Abstract
The marked increase in skeletal muscle mass during the neonatal period is largely due to a high rate of postprandial protein synthesis that is modulated by an enhanced sensitivity to insulin and amino acids. The amino acid signaling pathway leading to the stimulation of protein synthesis has not been fully elucidated. Among the amino acids, leucine is considered to be a principal anabolic agent that regulates protein synthesis. mTORC1, which controls protein synthesis, has been implicated as a target for leucine. Until recently, there have been few studies exploring the role of amino acids in enhancing muscle protein synthesis in vivo. In this review, we discuss amino acid-induced protein synthesis in muscle in the neonate, focusing on current knowledge of the role of amino acids in the activation of mTORC1 leading to mRNA translation. The role of the amino acid transporters, SNAT2, LAT1, and PAT, in the modulation of mTORC1 activation and the role of amino acids in the activation of putative regulators of mTORC1, i.e., raptor, Rheb, MAP4K3, Vps34, and Rag GTPases, are discussed.
Keywords: Mammalian target of rapamycin, Protein synthesis, Amino acids, Nutrient signaling, Skeletal muscle, Neonate, Raptor, Rheb, Translation initiation, Review
2. INTRODUCTION
The rate of growth is higher during the neonatal period than at any other stage of postnatal development. The more rapid gain in skeletal muscle mass than in the body as a whole during this period (1) results in a substantial increase in the proportion of the body protein pool that is muscle protein. The balance between protein synthesis and degradation is an important determinant of growth. During the neonatal period, the fractional rate of protein synthesis of newly formed skeletal muscle is much higher than that of protein degradation (2). Consequently, the high growth rate of neonatal muscle is mainly due to the high rate of protein synthesis (3). Our previous studies using neonatal pigs, a well-recognized animal model for the human neonate, showed that the fractional rate of protein synthesis in skeletal muscle is very high at birth and declines with age, particularly during the first month of life (4). This is achieved by an elevated capacity for protein synthesis which is driven by both the high ribosome content and an increased efficiency of the translation process in skeletal muscle (5, 6).
During periods of rapid growth, dietary amino acids are used for protein deposition with exceptionally high efficiency (6). In our previous studies in neonatal pigs, we demonstrated that feeding stimulates protein synthesis in all tissues, however, the highest response occurs in skeletal muscle (7, 8). To understand the mechanism for this response, we examined potential mediators for the feeding-induced stimulation of protein synthesis in skeletal muscle (9). There were five candidates that allegedly play a role, four hormones/growth factors (insulin, insulin-like growth factor-I (IGF-I), growth hormone, and glucagon), amino acids, and glucose. Due to their inability to both stimulate protein synthesis and increase rapidly after food ingestion, three candidates (IGF-I, growth hormone, and glucagon) were rejected (2, 9, 10). From our studies using the pancreatic-substrate clamp technique, which we developed to identify the independent effects of these factors in the regulation of protein synthesis (11), we concluded that insulin and amino acids (especially leucine) are the principal mediators of the postprandial rise in protein synthesis in skeletal muscle of the neonate (4, 11, 12).
Protein synthesis, one of the metabolic processes that is crucial for life, is complex. Translation of mRNA consists of three distinct stages: 1) Initiation – initiator tRNA binds to the start signal on mRNA; 2) Elongation – begins when an aa-tRNA binds to the ribosome A (amino acyl) site; and 3) Termination – occurs when one of three stop codon is encountered and the ribosomal subunits then break apart (reviewed in Reference 13). A strong body of evidence has shown that the postprandial (acute) regulation of protein synthesis is achieved in part through changes in the rate of mRNA translation via alterations in peptide-chain initiation, a process that involves enhanced binding of both mRNA and initiator methionyl-tRNA (met-tRNA) to the 40S ribosomal subunit (reviewed in Reference 14). Both insulin and amino acids (particularly leucine) independently affect the activation of the initiation phase of mRNA translation (15–18). While components in the insulin signaling pathway leading to this metabolic process have been intensively studied and elucidated, amino acid signaling toward protein synthesis, especially involving the upstream pathway of mTOR, are complex and less well understood when compared to the insulin signaling pathway.
In this review, we will summarize the role of amino acids in the regulation of protein synthesis in skeletal muscle of neonatal pigs. This review also aims to outline current knowledge of the effect of amino acids/leucine on the activation of mTORC1 leading to mRNA translation. We will also discuss our findings on the mechanism by which amino acids/leucine regulate signaling components of mRNA translation under physiological conditions in skeletal muscle of the neonate.
3. REGULATION OF PROTEIN SYNTHESIS IN SKELETAL MUSCLE OF NEONATES
During the neonatal period, all tissues presumably undergo rapid growth, however, skeletal muscle comprises a majority of the mass increase, resulting in a substantial increase in the proportion of the body protein pool that is muscle protein (approximately 45% of body mass) (2). Our early works suggest that the fractional rate of protein synthesis in skeletal muscle is elevated at birth and declines sharply with development (11). Feeding markedly stimulates protein synthesis, particularly in skeletal muscle. This enhanced rate of protein synthesis in skeletal muscle in response to feeding, thus, supports the rapid growth of the neonate (2). To elucidate the feeding components responsible for the postprandial rise in protein synthesis, we utilized our pancreatic-substrate clamp technique and found that insulin and amino acids independently stimulate protein synthesis. This response to insulin and amino acids, like the response to feeding, decreases with age (11, 19).
3.1. Effect of amino acids on protein synthesis in skeletal muscle of neonates
Amino acids are the building blocks of protein in the body, and thus amino acids are critical elements of the diet. During the neonatal period, muscles grow at a rate that is significantly greater than that of the body as a whole, and thus, a sufficient amino acid supply for protein synthesis is crucial (20). Results from in vitro as well as in vivo studies clearly show that amino acids alone can stimulate protein synthesis (12, 21–23). Studies using perfused muscle indicate that amino acids are as effective as insulin in stimulating protein synthesis (24). However, the concentration of amino acids that was used in cell culture and perfused muscle studies was typically 5 to 10 times higher than physiological levels, arguing against the physiological relevance of these studies. Furthermore, even if physiological concentrations of amino acids were used, they were compared to the absence of amino acids in the medium, a situation that is not physiologically possible (25). Most of the findings regarding the amino acid-induced stimulation of muscle protein synthesis in vivo were generated from studies in adult humans or mature animals (26–27). Our laboratory is one of the few laboratories that study the role of amino acids in the regulation of protein synthesis in skeletal muscle during the neonatal period.
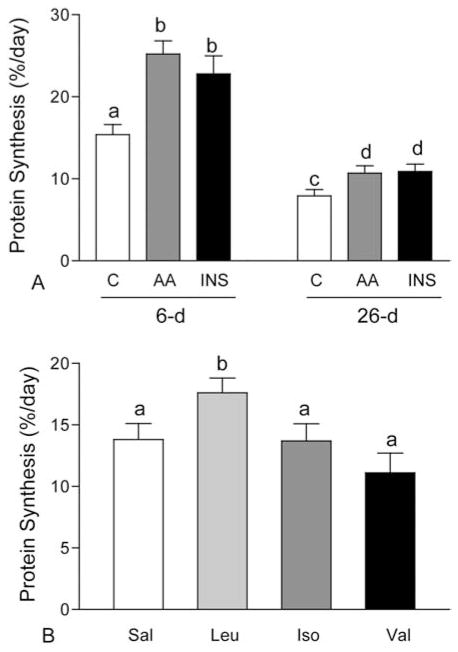
To examine the role of amino acids in the regulation of muscle protein synthesis, independent of changes in the circulating level of insulin, we developed a novel pancreatic-substrate clamp technique (11). This technique overcomes the confounding effects of insulin by blocking insulin secretion with somatostatin, while glucose and glucagon are maintained at fasting levels and insulin is maintained at any given level including undetectable (below fasting), fasting, intermediate, fed, or supraphysiological levels (12). We showed that infusing a balanced mixture of amino acids into fasting neonatal pigs to mimic levels present in the fed state increases protein synthesis in skeletal muscle to rates comparable to those of fed neonatal pigs (11, 12, 19). The stimulation of protein synthesis by amino acids occurs in the presence of fasting levels of insulin, at insulin concentrations that are intermediate between the fasting and the fully fed state, and when insulin concentrations are reduced to undetectable levels using somatostatin to block insulin secretion. The response to amino acids decreases with age (Figure 1A). The results provide strong evidence of an independent role of amino acids in the regulation of muscle protein synthesis and suggest that adequate amounts of amino acids are essential for muscle growth during the neonatal period.
Figure 1.
Fractional rates of protein synthesis in longissimus dorsi (LD) muscle of 6- and 26-day-old pigs in response to euinsulinemic-euglycemic-euaminoacidemic conditions (C), euinsulinemic-euglycemic-hyperaminoacidemic clamps (AA), and hyperinsulinemic-euglycemic-euaminoacidemic clamps (INS) (panel A). Fractional rates of protein synthesis in LD muscle of 6-day-old pigs in response to individual infusion of the branched-chain amino acids, leucine (Leu), isoleucine (Iso) and valine (Val), in comparison with saline (sal)/control (panel B). Values are means ± SEM, means with different letters differ at P < 0.05.
Several lines of evidence have demonstrated that leucine is the most effective single amino acid to activate protein synthesis in skeletal muscle. Leucine administration either orally or by infusion in adult humans or mature animals has been shown to enhance protein synthesis in skeletal muscle (23, 28). Recently, we demonstrated that infusion of physiological levels of leucine acutely stimulates protein synthesis in skeletal muscle of neonatal pigs (Figure 1B). This anabolic effect appears to be unique for leucine, as increasing circulating concentrations of the other branched-chain amino acids, isoleucine and valine, to fed levels does not increase muscle protein synthesis (29). This effect of leucine on neonatal muscle protein synthesis can be sustained for at least 24 h when the circulating levels of other amino acids are maintained and the leucine-induced drop in the concentrations of other amino acids is prevented (30, 31). Similarly, our recent data indicate that leucine supplementation of a low protein diet can stimulate muscle protein synthesis to a level that is comparable to that of a high protein diet (Unpublished data).
3.2. Amino acid-induced activation of signaling components leading to mRNA translation in skeletal muscle of neonates
Several lines of evidence indicate that amino acids are not only used as substrates for protein synthesis, they also serve as nutrient signals that regulate protein synthesis (32). Work in vitro and in cell cultures has clearly established that even without stimulation by anabolic hormones, such as insulin, amino acids themselves can stimulate mRNA translation (33). This suggests that amino acids act as anabolic agents that through nutrient signals can up-regulate the protein synthetic machinery, resulting in an enhanced rate of protein synthesis.
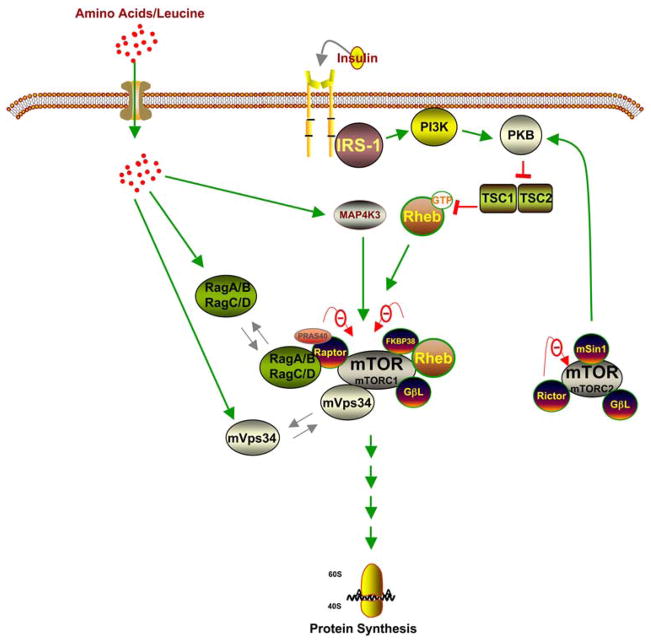
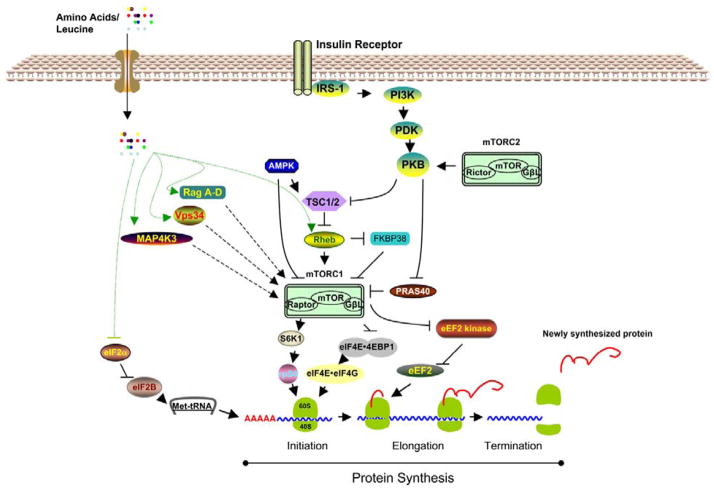
The underlying mechanisms by which amino acids regulate the activation of signaling components leading to mRNA translation are unclear (Figure 2). While the amino acid signaling pathway is critical to the cellular regulation of growth, the insulin signaling pathway senses and coordinates general nutrient status (34). For the sake of comparison, in this review we have included the insulin signaling pathway in this simple signaling schematic (Figure 2). The insulin signaling pathway leading to mRNA translation is well characterized (35–37). Briefly, upon the binding of insulin to its receptor, several downstream components are activated. The activated insulin receptor triggers the tyrosine phosphorylation of insulin receptor substrate (IRS) -1/2, followed by the activation of phosphoinositide 3-kinase (PI 3-kinase). PI 3-kinase then activates phosphoinositide-dependent kinase-1 (PDK-1), followed by the activation of protein kinase B (PKB). PKB, in turn, phosphorylates and inactivates tuberous sclerosis complex (TSC) -1/2, resulting in the activation of mTOR (see further assessment of mTOR regulation below). Activated mTOR stimulates mRNA translation by phosphorylating its principal effectors, ribosomal protein S6 kinase (S6K) and eIF4E-binding protein 1 (4E-BP1). Despite overwhelming evidence to support the assertion that amino acids utilize similar signaling components downstream of mTOR, the exact mechanisms by which amino acids relay their signal from the cell membrane to mTOR is still under debate (38, 39).
Figure 2.
Recent concepts of the insulin- and amino acid-induced activation of mTORC1.
Our recent studies have shown that either a balanced amino acid mixture or leucine alone up-regulates muscle protein synthesis in part by stimulating the phosphorylation of 4E-BP1 and S6K1 (11, 22). These findings are consistent with the reports from other in vivo studies (23). Most of the studies that have described the mechanisms of amino acid sensing to mTOR were generated from in vitro studies or cell cultures. It is important to note that several studies indicate that amino acids do not activate mTOR through PKB (39). Using our pancreatic-substrate clamp technique, we clearly showed that insulin, but not amino acids/leucine, induce PKB activation (40). There are conflicting data as to whether amino acids regulate mTOR by inhibiting TSC 1/2 (41). Our data and others support the notion that TSC 1/2 activation is not affected by amino acids/leucine (40). Thus, several lines of evidence indicate that the sensors for amino acids are located downstream of TSC 1/2 (39).
4. AMINO ACID SENSING MECHANISMS THAT REGULATE MTORC1
Target of rapamycin (TOR) is an important protein kinase that regulates cell growth and is highly conserved from yeast to humans (42–46). In fact, it is so crucial for life that genetic studies show that without this protein kinase, an organism cannot survive (46). Consequently, for decades scientists have been studying the nature of mTOR, hoping to find effective methods to kill cancer cells (47). Today, thanks to those studies, we are closer to understanding the molecular mechanisms by which anabolic factors (ranging from hormones to nutrients) regulate the activation of mTOR (48). Thus, mTOR plays a central role in the growth stimulating effect of nutrition on skeletal muscle accretion.
mTOR acts as a “hub” that conveys different signals that regulate cellular processes such as metabolism, gene expression, and protein synthesis (49). mTOR is comprised of two independently regulated complexes: mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2) (Figure 2). mTORC1 consists of mTOR, raptor, and the G-protein beta-like protein (GbetaL/LST8) while mTORC2 is composed of mTOR, rictor, GbetaL, and mSIN1. Since mTORC1, but not mTORC2, is involved in nutrient/amino acid sensing, in this review we will focus on mTORC1.
4.1 Role of amino acid transporters in the modulation of mTORC1 activation
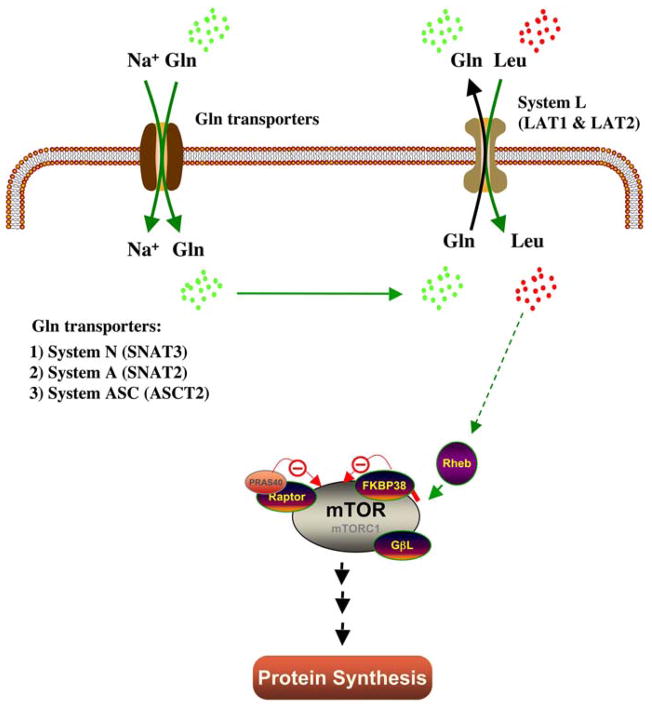
Unlike hormones or growth factors that have distinct receptors to initiate their signal, the biological nature of amino acid sensing in the cell membrane is highly debatable. Although yeast possess amino acid sensors that detect external amino acid levels (Ssy1P) (50), this mechanism in mammalian cells has not been established. Early studies using in vitro methods tried to elucidate whether amino acids initiate their signal in the plasma membrane (51). Nontransportable peptide (Leu8MAP which consist of 8 leucine molecules) treatment was used in isolated rat hepatocytes and evidence of the existence of a cell-surface receptor for leucine was shown. However, Lynch et al. (52) failed to detect any effect of Leu8MAP in isolated rat adipocytes suggesting that more studies are needed to address this hypothesis. Beugnet et al. (53) offered a more compelling study which showed that increases in the intracellular amino acid pool up-regulates mTOR signaling, resulting in an increased rate of protein synthesis and a reduction in protein degradation. More recently, an elegant study has shown that in order for leucine to activate mTORC1, leucine has to be transported into the cell by specific amino acid transporters (Figure 3) (54). In this model, intracellular glutamine accumulation is needed to facilitate leucine transport by the system L amino transporter.
Figure 3.
The regulation of leucine transport by glutamine leading to the activation of mTORC1 and protein synthesis.
Since all amino acids are required for cellular protein synthesis, it is apparent that cells need all amino acid transporters to be working properly. Genetic and biochemical approaches have been used to elucidate the role of amino acid transporters that are involved in the regulation of mTORC1 activation leading to mRNA translation (55). From these studies, only a handful of amino acid transporters have been implicated in mTORC1 activation (55). Thus, it is beyond the scope of this review to discuss all of the amino acid transporters.
4.1.1. SNAT2
Sodium-coupled neural amino acid transporter 2 (SNAT2) is a principal isoform of the system A amino acid transporter and is expressed in most extraneural tissues including skeletal muscle (56). In skeletal muscle, SNAT2 mediates the Na+ -dependent transport of glutamine (57). Among the first evidence to show the crucial role of SNAT2 in the amino acid-induced activation of mTORC1 was work performed in cell culture (58). In that study, down-regulation of SNAT2 abundance and activity by ceramide caused a marked reduction in protein synthesis due to reduced activation of translation initiation factors downstream of mTORC1. Evans et al. (59) used a different approach by silencing SNAT2 expression in L6 muscle cells. SNAT2 inhibition caused the depletion of cellular glutamine, resulting in depletion of other amino acids, notably leucine. Furthermore, lack of SNAT2 activity strongly reduced mTORC1 activation, leading to impairment of protein synthesis. These findings are consistent with the current model suggested by Nicklin et al. (54) (Figure 3).
SNAT2 is also considered a classical example of an amino acid transceptor (60). Transceptors can be defined as transporters that exhibit dual functions as transporters and sensors and their activity has been well established in yeast (60). Although the mechanistic mode of action is unclear, SNAT2 is capable of sensing and signaling amino acid availability to the mTORC1 signaling pathway, possibly through a PI 3-kinase dependent mechanism (59). Another indirect effect of SNAT2 activation is intracellular amino acid accumulation, which induces cell swelling due to osmotic water movement (61). In skeletal muscle, cell swelling stimulates the action of system A amino acid transport and the activation of the mTOR pathway (61). Although in vivo studies regarding the role of SNAT2 are lacking, a recent study reported that the placental expression of SNAT2 was markedly reduced in dams that consumed a low protein diet (62). Furthermore, a recent study showed that SNAT2 protein abundance was up-regulated in human skeletal muscle 2–3 hours after ingestion of essential amino acids (63). Our work (unpublished data) also suggests that in skeletal muscle of neonatal pigs, SNAT2 protein abundance is developmentally regulated, consistent with the developmental decline in the activation of mTORC1 (64). Considering that SNAT2 is not the only glutamine transporter in skeletal muscle, the notion that SNAT2 is crucial for mTORC1 activation is intriguing. To further understand the important contribution of SNAT2 to the regulation of protein synthesis in whole animals, more studies are needed.
4.1.2. LAT1
System L amino acid transporter 1 (LAT 1) is a Na+-independent amino acid transporter that facilitates the transport of large neutral amino acids such as phenylalanine, tyrosine, leucine, and tryptophan (65). To function, LAT1 requires the formation of a heterodimer complex with 4F2hc/CD98 (a heavy chain of the cell surface antigen) (66). Using a Xenopus oocytes expression system, several studies showed that LAT1 is an obligatory exchanger with a 1:1 stoichiometry and its function is controlled by intracellular amino acids (65). As shown in Figure 3, intracellular glutamine is crucial for cellular uptake of leucine through LAT1. LAT1 is expressed in various tissues including skeletal muscle (67). Interestingly, LAT1 expression is significantly up-regulated in various cancer cells, making it an excellent target for cancer therapy (68). Recent studies showed that a LAT1 inhibitor blocked the transport of leucine, resulting in the suppression of cancer cell growth (68). While the role of LAT1 in vivo is less well studied, it is interesting to find that LAT1 abundance in skeletal muscle of pigs is significantly higher during the neonatal period than in older pigs and is positively correlated with the activation of mTOR (unpublished data). In human skeletal muscle, Dummond et al. (63) recently showed that LAT1 protein abundance was significantly increased 3 hours after the ingestion of essential amino acids.
4.1.3. PAT
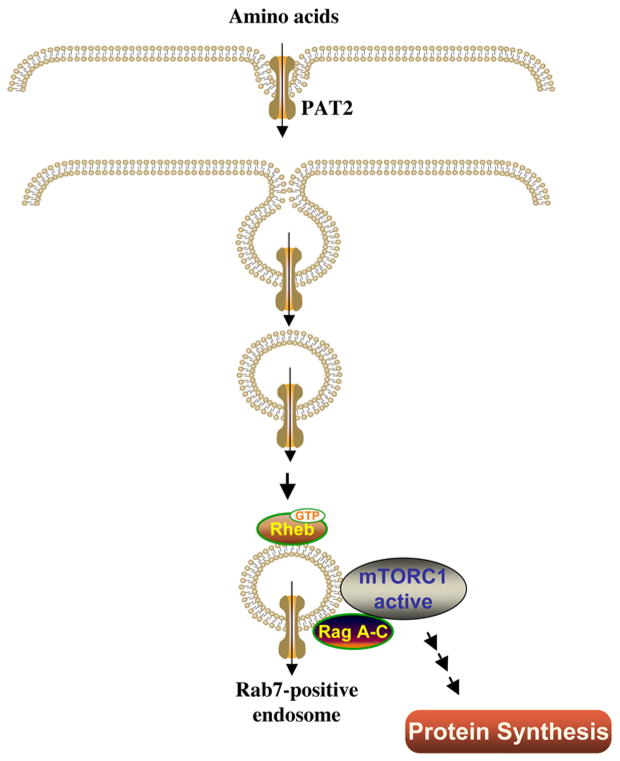
Genetic study in Drosophila highlighted the proton-assisted amino acid transporter (PAT) family as important transporters that have potent effects on growth (69). In general, PAT facilitates the transport of simple amino acids such as alanine, glycine, and proline. However, a recent study in Xenopus oocytes suggested that the PATs act as transceptors (55). PATs use a signaling- rather than a transport-dependent mechanism to regulate mTOR activation (69). In this model (Figure 4), upon activation, PAT undergoes internalization in the form of endosomes. These PAT-activated endosomes then interact with the mTORC1-rheb-rag complex, resulting in the activation of mTORC1 (69). Interestingly, several lines of compelling evidence suggest that PAT-activated endosomes are not only crucial for amino acid-induced activation of mTORC1, but also important for insulin-activated mTORC1, as well (69). At this point, the study of the PATs in mammalian species is very limited. A recent cell culture study indicated that the PAT isoforms, PAT1 and PAT4, are regulators of the amino-acid-dependent activation of mTORC1 in human MCF-7 breast cancer and HEK-293 cell lines (70). In addition, a recent human study showed that the mRNA of PAT1 in skeletal muscle was induced 1 hour after ingestion of essential amino acids (63). Our observations (64) also indicate that the protein abundance of another PAT isoform (PAT2) was highly expressed in skeletal muscle of neonatal pigs and was developmentally regulated. These early observations on the role of PATs in mTORC1 activation in cell cultures and on the abundance in skeletal muscle of mammalian species have shed new light regarding the role of PAT in mTORC1 activation.
Figure 4.
The molecular model of amino acid-induced activation of mTORC1 and protein synthesis by PAT2.
4.2. Role of amino acids in the activation of putative intracellular regulators of mTORC1
For more that a decade, investigators have studied the molecular mechanism by which amino acids activate mTORC1 leading to the stimulation of protein synthesis (39). Control of mTORC1 regulation by amino acids is complex and has not been completely elucidated. Understanding the exact role of amino acids in triggering the activation of mTORC1 has been challenging due to multiple targets in various cell types (75). There are several regulators of mTORC1 that potentially can be regulated by amino acids, and these include raptor, Rheb, MAP4K3, Vps34, and Rag GTPases (39) (Figure 2).
4.2.1. Raptor
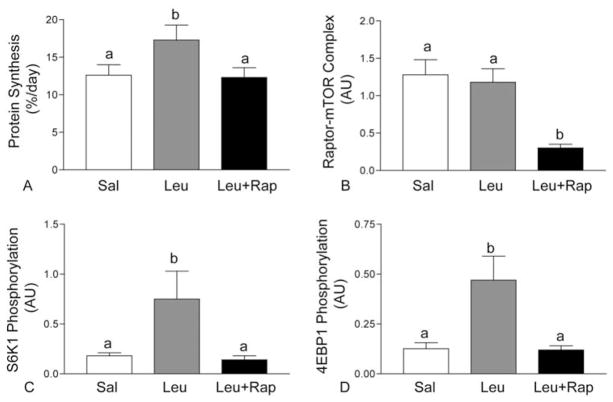
Regulatory associated protein of mTOR (raptor) is a 150 kDa binding protein and is a critical component of mTORC1 that regulates cell growth in response to nutrient levels by associating with mTOR (71). Genetic studies have shown that raptor deletion is embryonic lethal in mice, suggesting that raptor is an important component of mTORC1 (72). Raptor binding to mTOR is essential for mTOR to effectively phosphorylate downstream substrates such as S6K1 and 4EBP1 (73). The nature of raptor-mTOR interaction, in connection with mTORC1 activation, is unclear. Cell culture studies indicate that the interaction of mTOR and raptor is partly responsible for the activation of mTORC1 (71). However, our in vivo studies (74, 31) and others (75) show that neither feeding, amino acid infusion, nor leucine infusion affects the interaction of mTOR with raptor. Nevertheless, it is clear that raptor-mTOR association plays a crucial role in mTORC1 activation, since the disruption of this complex by rapamycin severely inhibits the activation of mTORC1 (76). To examine the importance of the raptor-mTOR association in vivo, we conducted a study on the effect of rapamycin on the leucine-induced stimulation of protein synthesis and the leucine-regulated activation of translation initiation factors in skeletal muscle of neonatal pigs (74). Rapamycin treatment completely blocked the leucine-induced stimulation of muscle protein synthesis (Figure 5A), which was partly due to a severe reduction in the raptor-mTOR interaction (Figure 5B). As a consequence of raptor-mTOR complex inhibition, the phosphorylation of S6K1 and 4EBP1 was significantly reduced (Figure 5C and 5D, respectively). Taken together, our study demonstrates that raptor-mTOR interaction in vivo is crucial for the mTORC1 pathway leading towards protein synthesis.
Figure 5.
Fractional rates of protein synthesis (A), raptor-mTOR interaction (B), S6K1 phosphorylation (C) and 4E-BP1 phosphorylation in longissimus dorsi (LD) muscle of 7-day-old pigs after 60 min of infusion of saline (Sal), 400 μmol·kg−1·h−1 leucine (Leu), and leucine with rapamycin (Leu+Rap). Values are means ± SEM; means with different letters differ at P < 0.05.
4.2.2. Rheb
The protein, Ras homolog enriched in brain (Rheb), plays a key role as a positive regulator of the mTORC1 pathway in integrating inputs from nutrient and growth factors for cell growth (77). An early observation concerning Rheb function revealed that this small GTPase is located downstream of TSC 1/2 and is very crucial for TOR activation (78). While it is clear that in order to activate mTORC1, insulin must inactivate TSC 1/2 (a GTPase-activator complex for Rheb), and that Rheb-GTP activates mTORC1, the amino acid pathways that lead to Rheb-induced activation of mTORC1 are more complicated (79). Although it is becoming apparent that amino acids do not regulate Rheb through TSC 1/2, the amino acid sensing mechanisms toward mTORC1 seems to be more complicated than initially anticipated (79). Early evidence from cell culture work suggests that Rheb binds directly to the amino-terminal lobe of the mTOR catalytic domain and activates mTORC1 in a GTP dependent manner (80). The authors further showed that withdrawal of all extracellular amino acids or just leucine from the culture medium inhibits Rheb-mTORC1 interaction. While the interaction of Rheb with mTORC1 appears crucial for mTORC1 activation, it does not induce autophosphorylation of mTOR (81). We have tried to detect Rheb-mTORC1 interaction in vivo but without success. This is probably due to the transient nature of the Rheb-mTORC1 interaction present during in vivo conditions. Furthermore, when we determined the protein abundance of Rheb in neonatal pigs, we did not find any developmental effect, suggesting that more studies are needed to elucidate the role of Rheb in the regulation of mTORC1 in skeletal muscle of neonates.
More recently, a new mode of action of Rheb in the regulation of mTORC1 was revealed by Bai et al. (82). In this model, it was proposed that Rheb regulates mTOR through an inhibitor called FK506-biding protein 38 (FKBP38). Under amino acid-rich conditions, Rheb directly interacts with FKBP38 and prevents its association with mTOR in a GTP-dependent manner. In this experimental condition, however, it was not clear whether the presence of subpopulations of Rheb that interacted with mTOR were taken into account. Furthermore, several findings from in vitro and cell culture studies appear to be unsupportive of this model. Using HEK293 cells, Wang et al. (83) confirmed that Rheb binds to FKBP38, but this binding was not affected by amino acids and had no effect on mTORC1 activation. In a more recent study, Uhlenbrock et al. (84) conducted a comprehensive biochemical characterization of the Rheb-FKBP38 interaction and found no detectable interaction between Rheb and FKBP38. Our efforts to determine an effect of amino acids on the Rheb-FKBP38 interaction in skeletal muscle of neonatal pigs resulted in no detection of an interaction (64). However, we were able to detect the presence of FKBP38 in skeletal muscle of neonatal pigs, although its protein abundance was not affected by age (64).
In another model, Sun et al. (85) suggested that phospholipase D1 (PLD1) plays a key role in the Rheb-induced activation of mTOR1. They found that upon growth factor or amino acid induction, Rheb binds and activates PLD1. Activated PLD1 generates the production of phosphatidic acid (PA), a lipid second messenger that can mediate the activation of mTORC1 (86). Whether PLD1 will be identified as the long sought after Rheb effector linking Rheb to mTORC1 is still unclear. Interestingly, we recently showed that Rheb forms a protein-protein complex with PLD1 and the abundance of this complex is developmentally regulated, consistent with the activation of mTORC1 in skeletal muscle of neonatal pigs. Nevertheless, more in vivo studies are needed to determine the role of the Rheb-PLD1 pathway in the amino acid-induced activation of mTORC1.
4.2.3. MAP4K3
Mitogen-activated protein kinase kinase kinase kinase-3 (MAP4K3) is a new component that has been implicated as part of the amino-acid sensing pathway toward mTORC1 activation (87). MAP4K3 was recognized as a Drosophila protein kinase responsible for the activation of dS6K, an effector of mTOR (87). Further tests using mammalian cell lines provided convincing evidence that MAP4K3 serves as an amino acid sensor responsible for activating mTORC1. Unlike Rheb, MAP4K3 is regulated by amino acids, but not by insulin (88), although the exact molecular mechanism by which activated MAP4K3 promotes mTORC1 activation is not well understood. However, recent observations from this same group identified a transautophosphorylation site in the MAP4K3 kinase activation segment (Ser170) that is required for MAP4K3 activity (89). The study showed that when amino acids were removed from the cell culture media, Ser170 of MAP4K3 is dephosphorylated by protein phosphatase 2 A (PP2A). Nonetheless, how amino acid sufficiency triggers the phosphorylation of MAP4K3 at Ser170 is unknown. A recent genetic study in Drosophila revealed that MAP4K3 is a regulator of animal growth and metabolism (90). MAP4K3 mutants display phenotypes characteristic of low mTOC1 activity, i.e., reduced growth rate, small body size, and low lipid reserves (90). The possible role of MAP4K3 in the regulation of animal growth in mammalian species is yet to be studied in detail.
4.2.4. Vps34
The class III PI3K, vacuolar protein sorting 34 (Vps34), is the primordial member of the PI 3 kinase family involved in vesicular trafficking, nutrient signaling, and autophagy (91). In 2005, two independent groups discovered almost simultaneously that human Vps34 (hVps34) acts as sensor that mediates amino acid sensing to mTOR, resulting in activation of mTORC1 (91, 92). The molecular mechanism by which hVps34 regulates mTORC1 activation is largely unknown and highly controversial. Early findings showed that introduction of amino acids to culture media induces a rise in intracellular Ca2+, leading to enhanced binding of calmodulin (CaM) to hVps34, resulting in the activation of mTORC1 (92). However, this hypothesis was strongly disputed by Backer’s group (93). In their hands, hVps34 activity toward mTORC1 is independent of Ca2+/CaM. Instead, they proposed that hVps34 activity is regulated through its interactions with hVps15 (93). Since both of the groups used mammalian cell lines in their studies, the exact mechanism of amino acid-induced activation of mTORC1 via hVps34 in the whole animal (mammalian species) still needs to be elucidated. Interestingly, deletion of Vps34 gene in Drosophila had no effect on TOR signaling. Furthermore, unlike mTORC1 activation, the protein abundance of Vps34 in skeletal muscle of neonatal pigs is not affected by age (64). Since Vps34 is considered to be one of the “true” amino acid sensors (i.e., is not affected by hormones/growth factors), in vivo studies are warranted to evaluate the role of Vps34 in protein synthesis.
4.2.4. Rag GTPases
Two independent investigators with different research approaches have reported that Rag GTPases are important for amino acid-induced mTORC1 activation (94, 95). In mammals, the Rag subfamily of Ras small GTPases consist of four members; RagA, RagB, RagC, and RagD (94, 95). They exist in a heterodimer of RagA or RagB with RagC or RagD (94, 95). Using HEK293 cells, Sancak et al. (94) showed that the introduction of expressed mutant Rag GTPases (permanently bound to GDP) induced a reduction in the activation of mTORC1. Conversely, the introduction of other mutant Rag GTPases resulted in increased mTORC1 activation and exhibited resistance to inhibition by amino acid removal. Convincing evidence has shed new light on the molecular mechanism by which Rag GTPases activate mTORC1 (94,95). These studies suggest that in order to activate mTORC1, first, Rag GTPases have to directly interact with raptor, which is facilitated by amino acids. However, an in vitro study also showed that Rag GTPases-raptor association does not stimulate mTORC1 kinase activity, but that Rheb is necessary for Rag GTPase-induced activation of mTOR. Thus, the recent model is as follows: an amino acid signal induces RagA or RagB-GTP binding, resulting in their interaction with raptor. This interaction then promotes the relocation of mTORC1 to vesicles containing Rheb. By an as yet unknown mechanism, these protein-protein interactions initiate the activation of mTORC1. To the best of our knowledge, to date there has been no study that has examined this hypothesis in vivo. Interestingly, in our initial observations we detected a RagB-raptor association in skeletal muscle of neonatal pigs. Consistent with the activation of mTORC1, RagB-raptor association also decreased with age (64). Although these preliminary data are convincing, more in vivo studies are needed to elucidate the role of Rag GTPases in the regulation of protein synthesis in skeletal muscle.
5. AMINO ACID SENSING MECHANISMS THAT REGULATE TRANSLATION INITIATION AND ELONGATION
Principally, protein synthesis is controlled at the level of the translation initiation stage which allows the rapid and reversible control of gene expression (96, 97). However, the process of translation elongation, which requires a substantial amount of metabolic energy, is also important for protein synthesis (98). There is a large body of evidence indicating the involvement of amino acids in the regulation of these two metabolically important processes (17).
Even though translation initiation in eukaryotes involves multiple steps, two are predominantly essential steps for the overall process: the binding of initiator methionyl-tRNA (met-tRNAi) to the 40S ribosomal subunit to form the 43S preinitiation complex, and the binding of mRNA to the 43S preinitiation complex (99). In the first step, eIF2 binds GTP and met-tRNAi, resulting in the ternary complex that binds to the 40S ribosomal subunit (reviewed in Reference 100). In the next step, the GTP bound to eIF2 is hydrolyzed to GDP, and the eIF2-GDP binary complex is detached from the ribosome. In this process, in order for eIF2 to bind to tRNAi and reform the active ternary complex, the GDP bound to eIF2 must be exchanged for GTP by the action of eIF2B. Thus, eIF2B catalyzes the guanine nucleotide exchange reaction to refurnish the eIF2-GTP pool (96). Cells contain a limited amount of eIF2B compared with eIF2, therefore, eIF2B is considered a main regulator of the overall rate of translation initiation (101). There are several ways to regulate the activity of eIF2B. The most recognized mechanism is the inhibition of eIF2B activity through the phosphorylation of the alpha-subunit of eIF2 (100) (Figure 6). The phosphorylation state of eIF2alpha transforms this signaling component into a potent inhibitor for eIF2B, resulting in the sequestering eIF2B into a nonproductive complex that prevents further the initiation process (100). Considering that all mRNA translation begins with met-tRNAi, the state of eIF2alpha phosphorylation is crucial for the up- or down-regulation of the synthesis of almost all proteins (100). Under various stress conditions, such as fasting or starvation, eIF2B activity is decreased, resulting in the reduction of protein synthesis (101). Interestingly, the results from our study (102) and others (103) showed that refeeding the animals after an overnight fast does not alter eIF2B activity. Conversely, in young humans, ingestion of essential amino acids after a bout of resistance exercise reduced eIF2alpha phosphorylation which positively correlated with the increase in muscle protein synthesis (26). In an acute study, orally administration of leucine had no effect on the eIF2alpha phosphorylation in rat skeletal muscle (23). However, in a relatively chronic study in our laboratory, 24 hour leucine infusion reduced eIF2alpha phosphorylation in parallel with an increase in muscle protein synthesis in neonatal pigs (31).
Figure 6.
The regulation of protein synthesis by amino acids and insulin.
The second predominant step of translation initiation, the binding of mRNA to the initiation complex is facilitated by a group of initiation factors called eIF4F complex (reviewed in Reference 99). The eIF4F is comprised of eIF4A, an RNA helicase, eIF4E, the protein that binds the m7GTP cap at the 5′-end on the mRNA, and eIF4G, a scaffolding protein that binds to eIF4A and eIF4E and also binds to the 43S preinitiation complex. The eIF4E-eIF4G complex, which is highly regulated by 4E-BP1, is crucial for the 43S preinitiation complex binding with mRNA. 4EBP1 acts as a potent inhibitor of translation initiation by partnering with eIF4E. Thus, when unphosphorylated, 4E-BP1 forms an inactive complex with eIF4E, resulting in the inhibition of translation initiation. On the contrary, in the hyperphosphorylated state, 4E-BP1 is no longer able to bind to eIF4E; this allows eIF4E and eIF4G to form an active complex (Figure 6). Both insulin and amino acids can induce phosphorylation of 4E-BP1 in an mTORC1-dependent fashion (17). Our studies using neonatal pigs clearly showed that either amino acid infusion or infusion of leucine alone significantly enhances the phosphorylation of 4E-BP1 in skeletal muscle (17, 22). Our results are in agreement with those of Anthony et al. (23) who showed that leucine stimulates phosphorylation of 4E-BP1 in skeletal muscle of mature rats.
Another mechanism that regulates the selection of mRNAs for translation (reviewed in Reference 99) involves modulation of the activity of the 70 kDa ribosomal protein (rp)S6 kinase (S6K1). Its target, rpS6 (Figure 6), is associated with the regulation of translation of mRNAs that contain an uninterrupted stretch of pyrimidine residues adjacent to the 5′-cap structure. These mRNAs are called terminal oligopyrimidine (TOP) mRNAs. Several pieces of evidence suggest that TOP mRNAs are coded for proteins involved in the protein synthetic apparatuses, such as ribosomal proteins, elongation factors, and poly (A) binding proteins. Thus, it is suggested that the activation of S6K1 is closely related with increasing the protein synthetic capacity of cells. Interestingly, our studies (17, 22) and others (23) show that amino acids as well as leucine significantly induce the activation of S6K1 by enhancing its phosphorylation.
During the elongation process, eukaryotic elongation factor 2 (eEF2) plays a crucial role in protein synthesis by catalyzing the translocation of two tRNAs and the mRNA after peptidyl transfer on the 80 S ribosome (104). Thus, eEF2 is responsible for mediating the translocation of the ribosome relative to the mRNA after addition of each amino acid to the nascent chain (98). The activity of eEF2 is regulated by reversible phosphorylation at Thr56, located within the GTP-binding domain of the protein (105). The kinase that catalyzes the phosphorylation of eEF2 is eEF2 kinase (98). Thus, phosphorylation of eEF2 by eEF2 kinase causes inactivation of eEF2, resulting in inhibition of the elongation process (98). Both insulin and amino acids have been shown to positively regulate the activation of eEF2 by inhibiting eEF2 kinase activation in an mTORC1-dependent mechanism (Figure 6) (98). Several in vivo studies (26, 106) including ours (74) indicate that administration of amino acids or leucine does not affect the phosphorylation of eEF2, suggesting that under normal physiological conditions, eEF2 is continuously active and the elongation process is not limiting for protein synthesis.
6. SUMMARY AND PERSPECTIVES
Using neonatal pigs, a widely accepted model for human infants, we have made significant progress in determining the effect of feeding on the rate of protein synthesis in skeletal muscle, a marker for neonatal growth. We have revealed that both insulin and amino acids independently regulate protein synthesis in skeletal muscle of neonates. Furthermore, we have also demonstrated that the high postprandial rate of protein synthesis in neonates is in part due to the enhanced activation of insulin/amino acid signaling components leading to mRNA translation. Unlike the insulin signaling pathway, the amino acid signaling pathway that controls protein synthesis is less well understood. Since the contribution of amino acids in supporting optimum growth in the infant is evident, the understanding of how amino acids regulate protein synthesis in skeletal muscle is essential.
For decades, scientists have used biochemical and genetic tools to unravel the complexities of the amino acid signaling pathways. The initial characterization of these pathways has shown that TOR is a vital target for amino acid sensing. While TOR downstream pathways that regulate protein synthesis are well characterized, the upstream pathways consisting of amino acid sensing components are not well understood at present. It is becoming clear that amino acid transporters serve as initial responders for amino acid sensing. Additionally, amino acid transceptors are also involved in the amino acid pathways that activate TORC1 activation, as least in the Drosophila model. Many other amino acid components/sensors have also been found to facilitate amino acid sensing, including raptor, Rheb, MAP4K4, Vps34, and Rag GTPases. Although each of these components has its own distinct pathway, the overall understanding of how amino acids regulate mTORC1 activation is still sketchy. Moreover, there are discrepancies regarding different models of the amino acid sensing pathways, which are probably due in part to the use of different cell lines or laboratory conditions. More importantly, the roles of amino acid transporter/transceptors and amino acid sensing components have not been fully studied in whole animals. On the contrary, our understanding on the effect of amino acids on the activation of signaling components down stream of mTORC1 seems to be much more settled. Taken together, increased knowledge of the molecular mechanisms that regulate amino acid signaling in vivo may provide important new information that will lead to the development of nutritional interventions to enhance the growth of low birth weight infants.
Acknowledgments
This work was supported in part by the National Institute of Arthritis and Musculoskeletal and Skin Diseases Grant AR-44474 (T.A. Davis) and by the USDA/ARS under Cooperative Agreement no. 6250510000-33 (T.A. Davis). This work is a publication of the United States Department of Agriculture/Agricultural Research Service (USDA/ARS) Children’s Nutrition Research Center, Department of Pediatrics, Baylor College of Medicine. The contents of this publication do not necessarily reflect the views or politics of the U.S. Department of Agriculture, nor does the mention of trade names, commercial products or organizations imply endorsement by the U.S. Government. We thank L.F. Weiser for secretarial assistance.
Abbreviations
- mTORC1/2
mammalian target of rapamycin complex 1/2
- IGF-I
insulin-like growth factorI
- met-tRNA
methionyl-tRNA
- PI 3-kinase
phosphoinositide 3-kinase
- PDK-1
phosphoinositide-dependent kinase-1
- PKB
protein kinase B
- TSC1/2
tuberous sclerosis complex
- S6K
protein S6 kinase
- 4E-BP1
eIF4E-binding protein 1
- GbetaL
G-protein beta-like protein
- SNAT2
sodium-coupled neural amino acid transporter 2
- LAT 1
system L amino acid transporter 1
- PAT
proton-assisted amino acid transporter
- raptor
regulatory associated protein of mTOR
- Rheb
Ras homolog enriched in brain
- FKBP38
FK506-biding protein 38
- PLD1
phospholipase D1
- MAP4K3
mitogen-activated protein kinase kinase kinase kinase-3
- Vps34
class III PI3K vacuolar protein sorting 34
- CaM
calmodulin
- met-tRNAi
methionyl-tRNA
- eIF
eukaryotic initiation factor
- rpS6
ribosomal protein S6
- eEF2
eukaryotic elongation factor 2
References
- 1.Young Vernon. The role of skeletal and cardiac muscle in the regulation of protein metabolism. In: Munro HN, editor. Mammalian protein metabolism. Academic; NY: 1991. [Google Scholar]
- 2.Davis Teresa, Fiorotto Marta. Regulation of muscle growth in neonates. Curr Opin Clin Nutr Metab Care. 2009;2:78–85. doi: 10.1097/MCO.0b013e32831cef9f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis Teresa, Fiorotto Marta, Nguyen Hanh, Reeds Peter. Protein turnover in skeletal muscle of suckling rats. Am J Physiol Regul Integr Comp Physiol. 1989;257:R1141–R1146. doi: 10.1152/ajpregu.1989.257.5.R1141. [DOI] [PubMed] [Google Scholar]
- 4.Davis Teresa, Fiorotto Marta, Beckett Philip, Burrin Douglas, Reeds Peter, Wray-Cahen Diane, Nguyen Hanh. Differential effects of insulin on peripheral and visceral tissue protein synthesis in neonatal pigs. Am J Physiol. 2001;280:E770–E779. doi: 10.1152/ajpendo.2001.280.5.E770. [DOI] [PubMed] [Google Scholar]
- 5.Fiorotto Marta, Davis Teresa, Reeds Peter. Regulation of myofibrillar protein turnover during maturation in normal and undernourished rat pups. Am J Physiol. 2000;278:845–854. doi: 10.1152/ajpregu.2000.278.4.R845. [DOI] [PubMed] [Google Scholar]
- 6.Davis Teresa, Fiorotto Marta, Reeds Peter. Amino acid compositions of body and milk protein change during the suckling period in rats. J Nutr. 1993;123:947–956. doi: 10.1093/jn/123.5.947. [DOI] [PubMed] [Google Scholar]
- 7.Burrin Douglas, Davis Teresa, Ebner Sylvia, Schoknecht Patricia, Fiorotto Marta, Reeds Peter, McAvoy Susan. Nutrient-independent and nutrient-dependent factors stimulate protein synthesis in colostrum-fed newborn pigs. Pediatr Res. 1995;37:593–599. doi: 10.1203/00006450-199505000-00006. [DOI] [PubMed] [Google Scholar]
- 8.Davis Teresa, Burrin Douglas, Fiorotto Marta, Nguyen Hanh. Protein synthesis in skeletal muscle and jejunum is more responsive to feeding in 7- than 26-day-old pigs. Am J Physiol. 1996;270:E802–E809. doi: 10.1152/ajpendo.1996.270.5.E802. [DOI] [PubMed] [Google Scholar]
- 9.Davis Teresa, Fiorotto Marta, Nguyen Hanh, Reeds Peter. Enhanced response of muscle protein synthesis and plasma insulin to food intake in suckled rats. Am J Physiol. 1993;265:R334–R340. doi: 10.1152/ajpregu.1993.265.2.R334. [DOI] [PubMed] [Google Scholar]
- 10.Davis Teresa, Burrin Douglas, Fiorotto Marta, Reeds Peter, Jahoor Farook. Roles of insulin and amino acids in the regulation of protein synthesis in the neonate. J Nutr. 1998;128:347S–350S. doi: 10.1093/jn/128.2.347S. [DOI] [PubMed] [Google Scholar]
- 11.Wray-Cahen Diane, Nguyen Hanh, Burrin Douglas, Beckett Philip, Fiorotto Marta, Reeds Peter, Wester Timothy, Davis Teresa. Response of skeletal muscle protein synthesis to insulin in suckling pigs decreases with development. Am J Physiol. 1998;275:E602–E609. doi: 10.1152/ajpendo.1998.275.4.E602. [DOI] [PubMed] [Google Scholar]
- 12.O’Connor Pamela, Bush Jill, Suryawan Agus, Nguyen Hanh, Davis Teresa. Insulin and amino acids independently stimulate skeletal muscle protein synthesis in neonatal pigs. Am J Physiol. 2003;284:E110–E119. doi: 10.1152/ajpendo.00326.2002. [DOI] [PubMed] [Google Scholar]
- 13.Marintchev Assen, Wagner Gerhard. Translation initiation: structures, mechanisms and evolution. Q Rev Biophys. 2004;37:197–284. doi: 10.1017/S0033583505004026. [DOI] [PubMed] [Google Scholar]
- 14.Kimball Scot. Regulation of translation initiation by amino acids in eukaryotic cells. Prog Mol Subcell Biol. 2001;26:155–184. doi: 10.1007/978-3-642-56688-2_6. [DOI] [PubMed] [Google Scholar]
- 15.Kimball Scot, Jefferson Leonard. Regulation of protein synthesis by branched-chain amino acids. Curr Opin Clin Nutr Metab Care. 2001;4:39–43. doi: 10.1097/00075197-200101000-00008. [DOI] [PubMed] [Google Scholar]
- 16.Kimball Scot, Jefferson Leonard. Control of protein synthesis by amino acid availability. Curr Opin Clin Nutr Metab Care. 2002;5:63–67. doi: 10.1097/00075197-200201000-00012. [DOI] [PubMed] [Google Scholar]
- 17.Proud Christopher. Role of mTOR signaling in the control of translation initiation and elongation by nutrients. Curr Top Microbiol Immunol. 2004;279:215–244. doi: 10.1007/978-3-642-18930-2_13. [DOI] [PubMed] [Google Scholar]
- 18.Proud Christopher. Regulation of protein synthesis by insulin. Biochem Soc Trans. 2006;34:213–216. doi: 10.1042/BST20060213. [DOI] [PubMed] [Google Scholar]
- 19.Suryawan Agus, Orellana Renan, Nguyen Hanh, Jeyapalan Asumthia, Fleming Jillian, Davis Teresa. Activation by insulin and amino acids of signaling components leading to translation initiation in skeletal muscle of neonatal pigs is developmentally regulated. Am J Physiol Endocrinol Metab. 2007;293:E1597–E1605. doi: 10.1152/ajpendo.00307.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reeds Peter, Burrin Douglas, Davis Teresa, Fiorotto Marta. Postnatal growth of gut and muscle: competitors or collaborators. Proc Nutr Soc. 1993;52:57–67. doi: 10.1079/pns19930037. [DOI] [PubMed] [Google Scholar]
- 21.Iresjö Britt-Marie, Svanberg Elisabeth, Lundholm Kent. Reevaluation of amino acid stimulation of protein synthesis in murine- and human-derived skeletal muscle cells assessed by independent techniques. Am J Physiol Endocrinol Metab. 2005;288:E1028–E1037. doi: 10.1152/ajpendo.00295.2004. [DOI] [PubMed] [Google Scholar]
- 22.Escobar Jeffery, Frank Jason, Suryawan Agus, Nguyen Hanh, Kimball Scot, Jefferson Leonard, Davis Teresa. Physiological rise in plasma leucine stimulates muscle protein synthesis in neonatal pigs by enhancing translation initiation factor activation. Am J Physiol Endocrinol Metab. 2005;288:E914–E921. doi: 10.1152/ajpendo.00510.2004. [DOI] [PubMed] [Google Scholar]
- 23.Anthony Joshua, Anthony Tracy, Kimball Scot, Vary Tom, Jefferson Leonard. Orally administered leucine stimulates protein synthesis in skeletal muscle of postabsorptive rats in association with increased eIF4F formation. J Nutr. 2000;130:139–145. doi: 10.1093/jn/130.2.139. [DOI] [PubMed] [Google Scholar]
- 24.Jefferson Leonard, Li Jeanne, Rannels Stephen. Regulation by insulin of amino acid release and protein turnover in the perfused rat hemicorpus. J Biol Chem. 1977;252:1476–1483. [PubMed] [Google Scholar]
- 25.Dardevet Dominique, Sornet Claire, Balage Michele, Grizard Jean. Stimulation of in vitro rat muscle protein synthesis by leucine decreases with age. J Nutr. 2000;130:2630–2635. doi: 10.1093/jn/130.11.2630. [DOI] [PubMed] [Google Scholar]
- 26.Drummond Micah, Bell Jill, Fujita Satoshi, Dreyer Hans, Glynn Erin, Volpi Elena, Rasmussen Blake. Amino acids are necessary for the insulin-induced activation of mTOR/S6K1 signaling and protein synthesis in healthy and insulin resistant human skeletal muscle. Clin Nutr. 2008;27:447–456. doi: 10.1016/j.clnu.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pruznak Anne, Kazi Abid, Frost Robert, Vary Thomas, Lang Charles. Activation of AMP-activated protein kinase by 5-aminoimidazole-4-carboxamide-1-beta-D-ribonucleoside prevents leucine-stimulated protein synthesis in rat skeletal muscle. J Nutr. 2008;138:1887–1894. doi: 10.1093/jn/138.10.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujita Satoshi, Dreyer Hans, Drummond Micah, Glynn Erin, Volpi Elena, Rasmussen Blake. Essential amino acid and carbohydrate ingestion before resistance exercise does not enhance postexercise muscle protein synthesis. J Appl Physiol. 2009;106:1730–1739. doi: 10.1152/japplphysiol.90395.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Escobar Jeffery, Frank Jason, Suryawan Agus, Nguyen Hanh, Kimball Scot, Jefferson Leonard, Davis Teresa. Regulation of cardiac and skeletal muscle protein synthesis by individual branched-chain amino acids in neonatal pigs. Am J Physiol Endocrinol Metab. 2006;290:E612–E621. doi: 10.1152/ajpendo.00402.2005. [DOI] [PubMed] [Google Scholar]
- 30.Escobar Jeffery, Frank Jason, Suryawan Agus, Nguyen Hanh, Davis Teresa. Amino acid availability and age affect the leucine stimulation of protein synthesis and eIF4F formation in muscle. Am J Physiol Endocrinol Metab. 2007;293:E1615–E1621. doi: 10.1152/ajpendo.00302.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson Fiona, Suryawan Agus, Gazzaneo Maria, Orellana Renan, Nguyen Hanh, Davis Teresa. Stimulation of muscle protein synthesis by prolonged parenteral infusion of leucine is dependent on amino acid availability in neonatal pigs. J Nutr. 2010;140:264–270. doi: 10.3945/jn.109.113621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dechant Reinhard, Peter Matthias. Nutrient signals driving cell growth. Curr Opin Cell Biol. 2008;20:678–687. doi: 10.1016/j.ceb.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 33.Talvas Jérémie, Obled Alain, Fafournoux Pierre, Mordier Sylvie. Regulation of protein synthesis by leucine starvation involves distinct mechanisms in mouse C2C12 myoblasts and myotubes. J Nutr. 2006;136:1466–1471. doi: 10.1093/jn/136.6.1466. [DOI] [PubMed] [Google Scholar]
- 34.Hietakangas Ville, Cohen Stephen. Regulation of tissue growth through nutrient sensing. Annu Rev Genet. 2009;43:389–410. doi: 10.1146/annurev-genet-102108-134815. [DOI] [PubMed] [Google Scholar]
- 35.White Morris, Kahn Ronald. The insulin-signaling system. J Biol Chem. 1994;269:1–4. [PubMed] [Google Scholar]
- 36.Pende Mario. mTOR, Akt, S6 kinases and the control of skeletal muscle growth. Bull Cancer. 2006;93:E39–E43. [PubMed] [Google Scholar]
- 37.Wang Xuemin, Beugnet Anne, Murakami Mirei, Yamanaka Shinya, Proud Christopher. Distinct signaling events downstream of mTOR cooperate to mediate the effects of amino acids and insulin on initiation factor 4E-binding proteins. Mol Cell Biol. 2005;25:2558–2572. doi: 10.1128/MCB.25.7.2558-2572.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Proud Christopher. Regulation of mammalian translation factors by nutrients. Eur J Biochem. 2002;206:5338–5348. doi: 10.1046/j.1432-1033.2002.03292.x. [DOI] [PubMed] [Google Scholar]
- 39.Avruch Joseph, Long Xiaomeng, Ortiz-Vega Sara, Rapley Joseph, Papageorgiou Angela, Dai Ning. Amino acid regulation of TOR complex 1. Am J Physiol Endocrinol Metab. 2009;296:E592–E602. doi: 10.1152/ajpendo.90645.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suryawan Agus, O’Connor Pamela, Kimball Scot, Bush Jill, Nguyen Hanh, Jefferson Leonard, Davis Teresa. Amino acids do not alter the insulin-induced activation of the insulin signaling pathway in neonatal pigs. J Nutr. 2004;134:24–30. doi: 10.1093/jn/134.1.24. [DOI] [PubMed] [Google Scholar]
- 41.Jozwiak Jaroslaw, Jozwiak Sergiusz, Grzela Tomasz, Maciej Lazarczyk M. Positive and negative regulation of TSC2 activity and its effects on downstream effectors of the mTOR pathway. Neuromolecular Med. 2005;7:287–296. doi: 10.1385/NMM:7:4:287. [DOI] [PubMed] [Google Scholar]
- 42.Yonezawa Kazuyoshi, Yoshino Ken-ichi, Tokunaga Chiharu, Kenta Hara K. Kinase activities associated with mTOR. Curr Top Microbiol Immunol. 2004;279:271–282. doi: 10.1007/978-3-642-18930-2_16. [DOI] [PubMed] [Google Scholar]
- 43.Tokunaga Chiharu, Yoshino Ken-ichi, Yonezawa Kazuyoshi. mTOR integrates amino acid- and energy-sensing pathways. Biochem Biophys Res Commun. 2004;313:443–446. doi: 10.1016/j.bbrc.2003.07.019. [DOI] [PubMed] [Google Scholar]
- 44.Schmelzle Tobias, Hall Michael. TOR, a central controller of cell growth. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- 45.Inoki Ken, Ouyang Hongjiau, Li Yong, Guan Kun-Liang. Signaling by target of rapamycin proteins in cell growth control. Microbiol Mol Biol Rev. 2005;69:79–100. doi: 10.1128/MMBR.69.1.79-100.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dann Stephen, Thomas George. The amino acid sensitive TOR pathway from yeast to mammals. FEBS Lett. 2006;580:2821–2829. doi: 10.1016/j.febslet.2006.04.068. [DOI] [PubMed] [Google Scholar]
- 47.Memmott Regan, Dennis Phillip. Akt-dependent and -independent mechanisms of mTOR regulation in cancer. Cell Signal. 2009;21:656–664. doi: 10.1016/j.cellsig.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tremblay Frederic, Jacques Helene, Marette Andre. Modulation of insulin action by dietary proteins and amino acids: role of the mammalian target of rapamycin nutrient sensing pathway. Curr Opin Clin Nutr Metab Care. 2005;8:457–462. doi: 10.1097/01.mco.0000172589.55434.03. [DOI] [PubMed] [Google Scholar]
- 49.Soliman Ghada. The mammalian target of rapamycin signaling network and gene regulation. Curr Opin Lipidol. 2005;16:317–323. doi: 10.1097/01.mol.0000169352.35642.06. [DOI] [PubMed] [Google Scholar]
- 50.Iraqui Ismail, Vissers Stephan, Bernard Florent, Craene Johan-Owen de, Boles Eckhard, Urrestarazu Antonio, André Bruno. Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-Box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol Cell Biol. 1999;19:989–1001. doi: 10.1128/mcb.19.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Sluijters Daphne, Dubbelhuis Peter, Blommaart Edward, Meijer Alfred. Amino-acid-dependent signal transduction. Biochem J. 2000;351:545–550. doi: 10.1042/0264-6021:3510545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lynch Christopher, Fox Heather, Vary Thomas, Jefferson Leonard, Kimball Scot. Regulation of amino acid-sensitive TOR signaling by leucine analogues in adipocytes. J Cell Biochem. 2000;77:234–251. doi: 10.1002/(sici)1097-4644(20000501)77:2<234::aid-jcb7>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 53.Beugnet Anne, Tee Andrew, Taylor Peter, Proud Christopher. Regulation of targets of mTOR (mammalian target of rapamycin) signaling by intracellular amino acid availability. Biochem J. 2003;372:555–566. doi: 10.1042/BJ20021266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nicklin Paul, Bergman Philip, Zhang Bailin, Triantafellow Ellen, Wang Henry, Nyfeler Beat, Yang Haidi, Hild Marc, Kung Charles, Wilson Christopher, Myer Vic, MacKeigan Jeffrey, Porter Jeffrey, Wang Karen, Cantley Lewis, Finan Peter, Murphy Leon. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2000;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taylor Peter. Amino acid transporters: éminences grises of nutrient signaling mechanisms? Biochem Soc Trans. 2009;37:237–341. doi: 10.1042/BST0370237. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Zhou, Grewer Christof. The sodium-coupled neutral amino acid transporter SNAT2 mediates an anion leak conductance that is differentially inhibited by transported substrates. Biophys J. 2007;92:2621–2632. doi: 10.1529/biophysj.106.100776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Evans Kate, Nasim Zeerak, Brown Jeremy, Clapp Emma, Amin Amin, Yang Bin, Herbert Terence, Bevington Alan. Inhibition of SNAT2 by metabolic acidosis enhances proteolysis in skeletal muscle. J Am Soc Nephrol. 2008;19:2119–2129. doi: 10.1681/ASN.2007101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hyde Russell, Hajduch Eric, Powell Darren, Taylor Peter, Hundal Harinder. Ceramide down-regulates System A amino acid transport and protein synthesis in rat skeletal muscle cells. FASEB J. 2005;19:461–463. doi: 10.1096/fj.04-2284fje. [DOI] [PubMed] [Google Scholar]
- 59.Evans Kate, Nasim Zeerak, Brown Jeremy, Butler Heather, Kauser Samira, Varoqui Helene, Erickson Jeffrey, Herbert Terence, Bevington Alan. Acidosis-sensing glutamine pump SNAT2 determines amino acid levels and mammalian target of rapamycin signaling to protein synthesis in L6 muscle cells. J Am Soc Nephrol. 2007;18:1426–1436. doi: 10.1681/ASN.2006091014. [DOI] [PubMed] [Google Scholar]
- 60.Hyde Russell, Cwiklinski Emma, MacAulay Katrina, Taylor Peter, Hundal Herinder. Distinct sensor pathways in the hierarchical control of SNAT2, a putative amino acid transceptor, by amino acid availability. J Biol Chem. 2007;282:19788–19798. doi: 10.1074/jbc.M611520200. [DOI] [PubMed] [Google Scholar]
- 61.Franchi-Gazzola Renata, Dall’Asta Valeria, Sala Roberto, Visigalli Rossana, Bevilacqua Elene, Gaccioli Francesca, Gazzola Gian, Bussolati Ovidio. The role of the neutral amino acid transporter SNAT2 in cell volume regulation. Acta Physiol. 2006;187:273–283. doi: 10.1111/j.1748-1716.2006.01552.x. [DOI] [PubMed] [Google Scholar]
- 62.Jansson Nina, Pettersson Jassica, Haafiz Allah, Ericsson Anette, Palmberg Isabelle, Tranberg Mattias, Ganapathy Vadivel, Powell Theresa, Jansson Thomas. Down-regulation of placental transport of amino acids precedes the development of intrauterine growth restriction in rats fed a low protein diet. J Physiol. 2006;576:935–946. doi: 10.1113/jphysiol.2006.116509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Drummond Micah, Glynn Erin, Fry Christopher, Timmerman Kyle, Volpi Elena, Rasmussen Blake. An increase in essential amino acid availability upregulates amino acid transporter expression in human skeletal muscle. Am J Physiol Endocrinol Metab. 2010;298:E1011–1018. doi: 10.1152/ajpendo.00690.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suryawan Agus, Davis Teresa. Abundance and activation of mTORC1 regulators in skeletal muscle of neonatal pigs are modulated by insulin, amino acids, and age. J Appl Physiol. 2010 doi: 10.1152/japplphysiol.00428.2010. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Uchino Hiroshi, Kanai Yoshikatsu, Kim Do Kyung, Wempe Michael, Chairoungdua Arthit, Morimoto Emiko, Anders MW, Endou Hitoshi. Transport of amino acid-related compounds mediated by L-type amino acid transporter 1 (LAT1): insights into the mechanisms of substrate recognition. Mol Pharmacol. 2002;61:729–737. doi: 10.1124/mol.61.4.729. [DOI] [PubMed] [Google Scholar]
- 66.Prasad Puttur, Wang Haiping, Huang Wei, Kekuda Ramesh, Rajan Deva, Leibach Frederick, Ganapathy Vadivel. Human LAT1, a subunit of system L amino acid transporter: molecular cloning and transport function. Biochem Biophys Res Commun. 1999;255:283–288. doi: 10.1006/bbrc.1999.0206. [DOI] [PubMed] [Google Scholar]
- 67.del Amo Eva, Urtti Arto, Yliperttula Marjo. Pharmacokinetic role of L-type amino acid transporters LAT1 and LAT2. Eur J Pharm Sci. 2008;35:161–174. doi: 10.1016/j.ejps.2008.06.015. [DOI] [PubMed] [Google Scholar]
- 68.Kim Chun Hee, Park Hyung Je, Park Jin Kyoo, Kanai Jae Yong, Endou Beon Ku, Park Tae Sub, Kim Do. The RNA interference of amino acid transporter LAT1 inhibits the growth of KB human oral cancer cells. Anticancer Res. 2006;26:2943–2948. [PubMed] [Google Scholar]
- 69.Goberdhan Deborah, Meredith David, Boyd Richard, Wilson Clive. PAT-related amino acid transporters regulate growth via a novel mechanism that does not require bulk transport of amino acids. Development. 2005;132:2365–2375. doi: 10.1242/dev.01821. [DOI] [PubMed] [Google Scholar]
- 70.Heublein S, Kazi Shubana, Ogmundsdóttir Margret, Attwood EV, Kala Sanjay, Boyd Richard, Wilson Clive, Goberdhan Deborah. Proton-assisted amino-acid transporters are conserved regulators of proliferation and amino-acid-dependent mTORC1 activation. Oncogene. 2010;29:4068–4079. doi: 10.1038/onc.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim Do-Hyung, Sabatini David. Raptor and mTOR: subunits of a nutrient-sensitive complex. Curr Top Microbiol Immunol. 2004;279:259–270. doi: 10.1007/978-3-642-18930-2_15. [DOI] [PubMed] [Google Scholar]
- 72.Guertin David, Stevens Deanna, Thoreen Carson, Burds Aurora, Kalaany Nada, Moffat Jason, Brown Michael, Fitzgerald Kevin, Sabatini David. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 73.Yonezawa Kazuyoshi, Tokunaga Chiharu, Oshiro Noriko, Yoshino Ken-ichi. Raptor, a binding partner of target of rapamycin. Biochem Biophys Res Commun. 2004;313:437–441. doi: 10.1016/j.bbrc.2003.07.018. [DOI] [PubMed] [Google Scholar]
- 74.Suryawan Agus, Jeyapalan Asumthia, Orellana Renan, Wilson Fiona, Nguyen Hanh, Davis Teresa. Leucine stimulates protein synthesis in skeletal muscle of neonatal pigs by enhancing mTORC1 activation. Am J Physiol Endocrinol Metab. 2008;295:E868–675. doi: 10.1152/ajpendo.90314.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vary Thomas, Gina Deiter G, Lynch Christopher. Rapamycin limits formation of active eukaryotic initiation factor 4F complex following meal feeding in rat hearts. J Nutr. 2007;137:1857–1862. doi: 10.1093/jn/137.8.1857. [DOI] [PubMed] [Google Scholar]
- 76.Oshiro Noriko, Yoshino Ken-ichi, Hidayat Sujuti, Tokunaga Chiharu, Hara Kenta, Eguchi Satoshi, Avruch Joseph, Yonezawa Kazuyoshi. Dissociation of raptor from mTOR is a mechanism of rapamycin-induced inhibition of mTOR function. Genes Cells. 2004;9:359–366. doi: 10.1111/j.1356-9597.2004.00727.x. [DOI] [PubMed] [Google Scholar]
- 77.Saucedo Leslie, Gao Xinsheng, Chiarelli Dominic, Li Ling, Pan Duoija, Edgar Bruce. Rheb promotes cell growth as a component of the insulin/TOR signaling network. Nat Cell Biol. 2003;5:566–571. doi: 10.1038/ncb996. [DOI] [PubMed] [Google Scholar]
- 78.Stocker Hugo, Radimerski Thomas, Schindelholz Benno, Wittwer Franz, Belawat Priyanka, Daram Pierre, Breuer Sebastian, Thomas George, Hafen Ernst. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat Cell Biol. 2003;5:559–565. doi: 10.1038/ncb995. [DOI] [PubMed] [Google Scholar]
- 79.Proud Christopher. Amino acids and mTOR signaling in anabolic function. Biochem Soc Trans. 2007;35:1187–1190. doi: 10.1042/BST0351187. [DOI] [PubMed] [Google Scholar]
- 80.Long Xiameng, Ortiz-Vega Sara, Lin Yenshou, Avruch Joseph. Rheb binding to mammalian target of rapamycin (mTOR) is regulated by amino acid sufficiency. J Biol Chem. 2005;280:23433–23436. doi: 10.1074/jbc.C500169200. [DOI] [PubMed] [Google Scholar]
- 81.Sato Tatsuhiro, Nakashima Akio, Guo Lea, Tamanoi Fuyuhiko. Specific activation of mTORC1 by Rheb G-protein in vitro involves enhanced recruitment of its substrate protein. J Biol Chem. 2009;284:12783–12791. doi: 10.1074/jbc.M809207200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bai Xiaochun, Ma Dongzhu, Liu Anling, Shen Xiaoyun, Wang Qiming J, Liu Yongjian, Jiang Yu. Rheb activates mTOR by antagonizing its endogenous inhibitor, FKBP38. Science. 2007;318:977–980. doi: 10.1126/science.1147379. [DOI] [PubMed] [Google Scholar]
- 83.Wang Xuemin, Fonseca Bruno, Tang Hua, Liu Rui, Elia Androulla, Clemens Michael, Bommer Ulrich-Axel, Proud Christopher. Re-evaluating the roles of proposed modulators of mammalian target of rapamycin complex 1 (mTORC1) signaling. J Biol Chem. 2008;283:30482–30492. doi: 10.1074/jbc.M803348200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Uhlenbrock Katharina, Weiwad Matthias, Wetzker Reinhard, Fischer Guter, Wittinghofer Alfred, Rubio Ignacio. Reassessment of the role of FKBP38 in the Rheb/mTORC1 pathway. FEBS Lett. 2009;583:965–970. doi: 10.1016/j.febslet.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 85.Sun Yan, Fang Yimin, Yoon Mee, Zhang Chongben, Roccio Marta, Zwartkruis Fried, Armstrong Miles, Brown Alex, Chen Jie. Phospholipase D1 is an effector of Rheb in the mTOR pathway. Proc Natl Acad Sci U S A. 2008;105:8286–8291. doi: 10.1073/pnas.0712268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sun Yuting, Jie Chen J. mTOR signaling: PLD takes center stage. Cell Cycle. 2008;7:3118–3123. doi: 10.4161/cc.7.20.6881. [DOI] [PubMed] [Google Scholar]
- 87.Findlay Greg, Yan Lijun, Procter Julia, Mieulet Virginie, Lamb Richard. A MAP4 kinase related to Ste20 is a nutrient-sensitive regulator of mTOR signaling. Biochem J. 2007;403:13–20. doi: 10.1042/BJ20061881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lam David, Martins Miguel. MAP4K3 enhances the expression of the BH3-only protein BID. Cell Cycle. 2009;8:3248–3249. doi: 10.4161/cc.8.20.9572. [DOI] [PubMed] [Google Scholar]
- 89.Yan Lijun, Mieulet Virginie, Burgess Darren, Findlay Greg, Sully Katherine, Procter Julia, Goris Jozef, Janssens Veerle, Morrice Nick, Lamb Richard. PP2A T61 epsilon is an inhibitor of MAP4K3 in nutrient signaling to mTOR. Mol Cell. 2010;37:633–642. doi: 10.1016/j.molcel.2010.01.031. [DOI] [PubMed] [Google Scholar]
- 90.Bryk Boris, Hahn Katrin, Cohen Stephen, Teleman Aurelio. MAP4K3 regulates body size and metabolism in Drosophila. Dev Biol. 2010;344:150–157. doi: 10.1016/j.ydbio.2010.04.027. [DOI] [PubMed] [Google Scholar]
- 91.Yan Ying, Backer Jonathan. Regulation of class III (Vps34) PI3Ks. Biochem Soc Trans. 2007;35:239–241. doi: 10.1042/BST0350239. [DOI] [PubMed] [Google Scholar]
- 92.Gulati Pawan, Thomas George. Nutrient sensing in the mTOR/S6K1 signaling pathway. Biochem Soc Trans. 2007;35:236–238. doi: 10.1042/BST0350236. [DOI] [PubMed] [Google Scholar]
- 93.Yan Ying, Flinn Rory, Wu Haiyan, Schnur Rachel, Backer Jonathan. hVps15, but not Ca2+/CaM, is required for the activity and regulation of hVps34 in mammalian cells. Biochem J. 2009;417:747–755. doi: 10.1042/BJ20081865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sancak Yasemin, Peterson Timothy, Shaul Yoav, Lindquist Robert, Thoreen Carson, Bar-Peled Liron, Sabatini David. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim Eunjung, Goraksha-Hicks Pankuri, Li Li, Neufeld Thomas, Guan Kun-Liang. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sarre Thomas. The phosphorylation of eukaryotic initiation factor 2: a principle of translational control in mammalian cells. Biosystems. 1989;22:311–325. doi: 10.1016/0303-2647(89)90053-1. [DOI] [PubMed] [Google Scholar]
- 97.Jackson Richard, Hellen Christopher, Pestova Tatyana. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Browne Gareth, Proud Christopher. Regulation of peptide-chain elongation in mammalian cells. Eur J Biochem. 2002;269:5360–5368. doi: 10.1046/j.1432-1033.2002.03290.x. [DOI] [PubMed] [Google Scholar]
- 99.Kimball Scot, Jefferson Leonard. Regulation of global and specific mRNA translation by oral administration of branched-chain amino acids. Biochem Biophys Res Commun. 2004;313:423–427. doi: 10.1016/j.bbrc.2003.07.014. [DOI] [PubMed] [Google Scholar]
- 100.Kimball Scot. Regulation of global and specific mRNA translation by amino acids. J Nutr. 2002;132:883–886. doi: 10.1093/jn/132.5.883. [DOI] [PubMed] [Google Scholar]
- 101.Kleijn Miranda, Scheper Gert, Voorma Harry, Thomas Adri. Regulation of translation initiation factors by signal transduction. Eur J Biochem. 1998;253:531–544. doi: 10.1046/j.1432-1327.1998.2530531.x. [DOI] [PubMed] [Google Scholar]
- 102.Davis Teresa, Nguyen Hanh, Suryawan Agus, Bush Jill, Jefferson Leonard, Kimball Scot. Developmental changes in the feeding-induced stimulation of translation initiation in muscle of neonatal pigs. Am J Physiol Endocrinol Metab. 2000;279:E1226–1234. doi: 10.1152/ajpendo.2000.279.6.E1226. [DOI] [PubMed] [Google Scholar]
- 103.Yoshizawa Fumiaki, Kimball Scot, Jefferson Leonard. Modulation of translation initiation in rat skeletal muscle and liver in response to food intake. Biochem Biophys Res Commun. 1997;240:825–831. doi: 10.1006/bbrc.1997.7652. [DOI] [PubMed] [Google Scholar]
- 104.Jørgensen Rene, Merrill A Rod, Andersen Gregers. The life and death of translation elongation factor 2. Biochem Soc Trans. 2006;34:1–6. doi: 10.1042/BST20060001. [DOI] [PubMed] [Google Scholar]
- 105.Redpath Nicholas, Price Nigel, Severinov Konstantin, Proud Christopher. Regulation of elongation factor-2 by multisite phosphorylation. Eur J Biochem. 1993;213:689–699. doi: 10.1111/j.1432-1033.1993.tb17809.x. [DOI] [PubMed] [Google Scholar]
- 106.Glover Elisa, Phillips Stuart, Oates Bryan, Tang Jason, Tarnopolsky Mark, Selby Anna, Smith Kenneth, Rennie Michael. Immobilization induces anabolic resistance in human myofibrillar protein synthesis with low and high dose amino acid infusion. J Physiol. 2008;586:6049–6061. doi: 10.1113/jphysiol.2008.160333. [DOI] [PMC free article] [PubMed] [Google Scholar]