Abstract
The cluster of differentiation 44 (CD44) signaling pathway is crucial in cancer-cell growth, invasion, proliferation and metastasis. CD44 is a transmembrane receptor for hyaluronan and osteopontin, and has recently attracted attention as a gastric cancer stem cell marker. Previous studies showed that polymorphisms in the CD44 gene can influence both human cancer survival and determine cellular response to cytotoxic chemotherapeutics. In addition, CD44 protein overexpression has been associated with poor prognosis in gastric adenocarcinoma (GA). We tested the hypothesis whether polymorphisms involved in the CD44 pathway will predict clinical outcome in patients with localized GA. Either blood or formalin-fixed paraffin-embedded (FFPE) tissues were obtained from 137 patients with localized GA at University of Southern California and Memorial Sloan-Kettering Cancer Center medical facilities. DNA was isolated and polymorphisms within the CD44 pathway were determined by PCR-RFLP technique. In univariate analysis CD44 rs187116 and CD44 rs7116432 were significantly associated with time to tumor recurrence (TTR) and overall survival (OS). After adjusting for covariates, patients harboring at least one G allele of CD44 rs187116 remained significantly associated with TTR (adjusted p=0.009) and OS (adjusted p=0.045). Further, patients harboring CD44 T-A haplotype were at the lowest risk of developing tumor recurrence (HR: 0.255; 95% CI: 0.11–0.591; adjusted p=0.001) and death (HR 0.198; 95% CI: 0.07–0.563; adjusted p=0.002). These results provide the first evidence that CD44 polymorphisms predict clinical outcome in patients with localized GA. This may help to identify localized GA patients at high risk for tumor recurrence.
Keywords: CD44, gastric adenocarcinoma, outcome, polymorphisms
Introduction
Worldwide, approximately 934,000 cases of GA are diagnosed annually 1. In 2010, an estimated 21,000 new cases of GA will be diagnosed in the United States, and 10,570 patients will succumb to their disease 2. Despite recent improvements in treatment options through the integration of targeted therapy in combination with cytotoxic chemotherapy for patients with HER2 positive advanced GA, the median overall survival (OS) is less than 14 months 3. The mortality for patients diagnosed with localized GA is still high, with five-year OS rates of 30% 4. In addition, of those patients who are fortunate enough to undergo standard combined radio-chemotherapy following complete gastric resection, an alarmingly high rate of 40–60% will develop recurrence 5. GA is a heterogeneous disease with two distinct histological subtypes (diffuse and intestinal type, classified by Laurén 6) which are characterized by different molecular alterations 7. However, GA is commonly treated uniformly, independent of histologic and molecular subtypes. Pathologic differentiation, depth of tumor invasion (T stage), and number of tumor infiltrated lymph nodes (N stage)represent the only prognostic markers. Therefore, it is critical to establish molecular markers that predict outcome to individualize therapeutic strategies by maximizing treatment efficacy and minimizing side effects.
CD44 is a major cell adhesion molecule playing distinct roles in a variety of physiological processes including cellular adhesion, migration and regulation of growth and lymphocyte homing 8. The CD44 gene is complex, comprising of 20 exons, 10 of which are expressed in most untransformed non-activated cells as standard CD44 (CD44s). The 10 remaining exons are incorporated in an extremely large number of CD44 family splice variants (CD44v) containing inserts of varying sizes in the extracellular portion of the molecule. These CD44v are found on the surface of tumor cells, dividing epithelial cells, and activated lymphocytes and possess distinct functional significance from CD44s 8. The major ligands of CD44 are hyaluronan and osteopontin 8, 9. CD44 signaling is crucial in cancer-cell growth, invasion, proliferation and metastasis. Activation of CD44 as a result of ligand binding, promotes proliferation and apoptosis resistance via phosphatidylinositol 3-kinase (PI3K)/Akt pathway 10. In addition, CD44 promotes tumor cell proliferation in co-operation with additional membrane-bound receptor tyrosine kinases including c-erbB-2 (HER2/neu) and c-Src 11. Another critical function of CD44 is the regulation of tumor cell adhesion and motility. In it’s active state, the cytoplasmic domain of CD44 can interact with the actin-based cytoskeletons via ezrin, radixin, and moesin (ERM family), thereby promoting membrane motility and tumor cell migration 12–14. Notably, CD44 has well documented tumor promoting activities that include stimulating cell growth and promoting metastasis in colorectal and GA 15, 16. Aberrant expression of CD44 and CD44v has also been reported to be associated with lymph node metastasis, invasion and survival in GA 17, 18. In addition CD44 positive gastric cancer cells were associated with chemo- (5-Fluorouracil and etoposide) and radiotherapy resistance which likely account, at least in part, for treatment resistance and subsequently early tumor recurrence 19. Consistent with this, genetic variants in the CD44 gene have been identified to affect both the cellular responses to chemotherapeutics 20, 21, as well as human cancer incidence and survival 22. A striking development in recent years is the emergence of CD44 as gastric cancer stem cell (CSC) marker. CD44 positive human gastric cancer cell lines have been reported to possess the capacity for self-renewal, longevity and multipotency 19
Considering the expanding body of evidence implicating the role of CD44 in promoting a variety of tumorigenic processes and the complexity of the CD44 gene and its splicing events, it is entirely plausible that the CD44 gene and signaling pathway could harbour functional genetic variants which may help further define GA sub-populations at high risk for early tumor recurrence. Therefore, we aimed to examine whether polymorphisms within the CD44 signaling pathway have potential signficance as molecular prognostic markers for localized GA.
Patients and Methods
Patients
A total of 137 patients with localized (stage Ib – IV) GA were included in this study. All patients were treated with surgery alone or surgery and adjuvant (radio)-chemotherapy, at the University of Southern California/Norris Comprehensive Cancer Center (USC/NCCC), the Los Angeles County/University of Southern California Medical Center, or the Memorial Sloan-Kettering Cancer Center/Cornell University from 1992 to 2008. Patient data were collected retrospectively through chart review. Study approval was obtained by the Institutional Review Boards of the University of Southern California and Memorial Sloan-Kettering Cancer Center. All participants signed informed consent for the analysis of molecular correlates.
Genotyping
Either blood or formalin-fixed paraffin-embedded (FFPE) normal gastric tissue specimens were obtained and genomic DNA was extracted using the QIAamp extraction kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol. The samples were tested either by PCR-based restriction fragment length polymorphism (PCR-RFLP) analysis or direct sequencing. The genes, reference SNP identification numbers, SNP location, function, forward and reverse primer and restriction enzymes are summarized in Table 1.
Table 1.
Analyzed polymorphisms within the CD44 signaling pathway and their functional significance, Primer Sequences, and Restriction Enzymes.
| Genes (rs number) |
Location of Polymorphisms |
Function of Polymorphisms |
Forward-Primer (5´-3´) | Reverse-Primer (5´-3´) | Enzyme |
|---|---|---|---|---|---|
|
CD44 (rs8193) |
3´UTR *438C>T |
T allele is associated with increased risk of adverse skin reactions after radiotherapie in breast cancer patients 40. |
CAGGGTTAATAGGGCCTGGT | GAAAAATTTCTAGAGGGGGTCTG | BsrDI |
|
CD44 (rs187115) |
Intron 1 +15242T>C |
C/C genotype is associated with increased risk for tumor- related death and lower drug sensitivity 21. |
CTCTGTCTCTCCTGCCCAAT | GCTAATTCAAATGCTTGGTTG | n.a* |
|
CD44 (rs187116) |
Intron 1 +4883G>A |
G allele is associated with increased risk of adverse skin reactions after radiotherapie in breast cancer patients 40. |
AGGTGGTTGGAGATCACCTG | CTTTCGCAAGAACCACTTCC | MspI |
| CD44 (rs4755392) | 3´UTR 98670T>A |
n.a. | TGGGTAATTTAGAGGAACAAAGTCA | ACACATCACTCATAGAAAACCAGA | n.a* |
|
CD44 (rs7116432) |
Exon 19 +779A>G |
G/G genotype is associated with Carboplatin sensitivity 20. |
CATCGTCTTCTTGCTGTTAGGA | GGTCTTGGTTCAGGTAGGGAGA | NlaIII |
|
OPN (rs1126616) |
Exon 8 708C>T |
T allele associated with higher risk for systemic lupus erythemtosus 41. |
TGAAACGAGTCAGCTGGATG | CTGTGGAATTCACGGCTGA | n.a* |
|
OPN (rs9138) |
Exon 8 *294A>C |
C allele associated with higher risk for systemic lupus erythemtosus 41. |
GAAGAAATGCAAACTATCACTGTATT | CCACAAAAAGATAATCACAACAAAA | Hpy166II |
|
HAS2 (rs4123220) |
5´UTR -173A>T |
n.a | CAAGGCTGGGTAGAGTTCGT | TTTCCTCAACAGTGCCACCT | n.a* |
|
HAS2 (rs1057308) |
Exon 1 .-155T>C |
n.a | AATAAGAGATCAGATGAATTTGAGACG | GCAACGGAAACATAAAGAGAA | n.a* |
direct sequencing
Abbreviations: CD44, cluster of differentiation 44; OPN, Osteopontin; HAS2, hyaluronan synthase 2; UTR, untranlsated region; n.a., not available;
Candidate polymorphisms
Common and potentially functional polymorphisms within the CD44 signaling pathway were selected by using the HapMap Project database (www.hapmap.org). We used the following criteria to select the candidate gene polymorphisms: (a) a minor allele frequency (MAF) ≥10% in Caucasians; (b) located in the 3´UTR, 5´UTR and coding regions of the tested genes and/or were shown to be of biological significance according to the location within the gene or according to literature review; (c) were associated with resistance to chemotherapeutic agents in literature review.
Statistical analysis
The primary endpoints of the analyses of germline polymorphisms within the CD44 signaling pathway in localized GA patients treated with surgery alone or surgery and adjuvant (radio)-chemotherapy were time to recurrence (TTR) and overall survival (OS). The TTR was calculated from the date of diagnosis of the disease to the date of first observation of tumor recurrence or until last follow-up if the patient was recurrence-free at that time. The OS was defined as the period from diagnosis to death from any cause or the last contact if the patient was alive.
The distributions of polymorphisms across baseline demographic, clinical and pathological characteristics were examined using Fisher’s exact test. The adjusted curves for TTR and OS of CD44 haplotypes were computed based on the multivariable Cox model 23. While the genetic model of inheritance for these polymorphisms was unknown, we considered the dominant, recessive, co-dominant, or additive model whenever appropriate.
Allelic distribution of all polymorphisms in each race/ethnic group was tested for deviation from Hardy-Weinberg equilibrium (HWE) using a chi-square test with 1 degree of freedom. Linkage disequilibrium among polymorphisms in CD44 was assessed using D’ and r2 values, and the haplotype frequencies were inferred using HaploView version 4.1 (www.broad.mit.edu/mpg/haploview).
The Cox proportional hazards regression model including T and N categories as covariates and race and type of adjuvant chemotherapy as stratum variables was fitted to re-evaluate the association between CD44 polymorphisms and TTR and OS considering the imbalances in the distributions of baseline characteristics.
All statistical tests were 2-sided and performed using the SAS statistical package version 9.2 (SAS Institute Inc. Cary, North Carolina, USA).
Results
A total of 137 patients with GA were enrolled in the study, and the median follow up time was 3.3 years. Of those patients, 61 (45%) had tumor recurrence, with a probability of 3-year recurrence of 0.52 ± 0.05. Fifty-five of 137 (40%) patients had recurrent disease within the first 3 years after surgery and the median time to recurrence (TTR) was 2.8 years (95% CI, 2.1–7.0 years). Of the 137 patients, forty-five (33%) have died and the median overall survival of the cohort is 4.7 years (95% CI, 3.8–7.3 years). T category (p=0.013), N-category (p=0.004), and type of chemotherapy (p=0.003) were significantly associated with TTR. The characteristics of the patients have been described in detail and are summarized in Table 2. No statistical significant association between tested genetic variations and Lauren classification was observed (p>0.05). The allelic frequencies observed were within the probability limits of HWE (p>0.05, exact test for HWE).
Table 2.
Baseline demographic and clinical characteristic and clinical outcome in patients with localized GA.
| Time to recurrence | Overall Survival | ||||||
|---|---|---|---|---|---|---|---|
| N | Median time to recurrence, yrs (95% CI) |
Relative risk (95% CI) |
P value* | Median overall survival, yrs (95% CI) |
Relative risk (95% CI) |
P value* | |
| Age, years | 0.42 | 0.65 | |||||
| < 60 | 80 | 2.2 (1.5, 14.5+) | 1 | 4.7 (3.8, 14.6+) | 1 | ||
| ≥ 60 | 57 | 3.7 (2.1, 12.3+) | 0.81 (0.48, 1.36) | 4.5 (3.3, 7.3) | 1.14 (0.64, 2.05) | ||
| Sex | 0.85 | 0.32 | |||||
| Male | 83 | 2.3 (1.8, 7.0) | 1 | 4.1 (3.3, 7.3) | 1 | ||
| Female | 54 | 7.0 (1.5, 8.3+) | 0.95 (0.56, 1.63) | 7.3 (3.8, 8.3+) | 0.72 (0.37, 1.39) | ||
| Race | 0.085 | 0.040 | |||||
| White | 63 | 1.7 (1.2, 4.4) | 1 | 3.8 (2.7, 5.5) | 1 | ||
| African American | 1 | 0.5+ | — | 0.5+ | — | ||
| Asian | 28 | 7.0 (2.3, 14.5+) | 0.45 (0.23, 0.91) | 7.3 (3.3, 14.6+) | 0.45 (0.20, 1.03) | ||
| Hispanic | 45 | 3.7 (2.1, 10.7+) | 0.63 (0.34, 1.17) | 10.7+ (3.6, 10.7+) | 0.36 (0.15, 0.85) | ||
| Stage | 0.030 | ||||||
| I | 12 | 4.3+ (2.2, 4.3+) | 1 | 4.4+ | — | 0.32 | |
| II | 36 | 7.0 (2.9, 10.7+) | 1.56 (0.35, 6.98) | 5.4 (4.1, 10.7+) | 1 | ||
| III | 71 | 1.8 (1.4, 2.8) | 3.24 (0.78, 13.5) | 3.8 (2.8, 7.3) | 1.31 (0.69, 2.50) | ||
| IV | 18 | 1.6 (1.2, 3.8+) | 4.00 (0.86, 18.5) | 7.3+ (1.4, 7.3+) | 1.33 (0.43, 4.09) | ||
| Tumor stage | 0.013 | 0.30 | |||||
| T1 † | 4 | ||||||
| T2 † | 44 | 8.3+ (2.9, 8.3+) | 1 | 5.4 (4.1, 8.3+) | 1 | ||
| T3 ‡ | 79 | 1.7 (1.4, 4.4) | 2.04 (1.14, 3.67) | 4.5 (3.3, 7.3) | 1.40 (0.73, 2.68) | ||
| T4 ‡ | 10 | ||||||
| N | 0.004 | 0.088 | |||||
| Negative | 27 | 7.0 (1.8, 10.7+) | 1 | 7.3 (3.4, 10.7+) | 1 | ||
| N1 | 64 | 4.4 (2.2, 14.5+) | 0.99 (0.47, 2.11) | 5.5 (4.1, 14.6+) | 1.07 (0.46, 2.47) | ||
| N2 | 31 | 1.3 (1.1, 2.3) | 2.62 (1.15, 5.94) | 3.3 (2.0, 5.7+) | 2.27 (0.85, 6.07) | ||
| N3 | 15 | 1.6 (1.0, 3.8+) | 1.96 (0.73, 5.32) | 2.4 (1.1, 3.8+) | 2.35 (0.66, 8.41) | ||
| ECOG performance status | 0.77 | 0.25 | |||||
| 0 | 62 | 2.5 (1.7, 10.7+) | 1 | 4.5 (2.8, 10.7+) | 1 | ||
| 1 | 65 | 2.3 (1.6, 14.5+) | 1.00 (0.60, 1.68) | 5.7 (3.8, 14.6+) | 0.68 (0.37, 1.25) | ||
| 2 | 10 | 2.2+ (1.7, 2.2+) | 0.60 (0.14, 2.53) | 2.2+ (1.2, 2.2+) | 1.67 (0.36, 7.75) | ||
| Differentiation | 0.15 | 0.098 | |||||
| Moderate | 27 | 2.1 (1.1, 3.8+) | 1 | 4.1 (2.7, 4.7) | 1 | ||
| Poor/Moderate ‡ | 10 | 3.7 (2.1, 14.5+) | 0.65 (0.36, 1.19) | 7.3 (3.8, 14.6+) | 0.59 (0.31, 1.13) | ||
| Poor ‡ | 97 | ||||||
| Lauren | 0.87 | 0.74 | |||||
| Diffuse | 40 | 3.7 (1.8, 8.9+) | 1 | 7.3 (5.4, 8.9+) | 1 | ||
| Intestinal | 50 | 7.0 (2.1, 14.5+) | 0.87 (0.45, 1.67) | 5.7 (3.8, 14.6+) | 1.10 (0.48, 2.51) | ||
| Mixed | 21 | 12.3+ (1.7, 12.3+) | 1.04 (0.45, 2.41) | 3.6 (1.9, 12.3+) | 1.52 (0.51, 4.58) | ||
| Type of chemotherapy | 0.003 | 0.004 | |||||
| 5-FU/LV | 70 | 7.0 (2.8, 10.6+) | 1 | 7.3 (4.1, 10.6+) | 1 | ||
| 5-FU/LV/oxaliplatin | 19 | 1.6 (1.1, 2.9) | 2.66 (1.23, 5.76) | 2.4 (1.2, 4.5+) | 4.24 (1.69, 10.6) | ||
| 5-FU, cisplatin, CPT-11 | 25 | 1.7 (1.2, 14.5+) | 1.46 (0.71, 3.01) | 4.1 (2.2, 14.6+) | 1.36 (0.59, 3.12) | ||
| None | 23 | 2.1 (0.8, 2.5) | 2.80 (1.48, 5.27) | 4.5 (2.8, 5.5) | 2.27 (1.09, 4.74) | ||
| Radiation | 0.92 | 0.68 | |||||
| Yes | 88 | 2.5 (1.8, 14.5+) | 1 | 4.5 (3.3, 14.6+) | 1 | ||
| No | 48 | 3.7 (1.7, 12.3+) | 1.03 (0.60, 1.76) | 5.4 (3.8, 12.3+) | 0.89 (0.48, 1.63) | ||
Based on log-rank test
Estimates were not reached.
No events occurred and estimates were not obtained.
Grouped together for the estimates of relative risk
Abbreviations: CI, confidence interval; ECOG, Eastern Cooperative Oncology Group; yrs, years; 5-FU, 5-fluorouracil; LV, leucovorin; CPT11, Irinotecan; —, no events occurred and estimates were not obtained.
CD44 rs187116 and clinical outcome in localized GA
Genotyping for CD44 rs187116 was successful in 124 (91%) of 137 cases. In 13 patients (9%), genotyping was not successful, because of limited quantity and quality of extracted genomic DNA. Patients harboring at least one G allele (A/G or G/G genotype) had a median TTR of 2.1 years (95%CI, 1.5–2.5 years) compared with 7.0 years (95% CI, 4.4–10.6+ years) in patients homozygous for A allele (p=0.022, log-rank test). In addition, patients with the CD44 rs187116 G allele reached borderline significance with median overall survival of 4.1 years (95% CI, 3.3–5.4 years) compared to 7.3 years (95% CI, 3.8–10.6+ years) for patients homozygous for A allele (p=0.079, log-rank test; Table 3)
Table 3.
CD44 signaling pathway polymorphisms and TTR and OS in patients with localized GA.
| Time to recurrence | Overall survival | ||||||
|---|---|---|---|---|---|---|---|
| N | Median time to recurrence, yrs (95% CI) |
Hazard Ratio (95% CI) | P value * | Median overall survival, yrs (95% CI) |
Hazard Ratio (95% CI) | P value * | |
| CD44 rs8193 | 0.95 | 0.39 | |||||
| C/C | 45 | 2.2 (1.2, 10.7+) | 1 (Reference) | 4.1 (2.0, 10.7+) | 1 (Reference) | ||
| C/T | 62 | 2.3 (1.7, 7.0) | 0.97 (0.54, 1.76) | 5.5 (3.8, 12.3+) | 0.65 (0.32, 1.31) | ||
| T/T | 21 | 2.9 (1.7, 7.0+) | 0.88 (0.39, 1.95) | 4.5 (2.7, 5.7) | 0.95 (0.40, 2.25) | ||
| C/T, T/T | 83 | 2.3 (1.7, 7.0) | 0.95 (0.54, 1.66) | 0.85 | 4.7 (3.8, 7.3) | 0.72 (0.37, 1.39) | 0.32 |
| CD44 rs187115 | 0.79 | 0.75 | |||||
| T/T | 58 | 2.5 (1.7, 12.3+) | 1 (Reference) | 4.1 (3.3, 12.3+) | 1 (Reference) | ||
| C/T | 52 | 2.2 (1.7, 7.0) | 1.21 (0.68, 2.15) | 5.5 (3.4, 7.3) | 1.04 (0.53, 2.01) | ||
| C/C | 15 | 7.0+ (1.1, 7.0+) | 1.01 (0.41, 2.50) | 3.8 (1.2, 7.0+) | 1.41 (0.55, 3.60) | ||
| CD44 rs187116 | 0.033 | 0.096 | |||||
| A/A | 30 | 7.0 (4.4, 10.6+) | 1 (Reference) | 7.3 (3.8, 10.6+) | 1 (Reference) | ||
| A/G | 56 | 2.2 (1.5, 7.0) | 2.07 (0.95, 4.51) | 4.5 (3.3, 7.3) | 1.78 (0.76, 4.16) | ||
| G/G | 38 | 1.7 (1.1, 2.3) | 2.90 (1.25, 6.72) | 2.8 (2.4, 4.5) | 2.65 (1.05, 6.70) | ||
| A/G,G/G | 94 | 2.1 (1.5, 2.5) | 2.32 (1.10, 4.91) | 0.022 | 4.1 (3.3, 5.4) | 2.02 (0.90, 4.57) | 0.079 |
| CD44 rs4755392 | 0.59 | 0.77 | |||||
| T/T | 43 | 3.2 (1.2, 5.9+) | 1 (Reference) | 3.8 (2.8, 7.3+) | 1 (Reference) | ||
| A/T | 53 | 2.1 (1.6, 14.5+) | 1.05 (0.56, 1.99) | 4.1 (3.6, 14.6+) | 0.76 (0.35, 1.66) | ||
| A/A | 34 | 3.7 (2.1, 7.0) | 0.76 (0.37, 1.57) | 5.4 (2.8, 7.3) | 0.80 (0.36, 1.81) | ||
| A/T,A/A | 87 | 2.3 (1.8, 7.0) | 0.93 (0.51, 1.69) | 0.80 | 4.7 (3.8, 7.3) | 0.78 (0.38, 1.60) | 0.48 |
| CD44 rs7116432 | 0.10 | 0.024 | |||||
| G/G | 36 | 7.0 (2.1, 12.3+) | 1 (Reference) | 7.3 (5.4, 12.3+) | 1 (Reference) | ||
| A/G | 60 | 2.3 (1.3, 7.0) | 1.77 (0.90, 3.47) | 4.1 (3.3, 7.3) | 2.16 (0.97, 4.83) | ||
| A/A | 31 | 1.7 (1.0, 5.7+) | 2.19 (1.02, 4.71) | 3.8 (2.0, 5.7+) | 3.32 (1.32, 8.40) | ||
| A/G,A/A | 91 | 2.2 (1.4, 4.4) | 1.89 (1.00, 3.60) | 0.045 | 3.8 (2.8, 4.7) | 2.44 (1.13, 5.27) | 0.018 |
| OPN rs1126616 | 0.79 | 0.80 | |||||
| C/C | 59 | 2.2 (1.2, 12.3+) | 1 (Reference) | 4.1 (3.3, 12.3+) | 1 (Reference) | ||
| C/T | 42 | 2.3 (1.7, 7.0) | 0.96 (0.53, 1.75) | 4.7 (2.8, 7.3) | 0.81 (0.41, 1.62) | ||
| T/T | 24 | 7.0 (1.8, 7.0+) | 0.78 (0.37, 1.63) | 4.5 (3.8, 7.3+) | 0.81 (0.36, 1.86) | ||
| OPN rs9138 | 0.70 | 0.94 | |||||
| A/A | 61 | 2.2 (1.3, 12.3+) | 1 (Reference) | 5.5 (2.5, 12.3+) | 1 (Reference) | ||
| A/C | 43 | 2.3 (1.7, 7.0) | 0.99 (0.55, 1.77) | 4.7 (2.8, 7.3) | 0.92 (0.47, 1.82) | ||
| C/C | 27 | 7.0 (1.8, 7.0+) | 0.75 (0.37, 1.52) | 4.5 (3.8, 7.3+) | 0.88 (0.40, 1.94) | ||
| HAS2 rs4123220 | 0.38 | 0.25 | |||||
| A/A | 60 | 1.7 (1.2, 7.0) | 1 (Reference) | 4.1 (2.8, 5.7) | 1 (Reference) | ||
| A/T | 49 | 2.9 (1.8, 12.3+) | 0.68 (0.38, 1.21) | 4.5 (2.8, 12.3+) | 0.98 (0.52, 1.85) | ||
| T/T | 18 | 2.3 (1.5, 5.9+) | 0.72 (0.33, 1.59) | 7.3+ (2.8, 7.3+) | 0.38 (0.11, 1.28) | ||
| HAS2 rs1057308 | 0.29 | 0.17 | |||||
| A/A | 60 | 1.7 (1.5, 7.0) | 1 (Reference) | 4.1 (2.8, 5.7) | 1 (Reference) | ||
| A/G | 52 | 2.9 (1.8, 12.3+) | 0.66 (0.37, 1.16) | 4.5 (3.3, 12.3+) | 0.83 (0.45, 1.55) | ||
| G/G | 19 | 5.9+ (1.5, 5.9+) | 0.68 (0.31, 1.49) | 7.3+ (3.8, 7.3+) | 0.33 (0.10, 1.11) | ||
Based on the log-rank test
Abbreviations: CD44, cluster of differentiation 44; OPN, Osteopontin; HAS2, hyaluronan synthase 2
CD44 rs7116432 and clinical outcome in localized GA
Genotyping for CD44 rs7116432 was successful in 127 (93%) of 137 cases. In 10 patients (7%), genotyping was not successful, because of limited quantity and quality of extracted genomic DNA. Patients with A/G or A/A genotype had a median TTR of 2.2 years (95%CI, 1.4–4.4 years) compared with 7 years (95% CI, 2.1–12.3+) for G/G genotype (p=0.045, log-rank test) and a median OS of 3.8 years (95% CI, 2.8–4.7 years) versus 7.3 years (95% CI, 5.4–12.3+ years; p=0.018, log-rank test), respectively (Table 3).
Multivariable analysis of CD44 rs187116 and CD44 rs7116432
Multivariable analysis for CD44 rs187116 and CD44 rs7116432 included T category, N category as covariates and was stratified by race and type of chemotherapy. CD44 rs187116 remained significantly associated with TTR (HR: 3.59, 95% CI, 1.37–9.40; adjusted p=0.009) and OS (HR: 2.98, 95% CI, 1.02–8.68; adjusted p=0.045, Table 4).
Table 4.
Multivariate and haplotype analysis of CD44 polymorphisms and TTR and OS in patients with localized GA.
| Time to recurrence | Overall survival | ||||
|---|---|---|---|---|---|
| N* | Hazard Ratio (95% CI) † | P value † | Hazard Ratio (95% CI) † | P value † | |
| CD44 rs187116 | |||||
| A/A | 30 | 1 (Reference) | 1 (Reference) | ||
| A/G,G/G | 92 | 3.59 (1.37, 9.40) | 0.009 | 2.98 (1.02, 8.68) | 0.045 |
| CD44 rs7116432 | |||||
| G/G | 34 | 1 (Reference) | 1 (Reference) | ||
| A/G,A/A | 88 | 1.24 (0.56, 2.76) | 0.60 | 2.34 (0.83, 6.59) | 0.11 |
| Haplotype analysis of CD44 rs187115 and rs187116 |
Haplotype Frequency |
||||
| T-G | 0.505 | 1 (Reference) | |||
| C-A | 0.298 | 0.639 (0.379, 1.077) | 0.093 | 0.887 (0.477, 1.647) | 0.70 |
| T-A | 0.171 | 0.255 (0.110, 0.591) | 0.001 | 0.198 (0.070, 0.563) | 0.002 |
Patients with incomplete CD44 rs187116 or CD44 rs7116432 were excluded in the multivariate analysis
Wald test in Cox Proportional hazards model including T category, N category as covariates and stratified by race and type of chemotherapy
Haplotype Analysis
CD44 rs187115 and CD44 rs187116 polymorphisms were in linkage disequilibrium in our study population (D’=0.85, 95% CI, 0.71–0.93; r2=0.39). Haplotypes were constructed from these two polymorphisms. Patients harboring the T-A haplotype were at lowest risk to develop tumor recurrence (HR: 0.255; 95% CI, 0.11–0.591) compared with patients with the most prevalent T-G haplotype (reference; adjusted p=0.001). In addition, patients with T-A haplotype were at lowest risk of death (HR 0.198; 95% CI, 0.07–0.563) compared with patients harboring T-G haplotype (reference; adjusted p-value=0.002; Table 4 and Figure 1A and 1B). The other polymorphisms tested did not show any linkage disequilibrium.
Figure 1.
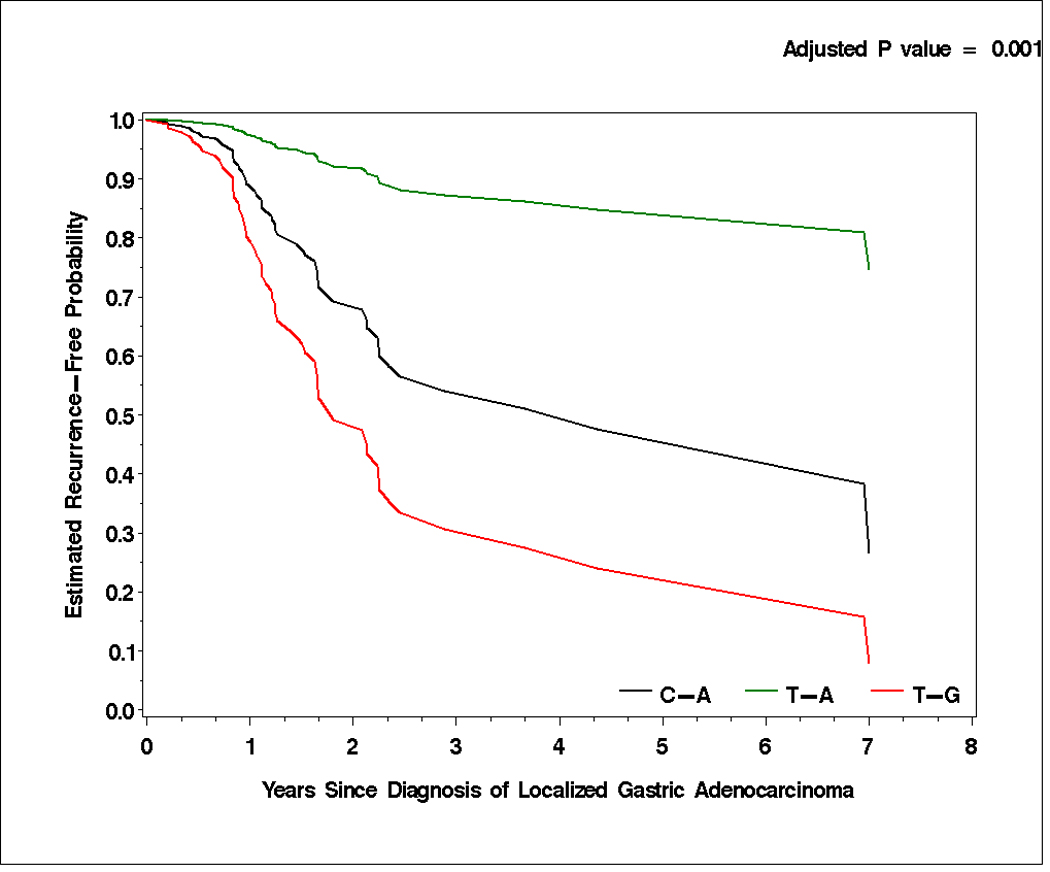
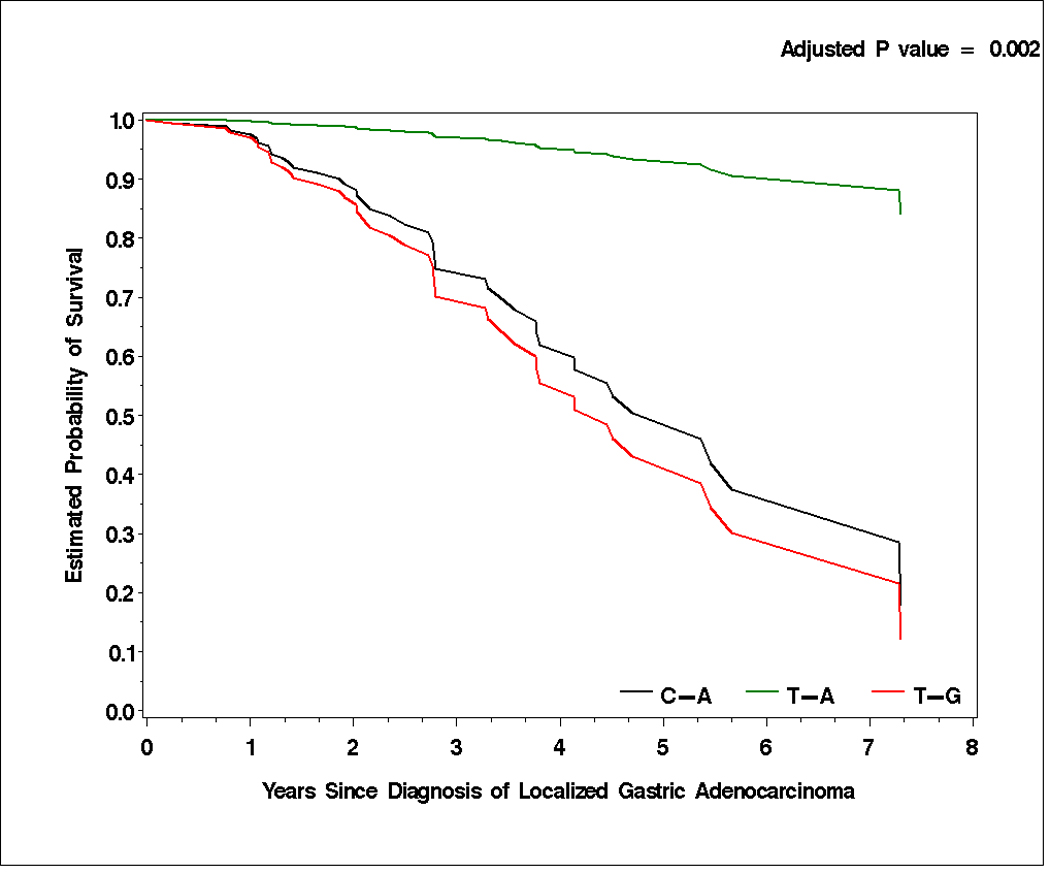
Time to tumor recurrence (A) and overall survival (B) by T-A, C-A, and T-G haplotypes of CD44 rs187115 and rs187116 polymorphisms in localized gastric adenocarcinoma patients. All censored patients and those who showed tumor recurrence are accounted for.
Analysis of other germline polymorphisms involved in the CD44 signaling pathway
None of the other tested germline polymorphisms demonstrated a statistically significant association with TTR and OS (Table 3).
Discussion
CD44 and its activating ligands hyaluronan and osteopontin interact within a signaling network to collectively drive numerous tumorigenic processes including the regulation of growth, survival, differentiation and motility. In addition, accumulating evidence in recent years strongly support CD44 as a gastric CSC marker. The results of the present research indicates that CD44 polymorphisms, alone or in combination, are significantly associated with both TTR and OS in localized GA patients who were treated with surgery alone or surgery and adjuvant (radio)-chemotherapy. These statistical associations retained their significance after adjusting for other potential predictors of patients’ outcome and were independent of ethnicity, tumor stage, lymph node involvement, and type of adjuvant therapy. To the best of our knowledge, this is the first study to show CD44 genetic variations as potential prognostic markers for localized GA.
Previous studies showed that CD44 protein overexpression is associated with poor prognosis in colorectal carcinoma and GA 24, 25. Although CD44 arises from a single gene, numerous splice variants are formed by alternative splicing and posttranscriptional modifications 26, 27. CD44 gained considerable interest, when it was described that certain CD44v facilitate to confer a metastatic phenotype in a variety of tumors, including GA 28–32. It is not yet clear whether particular modifications directly affect the binding affinity of CD44, or whether the structure of certain splice forms favors oligomerization, thereby conferring an increased affinity of CD44 for its ligands hyaluronan and osteopontin 8. Recent findings suggests that intronic germline variations can affect splicing regulatory elements, leading to aberrant splicing 33. Further, Narla et al. provided evidence of a link between a relatively common intronic polymorphism and KLF6, a Kruppel-like zinc finger transcription factor, through alternative splicing in prostate cancer 34.
Recently, Vazquez and colleagues showed that the C/C genotype of intronic CD44 rs187115 germline variation was significantly associated with decreased cellular response to cytotoxic chemotherapeutics including doxorubicin, carboplatin, RNA/DNA and DNA antimetabolites in vitro strongly suggesting a functionally significant role for this SNP in tumor cells 21. In addition, the functional significance of this polymorphism was further supported in an analysis of 129 soft-tis2sue sarcoma patients where patients homozygous for the C-allele were at a 2.1-fold increased risk for tumor related death compared with individuals harboring C/T or T/T genotype (p=0.041) 21. Interestingly, in our study, patients with localized GA harboring C/C genotype of CD44 rs187115 genetic variation had a 1.41-fold (95% CI, 0.55–3.60) increased risk of death compared with patients harboring T/T genotype but did not reach statistical significance in single marker analysis (log-rank p=0.75). However, haplotype analysis of polymorphisms rs187115 and rs187116 provided evidence for associaction with tumor recurrence. These polymorphisms were in LD (r2=0.39), which can mask or change the genetic effects of those loci in the association analysis. This may explain why rs187115 was not significantly associated with tumor recurrence in single marker analysis. The unvaforable T-G haplotype contains the protective T allele at rs187115 and the adverse G allele at rs187116. This suggests that the rs187116 unfavorable allele outweighs the rs187115 protective allele, and the overall effect of this haplotype significantly increases risk of tumor recurrence (adjusted p=0.001) and death (adjusted p=0.002; Table 4) compared to the T-A haplotype. As a result, the protective effect of rs187115 at haplotype T-G was masked by rs187116, and therefore its relation with tumor recurrence or death was not detected in single-marker analysis. In addition, in the current study we report for the first time, two CD44 intron genetic variations (rs187116 and rs7116423) which were significantly associated with OS and/or TTR in localized GA patients in single marker analysis (Table 3).
Although the precise functional and biological significance of these polymorphisms remain unknown, it is plausible that CD44 intronic polymorphisms may have a direct impact on the regulation of gene splicing events. The subsequent expression of alternate transcripts and CD44 receptor variants are reported to promote tumorigenic processes. To our knowledge, there have been no published reports characterizing the functional role of the CD44 intron polymorphisms tested in our study. Hence, the molecular mechanisms by which these germline variants exert their biological effects warrants further experimental investigation. However, these data strongly support a functionally and clinically important role of CD44 and its genetic variants in localized GA.
Our data may also have clinical-translational relevance. Early clinical trials using antibodies against CD44 that inhibit ligand binding have shown some promise by inhibiting tumor growth and metastasis 35–37, although some studies reported complications by various toxicities including the skin, a site in the body with a high level of CD44 38, 39. Therefore, the challenge is to improve these antibodies by taregeting them to specific CD44v that are uniquely expressed on the cancer in question. Future clinical trials should consider the genetic variations within the CD44 gene that we tested in an effort to further enhance tailored treatment strategies. In conclusion, the results of this study provide the first preliminary evidence that CD44 polymorphisms, alone or in combination, predict GA patients with early tumor recurrence. This may help to select patients at high risk for tumor recurrence who might benefit from more aggressive treatment strategies. Biomarker-embedded clinical trials are needed to validate our preliminary hypothesis-generating findings.
Acknowledgements
TW as supported in part by a Research Grant of the Austrian Society of Hematology and Oncology and the „Kurt und Senta-Herrmann foundation. PH was supported in part by a 2009 research grant from Cancer & Solidarité Foundation, Genève, Switzerland. AG was supported in part by a Research Grant of the Austrian Society of Hematology and Oncology and Bank Austria Visiting Scientists program.
This work was funded by the NIH grant 5 P30CA14089-27I and the Dhont Family Foundation. It was performed in the Sharon A. Carpenter Laboratory at USC/Norris Comprehensive Cancer Center and in memory of David Donaldson.
Footnotes
Previous Presentation: 2010 Gastrointestinal Cancers Symposium (oral presentation General Session II), ASCO 2010 (abstract #4047)
Conflicts of interest: There are no potential conflicts of interest
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Xu J, Ward E. Cancer Statistics, 2010. CA Cancer J Clin. 2010 Jul 7; doi: 10.3322/caac.20073. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 3.Van Cutsem E, Kang Y, Chung H, Shen L, Sawaki A, Lordick F, Hill J, Lehle M, Feyereislova A, Bang Y. Efficacy results from the ToGA trial: A phase III study of trastuzumab added to standard chemotherapy (CT) in first-line human epidermal growth factor receptor 2 (HER2)-positive advanced gastric cancer (GC). In ASCO 2009. J Clin Oncol. 2009;27(18s) (suppl; abstr LBA4509) [Google Scholar]
- 4.Macdonald JS. Treatment of localized gastric cancer. Semin Oncol. 2004;31:566–573. doi: 10.1053/j.seminoncol.2004.04.022. [DOI] [PubMed] [Google Scholar]
- 5.D'Angelica M, Gonen M, Brennan MF, Turnbull AD, Bains M, Karpeh MS. Patterns of initial recurrence in completely resected gastric adenocarcinoma. Ann Surg. 2004;240:808–816. doi: 10.1097/01.sla.0000143245.28656.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lauren P. The Two Histological Main Types of Gastric Carcinoma: Diffuse and So-Called Intestinal-Type Carcinoma. An Attempt at a Histo-Clinical Classification. Acta Pathol Microbiol Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- 7.Keller G, Hofler H, Becker KF. Molecular medicine of gastric adenocarcinomas. Expert Rev Mol Med. 2005;7:1–13. doi: 10.1017/S1462399405009592. [DOI] [PubMed] [Google Scholar]
- 8.Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 9.Weber GF, Ashkar S, Glimcher MJ, Cantor H. Receptor-ligand interaction between CD44 and osteopontin (Eta-1) Science. 1996;271:509–512. doi: 10.1126/science.271.5248.509. [DOI] [PubMed] [Google Scholar]
- 10.Lin YH, Yang-Yen HF. The osteopontin-CD44 survival signal involves activation of the phosphatidylinositol 3-kinase/Akt signaling pathway. J Biol Chem. 2001;276:46024–46030. doi: 10.1074/jbc.M105132200. [DOI] [PubMed] [Google Scholar]
- 11.Bourguignon LY, Zhu H, Chu A, Iida N, Zhang L, Hung MC. Interaction between the adhesion receptor, CD44, and the oncogene product, p185HER2, promotes human ovarian tumor cell activation. J Biol Chem. 1997;272:27913–27918. doi: 10.1074/jbc.272.44.27913. [DOI] [PubMed] [Google Scholar]
- 12.Bourguignon LY, Zhu H, Shao L, Zhu D, Chen YW. Rho-kinase (ROK) promotes CD44v(3,8-10)-ankyrin interaction and tumor cell migration in metastatic breast cancer cells. Cell Motil Cytoskeleton. 1999;43:269–287. doi: 10.1002/(SICI)1097-0169(1999)43:4<269::AID-CM1>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 13.Lesley J, Englis NM, Gal I, Mikecz K, Day AJ, Hyman R. Hyaluronan binding properties of a CD44 chimera containing the link module of TSG-6. J Biol Chem. 2002;277:26600–26608. doi: 10.1074/jbc.M201068200. [DOI] [PubMed] [Google Scholar]
- 14.Tsukita S, Oishi K, Sato N, Sagara J, Kawai A. ERM family members as molecular linkers between the cell surface glycoprotein CD44 and actin-based cytoskeletons. J Cell Biol. 1994;126:391–401. doi: 10.1083/jcb.126.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gotley DC, Fawcett J, Walsh MD, Reeder JA, Simmons DL, Antalis TM. Alternatively spliced variants of the cell adhesion molecule CD44 and tumour progression in colorectal cancer. Br J Cancer. 1996;74:342–351. doi: 10.1038/bjc.1996.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naor D, Sionov RV, Ish-Shalom D. CD44: structure, function, and association with the malignant process. Adv Cancer Res. 1997;71:241–319. doi: 10.1016/s0065-230x(08)60101-3. [DOI] [PubMed] [Google Scholar]
- 17.Liu YJ, Yan PS, Li J, Jia JF. Expression and significance of CD44s, CD44v6, and nm23 mRNA in human cancer. World J Gastroenterol. 2005;11:6601–6606. doi: 10.3748/wjg.v11.i42.6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoo CH, Noh SH, Kim H, Lee HY, Min JS. Prognostic significance of CD44 and nm23 expression in patients with stage II and stage IIIA gastric carcinoma. J Surg Oncol. 1999;71:22–28. doi: 10.1002/(sici)1096-9098(199905)71:1<22::aid-jso5>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 19.Takaishi S, Okumura T, Tu S, Wang SS, Shibata W, Vigneshwaran R, Gordon SA, Shimada Y, Wang TC. Identification of gastric cancer stem cells using the cell surface marker CD44. Stem Cells. 2009;27:1006–1020. doi: 10.1002/stem.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shukla SJ, Duan S, Wu X, Badner JA, Kasza K, Dolan ME. Whole-genome approach implicates CD44 in cellular resistance to carboplatin. Hum Genomics. 2009;3:128–142. doi: 10.1186/1479-7364-3-2-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vazquez A, Grochola LF, Bond EE, Levine AJ, Taubert H, Muller TH, Wurl P, Bond GL. Chemosensitivity profiles identify polymorphisms in the p53 network genes 14-3-3tau and CD44 that affect sarcoma incidence and survival. Cancer Res. 2010;70:172–180. doi: 10.1158/0008-5472.CAN-09-2218. [DOI] [PubMed] [Google Scholar]
- 22.Zhou J, Nagarkatti PS, Zhong Y, Creek K, Zhang J, Nagarkatti M. Unique SNP in CD44 intron 1 and its role in breast cancer development. Anticancer Res. 2010;30:1263–1272. [PMC free article] [PubMed] [Google Scholar]
- 23.Neuberger J, Altman DG, Christensen E, Tygstrup N, Williams R. Use of a prognostic index in evaluation of liver transplantation for primary biliary cirrhosis. Transplantation. 1986;41:713–716. doi: 10.1097/00007890-198606000-00009. [DOI] [PubMed] [Google Scholar]
- 24.Ghaffarzadehgan K, Jafarzadeh M, Raziee HR, Sima HR, Esmaili-Shandiz E, Hosseinnezhad H, Taghizadeh-Kermani A, Moaven O, Bahrani M. Expression of cell adhesion molecule CD44 in gastric adenocarcinoma and its prognostic importance. World J Gastroenterol. 2008;14:6376–6381. doi: 10.3748/wjg.14.6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huh JW, Kim HR, Kim YJ, Lee JH, Park YS, Cho SH, Joo JK. Expression of standard CD44 in human colorectal carcinoma: association with prognosis. Pathol Int. 2009;59:241–246. doi: 10.1111/j.1440-1827.2009.02357.x. [DOI] [PubMed] [Google Scholar]
- 26.Screaton GR, Bell MV, Jackson DG, Cornelis FB, Gerth U, Bell JI. Genomic structure of DNA encoding the lymphocyte homing receptor CD44 reveals at least 12 alternatively spliced exons. Proc Natl Acad Sci U S A. 1992;89:12160–12164. doi: 10.1073/pnas.89.24.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skelton TP, Zeng C, Nocks A, Stamenkovic I. Glycosylation provides both stimulatory and inhibitory effects on cell surface and soluble CD44 binding to hyaluronan. J Cell Biol. 1998;140:431–446. doi: 10.1083/jcb.140.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barbour AP, Reeder JA, Walsh MD, Fawcett J, Antalis TM, Gotley DC. Expression of the CD44v2-10 isoform confers a metastatic phenotype: importance of the heparan sulfate attachment site CD44v3. Cancer Res. 2003;63:887–892. [PubMed] [Google Scholar]
- 29.Gunthert U, Hofmann M, Rudy W, Reber S, Zoller M, Haussmann I, Matzku S, Wenzel A, Ponta H, Herrlich P. A new variant of glycoprotein CD44 confers metastatic potential to rat carcinoma cells. Cell. 1991;65:13–24. doi: 10.1016/0092-8674(91)90403-l. [DOI] [PubMed] [Google Scholar]
- 30.Heider KH, Dammrich J, Skroch-Angel P, Muller-Hermelink HK, Vollmers HP, Herrlich P, Ponta H. Differential expression of CD44 splice variants in intestinal- and diffuse-type human gastric carcinomas and normal gastric mucosa. Cancer Res. 1993;53:4197–4203. [PubMed] [Google Scholar]
- 31.Klingbeil P, Marhaba R, Jung T, Kirmse R, Ludwig T, Zoller M. CD44 variant isoforms promote metastasis formation by a tumor cell-matrix cross-talk that supports adhesion and apoptosis resistance. Mol Cancer Res. 2009;7:168–179. doi: 10.1158/1541-7786.MCR-08-0207. [DOI] [PubMed] [Google Scholar]
- 32.Wielenga VJ, Heider KH, Offerhaus GJ, Adolf GR, van den Berg FM, Ponta H, Herrlich P, Pals ST. Expression of CD44 variant proteins in human colorectal cancer is related to tumor progression. Cancer Res. 1993;53:4754–4756. [PubMed] [Google Scholar]
- 33.Pagani F, Baralle FE. Genomic variants in exons and introns: identifying the splicing spoilers. Nat Rev Genet. 2004;5:389–396. doi: 10.1038/nrg1327. [DOI] [PubMed] [Google Scholar]
- 34.Narla G, Difeo A, Reeves HL, Schaid DJ, Hirshfeld J, Hod E, Katz A, Isaacs WB, Hebbring S, Komiya A, McDonnell SK, Wiley KE, et al. A germline DNA polymorphism enhances alternative splicing of the KLF6 tumor suppressor gene and is associated with increased prostate cancer risk. Cancer Res. 2005;65:1213–1222. doi: 10.1158/0008-5472.CAN-04-4249. [DOI] [PubMed] [Google Scholar]
- 35.Borjesson PK, Postema EJ, Roos JC, Colnot DR, Marres HA, van Schie MH, Stehle G, de Bree R, Snow GB, Oyen WJ, van Dongen GA. Phase I therapy study with (186)Re-labeled humanized monoclonal antibody BIWA 4 (bivatuzumab) in patients with head and neck squamous cell carcinoma. Clin Cancer Res. 2003;9:3961S–3972S. [PubMed] [Google Scholar]
- 36.Colnot DR, Ossenkoppele GJ, Roos JC, Quak JJ, de Bree R, Borjesson PK, Huijgens PC, Snow GB, van Dongen GA. Reinfusion of unprocessed, granulocyte colony-stimulating factor-stimulated whole blood allows dose escalation of 186Relabeled chimeric monoclonal antibody U36 radioimmunotherapy in a phase I dose escalation study. Clin Cancer Res. 2002;8:3401–3406. [PubMed] [Google Scholar]
- 37.Marangoni E, Lecomte N, Durand L, de Pinieux G, Decaudin D, Chomienne C, Smadja-Joffe F, Poupon MF. CD44 targeting reduces tumour growth and prevents post-chemotherapy relapse of human breast cancers xenografts. Br J Cancer. 2009;100:918–922. doi: 10.1038/sj.bjc.6604953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rupp U, Schoendorf-Holland E, Eichbaum M, Schuetz F, Lauschner I, Schmidt P, Staab A, Hanft G, Huober J, Sinn HP, Sohn C, Schneeweiss A. Safety and pharmacokinetics of bivatuzumab mertansine in patients with CD44v6-positive metastatic breast cancer: final results of a phase I study. Anticancer Drugs. 2007;18:477–485. doi: 10.1097/CAD.0b013e32801403f4. [DOI] [PubMed] [Google Scholar]
- 39.Tijink BM, Buter J, de Bree R, Giaccone G, Lang MS, Staab A, Leemans CR, van Dongen GA. A phase I dose escalation study with anti-CD44v6 bivatuzumab mertansine in patients with incurable squamous cell carcinoma of the head and neck or esophagus. Clin Cancer Res. 2006;12:6064–6072. doi: 10.1158/1078-0432.CCR-06-0910. [DOI] [PubMed] [Google Scholar]
- 40.Suga T, Ishikawa A, Kohda M, Otsuka Y, Yamada S, Yamamoto N, Shibamoto Y, Ogawa Y, Nomura K, Sho K, Omura M, Sekiguchi K, et al. Haplotype-based analysis of genes associated with risk of adverse skin reactions after radiotherapy in breast cancer patients. Int J Radiat Oncol Biol Phys. 2007;69:685–693. doi: 10.1016/j.ijrobp.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 41.Han S, Guthridge JM, Harley IT, Sestak AL, Kim-Howard X, Kaufman KM, Namjou B, Deshmukh H, Bruner G, Espinoza LR, Gilkeson GS, Harley JB, et al. Osteopontin and systemic lupus erythematosus association: a probable gene-gender interaction. PLoS One. 2008;3:e0001757. doi: 10.1371/journal.pone.0001757. [DOI] [PMC free article] [PubMed] [Google Scholar]


