Abstract
Enteropathogenic and enterohaemorrhagic Escherichia coli are related pathotypes of bacteria that cause acute watery diarrhoea and haemorrhagic colitis, respectively, and enterohaemorrhagic E. coli can lead to a serious complication known as haemolytic uraemic syndrome. In both bacteria the global regulatory protein Ler controls virulence. The ler gene is found within the locus of enterocyte effacement, or LEE, encoding a type III secretion system necessary for injecting effector proteins into intestinal epithelial cells and causing net secretory diarrhoea. The nucleoid-associated protein H-NS silences, whereas Ler serves as an anti-silencer of, multiple LEE operons. Although Ler has a higher affinity for DNA than does H-NS, the precise molecular mechanism by which Ler increases LEE transcription remains to be determined. In this report we investigate the oligomerization activity of Ler. In solution, Ler forms dimers and soluble aggregates of up to 5000 kDa molecular mass, and appears to oligomerize more readily than the related protein H-NS. An insertional mutation into the Ler linker region diminished oligomerization activity. Despite being proteins of similar mass and having homologous DNA-binding domains, Ler and H-NS complexed to DNA migrated to distinct locations, as determined by an electrophoretic mobility shift assay, implying that the related proteins form different 3D shapes in the presence of DNA. Lastly, we present electron microscopy images of toroidal Ler–DNA structures that are predicted to be involved in stimulating gene expression.
INTRODUCTION
Enteropathogenic and enterohaemorrhagic Escherichia coli, EPEC and EHEC, respectively, are related bacteria that cause enteric disease. EPEC is a leading cause of acute infantile diarrhoea in developing countries, while EHEC infection results in haemorrhagic colitis, a disease found more commonly in developed nations (Clarke et al., 2003; Kaper et al., 2004). Several EHEC serotypes, including O157 : H7 and O111 : H−, have been implicated in disease outbreaks in the UK, Western Europe, the USA and Japan. EPEC infections are a significant cause of morbidity and mortality of children under the age of 2 years living in the developing world, and EHEC, particularly serotype O157 : H7, remains an important public health risk in many nations.
Although their sites of infection are different, EPEC infecting the small intestine and EHEC colonizing the large bowel, the two E. coli pathotypes share genetic relatedness and a similar mode of infection. Both organisms form attaching and effacing (A/E) intestinal lesions, a hallmark of their ability to cause disease (Moon et al., 1983). All of the genes necessary for EPEC to form A/E lesions reside in a pathogenicity island termed the locus of enterocyte effacement, or LEE (Campellone et al., 2004; Elliott et al., 1999; Garmendia et al., 2004). In addition to the LEE in EHEC strain EDL933, serotype O157 : H7, the EspFu protein encoded within the cryptic prophage CP-933U is also required for A/E lesion formation (Campellone et al., 2004; Elliott et al., 1999). The LEE pathogenicity islands of these bacteria encode type III secretion systems (T3SSs): molecular syringes that inject effector molecules into host cells, subverting signalling events and leading to net secretory diarrhoea. EHEC injects at least 39 effector molecules, many of which are encoded within cryptic prophages, into the host cytosol (Garmendia et al., 2005; Tobe et al., 2006), while EPEC is thought to inject fewer effectors. The LEE islands of the prototypical EPEC strain E2348/69, serotype O127 : H6, and EHEC strain EDL933 reside at the selC, tRNA locus, and these genetic elements were acquired by horizontal gene transfer (Schmidt et al., 2001; Sperandio et al., 1998).
Genetic elements acquired by horizontal transfer must be properly regulated in order to provide the bacterium with a selective advantage. For members of the Enterobacteriaceae, foreign DNA can be distinguished by its low G+C or high A+T content compared with the average base content across the genome. The nucleoid-associated protein H-NS binds to the minor groove of high-A+T- content DNA, and a major function of this 15.5 kDa protein is to silence horizontally acquired genetic elements (Navarre et al., 2006). Once an E. coli pathotype is within a host, co-ordinate gene expression must commence, i.e. silencing of virulence-associated genes must be relieved. In E. coli and Salmonella enterica serovar Typhimurium, a number of mechanisms of relieving silencing by H-NS have been described (reviewed by Stoebel et al., 2008). These include altering the secondary structure of DNA in response to changing environmental conditions, binding of regulatory proteins that can disrupt H-NS-mediated silencing and/or directly stimulate transcription by interacting with RNA polymerase, and the formation of heteromeric complexes between H-NS and H-NS paralogues, which lack DNA-binding motifs and disrupt H-NS function.
In EPEC and EHEC, H-NS silences expression of the T3SS, and the Ler protein encoded within the LEE is an important anti-silencer of H-NS (for a review, see Mellies et al., 2007). Ler is a member of the H-NS family of nucleoid-associated proteins, sharing 24 % amino acid identity and 44 % similarity with H-NS of Salmonella (Sperandio et al., 2000). Ler and H-NS share greatest identity over their C-terminal DNA-binding domains, while the N-terminal α-helical domain and the central linker region connecting the two domains are more divergent. These differences are thought, in part, to account for the ability of Ler to increase transcriptional activity. To date, Ler is the only H-NS-like molecule known to increase gene expression directly (Elliott et al., 2000; Mellies et al., 1999).
At the H-NS-silenced LEE5 operon of EPEC, Ler binds over an extended region from positions −221 to −60 in relation to the transcriptional start site (Haack et al., 2003). LEE5 encodes the proteins intimin and Tir, necessary for intimate attachment to the host epithelium and A/E lesion formation, and the Tir chaperone CesT. Genetic and biochemical data indicate that H-NS binds both upstream and downstream of the LEE5 promoter (Haack et al., 2003; our unpublished results). Ler relieves silencing by binding with higher affinity than H-NS (Umanski et al., 2002) to the same region upstream of LEE5, disrupting H-NS-containing nucleoprotein complexes. The LEE2 operon, encoding membrane components of the T3SS, is controlled by a similar mechanism (Bustamante et al., 2001; Sperandio et al., 2000).
Although the genetic role of Ler is now well established, little is known about the molecular mechanisms underlying its function. In this report we demonstrate that Ler forms dimers, and higher-order oligomers in solution. We also show that the interactions of Ler and H-NS with DNA are fundamentally different, and present images of toroidal Ler–DNA complexes reminiscent of the manner in which the Fis nucleoid-associated protein binds to DNA. These experiments provide a clearer understanding of how Ler stimulates transcriptional activity.
METHODS
Protein purification.
The Ler protein was expressed from pVS45 (Table 1), a derivative of the parent plasmid pBADMycHisA (Invitrogen) in the E. coli laboratory strain DH5α [supE44 Δ(argF-lac)U169 (ϕ80dlacΔ(Z)M15) deoR hsdR17 recA1 endA1 gyrA96 thi-1 relA1]. Ler expression was induced by the addition of arabinose, and the protein was purified by nickel chromatography, performed as described by Sperandio et al. (2000). In a parallel approach, the E. coli gene encoding H-NS was amplified by PCR with primers 5′-hns and 3′-hns (Table 2), using genomic DNA from strain MG1655 (F− λ− ilv rfb-50 rph-1). The PCR fragment was directionally cloned into pBADMycHisA by using the NcoI and EcoRI restriction sites, and the plasmid was designated pBAD-hns-6Xhis (Table 1). The plasmid construct necessary for expressing the mutated Ler protein containing the H-NS-derived linker sequence (ELLNSLAAVKS) inserted at position 67 was created by using the following strategy. Oligonucleotide primers K1575 and K1576 (Table 2) were used to amplify the mutated ler gene from pFL08 (Mellies et al., 2008), and the amplicon was ligated into the intermediate vector pCR2.1-TOPO (Invitrogen). The insert was excised by using the restriction enzymes KpnI and EcoRI and directionally ligated into the expression vector pBADMycHisA (Invitrogen) digested with the same enzymes. The plasmid was designated pBADHISFL08 (Table 1). The histidine-tagged proteins were purified by nickel affinity chromatography following the manufacturer's instructions (Invitrogen).
Table 1.
Plasmids used in this study
| Plasmid | Genotype or description | Reference or source |
|---|---|---|
| pVS45 | ler in pBADMycHisA | Sperandio et al. (2000) |
| pBADMycHisA | Arabinose-inducible expression vector | Invitrogen |
| pBAD-hns-6Xhis | hns in pBADMycHisA | This study |
| pBADHISFL08 | H-NS-derived residues ELLNSLAAVKS inserted at Ler position 67 in pBADMycHisA | This study |
| pRS551 | Promoterless lacZ reporter fusion vector | Simons et al. (1987) |
| pKMTIR3 | LEE5–lacZ (−303 to +172) in pRS551 | Haack et al. (2003) |
| pKHpro1 | proU–lacZ (−98 to +404) in pRS551 | Mellies et al. (2008) |
Table 2.
Oligonucleotides used in this study
| Primer | Sequence (5′ to 3′)* |
|---|---|
| 5′-hns | CCATGGGTATGAGCGAAGCACTTAAAATTCT |
| 3′-hns | CCGGAATTCCGTTGCTTGATCAGGAAATCGTC |
| K1575 | CGGGGTACCAATATGGAAACTAATTCACAT |
| K1576 | CCGGAATTCCGAATATTTTTCAGCGGTATTAT |
| proU-230 | TATCACGCAAATAATTTGTGGT |
| proU+193 | CGCCAAGCGATAGCCCAG |
| proUregF | CCGGAATTCCTTTCGCCGAATGCCGAT |
| proUregR | CGCGGATCCAGGACTGGAAGACCATCGCA |
*Restriction sites used in cloning are underlined.
DNA sequencing analysis.
All plasmid constructs were confirmed to be correct by DNA sequencing analysis performed at the Vollum Institute at the Oregon Health & Science University.
Oligomerization assays.
Purified Ler at 0.3 mg ml−1 final protein concentration in PBS (100 mM NaCl, 10 mM KCl, 10 mM Na2HPO4, pH 7.2) was cross-linked by using 0.4 % formaldehyde. Reactions were allowed to proceed for 220 min at room temperature. Samples were taken at 0, 1, 10, 30, 60, 110, 170 and 220 min and quenched by 1 : 4 addition of 1.5 M Tris/HCl, pH 8.8. Samples were then analysed by 15 % SDS-PAGE. Proteins were stained by Coomassie blue and imaged using a Kodak EDAS imaging system.
Size-exclusion chromatography with in-line multi-angle laser light scattering (SEC-MALS).
To investigate oligomerization, protein samples were concentrated to 0.2–1.0 mg ml−1 in chromatography buffer (20 mM NaH2PO4, 100 mM KCl, 1 mM EDTA, 1 mM DTT, pH 7.0) and injected onto a SEC-MALS system with in-line refractive index detector (HELEOS and Opti-Lab instruments, Wyatt Technology, Inc.). All experiments were done on a Superose 6 column (GE Healthcare) with a bed volume of 24 ml at a flow rate of 0.4 ml min−1. Molecular masses were calculated from the intensity of scattered light at up to 18 different angles according to Rayleigh light-scattering principles using astra software provided by the manufacturer.
Electrophoretic mobility shift assay (EMSA).
The LEE5 regulatory fragment (−303 to +172) was amplified with primers TIR1 and TIR3 by PCR using plasmid pKMTIR3 as template (Haack et al., 2003). The proU regulatory fragment (−230 to +193; Fig. 5b) was PCR-amplified with primers proU-230 and proU+193 using pKHpro1 (Table 1) as template (Mellies et al., 2008). A proU fragment with regulatory DNA between positions −398 and +404 amplified using primers proUregF and proUregR was used for determining KD values for Ler and H-NS at proU (Fig. 6). DNA fragments were separated by 1.0 % agarose gel electrophoresis, stained with ethidium bromide, excised and purified using a QIAQuick Gel Extraction kit (Qiagen).
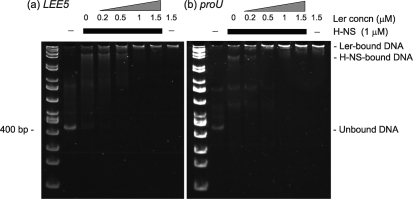
Fig. 5.
Ler can compete H-NS off LEE5 and proU DNA. EMSA was used to assess Ler and H-NS binding to (a) LEE5 (−303 to +172) and (b) proU (−230 to +193) regulatory DNA. DNA (100 ng) was combined with H-NS where indicated. Ler protein was then added at increasing concentrations, and incubation proceeded at room temperature for 15 min. Complexes were separated in a non-denaturing 5 % polyacrylamide gel at 4 °C. H-NS (1 μM) addition is indicated by the thick black bar, and increasing Ler concentrations are indicated by a triangle at a given concentration. Unbound DNA, and H-NS-dependent and Ler-dependent protein–DNA complexes are marked.
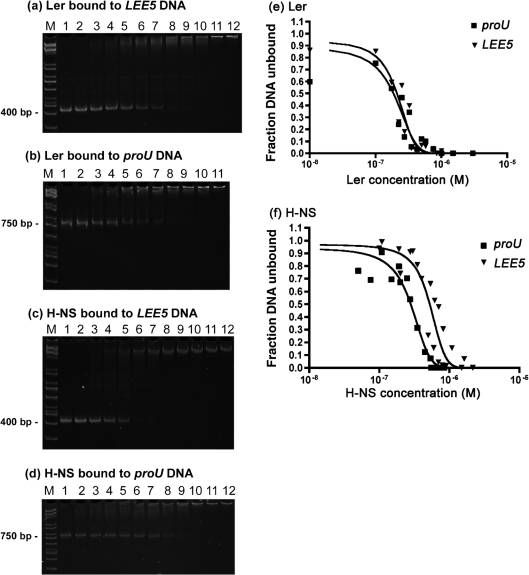
Fig. 6.
Ler binds with the same affinity to LEE5 and proU DNA. Dissociation constants (KD) were determined for Ler binding at LEE5 (a) and proU (b), and H-NS at LEE5 (c) and proU (d). PCR-amplified DNA (100 ng), containing LEE5 (−303 to +172) and the proU (−398 to +404) regulatory DNA, was incubated with Ler at the following protein concentrations (a, b): lane 1, 0 nM; lane 2, 1 nM; lane 3, 10 nM; lane 4, 100 nM; lane 5, 175 nM; lane 6, 225 nM; lane 7, 275 nM; lane 8, 350 nM; lane 9, 425 nM; lane 10, 500 nM; lane 11, 750 nM; lane 12, 850 nM. Similarly, 100 ng PCR-amplified DNA, containing the LEE5 and proU regulatory DNA fragments, was incubated with H-NS at the following protein concentrations (c, d): lane 1, 0 nM; lane 2, 50 nM; lane 3, 75 nM; lane 4, 100 nM; lane 5, 150 nM; lane 6, 200 nM; lane 7, 250 nM; lane 8, 350 nM; lane 9, 450 nM; lane 10, 550 nM; lane 11, 650 nM; lane 12, 750 nM. Complexes were separated electrophoretically in non-denaturing 5 % polyacrylamide gels at 4 °C to detect DNA binding. The ethidium bromide-stained DNA fragments were imaged, and exponential decay curves for Ler (e) and H-NS (f) were fitted to the data using Quantity One software. KD values are defined as 50 % free DNA. Data presented are representative of at least two replicate assays.
EMSA-based competition to assess Ler and H-NS binding to LEE5 and proU regulatory DNA was performed by using non-denaturing 5 % polyacrylamide gels. Polyacrylamide gels were prepared with a 37.5 : 1 acrylamide/bisacrylamide solution (Bio-Rad) following a standard protocol. Binding reaction mixtures containing 100 ng DNA, EMSA buffer (10 mM Tris, pH 7.4, 1 mM EDTA, 5 mM NaCl, 50 mM KCl, 50 μg BSA ml−1), and Ler and H-NS at the indicated concentrations were incubated at room temperature for 15 min. After the addition of glycerol to a concentration of 2.5 % (v/v), samples were separated by electrophoresis at 4 °C overnight at 35 V. Gels were stained with ethidium bromide and imaged using a Bio-Rad Fluor-S MultiImager.
For calculating KD values, the concentration at which half-maximal binding occurs, gels were imaged with the Bio-Rad Fluor-S MultiImager, and bands were quantified with Quantity One software. Apparent dissociation constants (KD) for Ler and H-NS were calculated by a modification of a published method (Carey, 1988; Umanski et al., 2002). The half-maximal binding point of each protein, which should correspond to its KD, was found by plotting the decrease in free DNA against increasing protein concentration.
Transmission electron microscopy (TEM).
For staining samples by rotary shadowing, a 30 μl sample containing 100 ng DNA (plasmid pKHpro1 or pKMTIR3 linearized by restriction digestion with BamHI) and/or Ler (0.5 μM) or H-NS (1 μM) protein in 10 mM Tris, pH 8.0, 5 mM NaCl and 50 mM KCl was incubated at room temperature for 15 min, diluted to 100 μg DNA ml−1, and mixed with 70 μl glycerol and nebulized, by using an airbrush, onto freshly cleaved mica. The sample was dried in a vacuum and rotary-shadowed by using a Pt-C electron beam gun angled at ° relative to the mica surface within a Balzers BAE 250 evaporator. The replica was backed with carbon, floated onto distilled water and picked up onto 600-mesh grids. Digital micrographs were collected by using an AMT 2k×2k camera mounted on an FEI G2 transmission electron microscope operated at 120 kV.
For gold sputtering, 100 ng of the 475 bp purified LEE5 fragment, excised as an EcoRI/BamHI fragment from plasmid pKMTIR3, was combined with 0.5 μM Ler or 1.0 μM H-NS and 0.5 μM Ler in 1× EMSA buffer, as indicated, and incubated at room temperature for 15 min. Samples were diluted 20-fold in 200 mM ammonium acetate, pH 7.4, containing 15 % (v/v) glycerol. Copper 300-mesh electron microscopy grids coated with parlodian were placed on 40 μl drops for 5 min, and washed for 2 min and then for 3 min by placing onto 500 μl drops of 50 mM ammonium acetate containing 15 % (v/v) glycerol. Grids containing samples were then dried for 1 min under a heat lamp. Grids were stained by placing a 3 mm-wide shield 1 mm above the electron microscopy grid (Colquhoun, 1984), and were sputter-coated with gold (Cressington Sputter Coater, model 108 Auto) under vacuum. Samples were observed and photographed under a JEOL 100CX transmission electron microscope at 100 kV.
RESULTS
The Ler predicted coiled-coil motif is shorter than that of H-NS
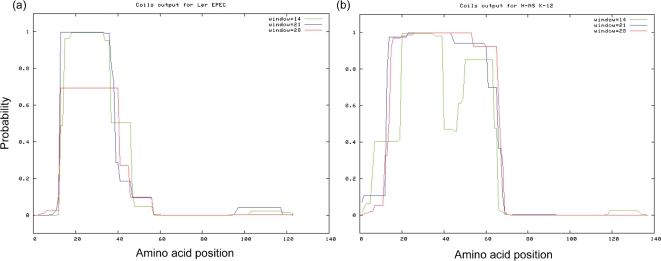
Ler and H-NS amino acid sequences vary considerably over their N-terminal α-helical domains. Previous analyses have demonstrated that Ler is predicted to have two α-helices in the N-terminal region, whereas H-NS possesses three α-helices between amino acid positions 1 and 70 (Arold et al., 2010; Bloch et al., 2003; Esposito et al., 2002; Mellies et al., 2008). The amino acid substitution I26R within the predicted coiled-coil domain of Ler eliminates Ler binding to regulatory DNA and the ability to increase transcriptional activity from a LEE2–lacZ fusion in vivo (Sperandio et al., 2000). Similarly, substitution of the leucine with a proline, L33P, in the coiled-coil motif eliminates DNA binding to curved DNA and dimer formation of H-NS (Ueguchi et al., 1997). Due to the differences in α-helical content of the N-terminal regions of the two proteins, we determined whether there were also differences in the predicted coiled-coil motifs. We therefore used the coils program (http://www.ch.embnet.org/software/COILS_form.html), and found that the Ler N-terminal coiled-coil domain was shorter than the coiled-coil of H-NS (Fig. 1). By this analysis, Ler contains a coiled-coil that extends from amino acids 10 to 40, while the predicted coiled-coil of H-NS extends from amino acids 10 to 64.
Fig. 1.
Ler protein is predicted to have a shorter N-terminal coiled-coil motif than H-NS. The amino acid sequences of Ler and H-NS were subjected to analysis by the coils program for predicting coiled-coils. The y axes indicate the probability of forming a left-handed coiled-coil, and the x axes are the amino acid positions in the proteins. Coiled-coil predictions for scanning windows of 14, 21 and 28 residues are presented. (a) Ler was predicted to have a coiled-coil extending from amino acid position 10 to position 40, whereas (b) the predicted H-NS coiled-coil extended from position 10 to position 64.
Ler forms dimers and higher-order oligomers in solution
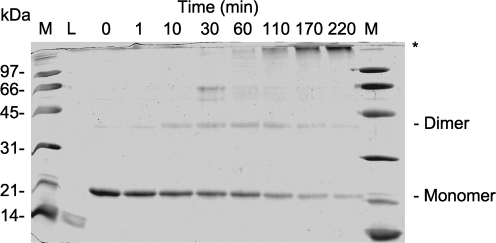
Full-length H-NS forms dimers in solution, as does a fragment containing the coiled-coil domain (Bloch et al., 2003; Esposito et al., 2002). Because the predicted coiled-coil domain of Ler is shorter than that of H-NS, we wanted to demonstrate clearly that Ler also forms dimers in solution. Ler was purified by using nickel affinity chromatography, subjected to cross-linking with formaldehyde, and species were separated by SDS-PAGE (Fig. 2). We observed a Ler monomer migrating to ∼22 kDa. In the presence of formaldehyde, a species migrating to ∼44 kDa appeared, corresponding to the Ler dimer. As the time of incubation increased beyond 120 min, a high-molecular-mass species of >100 kDa that was formaldehyde-dependent appeared, while the quantity of the Ler dimer diminished (Fig. 2). Thus, even though Ler contains a predicted coiled-coil shorter than that of H-NS, Ler forms dimers in solution, as well as higher-order oligomers.
Fig. 2.
Ler forms dimers and higher-order multimers in solution. Purified Ler protein (0.3 mg ml−1) was cross-linked by using 0.4 % formaldehyde in PBS, pH 7.2. Samples were taken at the indicated times, separated by SDS-PAGE and stained with Coomassie blue. Molecular mass standards (lane M) are indicated, and lysozyme, a 14 kDa monomer, was run as a control (lane L). Ler monomers and formaldehyde-dependent dimers are indicated. Formaldehyde-dependent, higher-order oligomers are marked with an asterisk.
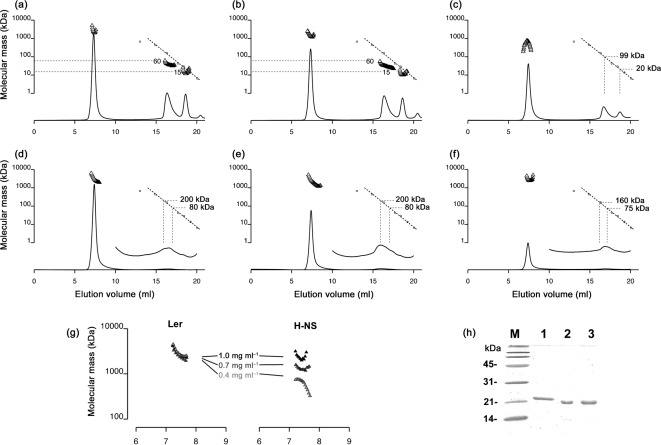
Extending the analysis, we compared oligomerization in solution of Ler with that of H-NS using SEC-MALS. Prior to injection, the purity of the samples was verified by SDS-PAGE (Fig. 3h). For H-NS, we observed an elution peak corresponding to the monomer (∼15–20 kDa) and a broader peak of mean molecular mass 60–100 kDa, probably an unresolved mixture of small multimers (Fig. 3a–c). In addition, about half the injected protein eluted in the void volume of the column as an aggregate with a molecular mass of 1000–2000 kDa (open triangles in Fig. 3a–c), measured by light scattering and corresponding to 50–100 copies of H-NS. The mean mass of the H-NS oligomers increased as a function of the concentration over the range tested, 0.4, 0.7 and 1.0 mg ml−1 (Fig. 3g). We noted that, despite its tendency to form large oligomers, H-NS remained soluble with no signs of precipitation at concentrations several times the highest used in the SEC-MALS experiments.
Fig. 3.
Oligomerization of Ler compared with that of H-NS. One hundred microlitres of either H-NS (a–c) or Ler (d–f) at a concentration of 1.0 mg ml−1 (a, d), 0.7 mg ml−1 (b, e) or 0.4 mg ml−1 (c, f) was injected on a Superose 6 size-exclusion column. As the protein eluted, concentration versus elution volume was measured by in-line UV absorbance detection, and mean molecular mass was measured simultaneously by in-line MALS. UV chromatograms are shown as solid lines. For Ler (d–f), a 10-fold vertical expansion of the chromatogram is also shown over the region 10–20 ml. Calibration standards (molecular mass versus elution volume) for the size-exclusion column employed are shown as open circles along with a calibration standard curve (dotted line). Where eluting protein peaks are strong enough to allow determination of molecular mass by light scattering, the calculated mean molecular mass versus elution time is shown above the chromatogram (triangles). Molecular mass estimates of the lower-order Ler multimers were determined by using the standard curve for the UV chromatogram, not from MALS (d–f). The y axis applies to both the molecular mass standards (dotted lines) and the molecular mass calculated from MALS data (triangles). (g) The mass of the H-NS oligomers eluting between 7 and 8 ml was found to be concentration-dependent, whereas Ler oligomers formed in a concentration-independent manner over the range tested, 0.4, 0.7 and 1.0 mg ml−1. (h) Purified protein used in SEC-MALS experiments. SDS-PAGE and Coomassie blue staining of purified Ler-FL08 (lane 1), H-NS (lane 2) and wild-type Ler (lane 3). Molecular mass standards are indicated.
The behaviour of Ler was distinctly different from that of H-NS, with a greater tendency to form higher-order oligomers (Fig. 3d–f). Less than 10 % of the injected Ler eluted as monomers, dimers or other low-copy-number multimers. We observed no sharp peak corresponding to these species, but rather a weak, broad peak eluting between 15 and 18 ml. Although the light scattering from this broad peak was not strong enough to allow molecular mass determination by multi-angle laser light scattering, the elution time, or UV chromatogram, matched that expected for solutes of 75–200 kDa, corresponding to Ler oligomers of 2–10 units. The majority of the protein – over 90 % of the injected sample – eluted in the void volume as a soluble aggregate with a mean molecular mass of 2000–5000 kDa (open triangles in Fig. 3d–f), corresponding to 100–250 monomers. The mean molecular mass differences between Ler and H-NS were particularly apparent for the protein concentration 0.4 mg ml−1 (Fig. 3g). Unlike H-NS, the mean mass of the Ler oligomers did not increase as a function of concentration over the range tested (Fig. 3g). However, like H-NS, Ler remained soluble, with no sign of precipitation at concentrations several times higher than the highest used in the SEC-MALS experiments.
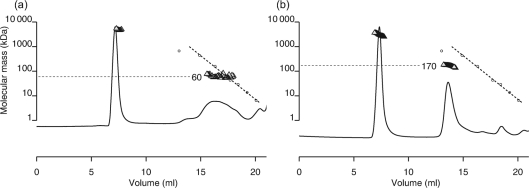
Because the flexible linker region connecting the N-terminal and C-terminal domains of Ler is important for proper function and has been implicated in oligomerization activity (Mellies et al., 2008; J. Levine and J. Kaper, personal communication), we compared the oligomerization of wild-type Ler with that of a mutated protein, termed Ler-FL08, in which an 11 aa sequence taken from the H-NS linker was inserted into the linker region of Ler (Fig. 4). In a SEC-MALS experiment with an injected concentration of 0.2 mg ml−1, most of the Ler-FL08 protein eluted as a soluble aggregate with a mean molecular mass matching that observed with wild-type Ler. However, a portion of the sample also eluted as a broad peak with a molecular mass of 30–70 kDa, corresponding to low-copy-number multimers (Fig. 4a). Under the same conditions, wild-type Ler eluted mostly as a soluble aggregate of 2000–5000 kDa and a smaller peak of ∼170 kDa. Thus, the Ler protein containing the 11 aa, H-NS-derived linker sequence was impaired in its ability to form higher-order multimers compared with wild-type Ler.
Fig. 4.
Ler protein containing an 11 aa insertion of the H-NS linker region shows diminished oligomerization activity. Oligomerization of mutated Ler protein containing the H-NS-derived linker amino acid sequence ELLNSLAAVKS, Ler-FL08 (a), is compared with that of wild-type Ler (b). MALS experiments were performed with 250 μl injections of 0.2 mg protein ml−1, as described in Fig. 3. The y axis applies to both the molecular mass standards (dotted lines) and the molecular mass calculated from MALS data (triangles).
Ler and H-NS form distinct species when bound to regulatory DNA
Due to the different oligomerization activity of Ler compared with H-NS, we hypothesized that Ler would form protein–DNA complexes that would migrate differently from H-NS–DNA complexes by EMSA. To test this hypothesis, we incubated purified Ler and H-NS proteins with LEE5 and proU regulatory DNA fragments and separated the species by non-denaturing PAGE. The proU regulatory fragment was used as a control because in vivo this operon is regulated by H-NS, but not by Ler (Elliott et al., 2000; Mellies et al., 2008). We also determined whether Ler could compete H-NS off the LEE5 and proU fragments by binding of H-NS followed by the addition of increasing concentrations of Ler.
Pre-bound H-NS at 1 μM impeded migration of both the LEE5 and proU DNA by EMSA, indicating that H-NS was able to bind to both of these regulatory fragments (Fig. 5). By adding increasing concentrations of Ler protein to pre-formed H-NS-bound DNA we observed Ler-dependent DNA complexes for both the LEE5 and proU fragments that were distinct from the H-NS–DNA complexes. The Ler-bound complexes were clearly visible upon addition of 1 μM Ler, the concentration of pre-bound H-NS (Fig. 5), consistent with the higher affinity of Ler, or lower KD for DNA than those of H-NS (Umanski et al., 2002). The LEE5 and proU DNA species pre-bound to 1 μM H-NS with 1.0 and 1.5 μM added Ler co-migrated with those complexes formed in the presence of Ler alone (Fig. 5a, b). By EMSA, we concluded that Ler competed pre-bound H-NS off LEE5 and proU when the proteins were at identical concentrations, and that Ler and H-NS formed distinct protein–DNA complexes.
We then determined the KD of Ler and H-NS binding at the LEE5 and proU operons. Increasing concentrations of purified Ler and H-NS proteins were bound to the regulatory DNA fragments, separated by non-denaturing PAGE and stained with ethidium bromide (Fig. 6a–d), and bands were quantified using GraphPad Prism software. For Ler, the KD values were 211 and 192 nM for binding to the LEE5 and proU fragments, respectively (Fig. 6e). For H-NS, the KD values were 525 and 281 nM for binding to the LEE5 and proU fragments, respectively (Fig. 6f). Thus, our data demonstrated that Ler bound with similar affinity to LEE5 and proU, with a KD of ∼0.2 μM, and that H-NS bound with approximately twofold higher KD to LEE5 than to proU. Because Ler increases LEE5 transcription ∼20-fold in vivo, while not affecting proU expression (Haack et al., 2003), we concluded that simple binding affinities are unlikely to explain the ability of Ler to differentially regulate LEE virulence genes.
Ler forms toroidal complexes when bound to DNA
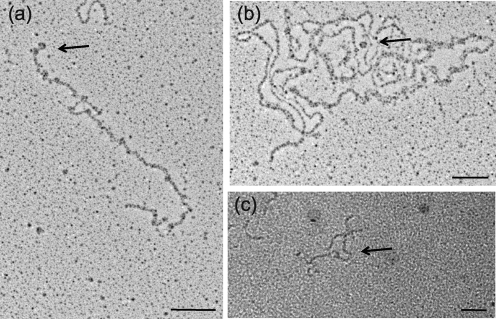
To investigate how Ler interacts with DNA, and to determine whether the interaction is different from that of H-NS–DNA complexes, we employed TEM, staining samples with platinum by rotary shadowing. Because Ler binds to DNA elements in vitro that it does not regulate in vivo, namely proU (Fig. 6a, b; Elliott et al., 2000; Mellies et al., 2008), while binding to LEE5 with the same affinity, we performed these assays using purified Ler and H-NS proteins, and a linearized pKHpro1 plasmid containing the 800 bp proU regulatory fragment in the reporter gene vector pRS551 (Table 1). Linearized pKHpro1 can be seen in Fig. 7(a). The protein-only controls can be seen in Fig. 7(b, d). Ler formed protein–DNA complexes that appeared toroidal, at somewhat regular intervals along the dsDNA strand (Fig. 7c). The toroidal structures were ∼25 nm in diameter, and appeared over a large extent of the 12.9 kb linearized plasmid, over which Ler exerts no regulatory control (Mellies et al., 2008). In contrast, H-NS formed protein–DNA complexes that were distinct from those formed by Ler (Fig. 7e). We observed projections extending from the DNA that were interpreted as bridges or loops, because it is known that H-NS is able to bridge DNA strands (Dame et al., 2005).
Fig. 7.
Ler–DNA complexes appear as toroidal structures ∼25 nm in diameter. Purified Ler and H-NS proteins, at 0.5 and 1 μM, respectively, were incubated with 0.1 μg pKHpro1 plasmid DNA linearized by restriction enzyme digestion with BamHI. BamHI was inactivated at 80 °C for 15 min prior to incubation. Plasmid pKHpro1 contains the proU regulatory region from positions −398 to +404 in the lac reporter gene vector pRS551 (Simons et al., 1987). After 15 min incubation at room temperature, samples were subjected to rotary shadowing and imaged by TEM as described in Methods. Samples are DNA only (a), Ler only (b), Ler plus DNA (c), H-NS only (d) and H-NS plus DNA (e). Bars, 100 nm.
We also demonstrated that Ler forms similar structures at the LEE5 operon using TEM with rotary shadowing and a gold-sputtering procedure (Fig. 8). Purified Ler protein was bound to the 475 bp DNA fragment extending from positions −303 to +172 in relation to the LEE5 transcriptional start site in plasmid pKMTIR3 (Haack et al., 2003) linearized with BamHI (Fig. 8a). We observed toroidal DNA structures consistent with images observed in Fig. 7(c), both in the presence of Ler alone (Fig. 8a) and in the presence of Ler and H-NS (Fig. 8b). The toroidal structure seen in Fig. 8(b) is consistent with the higher binding affinity of Ler compared with H-NS, and the ability of Ler to compete H-NS off regulatory DNA (Fig. 5; Torres et al., 2007). Lastly, we used a gold-sputtering procedure to image Ler bound to the 475 bp LEE5 DNA fragment extending from positions −303 to +172 (Haack et al., 2003; Fig. 8c). We observed toroidal DNA structures ∼25 nm in diameter, consistent with the structures observed in Figs 7(c) and 8(a, b). We did not observe the circular structures with the 475 bp LEE5 DNA fragment alone, in the absence of Ler protein (data not shown).
Fig. 8.
Ler bound to LEE5 DNA also forms circular structures. (a, b) Plasmid pKMTIR3 containing the LEE5 regulatory fragment (0.1 μg) with DNA from positions −303 to +172 was linearized by digestion with BamHI and then bound to purified Ler protein (0.5 μM) (a) or H-NS (1.0 μM) and Ler (0.5 μM) (b). After 15 min incubation at room temperature, samples were subjected to rotary shadowing and imaging by TEM as described in Methods. Bars, 100 nm. (c) The 475 bp LEE5 regulatory fragment (0.1 μg) containing DNA from positions −303 to +172 in relation to the transcriptional start site was bound to purified Ler protein at 0.5 μM, subjected to gold sputtering and viewed by TEM. Protein was not visible by gold sputtering. Arrows point to toroidal protein–DNA complexes. Bar, 50 nm.
DISCUSSION
The Ler protein, an important regulator of virulence in both EPEC and EHEC, forms dimers and higher-order oligomers in solution. Over the concentration range 0.2–1.0 mg ml−1, the primary species is a soluble aggregate with a molecular mass of 2000–5000 kDa, corresponding to 100–250 copies of Ler. This behaviour differs from that of H-NS, which at the same concentrations oligomerizes to some extent, but also exists in large part as monomers, dimers and other low-copy-number multimers. Over the range tested, we observed the mean mass of the H-NS oligomer to be concentration-dependent, whereas the mean mass of the Ler oligomers was not dependent upon concentration under the same conditions (Fig. 3g). These data were consistent with the observed concentration dependence of self-association of the truncated N-terminal H-NS1–83 protein (Arold et al., 2010; Leonard et al., 2009). Oligomerization of Ler was impaired in a chimeric protein containing an 11 aa portion of the H-NS linker inserted into the central region of the Ler molecule (Fig. 4), consistent with loss of in vivo activity and severely impaired DNA-binding ability of the Ler-FL08 protein in vitro (Mellies et al., 2008).
The Ler and H-NS amino acid sequences share 24 % identity overall, but are less similar in their N termini. The H-NS N-terminal domain contains three α-helices between amino acid positions 1 and 70 and dimerizes as a coiled-coil, as determined by NMR structural analysis (Arold et al., 2010; Bloch et al., 2003; Esposito et al., 2002). The Ler N-terminal domain contains only two predicted α-helices (Mellies et al., 2008) and a shorter coiled-coil motif than that of H-NS (Fig. 1). Along with the possession of inter-domain linkers of different lengths (Mellies et al., 2008), it is natural to speculate that these structural differences in the N termini underlie the observed differences in oligomerization and function. Interestingly, placing α-helices 1 and 2 of H-NS onto the N terminus of Ler had no effect on the ability of Ler to stimulate LEE transcription (Mellies et al., 2008). Although Ler, like H-NS, forms dimers and oligomers in solution, it is currently unknown whether Ler forms a coiled-coil structure similar to that of H-NS. More recent structural information, in addition to previously published reports, strongly suggests that H-NS forms anti-parallel dimers in solution (Arold et al., 2010; Bloch et al., 2003), and our preliminary structural data suggest that Ler also forms dimers in the same orientation, mediated by the N-terminal coiled-coil motif (data not shown).
As predicted from differing oligomerization activities, Ler and H-NS complexed to regulatory DNA migrated to distinct locations, as revealed by EMSA (Fig. 5). These data are consistent with results observed for Ler and H-NS competition assays at the lpf1 operon of EHEC (Torres et al., 2007). Ler and H-NS complexed to DNA also migrate to distinct locations, as shown by EMSA, with the Ler/lpf1 complex migrating more slowly through the gel than the H-NS/lpf1 complex (Torres et al., 2007). Both Ler and H-NS form protein–DNA complexes that migrate poorly into a non-denaturing polyacrylamide gel (Fig. 5; Haack et al., 2003; Torres et al., 2007). Under certain binding conditions, H-NS forms rigid filaments in the presence of DNA (R. Dame, personal communication), and we have observed similar structures by TEM, as well as more fibrous, higher-order protein–DNA complexes in the presence of Ler (G. Coriz and J. Mellies, unpublished data). These observations perhaps explain why the complexes are drastically shifted away from the unbound DNA (Fig. 5). Experiments performed using tagged and non-tagged Ler proteins have definitively demonstrated that Ler and H-NS do not form heterodimers in vivo (Yerushalmi et al., 2008), but we cannot rule out the possibility that Ler and H-NS dimers or higher-order oligomers bind simultaneously to regulatory DNA fragments.
Our most significant finding is that Ler forms toroidal structures in the presence of DNA (Figs 7c and 8). The image of the H-NS–DNA complexes (Fig. 7e) is consistent not only with H-NS being able to bridge DNA (Dame et al., 2005), but also with the observed helical scaffold formed by H-NS1–83 that is predicted to compact DNA by forming plectonemic nucleoprotein complexes (Arold et al., 2010). As predicted from Ler binding only in the upstream location at LEE5 (Haack et al., 2003), as opposed to both upstream and downstream of the promoter typical for H-NS, to date we have found no evidence that Ler is able to bridge dsDNA segments.
Although the stoichiometry of the Ler–DNA structures, i.e. the number of Ler monomers within each of the toroids, is unknown, as mentioned above we have observed that Ler in the absence of DNA forms soluble aggregates of molecular mass 2000–5000 kDa, corresponding to 100–250 copies of Ler. Assuming a Matthews coefficient of 2.0 for a well-packed protein complex with moderate solvent content, a species of this mass packed as a solid sphere would have an expected diameter of 20–27 nm. Therefore, it is plausible that the soluble aggregates observed in SEC-MALS experiments are equivalent to the toroids of ∼25 nm diameter observed by TEM.
Extending our analysis, we wanted to determine whether the mass of the Ler oligomer in solution and the diameter of the toroids were congruous with Ler binding at the LEE5 operon defined by DNase I protection assay (Haack et al., 2003). The observed diameter of the Ler–DNA toroids is consistent with the region of LEE5 to which Ler binds, previously determined to be from positions −221 to −60, in an uninterrupted or continuous footprint of ∼160 bp. The length of the DNA segment bound by Ler is 160 bp×0.3 nm/bp=∼48 nm. If the toroids are between 20 and 27 nm in diameter then their circumference (C=πr) would be between 63 and 85 nm. Coating of the sample with platinum for imaging enhances the size of the object so it is not unreasonable to suggest that the extent of the established footprint, wrapping of the DNA around a single Ler oligomer, and the diameter of the toroid are in agreement. We currently have no evidence to suggest that these large aggregates, or observed Ler–DNA complexes, form in vivo or that they are specifically necessary for Ler function, although the existence of such structures clearly demonstrates the oligomerization activity of Ler, and we predict them to be central to the ability of Ler to stimulate transcription.
To date, LEE5 is the only operon for which Ler can increase transcriptional activity in the absence of H-NS (Haack et al., 2003; Sánchez-SanMartín et al., 2001; Umanski et al., 2002). We suggest this is in part due to the location of Ler binding, between positions −221 and −60, and thus it is probable that this regulatory protein interacts with RNA polymerase, which binds to σ70-dependent promoter regions extending from positions ∼−55 to +20 in relation to the start of transcription (Craig et al., 1995). Perhaps consistent with this prediction, we have been unable to identify an additional negative-acting factor for LEE5 by using genetic approaches (data not shown). Furthermore, the toroidal structures formed by Ler in the presence of DNA are reminiscent of those formed by the nucleoid-associated protein Fis (Maurer et al., 2006, 2009). Fis is able to stimulate transcriptional activity either by altering DNA topology adjacent to promoter regions or by directly interacting with RNA polymerase (Aiyar et al., 2002; McLeod et al., 2002). Future experimentation will address whether Ler acts to stimulate LEE5 transcription by enhancing RNA polymerase binding, open complex formation, or both, and whether Ler interacts directly with RNA polymerase.
Acknowledgments
This work was supported by National Institutes of Health (NIH) AREA grant 2 R15 AI047802-03A1 to J. L. M. Funding for training for J. L. M. on the JEOL 100CX transmission electron microscope at Portland State University was provided by the National Science Foundation, as a Research Opportunity Award (ROA) Supplement to investigate protein–DNA complexes associated with DNA damage repair with Dr Justin Courcelle (summer 2010). Funding for light-scattering equipment in the laboratory of Dr Kirsten Lampi at Oregon Health & Sciences University was provided by NIH grant EY012239. We gratefully acknowledge Doug Keene for processing samples for rotary shadowing and imaging by TEM at Oregon Health & Science University.
Abbreviations
A/E, attaching and effacing
EHEC, enterohaemorrhagic Escherichia coli
EMSA, electrophoretic mobility shift assay
EPEC, enteropathogenic Escherichia coli
LEE, locus of enterocyte effacement
SEC-MALS, size-exclusion chromatography with in-line multi-angle laser light scattering
T3SS, type III secretion system
TEM, transmission electron microscopy
References
- Aiyar, S. E., McLeod, S. M., Ross, W., Hirvonen, C. A., Thomas, M. S., Johnson, R. C. & Gourse, R. L. (2002). Architecture of Fis-activated transcription complexes at the Escherichia coli rrnB P1 and rrnE P1 promoters. J Mol Biol 316, 501–516. [DOI] [PubMed] [Google Scholar]
- Arold, S. T., Leonard, P. G., Parkinson, G. N. & Ladbury, J. E. (2010). H-NS forms a superhelical protein scaffold for DNA condensation. Proc Natl Acad Sci U S A 107, 15728–15732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch, V., Yang, Y., Margeat, E., Chavanieu, A., Augé, M. T., Robert, B., Arold, S., Rimsky, S. & Kochoyan, M. (2003). The H-NS dimerization domain defines a new fold contributing to DNA recognition. Nat Struct Biol 10, 212–218. [DOI] [PubMed] [Google Scholar]
- Bustamante, V. H., Santana, F. J., Calva, E. & Puente, J. L. (2001). Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol Microbiol 39, 664–678. [DOI] [PubMed] [Google Scholar]
- Campellone, K. G., Robbins, D. & Leong, J. M. (2004). EspFU is a translocated EHEC effector that interacts with Tir and N-WASP and promotes Nck-independent actin assembly. Dev Cell 7, 217–228. [DOI] [PubMed] [Google Scholar]
- Carey, J. (1988). Gel retardation at low pH resolves trp repressor–DNA complexes for quantitative study. Proc Natl Acad Sci U S A 85, 975–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, S. C., Haigh, R. D., Freestone, P. P. & Williams, P. H. (2003). Virulence of enteropathogenic Escherichia coli, a global pathogen. Clin Microbiol Rev 16, 365–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquhoun, W. R. (1984). Sputter shadowing. J Ultrastruct Res 87, 97–105. [DOI] [PubMed] [Google Scholar]
- Craig, M. L., Suh, W. C. & Record, M. T. J., Jr (1995). HO. and DNase I probing of E sigma 70 RNA polymerase–lambda PR promoter open complexes: Mg2+ binding and its structural consequences at the transcription start site. Biochemistry 34, 15624–15632. [DOI] [PubMed] [Google Scholar]
- Dame, R. T., Luijsterburg, M. S., Krin, E., Bertin, P. N., Wagner, R. & Wuite, G. J. (2005). DNA bridging: a property shared among H-NS-like proteins. J Bacteriol 187, 1845–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott, S. J., Yu, J. & Kaper, J. B. (1999). The cloned locus of enterocyte effacement from enterohemorrhagic Escherichia coli O157 : H7 is unable to confer the attaching and effacing phenotype upon E. coli K-12. Infect Immun 67, 4260–4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott, S. J., Sperandio, V., Girón, J. A., Shin, S., Mellies, J. L., Wainwright, L., Hutcheson, S. W., McDaniel, T. K. & Kaper, J. B. (2000). The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun 68, 6115–6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito, D., Petrovic, A., Harris, R., Ono, S., Eccleston, J. F., Mbabaali, A., Haq, I., Higgins, C. F., Hinton, J. C. & Driscoll, P. C. (2002). H-NS oligomerization domain structure reveals the mechanism for high order self-association of the intact protein. J Mol Biol 324, 841–850. [DOI] [PubMed] [Google Scholar]
- Garmendia, J., Phillips, A. D., Carlier, M. F., Chong, Y., Schüller, S., Marches, O., Dahan, S., Oswald, E., Shaw, R. K. & other authors (2004). TccP is an enterohaemorrhagic Escherichia coli O157 : H7 type III effector protein that couples Tir to the actin-cytoskeleton. Cell Microbiol 6, 1167–1183. [DOI] [PubMed] [Google Scholar]
- Garmendia, J., Frankel, G. & Crepin, V. F. (2005). Enteropathogenic and enterohemorrhagic Escherichia coli infections: translocation, translocation, translocation. Infect Immun 73, 2573–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haack, K. R., Robinson, C. L., Miller, K. J., Fowlkes, J. W. & Mellies, J. L. (2003). Interaction of Ler at the LEE5 (tir) operon of enteropathogenic Escherichia coli. Infect Immun 71, 384–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaper, J. B., Nataro, J. P. & Mobley, H. L. (2004). Pathogenic Escherichia coli. Nat Rev Microbiol 2, 123–140. [DOI] [PubMed] [Google Scholar]
- Leonard, P. G., Ono, S., Gor, J., Perkins, S. J. & Ladbury, J. E. (2009). Investigation of the self-association and hetero-association interactions of H-NS and StpA from Enterobacteria. Mol Microbiol 73, 165–179. [DOI] [PubMed] [Google Scholar]
- Maurer, S., Fritz, J., Muskhelishvili, G. & Travers, A. (2006). RNA polymerase and an activator form discrete subcomplexes in a transcription initiation complex. EMBO J 25, 3784–3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer, S., Fritz, J. & Muskhelishvili, G. (2009). A systematic in vitro study of nucleoprotein complexes formed by bacterial nucleoid-associated proteins revealing novel types of DNA organization. J Mol Biol 387, 1261–1276. [DOI] [PubMed] [Google Scholar]
- McLeod, S. M., Aiyar, S. E., Gourse, R. L. & Johnson, R. C. (2002). The C-terminal domains of the RNA polymerase alpha subunits: contact site with Fis and localization during co-activation with CRP at the Escherichia coli proP P2 promoter. J Mol Biol 316, 517–529. [DOI] [PubMed] [Google Scholar]
- Mellies, J. L., Elliott, S. J., Sperandio, V., Donnenberg, M. S. & Kaper, J. B. (1999). The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol Microbiol 33, 296–306. [DOI] [PubMed] [Google Scholar]
- Mellies, J. L., Barron, A. M. & Carmona, A. M. (2007). Enteropathogenic and enterohemorrhagic Escherichia coli virulence gene regulation. Infect Immun 75, 4199–4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellies, J. L., Larabee, F. J., Zarr, M. A., Horback, K. L., Lorenzen, E. & Mavor, D. (2008). Ler interdomain linker is essential for anti-silencing activity in enteropathogenic Escherichia coli. Microbiology 154, 3624–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, H. W., Whipp, S. C., Argenzio, R. A., Levine, M. M. & Giannella, R. A. (1983). Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect Immun 41, 1340–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre, W. W., Porwollik, S., Wang, Y., McClelland, M., Rosen, H., Libby, S. J. & Fang, F. C. (2006). Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313, 236–238. [DOI] [PubMed] [Google Scholar]
- Sánchez-SanMartín, C., Bustamante, V. H., Calva, E. & Puente, J. L. (2001). Transcriptional regulation of the orf19 gene and the tir-cesT-eae operon of enteropathogenic Escherichia coli. J Bacteriol 183, 2823–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, H., Zhang, W. L., Hemmrich, U., Jelacic, S., Brunder, W., Tarr, P. I., Dobrindt, U., Hacker, J. & Karch, H. (2001). Identification and characterization of a novel genomic island integrated at selC in locus of enterocyte effacement-negative, Shiga toxin-producing Escherichia coli. Infect Immun 69, 6863–6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, R. W., Houman, F. & Kleckner, N. (1987). Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53, 85–96. [DOI] [PubMed] [Google Scholar]
- Sperandio, V., Kaper, J. B., Bortolini, M. R., Neves, B. C., Keller, R. & Trabulsi, L. R. (1998). Characterization of the locus of enterocyte effacement (LEE) in different enteropathogenic Escherichia coli (EPEC) and Shiga-toxin producing Escherichia coli (STEC) serotypes. FEMS Microbiol Lett 164, 133–139. [DOI] [PubMed] [Google Scholar]
- Sperandio, V., Mellies, J. L., Delahay, R. M., Frankel, G., Crawford, J. A., Nguyen, W. & Kaper, J. B. (2000). Activation of enteropathogenic Escherichia coli (EPEC) LEE2 and LEE3 operons by Ler. Mol Microbiol 38, 781–793. [DOI] [PubMed] [Google Scholar]
- Stoebel, D. M., Free, A. & Dorman, C. J. (2008). Anti-silencing: overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology 154, 2533–2545. [DOI] [PubMed] [Google Scholar]
- Tobe, T., Beatson, S. A., Taniguchi, H., Abe, H., Bailey, C. M., Fivian, A., Younis, R., Matthews, S., Marches, O. & other authors (2006). An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc Natl Acad Sci U S A 103, 14941–14946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, A. G., López-Sánchez, G. N., Milflores-Flores, L., Patel, S. D., Rojas-López, M., Martínez de la Peña, C. F., Arenas-Hernández, M. M. & Martínez-Laguna, Y. (2007). Ler and H-NS, regulators controlling expression of the long polar fimbriae of Escherichia coli O157 : H7. J Bacteriol 189, 5916–5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi, C., Seto, C., Suzuki, T. & Mizuno, T. (1997). Clarification of the dimerization domain and its functional significance for the Escherichia coli nucleoid protein H-NS. J Mol Biol 274, 145–151. [DOI] [PubMed] [Google Scholar]
- Umanski, T., Rosenshine, I. & Friedberg, D. (2002). Thermoregulated expression of virulence genes in enteropathogenic Escherichia coli. Microbiology 148, 2735–2744. [DOI] [PubMed] [Google Scholar]
- Yerushalmi, G., Nadler, C., Berdichevski, T. & Rosenshine, I. (2008). Mutational analysis of the locus of enterocyte effacement-encoded regulator (Ler) of enteropathogenic Escherichia coli. J Bacteriol 190, 7808–7818. [DOI] [PMC free article] [PubMed] [Google Scholar]