Abstract
The cyanobacterial gene cluster kaiABC encodes three essential circadian clock proteins: KaiA, KaiB and KaiC. The KaiB and KaiC protein levels are robustly rhythmical, whereas the KaiA protein abundance undergoes little if any circadian oscillation in constant light. The level of the KaiC protein is crucial for correct functioning of the clock because induction of the protein at phases when the protein level is normally low elicits phase resetting. Titration of the effects of the inducer upon phase resetting versus KaiC level shows a direct correlation between induction of the KaiC protein within the physiological range and significant phase shifting. The protein synthesis inhibitor chloramphenicol prevents the induction of KaiC and blocks phase shifting. When the metabolism is repressed by either translational inhibition or constant darkness, the rhythm of KaiC abundance persists; therefore, clock protein expression has a preferred status under a variety of conditions. These data indicate that rhythmic expression of KaiC appears to be a crucial component of clock precession in cyanobacteria.
Keywords: circadian/clock/cyanobacteria/kai
Introduction
Organisms keep time on a daily cycle using a biochemical oscillator. These circadian oscillators are found almost ubiquitously among the eukaryotes. While it was previously thought that circadian clocks were exclusively a property of eukaryotic organization, in the past decade it has been recognized that at least one group of prokaryotes, the eubacterial cyanobacteria, are also circadian time-keepers (Johnson and Golden, 1999). These timers elaborate a temporal program that enhances the fitness of cyanobacteria (Ouyang et al., 1998). The fundamental properties of circadian clocks in eukaryotes and cyanobacteria are the same: surprisingly precise self-sustained oscillations with an ∼24 h period that are temperature compensated and entrainable by environmental cycles. The mechanism that enables a precise oscillation with such a long time constant and that is relatively unaffected by the ambient temperature is of considerable interest. Are the mechanisms that accomplish this timing similar in diverse organisms, or has evolution ‘designed’ a series of solutions to the challenge of circadian timekeeping? The comparison of the molecular bases of circadian clocks among cyanobacteria and eukaryotes can address this question.
In eukaryotes, a web of evidence supports a model that proposes autoregulatory feedback loops of central clock gene expression (Hardin et al., 1990; Young, 1998; Dunlap, 1999). In Drosophila and Neurospora, where the model is most highly developed, there is an ensemble of different molecular components that are believed to participate in the feedback loop. The protein and mRNA levels of at least three of these genes, period (per), timeless (tim) and dclk (dclock), are rhythmic. In the evening, PER and TIM accumulate and associate in the cytoplasm, move to the nucleus, and repress the transcription of their own genes in a negative feedback loop (Rosbash and Hall, 1989; Sehgal et al., 1995; Darlington et al., 1998; Young, 1998). CYCLE and dCLK associate as positive regulatory elements to stimulate transcription at E-box sequences of the per and tim promoters (Hao et al., 1997; Darlington et al., 1998). In the other well characterized circadian clockwork, that in Neurospora, the gene frequency encodes a protein (FRQ) that is rhythmically abundant and represses its own transcription (Aronson et al., 1994; Dunlap, 1999). Expression of FRQ or PER proteins from inducible promoters at phases where they are normally low changes the phase of the clock (Aronson et al., 1994; Edery et al., 1994). Altering the stability of the clock proteins would be expected to change the period of the clock, and this is the interpretation of the influence of PER and FRQ phosphorylation on period (Price et al., 1998; Liu et al., 2000).
Less is known about the circadian clockwork in plants and mammals. There are two plant genes that have been cloned and shown to disrupt the expression of multiple circadian outputs when either constitutively overexpressed or mutated, namely the Myb-like transcriptional factors CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) (Schaffer et al., 1998; Wang and Tobin, 1998). CCA1 and LHY are currently thought to either be essential clock components in plants, or at least acting on an output path very close to the central oscillator. In the mouse, the emerging clockwork story is similar to that found in Drosophila. Three separate PER homologs (mPER1, mPER2 and mPER3) have been discovered in mice (Sun et al., 1997; Tei et al., 1997; Zylka et al., 1998). Other putative components of the mouse clockwork are the mammalian homologs of CRYPTOCHROME, mCRY1 and mCRY2 (Griffin et al., 1999; Kume et al., 1999). mRNA abundances within the suprachiasmatic nuclei are rhythmic for mPer1, mPer2, mPer3, MOP3 (BMAL1) and mCry1 (Sun et al., 1997; Tei et al., 1997; Zylka et al., 1998). The proteins encoded by at least some of these genes (as well as that for mCry2, whose mRNA is not rhythmic) are known to march in time with their mRNAs, further supporting the theme of rhythmic clock protein expression (Hastings et al., 1999; Kume et al., 1999).
In the prokaryotic cyanobacterium, Synechococcus sp. strain PCC 7942, a mutational analysis discovered a cluster of three circadian clock genes, named kaiA, kaiB and kaiC, that encode essential circadian clock components (Ishiura et al., 1998). Deletion of any of the three kai genes (or of the entire cluster) will cause arhythmicity of circadian outputs. Nineteen mutations have been mapped by genetic rescue and DNA sequencing to the three kai genes. All are missense mutants resulting from single nucleotide exchanges. Promoter activities were found in the upstream regions of both the kaiA and kaiB genes. The kaiA promoter gives rise to a monocistronic kaiA mRNA, whereas the kaiB promoter produces a dicistronic kaiBC mRNA (Ishiura et al., 1998).
Even though there are no sequence similarities between the kai genes and any known eukaryotic clock genes, some features of kai gene regulation are reminiscent of PER and FRQ regulation. Both the monocistronic kaiA and dicistronic kaiBC transcripts are rhythmically abundant, implying that the abundance of their proteins might be rhythmic. Continuous overexpression of kaiC repressed the kaiBC promoter (negative feedback) and eliminated rhythmicity, whereas kaiA overexpression enhanced the total activity of kaiBCp (positive feedback), dampening the amplitude of the oscillation without eliminating it. Moreover, overexpression of the kaiC gene for a few hours resets the phase of the rhythms, but KaiA overexpression did not affect the phase or period of the rhythm. Taken together, these results were used to propose a model wherein there is negative feedback control of kaiBC expression by the KaiC protein to generate a circadian oscillation and KaiA sustains the oscillation by enhancing kaiC expression (Ishiura et al., 1998). The cyanobacterial clock model implies that the Kai protein levels are rhythmic and that changes in Kai protein levels within the physiological range will elicit phase resetting, but these tantalizing similarities between the circadian clockwork models in eukaryotes versus cyanobacteria need to be tested explicitly. The studies in this paper provide insights into the biochemical mechanism of the cyanobacterial clockwork by testing and confirming those predictions.
Results
KaiB and KaiC proteins cycle in constant conditions
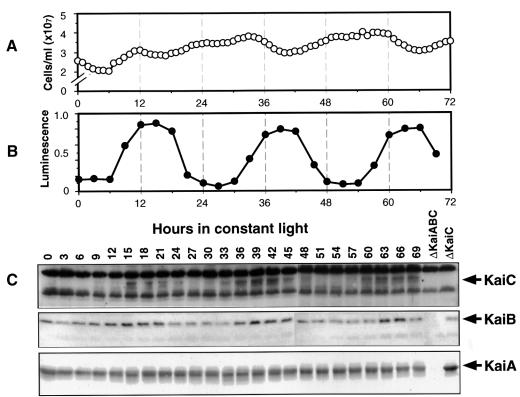
The promoter activities of both kaiA and kaiBC are rhythmic in constant light (LL), and there also appear to be rhythms of kaiA and kaiBC mRNA abundances (Ishiura et al., 1998; Iwasaki et al., 2000). If the turnover rate of the corresponding proteins is sufficiently high, these results predict rhythms of the Kai protein levels. To test this prediction, we assayed Kai proteins on immunoblots of cyanobacterial extracts. The cell cultures used for these experiments were in continuous log phase because we found that cells from this type of culture were the most homogeneous in terms of cell size and chlorophyll fluorescence, which is an indicator of metabolic status. Cells from these cultures exhibit a high amplitude rhythm of psbAI promoter activity (reported by luminescence), and divide rhythmically (Figure 1; Mori et al., 1996).
Fig. 1. Rhythms of Kai proteins for 3 days in LL from log-phase cultures that are continuously diluted to maintain an approximately equal cell concentration (30°C; light intensity = 125 µE/m2/s; strain = AMC149). In this experiment, the average generation time was 9.45 h. (A) Cell counts of culture showing the approximately equal cell numbers at all phases as in Mori et al. (1996). (B) Luminescence of the culture that reports the circadian rhythm of the psbAI promoter activity. (C) Immunoblots of protein extracts collected at the various phases. The blots were treated with antibodies to KaiC (top strip), KaiB (middle strip) and KaiA (bottom strip). The Kai-specific bands are indicated by the arrows to the right of the panels; note the ΔKaiABC and ΔKaiC lanes that are the controls to indicate non-specific bands.
Our antisera to KaiB and KaiA were specific, with few cross-reacting bands. Our antiserum to KaiC, however, cross-reacted with several non-specific proteins of high abundance that bracket the specific KaiC band on immunoblots (pre-immune sera did not react with any of these bands). Attempts to eliminate cross-reacting bands by adsorption of antiserum with extracts of ΔkaiABC cells or by using antibodies purified from immunoblots of purified KaiC (Harlow and Lane, 1999) were only partially successful. There may be an epitope that is common to KaiC and several high abundance proteins. We therefore routinely immunoblotted extracts from ΔkaiABC and ΔkaiC strains of cyanobacteria to confirm which bands were specific and which were not (Figure 1C, rightmost lanes). The non-specific reacting bands of anti-KaiC provided a separate internal control for the equal loading of total protein in each lane. The molecular weights of KaiA and KaiB estimated from their sequences should be 32.6 and 11.4 kDa, respectively, while those estimated from mobility on the immunoblots are 31.2 and 10.3 kDa. There are two KaiC-specific bands, one of which may be a phosphorylated form (Nishiwaki et al., 2000). From its sequence, KaiC is predicted to be 58.0 kDa; the two bands on immunoblots are estimated as 57.1 and 55.4 kDa. Therefore, there is an excellent correspondence between the molecular weights of the Kai proteins estimated from their sequences and from the specific bands on immunoblots.
Figure 1C shows that the abundances of KaiB and KaiC display high amplitude rhythms of abundance for at least 3 days in LL in continuous cultures. The KaiB and KaiC rhythms are roughly in phase with a peak at approximately circadian time (CT) 15 (CT0 = subjective dawn, CT12 = subjective dusk). Since KaiB and KaiC are co-transcribed, the phasing of the protein rhythms suggests that their turnover rates are similar. The peak phase of the KaiB and KaiC rhythms corresponds with the phase where the cell division rate begins to slow (Mori et al., 1996) and slightly phase-leads the peak phase of the psbAI promoter activity rhythm (reported by luminescence). The KaiC protein rhythm is not dependent on rhythmic cell division, as will be demonstrated later by the data depicted in Figures 4 and 6.
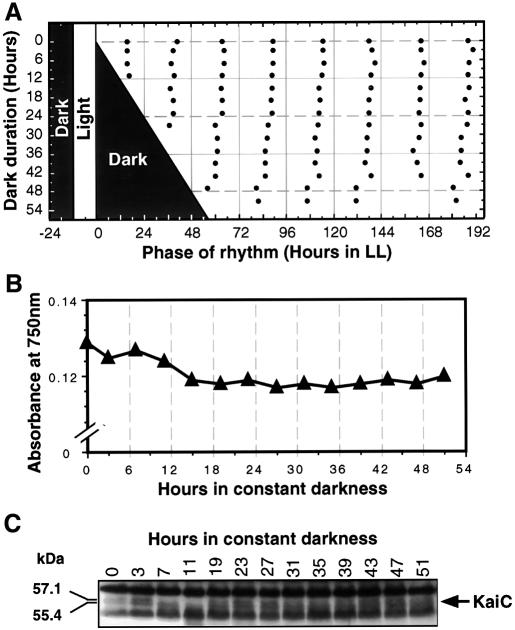
Fig. 4. The cyanobacterial clock and the KaiC oscillation continues to run in constant darkness in the AMC149 strain. Cells were grown for 4 days in an LD 12:12 cycle until the OD750 was 0.13. At that time, cultures were transferred to DD. (A) The clock continues to run in DD, as assessed by returning samples to LL after various durations of exposure to DD. Shaded areas indicate time of darkness; filled circles indicate the phases of the troughs of the luminescence rhythm during the subsequent free run in LL. (B) Cultures stop growing immediately in DD (measured by OD750). (C) KaiC immunoblot of cultures in DD (note that there is no sample for hour 17). To compare the phasing of the KaiC abundance rhythm in DD with that in LL, note that hour 0 in Figure 4 corresponds to hour 12 in Figure 1 (both are CT12). Molecular weights of the KaiC bands were calculated on the basis of migration.
KaiA levels are relatively high at all phases, displaying a possible rhythm of low peak-to-trough amplitude, even though kaiA mRNA is rhythmic (Ishiura et al., 1998; Iwasaki et al., 2000). Our data clearly show that a rhythm of mRNA abundance will not necessarily lead to a rhythm in the corresponding protein, and that post-transcriptional regulation is important in kai gene expression. The lack of a robust KaiA rhythm might be due to a slow turnover rate for this protein.
Expression of KaiC within the physiological range elicits phase resetting
If the rhythm of KaiC abundance is important for circadian clock precession in cyanobacteria, induction of KaiC expression at phases when it is normally low would be expected to reset the phase of the clock. Using inducible promoter constructs, expression of FRQ in Neurospora, PER in Drosophila and KaiC in cyanobacteria set the phase of circadian rhythms, and these results have been interpreted to mean that the oscillations of FRQ, PER or KaiC levels constitute a biochemical correlate of a state variable of the circadian clock (Aronson et al., 1994; Edery et al., 1994; Ishiura et al., 1998). In all those studies, however, the protein level that is necessary to achieve significant phase resetting was not measured (for FRQ, the level necessary to repress its own promoter has been quantified, but phase shifting was not measured in those experiments; Merrow et al., 1997). It is possible that the procedure induced the protein to levels far beyond the maximum levels normally reached during the circadian cycle. If so, the phase shifting by induction might be an artifact of expression beyond the physiological range.
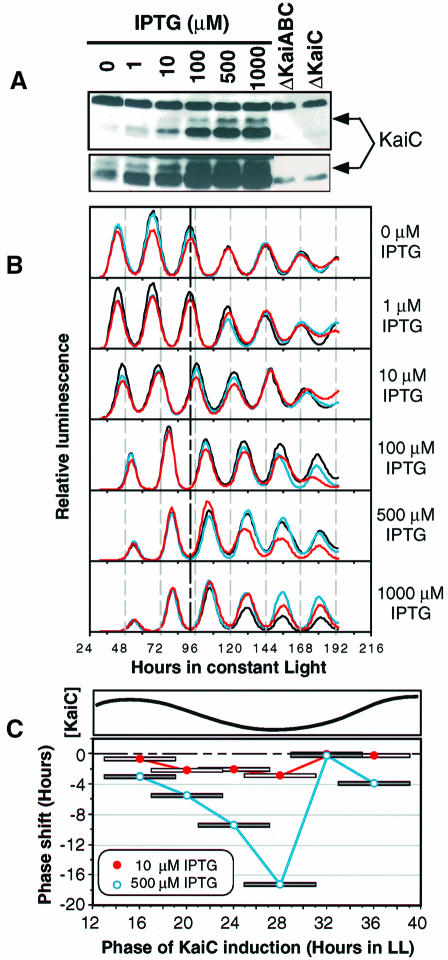
Therefore, we set out to test the dose response of expression of KaiC on the phase resetting of the psbAIp activity rhythm. We used a cyanobacterial strain harboring a lacIq-trcp::kaiC construct that is derepressed by the inducer isopropyl-β-d-thiogalactopyranoside (IPTG) (Ishiura et al., 1998). Figure 2A shows the induction of KaiC in the trcp::kaiC strain by 6 h exposures to various concentrations of IPTG (in wild-type cells without the trcp::kaiC construct, IPTG does not affect KaiC levels). A 1 µM IPTG concentration does not induce KaiC beyond basal (trough) levels, but 10 µM IPTG induces KaiC to levels above basal and which approximately correspond to sub-peak levels shown in Figure 1C. One hundred micromolar concentrations of IPTG induce levels of KaiC that are slightly beyond the normal physiological range (i.e. above peak levels), and higher concentrations of IPTG induce KaiC to levels far beyond normal. Treatment with IPTG also induced an antiserum-reacting protein with a molecular weight that is less than that predicted for KaiC. A band of the same mobility was also detected in the ΔkaiABC and ΔkaiC extracts at lower levels (Figure 2A). It is possible that this band is either a breakdown product of KaiC or that it is a completely unrelated protein. For the purpose of this dose–response experiment, only the bona fide KaiC band will be considered.
Fig. 2. Dose response of KaiC induction and clock resetting (strain = trcp::kaiC). Cells came from a continuously diluted culture grown as in Figure 1. (A) Induction of KaiC by 6 h treatments of IPTG given at the same CT as in the rhythm experiment shown in (B). The KaiC-specific band is indicated by the arrow to the right of the panel. The exposure time of the upper part is comparable to the exposure of the KaiC blot in Figure 1C, whereas the lower part is the relevant area of the same blot that was exposed longer to visualize the weaker bands. (B) Resetting of the luminscence rhythm by 6 h pulses of various concentrations of IPTG. After a 12 h synchronizing dark pulse, cyanobacterial cultures harboring the trcp::kaiC construct were placed in LL. The 6 h IPTG pulse was begun 21 h after the beginning of LL. Triplicate samples for each concentration are shown in different colors. The heavy vertical dashed line at hour 92 shows the control phase for comparison. (C) Phase-dependent resetting by KaiC induction in the trcp::kaiC strain. Upper part, the rhythm of KaiC abundance abstracted from the data depicted in Figure 1C. Lower part, phase resetting of the luminescence rhythm by IPTG induction of KaiC. Six hour pulses of IPTG at either 10 or 500 µM were administered to liquid cultures of cyanobacteria at different phases in LL. The midpoints of the 6 h pulses are connected by the lines. The phase shifts shown are the averages of duplicates for each point. The phase response curves are plotted monotonically as all-delay phase shifts (Johnson, 1999).
Cyanobacterial cultures were treated with 6 h pulses of IPTG from hour 21 to hour 27 after the beginning of LL. This phase was chosen because at hour 21, KaiC levels are declining, and hours 24–27 are essentially the trough of the KaiC abundance rhythm (Figure 1C). Expression of KaiC to peak levels in these trough phases should result in significant phase shifting if the rhythm of KaiC abundance is essential for clock function. As shown in Figure 2B, 1 µM IPTG does not elicit phase resetting, but 10 µM IPTG evokes significant phase shifts, and 100 µM IPTG gives essentially a saturated response (IPTG does not affect the rhythm in cyanobacteria that do not harbor the trcp::kaiC construct; data not shown). Therefore, there is an excellent correlation between the doses of IPTG that induce KaiC levels within the physiological range and phase resetting.
There is also a correlation between the phase of the endogenous KaiC abundance rhythm and the magnitude of clock resetting by IPTG induction of KaiC. This correlation is shown in the phase response curves depicted in Figure 2C. Phase-dependent resetting was measured for 6 h pulses of 10 and 500 µM IPTG, respectively. Phase shifts increase in magnitude during the phases of the clock when endogenous levels of KaiC are declining. Maximal phase shifts were obtained for IPTG pulses administered at the trough phase of KaiC. During phases of the KaiC rhythm when levels are rising, further increases in KaiC levels do not provoke significant phase resetting.
Inhibition of protein synthesis blocks KaiC induction and phase resetting
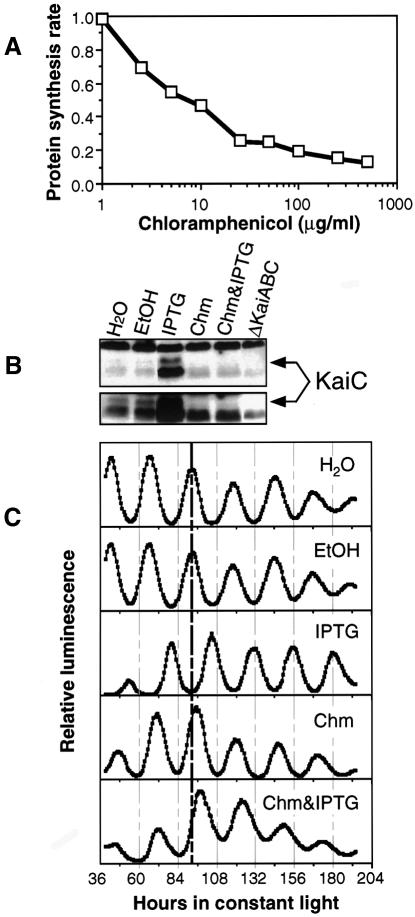
If IPTG treatment of cells containing the trcp::kaiC construct is phase shifting the circadian clock via new synthesis of KaiC, then prevention of KaiC induction using protein synthesis inhibitors should block the IPTG-induced phase resetting. Figure 3A shows that total protein synthesis is inhibited by chloramphenicol at 50 µg/ml by at least 75%, and further increases in chloramphenicol only slightly increase the amount of inhibition to 87%. It is likely that this residual 13% represents a background of label incorporation by metabolic processes other than protein synthesis, in which case the inhibition of protein synthesis by 50 µg/ml chloramphenicol is >75% (we used [35S]l-methionine and [35S]l-cysteine for measuring protein synthetic rate). Figure 3B shows that 50 µg/ml chloramphenicol appears totally to inhibit the induction of KaiC by 1 mM IPTG in the trcp::kaiC strain. Moreover, the data in Figure 3C show that simultaneous treatment with 50 µg/ml chloramphenicol inhibited the phase shift elicited by 1 mM IPTG. Chloramphenicol alone causes a very small phase delay that is the same as the combined treatment with chloramphenicol and IPTG, and clearly different from the large phase shift evoked by IPTG alone. Therefore, inhibition of protein synthesis prevents clock resetting by IPTG in the trcp::kaiC strain in concert with its inhibition of KaiC induction.
Fig. 3. Inhibition of KaiC induction and clock resetting by chloramphenicol in the trcp::kaiC strain. Cells were grown as in Figure 1 until the time of IPTG and/or chloramphenicol addition. (A) Inhibition of total protein synthesis by chloramphenicol. Protein synthetic rate is expressed as percent incorporation (incorporation/uptake) of [35S]l-methionine and [35S]l-cysteine with the control normalized to 1.0 (chloramphenicol concentration expressed as log concentration). Chloramphenicol was dissolved in ethanol; the final concentration of ethanol in all samples was 0.2%. (B) Inhibition of KaiC induction by chloramphenicol assayed by immunoblotting. Concentrations: 50 µg/ml chloramphenicol (Chm) in 0.2% ethanol; 1 mM IPTG (dissolved in dH2O); ethanol, 0.2%. The lower panel is the relevant region of the blot overexposed to visualize the expression level in the control sample. (C) Inhibition of IPTG-induced phase resetting by 50 µg/ml chloramphenicol. Six hour pulses of water, 0.2% ethanol, 1 mM IPTG, chloramphenicol (50 µg/ml in 0.2% ethanol) or IPTG plus chloramphenicol were initiated at hour 21 of LL as in Figure 2B.
KaiC is rhythmic in constant darkness
The data of Figure 1 show robust rhythms of KaiC levels in LL. Does this oscillator also operate in constant darkness? For obligate photoautotrophs like Synechococcus, the night is a time of shutdown for which these cells, and their internal clock, must be ready every day. It has long been recognized that cyanobacterial metabolism becomes largely (but not totally) quiescent in the dark (Doolittle, 1979). In many photoautotrophs, including eukaryotic plants, most output circadian rhythms are not expressed in continuous darkness (DD), although their central oscillators continue to tick (Johnson et al., 1998). In cyanobacteria that have been adapted to allow metabolism of exogenous carbon sources, circadian output rhythms persist when the carbon source is present (Aoki et al., 1997; Schneegurt et al., 1997).
In Synechococcus sp. strain PCC 7942, there has been no definitive evidence that the central clockwork continues to oscillate in DD. Therefore, we assayed clock precession and the expression of KaiC in DD. As shown in Figure 4A, the phase was set by the prior light/dark (LD) cycle, and dark treatment that begins at the normal time of lights-off does not cause major resetting of the clock even when the dark is extended for at least 52 h (if the phase of the clock had been strongly reset by the long dark exposures, the trough phases would have lined up along a diagonal that parallels the end of dark). Figure 4B shows that cell growth and division immediately and completely halt when cultures are transferred to DD. Figure 4C illustrates that KaiC abundance continues to oscillate in DD with an amplitude that is lower than that in LL, but still clearly rhythmic. These data demonstrate that the circadian clock in Synechococcus sp. strain PCC 7942 continues to run in constant darkness for at least 52 h.
KaiC rhythms persist even when translation is mostly inhibited
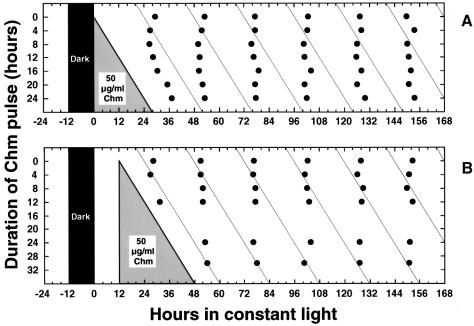
The intriguing results of Figure 4 encouraged us to try another condition of metabolic repression, namely by inhibiting protein synthesis with chloramphenicol. Similar to the results with DD, Figure 5 shows results from two different experiments in which long exposures to 50 µg/ml chloramphenicol do not cause steady-state phase shifts of the clock (if they had, the phase of the rhythm would have been set to the diagonal lines that parallel the chloramphenicol wash-out). The exposures to chloramphenicol began either immediately after the dark treatment (Figure 5A) or 12 h later (Figure 5B). We then assayed the expression of KaiC during long exposures to chloramphenicol. Growth is not immediately halted by 50 µg/ml chloramphenicol (Figure 6A), when protein synthesis is inhibited by at least 75–80% (Figure 3A). For the first 24 h in chloramphenicol, cell growth and division continue, but thereafter growth stops. After ∼48 h in chloramphenicol, cell numbers decline, and most of the cells are no longer able to recover from the chloramphenicol treatment. Nevertheless, the rhythms of the cells that do recover from extended chloramphenicol treatments are not phase shifted by the drug treatment. Therefore, a window of between 24 and 48 h in 50 µg/ml chloramphenicol exists during which growth has stopped, cells can recover after wash-out, and the clock is running.
Fig. 5. Chloramphenicol treatment does not set the phase of the circadian clock (strain = AMC149). Cells were grown in batch culture prior to addition of chloramphenicol. Chloramphenicol (50 µg/ml) was applied to cyanobacterial cultures for various durations, then removed. Filled circles indicate the phases of the troughs of the luminescence rhythm after wash-out of chloramphenicol. (A) Chloramphenicol treatment beginning immediately after the synchronizing dark treatment. (B) Chloramphenicol treatment beginning 12 h after the synchronizing dark treatment.
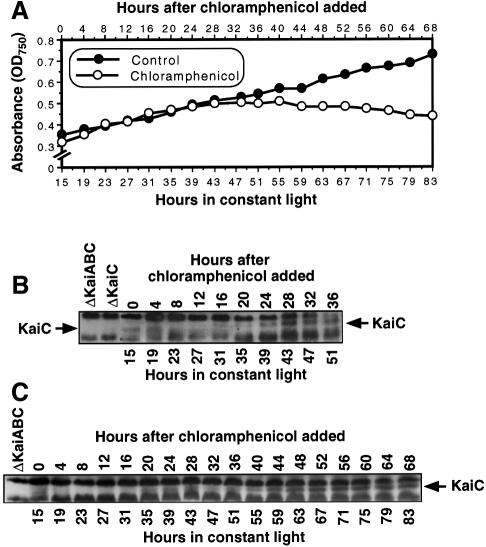
Fig. 6. KaiC abundance is still rhythmic in the presence of chloramphenicol. The cell culture for this experiment was a batch culture grown to an OD750 of 0.3–0.4, then 50 µg/ml chloramphenicol was added to cultures 15 h after the onset of LL (strain = AMC149). (A) In the presence of 50 µg/ml chloramphenicol, cell growth and division continue for ∼24 h in chloramphenicol, then stop, whereas the control culture continues to grow (growth measured by OD750). (B) KaiC immunoblot for a culture in chloramphenicol for 36 h (experiment 1). (C) KaiC immunoblot for a culture in chloramphenicol for 68 h (experiment 2).
What is happening to KaiC expression during this window? Even though the KaiC signal is not as strong as in control cultures, the rhythm of KaiC abundance continues for at least 48–60 h in the presence of chloramphenicol. Figure 6B and C shows two separate experiments in which KaiC continues to oscillate (two cycles in Figure 6B, three cycles in Figure 6C). However, as the cells enter the growth mode from which they cannot recover from chloramphenicol, the KaiC levels increase to peak levels and do not oscillate thereafter. These data indicate that the KaiC abundance rhythm persists in non-growing cells in which translation has been slowed to a trickle. The level of expression of KaiC is lower in DD (Figure 4) and under translational repression (Figure 6) than in LL (Figure 1). This lower level of KaiC might have been expected to interfere with clock expression, but the cyanobacterial clock has been found to run in a SasA-deletion mutant in which Kai levels are similarly reduced (Iwasaki et al., 2000). Finally, the data from non-dividing cells in DD or in the presence of chloramphenicol demonstrate that the KaiC rhythm is a circadian oscillation that is not linked obligatorily to cell growth and division.
Discussion
The abundance of KaiB and KaiC proteins oscillates in constant conditions. Since KaiB and KaiC are co-transcribed (Ishiura et al., 1998), the similar phase of the protein oscillations implies that KaiB and KaiC have similar rates of turnover. Even though the kaiA gene is rhythmically transcribed (Ishiura et al., 1998; Iwasaki et al., 2000), the abundance of its protein undergoes a very low amplitude oscillation. One explanation for this result is that the rate of KaiA turnover is significantly slower than that for KaiB or KaiC. The roles of the KaiA and KaiB proteins in the cyanobacterial clockwork are not clear. Overexpression of KaiA stimulates the activity of the kaiBC promoter without dramatically affecting its period or phase (Ishiura et al., 1998). Since KaiA expression is already nearly constitutive (Figure 1), the overexpression result is likely to be due to a non-physiologically high level, and not to the disruption of a temporal pattern of KaiA expression. Because KaiA interacts with KaiC (Iwasaki et al., 1999) and KaiC represses the expression of its own promoter, a possible mechanism by which KaiA could boost the activity of kaiBCp would be to inhibit KaiC, thereby derepressing kaiBCp.
This study focused on the expression patterns and properties of KaiC because it clearly is a central component of the cyanobacterial clockwork, and most of the mutants displaying altered circadian properties map to kaiC. KaiC shows sequence similarities to bacterial helicases and recombinases (Leipe et al., 2000), so it might directly regulate gene expression, which is controlled globally in cyanobacteria (Liu et al., 1995). Our data show that KaiC is rhythmically expressed even under conditions of metabolic shutdown. At first we were surprised to discover that clock precession and rhythmic translation of KaiC continue in the presence of chloramphenicol concentrations that are sufficient to halt growth. However, when compared with the metabolic repression that must accompany darkness in obligately photoautotrophic cells such as Synechococcus sp. strain PCC 7942 (Doolittle, 1979), chloramphenicol treatment might mimic DD in some respects. It seems natural that these cells would have evolved a mechanism that operates in darkness to repress simultaneously the expression of most of their genes in order to conserve energy while preferentially maintaining the expression of a subset of genes whose products are needed. The timing mechanism is a natural candidate for a process that should be maintained ‘around the clock’ in light or dark.
The mechanism that selects between those genes that continue to be expressed under metabolic stress versus those that are repressed is not known, but one possibility is that some mRNAs are translated and others are not. The translational selectivity could be accomplished by different types of ribosomes; in a closely related cyanobacterium, Synechococcus sp. strain PCC 6301, there are two rRNA operons: rrnA and rrnB (Tomioka et al., 1981; Kumano et al., 1986). Perhaps the ribosomes constructed from the rRNAs encoded by rrnA versus rrnB can distinguish between different classes of mRNAs. For example, one set of ribosomes that translates the majority of messages could become inactive during metabolic repression, whereas the other set of ribosomes that recognizes the preferential subset of transcripts churns away under all but the most extreme conditions.
The rhythm of KaiC abundance appears to be crucial for circadian timekeeping since induced expression of KaiC within the physiological range provokes phase resetting. Since KaiC expression also represses the activity of kaiBCp (Ishiura et al., 1998), the results are consistent with a negative feedback of KaiC onto kaiBCp and the circadian phase. Overexpression experiments with FRQ in Neurospora and PER in Drosophila have led to the conclusion that these clock proteins may be acting as molecular correlates of state variables in the circadian limit cycle oscillator (Aronson et al., 1994; Edery et al., 1994). The same may be true of KaiC in cyanobacteria, but it should not be concluded that we envision KaiC abundance to be merely like the grains of sand in an hourglass, namely, that the oscillator works by counting the number of KaiC molecules up to a certain threshold, which then triggers the next event. Rather, we can easily conceive that the relevant aspect of KaiC abundance could be the establishment of a critical stoichiometry of KaiC to other molecules in a complex, and that rhythmic changes of KaiC activate/repress this complex cyclically. Overexpression of KaiC could reset the clock by disrupting the stoichiometry within this complex. KaiC interacts with KaiA and KaiB (Iwasaki et al., 1999) and there are likely to be other molecular partners for KaiC (Iwasaki et al., 2000). These proteins may form one or more types of protein complex that play a crucial role in circadian timekeeping, as is thought to be true for eukaryotic clock proteins (Dunlap, 1999).
Materials and methods
Strains and growth conditions
AMC149 was used as the wild-type reporter strain, in which the gene fusion psbAIp::luxAB was integrated into the genome of Synechococcus sp. strain PCC 7942 (Kondo et al., 1993). To create a kaiC-null strain (ΔKaiC), the 1.67 kb Hpa I–Stu I fragment [which contains the entire kaiC open reading frame (ORF)] within the 4 kb SmaI–Hind III segment from p30N (Ishiura et al., 1998) containing the kaiABC cluster and its flanking regions was replaced by a chloramphenicol resistance gene (Chmr). AMC149 cells were transformed with this construct and subjected to several rounds of selection with chloramphenicol. The kaiC ORF was confirmed to have been deleted from the chromosome by genomic PCR. The kaiABC-null (ΔKaiABC) and the inducible trcp::kaiC strains were described previously (Ishiura et al., 1998; trcp::kaiC = Ptrc::kaiC strain in that paper). For induction of KaiC, trcp::kaiC cells were incubated for 6 h with IPTG. Following treatment, IPTG was removed by centrifugation and washing with fresh medium three times (control samples were spun and washed equivalentally).
All strains were grown on modified BG-11 medium (Bustos and Golden, 1991) at 30°C under LD 12:12 cycles or in LL under cool-white fluorescent lamps (40–50 µE/m2/s). Depending upon the antibiotic resistance of each strain, 20 µg/ml spectinomycin, 5 µg/ml kanamycin and/or 7.5 µg/ml chloramphenicol were added to the medium. Cell densities of liquid cultures were monitored either by counting cell numbers with an electronic particle counter (Coulter) or by measuring the absorbance at 750 nm. Before releasing the cultures into free-running conditions of either LL or DD, the clocks of the cells within the cultures were synchronized with either three or four cycles of LD 12:12 (experiments depicted in Figures 1, 3, 4, 5 and 6) or a single 12 h of darkness (experiments depicted in Figure 2). Luminescence rhythms were measured from liquid cultures transformed with the psbAIp::luxAB reporter construct that were grown in either standard batch cultures or in continuous cultures as described previously (Kondo et al., 1993; Mori et al., 1996).
In standard batch liquid cultures, cell suspensions were bubbled with sterile air. Growth of continuously diluted cultures was modified from the prior protocol (Mori et al., 1996) to allow harvesting of cells for protein extraction. For instance, in the experiment shown in Figure 1, cyanobacterial cells were grown in 935 ml of modified BG-11 medium in 1 l bottles in LD 12:12 with air bubbling and stirring. After four cycles of LD (OD750 ∼0.1), the culture was released into LL (intensity = 125 µE/m2/s), and continuous dilution with fresh BG-11 medium at a flow rate of 60 ml/h was begun. Concomitant with dilution, 3.4 ml of cell suspension were withdrawn every 1 h for monitoring cell density, and 170 ml of cell suspension were withdrawn every 3 h for measuring in vivo luminescence (1 ml) and collecting cells for later immunoblotting (∼169 ml). In different experiments, culture conditions were basically the same as described above, but the culture volume and dilution rate were slightly different.
Antibody production
Constructs for expressing His6::KaiA and His6::KaiB fusion proteins were described previously (Xu et al., 1999). To make a construct for expressing His6::KaiC fusion proteins, the coding region for KaiC was amplified by using a pair of primers: 5′-(BglII) AGATCTATGACTTCCGCTGAGATGACT-3′ and 5′-(HindIII) AAGCTTCTA AGCGCGATCGCTGGCTAG-3′ (underlined sequences indicate the added restriction enzyme sites). The PCR fragments containing the coding regions for KaiA, KaiB, and KaiC were inserted into the BglII–HindIII site of the Xpress™ vector pRSET-B (Invitrogen). His6::KaiA (residues 39–284), His6::KaiB and His6::KaiC fusion proteins were expressed in Escherichia coli BL21 (DE3) under the control of the bacteriophage T7 promoter by 3 h of induction with 1 mM IPTG at room temperature. The fusion proteins were purified by immobilized metal affinity chromatography on His⋅Bind® resin (Novagen). Polyclonal antisera against KaiA, KaiB and KaiC were generated by immunizing rabbits with the purified Kai antigens at 3 week intervals (two rabbits for each antigen). Antisera were screened by ELISA and immunoblots; after three to five immunization cycles (depending upon the development of reactive antisera in each rabbit), antisera were collected. The antisera against KaiA and KaiB were used directly, but for anti-KaiC, the antiserum was partially purified from immunoblots of purified His6-tagged KaiC (Harlow and Lane, 1999).
Immunoblot analyses of Kai proteins
Cyanobacterial cells were collected at various phases or after various treatments, spun and washed once with cold TBS (50 mM Tris–HCl, 200 mM NaCl pH 7.5). The pellets were frozen in liquid nitrogen and stored at –80°C. For extraction, each frozen pellet was thawed in 100 µl of lysis buffer (0.1 M CHES, 4% SDS, 20% glycerol, pH 9.5, 20 mM Na3VO4, 50 mM NaF, 50 mM glycerophosphate, 10 µg/ml aprotinin, 5 µg/ml leupeptin and 1 mM phenylmethylsulfonyl fluoride). The suspension was put on ice and sonicated until 90–100% of the cells were broken. The extract was centrifuged, and an aliquot of the supernatant was removed for determination of protein concentration (Lowry et al., 1951). An equal volume of 10% β-mercaptoethanol with 0.01% (w/v) bromophenol blue was added to each extract and the total protein concentration was adjusted using 1× SDS loading buffer so that all samples had an equal protein concentration. After this adjustment, all samples were boiled and then stored at –80°C.
Equal quantities of total proteins were loaded in each well and separated by SDS–PAGE (acrylamide concentrations: 15% for KaiA, 20% for KaiB and 10% for KaiC gels). Following protein transfer to nitrocellulose membranes (Bio-Rad), blots were blocked with 1× PBS buffer containing 0.1% Tween-20 and 0.2% I-Block™ (Tropix Inc.). Membranes were then incubated overnight at 4°C with either the antisera against KaiA or KaiB (1:10 000) or the immunoblot-purified anti-KaiC antibody (1:1000). After washing, blots were incubated for 1 h at room temperature with a 1:7500 dilution of alkaline phosphatase-conjugated goat anti-rabbit IgG (Promega). Immunoblots were developed using the CDP-Star™ chemiluminescent detection system (Tropix, Inc.).
Chloramphenicol inhibition of protein synthesis
Two microliters of various concentrations of chloramphenicol (Sigma) dissolved in ethanol were added to 1 ml cultures of cyanobacteria and incubated for 30 min. Ten microcuries of Tran35S-label (ICN; includes [35S]l-methionine and [35S]l-cysteine) were added to the samples and incubated at 30°C for 2 h, then half of each sample was washed on a glass fiber filter with ice-cold BG-11 medium plus 1 mg/ml methionine and 1 mg/ml cysteine (= uptake sample). The remaining half of each sample was incubated with trichloroacetic acid (TCA) and SDS (final concentrations: 10% TCA, 1% SDS) for 1 h at room temperature, then filtered onto glass fiber filters and washed three times with 5 ml of 10% TCA, followed by 5 ml of acetone (= incorporation sample). The radioactivity of the samples was measured by liquid scintillation counting and quench corrected. The protein synthetic rate was expressed as the incorporation divided by the uptake to compensate for changes in label uptake among the samples.
Acknowledgments
Acknowledgements
We thank Drs Shinsuke Kutsuna, Masahiro Ishiura and Takao Kondo for graciously providing the trcp::kaiC and ΔkaiABC strains. We appreciate the excellent assistance of Dr Heping Yan with antibody production, and are grateful to Dr Susan Golden for suggestions and discussion. This research was supported by the NSF (grant No. MCB-9874371 to C.H.J.) and the NIMH (grant No. K02 MH01179 to C.H.J.).
References
- Aoki S., Kondo,T., Wada,H. and Ishiura,M. (1997) Circadian rhythm of the cyanobacterium Synechocystis sp. strain PCC 6803 in the dark. J. Bacteriol., 179, 5751–5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson B., Johnson,K., Loros,J.J. and Dunlap,J.C. (1994) Negative feedback defining a circadian clock: autoregulation in the clock gene frequency. Science, 263, 1578–1584. [DOI] [PubMed] [Google Scholar]
- Bustos S.A. and Golden,S.S. (1991) Expression of the psbDII gene in Synechococcus sp. strain PCC 7942 requires sequences downstream of the transcription start site. J. Bacteriol., 173, 7525–7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington T.K., Wager-Smith,K., Ceriani,M.F., Staknis,D., Gekakis,N., Steeves,T.D.L., Weitz,C.J., Takahashi,J.S. and Kay,S.A. (1998) Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science, 280, 1599–1603. [DOI] [PubMed] [Google Scholar]
- Doolittle W.F. (1979) The cyanobacterial genome, its expression and the control of that expression. Adv. Microb. Physiol., 20, 1–102. [DOI] [PubMed] [Google Scholar]
- Dunlap J.C. (1999) Molecular bases for circadian clocks. Cell, 96, 271–290. [DOI] [PubMed] [Google Scholar]
- Edery I., Rutila,J.E. and Rosbash,M. (1994) Phase shifting of the circadian clock by induction of the Drosophila period protein. Science, 263, 237–240. [DOI] [PubMed] [Google Scholar]
- Griffin E.A. Jr, Staknis,D. and Weitz,C.J. (1999) Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science, 286, 765–768. [DOI] [PubMed] [Google Scholar]
- Hao H., Allen,D.L. and Hardin,P.E. (1997) A circadian enhancer mediates PER-dependent mRNA cycling in Drosophila melanogaster. Mol. Cell. Biol., 17, 3687–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin P.E., Hall,J.C. and Rosbash,M. (1990) Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature, 343, 536–540. [DOI] [PubMed] [Google Scholar]
- Harlow E. and Lane,D. (1999) Using Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Hastings M.H., Field,M.D., Maywood,E.S., Weaver,D.R. and Reppert,S.M. (1999) Differential regulation of mPER1 and mTIM proteins in the mouse suprachiasmatic nuclei: new insights into a core clock mechanism. J. Neurosci., 19, RC11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiura M., Kutsuna,S., Aoki,S., Iwasaki,H., Andersson,C.R., Tanabe,A., Golden,S.S., Johnson,C.H. and Kondo,T. (1998) Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science, 281, 1519–1523. [DOI] [PubMed] [Google Scholar]
- Iwasaki H., Taniguchi,Y., Ishiura,M. and Kondo,T. (1999) Physical interactions among circadian clock proteins KaiA, KaiB and KaiC in cyanobacteria. EMBO J., 18, 1137–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H., Williams,S.B., Kitayama,Y., Ishiura,M., Golden,S.S. and Kondo,T. (2000) A KaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell, 101, 223–233. [DOI] [PubMed] [Google Scholar]
- Johnson C.H. (1999) Forty years of PRCs—what have we learned? Chronobiol. Int., 16, 711–743. [DOI] [PubMed] [Google Scholar]
- Johnson C.H. and Golden,S.S. (1999) Circadian programs in cyanobacteria: adaptiveness and mechanism. Annu. Rev. Microbiol., 53, 389–409. [DOI] [PubMed] [Google Scholar]
- Johnson C.H., Knight,M., Trewavas,A. and Kondo,T. (1998) A clockwork green: circadian programs in photosynthetic organisms. In Lumsden,P. and Millar,A. (eds), Biological Rhythms and Photoperiodism in Plants. BIOS Scientific, Oxford, UK, pp. 1–34. [Google Scholar]
- Kondo T., Strayer,C.A., Kulkarni,R.D., Taylor,W.R., Ishiura,M., Golden,S.S. and Johnson,C.H. (1993) Circadian rhythms in prokaryotes: luciferase as a reporter of circadian gene expression in cyanobacteria. Proc. Natl Acad. Sci. USA, 90, 5672–5676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumano M., Tomioka,N., Shinozaki,K. and Sugiura,M. (1986) Analysis of the promoter region in the rrnA operon from a blue-green alga, Anacystis nidulans 6301. Mol. Gen. Genet., 202, 173–178. [Google Scholar]
- Kume K., Zylka,M., Sriram,S., Shearman,L.P., Weaver,D.R., Jin,X., Maywood,E.S., Hastings,M.H. and Reppert,S.M. (1999) mCRY1 and mCRY2 are essential components of the negative limb of the circadian feedback loop. Cell, 98, 193–205. [DOI] [PubMed] [Google Scholar]
- Leipe D.D., Aravind,L., Grishin,N.V. and Koonin,E.V. (2000) The bacterial replicative helicase DnaB evolved from a RecA duplication. Genome Res., 10, 5–16. [PubMed] [Google Scholar]
- Liu Y., Tsinoremas,N.F., Johnson,C.H., Lebedeva,N.V., Golden,S.S., Ishiura,M. and Kondo,T. (1995) Circadian orchestration of gene expression in cyanobacteria. Genes Dev., 9, 1469–1478. [DOI] [PubMed] [Google Scholar]
- Liu Y., Loros,J. and Dunlap,J.C. (2000) Phosphorylation of the Neurospora clock protein FREQUENCY determines its degradation rate and strongly influences the period length of the circadian clock. Proc. Natl Acad. Sci. USA, 97, 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry O.H., Rosebrough,N.J., Farr,A.L. and Randall,R.J. (1951) Protein measurement with the Folin phenol reagent. J. Biol. Chem., 193, 265–275. [PubMed] [Google Scholar]
- Merrow M.W., Garceau,N.Y. and Dunlap,J.C. (1997) Dissection of a circadian oscillation into discrete domains. Proc. Natl Acad. Sci. USA, 94, 3877–3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T., Binder,B. and Johnson,C.H. (1996) Circadian gating of cell division in cyanobacteria growing with average doubling times of less than 24 h. Proc. Natl Acad. Sci. USA, 93, 10183–10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiwaki T., Iwasaki,H., Ishiura,M. and Kondo,T. (2000) Nucleotide binding and autophosphorylation of the clock protein KaiC as a circadian timing process of cyanobacteria. Proc. Natl Acad. Sci. USA, 97, 495–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Y., Andersson,C.R., Kondo,T., Golden,S.S. and Johnson,C.H. (1998) Resonating circadian clocks enhance fitness in cyanobacteria. Proc. Natl Acad. Sci. USA, 95, 8660–8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J.L., Blau,J., Rothenfluh,A., Abodeely,M., Kloss,B. and Young,M.W. (1998) Double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell, 94, 83–95. [DOI] [PubMed] [Google Scholar]
- Rosbash M. and Hall,J.C. (1989) The molecular biology of circadian rhythms. Neuron, 3, 387–398. [DOI] [PubMed] [Google Scholar]
- Schaffer R., Ramsay,N., Samach,A., Corden,S., Putterill,J., Carré,I.A. and Coupland,G. (1998) LATE ELONGATED HYPOCOTYL, an Arabidopsis gene encoding a MYB transcription factor, regulates circadian rhythmicity and photoperiodic responses. Cell, 93, 1219–1229. [DOI] [PubMed] [Google Scholar]
- Schneegurt M.A., Sherman,D.M. and Sherman,L.A. (1997) Growth, physiology and ultrastructure of a diazotrophic cyanobacterium, Cyanothece sp. strain ATCC 51142, in mixotrophic and chemoheterotrophic cultures. J. Phycol., 33, 632–642. [Google Scholar]
- Sehgal A., Rothenfluh-Hilfiker,A., Hunter-Ensor,M., Chen,Y., Myers,M.P. and Young,M.W. (1995) Rhythmic expression of timeless: a basis for promoting circadian cycles in period gene autoregulation. Science, 270, 808–810. [DOI] [PubMed] [Google Scholar]
- Sun Z.S., Albrecht,U., Zhuchenko,O., Bailey,J., Eichele,G. and Lee,C.C. (1997) Rigui [mper1], a putative mammalian ortholog of the Drosophila period gene. Cell, 90, 1003–1011. [DOI] [PubMed] [Google Scholar]
- Tei H., Okamura,H., Shigeyoshi,Y., Fukuhara,C., Ozawa,R., Hirose,M. and Sakaki,Y. (1997) Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature, 389, 512–516. [DOI] [PubMed] [Google Scholar]
- Tomioka N., Shinozaki,K. and Sugiura,M. (1981) Molecular cloning and characterization of ribosomal RNA genes from a blue-green alga, Anacystis nidulans.Mol. Gen. Genet., 184, 359–363. [DOI] [PubMed] [Google Scholar]
- Wang Z.-Y. and Tobin,E.M. (1998) Constitutive expression of the circadian clock associated 1 (CCA-1) gene disrupts circadian rhythms and suppresses its own expression. Cell, 93, 1207–1217. [DOI] [PubMed] [Google Scholar]
- Xu Y., Piston,D.W. and Johnson,C.H. (1999) A bioluminescence resonance energy transfer (BRET) system: application to interacting circadian clock proteins. Proc. Natl Acad. Sci. USA, 96, 151–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.W. (1998) The molecular control of circadian behavioral rhythms and their entrainment in Drosophila. Annu. Rev. Biochem., 67, 135–152. [DOI] [PubMed] [Google Scholar]
- Zylka M., Shearman,L., Weaver,D. and Reppert,S. (1998) Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside the brain. Neuron, 20, 1103–1110. [DOI] [PubMed] [Google Scholar]