Summary
Spinocerebellar ataxia type 7 (SCA7) is a neurodegenerative disorder caused by CAG / polyglutamine repeat expansions in the ataxin-7 gene. Ataxin-7 is a component of two different transcription co-activator complexes, and recent work indicates that disease protein normal function is altered in polyglutamine neurodegeneration. Given this, we studied how ataxin-7 gene expression is regulated. The ataxin-7 repeat and translation start site are flanked by binding sites for CTCF, a highly conserved multi-functional transcription regulator. When we analyzed this region, we discovered an adjacent alternative promoter and a convergently transcribed antisense non-coding RNA, SCAANT1. To understand how CTCF regulates ataxin-7 gene expression, we introduced ataxin-7 mini-genes into mice, and found that CTCF is required for SCAANT1 expression. Loss of SCAANT1 de-repressed ataxin-7 sense transcription in a cis-dependent fashion, and was accompanied by chromatin remodeling. Discovery of this pathway underscores the importance of altered epigenetic regulation for disease pathology at repeat loci exhibiting bidirectional transcription.
Introduction
Spinocerebellar ataxia type 7 (SCA7) is an inherited neurological disorder characterized by cerebellar and retinal degeneration (Martin et al., 1994). SCA7 is caused by a CAG / polyglutamine (polyQ) repeat expansion in the ataxin-7 gene, and is therefore one of nine polyQ neurodegenerative disorders (La Spada and Taylor, 2010). Included in the CAG / polyQ repeat disease category are spinobulbar muscular atrophy (SBMA), Huntington’s disease (HD), dentatorubral-pallidoluysian atrophy (DRPLA), and five other forms of spinocerebellar ataxia (SCA1, 2, 3, 6 & 17). Numerous lines of investigation in the polyQ disease field suggest that expansion of the glutamine tract is a gain-of-function mutation, and that the initiating event in disease pathogenesis is transition of the polyQ expansion tract to an altered conformation (Paulson et al., 2000; Ross, 1997). However, as each polyQ disease displays distinct patterns of neuropathology despite overlapping patterns of disease gene expression, it is likely that the normal function, activities, and interactions of the polyQ disease protein determine the cell-type specificity in each disorder (La Spada and Taylor, 2003). Ataxin-7, the causal protein in SCA7, contains a polyQ tract that ranges in size from 4 – 35 glutamines in normal individuals, but expands to 37 – >400 glutamines in affected patients (David et al., 1997; Stevanin et al., 2000). The glutamine tract is located in the amino-terminus of ataxin-7, beginning at position #30. SCA7 is unique among the CAG / polyQ repeat diseases, as patients with this disorder develop a retinal degeneration phenotype, classified as a cone-rod dystrophy (Ahn et al., 2005; To et al., 1993). To understand the basis of SCA7 retinal degeneration and the reason for the selective loss of photoreceptor cells in this disease, we, and others, produced transgenic mice that recapitulated the SCA7 cone-rod dystrophy phenotype, and found that SCA7 retinal degeneration results from altered transcription regulation (Helmlinger et al., 2006; La Spada et al., 2001; Yoo et al., 2003).
As the vast majority of CAG / polyQ disease proteins are well-known transcription factors or can function as transcription co-regulators (Riley and Orr, 2006), a role for transcription dysregulation in SCA7 is consistent with an emerging view of these disorders as “transcriptionopathies” (La Spada and Taylor, 2003). The existence of an interaction between ataxin-7 and a retinal transcription factor, known as CRX, suggested that ataxin-7 is a transcription factor (La Spada et al., 2001), and this was supported by demonstration of a functional nuclear localization signal in ataxin-7 (Chen et al., 2004). When studies of the yeast orthologue of ataxin-7, Sgf73, indicated that Sgf73 is part of the SAGA complex (Sanders et al., 2002), we, and others, found that ataxin-7 is a core component of the analogous co-activator complex in mammals, known as the STAGA (Spt3-Taf9-Ada-Gcn5-Acetyltransferase) complex (Helmlinger et al., 2004; Palhan et al., 2005). STAGA is a transcriptional co-activator complex with histone acetyltransferase (HAT) activity (Martinez et al., 2001). In addition to being part of the STAGA complex, yeast Sgf73 and mammalian ataxin-7 are respectively components of the Ubp8 / USP22 deubiquitination complex (Kohler et al., 2008; Zhao et al., 2008). While the role of altered STAGA and USP22 deubiquitination complex function in SCA7 disease pathogenesis is unclear, recent studies of the related polyQ disorder SCA1 indicate that the polyQ expansion in ataxin-1 attenuates the formation and function of the Capicua transcription factor complex, contributing to SCA1 disease pathogenesis through a partial loss-of-function mechanism (Chen et al., 2003; Lim et al., 2008). Hence, polyQ disease may result from an alteration of normal function, combined with a novel gain-of-function, and in the case of SCA7, the native protein function of ataxin-7 appears critically important for chromatin remodeling at the level of histone acetylation and deubiquitination.
In addition to the CAG / polyQ repeat diseases, at least three other subclasses of repeat expansion disease are recognized: loss-of-function repeat expansion diseases, RNA gain-of-function repeat disorders, and polyalanine gain-of-function repeat expansion diseases (La Spada and Taylor, 2010). Although the four repeat expansion disease subcategories are based upon presumed differences in their mechanisms of repeat mutation toxicity, recent studies have revealed a number of shared genomic features for repeat disease loci, irrespective of repeat disease category, including: 1) genetic instability characterized by a strong tendency for repeats to further expand upon germ line transmission (Pearson et al., 2005); 2) binding sites for the multivalent transcription regulatory factor CTCF (Ohlsson et al., 2001), within close proximity of the repeats (Filippova et al., 2001); and 3) bidirectional transcription typically encompassing the repeat itself (Batra et al., 2010). These features suggest that certain epigenetic processes and chromatin regulatory pathways may be shared in common between different repeat diseases. As the SCA7 CAG repeat is the most unstable of all the CAG / polyQ repeat loci, and the SCA7 CAG repeat is closely flanked by two functional CTCF binding sites, the SCA7 CAG repeat is among the repeat disease loci likely to display this constellation of genomic features.
In light of the importance of ataxin-7 normal function for SCA7 disease pathogenesis and potentially for global transcription regulation, we initiated a series of studies aimed at understanding how ataxin-7 gene expression is regulated. The ataxin-7 CAG repeat tract and the start site of translation are both located in exon 3, which is flanked by two functional CTCF binding sites (Filippova et al., 2001). CTCF is a highly conserved 11 zinc-finger protein that mediates a variety of transcription regulatory functions, including transcription activation, transcription repression, insulator-boundary domain formation, and genomic imprinting (Phillips and Corces, 2009). When we analyzed the ataxin-7 repeat region, we discovered evidence for an alternative promoter just 5′ to exon 3, and identified an antisense non-coding RNA, SCAANT1 (for SpinoCerebellar Ataxia-7 Antisense Non-coding Transcript 1) that is convergently transcribed across exon 4, exon 3, and the alternative promoter. To understand the role of CTCF in regulating ataxin-7 transcription, we introduced ataxin-7 mini-genes, containing the ataxin-7 repeat region with a CAG repeat expansion, into transgenic mice. Studies of these transgenic mice and of human retinoblastoma cell lines revealed that CTCF binding is required for production of SCAANT1, and that loss of SCAANT1 expression de-repressed ataxin-7 sense transcription from the alternative promoter. Although SCAANT1 expression in trans did not reduce ataxin-7 alternative sense promoter activity in vitro or in vivo, convergent transcription of SCAANT1 in cis led to repression that was accompanied by post-translational modification of histones. Our studies reveal a novel regulatory pathway, linking CTCF transactivation of antisense non-coding RNA with repression of the corresponding sense transcript. The likely contribution of this pathway to SCA7 disease pathogenesis underscores the potential importance of altered epigenetic regulation for disease pathology at repeat loci characterized by bidirectional transcription.
Results
Identification of overlapping sense and antisense transcripts at the ataxin-7 locus
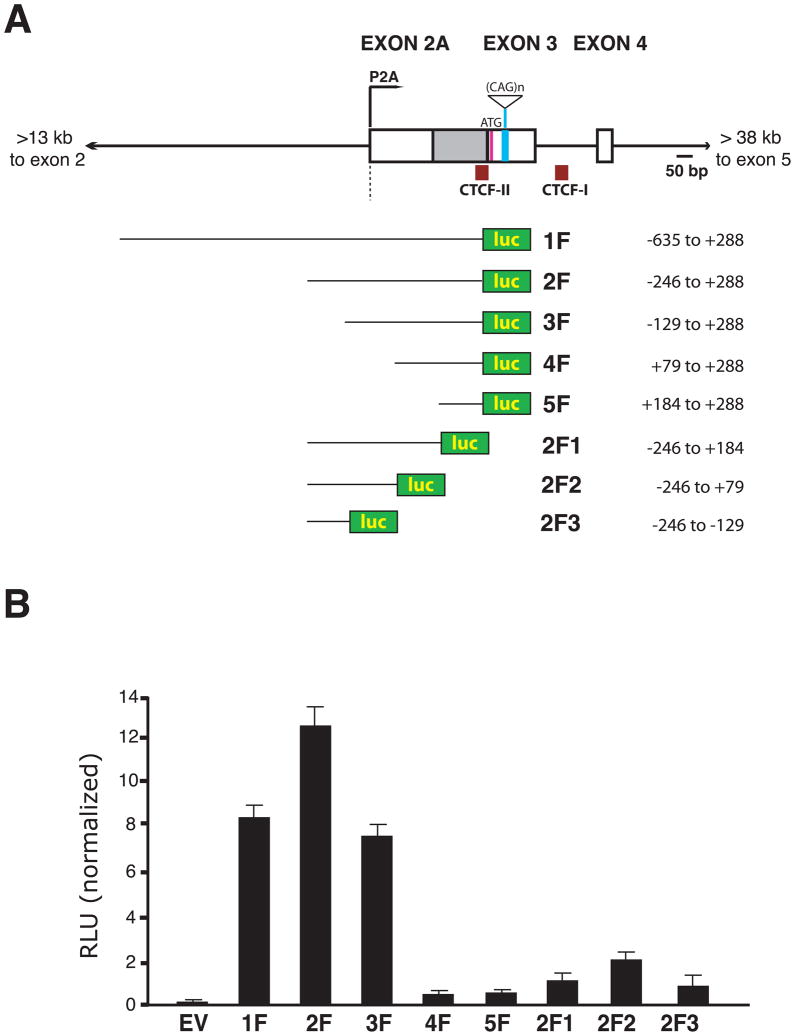
The ataxin-7 CAG / polyglutamine (polyQ) repeat is located in the first coding exon (i.e. exon 3), and is flanked by two CTCF binding sites (Filippova et al., 2001). To determine the basis of ataxin-7 gene expression regulation, we surveyed ataxin-7 human genomic sequence, including exon 3 and the CAG repeat region, with the UCSC genome browser <genome.ucsc.edu>. Bioinformatics analysis of this region, using FirstEF (Davuluri et al., 2001), revealed evidence for an alternative promoter in the intron 5′ to exon 3. The presence of a strong peak for the promoter-associated H3K4me3 modification at this location strongly supported the existence of this alternative promoter (Figure S1). When we analyzed mouse ataxin-7 genomic sequence, we found that the transcription start site (TSS) for the murine ataxin-7 gene is located in the ataxin-7 repeat region in close proximity to the alternative promoter predicted for the human gene. Interestingly, the previously defined human ataxin-7 TSS, which is located >40 kb 5′ to this region, annotated on the UCSC genome browser, and validated by RLM-RACE (Figure S1), is not predicted as a promoter or TSS in mouse <genome.ucsc.edu>. To evaluate this prediction, we performed RLM-RACE on murine RNA samples, and identified a cluster of TSSs located 85, 100, and 255 nucleotides 5′ to the annotated mouse ataxin-7 TSS. However, unlike in the human, there is no distant upstream ataxin-7 TSS in mice. Thus, the human ataxin-7 gene contains two promoters: the standard, upstream ‘P1’ promoter and the alternative ‘P2A’ promoter (Figure S2), while the mouse ataxin-7 gene contains only one promoter – P2A.
Although computational algorithms for identification of promoters and TSSs are powerful approaches for defining regulatory regions, experimental validation is necessary. To confirm the promoter calls, and to define the location of regulatory elements in this region, we cloned a series of human ataxin-7 CAG10 genomic fragments into a luciferase reporter (Figure 1A), and transfected the different ataxin-7 genomic fragment – luciferase constructs into primary cerebellar astrocytes. When we measured relative luciferase activity, we detected robust luciferase transactivation for ataxin-7 genomic fragments containing sequences just 5′ to the newly discovered TSS (Figure 1B). To define the alternative promoter, which we labeled ‘P2A’, we performed 5′ RACE and found that the alternative ataxin-7 TSS is located at the 3′ end of intron 2. Sequencing of 5′ RACE clones revealed that transcripts initiating from P2A contain a single first exon comprising the last ~400 bp of intron 2 and exon 3, or instead undergo splicing to join a shorter exon (“2A”) with exon 3 (Figure 1A).
Figure 1. The ataxin-7 gene contains an alternative sense promoter.
A) Diagram of the genomic region of the human ataxin-7 gene and constructs generated to map the alternative sense promoter. Boxes represent exons, while solid lines correspond to introns. The alternative promoter is indicated as P2A and is predicted to produce a transcript that contains an alternatively spliced intron (gray box). Various genomic fragments, containing 10 CAG/CTG repeats, were cloned into a firefly luciferase vector to yield the different luciferase expression constructs shown here.
B) Transactivation assay results for human ataxin-7 genomic fragments. The luciferase expression constructs shown in panel A were transfected into primary astrocytes, along with Renilla luciferase constructs for normalization, and relative luminescence unit (RLU) measurements were obtained. EV corresponds to empty vector, all experiments were performed in quadruplicate, and errors bars = s.e.m. RLU activity obtained for the vector 1F construct corresponded to ~10% of a positive control CMV-eGFP-IRES-luciferase vector (not shown).
See Figure S1.
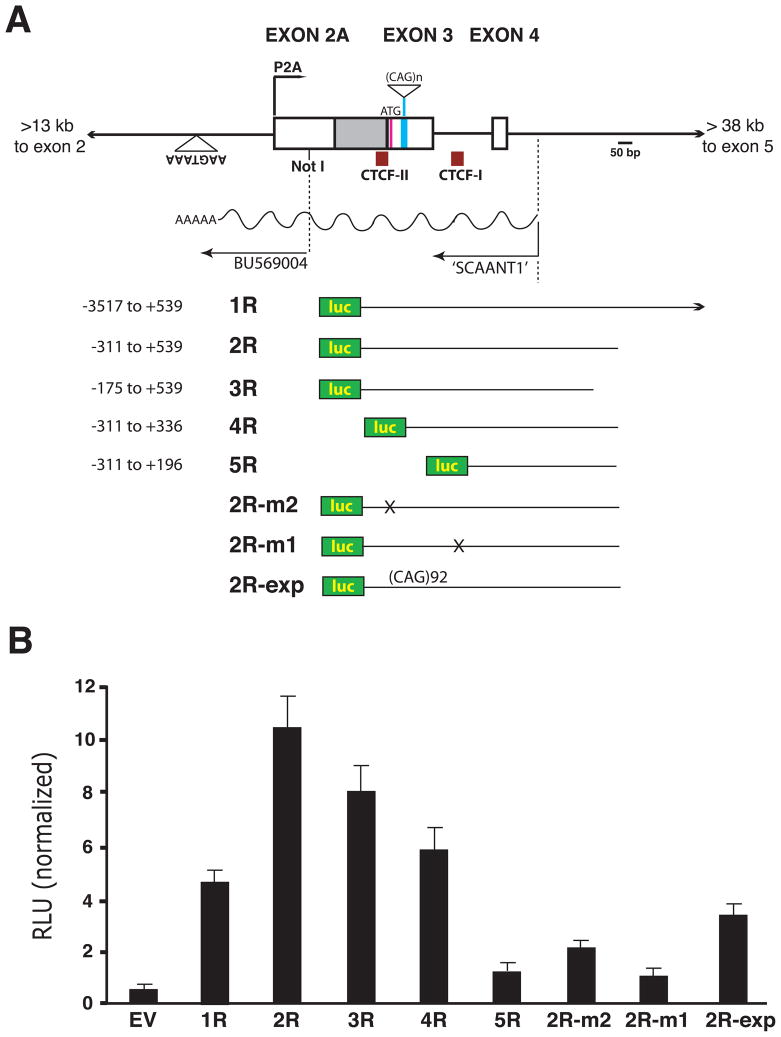
In the course of inventorying ESTs in the ataxin-7 repeat region, we discovered an EST (BU569004) in antisense orientation. Analysis of the sequence in this region revealed that the 5′ end of this EST corresponded to a Not I restriction site, and that the 3′ end of this EST co-localized with a putative polyadenylation signal sequence in antisense orientation (Figure 2A). As Not I digests were typically used during the former era of EST discovery, the 5′ end of this transcript was likely created during its cloning. To identify the extent of the antisense transcript, we performed strand-specific RT-PCR and 5′ RACE, and mapped the TSS to intron 4 (Figure 2A). We named this antisense transcript SCAANT1 for ‘SpinoCerebellar Ataxia-7 Antisense Non-coding Transcript 1’. To delineate the regulatory region responsible for transcription of SCAANT1, we cloned a series of human ataxin-7 CAG10 genomic fragments into a luciferase reporter construct in antisense orientation (Figure 2A), and transfected the different ataxin-7 antisense genomic fragment – luciferase constructs into primary cerebellar astrocytes. We noted that a short stretch of DNA 5′ to the SCAANT1 TSS was required for transactivation, while a sizable sequence 3′ to the SCAANT1 TSS was needed to achieve robust transactivation (Figure 2B). As the two CTCF binding sites lie within the regulatory domain mapped by the luciferase reporter assays, we derived another set of luciferase reporter constructs, based upon our most potent construct 2R, in which we mutated either of the CTCF binding sites (Figure 2A). When we measured the transactivation competence of the 2R-m2 and 2R-m1 constructs, we observed marked reductions in luciferase activity (Figure 2B), suggesting that CTCF binding site integrity is required for maximal SCAANT1 expression. We also derived an ataxin-7 antisense construct carrying a CAG92 repeat expansion (2R-exp), and when we measured its transactivation competence, we documented a significant reduction in luciferase activity (Figure 2B).
Figure 2. Identification and mapping of an antisense ncRNA at the human ataxin-7 locus.
A) Diagram of the genomic region of the human ataxin-7 gene, showing the human ataxin-7 antisense ncRNA (SCAANT1), and the constructs generated to map the SCAANT1 promoter. Boxes represent exons, while solid lines correspond to introns. The location of BU569004 – an antisense EST, a predicted polyadenylation signal sequence, the transcriptional domain for SCAANT1, and two CTCF binding sites are shown. Various genomic fragments in antisense orientation, all containing 10 CAG/CTG repeats except for 2R-exp, were cloned into a firefly luciferase vector to produce the different expression constructs shown here.
B) Transactivation assay results for human ataxin-7 antisense genomic fragments. The luciferase expression constructs shown in panel A were transfected into primary astrocytes, along with Renilla luciferase constructs for normalization, and relative luminescence unit (RLU) measurements were obtained. Construct 2R-m2 contains a mutated CTCF-II binding site, construct 2R-m1 contains a mutated CTCF-I binding site, and construct 2R-exp contains a 92 CAG/CTG repeat. EV corresponds to empty vector, experiments were performed in quadruplicate, and errors bars = s.e.m.
See Figure S2.
An ataxin-7 genomic fragment yields a SCA7 phenotype in transgenic mice upon mutation of the 3′ CTCF binding site
The existence of an ~1.4 kb antisense non-coding transcript overlapping a potentially strong sense promoter at the human ataxin-7 locus suggested that their transcription regulation might be linked. As CTCF binding site integrity was required for SCAANT1 transcription, we derived two ataxin-7 mini-gene constructs that contain the sense P2A promoter and SCAANT1, flanked by ~5 kb of DNA 5′ to this region, and ~8 kb of DNA 3′ to this region (Figure S2). Within this 13.5 kb human ataxin-7 genomic fragment reside two CTCF binding sites, known as CTCF-I and CTCF-II. To understand the regulatory relationship between SCAANT1 and ataxin-7 transcription from promoter P2A, we introduced an 11-nucleotide substitution mutation at the 3′ CTCF-I binding site (Figure S2). The location of the mutation was based upon DNA footprinting analysis, and validation of abrogated CTCF binding was achieved by electrophoretic mobility shift assays, as we have shown (Libby et al., 2008). In this way, we derived two distinct ataxin-7 genomic fragment constructs with an expanded CAG repeat tract: SCA7-CTCF-I-wt and SCA7-CTCF-I-mut (Figure S2). To determine the role of CTCF binding in regulating ataxin-7 repeat instability and transcription, we used these constructs to produce lines of transgenic mice. We have shown that mutation of the CTCF-I binding site significantly diminishes CTCF occupancy in vivo in the SCA7-CTCF-I-mut mice by ChIP analysis, and found that mutation of the CTCF-I binding site leads to increased repeat instability in the germ line and somatic tissues (Libby et al., 2008). Further studies of these mice also revealed that SCA7-CTCF-I-mut mice become tremulous, display weight loss, and develop an unsteady gait at 5 – 9 months of age (Movie S1). This phenotype, which is observed in both SCA7-CTCF-I-mut transgenic lines [(1) and (2)], progresses to become a prominent gait ataxia until the mice die prematurely at 8 – 14 months of age, with the SCA7-CTCF-I-mut-(2) line exhibiting a more rapidly progressive and severe phenotype. In contrast, four independent lines of SCA7-CTCF-I-wt mice did not exhibit any physical or neurological abnormalities, and have a normal lifespan.
As SCA7-CTCF-I-mut transgenic mice develop a pronounced ataxia, reminiscent of the gait difficulties seen in SCA7 patients and in other lines of SCA7 transgenic mice (La Spada et al., 2001; Yoo et al., 2003), we performed histopathology studies and behavioral testing. SCA7 patients develop a cone-rod dystrophy retinal degeneration, characterized by dramatic loss of cone photoreceptors and visual dysfunction (Ahn et al., 2005; To et al., 1993). To determine if SCA7-CTCF-I-mut mice recapitulate this phenotype, we immunostained retinal whole mounts from age-matched SCA7-CTCF-I-mut and SCA7-CTCF-I-wt mice, and observed a marked dropout of cone photoreceptors in SCA7-CTCF-I-mut mice (Figure 3A). Electroretinogram testing corroborated this finding, as SCA7-CTCF-I-mut mice went blind with a degradation of cone responses ahead of rod responses (Figure S3). The visible ataxia phenotype in affected SCA7-CTCF-I-mut mice led us to compare cerebellar sections from age-matched SCA7-CTCF-I-mut mice and SCA7-CTCF-I-wt mice. This analysis revealed dramatic Purkinje cell degeneration, as well as ataxin-7 positive aggregates in Purkinje cells in SCA7-CTCF-I-mut mice (Figure 3B). These findings confirm that mutation of the 3′ CTCF binding site, within a human ataxin-7 mini-gene lacking the canonical ataxin-7 TSS at exon 1, is sufficient to recapitulate the SCA7 phenotype in independent lines of transgenic mice.
Figure 3. SCA7-CTCF-I-mut mice develop retinal and cerebellar degeneration, produce polyQ-expanded protein, and suffer loss of ataxin-7 antisense ncRNA expression.
A) Confocal microscopy of retinal whole mounts from 11 month-old SCA7-CTCF-I-mut (mut) and SCA7-CTCF-I-wt (wt) mice. Retinas immunostained with antibodies against red/green cone photoreceptors (red) and rod photoreceptors (green) reveal dramatic loss of cone photoreceptors and more moderate, but nonetheless marked, drop-out of rods in SCA7-CTCF-I-mut mice.
B) Confocal microscopy of cerebellar sections from 11 month-old SCA7-CTCF-I-mut (mut) and SCA7-CTCF-I-wt (wt) mice. Sections immunostained with antibodies against calbindin (green) and ataxin-7 (magenta), and counterstained with DAPI (blue) reveal marked Purkinje cell degeneration, loss, and protein aggregate formation (arrows) in SCA7-CTCF-I-mut mice.
C) Western blot analysis of cortex protein lysates obtained from six week-old SCA7-CTCF-I-wt mice, two independent lines of SCA7-CTCF-I-mut mice, and a non-transgenic control (Ntg) immunoblotted with 1C2 antibody that detects polyQ-expanded protein. Both lines of SCA7-CTCF-I-mut mice display a ~ 47 kDa band (arrow) that is absent from the Ntg and barely visible in the SCA7-CTCF-I-wt mouse. The asterisk indicates a non-specific background band that confirmed equivalent loading.
D) Reciprocal expression of sense and antisense ataxin-7 RNAs in three month-old SCA7-CTCF-I-wt and SCA7-CTCF-I-mut mice. We performed strand-specific RT-PCR, and quantified RNA expression relative to 18S RNA. Results for three sets of brain RNAs per line are shown, with ataxin-7 sense RNA expression arbitrarily set to 1 for SCA7-CTCF-I-wt mice, and ataxin-7 antisense (SCAANT1) RNA expression arbitrarily set to 1 for SCA7-CTCF-I-mut mice. Comparable results were obtained when we normalized to GAPDH, and absolute values indicated that ataxin-7 antisense transcript levels are ~2.1-fold higher than ataxin-7 sense levels in SCA7-CTCF-I-wt mice (not shown). Assays were performed in triplicate, and error bars = s.e.m.
E) In situ hybridization of SCAANT1 in three month-old SCA7 transgenic mice. Prominent signal in the Purkinje cell layer of SCA7-CTCF-I-wt mice (wt) is detected with the ataxin-7 antisense riboprobe; however, minimal labeling in the Purkinje cell layer of SCA7-CTCF-I-mut mice (mut) is observed with this riboprobe.
See Figure S3.
Recapitulation of the SCA7 phenotype in SCA7-CTCF-I-mut mice, together with the observation of ataxin-7-positive inclusions in cerebellar Purkinje cells, suggested that mutation of the 3′ CTCF binding site had resulted in the initiation of sense transcription within the ataxin-7 mini-gene construct. To test this hypothesis, we performed RT-PCR analysis on SCA7-CTCF-I-mut mice and detected expression of the ataxin-7 first coding exon in RNA samples from cerebellum and cortex (data not shown). As SCA7 disease pathogenesis typically involves production of a misfolded polyQ-expanded ataxin-7 protein, we performed Western blot analysis with the 1C2 antibody that is specific for expanded polyQ tracts (Trottier et al., 1995), and observed an ~ 47 kDa protein in brain lysates from each SCA7-CTCF-I-mut transgenic line (Figure 3C). We noted higher expression in the SCA7-CTCF-I-mut-(2) line, consistent with its more severe phenotype. Low level expression of the ~ 47 kDa protein was detected in SCA7-CTCF-I-wt mice (Figure 3C), and this ~ 47 kDa protein product corresponds to an open reading frame starting at the initiator ATG codon in exon 3 and continuing through exon 4 until the first nonsense codon in intron 4. The production of a protein product and disease phenotype in the SCA7-CTCF-I-mut mice is reminiscent of the R6/2 mouse model of HD, in which a small fragment from the htt gene was introduced into mice to model repeat instability, but also yielded a truncated protein product resulting in a HD-like phenotype – despite the fact that the construct lacked a 3′ polyadenylation site or characterized promoter (Mangiarini et al., 1996).
Loss of SCAANT1 expression is accompanied by de-repression of sense transcription
Our findings indicate that mutation of the 3′ CTCF binding site is responsible for initiation of robust sense transcription in SCA7-CTCF-mut-I mice, as SCA7-CTCF-I-wt mice carrying an ataxin-7 genomic fragment with an intact 3′ CTCF binding site express low levels of ataxin-7 mRNA and protein. To determine if the levels of ataxin-7 sense and antisense transcription within the repeat region domain correlate in SCA7-CTCF-I-wt and SCA7-CTCF-I-mut mice, we performed quantitative strand-specific RT-PCR amplification, and detected ataxin-7 sense and antisense transcripts in each line. We found that ataxin-7 sense transcript levels were elevated ~370-fold in the brains of SCA7-CTCF-I-mut mice compared to SCA7-CTCF-I-wt mice, and this was accompanied by an ~140-fold decrease in SCAANT1 expression (Figure 3D). In situ hybridization analysis confirmed robust expression of SCAANT1 in the cerebellum of SCA7-CTCF-I-wt mice, but did not detect strong SCAANT1 expression in SCA7-CTCF-I-mut mice (Figure 3E). In situ hybridization analysis indicated moderate to strong expression of SCAANT1 in SCA7-CTCF-I-wt mice throughout the brain (Figure S4A). Correspondingly, in situ hybridization did not yield evidence for much SCAANT1 expression in the brain of SCA7-CTCF-I-mut mice (Figure S4B). Taken together, these findings show that reduced SCAANT1 expression correlates with increased P2A promoter activity, resulting in increased sense expression of the ataxin-7 gene.
Ataxin-7 sense expression and SCAANT1 expression are inversely correlated in human tissues and SCA7 patients
Our studies of the SCA7-CTCF-I-wt and SCA7-CTCF-I-mut mice suggested that expression of the ataxin-7 sense transcript inversely correlates with expression of SCAANT1. To determine if this reciprocal expression relationship exists in normal human tissues, we performed qRT-PCR analysis on a panel of human tissue RNAs. While we documented high levels of ataxin-7 sense transcript in cortex, cerebellum, striatum, and liver, we found much lower levels of ataxin-7 in lung and kidney (Figure 4A). Interrogation of SCAANT1 expression levels revealed an opposite pattern, as SCAANT1 was much higher in the lung and kidney than in cortex, cerebellum, striatum, or liver (Figure 4B). As the presence of the CAG repeat expansion decreased SCAANT1 promoter activity in our luciferase reporter assays (Figure 2), we tested if the diametrically opposed expression of ataxin-7 sense transcript and SCAANT1 might occur in the context of SCA7 disease. To test this hypothesis, we performed RT-PCR analysis of a SCA7 patient fibroblast cell line, and while we could amplify both the normal and expanded repeat alleles for the ataxin-7 sense transcript, we could not detect antisense SCAANT1 transcript expression from the expanded 55Q allele (Figure 4C). We then performed quantitative RT-PCR analysis of ataxin-7 sense expression on fibroblasts obtained from two SCA7 patients, one with a moderately sized disease repeat (55Q), and one with a severely expanded repeat (150Q). With increasing expansion size, we observed significantly increased ataxin-7 sense transcript levels (Figure 4D), indicating that expansion of the CAG repeat at the ataxin-7 locus yields increased levels of ataxin-7 transcript in association with reduced expression of SCAANT1. We also obtained a set of peripheral blood samples from three additional SCA7 patients, isolated RNA from their lymphocytes, and performed RT-PCR analysis. Antisense SCAANT1 transcript expression could not be detected from the expanded allele of the SCA7 samples, and all three SCA7 patients exhibited significantly increased ataxin-7 sense transcript levels (Figure 4E).
Figure 4. Reciprocal ataxin-7 sense and antisense expression in human tissues and SCA7.
A) Results of qRT-PCR analysis of ataxin-7 sense transcript. Expression of ataxin-7 sense transcript was significantly less in lung and kidney (p < .01 and p < .001 by ANOVA with post-hoc Tukey test). Assays were performed in triplicate, and error bars = s.e.m.
B) Results of qRT-PCR analysis of ataxin-7 antisense ncRNA SCAANT1. Expression of SCAANT1 was significantly elevated in lung and kidney (p < .001 and p < .01 by ANOVA with post-hoc Tukey test). Assays were performed in triplicate, and error bars = s.e.m.
C) RT-PCR analysis of SCA7 patient fibroblast RNAs. RT-PCR amplification reveals the ataxin-7 antisense ncRNA SCAANT1 from the normal allele in a control fibroblast line (15Q) and SCA7 patient fibroblast line (55Q), but not from the expanded allele for the SCA7 patient. RT-PCR amplification of the ataxin-7 sense transcript, however, indicates strong expression from the normal and expanded allele in SCA7 fibroblasts.
D) Quantification of ataxin-7 RNA levels in SCA7 patient fibroblasts. Significant increases in ataxin-7 sense expression were present in SCA7 patients carrying either 55Q or 150Q alleles (p < 0.01 by ANOVA). Experiments were performed in triplicate, and error bars = s.e.m.
E) Quantification of ataxin-7 RNA levels in SCA7 patient lymphocytes. Significant increases in ataxin-7 sense expression were present in SCA7 patients carrying 51Q, 59Q, or 66Q alleles (p < 0.01 by ANOVA). Experiments were performed in triplicate, and error bars = s.e.m.
See Figure S4.
CTCF regulates SCAANT1 expression and ataxin-7 alternative sense transcription
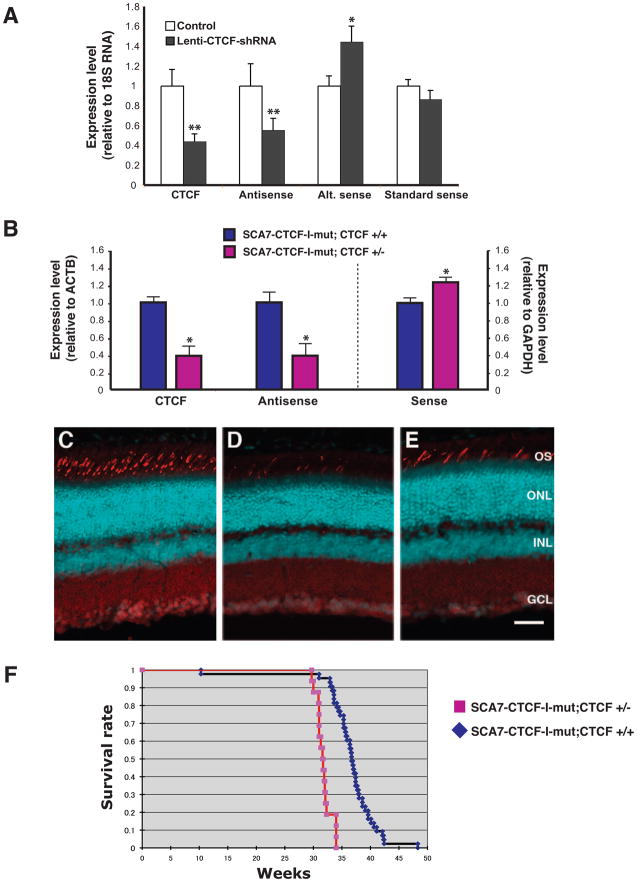
As mutation of the 3′ CTCF binding site reduced the activity of the SCAANT1 promoter while de-repressing ataxin-7 sense expression from promoter P2A, we hypothesized that CTCF modulates ataxin-7 sense expression from this promoter by driving the expression of SCAANT1. To test this hypothesis, we validated two different CTCF shRNA’s and derived a dual CTCF knock-down vector. After subcloning the CTCF dual shRNA knock-down fragment into a lentiviral construct with a linked eGFP expression cassette, we infected human Y-79 retinoblastoma cells and isolated RNA from flow-sorted GFP-positive Y-79 cells. Real-time RT-PCR analysis confirmed CTCF knock-down, and revealed a significant reduction in the expression level of SCAANT1 (Figure 5A). Significant reduction in SCAANT1 expression was accompanied by a marked increase in the ataxin-7 sense transcript from the P2A promoter, but not from the previously defined ‘standard’ ataxin-7 sense promoter, located >40 kb 5′ to the repeat region (Figure 5A). Although direct, physiological comparison of the standard P1 promoter and P2A promoter is complicated by the co-existence of SCAANT1 transcription, analysis of ataxin-7 alternative sense and standard sense expression at baseline in Y-79 cells revealed only modestly (i.e. ~2.5-fold) higher levels of the ataxin-7 standard sense transcript, consistent with the similar degree of H3K4me3 enrichment noted for their respective TSSs (Figure S1). To examine the trans contribution of CTCF protein levels to the regulation of ataxin-7 gene expression in the SCA7-CTCF-I-mut mice, we generated CTCF heterozygous knock-out mice (G.N. Filippova et al., unpublished results). ChIP analysis has indicated that reduced CTCF occupancy at the mutated 3′ CTCF binding site occurs in the SCA7-CTCF-I-mut mice (Libby et al., 2008), and this may account for the de-repression of ataxin-7 sense expression from promoter P2A. To test if the effect of this cis mutation could be compounded by reduction of CTCF expression in trans, we crossed SCA7-CTCF-I-mut mice with CTCF heterozygous null mice, and compared the resulting SCA7-CTCF-I-mut; CTCF +/− mice with their SCA7-CTCF-I-mut; CTCF +/+ littermates. We confirmed reduced dosage of CTCF expression in the SCA7-CTCF-I-mut; CTCF +/− mice, and observed significantly reduced expression of the antisense SCAANT1 transcript (Figure 5B). This was accompanied by increased ataxin-7 sense expression (Figure 5B), which yielded a more rapidly progressive retinal phenotype in SCA7-CTCF-I-mut; CTCF +/− mice (Figure 5C–E). The worsened phenotype was also reflected by a significantly shortened lifespan (Figure 5F). Hence, decreased expression of CTCF agonized the SCA7 phenotype in SCA7-CTCF-I-mut mice by further de-repressing ataxin-7 P2A promoter activity. As cohesin may play a role in CTCF insulator formation (Parelho et al., 2008; Stedman et al., 2008; Wendt et al., 2008), we performed ChIP analysis for two cohesin subunits in the SCA7 transgenic mice, and observed reduced occupancy of SMC1 and SMC3 at the 3′ CTCF binding site in the cerebellum of SCA7-CTCF-I-mut mice (Figure S5), suggesting that cohesin may also participate in CTCF-mediated transcription regulation at the ataxin-7 locus.
Figure 5. CTCF regulates promoter usage for ataxin-7 sense and antisense transcripts.
A) CTCF knock-down in Y-79 retinoblastoma cells leads to decreased ataxin-7 antisense RNA expression and increased exon 2A transcript expression. Human Y-79 retinoblastoma cells were infected with a lentiviral expression construct containing two tandem shRNAs targeting CTCF (lenti-CTCF-shRNA) or with empty vector (Control), and expression levels were measured by qPCR. Marked reductions in expression of CTCF and ataxin-7 antisense SCAANT1 transcripts were noted (p < 0.01 by ANOVA), and were accompanied by a significant increase in the ataxin-7 alternative sense transcript (p < .0.05 by ANOVA). Knock-down of CTCF did not affect expression of the ataxin-7 transcript initiated from a human-specific ataxin-7 promoter located >40 kb 5′ to the alternative promoter. Experiments were performed in triplicate, the expression level for each transcript upon CTCF knock-down is normalized to its respective control, and error bars = s.e.m.
B) Decreased CTCF dosage reduces SCAANT1 expression in SCA7-CTCF-I-mut mice. We crossed SCA7-CTCF-I-mut mice with CTCF +/− mice and derived SCA7-CTCF-I-mut mice on a CTCF heterozygous null background (CTCF +/−) or a wild-type background (CTCF +/+). We performed qRT-PCR analysis of CTCF in the brain of three month-old mice, and confirmed reduced expression of CTCF in SCA7-CTCF-I-mut; CTCF+/− mice (p < .0.05 by ANOVA). When we measured brain expression of SCAANT1 (Antisense) transcript and ataxin-7 sense transcript by qRT-PCR, we noted a marked reduction in antisense expression and a significant increase in sense expression in SCA7-CTCF-I-mut; CTCF+/− mice (p < .0.05 by ANOVA). Experiments were performed in triplicate, the expression level for each transcript is normalized to the SCA7-CTCF-I-mut; CTCF+/+ level, and error bars = s.e.m.
C–E) Reduced CTCF dosage enhances SCA7 retinal degeneration in SCA7-CTCF-I-mut mice. Confocal microscopy of retinal sections from six month-old non-transgenic controls (C), SCA7-CTCF-I-mut; CTCF +/− (D), and SCA7-CTCF-I-mut; CTCF +/+ (E), reveals more severe cone photoreceptor drop-out in SCA7-CTCF-I-mut mice on a CTCF heterozygous null background. Red/green pigment antibody = cyan; propidium iodide = red. OS = outer segments; ONL = outer nuclear layer; INL = inner nuclear layer; GCL = ganglion cell layer. Scale bar corresponds to 50 μM.
F) Kaplan-Meier survival analysis of SCA7-CTCF-I-mut; CTCF+/− mice. We recorded lifespan for cohorts of SCA7-CTCF-I-mut; CTCF+/− mice (n = 28) and SCA7-CTCF-I-mut; CTCF+/+ mice (n = 40), and observed a significant reduction in lifespan for SCA7-CTCF-I-mut mice on a CTCF heterozygous null background (p < 0.05 by log-rank test).
See Figure S5.
SCAANT1 mediates repression of the corresponding ataxin-7 sense transcript in cis
CTCF binding regulates sense and antisense transcription at the ataxin-7 locus, and expression levels of the ataxin-7 sense transcript and antisense SCAANT1 message are inversely correlated. Hence, a key question is whether SCAANT1 transcription is coincident, or required for de-repression of ataxin-7 sense promoter P2A. To determine if SCAANT1 transcription is necessary for the regulation of ataxin-7 sense expression, and to distinguish between a cis or trans regulatory mechanism, we developed a CMV-SCAANT1 expression construct. We then co-transfected astrocytes with a highly active ataxin-7 genomic fragment – luciferase reporter construct and the CMV-SCAANT1 expression construct, and tested if enforced expression of SCAANT1 would down-regulate ataxin-7 sense P2A promoter activity, but we observed no effect (Figure 6A). This in vitro experiment was paralleled by an in vivo study in which we crossed SCA7-CTCF-I-mut mice with SCA7-CTCF-I-wt mice, reasoning that the dramatically elevated levels of SCAANT1 antisense RNA in SCA7-CTCF-I-wt mice would reduce ataxin-7 sense expression in SCA7-CTCF-I-mut mice, if SCAANT1 regulation of ataxin-7 sense expression is occurring in trans. However, behavioral analysis revealed further worsening in rotarod performance in SCA7-CTCF-mut-I; SCA7-CTCF-I-wt mice in comparison to SCA7-CTCF-I-mut mice (Figure S6). Furthermore, SCA7-CTCF-I-mut; SCA7-CTCF-I-wt bigenic mice displayed more severe Purkinje cell degeneration than their singly transgenic SCA7-CTCF-I-mut littermates (Figure 6B-D). Thus, greater expression of SCAANT1 in SCA7-CTCF-I-mut; SCA7-CTCF-I-wt mice did not reduce ataxin-7 sense transcription; instead, low levels of ataxin-7 sense protein product from the SCA7-CTCF-I-wt mice (Figure 3C) enhanced the SCA7 phenotype in bigenic SCA7-CTCF-I-mut; SCA7-CTCF-I-wt mice.
Figure 6. The ataxin-7 antisense ncRNA, SCAANT1, regulates ataxin-7 alternative promoter activity in cis, but not in trans.
A) Effect of enforced expression of SCAANT1 upon transactivation competence of ataxin-7 promoter P2A. The human ataxin-7 genomic fragment luciferase expression construct (1F; see Figure 1A) was transfected into primary astrocytes, along with Renilla luciferase, and a CMV expression construct containing SCAANT1 (CMV-SCAANT1). Relative luminescence unit (RLU) measurements were obtained. EV corresponds to the empty luciferase vector, CMV-empty refers to the empty CMV expression construct, all experiments were performed in quadruplicate, and errors bars = s.e.m.
B–D) Effect of over-expression of SCAANT1 upon the de-repressed ataxin-7 alternative promoter in vivo. We crossed the SCA7-CTCF-I-mut mice with SCA7-CTCF-I-wt mice, and compared cerebellar histopathology between littermates of the following genotypes: SCA7-CTCF-I-wt (B), SCA7-CTCF-I-mut (C), and SCA7-CTCF-I-mut; SCA7-CTCF-I-wt (D). Here we see the results of confocal microscopy of cerebellar sections immunostained with antibodies against calbindin (green) and ataxin-7 (magenta), and counterstained with DAPI (blue) for mice at 7 months of age. Strong calbindin immunoreactivity is observed in SCA7-CTCF-I-wt mice (B), while an obvious reduction in calbindin immunoreactivity is apparent in the SCA7-CTCF-I-mut mice (C). Calbindin immunoreactivity is further reduced in SCA7-CTCF-I-mut; SCA7-CTCF-I-wt bigenic mice, and this is accompanied by an increased accumulation of aggregated ataxin-7 protein (magenta puncta) (D), indicating that enforced in vivo expression of SCAANT1 does not promote degradation of the complementary ataxin-7 sense transcript. Rather, crossing with SCA7-CTCF-I-wt mice actually worsens the phenotype, since SCA7-CTCF-I-wt mice produce modest levels of polyQ-expanded ataxin-7 protein (see Figure 3C).
E) Diagram of the ataxin-7 antisense premature termination construct. We replaced the ataxin-7 antisense ncRNA promoter with a tet-responsive element – minimal CMV promoter (TRE) and inserted Renilla luciferase (R-luc) in antisense orientation to quantify the activity of the TRE promoter. The IRES-luc segment is cloned in sense orientation to permit normalization. The poly-A transcription termination cassette was cloned into exon 3 in antisense orientation (asterisk), and we prepared two versions of this construct: either without the poly-A cassette (TRE-only) or with the poly-A cassette (TRE-poly-A-trap).
F) Validation of poly-A transcription termination activity. The TRE-only construct or the TRE-poly-A-trap construct was transfected into primary astrocytes, along with a CMV-tet-activator expression construct, and Renilla luciferase (R-luc) and luciferase (luc) activities were measured. A marked reduction in the R-luc / luc ratio in the TRE-poly-A-trap construct was noted (p < .01, ANOVA).
G) Convergent transcription is required for repression of ataxin-7 alternative promoter by the antisense ncRNA. The TRE-only construct or the TRE-poly-A-trap construct was transfected into primary astrocytes, along with tet-activator, and ataxin-7 sense transcript levels were measured by qPCR. Ataxin-7 sense expression was dramatically increased in cells transfected with the TRE-poly-A-trap construct (p < .01, ANOVA).
See Figure S6.
To test if SCAANT1 transcription regulates promoter P2A in cis, we engineered ataxin-7 P2A – exon 3(CAG10) – exon 4 genomic fragment constructs with a 3′ IRES-luciferase (luc) in sense orientation, and a Renilla luciferase (Rluc) in antisense orientation (Figure 6E). We replaced the antisense SCAANT1 promoter with a tet-regulatable element (TRE) to yield the “TRE-only” ataxin-7 genomic fragment construct, and then created a second version by cloning a polyA transcription termination signal (“polyA trap”) in antisense orientation into exon 3 (Figure 6E). To confirm the integrity of the polyA trap, we transfected astrocytes with either the TRE-only or TRE-polyA-trap ataxin-7 vector, induced with doxycycline, and observed marked reduction of Rluc/luc for the TRE-polyA-trap ataxin-7 vector (Figure 6F). When we measured ataxin-7 expression by qRT-PCR, we observed significant de-repression of ataxin-7 sense expression in TRE-polyA-trap ataxin-7 transfected cells (Figure 6G). Hence, premature termination of SCAANT1 transcription released repression of ataxin-7 P2A promoter activity, indicating that SCAANT1 regulates ataxin-7 sense expression in cis by convergent transcription.
Ataxin-7 antisense expression yields repressive chromatin modifications in transgenic mice
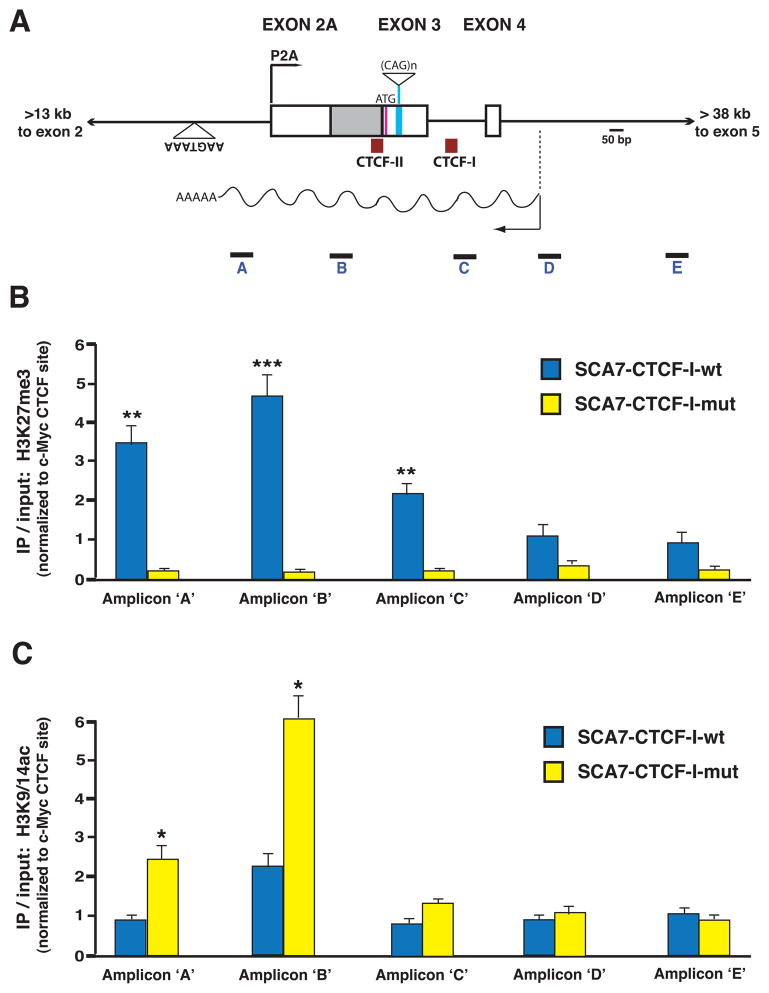
Silencing of ataxin-7 sense P2A promoter activity by convergent transcription of the SCAANT1 RNA raised a number of questions as to the mechanism of repression. We hypothesized that one possibility might be chromatin-dependent gene silencing, and proceeded to evaluate the status of covalent histone modifications known to correlate with transcription activation and repression in SCA7 CTCF-I-mut and SCA7-CTCF-I-wt mice. To do this, we performed ChIP analysis of histone marks, and interrogated covalent histone modification status at amplicons spanning the ataxin-7 repeat region, including the transcription domains and start sites for the sense P2A promoter and SCAANT1 (Figure 7A). Quantitative PCR analysis revealed highly enriched levels of the repressive H3K27me3 mark at amplicons 5′ and 3′ to the P2A TSS, and also showed low levels of the active H3K9/14ac mark in SCA7-CTCF-I-wt mice (Figure 7B–C). However, ChIP analysis revealed a reversal of this pattern in the SCA7-CTCF-I-mut mice, as amplicons adjacent to promoter P2A exhibited very low levels of H3K27me3 associated with elevated levels of H3K9/14ac (Figure 7B–C), consistent with active transcription at promoter P2A. Hence, our survey of covalent histone modifications in the ataxin-7 mini-gene mice supported a role for chromatin-dependent gene silencing by SCAANT1.
Figure 7. ChIP analysis of histone modification status at the ataxin-7 alternative promoter region in SCA7 transgenic mice.
A) Diagram of the genomic region of the human ataxin-7 gene, where the alternative promoter (P2A), first coding exon, CTCF binding sites, and antisense ncRNA, SCAANT1, are located. Boxes represent exons, while solid lines correspond to introns. The positions of the five amplicons employed in the ChIP analysis are indicated.
B–C) Results of ChIP analysis of histone marks in the cerebellum of SCA7 transgenic mice. ChIP was performed with antibodies against a repressive mark (H3K27me3) and activated transcription mark (H3K9/14Ac) on cerebellar DNAs isolated from six-month old SCA7-CTCF-I-wt and SCA7-CTCF-I-mut mice. B) In SCA7-CTCF-I-wt mice that express minimal amounts of ataxin-7 sense RNA from P2A, there was significant enrichment of H3K27me3 extending from the alternative promoter region into intron 3 of the ataxin-7 gene (p < .01 to .001, ANOVA). C) This pattern was reversed in SCA7-CTCF-I-mut mice that display robust activation of the P2A promoter, as significant enrichment of the H3K9/14Ac mark was present in the ataxin-7 sense P2A region and initial transcript region (p < 0.05, ANOVA). Results represent three independent experiments quantifying histone antibody ChIP normalized to the c-Myc promoter region. Error bars = s.e.m.
Discussion
Bidirectional transcription at repeat loci is emerging as an important theme in repeat expansion diseases, including myotonic dystrophy 1 (DM1), spinocerebellar ataxia type 8 (SCA8), the fragile X syndrome of mental retardation, Friedreich’s ataxia (FRDA), Huntington’s disease (HD), and Huntington’s disease-like 2 (HDL2) (Mirkin, 2007). At the same time, a role for CTCF in regulating chromatin structure and transcription at such repeat disease loci is being recognized (La Spada and Taylor, 2010). At the SCA7 locus, the significance of CTCF for regulating repeat instability was recently demonstrated, and shown to involve epigenetic processes (Libby et al., 2008). In this study, we examined the ataxin-7 repeat region where the CTCF binding sites reside, and discovered that ataxin-7 gene expression is governed by a novel antisense ncRNA transcript. This transcript, which we named ‘SCAANT1’, appears to regulate a previously unrecognized ataxin-7 sense promoter by convergent transcription that overlaps the ataxin-7 repeat and the adjacent P2A sense promoter. Our studies thus reveal a novel pathway for regulating ataxin-7 gene expression at this promoter, and link CTCF transactivation of SCAANT1 with repression of the convergently transcribed sense domain (Figure 8).
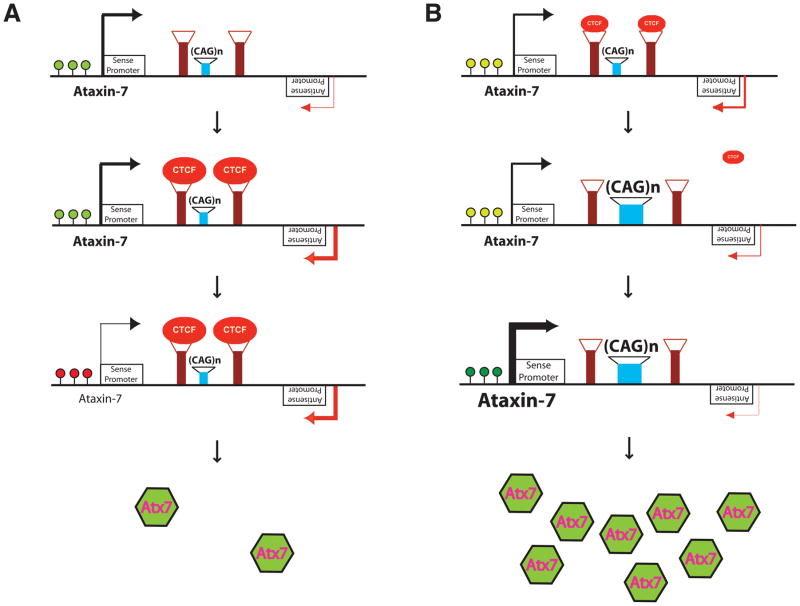
Figure 8. Model for transcription regulation at the ataxin-7 locus.
A) CTCF-induced, SCAANT1-mediated, Dicer-dependent repression of ataxin-7 sense transcription. The ataxin-7 CAG repeat is flanked by CTCF binding sites. When the CTCF binding sites are unoccupied, the adjacent ataxin-7 promoter is active, and covalent histone modification at the promoter region is consistent with active transcription (light green dots). When CTCF levels increase and CTCF binds, the antisense promoter for the non-coding RNA SCAANT1 is activated, yielding convergent transcription. This is accompanied by a transition from an active chromatin conformation to a repressed chromatin state (red dots), silencing ataxin-7 expression from sense promoter P2A.
B) CAG repeat expansion or CTCF reduction can de-repress ataxin-7 sense expression. Under conditions of modest CTCF expression, the ataxin-7 sense promoter is moderately active (light green dots). However, if the CAG repeat is expanded, or CTCF levels dwindle, the flanking CTCF binding sites become unoccupied, resulting in reduced activity of the SCAANT1 promoter. Under these conditions, the sense promoter is fully de-repressed, with a chromatin conformation consistent with highly active transcription (green dots), yielding robust expression of ataxin-7.
Repeat tracts can greatly influence chromatin structure, especially if they are CG-rich (Wang et al., 1996). The mapping of CTCF binding sites in close proximity to such repeats suggested the need to insulate surrounding DNA from the potentially untoward effects of repeat-induced changes upon chromatin structure. CTCF is a multivalent transcription regulatory factor, known to possess enhancer-blocking activity (Phillips and Corces, 2009). CTCF may also prevent inactivation of gene expression, as CTCF can restrict the spread of X-inactivation, thereby preserving the transcriptional activity of “escape” genes (Filippova et al., 2005). Previous studies of repeat disease loci have shown that CTCF can prevent epigenetic changes associated with heterochromatin formation and gene inactivation by constraining antisense transcription (Cho et al., 2005; De Biase et al., 2009; Filippova et al., 2005). We evaluated the role of CTCF in regulating ataxin-7 gene expression from an adjacent alternative promoter (P2A) by introducing two different ataxin-7 mini-genes into mice. These mini-genes are ~13.5 kb ataxin-7 genomic fragments that contain the P2A promoter, the SCAANT1 domain, the ataxin-7 start site of translation, and the CAG repeat tract. The two mini-genes were identical except for the presence of a substitution mutation in the ‘SCA7-CTCF-I-mut’ construct at the 3′ CTCF binding site. Importantly, this substitution mutation had been shown to completely abrogate CTCF binding (Libby et al., 2008). The SCA7-CTCF-I-wt construct yielded four independent lines of transgenic mice that did not develop a phenotype, despite possessing a CAG92 repeat tract in the fully intact ataxin-7 mini-gene. Instead, two independent lines of SCA7-CTCF-I-mut mice developed a SCA7-like phenotype, characterized by cone-rod dystrophy retinal degeneration and cerebellar atrophy. Further studies indicated that loss of CTCF binding results in dramatically reduced expression of SCAANT1 in association with high-level ataxin-7 expression from the newly discovered alternative sense promoter. Our findings thus reveal that CTCF does regulate ataxin-7 gene expression; however, instead of preventing transcription repression, CTCF supports it. Furthermore, rather than restricting antisense expression, CTCF promotes it.
Surveys of mammalian transcriptomes are uncovering tremendous numbers and varieties of non-coding RNAs, and the production of antisense transcripts appears to be a pervasive feature of the human and mouse transcriptomes (He et al., 2008; Kapranov et al., 2007; Okazaki et al., 2002). When we discovered that SCAANT1 expression levels inversely correlate with ataxin-7 sense expression in both SCA7 transgenic mice and human tissues, we considered the possibility that SCAANT1 might be regulating the expression of its sense counterpart, as reciprocal expression of sense and antisense transcripts has been reported for a number of human and mouse genes (Katayama et al., 2005). Indeed, at the human p15 locus, gene silencing of sense expression by an antisense RNA has been documented, and can be achieved by enforced expression of the antisense transcript (Yu et al., 2008). We tested if SCAANT1 expression in trans can down-regulate ataxin-7 alternative sense promoter activity in luciferase reporter assay experiments, and by crossing SCA7-CTCF-I-mut mice with SCA7-CTCF-I-wt mice, as the latter exhibit high-level SCAANT1 expression. However, SCAANT1 transcript elevation had no effect upon ataxin-7 alternative sense expression in vitro or in vivo. Studies of antisense transcripts in mice and humans, as well as other eukaryotes such as yeast, have revealed evidence for inhibition of transcription by virtue of actual transcription interference, when RNA polymerases moving in opposite directions collide with one another (Osato et al., 2007; Shearwin et al., 2005). To test if SCAANT1 regulates sense expression in cis, we engineered an ataxin-7 genomic fragment construct with a transcription terminator positioned in the antisense orientation, and placed antisense transcription under the control of an inducible promoter. After validating the efficiency of the transcription terminator, we measured the effect of premature transcription termination upon SCAANT1’s ability to repress ataxin-7 sense expression, and we noted a dramatic de-repression of sense transcription, when antisense transcription was prematurely terminated. These results confirmed that SCAANT1 is directly regulating ataxin-7 alternative sense expression and demonstrated that the regulation occurs in cis.
One obvious question, based upon our results is: Why does the cell need such a complicated pathway for adjusting ataxin-7 expression? Ataxin-7 is a core, and likely essential, component of different ubiquitously expressed transcription co-activator complexes (Helmlinger et al., 2004; Palhan et al., 2005; Zhao et al., 2008). As deletion of the ataxin-7 orthologue Sgf73 eliminates Ubp8-mediated histone deubiquitination in yeast (Kohler et al., 2008), and knock-down of ataxin-7 results in disassembly of the STAGA complex in mammals (Palhan et al., 2005), tight regulation of ataxin-7 expression could be a mechanism for controlling the activity of these co-activators. CTCF is a master regulator of transcription, and its expression cannot be significantly adjusted without killing the cell. However, minor changes in CTCF levels, or binding activity, could have a dramatic impact upon the transcriptional activity of the cell through its regulation of ataxin-7 expression, since ataxin-7 expression alterations would be amplified by affecting the stability and function of entire co-activator complexes. Thus, CTCF control of ataxin-7 levels could serve as a rheostat for setting global transcription activity status for the cell.
Another important implication of our work is its relevance to SCA7 disease pathogenesis and repeat disease biology. As we have shown, expansion of the ataxin-7 CAG repeat tract reduces SCAANT1 promoter activity, resulting in minimally detectable levels of SCAANT1 from the expanded allele in SCA7 patient fibroblasts. This reduction in SCAANT1 expression de-represses the ataxin-7 alternative promoter, and significantly boosts the level of ataxin-7, creating a feed forward effect that agonizes the SCA7 disease pathway by favoring increased production of mutant ataxin-7 protein (Figure 8). Although we can not exclude a role for altered transcript stability in this process, a survey of histone post-translational modifications in our SCA7 transgenic mice revealed repressive chromatin modifications in the alternative promoter when SCAANT1 transcription is robust, indicating that transcriptional activity is likely more important than transcript stability in controlling ataxin-7 sense expression. As bidirectional transcription in association with CTCF binding is emerging as a common feature of repeat disease loci, our findings could be applicable at other repeat disease loci, including loci that have not been carefully screened for antisense transcripts. Furthermore, the existence of regulatory bidirectional transcription may offer an entry point for therapeutically modulating ataxin-7 expression at the RNA level. In addition to providing a window into how gene transcription is regulated, convergent transcription may dictate which repeat loci are subject to dramatic genetic instability, as epigenetic modifications and chromatin alterations likely lie at the heart of the instability process. Hence, regulatory bidirectional transcription at repeat loci, coupled with CTCF-mediated epigenetic processes in the germ line, may explain why certain repeats exhibit dramatic parent-of-origin instability, while other repeats are relatively stable and not subject to parent-of-origin effects (La Spada, 1997; Pearson et al., 2005). Understanding the roles and regulation of convergent, bidirectional transcription at repeat loci should provide valuable clues as to how global transcriptional processes and epigenetic pathways work.
Experimental Procedures
Characterization of transgenic mice
The generation of the SCA7 transgenic constructs, and the production of the SCA7-CTCF-I-wt and SCA7-CTCF-I-mut mice have been previously described (Libby et al., 2008). ERG analysis, immunostaining analysis, RT-PCR studies, Western blot analysis, and behavioral studies were performed as previously described (Garden et al., 2002; La Spada et al., 2001). All experiments were approved by the University of Washington IACUC and UCSD IACUC. Please see Supplementary Material for detailed protocols for chromatin immunoprecipitation and in situ hybridization.
RNA Isolation and Analysis
Cell line, patient, and mouse tissue RNAs were isolated using Trizol and treated 2–3 times with DNase I (NEB), then precipitated for analysis. The integrity of RNA was determined by nested RT-PCR of cDNA generated with or without reverse transcriptase, using primers to at least 3 distinct genomic regions. For RT–PCR, cDNA was generated using gene-specific primers, which had a linker (LK) sequence, LK 5′-CGACTGGAGCACGAGGACACTGA-3′ attached to the 5′ end. Otherwise cDNA was generated with random decamers (Ambion). cDNA was generated with 1 – 3 μg of RNA and Superscript III (Invitrogen) at 50°C. PCR amplification was performed using two gene specific primers or with a primer to the Linker sequence and a gene-specific reverse primer for strand-specific analysis (35 cycles at 94°C for 30 s, 55°C for 30 s and 68°C for 1 min). The PCR products were cloned into the pCR®4-TOPO vector (Invitrogen) and sequenced. Strand-specific RT-PCR was essentially performed as previously described (Ladd et al., 2007). Please see the Supplementary Material for details of construct generation.
Statistical analysis
All data were prepared for analysis with standard spread sheet software (Microsoft Excel). Statistical analysis was done using Microsoft Excel, Prism 4.0 (Graph Pad), or the VassarStats website < http://faculty.vassar.edu/lowry/VassarStats.html>. For ANOVA, if statistical significance (p < 0.05) was achieved, we performed post-test analysis to account for multiple comparisons. The level of significance (alpha) was always set at 0.05.
Supplementary Material
Figure S1. Genome browser analysis of the ataxin-7 gene and repeat region.
This UCSC Genome Browser view of the ataxin-7 gene reveals the 5 ′ region of the ataxin-7 gene, with the Promoter H3K4me3, Transcription Factor ChIP track, CpG islands track, and some comparative sequence tracks. Both the previously predicted 5 ′ promoter and an alternative promoter at the repeat region contain strong peaks for tracks associated with promoter activity.
Figure S2. Diagram of ataxin-7 gene and the repeat region used in mini-gene constructs.
Across the top of this panel, the UCSC genome browser exon-intron prediction for the first half of the human ataxin-7 gene is shown. The previously described transcription start site at exon 1, adjacent to the predicted promoter (P1), is indicated. The purple rectangle overlies the genomic region used for the ataxin-7 mini-gene transgenic constructs, SCA7-CTCF-I-wt and SCA7-CTCF-I-mut. Below the purple rectangle, an expanded map of this region is shown. The 11-nucleotide substitution mutation introduced to create the ataxin-7 mini-gene construct for SCA7-CTCF-I-mut is given.
Figure S3. SCA7-CTCF-I-mut mice develop cone-rod dystrophy retinal degeneration.
Representative rod-initiated and cone-initiated electroretinogram (ERG) responses for 8 month-old SCA7-CTCF-I-wt and SCA7-CTCF-I-mut (line 1245) mice are shown. Each ERG was performed at a range of light intensities from dimmest (top) to brightest (bottom), and the extent of the response was monitored over time, with scale bars for the intensity of the response (in μV) and the time (in ms) shown respectively for the rod responses and cone responses. Note that rod-initiated responses are modestly reduced in the SCA7-CTCF-I-mut mouse compared to the SCA7-CTCF-I-wt mouse; however, cone-initiated responses are markedly reduced in the SCA7-CTCF-I-mut mouse compared to the SCA7-CTCF-I-wt mouse. Degradation of cone responses ahead of rod responses is consistent with a cone-rod dystrophy form of retinal degeneration in the SCA7-CTCF-I-mut mice.
Figure S4. In situ hybridization of SCAANT1 in SCA7 transgenic mice reveals a broad pattern of diametrically opposed antisense ataxin-7 expression.
A) When we probed sagittal brain sections from three month-old SCA7-CTCF-I-wt mice with the ataxin-7 antisense riboprobe, we noted prominent signal throughout the neuronal regions of the entire brain, including the cerebellum.
B) When we probed sagittal brain sections from three month-old SCA7-CTCF-I-mut mice with the ataxin-7 antisense riboprobe, we noted minimal signal throughout the neuronal regions of the entire brain.
Figure S5. Cohesin binding is reduced in the SCA7-CTCF-I-mut mice.
A) Diagram of the genomic region of the human ataxin-7 gene, where the CTCF binding sites and region of bidirectional transcription are located. Boxes represent exons, while solid lines correspond to introns. The position of the amplicon employed in the ChIP analysis is indicated as a black rectangle.
B) Results of ChIP analysis of SMC1 occupancy at the ataxin-7 CTCF binding site region in the cerebellum of six month-old SCA7 transgenic mice. There was a significant reduction in SMC1 occupancy at this amplicon in SCA7-CTCF-I-mut mice (p < .05, ANOVA). Results represent three independent experiments quantifying SMC1 antibody ChIP normalized to the c-Myc promoter region, and an amplicon 3′ to the CTCF-I binding site was used to diminish detection of SMC1 occupancy at the CTCF-II site. Error bars correspond to s.e.m.
C) Results of ChIP analysis of SMC3 occupancy at the ataxin-7 CTCF binding site region in the cerebellum of six month-old SCA7 transgenic mice. There was a significant reduction in SMC1 occupancy at this amplicon in SCA7-CTCF-I-mut mice (p < .05, ANOVA). Results represent three independent experiments quantifying SMC3 antibody ChIP normalized to the c-Myc promoter region, and an amplicon 3′ to the CTCF-I binding site was used to diminish detection of SMC3 occupancy at the CTCF-II site. Error bars correspond to s.e.m.
Figure S6. SCA7-CTCF-I-mut; SCA7-CTCF-I-wt bigenic mice perform worse than SCA7-CTCF-I-mut mice on the rotarod.
Groups of five to seven mice (littermates, all at 7 months of age) were tested over the course of four days on an accelerating rotarod. Mean latency to fall (in seconds) was obtained for each group and compared statistically by ANOVA with post-hoc Tukey test. On three of the four trial days, the SCA7-CTCF-I-mut; SCA7-CTCF-I-wt bigenic mice performed significantly worse than both the SCA7-CTCF-I-wt mice and the SCA7-CTCF-I-mut mice (p < .05). Error bars correspond to s.e.m.
Here we see two different transgenic mice, a SCA7-CTCF-I-wt mouse (light brown fur) and a SCA7-CTCF-I-mut line 1245 mouse (dark brown fur), at six months of age. The SCA7-CTCF-I-wt mouse of is average size and weight for the C57BL/6J strain background, and exhibits normal exploratory behavior without evidence of gait abnormality or tremor. However, the SCA7-CTCF-I-mut individual is much smaller in size and weight, though it is the same age and on the identical strain background. Furthermore, the SCA7-CTCF-I-mut mouse displays a moderate gait ataxia upon ambulation, and tremulousness while stationary.
Acknowledgments
The authors wish to thank A. Smith, S. Baccam, K. Takushi, J. Huang, and D. Possin for outstanding technical assistance, and B. Ren for critical reading of the manuscript. This work was supported by funding from the NIH (GM59356, EY14061, and ARRA award GM59356-09-S1 to A.R.L., and EY01730 [Vision Research Core] to UWMC). S.M.S. is supported by the Allen Institute for Brain Science, founded by Paul G. Allen and Jody Patton.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn JK, Seo JM, Chung H, Yu HG. Anatomical and functional characteristics in atrophic maculopathy associated with spinocerebellar ataxia type 7. Am J Ophthalmol. 2005;139:923–925. doi: 10.1016/j.ajo.2004.10.055. [DOI] [PubMed] [Google Scholar]
- Batra R, Charizanis K, Swanson MS. Partners in crime: bidirectional transcription in unstable microsatellite disease. Hum Mol Genet. 2010;19:R77–82. doi: 10.1093/hmg/ddq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HK, Fernandez-Funez P, Acevedo SF, Lam YC, Kaytor MD, Fernandez MH, Aitken A, Skoulakis EM, Orr HT, Botas J, et al. Interaction of Akt-phosphorylated ataxin-1 with 14-3-3 mediates neurodegeneration in spinocerebellar ataxia type 1. Cell. 2003;113:457–468. doi: 10.1016/s0092-8674(03)00349-0. [DOI] [PubMed] [Google Scholar]
- Chen S, Peng GH, Wang X, Smith AC, Grote SK, Sopher BL, La Spada AR. Interference of Crx-dependent transcription by ataxin-7 involves interaction between the glutamine regions and requires the ataxin-7 carboxy-terminal region for nuclear localization. Hum Mol Genet. 2004;13:53–67. doi: 10.1093/hmg/ddh005. [DOI] [PubMed] [Google Scholar]
- Cho DH, Thienes CP, Mahoney SE, Analau E, Filippova GN, Tapscott SJ. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol Cell. 2005;20:483–489. doi: 10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- David G, Abbas N, Stevanin G, Durr A, Yvert G, Cancel G, Weber C, Imbert G, Saudou F, Antoniou E, et al. Cloning of the SCA7 gene reveals a highly unstable CAG repeat expansion. Nat Genet. 1997;17:65–70. doi: 10.1038/ng0997-65. [DOI] [PubMed] [Google Scholar]
- Davuluri RV, Grosse I, Zhang MQ. Computational identification of promoters and first exons in the human genome. Nat Genet. 2001;29:412–417. doi: 10.1038/ng780. [DOI] [PubMed] [Google Scholar]
- De Biase I, Chutake YK, Rindler PM, Bidichandani SI. Epigenetic silencing in Friedreich ataxia is associated with depletion of CTCF (CCCTC-binding factor) and antisense transcription. PLoS One. 2009;4:e7914. doi: 10.1371/journal.pone.0007914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova GN, Cheng MK, Moore JM, Truong JP, Hu YJ, Nguyen DK, Tsuchiya KD, Disteche CM. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev Cell. 2005;8:31–42. doi: 10.1016/j.devcel.2004.10.018. [DOI] [PubMed] [Google Scholar]
- Filippova GN, Thienes CP, Penn BH, Cho DH, Hu YJ, Moore JM, Klesert TR, Lobanenkov VV, Tapscott SJ. CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nat Genet. 2001;28:335–343. doi: 10.1038/ng570. [DOI] [PubMed] [Google Scholar]
- Garden GA, Libby RT, Fu YH, Kinoshita Y, Huang J, Possin DE, Smith AC, Martinez RA, Fine GC, Grote SK, et al. Polyglutamine-expanded ataxin-7 promotes non-cell-autonomous purkinje cell degeneration and displays proteolytic cleavage in ataxic transgenic mice. J Neurosci. 2002;22:4897–4905. doi: 10.1523/JNEUROSCI.22-12-04897.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmlinger D, Hardy S, Abou-Sleymane G, Eberlin A, Bowman AB, Gansmuller A, Picaud S, Zoghbi HY, Trottier Y, Tora L, et al. Glutamine-expanded ataxin-7 alters TFTC/STAGA recruitment and chromatin structure leading to photoreceptor dysfunction. PLoS Biol. 2006;4:e67. doi: 10.1371/journal.pbio.0040067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmlinger D, Hardy S, Sasorith S, Klein F, Robert F, Weber C, Miguet L, Potier N, Van-Dorsselaer A, Wurtz JM, et al. Ataxin-7 is a subunit of GCN5 histone acetyltransferase-containing complexes. Hum Mol Genet. 2004;13:1257–1265. doi: 10.1093/hmg/ddh139. [DOI] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Kohler A, Schneider M, Cabal GG, Nehrbass U, Hurt E. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat Cell Biol. 2008;10:707–715. doi: 10.1038/ncb1733. [DOI] [PubMed] [Google Scholar]
- La Spada AR. Trinucleotide repeat instability: Genetic features and molecular mechanisms. Brain Path. 1997;7:943–963. doi: 10.1111/j.1750-3639.1997.tb00895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Spada AR, Fu Y, Sopher BL, Libby RT, Wang X, Li LY, Einum DD, Huang J, Possin DE, Smith AC, et al. Polyglutamine-expanded ataxin-7 antagonizes CRX function and induces cone-rod dystrophy in a mouse model of SCA7. Neuron. 2001;31:913–927. doi: 10.1016/s0896-6273(01)00422-6. [DOI] [PubMed] [Google Scholar]
- La Spada AR, Taylor JP. Polyglutamines placed into context. Neuron. 2003;38:681–684. doi: 10.1016/s0896-6273(03)00328-3. [DOI] [PubMed] [Google Scholar]
- La Spada AR, Taylor JP. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat Rev Genet. 2010;11:247–258. doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd PD, Smith LE, Rabaia NA, Moore JM, Georges SA, Hansen RS, Hagerman RJ, Tassone F, Tapscott SJ, Filippova GN. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum Mol Genet. 2007;16:3174–3187. doi: 10.1093/hmg/ddm293. [DOI] [PubMed] [Google Scholar]
- Libby RT, Hagerman KA, Pineda VV, Lau R, Cho DH, Baccam SL, Axford MM, Cleary JD, Moore JM, Sopher BL, et al. CTCF cis-regulates trinucleotide repeat instability in an epigenetic manner: a novel basis for mutational hot spot determination. PLoS Genet. 2008;4:e1000257. doi: 10.1371/journal.pgen.1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Crespo-Barreto J, Jafar-Nejad P, Bowman AB, Richman R, Hill DE, Orr HT, Zoghbi HY. Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature. 2008;452:713–718. doi: 10.1038/nature06731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- Martin JJ, Van Regemorter N, Krols L, Brucher JM, de Barsy T, Szliwowski H, Evrard P, Ceuterick C, Tassignon MJ, Smet-Dieleman H, et al. On an autosomal dominant form of retinal-cerebellar degeneration: an autopsy study of five patients in one family. Acta Neuropathol. 1994;88:277–286. doi: 10.1007/BF00310370. [DOI] [PubMed] [Google Scholar]
- Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, Chait BT, Roeder RG. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol Cell Biol. 2001;21:6782–6795. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- Osato N, Suzuki Y, Ikeo K, Gojobori T. Transcriptional interferences in cis natural antisense transcripts of humans and mice. Genetics. 2007;176:1299–1306. doi: 10.1534/genetics.106.069484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palhan VB, Chen S, Peng GH, Tjernberg A, Gamper AM, Fan Y, Chait BT, La Spada AR, Roeder RG. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc Natl Acad Sci U S A. 2005;102:8472–8477. doi: 10.1073/pnas.0503505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Paulson HL, Bonini NM, Roth KA. Polyglutamine disease and neuronal cell death. Proc Natl Acad Sci U S A. 2000;97:12957–12958. doi: 10.1073/pnas.210395797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley BE, Orr HT. Polyglutamine neurodegenerative diseases and regulation of transcription: assembling the puzzle. Genes Dev. 2006;20:2183–2192. doi: 10.1101/gad.1436506. [DOI] [PubMed] [Google Scholar]
- Ross CA. Intranuclear neuronal inclusions: a common pathogenic mechanism for glutamine-repeat neurodegenerative diseases? Neuron. 1997;19:1147–1150. doi: 10.1016/s0896-6273(00)80405-5. [DOI] [PubMed] [Google Scholar]
- Sanders SL, Jennings J, Canutescu A, Link AJ, Weil PA. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol Cell Biol. 2002;22:4723–4738. doi: 10.1128/MCB.22.13.4723-4738.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearwin KE, Callen BP, Egan JB. Transcriptional interference--a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. Embo J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevanin G, Durr A, Brice A. Clinical and molecular advances in autosomal dominant cerebellar ataxias: from genotype to phenotype and physiopathology. Eur J Hum Genet. 2000;8:4–18. doi: 10.1038/sj.ejhg.5200403. [DOI] [PubMed] [Google Scholar]
- To KW, Adamian M, Jakobiec FA, Berson EL. Olivopontocerebellar atrophy with retinal degeneration. An electroretinographic and histopathologic investigation. Ophthalmology. 1993;100:15–23. doi: 10.1016/s0161-6420(93)31702-1. [DOI] [PubMed] [Google Scholar]
- Trottier Y, Lutz Y, Stevanin G, Imbert G, Devys D, Cancel G, Saudou F, Weber C, David G, Tora L, et al. Polyglutamine expansion as a pathological epitope in Huntington’s disease and four dominant cerebellar ataxias. Nature. 1995;378:403–406. doi: 10.1038/378403a0. [DOI] [PubMed] [Google Scholar]
- Wang YH, Gellibolian R, Shimizu M, Wells RD, Griffith J. Long CCG triplet repeat blocks exclude nucleosomes: a possible mechanism for the nature of fragile sites in chromosomes. J Mol Biol. 1996;263:511–516. doi: 10.1006/jmbi.1996.0593. [DOI] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- Yoo SY, Pennesi ME, Weeber EJ, Xu B, Atkinson R, Chen S, Armstrong DL, Wu SM, Sweatt JD, Zoghbi HY. SCA7 knockin mice model human SCA7 and reveal gradual accumulation of mutant ataxin-7 in neurons and abnormalities in short-term plasticity. Neuron. 2003;37:383–401. doi: 10.1016/s0896-6273(02)01190-x. [DOI] [PubMed] [Google Scholar]
- Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Genome browser analysis of the ataxin-7 gene and repeat region.
This UCSC Genome Browser view of the ataxin-7 gene reveals the 5 ′ region of the ataxin-7 gene, with the Promoter H3K4me3, Transcription Factor ChIP track, CpG islands track, and some comparative sequence tracks. Both the previously predicted 5 ′ promoter and an alternative promoter at the repeat region contain strong peaks for tracks associated with promoter activity.
Figure S2. Diagram of ataxin-7 gene and the repeat region used in mini-gene constructs.
Across the top of this panel, the UCSC genome browser exon-intron prediction for the first half of the human ataxin-7 gene is shown. The previously described transcription start site at exon 1, adjacent to the predicted promoter (P1), is indicated. The purple rectangle overlies the genomic region used for the ataxin-7 mini-gene transgenic constructs, SCA7-CTCF-I-wt and SCA7-CTCF-I-mut. Below the purple rectangle, an expanded map of this region is shown. The 11-nucleotide substitution mutation introduced to create the ataxin-7 mini-gene construct for SCA7-CTCF-I-mut is given.
Figure S3. SCA7-CTCF-I-mut mice develop cone-rod dystrophy retinal degeneration.
Representative rod-initiated and cone-initiated electroretinogram (ERG) responses for 8 month-old SCA7-CTCF-I-wt and SCA7-CTCF-I-mut (line 1245) mice are shown. Each ERG was performed at a range of light intensities from dimmest (top) to brightest (bottom), and the extent of the response was monitored over time, with scale bars for the intensity of the response (in μV) and the time (in ms) shown respectively for the rod responses and cone responses. Note that rod-initiated responses are modestly reduced in the SCA7-CTCF-I-mut mouse compared to the SCA7-CTCF-I-wt mouse; however, cone-initiated responses are markedly reduced in the SCA7-CTCF-I-mut mouse compared to the SCA7-CTCF-I-wt mouse. Degradation of cone responses ahead of rod responses is consistent with a cone-rod dystrophy form of retinal degeneration in the SCA7-CTCF-I-mut mice.
Figure S4. In situ hybridization of SCAANT1 in SCA7 transgenic mice reveals a broad pattern of diametrically opposed antisense ataxin-7 expression.
A) When we probed sagittal brain sections from three month-old SCA7-CTCF-I-wt mice with the ataxin-7 antisense riboprobe, we noted prominent signal throughout the neuronal regions of the entire brain, including the cerebellum.
B) When we probed sagittal brain sections from three month-old SCA7-CTCF-I-mut mice with the ataxin-7 antisense riboprobe, we noted minimal signal throughout the neuronal regions of the entire brain.
Figure S5. Cohesin binding is reduced in the SCA7-CTCF-I-mut mice.
A) Diagram of the genomic region of the human ataxin-7 gene, where the CTCF binding sites and region of bidirectional transcription are located. Boxes represent exons, while solid lines correspond to introns. The position of the amplicon employed in the ChIP analysis is indicated as a black rectangle.
B) Results of ChIP analysis of SMC1 occupancy at the ataxin-7 CTCF binding site region in the cerebellum of six month-old SCA7 transgenic mice. There was a significant reduction in SMC1 occupancy at this amplicon in SCA7-CTCF-I-mut mice (p < .05, ANOVA). Results represent three independent experiments quantifying SMC1 antibody ChIP normalized to the c-Myc promoter region, and an amplicon 3′ to the CTCF-I binding site was used to diminish detection of SMC1 occupancy at the CTCF-II site. Error bars correspond to s.e.m.
C) Results of ChIP analysis of SMC3 occupancy at the ataxin-7 CTCF binding site region in the cerebellum of six month-old SCA7 transgenic mice. There was a significant reduction in SMC1 occupancy at this amplicon in SCA7-CTCF-I-mut mice (p < .05, ANOVA). Results represent three independent experiments quantifying SMC3 antibody ChIP normalized to the c-Myc promoter region, and an amplicon 3′ to the CTCF-I binding site was used to diminish detection of SMC3 occupancy at the CTCF-II site. Error bars correspond to s.e.m.
Figure S6. SCA7-CTCF-I-mut; SCA7-CTCF-I-wt bigenic mice perform worse than SCA7-CTCF-I-mut mice on the rotarod.
Groups of five to seven mice (littermates, all at 7 months of age) were tested over the course of four days on an accelerating rotarod. Mean latency to fall (in seconds) was obtained for each group and compared statistically by ANOVA with post-hoc Tukey test. On three of the four trial days, the SCA7-CTCF-I-mut; SCA7-CTCF-I-wt bigenic mice performed significantly worse than both the SCA7-CTCF-I-wt mice and the SCA7-CTCF-I-mut mice (p < .05). Error bars correspond to s.e.m.
Here we see two different transgenic mice, a SCA7-CTCF-I-wt mouse (light brown fur) and a SCA7-CTCF-I-mut line 1245 mouse (dark brown fur), at six months of age. The SCA7-CTCF-I-wt mouse of is average size and weight for the C57BL/6J strain background, and exhibits normal exploratory behavior without evidence of gait abnormality or tremor. However, the SCA7-CTCF-I-mut individual is much smaller in size and weight, though it is the same age and on the identical strain background. Furthermore, the SCA7-CTCF-I-mut mouse displays a moderate gait ataxia upon ambulation, and tremulousness while stationary.