Abstract
The complex retrovirus Human T-cell Leukemia Virus type I (HTLV-1) is the causative agent of adult T-cell leukemia (ATL). Deregulation of cellular transcription is thought to be an important step for T-cell transformation caused by viral infection. HTLV-1 basic leucine zipper factor (HBZ) is one of the viral proteins believed to be involved in this process as it deregulates the expression of numerous cellular genes. In the context of the provirus, HBZ represses HTLV-1 transcription, in part, by binding to the homologous cellular coactivators p300 and CBP. These coactivators play a central role in transcriptional regulation. In this study we determined that HBZ binds with high affinity to the KIX domain of p300/CBP. This domain contains two binding surfaces that are differentially targeted by multiple cellular factors. We show that two ϕXXϕϕ motifs in the activation domain of HBZ mediate binding to a single surface of the KIX domain, the mixed-lineage leukemia (MLL)-binding surface. Formation of this interaction inhibits binding of MLL to the KIX domain while enhancing the binding of the transcription factor, c-Myb to the opposite surface of KIX. Consequently, HBZ inhibits transcriptional activation mediated by MLL and enhances activation mediated by c-Myb. CREB, which binds the same surface of KIX as c-Myb, also exhibited an increase in activity through HBZ. These results indicate that HBZ is able to alter gene expression by competing with transcription factors for occupancy of one surface of KIX while enhancing the binding of factors to the other surface.
Keywords: Coactivators, activation domain, MLL, CREB, c-Myb
INTRODUCTION
Human T-cell Leukemia Virus type 1 (HTLV-1) is a complex retrovirus that is the causative agent of adult T-cell leukemia (ATL), an aggressive and fatal form of leukemia 1; 2; 3. The molecular mechanisms that lead to ATL have not been fully determined, but they were initially believed to predominantly involve pleiotropic activities of the viral protein Tax. However, the most recently discovered HTLV-1-encoded protein, HTLV-1 basic leucine zipper factor (HBZ), is now also implicated in development of this disease 4. Indeed, HBZ has been found to enhance T-cell proliferation, and among the viral proteins, HBZ is the only one consistently expressed in ATL cells from patients 5; 6.
HBZ is believed to contribute to the development of ATL by disrupting cellular gene expression 5; 7; 8; 9. HBZ contains three central basic regions that mediate its nuclear import 10. Within the nucleus HBZ is able to dimerize with a subset of cellular basic leucine zipper (bZIP) transcription factors through its C-terminal leucine zipper 11; 12; 13; 14; 15; 16. Since the amino acid sequence of the basic region of HBZ in proximity to its ZIP diverges from corresponding basic regions of cellular bZIP factors, interactions with HBZ generally prevent the cellular factors from binding their consensus DNA sequences and activating transcription. Although HBZ has not been shown to bind DNA, a recent report indicates that it forms a complex with Maf on the MARE binding site 15. However, it is unclear whether HBZ contacts the DNA in this context. HBZ additionally contains an N-terminal activation domain (AD) that directly contacts the KIX domain of the homologous coactivators p300 and CBP 17.
p300/CBP play an important role in the regulation of gene expression. Each coactivator contains a histone acetyltransferase (HAT) domain that acetylates specific lysine residues on histones. These modifications lead to an open chromatin structure and facilitate recruitment of specific transcriptional regulators directly to the chromatin 18. This enzymatic activity also targets certain transcription factors to modulate their activity 18. In addition to their HAT activity, p300/CBP bridge specific transcription factors to the general transcriptional machinery and are also able to recruit additional regulatory proteins to the DNA 18. Currently, p300/CBP are known to bind over 400 proteins 19.
The KIX domain is highly conserved between the coactivators 20; 21, encompassing amino acids (aa) 566-652 of p300 and 586-672 of CBP 22. This domain serves as a docking site for at least a dozen transcription factors 23. The structure of KIX consists of a bundle of three amphipathic α-helices with a hydrophobic core that contains two distinct protein binding surfaces 20. Interactions involving the activation domain of c-Myb and the phosphorylated kinase-inducible domain (pKID) of CREB have been structurally defined on one surface 20; 24. On the other surface, an interaction with the activation domain of mixed-lineage leukemia protein (MLL) has been well-characterized 22; 25; 26. The MLL-binding surface is targeted by a number of other proteins, including Tax 27; 28. Interestingly, certain combinations of proteins that contact the separate surfaces have been shown to bind simultaneously and cooperatively to the KIX domain 22; 25; 26; 29.
Proteins that bind to the KIX domain have been consistently found to carry a ϕXXϕϕ motif that mediates the interaction, with ϕ denoting a hydrophobic amino acid and X denoting any amino acid 30. These motifs are found within amphipathic α-helices that are thought to form upon binding to the KIX domain 20; 22; 24; 25; 27; 31. Interestingly, the ϕXXϕϕ motifs contribute to binding regardless of which KIX surface is contacted by a specific factor.
We have shown that HBZ represses HTLV-1 transcription through competition with Tax for the KIX domain, which inhibits the recruitment of p300/CBP to Tax:CREB complexes at the viral promoter 17. Based on this observation, we hypothesized that HBZ also alters interactions of cellular factors with KIX, an effect that may contribute to deregulation of cellular gene expression by the viral protein. In this study, we further characterized the HBZ:KIX complex and determined the effects of HBZ on the binding of a select group of transcription factors to the KIX domain. Using a unique electrophoretic mobility shift assay (EMSA), we found that HBZ exhibits a high affinity for the KIX domain, with an apparent Kd of 3 ± 0.5 nM. We show that this affinity is dependent on two ϕXXϕϕ motifs in the activation domain of HBZ. Computational analyses predict that each ϕXXϕϕ motif in HBZ-AD lies within an amphipathic α-helix, consistent with the characteristics of other transcription factors that bind KIX. Also consistent with other proteins that bind KIX, we show that the HBZ:KIX complex adopts a 1:1 stoichiometry. Using site-directed mutagenesis to compromise one or the other binding surface of KIX, we determined that HBZ contacts the MLL-binding surface and effectively competes with MLL for occupancy of this site. In addition, we demonstrated that HBZ enhances the binding of c-Myb to the opposite surface of KIX. Finally, we found that HBZ increased transcriptional activation through the pKID domain of CREB within cells.
RESULTS
HBZ binds with high affinity to the KIX domain of p300/CBP
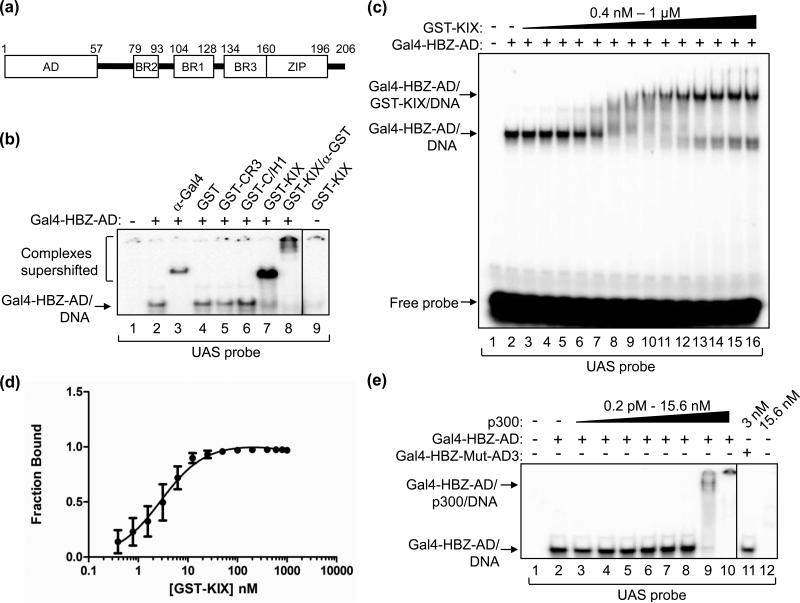
We previously reported that the AD of HBZ binds directly to the KIX domain of p300/CBP and is able to displace Tax from this domain, repressing transcription from the HTLV-1 promoter 17. In order to gauge the potential for HBZ to disrupt functions of other transcription factors whose activities depend on interactions with p300/CBP through the KIX domain, we first designed an EMSA system to quantify the HBZ:KIX interaction. The HBZ polypeptides we used were derived from the major alternative splice variant that produces the 206-amino acid protein, which differs from the other two variants by a small group of amino acids at the N-terminus 5; 32; 33. Since HBZ is not known to bind DNA by itself, our approach was to fuse the DNA-binding domain (DBD) of the yeast transcription factor Gal4 (aa 1-147) to the N-terminus of the HBZ-AD (aa 1-77)(Fig. 1A), allowing HBZ to be indirectly tethered to a 32P-labeled DNA probe containing the Gal4-DBD-binding site (Fig. 1B, lane 2). This DNA:protein complex was supershifted with an antibody recognizing the DBD of Gal4 (lane 3). GST-KIX (aa 588-683) efficiently supershifted Gal4-HBZ-AD and was confirmed to be associated with the complex by addition of an antibody directed against GST, which further retarded migration of the complex (lanes 7 and 8). No supershift was detected with GST alone (lane 4), demonstrating that the GST portion of GST-KIX was not involved in the interaction. Two other domains of p300/CBP that do not interact with HBZ, GST-CR3 (aa 2221-2441) and GST-C/H1 (aa 302-451) 17, also did not supershift Gal4-HBZ-AD (lanes 5 and 6), corroborating the specificity of the interaction. Finally, GST-KIX alone did not bind the probe (lane 9).
Figure 1. HBZ-AD displays high affinity-binding to the KIX domain of p300/CBP.
(a) The schematic representation of HBZ shows the N-terminal activation domain (AD), three basic regions (BR2, BR1 and BR3) and the C-terminal leucine zipper (ZIP). (b) Gal4-HBZ-AD interacts specifically with the KIX domain. EMSA reactions contained the UAS DNA probe (5 nM) and, where indicated, Gal4-HBZ-AD (1 nM). Reactions were supplemented with either anti-Gal4-DBD antibody (5 ng), GST (50 nM), GST-CR3 (50 nM), GST-C/H1 (50 nM), GST-KIX (50 nM) or anti-GST antibody (3.1 μg) as indicated. (c) EMSAs were used to determine the apparent Kd for the Gal4-HBZ-AD:GST-KIX complex. Reactions contained UAS probe (2.5 nM), Gal4-HBZ-AD (1 nM; lanes 2-16) and increasing concentrations of GST-KIX (0.4, 0.8, 1.6, 3.2, 6.25, 12.5, 25, 50, 100, 200, 400, 600, 800, and 1000 nM; lanes 3-16). (d) The binding curve, generated by nonlinear regression analysis, yields an apparent Kd of 3 ± 0.5 nM. The graph shows the fraction of Gal4-HBZ-AD bound to GST-KIX versus the concentration of GST-KIX (nM) as determined by quantification of three independent EMSAs. (E) Gal4-HBZ-AD binds strongly to full-length p300. EMSA reactions contained UAS probe (2.5 nM), Gal4-HBZ-AD (2.5 nM; lanes 2-10) and increasing concentrations of p300 (0.2 pM, 1 pM, 5 pM, 25 pM, 125 pM, 625 pM, 3 nM, and 15.6 nM; lanes 3-10). Lane 11 contained Gal4-HBZ-Mut-AD3 (2.5 nM) and p300 (3 nM); lane 12 contained only p300 (15.6 nM) and the UAS probe. Lanes shown are from the same gel. Vertical lines show where the gel was cropped.
To determine the affinity of the interaction between HBZ-AD and KIX, GST-KIX was titrated into binding reactions with Gal4-HBZ-AD (Fig. 1C, lanes 3-16). The apparent equilibrium dissociation constant (Kd) was calculated according to the appearance of the supershifted complex containing GST-KIX. The apparent Kd was determined to be 3 ± 0.5 nM based on nonlinear regression analysis of the quantified band densities from three independent experiments (Fig. 1D), indicating that HBZ-AD binds strongly to KIX. We additionally used Gal4-HBZ full-length (aa 1-206) with GST-KIX and obtained a similar apparent Kd (data not shown). These results confirm that HBZ-AD is sufficient for the strong affinity between the two proteins.
The KIX domain has been shown to fold and function independently of other domains in p300/CBP 19; 20; 23; 31; 34. To confirm that KIX remains accessible for binding by HBZ-AD within full-length p300, we performed the same experiment using p300 in place of GST-KIX. Figure 1E shows a similar high affinity to that observed using GST-KIX, with the Gal4-HBZ-AD:DNA complex almost completely shifted between 0.625 and 3 nM of p300 (lanes 8 and 9). In control reactions, p300 alone did not bind the probe (lane 12).
The two ϕXXϕϕ motifs in HBZ-AD are necessary for high-affinity KIX-binding
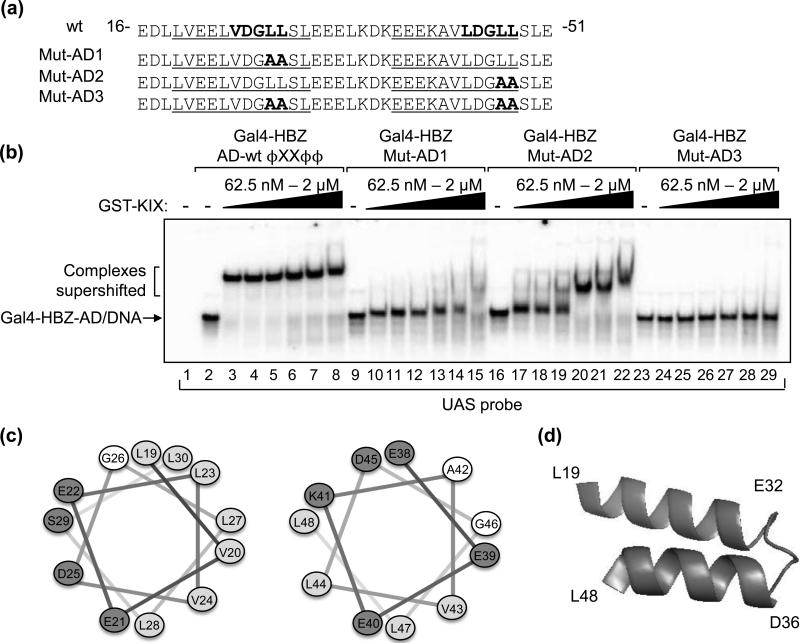
We previously showed that HBZ binds KIX via two ϕXXϕϕ motifs, VDGLL and LDGLL, located at positions 24-28 and 44-48 in HBZ, respectively (Fig. 2A)17. To determine the relative contribution of each ϕXXϕϕ motif to KIX-binding, we mutated the first motif (VDGLL to VDGAA, Gal4-HBZ-Mut-AD1) or the second motif (LDGLL to LDGAA, Gal4-HBZ-Mut-AD2) or both motifs (Gal4-HBZ-Mut-AD3) in the Gal4-HBZ-AD construct (Fig. 2A). We then performed EMSA experiments with each mutant using equivalent GST-KIX titrations and found that these mutants exhibit a reduced capacity to bind GST-KIX (Fig. 2B). In separate experiments, titrations of GST-KIX were carried to higher concentrations than those shown in figure 2B in order to obtain the apparent Kd values for the mutant interactions (data not shown). Mutation of either motif decreased the affinity of the HBZ-AD:KIX interaction, as complexes for Gal4-HBZ-Mut-AD1 and Gal4-HBZ-Mut-AD2 exhibited Kd values of 800 nM and 180 nM, respectively. No supershift was observed for the Gal4-HBZ-Mut-AD3 in figure 2B (lanes 23-29). This mutant also failed to interact with full-length p300 at the concentrations used to supershift Gal4-HBZ-AD wt (Fig. 1D, lane 11). These results indicate that the high-affinity interaction with p300 is modulated through the ϕXXϕϕ motifs in HBZ-AD.
Figure 2. Two ΦXXΦΦ motifs within predicted amphipathic α-helices of HBZ-AD mediate binding to KIX.
(a) Amino acids 16 to 51 of HBZ are shown. Indicated in bold are the two ΦXXΦΦ motifs in the wild-type (wt) sequence and amino acid substitutions in HBZ-Mut-AD1, HBZ-Mut-AD2 and HBZ-Mut-AD3. Sequences predicted to form amphipathic α-helices are underlined. (b) Each ΦXXΦΦ motif contributes to the affinity of Gal4-HBZ-AD for GST-KIX. EMSA reactions contained UAS probe (3.25 nM) and 1nM Gal4-HBZ-AD wt, Gal4-HBZ-Mut-AD1, Gal4-HBZ-Mut-AD2 or Gal4-HBZ-Mut-AD3 as indicated. GST-KIX was titrated into reactions (62.5 nM, 125 nM, 250 nM, 500 nM, 1 μM, and 2 μM) as indicated. (c) Helical wheels show uncharged, hydrophobic, and charged amino acids as unshaded, lightly shaded and darkly shaded, respectively. (d) The schematic shows that the two amphipathic α-helices in HBZ-AD are predicted to form a hairpin structure.
Transcription factors that have well-characterized interactions with the KIX domain contain ϕXXϕϕ motifs within amphipathic α-helices, which mediate the binding 20; 22; 24; 25; 27. Analysis of the HBZ-AD amino acid sequence using secondary structure prediction programs showed a high probability of α-helical formation in each of the two regions containing a ϕXXϕϕ motif, with a break predicted to occur between the helices. Using helical wheel analysis, each predicted helix was found to be amphipathic (Fig. 2C). Therefore, the motifs in HBZ responsible for high-affinity KIX-binding appear similar to those of other transcription factors that contact KIX. Interestingly, the two α-helices of HBZ-AD are predicted to form a helix-loop-helix hairpin tertiary structure (Fig. 2D).
The stoichiometry of the HBZ-AD:KIX complex is 1:1
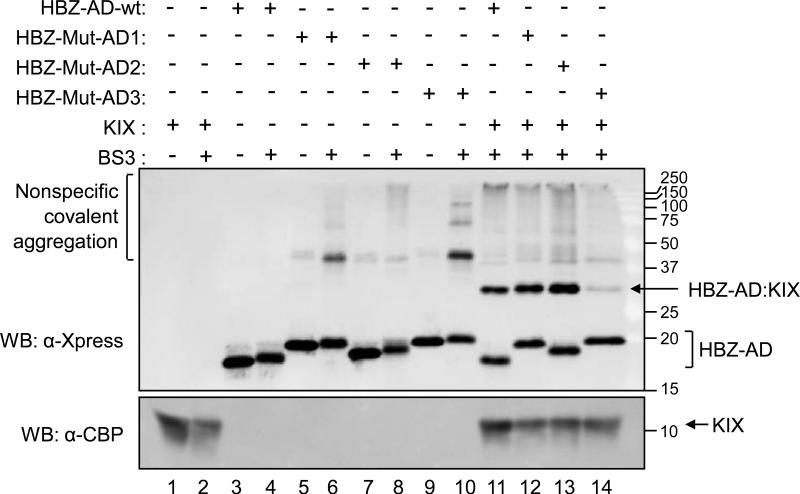
The binding of cellular transcription factors to KIX is commonly mediated by a single ϕXXϕϕ motif. Because HBZ carries two such motifs, it was possible that the HBZ:KIX complex was composed of a 1:2 stoichiometric ratio of HBZ:KIX. To test this possibility, we utilized HBZ-AD wt and each HBZ-AD mutant without the Gal4-DBD fusion. These proteins were individually combined with untagged KIX (aa 587-679) and the crosslinking reagent bis(sulfosuccinimidyl) suberate (BS3). BS3 crosslinks primary amine groups in close proximity to one another, creating covalent bonds between interacting protein species. Crosslinked protein complexes were resolved using SDS-PAGE, and the stoichiometry was deduced based on the apparent molecular weight of the complexes compared to the additive molecular weight of the individual proteins (Fig. 3). Untagged KIX alone migrated at 12 kDa (lane 1). In the absence of KIX, the HBZ-AD polypeptides were detected between 18-20 kDa (lanes 3-10). Although each HBZ-AD construct consists of amino acids 1-77 plus a 6xHis tag and linker (119 aa total), verified by DNA sequencing, their migrations by SDS-PAGE were not equivalent. This phenomenon was reported in a study using mutants of the α-helical hairpin polypeptide in the human cystic fibrosis transmembrane conductance regulator35. In the previous study, variations in gel migration were thought to arise from a relationship between SDS-binding, hydrophobicity and robustness of the tertiary structure. Weak non-specific aggregation was observed with each HBZ-AD construct in the presence of BS3 (lanes 4, 6, 8 and 10). However, in the presence of KIX, the specificity of the HBZ-KIX interaction abrogated these effects (lanes 11-14).
Figure 3. The stoichiometry of the HBZ-AD:KIX complex is 1:1.
Binding reactions contained 750 nM HBZ-AD wt, HBZ-Mut-AD1, HBZ-Mut-AD2 or HBZ-Mut-AD3 as indicated. Reactions additionally contained untagged KIX (27 μM) and/or BS3 crosslinking reagent (1.8 mM) as indicated. Proteins were resolved by SDS-PAGE and detected by Western blot (WB) using the specified antibodies. Molecular weights are shown in kilodaltons.
HBZ-AD wt incubated with untagged KIX and BS3 resulted in a covalently-linked complex formed between KIX and HBZ-AD wt that migrated at 31 kDa, consistent with a 1:1 stoichiometry between the two proteins (lane 11). Interestingly, HBZ-Mut-AD1 and HBZ-Mut-AD2 also formed complexes with KIX in the presence of BS3 (lanes 12 and 13), confirming that these mutants retain the ability to interact with KIX. In this assay, the band intensities of the complexes do not reflect the different affinities of the HBZ-AD constructs for KIX, as higher concentrations of all proteins were used relative to the EMSA experiments.
HBZ-AD interacts with the MLL-binding surface of KIX
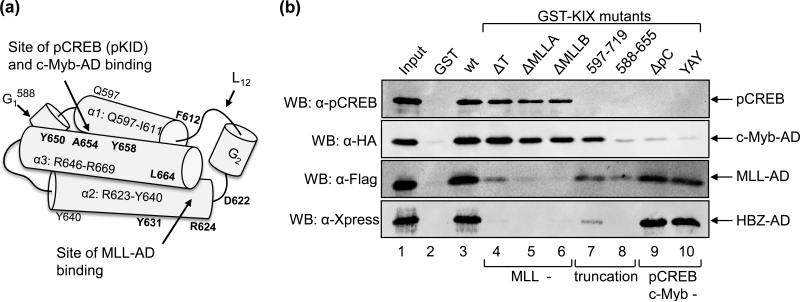
NMR studies have demonstrated that the KIX domain of p300/CBP contains two transcription factor-binding surfaces (Fig. 4A) 20; 22. The first binding surface is a shallow groove formed between α-helices 1 and 3, and binds the pKID domain of pCREB and the AD of c-Myb. The second binding surface is a larger groove located between α-helices 2 and 3. This surface incorporates residues from the C-terminus of α-helix 1 as well as the loop L12 and small 310 helix G2 that connect α-helices 1 and 2. This surface binds a variety of transcription factors, including MLL.
Figure 4. HBZ-AD binds the MLL-binding surface of KIX.
(a) A schematic representation of the KIX domain. The long cylinders represent α-helices 1, 2 and 3 as indicated with their amino acid boundaries (relative to CBP). Short cylinders represent 310 helices. L12 denotes the loop connecting α-helix 1 and the 310 helix G2. Mutated residues in GST-KIX constructs are shown in bold at their approximate locations. The two protein-binding surfaces are identified by arrows. (b) Mutations in the MLL-binding surface of KIX disrupt the HBZ interaction in GST pull-down assays. Binding reactions for pCREB, MLL-AD and HBZ-AD contained 500 nM GST, GST-KIX (aa 588-683) or individual GST-KIX mutants ΔT (F612A, D622A, R624A), ΔMLLA (Y631A, L664A), ΔMLLB (D622A, R624A, Y631A, L664A), GST-KIX597-719, GST-KIX588-655, ΔpC (Y658A), and YAY (Y650A, A654Q, Y658A). Reactions for c-Myb-AD contained 350 nM GST or GST-KIX fusion protein. The concentrations of pCREB, c-Myb-AD, MLL-AD and HBZ-AD used in reactions were 6.5 nM, 5 μM, 83 μM and 150 nM, respectively. Bound proteins were resolved by SDS-PAGE and detected by Western blot (WB) using the antibodies indicated. A fraction of the input is shown in lane 1: pCREB (100%), c-Myb-AD (0.2%), MLL-AD (0.1%), and HBZ-AD (80%).
Experimental evidence indicates that Tax contacts the MLL-binding surface of KIX 27; 28. We previously reported that HBZ competes with Tax for KIX-binding, leading to partial repression of HTLV-1 transcription 17. Based on these results, it was possible that HBZ also binds the MLL-binding surface. Alternatively, HBZ could contact both surfaces based on our findings that HBZ binds KIX with a 1:1 stoichiometry but carries two ϕXXϕϕ motifs that participate in the interaction. With respect to this hypothesis, p53 was recently shown to contain two ϕXXϕϕ motifs located in separate α-helices within its AD, with each motif able to bind simultaneously and synergistically to both surfaces 30. In order to determine whether HBZ-AD contacts a specific surface or both surfaces of KIX, we performed a series of GST pull-down assays using GST-KIX mutants with disruptions in either the pCREB/c-Myb-binding surface or the MLL-binding surface (Table 1; Fig. 4A). All GST-KIX point mutations were introduced into GST-KIX588-683. Figure 4B shows results from GST pull-down assays using the GST-KIX mutants in binding reactions with either pCREB, c-Myb-AD (aa 186-325), MLL-AD (aa 2829-2883) or HBZ-AD. All purified proteins bound specifically to KIX wt (lane 3) since they did not bind to GST alone (lane 2). As expected, pCREB and c-Myb-AD bound GST-KIXΔT (lane 4), GST-KIXΔMLL-A (lane 5) and GST-KIXΔMLL-B (lane 6), which are defective specifically for MLL-binding. Like MLL, HBZ-AD failed to bind these mutants (lanes 4-6). However, HBZ-AD did interact with GST-KIXΔpC (lane 9) and GST-KIXYAY (lane 10), which are defective for pCREB and c-Myb-binding but not MLL-binding. None of the factors bound to GST-KIX588-655 (lane 8), which is structurally disrupted in both surfaces. Finally, all factors, except c-Myb, exhibited very weak or undetectable binding to GST-KIX597-719 (lane 7). Binding of c-Myb-AD to this mutant was unexpected, since c-Myb binds the same surface of KIX as pCREB. The difference in c-Myb and pCREB binding to this mutant is potentially due to reported variations in their mechanisms of binding to KIX 20; 24. Overall, the pattern that emerges from the GST-KIX mutagenesis data indicates that HBZ-AD contacts only the MLL-binding surface.
Table 1.
GST-KIX mutants.
| Mutants name | Factors affected | Mutations | References |
|---|---|---|---|
| ΔT | Tax | F612A, D622A, R624A | 28 a |
| ΔMLL-A | MLL | Y631A, L664A | 22b, 26b |
| ΔMLL-B | MLL | D622A, R624A, Y631A, L664A | 22b, 26b |
| 597-719 | Tax, pCREB | Truncation | 53a, 65c |
| 588-655 | c-Myb, MLL | Truncation | 22d, 65c, 54c |
| ΔpC | pCREB | Y658A | 20a, 28a |
| YAY | c-Myb | Y650A, A654Q, Y658A | 66a,45a |
Reference showing the mutant was defective for the interaction with the denoted factor(s)
Reference used to design the mutant in order to disrupt the interaction with the denoted factor
Reference in which the mutant was used to disrupt the binding of additional factors to KIX
Reference indicating that the mutant contains disruptions in both binding surfaces.
HBZ-AD competes with MLL for KIX-binding
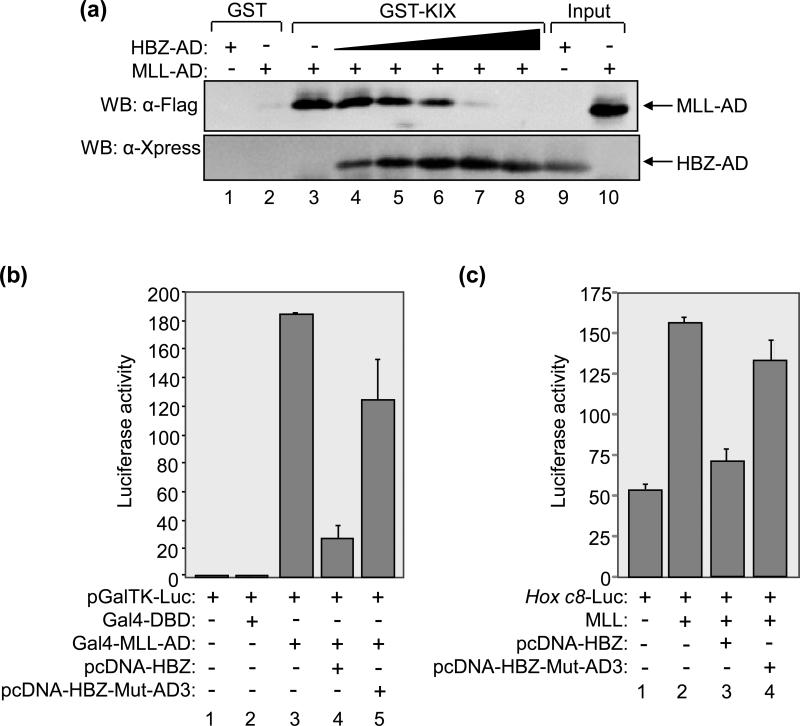
Our findings that HBZ-AD interacts with the MLL-binding surface of KIX suggested that, like Tax, HBZ competes with MLL for KIX-binding. To test this hypothesis, we performed GST pull-down assays in which we titrated HBZ-AD into reactions containing MLL-AD and GST-KIX (Fig. 5A). In these competition binding assays, HBZ-AD bound to GST-KIX and abrogated the interaction between MLL-AD and GST-KIX, indicating that HBZ effectively competes with MLL for KIX-binding (lanes 4-8). Due to the high affinity of the HBZ-AD:KIX interaction, a 15-fold lower concentration of HBZ-AD relative to the concentration of MLL-AD was sufficient to completely displace MLL-AD from GST-KIX (lane 7). Neither protein bound GST alone (lanes 1 and 2).
Figure 5. HBZ-AD competes with MLL-AD for KIX-binding.
(a) HBZ-AD competes with MLL-AD for KIX-binding in GST pull-down assays. Competition binding reactions contained GST-KIX (500 nM) and Flag-MLL-AD (15 μM) (lanes 3-8) and increasing concentrations of HBZ-AD (50 nM, 100 nM, 200 nM, 1 μM, and 5 μM, lanes 4-8). Control binding reactions contained GST (500 nM) with HBZ-AD (5 μM, lane 1) or Flag-MLL-AD (15 μM, lane 2). Fractions of HBZ-AD (2%) and Flag-MLL-AD (0.5%) inputs are shown in lanes 9 and 10, respectively. Bound proteins were resolved by SDS-PAGE and analyzed by Western blot (WB) using the antibodies indicated. (b) HBZ reduces transcriptional activation by MLL-AD in cells. Jurkat cells were transfected with pGalTK-Luc (100 ng) alone or in combination with Gal4-DBD (50 ng), Gal4-MLL-AD (50 ng), pcDNA-HBZ (800 ng) or pcDNA-HBZ-Mut-AD3 (800 ng) as indicated. Luciferase assays were performed 24 hours after transfection. The reported values are the average luminescence ± S.E. from one experiment performed in duplicate and are representative of three independent experiments. (c) HBZ reduces MLL-mediated transcriptional activation from the Hox c8 promoter in cells. 293T/T7 cells were transfected with Hox c8-Luc (200 ng) alone or in combination with Flag-MLL (100 ng), pcDNA-HBZ (700 ng) or pcDNA-HBZ-Mut-AD3 (700 ng) as indicated. Luciferase assays were performed 24 hours after transfection. The reported values are the average luminescence ± S.E. from one experiment performed in triplicate and are representative of three independent experiments.
To determine whether HBZ also competes with MLL for KIX-binding in cells, luciferase assays were performed using Jurkat T-cells. Cells were co-transfected with a luciferase reporter plasmid containing five Gal4 binding sites (pGalTK-Luc)36 and an expression plasmid for MLL-AD (aa 2829 to 2883)25 fused to the DBD of Gal4. A previous study correlated transcriptional activation of this reporter plasmid with binding of MLL-AD to endogenous p300/CBP and recruitment of the coactivator to the promoter 25. Consistent with this report, we obtained strong luciferase activity induced by expression of Gal4-MLL-AD compared to that of Gal4-DBD alone (Fig. 5B, lanes 2 and 3). Additional co-transfection of an expression plasmid for full-length HBZ17 led to a greater than 6-fold reduction in luciferase activity (lane 4). In contrast, only a slight decrease in luciferase activity (<2-fold) was obtained by expression of HBZ carrying mutations in both ϕXXϕϕ motifs (lane5)17.
To determine whether HBZ competes with MLL for KIX-binding at a natural promoter, we used the Hox c8 promoter, which was previously shown to be activated by MLL 37. In the luciferase assay shown in Figure 5C, HBZ repressed activation from the Hox c8 promoter mediated by MLL, while HBZ carrying mutations in both ϕXXϕϕ motifs did not. These results show that HBZ competes effectively with MLL for KIX-binding in vivo.
HBZ-AD enhances the binding of c-Myb-AD to KIX
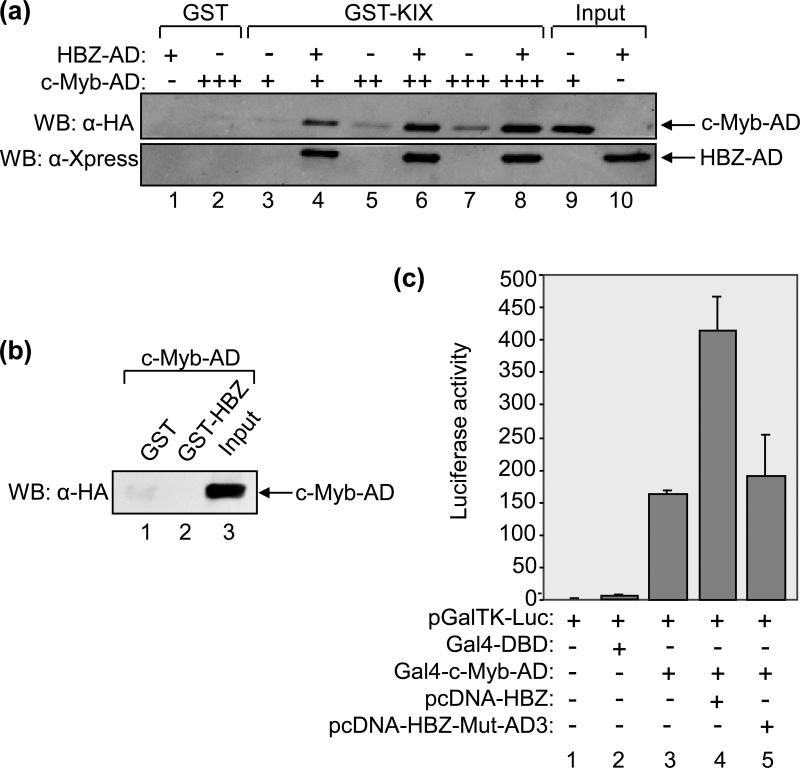
The interaction of MLL-AD with its KIX-binding surface was found to induce cooperative binding of either pCREB or c-Myb-AD to the opposite surface 22; 25; 26. We were therefore interested in determining whether HBZ similarly enhances the binding of these transcription factors to KIX. To test for a potential effect on c-Myb-binding, we performed GST pull-down assays in which we incubated increasing amounts of c-Myb-AD with GST-KIX in the absence or presence of a constant amount of HBZ-AD sufficient to saturate the MLL-binding surface (Fig. 6A, lanes 3-8). In reactions containing HBZ, both the viral protein and c-Myb bound to GST-KIX. Strikingly, the presence of HBZ-AD induced a large increase in the amount of c-Myb-AD bound to GST-KIX (lanes 4, 6 and 8). Quantification of the signals obtained by Western blot (lanes 5 and 6 compared) shows a 6-fold increase in c-Myb-binding. To confirm that this effect was not due to a direct interaction between c-Myb and HBZ, we performed a GST pull-down assay using the equivalent concentration of GST-HBZ and the maximum concentration of c-Myb-AD from the experiment. Figure 6B shows that the proteins did not directly interact (lane 2), confirming that the effect was specifically due an enhanced binding of c-Myb to GST-KIX.
Figure 6. HBZ-AD enhances the binding of c-Myb-AD to KIX.
(a) HBZ-AD enhances the binding of c-Myb-AD to KIX in GST pull-down assays. Binding reactions contained GST-KIX (250 nM, lanes 3-8) and increasing concentrations of c-Myb-AD (0.1 μM, lanes 3 and 4; 0.25 μM, lanes 5 and 6; 0.5 μM; lanes 7 and 8). Lanes 4, 6 and 8 contained 7.5 μM HBZ-AD. Control reactions contained GST (250 nM) with HBZ-AD (7.5 μM, lane 1) or c-Myb-AD (0.5 μM, lane 2) as indicated. Fractions of c-Myb-AD (1%) and HBZ-AD (0.8%) inputs are shown in lanes 9 and 10, respectively. Bound proteins were resolved by SDS-PAGE and analyzed by Western blot (WB) using the antibodies indicated. (b) c-Myb-AD does not bind GST-HBZ in GST pull-down assays. Binding reactions contained c-Myb-AD (0.5 μM) and GST (7.5 μM, lane 1) or GST-HBZ (7.5 μM, lane 2). A fraction of the input of c-Myb-AD (1%) is shown in lane 3. (c) HBZAD increases transcriptional activation by c-Myb-AD in cells. Jurkat cells were transfected with pGalTK-Luc (100 ng) alone or in combination with Gal4-DBD (50 ng), Gal4-c-Myb-AD (50 ng), pcDNA-HBZ (100 ng) or pcDNA-HBZ-Mut-AD3 (100 ng) as indicated. Luciferase assays were performed 24 hours after transfection. The reported values are the average luminescence ± S.E. from one experiment performed in duplicate and are representative of three independent experiments.
To determine whether HBZ increases c-Myb-binding to p300/CBP in cells, we used the luciferase assay system described for figure 5B. c-Myb-AD (aa 186-325) was fused with the DBD of Gal4 21, and Jurkat T-cells were co-transfected with an expression plasmid for this construct and with pGalTK-Luc. As reported for Gal4-MLL-AD, the interaction of Gal4-c-Myb-AD with endogenous p300/CBP has been shown to activate transcription from the reporter plasmid 21. As expected, we found that expression of Gal4-c-Myb-AD caused a large increase in luciferase activity, while Gal4-DBD alone did not produce such an effect (Fig. 6C, lanes 2 and 3). Interestingly, expression of HBZ augmented transcriptional activation by Gal4-c-Myb-AD greater than 2-fold (lane 4), suggesting that HBZ enhances the interaction between p300/CBP and c-Myb in vivo. However, HBZ-Mut-AD3, which contains mutations in both ϕXXϕϕ motifs, failed to significantly enhance Gal4-c-Myb-AD transcriptional activity (lane 5). These results support the in vitro data and further suggest that HBZ enhances binding of c-Myb to the endogenous cellular coactivators.
HBZ-AD affects the binding of CREB to KIX
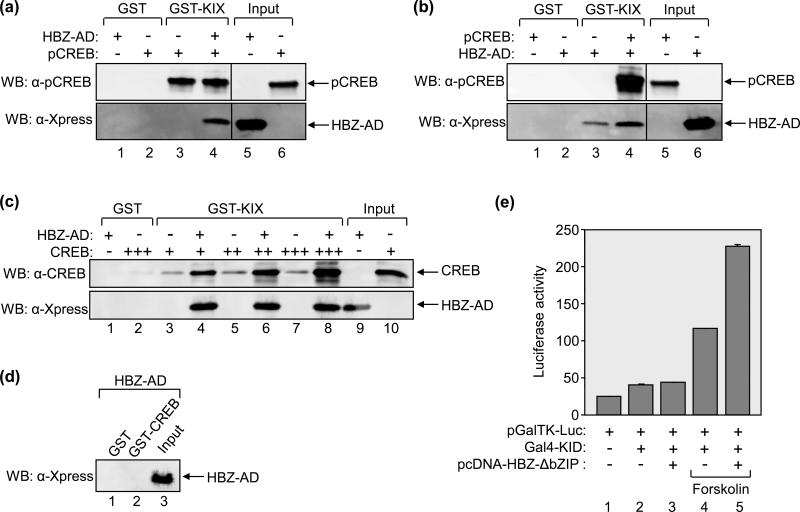
We also tested whether HBZ similarly enhances the pCREB:KIX interaction. Using a GST pull-down assay, we compared the binding of pCREB to GST-KIX in the absence or presence of HBZ-AD at a concentration sufficient to saturate the MLL-binding surface (Fig. 7A). Unlike the results obtained with c-Myb, HBZ did not increase the amount of pCREB bound to KIX in this assay (lanes 3 and 4). To investigate whether pCREB increases HBZ-AD binding to GST-KIX, we performed the reverse experiment in which HBZ was combined with a saturating concentration of pCREB. As shown in figure 7B, pCREB induced a 2-fold increase in HBZ-AD binding to KIX (lanes 3 and 4). We also investigated whether HBZ-AD enhances the binding of unphosphorylated CREB to KIX. In its unphosphorylated form, CREB has been shown to exhibit a low affinity for KIX, with a Kd of 108 ± 5 μM 38. To determine whether HBZ-AD enhanced the binding of unphosphorylated CREB to KIX, we performed GST pull-down assays in which three concentrations of unphosphorylated CREB were bound to GST-KIX in the absence or presence of HBZ-AD (Fig. 7B). As expected, very little CREB bound to GST-KIX in the absence of HBZ-AD (lanes 3, 5 and 7). However, when HBZ-AD was present, we observed a large increase in the amount of CREB bound to KIX (lanes 4, 6 and 8). Quantification of the signals obtained by Western blot (lanes 5 and 6 compared) shows a 6.5-fold increase in CREB-binding. Because HBZ-AD lacks its bZIP domain, it should not bind to CREB as previously reported 14. Indeed, HBZ-AD did not bind to GST-CREB in a GST pull-down assay in which the concentrations of HBZ-AD and GST-CREB were identical to the concentration of HBZ-AD and the highest concentration of CREB used in figure 7B (Fig. 7C). These data provide evidence that HBZ may facilitate an interaction between CREB and p300/CBP in the absence of S133 phosphorylation of CREB.
Figure 7. HBZ-AD affects the binding of CREB to KIX.
(a) HBZ-AD does not enhance pCREB-binding to KIX in GST pull-down assays. Binding reactions contained GST-KIX (25 nM, lanes 3 and 4) and pCREB (25 nM, lanes 3 and 4) in the absence or presence of HBZ-AD (1 μM, lane 4). Control reactions contained GST (25 nM) with HBZ-AD (1 μM, lane 1) or pCREB (25 nM, lane 2). Fractions of HBZ-AD (3.3%) and pCREB (24%) inputs are shown in lanes 5 and 6, respectively. Lanes shown are from the same membrane. Vertical lines show where the membrane was cropped. (b) pCREB enhances HBZ-AD-binding to KIX in GST pull-down assays. Binding reactions contained GST-KIX (25 nM, lanes 3 and 4) and HBZ-AD (25 nM, lanes 3 and 4) in the absence or presence of pCREB (100 nM, lane 4). Control reactions contained GST (25 nM) with pCREB (100 nM, lane 1) or HBZ-AD (25 nM, lane 2). Inputs of pCREB (6%) and HBZ-AD (130%) are shown in lanes 5 and 6, respectively. Lanes shown are from the same membrane. Vertical lines show where the membrane was cropped. (c) HBZ-AD enhances the binding of unphosphorylated CREB to KIX in vitro. Binding reactions contained GST-KIX (500 nM, lanes 3-8) and increasing concentrations of CREB (50 nM, lanes 3 and 4; 100 nM, lanes 5 and 6; 200 nM CREB, lanes 7 and 8). Lanes 4, 6 and 8 contained 5 μM HBZ-AD. Control reactions contained GST (500 nM) with HBZ-AD (5 μM, lane 1) or CREB (200 nM, lane 2). A fraction of HBZ-AD (2%) and CREB (12%) inputs are shown in lanes 9 and 10, respectively. (d) CREB does not bind HBZ-AD in GST pull-down assays. Binding reactions contained 5 μM HBZ-AD with either GST (200 nM, lane 1) or GST-CREB (200 nM, lane 2). A fraction of the input of HBZ-AD (2%) is shown in lane 3. (e) HBZ-AD increases transcriptional activation by pKID in cells. 293T/17 cells were transfected with pGalTK-Luc (100 ng) alone or in combination with Gal4-KID (100 ng), pcDNA-HBZ-ΔbZIP (100 ng) as indicated. Cells were treated with forskolin (2 μM) for 5h where indicated before the assays were performed. Luciferase assays were performed 24 hours after transfection. The reported values are the average luminescence ± S.E. from one experiment performed in duplicate and are representative of three independent experiments.
To determine whether HBZ-AD increases pCREB and CREB activity in cells, we fused the CREB activation domain, KID (aa 102-160), to the Gal4-DBD. Luciferase assays were then performed using 293T/17 cells co-transfected with the expression plasmid for Gal4-KID and the pGalTK-Luc reporter plasmid. As previously reported, Gal4-KID in its unphosphorylated state did not activate transcription ( Fig. 7E, lane 2) 39. However, treatment of cells with forskolin to stimulate protein kinase A-dependent S133 phosphorylation of KID led to a 2.8-fold increase in luciferase activity (compare lanes 2 and 4) 39. Surprisingly, co-transfection of HBZ-AD did not increase Gal4-KID activity without forskolin treatment (compare lanes 2 and 3). In contrast to the GST pull-down data, these results suggest that HBZ does not provide a sufficient increase in affinity between unphosphorylated CREB and KIX for p300/CBP recruitment in vivo. Interestingly, expression of HBZ-AD in cells treated with forskolin led to an almost 2-fold increase in Gal4-KID activity compared to Gal4-KID activity in untreated cells (compare lanes 4 and 5). These results support an effect of HBZ in augmenting pCREB affinity for the KIX domain that was not observed using GST pull-down assays.
DISCUSSION
We previously showed that HBZ, through its AD, binds to the KIX domain of p300/CBP 17. In this study, we determined that the apparent Kd of this interaction is 3 ± 0.5 nM. To our knowledge, this is one of the strongest interactions between a single protein and the KIX domain reported, as Kd values involving other proteins are in the micromolar range (pKID, 1μM; c-Myb, 10 μM; MLL, 2.8 μM; and p53, 19 μM)26; 30. Therefore, expression of HBZ in HTLV-1-infected cells is likely to have a significant impact on p300/CBP function. It is believed that some oncogenic properties of other viral proteins, such as Tax and the adenoviral protein E1A, stem from their interactions with these coactivators 40; 41. Consequently, the role of HBZ in ATL may partially relate to its interaction with p300/CBP.
Proteins that bind the KIX domain contain a ϕXXϕϕ motif within an amphipathic helix that directly contacts one of the KIX-binding surfaces 20; 22; 24; 26; 27; 30; 42; 43; 44; 45; 46. Interestingly, HBZ carries two such motifs in its AD, both of which contribute to the high-affinity interaction with the KIX domain. The first motif contributes to KIX-binding to a larger extent than the second motif, as mutations in the first motif result in a 267-fold increase in the Kd compared to a 60-fold increase caused by mutations in the second motif.
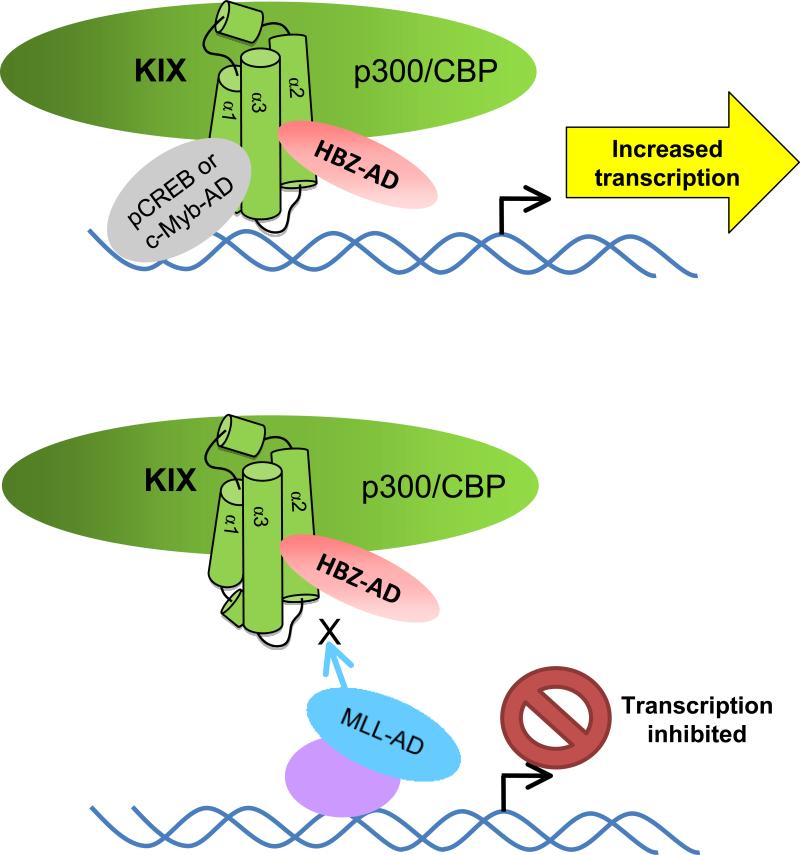
We found that HBZ targets the MLL-binding surface of KIX, but not the surface contacted by pCREB and c-Myb. The predicted hairpin structure of HBZ-AD could potentially allow both ϕXXϕϕ motifs to contact the MLL-binding surface. This surface is known to be larger and more accommodating to a variety of transcription factors than the pCREB/c-Myb-binding surface 22, which may permit for the simultaneous binding of both predicted amphipathic α-helices in HBZ-AD. However, it is alternatively possible that one helix stabilizes the tertiary structure of the AD without directly contacting the KIX domain.
In targeting the MLL-binding surface, we found that HBZ competes with MLL for KIX-binding. This result extends our previous finding that HBZ disrupts the interaction between Tax and KIX, as experimental evidence indicates that Tax also contacts the MLL-binding surface 27; 28. MLL was shown to induce positive cooperativity in the binding of c-Myb and pCREB to KIX 22; 25; 26. We found that HBZ-AD enhanced the binding of c-Myb to KIX. While HBZ-AD did not enhance the interaction between pCREB and KIX in GST pull-down assays, it did enhance the transcriptional activity of pKID, which is directly linked to p300/CBP recruitment. It is possible that certain limitations of the GST pull-down assay prevent detection of an increase in the already high-affinity interaction between pCREB and KIX. The combined results that HBZ enhances pKID activity in vivo and pCREB enhances the binding of HBZ to KIX in vitro suggest that these proteins do bind cooperatively to KIX. Interestingly, HBZ increased the affinity of unphosphorylated CREB for KIX. However, this interaction did not produce transcriptional activity of KID in cells, indicating that HBZ does not enhance binding of unphosphorylated CREB to KIX to the same level induced by phosphorylation of CREB.
The fact that HBZ alters the binding of transcription factors to both surfaces of the KIX domain may have important implications with regard to mechanisms leading to deregulated gene expression in HTLV-1-infected cells. Since transcription factors besides those included in this study have been reported to bind the KIX domain, HBZ is likely to modulate transcriptional activity of KIX-binding proteins beyond MLL, c-Myb and CREB. Regarding the deregulation of MLL, c-Myb and CREB activity, each has been linked to leukemia and/or T-cell proliferation 47; 48. MLL translocations are found in infant acute lymphoblastic and myeloid leukemias 49, while dysfunction in c-Myb activity is associated with T-cell acute lymphoblastic leukemia 47; 50. Furthermore, c-Myb activity is essential for the continued proliferation and viability of acute myeloid and chronic myeloid leukemia cells 50. CREB controls diverse functions including cell survival, proliferation and hematopoiesis and is implicated in various acute leukemias 48. Therefore, downstream effects of the interaction between HBZ and the KIX domain may play a role in ATL. Finally, like HBZ, Tax contacts the MLL-binding surface of KIX. Tax is oncogenic and believed to play an essential role in triggering early events that lead to ATL, some of which may involve its interaction with p300/CBP 40. Consequently, it is possible that HBZ complements Tax-function in binding p300/CBP, as Tax expression is frequently lost during the course of the disease 4.
MATERIALS AND METHODS
Bacterial expression vectors and cloning
Full-length HBZ splice 1 (aa 1-206) was PCR-amplified from pcDNA3.1-MycHis-HBZ 17 and cloned into the EcoRI and XbaI sites of pSG42451, containing the Gal4-DBD (aa 1-147), to create the fusion protein Gal4-HBZ (pSG424-HBZ). pRSET-Gal4-HBZ was constructed by amplifying Gal4-HBZ from pSG424-HBZ and cloning the product into the BamHI and NcoI sites of pRSET-A (Invitrogen). pRSET-Gal4-HBZ-AD was constructed by amplifying Gal4-HBZ-AD (aa 1-77) from pRSET-Gal4-HBZ and cloning the product into the BamHI and NcoI sites of pREST-A. Two clones of pRSET-Gal4-HBZ-AD were used: the one used in figure 1 did not have any mutations, while the one used in Figure 2B had an A to T substitution at aa 3 of HBZ. We tested the affinities of both constructs for GST-KIX and found they were the same (data not shown). pRSET-Gal4-HBZ-AD mutants were constructed by amplifying HBZ-AD from the HBZ mutant templates with L27A/L28A, L47A/L48A, or L27A/L28A/L47A/L48 substitutions (pcDNA3.1-MycHis-HBZ)17. Products were cloned into the EcoRI site of pRSET-Gal4-HBZ-AD after removal of the HBZ-AD wt sequence by digestion with EcoRI. pRSET-HBZ-AD was constructed by amplifying HBZ-AD from pRSET-Gal4-HBZ and cloning the product into the EcoRI site of pRSET-B (Invitrogen). pRSET-HBZ-AD mutants were constructed by cloning the above HBZ-AD mutant PCR products into the EcoRI site of pRSET-B.
Escherichia coli expression plasmids for untagged KIX (aa 587-679, from mouse CBP)52, GST-C/H1 (aa 302-451, from mouse CBP), GST-CR3 (aa 2221-2441, from mouse CBP), GST-KIX (aa 588-683, from mouse CBP) and GST-KIX deletion mutants (aa 588-655 and 597-719, from mouse CBP) were previously described 53; 54. GST-KIX588-683 point mutants ΔT (F612A, D622A, R624A), ΔMLL-A (Y631A, L664A), ΔMLL-B (D622A, R624A, Y631A, L664A), ΔpC (Y658A), and YAY (Y650A, A654Q, Y658A) were made using the QuikChange II site-directed mutagenesis kit (Stratagene) with the GST-KIX expression plasmid template. GST-HBZ and GST-CREB E. coli expression plasmids were described previously 17; 55. The E. coli expression plasmid for CREB was described previously 56.
pRSET-Flag-MLL-AD (aa 2829-2883) was constructed by PCR-amplifying MLL-AD from pF-MLL 37 with the forward primer containing the Flag sequence, and cloning the product into the BamHI and EcoRI sites of pRSET-A. HA-c-Myb-AD (aa 186-325) was constructed by amplifying c-Myb-AD from pM2 Gal4-c-Myb (aa 186-325)21 with the forward primer containing the HA sequence, and cloning the product into the BamHI and EcoRI sites of pRSET-A. All plasmids were sequenced. The original HA-c-Myb-AD construct has a V to A substitution at aa 267, which is outside the KIX-binding domain and does not contribute to the interaction 57.
Expression and purification of recombinant proteins
E. coli BL21(DE3) pLysS cells (Agilent Technologies) were used for expression of Gal4-HBZ-AD wt and mutants, HBZ-AD wt and mutants, HA-c-Myb-AD, GST-CREB, GST-KIX, untagged KIX and CREB. BL21-codon plus (DE3) cells (Agilent Technologies) were used for expression of Gal4-HBZ, Flag-MLL-AD and GST-HBZ. The LB medium for HBZ-AD and Flag-MLL-AD cultures was supplemented with 1% glucose. Expression and purification of GST fusion proteins were performed as described previously 58, except that proteins were dialyzed in HM 0.1 buffer (50 mM HEPES [pH 7.9], 100 mM KCl, 12.5 mM MgCl2, 1 mM EDTA, 20% [vol/vol] glycerol, 0.025% [vol/vol] Tween 20 and 1 mM dithiothreitol [DTT]) overnight at 4°C. Dialyzed proteins were aliquoted and stored at -80°C. Proteins with a 6XHis tag were purified using Ni-NTA agarose (QIAGEN) as described by the manufacturer and subsequently supplemented with 15% glycerol and 5 mM DTT, aliquoted and stored at -80°C. p300 was expressed from a recombinant baculovirus in Sf9 cells and purified as previously described 59. Untagged KIX was purified using Q-sepharose and SP-sepharose ion exchange columns (Amersham) as described previously 28. CREB was purified as described previously 60 with additional heparin-agarose chromatography 61 and dialyzed overnight at 4°C against TM 0.1 (50 mM Tris [pH 7.9], 100mM KCl, 12.5 mM MgCl2, 1 mM EDTA [pH 8.0], 20% [vol/vol] glycerol, 0.025% [vol/vol] Nonidet P40 and 1 mM DTT). Purity and concentration of proteins were assessed by SDS-PAGE using BSA for standard curves. Protein gels were stained with SYPRO Ruby protein gel stain (Invitrogen) or Coomassie Brilliant Blue. CREB (1 μM) was phosphorylated using 50 units of the catalytic subunit of protein kinase A (P2645, Sigma) in 25 mM potassium phosphate buffer [pH 6.6], containing 20 μM ATP and 5 mM MgCl2 at 30°C.
Electrophoretic mobility shift assays (EMSA)
Reactions (20 μl) contained 0.5x TM 0.1, 5 μM ZnSO4, 5 mM DTT, and concentrations of protein and 32P-end-labeled DNA probe indicated in the figure legends. The Gal4-DBD DNA probe sequence was TTTGCATCGGAGGACTGTCCTCCGATGC. Antibodies used to supershift complexes were anti-GST (G7781, Sigma) and anti-Gal4-DBD (sc-510, Santa Cruz). Reactions were incubated at 20°C for 2 hours to ensure that equilibrium binding conditions were reached and then resolved on a non-denaturing polyacrylamide gel containing 40 mM Tris [pH 8.5] and 306 mM glycine. Gels ranged in acrylamide concentration from 5-7% (49:1 bis) and were run at 230V and 4°C for 2-3 hours in 40 mM Tris [pH 8.5] and 306 mM glycine. Following electrophoresis, gels were dried and visualized by PhosphorImager analysis with a Typhoon 9410 (GE Healthcare). Band intensities were quantified using ImageQuant TL (GE Healthcare). Kd values were determined using the same method described by Giebler et al. 58, with the formula: Kd = [GST-KIX]free[Gal4-HBZ-AD]free/[GST-KIX:Gal4-HBZ-AD]. The fraction bound was determined based on the appearance of the shifted band with the formula: fraction bound = [Gal4-HBZ-AD]bound/[Gal4-HBZ-AD]total. Prism® (GraphPad Software) was used to plot the data using nonlinear regression to a hyperbolic one-site binding model with the equation y = (Bmax* x)/(x + Kd), where Bmax is the maximum fraction bound. Gibbs free-energy values were calculated from the Kd values using the equation ΔG = -RTln(1/Kd), where R = 0.001987 kcal mol-1 K-1, and T = 293 K (20°C).
Crosslinking assays
Binding reactions (15 μl) for bis(sulfosuccinimidyl) suberate (BS3, Pierce) crosslinking assays were performed in 0.25x HM 0.1 buffer, with concentrations of recombinant proteins indicated in the figure legend. Reactions were incubated for approximately 2 hours at 20°C without BS3, then 1.8 mM BS3 was added, and reactions were further incubated at room temperature for 30 minutes. Crosslinking was quenched by addition of Tris (pH 7.5) to a final concentration of 50 mM followed by incubation at room temperature for 15 minutes. Proteins were resolved by SDS-PAGE and analyzed by Western blot analysis using the anti-Xpress antibody (R910-25, Invitrogen) to detect the Xpress epitope for pRSET vectors (HBZ-AD) and anti-CBP (sc-1211, Santa Cruz) to detect untagged KIX. Blots were developed using ECL Plus chemiluminescence (GE Healthcare) and visualized with a Typhoon 9410 (GE Healthcare).
GST pull-down assays
GST fusion proteins (concentrations indicated in the figure legends) were bound at 4°C for 1 hour to 20 μl of glutathione-agarose beads (Sigma) that were equilibrated with 0.5x Superdex buffer (12.5 mM HEPES [pH 7.9], 75 mM KCl, 6.25 mM MgCl2, 5 μM ZnSO4, 20% [vol/vol] glycerol, 0.05% [vol/vol] Nonidet P40, 1 mM EDTA and 1 mM DTT). Beads were then washed twice with 500 μl 0.5x Superdex buffer, combined with the second protein (concentrations indicated in the figure legends) in a final volume of 50 μl 0.5x Superdex buffer for all reactions except those with MLL-AD, which were incubated in JM buffer (20 mM HEPES [pH 7.9], 0.5 mM EDTA, 10% glycerol, 0.05% Nonidet P40, 5 μM ZnSO4, 2.5 mM MgCl2, 25 mM KCl and 1 mM DTT). Reactions were incubated at 4°C overnight, and beads were then washed four times with 500 μl 0.5x Superdex buffer (or JM buffer for MLL-AD reactions). Proteins retained on the beads were resolved by SDS-PAGE and detected by Western blot analysis using the following antibodies: anti-X-press (R910-25) from Invitrogen; anti-CREB-1 (sc-186), anti-pCREB-1 (sc-7978) from Santa-Cruz; anti-Flag M2 (F3165), anti-HA (H3663) and anti-GST (G7781) from Sigma. Blots were developed using ECL Plus chemiluminescence (GE Healthcare) and visualized with a Typhoon 9410 (GE Healthcare).
Cell culture, transient cotransfection assays and luciferase assays
Jurkat cells and 293T/17 cells (ATCC) were cultured in Iscove's modified Dulbecco's medium supplemented with 10% fetal bovine serum, glutamine, and penicillin/streptomycin. For transient cotransfection assays, cells were plated at 4×105 cells per well for Jurkat and 6×104 cells per well for 293T/17 cells. Jurkat and 293T/17 cells were transfected with Lipofectamine (Invitrogen) and TurboFect (MBI), respectively, and a constant amount of DNA (1 μg). After 24 hours, cells were harvested and lysed, and luciferase activity was measured using the Dual-Luciferase reporter assay system (Promega) with a Turner Designs model TD 20-e luminometer. Luciferase activity was normalized to Renilla luciferase from the herpes simplex virus thymidine kinase promoter (pRL-TK, Promega). Mammalian expression plasmids pGal-TK-Luc 36, Gal4-c-Myb (aa 186-325)21, pBXG1-MLL-AD (aa 2829 to 2883)25, Hox c8-Luc (Hox c8 promoter) and pF-MLL (full-length MLL)37 were previously described. Gal4-KID was constructed by PCR-amplifying KID (aa 102-160) from CREB, and cloning the product into the EcoRI and XbaI sites of pSG424 51 (pSG424-KID). pcDNA-HBZ, pcDNA-HBZ-MutAD3 and pcDNA-HBZ-ΔbZIP were previously described 9,17.
Protein structure prediction and helical wheel projection
Secondary and tertiary prediction programs used were PSIPRED 62, http://bioinf.cs.ucl.ac.uk/psipred/); NetSurfP 63, http://ca.expasy.org/tools/); and I-TASSER 64, http://zhanglab.ccmb.med.umich.edu/I-TASSER/). Helical wheel projection programs used were located at http://rzlab.ucr.edu/scripts/wheel/wheel.cgi and EMBOSS Pepwheel at http://www.tcdb.org/progs/pepwheel.php.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the National Institutes of Health (R01CA128800 to I.L.); and the American Heart Association (09BGIA2050059 to I.L.). We would like to thank Dr. F.E. Bertrand, Dr. P. Brindle, Dr. P. Ernst and Dr. J. Hess for the generous gift of plasmids; and Dr. C. Hardin for helpful discussion during the course of the studies.
Abbreviations
- HTLV-1
human T-cell leukemia virus type 1
- HBZ
HTLV-1 basic leucine zipper factor
- AD
activation domain
- bZIP
basic leucine zipper
- DBD
DNA-binding domain
- WB
Western blot
- ATL
adult T-cell leukemia
- MLL
mixed-lineage leukemia
- aa
amino acid residue(s)
- GST
glutathione-S-transferase
- EMSA
electrophoretic mobility shift assay
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Uchiyama T, Yodoi J, Sagawa K, Takatsuki K, Uchino H. Adult T-cell leukemia: clinical and hematologic features of 16 cases. Blood. 1977;50:481–492. [PubMed] [Google Scholar]
- 2.Poiesz BJ, Ruscetti FW, Gazdar AF, Bunn PA, Minna JD, Gallo RC. Detection and isolation of type C retrovirus particle from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. USA. 1980;77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hinuma Y, Nagata K, Hanaoka M, Nakai M, Matsumoto T, Kinoshita KI, Shirakawa S, Miyoshi I. Adult T-cell leukemia: antigen in an ATL cell line and detection of antibodies to the antigen in human sera. Proc. Natl. Acad. Sci. USA. 1981;78:6476–6480. doi: 10.1073/pnas.78.10.6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsuoka M, Green PL. The HBZ gene, a key player in HTLV-1 pathogenesis. Retrovirology. 2009;6:71. doi: 10.1186/1742-4690-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Satou Y, Yasunaga J, Yoshida M, Matsuoka M. HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proc Natl Acad Sci U S A. 2006;103:720–725. doi: 10.1073/pnas.0507631103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnold J, Zimmerman B, Li M, Lairmore MD, Green PL. Human T-cell leukemia virus type-1 antisense-encoded gene, Hbz, promotes T-lymphocyte proliferation. Blood. 2008;112:3788–3797. doi: 10.1182/blood-2008-04-154286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuhlmann AS, Villaudy J, Gazzolo L, Castellazzi M, Mesnard JM, Duc Dodon M. HTLV-1 HBZ cooperates with JunD to enhance transcription of the human telomerase reverse transcriptase gene (hTERT). Retrovirology. 2007;4:92. doi: 10.1186/1742-4690-4-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao T, Yasunaga J, Satou Y, Nakao M, Takahashi M, Fujii M, Matsuoka M. Human T-cell leukemia virus type 1 bZIP factor selectively suppresses the classical pathway of NF-kappaB. Blood. 2009;113:2755–2764. doi: 10.1182/blood-2008-06-161729. [DOI] [PubMed] [Google Scholar]
- 9.Polakowski N, Gregory H, Mesnard JM, Lemasson I. Expression of a protein involved in bone resorption, Dkk1, is activated by HTLV-1 bZIP factor through its activation domain. Retrovirology. 2010;7:61. doi: 10.1186/1742-4690-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hivin P, Frederic M, Arpin-Andre C, Basbous J, Gay B, Thebault S, Mesnard JM. Nuclear localization of HTLV-I bZIP factor (HBZ) is mediated by three distinct motifs. J Cell Sci. 2005;118:1355–1362. doi: 10.1242/jcs.01727. [DOI] [PubMed] [Google Scholar]
- 11.Gaudray G, Gachon F, Basbous J, Biard-Piechaczyk M, Devaux C, Mesnard JM. The complementary strand of the human T-cell leukemia virus type 1 RNA genome encodes a bZIP transcription factor that down-regulates viral transcription. J. Virol. 2002;76:12813–12822. doi: 10.1128/JVI.76.24.12813-12822.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basbous J, Arpin C, Gaudray G, Piechaczyk M, Devaux C, Mesnard JM. The HBZ factor of human T-cell leukemia virus type I dimerizes with transcription factors JunB and c-Jun and modulates their transcriptional activity. J. Biol. Chem. 2003;278:43620–46327. doi: 10.1074/jbc.M307275200. [DOI] [PubMed] [Google Scholar]
- 13.Thebault S, Basbous J, Hivin P, Devaux C, Mesnard JM. HBZ interacts with JunD and stimulates its transcriptional activity. FEBS Lett. 2004;562:165–170. doi: 10.1016/S0014-5793(04)00225-X. [DOI] [PubMed] [Google Scholar]
- 14.Lemasson I, Lewis MR, Polakowski N, Hivin P, Cavanagh MH, Thebault S, Barbeau B, Nyborg JK, Mesnard JM. Human T-cell leukemia virus type 1 (HTLV-1) bZIP protein interacts with the cellular transcription factor CREB to inhibit HTLV-1 transcription. J Virol. 2007;81:1543–1553. doi: 10.1128/JVI.00480-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reinke AW, Grigoryan G, Keating AE. Identification of bZIP interaction partners of viral proteins HBZ, MEQ, BZLF1, and K-bZIP using coiled-coil arrays. Biochemistry. 2010;49:1985–1997. doi: 10.1021/bi902065k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohshima T, Mukai R, Nakahara N, Matsumoto J, Isono O, Kobayashi Y, Takahashi S, Shimotohno K. HTLV-1 basic leucine-zipper factor, HBZ, interacts with MafB and suppresses transcription through a Maf recognition element. J Cell Biochem. 2010;111:187–194. doi: 10.1002/jcb.22687. [DOI] [PubMed] [Google Scholar]
- 17.Clerc I, Polakowski N, Andre-Arpin C, Cook P, Barbeau B, Mesnard JM, Lemasson I. An interaction between the human T cell leukemia virus type 1 basic leucine zipper factor (HBZ) and the KIX domain of p300/CBP contributes to the down-regulation of tax-dependent viral transcription by HBZ. J Biol Chem. 2008;283:23903–23913. doi: 10.1074/jbc.M803116200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell. Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- 19.Bedford DC, Kasper LH, Fukuyama T, Brindle PK. Target gene context influences the transcriptional requirement for the KAT3 family of CBP and p300 histone acetyltransferases. Epigenetics. 2010;5:9–15. doi: 10.4161/epi.5.1.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radhakrishnan I, Perez-Alvarado GC, Parker D, Dyson HJ, Montminy MR, Wright PE. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 21.Parker D, Rivera M, Zor T, Henrion-Caude A, Radhakrishnan I, Kumar A, Shapiro LH, Wright PE, Montminy M, Brindle PK. Role of secondary structure in discrimination between constitutive and inducible activators. Mol Cell Biol. 1999;19:5601–5607. doi: 10.1128/mcb.19.8.5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Guzman RN, Goto NK, Dyson HJ, Wright PE. Structural basis for cooperative transcription factor binding to the CBP coactivator. J Mol Biol. 2006;355:1005–1013. doi: 10.1016/j.jmb.2005.09.059. [DOI] [PubMed] [Google Scholar]
- 23.Lee LW, Mapp AK. Transcriptional switches: chemical approaches to gene regulation. J Biol Chem. 2010;285:11033–11038. doi: 10.1074/jbc.R109.075044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zor T, De Guzman RN, Dyson HJ, Wright PE. Solution structure of the KIX domain of CBP bound to the transactivation domain of c-Myb. J Mol Biol. 2004;337:521–534. doi: 10.1016/j.jmb.2004.01.038. [DOI] [PubMed] [Google Scholar]
- 25.Ernst P, Wang J, Huang M, Goodman RH, Korsmeyer SJ. MLL and CREB bind cooperatively to the nuclear coactivator CREB-binding protein. Mol Cell Biol. 2001;21:2249–2258. doi: 10.1128/MCB.21.7.2249-2258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goto NK, Zor T, Martinez-Yamout M, Dyson HJ, Wright PE. Cooperativity in transcription factor binding to the coactivator CREB-binding protein (CBP). The mixed lineage leukemia protein (MLL) activation domain binds to an allosteric site on the KIX domain. J Biol Chem. 2002;277:43168–43174. doi: 10.1074/jbc.M207660200. [DOI] [PubMed] [Google Scholar]
- 27.Vendel AC, McBryant SJ, Lumb KJ. KIX-mediated assembly of the CBP-CREB-HTLV-1 tax coactivator-activator complex. Biochemistry. 2003;42:12481–7. doi: 10.1021/bi0353023. [DOI] [PubMed] [Google Scholar]
- 28.Ramirez JA, Nyborg JK. Molecular characterization of HTLV-1 Tax interaction with the KIX domain of CBP/p300. J Mol Biol. 2007;372:958–969. doi: 10.1016/j.jmb.2007.06.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Q, Vo N, Goodman RH. Histone binding protein RbAp48 interacts with a complex of CREB binding protein and phosphorylated CREB. Mol Cell Biol. 2000;20:4970–4978. doi: 10.1128/mcb.20.14.4970-4978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee CW, Arai M, Martinez-Yamout MA, Dyson HJ, Wright PE. Mapping the Interactions of the p53 Transactivation Domain with the KIX Domain of CBP (dagger). Biochemistry. 2009;48:2115–2124. doi: 10.1021/bi802055v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 32.Cavanagh M, Landry S, Audet B, Arpin-Andre C, Hivin P, Pare M-E, Thete J, Wattel E, Marriott SJ, Barbeau B, Mesnard J-M. HTLV-I antisense transcripts initiate in the 3’LTR and are alternatively spliced and polyadenylated. Retrovirology. 2006;3:15. doi: 10.1186/1742-4690-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murata K, Hayashibara T, Sugahara K, Uemura A, Yamaguchi T, Harasawa H, Hasegawa H, Tsuruda K, Okazaki T, Koji T, Miyanishi T, Yamada Y, Kamihira S. A novel alternative splicing isoform of human T-cell leukemia virus type 1 bZIP factor (HBZ-SI) targets distinct subnuclear localization. J Virol. 2006;80:2495–2505. doi: 10.1128/JVI.80.5.2495-2505.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Radhakrishnan I, Perez-Alvarado GC, Parker D, Dyson HJ, Montminy MR, Wright PE. Structural analyses of CREB-CBP transcriptional activator-coactivator complexes by NMR spectroscopy: implications for mapping the boundaries of structural domains. J Mol Biol. 1999;287:859–865. doi: 10.1006/jmbi.1999.2658. [DOI] [PubMed] [Google Scholar]
- 35.Rath A, Glibowicka M, Nadeau VG, Chen G, Deber CM. Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc Natl Acad Sci U S A. 2009;106:1760–1765. doi: 10.1073/pnas.0813167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dobner T, Horikoshi N, Rubenwolf S, Shenk T. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science. 1996;272:1470–1473. doi: 10.1126/science.272.5267.1470. [DOI] [PubMed] [Google Scholar]
- 37.Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 38.Zor T, Mayr BM, Dyson HJ, Montminy MR, Wright PE. Roles of phosphorylation and helix propensity in the binding of the KIX domain of CREB-binding protein by constitutive (c-Myb) and inducible (CREB) activators. J Biol Chem. 2002;277:42241–42248. doi: 10.1074/jbc.M207361200. [DOI] [PubMed] [Google Scholar]
- 39.Brindle P, Linke S, Montminy M. Protein-kinase-A-dependent activator in transcription factor CREB reveals new role for CREM repressors. Nature. 1993;364:821–824. doi: 10.1038/364821a0. [DOI] [PubMed] [Google Scholar]
- 40.Van Orden K, Nyborg JK. Insight into the tumor suppressor function of CBP through the viral oncoprotein Tax. Gene Expr. 2000;9:29–36. doi: 10.3727/000000001783992678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 42.Campbell KM, Lumb KJ. Structurally distinct modes of recognition of the KIX domain of CBP by Jun and CREB. Biochemistry. 2002;41:13956–13964. doi: 10.1021/bi026222m. [DOI] [PubMed] [Google Scholar]
- 43.Liu YP, Chang CW, Chang KY. Mutational analysis of the KIX domain of CBP reveals residues critical for SREBP binding. FEBS Lett. 2003;554:403–409. doi: 10.1016/s0014-5793(03)01200-6. [DOI] [PubMed] [Google Scholar]
- 44.Vendel AC, Lumb KJ. NMR mapping of the HIV-1 Tat interaction surface of the KIX domain of the human coactivator CBP. Biochemistry. 2004;43:904–908. doi: 10.1021/bi035612l. [DOI] [PubMed] [Google Scholar]
- 45.Bayly R, Murase T, Hyndman BD, Savage R, Nurmohamed S, Munro K, Casselman R, Smith SP, LeBrun DP. Critical role for a single leucine residue in leukemia induction by E2A-PBX1. Mol Cell Biol. 2006;26:6442–6452. doi: 10.1128/MCB.02025-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang F, Marshall CB, Li GY, Yamamoto K, Mak TW, Ikura M. Synergistic interplay between promoter recognition and CBP/p300 coactivator recruitment by FOXO3a. ACS Chem Biol. 2009;4:1017–1027. doi: 10.1021/cb900190u. [DOI] [PubMed] [Google Scholar]
- 47.O'Neil J, Look AT. Mechanisms of transcription factor deregulation in lymphoid cell transformation. Oncogene. 2007;26:6838–6849. doi: 10.1038/sj.onc.1210766. [DOI] [PubMed] [Google Scholar]
- 48.Shankar DB, Cheng JC, Sakamoto KM. Role of cyclic AMP response element binding protein in human leukemias. Cancer. 2005;104:1819–1824. doi: 10.1002/cncr.21401. [DOI] [PubMed] [Google Scholar]
- 49.Slany RK. The molecular biology of mixed lineage leukemia. Haematologica. 2009;94:984–993. doi: 10.3324/haematol.2008.002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat Rev Cancer. 2008;8:523–534. doi: 10.1038/nrc2439. [DOI] [PubMed] [Google Scholar]
- 51.Sadowski I, Ptashne M. A vector for expressing GAL4(1-147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mestas SP, Lumb KJ. Electrostatic contribution of phosphorylation to the stability of the CREB-CBP activator-coactivator complex. Nat Struct Biol. 1999;6:613–614. doi: 10.1038/10655. [DOI] [PubMed] [Google Scholar]
- 53.Yan JP, Garrus JE, Giebler HA, Stargell LA, Nyborg JK. Molecular interactions between the coactivator CBP and the human T-cell leukemia virus Tax protein. J. Mol. Biol. 1998;281:395–400. doi: 10.1006/jmbi.1998.1951. [DOI] [PubMed] [Google Scholar]
- 54.Van Orden K, Yan JP, Ulloa A, Nyborg JK. Binding of the human T-cell leukemia virus Tax protein to the coactivator CBP interferes with CBP-mediated transcriptional control. Oncogene. 1999;18:3766–3772. doi: 10.1038/sj.onc.1202703. [DOI] [PubMed] [Google Scholar]
- 55.Harrod R, Tang Y, Nicot C, Lu HS, Vassilev A, Nakatani Y, Giam CZ. An exposed KID-like domain in human T-cell lymphotropic virus type 1 Tax is responsible for the recruitment of coactivators CBP/p300. Mol. Cell. Biol. 1998;18:5052–5061. doi: 10.1128/mcb.18.9.5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Franklin AA, Kubik MF, Uittenbogaard MN, Brauweiler A, Utaisincharoen P, Matthews MA, Dynan WS, Hoeffler JP, Nyborg JK. Transactivation by the human T-cell leukemia virus Tax protein is mediated through enhanced binding of activating transcription factor-2 (ATF-2) ATF-2 response and cAMP element-binding protein (CREB). J. Biol. Chem. 1993;268:21225–21231. [PubMed] [Google Scholar]
- 57.Wilczek C, Chayka O, Plachetka A, Klempnauer KH. Myb-induced chromatin remodeling at a dual enhancer/promoter element involves non-coding rna transcription and is disrupted by oncogenic mutations of v-myb. J Biol Chem. 2009;284:35314–35324. doi: 10.1074/jbc.M109.066175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Giebler HA, Loring JE, Van Orden K, Colgin MA, Garrus JE, Escudero KW, Brauweiler A, Nyborg JK. Anchoring of CREB binding protein to the human T-cell leukemia virus type 1 promoter: a molecular mechanism of Tax transactivation. Mol. Cell. Biol. 1997;17:5156–5164. doi: 10.1128/mcb.17.9.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kraus WL, Manning ET, Kadonaga JT. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol. Cell. Biol. 1999;19:8123–8135. doi: 10.1128/mcb.19.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoeffler JP, Lustbader JW, Chen C-Y. Identification of multiple nuclear factors that inter-act with Cyclic Adenosine 3',5'-Monophosphate Response Element-Binding Protein and Activating Transcription Factor-2 by protein-protein interactions. Mol. Endocrin. 1991;5:256–266. doi: 10.1210/mend-5-2-256. [DOI] [PubMed] [Google Scholar]
- 61.Lopez DI, Mick JE, Nyborg JK. Purification of CREB to apparent homogeneity: removal of truncation products and contaminating nucleic acid. Protein Expr Purif. 2007;55:406–418. doi: 10.1016/j.pep.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buchan DW, Ward SM, Lobley AE, Nugent TC, Bryson K, Jones DT. Protein annotation and modelling servers at University College London. Nucleic Acids Res. 2010;38(Web Server issue):W563–568. doi: 10.1093/nar/gkq427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Petersen B, Petersen TN, Andersen P, Nielsen M, Lundegaard C. A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct Biol. 2009;9:51. doi: 10.1186/1472-6807-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van orden K, Giebler HA, Lemasson I, Gonzales M, Nyborg JK. Binding of p53 to the KIX domain of CREB binding protein. A potential link to human T-cell leukemia, type I-associated leukemogenesis. J. Biol Chem. 1999;274:26321–26328. doi: 10.1074/jbc.274.37.26321. [DOI] [PubMed] [Google Scholar]
- 66.Kasper LH, Boussouar F, Ney PA, Jackson CW, Rehg J, van Deursen JM, Brindle PK. A transcription-factor binding surface of coactivator p300 is required for haematopoiesis. Nature. 2002;419:738–743. doi: 10.1038/nature01062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.