Abstract
The ability of animals to regenerate missing parts is a dramatic and poorly understood aspect of biology. The sources of new cells for these regenerative phenomena have been sought for decades. Recent advances involving cell fate tracking in complex tissues have shed new light on the cellular underpinnings of regeneration in Hydra, planarians, zebrafish, Xenopus, and Axolotl. Planarians accomplish regeneration with use of adult pluripotent stem cells, whereas several vertebrates utilize a collection of lineage-restricted progenitors from different tissues. Together, an array of cellular strategies—from pluripotent stem cells to tissue-specific stem cells and dedifferentiation—are utilized for regeneration.
Sources of new cells in animal regeneration
The ability to regenerate is widespread in the animal kingdom, with representatives from most animal phyla displaying the ability to regrow missing body parts (Brockes et al., 2001; Sánchez Alvarado, 2000). Prominent examples include cnidarians such as Hydra, annelids, molluscs, nemertean worms, platyhelminthes such as planarians, and chordates including vertebrates. The regenerative capacities of these animals vary. Planarians, for instance, are capable of regenerating missing heads or entire bodies from small fragments, whereas salamanders are capable of regrowing missing limbs. In this review we discuss work in classic animal regeneration model systems, which are capable of regenerating large missing parts of their bodies.
Experimentation with regeneration dates back to the 1700s and the experiments of Abraham Trembley with Hydra (Lenhoff and Lenhoff, 1986). There are many questions that have captured the imagination of the generations of biologists who have since seen new heads and limbs growing from injured animals. How does the process start? How do the wounded tissues specify what to make? Where do the new cells come from? Recently, significant progress clarifying the source of new cells for regeneration has been made in multiple different regenerative contexts and is therefore the focus of this review.
Regenerative phenomena in the animal kingdom involve differences in the number of cell types to be made, ranging from replacing a single cell type (such as in the case of the salamander lens) to replacing all the cells within a region of the body (such as in the case of planarian regeneration). In the case of the salamander lens, the dorsal iris normally regenerates the missing lens. Because a dorsal iris placed into a regenerating limb still regenerates a lens, the regenerative potential of the dorsal iris appears to be restricted and unipotent (Reyer et al., 1973; Tsonis et al., 2004; Wolff, 1895). By contrast, at the tissue-scale level, a small piece of planarian tissue can be considered pluripotent because it can regenerate all cell types of the entire organism, including cell types typically made in the embryo from the three embryonic germ layers (Reddien and Sánchez Alvarado, 2004). A crucial issue for understanding planarian regeneration, however, is how this capacity of tissues to regenerate all adult cell types is achieved at the level of individual cells (Figure 1). The regenerative pluripotency at the tissue scale could be achieved either by the action of pluripotent cells that, as individual cells, have the potential to produce all cells of the body. Alternatively, tissue-level pluripotency could be attained via the collective action of multiple cell types that each has different, restricted potential.
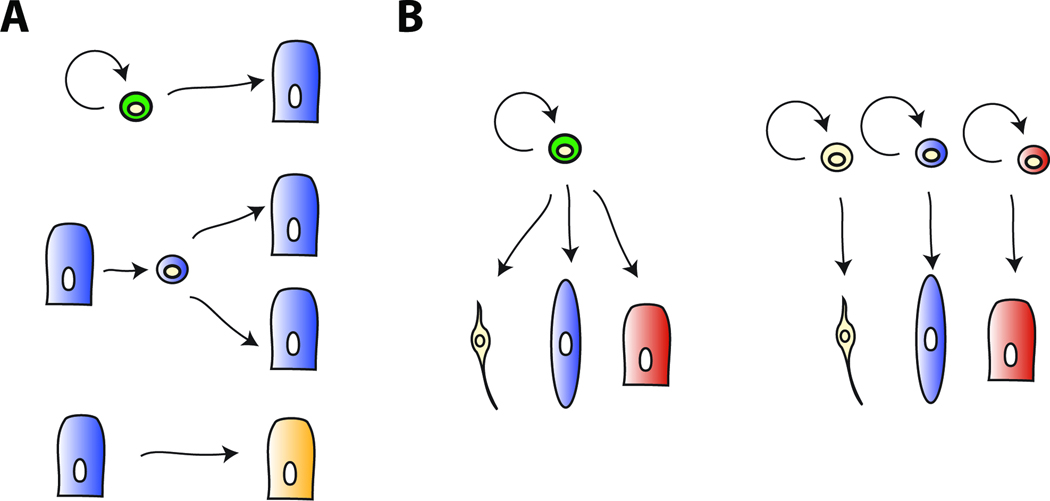
Figure 1. Sources for new cells in regeneration.
A. Top, stem cells self-renew and produce one or more differentiated cells. Middle, dedifferentiation is the process by which a cell loses differentiated character to produce a progenitor cell that can divide to produce more differentiated cells. Bottom, transdifferentiation involves the change of one cell type into others. This could occur without division, or following de-differentiation of one cell type into a progenitor for additional cell types. B. Distinct ways for accomplishing tissue-level pluripotency. Left, a pluripotent progenitor cell (a stem cell is depicted) produces differentiated progeny cells spanning multiple germ layers. There could exist multiple, and/or self-renewing intermediates along different lineage paths. Right, different lineage-restricted progenitor cells (stem cell types are depicted) each produce different differentiated cells. Each different tissue separately generates or harbors a restricted stem cell. These stem cells together can reconstitute the three different tissues, while any individual on its own is not sufficient.
There are multiple possible means by which injured tissues could provide new cells for regeneration (Figure 1A). First, new cell types could be produced by resident stem cells. Stem cells are a type of cell that self renews (dividing to produce more cells like itself) and can produce one or more differentiated cell types (Weissman et al., 2001). Second, new cells could be produced through de-differentiation – loss of the differentiated character of a cell type – to produce a dividing cell that acts as a progenitor cell (Jopling et al., 2011). Finally, new cell types could arise as a result of transdifferentiation, or a change in state of one cell type into another (Jopling et al., 2011; Selman and Kafatos, 1974). Transdifferentiation could happen without cell division, or via a progenitor cell produced by de-differentiation. Multiple of these candidate sources of new cells could in principle act in concert to allow regeneration of a complex tissue. For any specific cell type that acts as a source for new cells, whether it functions as a stem cell or through de-differentiation to a progenitor state, it is important to determine the developmental potential of that cell type in regeneration (unipotent, multipotent, or pluripotent).
Determination of the source of new cell types in regeneration connects the trait of regeneration at the organismal scale to cellular behaviors that can be studied on a molecular, mechanistic level. Only a fraction of the cells at the injury site may represent the source cells for the regenerating tissue. Important work over recent years has identified tissue interactions and signaling molecules associated with injured tissues that are required for proper regeneration (for reviews see (Adell et al., 2010; Antos and Tanaka, 2010; Forsthoefel and Newmark, 2009; Poss, 2010; Reddien, 2011; Stoick-Cooper et al., 2007; Yokoyama, 2008)). Mechanistic analyses of molecules involved in regeneration have been hampered by the inability to precisely identify the source cells for regeneration, and to follow how they undergo proliferation, patterning, and differentiation. An important next step in the regeneration field will be to define how the molecular changes that occur upon tissue removal control the biology of progenitor cells for the regenerating tissue.
Identifying the cellular underpinnings of regeneration has historically been a difficult challenge. Various models and hypotheses for the cellular basis of regeneration have been posited and debated for decades. The lack of clarity can in part be explained by limitations in tools available for cell-lineage experiments. Development of cellular and molecular tools for study of highly regenerative animals has lagged far behind the case for other organisms that have been the workhorses of molecular and developmental biology. However, new tools are rapidly emerging that allow a new generation of experiments to address the fundamental questions of regeneration. Recent work in several classic regenerative organisms has thus begun to shed light on the central and long-standing topic of the cellular explanation for regeneration.
cNeoblasts: Adult pluripotent stem cells and the source of new cells in planarian regeneration
Planarians are flatworms and one of the classic model systems for the study of regeneration. Planarians are capable of re-growing new heads, tails, sides, or even entire organisms from tiny body fragments (Morgan, 1898; Randolph, 1897). Planarians possess bilateral symmetry and complex internal anatomy, including nervous system, musculature, excretory system, epidermis, eyes, and intestine (Hyman, 1951; Reddien and Sánchez Alvarado, 2004). Planarian regeneration involves changes in pre-existing tissues and formation of an outgrowth at wounds called a blastema, in which missing tissues are produced. Because small fragments of tissue can regenerate new animals, all of the various organ systems and cell types of the body can be produced in the adult. Therefore, pluripotency at the tissue-scale level exists in adult planarians. Planarians of the species Schmidtea mediterranea come in two types: sexual animals that are crossfertilizing hermaphrodites, and asexual animals that reproduce by transverse fission and regeneration. Because entire adult strains of animals can be generated by amputation and regeneration, including animals capable of sexual reproduction, adult planarian tissues could be considered to possess totipotency (for adult cell types). Furthermore, because any planarian body region containing roughly 10,000 or more cells (Montgomery and Coward, 1974) (with the exception of the tip of the head and the pharynx) can regenerate an entire animal, this attribute of tissue-scale pluripotency is spread throughout the planarian body. How this widespread, adult tissue pluripotency in planarians is explained at the level of cells has been explored for over a century.
Planarian regeneration requires a proliferative cell population
A population of adult dividing cells, called "neoblasts", has long been prominent in planarian regeneration research. In the late 1800s, dividing cells with simple morphology were described to exist in the bodies of flatworms (Curtis, 1902; Keller, 1894; Lehnert, 1891; Wagner, 1890). These cells have gone by multiple names, such as Stammzellen and formative cells (Wolff, 1962), but eventually the name neoblast became affixed to these cells (Buchanan, 1933; Dubois, 1949; Wolff, 1962). Providing one name to a cell population sometimes led to the perception that all adult dividing planarian cells are the same (which would imply all neoblasts are pluripotent). However, the term neoblast, by contrast, has historically described all adult somatic planarian cells that are dividing and not necessarily a specific single cell type. This distinction is important, because dividing cells in many cell populations are frequently very heterogeneous (Spangrude et al., 1988), and neoblasts could therefore consist of a collection of very different cell types. Hereafter in this review, the word neoblast is used strictly to refer to all somatic dividing cells in adult planarians.
Dividing somatic cells (neoblasts) are distributed throughout the planarian body in a tissue region called the parenchyma, which is beneath the basement membrane and body wall musculature, and surrounds the intestine and nervous system (Figure 2A) (Hyman, 1951; Pedersen, 1961). Dividing cells are absent from the tip of the animal head and the animal pharynx, providing a candidate explanation for the inability of these two regions to regenerate other parts of the body in isolation (Newmark and Sánchez Alvarado, 2000; Reddien and Sánchez Alvarado, 2004). Evidence that proliferating cells (neoblasts) contribute to blastema formation originally came from irradiation experiments. Irradiation is commonly used to kill dividing cells, and was observed to cause neoblast degeneration and block regeneration in planarians (Bardeen and Baetjer, 1904; Curtis and Hickman, 1926; Dubois, 1949; Lange, 1968; Wolff, 1962; Wolff and Dubois, 1948). These observations correlate the absence of cell division with the lack of regenerative capacity, although it remained possible that irradiation blocked some other process. Analysis using molecular markers that label dividing cells have since confirmed that irradiation can effectively and largely specifically eliminate the dividing cells and their immediate descendent cells (Eisenhoffer et al., 2008; Newmark and Sánchez Alvarado, 2000; Reddien et al., 2005b).
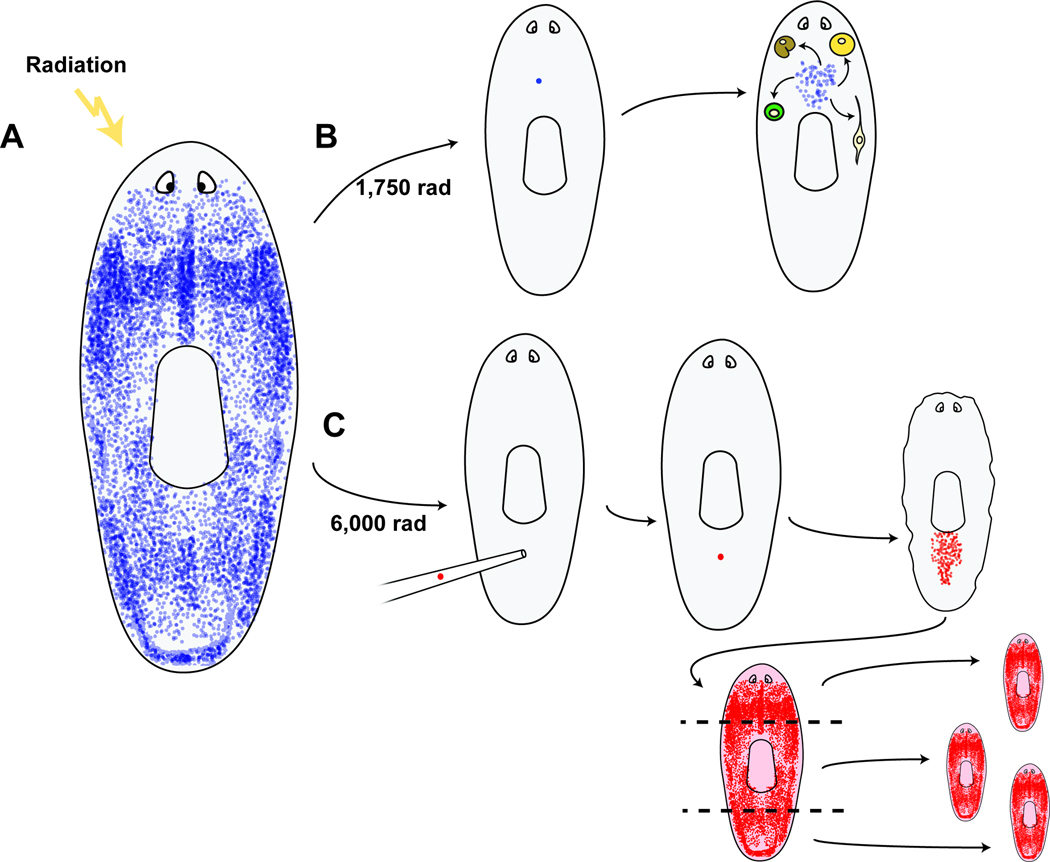
Figure 2. Planarian regeneration is accomplished with pluripotent stem cells called cNeoblasts.
A. Neoblasts (blue), are the somatic dividing cells of planarians and are depicted in blue. Neoblasts are scattered throughout the body, but restricted to behind the eyes and absent from the pharynx (centrally located). B. Irradiation with 1,750 rad can result in animals with a single surviving neoblast. This single cell can divide and produce a colony of neoblasts, ultimately producing differentiated cells spanning germ layers (Wagner et al., 2011). For example, individual neoblasts can generate both neurons and intestine cells, as well as defined neoblast progeny populations. C. Irradiation with 6,000 rad eliminates all neoblasts. Transplant of a single cNeoblast from a donor strain (red) results in clonogenic growth and, ultimately, the restored capacity for regeneration.
Whether or not planarian regeneration is explained entirely by a dividing, self-renewing cell population or involves other types of cellular changes, such as dedifferentiation has been explored. Some experiments have suggested that differentiated cells can contribute to new cell formation in planarian regeneration. These observations included those made using histological approaches and electron microscopy (EM) (Flickinger, 1964; Hay, 1966; Woodruff and Burnett, 1965), as well as those made with the use of vital dyes for lineage-tracing (Rose and Shostak, 1968). Additional support for de-differentiation came from observations involving planarian species containing cells with different ploidy; cells with the ploidy of germ cells were observed in regeneration blastemas following amputation through gonads (Gremigni and Miceli, 1980; Gremigni et al., 1980; Gremigni et al., 1982). Overall the possibility of dedifferentiation was controversial because of multiple potential interpretations for some of the data and because other EM investigations did not observe de-differentiation (Baguñà, 1998; Baguñà et al., 1989; Reddien and Sánchez Alvarado, 2004).
Several transplant experiments have been performed that support the idea that a renewing population of dividing cells are the primary contributors to planarian regeneration. First, transplant of normal tissue into irradiated hosts can rescue the capacity for regeneration (Dubois, 1949; Lange, 1968) and contribute [3H]uridine-labeled tissues to the host (Lender, 1962; Lender and Gabriel, 1965). Second, transplant of a population of small cells, enriched for small cells, restored regeneration and changed animal behavior from sexual (the host) to asexual (the donor), whereas transplant of differentiated cells showed no effect (Baguñà et al., 1989). Furthermore, BrdU-labeling experiments demonstrate that dividing cells contribute new cells to blastemas, and that cells with simple morphology (rather than differentiated morphology) are the first cells labeled following a BrdU pulse (Newmark and Sánchez Alvarado, 2000). Together, these observations are consistent with the existence of a population of adult dividing cells responsible for new tissue formation in planarian regeneration; however, they do not exclude the possibility of de-differentiation or transdifferentiation as candidate contributing sources for dividing cells or other cells in regeneration.
Clonal analysis of dividing planarian cells identify pluripotent stem cells
Taken together, prior data described above indicated dividing cells contribute to regeneration, but do not distinguish between multiple possible models for how these dividing cells participate in regeneration (Figure 1B). Whether the dividing cell population possesses a pluripotent cell type or whether the dividing cells consist of many different cell types with each possessing more restricted potential is a critical distinction for understanding planarian regeneration and has only recently been determined (Wagner et al., 2011). Dividing cells (neoblasts) can be isolated with flow cytometry, based simply on their possession of greater than 2N DNA content during replication and mitosis (Hayashi et al., 2006). A variety of genes are expressed broadly in this population of dividing cells (Aboobaker, 2011; Reddien et al., 2005b; Shibata et al., 2010), with the commonly used neoblast marker gene smedwi-1 expressed in all dividing cells (Wagner et al., 2011). However, heterogeneity in gene expression does in fact exist within this population of dividing cells (Hayashi et al., 2010). Regardless of potential heterogeneity within the dividing cells, some subset of the proliferating cells could be the ultimate source of new cells and be pluripotent at the single cell level.
To determine whether pluripotent cells explain planarian regeneration, the potential of single dividing cells was examined (Wagner et al., 2011). Some cells were observed to have clonogenic potential, that is, the capacity to produce a large number of descendent cells through the process of cell division. Two different and complementary assays were developed and used to obtain these data (Figure 2B, C). The first assay relies upon an irradiation dose that leaves a small number of dividing cells that survived irradiation behind. This method was made possible by the recent development of markers for dividing cells and their progeny (Eisenhoffer et al., 2008; Reddien et al., 2005b; Wagner et al., 2011). A strength of this method is that remaining dividing cells reside in their original tissue environment. A second assay was developed that involved transplant of single cells from a donor into a lethally irradiated host lacking all dividing cells. A strength of this more technically challenging method is that cells of different genotypes can be transplanted into hosts, and the location of the cell that will divide is known.
Following sublethal irradiation, some surviving dividing cells were capable of producing large numbers of clonally derived descendent cells. These colonies presented the first opportunity to analyze the differentiation potential of single cells from within the neoblast population, for example, by using established markers for immediate, nondividing neoblast progeny cell types (Eisenhoffer et al., 2008). The clonally derived descendent cells in colonies included cells that differentiated into intestine, neurons, and all other known neoblast progeny cell types (Figure 2B) (Wagner et al., 2011). The capacity to make these cell types existed in colonies derived from throughout the body. Cells from within the dividing cell population, therefore, have the capacity to produce differentiated progeny spanning multiple germ layers, displaying pluripotency at the single cell level. Cells with these attributes were defined as "cNeoblasts," for clonogenic neoblasts. Adult planarians constantly turnover their tissues, with the progeny of dividing cells replacing aging differentiated cells (Pellettieri and Sánchez Alvarado, 2007; Reddien and Sánchez Alvarado, 2004). As a consequence, irradiated animals are incapable of long-term survival without cell division. As few as 3 to 5 cNeoblasts surviving irradiation in asexual animals were sufficient to lead to survival and to restore regenerative capacity (Wagner et al., 2011).
The capacity of cNeoblasts to produce large numbers of dividing cells, to repopulate planarian tissues with dividing somatic cells, and to produce cells spanning germ layers was confirmed with transplantation experiments (Wagner et al., 2011). In the transplantation experiments, individual cells from asexual animals were transplanted into lethally irradiated sexual hosts, which lacked any other source for dividing cells. These experiments were made possible by the development of a cell-sorting procedure for isolation of individual cNeoblasts involving flow cytometry followed by morphological identification of cells. Also important was the development of a transplant procedure yielding high frequency engraftment of single cells and the identification of sequence polymorphisms for genotyping. Optimization of this procedure relied upon recently identified molecular markers for detection of proliferating cells. Remarkably, single transplanted cells were sufficient to lead to survival and a restored capacity for regeneration in some of these sexual hosts (Figure 2C). In order for enough time to elapse for repopulation of tissue with proliferating cells, transplant recipients needed to survive for 40–50 days without other sources of cell division, and 7/130 animals recovered. These rescued animals displayed asexual behavior, and importantly, the asexual genotype. Therefore, the single transplanted cell slowly but surely replaced host tissues, with the animal becoming a genetic clone of the donor. This experiment confirms that the transplanted cell had the capacity to make the essential tissues of planarians, and provides candidate cellular explanation for planarian regeneration: the persistence into adulthood of a pluripotent stem cell type that is widespread throughout the body.
The origin of a clonogenic neoblast has not yet been characterized in detail; however, several observations support the idea that cNeoblasts self-renew. First, cNeoblasts produce large, growing colonies of dividing cells, in fact, repopulating the animal with dividing cells. The restoration of regenerative potential (and even entire strains of clonal animals) from animals harboring from one or a few cNeoblasts further suggests that capacity for tissue pluripotency expanded in these animals. Therefore, self-renewal appears to be an attribute of cNeoblasts. cNeoblasts are scattered along the head-to-tail axis in the animal parenchyma. However, the percentage of dividing cells that function as cNeoblasts is unknown—it could be that the vast majority of dividing cells have this clonogenic potential and pluripotency, or there could be more complexity in the dividing cell population (neoblasts) than was previously imagined. Many possibilities exist for the behavior of cNeoblast descendant cells—from rapid differentiation, to the existence of long-lived transit amplifying cells. It will be of great interest to understand the processes by which so many different cell types emerge from a single adult cell type, and whether this requires slow-and-steady lineage restriction through many rounds of division, or dramatic and rapid steps to final cell states.
An expanding set of molecular tools are emerging for the study of genes in planarian regeneration (Newmark and Sánchez Alvarado, 2002; Saló et al., 2009; Sánchez Alvarado, 2006), including efficient inhibition of gene function with RNAi (Newmark et al., 2003; Reddien et al., 2005a; Sánchez Alvarado and Newmark, 1999), labeling of dividing cells with RNA probes, BrdU, or antibodies (Guo et al., 2006; Newmark and Sánchez Alvarado, 2000; Reddien et al., 2005a), isolation of dividing cells using flow cytometry (Hayashi et al., 2006), and assessment of differentiation of progenitor cells (Eisenhoffer et al., 2008; Guo et al., 2006; Newmark and Sánchez Alvarado, 2000; Scimone et al., 2010; Wagner et al., 2011; Wenemoser and Reddien, 2010). Many genes have already been identified that impact biology of the neoblast population (Fernandez-Taboada et al., 2010; Guo et al., 2006; Hayashi et al., 2010; Oviedo and Levin, 2007; Pearson and Sánchez Alvarado, 2010; Reddien et al., 2005a; Reddien et al., 2005b; Scimone et al., 2010; Solana et al., 2009; Wenemoser and Reddien, 2010). cNeoblasts therefore present the opportunity for molecular genetic dissection of maintenance, differentiation, and deployment for regeneration of a pluripotent stem cell in vivo.
Regenerative cells must respond to wound signals for the initiation of regeneration. Furthermore, to regenerate, the identity of missing tissues must be specified. For example, recent work has identified Wnt signaling as important for controlling head-versus-tail regeneration decisions at transverse wounds and serves as a paradigm for study of the specification of regeneration programs (Gurley et al., 2008; Iglesias et al., 2008; Petersen and Reddien, 2008, 2011, Adell et al., 2009; Gurley et al., 2010; Petersen and Reddien, 2009; Reddien, 2011). The identity of these wound and missing tissue identity signals, and whether they act on the cNeoblasts or lineage-committed cNeoblast progeny, will be important research directions for understanding the regulatory logic of regeneration.
Hydra regeneration and stem cells
Hydra, like planarians, display some of the most dramatic regenerative feats known to occur in the animal kingdom (Galliot and Schmid, 2002). Hydra are cnidarians that live as freshwater polyps with a polarized, primary body axis. Cnidarians are metazoans that possess two germ layers and represent an outgroup to the Bilateria (Adoutte et al., 2000; Putnam et al., 2007). The Hydra body axis contains two poles separated by a body column (Figure 3A). The oral pole, or head, contains tentacles and hypostome (mouth) and the aboral pole, or foot, contains the basal disc. These animals are composed of two tissue layers, each a single cell thick: an outer layer of ectodermal myoepithelial cells and an inner layer of endodermal myoepithelial cells, arranged together in a tube ending in tentacles (Figure 3B). Hydra were the first subjects of described regeneration experimentation (Lenhoff and Lenhoff, 1986) and are capable of regenerating entire polyps from tiny body fragments (Bosch, 2007; Holstein et al., 2003). In a remarkable display of body organization capacity, dissociated Hydra cells can even be re-aggregated and produce a new Hydra polyp (Gierer et al., 1972; Noda, 1971; Technau et al., 2000). Because of the ability of small body fragments to regenerate Hydra polyps, tissue pluripotency is spread broadly in these animals. The current model, described below, for tissue pluripotency in Hydra does not involve a single cNeoblast-like cell type, but instead involves action of multiple different stem cell types.
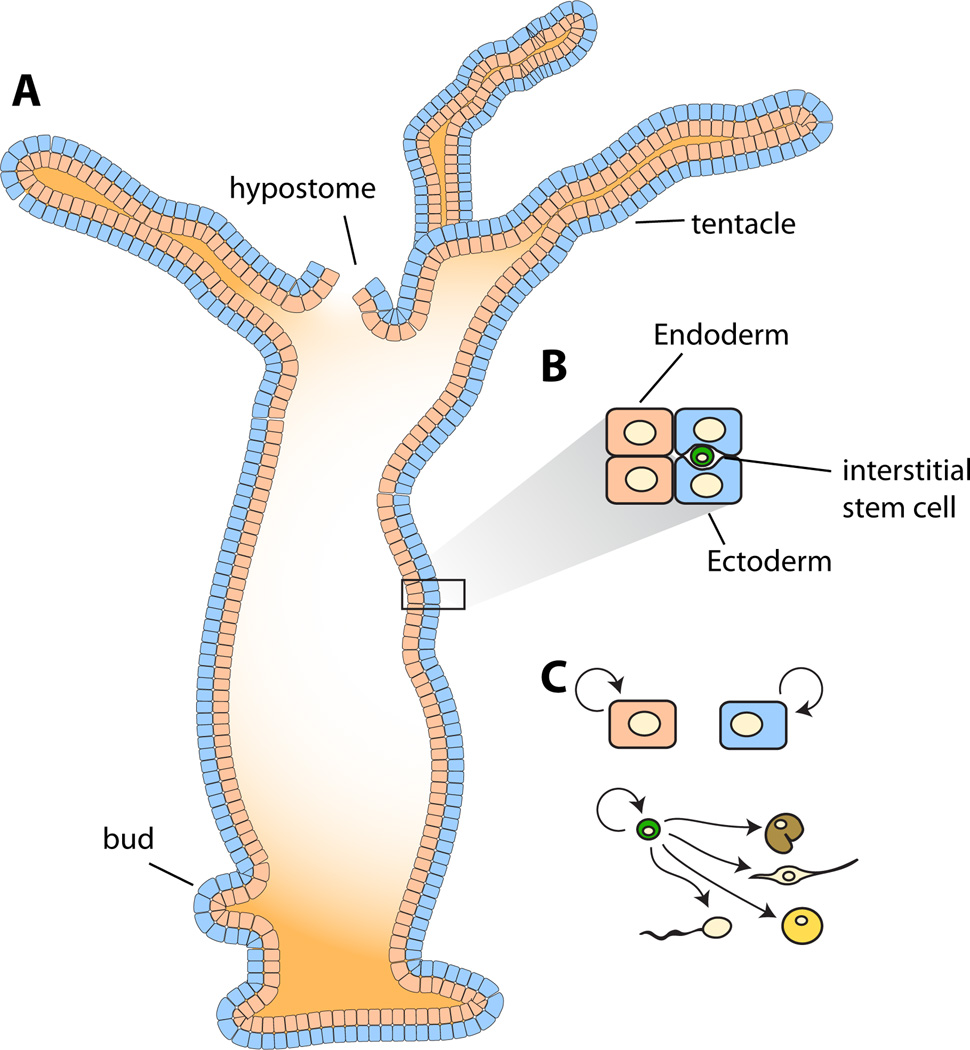
Figure 3. Hydra regeneration is accomplished with three different stem cell populations.
A. Hydra are cnidarians with a primary body axis containing a hypostome (or head) at one end and a foot at the other. Cell proliferation in the body column continually pushes cells to the poles of the body. Asexual reproduction is accomplished by budding. B. The body wall contains two epithelial cell layers, ectodermal and endodermal epithelial cells. Interstitial stem cells exist within the ectodermal epithelial cell layer. C. The ectodermal and endodermal epithelial cells proliferate continuously to maintain these tissue layers, producing differentiated epithelial cells, and are therefore considered to be distinct stem cells. A third stem cell type, the multipotent, interstitial stem cell can self-renew and produce neurons, nematocytes, secretory cells, and gametes.
Regeneration in Hydra can be accomplished by changes in tissue morphology involving existing cells in the absence of cell proliferation (often referred to, for Hydra, as morphallaxis) (Bosch, 2007; Cummings and Bode, 1984; Galliot and Ghila, 2010; Holstein et al., 1991; Marcum and Campbell, 1978a, b; Park et al., 1970; Wittlieb et al., 2006). Despite regeneration being capable of occurring without significant cell division, new cells in Hydra are normally continuously made for maintenance of the polyp. Small body fragments can regenerate polyps through morphogenetic changes, which can then feed and grow to produce an individual similar to the original (Bode and Bode, 1980). Furthermore, regenerated individuals can reproduce long-term through budding. Therefore, sources for massive cell production exist in the adult Hydra. Finally, following mid-gastric amputation (but not after decapitation close to the head), stimulation of interstitial cell proliferation occurs and contributes to regeneration (Chera et al., 2009). What is the source of new materials?
The epithelial cells in the Hydra body column continuously proliferate and replace differentiated epithelial cells at the poles of the polyp, with older cells sloughed off at the tentacle and foot (Campbell, 1967; Dubel et al., 1987). These dividing epithelial cells of the body column carry out differentiated tasks, such as osmoregulation (ectoderm), food digestion (endoderm), and muscle-like contraction (both layers) (Bode, 1996). These two cell layers in the body column are broadly considered to each possess cells that act as separate epithelial stem cell types (Figure 3C) (Bosch, 2007). Understanding the division and differentiation behavior of these cells could be enhanced with single cell-based lineage studies.
In addition to proliferation occurring in the myoepithelial cells, highly proliferative interstitial cells exist within the epithelial cell layers (Figure 3B) (Bode, 1996). To investigate the potential of individual interstitial cells, clonal analyses were performed taking advantage of the capacity of Hydra to re-aggregate following dissociation to a cell suspension (Bosch and David, 1987; David and Murphy, 1977). Animals lacking the interstitial cell lineage (following treatment with nitrogen-mustard or using a strain with temperature-sensitive interstitial cells) were dissociated and re-aggregated together with a small number of cells from normal animals. Clones of growing interstitial cells emerged in these chimeras, with clonal growth having occurred from donor-derived cells, demonstrated using [3H]thymidine-labeled donor cells. A statistical approach was used to indicate the majority of clones analyzed arose from single cells. From these experiments, it was shown that interstitial cells are multipotent stem cells that can generate neurons, nematocytes, secretory cells, and gametes but not the epithelial layers (Figure 3C) (Bosch and David, 1987; David and Murphy, 1977). Similar chimera experiments demonstrate that interstitial cells with clonogenic capacity, deemed to be stem cells, are distributed throughout the body column (David and Plotnick, 1980) in the ectodermal epithelial layer (Smid and Tardent, 1986) with descendant cells present in both epithelial layers.
Because under starvation conditions, regeneration in Hydra can occur by tissue morphogenesis without new cell production, some differentiated cells will initially find themselves in inappropriate areas. Some observations have led to proposals that certain differentiated cells can change state. For example, zymogen gland cells (ZGC) of the body column can become found in the regenerating head region following amputation (Siebert et al., 2008). Histological studies of cells with intermediate appearance between body column (ZGC) and head mucous gland cells (MGC), suggest that ZGCs can transdifferentiate to become MGCs (Siebert et al., 2008). Some experiments involving elimination of neuron precursors (interstitial cells) followed by amputation and assessment of maintenance or change in neuron cell state led to similar proposals that pre-existing neurons might be capable of changing type during regeneration (Bode, 1992, 1996). By contrast, other experiments have suggested that peduncle neurons arise largely, if not entirely, from interstitial cell-derived precursors (Technau and Holstein, 1996). Lineage-tracing experiments will therefore be important for investigating the possibility of changes in the state of differentiated cells in Hydra regeneration more definitively.
In summary, tissue pluripotency in Hydra involves three stem cell types (ectodermal and endodermal epithelial cells, and interstitial stem cells) that enable continual new tissue production (Bosch, 2007). Transgenic Hydra have now been generated that allow observation of individual cells during regeneration (Khalturin et al., 2007; Wittlieb et al., 2006). This innovation will in principle enable a new suite of chimera and transplantation experiments that allow observation and lineage tracing of individual cells. A limited number of labeled epithelial cells have already been observed to expand in number and populate the entire epithelial layer (Wittlieb et al., 2006). Transplantation of single epithelial cells has been described as possible (Wittlieb et al., 2006), and it will be of interest to observe the behavior of such single cells. These transgenic and chimera methods should prove powerful for evaluating the potential of individual proliferative cells, the lineage decisions between stem cells and differentiated cells, and candidate transdifferentiation events that occur during Hydra regeneration. Numerous factors have been identified that regulate the process of Hydra regeneration, including Wnt signaling and the MAP kinase-CREB pathway (Bosch, 2007; Galliot and Chera, 2010). Understanding Hydra regeneration requires combining knowledge of the cellular sources for new tissue with knowledge of wound signaling and other molecular mechanisms that specify the identity of new tissues in regeneration (Bosch, 2007; Galliot and Chera, 2010). Therefore how regulatory molecules control the division and differentiation behavior of Hydra stem cells and stem cell-progeny cells is an important area of investigation.
How do vertebrates regenerate all of the missing cells?
While regenerative vertebrates such as salamanders, frogs, and fish, do not show full body regeneration, they can regrow substantial parts of the body. For example, tissue or cell removal from internal organs such as the heart, the brain, and the kidney in these animals result in a regeneration response (Kirsche and Kirsche, 1963; Oberpriller and Oberpriller, 1974; Parish et al., 2007; Poss et al., 2002). In many of these cases, the correct cell types and tissue mass are restored but exact organ form is not always replicated. By contrast, amputation of appendages results in restoration of the correct cell types and form. The salamander limb faithfully regenerates the missing limb segments, upper limb, lower limb or foot/hand, when amputated anywhere along the limb axis. Vertebrate appendages are composed of intricately patterned tissues originating from multiple germ layers. For example, the vertebrate limb consists of epidermis and peripheral nervous tissue deriving from ectoderm, and other internal tissues such as muscle, bone, dermis and blood vessels that derive from mesoderm. Therefore the same conceptual issues arise as for invertebrate regeneration—how is the full spectrum of cell types and pattern re-formed after tissue removal? Does, for example, a resident, pluripotent cNeoblast exist in vertebrate tissues that executes regeneration?
A brief history of blastema cell origin and potency
During limb regeneration, a zone of seemingly homogeneous, undifferentiated progenitor cells, called the blastema, forms at the amputation site. The blastema consists of mesenchymal blastema cells encased by a simple, wound epidermis. The cellular sources of the limb blastema and the potency of blastema cells have been investigated by excellent researchers over many years, but, until recently, many questions remained unresolved and a diversity of conflicting conclusions had arisen because of the lack of satisfactory lineage-tracing tools and molecular markers to address these questions. In the salamander limb system, where much of this work has been performed, two primary classes of experiments were historically used to examine blastema cell potency and tissue origin. The first involved grafting limb blastemas to ectopic sites such as the fin that support blastema growth and differentiation in order to reveal the intrinsic differentiation capacities of blastema cells (Pietsch, 1961; Stocum, 1968). The diversity of cell types and the completeness of limb segments that formed in such experiments varied widely, leading some researchers to propose that blastema cells had limited potential and others to speculate that blastema cells were pluripotent (Holtzer, 1969; Pietsch, 1961; Steen, 1970).
Many researchers then attempted to directly track the fate of cells coming from specific cell types by transplanting triploid or tritiated thymidine-marked tissues into normal or irradiated regenerating host limbs. Cartilage was recognized as one tissue where grafts consisting purely of cartilage cells could be isolated. Steen transplanted triploid and tritiated thymidine-labeled cartilage pieces into normal regenerating limbs and found that labeled cells contributed to the blastema with the vast majority of cells reforming cartilage (Steen, 1968). These results led Steen to conclude that during normal regeneration, cartilage cells only form cartilage. Interestingly, when Namenwirth (1974) performed similar experiments transplanting cartilage into irradiated limbs, the labeled cells formed not only cartilage, but also perichondrium and soft connective tissues in the joints and dermis (Namenwirth, 1974). It was proposed that under conditions where host cells cannot contribute to blastema formation, a broader potential of cartilage to form soft tissue connective cells was revealed. Given that cartilage and soft connective tissue cells arise from a common progenitor during limb development, this hypothesis is plausible (Pearse et al., 2007). However, it is also possible that the transplanted cartilage pieces in the separate experiments were of differing purity. Because the perichondrium that encases the limb cartilage is a likely source of cells with soft connective tissue potential, any graft that had not sufficiently removed this layer could also have given this result. Therefore, whether or not cartilage can contribute to soft connective tissue regeneration remains unresolved. Wallace and colleagues performed a similar irradiation rescue experiment using unlabelled cartilage and found regenerated limbs possessing cartilage and muscle (Wallace et al., 1974). These investigators concluded that cartilage cells have the potential to form all limb tissue types, including muscle. Due to lack of any lineage-tracing markers in this latter experiment, it was unclear if the muscle tissue arose from the host or the graft; and the purity of the grafted piece was also a consideration. In summary, many possible interpretations of cartilage cell potential have historically been proposed.
Tracking of the fate of other limb tissues during regeneration, such as muscle, Schwann cells and dermis, had yielded uninterpretable results due to the complexity of these tissues. When Steen transplanted labeled muscle tissue he observed efficient contribution to regenerating cartilage (Steen, 1968). He wisely noted that since muscle is actually a complex tissue consisting of muscle fibers, muscle satellite cells, connective tissue and vessels, the cellular source of this regenerated cartilage, and thus the true regenerative potential of muscle cells, remained unclear. Similarly, the potential of dermis to form muscle was unclear based on sporadic muscle labeling in graft experiments. When Namenwirth rescued irradiated limbs with skin transplants that included dermis, patterned limbs formed, with some, but limited muscle tissue formation (Dunis and Namenwirth, 1977). The muscle formation observed was ascribed to contamination of skin transplants with some muscle cells. Finally, the regenerative potential of nerve cells was studied by Wallace, who rescued amputated, irradiated limbs with unirradiated nerve grafts (Wallace and Wallace, 1973). Because such samples generated limbs consisting of all tissue types, Wallace concluded that cells in nerve grafts can dedifferentiate and ultimately form other cell types, such as muscle and connective tissue. Whether the source of cells was Schwann cells or accompanying connective tissue cells was unresolved. In summary, the study of the fate of internal limb tissues during regeneration was largely obscured by the inability to label defined cell types within a given tissue and therefore the true potency of blastema cells coming from different cell types was unresolved.
Vertebrate appendage regeneration implements lineage-restricted progenitors
Recent advances in generating GFP-expressing transgenic frogs, salamanders, and fish, combined with molecular marker analysis, have allowed in vivo tracking of cells with higher precision to resolve many of the questions from the previous studies. In this recent work, limited cell potential in regeneration has been found for all examined tissue lineages in frog, salamander, and fish. Each different tissue provides a distinct progenitor cell pool to the regeneration blastema indicating that from the very outset, the vertebrate blastema is not generated from or comprised of cells of a single type. By contrast, the blastema as a whole is a mixture of cells with different, restricted potentials and tissue origins that together coordinately regenerate the complex appendage. In other words, vertebrate appendages do not harbor a pluripotent cNeoblast-like cell as found in planaria.
Limb of newt and tail of frog
The first demonstration in a vertebrate that different tissues such as muscle and nerve are regenerated from distinct progenitor cell pools came from investigation of Xenopus tadpole tail regeneration. Embryonic grafts of posterior neural plate, posterior presomitic mesoderm, or posterior axial mesoderm from GFP-transgenic donors into unlabeled hosts were used to generate animals, each having one of the three major tissues (spinal cord, muscle, or notochord) labeled in the Xenopus tail (Figure 4A) (Gargioli and Slack, 2004). Amputation of the differently labeled tails revealed that each tissue layer regenerated separately and did not contribute to the other. These studies also addressed whether muscle regeneration occurred via dedifferentiation or recruitment of stem cells. Vertebrate skeletal muscle harbors a population of stem cells called satellite cells, which lie adjacent to mature muscle fibers and are activated by injury to proliferate and then differentiate and fuse into muscle fibers during repair (for review see Le Grand and Rudnicki, 2007). Tissue grafts of early, medial presomitic mesoderm yielded labeling only of tail muscle fibers but not satellite cells. Amputation of these tails showed no GFP+ cells in the regenerate. In contrast, later stage presomitic mesoderm grafts that produced labeling of both muscle fibers and satellite cells did regenerate GFP+ muscle. These results indicated that stem cell activation rather than dedifferentiation was the major mechanism of muscle regeneration in Xenopus tail regeneration.
Figure 4. Cell tracking of GFP-labeled cells in amphibians shows that vertebrate appendage regeneration occurs by producing lineage-restricted progenitors in the Xenopus tail and Axolotl limb blastema.
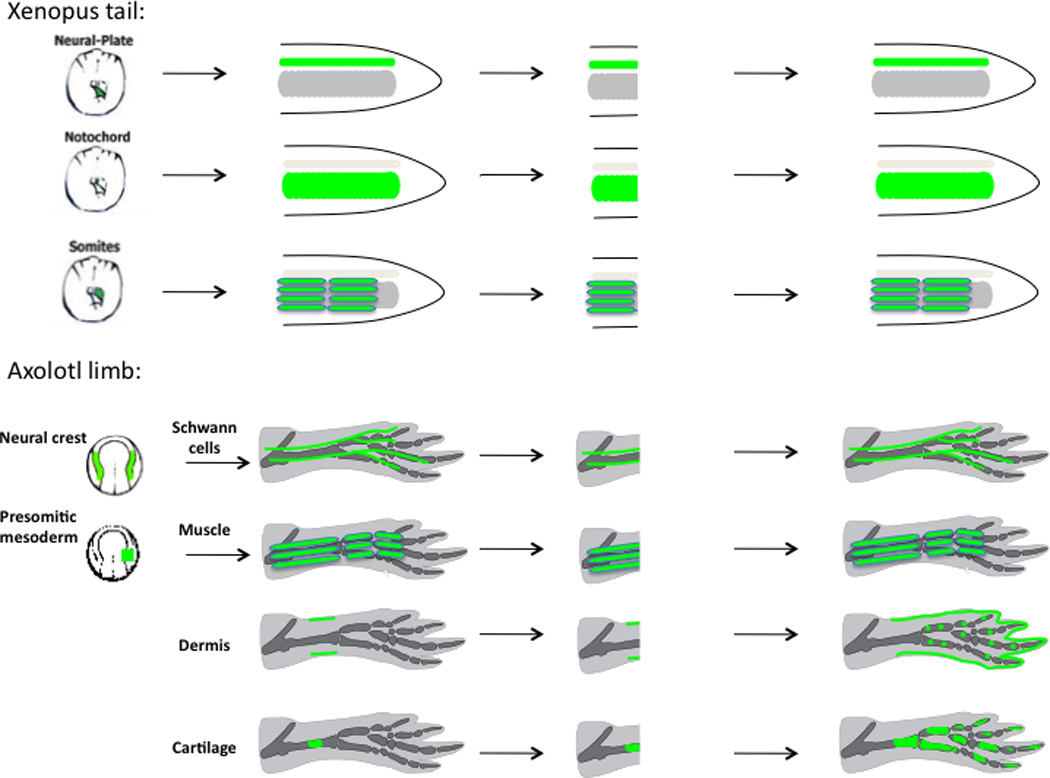
Cell labeling was primarily achieved via grafting of embryonic tissues during the neurula stage from GFP-expressing donors to normal hosts. (Top) Xenopus: posterior neural plate, presomitic mesoderm, and notochord were transplanted to label tail spinal cord, muscle, and notochord, respectively. After tail amputation, the labeled tissues regenerated the same tissue type as prior to amputation (Gargioli and Slack, 2004). (Bottom) Axolotl limb Schwann cells and muscle were labeled by embryonic presomitic mesoderm and neural crest transplantation (Kragl et al., 2009). Dermis and cartilage were labeled by direct tissue transplantation in the limb, as well as embryonic tissue grafts (Kragl et al., 2009). After limb amputation, labeled Schwann cells regenerated Schwann cells only. Muscle regenerated muscle and no cartilage. Dermis regenerated dermis, cartilage and connective tissues (also described by Dunis and Namenwirth, 1977), while cartilage regenerated cartilage (also described by Steen, 1968).
Since Xenopus loses regeneration ability during metamorphosis when the tail is resorbed, it was unclear whether these results were generalizable to all appendage regeneration, especially to animals that display lifelong limb and tail regeneration, such as salamanders. This was an important consideration in light of contrasting results obtained by following the fate of electroporated spinal cord cells in the salamander species, Axolotl, which indicated contribution to muscle and cartilage (Echeverri and Tanaka, 2002). Further transgenic labeling methods are currently being used in Axolotl to resolve the fate of the cells exiting the regenerating Axolotl spinal cord (McHedlishvili et al., 2007).
Limb regeneration in salamanders, such as Axolotl and newts, represents the canonical example of complex vertebrate regeneration, where previous lineage-tracing studies had raised many unanswered questions. Recently, many long-standing issues were resolved using transgenic animals. Transgenic Axolotls that constitutively express a GFP transgene were used as donors in embryonic tissue grafts of prospective limb forming regions to specifically label limb epidermis, muscle, Schwann cells, or connective tissue (Figure 4B) (Kragl et al., 2009). GFP labeling was used to sensitively detect and eliminate samples with labeling of undesired cell types prior to initiation of the regeneration experiment. Furthermore, by using embryonic grafts rather than limb tissue grafts, defined cell populations could be labeled. Limbs of animals with different GFP-labeled tissue types were amputated and the identity of GFP+ regenerated tissues was determined. These experiments answered three of the major issues raised in previous studies in salamanders. First, they showed that labeled muscle (muscle fibers and satellite cells) did not contribute to cartilage or epidermis but gave rise primarily to muscle. Because the muscle labeling experiments involved grafts of presomitic mesoderm, blood vessels (which have a common origin in somitic mesoderm with muscle) were also unavoidably labeled. Therefore, it is still unresolved whether muscle can potentially contribute to endothelial cells and vice versa. Second, the embryonic lateral plate mesoderm as well as adult skin grafting studies showed that dermis cells contributed to cartilage and connective tissue, but did not give rise to muscle – neither Pax7+ muscle satellite cells nor mature muscle fibers. Third, the question of whether or not irradiation rescue causes cells to display broader cell potency was addressed by combining nucCherry-expressing transgenics as irradiated hosts with nerve grafts derived from GFP transgenics, or from GFP-Schwann cell-labeled animals. If the irradiated animals were rescued with nerve tissue where all the cells were GFP+, then all the regenerated cartilage was GFP+. However, if nerve grafts – where only Schwann cells were GFP+ – were used to rescue nucCherry hosts, the cartilage in the regenerated host was negative for both transgenes. This indicated that Schwann cells had not acquired cartilage regenerative potential during irradiation rescue. By contrast, Schwann cells only reformed Schwann cells, whereas the regenerated cartilage and connective tissue derived from an accompanying cell—presumably connective tissue fibroblasts that are closely intertwined with the Schwann cell in nerve sheaths. An important dimension of the irradiation experiments was that since all cells in the regenerates were either transgenic GFP+ or nucCherry+, the origin of the different tissues could be quantitatively assessed. As a whole, in these GFP embryonic labeling experiments of Schwann cells, muscle, and cartilage/connective tissue, a large majority of the given tissue type could be labeled, allowing the conclusion that the observed lineage restrictions reflects the behavior of the vast majority of the cells.
While the Axolotl limb experiments resolved the overall lineage restrictions of the Axolotl limb blastema, they did not address whether the cellular mechanisms involved in producing blastema cells from the different tissues involves activation of a resident tissue-specific stem/progenitor cell, or involves the dedifferentiation of a post-mitotic cell. This is an important issue, discussed further below, because the concept and occurrence of muscle dedifferentiation has been a major theme in salamander limb regeneration studies (for review see Straube and Tanaka, 2006).
Lineage-restrictions during zebrafish fin development and regeneration
The fish caudal fin is another major experimental system for investigation of vertebrate appendage regeneration. The caudal fin is an innervated structure consisting of segmented bony fin rays that surround fibroblasts that together are encased in epidermis; no muscle is present in the region of the fin that regenerates (for review see Akimenko et al., 2003). Upon amputation, the tip of each fin ray forms a growth zone called the blastema, which independently grows to elongate the missing fin ray. Until present, it was unknown if the different fin tissues each supplied distinct, lineage-restricted progenitors to the blastema, or whether blastema cells represented a single cell type that had the potential to form all cell types of the fin. Two recent studies that employed different cell-tracking methods to follow the fate of fin cells in regeneration both came to the conclusion that cells show lineage restriction. The Johnson group used sporadic Tol2 transgene insertion to follow cells during fin development and regeneration while the Weidinger group, as described in the next section, used Cre/loxP-marking technology to follow the fate of osteoblasts during regeneration. Tu and Johnson (2011) generated mosaically labeled fish fins by injecting embryos with limiting amounts of plasmid that insert a XenEF1a:GFP transgene by Tol2 transposition. By analyzing coherent cell groups that label a given tissue type as clones, the authors propose that the fin is built in a highly mosaic fashion from 9 different cell lineages—vessel/artery, osteoblast, fibroblast, glial, melanophore/xanthophore, iridiphore, epidermis, and lateral line. Since most fins were apparently labeled with more than one clone, the authors use co-occurrence analysis to infer that most lineages, by the time cells form the fin bud, were unipotent except the bipotent vein/artery lineage and the melanophore/xanthophore lineage. Upon fin amputation through labeled cell patches, the labeled cells regenerated the same cell type and did not contribute to other cell types. The authors point out that in their work the dermal and osteoblast lineages remained separate during both fin development and regeneration. These results are quite distinct to results from the development of other vertebrate appendages. For example, clonal analyses in the mouse and chicken limb bud showed that single progenitor cells contribute to dermis, cartilage, tendon, and connective tissue (Arques et al., 2007; Pearse et al., 2007). During salamander limb regeneration, transplantation of dermis-containing skin onto an amputated host limb results in dermis cells contributing to, and even fully regenerating, patterned bone (Dunis and Namenwirth, 1977; Kragl et al., 2009). Biological and/or technical reasons may be responsible for the divergent dermis/bone tracking results between the fish fin and other vertebrate appendages. First, it is likely, though not established, that bone origin and formation in the fish fin differs significantly from that in vertebrate limbs. The vertebrate limb consists of endochondral bone with a clear origin in lateral plate mesoderm that differentiates through a cartilage intermediate (for reviews see Goldring et al., 2006; Tuan, 2004). In contrast, the zebrafish fin consists of dermal bone that may derive from neural crest and that appears to ossify directly from progenitors without a cartilage intermediate (Smith et al., 1994). Therefore, the lineage relationship between bone and connective tissue may be different in these two contexts. Furthermore, because many of the cell types described in the fish fin study may be of neural crest origin, the results may largely reflect the diversification and commitment of neural crest during fin development and regeneration. On the other hand, because the method of clonal analysis did not address whether the labeling was representative of all fin cells, it is still possible that other, less restricted, clone behaviors may be occurring in fish fin development and regeneration.
The fish results emphasized that during regeneration there is no crossing between neural crest subtypes such as glia and melanophore/xanthophore lineages. This is interesting in light of cell culture work characterizing the ability of chick and quail glia to dedifferentiate and form melanocytes, and the ability of clonally cultured melanocytes to dedifferentiate to a progenitor state that can differentiate into myofibroblasts and glial cells (Dupin et al., 2003; Real et al., 2006). It should be investigated whether these differences reflect organism-specific traits or whether long-term clonal culture conditions impart a broader potential onto cells than they normally have in vivo.
Dedifferentiation versus stem cells
With the exception of the Xenopus studies, the lineage-tracing results left open whether blastema formation occurs via the activation of resident tissue stem cells, or via dedifferentiation. When taking all studies across different tissues into account, it is likely that both processes contribute to regeneration by differing amounts for different tissues. In the context of newt limb, tail, and heart regeneration, a number of studies, mostly employing non-genetic cell lineage tracers, previously suggested that skeletal and cardiac muscle cells can dedifferentiate and become proliferative during regeneration (Echeverri et al., 2001; Kumar et al., 2000; Laube et al., 2006; Lo et al., 1993). Nongenetic lineage tracers such as fluorescent lipidic molecules or fluorescent cytoplasmic tracers used in these experiments, however, are not ideal because there is always the possibility of transfer to other cell types. Most recently, three studies in zebrafish, two in the heart and the other in the fish fin, used genetic-labeling-based fate mapping to establish that cardiomyocytes and osteoblast cells do dedifferentiate and proliferate during heart and fin regeneration (Jopling et al., 2010; Kikuchi et al., 2010; Knopf et al., 2011).
Zebrafish and salamanders regenerate heart tissue after resection of the ventricle. To address whether resident differentiated cardiomyocytes contribute to this regeneration, Jopling et al. and Kikuchi et al. used Cre/loxP-based genetic marking to track cardiomyocytes (Figure 5A). Both studies implemented double-transgenic animals harboring a cardiac myosin light chain (cmlc2) promoter driving a tamoxifen-inducible CreER gene as well as a loxP-reporter transgene where a cardiac-restricted promoter drove a floxed STOP cassette followed by the GFP gene (Figure 5A). To obtain cardiomyocyte-specific GFP expression, Jopling et al. treated double-transgenic embryos with tamoxifen, while Kikuchi et al. injected tamoxifen into adults prior to regeneration. These treatments caused Cre-mediated excision of the STOP cassette and GFP expression specifically in embryonic and adult cardiomyocytes respectively. After adult heart transection essentially all of the newly made cardiomyocytes were derived from these pre-labeled, GFP-expressing cardiomyocytes, indicating that the primary cell source for heart regeneration was differentiated cardiomyocytes that had proliferated rather than an unlabeled stem cell that had differentiated after transection. Interestingly, Kikuchi and colleagues showed that the proliferating cardiomyocytes reactivate a transgenic reporter for the heart developmental regulator, GATA4. These results indicate that cardiomyocytes proliferate to restore heart mass in zebrafish heart regeneration. Since a cardiomyocyte-specific promoter was used to drive expression of the loxP reporter, no conclusions on whether the labeled cardiomyocytes contribute to other lineages were made.
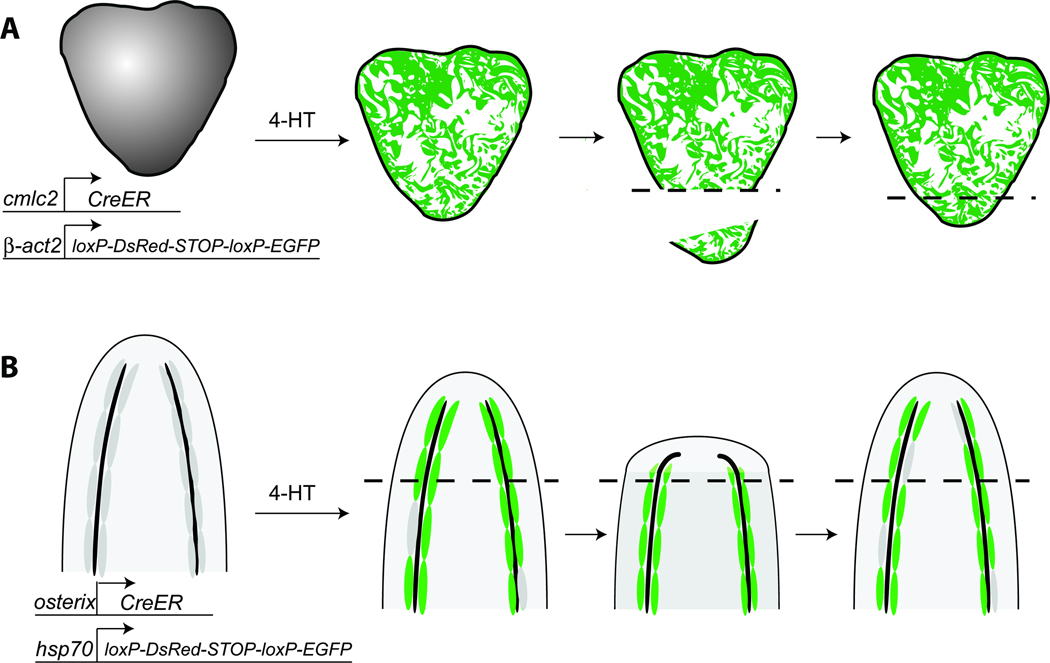
Figure 5. Cre/loxP-based cell fate mapping establishes that dedifferentiation occurs during zebrafish heart and fin regeneration.
A. Prior to heart resection, cardiomyocytes were labeled via a cardiomyocyte-specific promoter driving the CreER sequence. CreER acted on a cardiomyocyte-specific loxP reporter where a floxed STOP cassette was excised by CreER, which is active in the presence of 4-HT (4-hydroxytamoxifen), to induce GFP expression. Newly regenerated cardiomyocytes (right, below dotted line) express GFP, indicating that they derived from cardiomyocytes in the injured heart tissue (Kikuchi et al, 2010; Jopling et al, 2010). B. Tracking of osteoblasts during caudal fin regeneration demonstrates that they contribute to the regenerated fin and remain restricted to an osteoblast identity. osterix:Cre-ERT2 acting on the loxP reporter; upon Cre-mediated excision of a STOP cassette, the hsp70 promoter drives expression of GFP. GFP expression was induced prior to fin amputation leading to sporadic cell labeling. GFP-expressing cells generate osteoblasts in the regenerated fin, indicating that osteoblasts de-differentiated and divided to produce more osteoblasts, remaining restricted to the osteoblast fate during regeneration (Knopf et al, 2011).
It will be important to further map the fate of cardiomyocytes and other cardiac cell lineages to gain a complete picture of cell fates during zebrafish heart regeneration. Recently, Kikuchi and colleagues investigated the fate of another major cell type in the heart, the epicardium (Kikuchi et al., 2011). The authors identified tcf21 as a gene that is specifically expressed in the developing and adult zebrafish epicardium. Using a similar Cre/loxP-based cell tracing strategy as described above, the authors showed that driving CreER by the tcf21 promoter yielded no GFP-positive cardiomyocytes. To track the potential contribution of epicardium to other heart cell types, tcf21:CreER transgenic animals were crossed to the loxP reporter gata5:RnG that drives reporter expression in all heart cells. When tamoxifen was administered in growing larvae that were later examined as adults, GFP+ cells were found not only throughout the adult epicardium but also in some MLCK+ smooth muscle cells of the bulbus arteriosus and coronary vessels. Resection of these adult hearts was followed by the appearance of GFP+ perivascular cells in the regenerated heart. Similar results were found when tamoxifen was administered in the adult stage prior to regeneration. These results indicate that epicardial cells do not form cardiomyocytes during heart regeneration and appear to show limited flexibility to form perivascular cells.
Similarly, Knopf et al. implemented Cre/loxP-based fate mapping of osteogenic populations during fish fin regeneration, and found that differentiated osteoblasts temporarily dedifferentiate, enter into the fin blastema and then redifferentiate into osteoblasts. In the fish fin bones, an osterix:GFP reporter labels pre-osteoblast precursors as well as mature osteoblasts, while the osteocalcin:GFP reporter labels mature osteoblasts. Amputation of the fish fin caused proliferation as well as downregulation of both osterix:GFP and osteocalcin:GFP in osteoblasts at the amputation plane and upregulation of the transcription factor, Runx2, which is expressed in osteoblast precursors. To show that osteoblasts enter the fin blastema, cells that remained transiently GFP+ in the osteocalcin:GFP fish were followed into the early blastema. This participation of osteoblasts in fin regeneration was confirmed by Cre/loxP-mediated genetic fate mapping implementing an osterix:CreERT2 transgenic in conjunction with a loxP reporter where the heat shock promoter drove a floxed DsRedStop cassette followed by nucGFP (Figure 5B). Injection of tamoxifen into double transgenic animals caused excision of the DsRed cassette, and expression of nucGFP from the heat shock promoter in osteoblasts and their immediate precursors. NucGFP-expressing cells were observed to enter the regeneration blastema, and form newly regenerated osteoblasts, and did not appear to significantly contribute to other lineages. Taken together, these results indicate that during zebrafish fin regeneration, osteoblasts dedifferentiate, proliferate, and re-differentiate into osteoblasts. Since the Cre-based labeling was sporadic and did not include all osteoblasts, it is still not known if osteoblast dedifferentiation accounts for the major cell type that regenerates the bony fin ray or whether other cell types also contribute. In summary, heart and fin regeneration results in zebrafish both demonstrate that limited dedifferentiation occurs resulting in expansion and redifferentiation to the original cell type.
Amphibian appendages harbor skeletal muscle, and this situation is likely more complex than the cardiomyocyte and fin osteoblast situation described above. Vertebrate skeletal muscle harbors a population of stem cells called satellite cells, which lie adjacent to mature muscle fibers and are activated by injury to proliferate and then differentiate and fuse into muscle fibers during repair (Le Grand and Rudnicki, 2007). Pax7+ satellite cells were shown to reside in salamander limb skeletal muscle (Kragl et al., 2009; Morrison et al., 2006). Four days after newt limb amputation, the blastema was shown to contain Pax7+ cells that presumably derived from muscle satellite cells but with the absence of cell-tracing data, other sources of the Pax7+ cells were not excluded. Furthermore clonally-expanded newt muscle satellite cells from cell culture transplanted into regenerating newt limbs contributed to both regenerated muscle and cartilage (Morrison et al., 2010). These results indicate that, as was the case in frog, muscle stem cells contribute to the newt regeneration blastema. Considering that the in vivo muscletracking experiments in Axolotl showed no contribution to cartilage, it needs to be resolved whether this difference in muscle tissue potency reflects a biological difference between two different salamander species (Axolotl versus newt) or is due to satellite cells acquiring an increased potency upon extensive culturing.
On the other hand, a number of experiments tracking in vivo or implanted muscle cells suggest that differentiated muscle cells may fragment, dedifferentiate and proliferate during salamander but not frog appendage regeneration (Echeverri et al., 2001; Kumar et al., 2000; Lo et al., 1993). TenascinC, an extracellular matrix protein consisting of 14 EGF-like repeats, and at least eight fibronectin-III domains, has been implicated in the fragmentation process while msx1 has been proposed to be important for the dedifferentiation process (Calve et al., 2010; Odelberg et al., 2000). Since the studies have focused on tracking a small number of in vivo or implanted myotubes, the true contribution of these cells to regenerating muscle has not been evaluated. Cre/loxP-based lineage tracing of skeletal muscle fibers versus satellite cells will be critical to evaluate the significance of muscle dedifferentiation versus stem cell activation in this lineage. Similarly, regeneration from the dermal compartment is widely assumed to derive from the dedifferentiation of fibroblasts to a lateral plate mesoderm-like cell, but the possibility of a resident stem cell taking on most of the regenerative role has not yet been excluded (Dunis and Namenwirth, 1977; Weiss, 1925).
Implications of the mosaic composition of the blastema
The vertebrate studies have shown that regeneration occurs by each tissue providing a separate pool of progenitor cells, with each having limited, if any, flexibility to form other tissue types. This information has several important implications. First, it suggests that some of the stem/progenitor cells utilized in appendage regeneration, for example the muscle satellite cell are similar to cognate stem/progenitor cells used during tissue repair in mammals. How limb amputation in salamanders can induce such progenitor cells to build an entire limb and why this does not occur in mammals is an enduring question. During limb regeneration, cut nerves and the convergence of skin cells from around the amputated limb are two crucial events required to signal the accumulation of blastema cells that can collectively build the missing limb structure. The work investigating these phenomena and the associated molecular knowledge have recently been reviewed (Nacu and Tanaka, 2011; Yokoyama, 2008). Second, cells deriving from the different tissues may respond to injury and regeneration cues differently. A host of molecular signaling factors including WNTs, BMP/TGF-βs, IGFs, and FGFs, have been identified as involved in appendage regeneration based on the inhibition of regeneration upon their inactivation, but detailed analysis of these phenotypes has been limited (for reviews see: Antos and Tanaka, 2010; Poss, 2010; Stoick-Cooper et al., 2007; Yokoyama, 2008). Precisely how these pathways affect cells from each different tissue is crucially lacking. Third, the similarity in lineage restriction between limb blastema cells and progenitors found in the developing limb bud suggests that morphogenesis and patterning events occurring during limb development and regeneration may be more similar than previously appreciated (Nacu and Tanaka, 2011). Finally, it is interesting that in vertebrate limb regeneration, blastema cells show restriction not only in their tissue fates, but also in the positional identity they can adopt along the proximal distal axis (Butler, 1955; Kragl et al., 2009). Understanding the molecular basis of positional identity and its restriction during regeneration is also an important future goal (Tamura et al., 2010).
Concluding remarks
The recent advances in identifying the cell sources for regeneration in several invertebrate and vertebrate model organisms have revealed a diversity of ways by which injured tissues provide progenitor cells for regeneration. Hydra and planarians, which regenerate whole body structures from small animal pieces, display distinct modes of achieving tissue-level pluripotency in the adult. Hydra appears to employ several separate, restricted stem cell pools, whereas planarians utilize a clonogenic, pluripotent stem cell. Among vertebrates, the blastema used for appendage regeneration is a mosaically built structure made up of several distinct, restricted progenitor cell pools that act in concert. However, the types of cellular mechanisms involved in new tissue production can be varied. Dedifferentiation and stem cell activation both appear to be contributing mechanisms for producing proliferating progenitors for regeneration, whereas regeneration of tissues such as the lens occurs via transdifferentiation (Eguchi, 1986).
These results demonstrate the importance of studying regeneration in many different animal and tissue contexts, as each system contributes distinct concepts for understanding tissue regeneration biology. The pluripotent planarian cNeoblast provides a unique system for studying how a dispersed population of pluripotent stem cells can be maintained in an adult tissue context and how pluripotent cells can be directed along different lineages to regenerate complex tissues and organs. The vertebrate models provide an arena for study of how tissue-restricted stem cells are implemented towards functional regeneration instead of imperfect tissue repair. Finally, the ability to track a differentiated cell through dedifferentiation toward regeneration events opens the path for dissection of the molecular control of in vivo dedifferentiation. In summary, the cell-tracking results described in this review go beyond the achievement of addressing longstanding, fundamental questions of regeneration biology and evolution, to opening exciting new opportunities to delve into molecular mechanisms of regeneration.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aboobaker AA. Planarian stem cells: a simple paradigm for regeneration. Trends Cell Biol. 2011 doi: 10.1016/j.tcb.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Adell T, Cebria F, Saló E. Gradients in planarian regeneration and homeostasis. Cold Spring Harb Perspect Biol. 2010;2 doi: 10.1101/cshperspect.a000505. a000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adell T, Saló E, Boutros M, Bartscherer K. Smed-Evi/Wntless is required for beta-catenin-dependent and -independent processes during planarian regeneration. Development. 2009;136:905–910. doi: 10.1242/dev.033761. [DOI] [PubMed] [Google Scholar]
- Adoutte A, Balavoine G, Lartillot N, Lespinet O, Prud'homme B, de Rosa R. The new animal phylogeny: reliability and implications. Proc Natl Acad Sci USA. 2000;97:4453–4456. doi: 10.1073/pnas.97.9.4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimenko MA, Mari-Beffa M, Becerra J, Geraudie J. Old questions, new tools, and some answers to the mystery of fin regeneration. Dev Dyn. 2003;226:190–201. doi: 10.1002/dvdy.10248. [DOI] [PubMed] [Google Scholar]
- Antos CL, Tanaka EM. Vertebrates that regenerate as models for guiding stem cells. Adv Exp Med Biol. 2010;695:184–214. doi: 10.1007/978-1-4419-7037-4_13. [DOI] [PubMed] [Google Scholar]
- Arques CG, Doohan R, Sharpe J, Torres M. Cell tracing reveals a dorsoventral lineage restriction plane in the mouse limb bud mesenchyme. Development. 2007;134:3713–3722. doi: 10.1242/dev.02873. [DOI] [PubMed] [Google Scholar]
- Baguñà J. Planarians. In: Ferretti P, Géraudie J, editors. Cellular and Molecular Basis of Regeneration: From Invertebrates to Humans. Chichester: John Wiley & Sons Ltd.; 1998. pp. 135–165. [Google Scholar]
- Baguñà J, Saló E, Auladell C. Regeneration and pattern formation in planarians. III. Evidence that neoblasts are totipotent stem cells and the source of blastema cells. Development. 1989;107:77–86. [Google Scholar]
- Bardeen CR, Baetjer FH. The inhibitive action of the Roentgen rays on regeneration in planarians. J Exp Zool. 1904;1:191–195. [Google Scholar]
- Bode HR. Continuous conversion of neuron phenotype in hydra. Trends Genet. 1992;8:279–284. doi: 10.1016/0168-9525(92)90254-2. [DOI] [PubMed] [Google Scholar]
- Bode HR. The interstitial cell lineage of hydra: a stem cell system that arose early in evolution. J Cell Sci. 1996;109:1155–1164. doi: 10.1242/jcs.109.6.1155. [DOI] [PubMed] [Google Scholar]
- Bode PM, Bode HR. Formation of pattern in regenerating tissue pieces of hydra attenuata. I. Head-body proportion regulation. Dev Biol. 1980;78:484–496. doi: 10.1016/0012-1606(80)90348-6. [DOI] [PubMed] [Google Scholar]
- Bosch TC. Why polyps regenerate and we don't: towards a cellular and molecular framework for Hydra regeneration. Dev Biol. 2007;303:421–433. doi: 10.1016/j.ydbio.2006.12.012. [DOI] [PubMed] [Google Scholar]
- Bosch TC, David CN. Stem cells of Hydra magnipapillata can differentiate into somatic cells and germ line cells. Dev Biol. 1987;121:182–191. [Google Scholar]
- Brockes JP, Kumar A, Velloso CP. Regeneration as an evolutionary variable. J Anat. 2001;199:3–11. doi: 10.1046/j.1469-7580.2001.19910003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan Regeneration in Phagocata gracilis (Leidy) Phys Zool. 1933;6:185–204. [Google Scholar]
- Butler EG. Regeneration of the urodele forelimb after reversal of its proximodistal axis. J Morphol. 1955;96:265–281. [Google Scholar]
- Calve S, Odelberg SJ, Simon HG. A transitional extracellular matrix instructs cell behavior during muscle regeneration. Dev Biol. 2010;344:259–271. doi: 10.1016/j.ydbio.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell R. Tissue dynamics of steady state growth in Hydra littoralis. I. Patterns of cell division. Dev Biol. 1967;15:487–502. doi: 10.1016/0012-1606(67)90039-5. [DOI] [PubMed] [Google Scholar]
- Chera S, Ghila L, Dobretz K, Wenger Y, Bauer C, Buzgariu W, Martinou JC, Galliot B. Apoptotic cells provide an unexpected source of Wnt3 signaling to drive hydra head regeneration. Dev Cell. 2009;17:279–289. doi: 10.1016/j.devcel.2009.07.014. [DOI] [PubMed] [Google Scholar]
- Cummings S, Bode H. Head regeneration and polarity reversal in Hydra attenuata can occur in the absence of DNA synthesis. Roux's Arch Dev Biol. 1984;194:79–86. doi: 10.1007/BF00848347. [DOI] [PubMed] [Google Scholar]
- Curtis WC. The life history, the normal fission, and the reproductive organs of Planaria maculata. Proc Boston Soc Nat Hist. 1902;30:515–559. [Google Scholar]
- Curtis WC, Hickman J. Effects of X-rays and radium upon regeneration in planarians. Anat Rec. 1926;34:145–146. [Google Scholar]
- David CN, Murphy S. Characterization of interstitial stem cells in hydra by cloning. Dev Biol. 1977;58:372–383. doi: 10.1016/0012-1606(77)90098-7. [DOI] [PubMed] [Google Scholar]
- David CN, Plotnick I. Distribution of interstitial stem cells in Hydra. Dev Biol. 1980;76:175–184. doi: 10.1016/0012-1606(80)90370-x. [DOI] [PubMed] [Google Scholar]
- Dubel S, Hoffmeister SA, Schaller HC. Differentiation pathways of ectodermal epithelial cells in hydra. Differentiation. 1987;35:181–189. doi: 10.1111/j.1432-0436.1987.tb00167.x. [DOI] [PubMed] [Google Scholar]
- Dubois F. Contribution á l 'ètude de la migration des cellules de règènèration chez les Planaires dulcicoles. Bull Biol Fr Belg. 1949;83:213–283. [Google Scholar]
- Dunis DA, Namenwirth M. The role of grafted skin in the regeneration of x-irradiated axolotl limbs. Dev Biol. 1977;56:97–109. doi: 10.1016/0012-1606(77)90157-9. [DOI] [PubMed] [Google Scholar]
- Dupin E, Real C, Glavieux-Pardanaud C, Vaigot P, Le Douarin NM. Reversal of developmental restrictions in neural crest lineages: transition from Schwann cells to glial-melanocytic precursors in vitro. Proc Natl Acad Sci U S A. 2003;100:5229–5233. doi: 10.1073/pnas.0831229100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri K, Clarke JD, Tanaka EM. In vivo imaging indicates muscle fiber dedifferentiation is a major contributor to the regenerating tail blastema. Dev Biol. 2001;236:151–164. doi: 10.1006/dbio.2001.0312. [DOI] [PubMed] [Google Scholar]
- Echeverri K, Tanaka EM. Ectoderm to mesoderm lineage switching during axolotl tail regeneration. Science. 2002;298:1993–1996. doi: 10.1126/science.1077804. [DOI] [PubMed] [Google Scholar]
- Eguchi G. Instability in cell commitment of vertebrate pigmented epithelial cells and their transdifferentiation into lens cells. Curr Top Dev Biol. 1986;20:21–37. doi: 10.1016/s0070-2153(08)60652-3. [DOI] [PubMed] [Google Scholar]
- Eisenhoffer GT, Kang H, Sánchez Alvarado A. Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell. 2008;3:327–339. doi: 10.1016/j.stem.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Taboada E, Moritz S, Zeuschner D, Stehling M, Scholer HR, Saló E, Gentile L. Smed-SmB, a member of the LSm protein superfamily, is essential for chromatoid body organization and planarian stem cell proliferation. Development. 2010;137:1055–1065. doi: 10.1242/dev.042564. [DOI] [PubMed] [Google Scholar]
- Flickinger RA. Isotopic evidence for a local origin of blastema cells in regenerating planarians. Exp Cell Res. 1964;34:403–406. [Google Scholar]
- Forsthoefel DJ, Newmark P. Emerging patterns in planarian regeneration. Current Opinion in Genetics and Development. 2009;19:412–420. doi: 10.1016/j.gde.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galliot B, Chera S. The Hydra model: disclosing an apoptosis-driven generator of Wnt-based regeneration. Trends Cell Biol. 2010;20:514–523. doi: 10.1016/j.tcb.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Galliot B, Ghila L. Cell plasticity in homeostasis and regeneration. Mol Reprod Dev. 2010;77:837–855. doi: 10.1002/mrd.21206. [DOI] [PubMed] [Google Scholar]
- Galliot B, Schmid V. Cnidarians as a model system for understanding evolution and regeneration. Int J Dev Biol. 2002;46:39–48. [PubMed] [Google Scholar]
- Gargioli C, Slack JM. Cell lineage tracing during Xenopus tail regeneration. Development. 2004;131:2669–2679. doi: 10.1242/dev.01155. [DOI] [PubMed] [Google Scholar]
- Gierer A, Berking S, Bode H, David C, Flick K, Hansmann G, Schaller H, Trenkner E. Regeneration of hydra from reaggregated cells. Nat New Biol. 1972;239:98–101. doi: 10.1038/newbio239098a0. [DOI] [PubMed] [Google Scholar]
- Goldring MB, Tsuchimochi K, Ijiri K. The control of chondrogenesis. J Cell Biochem. 2006;97:33–44. doi: 10.1002/jcb.20652. [DOI] [PubMed] [Google Scholar]
- Gremigni V, Miceli C. Cytophotometric evidence for cell 'transdifferentiation' in planarian regeneration. Wilhelm Roux's Archives. 1980;188:107–113. doi: 10.1007/BF00848801. [DOI] [PubMed] [Google Scholar]
- Gremigni V, Miceli C, Picano E. On the role of germ cells in planarian regeneration. II. Cytophotometric analysis of the nuclear Feulgen-DNA content in cells of regenerated somatic tissues. J Embryol Exp Morphol. 1980;55:65–76. [PubMed] [Google Scholar]
- Gremigni V, Nigro M, Puccinelli I. Evidence of male germ cell redifferentiation into female germ cells in planarian regeneration. J Embryol Exp Morphol. 1982;70:29–36. [PubMed] [Google Scholar]
- Guo T, Peters AH, Newmark PA. A Bruno-like gene is required for stem cell maintenance in planarians. Dev Cell. 2006;11:159–169. doi: 10.1016/j.devcel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Gurley KA, Elliott SA, Simakov O, Schmidt HA, Holstein TW, Sánchez Alvarado A. Expression of secreted Wnt pathway components reveals unexpected complexity of the planarian amputation response. Dev Biol. 2010;347:24–39. doi: 10.1016/j.ydbio.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley KA, Rink JC, Sánchez Alvarado A. Beta-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science. 2008;319:323–327. doi: 10.1126/science.1150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay ED. Regeneration. New York, Holt, Rinehart and Winston: 1966. [Google Scholar]
- Hayashi T, Asami M, Higuchi S, Shibata N, Agata K. Isolation of planarian X-ray-sensitive stem cells by fluorescence-activated cell sorting. Dev Growth Differ. 2006;48:371–380. doi: 10.1111/j.1440-169X.2006.00876.x. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Shibata N, Okumura R, Kudome T, Nishimura O, Tarui H, Agata K. Single-cell gene profiling of planarian stem cells using fluorescent activated cell sorting and its "index sorting" function for stem cell research. Dev Growth Differ. 2010;52:131–144. doi: 10.1111/j.1440-169X.2009.01157.x. [DOI] [PubMed] [Google Scholar]
- Holstein TW, Hobmayer E, David CN. Pattern of epithelial cell cycling in hydra. Dev Biol. 1991;148:602–611. doi: 10.1016/0012-1606(91)90277-a. [DOI] [PubMed] [Google Scholar]
- Holstein TW, Hobmayer E, Technau U. Cnidarians: an evolutionarily conserved model system for regeneration? Dev Dyn. 2003;226:257–267. doi: 10.1002/dvdy.10227. [DOI] [PubMed] [Google Scholar]
- Holtzer H. In: Mauro A, Shafiq SA, Milhorat AT, editors. Regeneration of striated muscle, and myogenesis; proceedings of the international conference convened by Muscular Dystrophy Associations of America at the Institute for Muscle Disease; March 28–29, 1969; New York. Amsterdam: Excerpta Medica; 1969. [Google Scholar]
- Hyman LH. The Invertebrates: Platyhelminthes and Rhynchocoela The acoelomate bilateria. Vol II. New York: McGraw-Hill Book Company Inc.; 1951. [Google Scholar]
- Iglesias M, Gomez-Skarmeta JL, Saló E, Adell T. Silencing of Smed-betacatenin1 generates radial-like hypercephalized planarians. Development. 2008;135:1215–1221. doi: 10.1242/dev.020289. [DOI] [PubMed] [Google Scholar]
- Jopling C, Boue S, Izpisua Belmonte JC. Dedifferentiation, transdifferentiation and reprogramming: three routes to regeneration. Nat Rev Mol Cell Biol. 2011;12:79–89. doi: 10.1038/nrm3043. [DOI] [PubMed] [Google Scholar]
- Jopling C, Sleep E, Raya M, Marti M, Raya A, Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller J. Die ungeschlechtliche Fortpflanzungder Süsswasser-Turbellarien. Jen Zeit Naturw. 1894;28:370–407. [Google Scholar]
- Khalturin K, Anton-Erxleben F, Milde S, Plotz C, Wittlieb J, Hemmrich G, Bosch TC. Transgenic stem cells in Hydra reveal an early evolutionary origin for key elements controlling self-renewal and differentiation. Dev Biol. 2007;309:32–44. doi: 10.1016/j.ydbio.2007.06.013. [DOI] [PubMed] [Google Scholar]
- Kikuchi K, Gupta V, Wang J, Holdway JE, Wills AA, Fang Y, Poss KD. tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development. 2011 doi: 10.1242/dev.067041. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, Poss KD. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopf F, Hammond C, Chekuru A, Kurth T, Hans S, Weber CW, Mahatma G, Fisher S, Brand M, Schulte-Merker S, et al. Bone Regenerates via Dedifferentiation of Osteoblasts in the Zebrafish Fin. Dev Cell. 2011;20:713–724. doi: 10.1016/j.devcel.2011.04.014. [DOI] [PubMed] [Google Scholar]
- Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, Tanaka EM. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460:60–65. doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- Kumar A, Velloso CP, Imokawa Y, Brockes JP. Plasticity of retrovirus-labelled myotubes in the newt limb regeneration blastema. Dev Biol. 2000;218:125–136. doi: 10.1006/dbio.1999.9569. [DOI] [PubMed] [Google Scholar]
- Lange C. Studies on the cellular basis of radiation lethality. I. The pattern of mortality in the whole-body irradiated planarian (Tricladida, Paludicola) Int J Radiat Biol Relat Stud Phys Chem Med. 1968;13:511–530. doi: 10.1080/09553006814550581. [DOI] [PubMed] [Google Scholar]
- Laube F, Heister M, Scholz C, Borchardt T, Braun T. Reprogramming of newt cardiomyocytes is induced by tissue regeneration. J Cell Sci. 2006;119:4719–4729. doi: 10.1242/jcs.03252. [DOI] [PubMed] [Google Scholar]
- Le Grand F, Rudnicki MA. Skeletal muscle satellite cells and adult myogenesis. Curr Opin Cell Biol. 2007;19:628–633. doi: 10.1016/j.ceb.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnert GH. Beobachtung an Landplanarien. Arch Naturgech. 1891;1:306–350. [Google Scholar]
- Lender T. Factors in morphogenesis of regenerating fresh-water planaria. In: Abercrombie M, Brachet J, editors. Advances in Morphogenesis. New York: Academic Press, Inc.; 1962. pp. 305–331. [Google Scholar]
- Lender T, Gabriel A. Les néoblasts marqués par l'uridine tritiée migrent et édifient le blastème des planaires d'eau douce. C R Acad Sc Paris. 1965;260:4095–4097. [PubMed] [Google Scholar]
- Lenhoff SG, Lenhoff HM. Hydra and the Birth of Experimental Biology, 1744: Abraham Trembley's Memoirs Concerning the Natural History of a Type of Freshwater Polyp with Arms Shaped like Horns. Pacific Grove, California: Boxwood Press; 1986. [Google Scholar]
- Lo DC, Allen F, Brockes JP. Reversal of muscle differentiation during urodele limb regeneration. Proc Natl Acad Sci U S A. 1993;90:7230–7234. doi: 10.1073/pnas.90.15.7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcum BA, Campbell RD. Development of Hydra lacking nerve and interstitial cells. J Cell Sci. 1978a;29:17–33. doi: 10.1242/jcs.29.1.17. [DOI] [PubMed] [Google Scholar]
- Marcum BA, Campbell RD. Developmental roles of epithelial and interstitial cell lineages in hydra: analysis of chimeras. J Cell Sci. 1978b;32:233–247. doi: 10.1242/jcs.32.1.233. [DOI] [PubMed] [Google Scholar]
- McHedlishvili L, Epperlein HH, Telzerow A, Tanaka EM. A clonal analysis of neural progenitors during axolotl spinal cord regeneration reveals evidence for both spatially restricted and multipotent progenitors. Development. 2007;134:2083–2093. doi: 10.1242/dev.02852. [DOI] [PubMed] [Google Scholar]
- Montgomery J, Coward S. On the minimal size of a planarian capable of regeneration. Trans Am Microsc Soc. 1974;93:386–391. [PubMed] [Google Scholar]
- Morgan TH. Experimental studies of the regeneration of Planaria maculata. Arch Entw Mech Org. 1898;7:364–397. [Google Scholar]
- Morrison JI, Borg P, Simon A. Plasticity and recovery of skeletal muscle satellite cells during limb regeneration. FASEB J. 2010;24:750–756. doi: 10.1096/fj.09-134825. [DOI] [PubMed] [Google Scholar]
- Morrison JI, Loof S, He P, Simon A. Salamander limb regeneration involves the activation of a multipotent skeletal muscle satellite cell population. J Cell Biol. 2006;172:433–440. doi: 10.1083/jcb.200509011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacu E, Tanaka EM. Limb regeneration: a new development? Ann Rev Cell Dev Biol. 2011 doi: 10.1146/annurev-cellbio-092910-154115. in press. [DOI] [PubMed] [Google Scholar]
- Namenwirth M. The inheritance of cell differentiation during limb regeneration in the axolotl. Dev Biol. 1974;41:42–56. doi: 10.1016/0012-1606(74)90281-4. [DOI] [PubMed] [Google Scholar]
- Newmark P, Sánchez Alvarado A. Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev Biol. 2000;220:142–153. doi: 10.1006/dbio.2000.9645. [DOI] [PubMed] [Google Scholar]
- Newmark PA, Reddien PW, Cebria F, Sánchez Alvarado A. Ingestion of bacterially expressed double-stranded RNA inhibits gene expression in planarians. Proc Natl Acad Sci. 2003;100:11861–11865. doi: 10.1073/pnas.1834205100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmark PA, Sánchez Alvarado A. Not your father's planarian: a classic model enters the era of functional genomics. Nature Reviews Genetics. 2002;3:210–219. doi: 10.1038/nrg759. [DOI] [PubMed] [Google Scholar]
- Noda K. Reconstitution of dissociated cells of hydra. Zool Mag. 1971;80:99–101. [Google Scholar]
- Oberpriller JO, Oberpriller JC. Response of the adult newt ventricle to injury. J Exp Zool. 1974;187:249–253. doi: 10.1002/jez.1401870208. [DOI] [PubMed] [Google Scholar]
- Odelberg SJ, Kollhoff A, Keating MT. Dedifferentiation of mammalian myotubes induced by msx1. Cell. 2000;103:1099–1109. doi: 10.1016/s0092-8674(00)00212-9. [DOI] [PubMed] [Google Scholar]
- Oviedo NJ, Levin M. smedinx-11 is a planarian stem cell gap junction gene required for regeneration and homeostasis. Development. 2007;134:3121–3131. doi: 10.1242/dev.006635. [DOI] [PubMed] [Google Scholar]
- Parish CL, Beljajeva A, Arenas E, Simon A. Midbrain dopaminergic neurogenesis and behavioural recovery in a salamander lesion-induced regeneration model. Development. 2007;134:2881–2887. doi: 10.1242/dev.002329. [DOI] [PubMed] [Google Scholar]
- Park HD, Ortmeyer AB, Blankenbaker DP. Cell division during regeneration in Hydra. Nature. 1970;227:617–619. doi: 10.1038/227617a0. [DOI] [PubMed] [Google Scholar]
- Pearse RV, 2nd, Scherz PJ, Campbell JK, Tabin CJ. A cellular lineage analysis of the chick limb bud. Dev Biol. 2007;310:388–400. doi: 10.1016/j.ydbio.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson BJ, Sánchez Alvarado A. A planarian p53 homolog regulates proliferation and self-renewal in adult stem cell lineages. Development. 2010;137:213–221. doi: 10.1242/dev.044297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen KJ. Studies on the nature of planarian connective tissue. Z Zellforsch Mikrosk Anat. 1961;53:569–608. [Google Scholar]
- Pellettieri J, Sánchez Alvarado A. Cell turnover and adult tissue homeostasis: from humans to planarians. Annu Rev Genet. 2007;41:83–105. doi: 10.1146/annurev.genet.41.110306.130244. [DOI] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW. Smed-betacatenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science. 2008;319:327–330. doi: 10.1126/science.1149943. [DOI] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW. A wound-induced Wnt expression program controls planarian regeneration polarity. Proc Natl Acad Sci U S A. 2009;106:17061–17066. doi: 10.1073/pnas.0906823106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW. Polarized notum activation at wounds inhibits Wnt function to promote planarian head regeneration. Science. 2011;332:852–855. doi: 10.1126/science.1202143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietsch P. Differentiation in regeneration. I. The development of muscle and cartilage following deplantation of of regenerating limb blastemata of Amblystoma larvae. Dev Biol. 1961;3:255–264. doi: 10.1016/0012-1606(61)90046-x. [DOI] [PubMed] [Google Scholar]
- Poss KD. Advances in understanding tissue regenerative capacity and mechanisms in animals. Nat Rev Genet. 2010;11:710–722. doi: 10.1038/nrg2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- Randolph H. Observations and experiments on regeneration in planarians. Arch Entw Mech Org. 1897;5:352–372. [Google Scholar]
- Real C, Glavieux-Pardanaud C, Le Douarin NM, Dupin E. Clonally cultured differentiated pigment cells can dedifferentiate and generate multipotent progenitors with self-renewing potential. Dev Biol. 2006;300:656–669. doi: 10.1016/j.ydbio.2006.09.032. [DOI] [PubMed] [Google Scholar]
- Reddien PW. Constitutive gene expression and specification of tissue identity in adult planarian biology. Trends in Genetics. 2011 doi: 10.1016/j.tig.2011.04.004. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sánchez Alvarado A. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev Cell. 2005a;8:635–649. doi: 10.1016/j.devcel.2005.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sánchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells for regeneration and homeostasis. Science. 2005b;310:1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Sánchez Alvarado A. Fundamentals of planarian regeneration. Ann Rev Cell Dev Bio. 2004;20:725–757. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- Reyer RW, Woolfitt RA, Withersty LT. Stimulation of lens regeneration from the newt dorsal iris when implanted into the blastema of the regenerating limb. Dev Biol. 1973;32:258–281. doi: 10.1016/0012-1606(73)90240-6. [DOI] [PubMed] [Google Scholar]
- Rose C, Shostak S. The transformation of gastrodermal cells to neoblasts in regenerating Phagocata gracilis (Leidy) Exp Cell Res. 1968;50:553–561. doi: 10.1016/0014-4827(68)90418-7. [DOI] [PubMed] [Google Scholar]
- Saló E, Abril JF, Adell T, Cebria F, Eckelt K, Fernandez-Taboada E, Handberg-Thorsager M, Iglesias M, Molina MD, Rodriguez-Esteban G. Planarian regeneration: achievements and future directions after 20 years of research. Int J Dev Biol. 2009;53:1317–1327. doi: 10.1387/ijdb.072414es. [DOI] [PubMed] [Google Scholar]
- Sánchez Alvarado A. Regeneration in the Metazoans: Why does it happen? BioEssays. 2000;22:578–590. doi: 10.1002/(SICI)1521-1878(200006)22:6<578::AID-BIES11>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Sánchez Alvarado A. Planarian regeneration: its end is its beginning. Cell. 2006;124:241–245. doi: 10.1016/j.cell.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Sánchez Alvarado A, Newmark PA. Double-stranded RNA specifically disrupts gene expression during planarian regeneration. Proc Natl Acad Sci. 1999;96:5049–5054. doi: 10.1073/pnas.96.9.5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone ML, Meisel J, Reddien PW. The Mi-2-like Smed-CHD4 gene is required for stem cell differentiation in the planarian Schmidtea mediterranea. Development. 2010:1231–1241. doi: 10.1242/dev.042051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman K, Kafatos FC. Transdifferentiation in the labial gland of silk moths: is DNA required for cellular metamorphosis? Cell Differ. 1974;3:81–94. doi: 10.1016/0045-6039(74)90030-x. [DOI] [PubMed] [Google Scholar]
- Shibata N, Rouhana L, Agata K. Cellular and molecular dissection of pluripotent adult somatic stem cells in planarians. Dev Growth Differ. 2010;52:27–41. doi: 10.1111/j.1440-169X.2009.01155.x. [DOI] [PubMed] [Google Scholar]
- Siebert S, Anton-Erxleben F, Bosch TC. Cell type complexity in the basal metazoan Hydra is maintained by both stem cell based mechanisms and transdifferentiation. Dev Biol. 2008;313:13–24. doi: 10.1016/j.ydbio.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Smid I, Tardent P. The potentialities of endoderm interstitial cells in Hydra attenuata Pall. Dev Biol. 1986;117:672–675. [Google Scholar]
- Smith M, Hickman A, Amanze D, Lumsden A, Thorogood P. Trunk neural crest origin of caudal fin mesenchyme in the zebrafish Brachydanio rerio. Proc R Soc Lond. 1994;256:137–145. [Google Scholar]
- Solana J, Lasko P, Romero R. Spoltud-1 is a chromatoid body component required for planarian long-term stem cell self-renewal. Dev Biol. 2009;328:410–421. doi: 10.1016/j.ydbio.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- Steen TP. Stability of chondrocyte differentiation and contribution of muscle to cartilage during limb regeneration in the axolotl (Siredon mexicanum) J Exp Zool. 1968;167:49–78. doi: 10.1002/jez.1401670105. [DOI] [PubMed] [Google Scholar]
- Steen TP. Origin and differentiative capacities of cells in the blastema of the regenerating salamander limb. Am Zool. 1970;10:119–132. doi: 10.1093/icb/10.2.119. [DOI] [PubMed] [Google Scholar]
- Stocum DL. The urodele limb regeneration blastema: a self-organizing system. I. Morphogenesis and differentiation of autografted whole and fractional blastemas. Dev Biol. 1968;18:457–480. doi: 10.1016/0012-1606(68)90052-3. [DOI] [PubMed] [Google Scholar]
- Stoick-Cooper CL, Moon RT, Weidinger G. Advances in signaling in vertebrate regeneration as a prelude to regenerative medicine. Genes Dev. 2007;21:1292–1315. doi: 10.1101/gad.1540507. [DOI] [PubMed] [Google Scholar]
- Straube WL, Tanaka EM. Reversibility of the differentiated state: regeneration in amphibians. Artif Organs. 2006;30:743–755. doi: 10.1111/j.1525-1594.2006.00296.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Ohgo S, Yokoyama H. Limb blastema cell: a stem cell for morphological regeneration. Dev Growth Differ. 2010;52:89–99. doi: 10.1111/j.1440-169X.2009.01144.x. [DOI] [PubMed] [Google Scholar]
- Technau U, Cramer von Laue C, Rentzsch F, Luft S, Hobmayer B, Bode HR, Holstein TW. Parameters of self-organization in Hydra aggregates. Proc Natl Acad Sci U S A. 2000;97:12127–12131. doi: 10.1073/pnas.97.22.12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Technau U, Holstein TW. Phenotypic maturation of neurons and continuous precursor migration in the formation of the peduncle nerve net in Hydra. Dev Biol. 1996;177:599–615. doi: 10.1006/dbio.1996.0189. [DOI] [PubMed] [Google Scholar]
- Tsonis PA, Madhavan M, Tancous EE, Del Rio-Tsonis K. A newt's eye view of lens regeneration. Int J Dev Biol. 2004;48:975–980. doi: 10.1387/ijdb.041867pt. [DOI] [PubMed] [Google Scholar]
- Tu S, Johnson SL. Fate restriction in the growing and regenerating zebrafish fin. Dev Cell. 2011;20:725–732. doi: 10.1016/j.devcel.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuan RS. Biology of developmental and regenerative skeletogenesis. Clin Orthop Relat Res. 2004:S105–S117. doi: 10.1097/01.blo.0000143560.41767.ee. [DOI] [PubMed] [Google Scholar]
- Wagner DE, Wang IE, Reddien PW. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science. 2011;332:811–816. doi: 10.1126/science.1203983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner F. Zur Kenntnis der ungeschlechtlichen Fortpflanzung von Microstoma nebst allegemeinen Bemerkungen über Teilung und Knospung im Tierreich. Z Jahrb. 1890;4:349–423. [Google Scholar]
- Wallace BM, Wallace H. Participation of grafted nerves in amphibian limb regeneration. J Embryol Exp Morphol. 1973;29:559–570. [PubMed] [Google Scholar]
- Wallace H, Maden M, Wallace BM. Participation of cartilage grafts in amphibian limb regeneration. J Embryol Exp Morphol. 1974;32:391–404. [PubMed] [Google Scholar]
- Weiss P. Unabhängigkeit der Extremitätenregenerationvom Skelett (bei Triton cristatus) Wilhelm Roux’ Arch Entwicklungsmech Org. 1925;104:359–394. [Google Scholar]
- Weissman IL, Anderson DJ, Gage F. Stem and progenitor cells: origins, phenotypes, lineage commitments, and transdifferentiations. Annu Rev Cell Dev Biol. 2001;17:387–403. doi: 10.1146/annurev.cellbio.17.1.387. [DOI] [PubMed] [Google Scholar]
- Wenemoser D, Reddien PW. Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev Biol. 2010;344:979–991. doi: 10.1016/j.ydbio.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittlieb J, Khalturin K, Lohmann JU, Anton-Erxleben F, Bosch TC. Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. Proc Natl Acad Sci U S A. 2006;103:6208–6211. doi: 10.1073/pnas.0510163103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff E. Recent researches on the regeneration of planaria. In: Rudnick D, editor. Regeneration 20th Growth Symposium. New York: The Ronald Press Co.; 1962. pp. 53–84. [Google Scholar]
- Wolff E, Dubois F. Sur la migration des cellules de régénération chez les planaires. Revue Swisse Zool. 1948;55:218–227. [Google Scholar]
- Wolff G. Entwicklungsphysiologische Studien. I. Die Regeneration der Urodelenlinse. Wilhelm Roux Arch EntwMech Org. 1895;1:380–390. [Google Scholar]
- Woodruff L, Burnett AL. The origin of the blastemal cells in Dugesia tigrina. Exp Cell Res. 1965;38:295–305. doi: 10.1016/0014-4827(65)90405-2. [DOI] [PubMed] [Google Scholar]
- Yokoyama H. Initiation of limb regeneration: the critical steps for regenerative capacity. Dev Growth Differ. 2008;50:13–22. doi: 10.1111/j.1440-169X.2007.00973.x. [DOI] [PubMed] [Google Scholar]