Abstract
A ubiquitin-like modifier, NEDD8, is covalently attached to cullin-family proteins, but its physiological role is poorly understood. Here we report that the NEDD8-modifying pathway is essential for cell viability and function of Pcu1 (cullin-1 orthologue) in fission yeast. Pcu1 assembled on SCF ubiquitin-ligase was completely modified by NEDD8. Pcu1K713R defective for NEDD8 conjugation lost the ability to complement lethality due to pcu1 deletion. Forced expression of Pcu1K713R or depletion of NEDD8 in cells resulted in impaired cell proliferation and marked stabilization of the cyclin-dependent kinase inhibitor Rum1, which is a substrate of the SCF complex. Based on these findings, we propose that covalent modification of cullin-1 by the NEDD8 system plays an essential role in the function of SCF in fission yeast.
Keywords: cullin/NEDD8/Rub1/SCF/ubiquitin
Introduction
Ubiquitin (Ub), an 8.6 kDa highly conserved protein, is distributed widely in all eukaryotic cells and has become a landmark molecule in a post-translational protein-modifying system that plays a central role in intracellular protein breakdown (Hochstrasser, 1996; Hershko and Ciechanover, 1998; Kornitzer and Ciechanover, 2000). Ub is covalently attached to target proteins via an isopeptide linkage between the C-terminal glycine of Ub and the ε-NH2 group of the lysine residue of the acceptor substrate. This protein ubiquitylation is mediated by a cascade of three enzymes, termed E1 (Ub-activating), E2 (Ub-conjugating) and E3 (Ub-ligating) enzymes, to form a poly-Ub chain, which serves as the degradation signal for proteolytic attack by the 26S proteasome, a eukaryotic ATP-dependent protease complex (Coux et al., 1996; Baumeister et al., 1998; Bochtler et al., 1999; Voges et al., 1999).
The progress in the study of the Ub system has revealed the existence of multiple molecules that are structurally related to Ub (reviewed by Hochstrasser, 1998). Of these, the gene encoding NEDD8 (a mammalian orthologue of the budding yeast Rub1, a modifier related to Ub 1) was identified as one of the neural precursor cell-expressed developmentally down-regulated genes in mice (Kumar et al., 1993). NEDD8 has the highest homology to Ub, displaying an overall identity of 57%, among an expanding family of Ub-like proteins in eukaryotes. Recently, we found that human NEDD8 was covalently linked to target proteins via the C-terminal glycine residue in a manner analogous to ubiquitylation, where the reaction is catalysed by the APP-BP1–hUba3 heterodimer complex and hUbc12 as the E1- and E2-like enzymes, respectively (Osaka et al., 1998; Gong and Yeh, 1999). A similar Ub-related conjugation pathway for yeast Rub1 is known to operate in Saccharomyces cerevisiae (Liakopoulos et al., 1998), indicating that the NEDD8/Rub1-modifying system has been widely conserved during evolution.
Yeast Cdc53 is structurally related to Cul-1, one member of human cullin (abbreviated Cul)-family proteins, and was first found to be modified by Rub1 (Lammer et al., 1998). To date, it has become clear that Cdc53/Cul-1 functions as a common subunit of the growing family of Ub–protein ligases termed SCF (Skp1, Cul-1 or Cdc53, F-box protein), consisting of the core subunits Skp1, Cul-1/Cdc53 and Roc1/Rbx1/Hrt1, and substrate recognition adaptors called F-box proteins (Kamura et al., 1999a; Ohta et al., 1999; Seol et al., 1999; Skowyra et al., 1999; Tan et al., 1999), which are responsible for ubiquitylation of a variety of regulatory factors involved in the cell cycle and signal transduction (reviewed by Hershko and Ciechanover, 1998; Patton et al., 1998a; Deshaies, 1999; Harper and Elledge, 1999). Recently, it was reported that NEDD8 attaches to human Cul-2, which assembles with the von Hippel–Lindau tumour suppressor protein pVHL and elongin B/C, forming an SCF-like protein complex, CBCVHL (Liakopoulos et al., 1999; Wada et al., 1999a). Quite recently, the Rbx1 subunit of SCF was found to activate Rub1 modification of cullins Cdc53 and Cul-2 (Kamura et al., 1999b). We reported covalent modification of Cul-4A by NEDD8 in rabbit reticulocyte lysates (Osaka et al., 1998) and subsequently found that other human cullins, Cul-1, Cul-2, Cul-3, Cul-4B and Cul-5, are targets of the NEDD8-ligating pathway (Hori et al., 1999). It is notable that mutation of the Rub1-ligating system resulted in impairment of the auxin response in Arabidopsis (Pozo et al., 1998) and cell cycle progression in budding yeast (Lammer et al., 1998), although it is not essential in the latter organisms (Lammer et al., 1998; Liakopoulos et al., 1998). These observations imply a fundamentally important role for the modification of Cul-family proteins by NEDD8, which perhaps functions as a new modulator of SCF Ub–protein ligases. However, the role of the NEDD8/Rub1 conjugation pathway at the physiological level remains obscure.
We report here that disruptions of the genes encoding NEDD8 and the ligating E1 and E2 enzymes had a lethal effect in Schizosaccharomyces pombe. Moreover, we show that NEDD8 modification plays an indispensable role in the functions of Cul-family proteins Pcu1 and Pcu4 [Cul-1 and Cul-4A or Cul-4B orthologues, respectively (Kominami et al., 1998)] in fission yeast, based on the findings that Pcu1K713R and Pcu4K680R (Lys713 and Lys680 in Pcu1 and Pcu4, respectively, replaced by arginine), which are defective for modification by NEDD8, have completely lost their functions in vivo.
Results
The NEDD8 system is essential in fission yeast
Computer-assisted homology analysis revealed the existence of three genes, ned8+, uba3+ and ubc12+ encoding SpNEDD8, SpUba3 and SpUbc12, respectively. These genes are thought to be actual components of the NEDD8-ligating pathway in S.pombe, judging from their high overall similarities to their human homologues (Figure 1A). SpNEDD8, SpUba3 and SpUbc12 display 81, 40 and 55% identity to their human counterparts, respectively. SpUba3 and SpUbc12 have a presumptive active cysteine residue that forms a thioester linkage with NEDD8, which resembles a conserved cysteine residue in E1 and E2 of the Ub pathway.
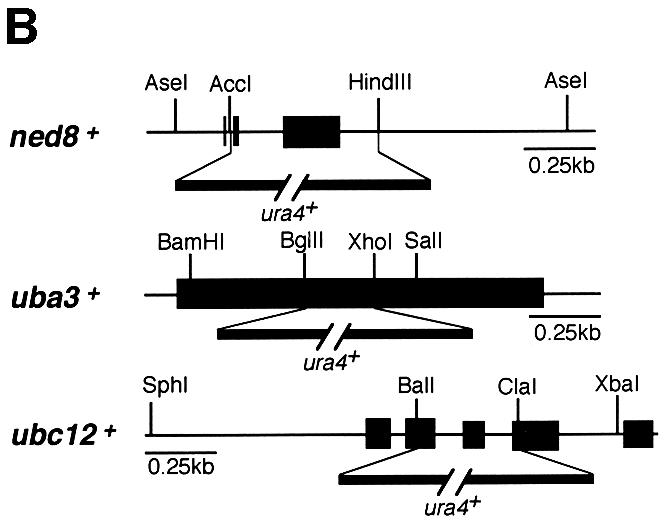
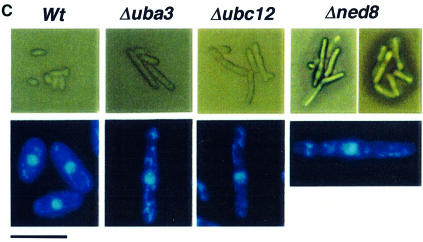
Fig. 1. Functional analysis of the NEDD8-ligating system in fission yeast. (A) Amino acid comparison of S.pombe (Sp) NEDD8 (DDBJ/EMBL/GenBank accession No. AJ003818), Uba3 (SWISS-PROT accession No. Q09765) and Ubc12 (DDBJ/EMBL/GenBank accession No. AL031532, SPC777.10C) with their human homologues (Hs). The identical amino acids are boxed in black. The asterisks in Uba3 and Ubc12 indicate the putative active cysteine residues. The arrow in NEDD8 indicates the processing site. (B) Disruption of ned8+, uba3+ and ubc12+ genes. The coding sequences in exons of the ned8+, uba3+ and ubc12+ genes are shown by solid boxes. For gene disruption, the indicated DNA fragments were replaced by a PCR-generated fragment containing the ura4+ marker gene. (C) After tetrad analysis of diploids heterozygous for ned8, uba3 and ubc12, inviable Ura+ cells that had germinated from each of the spores were observed under a phase-contrast microscope (upper panel). Note that in SpUba3 and SpUbc12, a part of the cells derived from a microcolony is shown. To observe the defective phenotype by DAPI staining, heterozygous diploids were sporulated and treated with glusulase to kill non-sporulating cells. Spores were grown in minimal medium lacking uracil at 30°C for 24 h then stained with DAPI (lower panel). Bar, 35 µm (upper panels) and 10 µm (lower panels).
To explore the function of the NEDD8-ligating pathway, null alleles of the ned8+, uba3+ or ubc12+ genes were created by replacing the majority of their open reading frames (ORFs) with the ura4+ marker (Figure 1B). Tetrad analyses of all these strains resulted in only two viable Ura– colonies from each ascus (data not shown), indicating that ned8+, uba3+ and ubc12+ are essential for cell viability. The lethality of Δubc12 was complemented by human Ubc12 (data not shown). Microscopic observation showed that Δned8 spores germinated and stopped growing after two or three cycles of cell division with an elongated cell shape (Figure 1C, upper panel). Δuba3 and Δubc12 spores germinated, divided several times into a microcolony and ceased division after forming a number of elongated cells. 4′,6-Diamidino-2-phenylindole (DAPI) staining revealed that most of the elongated cells in Δned8, Δuba3 or Δubc12 strains had single nuclei and decondensed chromosomes (Figure 1C, lower panel). These results strongly indicate that the NEDD8-modifying system in fission yeast is essential for cell cycle progression.
Covalent attachment of NEDD8 to Pcu1 and Pcu4
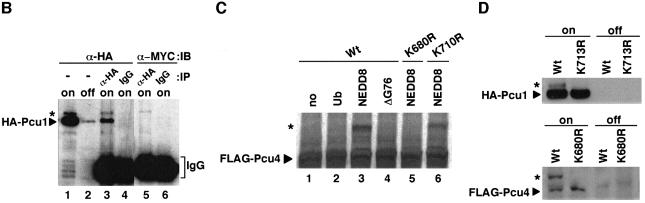
It is interesting to determine why disruption of the NEDD8 pathway in fission yeast is lethal. Presumably, modification of target protein(s) by NEDD8 (abbreviated NEDD8-ylation) may play an indispensable role in maintaining cell viability. Recently, we found that NEDD8 is covalently attached to all six known members of a family of human cullins in vitro (Osaka et al., 1998; Hori et al., 1999), suggesting that NEDD8-ylation of cullins plays a universal role in cells. Here, we first investigated whether or not S.pombe cullins are modified by NEDD8. For this purpose, Myc-NEDD8 and haemagglutinin (HA)-Pcul under the control of the thiamine-repressible promoter (nmt1 promoter) were co-expressed in cells. As shown in Figure 2B, when HA-Pcu1 was immunoprecipitated and used for western blotting, two Pcu1 bands were observed by anti-HA antibody staining, whereas only the upper band was stained with anti-Myc antibody, indicating that the HA-Pcul with a slow electrophoretic mobility is actually modified by Myc-NEDD8 in vivo.
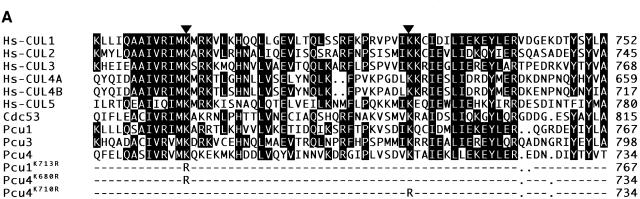
Fig. 2. Identification of a lysine residue modified by NEDD8 in Pcu1 and Pcu4 in S.pombe. (A) Amino acid sequences of the C-terminal region of six members of the human cullin family, Hs-Cul-1, -2, -3, -4A, -4B and -5, three members of the fission yeast cullin family, Pcu1, Pcu3 and Pcu4, and the budding yeast Cdc53 are shown. The numbers shown are the last residue numbers of the peptides in the respective proteins. Gaps (shown by dots) are inserted to maximize sequence homologies. Amino acids conserved in half or more of these cullins are boxed in black. The arrowheads indicate two lysine residues conserved in all members of the cullin family proteins. Pcu1K713R, Pcu4K680R and Pcu4K710R indicate that lysine at positions 713, 680 and 710 in Pcu1 and Pcu4, respectively, was replaced by arginine. (B) Wild-type cells containing pREP41-HA-pcu1+ and pREP81-Myc-ned8+ were grown in minimal medium without (de-repressed, lanes 1 and 3–6) or with (repressed, lane 2) thiamine. Cell extracts were immunoprecipitated with anti-HA antibody or non-immune mouse IgG. Cell extracts (lanes 1 and 2) and immunoprecipitates (lanes 3–6) were separated by SDS–PAGE, followed by immunoblotting using anti-HA or anti-Myc antibody. An asterisk indicates HA-Pcu1 ligated by Myc-NEDD8. (C) After [35S]FLAG-Pcu4 was synthesized in a reticulocyte lysate transcription/translation system in the presence or absence of unlabeled GST–Ub, GST–NEDD8 or GST–NEDD8ΔG76 (ΔG76) lacking the C-terminal glycine residue, samples of the resultant translational products were subjected directly to SDS–PAGE in the presence of DTT and autoradiographed (lanes 1–4). Two Pcu4 variants, FLAG-Pcu4K680R and FLAG-Pcu4K710R, were synthesized in vitro in the presence of GST–NEDD8 and analysed by the same method (lanes 5 and 6). An asterisk indicates [35S]FLAG-Pcu4 ligated by GST–NEDD8. (D) Wild-type cells containing pREP41-HA-pcu1+ (Wt) or pREP41-HA-pcu1K713R (K713R) (upper panel), or pREP81-FLAG-pcu4+ (Wt) or pREP81-FLAG-pcu4K680R (K680R) (lower panel) were grown in minimal medium with or without thiamine. Proteins were detected by immunoblotting using anti-HA or anti-FLAG antibody. Asterisks indicate FLAG-Pcu4 or HA-Pcu1 ligated by NEDD8. Note that the nmt1 promoter on pREP41 is ∼12-fold more active than that on pREP81 and that the expressed levels of the mutants FLAG-Pcu4K680R and Myc-Pcu1K713R, which did not form a linkage to NEDD8, were roughly similar to those of their wild types.
To ascertain the role of NEDD8-ylation in cullins, we next determined the site undergoing NEDD8-ylation in Pcu1 and Pcu4, which are S.pombe homologues of a human Cul-1 and Cul-4A or Cul-4B, respectively (Kominami et al., 1998). Previously, we found that the 171 C-terminal amino acids of human Cul-4A were sufficient for ligation by NEDD8 in vitro (Osaka et al., 1998) and, accordingly, we predicted that the NEDD8-ylation site exists in the C-terminal region of cullins. Comparing amino acid sequences among various cullins, we found two lysine residues within 69–71 C-terminal amino acids that have been conserved in all species (Figure 2A). To determine which lysine residue is modified by NEDD8, we synthesized two Pcu4 variants, FLAG-Pcu4K680R (Lys680 in Pcu4 was replaced by arginine) or FLAG-Pcu4K710R, in rabbit reticulocyte lysate (Figure 2A). [35S]FLAG-Pcu4 formed a linkage with GST–NEDD8, but not with GST–Ub or GST–NEDD8ΔG76, in which Gly76 had been deleted (Figure 2C). However, [35S]FLAG-Pcu4K680R did not form a linkage with GST–NEDD8, although [35S]FLAG- Pcu4K710R did, like wild-type Pcu4 (Figure 2C), indicating that Lys680 in Pcu4 is a presumptive acceptor site for covalent attachment to NEDD8 in vitro.
Subsequently, we investigated whether or not FLAG-Pcu4K680R and Myc-Pcu1K713R (note that Lys713 in Pcu1 corresponds to Lys680 in Pcu4; see Figure 2A) can form a linkage with NEDD8 in vivo. When the epitope-tagged Myc-Pcu1 or FLAG-Pcu4 under the control of the nmt1 promoter were overexpressed, two close bands with different electrophoretic mobilities for Myc-Pcu1 and FLAG-Pcu4 were detected by western blot analysis (Figure 2D). However, when the mutated cullins, Myc-Pcu1K713R or FLAG-Pcu4K680R, were overexpressed instead of their wild types, both upper bands disappeared completely, indicating that the larger forms were FLAG-Pcu4 and Myc-Pcu1 ligated by NEDD8. From these in vitro and in vivo experiments, it was concluded that the conserved lysine residue at positions 713 or 680 near the C-terminal region of Pcu1 or Pcu4, respectively, is the site required for NEDD8-ylation. Recently, consistent with these findings, Lys689 of human Cul-2 (Wada et al., 1999b) and Lys720 of human Cul-1 (Read et al., 2000) were reported to be the sites that undergo NEDD8-ylation.
Modification of the SCF complex by NEDD8 in vivo
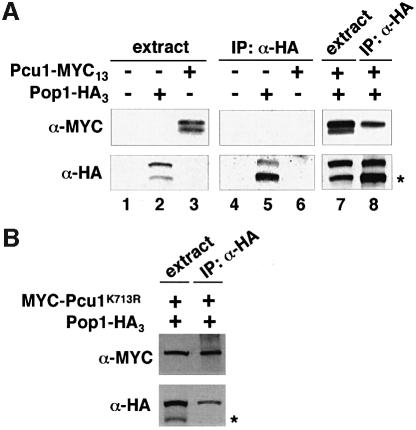
We next attempted to clarify whether Pcu1 complexed with Pop1, an S.pombe F-box protein (Kominami and Toda, 1997), is modified by NEDD8 in vivo. To investigate the NEDD8-ylation of Pcu1 in the SCFPop1 complex under physiological conditions, we replaced the chromosomal pcu1+ and pop1+ genes by their epitope-tagged genes, pcu1+-Myc13 and pop1+-HA3, respectively. Next, SCFPop1–HA was immunoprecipitated by anti-HA antibody from the resultant extracts, and subjected to western blotting with anti-HA and anti-Myc antibodies. As previously reported (Kominami et al., 1998), Pcu1-Myc13 was co-immunoprecipitated with Pop1-HA3 (Figure 3A, lane 8). Western blotting with anti-Myc antibody revealed two Pcu1 bands in the cell extracts (Figure 3A, lane 3 or 7), whereas only the upper band was immunoprecipitated by Pop1-HA3 (Figure 3A, lane 8). These results indicate that Pcu1 assembled into the SCFPop1 complex is modified preferentially by NEDD8.
Fig. 3. Preferential modification of Pcu1 by NEDD8 in the SCFPop1 complex. (A) Wild-type cells (lanes 1 and 4) or cells in which chromosomal pcu1+ and pop1+ had been replaced by the epitope-tagged genes pop1+-HA3 (lanes 2 and 5), pcu1+-Myc13 (lanes 3 and 6) or pop1+-HA3 and pcu1+-Myc13 (lanes 7 and 8) were grown in YE. Cell extracts were immunoprecipitated (IP) with anti-HA antibody. The cell extracts (lanes 1–3 and 7) and IP-proteins (lanes 4–6 and 8) were separated by SDS–PAGE followed by immunoblotting with anti-HA, anti-Myc antibody. (B) Fission yeast cells containing the integrated pop1+-HA3 and pREP41-Myc-Pcu1K713R were grown in minimal medium without thiamine for 20 h at 32°C. Cell extracts were immunoprecipitated (IP) with anti-HA antibody. The cell extracts and immunoprecipitated proteins were separated by SDS–PAGE and followed by immunoblotting with anti-HA, anti-Myc antibody. Asterisks indicate truncated Pop1, which may be produced by proteolytic digestion or synthesized from a different translation initiation site.
To test whether NEDD8-ylation of Pcu1 is needed for assembly of the SCFPop1 complex, the same immunoprecipitation with anti-HA antibody was performed for the lysates of cells containing pREP41-Myc-pcu1K713R, in which chromosomal pop1+ had been replaced by pop1+-HA3. When the immunoblotting with anti-Myc antibody was carried out for the Pop1-HA3 immunoprecipitates, Pcu1K713R was found to be incorporated into the SCFPop1 complex (Figure 3B), indicating that NEDD8-ylation of Pcu1 is not essential for the formation of the SCFPop1 complex. Furthermore, both Pcu1K713R and wild-type Pcu1 modified with or without NEDD8 were fractionated similarly at ∼400 kDa by molecular sieve chromatography, indicating that NEDD8 modification does not have an apparent effect on complex formation nor oligomerization of SCF (data not shown).
Impairment of SCF function by Pcu1K713R
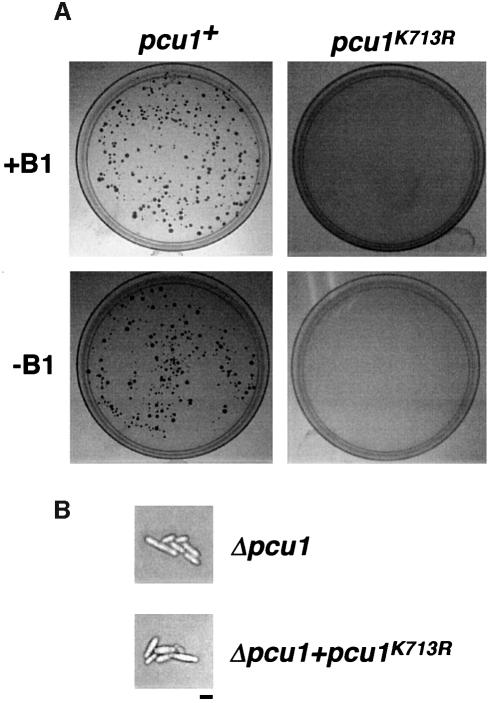
Since S.pombe pcu1+ is essential for cell viability (Kominami et al., 1998), we examined whether or not the NEDD8-ylation-defective mutant Myc-pcu1K713R can complement the loss of pcu1. As shown in Figure 4A (lower panel), the lethality of Δpcu1 cells was not complemented by overexpression of Myc-pcu1K713R from the nmt1 promoter, unlike the wild-type Myc-pcu1+, indicating that modification of Pcu1 by NEDD8 plays a pivotal role in the proliferation of S.pombe cells. Intriguingly, the weak expression of Myc-pcu1+, but not Myc-pcu1K713R under repression of the nmt1 promoter, was also able to complement the lethality of Δpcu1 cells (Figure 4A, upper panel). It is noteworthy that this weak expression of Myc-pcu1K713R did not cause any defective phenotypes in wild-type cells, unlike the growth-suppressive effect caused by overexpression of Myc-pcu1K713R (see Figure 5). Accordingly, it is unlikely that the lack of complementation by Myc-pcu1K713R is the toxic effect due to its overexpression (Figure 4A, upper panel). Furthermore, Δpcu1 spores expressing Pcu1K713R weakly from the repressed nmt1 promoter were germinated and stopped growing after two or three cycles of cell division, similarly to the Δpcu1 spores (Figure 4B).
Fig. 4. pcu1K713R loses the ability to complement the lethality of Δpcu1 cells. (A) Spores (5 × 103) derived from the IO100 (pcu1::ura4+/pcu1+) diploid strain containing [pREP41-Myc-pcu1+ LEU2] or [pREP41-Myc-pcu1K713R LEU2] were germinated and grown on minimal medium plates lacking uracil and leucine in the presence (upper panel, repressed) or absence (lower panel, de-repressed) of thiamine (B1). (B) Δpcu1 cells and Δpcu1 cells expressing Myc-Pcu1K713R, germinated from spores, are shown. Bar, 10 µm.
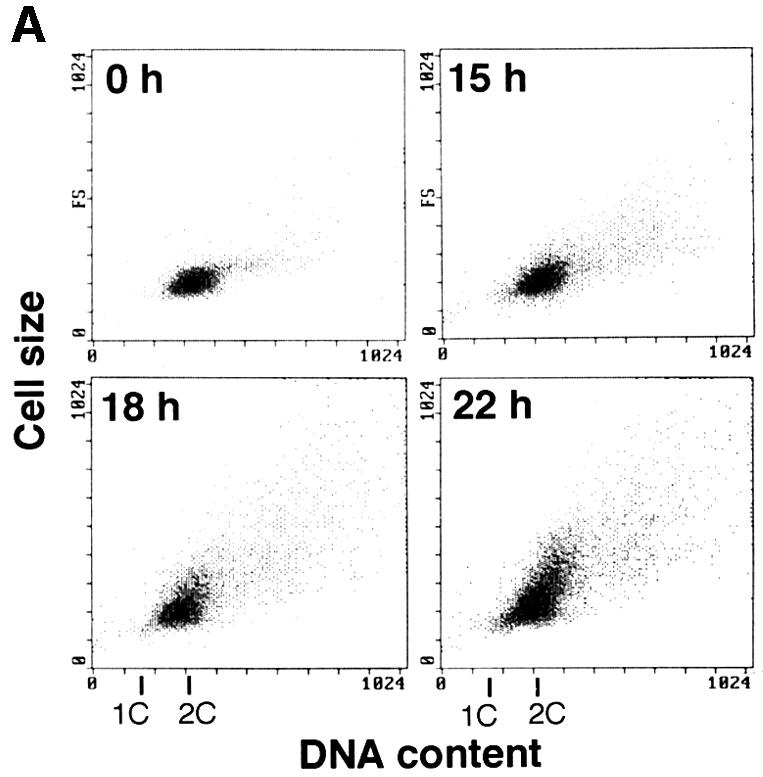
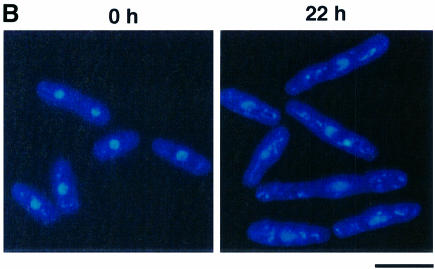
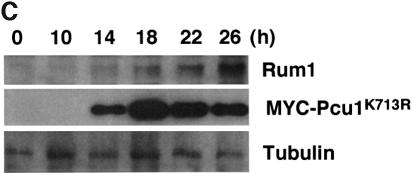
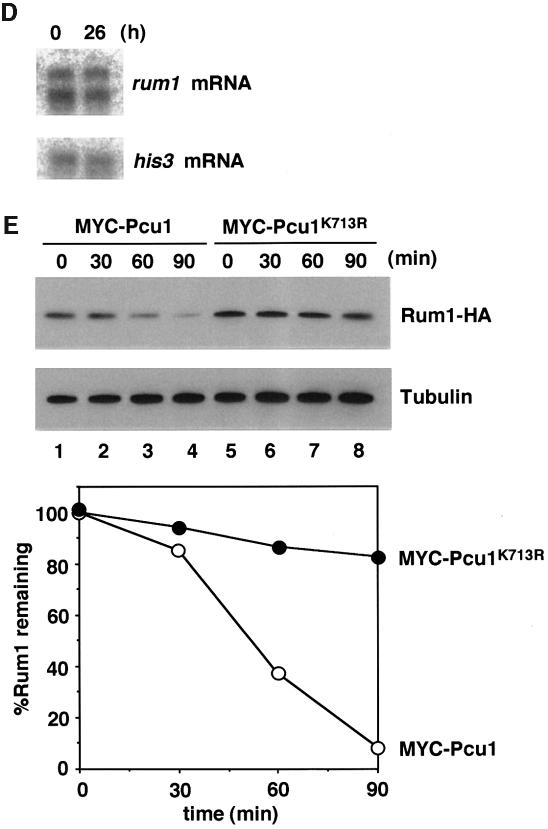
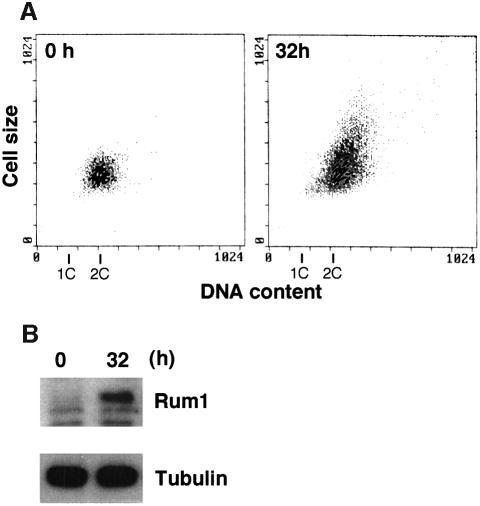
Fig. 5. Stabilization of Rum1 in the Pcu1K713R-overexpressed cells. Fission yeast cells (SKP401) containing pREP41-Myc-pcu1K713R were grown in minimal medium without thiamine for the indicated time at 32°C. (A) DNA content and cell size were measured by a flow cytometer. (B) Cells were stained with DAPI. Bar, 10 µm. (C) Cell extracts were separated by SDS–PAGE, followed by immunoblotting using anti-Myc, anti-Rum1 or anti-tubulin antibody. (D) Northern blot analysis was carried out using rum1 or his3 cDNA as a probe. (E) Wild-type cells containing pREP42-rum1+-HA/His6 and pREP41-Myc-pcu1+ (lanes 1–4), or pREP42-rum1+-HA/His6 and pREP41-Myc-pcu1K713R (lanes 5–8) were grown in minimal medium without thiamine for 22 h. At time 0, thiamine was added to repress the nmt1 promoter. After the indicated times, cell extracts were prepared, separated by SDS–PAGE, then subjected to immunoblotting using anti-HA or anti-tubulin antibody (upper panels). The levels of Rum1 were quantified densitometrically, and the relative amounts of remaining Rum1 are shown (lower panel).
We next tested the effect of forced expression of the Myc-pcu1K713R mutant in yeast cells. At 18 h after induction of Myc-pcu1K713R, a number of cells started to elongate (Figure 5A and B) and cell division was severely inhibited (data not shown). The most elongated cells had a 2C DNA content, which is characteristic of haploid cells in the G2/M phase (Figure 5A), and decondensed chromosomes (Figure 5B), suggesting that cell cycle progression had been largely delayed at the G2 phase.
This effect may be due to dysfunction of SCF mediated by lack of NEDD8-ylation of Pcu1. It is notable that Pcu1 is complexed with two related F-box/WD40-repeat proteins, Pop1 and Pop2, forming SCFPop1 and/or SCFPop2, which ubiquitinylate the CDK (cyclin-dependent kinase) inhibitor Rum1 and the S phase regulator Cdc18 (Kominami and Toda, 1997; Jallepalli et al., 1998; Kominami et al., 1998; Maekawa et al., 1998). To ascertain the role of NEDD8-ylation of Pcu1 in SCF function, we measured the level of Rum1 (Moreno and Nurse, 1994) in the Myc-Pcu1K713R-overexpressing cells. As shown in Figure 5C, Rum1 accumulated after induction of the Myc-Pcu1K713R, which is consistent with the appearance of abnormality in cell morphology, although the level of Rum1 protein in exponentially growing wild-type cells was extremely low and barely detectable, as previously reported (Kominami et al., 1998). Accumulation of Myc-Pcu1K713R preceded that of Rum1, whereas no appreciable change in the tubulin level was observed. Northern blot analysis revealed no obvious change in the mRNA levels of Rum1 (Figure 5D), indicating that the increase in Rum1 is not due to the elevated level of transcription, but rather is due to the defective degradation of Rum1. Finally, we examined the stability of Rum1 in the cells. For this, HA/His6-tagged Rum1 was ectopically expressed from the nmt1 promoter, then the disappearance of Rum1-HA/His6 was monitored by immunoblotting after the addition of thiamine to repress the promoter. As shown in Figure 5E, Rum1 was degraded rapidly in the presence of wild-type Pcu1 after a short lag time, whereas it was stabilized greatly by overexpression of Pcu1K713R (upper panel). Their decay curves reveal that the apparent half-life of Rum1 was <45 min under the expression of the wild-type Pcu1, which is in accordance with the previously reported value (Benito et al., 1998), but no significant degradation of Rum1 was observed in the presence of excess NEDD8-ylation-defective Pcu1 mutant (Figure 5E, lower panel).
Since overexpression of Rum1 has been shown to induce various cell defects, including an increment of genome ploidy (Moreno and Nurse, 1994), it is likely that the deletion of rum1+ would cancel cellular impairments induced by Pcu1K713R. However, when the rum1+-deleted cells (PN1012) containing pREP41-Myc-pcu1K713R were grown in minimal medium in the absence of thiamine for 24 h at 32°C, the delay in cell cycle progression and abnormal morphology induced by Pcu1K713R were observed to the same extent as in rum1+ cells (data not shown), suggesting that accumulation of Rum1 caused by overexpression of Pcu1K713R was not directly responsible for the inhibition of cell cycle progression and that there is another unknown target(s) of SCF involved in normal cell proliferation (see Discussion).
We next tested whether or not Rum1 is stabilized in cells lacking the NEDD8 system by producing an S.pombe strain whose chromosomal ned8+ was deleted by ura4+ and NEDD8 expression can be controlled conditionally by the nmt1 promoter. At 24 h after repression of NEDD8, a number of cells started to elongate and cell growth was inhibited (data not shown). The most elongated cells had a 2C DNA content (Figure 6A) and decondensed chromosomes like those of Δned8 cells (data not shown). Western blotting revealed that the depletion of NEDD8 caused accumulation of Rum1 (Figure 6B). Based on these results, it was concluded that covalent modification of Pcu1 by NEDD8 is essential for the function of SCF Ub-ligase in fission yeast. It is noteworthy that the NEDD8-modified levels of both Pcu1 and Pcu4 did not change significantly during the cell cycle progression of the fission yeast (data not shown).
Fig. 6. Accumulation of Rum1 in NEDD8-depleted cells. Δned8+ cells containing pREP81- ned8+ were grown in minimal medium with thiamine for the indicated time at 32°C. (A) DNA content and cell size were measured by a flow cytometer. (B) Cell extracts were separated by SDS–PAGE, followed by immunoblotting using anti-Rum1 or anti-tubulin antibody.
NEDD8-ylation is essential for function of Pcu4
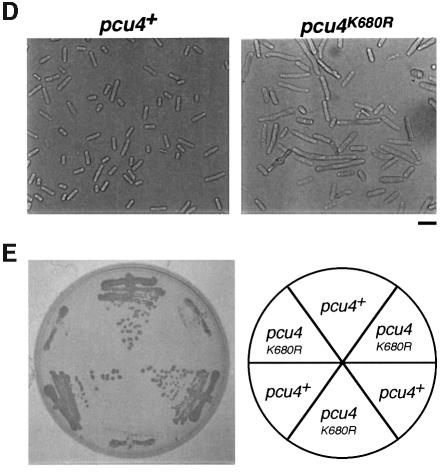
It is of interest to determine whether the modification of NEDD8 influences the functions of other cullins. As we first found that mammalian Cul-4A was covalently ligated by NEDD8 in reticulocyte lysates (Osaka et al., 1998), we analysed the role of NEDD8 modification of Pcu4, its possible counterpart in S.pombe. To examine the loss-of-function phenotypes of the pcu4+ gene, we replaced almost the entire ORF of the pcu4+ gene with the ura4+ marker (Figure 7A). Tetrad manipulation of a heterozygous diploid dissected out two normal sized and two tiny colonies (Figure 7B). In all cases, small colonies were Ura+, indicating that pcu4+ is important for normal growth, but not essential for cell viability. Microscopic observation showed that a number of Δpcu4 cells were extremely elongated with decondensed chromosomes (Figure 7C). Interestingly, all of the defective phenotypes observed in Δpcu4 cells, such as abnormal morphology (Figure 7D) and slow growth rate (Figure 7E), were complemented by FLAG-pcu4+, but not by FLAG-pcu4K680R. Overexpression of Pcu4K680 from the nmt1 promoter of pREP81 vector in the wild-type cells, however, did not show any significant abnormality, differing from that of Pcu1K713R (data not shown). This finding strongly indicates that FLAG-Pcu4K680R, which cannot be modified by NEDD8, completely loses the function of Pcu4 in vivo and that modification of Pcu4 by NEDD8 plays a pivotal role in the function of Pcu4.
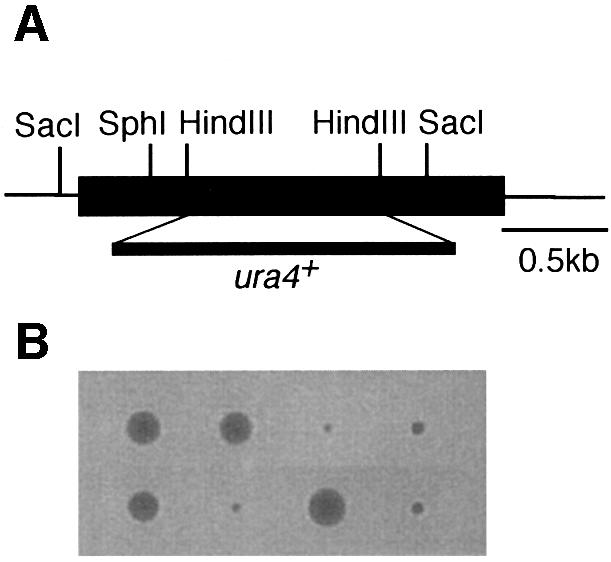
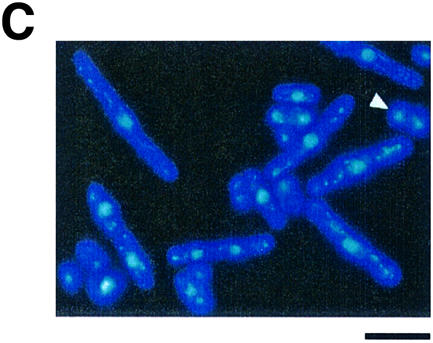
Fig. 7. pcu4K680R loses the ability to complement the defective phenotypes of Δpcu4 cells. (A) Disruption of the pcu4+ gene. Almost the entire ORF of the pcu4+ gene (DDBJ/EMBL/GenBank accession No. Z99260, SPC3A11.08) was deleted by use of a PCR-generated fragment containing the ura4+ marker. (B) Tetrad analysis of diploids heterozygous for pcu4. (C) Δpcu4 cells were stained by DAPI as described in Figure 1C. Ungerminated spores are marked with an arrow. Bar, 10 µm. (D) Δpcu4 cells containing pREP81-FLAG-pcu4+ or pREP81-FLAG-pcu4K680R were grown in minimal medium without thiamine, then observed under a phase contrast microscope. Bar, 10 µm. (E) Δpcu4 cells containing pREP81-FLAG-pcu4+ or pREP81-FLAG-pcu4K680R were grown on a minimal plate without thiamine for 4 days at 30°C.
Discussion
In the present study, we showed that the NEDD8-ligating pathway is essential in fission yeast, presumably due to the indispensable role of NEDD8-ylation of Pcu1 in the function of S.pombe SCF Ub-ligase. However, it may also affect pleiotropically various cellular functions, because NEDD8 modifies all Cul proteins (Osaka et al., 1998; Hori et al., 1999; Liakopoulos et al., 1999; Wada et al., 1999a,b) and possibly other as yet unknown target(s). Intriguingly, overexpression of the NEDD8-ylation-defective mutant Myc-Pcu1K713R resulted in inhibition of cell division at the G2 phase concomitant with abnormal accumulation of Rum1 capable of inhibiting the activity of Cdc2 mitotic kinase (Correa-Bordes and Nurse, 1995; Martin-Castellanos et al., 1996). Details of the mechanism by which forced expression of Pcu1K713R induces the G2 arrest remain elusive, but it is noteworthy that accumulation of Rum1 is not a major cause of induction of cell cycle abnormality, because disruption of the rum1+ gene did not cancel the cell abnormalities caused by overexpression of Pcu1K713R. Consistent with this result, we found that pcu1 deletion is still lethal in a rum1 deletion background (T.Toda and S.Katayama, unpublished result). Quite recently, it was reported that Cul-1, perhaps existing as an SCF complex, is present abundantly in centrosomes and that the centrosome-associated Cul-1 is modified mostly by NEDD8 (Freed et al., 1999). The NEDD8-ylation, therefore, may play a pivotal role in the centrosome function(s), which somehow influences the G2–M transition of the cell cycle, finally causing a cell cycle delay at the G2 phase.
It is notable that ectopic expression of Rum1 or pop1 mutation causes a high ploidy phenotype of the cell (Kominami et al., 1997), but here we found that overexpression of Pcu1K713R did not have a serious influence on induction of polyploidy, irrespective of Rum1 accumulation. This difference is presumably due to the fact that overexpression of Pcu1K713R may result in dysfunction of all species of SCF complexes, which cause the cell cycle arrest at multiple stages and a variety of abnormal phenotypes, whereas pop1 mutation causes the defect of the SCFpop1 ligase leading to abnormal accumulation of Rum1. Intriguingly, we also observed a slight, but significant accumulation of Cdc18, which is known to be another target of SCFPop1/2 in fission yeast (Kominami and Toda, 1997; Jallepalli et al., 1998; Kominami et al., 1998; Maekawa et al., 1998), which was consistent with the observation that deletions of pop1+ and/or pop2+ resulted in a relatively low accumulation of Cdc18 compared with the marked accumulation of Rum1 (Kominami et al., 1998). It remains to be discovered, however, why Myc-Pcu1K713R unmodified by NEDD8 completely loses SCFPop1/2 function in vivo. We found here that NEDD8-ylation of Pcu1 is not required for formation of the SCF complex, because Pcu1K713R, like wild- type Pcu1, was incorporated into the SCFPop1 complex. The observation that Pcu1K713R is capable of forming an SCF complex with an apparently normal size indicates that Pcu1K713R possesses considerable, if not total, functional integrity compared with the wild-type Pcu1, except for the linkage with NEDD8, although Pcu1K713R may not be structurally equivalent to the wild-type Pcu1. Therefore, although unlikely, modification of Pcu1 by NEDD8 may be necessary for interaction with Roc1/Rbx1/Hrt1, which has recently been identified as the fourth essential subunit of the SCF (Kamura et al., 1999a; Ohta et al., 1999; Seol et al., 1999; Skowyra et al., 1999; Tan et al., 1999), or for recruitment of the E2 enzyme. Alternatively, the possibility that NEDD8 is related to the stability or localization of the SCF complex cannot be ruled out. Further study is required to clarify this issue.
Similarly, a deletion of the S.cerevisiae ENR2 (equivalent to APP-BP1) is synthetic lethal in temperature-sensitive alleles of cdc34 and enhances the phenotypes of cdc4, cdc53 and skp1. This suggests that modification of Cdc53 by Rub1/NEDD8 affects the optimal assembly or functions of the SCF complex (Lammer et al., 1998). Accordingly, the Rub1/NEDD8 ligation pathway is closely linked to cell cycle regulation, although this pathway and the modification of Cdc53 by Rub1 do not seem to be strictly essential for the function of SCFCDC4 in budding yeast (Lammer et al., 1998; Liakopoulos et al., 1998), unlike in fission yeast. In Arabidopsis, AXR1 (APP-BP1 homologue) has been identified as one of the genes required for a normal auxin response (Leyser et al., 1993). In the axr1 mutant, it is known that ECR1 (Uba3 homologue) cannot form an efficient thiolester linkage with Rub1/NEDD8 (Pozo et al., 1998). Interestingly, an F-box protein mutant, tir1, identified from genetic analysis of auxin-response mutants, has a synergistic interaction with axr1, implying that modification of the SCFtir function by the NEDD8 ligation system is required for the normal auxin response (Ruegger et al., 1998; Gray et al., 1999). These observations are consistent with our conclusion that the NEDD8 ligation system regulates the function of the SCF complex. Taken together, these findings suggest that the NEDD8/Rub1-ligating pathway may play a critical role in various functions of the SCF complex in a variety of eukaryotic cells.
Budding yeast Cdc53 functions as a common component of distinct SCF Ub-ligases that regulate multiple cellular functions, such as the cell cycle (Feldman et al., 1997; Skowyra et al., 1997), gene expression (Li and Johnston, 1997) and methionine biosynthesis (Patton et al., 1998b), by replacing the appropriate F-box proteins. Consistent with this notion, the mutation of hamster SMC, encoding a protein nearly identical to APP-BP1, is responsible for cell cycle defects in the hamster ts41 cell line (Handeli and Weintraub, 1992; Chen et al., 2000). Moreover, mammalian SCFβTrCP has recently been found to act as a Ub-ligase for IκBα (Yaron et al., 1998; Suzuki et al., 1999, 2000; Winston et al., 1999), and β-catenin (Kitagawa et al., 1999; Maniatis, 1999; Winston et al., 1999), suggesting its involvement in signal transduction. Recently, it was also suggested that SCFSKP2 catalyses ubiquitylation of the CDK inhibitor p27Kip1 (Carrano et al., 1999; Tsvetkov et al., 1999) and E2F-1 (Marti et al., 1999). It was found recently that Cul-1 associated with phosphorylated IκBα and p27Kip1 was modified preferentially by NEDD8 in mammalian cells (Podust et al., 2000; Read et al., 2000; T.Kawakami, T.Chiba and K.Tanaka, in preparation), which seems to be in agreement with the present finding that Pcu1 linked to SCFPop1 was almost fully modified by NEDD8. Interestingly, the formation of NEDD8–Cul-2 conjugates is stimulated by pVHL, but not by a tumorigenic pVHL variant, suggesting that ligation of NEDD8 to Cul-2 presumably is linked to the pVHL tumour suppression function via CBCpVHL (Liakopoulos et al., 1999). Here we also provided evidence that modification of NEDD8 by Pcu4 plays a critical role in the function of Pcu4, although whether or not Pcu4 is incorporated into the SCF-like complex remains unknown. Quite recently, it was reported that the CBCpVHL complex containing Cul-2 functions as a Ub–protein ligase (Iwai et al., 1999; Lisztwan et al., 1999) and Cul-3 targets cyclin E for ubiquitylation (Singer et al., 1999), implying that all Cul-family proteins may act as components of the Ub–protein ligase complex. Thus, it is likely that NEDD8-ylation is a common and essential modification required not only for Cul-1 of the SCF Ub-ligase complex, but also for function(s) of other Cul-family proteins.
Materials and methods
Strains, media and genetic methods
In this study, the following strains were used: TP4-5A (h–leu1ura4ade6-M210), TP4-1D (h+leu1ura4his2ade6-M216), IO100 (h–/h+leu1/leu1ura4/ura4his7/his7ade6-M210/ade6-M216pcu1::ura4+/pcu1+), SKP401 (h–leu1pop2+-3HA-kan), SKP414-17 (h–leu1ura4pcu1+-13Myc-kan), SKP424-17 (h–leu1pcu1+-13Myc-kan1, pop1+-3HA-kan), SKP408-4 (h–leu1ura4pop1+-3HA-kan) and PN1012 (h90 leu1 ade6 ura4 rum1:: ura4+). Cells were grown in YE medium or minimal medium supplemented with amino acids, uracil and/or adenine (Moreno et al., 1991). To regulate the expression of genes under the thiamine-repressible promoter (nmt1 promoter), cells were grown in minimal medium with or without 10 µM thiamine (Maundrell, 1990). For gene disruption, a diploid strain crossed between TP4-5A and TP4-1D was used. Correct disruption of all heterozygous diploids was confirmed by both genomic Southern blotting and PCR analyses.
Immunological analyses
Cells were suspended in ESB buffer [2% SDS, 80 mM Tris pH 6.8, 10% glycerol, 1.5% dithiothreitol (DTT) and 0.1 mg/ml bromophenol blue], boiled for 3 min, followed by treatment with glass beads. Cell extracts were separated by SDS–PAGE, followed by immunoblotting using anti-Myc antibody (9E10 Boehringer Mannheim), anti-FLAG M2 antibody (IB13010 Kodak), anti-HA antibody (16B12 BAbCO), anti-Rum1 antibody (provided by S.Moreno) or anti-tubulin antibody (T5168 Sigma or YOL1/34 Harlan). For immunoprecipitation, cell extracts (2 mg) were prepared as described (Moreno et al., 1991) and mixed with anti-HA antibody, followed by AffiPrep Protein A (Bio-Rad). After washing with HB buffer (Moreno et al., 1991), the absorbed proteins were eluted by heating for 3 min in SDS–PAGE sample buffer.
Flow cytometry
Cells were fixed with ethanol, treated with DNase-free RNase A (100 µg/ml) and stained with 2.5 µg/ml propidium iodide. Samples of 30 000 cells were analysed with a flow cytometer (EPICS XL Beckman Coulter).
Acknowledgments
Acknowledgements
We are grateful to Drs S.Moreno and P.Nurse for the gift of the anti-Rum1 antibody and strains, respectively, and to S.Kobayashi for technical advice. K.K. and S.K. were supported by JSPS Postdoctoral Fellowships for Research Abroad. Part of this research was supported by the Human Frontier Science Program Research Grant (T.T.).
References
- Baumeister W., Walz,J., Zühl,F. and Seemüller,E. (1998) The proteasome: paradigm of a self-compartmentalizing protease. Cell, 92, 367–380. [DOI] [PubMed] [Google Scholar]
- Benito J., Martin-Castellanos,C. and Moreno,S. (1998) Regulation of the G1 phase of the cell cycle by periodic stabilization and degradation of the p25rum1 CDK inhibitor. EMBO J., 17, 482–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochtler M., Ditzel,L., Groll,M., Hartmann,C. and Huber,H. (1999) The proteasome. Annu. Rev. Biophys. Biomol. Struct., 28, 295–317. [DOI] [PubMed] [Google Scholar]
- Carrano A., Eytan,E., Hershko,A. and Pagano,M. (1999) Skp2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nature Cell Biol., 1, 193–199. [DOI] [PubMed] [Google Scholar]
- Chen Y., McPhie,D.L., Hirschberg,J. and Neve,R.L. (2000) The amyloid precursor protein-binding protein APP-BP1 drives the cell cycle through the S–M checkpoint and causes apoptosis in neurons. J. Biol. Chem., 275, 8929–8935. [DOI] [PubMed] [Google Scholar]
- Correa-Bordes J. and Nurse,P. (1995) p25rum1 orders S phase and mitosis by acting as an inhibitor of the p34cdc2 mitotic kinase. Cell, 83, 1001–1009. [DOI] [PubMed] [Google Scholar]
- Coux O., Tanaka,K. and Goldberg,A.L. (1996) Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem., 65, 801–847. [DOI] [PubMed] [Google Scholar]
- Deshaies R.J. (1999) SCF and Cullin/RING-H2-based ubiquitin-ligases. Annu. Rev. Cell Dev. Biol., 15, 435–467. [DOI] [PubMed] [Google Scholar]
- Feldman R.M., Correll C.C., Kaplan,K.B. and Deshaies,R.J. (1997) A complex of Cdc4p, Skp1p and Cdc53p/Cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell, 91, 221–230. [DOI] [PubMed] [Google Scholar]
- Freed E., Lacey,K.R., Huie,P., Lyapina,S.A., Deshaies,R.J., Stearns,T. and Jackson,P.K. (1999) Components of an SCF ubiquitin ligase localize to the centrosome and regulate the centrosome duplication cycle. Genes Dev., 13, 2242–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L. and Yeh,E.T.H. (1999) Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J. Biol. Chem., 274, 12036–12042. [DOI] [PubMed] [Google Scholar]
- Gray W.M. et al. (1999) Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana.Genes Dev., 13, 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handeli S. and Weintraub,H. (1992) The ts41 mutation in Chinese hamster cells leads to successive S phases in the absence of intervening G2, M and G1. Cell, 71, 599–611. [DOI] [PubMed] [Google Scholar]
- Harper J.W. and Elledge,S.J. (1999) Skipping into the E2F1-destruction pathway. Nature Cell Biol., 1, E5–E7. [DOI] [PubMed] [Google Scholar]
- Hershko A. and Ciechanover,A. (1998) The ubiquitin system. Annu. Rev. Biochem., 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. (1996) Ubiquitin-dependent protein degradation. Annu. Rev. Genet., 30, 405–439. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. (1998) There’s the Rub: a novel ubiquitin-like modification linked to cell cycle regulation. Genes Dev., 12, 901–907. [DOI] [PubMed] [Google Scholar]
- Hori T., Osaka,F., Chiba,T., Miyamoto,C., Okabayashi,K., Shimbara,N., Kato,S. and Tanaka,K. (1999) Covalent modification of all members of human cullin family proteins by NEDD8. Oncogene, 18, 6829–6834. [DOI] [PubMed] [Google Scholar]
- Iwai K., Yamanaka,K., Kamura,T., Minato,N., Conaway,R.C., Conaway,J.W., Klausner,R.D. and Pause A. (1999) Identification of the von Hippel–Lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc. Natl Acad. Sci. USA, 96, 12436–12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallepalli P.V., Tien,D. and Kelly,T.J. (1998) sud1+ targets cyclin-dependent kinase-phosphorylated Cdc18 and Rum1 proteins for degradation and stops unwanted diploidization in fission yeast. Proc. Natl Acad. Sci. USA, 95, 8159–8164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T. et al. (1999a) Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science, 284, 657–661. [DOI] [PubMed] [Google Scholar]
- Kamura T., Conrad,M.N., Yan,Q., Conaway,R.C. and Conaway,J.W. (1999b) The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev., 13, 2928–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M. et al. (1999) An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of β-catenin. EMBO J., 18, 2401–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominami K. and Toda,T. (1997) Fission yeast WD-repeat protein Pop1 regulates genome ploidy through ubiquitin–proteasome-mediated degradation of the CDK inhibitor Rum1 and the S-phase initiater Cdc18. Genes Dev., 11, 1548–1560. [DOI] [PubMed] [Google Scholar]
- Kominami K., Ochotorena,I. and Toda,T. (1998) Two F-box/WD-repeat proteins Pop1 and Pop2 form hetero- and homo-complexes together with cullin-1 in the fission yeast SCF (Skp1–Cullin-1–F-box) ubiquitin ligase. Genes Cells, 3, 721–735. [DOI] [PubMed] [Google Scholar]
- Kornitzer D. and Ciechanover,A. (2000) Modes of regulation of ubiquitin-mediated protein degradation. J. Cell Physiol., 182, 1–11. [DOI] [PubMed] [Google Scholar]
- Kumar S., Yoshida,Y. and Noda,M. (1993) Cloning of a cDNA which encodes a novel ubiquitin-like protein. Biochem. Biophys. Res. Commun., 195, 393–399. [DOI] [PubMed] [Google Scholar]
- Lammer D., Mathias,N., Laplaza,J.M., Jiang,W., Liu,Y., Callis,J., Goebl,M. and Estelle,M. (1998) Modification of yeast Cdc53p by the ubiquitin-related protein Rub1p affects function of the SCFCdc4 complex. Genes Dev., 12, 914–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser O.H.M., Lincoln,C.A., Timpte,C., Lammer,D., Turner,J. and Estelle,M. (1993) Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature, 364, 161–164. [DOI] [PubMed] [Google Scholar]
- Li F.N. and Johnston,M. (1997) Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: coupling glucose sensing to gene expression and the cell cycle. EMBO J., 16, 5629–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos D., Doenges,G., Matuschewski,K. and Jentsch,S. (1998) A novel protein modification pathway related to the ubiquitin system. EMBO J., 17, 2208–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos D., Busgen,T., Brychzy,A., Jentsch,S. and Pause,A. (1999) Conjugation of the ubiquitin-like protein NEDD8 to cullin-2 is linked to von Hippel–Lindau tumor suppressor function. Proc. Natl Acad. Sci. USA, 96, 5510–5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisztwan J., Imbert,G., Wirbelauer,C., Gstaiger,M. and Krek,W. (1999) The von Hippel–Lindau tumor suppressor protein is a component of an E3 ubiquitin–protein ligase activity. Genes Dev., 13, 1822–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa H., Kitamura,K. and Shimoda,C. (1998) The Ste16 WD-repeat protein regulates cell-cycle progression under starvation through the Rum1 protein in Schizosaccharomyces pombe. Curr. Genet., 33, 29–37. [DOI] [PubMed] [Google Scholar]
- Maniatis T. (1999) A ubiquitin ligase complex for the NF-κB, Wt/Wingless and Hedgehog signaling pathways. Genes Dev., 13, 505–510. [DOI] [PubMed] [Google Scholar]
- Marti A., Wirbelauer,C., Scheffner,M. and Krek,W. (1999) Interaction between the ubiquitin–protein ligase SCFskp2 and E2F-1 underlies the regulation of E2F-1 degradation. Nature Cell Biol., 1, 1–6. [DOI] [PubMed] [Google Scholar]
- Martin-Castellanos C., Labib,K. and Moreno,S. (1996) B-type cyclins regulate G1 progression in fission yeast in opposition to the p25rum1 CDK inhibitor. EMBO J., 15, 839–849. [PMC free article] [PubMed] [Google Scholar]
- Maundrell K. (1990) nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J. Biol. Chem., 265, 10857–10864. [PubMed] [Google Scholar]
- Moreno S. and Nurse,P. (1994) Regulation of progression through the G1 phase of the cell cycle by the rum1+ gene. Nature, 367, 236–242. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar,A. and Nurse,P. (1991) Molecular genetic analyses of fission yeast Schizosaccharomyces pombe. Methods Enzymol., 194, 795–823. [DOI] [PubMed] [Google Scholar]
- Ohta T., Jennifer,J., Michel,J.J., Schottelius,A.J. and Xiong,Y. (1999) ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell, 3, 535–541. [DOI] [PubMed] [Google Scholar]
- Osaka F., Kawasaki,H., Aida,N., Saeki,M., Chiba,T., Kawashima S., Tanaka,K. and Kato S. (1998) A new NEDD8-ligating system for cullin-4A. Genes Dev., 12, 2263–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton E.E., Willems,A.R. and Tyers,M. (1998a) Combinatorial control in ubiquitin-dependent proteolysis: don’t Skp the F-box hypothesis. Trends Genet., 14, 236–243. [DOI] [PubMed] [Google Scholar]
- Patton E.E., Willems,A.R., Sa,D., Kuras,L., Thomas,D., Craig,K.L. and Tyers,M. (1998b) Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev., 12, 692–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podust V.N., Brownell,J.E., Gladysheva,T.B., Luo,R.-S., Wang,C., Coggins,M.B., Pierce,J.W., Lightcap,E.S. and Chau,V. (2000) A Nedd8 conjugating pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proc. Natl Acad. Sci. USA, 97, 4579–4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo J.C., Timpte,C., Tan,S., Callis,J. and Estelle,M. (1998) The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science, 280, 1760–1763. [DOI] [PubMed] [Google Scholar]
- Read M. et al. (2000) Nedd8 modification of Cul-1 activates SCFβTrCP-dependent ubiquitination of IκBα. Mol. Cell. Biol., 20, 2326–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M., Deway,E., Gray,W.M., Hobbie,L., Turner,J. and Estella,M. (1998) The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev., 12, 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol J.H. et al. (1999) Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev., 13, 1614–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer J.D., Gurian-West,M., Clurman,B. and Roberts,J.M. (1999) Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev., 13, 2375–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D., Craig,K.L., Tyers,M., Elledge,S.J. and Harper,J.W. (1997) F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell, 91, 209–219. [DOI] [PubMed] [Google Scholar]
- Skowyra D., Koepp,D.M., Kamura,T., Conrad,M.N., Conaway,R.C., Conaway,J.W., Elledge,S.J. and Harper,J.W. (1999) Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science, 284, 662–665. [DOI] [PubMed] [Google Scholar]
- Suzuki H. et al. (1999) IκBα ubiquitination is catalyzed by an SCF-like complex containing Skp1, cullin-1 and two F-box/WD40-repeat proteins βTrCP1 and βTrCP2. Biochem. Biophys. Res. Commun., 256, 127–132. [DOI] [PubMed] [Google Scholar]
- Suzuki H., Chiba,T., Suzuki,T., Fujita,T., Ikenoue,T., Omata,M., Furuichi,K., Shikama,H. and Tanaka,K. (2000) Homodimer of two F-box proteins βTrCP1 or βTrCP2 binds to IκBα for signal-dependent ubiquitination. J. Biol. Chem., 275, 2877–2884. [DOI] [PubMed] [Google Scholar]
- Tan P., Fuchs,S.Y., Chen,A., Wu,K., Gomez,C., Ronai,Z. and Pan,Z.Q. (1999) Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of IκBα. Mol. Cell, 3, 527–533. [DOI] [PubMed] [Google Scholar]
- Tsvetkov L.M., Yeh,K.H., Lee,S.J., Sun,H. and Zhang,H. (1999) p27(Kip1) ubiquitination and degradation is regulated by the SCFSkp2 complex through phosphorylated Thr187 in p27. Curr. Biol., 9, 661–664. [DOI] [PubMed] [Google Scholar]
- Voges D., Zwickl,P. and Baumeister,W. (1999) The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem., 68, 1015–1068. [DOI] [PubMed] [Google Scholar]
- Wada H., Yeh,E.T.H. and Kamitani,T. (1999a) The von Hippel–Lindau tumor suppressor gene product promotes, but is not essential for, Nedd8 conjugation to cullin-2. J. Biol. Chem., 274, 36025–36029. [DOI] [PubMed] [Google Scholar]
- Wada H., Yeh,E.T.H. and Kamitani,T. (1999b) Identification of NEDD8-conjugating site in human cullin-2. Biochem. Biophys. Res. Commun., 257, 100–105. [DOI] [PubMed] [Google Scholar]
- Winston J.T., Strack,P., Beer-Romero,P., Chu,C.Y., Elledge,S.J. and Harper,J.W. (1999) The SCFβ-TRCP–ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev., 13, 270–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaron A. et al. (1998) Identification of the receptor component of the IκBα–ubiquitin-ligase. Nature, 396, 590–594. [DOI] [PubMed] [Google Scholar]