Abstract
Two percentage of the cat genome is a repetitive, feline-specific satellite sequence (FA-SAT) of 483 bp and 65% guanine-cytosine content. Previous chromosomal localization of the satellite has demonstrated the satellite’s presence on several discrete regions of the telomeres of chromosomes, predominately on the D, E, and F chromosome groups. The recent assembly of the 1.9× whole-genome shotgun (WGS) sequence of cat illustrates the challenge of the assembly of these large numbers of relatively short, similar sequences. Clones with paired end reads that include FA-SAT sequence have a high level of assembly discrepancies compared with clones with other types of repetitive elements, such as short interspersed nuclear elements (SINEs) and long interspersed nuclear elements (LINEs). The influence of the presence of FA-SAT but not SINEs and LINEs on genome assembly may likely reflect the evolutionary emergence of FA-SAT, which has lead to an excess of FA-SAT copies with identical sequence, which is less an issue with older, more diverse SINE and LINE sequences. The FA-SATs are restricted to a few hundred discrete regions of the cat genome, and associated errors in the assembly seem to be restricted to these loci. The findings regarding the feline-specific sequence should be considered in the pending 8x assembly of the cat genome.
Keywords: artifacts, FA-SAT, genome assembly, repetitive elements, satellite, whole-genome shotgun
Whole-genome shotgun (WGS) genome assembly entails fragmentation of the genome, cloning the resulting fragments in vectors of known insert length, and end sequencing of the clones using capillary Sanger sequencing. The sequence reads are then combined using assembly algorithms such as ARACHNE (Batzoglou et al. 2002), PHUSION (Mullikin and Ning 2003), or PCAP (Huang et al. 2003). Sequence similarity is used to combine end reads into contiguous sequences (contigs), whereas information about clone insert length, between paired end reads, is used to estimate the distances between contigs and place them, separated by gaps of unknown sequence, onto scaffolds. For 2× coverage of the genome, in which each nucleotide is represented on the average by 2 observations, the final assembly is highly fragmented (Kirkness et al. 2003; O'Brien and Murphy 2003; Green 2007; Pontius et al. 2007). Mammalian 2× genomes consist of hundreds of thousands of short contigs (<5 kb) placed onto scaffolds. This fragmented data make it difficult to glean the same level of information that can be derived from an 8× genome.
In the case of the 1.9× cat genome, the availability of a highly resolved dog genome (Lindblad-Toh et al. 2005), a feline radiation hybrid map (Murphy et al. 2007), and thorough annotations of other mammalian genomes such as human (Sayers et al. 2009) have enabled the cat assembly to be annotated with a variety of features including proposed feline orthologs for more than 17 000 human genes (Pontius et al. 2007). The availability of the generic genome browser (Stein et al. 2002) has allowed the assembly and its annotations to be organized online on a genome browser, GARFIELD (Pontius and O'Brien 2007), and has served as a useful resource for further studies of cat genetics, such as gene discovery (Fyfe et al. 2006; Ishida et al. 2006; Kehler et al. 2007; Menotti-Raymond et al. 2007).
The feline-specific satellite sequence (FA-SAT) (Fanning 1987) is a unique characteristic of the cat genome that has posed a challenge to genome assembly. The FA-SAT is a short sequence repeat (483 bp) that represents 2% of the genome, suggesting that there are more than 100 000 copies of the satellite in the cat genome. The satellite has low intrasequence variability due to its high G + C content of 64% (Figure 1) and presence, in each satellite, of 25 variants of the hexanucleotide TAACCC (Fanning 1987). Lastly, as the satellite occurs in tandem copies, end sequencing inevitably includes pairs of end reads that both represent the FA-SAT. These aspects of the FA-SAT present a challenge to the assembly of the regions of the genome that contain it and can lead to a variety of assembly errors (Phillippy et al. 2008).
Figure 1.
Histogram of C + G content for cat WGS sequences. The FA-SAT is distinguishable as a subpopulation on the histogram of percent G + C content in windows of 500 bp across the cat genome.
In a preliminary analysis of FA-SATs in the 1.9× cat genome, it became evident that many FA-SAT end reads were being combined in the course of the assembly by ARACHNE to 1 locus, even when they represented end reads from clones having insert sizes of 35 kb. In the case of a clone with 2 FA-SAT end reads, the combination of these identical sequences would generally have irrelevant consequences. However, the merging of 2 different clones based on their each having only one end read that is FA-SAT could lead to inadvertent misassembly and incorrect scaffold placement of the partner end reads.
To assess the possibility that the FA-SAT has led to assembly discrepancies, we analyzed paired end reads with respect to their content and their position in the assembly. We have found that, compared with other end reads, assembly discrepancies are elevated for clones that include the FA-SAT. We also found that the regions of the genome that have a large percentage of misplaced reads have a higher fraction of FA-SAT than in well-assembled regions.
Recently, GARFIELD (Pontius and O'Brien 2007) has been updated to include a display of regions that contain these irregularities. This addition helps users to exercise caution in interpretation of the data provided.
Materials and Methods
Assembly information for the ARACHNE 1.9× assembly of cat was taken from The Broad (ftp://ftp.broad.mit.edu/pub/assemblies/mammals/cat/felCat3/). Our analysis used only clones that have 2 end reads in the assembly. Each end read was classified with respect to 2 characteristics: repeat content and assembly irregularity.
End reads that represent repetitive elements were identified using REPEATMASKER (www.repeatmasker.org) and were categorized as being: LINE, SINE, FA-SAT, any combination of any of these, or NONE. Each clone was then assigned to a category based on the exact repeat content of its member reads (See Table 1). This allowed us to distinguish clones that may be entirely FA-STAT, for example, as indicated by both end reads being FA-STAT, from clones that may include a region flanking the FA-STAT introgression.
Each clone was also assigned to one of the following categories with regard to the inconsistency of the placement of its end reads in the final assembly:
Disparate scaffolds: 2 paired reads are not assembled on the same scaffold.
Too close: the midpoints of end reads are within 500 bp of one another.
Too distant: the midpoints of end reads are separated by a distance that is more than double the insert length of the clone.
Strand discrepancy: end reads are on the same scaffold and at a reasonable distance of separation, but on the same strand instead of opposite strand.
Well placed: any clone that is not included in the other categories.
With the clones thus categorized by both repeat content and assembly quality, we then tallied, for each repetitive element category, the total number of clones with assembly irregularities.
To analyze irregularities of the assembly across the genome as a whole, windows of 10 kb across the chromosome of the cat genome were analyzed. As with the individual clones, each window was categorized with respect to its repeat content and assembly irregularities, but with categories that differed from those used for individual clones. For each window, the fraction of end reads that were tagged by REPEATMASKER as being LINE, SINE, or FA-SAT were calculated. Assembly irregularities within the window were assessed based on the percentage of its end reads that were not “Well placed”; each window was assigned to 1 of 5 categories: 0–20% of assembly end reads with irregularities, 20–40%, 40–60%, 60–80%, or 80–100% irregularities.
This allowed each window to be categorized by both its repeat content and assembly quality. Then, for each repetitive element category, the total number of windows in the 5 groups of assembly irregularities was tallied.
Results and Discussion
A summary of the end read counts, their repeat content and assembly discrepancies is found in Table 1.
Table 1.
Paired end reads from clones included in the 1.9× feline assembly
| Repetitive element content of clone | Assembly discrepancy counts | Well-placed pairs | |||||
| End1 | End2 | Disparate scaffolds | Too far | Too close | Strand | Counts | Percentage of total |
| Plasmids | |||||||
| SINE | FA-SAT | 194 | 3 | 0 | 0 | 209 | 0.51 |
| LINE | FA-SAT | 360 | 12 | 0 | 0 | 437 | 0.54 |
| FA-SAT | NONE | 929 | 61 | 20 | 0 | 1341 | 0.57 |
| FA-SAT | FA-SAT | 28968 | 159 | 2441 | 2 | 18362 | 0.37 |
| LINE | LINE | 4764 | 442 | 1128 | 8 | 52269 | 0.89 |
| SINE | LINE | 2989 | 1472 | 132 | 13 | 139123 | 0.97 |
| SINE | SINE | 2183 | 1729 | 2098 | 26 | 157417 | 0.96 |
| LINE | NONE | 5497 | 2231 | 462 | 22 | 201132 | 0.96 |
| NONE | NONE | 13339 | 5584 | 5629 | 68 | 383155 | 0.94 |
| SINE | NONE | 6231 | 5126 | 1120 | 66 | 422861 | 0.97 |
| Total | 92759 | 23847 | 16724 | 310 | 2023650 | 0.94 | |
| Fosmids | |||||||
| FA-SAT | NONE | 551 | 0 | 7 | 0 | 142 | 0.20 |
| FA-SAT | FA-SAT | 2508 | 0 | 77 | 0 | 191 | 0.07 |
| LINE | LINE | 1510 | 17 | 317 | 2 | 6099 | 0.77 |
| SINE | SINE | 484 | 23 | 419 | 2 | 15640 | 0.94 |
| SINE | LINE | 1385 | 25 | 4 | 3 | 17962 | 0.93 |
| LINE | NONE | 2922 | 60 | 38 | 9 | 32069 | 0.91 |
| NONE | NONE | 2196 | 97 | 803 | 20 | 48816 | 0.94 |
| SINE | NONE | 1469 | 75 | 87 | 10 | 51534 | 0.97 |
| Total | 18015 | 368 | 2133 | 59 | 214037 | 0.91 | |
REPEATMASKER was used to categorize each read as to whether it includes LINEs, SINEs, or FA-SAT, and clones were categorized by the repeat content of their end reads as well as the assembly consistency of the 2 reads. Assembly discrepancies include the following: being placed on disparate scaffolds, being too distant (separated by more than 2× the length of the insert), being too close (less than 500 bp apart), and being placed on the same strand of the scaffold. All other end reads are considered well-placed.
The category of assembly irregularity with the most numbers was that of “Disparate scaffolds.” Generally, assembly algorithms generate contigs based on sequence similarity between reads, whereas the placing of contigs onto scaffolds is based on distance of separation of paired end reads as estimated by the insert size of their clone. When paired reads are not placed on the same scaffold, it could simply be the result of this information not being used. Alternatively, it could mean that taking into account such information would have been inconsistent with other aspects of the assembly and, as such, could mean that unpaired end reads are an indication of mistakes in the assembly. In the cat 1.9× assembly, the majority of paired end reads were placed on the same scaffold. Overall, more than 90% of the pairs are placed on consistent scaffolds. However, when one or both end reads represent the FA-SAT, the percentage of end reads that are assembled on the same scaffold is much lower: less than 60% of plasmid and less than 20% of fosmid end reads.
Being too close together is another assembly inconsistency that is elevated in the FA-SAT end reads. For end reads that are on the same scaffold, more than 10% of FA-SAT end reads are separated by less than 500 bp, whereas this reaches only 2% for LINEs, SINEs, and end reads with no repetitive elements. For fosmids, this reaches 28% for FA-SAT, whereas less than 3% for other categories of repetitive elements.
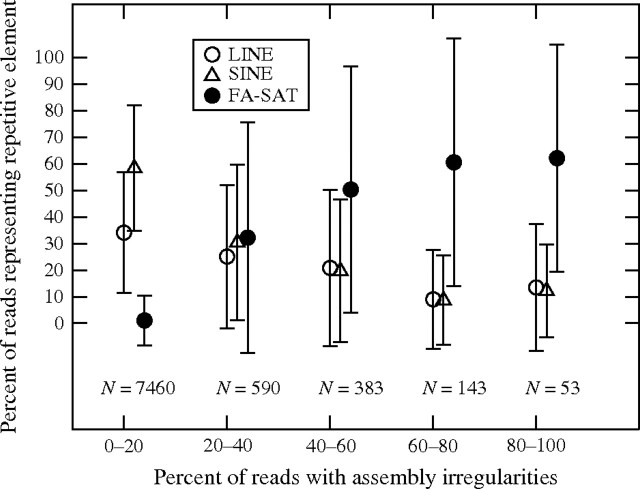
For the analysis of the genome as a whole, using windows of 10 kb, we found a strong correlation between FA-SAT content and assembly inconsistencies. On the whole, the majority of windows (7460/8628) have less than 20% irregularities, and these well-assembled windows also averaged 0% FA-SAT content. The percentage of FA-SAT end reads increases progressively with the increasing window irregularities (correlation coefficient = 0.935), reaching an average of 60% in the 53 windows that have 80–100% irregularities (Figure 2). On the other hand, the content of LINEs and SINEs is highest in well-assembled windows, decreasing progressively in windows with assembly irregularities (correlation coefficient of −0.920 and −0.902 for LINEs and SINEs, respectively). These results suggest that generally LINEs and SINEs were well assembled, whereas the FA-SAT posed a challenge to the assembly algorithm, resulting in windows that include FA-SAT having an excess of the assembly irregularities analyzed here.
Figure 2.
Percentage of misplaced end reads in windows of 10 kb along the cat genome compared with LINE, SINE, and FA-SAT content of the windows. Shown is the mean and standard deviation of the percentage of reads representing FA-SAT, LINEs, and SINEs as a function of their category of assembly irregularities, with the number of 10-kb windows that were analyzed. In windows with high assembly irregularities, about 50% of the end reads represent FA-SAT, whereas the level of LINEs and SINEs in these windows is lower than for well-assembled windows.
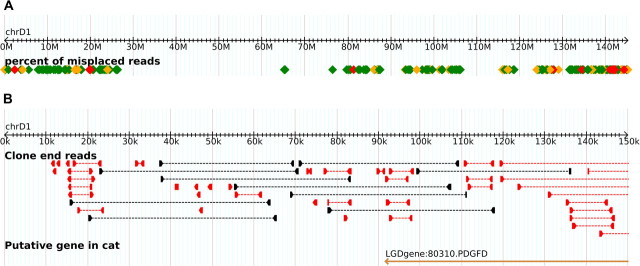
In order to allow a visual representation of these irregularities, the cat genome browser GARFIELD has been supplemented with a track displaying the misplaced reads (Figure 3a,b).
Figure 3.
FA-SATs at the telomeres of chromosome D1. The GARFIELD genome browser includes a track for showing regions of possible assembly artifacts. (a) The chromosome view includes the color-coded regions representing the percentage of misplaced reads in windows of 10 kb. Windows having a high percentage of outliers are represented in red and orange, whereas low, yet nonzero, levels are in green. (b) The detailed view of GARFIELD shows paired end reads of the assembly. Inconsistently placed end reads are displayed in red. Well-placed end reads (those that are 31.4–49.5 kb apart) are in black. Well-placed plasmids are so numerous that they are not included in the display.
In spite of the difficulty of assembly of the FA-SATs, the majority of those that could be placed were assigned to cat chromosomes E3, A3, D1, and X, and all these loci are consistent with published cytogenetic studies (Modi et al. 1988; Santos et al. 2004, 2006). The loci of FA-SAT assembled on chromosomes cat A3 could also be corroborated by known synteny between cat and the dog and human genomes: for clones with a single FA-SAT end read on A3 could be aligned to regions of the dog and human genomes that share known synteny between cat chromosome A3 and the dog and human genomes (Murphy et al. 2007; Pontius et al. 2007). Other FA-SAT end reads with undetermined loci on cat chromosomes include those from clones that share sequence similarity to chromosomes 25, 27, and 36 in dog and chromosome X in human.
In summary, the FA-SAT end reads of the cat genome are associated with unusual and likely erroneous distances of separation on the scaffolds of the 1.9× WGS assembly. This FA-SAT seems to have presented more of a challenge in assembly than other repetitive elements such as LINEs and SINEs. However, as these satellite sequences are restricted to very few regions of the genome, so are their associated errors in the assembly as a whole. The regions of the genome that include the FA-SAT will pose a challenge to the pending 8× assembly of the cat genome.
Funding
National Cancer Institute, National Institutes of Health, (N01-CO-12400); Intramural Research Program of the National Institutes of Health; National Cancer Institute; Center for Cancer Research.
Acknowledgments
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
References
- Batzoglou S, Jaffe DB, Stanley K, Butler J, Gnerre S, Mauceli E, Berger B, Mesirov JP, Lander ES. ARACHNE: a whole-genome shotgun assembler. Genome Res. 2002;12:177–189. doi: 10.1101/gr.208902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning TG. Origin and evolution of a major feline satellite DNA. J Mol Biol. 1987;197(4):627–634. doi: 10.1016/0022-2836(87)90469-4. [DOI] [PubMed] [Google Scholar]
- Fyfe JC, Menotti-Raymond M, David VA, Brichta L, Schäffer AA, Agarwala R, Murphy WJ, Wedenmeyer WJ, Drummond MC, Buzzell BG, et al. An ∼ 140 kb deletion associated with feline spinal muscular atrophy implies an essential LIX1 function for motor neuron survival. Genome Res. 2006;16(9):1084–1090. doi: 10.1101/gr.5268806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P. 2x genomes—does depth matter? Genome Res. 2007;17(11):1547–1549. doi: 10.1101/gr.7050807. [DOI] [PubMed] [Google Scholar]
- Huang X, Wang J, Aluru S, Yang SP, Hillier L. PCAP: a whole-genome assembly program. Genome Res. 2003;13(9):2164–2170. doi: 10.1101/gr.1390403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida Y, David VA, Eizirik E, Schäffer AA, Neelam BA, Roelke ME, Hannah SS, O'Brien SJ, Menotti-Raymond M. A homozygous single-base deletion in MLPH causes the dilute coat color phenotype in the domestic cat. Genomics. 2006;88(6):698–705. doi: 10.1016/j.ygeno.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Kehler J, David V, Schäffer AA, Eizirik E, Ryugo D, Hannah S, O'Brien SJ, Menotti-Raymond M. Four separate mutations in the feline fibroblast growth factor 5 gene are responsible for long hair in the domestic cat. J Hered. 2007;98(6):555–566. doi: 10.1093/jhered/esm072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkness EF, Bafna V, Halpern AL, Levy S, Remington K, Rusch DB, Delcher AL, Pop M, Wang W, Fraser CM, et al. The dog genome: survey sequencing and comparative analysis. Science. 2003;301(5641):1898–1903. doi: 10.1126/science.1086432. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ, 3rd, Zody MC, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438(7069):803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Menotti-Raymond M, David VA, Schäffer AA, Stephens R, Wells D, Kumar-Singh R, O'Brien SJ, Narfström K. Mutation in CEP290 discovered for cat model of human retinal degeneration. J Hered. 2007;98(3):211–220. doi: 10.1093/jhered/esm019. [DOI] [PubMed] [Google Scholar]
- Modi WS, Fanning TG, Wayne RK, O'Brien SJ. Chromosomal localization of satellite DNA sequences among 22 species of felids and canids (Carnivora) Cytogenet Cell Genet. 1988;48(4):208–213. doi: 10.1159/000132630. [DOI] [PubMed] [Google Scholar]
- Mullikin JC, Ning Z. The phusion assembler. Genome Res. 2003;13:81–90. doi: 10.1101/gr.731003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy WJ, Davis B, David VA, Agarwala R, Schäffer AA, Pearks Wilkerson AJ, Neelam B, O'Brien SJ, Menotti-Raymond M. A 1.5-Mb-resolution radiation hybrid map of the cat genome and comparative analysis with the canine and human genomes. Genomics. 2007;89:189–196. doi: 10.1016/j.ygeno.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien SJ, Murphy WJ. Genomics. A dog’s breakfast? Science. 2003;301(5641):1854–1855. doi: 10.1126/science.1090531. [DOI] [PubMed] [Google Scholar]
- Phillippy AM, Schatz MC, Pop M. Genome assembly forensics: finding the elusive mis-assembly. Genome Biol. 2008;9(3):R55. doi: 10.1186/gb-2008-9-3-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- doi: 10.1101/gr.6380007. Pontius JU, Mullikin JC, Smith DR, Agencourt Sequencing Team, Lindblad-Toh K, Gnerre S, Clamp M, Chang J, Stephens R, Neelam B, et al. 2007. Initial sequence and comparative analysis of the cat genome. Genome Res. 17(11):1675–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontius JU, O'Brien SJ. Genome annotation resource fields—GARFIELD: a genome browser for Felis catus. J Hered. 2007;98(5):386–389. doi: 10.1093/jhered/esm055. [DOI] [PubMed] [Google Scholar]
- Santos S, Chaves R, Adega F, Bastos E, Guedes-Pinto H. Amplification of the major satellite DNA family (FA-SAT) in a cat fibrosarcoma might be related to chromosomal instability. J Hered. 2006;97(2):114–118. doi: 10.1093/jhered/esj016. [DOI] [PubMed] [Google Scholar]
- Santos S, Chaves R, Guedes-Pinto H. Chromosomal localization of the major satellite DNA family (FA-SAT) in the domestic cat. Cytogenet Genome Res. 2004;107(1–2):119–122. doi: 10.1159/000079581. [DOI] [PubMed] [Google Scholar]
- Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37(Database issue):D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12(10):1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]