Abstract
Poly(dA)⋅poly(dT) tracts are common and often found upstream of genes in eukaryotes. It has been suggested that poly(dA)⋅poly(dT) promotes transcription in vivo by affecting nucleosome formation. On the other hand, in vitro studies show that poly(dA)⋅poly(dT) can be easily incorporated into nucleosomes. Therefore, the roles of these tracts in nucleosome organization in vivo remain to be established. We have developed an assay system that can evaluate nucleosome formation in yeast cells, and demonstrated that relatively longer tracts such as A15TATA16 and A34 disrupt an array of positioned nucleosomes, whereas a shorter A5TATA4 tract is incorporated in positioned nucleosomes of yeast minichromosomes. Thus, nucleosomes are destabilized by poly(dA)⋅poly(dT) in vivo in a length-dependent manner. Furthermore, in vivo UV footprinting revealed that the longer tracts adopt an unusual DNA structure in yeast cells that corresponds to the B′ conformation described in vitro. Our results support a mechanism in which a unique poly(dA)⋅ poly(dT) conformation presets chromatin structure to which transcription factors are accessible.
Keywords: chromatin/DNA structure/nucleosome positioning/poly(dA)⋅poly(dT)/yeast
Introduction
Gene expression in eukaryotic cells involves the interaction of proteins with DNA packaged into nucleosomes; the primary structural units of chromatin are composed of a histone octamer and 145 bp DNA. Since the nucleosome limits the access of trans-acting factors to their cognate DNA sites, it is widely accepted that the nucleosome acts as a barrier to transcription. Thus, an alteration of chromatin structure is an essential step for the initiation of transcription. Recent genetic and biochemical studies have shown that chromatin structure is altered by several mechanisms, including chromatin remodeling complexes and the acetylation–deacetylation of histones in living cells (for reviews see Kadonaga, 1998; Kornberg and Lorch, 1999; Travers, 1999; Wolffe and Hayes, 1999).
Nucleosomes are often located at distinct positions in promoter regions and close to cis-acting DNA elements. Nucleosome positioning appears to be influenced by a combination of several factors, including histone–DNA interaction, anisotropy of the DNA structure, boundary formation, chromatin folding and interaction of trans-acting factors with histones (for reviews see Simpson, 1991; Thoma, 1992). Since DNA structures are considered to act as determinants of nucleosome organization, the effects of DNA sequences on nucleosome formation have been extensively studied using in vitro reconstitution from purified histone octamers and DNA such as synthetic polymers, restriction fragments and plasmids. For example, earlier studies found that synthetic homopolymers such as poly(dA)⋅poly(dT) and poly(dG)⋅poly(dC) have inherent resistance to nucleosome formation (Rhodes, 1979; Simpson and Kunzler, 1979; Kunkel and Martinson, 1981). Since homopolymeric An⋅Tn sequences are common in most eukaryotes and are often found in promoters (Struhl, 1985; Schlapp and Rodel, 1990; Thiry-Blaise and Loppes, 1990; Karlin et al., 1993; Behe, 1995), the influence of An tracts on nucleosome organization has been studied in detail by several investigators.
In agreement with the earlier observations, a T14A11 tract blocks nucleosome formation over the human NF1-Alu element in vitro (Englander and Howard, 1996). On the other hand, Losa et al. (1990) found that an An⋅Tn tract, T3CCT6CT5GCT5CT7, is located in a nucleosome-free region in the yeast DED1 promoter in vivo, whereas it can be incorporated in the nucleosome core in vitro. Similarly, human genomic clones containing a longer An tract (31, 41 and 69 bp in length) with other mixed sequences readily wrap around histone octamers in vitro (Fox, 1992). Other studies also revealed that poly(dA)⋅poly(dT) sequences can be reconstituted into nucleosomes in vitro (Hayes et al., 1991; Puhl et al., 1991; Schieferstein and Thoma, 1996; Brown and Fox, 1998) and that the free energy of nucleosome formation is virtually identical to that of the nucleosome with a heterologous sequence DNA (Puhl et al., 1991). Furthermore, poly(dA)⋅poly(dT) forms nucleosomes more strongly as the temperature is increased (Puhl and Behe, 1995; Mahloogi and Behe, 1997), suggesting that the DNA conformation adopted at elevated temperatures facilitates nucleosome formation. Thus, the formation and stability of nucleosomes in poly(dA)⋅poly(dT) tracts remain to be elucidated.
Iyer and Struhl (1995) showed that an An tract of 17 bp or more in the yeast HIS3 promoter stimulates transcription by improving accessibility to the promoters in vivo, suggesting that An tracts affect local nucleosome formation. Consistent with the above results, Filetici et al. (1998) reported that the HIS3 upstream region containing the An tract (T4CAT11) and a Gcn4p binding site is nucleosome-free. Thiele and his colleagues (Zhu and Thiele, 1996; Koch and Thiele, 1999) showed that a naturally occurring A16 element in the Candida glabrata AMT1 gene distorts the nucleosomal DNA within a positioned nucleosome to facilitate transcription factor binding in vivo. These studies shed light on the functional importance of poly(dA)⋅poly(dT) in chromatin structure, but the roles of DNA structures in nucleosome organization in vivo are not yet well understood.
We have developed an assay system with which to examine the effects of DNA sequences on the formation and organization of nucleosomes in yeast minichromosomes in vivo. We found that longer An⋅Tn sequences disrupt an array of positioned nucleosomes, but shorter sequences are included. Furthermore, in vivo UV photofootprinting revealed that the longer An⋅Tn sequences adopt an unusual DNA conformation in yeast cells, supporting the notion that a unique DNA structure prevents wrapping around histone octamers in vivo.
Results
Experimental design
Previous studies indicate that the yeast α2/Mcm1p precisely positions nucleosomes adjacent to the α2 operator in genomic a-specific genes as well as in minichromosomes (Roth et al., 1990, 1992; Shimizu et al., 1991; Ganter et al., 1993), together with corepressors Ssn6p-Tup1p (Cooper et al., 1994). Tup1p directly interacts with deacetylated histones H3 and H4 (Edmondson et al., 1996), indicating that the mechanism of the positioning involves interaction of the multiple repressor complex with histones.
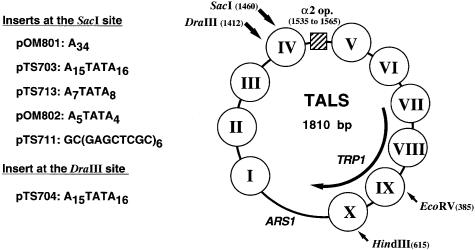
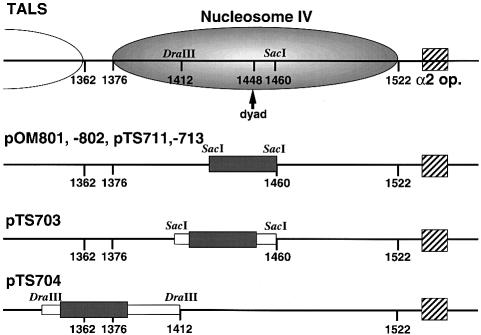
Roth et al. (1990) constructed the TALS plasmid by inserting 356 bp of the STE6 promoter, including the α2 operator, into the EcoRI site of the TRP1/ARS1 plasmid. Figures 1 and 2 show the chromatin structure of the TALS minichromosomes and DNA inserts used in this study. The α2 operator is located in the region of 1535–1565 m.u. (map units). Figure 2 indicates that the main location of nucleosome IV is between 1376 and 1522 m.u. and that the pseudo-dyad axis is mapped at 1448 m.u., as determined by high resolution mapping of micrococcal nuclease (MNase) cleavage sites (Shimizu et al., 1991; Kladde and Simpson, 1994). The SacI site is located at 1460 m.u., corresponding to 63 bp from the edge of nucleosome IV at 1522 m.u., whereas the DraIII site at 1412 m.u. corresponds to 111 bp from the edge. Thus, Figure 2 shows that when DNA sequences were inserted into the SacI site, the inserts lay around the dyad of nucleosome IV (pOM801, -802, pTS711, -713 and -703). On the other hand, when a sequence was inserted into the DraIII, it lay proximal to the linker region between nucleosomes III and IV (pTS704). These TALS derivatives were introduced into yeast α cells to assemble chromatin in vivo. The chromatin structure of the minichromosomes was examined by limited digestion of the isolated nuclei with MNase, which preferentially cuts the linker DNA region between neighboring nucleosomes.
Fig. 1. Chromatin structure of TALS minichromosomes in yeast α cells and DNA inserts used in this study. TALS contains the α2 operator (indicated by the hatched box) from the STE6 promoter at the EcoRI site of TRP1/ARS1 plasmid, and the location of nucleosomes in TALS was determined previously (Roth et al., 1990). DNA inserts were cloned into the SacI site of TALS, corresponding to near the pseudo-dyad axis of nucleosome IV, or into the DraIII site corresponding to the linker region between nucleosomes III and IV.
Fig. 2. Locations of the positioned nucleosome IV, α2 operator and poly(dA)⋅poly(dT) inserts in TALS and its derivative minichromosomes as described (Shimizu et al., 1991; Kladde and Simpson, 1994). The SacI and DraIII sites are at 1460 and 1412 m.u. (map units), respectively, in nucleosome IV as shown by the shaded ellipse. The α2 operator and poly(dA)⋅poly(dT) tracts are shown as hatched and shaded boxes, respectively. The polylinker attached with the A15TATA16 insert is indicated by white boxes (see Materials and methods).
High resolution mapping of MNase cleavage sites in the nucleosome IV region
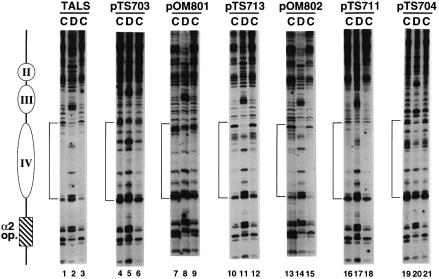
Figure 3 shows high resolution mapping of MNase cleavage sites in the region of nucleosome IV in TALS and their derivative minichromosomes. In the TALS chromatin, a region of ∼140 bp adjacent to the α2 operator (shown by a bracket in Figure 3, lanes 1 and 3) is largely protected against MNase digestion, compared with the digestion of naked DNA (lane 2), confirming the precise translational positioning of nucleosome IV (Shimizu et al., 1991).
Fig. 3. Primer extension analysis of MNase cleavage sites in the nucleosome IV region of TALS and its derivative minichromosomes. Plasmids pTS703, pOM801, pTS713, pOM802 and pTS711 contain A15TATA16, A34, A7TATA8, A5TATA4 and GC(GAGCTCGC)6, respectively, at the SacI site of TALS. Plasmid pTS704 contains A15TATA16 at the DraIII site. Chromatin (in isolated nuclei) (lanes labeled C) and naked DNA (lanes labeled D) were digested at 37°C for 10 min with various concentrations of micrococcal nuclease as follows: 2.5 U/ml (lanes 1, 4, 7, 10, 13, 16 and 19), 1.25 U/ml (lanes 3, 6, 9, 12,15,18 and 21) and 0.05 U/ml (lanes 2, 5, 8, 11, 14, 17 and 20), as described (Shimizu et al., 1991). The nucleosome IV regions are shown on the left of each gel by brackets. The locations of nucleosomes II to IV and the α2 operator are schematically shown to the left of the gel.
A mixed sequence, GC(GAGCTCGC)6 (50 bp), was inserted at the SacI site, near the pseudo-dyad axis of nucleosome IV (pTS711) for a control sequence. Figure 3 (lanes 16–18) shows that the nucleosome IV region in pTS711 chromatin was protected from MNase digestion, and that the MNase cleavage profile is very similar to that of vector TALS chromatin (lanes 1–3). We also analyzed the effect of (CA)17⋅(TG)17 (34 bp) insertion at the SacI site and found no alterations in the nucleosome positioning (data not shown). These results indicate that DNA insertions per se do not affect nucleosome formation. This is consistent with the fact that the nucleosomes are positioned independently of DNA sequences flanked by the α2 operator. At least seven such examples of precise positioning of nucleosomes adjacent to the α2 operator have been reported: three a-cell-specific genes and four minichromosomes (Roth et al., 1990; Simpson, 1990; Shimizu et al., 1991; Ganter et al., 1993; Simpson et al., 1993; Patterton and Simpson, 1994).
In contrast to the control insert, relatively longer tracts such as A15TATA16 and A34 (pTS703 and pOM801, respectively) remarkably affected nucleosome formation. That is, the nucleosome IV region became hypersensitive to MNase and the cleavage profile of chromatin was the same as that of naked DNA (Figure 3, lanes 4–6 and 7–9, respectively). This demonstrated that the insertion of longer An⋅Tn tracts disrupts nucleosome positioning by the α2 repressor in vivo. The MNase cleavage sites in the nucleosome IV region of pTS713 chromatin containing the A7TATA8 insert were partially protected, and the region near the tract became more accessible to MNase (Figure 3, lanes 10–12), compared with the vector TALS or the control pTS711. On the other hand, a shorter A5TATA4 sequence that matched the ARS (autonomously replicating sequence) consensus sequence (ACS) in Saccharomyces cerevisiae was incorporated into nucleosome IV (pOM802; Figure 3, lanes 13–15). These results demonstrate that An⋅Tn tracts disrupt or distort the nearby nucleosome structures and that a certain length is required to disrupt nucleosome positioning.
Furthermore, we examined the effect of the location of the destabilizing sequence (A15TATA16) on nucleosome formation by inserting the sequence into the DraIII site (pTS704; Figure 2) to cover the region corresponding to the linker between nucleosomes III and IV. High resolution mapping of pTS704 (Figure 3, lanes 19–21) revealed similar cleavage profiles between naked DNA and chromatin samples in the region of nucleosomes III and IV, indicating that nucleosome positioning is disrupted by the insert. This result implies that longer An tracts inserted in the linker region as well as in the center of the nucleosome prevent wrapping of histone octamers.
Nucleosome positioning in the minichromosomes analyzed by indirect end-labeling
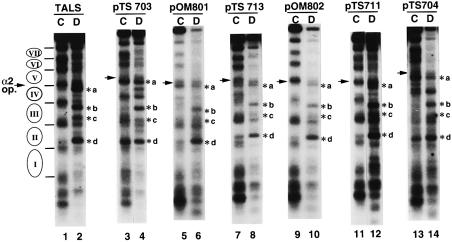
The chromatin structures of minichromosomes were also analyzed by indirect end-labeling and the results are shown in Figure 4. For the vector TALS, the four MNase cleavage sites (indicated by *a, *b, *c and *d) in the naked DNA (Figure 4, lane 2) were protected in the chromatin sample (lane 1). Also, the cleavage sites with equal spacing (140–150 bp) in TALS chromatin were enhanced (Figure 4, lane 1). This result is in good agreement with published reports (Roth et al., 1990, 1992), confirming that the TALS chromatin is assembled with precisely and stably positioned nucleosomes. The same profile of MNase cleavage sites was obtained for the pTS711 minichromosomes containing the control sequence GC(GAGCTCGC)6 (Figure 4, lane 11), indicating that DNA insertion per se does not affect nucleosome positioning in the minichromosomes.
Fig. 4. Chromatin structure of TALS and its derivatives analyzed by indirect end-labeling. Plasmids pTS703, pOM801, pTS713, pOM802 and pTS711 contain A15TATA16, A34, A7TATA8, A5TATA4 and GC(GAGCTCGC)6, respectively, at the SacI site of TALS. Plasmid pTS704 contains A15TATA16 at the DraIII site. Chromatin (in isolated nuclei) (lanes labeled C) and naked DNA (lanes labeled D) were digested at 37°C for 10 min with micrococcal nuclease as follows: 2.5 U/ml (odd-numbered lanes) and 0.05 U/ml (even-numbered lanes). The samples were digested with EcoRV and resolved by electrophoresis on 1.2% agarose gel, then the MNase cleavage sites were detected by indirect end-labeling using the EcoRV–HindIII fragment (385–615 m.u. in TALS; see Figure 1) as a probe (Roth et al., 1990). The locations of nucleosomes I to VII and the α2 operator are shown to the left of the gel. The four cleavage sites of TALS in naked DNA, but not in chromatin, are indicated by *a, *b, *c and *d.
In contrast, the two bands (*a and *d) that were protected in TALS chromatin were evident in the pTS703 chromatin (A15TATA16) (Figure 4, compare lanes 1 and 2 with lanes 3 and 4). Furthermore, the discrete bands in TALS chromatin that corresponded to the linker regions between nucleosomes II and IV became broader or disappeared in pTS703 chromatin (Figure 4, lane 3). Similarly, the band *a was present in pOM801 chromatin (A34; Figure 4, lane 5) and the bands corresponding to the linker regions between nucleosomes II and IV were broader or lost. These results indicate that nucleosome positioning is disrupted by the longer inserts, not only in nucleosome IV but also in nucleosomes II and III. For pTS713 containing an A7TATA8 insert (Figure 4, lanes 7 and 8), the MNase cleavage pattern of chromatin sample was similar to that of TALS chromatin, although the characteristic band *a in naked DNA was detected to some extent in the chromatin sample, and the band corresponding to the linker regions between positioned nucleosomes III and IV was broader than that in TALS chromatin. This implies that nucleosome positioning in pTS713 chromatin is less stable than that in TALS chromatin.
The MNase cleavage profile of the minichromosomes containing A5TATA4 (pOM802; Figure 4, lane 9) was similar to that of vector TALS (lane 1) and the control pTS711 (lane 11), indicating that the shorter tract did not significantly affect the formation of positioned nucleosomes.
The effect of inserting A15TATA16 into the linker DNA region (pTS704; Figure 4, lanes 13 and 14) was also analyzed by indirect end-labeling. The cleavage profile of pTS704 chromatin was quite different from that of TALS chromatin (Figure 4, compare lanes 1 and 13). Two characteristic bands, *a and *d, were clear, while no obvious MNase cut sites were observed in the region containing bands *b and *c in pTS704 chromatin (Figure 4, lane 13). Thus, an insertion into the linker region also disrupted the positioning of nucleosomes II to IV in the minichromosomes. These results are consistent with those of the high resolution mapping described above.
Nucleosome organization in minichromosomes analyzed by nucleosome repeat assay
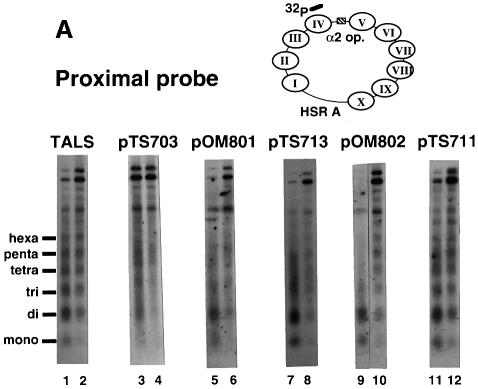
We further examined whether or not the regularity of the nucleosome array is disrupted by the An⋅Tn tracts using the nucleosome repeat assay (Southern blots of nucleosome ladders produced by MNase digestion followed by hybridization with a unique oligonucleotide probe). Figure 5A shows the results of nucleosome repeat analysis detected using a proximal probe with a complementary sequence (1502–1473 m.u.) flanked by the inserts in the nucleosome IV region. We examined MNase digests corresponding to the mono-nucleosome up to the hexa-nucleosomes, as observed for TALS chromatin (Figure 5A, lanes 1 and 2). The nucleosome ladders of pTS711 and pOM802, which contain the control GC(GAGCTCGC)6 insert and the shorter A5TATA4 tract, respectively (Figure 5A, lanes 9–12), were similar to that of the vector TALS (Figure 5A, lanes 1 and 2). Thus, this result indicates that these inserts were incorporated into the nucleosome array with regular spacing.
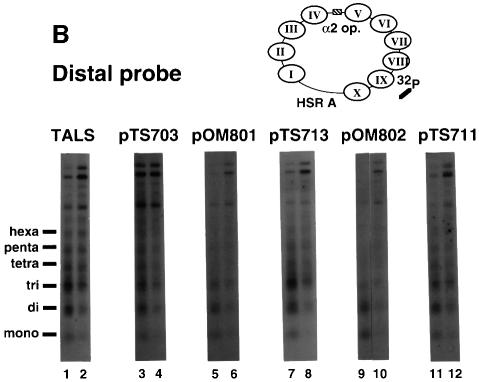
Fig. 5. Nucleosome repeat analysis of TALS and its derivative minichromosomes. Plasmids pTS703, pOM801, pTS713, pOM802 and pTS711 contain A15TATA16, A34, A7TATA8, A5TATA4 and GC(GAGCTCGC)6, respectively, at the SacI site of TALS. Chromatin (in isolated nuclei) was digested at 37°C for 10 min with micrococcal nuclease: 2.5 U/ml (odd-numbered lanes) and 1.25 U/ml (even-numbered lanes). The MNase digest samples were resolved by 1.3% agarose gel electrophoresis. (A) Southern blots of MNase-digested chromatin were probed using a 32P-labeled oligonucleotide with a complementary sequence adjacent to the SacI site in nucleosome IV (proximal probe, top of panel; see Materials and methods). (B) After stripping the proximal probe off, the blots were re-probed using a 32P-labeled oligonucleotide with a sequence complementary to the nucleosome IX region (distal probe, top of panel; see Materials and methods).
Nucleosome ladders for more than tri-nucleosomes were not clear for the pTS713 (A7TATA8) chromatin (Figure 5A, lanes 7 and 8), indicating less periodicity of nucleosome spacing compared with the vector TALS. Furthermore, the ladders became much more smeared for pTS703 (A15TATA16) and pOM801 (A34) (Figure 5A, lanes 3–6), indicating that the nucleosome IV regions near the inserts completely lost spacing periodicity. This implies that the regularity of the nucleosome array is disrupted by the longer An inserts.
After stripping the proximal probe off, the blots were re-hybridized with a distal probe that has a sequence complementary to a region within nucleosome IX (409–445 m.u.) (Figure 5B). Figure 5B shows that ladders from the minichromosomes appeared nearly identical to the canonical nucleosome ladders, demonstrating that the perturbation of nucleosome organization does not propagate into the distal region.
Unusual conformation of An⋅Tn tracts detected by in vivo UV photofootprinting
Poly(dA)⋅poly(dT) adopts at least two double-helical conformations (e.g. Premilat and Albiser, 1997), depending on conditions such as temperature and relative humidity. Lyamichev (1991) reported that the structural transition of An⋅Tn tracts from an unusual structure to a structure close to B-DNA can be detected in vitro by UV photofootprinting as a function of temperature. Thus, we used in vivo UV footprinting to determine whether or not the An⋅Tn tracts in minichromosomes adopt the same structure as that in vitro. Intact yeast cells harboring the TALS derivatives were irradiated with UV light at 254 nm, and thymine dimerization at An⋅Tn tracts was analyzed by primer extension mapping as described (Axelrod and Majors, 1989; Murphy et al., 1993; Shimizu et al., 1997, 1998).
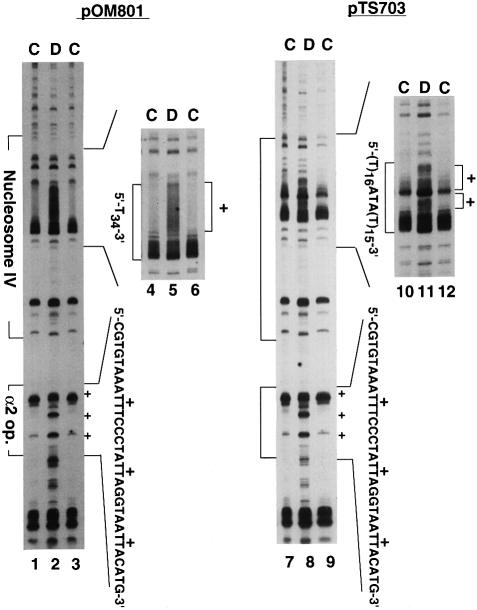
Figure 6 shows the in vivo UV photofootprints of inserts A34 (pOM801) and A15TATA16 (pTS703). Eight to ten thymine residues from the 3′-end exhibited high photoreactivity in the T34 tract, whereas thymine bases in the other 2/3 region of the tract were protected in pOM801 chromatin (Figure 6, lanes 1, 3, 4 and 6). Similarly, the photoreactivity of naked DNA decreased gradually from the middle of the tract to the 5′-end, but the degree of protection was much weaker (Figure 6, lanes 2 and 5) compared with samples of intact cells (lanes 1, 3, 4 and 6). The footprints were also similar for the A15TATA16 insert, as seen in lanes 7–12 of Figure 6. Yields of thymine dimer formation were maximal at regions near the 3′-end of both T16 and T15 tracts in the T16ATAT15 insert, whereas the 5′-half of each of the T16 and T15 tracts was protected. Thus, the ATA sequence at the center interrupted the protection associated with the Tn stretch. The UV foot prints in the yeast cells were in good agreement with the findings of Lyamichev (1991): the yield of thymine dimer formation is maximal at the 3′-ends of the Tn tracts, whereas the photoreactivity of thymine residues is repressed at the 5′-sides. Thus, A34 and A15TATA16 tracts predominantly form an unusual conformation in yeast cells. The unique protection at the 5′-side was not observed in vivo for a shorter insert, T4ATAT5 in pOM802 (data not shown), indicating that a certain length of An⋅Tn tract is required to stabilize the unusual conformation.
Fig. 6. In vivo UV photofootprinting of the T34 and T16ATAT15 strands in pOM801 and pTS703 minichromosomes. Intact cells (lanes labeled C) and naked DNA (lanes labeled D) were irradiated at 254 nm with different UV dosages as follows: 500 mJ (lanes 1, 4, 7 and 10), 750 mJ (lanes 3, 6, 9 and 12) and 60 mJ (lanes 2, 5, 8 and 11), as described (Shimizu et al., 1997, 1998). The nucleosome IV region and the α2 operator are shown by brackets on the left of the gel. The sequences of T34 and T16ATAT15 tracts and the α2 operator are shown to the side of gel. + indicates the sites protected from UV irradiation in cells.
It should be noted that the pyrimidine dimer formation was also protected in the α2 operator in intact cells, and its profile was identical to the in vivo UV footprints of α2/Mcm1p binding (Murphy et al., 1993), indicating that α2/Mcm1p proteins are present in these minichromosomes. Thus, destablization of nucleosomes by An tracts is not due to the loss of α2/Mcm1p binding. In other words, nucleosomes cannot be positioned near An tracts, even in the presence of α2/Mcm1p.
Discussion
We described an assay system that can evaluate nucleosome formation in vivo using well defined yeast minichromosomes (Roth et al., 1990; Shimizu et al., 1991). Similar approaches have examined the effects of transcription factor binding on nucleosomal DNA (Morse et al., 1992; Morse, 1993). The key advantages in this system are as follows. The effect of inserts at specific locations, either at the center or at linker regions in nucleosomes can be examined, and the constructed plasmids are physiologically and naturally assembled into chromatin structures in living yeast cells. Thus, potential artifacts of chromatin assembly arising from reconstitution conditions such as high concentrations of salts and/or urea etc. can be eliminated. We used this system to show the effects of poly(dA)⋅poly(dT) sequences on the formation of positioned nucleosome in vivo.
We examined the chromatin structures of minichromosomes using three different assays: (i) primer extension mapping of positioned nucleosome IV at base-pair resolution; (ii) nucleosome locations in minichromosomes by indirect end-labeling; and (iii) nucleosome spacing by nucleosome repeat analysis. These results were all consistent in terms of the effects of An⋅Tn tracts on nucleosome organization, and led to the following conclusions. Relatively longer tracts such as A15TATA16 and A34 excluded the positioned nucleosome and a moderate length of tract, A7TATA8, was incorporated into a positioned nucleosome, but the nucleosomal DNA near the insert appeared to become more accessible to MNase compared with the control minichromosomes. Finally, a shorter tract (A5TATA4) as well as a mixed control sequence [GC(GAGCTCGC)6] did not affect the formation of positioned nucleosomes. Because in vivo UV footprints indicated occupancy of the α2 operator, the changes of chromatin structure are not due to the loss of α2/Mcm1p binding in these minichromosomes.
Nucleosome exclusion by longer An⋅Tn tracts would be accounted for by occupancy of the tracts by a specific DNA binding protein, or by a non-canonical DNA structure that inhibits histone binding. In S.cerevisiae, ARS1 lies on the nucleosome-free region between two positioned nucleosomes in yeast chromosome IV (Diffley and Cocker, 1992) as well as in TRP1/ARS1 minichromosomes (Thoma et al., 1984; Thoma, 1986; see also Figure 1). The ACS is a 12 bp poly(dA)⋅poly(dT)-rich sequence [5′-(T/A)TTTA(C/T)(A/G)TTT(T/A)(T/C/G)-3′] and the ORC (origin recognition complex) binds to the ACS (for review see Rowley et al., 1994). However, we showed that the inserted ACS is incorporated into the positioned nucleosome in pOM802 chromatin. Thus, ACS alone does not establish a nucleosome-free region. Another candidate is Dat1p (or Datin protein), the only known yeast poly(dA)⋅poly(dT) binding protein, which recognizes at least consecutive An⋅Tn tracts of 10 bp and functions as a repressor (Winter and Varshavsky, 1989; Reardon et al., 1995). However, our in vivo UV footprints argue against this notion. The footprints for A15TATA16 and A34 indicated enhanced thymine dimer formation in the region near the 3′-end of the Tn tracts and protection in the other 5′-half. This asymmetry is the same as that of an unusual conformation of Tn tract at lower temperatures in vitro, as shown by Lyamichev (1991). Also, Iyer and Struhl (1995) reported that dA⋅dT tracts stimulate transcription from the yeast HIS3 promoter, and that several lines of evidence argue against the idea that specific proteins binding to poly(dA)⋅poly(dT) play a positive role in transcription due to increasing accessibility to promoters. Taken together, we conclude that the unusual conformation of An⋅Tn tracts excludes histones from binding, rather than the occupancy of specific binding proteins.
The transition of poly(dA)⋅poly(dT) DNA fiber proceeds from the B′ to the B* structure (two defined conformations of the double helix) at temperatures >30°C and a relative humidity of ∼80% (e.g. Premilat and Albiser, 1997). The B′ conformation is rigid and heteronomous with two sugar–phosphate geometries and has a narrow minor groove (Alexeev et al., 1987; Nelson et al., 1987; Park et al., 1987). On the other hand, the B* structure is a member of the B-DNA family with single sugar–phosphate geometry (Premilat and Albiser, 1997). Thus, the unusual conformation of the Tn tract detected by UV photofootprinting at lower temperatures (Lyamichev, 1991) is most likely to be the B′ conformation. The strong protections in the 5′-half may be attributed to the fact that the minor groove of An⋅Tn tracts narrows from the 3′ towards the 5′-end of Tn stretches (Alexeev et al., 1987; Nelson et al., 1987; Park et al., 1987). Schieferstein and Thoma (1996) analyzed cyclobutane pyrimidine dimer formation in a positioned nucleosome containing a Tn-rich sequence (T3CCT6CT5GCT5CT7) from the yeast DED1 promoter. They showed that most of the Tn stretches formed an unusual conformation in the DNA solution that disappeared upon folding in nucleosomes at 37°C in vitro. Thus, our results indicate that the predominant formation of the B′ conformation in living yeast cells prevents DNA wrapping around histone octamers. This is also consistent with the observation that poly(dA)⋅poly(dT) forms nucleosomes in vitro much more effectively at higher temperatures (Puhl and Behe, 1995), which then stabilize the B-DNA family conformation.
When the size of the An tract is moderate, the region near the A7TATA8 insert is more accessible to MNase within the positioned nucleosome. This result may correspond to the observations of Thiele and colleagues (Zhu and Thiele, 1996; Koch and Thiele, 1999). They showed that an A16 element distorted the DNA structure in a positioned nucleosome, facilitating nucleosomal access to transcription factors in the C.glabrata metal response promoter. We consider that the dynamic changes of the An⋅Tn structure between the unusual conformation and the B-form family may distort the local nucleosome structure.
Regarding the biological implications of the An⋅Tn tracts, analyses of S.cerevisiae chromosome III have shown that ∼25% of yeast genes contain An⋅Tn tracts n ≥ 10 bp (Karlin et al., 1993; Behe, 1995; Iyer and Struhl, 1995; Reardon et al., 1995). We extended this type of analysis, since a database of the completed yeast genome sequence is now available. Consistent with earlier reports, An⋅Tn tracts n ≥ 10 bp are found in 2495 sites in the yeast genome, among which 1760 are located within 500 bp of the untranslated region of the open reading frame (ORF) (6183 ORFs >100 amino acids long were reported). Longer An⋅Tn tracts (n ≥ 20 bp) are found in 130 sites in the genome, and 98 of these are located within 500 bp upstream of the ORF. Although many of the loci mapped are hypothetical ORFs, the longer An tracts have been mapped in many housekeeping genes such as metabolic enzymes and ribosomal proteins. This supports the notion that poly(dA)⋅poly(dT) modulates chromatin structures to make transcription factors accessible to the promoters of constitutively expressed genes.
In summary, we showed that yeast minichromosomes provide a unique system with which to examine the effect of specific DNA sequences and hence their intrinsic DNA structures on nucleosome organization in vivo. We found that poly(dA)⋅poly(dT) tracts disrupt or at least modulate nucleosome organization in yeast. Using this assay, we are currently extending these studies to other specific DNA sequences associated with non-B-DNA structures such as Z-DNA, cruciforms and triplet repeat sequences. The effects of these DNA structures on chromatin organization in vivo will be published elsewhere.
Materials and methods
Plasmids and strains
The TALS-pUC19 plasmid was provided by Drs S.Y.Roth and R.T.Simpson. To manipulate plasmid construction, TALS linearized by HindIII digestion was inserted into the HindIII site of pBR322ΔRI. Plasmid pBR322ΔRI was constructed as follows: pBR322 was digested with EcoRI, then the site was filled-in with Klenow fragment and dNTPs, and ligated. A set of oligonucleotides synthesized chemically was annealed and ligated with the vector TALS-pBR322ΔRI digested with SacI to construct pTS711, -713, pOM801 and -802. To construct pTS703, oligonucleotides A15TATA16 and T16ATAT15 were annealed and cloned into the SmaI site of a pRS424 derivative, forming pKB103, and then pKB103 was digested with BamHI and EcoRI to obtain the fragment containing A15TATA16. The 5′-overhang of the A15TATA16 fragment was filled-in, ligated with the SacI linker, digested with SacI and cloned into the SacI site of the TALS-pBR322ΔRI, forming pTS703. To construct pTS704, pKB103 was digested with EcoRI and NotI, ligated with a DraIII adapter and cloned into the DraIII site of TALS-pBR322ΔRI, forming pTS704. All inserts were verified by DNA sequencing. The location of the inserts is schematically shown in Figure 2.
TALS-pBR322ΔRI and its derivatives containing test DNA inserts were digested with HindIII to eliminate the pBR322 backbone. TALS or TALS derivative plasmids were then isolated, ligated and introduced into the yeast strain FY24 (MATα ura3-52 trp1Δ63 leuΔ21).
Analysis of chromatin structure
Yeast cells harboring plasmids were cultured in SD medium (0.67% yeast nitrogen base without amino acids, 2% glucose) supplemented with 0.02% uracil and 1% leucine until the OD600 reached ∼0.7. Nuclei were isolated and MNase digestion proceeded as described (Shimizu et al., 1991; see legends to Figures 3–5). Cleavage sites for MNase were analyzed by primer extension mapping using the primer MS-2 (1735–1701 m.u. in TALS) (Shimizu et al., 1991) and indirect end-labeling (Thoma et al., 1984; Thoma, 1986; Roth et al., 1990). Nucleosome spacing was analyzed using the nucleosome repeat assay (Cavalli and Thoma, 1993; Becker et al., 1994). Oligonucleotides 5′-GAAAACTGTTGCATTATTGCCGTTCATCATTTTCGA-3′ (1502–1473 m.u.) and 5′-GAGTCGTGGCAAGAATACCAAGAGTTCCTCGGTTTGC-3′ (409–445 m.u.) were used as proximal and distal probes, respectively.
In vivo UV photofootprinting
In vivo UV photofootprinting was performed as described (Axelrod and Majors, 1989; Shimizu et al., 1997, 1998). The sites of UV photoproducts were analyzed by primer extension using the primer MS-2 (Murphy et al., 1993).
Acknowledgments
Acknowledgements
We thank Drs R.T.Simpson and S.Y.Roth for providing the TALS plasmid and yeast strains. We also thank I.Kinebuchi-Nakamura, Y.Naganuma, Y.Fan and K.Sato-Kubo for plasmid construction. M.S. thanks Dr Susumu Hirose for helpful advice and his encouragement in the course of this work. This work was supported by a grant-in-aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan to M.S.
References
- Alexeev D.G., Lipanov,A.A. and Skuratovskii,I.Ya. (1987) Poly(dA)⋅ poly(dT) is a B-type double helix with a distinctively narrow minor groove. Nature, 325, 821–823. [DOI] [PubMed] [Google Scholar]
- Axelrod J.D. and Majors,J. (1989) An improved method for photofootprinting yeast genes in vivo using Taq polymerase. Nucleic Acids Res., 17, 171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker P.B., Tsukiyama,T. and Wu,C. (1994) Chromatin assembly extracts from Drosophila embryos. Methods Cell Biol., 44, 207–223. [DOI] [PubMed] [Google Scholar]
- Behe M.J. (1995) An overabundance of long oligopurine tracts occurs in the genome of simple and complex eukaryotes. Nucleic Acids Res., 23, 689–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P.M. and Fox,K.R. (1998) DNA triple-helix formation on nucleosome-bound poly(dA)⋅poly(dT) tracts. Biochem. J., 333, 259–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli G. and Thoma,F. (1993) Chromatin transitions during activation and repression of galactose-regulated genes in yeast. EMBO J., 12, 4603–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J.P., Roth,S.Y. and Simpson,R.T. (1994) The global transcriptional regulators, SSN6 and TUP1, play distinct roles in the establishment of a repressive chromatin structure. Genes Dev., 8, 1400–1410. [DOI] [PubMed] [Google Scholar]
- Diffley J.F. and Cocker,J.H. (1992) Protein–DNA interactions at a yeast replication origin. Nature, 357, 169–172. [DOI] [PubMed] [Google Scholar]
- Edmondson D.G., Smith,M.M. and Roth,S.Y. (1996) Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev., 10, 1247–1259. [DOI] [PubMed] [Google Scholar]
- Englander E.W. and Howard,B.H. (1996) A naturally occurring T14A11 tract blocks nucleosome formation over the human neurofibromatosis type 1 (NF1)-Alu element. J. Biol. Chem., 271, 5819–5823. [DOI] [PubMed] [Google Scholar]
- Filetici P., Aranda,C., Gonzalez,A. and Ballario,P. (1998) GCN5, a yeast tanscriptional coactivator, induced chromatin reconfiguration of HIS3 promoter in vivo. Biochem. Biophys. Res. Commun., 242, 84–87. [DOI] [PubMed] [Google Scholar]
- Fox K.R. (1992) Wrapping of genomic poly dA⋅poly dT tracts around nucleosome core particles. Nucleic Acids Res., 20, 1235–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganter B., Tan,S. and Richmond,T.J. (1993) Genomic footprinting of the promoter regions of STE2 and STE3 genes in the yeast Saccharomyces cerevisiae. J. Mol. Biol., 234, 975–987. [DOI] [PubMed] [Google Scholar]
- Hayes J.J., Bashkin,J., Tullius,T.D. and Wolffe,A.P. (1991) The histone core exerts a dominant constraint on the structure of DNA in a nucleosome. Biochemistry, 30, 8434–8440. [DOI] [PubMed] [Google Scholar]
- Iyer V. and Struhl,K. (1995) Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J., 14, 2570–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga J.T. (1998) Eukaryotic transcription: an interlaced network of transcription and chromatin-modifying machines. Cell, 92, 307–313. [DOI] [PubMed] [Google Scholar]
- Karlin S., Blaisdell,B.E., Sapolsky,R.J., Cardon,L. and Burge,C. (1993) Assessments of DNA inhomogeneties in yeast chromosome III. Nucleic Acids Res., 21, 703–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kladde M.P. and Simpson,R.T. (1994) Positioned nucleosomes inhibit Dam methylation in vivo. Proc. Natl Acad. Sci. USA, 91, 1361–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch K.A. and Thiele,D.J. (1999) Functional analysis of a homopolymeric (dA–dT) element that provides nucleosome access to yeast and mammalian transcription factors. J. Biol. Chem., 274, 23752–23760. [DOI] [PubMed] [Google Scholar]
- Kornberg R.D. and Lorch,Y. (1999) Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell, 98, 285–294. [DOI] [PubMed] [Google Scholar]
- Kunkel G.R. and Martinson,H.G. (1981) Nucleosomes will not form on double-stranded RNA or over poly(dA)⋅poly(dT) tracts in recombinant DNA. Nucleic Acids Res., 9, 6869–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losa R., Omari,S. and Thoma,F. (1990) Poly(dA)⋅poly(dT) rich sequences are not sufficient to exclude nucleosome formation in a constitutive yeast promoter. Nucleic Acids Res., 18, 3495–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyamichev V. (1991) Unusual conformation of (dA)n⋅(dT)n-tracts as revealed by cyclobutane thymine–thymine dimer formation. Nucleic Acids Res., 19, 4491–4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahloogi H. and Behe,M.J. (1997) Oligoadenosine tracts favor nucleosome formation. Biochem. Biophys. Res. Commun., 235, 663–668. [DOI] [PubMed] [Google Scholar]
- Morse R.H. (1993) Nucleosome disruption by transcription factor binding in yeast. Science, 262, 1563–1566. [DOI] [PubMed] [Google Scholar]
- Morse R.H., Roth,S.Y. and Simpson,R.T. (1992) A transcriptionally active tRNA gene interferes with nucleosome positioning in vivo. Mol. Cell. Biol., 12, 4015–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M.R., Shimizu,M., Roth,S.Y., Dranginis,A.M. and Simpson,R.T. (1993) DNA–protein interactions at the S.cerevisiae α2 operator in vivo. Nucleic Acids Res., 21, 3295–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson H.C.M., Finch,J.T., Luisi,B.F. and Klug,A. (1987) The structure of an oligo(dA)⋅oligo(dT) tract and its biological implications. Nature, 330, 221–226. [DOI] [PubMed] [Google Scholar]
- Park H.S., Arnott,S., Chandrasekaran,R., Millane,R.P. and Campagnari,F. (1987) Structure of the α-form of poly[d(A)]⋅poly[d(T)] and related polynucleotide duplexes. J. Mol. Biol., 197, 513–523. [DOI] [PubMed] [Google Scholar]
- Patterton H.-G. and Simpson,R.T. (1994) Nucleosome location of the STE6 TATA box and Matα2p-mediated repression. Mol. Cell. Biol., 14, 4002–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premilat S. and Albiser,G. (1997) X-ray fibre diffraction study of an elevated temperature structure of poly(dA)⋅poly(dT). J. Mol. Biol., 274, 64–71. [DOI] [PubMed] [Google Scholar]
- Puhl H.L. and Behe,M.J. (1995) Poly(dA)⋅poly(dT) forms very stable nucleosomes at higher temperature. J. Mol. Biol., 245, 559–567. [DOI] [PubMed] [Google Scholar]
- Puhl H.L., Satyanarayana,R., Gudibandem,R. and Behe,M.J. (1991) Poly[d(A⋅T)] and other synthetic polydeoxynucleotides containing oligoadenosine tracts form nucleosomes easily. J. Mol. Biol., 222, 1149–1160. [DOI] [PubMed] [Google Scholar]
- Reardon B.J., Gordon,D., Ballard,M.J. and Winter,E. (1995) DNA binding properties of the Saccharomyces cerevisiae DAT1 gene product. Nucleic Acids Res., 23, 4900–4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D. (1979) Nucleosome cores reconstituted from poly(dA–dT) and the octamer of histones. Nucleic Acids Res., 6, 1805–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S.Y., Dean,A. and Simpson,R.T. (1990) Yeast α2 repressor positions nucleosomes in TRP1/ARS1 chromatin. Mol. Cell. Biol., 10, 2247–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S.Y., Shimizu,M., Johnson,L., Grunstein,M. and Simpson,R.T. (1992) Stable nucleosome positioning and complete repression by the yeast α2 repressor are disrupted by amino-terminal disruptions in histone H4. Genes Dev., 6, 411–425. [DOI] [PubMed] [Google Scholar]
- Rowley A., Dowell,S.J. and Diffley,J.F. (1994) Recent developments in the initiation of chromosomal DNA replication: a complex picture emerges. Biochim. Biophys. Acta, 1217, 239–256. [DOI] [PubMed] [Google Scholar]
- Schieferstein U. and Thoma,F. (1996) Modulation of cyclobutane pyrimidine dimer formation in a positioned nucleosome containing poly(dA⋅dT) tracts. Biochemistry, 35, 7705–7714. [DOI] [PubMed] [Google Scholar]
- Schlapp T. and Rodel,G. (1990) Transcription of two divergently transcribed yeast genes initiates at a common oligo(dA–dT) tract. Mol. Gen. Genet., 223, 438–442. [DOI] [PubMed] [Google Scholar]
- Shimizu M., Roth,S.Y., Szent-Gyorgyi,C. and Simpson,R.T. (1991) Nucleosomes are positioned with base pair precision adjacent to the α2 operator in Saccharomyces cerevisiae. EMBO J., 10, 3033–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu M., Li,W., Shindo,H. and Mitchell,A.P. (1997) Transcriptional repression at a distance through exclusion of activator binding in vivo. Proc. Natl Acad. Sci. USA, 94, 790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu M., Li,W., Covitz,P.A., Hara,M., Shindo,H. and Mitchell,A.P. (1998) Genomic footprinting of the yeast zinc finger protein Rme1p and its roles in repression of the meiotic activator IME1. Nucleic Acids Res., 26, 2329–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R.T. (1990) Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature, 343, 387–389. [DOI] [PubMed] [Google Scholar]
- Simpson R.T. (1991) Nucleosome positioning: occurrence, mechanisms and functional consequences. Prog. Nucleic Acid Res. Mol. Biol., 40, 143–184. [DOI] [PubMed] [Google Scholar]
- Simpson R.T. and Kunzler,P. (1979) Chromatin and core particles formed from the inner histones and synthetic polydeoxyribo nucleotides of defined sequence. Nucleic Acids Res., 6, 1387–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R.T., Roth,S.Y., Morse,R.H., Patterton,H.-G., Cooper,J.P., Murphy,M.R., Kladde,M.P. and Shimizu,M. (1993) Nucleosome positioning and transcription. Cold Spring Harb. Symp. Quant. Biol., 58, 237–245. [DOI] [PubMed] [Google Scholar]
- Struhl K. (1985) Naturally occurring poly(dA–dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc. Natl Acad. Sci. USA, 82, 8419–8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiry-Blaise L.M. and Loppes,R. (1990) Deletion analysis of the ARG4 promoter of Saccharomyces cerevisiae: a poly (dAdT) stretch involved in gene transcription. Mol. Gen. Genet., 223, 474–480. [DOI] [PubMed] [Google Scholar]
- Thoma F. (1986) Protein–DNA interactions and nuclease-sensitive regions determine nucleosome positions on yeast plasmid chromatin. J. Mol. Biol., 190, 177–190. [DOI] [PubMed] [Google Scholar]
- Thoma F. (1992) Nucleosome positioning. Biochim. Biophys. Acta, 1130, 1–19. [DOI] [PubMed] [Google Scholar]
- Thoma F., Bergman,L.W. and Simpson,R.T. (1984) Nuclease digestion of circular TRP1ARS1 chromatin reveals positioned nucleosomes separated by nuclease-sensitive regions. J. Mol. Biol., 177, 715–733. [DOI] [PubMed] [Google Scholar]
- Travers A., (1999) Chromatin modification by DNA tracking. Proc. Natl Acad. Sci. USA, 96, 13634–13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter E. and Varshavsky,A. (1989) A DNA binding protein that recognizes oligo(dA)⋅oligo(dT) tracts. EMBO J., 8, 1867–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A.P. and Hayes,J.J. (1999) Chromatin disruption and modification. Nucleic Acids Res., 27, 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z. and Thiele,D.J. (1996) A specialized nucleosome modulates transcription factor access to a C.glabrata metal responsive promoter. Cell, 87, 459–470. [DOI] [PubMed] [Google Scholar]