like STGC events, expansion via an LTGC mechanism will result in conversion of the I-SceI site in the S2neo gene into an NcoI site and generate a neo+ gene (Figure 1A). Consistent with their G418 resistance, all of the expansion products were found to contain a neo+ gene by Southern blot analysis (data not shown).
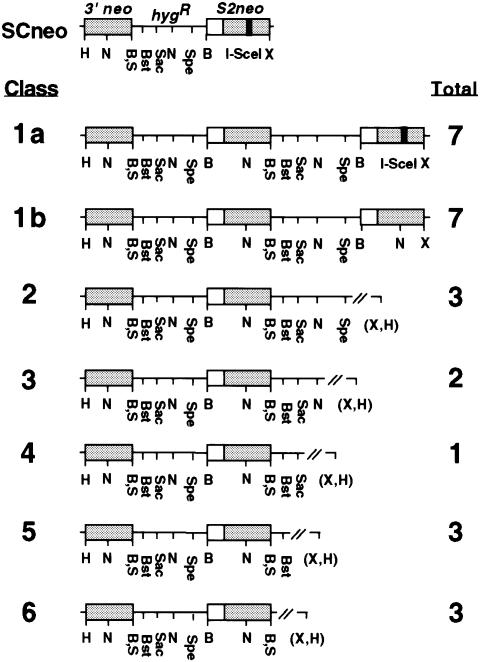
We next determined the extent of the conversion tract in the 29 expansion clones. In an LTGC event, the gene conversion tract can extend downstream of the NcoI site of the 3′ neo gene into hygR gene sequences prior to rejoining the end of what was the S2neo gene on the original chromatid. Consequently, the neo+ gene will be followed by a SalI site, rather than the XhoI site found after the S2neo gene (Figure 1A). Restriction analysis indicated that 26 of the 29 expansion products contained the SalI site, demonstrating that conversion extended at least 0.3 kb. These clones were examined further by Southern blot analysis to determine how far the gene conversion tract extended after the 3′ neo gene. For this, we used restriction endonucleases with sites present within the hygR and S2neo genes. Based on these digests, six different classes of clones were distinguished (Figure 4). Class 1 clones, which consisted of 14 clones, contained two hygR and three neo genes, representing a conversion of ∼3.3 kb. Classes 2–6, consisting of 12 clones, had shorter conversion tracts extending for varying distances into the 2.1 kb hygR gene. The gene conversion tracts in these recombinants ranged from ∼1.7 kb (class 2) to just over 0.3 kb (class 6). The variability of the conversion tracts suggests that the junction between the newly converted sequence and the chromatid involves NHEJ. It is possible that in the three clones in which the SalI site was not present, conversion extended just under 0.3 kb, i.e. past the stop codon of the 3′ neo gene but not as far as the SalI site.
Fig. 4. Structure of sister chromatid recombination products. To derive the structures, genomic DNA from each LTGC clone was digested with the indicated restriction enzymes and subjected to Southern blot analysis. The total number of events obtained for each class is indicated. H, HindIII; N, NcoI; S, SalI; B, BamHI; Bst, BstXI; Sac, SacI; Spe, SpeI; X, XhoI, (X,H), either a XhoI or HindIII site.
The class 1 clones were analyzed further to determine if the neo gene in the downstream repeat retained the I-SceI site (Figure 4). Seven of the clones (class 1a) maintained the I-SceI site, whereas the other seven (class 1b) had converted the downstream I-SceI site to an NcoI site. We also determined the structure of the 4.0 kb XhoI–HindIII fragment in these clones. In the six class 1a clones that also contained a 4.0 kb fragment, the 4.0 kb fragment had a parental structure, i.e. an S2neo gene with an intact I-SceI site (data not shown). In contrast, in the seven class 1b clones, the 4.0 kb fragment had the structure of an STGC event, i.e. a neo+ gene with an NcoI site. Therefore, all three full-length neo genes in the class 1b clones were neo+, whereas only one of the three in the class 1a clones was neo+. No clones were obtained with the intermediate genotype in which two full-length neo genes were neo+.
Discussion
Here we present evidence that homologous recombination (gene conversion) between sister chromatids is an important pathway for DSB repair in mammalian cells. A screen of unselected repair events revealed that gene conversion, as well as NHEJ and single strand annealing, can all make significant contributions to mammalian DSB repair. Gene conversion events involving the sister chromatid were not associated with crossing over, such that exchanges within the SCneo substrate could be estimated to be ≤3% (<1/29) of total sister chromatid repair events. The lack of crossing over suggests a major mechanistic difference from homologous recombination during meiosis, in which conversion frequently is associated with crossing over (Paques and Haber, 1999). This may reflect the different roles of homologous recombination during these two cell division cycles. In mitotically dividing cells, recombination is used primarily for DNA repair, with the major template for repair being the sister chromatid. During meiosis, homologs recombine at high frequency, with crossing over being essential for their proper segregation during the reductional division (Roeder, 1997).
In addition to the V79 and AA8 hamster cell lines used in this study, homologous repair, like NHEJ, was shown to be an important pathway for DSB repair in mouse embryonic stem (ES) cells (Moynahan et al., 1999) and another hamster cell line, CHO-K1 (Liang et al., 1998), using direct repeat reporter substrates. DSBs have also been shown to be potent inducers of gene conversion in human cell lines (A.J.Pierce and M.Jasin, unpublished data). Sister chromatid recombination was not assayed directly in these cases; however, results presented here, as well as in a recent study in ES cells using the SCneo substrate (Dronkert et al., 2000), suggest that sister chromatid recombination is an important DSB repair pathway in these cell lines as well. In light of organismal complexity, it will be interesting to determine how DSB repair pathways vary in adult tissues and at embryonic stages. Since Rad51 is an essential gene early in mouse development, homologous recombination is predicted to be an essential process at embryonic stages (Lim and Hasty, 1996; Tsuzuki et al., 1996). However, its role in adult tissues is probably restricted, as expression of Rad51 is limited to a subset of proliferative tissues (Shinohara et al., 1993). Another homologous repair protein, Rad54, was recently demonstrated to make different contributions to repair during embryonic and adult stages of the mouse (Essers et al., 2000). Considering the results presented here, the importance of homologous repair would be expected to diminish in cells that have exited the cell cycle, when sister chromatids are not available.
Derivation of the sister chromatid gene conversion events
A number of gene conversion products were obtained from repair of the DSB in the SCneo substrate. The major product was an STGC product in which conversion was limited to the 0.7 kb neo gene repeat. LTGC events were also frequent and could be divided into a number of classes depending on the extent of the conversion tract. DSB-promoted gene conversion is expected to be initiated in all of the events by invasion of a broken end(s) into the homologous neo repair template to prime repair synthesis (Figure 5). The structures of the fully homologous STGC and class 1 LTGC events are consistent with either one-ended or two-ended strand invasion. After repair synthesis is completed during a one-ended invasion, the newly synthesized strand would be expected to anneal to the non-invading end to complete the homologous repair event (Figure 5A). This has been proposed previously for gene conversion involving two chromosomes (Richardson et al., 1998). For two-ended invasion (not shown), the two newly synthesized strands would instead anneal to each other, rather than to a non-invading strand, to complete homologous repair. This is similar to the SDSA pathway proposed for D.melanogaster DSB repair events (Nassif et al., 1994).
Fig. 5. Model for the generation of sister chromatid gene conversion events. LTGC and LTGC/NHEJ events can be initiated by strand invasion of the broken S2neo gene into the 3′ neo gene on the sister chromatid, with repair synthesis primed by the invading end. (A) In an LTGC event, repair synthesis continues into the downstream S2neo gene and is followed by homologous annealing of the newly synthesized strand to the end of the broken chromatid. (B) In an LTGC/NHEJ event, repair synthesis terminates downstream of the neo+ gene and is followed by NHEJ of the newly synthesized strand to the end of the broken chromatid. See text for details.
Although both the STGC and class 1 LTGC events are fully homologous repair events, they differ from each other in the length of the conversion tract. This is due to differences in the extent of repair synthesis or, possibly, the position of annealing of the newly synthesized strand(s). In the STGC events, synthesis (or annealing) would be confined to the 0.7 kb neo gene repeat and involve the 3′ neo gene on the same chromatid or sister chromatid. For the class 1 LTGC events, repair synthesis would have continued into the non-homologous sequences downstream of the 3′ neo gene repeats (Figure 5A), leading to a conversion tract of ∼3.3 kb. An involvement of the sister chromatid is certain for the class 1a events, since repair synthesis must incorporate an I-SceI site to maintain an intact S2neo gene in the last repeat of the triplication (Figure 5A).
Class 1b events were likely generated by the same mechanism as class 1a events, with the exception that an additional STGC event occurred. If the STGC occurred on one sister chromatid prior to the LTGC event on the other chromatid, an NcoI site would have been copied into the last repeat of the triplication during the LTGC event, rather than an I-SceI site as in the class 1a events. Alternatively, two STGCs could have occurred independently after the LTGC event, leading to conversion of the I-SceI sites to NcoI sites. This seems less likely because it would have required three independent gene conversion events, and no clones were detected with the intermediate genotype of two full-length neo genes with NcoI sites and one with an I-SceI site. Since an I-SceI site was not retained in the final products, it is formally possible that the class 1b events were generated by strand invasion into the 3′ neo gene on the same chromatid, rather than the sister chromatid. However, we do not think intrachromatid invasion is likely for two reasons. First, every class 1b event is associated with a 4.0 kb XhoI–HindIII fragment that could function as a donor of sequence information, and intrachromatid events require no such association. Secondly, strand synthesis could have continued to generate additional repeats, rather than only forming a triplication, yet larger expansions were not observed. Considering that class 1b events are very similar to class 1a events, we believe it is more likely that they are sister chromatid rather than intrachromatid repair events.
The remaining classes of LTGC events, classes 2–6, can be classified more precisely as LTGC/NHEJ events since the LTGC event apparently was associated with an NHEJ event. Only one-ended strand invasions are expected to have initiated each of these events (Figure 5B), since none of these events gave products consistent with two-ended invasion (i.e. a neo gene triplication with the second hygR gene containing a deletion). In each of these classes, repair synthesis from the strand invasion would have continued downstream of the 3′ neo gene but terminated prior to the end of the S2neo gene. Since newly synthesized hygR gene sequences are not homologous to the other end of the broken chromosome, rejoining would have had to occur by NHEJ. Although these junctions have not been characterized at the sequence level, they are structurally similar to the NHEJ junctions observed in LTGC events between non-homologous chromosomes (Richardson et al., 1998; C.Richardson and M.Jasin, submitted). Possibly, these LTGC/NHEJ events only arise because they are products of recombination between short repeats on unequally positioned sister chromatids. Sister chromatid repair events normally would be expected to occur between sequences that are positioned equally and therefore have extensive homology. In this case, the complete identity between the newly synthesized DNA and the original chromatid might always promote homologous annealing. Also, the occurrence of LTGC/NHEJ events might be favored in the SCneo substrate because the donor chromatid may also have a DSB, blocking repair synthesis to the end of the S2neo gene.
Five LTGC events were obtained in which only a single band was present, without the presence of the donor chromatid. This was verified by examining subclones of each of these clones. One of these is a class 1a event (U3-537), which strongly suggests that a donor chromatid was involved in the LTGC event. Possibly, the donor chromatid underwent a DSB that was not repaired, leading to loss of the daughter cell. Alternatively, the daughter cell could have had a slightly slower cell division time, so that its progeny make up only a small portion of cells in the clone. An unequal contribution of daughter cells in some of the clones is indicated by the different band intensities (Figure 2).
Preference for the sister chromatid as a homologous repair template
Our results in mammalian cells are consistent with those previously obtained in S.cerevisiae, which demonstrated that sister chromatids are preferred templates for homologous repair (Kadyk and Hartwell, 1992). In yeast, when sister chromatids are not available (i.e. during the G1 phase), homologs efficiently substitute as templates for repair (Kadyk and Hartwell, 1992). As in yeast, homologs, as well as sequence repeats on heterologs, have been shown to serve as templates for DSB repair in mammalian cells (Moynahan and Jasin, 1997; Richardson et al., 1998). However, in contrast to yeast, the frequency of repair events that use a homolog or heterolog in mammalian cells is two to three orders of magnitude lower than those that use the sister chromatid (Moynahan and Jasin, 1997; Richardson et al., 1998).
An important difference between sister chromatids and homologs is their relative proximity. Sister chromatids are attached to each other by cohesion proteins that are thought to assemble during DNA replication and remain assembled until mitosis (Miyazaki and Orr-Weaver, 1994). Except during meiosis, homologs in mammalian cells generally are not any closer to each other in the nucleus than they are to other chromosomes (Ferguson and Ward, 1992). The smaller nuclear volume that sister chromatids occupy relative to homologs, or even the assemblage of cohesion proteins itself, may promote recombination between sister chromatids. When sister chromatids are not available in mammalian cells, NHEJ may supplant homolog recombination for the repair of DSBs.
DSB-promoted recombination is usually examined following the addition of DNA-damaging agents to cells. Nonetheless, homologous recombination evidently also plays an important role in the repair of spontaneously arising DNA damage. In Escherichia coli and yeast, homologous recombination is thought to be required to repair replication errors, e.g. to restart stalled replication forks (Symington, 1998; Marians, 2000; Rothstein et al., 2000). A similar role for homologous recombination in vertebrate cells is supported by the essential role of Rad51 for cell survival (Lim and Hasty, 1996; Tsuzuki et al., 1996; Sonoda et al., 1998), as well as by the analysis of mammalian homologous repair mutants (Johnson et al., 1999; Moynahan et al., 1999; Pierce et al., 1999), which exhibit a high frequency of chromosomal aberrations in the absence of exogenous DNA-damaging agents (Tebbs et al., 1995; Liu et al., 1998; Shen et al., 1998). Although I-SceI-generated DSBs may not precisely mimic damage that occurs during DNA replication, sister chromatids nevertheless may be expected to be important repair templates for both types of damage. Expansions observed in spontaneous direct repeat recombination have also been attributed to sister chromatid interactions (Bollag and Liskay, 1991), and, when compared, spontaneous recombination between direct repeats has been found to be significantly more frequent than homologous recombination between two chromosomes (Shulman et al., 1995).
Gene conversion unassociated with reciprocal exchange
Experiments analyzing plasmid DSB repair in yeast had initially indicated that mitotic gene conversion was associated with crossing over (Orr-Weaver and Szostak, 1983). However, a number of other studies have concluded that mitotic gene conversion is not usually associated with crossing over, whether in yeast (Jackson and Fink, 1981; Strathern et al., 1982; Aguilera and Klein, 1989; Paques et al., 1998), D.melanogaster (Gloor et al., 1991; Nassif et al., 1994), Ustilago maydis (Ferguson and Holloman, 1996) or mammalian cells (Bollag and Liskay, 1988; Richard et al., 1994; Richardson et al., 1998). This includes a study in yeast utilizing an experimental design similar to that presented here, in which non-crossover LTGC events predominated over reciprocal exchanges, although by only a 2:1 ratio (Kadyk and Hartwell, 1992). Thus, the bias against crossing over in mitotically growing cells appears to extend to sister chromatid recombination.
Despite the fact that SCEs were not detected in our study, SCEs that appear to reflect homologous recombination events (Sonoda et al., 1999; Dronkert et al., 2000) can be detected cytologically. Approximately six SCEs per cell are observed in untreated hamster cells (Pinkel et al., 1985), with the frequency of SCEs increasing in a dose-dependent manner following exposure to a variety of DNA-damaging agents (Perry and Evans, 1975; Carrano et al., 1978). Since agents that create strand breaks, including both ionizing radiation and restriction enzymes, are usually poor inducers of SCE (Perry and Evans, 1975; Solomon and Bobrow, 1975; Morgan et al., 1988), it initially had seemed contradictory that an endonuclease-generated DSB would be a potent inducer of homologous recombination (Rouet et al., 1994b; Liang et al., 1998; Johnson et al., 1999). These two observations are readily reconciled by the experiments presented here, in that most homologous repair events would not lead to SCE. Homologous recombination models that invoke a low level frequency of reciprocal exchange following DSB repair (e.g. Ferguson and Holloman, 1996) may account satisfactorily for the few SCEs that are detected cytologically.
The bias against crossing over observed during interchromosomal homologous recombination has been argued to be an important mechanism by which inappropriate genomic alterations, such as translocations (Richardson et al., 1998) and loss of heterozygosity (Moynahan and Jasin, 1997), are suppressed in mammalian cells. This bias appears to be stronger in D.melanogaster and mammals whose genomes contain a much greater abundance of repetitive elements than in yeast. For example in mammalian cells, crossovers between heterologous chromosomes occur in ≤1% of events and, as shown here for sister chromatids, in ≤3% of events. Given the fact that homologous recombination is a major DSB repair pathway (Liang et al., 1998), frequent reciprocal exchange would lead to genome instability. Although SCEs should not result in genetic alterations, a common mechanism for both chromatid and chromosome mitotic recombination may provide a safeguard against genomic alterations.
Materials and methods
Plasmids and DNA manipulations
The construction of the recombination reporter substrate SCneo was described previously (Johnson et al., 1999), as was the I-SceI expression vector, pCMV3xnls-I-SceI (Rouet et al., 1994a; Donoho et al., 1998). Southern blot analysis was performed using 8 µg of genomic DNA according to standard procedures, with a 1.2 kb XhoI–HindIII fragment containing the complete neo gene.
Construction of cell lines and cell transfections
Construction of the V79 (4-18) hamster cell line was described previously (Johnson et al., 1999). To construct the AA8 (10-4) cell line, the hamster cell line AA8 (ATCC) was electroporated at 250 V/960 µF with the SCneo substrate and plated in non-selective medium. Hygromycin (0.5 mg/ml) was added 24 h later and, after 11 days, colonies were isolated and expanded. The AA8 (10-4) cell line was determined by Southern blotting to contain a single, integrated copy of SCneo. In subsequent electroporations, 1.6 × 107 cells were suspended in phosphate-buffered saline with either 25 µg (frequency analysis) or 100 µg (random clone analysis) of uncut pCMV3xnls-I-SceI. For random clone analysis, single cells were seeded into 96-well tissue culture plates in non-selective media and the presence of a single cell was verified microscopically. Resulting colonies were analyzed by Southern blotting either immediately after colony expansion or after replica plating from 24-well plates ∼16 days after seeding to determine which clones had undergone a gene conversion event. Replicas were produced in non-selective media and in media containing 1 mg/ml G418, and clones that contained G418R cells were expanded from the replica plate containing non-selective media for further analysis. Segregation analysis was performed by plating single cells in non-selective media.
Acknowledgments
Acknowledgements
We thank members of the Jasin laboratory and Scott Keeney for helpful discussions. This work was supported by an NRSA fellowship (GM18640) to R.D.J. and an NIH (GM54688) grant to M.J.
References
- Aguilera A. and Klein,H.L. (1989) Yeast intrachromosomal recombination: long gene conversion tracts are preferentially associated with reciprocal exchange and require the RAD1 and RAD3 gene products. Genetics, 123, 683–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag R.J. and Liskay,R.M. (1988) Conservative intrachromosomal recombination between inverted repeats in mouse cells: association between reciprocal exchange and gene conversion. Genetics, 119, 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag R.J. and Liskay,R.M. (1991) Direct-repeat analysis of chromatid interactions during intrachromosomal recombination in mouse cells. Mol. Cell. Biol., 11, 4839–4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrano A.V., Thompson,L.H., Lindl,P.A. and Minkler,J.L. (1978) Sister chromatid exchange as an indicator of mutagenesis. Nature, 271, 551–553. [DOI] [PubMed] [Google Scholar]
- Colleaux L., d’Auriol,L., Gailbert,F. and Dujon,B. (1988) Recognition and cleavage site of the intron-encoded omega transposase. Proc. Natl Acad. Sci. USA, 85, 6022–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoho G., Jasin,M. and Berg,P. (1998) Analysis of gene targeting and intrachromosomal homologous recombination stimulated by genomic double-strand breaks in mouse embryonic stem cells. Mol. Cell. Biol., 18, 4070–4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dronkert M.L., Beverloo,H.B., Johnson,R.D., Hoeijmakers,J.H., Jasin,M. and Kanaar,R. (2000) Mouse RAD54 affects DNA double-strand break repair and sister chromatid exchange. Mol. Cell. Biol., 20, 3147–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers J., van Steeg,H., de Wit,J., Swagemakers,S.M., Vermeij,M., Hoeijmakers,J.H. and Kanaar,R. (2000) Homologous and non-homologous recombination differentially affect DNA damage repair in mice. EMBO J., 19, 1703–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson D.O. and Holloman,W.K. (1996) Recombinational repair of gaps in DNA is asymmetric in Ustilago maydis and can be explained by a migrating D-loop model. Proc. Natl Acad. Sci. USA, 93, 5419–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson M. and Ward,D.C. (1992) Cell cycle dependent chromosomal movement in pre-mitotic human T- lymphocyte nuclei. Chromosoma, 101, 557–565. [DOI] [PubMed] [Google Scholar]
- Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. American Society of Microbiology Press, Washington, DC. [Google Scholar]
- Gloor G.B., Nassif,N.A., Johnson-Schlitz,D.M., Preston,C.R. and Engels,W.R. (1991) Targeted gene replacement in Drosophila via P element-induced gap repair. Science, 253, 1110–1117. [DOI] [PubMed] [Google Scholar]
- Holmes A.M. and Haber,J.E. (1999) Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell, 96, 415–424. [DOI] [PubMed] [Google Scholar]
- Jackson J.A. and Fink,G.R. (1981) Gene conversion between duplicated genetic elements in yeast. Nature, 292, 306–311. [DOI] [PubMed] [Google Scholar]
- Johnson R.D., Liu,N. and Jasin,M. (1999) Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature, 401, 397–399. [DOI] [PubMed] [Google Scholar]
- Kadyk L.C. and Hartwell,L.H. (1992) Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics, 132, 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoma T. (1997) Stable DNA replication: interplay between DNA replication, homologous recombination and transcription. Microbiol. Mol. Biol. Rev., 61, 212–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasko D., Cavenee,W. and Nordenskjold,M. (1991) Loss of constitutional heterozygosity in human cancer. Annu. Rev. Genet., 25, 281–314. [DOI] [PubMed] [Google Scholar]
- Liang F., Han,M., Romanienko,P.J. and Jasin,M. (1998) Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl Acad. Sci. USA, 95, 5172–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim D.-S. and Hasty,P. (1996) A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol., 16, 7133–7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Lukacsovich,T. and Waldman,A.S. (1999) Multiple pathways of repair of DNA double-strand breaks in mammalian chromosomes. Mol. Cell. Biol., 19, 8353–8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N. et al. (1998) XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol. Cell, 1, 783–793. [DOI] [PubMed] [Google Scholar]
- Marians K. (2000) Replication and recombination intersect. Curr. Opin. Genet. Dev., 10, 151–156. [DOI] [PubMed] [Google Scholar]
- Miyazaki W.Y. and Orr-Weaver,T.L. (1994) Sister-chromatid cohesion in mitosis and meiosis. Annu. Rev. Genet., 28, 167–187. [DOI] [PubMed] [Google Scholar]
- Morgan W.F., Fero,M.L., Land,M.C. and Winegar,R.A. (1988) Inducible expression and cytogenetic effects of the EcoRI restriction endonuclease in Chinese hamster ovary cells. Mol. Cell. Biol., 8, 4204–4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan M.E. and Jasin,M. (1997) Loss of heterozygosity induced by a chromosomal double-strand break. Proc. Natl Acad. Sci. USA, 94, 8988–8993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan M.E., Chiu,J.W., Koller,B.H. and Jasin,M. (1999) Brca1 controls homology-directed repair. Mol. Cell, 4, 511–518. [DOI] [PubMed] [Google Scholar]
- Nassif N., Penney,J., Pal,S., Engels,W.R. and Gloor,G.B. (1994) Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol., 14, 1613–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T.L. and Szostak,J.W. (1983) Yeast recombination: the association between double-strand gap repair and crossing-over. Proc. Natl Acad. Sci. USA, 80, 4417–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F. and Haber,J.E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev., 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F., Leung,W.Y. and Haber,J.E. (1998) Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol. Cell. Biol., 18, 2045–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry P. and Evans,H.J. (1975) Cytological detection of mutagen–carcinogen exposure by sister chromatid exchange. Nature, 258, 121–125. [DOI] [PubMed] [Google Scholar]
- Pierce A.J., Johnson,R.D., Thompson,L.H. and Jasin,M. (1999) XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev., 13, 2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D., Thompson,L.H., Gray,J.W. and Vanderlaan,M. (1985) Measurement of sister chromatid exchanges at very low bromodeoxyuridine substitution levels using a monoclonal antibody in Chinese hamster ovary cells. Cancer Res., 45, 5795–5798. [PubMed] [Google Scholar]
- Richard M., Belmaaza,A., Gusew,N., Wallenburg,J.C. and Chartrand,P. (1994) Integration of a vector containing a repetitive LINE-1 element in the human genome. Mol. Cell. Biol., 14, 6689–6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C., Moynahan,M.E. and Jasin,M. (1998) Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev., 12, 3831–3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder G.S. (1997) Meiotic chromosomes: it takes two to tango. Genes Dev., 11, 2600–2621. [DOI] [PubMed] [Google Scholar]
- Rothstein R., Michel,B. and Gangloff,S. (2000) Replication fork pausing and recombination, or ‘gimme a break’. Genes Dev., 14, 1–10. [PubMed] [Google Scholar]
- Rouet P., Smih,F. and Jasin,M. (1994a) Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc. Natl Acad. Sci. USA, 91, 6064–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet P., Smih,F. and Jasin,M. (1994b) Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell. Biol., 14, 8096–8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent R.G., Brenneman,M.A. and Wilson,J.H. (1997) Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol. Cell. Biol., 17, 267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid C.W. (1996) Alu: structure, origin, evolution, significance and function of one-tenth of human DNA. Prog. Nucleic Acid Res. Mol. Biol., 53, 283–319. [DOI] [PubMed] [Google Scholar]
- Schwacha A. and Kleckner,N. (1995) Identification of double Holliday junctions as intermediates in meiotic recombination. Cell, 83, 783–791. [DOI] [PubMed] [Google Scholar]
- Shen S.X., Weaver,Z., Xu,X., Li,C., Weinstein,M., Chen,L., Guan,X.Y., Ried,T. and Deng,C.X. (1998) A targeted disruption of the murine Brca1 gene causes gamma-irradiation hypersensitivity and genetic instability. Oncogene, 17, 3115–3124. [DOI] [PubMed] [Google Scholar]
- Shinohara A., Ogawa,H., Matsuda,Y., Ushio,N., Ikeo,K. and Ogawa,T. (1993) Cloning of human, mouse and fision yeast recombination genes homologous to RAD51 and recA. Nature Genet., 4, 239–243. [DOI] [PubMed] [Google Scholar]
- Shulman M.J., Collins,C., Connor,A., Read,L.R. and Baker,M.D. (1995) Interchromosomal recombination is suppressed in mammalian somatic cells. EMBO J., 14, 4102–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon E. and Bobrow,M. (1975) Sister chromatid exchanges—a sensitive assay of agents damaging human chromosomes. Mutat. Res., 30, 273–278. [PubMed] [Google Scholar]
- Sonoda E., Sasaki,M.S., Buerstedde,J.M., Bezzubova,O., Shinohara,A., Ogawa,H., Takata,M., Yamaguchi-Iwai,Y. and Takeda,S. (1998) Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J., 17, 598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E., Sasaki,M.S., Morrison,C., Yamaguchi-Iwai,Y., Takata,M. and Takeda,S. (1999) Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell. Biol., 19, 5166–5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern J.N., Klar,A.J., Hicks,J.B., Abraham,J.A., Ivy,J.M., Nasmyth,K.A. and McGill,C. (1982) Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell, 31, 183–192. [DOI] [PubMed] [Google Scholar]
- Symington L.S. (1998) Homologous recombination is required for the viability of rad27 mutants. Nucleic Acids Res., 26, 5589–5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J.W., Orr-Weaver,T.L., Rothstein,R.J. and Stahl,F.W. (1983) The double-strand-break repair model for recombination. Cell, 33, 25–35. [DOI] [PubMed] [Google Scholar]
- Tebbs R.S., Zhao,Y., Tucker,J.D., Scheerer,J.B., Siciliano,M.J., Hwang, M., Liu,N., Legerski,R.J. and Thompson,L.H. (1995) Correction of chromosomal instability and sensitivity to diverse mutagens by a cloned cDNA of the XRCC3 DNA repair gene. Proc. Natl Acad. Sci. USA, 92, 6354–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuzuki T., Fujii,Y., Sakumi,K., Tominaga,Y., Nakao,K., Sekiguchi,M., Matsushiro,A., Yoshimura,Y. and Morita,T. (1996) Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl Acad. Sci. USA, 93, 6236–6340. [DOI] [PMC free article] [PubMed] [Google Scholar]