Abstract
Two ubiquitin-conjugating enzymes, RAD6 and the heteromeric UBC13–MMS2 complex, have been implicated in post-replicative DNA damage repair in yeast. Here we provide a mechanistic basis for cooperation between the two enzymes. We show that two chromatin-associated RING finger proteins, RAD18 and RAD5, play a central role in mediating physical contacts between the members of the RAD6 pathway. RAD5 recruits the UBC13–MMS2 complex to DNA by means of its RING finger domain. Moreover, RAD5 association with RAD18 brings UBC13–MMS2 into contact with the RAD6–RAD18 complex. Interaction between the two RING finger proteins thus promotes the formation of a heteromeric complex in which the two distinct ubiquitin-conjugating activities of RAD6 and UBC13–MMS2 can be closely coordinated. Surprisingly, UBC13 and MMS2 are largely cytosolic proteins, but DNA damage triggers their redistribution to the nucleus. These findings suggest a mechanism by which the activity of this DNA repair pathway could be regulated.
Keywords: chromatin/DNA repair/RING finger/SNF2/SWI2 homolog/ubiquitin-conjugating enzyme
Introduction
Accurate maintenance of a cell’s genetic information is endangered by the fact that DNA is a rather unstable molecule that is constantly subject to damage by both environmental influences and spontaneous decay. It is therefore not surprising that cells support a complex enzymatic machinery for the maintenance and repair of their genomes. In yeast and higher eukaryotes DNA damage repair is executed mainly by three groups of enzymes corresponding to three mechanistically distinct repair systems (for reviews see Prakash et al., 1993; Friedberg et al., 1995). Nucleotide excision repair is responsible for recognizing and removing UV-induced pyrimidine dimers and other damaged or chemically modified bases and nucleotides. Double strand breaks are repaired largely via homologous recombination. Finally, the third system, which is collectively called post-replicative repair, is believed to act during and after DNA synthesis when unrepaired lesions in the template strand cause a stalling of the replication machinery and thereby lead to gaps in the newly synthesized strand. This repair system comprises a rather heterogeneous group of enzymes that fall into at least two branches, an error-prone and an error-free pathway (Lawrence, 1994). While error-prone repair entails a mutagenic bypass of damaged sites by a specialized polymerase capable of translesion synthesis, the mechanism of the error-free repair system remains poorly understood. The principal member of the post-replication repair group, mediating both the error-free and the error-prone branch, is RAD6. Consequently, mutations in RAD6 confer a high degree of sensitivity towards various DNA-damaging agents and an enhanced spontaneous mutation rate, but a defect in damage-induced mutagenesis (Lawrence et al., 1974; Prakash, 1974, 1981; Montelone et al., 1981).
The RAD6 gene encodes a ubiquitin-conjugating enzyme (UBC) (Jentsch et al., 1987). UBCs are a class of structurally related proteins that catalyze the transfer of the small protein ubiquitin to a substrate protein (for reviews see Hochstrasser, 1996; Scheffner et al., 1998). Further conjugation of ubiquitin, usually to Lys48 of the previous ubiquitin moiety (Chau et al., 1989; Finley et al., 1994), results in the formation of multiubiquitin chains that label the substrate for selective degradation by the 26S proteasome (Coux et al., 1996). In other cases, (mono-) ubiquitylation may serve as a targeting or localization signal (Strous, 1999). The UBC RAD6 is involved not only in DNA damage repair, but also in sporulation (Cox and Parry, 1968), retrotransposition (Picologlu et al., 1990), silencing (Huang et al., 1997) and the degradation of several short-lived proteins (Dohmen et al., 1991; Sung et al., 1991; Kornitzer et al., 1994; Madura and Varshavsky, 1994). Its catalytic activity as a UBC is essential for these functions (Sung et al., 1990). In the context of the protein degradation pathway via the N-end rule, RAD6 in complex with the ubiquitin ligase UBR1 catalyzes the assembly of Lys48-linked multiubiquitin chains that target the ubiquitylated substrate protein to proteasomal degradation (Chau et al., 1989; Dohmen et al., 1991; Sung et al., 1991; Madura et al., 1993). For DNA repair, RAD6 forms an alternative complex with the single-stranded DNA-dependent ATPase RAD18 (Bailly et al., 1994, 1997a). However, the proteins relevant to DNA repair that are ubiquitylated by RAD6 remain unknown.
Recently, another ubiquitin-conjugating activity has been implicated in the RAD6 pathway. Hofmann and Pickart (1999) have shown that a stable complex of the conjugating enzyme UBC13 and a non-canonical UBC variant, MMS2, is capable of assembling an alternative type of multiubiquitin chain, in which the ubiquitin moieties are linked via Lys63. This type of chain previously had been associated with post-replication DNA repair, since yeast cells depending on a mutated form of ubiquitin in which Lys63 is replaced by arginine display a UV-sensitive phenotype that falls into the RAD6 group (Spence et al., 1995). Moreover, the MMS2 gene was cloned by functional complementation of a mutant sensitive to the alkylating agent methyl methanesulfonate (MMS) and was classified as a member of the error-free RAD6 pathway (Broomfield et al., 1998; Xiao et al., 1999). Both UBC13 and MMS2 have homologs in higher eukaryotes, and human MMS2 complements the UV sensitivity of the yeast mms2 mutant (Xiao et al., 1998), suggesting that this conjugation system may be highly conserved.
Here we show that the function of UBC13 and MMS2 in DNA damage repair is mediated by the chromatin-associated RING finger protein RAD5, another component of the error-free repair system. RAD5 recruits the UBC13–MMS2 complex to DNA by means of its RING finger domain. Moreover, we found that RAD5 acts as a bridging factor to bring UBC13 and MMS2 into contact with the RAD6–RAD18 complex, thereby providing a means to coordinate the distinct ubiquitin-conjugating activities of RAD6 and UBC13–MMS2. UBC13 and MMS2 are normally cytoplasmic proteins, but are redistributed to the nucleus in response to DNA-damaging agents. This damage-induced recruitment of UBC13–MMS2 suggests a mechanism by which the activity of error-free post-replicative DNA repair can be modulated.
Results
The function of UBC13–MMS2 in DNA repair is dependent on RAD5
By double mutant analysis based on the UV sensitivities of the respective mutants, the MMS2 gene had been assigned to the post-replicative repair group defined by RAD6 (Broomfield et al., 1998). Hofmann and Pickart (1999) have implicated UBC13 in the same pathway, based on its cooperation with MMS2 in Lys63-linked ubiquitin chain synthesis and because the ubc13 mms2 double mutant shows a UV sensitivity identical to that of the ubc13 and mms2 single mutants. Using deletion mutants representative of the three major epistasis groups of DNA repair genes, rad2, rad52 and rad6, we similarly confirmed that the function of UBC13, like that of MMS2, is specific for the RAD6 pathway, consistent with the notion that the two proteins function cooperatively and exclusively in the post-replication repair pathway, and that the activity of one component depends on the presence of the other (data not shown).
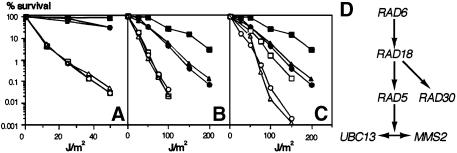
MMS2 had been assigned to the error-free branch of the RAD6 pathway and was shown to be dependent on RAD18 (Broomfield et al., 1998; Xiao et al., 1999). We found the same phenotype for ubc13 deletion mutants (Figure 1A). We therefore analyzed the relationship of MMS2 and UBC13 to the other known components of error-free repair, RAD5 and RAD30, which are both dependent on RAD18, but act independently of each other (Johnson et al., 1992; McDonald et al., 1997). UV sensitivity assays of the double mutants mms2 rad5 and ubc13 rad5 show that RAD5 is epistatic to both MMS2 and UBC13 (Figure 1B). On the other hand, the effects of rad30 and mms2 or ubc13 deletions, respectively, are additive (Figure 1C). In summary, our genetic analysis suggests that the function of UBC13–MMS2 in the RAD6 pathway is mediated by RAD5. The repair system comprising the five components RAD6, RAD18, RAD5, UBC13 and MMS2 thus represents a sub-branch of the RAD6 pathway that seems distinct from the branch represented by the epistasis group RAD6–RAD18–RAD30 (Figure 1D).
Fig. 1. Genetic interactions between the members of the RAD6 pathway. Survival after irradiation (254 nm) is plotted against the applied UV dosage. Experiments were performed in duplicate or triplicate. Filled square, wild-type; filled triangle, ubc13; filled circle, mms2. (A) Open square, rad18; open triangle, rad18 ubc13; open circle, rad18 mms2. (B) Open square, rad5; open triangle, rad5 ubc13; open circle, rad5 mms2. (C) Open square, rad30; open triangle, rad30 ubc13; open circle, rad30 mms2. (D) Schematic representation of the epistatic relationships within error-free post-replicative DNA repair.
RAD5 interacts with UBC13 and RAD18
Stable physical interactions have been demonstrated between RAD6 and RAD18 (Bailly et al., 1994) as well as between UBC13 and MMS2 (Hofmann and Pickart, 1999). We used the two-hybrid system to identify possible additional interactions within the RAD6 pathway. The yeast strain used here carries two reporter genes, HIS3 and ADE2, for growth assays on selective media with different stringencies, with ADE2 being more stringent than HIS3 (James et al., 1996). Figure 2A shows an array examining all possible interactions between the five components, RAD6, RAD18, RAD5, UBC13 and MMS2. Both previously known interactions, those between RAD6 and RAD18 and between UBC13 and MMS2, were confirmed in this experiment and gave strong signals on both selective media. In addition, we observed that both RAD18 and RAD5 are capable of self-interaction in the two-hybrid system. Moreover, RAD5 showed interactions with both RAD18 and UBC13, resulting in a series of associations that reflects the order established by genetic analysis.
Fig. 2. Physical interactions between the members of the RAD6 pathway. (A) Two-hybrid assays. The open reading frames were expressed in PJ69-4A as a fusion to the GAL4 activation domain (AD) in combination with a fusion to the GAL4 DNA-binding domain (BD), as indicated to the left of and above the panels. The AD and BD vectors without inserts (pGAD424 and pGBT9) are represented as ‘–’. Transformants were spotted onto selective media as indicated. Positive interactions result in His or, in case of stronger interactions, His and Ade prototrophy. The absence of growth on selective media is not likely to be due to a failure to express the corresponding proteins, as all constructs give rise to positive signals when combined with the appropriate partners. (B) Co-immunoprecipitations. Immunoprecipitations were performed from doubly tagged strains as indicated using a rabbit polyclonal anti-myc antibody (+) or an unrelated rabbit antibody (–). Interacting proteins were detected on western blots with a mouse monoclonal anti-HA or anti-VSV antibody. The ‘input’ samples represent 1% of the crude extract; 30% of the precipitated material was loaded in the ‘+’ and ‘–’ samples.
In order to confirm the newly identified interactions by an independent method, we performed co-immunoprecipitations using extracts of yeast strains that carried epitope-tagged versions of the respective proteins (Figure 2B). The tagged proteins were expressed from their own promoters in the original genetic context, with the exception of VSVRAD5, which was overexpressed at least 10-fold from an integrative plasmid using the GAL1-10 promoter in order to facilitate its detection (our unpublished results). The in vivo functionality of the tagged proteins was verified by examining the UV sensitivity of the modified strains (data not shown). Proteins were precipitated using polyclonal anti-myc antibodies, and associated proteins were detected subsequently by western blots using a monoclonal anti-hemagglutinin (HA) or, in the case of RAD5, anti-vesicular stomatitis virus (VSV) antibody. A diploid strain carrying two differently tagged RAD18 alleles was used to examine the self-association of RAD18, and plasmid-derived VSVRAD5 was co-precipitated with the genomically encoded RAD5myc protein. Using this combination of epitopes, we were able to confirm all pairwise interactions found in the two-hybrid system. We conclude that the involvement of RAD18, RAD5 and UBC13–MMS2 in the error-free branch of the RAD6 pathway is due to direct physical associations. Consistent with the results of the two-hybrid analysis, the low ratio of co-precipitated to input material in the assays involving RAD5 as well as the RAD18 self-association indicates that these interactions are rather weak or transient and thus of a different nature to that of the stable RAD6–RAD18 complex. Preliminary experiments indicate that the strength of these interactions does not increase when DNA damage is induced, indicating that the observed affinities are inherent in the proteins (unpublished data).
Domains responsible for interactions between RAD18 and RAD5 are distinct from those mediating UBC binding
To address the question of whether RAD18 and RAD5 may be capable of mediating higher order assemblies involving more than two of the factors at a time, we designed a series of truncation constructs in order to identify the domains responsible for each of the newly found interactions. Both RAD18 and RAD5 are multidomain proteins bearing several structural motifs that allow their classification into a number of protein families (Figure 3A). RAD18 was identified as an ATPase based on a Walker type A nucleotide-binding motif (Bailly et al., 1997a). At its N-terminus, RAD18 carries a RING finger domain (residues 28–65), and a second cysteine-rich, possibly zinc-binding motif of the C2HC type is located between amino acids 190 and 210. RAD5 is a member of the SNF2/SWI2 superfamily of ATPases containing seven helicase consensus motifs (Johnson et al., 1992, 1994). The helicase-like domain comprises the C-terminal half of the protein (amino acids 526–1170) and is interrupted by a centrally positioned RING finger.
Fig. 3. Interactions between the members of the RAD6 pathway: domain analyses of RAD18 and RAD5. (A) Schematic representation of the domains present in RAD5 and RAD18 and the constructs used for two-hybrid assays. Nucleotide-binding domain motifs are indicated in black. Characteristic protein domains of RAD5 and RAD18 and positively (+) and negatively (–) charged regions of RAD18 are indicated. Results of the interaction analysis are summarized on the right. (B–F) Two-hybrid assays were performed as in Figure 2. The top panels represent non-selective growth, while the bottom panels show positive interactions on medium lacking histidine. Negative controls (AD or BD vector without insert) are included in each assay (–). (B) Interactions of RAD18 truncations with full-length RAD18 and RAD18(1–248). (C) Interactions of RAD5 constructs with full-length RAD5 and UBC13. (D) Interactions of RAD5 constructs with RAD5(1–556) and RAD5(C914S). (E) Interactions of RAD5 constructs with full-length RAD18 and RAD18(1–248). (F) Interactions of RAD18 truncations with RAD5(1–556).
In order to delimit the region responsible for the RAD18 self-association, we examined the ability of truncated versions of RAD18 to interact with the full-length protein in the two-hybrid system. Figure 3B shows that the RING finger as well as the C-terminal half of RAD18, including the nucleotide-binding motif, are dispensable for this interaction. Thus, the region of RAD18 which comprises the C2HC motif [i.e. the variant RAD18(83–248)] is sufficient for self-association. Notably, this self-association of RAD18 is independent of its interaction with RAD6, which was localized to a basic region comprising amino acids 371–410 between the nucleotide-binding site and a C-terminal more acidic domain (Bailly et al., 1997b). Self-association of RAD5 is also mediated by its N-terminal half, since deletion of the C-terminal helicase domain or inactivation of the RING finger domain by a point mutation in one of its conserved cysteines (C914S) did not prevent the interaction (Figure 3C and D). In contrast to the self-association, however, interaction with UBC13 requires an intact RING domain, as truncation of RAD5 N-terminally of the RING finger or selective inactivation by the C914S mutation abolished growth on selective media (Figure 3C). Constructs containing the RING finger but lacking the N-terminal half of RAD5 [i.e. RAD5(598–984) and RAD5(801–984)] showed no interaction with UBC13, indicating that N-terminal regions of RAD5 may be required for a productive interaction with UBC13. Alternatively, these constructs may be folded incorrectly or poorly expressed. Thus, the RING finger of RAD5 is necessary, but may not be sufficient for interaction with UBC13.
Figure 3E and F shows an analysis of the RAD18–RAD5 interaction. Full-length RAD18 and the variant RAD18(1–248) interact with RAD5 independently of the RAD5 RING finger and helicase domains (Figure 3E). Because of the autoactivating nature of RAD5(1–556) and to some extent RAD5(1–800) as fusions to the DNA-binding domain (Figure 3D and E), we used the reverse combination to demonstrate that the N-terminal half of RAD5 is sufficient for an interaction with RAD18 (Figure 3F). Similarly, the RAD18 RING finger as well as its nucleotide-binding and RAD6 interaction domains are dispensable for the RAD18–RAD5 interaction.
In summary, the regions responsible for self-association of both RAD18 and RAD5 coincide with those mediating the contact between the two proteins, suggesting that self-association and RAD18–RAD5 interaction may be competing processes. At the same time, these regions are distinct from the domains required for interaction with the corresponding UBCs. Both RING finger proteins, RAD18 and RAD5, should thus be capable of a simultaneous interaction with their corresponding UBC, RAD6 or UBC13, and another molecule of RAD5 or RAD18.
RAD18 and RAD5 can coordinate multimeric UBC complexes
If dimerization of RAD5 with RAD18 via its N-terminal domain is indeed independent of the RING finger-mediated interaction with UBC13, RAD5 should be capable of bringing UBC13–MMS2 into contact with the RAD6–RAD18 complex. In order to obtain direct evidence for such a bridging function, we re-examined the interaction between RAD18 and UBC13 in the three-hybrid system, overexpressing RAD5 from an episomal plasmid. Figure 4A shows that RAD5 can in fact mediate an indirect interaction between RAD18 and UBC13. This bridging function was dependent on the RAD5 RING finger, as the C914S mutation in the RING finger or a truncated construct lacking this domain abolished the interaction. Similarly, overexpression of UBC13 conveyed an indirect association between RAD5 and MMS2, indicating that the entire UBC13–MMS2 dimer contacts RAD5 (Figure 4B). Again, this interaction was dependent on an intact RAD5 RING finger.
Fig. 4. Higher order interactions between the DNA repair proteins. (A) Three-hybrid interaction of RAD18 and UBC13, promoted by overexpression of RAD5 from an episomal plasmid. Empty vectors are designated as ‘–’, the variant RAD5(C914S) as CS, and RAD5(1–800) as (1–800). (B) Three-hybrid interaction of RAD5 and MMS2, promoted by overexpression of UBC13 from an episomal plasmid. Variants are labeled as in (A). (C) Co-immunoprecipitation of RAD5 and UBC13 with RAD18. Immunoprecipitations were performed from the strain RAD18HA UBC13myc overexpressing VSVRAD5 (labeled wt) or VSVRAD5(C914S) (labeled CS) using mouse monoclonal anti-HA antibody 12CA5 (+) or an unrelated mouse antibody (–). Proteins were detected on western blots with anti-HA, anti-VSV and anti-myc antibodies as indicated. The ‘input’ samples represent 1% of the crude extract; 30% of the precipitated material was loaded in the ‘+’ and ‘–’ samples. (D) RAD18 mediates association between multiple molecules of RAD6. Immunoprecipitations and western blots were performed as described for (C) from diploid RAD6myc/RAD6HA strains that were either RAD18 or rad18.
Independent evidence for the bridging function of RAD5 was obtained by co-immunoprecipitation. For this purpose, VSVRAD5 was overexpressed in a rad5Δ strain carrying genomically encoded RAD18HA and UBC13myc. Both VSVRAD5 and UBC13myc were detectable in immunoprecipitates of RAD18HA (Figure 4C). However, when VSVRAD5 was replaced by the C914S RING finger mutant, VSVRAD5(C914S) still co-precipitated with RAD18HA, but UBC13myc was no longer detectable, indicating that the RAD5 RING domain is indeed responsible for recruitment of UBC13, but does not affect the association between RAD5 and RAD18. Since Bailly et al. (1997a) have presented evidence that RAD18 exists quantitatively in a stable complex with RAD6, and its RAD6-binding domain is independent of the domain responsible for interaction with RAD5 (see above), we conclude that RAD5 indeed promotes an association of the two UBCs, RAD6 and UBC13–MMS2, in a multimeric complex.
On the other hand, di- or oligomerization of RAD18 similarly should lead to an association of multiple molecules of RAD6. We tested this hypothesis by co-immunoprecipitation, using diploid strains carrying two differently tagged RAD6 alleles. RAD6myc co-precipitated with RAD6HA only in the wild-type, but not in a rad18/rad18 deletion strain (Figure 4D). These results are consistent with the notion that individual RAD6–RAD18 dimers can assemble into larger complexes by means of RAD18 self-association.
UBC13 and MMS2 relocalize from the cytoplasm to the nucleus in response to DNA damage
Although it is generally believed that the components of post-replicative DNA repair reside in the nucleus, their in vivo localization has never been demonstrated experimentally. In order to visualize the members of the UBC13–MMS2 pathway directly, we performed immunofluorescence microscopy using the the epitope-tagged alleles. Figure 5A shows that both RAD18 and RAD5 are indeed nuclear proteins, while RAD6 is present in the nucleus as well as the cytoplasm, as might be expected based on its nuclear and cytoplasmic functions. To demonstrate a chromatin association of RAD18 and RAD5, we visualized the proteins on chromatin spreads. Genomic epitope-tagged RAD5myc was used for these experiments in order to avoid any mislocalization due to overexpression. Both proteins localized to the chromatin in a punctate pattern (Figure 5B). Their associations with chromatin were independent of each other, as RAD18 was detectable on the spreads in the absence of RAD5 and vice versa (Figure 5B). Double labeling of RAD18 and RAD5 showed significant co-localization of the two proteins (Figure 5C), implying that the interactions demonstrated above reflect the physiological situation in the context of chromatin.
Fig. 5. In vivo localization of DNA repair proteins. (A) Localization of RAD6myc, RAD18myc and VSVRAD5. Tagged proteins were detected as described in Materials and methods. A Cy3 conjugate served as secondary antibody. The untagged control strain is designated as wt. Images show focal planes across the nuclei. Lysed spheroplasts are shown for VSVRAD5, as the protein was not detectable in intact cells. (B) Localization of RAD18myc and RAD5myc on spread chromatin. Cells harboring tagged alleles of RAD18 or RAD5 were spread onto microscope slides. The DNA was stained with DAPI, and proteins were visualized by immunofluorescence. (C) Co-localization of RAD18HA and RAD5myc on the chromatin in a doubly tagged strain, using Cy3 and Cy5 conjugates as secondary antibodies. Colors were assigned for visualization of the superpositions. (D) Staining controls performed on spreads of a strain bearing no tagged alleles (wt). (E–H) DNA damage-dependent localization of UBC13myc and MMS2myc. Treatment of the cells with UV light (+ UV) or MMS (+ MMS) before fixation is indicated on the left. Intact cells are depicted in (E) and (F), while (G) and (H) show images of lysed speroplasts. (E and G) UBC13myc; (F and H) MMS2myc.
UBC13 and MMS2 were predicted to reside in the nucleus based on putative nuclear localization signals (Hofmann and Pickart, 1999). To our surprise, however, we found that both UBC13 and MMS2 appear to be cytoplasmic proteins. In fact, they seemed to be largely excluded from the nucleus under normal circumstances (Figure 5E and F). Interestingly, this exclusion was abolished when the cells were exposed to DNA damage, either by UV irradiation or by treatment with the alkylating agent MMS, before fixation. This change in localization was even more obvious in partially lysed spheroplasts, which allow better visualization of the nucleus (Figure 5G and H). Despite its presence in the nucleus, however, UBC13 was not detectable on chromatin spreads even after MMS treatment (data not shown). This finding suggests that the UBC13–MMS2 complex enters the nucleus upon DNA damage, but the RAD5–UBC13 interaction is not stable enough to withstand the chromatin fixation procedure, again arguing for a dynamic association of the two proteins.
RAD5 recruits UBC13–MMS2 to chromatin in response to DNA damage
The ability of UBC13 to co-precipitate with RAD5 in a cell extract prepared under non-damage conditions may seem contradictory in light of the fact that these proteins normally reside in different cellular compartments. In order to prove that this interaction reflects an inherent affinity of the two proteins for each other and is indeed relevant in vivo in the context of chromatin, we made use of the formaldehyde cross-linking procedure first devised by Solomon and Varshavsky (1985). Treatment of live cells with formaldehyde results in reversible cross-links bridging ∼2 Å between proteins and DNA, thus providing a means to ‘freeze’ a particular state of protein–protein and protein–DNA complexes and to prevent a redistribution of the components during purification and analysis. We asked whether this method could be used to preserve a transient association of UBC13 with RAD5 on the chromatin for detection. We subjected extracts of cross-linked cells expressing myc-tagged UBC13 to isopycnic centrifugation in a CsCl gradient in order to separate the chromatin and associated components from bulk protein. Figure 6 (left column) shows the distribution of UBC13myc along the gradient for extracts prepared under several different conditions, as well as the DNA contents of the respective fractions (indicated at the bottom of the figure). Extracts of a rad5 deletion mutant (first row) or cells not treated with MMS (second row) showed a low background of material in fractions 3–5, which correspond to the chromatin peak. This amount was somewhat increased after MMS treatment (third row). Overexpression of VSVRAD5 strikingly increased the fraction of UBC13myc associated with the chromatin (fourth row). No UBC13myc was detectable in the corresponding fractions if the cross-linking step was omitted (fifth row). We conclude that the UBC13–MMS2 complex is recruited to the chromatin by RAD5 upon its accumulation in the nucleus in response to DNA damage.
Fig. 6. Recruitment of UBC13 to chromatin by RAD5. Extracts prepared from formaldehyde-cross-linked cells of the indicated strains were subjected to isopycnic centrifugation on a CsCl gradient, and fractions were analyzed by SDS–PAGE and western blot after reversal of the cross-links in SDS sample buffer. The distribution of the vacuolar enzyme carboxypeptidase Y served as a control, representing a protein that should not be cross-linked to chromatin. DNA contents of the individual fractions were visualized by spotting on ethidium bromide-containing agarose. Fractions are numbered from the highest to lowest density. ‘Input’ represents an aliquot of each extract not subjected to CsCl gradient centrifugation. Treatment of the cells with MMS before cross-linking (+ MMS) and omission of the formaldehyde treatment step (– x-link) are indicated.
Discussion
Evidence for alternative UBC complexes in DNA repair
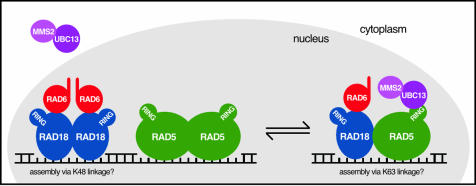
We have provided evidence for a role for RAD18 and RAD5 in mediating the assembly of repair complexes involving different combinations of UBCs. Self-association of RAD18 can promote the assembly of multiple molecules of RAD6, while a heteromeric association with RAD5 results in a complex comprising the two distinct UBCs RAD6 and UBC13–MMS2. Figure 7 shows a model of these repair complexes. While at this time we cannot rigorously exclude the alternative scenario that RAD18 can interact with itself and with RAD5 simultaneously, resulting in the formation of large, multimeric complexes, based on our domain analysis of RAD18 and RAD5 we favor the notion that homo- and heterodimerization are mutually exclusive. A prerequisite for this model is in fact given by the weakness of the interactions between RAD18, RAD5 and UBC13. Dynamic associations and dissociations would favor an equilibrium between the alternative complexes and thus provide the basis for a potential mechanism of regulation of RAD6 function in DNA repair. The heteromeric complex could function in the recruitment of UBC13–MMS2 for cooperation with RAD6 in error-free repair, resulting in the formation of Lys63-linked multiubiquitin chains, while homodimers of RAD18 could target RAD6 independently of UBC13–MMS2 to a different set of RAD6 substrates, for example in the error-prone or the RAD30-dependent sub-pathway. Ubiquitin conjugation in this case would probably involve mono-ubiquitylation or the Lys48-linked chains for which RAD6 is known (Jentsch et al., 1987; Chau et al., 1989). In addition, the RAD5 ATPase may also act independently of UBC13–MMS2. An equilibrium between the alternative complexes might be controlled by the abundance of UBC13–MMS2 in the nucleus. Its redistribution within the cell from the cytoplasm to the nucleus in response to DNA damage supports this notion.
Fig. 7. Model for alternative repair complexes within the RAD6 pathway of DNA repair. A schematic representation of the DNA repair proteins shows the distribution of RING finger–UBC complexes that would result from a competition between RAD18 and RAD5 homo- and heterodimerization. Homodimerization of RAD18 would lead to assemblies comprising RAD6 as the sole UBC, whereas heterodimerization with RAD5 would promote the recruitment of UBC13–MMS2 for the assembly of lysine (K) 63-linked multiubiquitin chains.
RING finger domains for recruitment of UBCs to chromatin
RING finger proteins are now emerging as a new class of members of the ubiquitin system. While this conserved zinc-binding motif is present in a multitude of proteins involved in diverse cellular processes (Saurin et al., 1996), an increasing number of RING finger proteins are found associated with the ubiquitin conjugation machinery (Freemont, 2000; Tyers and Jorgensen, 2000). In many cases, ubiquitin ligase activity of the RING finger protein itself or the associated protein complex has been demonstrated directly, with the RING finger being responsible for mediating contact with the UBC. Lorick et al. (1999) have even proposed a general function in UBC-dependent ubiquitin ligation for the RING domain of most if not all RING finger proteins.
We have provided evidence that two chromatin-associated RING finger proteins, RAD18 and RAD5, mediate the UBC13–MMS2-dependent sub-branch of the RAD6 pathway by physical interaction with a UBC. Thus, even though the lack of a physiological substrate precludes experiments that would allow the demonstration of ubiquitin ligase activity, a targeting function of RAD18 and RAD5 for the UBCs to their sites of action is evident. The importance of the RING domains for post-replicative DNA repair is underscored further by the fact that neither RAD18 nor RAD5 RING finger mutants can rescue the UV sensitivities of the respective deletion strains (our unpublished results). Considering that the RING finger of RAD18 is dispensable for interaction with RAD6, these observations in fact point to a catalytic function of this domain, as in the case of the RAD6-dependent ubiquitin ligase UBR1 (Xie and Varshavsky, 1999).
RAD5 is the first example of a member of the ubiquitin system in which the RING domain is embedded in another conserved structural motif, the helicase-like domain of the SNF2/SWI2 superfamily. RAD5 shares this arrangement with several related proteins which so far have not been implicated in ubiquitin conjugation, such as yeast RAD16 and DIS1, Schizosaccharomyces pombe RAD8 (Doe et al., 1993), and mammalian HIP116 (Sheridan et al., 1995) and RUSH-1 (Hayward-Lester et al., 1996). Together, these proteins constitute a distinct subfamily within the SNF2/SWI2 superfamily, which comprises DNA and RNA helicases as well as chromatin remodeling factors and sequence-specific transcription factors (Eisen et al., 1995; Pazin et al., 1997). We imagine that the RING fingers could act as UBC recruitment domains in the other members of this protein family as well, thus combining their role in chromatin metabolism with a function in ubiquitin conjugation.
Mechanism of RAD6 and UBC13–MMS2 cooperation in DNA repair
The RAD5-mediated recruitment of UBC13–MMS2 to the RAD6–RAD18 DNA repair complex reported here implies a close cooperation between the UBCs in ubiquitin conjugation and raises the question of how the two enzymes might coordinate their activities towards a common substrate protein. A possible answer is suggested by the observation that UBC13–MMS2 assembles unanchored multiubiquitin chains in the absence of a substrate (Hofmann and Pickart, 1999), while RAD6 is known to attach ubiquitin directly to a substrate protein either with or without the help of a ubiquitin ligase (Jentsch et al., 1987; Dohmen et al., 1991; Sung et al., 1991). Based on these notions, we can envisage two scenarios for a cooperation of the two UBCs in DNA repair. One possibility is that RAD6 might use multiubiquitin chains pre-assembled by UBC13–MMS2 for their attachment to a substrate protein. Alternatively, RAD6 may serve to conjugate a single ubiquitin moiety to a substrate, which can then be used for further elongation by the UBC13–MMS2 complex directed to the correct location by means of its association with RAD5.
Nevertheless, the biological function of Lys63-linked multiubiquitin chains in DNA repair remains enigmatic. While Prakash (1994) and others have invoked protein degradation for RAD6-dependent DNA repair, Hofmann and Pickart (1999) have suggested a role in signaling distinct from the degradation signal of Lys48-linked chains for recognition by the 26S proteasome. The unusual type of chains assembled by UBC13–MMS2 would thus serve in the recruitment of as yet unidentified downstream factors to sites of DNA damage. This model is supported by the fact that the general turnover of short-lived proteins is not perturbed in a ubiquitin mutant strain lacking Lys63 (Spence et al., 1995), and the finding that mutations affecting the catalytic function of the proteasome do not result in UV sensitivity (Dor et al., 1996). Identification of the ubiquitylation substrates of RAD6 and UBC13–MMS2 as well as a putative receptor for Lys63-linked multiubiquitin chains will be crucial to our understanding of post-replicative DNA damage repair.
Materials and methods
Yeast strains and media
Standard protocols were followed for the preparation of yeast media, transformation, sporulation and tetrad dissection (Guthrie and Fink, 1991; Ausubel et al., 1994). With the exception of the two-hybrid assay strain PJ69-4A, provided by P.James, all strains are derivatives of the diploid wild-type DF5 (Finley et al., 1985) and its haploids (his3-Δ200 leu2-3,112 lys2-801 trp1-1 ura3-52). They are designated by their relevant genotypes in the text and figures. The rad6Δ::HIS3 and ubc13Δ::HIS3 strains were provided by W.Seufert and K.Matuschewski, respectively (unpublished). All other deletion strains were prepared by transformation with the respective deletion constructs described below. Deletions were confirmed by an assay of the expected phenotype (UV sensitivity) in combination with PCR and/or Southern blot analysis. Double mutants were constructed by standard crossing. Yeast strains bearing 9myc- or 6HA-tagged alleles of RAD6, RAD18, RAD5, UBC13 and MMS2 were constructed by a PCR strategy using in vivo recombination (Knop et al., 1999). Functionality of the tagged alleles was confirmed by assaying the strains for UV sensitivity (see below).
Construction of plasmids
Plasmid mms2Δ::HIS3 was constructed by amplifying 511 bp of the upstream and 420 bp of the downstream flanking regions of MMS2 by PCR from genomic DNA and combining the fragments with a HIS3 cassette in pBluescript-SK(+) (Stratagene). rad18Δ::TRP1 contains 464 bp of the RAD18 upstream flanking region and the C-terminal part of the open reading frame (ORF; 520 bp) interrupted by the TRP1 locus in pBluescript-SK(+). rad30Δ::HIS3 was constructed by inserting a 3076 bp PCR fragment containing the RAD30 gene with flanking regions into pBluescript-SK(+) and replacing the StuI–EheI fragment, containing the entire ORF, with the HIS3 marker. rad5Δ::HIS3 was constructed similarly by amplification of the RAD5 ORF, insertion into pBluescript-SK(+) and replacement of the internal BclI–BglII fragment with HIS3. The complete ORFs of RAD18, RAD5, UBC13 and MMS2 were cloned into the two-hybrid vectors pGAD424 and pGBT9 (Clontech) by PCR from genomic DNA. RAD6 was cloned into pAS2-1 and a derivative of pGAD GH (Clontech). Truncations of RAD18 and RAD5 were constructed using internal restriction sites. The point mutation in the RING finger of RAD5 was created by PCR mutagenesis. The constructs for VSVRAD5 overexpression are based on the integrative plasmid YIplac211 carrying the URA3 marker (Gietz and Sugino, 1988), the GAL1-10 promoter, a VSV epitope and a transcription terminator derived from pGAD424. Plasmids YEp-RAD5, YEp-RAD5(1–800) and YEp-RAD5(C914S) were constructed similarly under the control of the ADH1 promoter derived from pAS2-1 in the episomal vector YEplac195, carrying the URA3 marker (Gietz and Sugino, 1988). Sequence maps of all constructs are available on request.
UV sensitivity assays
Yeast cultures grown to logarithmic phase in YPD medium were spread onto YPD plates at appropriate dilutions and irradiated with UV light (254 nm) at 0.83 J/m2/s for varying times to apply the specified dosage. Plates were incubated in the dark and colonies were counted after 3 days. All experiments were performed in duplicate or triplicate, and relative survival was averaged.
Two-hybrid assays
Yeast two-hybrid strain PJ69-4A (James et al., 1996) was transformed simultaneously with combinations of activation (AD) and binding domain (BD) vectors and plated onto SC –Leu/Trp. Colonies were resuspended in water at equal densities, spotted onto SC –Leu/Trp, SC –His/Leu/Trp and SC –Ade/His/Leu/Trp, and scored for differential growth on the selective media after 3–5 days. The three-hybrid assays were performed similarly by transformation of PJ69-4A with the episomal YEp-RAD5 constructs or the empty vector YEplac195 in combination with the respective AD and BD constructs and omission of uracil in all media.
Immunoprecipitations
Cells from an exponential culture were disrupted by French press lysis or by agitation with glass beads in protein buffer (20 mM Tris–HCl pH 8.0, 150 mM NaCl) supplemented with Complete™ protease inhibitors (Roche), 5 mM benzamidine and 2 mM phenylmethylsulfonyl fluoride (PMSF). Extracts were made 0.05–0.2% in Triton X-100 and clarified by centrifugation (20 000 g, 20 min). The supernatant was incubated at 4°C with 1 or 2 µg of antibody for 2 h. Protein G–agarose (Roche) was added and incubation was continued for 1 h. After washing the beads with protein buffer + 0.05–0.2% Triton X-100, proteins were eluted by incubation in SDS sample buffer at 95°C for 10 min, and aliquots were subjected to SDS–PAGE and western blot analysis. Antibodies used for immunoprecipitation and western blots were mouse monoclonal anti-myc antibody 9E10, anti-HA 12CA5, anti-VSV (Roche), and rabbit polyclonal anti-myc and anti-HA antibodies (Santa Cruz Biotechnology). Unrelated antibodies from the same species served as negative controls during immunoprecipitations.
Immunofluorescence studies
Exponential cultures of diploid strains were grown in YPD liquid medium at 30°C. For UV treatment, cells were spread onto YPD plates and irradiated with a dose corresponding to ∼50% survival. The cells were then washed off the plates and incubated in liquid culture for another 90 min at 30°C to allow induction of the repair process. For induction of chemical DNA damage, 0.1% MMS was added directly to the culture medium, and incubation was continued for 2 h. The cells were then fixed with 4% formaldehyde and prepared for immunofluorescence as described (Spector et al., 1998). Tagged proteins were detected with monoclonal anti-myc, anti-VSV or rat anti-HA antibody (Roche). The secondary antibodies were Cy3 or Cy5 conjugates (Dianova). DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI). Pictures were captured on a Leica DM RXA fluorescence microscope equipped with a digital camera, and images corresponding to the focal planes of the nuclei were generated using the deconvolution procedure included in the ‘Open Lab’ software (Improvision). Chromatin spreads were performed as described by Klein et al. (1992). Briefly, spheroplasts were lysed on microscope slides by sequential additions of fixative (4% paraformaldehyde, 3.4% sucrose) and detergent (1% Lipsol), and the mixture was spread out on the slides with a glass rod. Slides were air-dried, washed with phosphate-buffered saline (PBS), and immunofluorescence was performed as described.
Formaldehyde cross-linking experiments
The in vivo cross-linking procedure was adapted from Orlando et al. (1997) and Hecht et al. (1999). Cells grown in synthetic complete medium at 30°C to exponential phase were treated with 0.1% MMS for 2 h where indicated. Formaldehyde was added to 1% directly to the growth medium, and the cultures were incubated with shaking at room temperature for 20 min, followed by addition of 125 mM glycine and further incubation for 5 min. Cells (200 OD600) were harvested, washed twice with PBS, resuspended in lysis buffer (50 mM HEPES–KOH pH 7.5, 140 mM NaCl, 1 mM EDTA) with Complete™ protease inhibitors (Roche) and broken by agitation with glass beads. The chromatin was sheared by sonication, and detergents were added (1% Triton X-100, 0.1% sodium deoxycholate). After centrifugation, cesium chloride was added to the clarified lysate to a final density of 1.42 g/ml, adjusting the final volume to 5 ml. Isopycnic centrifugation was performed in a Beckmann SW 55Ti rotor at 195 000 g, 20°C, for 72 h. Fractions of 0.5 ml each were collected from the bottom of the tube. Aliquots were subjected to trichloroacetic acid precipitation, and pellets were resuspended in SDS sample buffer containing 0.5 M β-mercaptoethanol and incubated for 30 min at 95°C. Samples were subjected to SDS–PAGE and western blot analysis, using anti-myc antibody for detection of UBC13myc and monoclonal antibody 10A5-B5 (Molecular Probes) for carboxypeptidase Y. DNA was analyzed by agarose gel electrophoresis as described (Orlando et al., 1997) or by spotting aliquots onto ethidium bromide-containing agarose plates.
Acknowledgments
Acknowledgements
We thank K.Matuschewski for the isolation of UBC13 and the construction of the ubc13 knock out mutant, P.James, M.Knop and W.Seufert for gifts of plasmids and/or strains, V.Orlando for advice on formaldehyde cross-linking, and M.Knop for advice on microscopy. We are grateful to S.Thalmeir and E.Löser for excellent technical assistance, and we thank J.Höhfeld, C.Koch and G.Pyrowolakis for critical comments on the manuscript. H.D.U. was supported by a fellowship from the American Cancer Society, and S.J. by grants from the Deutsche Forschungsgemeinschaft and the European TMR ubiquitin network.
References
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1994) Current Protocols in Molecular Biology. Greene Publishing and John Wiley, New York, NY. [Google Scholar]
- Bailly V., Lamb,J., Sung,P., Prakash,S. and Prakash,L. (1994) Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev., 8, 811–820. [DOI] [PubMed] [Google Scholar]
- Bailly V., Lauder,S., Prakash,S. and Prakash,L. (1997a) Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J. Biol. Chem., 272, 23360–23365. [DOI] [PubMed] [Google Scholar]
- Bailly V., Prakash,S. and Prakash,L. (1997b) Domains required for dimerization of yeast Rad6 ubiquitin-conjugating enzyme and Rad18 DNA binding protein. Mol. Cell. Biol., 17, 4536–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broomfield S., Chow,B.L. and Xiao,W. (1998) MMS2, encoding a ubiquitin-conjugating-enzyme-like protein, is a member of the yeast error-free postreplication repair pathway. Proc. Natl Acad. Sci. USA, 95, 5678–5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau V., Tobias,J.W., Bachmair,A., Marriott,D., Ecker,D.J., Gonda,D.K. and Varshavsky,A. (1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science, 243, 1576–1583. [DOI] [PubMed] [Google Scholar]
- Coux O., Tanaka,K. and Goldberg,A.L. (1996) Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem., 65, 801–847. [DOI] [PubMed] [Google Scholar]
- Cox B.S. and Parry,J.M. (1968) The isolation, genetics and survival characteristics of ultraviolet light sensitive mutants in yeast. Mutat. Res., 6, 37–55. [DOI] [PubMed] [Google Scholar]
- Doe C.L., Murray,J.M., Shayeghi,M., Hoskins,M., Lehmann,A.R., Carr,A.M. and Watts,F.Z. (1993) Cloning and characterization of the Schizosaccharomyces pombe rad8 gene, a member of the SNF2 helicase family. Nucleic Acids Res., 21, 5964–5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen J.R., Madura,K., Bartel,B. and Varshavsky,A. (1991) The N-end rule is mediated by the UBC2(RAD6) ubiquitin-conjugating enzyme. Proc. Natl Acad. Sci. USA, 88, 7351–7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dor Y, Raboy,B. and Kulka,R.G. (1996) Role of the conserved carboxy-terminal α-helix of Rad6p in ubiquitination and DNA repair. Mol. Microbiol., 21, 1197–1206. [DOI] [PubMed] [Google Scholar]
- Eisen J.A., Sweder,K.S. and Hanawalt,P.C. (1995) Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res., 23, 2715–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D., Özkaynak,E. and Vashavsky,A. (1985) The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell, 48, 1035–1046. [DOI] [PubMed] [Google Scholar]
- Finley D., Sadis,S., Monia,B.P., Boucher,P., Ecker,D.J., Crooke,S.T. and Chau,V. (1994) Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol. Cell. Biol., 14, 5501–5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freemont P.S. (2000) Ubiquitination: RING for destruction? Curr. Biol., 10, R84–R87. [DOI] [PubMed] [Google Scholar]
- Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. American Society for Microbiology Press, Washington, DC. [Google Scholar]
- Gietz R.D. and Sugino,A. (1988) New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene, 74, 527–534. [DOI] [PubMed] [Google Scholar]
- Guthrie C. and Fink,G.R. (1991) Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego, CA. [Google Scholar]
- Hayward-Lester A., Hewetson,A., Beale,E.G., Oefner,P.J., Doris,P.A. and Chilton,B.S. (1996) Cloning, characterization, and steroid-dependent posttranscriptional processing of RUSH-1 α and β, two uteroglobin promoter-binding proteins. Mol. Endocrinol., 10, 1335–1349. [DOI] [PubMed] [Google Scholar]
- Hecht A., Strahl-Bolsinger,S. and Grunstein,M. (1999) Mapping DNA interaction sites of chromosomal proteins: crosslinking studies in yeast. In Becker,P. (ed.), Methods in Molecular Biology—Chromatin Protocols. Humana Press, Totowa, NJ, pp. 467–479. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. (1996) Ubiquitin-dependent protein degradation. Annu. Rev. Genet., 30, 405–439. [DOI] [PubMed] [Google Scholar]
- Hofmann R.M. and Pickart,C.M. (1999) Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell, 96, 645–653. [DOI] [PubMed] [Google Scholar]
- Huang H., Kahana,A., Gottschling,D., Prakash,L. and Liebman,S. (1997) The ubiquitin-conjugating enzyme Rad6 (Ubc2) is required for silencing in Saccharomyces cerevisiae. Mol. Cell. Biol., 17, 6693–6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Halladay,J. and Craig,E.A. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics, 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S., McGrath,J.P. and Varshavsky,A. (1987) The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature, 329, 131–134. [DOI] [PubMed] [Google Scholar]
- Johnson R.E., Henderson,S.T., Petes,T.D., Prakash,S., Bankmann,M. and Prakash,L. (1992) Saccharomyces cerevisiae RAD5-encoded DNA repair protein contains DNA helicase and zinc-binding sequence motifs and affects the stability of simple repetitive sequences in the genome. Mol. Cell. Biol., 12, 3807–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.E., Prakash,S. and Prakash,L. (1994) Yeast DNA repair protein RAD5 that promotes instability of simple repetitive sequences is a DNA-dependent ATPase. J. Biol. Chem., 269, 28259–28262. [PubMed] [Google Scholar]
- Klein F., Laroche,T., Cardenas,M.E., Hofmann,J.F.-X., Schweizer,D. and Gasser,S.M. (1992) Localization of RAP1 and topoisomerase II in nuclei and meiotic chromosomes of yeast. J. Cell Biol., 117, 935–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Siegers,K., Pereira,G., Zachariae,W., Winsor,B., Nasmyth,K. and Schiebel,E. (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast, 15, 963–972. [DOI] [PubMed] [Google Scholar]
- Kornitzer D., Raboy,B., Kulka,R.G. and Fink,G.R. (1994) Regulated degradation of the transcription factor Gcn4. EMBO J., 13, 6021–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C. (1994) The RAD6 DNA repair pathway in Saccharomyces cerevisiae: what does it do, and how does it do it? BioEssays, 16, 253–258. [DOI] [PubMed] [Google Scholar]
- Lawrence C.W., Steward,J.W., Sherman,F. and Christensen,R. (1974) Specificity and frequency of ultraviolet-induced reversion of an iso-1-cytochrome c ochre mutant in radiation-sensitive strains of yeast. J. Mol. Biol., 85, 137–162. [DOI] [PubMed] [Google Scholar]
- Lorick K.L., Jensen,J.P., Fang,S., Ong,A.M., Hatakeyama,S. and Weissman,A.M. (1999) RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl Acad. Sci. USA, 96, 11364–11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madura K. and Varshavsky,A. (1994) Degradation of Gα by the N-end rule pathway. Science, 265, 1454–1458. [DOI] [PubMed] [Google Scholar]
- Madura K., Dohmen,R.J. and Varshavsky,A. (1993) N-recognin/Ubc2 interactions in the N-end rule pathway. J. Biol. Chem., 268, 12046–12054. [PubMed] [Google Scholar]
- McDonald J.P., Levine,A.S. and Woodgate,R. (1997) The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics, 147, 1557–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montelone B.A., Prakash,S. and Prakash,L. (1981) Recombination and mutagenesis in rad6 mutants of Saccharomyces cerevisiae: evidence for multiple functions of the RAD6 gene. Mol. Gen. Genet., 184, 410–415. [DOI] [PubMed] [Google Scholar]
- Orlando V., Strutt,H. and Paro,R. (1997) Analysis of chromatin structure by in vivo formaldehyde-crosslinking. Methods, 11, 205–214. [DOI] [PubMed] [Google Scholar]
- Pazin M.J. and Kadonaga,J.T. (1997) SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein–DNA interactions. Cell, 88, 737–740. [DOI] [PubMed] [Google Scholar]
- Picologlou S., Brown,N. and Liebman,S.W. (1990) Mutations in RAD6, a yeast gene encoding a ubiquitin-conjugating enzyme, stimulate retrotransposition. Mol. Cell. Biol., 10, 1017–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash L. (1974) Lack of chemically induced mutation in repair-deficient mutant of yeast. Genetics, 78, 1101–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash L. (1981) Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3, and rad52 mutations. Mol. Gen. Genet., 184, 471–478. [DOI] [PubMed] [Google Scholar]
- Prakash L. (1994) The RAD6 gene and protein of Saccharomyces cerevisiae. Ann. N Y Acad. Sci., 726, 267–273. [DOI] [PubMed] [Google Scholar]
- Prakash S., Sung,P. and Prakash,L. (1993) DNA repair genes and proteins of Saccharomyces cerevisiae. Annu. Rev. Genet., 27, 33–70. [DOI] [PubMed] [Google Scholar]
- Saurin A.J., Borden,K.L.B., Boddy,M.N. and Freemont,P.S. (1996) Does this have a familiar RING? Trends Biochem. Sci., 21, 208–214. [PubMed] [Google Scholar]
- Scheffner M., Smith,S. and Jentsch,S. (1998) The ubiquitin-conjugation system. In Peters,J.-M., Harris,J.R. and Finley,D. (eds), Ubiquitin and the Biology of the Cell. Plenum Press, New York, pp. 65–98. [Google Scholar]
- Sheridan P.L., Schorpp,M., Voz,M.L. and Jones,K.A. (1995) Cloning of an SNF2/SWI2-related protein that binds specifically to the SPH motifs of the SV40 enhancer and to the HIV-1 promoter. J. Biol. Chem., 270, 4575–4587. [DOI] [PubMed] [Google Scholar]
- Solomon M.J. and Varshavsky,A. (1985) Formaldehyde-mediated DNA–protein crosslinking: a probe for in vivo chromatin structures. Proc. Natl Acad. Sci. USA, 82, 6470–6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D.L., Goldman,R.D. and Leinwand,L.A. (1998) Cells: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Spence J., Sadis,S., Haas,A.L. and Finley,D. (1995) A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol. Cell. Biol., 15, 1265–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strous G.J. and Govers,R. (1999) The ubiquitin–proteasome system and endocytosis. J. Cell Sci., 112, 1417–1423. [DOI] [PubMed] [Google Scholar]
- Sung P., Prakash,S. and Prakash,L. (1990) Mutation of cysteine-88 in the Saccharomyces cerevisiae RAD6 protein abolishes its ubiquitin-conjugating activity and its various biological functions. Proc. Natl Acad. Sci. USA, 87, 2685–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P., Berleth,E., Pickart,C., Prakash,S. and Prakash,L. (1991) Yeast RAD6 encoded ubiquitin conjugating enzyme mediates protein degradation dependent on the N-end-recognizing E3 enzyme. EMBO J., 10, 2187–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M. and Jorgensen P. (2000) Proteolysis and the cell cycle: with this RING I do thee destroy. Curr. Opin. Genet. Dev., 10, 54–64. [DOI] [PubMed] [Google Scholar]
- Xiao W., Lin,S.L., Broomfield,S., Chow,B.L. and Wei,Y.-F. (1998) The products of the yeast MMS2 and two human homologs (hMMS2 and CROC-1) define a structurally and functionally conserved Ubc-like protein family. Nucleic Acids Res., 26, 3908–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W., Chow,B.L., Fontanie,T., Ma,L., Bacchetti,S., Hryciw,T. and Broomfield,S. (1999) Genetic interactions between error-prone and error-free postreplication repair pathways in Saccharomyces cerevisiae. Mutat. Res., 435, 1–11. [DOI] [PubMed] [Google Scholar]
- Xie Y. and Varshavsky,A. (1999) The E2–E3 interaction in the N-end rule pathway: the RING-H2 finger of E3 is required for the synthesis of multiubiquitin chains. EMBO J., 18, 6832–6844. [DOI] [PMC free article] [PubMed] [Google Scholar]