Abstract
Chromatin remodelling complexes containing the nucleosome-dependent ATPase ISWI were first isolated from Drosophila embryos (NURF, CHRAC and ACF). ISWI was the only common component reported in these complexes. Our purification of human CHRAC (HuCHRAC) shows that ISWI chromatin remodelling complexes can have a conserved subunit composition in completely different cell types, suggesting a conserved function of ISWI. We show that the human homologues of two novel putative histone-fold proteins in Drosophila CHRAC are present in HuCHRAC. The two human histone-fold proteins form a stable complex that binds naked DNA but not nucleosomes. HuCHRAC also contains human ACF1 (hACF1), the homologue of Acf1, a subunit of Drosophila ACF. The N-terminus of mouse ACF1 was reported as a heterochromatin-targeting domain. hACF1 is a member of a family of proteins with a related domain structure that all may target heterochromatin. We discuss a possible function for HuCHRAC in heterochromatin dynamics. HuCHRAC does not contain topoisomerase II, which was reported earlier as a subunit of Drosophila CHRAC.
Keywords: bromodomain/nucleosome/PHD finger/topoisomerase II/WSTF
Introduction
Recent discoveries have highlighted the interrelationship of the packaging of the eukaryotic genome in chromatin and regulated processes such as transcription. Alterations in chromatin structure are often a prerequisite for transcriptional regulation. The nucleosome, the basic subunit of chromatin, is a major target for such alterations. Various enzymes change the nucleosome structure and mediate transcriptional regulation via ‘chromatin remodelling’. These enzymes include histone acetyltransferases and histone deacetylases, histone kinases and ATP-dependent nucleosome remodelling factors (reviewed in Workman and Kingston, 1998; Kingston and Narlikar, 1999). ATP-dependent nucleosome remodelling factors function as complexes, ranging from heterodimers to complexes of 12 or more subunits. Central to their function are ATPases with a conserved domain structure. Some of these ATPases are similar to the yeast SWI2/SNF2 ATPase, others are of the CHD type or of the ISWI-type (Kingston and Narlikar, 1999).
Three different chromatin remodelling complexes, isolated from Drosophila embryos, contain the ISWI ATPase: (i) nucleosome remodelling factor (NURF; Tsukiyama and Wu, 1995); (ii) ATP-utilizing chromatin assembly and remodelling factor (ACF; Ito et al., 1997); and (iii) chromatin accessibility complex (CHRAC; Varga-Weisz et al., 1997). Apart from the shared ISWI, no common subunits were reported for these complexes. Yeast Saccharomyces cerevisiae contains two highly similar genes with homology to Drosophila ISWI. Nevertheless, their gene products form functionally and biochemically different complexes (Tsukiyama et al., 1999). In human, two very similar ISWI homologues, hSNF2L (Okabe et al., 1992) and hSNF2H (Aihara et al., 1998), have been described. A heterodimeric complex termed RSF (remodelling and spacing factor) containing hSNF2H recently was purified from HeLa cell nuclear pellet (LeRoy et al., 1998). The other subunit of RSF is a novel 325 kDa protein. RSF facilitates regular spacing between nucleosomes and allows transcription initiation from a nucleosomal promoter.
The finding of three different ISWI-containing complexes in Drosophila early embryos raises the question of whether they represent a limited number of functional units with an evolutionarily conserved subunit composition, or whether they are just a small sample of a much larger number of complexes that form as a result of promiscuous interaction with ISWI. The necessity for rapid replication-associated chromatin assembly in Drosophila embryos may influence the abundance and composition of ISWI-containing chromatin remodelling factors as compared with, for example, much slower dividing HeLa cells. The subunit composition and biochemistry of ISWI complexes in cells other than those from Drosophila embryos may provide clues about the in vivo function of ISWI.
Here we report the characterization of a human ISWI-containing complex, human CHRAC (HuCHRAC), purified from HeLa cell nuclear extracts. HuCHRAC contains subunits that are conserved in Drosophila ISWI complexes: hACF1, the human homologue of Drosophila Acf1, a subunit of ACF, and the human counterparts of two novel histone-fold proteins that are part of Drosophila CHRAC. This demonstrates conservation of ISWI complexes between Drosophila embryo cells and human cells, despite their physiological differences, and indicates a conserved function for ISWI. The N-terminal part of mouse ACF1 recently was shown to target pericentric heterochromatin upon cell differentiation (Tate et al., 1998), and we discuss potential implications of this finding concerning the function of HuCHRAC.
Results
Purification of a human ISWI complex from HeLa nuclear extracts
The Drosophila ISWI complexes have all been identified with chromatin remodelling assays. However, crude HeLa cell nuclear extract or crude fractions thereof showed no chromatin remodelling activities in such assays (data not shown). This possibly is due to inhibitors of activity. We therefore decided to purify human ISWI by following its immunoreactivity with western blot analysis. We generated a rabbit polyclonal antibody against a part of human ISWI isoform hSNF2L that is highly conserved in the other isoform of human ISWI, hSNF2H (see Materials and methods). The antibody recognizes the recombinant epitope and shows one predominant band of ∼130 kDa in a western analysis of HeLa nuclear extract (data not shown). Since the antibody does not distinguish between the hSNF2H and hSNF2L isoforms of human ISWI (data not shown), we refer to the antibody-recognized protein as ‘human ISWI’ (hISWI).
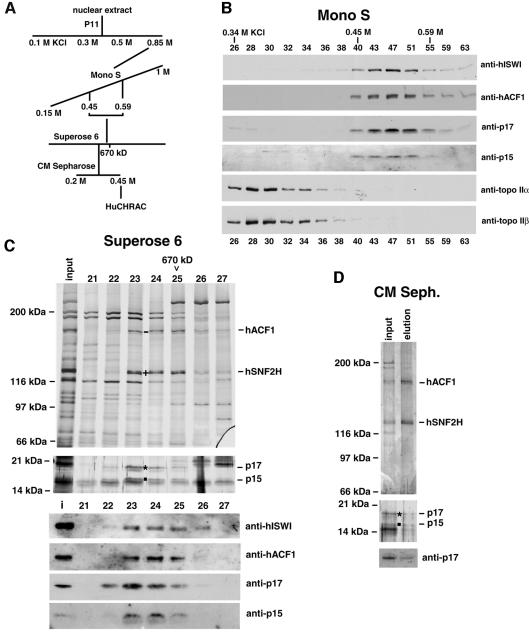
To purify hISWI complexes, we followed chromatographic fractions that contained the highest concentration of ISWI. After two purification steps (see Figure 1A for purification scheme), a gel filtration analysis was performed. A 130 kDa protein (Figure 1C, upper panel, indicated by a cross), which was recognized by the anti-hISWI antibody (Figure 1C, lower panel), co-purified with a 185 kDa protein (indicated by a bar in the upper panel) at an apparent mol. wt of ∼800 kDa. Two small proteins, in apparently stoichiometric amounts to each other, with a mol. wt of 15 and 17 kDa, also co-purified in the same fractions, with the earliest fraction (23) being more enriched for both proteins (Figure 1C, middle panel).
Fig. 1. Human ISWI co-purifies with human ACF1 and histone-fold proteins p17 and p15. (A) Purification scheme for HuCHRAC. (B) Co-fractionation of hISWI with hACF1, p17 and p15, but separation from topoisomerase IIα and IIβ in Mono S chromatography. Proteins in the input and indicated fractions were visualized by western blot analysis with the indicated antibodies. (C) Co-fractionation of hISWI with hACF1, p17 and p15 in Superose 6 gel filtration chromatography. Proteins in the input and indicated fractions (0.5 ml) were visualized by silver staining (two upper panels) or western blot analysis (four lower panels) with the indicated antibodies. ‘–’ indicates hACF1, identified by mass spectroscopy (see text), ‘+’ indicates hISWI isoform hSNF2H, identified by mass spectroscopy (see text), * indicates protein recognized by anti-p17, and the filled square indicates protein recognized by anti-p15. The running position of a molecular size standard (thyroglobulin) is indicated at the top of the panel. (D) Co-fractionation of hSNF2H, hACF1, p17 and p15 in the final CM-Sepharose chromatography. The input and elution fraction were visualized by silver staining (upper two panels). The lower panel represents a western blot analysis with anti-p17 antibodies.
The hISWI-containing gel filtration fractions were purified further with CM-Sepharose. The eluate contains two large proteins of 130 and 185 kDa, in an apparent stoichiometric ratio (Figure 1D). The 185 kDa band is the protein that co-purified with hISWI on the gel filtration column (Figure 1C). The 15 and 17 kDa proteins also co-purified with hISWI over this last purification step (Figure 1D, lower panels), although in reduced amounts.
When we fractionated a crude HeLa nuclear extract on a gel filtration column, the majority of hISWI runs with an apparent mol. wt of ∼ 800 kDa; a minor fraction runs at ∼300 kDa (data not shown). This suggests that the majority of the total hISWI protein in the nuclear extract is part of the hISWI complex presented here. We cannot, however, exclude that another ISWI complex in the crude extract is running in the same gel filtration column fractions.
We estimated, by staining comparison with a protein standard, that the amount of ‘hISWI complex’ on the gel filtration column, defined as hISWI and the co-purifying proteins of 185, 17 and 15 kDa (Figure 1C), is ∼50 µg, starting from 140 mg of nuclear extract.
ISWI homologue hSNF2H and a protein with homology to Acf1 are part of the hISWI complex
Tryptic peptides from the 130 and 185 kDa proteins in the purified hISWI complex were analysed by ion trap mass spectrometry. From the 130 kDa band, the sequences of 38 peptides were identified. Thirty six were specific for hSNF2H and two were specific for hSNF2L (data not shown), indicating that the hISWI band contains predominantly the hSNF2H ISWI isoform. The minor amount of hSNF2L that co-purifies with hSNF2H may suggest that at least a fraction of hSNF2L is present in a complex similar to the hSNF2H complex described here.
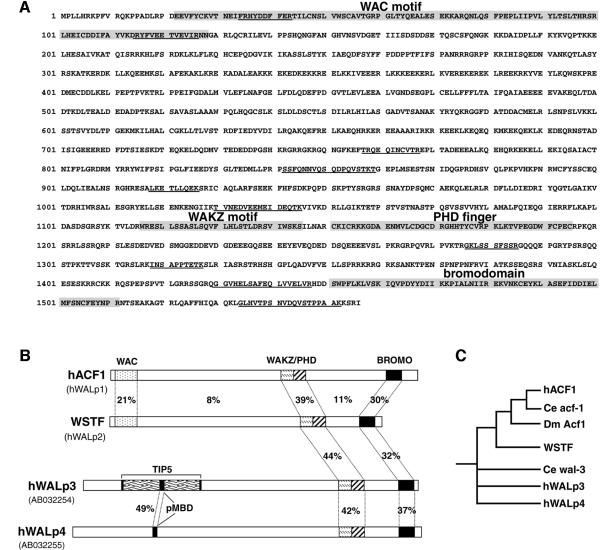
Eleven peptides were sequenced from the 185 kDa band, nine of which identified a human partial cDNA (DDBJ/EMBL/GenBank accession No. AL050089); the other two peptide sequences identified the partial mouse cDNA from Ct146 (Tate et al., 1998). The full-length sequence of the human cDNA encoding the 185 kDa protein was determined by combining sequences derived from human expressed sequence tags (ESTs) that matched the identified cDNAs, and sequences derived from reverse transcription of HeLa RNA (see Materials and methods). The protein sequence encoded by the full-length cDNA is shown in Figure 2A. We call this protein human ACF1 (hACF1) because of its high homology to Drosophila Acf1 (Dm Acf1, Figure 2C). Acf1 is a subunit of the chromatin remodelling complex ACF (Ito et al., 1997) and co-operates with ISWI in the ATP-dependent catalysis of chromatin assembly by ACF (Ito et al., 1999). We created an antiserum against a 17 amino acid peptide specific to hACF1. This antiserum recognizes a 185 kDa protein in HeLa nuclear extracts. Western blot analysis of the Mono S and gel filtration fractions with the anti-hACF1 antiserum confirms the co-purification of hACF1 and hISWI (Figure 1C, lower panel, and Figure 1B). Topoisomerases II (topo II) α and β were clearly separated from hISWI and hACF1 on the Mono S column (Figure 1B). Furthermore, the antiserum against hISWI co-immunoprecipitates hACF1 (Figure 4). We conclude that the hISWI isoform hSNF2H is in a complex with hACF1.
Fig. 2. Human ACF1 is a member of a family of WAL proteins. (A) Predicted amino acid sequence of hACF1. The WAC and WAKZ domains, PHD finger and bromodomain in hACF1 are indicated by shaded boxes. Underlined amino acids indicate tryptic peptides whose sequences were identified by ion trap tandem mass spectrometry. (B) Known human members of a family of WAL (WSTF/Acf1-like) proteins. The percentage amino acid identity between the indicated domains is shown. The human (h)WALp3 sequence was derived from the AB032254 cDNA. hWALp3 contains the TIP5 (TTF1 interaction peptide 5, EST AF000422; Jansa et al., 1998). The hWALp4 sequence was derived from AB032255 cDNA. The TIP5 peptide in hWALp3 and the N-terminal part of hWALp4 contain a putative methyl-binding domain (pMBD). (C) Phylogeny tree of the WAL proteins. Ce acf-1 is encoded by H20J04.2 cDNA, Ce wal-3 is encoded by ZK783.4 cDNA.
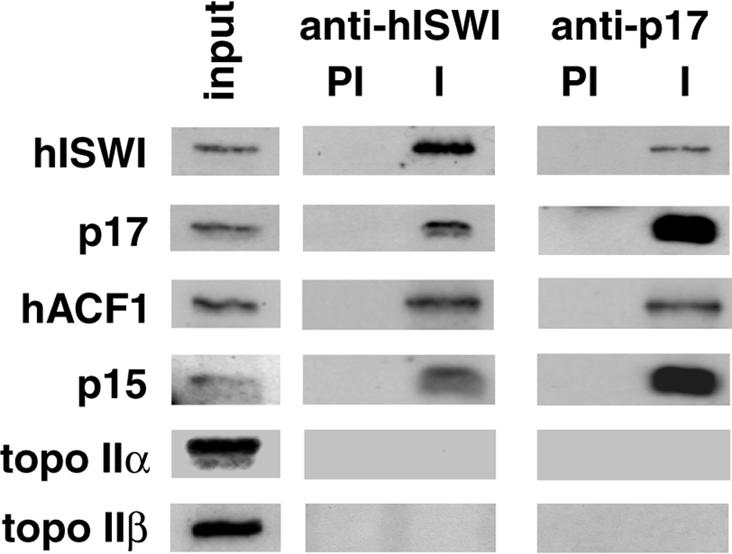
Fig. 4. Human ISWI co-immunoprecipitates with the p15 and p17 histone-fold proteins and hACF1. Immunoprecipitations from HeLa nuclear extracts with anti-hISWI and anti-p17 antibodies. Immuno precipitates with pre-immune (PI) or immune (I) sera and 5% input were western blotted and probed with antibodies against hISWI, p17, ACF1, p15, topo IIα and β, as indicated.
The N-terminal part of hACF1 is >90% identical with the protein sequence of the Ct146 mouse cDNA clone (data not shown). The hACF1 gene was localized to chromosome 14q11.2 by fluorescent in situ hybridization (FISH) analysis (data not shown). Ct146 maps to mouse chromosome 12, band C, which is in conserved synteny with the region on human chromosome 14 to which hACF1 localizes (www.informatics.jax.org). This strongly suggests that Ct146 encodes the N-terminal part of mouse ACF1, the mouse homologue of hACF1. The product of Ct146 was identified recently in a gene-trap screen as a protein concentrated in pericentric heterochromatin at interphase and metaphase in differentiated mouse cells (Tate et al., 1998; see Discussion).
Human ACF1 and WSTF are members of a novel family of WAL proteins
Analysis of the DDBJ/EMBL/GenBank databases revealed three human genes that encode proteins highly related to hACF1 (Figure 2B). One gene encodes the WSTF protein (Williams syndrome transcription factor) and is one of (at least) 15 genes that are part of a 1 Mb deletion of human chromosome 7 associated with Williams syndrome, a developmental disorder (Lu et al., 1998; Peoples et al., 1998). Two hACF-related genes were characterized as ‘bromodomain adjacent to zinc finger domain 2A and 2B’ (BAZ2A and BAZ2B; Jones et al, 2000). This suggests the existence of a family of WSTF/ACF1-related proteins, which we have named the WAL (WSTF-, ACF1-like) family of proteins. Members of the WAL protein family are characterized by a conserved WAKZ motif (Ito et al., 1999) followed directly by a PHD finger motif (Aasland et al., 1995) and a bromodomain (reviewed in Winston and Allis, 1999) near the C-terminus of the proteins (Figure 2B). hACF1 and WSTF, in the upper branch of the WAL family tree (Figure 2C), contain a WAC motif near their N-terminus (Ito et al., 1999).
Interestingly, hWALp3, hWALp4 and their Caenorhabditis elegans homologue Ce wal-3 (Figure 2C), which are more distant from hACF1 and WSTF, do not contain an N-terminal WAC motif. Instead, hWALp3 contains a sequence named TIP5 (see Discussion; Jansa et al., 1998; and Figure 2B), in the centre of which we found a 70 amino acid motif (Figure 2B; pMBD) that is conserved in the methyl-binding domains (MBD) of human methyl-CpG-binding proteins such as MBD1-4 and MeCP2 (Hendrich and Bird, 1998; G.Dellaire and W.A.Bickmore, unpublished data). This domain is highly conserved between hWALp3 and hWALp4 (49% identity; Figure 2B, and data not shown). Moreover, the TIP5 sequence is conserved between hWALp3 and Ce wal-3 only in this MBD-homologous domain (39% identity; data not shown).
A set of novel histone-fold proteins interacts with hISWI: evidence for co-purification with the human ISWI complex
Two peptides of 15 and 17 kDa co-purified with hISWI (Figure 1C and D), similar to the co-purification of two small peptides in the Drosophila CHRAC (Varga-Weisz et al., 1997). Homology searches with the sequences of these two Drosophila peptides, named CHRAC-14 and CHRAC-16 (Corona et al., 2000), revealed human ESTs encoding two novel proteins. The peptide sequence of CHRAC-14 showed close homology to one unknown human 17 kDa protein of 148 amino acids. We call this protein HuCHRAC-17, abbreviated here as p17 (Figure 3). CHRAC-16 showed close homology to one 131 amino acid, 15 kDa protein. There was no in-frame stop codon before the first ATG of the open reading frame (ORF) in the EST. Therefore, we cannot exclude the possibility that the actual ORF is larger. However, comparison with the Drosophila homologue suggests that the 131 amino acids represent the full-length ORF of a protein that we call HuCHRAC-15, abbreviated as p15 (Figure 3). Comparison with putative histone-fold domains of other proteins indicated that p17 and p15, like their Drosophila homologues, contain a putative histone-fold domain (indicated in Figure 3; Baxevanis et al., 1995; genome. nhgri.nih.gov/histones/fold.html, see also below).
Fig. 3. HuCHRAC-15 and HuCHRAC-17 are the human homologues of Drosophila CHRAC-16 and CHRAC-14, respectively. The predicted amino acid sequences of HuCHRAC-15 and HuCHRAC-17, abbreviated as p15 and p17, were aligned with the sequences of CHRAC-16 and CHRAC-14, respectively. Identical amino acids are assigned; similar amino acids are indicated by +. Putative histone-fold domains are indicated by shaded boxes. Histone-fold domains for p15 and p17 homologues were assigned by homology to the putative histone-fold domains of CBF-C and HAP5 (p15) or CBF-A and HAP3 (p17).
We expressed recombinant p17 and p15 in Escherichia coli and used the recombinant protein to raise antibodies. A rabbit polyclonal antibody against recombinant p17 specifically recognized and efficiently immunoprecipitated a protein of 17 kDa from nuclear extract (Figure 4). hISWI co-immunoprecipitated in these experiments (Figure 4) and the anti-hISWI antibody co-immunoprecipitated p17 (Figure 4). Both anti-hISWI and the anti-p17 antibodies co-immunoprecipitated hACF1 and p15, but not topo II α and β (Figure 4). Furthermore, western blot analysis shows the co-purification of p17 and p15 with hISWI and hACF1 in the Mono S and in the gel filtration chromatography (Figure 1B, C and D). p17 was also present in the CM-Sepharose-purified complex (Figure 1D, lowest panel). The p15 antiserum was too weak to prove the presence of p15 in the CM-Sepharose fraction, although the silver stain analysis suggests that it co-purifies in equimolar amounts to p17 (Figure 1D, middle panel). We conclude that hISWI, hACF1, p17 and p15 are interacting in one complex. In reference to the presence in our hISWI complex of the two histone-fold proteins that were also found in Drosophila CHRAC (Corona et al., 2000), we call our ISWI complex human CHRAC (HuCHRAC).
HuCHRAC is a nucleosome remodelling factor
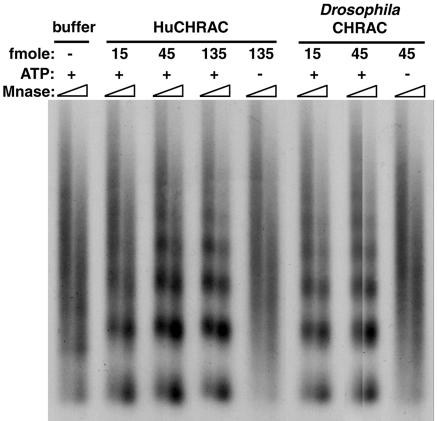
We tested whether HuCHRAC could mobilize nucleosomes in an ATP-dependent manner into a regularly spaced nucleosomal array, as was shown for other ISWI-containing chromatin remodelling factors such as Drosophila CHRAC (Varga-Weisz et al., 1997) and human RSF (LeRoy et al., 1998). Plasmid DNA was assembled into ‘irregular’ chromatin using Drosophila assembly extracts in the absence of ATP (Varga-Weisz et al., 1997). Endogenous chromatin remodelling activity was stripped from the chromatin by sarcosyl wash. Subsequently, HuCHRAC or the Drosophila CHRAC complex were added to the chromatin, followed by incubation in the presence or absence of ATP. The chromatin was digested with micrococcal nuclease, which cleaves preferentially in linker DNA but less in nucleosomal DNA. The purified DNA was analysed by Southern blot hybridization. Figure 5 shows that addition of HuCHRAC created a more regularly spaced nucleosomal array, as compared with the control. The nucleosome spacing activity of HuCHRAC is strictly ATP dependent and is qualitatively and quantitatively similar to the remodelling activity of Drosophila CHRAC. We conclude that HuCHRAC is a chromatin remodelling factor.
Fig. 5. HuCHRAC has nucleosome spacing activity. The indicated amounts of HuCHRAC or Drosophila CHRAC were added to sarkosyl-stripped chromatin DNA, assembled in Drosophila embryo extracts without ATP. Addition of ATP was as indicated. After the chromatin remodelling reaction, DNA was digested for 30 or 60 s with micrococcal nuclease, separated on a 1.3% agarose gel, Southern blotted and probed with random labelled input DNA.
HuCHRAC has DNA-dependent ATPase activity, which is enhanced >3-fold when the DNA is packaged into nucleosomes (data not shown). This was to be expected since recombinant Drosophila ISWI is a nucleosome-dependent ATPase by itself (Corona et al., 1999). Addition of recombinant p15–p17 complex (see below) to HuCHRAC did not have a significant effect on the chromatin-dependent ATPase activity or the nucleosome spacing activity of HuCHRAC (data not shown).
Recombinant p15 and p17 form a complex that binds DNA but not nucleosomes
The histone fold is a conserved protein structural motif which allows dimerization of two proteins carrying this motif (Arents and Moudrianakis, 1995; Baxevanis et al., 1995). A putative histone-fold domain in p15 (indicated in Figure 3) has similarity to the suggested histone-fold domains of CBF-C, a subunit of the CCAAT-binding factor, yeast activator HAP5 and NC2α, the dimerization partner of NC2β (Zemzoumi et al., 1999). The putative histone-fold motifs of CBF-C, HAP5 and NC2α are of the histone H2A type. The putative p17 histone-fold domain is similar to the suggested histone-fold domains of CBF-A, yeast transcriptional activator HAP3 and NC2β (Maity and de Crombrugghe, 1998), which are of the histone H2B type. This suggests that p15 and p17 can form a histone-fold complex of the H2A–H2B type.
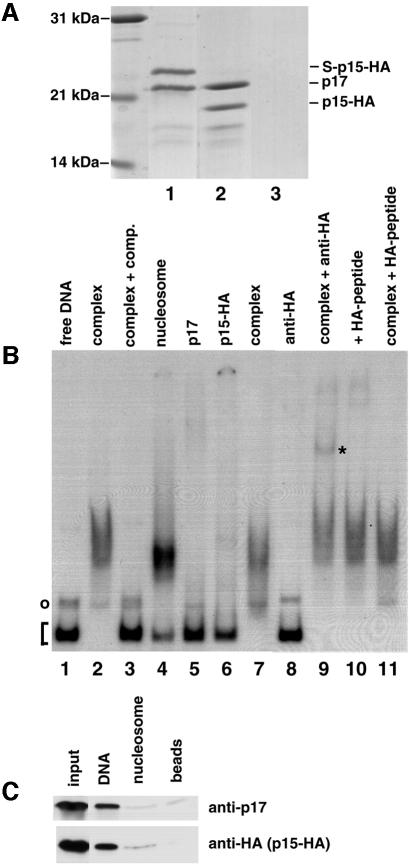
To test if the ISWI-associated peptides p15 and p17 interact directly with each other, we co-expressed both proteins in E.coli, each having different tags: GST with p17, and His tag, S tag and haemagglutinin (HA) tag with p15 (‘S-p15-HA’; see Materials and methods). The bacterial lysate was incubated with glutathione–Sepharose beads to bind GST–p17. p17 was released by thrombin protease cleavage. Figure 6A, lane 1 shows that the thrombin cleavage released two proteins. Western blot analysis with the anti-p17 antibody and an anti-HA antibody confirmed that the lower band was p17 and the upper band was S-p15-HA (data not shown; thrombin also cleaves off the His tag from S-p15-HA). A lysate of bacteria expressing only p15 released no protein upon cleavage with thrombin (Figure 6A, lane 3). The p17–S-p15-HA complex was then bound to an S-protein resin. Cleavage by enterokinase-protease released the purified p17–p15-HA from the S-column (Figure 6A, lane 2). We conclude that p15 and p17 form a stable complex. Because histone-fold type proteins interact with each other via the histone-fold domains (Arents and Moudrianakis, 1995; Baxevanis et al., 1995), it is very likely that this is also true for the p15–p17 complex.
Fig. 6. Recombinant proteins p15 and p17 form a complex that binds to naked, but not nucleosomal DNA. (A) Coomassie-stained SDS–polyacrylamide gel showing the purification of the p15–p17 complex. Lane 1, elution of the S-p15-HA–p17 complex from glutathione–Sepharose by cleavage with thrombin protease; lane 2, elution of the p15-HA–p17 complex from S-protein–agarose by cleavage with enterokinase-protease; lane 3, control elution from glutathione–Sepharose loaded with lysate from cells expressing only His-S-p15-HA, showing that binding of p15 to this column is only possible via p17–GST. (B) Electrophoretic mobility shift analysis of DNA–p15–p17 complexes with a 150 bp DNA fragment. Lane 1, 32P-labelled DNA fragment without protein; lane 2, p15-HA–p17 complex (0.2 µg); lane 3, p15-HA–p17 complex (0.2 µg) + excess unrelated plasmid DNA; lane 4, nucleosomal DNA; lane 5, p17 (0.3 µg); lane 6, p15-HA (0.12 µg); lane 7, p15-HA–p17 complex (0.1 µg); lane 8, anti-HA-antibody; lane 9, as lane 2 + anti-HA-antibody (*indicates the supershifted p15-HA–p17–DNA complex); lane 10, as lane 9 +HA-peptide; lane 11, as lane 2 + HA-peptide. The bracket on the left side denotes uncomplexed DNA, the circle indicates single-stranded DNA, a PCR artificial product. (C) Binding of p15–p17 complex to DNA coupled to magnetic beads. p15–p17 complex was incubated with DNA coupled to magnetic beads, or nucleosomal DNA coupled to magnetic beads, or beads alone. Bound proteins were examined by western blot analysis with antibodies against p17, or anti-HA-antibodies (p15-HA).
To investigate a possible function of the histone-fold proteins within HuCHRAC, we tested the DNA-binding capacities of the recombinant p15–p17 complex. Addition of the p15–p17 complex to a randomly chosen 150 bp, 32P-end-labelled DNA fragment caused a defined shift of the DNA to lower mobility in acrylamide gels (Figure 6B, lanes 2 and 7). In contrast, addition of the same amounts of p15 or p17 alone did not cause a significant DNA shift (lanes 5 and 6). Also, when p15 and p17, both purified separately, were added together, no DNA shift occurred (data not shown), suggesting that p15 and p17 monomers do not bind each other in the reaction conditions used here. An excess amount of unrelated plasmid DNA out-competed binding of the p15–p17 complex to the 150 bp fragment (Figure 6B, lane 3), suggesting that the complex does not bind to a specific sequence in the 150 bp fragment. Addition of anti-HA-antibody to the p15-HA–p17–DNA complex supershifted part of the p15-HA–p17–DNA complex (lane 9, supershift indicated by *). Addition of excess HA-peptide avoided the supershift by competition with the HA tag of p15-HA (lane 10). Titration experiments revealed that the dissociation constant of the p15–p17–DNA complex is in the submicromolar range, indicating a high affinity of p15–p17 for DNA (data not shown). These data show that the p15–p17 complex, but not its components, binds strongly to DNA to form a complex that causes a distinct DNA shift. Pull-down experiments using magnetic beads coupled to the same 150 bp DNA fragment, either ‘naked’ or assembled into a nucleosome, confirmed the DNA-binding capacity of p15–p17, but indicated that the complex did not interact well with nucleosomes (Figure 6C).
The relative levels of p15 and p17 mRNA are correlated in different human tissues
If p15 and p17 are in a complex in vivo, one would predict that their relative expression levels in different tissues are related. We measured the levels of p15-coding mRNA and p17-coding mRNA in different human tissues by northern blot analysis, using labelled coding cDNA sequences for p15 and p17, respectively. As shown in Figure 7, p15 and p17 mRNA are expressed in all the tissues tested. p15 and p17 mRNA levels, standardized to β-actin mRNA levels, are variable but parallel in different tissues (Figure 7).
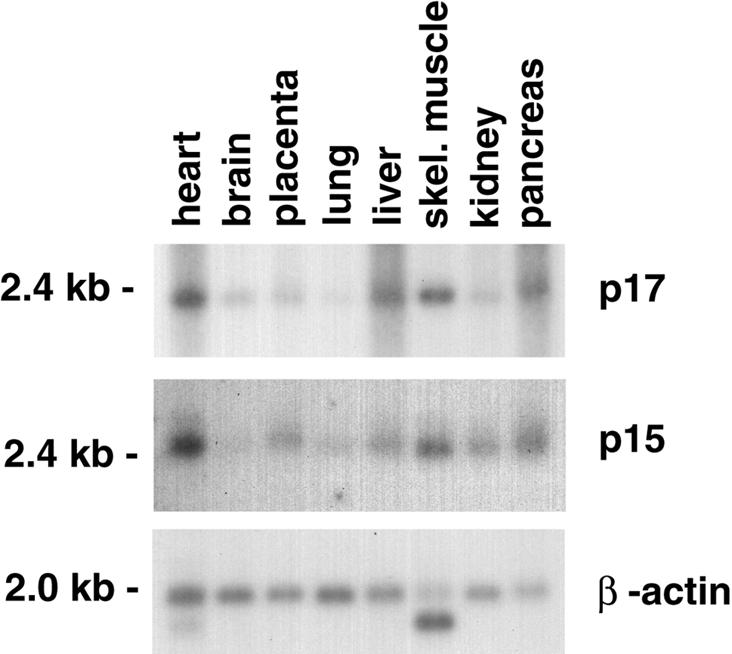
Fig. 7. Relative amounts of p15 and p17 mRNA in human tissues. Multiple tissue northern blot analysis of p15 and p17 mRNA, and β-actin mRNA as a loading control. The tissue and the position of size markers are indicated.
Discussion
HuCHRAC is a human chromatin remodelling complex with subunit homology to Drosophila chromatin remodelling complexes
We describe a novel chromatin remodelling complex from HeLa nuclear extract, HuCHRAC. HuCHRAC contains four subunits, all of which are homologous to proteins previously found in the Drosophila chromatin remodelling complexes ACF and/or CHRAC. HuCHRAC contains predominantly the hISWI isoform hSNF2H. We found minor amounts of the hSNF2L isoform, suggesting that at least a fraction of hSNF2L is present in a HuCHRAC-like complex. The 185 kDa subunit of HuCHRAC is hACF1, the human homologue of Drosophila Acf1, the large subunit of the chromatin remodelling complex ACF (Ito et al., 1999). Drosophila Acf1 is also the 185 kDa subunit of CHRAC (S.Ferrari, A.Eberharter, T.Straub, P.Varga-Weisz and P.B.Becker, in preparation). Drosophila Acf1 is required for optimal in vitro nucleosome assembly activity of ISWI (Ito et al., 1999). HuCHRAC also contains two putative histone-fold proteins, p15 and p17, which are the human homologues of novel proteins identified in the Drosophila CHRAC complex (Corona et al., 2000). However, HuCHRAC and Drosophila CHRAC seem to be different in one respect: whereas the Drosophila complex was reported to contain a dimer of topo II (Varga-Weisz et al., 1997), HuCHRAC does not contain topo II. We show that topo IIα and topo IIβ do not co-purify with HuCHRAC and do not co-immunoprecipitate with hISWI and p17. The absence of topo II does not alter the nucleosome spacing activity of HuCHRAC, as compared with Drosophila CHRAC. Also, the gel filtration behaviour of HuCHRAC is similar to that of its Drosophila counterpart (Varga-Weisz et al., 1997; this study). Recent observations on Drosophila CHRAC support our findings: topo II could be removed from CHRAC without affecting its gel filtration profile or catalytic activity (S.Ferrari, A.Eberharter, T.Straub, P.Varga-Weisz and P.B.Becker, in preparation).
The conservation of subunit composition in ISWI-containing chromatin remodelling factors from fly embryos to human carcinoma cells suggests that these factors have biological roles that are common between these divergent organisms at different developmental stages. In addition, it argues against a ‘promiscuous’ ISWI ATPase, and suggests that ISWI interacts with a limited set of proteins.
Recently, three other hSNF2H complexes have been described: RSF, a two-subunit complex containing RSF325, which is unrelated to any known protein (LeRoy et al., 1998); WCRF (Bochar et al., 2000); and human ACF (LeRoy et al., 2000). WCRF and hACF are both purified from HeLa nuclear extract as two-subunit complexes containing hACF1 (named as WCRF180 and BAZ1A, respectively). Although their reported subunit composition is identical, gel filtration analysis suggests that hACF has an apparent mol. wt of 300–400 kDa while WCRF runs as a 600–700 kDa complex (Bochar et al., 2000; LeRoy et al., 2000). HuCHRAC differs from WCRF/hACF in the presence of the p15–p17 putative histone-fold pair. An explanation for this discrepancy may be that p15–p17 are loosely associated with HuCHRAC and therefore easily lost, as happened partially in our final purification step (Figure 1D). We also find that p15–p17 are enriched in early hISWI-containing gel filtration fractions (Figure 1C, fraction 23). This may suggest loss of p15–p17 from a fraction of the hISWI complexes, presumably causing their later gel filtration elution due to a smaller size. Alternatively, WCRF and/or hACF do contain p15–p17, but they have remained undetected, due to their small size.
Human ACF1 is a member of a family of related proteins, which may target heterochromatin
We find three human proteins related to hACF1: WSTF and two uncharacterized proteins, which we named hWALp3 and hWALp4, being the third and fourth human members of a family of WAL proteins (Figure 2B and C). All human WAL family members have a WAKZ motif (Ito et al., 1999) followed directly by a PHD finger, and a C-terminal bromodomain. PHD fingers and bromodomains are found in many proteins that function in chromatin remodelling processes and that have been suggested to interact specifically with chromatin components (Aasland et al., 1995; Winston and Allis, 1999).
Human and Drosophila Acf1, and WSTF contain the WAC motif at their N-terminus, a novel protein domain of unknown function (Ito et al., 1999). Recently, the N-terminal 400 amino acids of the mouse homologue of hACF1 (encoded by Ct146) were isolated by a gene-trapping strategy (Tate et al., 1998). The ACF1 N-terminal peptide fused to a reporter localized to pericentric heterochromatin in differentiated mouse cells (Tate et al., 1998). Of the >100 nuclear peptides isolated with this strategy, only 2% are targeted to this location and, where tested, their specific nuclear localization faithfully reflected the localization of the endogenous protein (McDowell et al., 1999; W.A.Bickmore, unpublished data). The WAC motif is the only obvious conserved domain in this N-terminal sequence of mouse ACF1 (Ito et al., 1999; this study), suggesting that this domain may be sufficient for targeting to pericentric heterochromatin. By extrapolation, hACF1 may target HuCHRAC to heterochromatin, where its remodelling activity could play a role in the assembly, disassembly or maintenance of the local chromatin structure.
hWALp3, hWALp4 and their C.elegans homologue Ce wal-3 have a different amino acid sequence at their N-terminus. hWALp3 contains the TIP5 (TTF1-interacting peptide 5) sequence, which interacts with TTF-I, an RNA polymerase I (pol I) transcription factor (Jansa, 1998; Figure 2B). Nucleosome-bound TTF-I synergizes with ATP-dependent co-factors, present in Drosophila embryo extracts and mouse cell extracts, to position a nucleosome over the transcription start site, which is a prerequisite for transcription activation (Längst et al., 1998, and references therein). This opens up the possibility that a chromatin remodelling complex containing hWALp3 is targeted to pol I promoters by TTF-I via the TIP5 peptide in hWALp3.
Interestingly, hWALp3 and hWALp4 contain an amino acid motif that is homologous to the MBD of methylated DNA-binding proteins, such as MeCP2 (Lewis et al., 1992). Methylated DNA is found primarily in transcriptionally repressed chromatin and heterochromatin (for a review see Bird and Wolffe, 1999). MeCP2 is targeted specifically to methylated CpGs in the pericentric heterochromatin of mouse cells via its MBD (Nan et al., 1996). We suggest that all members of the WAL protein family have a heterochromatin-targeting domain: the WAC motif in ACF1 and WSTF and the putative MBD in WALp3 and WALp4.
Possible in vivo functions of HuCHRAC
Although a number of ISWI complexes have been purified from different organisms, the in vivo function of ISWI is unclear. Experiments with Drosophila ACF and NURF (Ito et al., 1997; Mizuguchi et al., 1997) and human RSF (LeRoy et al., 1998) have shown that these ISWI complexes can facilitate transcription initiation on a chromatin template in vitro. However, ISWI does not co-localize with RNA polymerase II on Drosophila polytene chromosomes (Deuring et al., 2000), suggesting that it is not generally required for transcription initiation in vivo. A characteristic feature of several ISWI-containing complexes is their capacity to assemble a spaced nucleosomal array. The finding that human ISWI co-purifies with a protein that has a heterochromatin-targeting domain raises the possibility that the nucleosome spacing activity of ISWI has a function in the formation or maintenance of heterochromatin, which is characterized by very regularly spaced nucleosomes (Cryderman et al., 1999). Mutations in the ISWI protein caused decondensation of the Drosophila male X chromosome, suggesting a function for ISWI in the maintenance of higher order chromatin structure (Deuring et al., 2000).
Alternatively, HuCHRAC components may be stored at sites of heterochromatin, but function elsewhere in the genome. Another mammalian protein, ATRX, the sequence of which suggests that it may be a chromatin remodelling factor, was found to be located predominantly at sites of constitutive heterochromatin in human and mouse cells (McDowell et al., 1999). However, the phenotype of individuals with mutations in ATRX demonstrates a role for this protein in the expression of genes located in euchromatin (e.g. α-globin).
The functional relevance of the two small histone-fold proteins in Drosophila and human CHRAC is unclear. Addition of extra, recombinant, p15–p17 complex to HuCHRAC did not improve or alter its chromatin remodelling activities in the assays we performed. This was not unexpected since Drosophila ACF, which lacks the histone-fold proteins, has remodelling activities similar to those described for HuCHRAC and Drosophila CHRAC (Ito et al., 1999). The DNA-binding properties of the p15–p17 complex possibly are relevant for incorporation of p15–p17 into chromatin, aided by the nucleosome remodelling activity of hSNF2H plus hACF1. The proposed targeting of HuCHRAC to heterochromatin via hACF1 may provide the specificity in this process.
Materials and methods
Antibodies
hSNF2L cDNA encoding amino acids 647–884 of the hSNF2L-2 protein (Okabe et al., 1992; cDNA provided by ATCC, IMAGE consortium clone ID 79559) was cloned into pGEX-4T-3 (Pharmacia) and expressed as a GST fusion protein in E.coli strain BL21 (DE3) (Harper and Speicher, 1997). The fusion protein was injected into rabbits as washed inclusion bodies (Harlow and Lane, 1988). Full-length p17 cDNA was amplified by PCR from a p17 cDNA-containing plasmid (IMAGE consortium clone ID 291522; Lennon et al., 1996). p17 cDNA was cloned into pGEX-4T-3 (Pharmacia) and expressed as a p17–GST fusion protein in BL21 (DE3) (Harper and Speicher, 1997). Soluble p17–GST was injected into rabbits for antibody production. The p15-encoding ORF in p15 cDNA (IMAGE consortium clone ID 40740) was amplified by PCR and inserted into vector pRK-3HA-10His. This places one HA tag at the C-terminus of p15. The p15 coding region together with the C-terminal HA epitope tag were amplified from pRK-p15-HA-His10 by PCR, inserted into the pET-30 Ek/LIC plasmid (Novagen) and expressed in BL21 (DE3). Anti-p15 antiserum was raised by injection of recombinant S-p15-HA-containing inclusion bodies into rabbits. Antiserum against hACF1 was raised by injection of the peptide RQKPPADLRPDEEVFYC (amino acids 10–26) coupled to keyhole limpet haemocyanin into rabbits. Polyclonal rabbit anti-topo IIα and β antibodies were from Biotrend, Cologne.
Purification of HuCHRAC
Nuclear extract from HeLa S3 cells was prepared as described by Dignam et al. (1983). A 20 ml nuclear extract (140 mg protein) in 100 mM KCl (BC-100): 20 mM HEPES pH 7.6, 20% glycerol, 100 mM KCl, 0.2 mM EDTA, 3 mM dithiothreitol (DDT), 0.2 mM phenylmethylsulfonyl fluoride (PMSF) was loaded onto a 20 ml P11 cellulose phosphate column (Whatman). The column was washed with BC-100 and step-eluted with BC-300, BC-500 and BC-850. The BC-850 fraction was diluted with BC-0 to 150 mM KCl and fractionated on an 8 ml Mono S column (Pharmacia) with a salt gradient from 150 mM to 1 M KCl in Mono S buffer: 20 mM HEPES pH 7.6, 1 mM MgCl2, 10% glycerol, 0.5 mM EGTA, 0.1 mM EDTA, 1 mM DTT. ISWI-containing fractions were size fractionated with a 25 ml Superose 6 gel filtration column (Pharmacia), as described (Varga-Weisz et al., 1997). Human ISWI-containing fractions (23 + 24) in Mono S-200 were loaded onto CM-Sepharose Fast Flow (Pharmacia, 0.2 ml). The column was washed with Mono S-200 and eluted with Mono S-450.
Tryptic digestion, nanoLC ion trap mass spectrometry and sequencing
SDS–PAGE excised bands were subjected to in-gel reduction, carboxyamido methylation and tryptic digestion (Promega). Multiple peptide sequences were determined in a single run by microcapillary reverse-phase chromatography directly coupled to a Finnigan LCQ DECA quadrupole ion trap mass spectrometer equipped with a custom nanoelectrospray source. The column was packed in-house with 5 cm of C18 support into a New Objective one-piece 75 µm i.d. column terminating in a 15 µm tip. The flow rate was 190 nl/min. The ion trap was programmed to acquire full scan MS surveying the range 395–1295 m/z, followed by successive sets of two data-dependent scans on the three most abundant ions in that survey. MS/MS spectra were acquired with a relative collision energy of 30%, an isolation width of 2.5 Da and recurring ions dynamically excluded. Interpretation of the resulting MS/MS spectra of the peptides was facilitated by database correlation with the algorithm SEQUEST (Eng et al., 1994) and programs developed in the Harvard Microchemistry Facility (Chittum et al., 1998).
Bioinformatic analysis
EMBL, GenBank and EST databases were queried with the BLAST 2.0 algorithm (www.ncbi.nlm.nih.gov; Altschul et al., 1997). Protein domain and motif analysis was done with PROSITE (www.isrec.isb-sib.ch) and the pileup analysis and phylogeny trees were constructed with the PILEUP and PAUPSEARCH programs (GCG 10 package; www.hgmp.mrc.ac.uk). Chromosomal localization was through cross-referencing matching ESTs with the radiation hybrid mapped ESTs found in the UniGene database (NCBI web server). The C.elegans WALp proteins were determined using the BLAST algorithm against the WORMPEP database (www.sanger.ac.uk).
Sequence analysis of hACF1
An RT–PCR (Superscript-One Step RT–PCR, Gibco-BRL) was set up with primers complementary to the 3′ end of EST AA765931 (the 3′ end of the human homologue of mouse Ct146, Tate et al., 1998), and the 5′ end of EST AL050089, and 1 µg of HeLa total RNA as template. The resulting band was gel purified (Qiagen) and used directly as a template for PCR cycle sequencing (Big-Dye, Perkin-Elmer Cetus). The full-length cDNA sequence for hACF1 was determined by joining ESTs with the sequences obtained using RT–PCR analysis. Experimental details are available on request.
Immunoprecipitations
A 60 µl aliquot of anti-hISWI, anti-p17 serum or pre-immune serum was added to 210 µl of HeLa nuclear extract in BC buffer with 300 mM NaCl + 5 µl of 50× Complete™ protease inhibitor (Roche) and incubated for 2 h on ice. Protein A–Sepharose CL-4B beads (Pharmacia) were washed twice with IP-400 (20 mM HEPES pH 7.6, 5 mM MgCl2, 400 mM KCl, 0.1 mM EDTA, 1 mM DDT, 0.1% NP-40, 20% glycerol), once with IP-400 + 1 mg/ml ovalbumin, and 25 µl of washed beads were added to the serum plus nuclear extract and mixed for 2 h at 4°C. Beads were washed four times with 400 µl of IP-400, once with 150 µl of IP-150 and boiled with 40 µl of Laemmli buffer. Supernatants were analysed by western blotting.
Recombinant p15 and p17 proteins
BL21 (DE3) cells containing pET-30-p15-HA (see above) and pGEX-p17 (see above) were induced at an OD600 of 0.5 with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and harvested after 3 h at 37°C. The p15–p17 complex was purified using glutathione–Sepharose 4B (Pharmacia; Harper and Speicher, 1997) and released from the resin with thrombin protease (Pharmacia, 60 U/ml resin), which cleaves between the GST tag and the fused protein (in this case p17). The p15–p17 complex was purified further with S-protein–agarose (Novagen) according to the manufacturer’s instructions. Ten units of enterokinase-protease (Novagen, which cuts between the S tag and the fusion protein) was added to 1 ml of resin in a total volume of 2 ml and mixed for 10 h at room temperature. Enterokinase was removed with 0.5 ml of Ekapture–agarose (Novagen). Protein expression and purification of p15-HA alone with S-protein–agarose was as described above followed by chromatography with a 1 ml Mono Q column (Pharmacia). p17 protein alone was purified from BL21 (DE3) cells expressing p17–GST (see above) by release from glutathione–Sepharose 4B resin with thrombin cleavage.
Gel shift analysis
A DNA fragment, 150 bp in size and randomly chosen (sequence available upon request), was generated by PCR with one 32P-end-labelled primer. Binding was in 20 mM HEPES pH 7.6, 0.5 mM EDTA, 0.05% NP-40, 10% glycerol, 60 µg/ml acetylated bovine serum albumin (BSA; Sigma) plus 4.4 ng of a 150 bp 32P-labelled DNA fragment and 17.5 ng or excess (1 µg) of unrelated, unlabelled, plasmid DNA of 5.2 kb and the indicated amounts of p17, p15-HA or p15-HA–p17 complex, in a reaction volume of 8 µl. Mobility shift analysis was performed by electrophoresis through a 4% acrylamide gel in 0.5× TBE at 4°C. The p15-HA–p17 complex was supershifted with 0.3 µg of monoclonal anti-HA antibody (Roche). The supershift was competed with 4 µg of HA-peptide (Roche). Mononucleosomes were assembled on the same 150 bp fragment with purified HeLa histones by the salt dialysis method (Varga-Weisz et al., 1999).
Binding studies with DNA and nucleosomes coupled to magnetic beads
The same 150 bp DNA fragment as used in the gel shift analysis (see above) was generated by PCR with one primer having a biotinylated nucleotide at its 5′ end. This fragment was either coupled directly to streptavidin-coated magnetic beads or first assembled into a nucleosome, as described (Varga-Weisz et al., 1999). A 100 ng aliquot of bead-coupled naked DNA, or nucleosomal DNA, or the equivalent amount of uncoupled beads was washed twice with 0.5 ml of washing buffer (20 mM HEPES pH 7.6, 0.5 mM EGTA, 0.05% NP-40, 10% glycerol, 1 mM DTT) and resuspended in 10 µl of binding buffer (20 mM HEPES pH 7.6, 0.5 mM EGTA, 0.1 mM EDTA, 0.2 mM MgCl2, 20 mM KCl, 0.04% NP-40, 10% glycerol, 1 mM DTT) containing 0.4 µg of p17–p15 complex. After mixing for 1 h at 4°C, the beads were washed four times with 20 µl of washing buffer. Bound proteins were extracted by boiling in Laemmli buffer and analysed by western blot with a monoclonal anti-HA antibody (Roche) against HA epitope-tagged p15, or the p17 antibody.
Northern blots
Northern blot analysis was with a Human Multiple Tissue Northern Blot from Clontech, California, with ∼2 µg of poly(A)+ RNA per lane. The blot was probed with 32P-labelled ORF cDNA of p15 or p17, or human β-actin cDNA, according to the manufacturer’s protocol. Labelling was with the High Prime DNA labelling kit (Roche).
Chromatin assays
The nucleosome spacing assay using HH4.8 plasmid DNA was essentially as described (Varga-Weisz et al., 1997, 1999). Nucleosome spacing reactions were assembled by adding 40 µl of HuCHRAC (Superose 6 fraction) or Drosophila CHRAC in Ex100 (20 mM HEPES pH 7.6, 100 mM KCl, 5 mM MgCl2, 0.5 mM EGTA, 10% glycerol, 1 mM DTT) to 10 µl of sarkosyl-stripped chromatin in Ex120 (contains ∼40 ng of DNA). Quantities of HuCHRAC or Drosophila CHRAC added to the reaction (in fmol) were estimated from silver-stained SDS–gels, using BSA as a standard. A 1 µl aliquot of 100 mM ATP in Ex120 or of Ex120 was added to the reaction and the mixture was incubated at 28°C for 1 h. The chromatin was digested at 28°C by addition of 70 µl of micrococcal nuclease (50 U/ml in Ex120/5 mM CaCl2), for 30 s or 1 min before reactions were stopped. Purified DNA was analysed by Southern blot (Varga-Weisz et al., 1995).
Accession numbers
The following nucleotide sequences have been deposited in the DDBJ/EMBL/GenBank database: hACF1, accession No. AF213467; HuCHRAC-15, accession No. AF226076; HuCHRAC-17, accession No. AF226077
Acknowledgments
Acknowledgements
We thank Custodia Garcia-Jimenez (Marie Curie Research Institute, Oxted) for plasmid constructs, extracts and discussion, William S.Lane (Harvard, Cambridge, MA) for mass spectrometry, and Thomas Oelgeschläger (Marie Curie Research Institute, Oxted) for HeLa extract fractions. Robert Roeder kindly provided an antiserum against hISWI. We thank Thomas Oelgeschläger, Peter O’Hare and Enrique Castano for critical reading of the manuscript. This research was funded by the Marie Curie Cancer Care, a grant from the Association for International Cancer Research (AICR), St Andrews, Scotland, to R.A.P. and a post-doctoral fellowship from the Medical Research Council of Canada to G.D.
References
- Aasland R., Gibson,T. and Stewart,A.F. (1995) The PHD-finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci., 20, 56–59. [DOI] [PubMed] [Google Scholar]
- Aihara T., Miyoshi,Y., Koyama,K., Suzuki,M., Takahashi,E., Monden,M. and Nakamura,Y. (1998) Cloning and mapping of SMARCA5 encoding hSNF2H, a novel human homologue of Drosophila ISWI. Cytogenet. Cell Genet., 81, 191–193. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden,T.L., Schaffen,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arents G. and Moudrianakis,E.N. (1995) The histone fold: a ubiquitous architectural motif utilised in DNA compaction and protein dimerization. Proc. Natl Acad. Sci. USA, 92, 11170–11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxevanis A.D., Arents,G., Moudrianakis,E.N. and Landsman,D. (1995) A variety of DNA-binding and multimeric proteins contain the histone fold motif. Nucleic Acids Res., 23, 2685–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A.P. and Wolffe,A.P. (1999) Methylation-induced repression—belts, braces, and chromatin. Cell, 99, 451–454. [DOI] [PubMed] [Google Scholar]
- Bochar D.A., Savard,J., Wang,W., Lafleur,D.,W., Moore,P., Côté,J. and Shiekhattar,R., (2000) A family of chromatin remodeling factors related to Williams syndrome transcription factor. Proc. Natl Acad. Sci. USA, 97, 1038–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittum H.S., Lane,W.S., Carlson,B.A., Roller,P.P., Lung,F.T., Lee,B.J. and Hatfield,D.L. (1998) β-Globin is extended beyond its UGA stopcodon by multiple suppressions and translational reading gaps. Biochemistry, 37, 10866–10870. [DOI] [PubMed] [Google Scholar]
- Corona D.F.V., Längst,G., Clapier,C.R., Bonte,E.J., Ferrari,S., Tamkun,J.W. and Becker,P.B. (1999) ISWI is an ATP-dependent nucleosome remodeling factor. Mol. Cell, 3, 239–245. [DOI] [PubMed] [Google Scholar]
- Corona D.F.V., Budde,A., Deuring,R., Ferrari,S., Varga-Weisz,P.D., Wilm,M., Tamkun,J.W. and Becker,P.B. (2000) Two histone fold proteins, CHRAC-14 and CHRAC-16, are developmentally regulated subunits of chromatin accessibility complex (CHRAC). EMBO J., 19, 3049–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryderman D.E., Tang,H., Bell,C., Gilmour,D.S. and Wallrath,L.L. (1999) Heterochromatin silencing of Drosophila heat shock genes acts at the level of promoter potentiation. Nucleic Acids Res., 16, 3364–3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuring R. et al. (2000) The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol. Cell, 5, 355–365. [DOI] [PubMed] [Google Scholar]
- Dignam J.D., Lebovitz,R.M. and Roeder,R.G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res., 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng J.K., McCormick,A.L. and Yates,J.R.,III (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom., 5, 976–989. [DOI] [PubMed] [Google Scholar]
- Harlow E. and Lane,D. (1988) Antibodies. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Harper S. and Speicher,D.W. (1997) Expression and purification of GST fusion proteins. In Coligan,J.E., Dunn,B.M., Ploegh,H.L., Speicher,D.W. and Wingfield,P.T. (eds), Current Protocols in Protein Science. John Wiley and Sons, New York, NY, pp. 6.6.1–6.6.21. [Google Scholar]
- Hendrich B. and Bird,A. (1998) Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol., 18, 6538–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Bulger,M., Pazin,M.J., Kobayashi,R. and Kadonaga,J.T. (1997) ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell, 90, 145–155. [DOI] [PubMed] [Google Scholar]
- Ito T., Levenstein,M.E., Fyodorov,D.V., Kutach,A.K., Kobayashi,R. and Kadonaga,J.T. (1999) ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev., 13, 1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansa P., Mason,S.W., Hoffmann-Rohrer,U. and Grummt,I. (1998) Cloning and functional characterization of PTRF, a novel protein which induces dissociation of paused ternary transcription complexes. EMBO J., 17, 2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.H., Hamana,N., Nezu,J. and Shimane,M. (2000) A novel family of bromodomain genes. Genomics, 63, 40–45. [DOI] [PubMed] [Google Scholar]
- Kingston R.E. and Narlikar,G.J. (1999) ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev., 13, 2339–2352. [DOI] [PubMed] [Google Scholar]
- Längst G., Becker,P.B. and Grummt,I. (1998) TTF-I determines the chromatin architecture of the active rDNA promoter. EMBO J., 17, 3135–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon G.G., Auffray,C., Polymeropoulos,M. and Soares,M.B. (1996) The IMAGE consortium: an integrated molecular analysis of genomes and their expression, Genomics, 33, 151–152. [DOI] [PubMed] [Google Scholar]
- LeRoy G., Orphanides,G., Lane,W.S. and Reinberg,D. (1998) Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science, 282, 1900–1904. [DOI] [PubMed] [Google Scholar]
- LeRoy G., Loyola,A., Lane,W.S. and Reinberg,D. (2000) Purification and characterization of a human factor that assembles and remodels chromatin. J. Biol. Chem., 275, 14787–14790. [DOI] [PubMed] [Google Scholar]
- Lewis J.D., Meehan,R.R., Henzel,W.J., Maurer-Fogy, Jeppesen,P., Klein,F. and Bird,A. (1992) Purification, sequence and cellular localisation of a novel chromosomal protein that binds to methylated DNA. Cell, 69, 905–914. [DOI] [PubMed] [Google Scholar]
- Lu X., Meng,X., Morris,C.A. and Keating,M.T. (1998) A novel human gene, WSTF, is deleted in Williams syndrome. Genomics, 54, 241–249. [DOI] [PubMed] [Google Scholar]
- Maity S.N. and de Crombrugghe,B. (1998) Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem. Sci., 23, 174–178. [DOI] [PubMed] [Google Scholar]
- McDowell T.L. et al. (1999) Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes. Proc. Natl Acad. Sci. USA, 96, 13983–13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G., Tsukiyama,T., Wisniewski,J. and Wu,C. (1997) Role of nucleosome remodeling factor NURF in transcriptional activation of chromatin. Mol. Cell, 1, 141–150. [DOI] [PubMed] [Google Scholar]
- Nan X., Tate,P., Li,E. and Bird,A. (1996) DNA methylation specifies chromosomal localization of MeCP2. Mol. Cell. Biol., 16, 414–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe I., Bailey,L.C., Attree,O., Srinivasan,S., Perkel,J.M., Laurent,B.C., Carlson,M., Nelson,D.L. and Nussbaum,R L. (1992) Cloning of human and bovine homologs of SNF2/SWI2: a global activator of transcription in yeast S.cerevisiae. Nucleic Acids Res., 20, 4649–4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peoples R.J., Cisco,M.J., Kaplan,P. and Francke,U. (1998) Identification of the WBSCR9 gene, encoding a novel transcriptional regulator, in the Williams–Beuren syndrome deletion at 7q11.23. Cytogenet. Cell Genet., 82, 238–246. [DOI] [PubMed] [Google Scholar]
- Tate P., Lee,M., Tweedie,S., Skarnes,W.C., and Bickmore,W.A. (1998) Capturing novel mouse genes encoding chromosomal and other nuclear proteins. J. Cell Sci., 111, 2575–2585. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T. and Wu,C. (1995) ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kD subunit of the nucleosome remodeling factor. Cell, 83, 1021–1026. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T., Palmer,J., Landel,C.C. and Shiloach,J. and Wu,C. (1999) Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev., 13, 686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga-Weisz P.D., Blank,T.A. and Becker,P.B. (1995) Energy-dependent chromatin accessibility and nucleosome mobility in a cell-free system. EMBO J., 14, 2209–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga-Weisz P.D., Wilm,M., Bonte,E., Dumas,K., Mann,M. and Becker,P.B. (1997) Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature, 388, 598–602. [DOI] [PubMed] [Google Scholar]
- Varga-Weisz P.D., Bonte,E.J. and Becker,P.B. (1999) Analysis of modulators of chromatin structure in Drosophila. Methods Enzymol., 304, 742–757. [DOI] [PubMed] [Google Scholar]
- Winston F. and Allis,C.D. (1999) The bromodomain: a chromatin-targeting module? Nature Struct. Biol., 7, 601–604. [DOI] [PubMed] [Google Scholar]
- Workman J.L. and Kingston,R.E. (1998) Alterations of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem., 67, 545–579. [DOI] [PubMed] [Google Scholar]
- Zemzoumi K., Frontini,M., Bellorini,M. and Mantovani,R. (1999) NF-Y histone fold α1 helices help impart CCAAT specificity. J. Mol. Biol., 286, 327–337. [DOI] [PubMed] [Google Scholar]