Abstract
Here, we report the identification and functional characterization of the Streptomyces globisporus 1912 gene lndYR, which encodes a GntR-like regulator of the YtrA subfamily. Disruption of lndYR arrested sporulation and antibiotic production in S. globisporus. The results of in vivo and in vitro studies revealed that the ABC transporter genes lndW–lndW2 are targets of LndYR repressive action. In Streptomyces coelicolor M145, lndYR overexpression caused a significant increase in the amount of extracellular actinorhodin. We suggest that lndYR controls the transcription of transport system genes in response to an as-yet-unidentified signal. Features that distinguish lndYR-based regulation from other known regulators are discussed.
INTRODUCTION
Streptomycetes are notable for their ability to produce secondary metabolites that are immensely diverse in terms of structure, biogenesis and the biological responses they elicit. The chromosomes of streptomycetes contain many gene clusters for secondary metabolite biosynthesis, although not all of them are expressed in wild-type strains under routine laboratory conditions (Nett et al., 2009). Those that are expressed usually produce only small quantities of the product, creating the need for extensive strain improvement programs for industrial purposes. These observations indicate that streptomycetes have evolved a complicated regulatory web that orchestrates their secondary metabolism. Indeed, available data underscore a great abundance of regulatory genes in actinomycete genomes.
GntR superfamily regulators, which constitute one of the biggest groups of bacterial regulators, contain a typical helix–turn–helix DNA-binding domain at their N terminus (Hoskisson & Rigali, 2009). This superfamily is named after the prototypical member, GntR, a repressor of the Bacillus subtilis gluconate operon. The Streptomyces coelicolor genome encodes 57 GntR-like regulators (Bentley et al., 2002; Hillerich & Westpheling, 2006), but only a few of these genes have been studied experimentally, namely whiH, devA, dasR and aglR3R. Genes whiH and devA control the early steps of sporulation (Flärdh & Buttner, 2009; Hoskisson et al., 2006), while the other two influence carbohydrate transport and antibiotic biosynthesis (Rigali et al., 2006; Hillerich & Westpheling, 2006). Also, the GntR-like genes korSA (korA) are known to control conjugal transfer of actinomycete plasmids, such as pIJ101 and pSAM2 (Sezonov et al., 2000). Except for these data and bioinformatic predictions (Rigali et al., 2002, 2004), little is known about the role(s) of many other members of the GntR family in streptomyces biology. Our interest in this family arose when we uncovered a new GntR-like gene, lndYR, in the vicinity of the landomycin E (LaE) biosynthesis gene (lnd) cluster. This cluster is an object of intense investigation in our laboratory, with an emphasis on the genetic regulation of LaE production. To date, two regulatory genes have been shown to control LaE production: one encoding streptomyces antibiotic regulatory protein-like regulator LndI and another that specifies putative proteinase Prx (Ostash et al., 2009). The lndYR gene is located between genes for a putative serine-threonine kinase (lndY) and a two-component ABC transporter system (lndW–lndW2; see Fig. 1). While the function of lndY remains unknown, lndW has been shown to confer increased LaE resistance under heterologous overexpression conditions (Ostash et al., 2008). Taking into account the physical linkage of lndYR to other lnd genes and the fact that GntR-like genes often control transport processes (Hoskisson & Rigali, 2009), we set out to investigate the possible functional relationships between lndYR, lndW–lndW2 and LaE production. Here, we report that LndYR influences sporulation and landomycin production in Streptomyces globisporus; significant changes in actinorhodin distribution are also seen in S. coelicolor overexpressing lndYR. As LndYR has numerous homologues in sequenced actinomycete genomes, future studies of this family of regulators may lead to valuable insights into morphogenesis and antibiotic production by members of the order Actinomycetales.
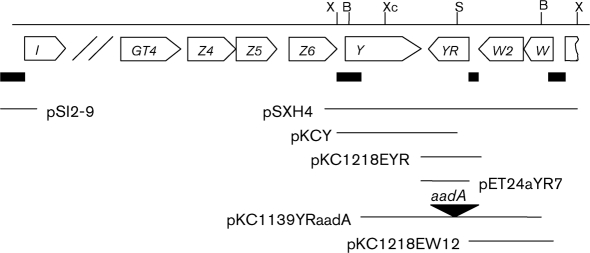
Fig. 1.
Genetic organization of the lnd cluster in the vicinity of the lndY–lndW fragment (not drawn to scale). Subclones used throughout the work are shown below. Restriction sites: X, XhoI; B, BamHI; Xc, XcmI; S, StuI. Black rectangles below gene symbols indicate promoter regions used in this work. The black triangle points to the site of insertion of the spectinomycin resistance cassette (aadA) into the lndYR coding sequence.
METHODS
Bacterial strains and culture conditions.
Bacterial strains and plasmids used in this work are listed in Table 1. Escherichia coli strains were grown at 37 °C in LB, 2×TY or TB for routine applications. Streptomyces strains were grown at 30 °C. Solid oatmeal medium (OM) (Luzhetskiǐ et al., 2001) or mannitol-soy medium (MS) (Kieser et al., 2000) was used for sporulation of streptomycetes and plating of E. coli–Streptomyces matings. For isolation of genomic and plasmid DNA, streptomycetes was grown in TSB for 48–60 h. LaE production by S. globisporus strains was assayed in liquid media SG (Rebets et al., 2003), TSB, R5A and on solid OM (Rebets et al., 2003), Bennett (Shirling & Gottlieb, 1966), MM, MS, R2YE, and MMGT (Kieser et al., 2000). Total RNA was isolated from S. globisporus cultures grown in TSB or R5A (Kieser et al., 2000). Actinorhodin (Act) and prodiginines (Red) production by S. coelicolor was determined in liquid medium YMPG (Ishizuka et al., 1992) as previously described (Kieser et al., 2000). Complex agar YMA (Ostash et al., 2007), MS, Bennett, R2YE, MMGT and chemically defined minimal medium (Kieser et al., 2000) and Evans medium (Evans et al., 1970), modified as described by Sun et al. (2001), were also used to analyse Act and Red production. Moenomycin production by Streptomyces ghanaensis (ATCC14672) was studied in TSB as described by Makitrynskyy et al. (2010). Siomycin production by Streptomyces sioyaensis was determined in medium SG1 as described by Myronovskyy et al. (2009). In all cases, the amount of antibiotic was normalized to equal amounts of biomass (dry weight) and the reported values are the means of three independent experiments. Antibiotic resistance was analysed by using the disc diffusion method.
Table 1.
Strains and plasmids used in this work
| Bacterial strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli DH5α | supE44 λlacU169(Δ80lacZλM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | MBI Fermentas |
| E. coli ET12567 (pUB307) | dam-13 : : Tn9(Cmr) dcm-6hsdM; harbours conjugative plasmid pUB307; Cmr, Kmr | C. P. Smith, UMIST, UK |
| E. coli BL21(DE3) | F−ompThsdS gal [dcm][lon]hsdSB ( |
Novagen |
| S. globisporus 1912 | Wild-type LaE producer | Rebets et al. (2005) |
| S. globisporus YR | A lndYR-disrupted mutant (lndY : : aadA) | This work |
| S. coelicolor M145 | Model strain, ACT and RED producer | M. Bibb, JIC |
| S. ghanaensis ATCC14672 | Wild-type moenomycin producer | ATCC |
| S. sioyaensis NRRL-B5408 | Wild-type siomycin producer | NRRL |
| pBluescriptIISK− | General purpose cloning vector; Apr | MBI Fermentas |
| pSTBlue-1 | E. coli T-cloning vector for PCR products cloning; Apr | Novagen |
| pET24a | Expression vector to purify His-tagged proteins; Kmr | Novagen |
| pKC1218E | E. coli/streptomyces shuttle vector with ermEp and SCP2* replicon, derivative of pKC1218; Amr | Rebets et al. (2005) |
| pKC1139 | E. coli/streptomyces shuttle vector, pSG5 replicon; Amr | Kieser et al. (2000) |
| pHP4 Ω | Plasmid carrying Ω interposon with spectinomycin resistance gene cassette aadA; Apr, Smr/Spr | Prentki & Krisch (1984) |
| pSXH4 | pBluescriptIISK− carrying 6 kb XhoI fragment of lnd cluster (lndY–lndW segment) | Fedorenko et al. (2000) |
| pKCYRaadA | pKC1139 carrying the disrupted allele of lndYR gene (lndYR : : aadA) | This work |
| pKC1218EYR | pKC1218E carrying lndYR behind ermEp | This work |
| pKCY | pKC1139 harbouring lndY gene | This work |
| pSI2-9 | pSET152 containing lndI gene with promoter | Rebets et al. (2005) |
| pET24aYR7 | pET24a carrying lndYR | This work |
| pKC1218EW12 | pKC1218E carrying lndW lndW2 behind ermEp | This work |
DNA manipulations.
Genomic and plasmid DNA from Streptomyces and plasmid DNA from E. coli was isolated using standard protocols (Kieser et al., 2000; Sambrook & Russell, 2001). E. coli transformation and intergenetic E. coli–Streptomyces matings were performed as described by Kieser et al. (2000) and Luzhetskiǐ et al. (2001). Molecular biology enzymes were purchased from standard commercial sources (MBI Fermentas, NEB, Amersham Biosciences, Invitrogen) and used according to the manufacturer's instructions. quantitative PCR (qPCR) experiments were done on a MyiQ single-colour real-time PCR detection system (Bio-Rad). PCRs were performed using thermal cyclers Mastercycler (Eppendorf) and PTC-100 (MJ Research). All amplicons used in this work were verified by sequencing. DNA sequencing was performed by using the Sanger method using an ABI3700xl instrument. Sequence assembly was carried out using Lasergene software (dnastar). The GenBank accession number for fragments containing genes lndY, lndYR, lndW2 and lndW is HM204451.
Bioinformatic analysis of sequences.
FramePlot 2.3.2 (Ishikawa & Hotta, 1999) was used to find open reading frames (ORFs) within a sequenced fragment of the lnd cluster. blast search tools (on the NCBI server) were used to identify homologues of lndY, lndYR and lndW2 genes. The CDD search engine (blast) and HHPred, Pfam and TMHMM on the ExPaSy proteomics server were utilized for in silico analysis. Phylogenetic trees were constructed by using an online phylogenetic server (http://www.phylogeny.fr; Dereeper et al., 2008). The LndYR tree was rooted using Bacillus pumilus ATCC 7061 YtrA protein as an outgroup. The topology of the resultant neighbour-joining tree was evaluated by an approximate-likelihood ratio test (Anisimova & Gascuel, 2006).
Plasmid construction
Sequences of the primers used throughout this work are given in Table 2.
Table 2.
Primers used in this study
| Purpose/name | Sequence (5′–3′) |
|---|---|
| To clone lndYR for pKC1218E | |
| Y2upXbaI | AAATCTAGACCACCGTGTTCGTCTGGGACG |
| Y2rpEcoRI | AAAGAATTCTCAGTCCGGCGCCTCGC |
| To clone lndW1W2 | |
| W12XbaF | ATTCTAGATGGAATCACGATCACGGATCA |
| W12EcIR | ATGAATTCCGCTCGATTGCCGACACTT |
| To clone lndYR for pET24a | |
| LdY2EF | TACATATGCGTAAAGTGTCGGCAATCG |
| LdY2ER | TACTCGAGGTCCGGCGCCTCGCCCG |
| RT-PCR analysis of lnd genes expression | |
| LdIF | ATACTTCGCACACCGTGTAG |
| LdIR | AGGTCGAGGAGAACTAGGT |
| LdEF | ACGCTGCGGTGATCGTTGT |
| LdER | TGGCCTTGCGTACTGGTCT |
| LdG4F | TCGAGTTCAACTCGAACCAG |
| LdG4R | AGCATGCCGTCCATCAGG |
| LdJF | CGTGGTCGCGATGTTACTC |
| LdJR | CGGAAGCCGATGAGCTGG |
| LdYF | CTTCCGTCGGGAGATCGA |
| LdYR | CGATGCCGAAGTCGATGAC |
| LdY2F | AAGGTGAAGTCCGTGGTCG |
| LdY2R | GCGTTCCAGCTCGCGGTA |
| LdWF | CTGTTCAAGCGGTTCACCG |
| LdWR | AGGACATCACCACCGTGGT |
| rrnAF | CACATGCAAGTCGAACGATG |
| rrnAR | GCTGCTGGCACGTAGTTAG |
| Promoter fragments for EMSA | |
| W12PR | TCGGTTCCGTGATCCGTGAT |
| W12PF | AGCGTGCCGTCGAAGTCGA |
| LdWF1 | GTGATTCCATGAATCAATTAATG |
| LdWF2 | AATGGAATCATGGAGTCGTGA |
| LdWF3 | GTGAGGGAATGACTGTCAAG |
| LdWF4 | CAGTGCCCAGTTGCCGGA |
| LdWR4 | CTTGACAGTCATTCCCTCAC |
| LdWF5 | CTGTCAAGCCGGGGTGGT |
| LdWF6 | TGAATCAATTAATGGAATCATGG |
| ldYRF | TTGCCGACACTTTACGTAGG |
| ldYRR | GTCAACGACGTGATCGAGTA |
| ldYPF | ATCCGCAGGAAGCGAGAGC |
| ldYPR | TCGTGCACCACCTTCACGG |
| ldIPF | CGAACCACCGGAAATTGATC |
| ldIPR | CTGCCGTCTCATTTACTACC |
| Intact fragment | |
| intact-up | TGACAGTCATTCCCTCACGACTCCATGATTCCATTAATTGATTCATGGAATCACGATCA |
| Mutant versions of intact fragment (only one strand is shown for each duplex) | |
| Δs1s2s3-up | TGACAGAAAAAACCTCACGACTCCAAAAAAACATTAATAAAAAAATGGAATCACGATCA |
| Δs1s2s3s4-up | TGACAGAAAAAACCTCACGACTCCAAAAAAACATTAATAAAAAAATGAAAAAACGATCA |
| Δs4-up | TGACAGTCATTCCCTCACGACTCCATGATTCCATTAATTGATTCATGAAAAAACGATCA |
| Δs1-up | TGACAGAAAAAACCTCACGACTCCATGATTCCATTAATTGATTCATGGAATCACGATCA |
| Δs2s3-up | TGACAGTCATTCCCTCACGACTCCAAAAAAACATTAATAAAAAAATGGAATCACGATCA |
For lndYR expression.
A 798 bp fragment containing lndYR and its 400 bp upstream region was amplified from pSXH4 (Fedorenko et al., 2000) with primers Y2upXbaI and Y2rpEcoRI. The resulting amplicon was digested with XbaI and EcoRI and cloned into the same sites of pKC1218E to give pKC1218EYR.
For lndY expression.
The lndY gene was amplified as a 4.5 kb XhoI–StuI fragment from pSXH4, treated with Klenow fragment and cloned into EcoRV-digested pKC1139 to give pKCY.
For lndW–lndW2 expression.
The genes lndW–lndW2 were amplified by using primers W12XbaF and W12EcIR, digested with XbaI and EcoRI and cloned into the respective sites of pKC1218E resulting in pKCEW12. In this construct, the cloned lndW–lndW2 segment contains its own ribosome-binding site (15 bp upstream of lndW) and its expression is driven only by ermEp.
For lndYR knockout.
A 3.5 kb BamHI fragment of pSXH4 (containing lndY, lndYR, lndW2 and lndW lacking its first 119 bp; Fig. 1) was cloned into BamHI-digested pBluescriptIISK− to give pBL35. The latter contains a unique StuI site within the lndYR coding region, which was utilized to clone SmaI-digested spectinomycin resistance cassette aadA to yield pBL35StuIaadA. The mutant lndYR : : aadA allele was moved as a 5.5 kb BamHI fragment from pBL35StuIaadA to pKC1139 to give the final construct pKCYRaadA.
For lndYR expression in E. coli.
The lndYR gene was amplified with primers LdY2EF and LdY2ER, digested with NdeI and XhoI and cloned into the respective sites of pET24a to give pET24aYR7.
RNA isolation and reverse transcriptase (RT)-PCR analysis.
Total RNA was isolated after 3 days of S. globisporus growth in TSB or R5A according to the Trizol manufacterer's instructions (Invitrogen). To avoid DNA contamination, samples were treated with RQ1 DNase (Promega). RNA concentration and purity were determined by measuring the ratio of OD260 : OD280, and an equal amount of RNA from each studied strain was used for the RT reactions. cDNA was obtained by using a cloned AMV first-strand synthesis kit (Invitrogen) and random hexanucleotide primers. PCR was performed using Taq DNA polymerase (NEB) and primer pairs specific to each individual lnd gene (Table 2). As a positive control, the rrnA primer pair specific to 16S rRNA of S. globisporus was used. Negative controls were done with rrnA primers to confirm the absence of contaminating DNA in the RNA preparations. The PCR parameters for quantitative analysis were as follows: initial denaturation for 5 min at 95 °C, then 35 extension cycles (30 s, 95 °C; 30 s, 53 °C; 30 s, 72 °C) and final denaturation with a gradient of 0.5 °C step every 10 s. CT was calculated to be 26.0 using the maximum curvation method, and the relative ratios of gene expression were determined. PCR products were analysed by electrophoresis on a 1.5 % agarose gel, and band intensities were measured by using the ImageJ1.36b software (NIH). For qPCR, the QuantiTect SYBR green PCR kit (Qiagen) was used. Data analysis was performed by using the comparative CT (2 –ΔΔCT) method using the 16S rRNA signal as an internal reference (Livak & Schmittgen, 2001).
LndYR purification.
To obtain C-terminally 6×His-tagged LndYR, an E. coli BL21(DE3) strain carrying plasmid pET24aYR7 was grown overnight at 37 °C. LB (400 ml) containing 50 μg kanamycin ml−1 was inoculated with 1 ml of overnight culture and incubated at 37 °C until OD600 0.6. LndYR expression was then induced with 0.4 mM IPTG. After incubation for an additional 6 h at 25 °C, the culture was harvested by centrifugation at 4200 r.p.m. for 10 min at 4 °C and lysed by treatment with BugBuster protein extraction reagent supplemented with Benzonase and rLysozyme (Novagen). Native purification of the LndYR protein on His-Tag binding resin was performed according to Novagen's instructions. LndYR was eluted with 200 mM imidazole and dialysed overnight at 4 °C against the storage buffer (50 mM Tris/HCl, pH 7.5, 100 mM KCl, 1 mM MgCl2, 10 % glycerol). Protein was stored at –80 °C.
Electrophoretic mobility shift assays (EMSAs).
Four gene fragments upstream of lndI (lndIp; 242 bp), lndY (lndYp; 293 bp), lndW (lndWp; 294 bp) and the lndYR–lndW2 intergenic region (lndYRp; 169 bp) were used in DNA binding assays. The DNA fragments were amplified from plasmid pSXH4 or pSI2-9 (Fig. 1) using primers described in Table 2. One DNA binding assay contained 40 pM DNA fragment and 0–250 pM LndYR protein in a total volume of 30 μl binding buffer [50 mM Tris/HCl, pH 7.5, 100 mM KCl, 2 mM MgCl2, 10 % glycerol, 0.5 μg poly(dA)-poly(dT)]. After incubation for 20 min at room temperature, protein-bound and free DNA were resolved by electrophoresis at 4 °C on a 5–8 % non-denaturing polyacrylamide gel in 0.5×TBE buffer. The gel was stained with ethidium bromide and analysed with a UV-imaging system (Fluorochem 5330). Short fragments of lndWp were also tested to map LndYR binding sites (see Results and Supplementary Fig. S1, available with the online version of this paper), and competitor was not included in these experiments.
RESULTS
Sequencing and in silico analysis of novel regulatory genes in the lnd cluster of S. globisporus 1912
Sequencing of a 4 kb segment between lndW and lndZ6 (Fig. 1) revealed three ORFs, referred to as lndY, lndYR and lndW2. Typically for actinomycetes, these genes are GC-rich (68–73 %) and extremely biased in GC usage in the third codon position (over 90 %). LndY (690 aa) showed local similarity to several known serine/threonine protein kinases from S. coelicolor M145 [SCO3621, SCO4487 (PkaG) and SCO4507; more than 60 % similarity over a 300 aa stretch] and Streptomyces granaticolor (68 % similarity over a 272 aa segment) (Vomastek et al., 1998). The 5′ end of lndW2 overlaps by 4 nt with the stop codon of lndW. LndW2 (331 aa) is homologous to putative transmembrane subunits of ABC transporters (Méndez & Salas, 2001), and is likely to cooperate with the ATPase subunit LndW to produce a functional two-component ABC transporter (Ostash et al., 2008). According to blast and CDD search results, lndYR encodes a GntR-superfamily transcriptional regulator, which belongs to the YtrA subfamily of GntR-like regulators. Like other members of this subfamily (Rigali et al., 2002), LndYR is small (144 aa) and contains a greatly reduced C-terminal effector binding/oligomerization domain. LndYR homologues are present in all sequenced genomes of members of the order Actinomycetales. Moreover, LndYR-like genes are part of a genomic segment showing good local synteny, e.g. they are located near putative ABC transporter genes, which are the usual targets of YtrA-like proteins (Hoskisson & Rigali, 2009). The YtrA subfamily remains the least studied GntR subgroup due to low sequence similarity and the scarcity of structural data for the operator sites. To our knowledge, no experimental work has been carried out to date on LndYR or any of its homologues. The dense location of lndW, lndW2 and lndYR suggests their transcriptional coupling.
We analysed phylogenetic relationships within a group of 18 LndYR homologues of streptomycete origin. The LndYR tree (see Supplementary Fig. S2, available with the online version of this paper) consists of two major clades, one of which contains LndYR and its orthologues (Streptomyces griseus SGR3238 and proteins found in partially sequenced genomes of Streptomyces roseosporus, Streptomyces albus J1074, Streptomyces flavogriseus, Streptomyces sp. C1, Streptomyces. sp. SPB8). The second, larger clade of LndYR paralogues includes S. coelicolor SCO0823 and 10 other proteins (Supplementary Fig. S2). Several genomes (S. griseus, S. roseosporus and S. albus) contain both paralogues and orthologues of LndYR, suggesting that gene duplications shaped the evolution of this group of regulators.
An S. globisporus lndYR mutant is deficient in antibiotic production and sporulation
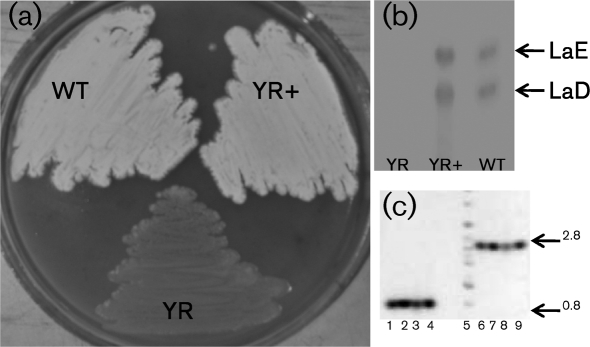
lndY, lndYR and lndW–lndW2 were overexpressed individually under the control of ermEp in S. globisporus, but did not exert any significant effect on landomycin production (in liquid and solid media), resistance to antibiotics or sporulation compared with a control strain carrying the empty vector (data not shown). The lndYR disruption was done via insertional inactivation (Kieser et al., 2000), using the temperature-sensitive plasmid pKCYRaadA. The spectinomycin-resistant and apramycin-sensitive (Spr Ams, respectively) strain S. globisporus YR, in which lndYR is replaced with the lndYR : : aadA allele, was selected, and the gene replacement event was verified by PCR amplification using primers Y2upXbaI and Y2rpEcoRI. A 0.8 kb DNA fragment corresponding to lndYR was amplified from strain 1912, whereas a 2.8 kb DNA fragment was amplified from strain YR (Fig. 2c), indicating that the wild-type copy of lndYR had been inactivated by insertion of aadA.
Fig. 2.
Phenotype of the S. globisporus lndYR mutant. (a) Lawns after 10 days growth of lndYR mutant (YR), wild-type (WT) and complemented YR (YR+) on solid OM. (b) Thin layer chromatography of ethyl acetate extracts from YR, YR+ and WT strains grown in liquid SG medium for 4 days. Arrows indicate spots corresponding to LaE and its disaccharide precursor, LaD. (c) Amplicons obtained with lndYR-specific primers from pKC1218EYR (lane 1), from WT genome (lanes 3–4, independent clones), pKC1139YRaadA (lane 6) and the YR genome (lanes 7–9, independent clones). Lane 5, DNA ladder (Bionexus); sizes given in kb.
The lndYR-deficient S. globisporus YR differed from the 1912 parental strain in both morphology and landomycin production (Fig. 2). YR did not accumulate LaE or its aglyca during submerged fermentation (Fig. 2b) or growth on agar plates. Poor aerial mycelia were formed after 10 days of growth on solid OM, and sporulation, confined to the edges of the lawn, was scant and sporadic (Fig. 2a). There were no differences in the antibiotic resistance profiles of 1912 and YR. Expression of an intact lndYR copy fused to ermEp (plasmid pKC1218EYR) in the YR strain restored its ability to sporulate and produce LaE (Fig. 2) to the levels of its parent strain, thus ruling out the possibility of polar effects in the YR mutant and unexpected rearrangements in the YR genome. We grew the YR strain on a variety of minimal and complex solid media, and none of them restored sporulation to the strain or stimulated LaE production (see Supplementary Fig. S3, available with the online version of this paper). Since the loss of lndYR function has a strong dual phenotype, affecting both morphology and antibiotic production, we conclude that LndYR is not simply a pathway-specific regulator.
Expression of lndW is strongly increased in the lndYR-deficient strain
We performed semiquantitative RT-PCR analysis of representative landomycin biosynthetic (lnd) genes from two main lnd operons: the oxygenase lndE operon (lndFABCD) and the glycosyltransferase lndGT4 operon (lndZ1Z3GT4Z4Z5). We monitored the proton-dependent landomycin antiporter lndJ, the ABC transporter lndW, and regulators lndI, lndY and lndYR in both the parent and lndYR mutant to determine the effect(s) of LndYR deficiency on the expression of those genes. The strains were grown in liquid TSB (supports low landomycin production) and R5A (supports high landomycin production). Despite the lack of landomycin production in the mutant, transcripts for all tested lnd genes were observed in this strain. However, there was a dramatic increase in lndW transcription in the YR strain (Supplementary Fig. S4, available with the online version of this paper). Hence, we focused on qPCR analysis of lndW and lndI gene expression to further assess this phenomenon. In TSB, the amount of lndW mRNA in the YR strain was 150±9 times higher than in 1912; in R5A, the difference was approximately 430-fold. There was no significant difference in lndI expression when YR and 1912 were cultivated in TSB. On R5A, lndI showed increased expression but the difference compared with the parent strain was not nearly as dramatic (two- to threefold; data not shown). These results imply that LndYR acts as repressor of lndW and all contranscribed genes, most likely lndW2 and lndYR. Although LndYR also affects lndI transcription, the latter does not seem to be a primary LndYR target. The cessation of landomycin production by the YR strain is not likely to be a direct result of disrupting repressive LndYR interactions with promoters of structural or regulatory lnd genes, since their expression levels are either unchanged or increased. Rather, it could stem from deregulated expression of lndW–lndW2 and/or other, as-yet-unknown LndYR-dependent genes. For example, LndYR may influence the expression of genes that control landomycin production at a post-transcriptional level (mRNA translation, post-translational Lnd protein modification through phosphorylation or proteolysis). The existence of such regulatory mechanisms is not inconceivable since we have previously shown the essentiality of the putative proteinase gene prx for LaE production (Dutko et al., 2006).
LndYR binds to the promoter regions of lndW and lndI
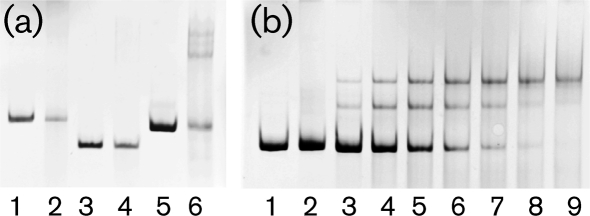
To confirm DNA binding activity and establish the specificity of LndYR, we purified the protein and performed in vitro binding assays using several DNA fragments (Fig. 1) containing the upstream regions of lndI (lndIp), lndY (lndYp), lndW (lndWp) and the lndYR–lndW2 intergenic region (lndYRp). No DNA shift was detected when using lndYp or lndYRp, whereas significant DNA retardation occurred when lndWp was used in the binding assay (Fig. 3); LndYR also interacted with lndIp (Fig. 3 and Supplementary Fig. S5, available with the online version of this paper). The presence of several discrete shifted bands in the EMSA assays may stem from the availability of several binding sites (as detailed further in this work) as well as the ability of LndYR (like many other GntR proteins), to bind DNA as both a mono- and an oligomer. In regard to the latter, we noted that even on denaturing gels, a portion of purified LndYR appeared to move as a dimer and, probably, a trimer (Supplementary Fig. S6); this observation was confirmed via FPLC and Western blot (data not shown). Several experiments demonstrated the specificity of LndYR binding to lndWp and lndIp. For instance, LndYR did not bind all of the four tested promoters, and increased amounts of the protein caused a more complete conversion of free (unbound) lndWp into the LndYR-bound form (Fig. 3). Randomly chosen DNA did not interact with LndYR or compete for binding (Supplementary Fig. S1).
Fig. 3.
EMSAs of DNA fragments containing different lnd promoters. (a) Binding of LndYR (250 pM) to lndYp (4 pM) lndYRp (40 pM) or lndIp (4 pM). Lanes: 1, lndYp; 2, lndYp+LndYR; 3, lndYRp; 4, lndYRp+LndYR; 5, lndIp; 6, lndIp+LndYR. (b) Retardation of lndWp DNA (6 pM) in the presence of increasing concentrations of LndYR. Lanes: 1, lndWp; 2, lndWp+cell lysate of BL21(DE3); 3–9, lndWp+LndYR 16, 40, 80, 110, 160, 200 and 250 pM, respectively.
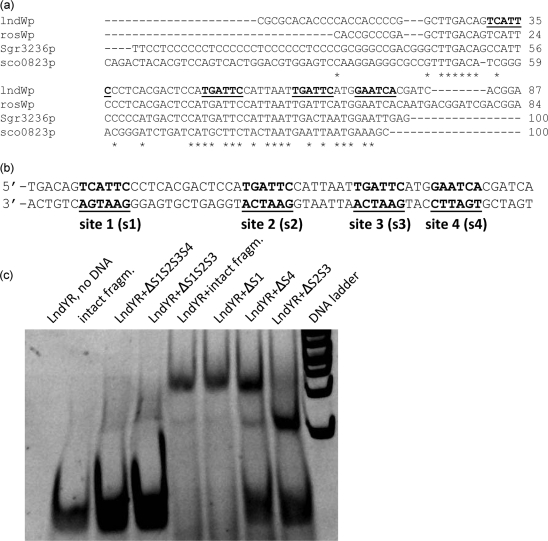
We performed multiple sequence alignments of DNA regions that are putative targets for LndYR or its homologues. Such regions are within promoters located upstream of lndW, its orthologues from S. griseus (sgr3236) and S. roseosporus (ZP_04710279), and upstream of the lndYR paralogue sco0823. All sequences aligned well and revealed the presence of a moderately conserved 80 bp region upstream of the lndW start codon (Fig. 4a). This region did not seem to contain the consensus sequence that is proposed to be recognized by YtrA-like proteins [N(x)GTN(y)TAN(z)TAN(y)ACN(x)]. Nevertheless, visual inspection of the alignment revealed several elements of symmetry. Particularly, an array of imperfect direct and inverted hexanucleotide repeats, embedded into an AT-rich sequence, is present in promoters from orthologous groups (Fig. 4a). Using small overlapping fragments of lndWp, we decided to map the LndYR cis-acting element within lndWp. Our strategy is outlined in Supplementary Fig. S7 (available with the online version of this paper). We found that a short lndWp segment (approx. 60 bp; located 25 bp upstream of the lndW start codon), shown in Fig. 4(b) and referred to as the ‘intact fragment’, contains all of the elements necessary to interact with LndYR and to form two discrete, electrophoretically retarded DNA species (Supplementary Fig. S7). Shorter fragments formed a single shifted DNA band implying that they lack one of the LndYR binding sites (Supplementary Fig. S7), while longer fragments yielded supershifted bands at higher LndYR concentrations (Supplementary Fig. S7c, d). It is possible that LndYR may have additional binding sites far upstream of lndW.
Fig. 4.
The lndW upstream region contains several LndYR binding sites. (a) Multiple alignment of DNA segments located immediately upstream of genes lndW, sgr3236, the lndW homologue from S. roseosporus (rosWp) and sco0823. Asterisks show positions conserved in all four sequences. Imperfect direct repeats are shown in bold and underlined. (b) Sequence of the minimal intact fragment of lndWp (in double-stranded form) that binds LndYR, as defined by EMSAs of lndWp subclones (see Methods and Supplementary Fig. S7). Repeats are marked as s1, s2, s3 and s4 in the intact fragment. (c) Retardation of the intact fragment and its mutant versions carrying the replacement of one or several repeats with hexaadenine sequence. For example, Δs1s2s3s4 is an intact fragment mutant where all four sites (s1, s2, s3, s4) have been replaced with adenines. In all experiments, LndYR was added in molar excess (50 pM, final concentration) with regard to DNA (350 fM).
Having defined a relatively short LndYR DNA target, we set out to determine the nature and importance of the direct and inverted repeats underpinned by bioinformatic analysis. For this purpose, we carried out EMSAs with the intact fragment and its mutant variants where repeats were replaced with adenines. To this end, we tested several combinations of repeat deletions, as shown on Fig. 4(c). While the removal of a single repeat had little effect on its ability to be recognized by LndYR, deletions of two, three or all four sites demonstrated their importance for LndYR binding. A detailed investigation of the intact fragment can now be performed to obtain more precise information about LndYR cis-acting elements.
lndYR affects antibiotic production in heterologous strains
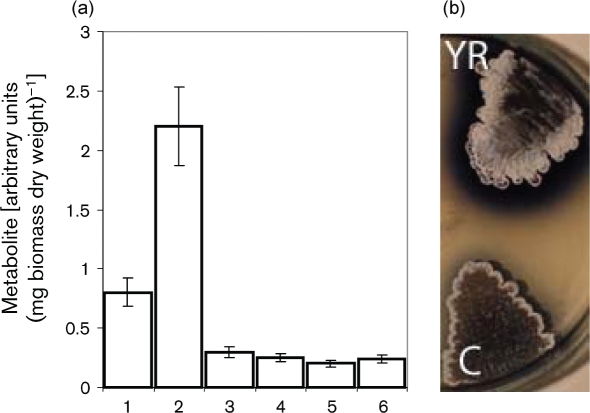
To test whether LndYR influences morphogenesis and secondary metabolism in other streptomycetes, we overexpressed it in the model strain S. coelicolor M145 and in two producers of industrially important antibiotics, S. ghanaensis ATCC14672, which produces moenomycins, and S. sioyaensis NRRL-B5408, which produces siomycins. S. coelicolor M145 carrying plasmid pKC1218EY (M145-YR) was grown in parallel with a control strain (carrying an empty vector) and the amount of Act and Red in the cells and spent medium was determined. On average, both strains yielded a similar amount of biomass, but M145-YR accumulated 2.5 times more Act in the supernatant than the control strain (Fig. 5). M145-YR and its control strain differed negligibly in cellular amounts of Act and Red. Overexpression of lndYR influenced Act production in S. coelicolor during growth on certain solid media. For example, on MMGT plates, strain M145-YR accumulated more Act than did M145 (Supplementary Fig. S8, available with the online version of this paper), probably because M145-YR entered the Act production phase earlier (Supplementary Fig. S8). On replete medium R2YE, M145-YR produced more Act over time (Supplementary Fig. S9). We also grew M145-YR on chemically defined Evans medium and found that on complete, carbon- and phosphate-limited agar plates, M145-YR produced more Act than did M145, while nitrogen-limited medium interfered with Act biosynthesis both in M145-YR and in the control strain (Supplementary Fig. S10). Hence, lndYR overexpression appears to affect the export of Act, and its regulatory function is dependent on nutritional status. As a further illustration of the complexity of biological effects caused by lndYR, we discovered that its overexpression in S. sioyaensis reduced total siomycin production [wild-type, 2.2±0.5 μg (mg dry weight)–1; lndYR+ strain, 1.0±0.2 μg (mg dry weight)–1], while it slightly increased moenomycin titres in S. ghanaensis [wild-type, 0.9±0.3 μg (mg dry weight)–1; lndYR+ strain, 1.5±0.2 μg (mg dry weight)–1].
Fig. 5.
Overexpression of lndYR in S. coelicolor M145. (a). Production of Act and Red by S. coelicolor strains after 5 days of incubation in YMPG. 1, 2, extracts of Act from spent medium of control and lndYR+ strains, respectively; 3, 4, cell extracts of Act from control and lndYR+ strains, respectively; 5, 6, extracts of Red from control and lndYR+ strains, respectively. (b) Patches of M145-YR (YR) and control (C) S. coelicolor strains grown for 5 days on MMGT.
DISCUSSION
We have identified and characterized a new gene, lndYR, encoding a GntR-like regulator (YtrA subfamily) from the LaE producer S. globisporus 1912. LndYR homologues are found in most actinomycete genomes, making LndYR an attractive research target. Our studies have revealed the pleiotropic nature of this gene since an S. globisporus lndYR mutant was deficient in both morphogenesis and antibiotic production, and these processes were also significantly changed in S. coelicolor overexpressing lndYR. Genes within the lndW operon (W-W2-YR) are targets of the LndYR repressor, but most likely not a single major one: the ability of LndYR to bind lndIp and to form lndWp supershifted bands indirectly supports this suggestion. Since increased transcriptional activity of lndW–lndW2 does not seem to be the immediate cause of the drastic changes in sporulation and landomycin production in the YR strain, other LndYR-regulated genes must exist in the genomes of S. globisporus and other species that carry lndYR orthologues. Their discovery will be expedited by precise mapping of LndYR operator sequences and further in silico screening for genes containing similar motifs. Limited experimental data and comparative genomic analyses point to the fact that YtrA proteins are repressors involved in the regulation of transport processes (Hoskisson & Rigali, 2009), and our experiments with S. globisporus and S. coelicolor adhere to this hypothesis. We suggest that, besides influencing the expression of adjacent genes lndW–lndW2 and maybe lndYR, LndYR also regulates the transcription of other, currently unidentified, transporter genes. Their inappropriate expression in the lndYR mutant may produce its phenotype.
In its influence on morphology and secondary metabolism, lndYR resembles gene agl3R (SCO7168), a GntR-type regulator that controls ABC transporter genes in S. coelicolor and responds to the presence of α-glucosides (e.g. trehalose and mellibiose). It is interesting to note that lndW orthologues in S. roseosporus and S. griseus genomes are situated near genes for trehalose metabolism. We tested several sugars, including trehalose, as potential LndYR ligands but they did not interact with LndYR (data not shown); more detailed screening for an LndYR ligand(s) is underway in our laboratories. Unlike Agl3R and most of the studied GntR-like regulators, LndYR does not bind the upstream region of its own gene, probably because lndWp is a single point of control of the entire lndW–lndW2–lndYR operon. At the same time, LndYR binds the lndI gene upstream region. The biological significance of this binding is unclear, as our RT-PCR analysis showed only slight changes in the transcription of lndI in the lndYR mutant compared with the parent strain. Nevertheless, the observed binding reinforces the idea that LndYR may regulate several genes scattered throughout the S. globisporus genome, one of which is directly connected to the expression of lnd genes.
Studies on lndW–lndW2 have provided valuable initial information about LndYR binding sequences. A consensus recognition sequence for the YtrA subfamily has been proposed [N(x)GTN(y)TAN(z)TAN(y)ACN(x); bases essential for binding/recognition are shown in bold] based on a small dataset (Rigali et al., 2002; Hoskisson & Rigali, 2009). Our EMSAs of lndWp fragments show that LndYR binding sites are located on a moderately long DNA stretch (approx. 60 bp) that contains several operators. According to sequence analysis, lndWp and several other upstream gene regions, also believed to be regulated by LndYR-like proteins, share a moderately conserved AT-rich motif. We suggest that, in lndWp, an array of direct (T1C/G2A3T4T5C6) and inverted (T1G2A3T4T5C6, G1A2A3T4C5A6) hexanucleotide repeats (see Fig. 4) serve as an LndYR binding site(s). We note that lndIp did not align with lndWp or its homologues, and yet it is bound by LndYR. It is possible that LndYR recognizes DNA elements showing no obvious similarity, which would not be unprecedented (McKenzie & Nodwell, 2007; Hoskisson & Rigali, 2009). Uncovering the variations in LndYR binding sites will be key to finding new members of the lndYR regulon that influence sporulation and secondary metabolism.
Overexpression of lndYR in S. coelicolor caused significant increases in Act production and extrusion, both in liquid medium YMPG and on relatively poor MMGT agar. Sporulation was faster on MMGT as well. In contrast, lndYR did not exert upregulatory effects when M145-YR was grown on rich medium R2YE. These results, as well as the expression of lndYR in moenomycin and siomycin producers, indirectly support the idea that lndYR controls gene expression in response to certain nutrients, as many other GntRs (most notably, agl3R) do. Hence, the use of different media may lead to different results, since additional regulatory pathways (often linked to both morphogenesis and antibiotic production) are turned on.
Acknowledgments
The authors thank Dr Jennifer Campbell (Department of Microbiology and Immunobiology, Harvard Medical School) for manuscript refinement. The work was supported by the Ministry of Education and Science of Ukraine grant Bg-01F (to V. F.) and by National Institutes of Health grant 5R01GM076710 (to S. W.).
Abbreviations
Act, actinorhodin
EMSA, electrophoretic mobility shift assay
LaE, landomycin E
qPCR, quantitative PCR
Red, prodiginines
RT, reverse transcriptase [PCR]
Footnotes
The GenBank/EMBL/DDBJ accession number for the fragments containing genes lndY, lndYR, lndW2 and lndW is HM204451.
Ten supplementary figures and a supplementary table are available with the online version of this paper.
References
- Anisimova, M. & Gascuel, O. (2006). Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol 55, 539–552. [DOI] [PubMed] [Google Scholar]
- Bentley, S. D., Chater, K. F., Cerdeño-Tárraga, A. M., Challis, G. L., Thomson, N. R., James, K. D., Harris, D. E., Quail, M. A., Kieser, H. & other authors (2002). Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147. [DOI] [PubMed] [Google Scholar]
- Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chevenet, F., Dufayard, J. F., Guindon, S., Lefort, V. & other authors (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36, W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutko, L., Rebets, Y., Ostash, B., Luzhetskyy, A., Bechthold, A., Nakamura, T. & Fedorenko, V. (2006). A putative proteinase gene is involved in regulation of landomycin E biosynthesis in Streptomyces globisporus 1912. FEMS Microbiol Lett 255, 280–285. [DOI] [PubMed] [Google Scholar]
- Evans, C. G. T., Herbert, D. & Tempest, D. W. (1970). The continuous culture of microorganisms. Construction of a chemostat. In Methods in Microbiology, vol. 2, pp. 277–327. Edited by Norris, J. R. & Ribbons, D. W.. London, UK. : Academic Press. [Google Scholar]
- Fedorenko, V., Basiliya, L., Pankevych, K., Dubitska, L., Ostash, B., Luzhetskyy, A., Gromyko, O. & Krugel, H. (2000). Genetic control of antitumor antibiotics-polyketides by actinomycetes. Bull Inst Agric Microbiol 8, 27–31. [Google Scholar]
- Flärdh, K. & Buttner, M. J. (2009). Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol 7, 36–49. [DOI] [PubMed] [Google Scholar]
- Hillerich, B. & Westpheling, J. (2006). A new GntR family transcriptional regulator in Streptomyces coelicolor is required for morphogenesis and antibiotic production and controls transcription of an ABC transporter in response to carbon source. J Bacteriol 188, 7477–7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskisson, P. A. & Rigali, S. (2009). Chapter 1: variation in form and function the helix-turn-helix regulators of the GntR superfamily. Adv Appl Microbiol 69, 1–22. [DOI] [PubMed] [Google Scholar]
- Hoskisson, P. A., Rigali, S., Fowler, K., Findlay, K. C. & Buttner, M. J. (2006). DevA, a GntR-like transcriptional regulator required for development in Streptomyces coelicolor. J Bacteriol 188, 5014–5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa, J. & Hotta, K. (1999). FramePlot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G+C content. FEMS Microbiol Lett 174, 251–253. [DOI] [PubMed] [Google Scholar]
- Ishizuka, H., Horinouchi, S., Kieser, H. M., Hopwood, D. A. & Beppu, T. (1992). A putative two-component regulatory system involved in secondary metabolism in Streptomyces spp. J Bacteriol 174, 7585–7594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F. & Hopwood, D. A. (2000). Practical streptomyces genetics. Norwich, UK. : The John Innes Foundation.
- Livak, K. J. & Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔCT) method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Luzhetskiǐ, A. N., Ostash, B. E. & Fedorenko, V. A. (2001). [Intergeneric conjugation of Escherichia coli–Streptomyces globisporus 1912 using integrative plasmid pSET152 and its derivatives]. Genetika 37, 1340–1347 (in Russian). [PubMed] [Google Scholar]
- Makitrynskyy, R., Rebets, Y., Ostash, B., Zaburannyi, N., Rabyk, M., Walker, S. & Fedorenko, V. (2010). Genetic factors that influence moenomycin production in streptomycetes. J Ind Microbiol Biotechnol 37, 559–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie, N. L. & Nodwell, J. R. (2007). Phosphorylated AbsA2 negatively regulates antibiotic production in Streptomyces coelicolor through interactions with pathway-specific regulatory gene promoters. J Bacteriol 189, 5284–5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez, C. & Salas, J. A. (2001). The role of ABC transporters in antibiotic-producing organisms: drug secretion and resistance mechanisms. Res Microbiol 152, 341–350. [DOI] [PubMed] [Google Scholar]
- Myronovskyy, M., Ostash, B., Ostash, I. & Fedorenko, V. (2009). A gene cloning system for the siomycin producer Streptomyces sioyaensis NRRL-B5408. Folia Microbiol (Praha) 54, 91–96. [DOI] [PubMed] [Google Scholar]
- Nett, M., Ikeda, H. & Moore, B. S. (2009). Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep 26, 1362–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostash, B., Saghatelin, A. & Walker, S. (2007). A streamlined metabolic pathway for the biosynthesis of moenomycin A. Chem Biol 14, 257–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostash, I., Ostash, B., Walker, S. & Fedorenko, V. (2007). Proton-dependent transporter gene lndJ confers resistance to landomycin E in Streptomyces globisporus. Genetika 43, 1032–1037. [PubMed] [Google Scholar]
- Ostash, I., Rebets, Y., Ostash, B., Kobylyanskyy, A., Myronovskyy, M., Nakamura, T., Walker, S. & Fedorenko, V. (2008). An ABC transporter encoding gene lndW confers resistance to landomycin E. Arch Microbiol 190, 105–109. [DOI] [PubMed] [Google Scholar]
- Ostash, B., Korynevska, A., Stoika, R. & Fedorenko, V. (2009). Chemistry and biology of landomycins, an expanding family of polyketide natural products. Mini Rev Med Chem 9, 1040–1051. [DOI] [PubMed] [Google Scholar]
- Prentki, P. & Krisch, H. M. (1984). In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29, 303–313. [DOI] [PubMed] [Google Scholar]
- Rebets, Y., Ostash, B., Luzhetskyy, A., Hoffmeister, D., Brana, A., Mendez, C., Salas, J. A., Bechthold, A. & Fedorenko, V. (2003). Production of landomycins in Streptomyces globisporus 1912 and S. cyanogenus S136 is regulated by genes encoding putative transcriptional activators. FEMS Microbiol Lett 222, 149–153. [DOI] [PubMed] [Google Scholar]
- Rebets, Y., Ostash, B., Luzhetskyy, A., Kushnir, S., Fukuhara, M., Bechthold, A., Nashimoto, M., Nakamura, T. & Fedorenko, V. (2005). DNA-binding activity of LndI protein and temporal expression of the gene that upregulates landomycin E production in Streptomyces globisporus 1912. Microbiology 151, 281–290. [DOI] [PubMed] [Google Scholar]
- Rigali, S., Derouaux, A., Giannotta, F. & Dusart, J. (2002). Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J Biol Chem 277, 12507–12515. [DOI] [PubMed] [Google Scholar]
- Rigali, S., Schlicht, M., Hoskisson, P., Nothaft, H., Merzbacher, M., Joris, B. & Titgemeyer, F. (2004). Extending the classification of bacterial transcription factors beyond the helix-turn-helix motif as an alternative approach to discover new cis/trans relationships. Nucleic Acids Res 32, 3418–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigali, S., Nothaft, H., Noens, E. E., Schlicht, M., Colson, S., Müller, M., Joris, B., Koerten, H. K., Hopwood, D. A. & other authors (2006). The sugar phosphotransferase system of Streptomyces coelicolor is regulated by the GntR-family regulator DasR and links N-acetylglucosamine metabolism to the control of development. Mol Microbiol 61, 1237–1251. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. & Russell, D. W. (2001). Molecular Cloning: a Laboratory Manual, 3rd edn. Cold Spring Harbor, NY. : Cold Spring Harbor Laboratory.
- Sezonov, G., Possoz, C., Friedmann, A., Pernodet, J. L. & Guérineau, M. (2000). KorSA from the Streptomyces integrative element pSAM2 is a central transcriptional repressor: target genes and binding sites. J Bacteriol 182, 1243–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirling, E. B. & Gottlieb, D. (1966). Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16, 313–340. [Google Scholar]
- Sun, J., Hesketh, A. & Bibb, M. (2001). Functional analysis of relA and rshA, two relA/spoT homologues of Streptomyces coelicolor A3(2). J Bacteriol 183, 3488–3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vomastek, T., Nádvorník, R., Janecek, J., Techniková, Z., Weiser, J. & Branny, P. (1998). Characterisation of two putative protein Ser/Thr kinases from actinomycete Streptomyces granaticolor both endowed with different properties. Eur J Biochem 257, 55–61. [DOI] [PubMed] [Google Scholar]