Abstract
Coxiella burnetii is the bacterial agent of Q fever in humans. Here, we describe a unique, ∼7.2 kDa, surface-exposed lipoprotein involved in metal binding which we have termed LimB. LimB was initially identified as a potential metal-binding protein on far-Western (FW) blots containing whole-cell lysate proteins when probed with nickel-coated horseradish peroxidase (Ni-HRP) and developed with a chemiluminescent HRP substrate. The corresponding identity of LimB as CBU1224a was established by matrix-assisted laser desorption ionization-tandem time-of-flight mass spectrometry. blast analyses with CBU1224a showed no significant similarity to sequences outside strains of C. burnetii. Additional in silico analyses revealed a putative 20 residue signal sequence with the carboxyl end demarcated by a potential lipobox (LSGC) whose Cys residue is predicted to serve as the N-terminal, lipidated Cys of mature LimB. The second residue of mature LimB is predicted to be Ala, an uncharged envelope localization residue. These features suggest that CBU1224a is synthesized as a prolipoprotein which is subsequently lipidated, secreted and anchored in the outer membrane. Mature LimB is predicted to contain 45 aa, of which there are 10 His and 5 Cys; both amino acids are frequently involved in binding transition metal cations. Recombinant LimB (rLimB) was generated and its Ni-HRP-binding activity demonstrated on FW blots. Ni-HRP binding by rLimB was inhibited by >95 % on FW blots done in the presence of EDTA, imidazole, Ni2+ or Zn2+, and roughly halved in the presence of Co2+ or Fe3+. The limB gene was maximally expressed at 3–7 days post-infection in Coxiella-infected Vero cells, coinciding with exponential phase growth. Two isoforms of LimB were detected on FW and Western blots, including a smaller (∼7.2 kDa) species that was the predominant form in small cell variants and a larger isoform (∼8.7 kDa) in large cell variants. LimB is Sarkosyl-insoluble, like many omps. The predicted surface location of LimB was verified by immunoelectron and immunofluorescence microscopy using anti-rLimB antibodies. Overall, the results suggest that LimB is a unique Coxiella lipoprotein that serves as a surface receptor for divalent metal cations and may play a role in acquiring at least one of these metals during intracellular growth.
INTRODUCTION
Coxiella burnetii is a gammaproteobacterium and the agent of Q fever in humans. C. burnetii is one of the most infectious pathogens known, with an ID50 of 1–10 bacteria in the guinea pig model (Moos & Hackstadt, 1987). Human infections with C. burnetii are generally zoonoses acquired by inhalation of contaminated aerosols. Q fever typically presents as an acute, self-limiting flu-like illness accompanied by pneumonia or hepatitis. In roughly 1 % of cases, a severe chronic infection can occur, in which endocarditis is the predominant manifestation (Maurin & Raoult, 1999). These attributes and its past use as a biological weapon component (Regis, 1999) were grounds for classifying Coxiella as an ‘HHS Select Agent’.
In nature, C. burnetii is an obligate intracellular pathogen and undergoes a developmental cycle that exhibits two distinct cell morphotypes. The infectious small cell variant (SCV) has been described as ‘spore-like’, and it is extremely resistant to environmental stressors, including UV light and desiccation. Shortly after entry into a eukaryotic phagolysosome-like compartment called a parasitophorous vacuole (pH ∼4.5), the SCV transforms into a ‘vegetative’ morphotype, termed a large cell variant (LCV) (McCaul & Williams, 1981). After 5–6 days of intracellular replication, the LCV transforms back to a SCV that is released from the host cell to complete the developmental cycle (Coleman et al., 2004; McCaul & Williams, 1981). In addition to distinct developmental forms, two separate phase variants of the bacterium have also been described. First, wild-type C. burnetii is typically ‘phase I’, in reference to serological reactivity against its ‘smooth’, long-chain LPS, and must be manipulated in a bio-safety level (BSL)-3 facility. Second, repeated passage in tissue culture or embryonated hens' eggs results in the spontaneous generation of attenuated ‘phase II’ (PII) organisms that synthesize a ‘rough’ LPS that can be caused by a chromosomal deletion (Hoover et al., 2002; Vodkin & Williams, 1986), and may be handled in a BSL-2 laboratory. Finally, a number of C. burnetii strains [such as ‘Nine Mile’ (NM)] have been isolated from different parts of the world and have been grouped according to their association with acute or chronic disease manifestations of Q fever (Beare et al., 2009; Seshadri & Samuel, 2005).
Despite several fascinating attributes, little is known regarding Coxiella's virulence determinants. In fact, LPS is the only bona fide virulence factor described to date (Moos & Hackstadt, 1987), and it has been shown to function as an immune-evasion shield (Shannon et al., 2005). One potential set of virulence factors that has received relatively little attention is the pathogen's considerable assemblage of lipoproteins. Lipoproteins have long been recognized as virulence determinants in a variety of bacterial pathogens, and a number of them have been developed into vaccine immunogens (Fortney et al., 2000; Hayashi & Wu, 1990; Schwan et al., 1995; Sha et al., 2004). Lipoproteins are anchored to the inner or outer membrane through a covalently linked lipid moiety (Babu et al., 2006; Hantke & Braun, 1973; Inouye et al., 1977). The genome of the NM strain of C. burnetii (Seshadri et al., 2003) is predicted to encode 25 lipoproteins (Babu et al., 2006; Flores-Ramírez et al., 2009), including DotC, a type IV secretion system protein (Zamboni et al., 2003). However, with the exception of DotC, the functions of Coxiella's lipoproteins are unknown. We hypothesize that some of these lipoproteins play critical roles in growth and virulence, and represent a set of subunit vaccine candidates and targets for antimicrobial therapy against C. burnetii.
It is widely accepted that metal acquisition is an essential process for bacterial survival and pathogenesis, and bacteria have evolved an impressive array of strategies and mechanisms to acquire, transport and store bio-reactive metals such as iron and zinc (Ratledge & Dover, 2000; Rosenzweig, 2002). This paper describes a unique C. burnetii outer-membrane protein and putative lipoprotein encoded by the CBU1224a open reading frame (Seshadri et al., 2003) which binds metal cations with high affinity; we have termed this LimB (lipoprotein involved in metal binding).
METHODS
Bacterial strains, cell lines and growth conditions.
NM PII C. burnetii (strain RSA 439; clone 4) was propagated in African green monkey kidney (Vero) epithelial cells (CCL-81; American Type Culture Collection), as described previously (Raghavan et al., 2008). SCVs were prepared and purified from 28-day-old cultures as described previously (Raghavan et al., 2008). SCVs were used to inoculate Vero cell monolayers to produce synchronous infections (Coleman et al., 2004). Mixed-cell populations of LCVs and SCVs were harvested from synchronous cultures as for SCVs, but at 10 days post-infection. LCVs were prepared from 72 h synchronous cultures using a digitonin-based protocol (Cockrell et al., 2008). Whole-cell lysates of Coxiella were obtained by using a freeze–thaw method (Samoilis et al., 2007).
SDS-PAGE and far-Western (FW) blots.
Protein concentrations were determined by using a BCA protein assay kit (Thermo Scientific). Bacterial suspensions were solubilized with Laemmli sample buffer (Laemmli, 1970). Samples were typically boiled for 10 min and centrifuged (1 min, 10 000 g) to remove insoluble material; the resulting supernatant was separated on an SDS-PAGE gel (Laemmli, 1970). Protein bands were visualized by staining gels with Coomassie brilliant blue [CBB; 0.1 % (w/v) in 50 % methanol, 7 % (v/v) acetic acid].
FW blots were prepared using whole-cell lysate proteins (20 μg) from SCVs, LCVs or mixed-cell populations, separated by SDS-PAGE, then transferred to supported nitrocellulose [45 μm pore; GE Osmonics (Towbin et al., 1979)]. Resulting blots were probed with nickel-coated horseradish peroxidase (Ni-HRP; Pierce) and developed with ECL reagents (SuperSignal West Pico; Pierce) according to the manufacturer's instructions. Blots were developed for 5 min in chemiluminescent horseradish peroxidase (HRP) substrate (SuperSignal West Pico; Pierce) according to the manufacturer's instructions. Blots were visualized using an LAS-3000 digital imaging system (Fujifilm).
Inhibition studies were done using recombinant LimB (rLimB) and FW blots were prepared as described above, except the probing solution contained a potential competitive inhibitor of binding, including EDTA (100 mM) or imidazole (500 mM). In some cases, HRP was used in place of Ni-HRP, at the same final concentration (1 μg ml−1), to demonstrate that LimB binding activity was dependent on the nickel moiety of Ni-HRP. Finally, to qualitatively analyse the relative affinity of LimB for various metal ions, modified FW blots were performed based on a previously published protocol (Zhao & Waite, 2006). First, rLimB (5 μg protein) was separated by SDS-PAGE, transferred to nitrocellulose, blocked and washed as above. Second, blots were incubated (1 h; 25 °C) in TBST containing chloride salts of individual metal ions (FeCl3·6H2O, CoCl2·6H2O, NiCl2·6H2O or ZnCl2) at a final concentration of 10 mM. Excess metal was subsequently removed by washing three times for 5 min with TBST containing 0.1 mM of the individual metal ion. Third, resulting FW blots were probed and washed as above except solutions contained 0.1 mM of the particular metal ion. Finally, blots were developed and visualized as above.
Mass spectrometry (MS).
A C. burnetii whole-cell lysate (20 μg protein) was separated by using SDS-PAGE [10–15 % acrylamide (w/v)] and stained with CBB, as above. Bands corresponding to Ni-HRP-binding activity on FW blots were excised from gels and subjected to trypsin digestion and MALDI-TOF peptide mass fingerprinting and MALDI-tandem TOF (MALDI-TOF/TOF) peptide sequencing. All MS work and database searches (NRDB1 database using Mascot version 2.2.03 software) were performed by Alphalyse.
Nucleic acid isolation and rLimB production.
Infected monolayers were harvested at specific time points (0, 3, 5, 6, 7 and 8 days) by replacing culture medium with 2.5 ml TRI reagent (Ambion), briefly incubating (5 min, 25 °C) and scraping. Harvested monolayers were stored (−80 °C) for subsequent nucleic acid isolation. RNA and DNA were isolated from each harvested monolayer sample essentially as described previously (Raghavan et al., 2008). RNA was isolated from this extract by the addition of 300 μl 100 % ethanol and use of a RiboPure kit (Ambion) according to the manufacturer's instructions, and then stored at −20 °C. DNA and RNA were removed from these samples by DNase treatment (TURBO DNase kit; Ambion) and an RNA clean-up step (RNeasy kit; Qiagen). Purified RNA was quantified spectrophotometrically (260 nm), and 1 μg RNA from each sample was converted to cDNA with an iScript cDNA kit (Bio-Rad), as instructed. Finally, cDNA was used to analyse limB transcript profiles by quantitative real-time PCR (qRT-PCR).
DNA was isolated using TRI Reagent DNA/protein isolation protocol (Ambion). DNA-containing supernatant was transferred to another microcentrifuge tube and 40 μl HEPES (0.1 M) was added to adjust the pH to 8.2. Isolated DNA was cleaned using a QIAquick PCR purification kit (Qiagen) as instructed and stored (−20 °C) until needed.
A portion of Coxiella limB encoding the predicted, mature LimB (i.e. Cys21 to His65) was cloned into pMAL-c5X (NEB) to create pJB202, a plasmid that encodes a LimB/maltose-binding protein (MBP) fusion. Briefly, primers (LimBF+XmnI, CTGCAGGAAGGCTTTCAGGTTGTGCTCCACGCAC; LimBR+BamHI, AAGGATCCTTATCAATGATGATTACAATGACGATG) were designed to amplify the CBU1224a ORF from the codon for Cys21 to the termination codon, and both primers contained restriction sites (underlined). PCR was done by using standard methods (Ausubel et al., 1995) and the resulting amplicon was purified (PCR Purification kit, Qiagen), digested with XmnI and BamHI, repurified, and ligated into the corresponding restriction sites of pMAL-c5x (NEB) using T4 DNA ligase. E. coli BL21(DE3) was transformed with pJB202 (Chung et al., 1989) and the resulting transformants were selected by plating on Luria–Bertani plates containing ampicillin (100 μg ml−1). Plasmid constructs of selected clones were verified by automated sequence analysis. Amylose resin was used according to the manufacturer's instructions (NEB) to isolate rLimB fusion protein from the supernatant. Fractions containing rLimB were identified by SDS-PAGE, pooled and dialysed against 20 mM Tris, 50 mM NaCl, 1 mM CaCl2 buffer (pH 6.5) using a Slide-a-Lyser cassette (Pierce; molecular weight cut-off, 3500 Da). rLimB was stored at −20 °C until needed.
qRT-PCR and quantitative PCR (qPCR).
Primers were designed using Beacon Designer Software (Premier Biosoft). Primers were synthesized (Integrated DNA Technologies) and used to quantify limB mRNA over the course of a synchronous infection by normalizing to the number of C. burnetii genomes, as described previously (Coleman et al., 2004; Raghavan et al., 2008). First, the number of genomes in each purified DNA sample at each time point (0, 3, 5, 6, 7 and 8 days) was determined in triplicate qPCR reactions (each triplicate experiment was performed twice, thus resulting in six independent determinations). This was accomplished using an iCycler single-colour real-time PCR detection system (Bio-Rad), SYBR Green supermix (Bio-Rad) and a dotA (CBU_1648) primer set (DOTA-F, GATAGCTGTGGAGCCGTGAC; DOTA-R, TCCTTTGCCATCGTGCATTTT) by normalization to a standard curve of 10-fold serially diluted (107–101) C. burnetii genomes. Second, cDNA from each time point was used to determine the starting quantity (SQ) mean, a value calculated by iCycler software (MyIQ, v1.0, Bio-Rad) by normalizing limB transcript numbers with a limB primer set (LIMB-F, GGTCCTGT TTGATAAACTACG; LIMB-R, GCTGTTATGATTTCAGTGATTG) to a standard curve generated by dotA qPCR of serially diluted C. burnetii genomes. Finally, the number of limB mRNAs per genome equals the SQ mean divided by the number of genomes × 1000.
Antibodies, Western blots, immunofluorescence and immunoelectron microscopy.
Anti-rLimB and anti-rCom1 polyclonal antibodies were generated in female New Zealand white rabbits, as described previously (Parrow et al., 2009), using 100 μg purified rLimB or rCom1 (Hicks et al., 2010) for both primary and booster injections. Rabbit anti-MBP polyclonal antibodies were purchased (NEB) and used as a negative control for the LimB fusion protein partner. Mouse monoclonal antibody against Coxiella elongation factor-Ts (EF-Ts) was used as a negative control for intracellular proteins that may have inadvertently leaked from the bacterium during handling and preparation for microscopy (a generous gift from Robert Heinzen).
Immunoblots were done by transferring un-fixed SDS-PAGE gels to supported nitrocellulose and probing overnight with anti-rLimB antibodies (1 : 5000 dilution) and peroxidase-conjugated goat anti-rabbit antibodies (Sigma), as described previously (Parrow et al., 2009), except ECL reagents were used during development according to the manufacturer's instructions (Pierce), and signals were visualized with an LAS-3000 digital imaging system (Fujifilm).
Immunofluorescence microscopy to identify surface-exposed proteins was done by the general methods described by Ge & Rikihisa (2007). Bacteria were cultured and harvested from Vero cells at 5 days post-infection, as above, pelleted and washed in PBS containing 0.25 M sucrose (PBSS; pH 7.4). This, and all subsequent steps were done at 25 °C. Bacteria were fixed for 45 min in 2 % (w/v) paraformaldehyde. After quenching in PBS containing 0.1 M glycine and subsequently washing in PBS, bacteria were incubated for 1 h with anti-LimB, anti-Com1, anti-EF-Ts, anti-MBP or pre-immune serum (PIS) at a 1 : 100 dilution in 0.2 % (w/v) gelatin in PBS (PG buffer). After washing with PG buffer, bacteria were labelled for 1 h with Alexa Fluor 488 goat anti-rabbit or mouse (for anti-EF-Ts) immunoglobulin G (Invitrogen) at a dilution of 1 : 100 in PG buffer. Bacteria were washed in PG buffer, suspended in PBS and imaged with a BX51 fluorescence microscope and digital acquisition software (DP2-BSW) (Olympus).
To conduct immunoelectron microscopy, bacteria were cultured and harvested at 5 days post-infection as described above, and diluted in PBSS to a concentration of ∼1×105 genomes μl−1. The suspension was combined with an equal volume (100 μl) of antiserum or PIS [diluted 1 : 100 in PBS-B (PBS containing 0.01 mg BSA ml−1)] and incubated for 1 h at 37 °C. Cells were then centrifuged (10 min at 10 000 g), the pellet was washed with 300 μl PBSS, and the pellet was suspended in 200 μl PBS-B. An aliquot (100 μl) of the cell suspensions was combined with an equal volume of protein A–gold conjugate [Electron Microscopy Sciences (EMS)] pre-diluted 1 : 20 in PBS-B. Following a brief incubation (50 min, 37 °C), cells were centrifuged and washed as above, and finally suspended in 30 μl PBS. Half the volume of these suspensions was placed on Formvar-coated TEM grids (EMS; 400 mesh) and incubated for 10 min at 23 °C to allow for adherence. Excess suspension was wicked from the grid surface by using blotting paper, replaced with 15 μl uranyl acetate [2 % (w/v) in H2O, J. T. Baker, Phillipsburg, NJ], and incubated for 3 min at 23 °C. Excess protein A–gold was washed from grids by successive immersions in three beakers containing 2 l dH2O. Finally, grids were air-dried and analysed by using a Hitachi 7100 transmission electron microscope.
Sarkosyl fractionation.
Omps of C. burnetii were enriched by Sarkosyl-fractionation of the bacterium. A whole-cell lysate from a mixed-cell population of Coxiella was prepared and the insoluble cell-wall fraction was pelleted by centrifugation. Briefly, the pellet was suspended in 300 μl PBS (pH 7.4), to which 38 ml 0.1 M sodium bicarbonate (pH 11) was added, then incubated on ice for 1 h (with rocking). The mixture was centrifuged (141 000 g for 80 min, 4 °C) and the resulting pellet was extracted with 4 ml sodium N-lauroylsarcosine (Sarkosyl; Sigma-Aldrich) solution (2 %; w/v). The extraction was done on ice for 30 min (with rocking). The Sarkosyl-insoluble (omp-rich) fraction was isolated by centrifuging the mixture (138 000 g for 1h, 4 °C). The Sarkosyl-soluble fraction (supernatant) was precipitated in 4 vols acetone for 1 h at −20 °C. The mixture was centrifuged (3000 g for 30 min at 4 °C) to obtain a pellet. Both fractions were analysed by SDS-PAGE and blotting.
Sequencing, in silico analyses and statistics.
Sequence data were obtained with an automated DNA sequencer (ABI3130x1) and a BigDye terminator cycle sequencing ready reaction kit (ABI). GenBank searches were done by using blast (http://www.ncbi.nlm.nih.gov/BLAST/). In silico tools available at the database of bacterial lipoproteins (DOLOP; http://www.mrc-lmb.cam.ac.uk/genomes/dolop/; Babu et al., 2006) were used to determine if a predicted protein sequence contained a prolipoprotein signal peptide. MacVector genetic software (Version 11.1.1) was used for protein sequence analysis as well as sequence manipulations. Comparative densitometry of FW blot signals was done using ImageJ 1.42q software (http://rsbweb.nih.gov/ij/). At least three independent determinations were used to calculate means and standard deviations for all numerical data. Statistical and graphical analyses were done using InStat3 (GraphPad) and/or Excel (Microsoft) software. Statistical significance was determined using Student's t test, where a P-value of <0.05 was considered significant.
RESULTS
FW blots reveal a ∼7.2 kDa component of Coxiella that binds Ni-HRP
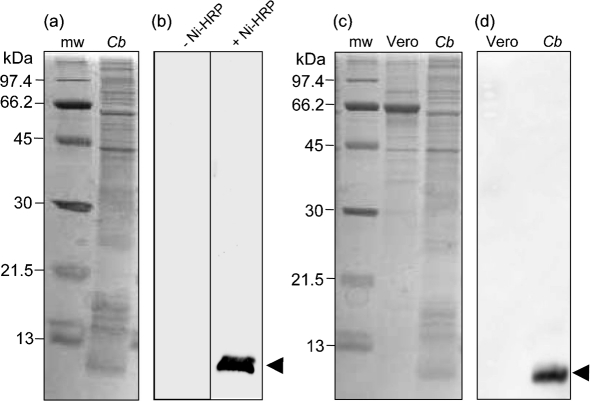
A prominent, chemiluminescent band of ∼7.2 kDa was discovered on Ni-HRP-probed FW blots containing whole-cell lysate proteins from a mixed-cell population (10-day-old culture) of C. burnetii LCVs and SCVs (Fig. 1a and b). We were curious to determine whether the signal was a result of Ni-HRP binding to a C. burnetii constituent or whether it was due to an endogenous peroxidase that was directly acting on the chemiluminescent HRP substrate. To address this question, we performed FW blots as above but without Ni-HRP. The resulting FW blots showed that even with prolonged exposure (5 min) a chemiluminescent band was not detectable Fig. 1(b) (−Ni-HRP), suggesting that the FW blot signal was due to Ni-HRP binding to a C. burnetii component, and not due to an endogenous peroxidase. To ensure that Ni-HRP was not binding a contaminating host-cell constituent, Vero cells were cultivated and harvested for Ni-HRP FW blot analyses (Fig. 1c). The resulting FW blots, prepared as above, showed no detectable chemiluminescent signal in Vero host cell lysate proteins (Fig. 1d). Finally, FW blots of C. burnetii were prepared and probed with HRP; however, a chemiluminescent signal was not produced in the absence of Ni (data not shown). From these results, we hypothesized that the Ni-HRP was binding to a C. burnetii component, that binding was dependent on the nickel moiety of the probe, and that a small, His-rich protein might be responsible.
Fig. 1.
An ∼7.2 kDa component of C. burnetii binds Ni-HRP and is not present in host cells. (a) CBB-stained SDS-PAGE gel (12–15 % acrylamide; w/v) showing the protein profile of a whole-cell lysate from a mixed-cell population (10 days) of C. burnetii (Cb; 20 μg protein). Molecular mass markers (mw) are shown. (b) Corresponding FW blots of Cb, as in (a), showing an intense chemiluminescent band of ∼7.2 kDa (arrowhead) when probed with Ni-HRP (+ Ni-HRP; exposure time 2 min), but which is absent without Ni-HRP (−Ni-HRP; exposure time 5 min). (c) CBB-stained SDS-PAGE gel (12–15 % acrylamide; w/v) showing Cb and mw as above plus the protein profile of uninfected Vero host cells (Vero; 20 μg protein). (d) FW blot corresponding to Vero and Cb, as in (c), showing that the Ni-HRP binding component is specific to C. burnetii and is not detected in Vero cells.
MS identifies the Ni-HRP binding protein as CBU1224a
To identify the C. burnetii protein responsible for Ni-HRP binding, MALDI-TOF peptide mass fingerprinting and MALDI-TOF/TOF peptide sequencing were employed; these identified two major peaks in the chromatogram that matched two of the tryptic peptide fragments calculated for GI-153209557, a C. burnetii ‘putative lipoprotein’. MS sequencing results represented 56 % coverage of the predicted mature, 45 aa lipoprotein. These data allowed us to presumptively identify the Ni-HRP-binding ligand as the protein encoded by the CBU1224a locus of C. burnetii (strain RSA 493).
In silico analysis of CBU1224a reveals a potential lipoprotein signal sequence, a ‘lipobox’ and a His-rich carboxyl-terminus
Several in silico analyses were performed on CBU1224a and its encoded protein. blast was used to search the GenBank database for similar sequences. Published genomic sequences of six C. burnetii strains (RSA 493, RSA 331, Q177, Q154, Q212 and 5J108-111) (Beare et al., 2009) (Seshadri & Samuel, 2005; Seshadri et al., 2003) possess ORFs that are 100 % identical to CBU1224a. In each strain, the 198 bp ORF is flanked by genes encoding haloalkane dehalogenase (111 bp upstream) and an RmuC family protein (105 bp downstream). RmuC (rearrangement mutator) family proteins are hypothesized to play a role in DNA structure/recombination (Slupska et al., 2000) and haloalkane dehalogenase (EC 3.8.1.5) is an enzyme involved in degradation of halocarbons. Based on considerable intergenic spacing, neither of the flanking genes appears to be transcriptionally linked to CBU1224a. Finally, blast searches of the non-redundant GenBank database using DNA, as well as the predicted 65 aa residue prolipoprotein and 45 aa mature lipoprotein sequences, did not yield significant similarities outside C. burnetii.
We also used in silico tools available on the DOLOP website (Babu et al., 2006) to investigate the putative lipoprotein encoded by CBU1224a. These analyses revealed a potential ‘lipobox’ and characteristic N, H and C prolipoprotein signal peptide regions, supporting the ‘lipoprotein’ annotation for the locus. Specifically, the characteristics of the lipobox are: (i) it consists of 4 aa with the consensus sequence [LVI][ASTVI][ASG][C] (abbreviated as LXXC), (ii) it is located within the first 40 residues of the prolipoprotein, and (iii) it is also called the Cys region (Fig. 2).
Fig. 2.
Predicted features of the CBU1224a-encoded lipoprotein involved in metal binding (LimB). In silico analyses reveal a predicted signal peptide (lower case) containing a potential lipobox (LSGC; blue box), a consensus peptide cleavage site (red line), and an N-terminal Cys (green) in mature LimB (upper case) that is predicted to be covalently modified with diacylglyceryl (DAG) and a fatty acid (FA), as observed in other Gram-negative bacteria. The uncharged Ala envelope localization residue (ELR) (circled) immediately following the N-terminal Cys suggests that mature LimB is translocated to the outer membrane, as reported for Gram-negative bacteria. Mature LimB contains ten His (red) and four Cys (orange) residues that may be involved in metal binding.
Based on studies with other Gram-negative bacteria, the mature CBU1224a lipoprotein (unlipidated) is predicted to be 45 aa in length (∼5.2 kDa), and would be located in the C. burnetii outer membrane. Several features and results support this hypothesis. First, the envelope localization residue is an Ala, and would therefore help direct the mature protein to the outer membrane (Yamaguchi et al., 1988). Second, two tryptic fragments identified by MALDI-TOF MS (Thr25–Lys41 and His53–Arg60) are peptides from the predicted mature lipoprotein and not the leader sequence.
The high-affinity binding of Ni-HRP observed in FW blots (Fig. 1) is probably explained by the high percentage of His residues located in the C-terminal portion of CBU1224a (Fig. 2). Specifically, the predicted, mature 45 aa lipoprotein contains ten His (22 % of the total amino acids), nine of which are located in the last 20 aa of the protein. Four Cys residues are also found in this region, an observation that is noteworthy for two reasons: (i) Cys residues could be partially responsible for Ni-HRP binding as both His and Cys are known to coordinate binding of transition metals, and (ii) intra- and/or inter-disulfide bonds could contribute to structure or multimerization of the protein, respectively.
Recombinant protein encoded by CBU1224a binds Ni-HRP
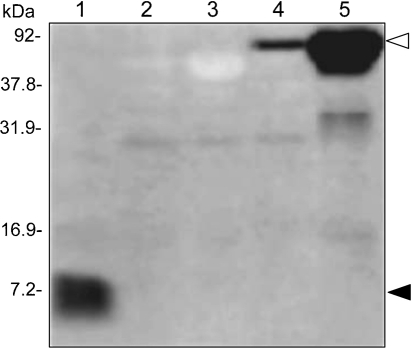
To unequivocally demonstrate that Ni-HRP binding on FW blots was due to the protein encoded by CBU1224a, a plasmid (pJB202) was constructed to produce a translational fusion with MBP. FW blots were prepared to compare Ni-HRP binding characteristics of E. coli BL21 (pJB202) versus E. coli BL21 (pMAL-c5X; empty cloning vector). Results clearly show that the recombinant protein binds Ni-HRP (Fig. 3, lanes 4 and 5), like the native protein (Fig. 3, lane 1). In addition, recombinant MBP (rLimB’s fusion partner) does not bind the probe (Fig. 3, lanes 2 and 3). Based upon these results and our in silico analyses, we designated CBU1224a as a lipoprotein involved in metal-binding (LimB).
Fig. 3.
rLimB binds Ni-HRP. FW blot prepared from a SDS-PAGE gel (12.5 % acrylamide; w/v) containing separated proteins (20 μg) of whole-cell lysates of C. burnetii (mixed-cell population; lane 1), E. coli (pMAL-c5x) (2 h, no IPTG; lane 2), E. coli (pMAL-c5x) (2 h, 1 mM IPTG; lane 3), E. coli (pJB202) (2 h, no IPTG; lane 4) and E. coli (pJB202) (2 h, 1 mM IPTG; lane 5). FW blot chemiluminescent signals from native LimB and rLimB binding of Ni-HRP are indicated by closed and open arrowheads, respectively. Molecular masses were determined from protein standards.
rLimB binds divalent cations, which involves His residues
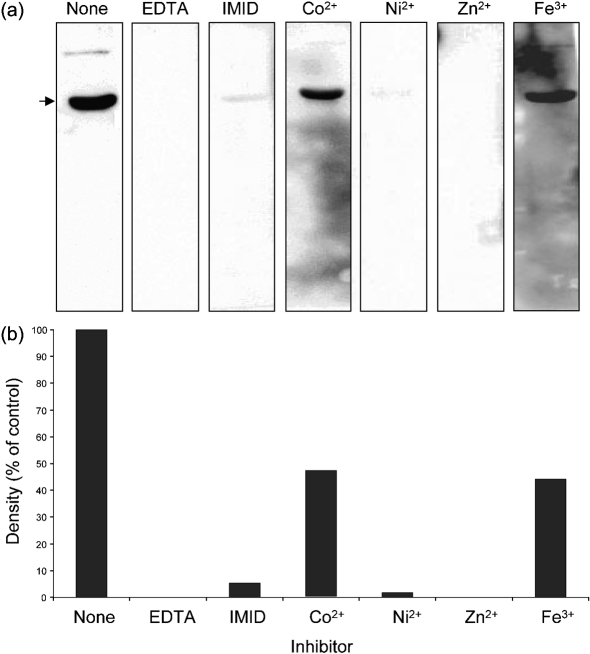
To further characterize Ni-HRP binding, rLimB was employed in FW blots done in the presence of inhibitors or divalent metal cations. First, when the metal chelator EDTA was added along with the Ni-HRP probe, a chemiluminescent signal could not be detected, in contrast with untreated controls done in parallel (Fig. 4, EDTA vs none). These results suggest that the Ni moiety of Ni-HRP mediates binding to LimB. Second, when the His analogue imidazole is added along with Ni-HRP, the luminescent signal was reduced by >95 % compared with untreated controls (Fig. 4, IMID vs none). These data suggest that LimB His residues are involved in Ni-HRP binding. However, since the signal was not completely inhibited, other residues, such as Cys, may also be involved. Third, we wanted to determine whether LimB was capable of binding other metals. We hypothesized that metal ions could fill LimB binding residues, and inhibit or ‘block’ the binding of Ni-HRP on FW blots. The relative competition capacity of free metal ions could therefore be analysed by FW blots, where a reduction in chemiluminescent signal is directly proportional to the affinity of that particular metal to LimB. To this end, immediately following transfer of proteins to nitrocellulose, we blocked FW blots by incubation in TBST containing chloride salts of four metal ions. FW blots blocked with nickel showed ∼95 % reduction in their Ni-HRP chemiluminescent signal relative to untreated controls (Fig. 4, Ni2+), further supporting previous results showing that the nickel moiety of Ni-HRP mediates binding to LimB. Interestingly, when blots were blocked with zinc (Fig. 4, Zn2+) the chemiluminescent signal was no longer detectable relative to untreated controls. These data suggest that rLimB binds Ni2+ and Zn2+ with high affinity. In contrast, when FW blots were blocked with cobalt (Co2+) or iron (Fe3+), the chemiluminescent signal was roughly half of untreated controls, suggesting that these metals bind LimB with relatively lower affinity than Ni2+ or Zn2+. Taken as a whole, the results demonstrate that LimB binds a variety of divalent metal cations with a higher affinity for Ni2+ and Zn2+ relative to Co2+ and Fe3+.
Fig. 4.
Ni-HRP binding by rLimB is inhibited by EDTA, imidazole and divalent metal cations. (a) FW blots were done in parallel and prepared from the same SDS-PAGE gel (12.5 % acrylamide; w/v) using whole-cell lysates of C. burnetii (mixed-cell population; 20 μg protein per lane). Blots were incubated in the presence of EDTA, imidazole (IMID) or divalent metal cations, as described in Methods. The resulting FW blots demonstrate a marked reduction in rLimB binding of Ni-HRP in the presence of Co2+ or Fe3+ (<50 % of control), as shown by the reduced chemiluminescent signal (arrow). Binding signal was not detectable on blots incubated with EDTA or Zn2+ and was only slightly detectable (<5 % of control) if imidazole (IMID) or Ni2+ were present. (b) Densitometric analysis of the blots in (a), shown as a percentage of the signal density of the untreated control (None). The assay is representative of two independent sets of experiments.
2-Mercaptoethanol (2-ME) treatment implicates the Cys residues of LimB in metal binding
We were curious whether the Cys residues of LimB also play a role in Ni-HRP binding. As mentioned above, four Cys residues are located in the C-terminal portion of LimB and might bind nickel and/or confer inter- or intra-LimB disulfide bonds. To address these possibilities, cell lysate proteins of C. burnetii were prepared with and without 2-ME, and FW blots were done as described above. Resulting blots showed a marked reduction in the chemiluminescent signal of LimB when samples were prepared without 2-ME compared with those with 2-ME (data not shown). Increased binding of Ni-HRP in the presence of 2-ME may be due to increased exposure of His and/or increased availability of thiol ligands as a result of disulfide reduction. Multiple LimB bands that would indicate disulfide-mediated inter-LimB multimerization were never observed; however, it is still possible that multimers occur, especially considering the low Ni-HRP binding efficiency observed in the absence of 2-ME.
limB expression is maximal during exponential phase
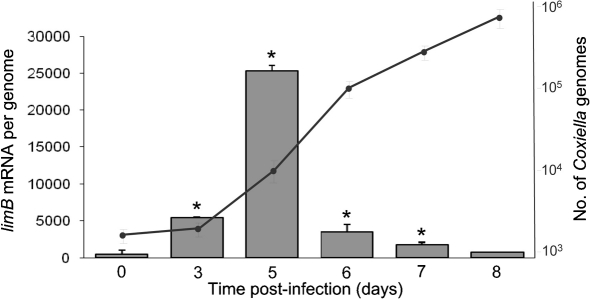
To determine whether expression of limB changes during the developmental cycle of C. burnetii, synchronous infections were initiated and the transcript quantity was analysed over an 8 day time-course using qRT-PCR. Results showed that limB mRNA was significantly higher (P<0.05) at 3–7 days post-infection, relative to the initial day (day 0). The limB transcripts were highest at 5 days post-infection (∼48-fold increase over the day 0 value). A corresponding analysis of Coxiella genomes in these samples shows that limB expression is highest during exponential phase growth of the bacterium (Fig. 5). Although transcription of limB is maximal during exponential phase growth (Fig. 5), the LimB protein can be detected in both LCVs and SCVs (Fig. 6). Interestingly, FW blots and immunoblots prepared from 12.5 % acrylamide gels revealed two distinct molecular mass forms of native LimB, with a larger (∼8.7 kDa) LimB species dominant in LCVs and an ∼7.2 kDa isoform of LimB in SCVs (28 days post-infection) (Fig. 6). Not surprisingly, mixed-cell populations of the bacterium possess both LimB isoforms (Fig. 6a and b, lane 1). Although the two LimB isoforms can be distinguished as a doublet or smear on blots prepared from gradient gels (e.g. Fig. 1b), resolution of the bands was much clearer when blots were prepared from 12.5 % acrylamide gels (Figs 6 and 7). The two LimB isoforms may reflect post-translational modification of LimB, and perhaps cleavage of LimB’s signal sequence and/or lipidation during the protein’s maturation.
Fig. 5.
limB expression is highest during exponential phase growth of C. burnetii in synchronous infections of cultured Vero cells. An analysis of limB transcript quantities (histogram) relative to the number of genomes (• and line) was determined by qRT-PCR and qPCR, respectively, over the course of an 8 day synchronous infection of Vero cells, as described in Methods. Values represent the means±sd from six independent determinations. Asterisks denote a statistically significant increase in limB mRNA relative to 0 days (P<0.05).
Fig. 6.
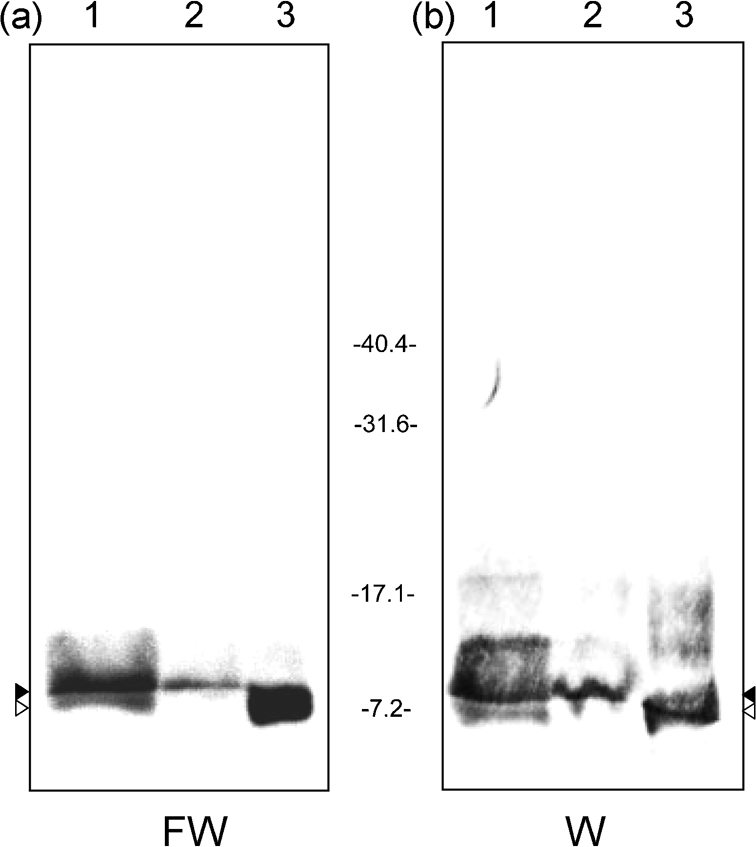
LimB is found in LCVs and SCVs and two isoforms of the protein exist. (a) FW blot prepared from an SDS-PAGE gel (12.5 % acrylamide; w/v) using whole-cell lysates of C. burnetii from a mixed-cell population (10 days) (lane 1), LCVs (5 days; lane 2) and SCVs (28 days; lane 3) (20 μg protein per lane). (b) Corresponding Western blot (W) from the other half of the gel, developed with anti-rLimB antibodies. The small (∼7.2 kDa) and large (∼8.7 kDa) isoforms of LimB are indicated by white and black arrowheads, respectively. Molecular masses were determined from pre-stained standards (Bio-Rad) and are shown in kDa.
Fig. 7.
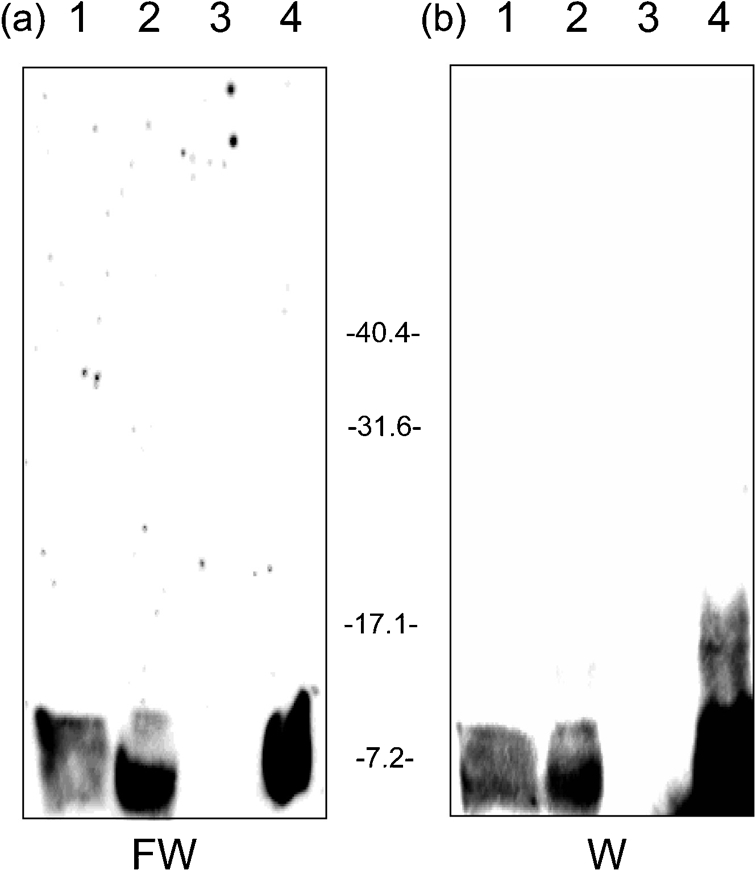
LimB is found in the Sarkosyl-insoluble fraction of Coxiella. (a) FW blot prepared from an SDS-PAGE gel (12.5 % acrylamide; w/v) using a mixed-cell population (lane 1), whole-cell lysates (lane 2), and Sarkosyl-soluble and Sarkosyl-insoluble fractions of the mixed-cell population (lanes 3 and 4, respectively) (20 μg protein per lane). (b) Corresponding Western blot (W) from the other half of the gel developed with anti-rLimB antibodies. Molecular masses were determined from pre-stained standards (Bio-Rad) and are shown in kDa.
LimB is a surface-exposed omp
The second residue of mature LimB is predicted to be Ala (Fig. 2). Previous work has shown that the second residue of mature lipoproteins plays an important role in coordinating transport of the protein to its final destination in the cell, thus it is referred to as an envelope localization residue (ELR) (Yamaguchi et al., 1988). Since lipoproteins with an uncharged ELR are translocated to the outer membrane of Gram-negative bacteria, we hypothesized that LimB was surface-exposed in C. burnetii. To address the hypothesis, we used three different methods. First, we prepared a Sarkosyl-insoluble fraction of Coxiella, a method commonly used to purify omps of Gram-negative bacteria, previously employed for this purpose with C. burnetii (Zhang et al., 2005). Analysis of Sarkosyl fractions of a mixed-cell population of Coxiella by FW blots and immunoblotting with anti-rLimB antibodies showed that: (i) both molecular mass forms of LimB were present in the insoluble fraction but absent in the Sarkosyl-soluble fraction, (ii) Ni-HRP was bound by both LimB isoforms and (iii) both protein isoforms were recognized by anti-LimB antibodies (Fig. 7). These data suggest that LimB is an omp.
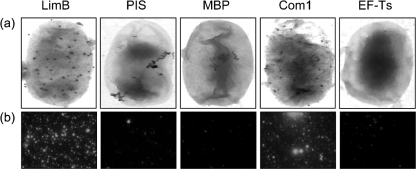
Second, we employed immunoelectron microscopy with anti-rLimB or control antibodies and live bacteria. Resulting electron micrographs showed uniform protein A–gold binding on the surface of Coxiella treated with either anti-LimB or anti-Com1 (positive control) antibodies (Fig. 8a). In contrast, treatment with anti-MBP antibodies (a control for the fusion partner of the rLimB immunogen), anti-EF-Ts (a control for leaked intracellular proteins) or rabbit PIS from the same animal used to generate anti-rLimB antibodies, showed minor and inconsistent protein A–gold binding to the bacteria (Fig. 8a).
Fig. 8.
Immunoelectron and immunofluorescence microscopy showing the surface location of LimB in Coxiella. (a) Typical immunoelectron micrographs showing uniform protein A–gold binding to the surface of intact C. burnetii treated with anti-LimB or anti-Com1 (positive control for a surface-exposed protein) antibodies. Protein A–gold binding was rarely observed when negative controls were employed, including PIS, anti-MBP or anti-EF-Ts antibodies. (b) Typical immunofluorescence micrographs showing intense fluorescence at the surface of paraformaldehyde-fixed C. burnetii treated with antibodies against LimB or Com1 versus relatively minor, sporadic fluorescence when negative controls, as in (a), were employed.
Finally, we employed these antibodies and PIS in immunofluorescence microscopy with Coxiella cells that had been fixed with paraformaldehyde. This procedure was previously employed to detect surface-exposed proteins of Ehrlichia and Anaplasma species (Ge & Rikihisa, 2007). Results of these experiments correlated with the immunoelectron microscopy data, and showed prominent immunofluorescence when Coxiella was treated with anti-rLimB or anti-Com1 antibodies, and minor, sporadic immunofluorescence when negative-control antibodies or PIS were used (Fig. 8b). Taken together, the Sarkosyl insolubility and detection of LimB on the surface of live cells by immunoelectron microscopy and on paraformaldehyde-fixed cells by immunofluorescence microscopy strongly suggest that LimB is a surface-exposed omp in Coxiella.
DISCUSSION
In this study, we present data on the discovery and partial characterization of Coxiella LimB, a surface-exposed omp that binds divalent metal cations, including nickel and zinc with high affinity, and cobalt and iron with relatively lower affinity. Although assigning specific roles to LimB requires additional investigation regarding the protein’s structure and ligand-binding characteristics, the results of this study allow us to formulate reasonable hypotheses. First, maximal expression of the limB gene occurs during exponential phase growth, suggesting that LimB and its ligand(s) may be important for replication. Second, LimB’s surface location suggests that its metal ligand is obtained extracellularly. Third, because the replication of Coxiella in nature is restricted to an acidified, lysosome-like compartment of the host cell, termed a parasitophorous vacuole (PV) (Voth & Heinzen, 2007), it is logical to assume that the ligand(s) is present in the PV niche. Fourth, it is plausible that LimB activity in the PV generates an active competition with the host cell, especially if the ligand is limiting in concentration. Based upon these results, we hypothesize that LimB is a lipoprotein that serves as a receptor for a divalent metal cation(s) at the surface of the pathogen during active replication within host cells. Considering that bio-reactive metal acquisition is often key to bacterial replication (Ratledge & Dover, 2000), LimB may play a role in the pathogenesis of C. burnetii.
Gram-negative bacteria employ a set of highly conserved proteins to process and transport lipoproteins (Babu et al., 2006; Hantke & Braun, 1973). Prolipoprotein precursors are synthesized in the cytosol and then translocated through the cytosolic membrane via a Sec translocon [or Tat for proteins with a type I signal sequence (Hutchings et al., 2009)]. Once across, a phosphatidylglycerol:prolipoprotein diacylglyceryl transferase (Lgt) catalyses addition of a DAG moiety to the thiol group of the lipobox Cys residue. Subsequently, a lipoprotein-specific signal peptidase (Lsp; also called signal peptidase II) cleaves the signal peptide at the N-terminal side of lipobox Cys. Finally, a phospholipid:apolipoprotein transacylase (Lnt) covalently attaches a fatty acid to the amide group of the N-terminal Cys to create the mature lipoprotein (Tokuda, 2009) (see Fig. 2). The ELR at position 2 of mature lipoproteins plays an important role in coordinating the protein’s transport to its final destination in the cell (Yamaguchi et al., 1988). Namely, mature lipoproteins that possess a charged ELR (especially Asp) are retained in the cytosolic membrane during transport, whereas those with an uncharged ELR (e.g. the Ala of LimB; Fig. 2) are translocated to the outer membrane with the assistance of Lol sorting-system proteins in the periplasm (Tokuda, 2009; Yamaguchi et al., 1988). Based upon characteristics of the predicted LimB protein (Fig. 2) we hypothesize that pro-LimB follows a typical Sec-dependent (type II signal peptide) pathway wherein its 20-residue type II signal sequence would be cleaved by an LspA-like protein, its resulting N-terminal Cys is lipidated by Lgt and Lnt homologues, and the Ala ELR helps coordinate translocation of mature LimB to the outer membrane with the assistance of a Lol-like sorting-system. In fact, the surface location determined for LimB (Figs 7 and 8) is in keeping with the hypothesized outer membrane location of LimB, as predicted from its Ala ELR (Fig. 2).
Although the exact role of LimB in the biology of Coxiella remains to be determined, its demonstrated surface exposure and potentially chimeric nature (a protein with lipids) would be attractive features for a subunit vaccine candidate. In addition, the biosynthetic pathway of LimB might also provide an alternative strategy for therapeutic intervention during chronic Q fever; specifically, lipoprotein secretion could conceivably be inhibited by drugs like globomycin. Globomycin, a cyclic pentapeptide antibiotic, has been shown to inhibit secretion of lipoproteins in a wide variety of pathogens by inhibiting Lsp function (Geukens et al., 2006; Kiho et al., 2004; Rahman et al., 2007). Further investigation of LimB and other surface-exposed metal-binding proteins of Coxiella is expected to yield clues regarding the poorly characterized pathogenesis of this highly infectious agent. An illustration of LimB lipoprotein maturation as well as several hypothetical functions are provided in Supplementary Fig. S1, available with the online version of this paper.
Acknowledgments
We are grateful to Michele McGuirl for helpful suggestions and assistance with experimental design. Assistance with LAS-3000 digital imaging was kindly provided by Laura Hall and Meghan Lybecker. Monoclonal antibodies against Coxiella EF-Ts were generously provided by Robert Heinzen. The technical assistance of Jim Driver, Rahul Raghavan and Kate Sappington is greatly appreciated. This work was supported by National Institutes of Health (NIH) Rocky Mountain Regional Center of Excellence (RMRCE) grant U54-AI065357-040023 to M. F. M. and an NIH COBRE (P20-RR020185) project to J. M. B.
Abbreviations
CBB, Coomassie brilliant blue
EF-Ts, elongation factor-Ts
ELR, envelope localization residue
FW, far-Western
HRP, horseradish peroxidase
LCV, large cell variant
MBP, maltose-binding protein
Ni-HRP, nickel-coated horseradish peroxidase
NM, Nine Mile
qRT-PCR, quantitative real-time PCR
PII, ‘phase II’
PIS, pre-immune serum
SCV, small cell variant
Footnotes
A supplementary figure, showing synthesis of and possible roles for LimB, is available with the online version of this paper.
References
- Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (editors) (1995). Current Protocols in Molecular Biology. New York. : Wiley.
- Babu, M. M., Priya, M. L., Selvan, A. T., Madera, M., Gough, J., Aravind, L. & Sankaran, K. (2006). A database of bacterial lipoproteins (DOLOP) with functional assignments to predicted lipoproteins. J Bacteriol 188, 2761–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beare, P. A., Unsworth, N., Andoh, M., Voth, D. E., Omsland, A., Gilk, S. D., Williams, K. P., Sobral, B. W., Kupko, J. J., III & other authors (2009). Comparative genomics reveal extensive transposon-mediated genomic plasticity and diversity among potential effector proteins within the genus Coxiella. Infect Immun 77, 642–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, C. T., Niemela, S. L. & Miller, R. H. (1989). One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci U S A 86, 2172–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockrell, D. C., Beare, P. A., Fischer, E. R., Howe, D. & Heinzen, R. A. (2008). A method for purifying obligate intracellular Coxiella burnetii that employs digitonin lysis of host cells. J Microbiol Methods 72, 321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, S. A., Fischer, E. R., Howe, D., Mead, D. J. & Heinzen, R. A. (2004). Temporal analysis of Coxiella burnetii morphological differentiation. J Bacteriol 186, 7344–7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Ramírez, G., Toman, R., Sekeyova, Z. & Skultety, L. (2009). In silico prediction and identification of outer membrane proteins and lipoproteins from Coxiella burnetii by the mass spectrometry techniques. Clin Microbiol Infect 15 (Suppl. 2), 196–197. [DOI] [PubMed] [Google Scholar]
- Fortney, K. R., Young, R. S., Bauer, M. E., Katz, B. P., Hood, A. F., Munson, R. S., Jr & Spinola, S. M. (2000). Expression of peptidoglycan-associated lipoprotein is required for virulence in the human model of Haemophilus ducreyi infection. Infect Immun 68, 6441–6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, Y. & Rikihisa, Y. (2007). Surface-exposed proteins of Ehrlichia chaffeensis. Infect Immun 75, 3833–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geukens, N., De Buck, E., Meyen, E., Maes, L., Vranckx, L., Van Mellaert, L., Anné, J. & Lammertyn, E. (2006). The type II signal peptidase of Legionella pneumophila. Res Microbiol 157, 836–841. [DOI] [PubMed] [Google Scholar]
- Hantke, K. & Braun, V. (1973). Covalent binding of lipid to protein. Diglyceride and amide-linked fatty acid at the N-terminal end of the murein-lipoprotein of the Escherichia coli outer membrane. Eur J Biochem 34, 284–296. [DOI] [PubMed] [Google Scholar]
- Hayashi, S. & Wu, H. C. (1990). Lipoproteins in bacteria. J Bioenerg Biomembr 22, 451–471. [DOI] [PubMed] [Google Scholar]
- Hicks, L. D., Raghavan, R., Battisti, J. M. & Minnick, M. F. (2010). A DNA-binding peroxiredoxin of Coxiella burnetii is involved in countering oxidative stress during exponential-phase growth. J Bacteriol 192, 2077–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover, T. A., Culp, D. W., Vodkin, M. H., Williams, J. C. & Thompson, H. A. (2002). Chromosomal DNA deletions explain phenotypic characteristics of two antigenic variants, phase II and RSA 514 (crazy), of the Coxiella burnetii nine mile strain. Infect Immun 70, 6726–6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchings, M. I., Palmer, T., Harrington, D. J. & Sutcliffe, I. C. (2009). Lipoprotein biogenesis in Gram-positive bacteria: knowing when to hold ‘em, knowing when to fold ‘em. Trends Microbiol 17, 13–21. [DOI] [PubMed] [Google Scholar]
- Inouye, S., Wang, S., Sekizawa, J., Halegoua, S. & Inouye, M. (1977). Amino acid sequence for the peptide extension on the prolipoprotein of the Escherichia coli outer membrane. Proc Natl Acad Sci U S A 74, 1004–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiho, T., Nakayama, M., Yasuda, K., Miyakoshi, S., Inukai, M. & Kogen, H. (2004). Structure–activity relationships of globomycin analogues as antibiotics. Bioorg Med Chem 12, 337–361. [DOI] [PubMed] [Google Scholar]
- Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Maurin, M. & Raoult, D. (1999). Q fever. Clin Microbiol Rev 12, 518–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaul, T. F. & Williams, J. C. (1981). Developmental cycle of Coxiella burnetii: structure and morphogenesis of vegetative and sporogenic differentiations. J Bacteriol 147, 1063–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moos, A. & Hackstadt, T. (1987). Comparative virulence of intra- and interstrain lipopolysaccharide variants of Coxiella burnetii in the guinea pig model. Infect Immun 55, 1144–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrow, N. L., Abbott, J., Lockwood, A. R., Battisti, J. M. & Minnick, M. F. (2009). Function, regulation, and transcriptional organization of the hemin utilization locus of Bartonella quintana. Infect Immun 77, 307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan, R., Hicks, L. D. & Minnick, M. F. (2008). Toxic introns and parasitic intein in Coxiella burnetii: legacies of a promiscuous past. J Bacteriol 190, 5934–5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman, M. S., Ceraul, S. M., Dreher-Lesnick, S. M., Beier, M. S. & Azad, A. F. (2007). The lspA gene, encoding the type II signal peptidase of Rickettsia typhi: transcriptional and functional analysis. J Bacteriol 189, 336–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratledge, C. & Dover, L. G. (2000). Iron metabolism in pathogenic bacteria. Annu Rev Microbiol 54, 881–941. [DOI] [PubMed] [Google Scholar]
- Regis, E. (1999). The biology of doom: The history of America’s secret germ warfare project. New York. : Henry Holt & Co.
- Rosenzweig, A. C. (2002). Metallochaperones: bind and deliver. Chem Biol 9, 673–677. [DOI] [PubMed] [Google Scholar]
- Samoilis, G., Psaroulaki, A., Vougas, K., Tselentis, Y. & Tsiotis, G. (2007). Analysis of whole cell lysate from the intercellular bacterium Coxiella burnetii using two gel-based protein separation techniques. J Proteome Res 6, 3032–3041. [DOI] [PubMed] [Google Scholar]
- Schwan, T. G., Piesman, J., Golde, W. T., Dolan, M. C. & Rosa, P. A. (1995). Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc Natl Acad Sci U S A 92, 2909–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri, R. & Samuel, J. (2005). Genome analysis of Coxiella burnetii species: insights into pathogenesis and evolution and implications for biodefense. Ann N Y Acad Sci 1063, 442–450. [DOI] [PubMed] [Google Scholar]
- Seshadri, R., Paulsen, I. T., Eisen, J. A., Read, T. D., Nelson, K. E., Nelson, W. C., Ward, N. L., Tettelin, H., Davidsen, T. M. & other authors (2003). Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc Natl Acad Sci U S A 100, 5455–5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha, J., Fadl, A. A., Klimpel, G. R., Niesel, D. W., Popov, V. L. & Chopra, A. K. (2004). The two murein lipoproteins of Salmonella enterica serovar Typhimurium contribute to the virulence of the organism. Infect Immun 72, 3987–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, J. G., Howe, D. & Heinzen, R. A. (2005). Virulent Coxiella burnetii does not activate human dendritic cells: role of lipopolysaccharide as a shielding molecule. Proc Natl Acad Sci U S A 102, 8722–8727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupska, M. M., Chiang, J. H., Luther, W. M., Stewart, J. L., Amii, L., Conrad, A. & Miller, J. H. (2000). Genes involved in the determination of the rate of inversions at short inverted repeats. Genes Cells 5, 425–437. [DOI] [PubMed] [Google Scholar]
- Tokuda, H. (2009). Biogenesis of outer membranes in Gram-negative bacteria. Biosci Biotechnol Biochem 73, 465–473. [DOI] [PubMed] [Google Scholar]
- Towbin, H., Staehelin, T. & Gordon, J. (1979). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A 76, 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodkin, M. H. & Williams, J. C. (1986). Overlapping deletion in two spontaneous phase variants of Coxiella burnetii. J Gen Microbiol 132, 2587–2594. [DOI] [PubMed] [Google Scholar]
- Voth, D. E. & Heinzen, R. A. (2007). Lounging in a lysosome: the intracellular lifestyle of Coxiella burnetii. Cell Microbiol 9, 829–840. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, K., Yu, F. & Inouye, M. (1988). A single amino acid determinant of the membrane localization of lipoproteins in E. coli. Cell 53, 423–432. [DOI] [PubMed] [Google Scholar]
- Zamboni, D. S., McGrath, S., Rabinovitch, M. & Roy, C. R. (2003). Coxiella burnetii express type IV secretion system proteins that function similarly to components of the Legionella pneumophila Dot/Icm system. Mol Microbiol 49, 965–976. [DOI] [PubMed] [Google Scholar]
- Zhang, G., To, H., Russell, K. E., Hendrix, L. R., Yamaguchi, T., Fukushi, H., Hirai, K. & Samuel, J. E. (2005). Identification and characterization of an immunodominant 28-kilodalton Coxiella burnetii outer membrane protein specific to isolates associated with acute disease. Infect Immun 73, 1561–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, H. & Waite, J. H. (2006). Proteins in load-bearing junctions: the histidine-rich metal-binding protein of mussel byssus. Biochemistry 45, 14223–14231. [DOI] [PMC free article] [PubMed] [Google Scholar]