Abstract
The avrPphF gene was cloned from Pseudomonas syringae pathovar phaseolicola (Pph) races 5 and 7, based on its ability to confer avirulence towards bean cultivars carrying the R1 gene for halo-blight resistance, such as Red Mexican. avrPphF comprised two open reading frames, which were both required for function, and was located on a 154 kb plasmid (pAV511) in Pph. Strain RW60 of Pph, lacking pAV511, displayed a loss in virulence to a range of previously susceptible cultivars such as Tendergreen and Canadian Wonder. In Tendergreen virulence was restored to RW60 by avrPphF alone, whereas subcloned avrPphF in the absence of pAV511 greatly accelerated the hypersensitive resistance reaction caused by RW60 in Canadian Wonder. A second gene from pAV511, avrPphC, which controls avirulence to soybean, was found to block the activity of avrPphF in Canadian Wonder, but not in Red Mexican. avrPphF also conferred virulence in soybean. The multiple functions of avrPphF illustrate how effector proteins from plant pathogens have evolved to be recognized by R gene products and, therefore, be classified as encoded by avirulence genes.
Keywords: avirulence/disease resistance/gene-for-gene interactions/hypersensitive reaction/virulence
Introduction
The bacteria and fungi that colonize living plants and cause disease have evolved the ability to overcome the innate resistance of their hosts to infection. Superimposed on this establishment of basic parasitism (or pathogenicity) is the phenomenon of cultivar (cv.)-specific resistance. A plant pathogen may, therefore, be fully virulent causing disease on certain cultivars of its host, but avirulent on others (De Wit, 1995; Alfano and Collmer, 1997; Grant and Mansfield, 1999). Avirulence is associated with activation of the hypersensitive reaction (HR), a form of rapid programmed cell death, which restricts colonization to the site of inoculation in resistant cultivars of the host plant (De Wit, 1995; Mansfield et al., 1997a). This article concerns the genetics of avirulence and virulence in the bean halo-blight bacterium Pseudomonas syringae pathovar (pv.) phaseolicola (hereafter Pph). Nine races of Pph have been differentiated based on their virulence to a range of bean cultivars, as summarized in Table I (Taylor et al., 1996).
Table I. Gene-for-gene relationship (based on five matching gene pairs) proposed from analysis of the reactionsa of bean cultivars to races of P.s. pv. phaseolicola (from Taylor et al., 1996).
| Races/avirulence (A) genes | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
||||||
| A1 | . | . | . | A1 | . | A1 | . | A1 | ||||||
| . | A2 | . | A2 | A2 | . | A2 | . | . | ||||||
| . | . | A3 | A3 | . | . | . | . | . | ||||||
| . | . | . | . | A4 | . | . | . | . | ||||||
| Differential cultivars | Resistance genes | A5 | . | . | . | . | . | A5 | A5 | |||||
| Canadian Wonder | . | . | . | . | . | + | + | + | + | + | + | + | + | + |
| A52 (ZAA54) | . | . | . | R4 | . | + | + | + | + | – | + | + | + | + |
| Tendergreen | . | . | R3 | . | . | + | + | – | – | + | + | + | + | + |
| Red Mexican | R1 | . | . | R4 | . | – | + | + | + | – | + | – | + | – |
| A53 (ZAA55) | . | . | R3 | R4 | . | + | + | – | – | – | + | + | + | + |
| A43 (ZAA12) | . | R2 | R3 | R4 | R5 | + | – | – | – | – | + | – | – | – |
a+, susceptible; –, resistant.
The gene-for-gene theory explaining cultivar-specific resistance to plant disease was developed by Flor (1942, 1971) based on his analysis of the differential reactions of flax to the rust fungus Melampsora lini. Person et al. (1962) summarized the essence of Flor’s hypothesis in the statement that ‘…for every resistance gene in flax there is a specific and related gene for virulence in those rusts to which it is susceptible’. The emphasis on susceptibility suggested that disease was the result of the positive function of cultivar-specific virulence determinants. The advances of molecular genetics have proved the gene-for-gene theory by the cloning not of virulence (vir) genes, but of avirulence (avr) genes from bacteria and fungi, and the matching resistance (R) genes from plant hosts (Dangl, 1994; De Wit, 1995; Jones and Jones, 1996; Vivian et al., 1997). The proteins encoded by avr and R genes are envisaged to interact either directly or indirectly to activate signalling cascades leading to resistance, which is expressed by the HR (Hammond-Kosack and Jones, 1996; Van den Ackerveken et al., 1996). Bacterial avr genes have been cloned by function through the transfer of genomic libraries from avirulent to virulent races of the pathogen (Staskawicz et al., 1987). Genomic clones causing changes in the virulence phenotype on certain cultivars have been isolated and genes for avirulence subsequently characterized, e.g. avrPphB and avrPphE from Pph, which correspond to A3 and A2, respectively, in Table I (Mansfield et al., 1994; Vivian et al., 1997). Similarly, genes for avirulence but not virulence have also been cloned, which operate at the level of host specificity (Kobayashi et al., 1989; Fillingham et al., 1992; Wood et al., 1994).
Clearly, the continued presence of genes that restrict host range in certain strains or races of plant pathogens is somewhat of a paradox, but some avr genes have been implicated in basic parasitism, e.g. avrRpm1 from P.s. pv. maculicola and avrE from P.s. pv. tomato. Mutations in avrRpm1 or avrE lead to greatly reduced ability to colonize host plants (Lorang et al., 1994; Ritter and Dangl, 1995). The emerging pattern is that plant pathogens may produce numerous proteinaceous virulence factors, which act synergistically to promote disease (Alfano and Collmer, 1997; Collmer, 1998). Some of these factors may become recognized by the products of R genes and, therefore, be identified as determinants of avirulence. Because of functional redundancy, however, the loss of what has become an avr gene rather than a vir gene does not always compromise the basic pathogenicity on which cultivar specificity is superimposed.
The vir gene concept has been strengthened by the identification of a gene, virPphA from Pph, based on its ability to restore virulence to strains of the bean pathogen cured of a 154 kb plasmid designated pAV511 (Jackson et al., 1999). Cured strains (such as that named RW60) lost virulence towards bean, causing hypersensitive reactions in all previously susceptible cultivars tested. A cluster of potential vir genes including virPphA was located to a region described as a pathogenicity island (PAI), on pAV511. The ability of RW60 to elicit the HR suggested that it contained avr genes, located on the chromosome or other plasmids, whose function was usually masked by virulence factors encoded by the PAI. Other avr genes, such as avrPphB or avrPphE, continued to function despite the operation of the plasmid-borne PAI in wild-type strains and were, therefore, classified as α avr genes. The genes detected only in the absence of the PAI were classified as β avr genes (Jackson et al., 1999).
The resistance responses activated by different avr–R gene combinations may all be classified as the HR, but they may be phenotypically different because of, for example, differential timing of plant cell collapse. Differences are clear in bean as the avrPphB–R3 interaction provokes a more rapid HR than avrPphE–R2 (Mansfield et al., 1994, 1997a,b). In many race–cultivar interactions, several different α avr genes and their matching R genes may be present in the pathogen and plant, respectively (Mansfield et al., 1997a). The potential for epistatic interaction between various avr–R gene combinations is apparent from analysis of Arabidopsis harbouring the RPM1 and RPS2 genes. Although the HR controlled by avrRpt2–RPS2 is slower than that activated by the avrRpm1–RPM1 interaction, the phenotypically slower response is observed when both avr genes are present (Reuber and Ausubel, 1996; Ritter and Dangl, 1996). It has been proposed that such epistasis involves direct interaction between avr and R gene products (Ritter and Dangl, 1996).
Here we describe molecular characterization of the avrPphF locus in Pph. The gene was found to comprise an operon with two open reading frames (ORFs), both of which were required for function. Although we identified avrPphF as an α avr gene that determined race structure in the halo-blight bacterium, it was also found to confer cultivar-specific virulence to cv. Tendergreen. Furthermore, in the absence of the plasmid pAV511, containing the PAI, avrPphF also acted as a β avr gene in cv. Canadian Wonder, which is susceptible to all known races of the halo-blight bacterium. The gene within pAV511 that masked the β type of avr activity of avrPphF to Canadian Wonder was found to be avrPphC, which was previously identified as an avr gene acting on soybean cultivars. In short, an intriguing web of interacting avirulence and virulence functions has been revealed for the avr genes from Pph. In the bean–P.syringae interaction, the gene-for-gene concept, although continuing to provide a useful framework for analysis of cultivar-specific resistance, is far more complex than is apparent from simple analysis of race structures and α avr–R gene interactions as outlined in Table I. It is proposed that similar complexity will emerge in other host–pathogen systems once the problem of the redundancy of multiple virulence factors has been overcome by the removal of certain PAIs.
Results
Cloning of avrPphF from race 5
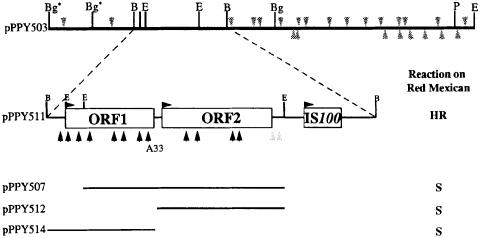
Based on the race structure proposed in Table I (Taylor et al., 1996), we sought to clone the putative A1 and A4 genes from race 5. A genomic library of race 5 strain 1375A, predicted to contain A1 and A4 and known to harbour A2 (cloned as avrPphE by Stevens et al., 1998), was screened for avr genes by mating into race 6 strain 1448A, which is virulent on all cultivars of bean used to define race structure (Mansfield et al., 1994). Transconjugants were initially tested for virulence on pods of cvs Red Mexican UI3 (proposed R1, R4) and A53 (R3, R4). Two genomic clones, pPPY501 and pPPY502, containing overlapping regions of insert DNA conferred avirulence on Red Mexican but none from the 1500 tested affected reactions on A53. Avirulence activity was retained by a 5.3 kb fragment, designated pPPY503. Transposon mutagenesis with Tn3gus located avrPphF to a 1.3 kb region flanked by BamHI and EcoRI restriction sites (Figure 1). The 1.8 kb BamHI fragment containing this region was cloned as pPPY511, and found to activate the HR in a cultivar-specific manner; avrPphF therefore acted as a typical α avr gene.
Fig. 1. Location of avrPphF within pPPY503 and the 1.8 kb of DNA sequenced. The direction of transcription is indicated by the horizontal arrowheads. Sites of insertion of Tn3gus in pPPY503 are indicated by vertical arrowheads; only those marked black in the expanded fragment abolished the avirulence activity of avrPphF. All transposon insertions were mapped from restriction digests, except A33, which was located by sequencing. The region found to have similarity to IS100 is indicated. Restriction sites for BamHI (B), BglII (Bg), EcoRI (E) and PstI (P) are marked; an asterisk indicates site in the vector pLAFR3. The reactions caused in bean cv. Red Mexican by transconjugants of race 6 harbouring subclones are noted. HR, hypersensitive reaction; S, susceptible response.
Segregation of the R1 resistance gene matching avrPphF was examined in 88 F2 progeny derived from a cross between Red Mexican and Tendergreen (R3). Progeny undergoing the HR to the transconjugant race 6(pPPY503) were also resistant to race 1 (A1). The pattern of incompatibility observed fitted that expected if avrPphF matched the R1 gene in Red Mexican, the numbers of resistant and susceptible plants (66:22) exactly fitting the 3:1 ratio expected if the R1 gene was dominant. The F2 progeny from the cross between Red Mexican and Tendergreen were also tested with a transconjugant of race 6 containing the clone pMS2330, which contains avrPphA isolated by Shintaku et al. (1989). Although race 6(pMS2330) did cause a weak HR on Red Mexican, reactions on pods from the F2 progeny of Red Mexican and Tendergreen showed that there was no co-segregation with race 1 (data not shown).
Multiplication of race 1 and race 6 was compared with race 6(pPPY503) in leaves of cv. Red Mexican. The HR determined by the presence of the cloned avrPphF gene was associated with the failure of bacteria to multiply. Following inoculation to introduce ∼0.2 × 106 c.f.u. per excised leaf disc, after 2 days race 6 had reached 3.7 × 106, whereas race 1 and race 6(pPPY503) were present at only 9.3 × 102 and 16 × 102 c.f.u. per disc, respectively.
Sequence analysis and expression of avrPphF
The nucleotide sequences of the functional 1.8 kb fragments from race 5, and as also recovered from race 7 containing avrPphF, were found to be identical (DDBJ/EMBL/GenBank accession Nos AF231452 and AF231453). The sequence revealed two ORFs with a single upstream promoter region containing the hrp box motif, indicating probable regulation by HrpL (Innes et al., 1993; Pirhonen et al., 1996). Both ORFs were preceded by purine-rich regions, which would be expected to act as ribosome binding sites (Singer and Berg, 1991) and they were separated by a 57 bp sequence. A rho-dependent transcription termination loop was located after the translation stop signal in ORF2. The avrPphF locus therefore had the structural features of an operon. The G+C content of avrPphF was 47%, with ORF1 40% and ORF2 52.5%, both significantly lower than the overall figures of 59–61% reported for pvs of P.syringae (De Ley, 1968).
Transposon insertions that compromised avirulence were located in both ORF1 and ORF2 of avrPphF, and further subcloning revealed that both ORFs were required for avr function (Figure 1). To examine further the role of the two ORFs, non-polar mutations were created for ORF1 and ORF2, resulting in race 7ΔORF1 and race 7ΔORF2, respectively; both mutants failed to elicit the HR in cv. Red Mexican.
Protein production from ORFs 1 and 2 was examined in Escherichia coli using constructs of avrPphF cloned for expression in pBluescript. Proteins of 17 and 24 kDa were detected on SDS–PAGE gels after Coomassie Blue staining, corresponding to the peptides predicted to be encoded by ORFs 1 and 2, i.e. 15.6 and 21.9 kDa, respectively (data not shown). Searches of the databases using the BLAST or Propsearch approaches (Hobohm and Sander, 1995; Altschul et al., 1997) failed to reveal significant homology with known protein sequences. Both peptides are predicted to be hydrophilic; they lack leader sequences or potentially membrane-spanning domains.
avrPphF is plasmid borne and present only in races expressing the A1 phenotype
The presence of homologues of avrPphF was examined in strains of Pph representing all nine races and also in other pathovars of P.syringae. Using specific probes for ORF1 and ORF2 in PCR or Southern analysis, signals identical to those found in race 5, the origin of the avrPphF clone, were present only in races 1, 7 and 9, but not in races 2, 3, 4, 6 or 8, which are virulent on Red Mexican (Table I; Figure 2A and B). However, when the entire 1.8 kb BamHI fragment containing avrPphF was used as a probe the pattern changed. Multiple signals were detected in all races of Pph (Figure 2C). Further probing revealed that multiple signals were due to the 0.5 kb EcoRI–BamHI fragment downstream of avrPphF (Figure 1), which was found to contain an incomplete ORF with predicted similarity to the IS100 transposase homologue from the PAI in Pph (Jackson et al., 1999). Sequences hybridizing to avrPphF were found in strains of P.s. pvs pisi and glycinea, but not in pvs coronafaciens, maculicola, tabaci, tomato or syringae, or in Pseudomonas cichorii. Interestingly, only P.s. pv. syringae and P.s. pv. pisi contained sequences hybridizing to the putative transposase, but, in contrast to Pph, only one band was observed on Southern blots (data not shown), indicating that it is highly specific to pv. phaseolicola.
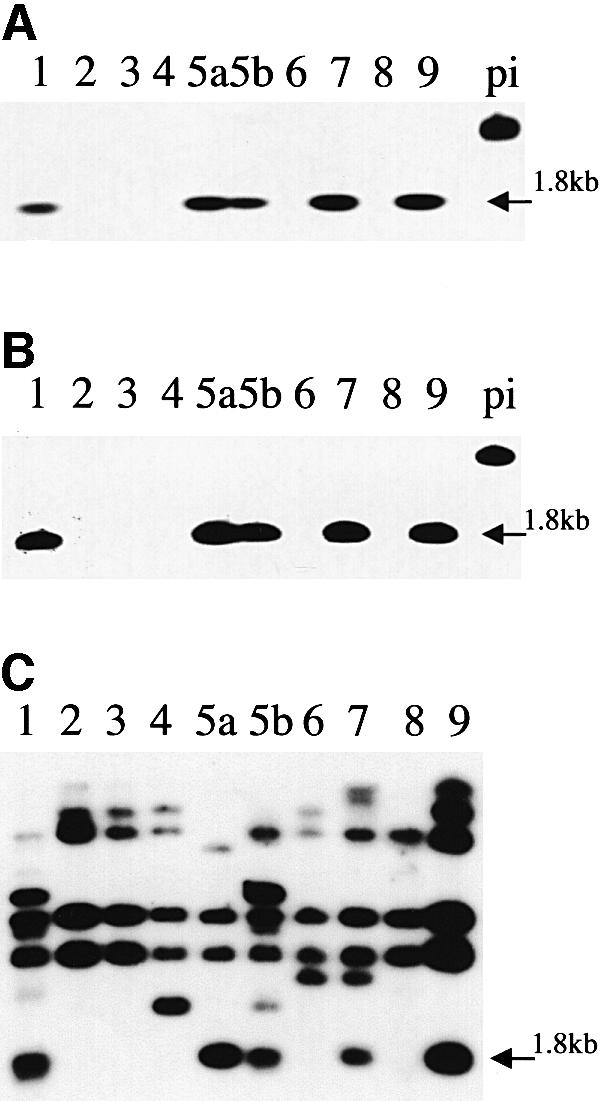
Fig. 2. Southern hybridization (high stringency) of BamHI-digested total DNA from different strains of P.syringae using (A) ORF1 and (B) ORF2 of avrPphF and (C) the 0.5 kb EcoRI–BamHI fragment with similarity to IS100 as probes. Digests from races 1, 2, 3, 4, 5 (a, strain 52A; b, strain 1375A), 6, 7, 8 and 9 of P.s. pv. phaseolicola, and P.s. pv. pisi (pi) are shown.
Hybridization experiments also located avrPphF to the 154 kb indigenous plasmid pAV511. avrPphF was positioned at the left of the PAI recently identified in Pph (Figure 3), close to avirulence genes avrPphC and a homologue of avrD, both of which were cloned as soybean interactors (Kobayashi et al., 1990; Yucel et al., 1994).
Fig. 3. Location of avrPphF in relation to avrD, virPphA, avrPphC and ORF4 in the 154 kb plasmid identified in race 7 strain 1449B (Jackson et al., 1999). The insert cloned in pAV520 (vector pLAFR3) is shown; the region present in pAV521 is underlined. Restriction sites for BamHI (B), EcoRI (E) and HindIII (H) are marked.
avrPphF acts as a gene for virulence in cv. Tendergreen
Although race 7 of Pph is virulent on cv. Tendergreen, the plasmid-cured strain RW60 causes an HR. Virulence towards Tendergreen was restored by the genomic clones pAV520 or pAV521 harbouring avrPphF recovered from race 7 strain 1449B (Jackson et al., 1999). A transposon insertion in pAV521, which reduced ability to restore virulence only to cv. Tendergreen, was located to avrPphF and we subsequently found that avrPphF alone (pPPY511) effectively restored water-soaking ability to RW60 in pods and leaves of Tendergreen, as well as the distinct avrPphF–R1 interaction phenotype of the HR in cv. Red Mexican (Table II; Figure 4A). The complementation achieved showed, therefore, that avrPphF acts as a cultivar-specific virulence gene on cv. Tendergreen. Transconjugants, RW60(pPPY514) and RW60 (pPPY512), separately harbouring ORF1 and ORF2 of avrPphF, respectively, were unable to restore virulence (Table II).
Table II. Reactionsa caused by strains of P.s. pv. phaseolicola on bean and soybean cultivars.
| Strainb | Bean cultivar |
Soybean cultivar |
|||
|---|---|---|---|---|---|
| Red Mexican | Tendergreen | Canadian Wonder | Osumi | Choska | |
| Race 7 | HR* | S | S | HR | S– |
| RW60 | HR | HR | hr | N | N |
| RW60(pAV520) | HR* | S | S | HR | S– |
| RW60(pPPY511) | HR* | S– | HR | S– | S– |
| RW60(pPPY512) | HR | HR | hr | N | N |
| RW60(pPPY514) | HR | HR | hr | N | N |
| RW60(pPPY511, pDAHR15) | HR* | S– | hr | nt | nt |
| Race 7::avrPphFTn3gus A33 | S | S | S | nt | nt |
| P.s. pv. glycinea | HR | HR | HR | S | S |
aS, fully susceptible water-soaked lesion; S–, slower development of lesions than S; HR, hypersensitive reaction; HR*, characteristic of the avrPphF–R1 interaction; hr, slow development of the HR; N, null reaction; nt, not tested.
bpAV520 is a genomic clone; plasmids encoding single ORFs or avr genes are pPPY511 (avrPphF), pPPY512 (avrPphF, ORF2), pPPY514 (avrPphF, ORF1) and pDAHR15 (avrPphC).
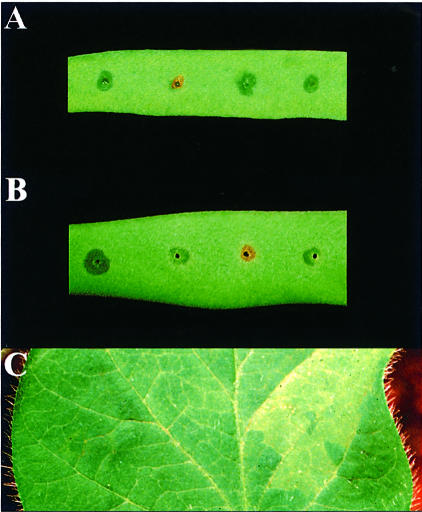
Fig. 4. (A) Reaction phenotypes in a pod of cv. Tendergreen 2 days after inoculation with (from left to right) race 7, RW60, RW60 (pAV520) containing the PAI and RW60(pPPY511) expressing avrPphF alone. (B) Reaction phenotypes in a pod of cv. Canadian Wonder 2 days after inoculation with (from left to right) race 7, RW60, RW60(pPPY511) expressing avrPphF, and RW60(pPPY511, pDAHR15) expressing both avrPphF and avrPphC. Note that the lesions caused by RW60 and RW60(pPPY511, pDAHR15) are sunken at this stage compared with the water-soaked susceptible response to race 7, whereas RW60(pPPY511) has already caused a brown lesion characteristic of the rapid HR induced. (C) Reactions at infiltration sites in soybean leaves (cv. Osumi) 3 days after inoculation with (left to right) RW60, which causes a null reaction, and the transconjugant RW60(pPPY511) expressing avrPphF, which causes a susceptible response recognized by yellowing as shown and later some water- soaking.
The increased virulence of transconjugants of RW60 harbouring avrPphF was also demonstrated by increases in bacterial populations in Tendergreen leaves (Figure 5). Interestingly, bacterial growth of the marker exchange avrPphF mutant, race 7::avrPphF Tn3gus A33 (Figure 1), was restricted compared with the wild type, but the symptoms produced were still classified as susceptible. The two non-polar mutants, race 7ΔORF1 and race 7ΔORF2, respectively, were also virulent in cv. Tendergreen. The continued presence of plasmid-borne virPphA, and possibly other vir genes in race 7 strains, may allow the avrPphF mutants to grow but not to reach wild-type levels.
Fig. 5. Bacterial multiplication in leaves of bean cv. Tendergreen inoculated with suspensions of 2 × 108 cells/ml of race 7 (open), RW60 (striped), RW60 (pPPY511) (light grey), RW60 (pAV521) (dark grey) and the avrPphF marker exchange mutant race 7::avrPphF Tn3gusA33 (black); bars, ± SEM.
avrPphF behaves as a masked avirulence gene in cv. Canadian Wonder
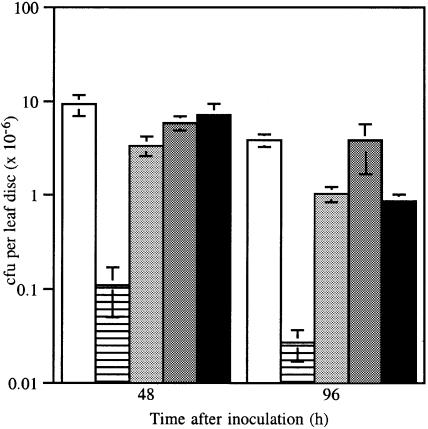
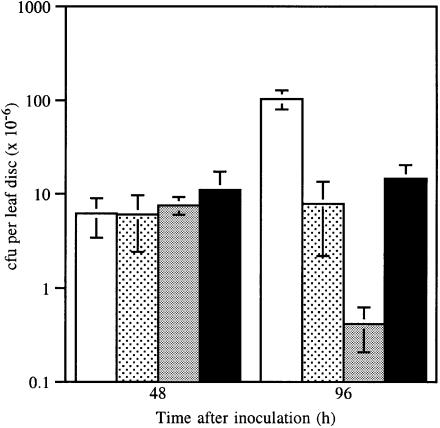
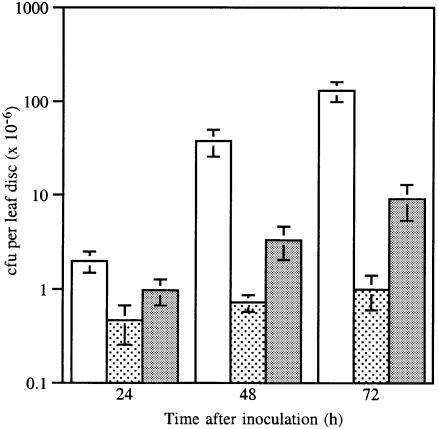
The plasmid-cured strain RW60 was found to cause a slow HR (subsequently designated hr) in cv. Canadian Wonder, whereas wild-type race 7 causes water soaking characteristic of a susceptible reaction. Unexpectedly, transconjugant RW60(pPPY511), expressing avrPphF, caused a very rapid HR on leaves and pods of cv. Canadian Wonder quite distinct from the hr caused by RW60 alone (Figure 4B). Transconjugants of RW60 containing pAV520 (which harbours avrPphF; Figure 3), like wild-type race 7, caused a susceptible reaction on cv. Canadian Wonder, indicating that a gene in pAV520 must suppress the avirulence function of avrPphF. Analysis of subcloned regions of pAV520 revealed that the gene suppressing the avr activity of avrPphF was the non-host avirulence gene avrPphC (Yucel et al., 1994). Transconjugants of RW60 containing both avrPphF and avrPphC (in pPPY511 and pDAHR15, respectively) gave a phenotype identical to that caused by RW60 (Table II; Figure 4B). Multiplication of race 7, RW60, RW60(pPPY511) and RW60(pPPY511, pDAHR15) in leaves was compared. The rapid HR determined by the presence of avrPphF in RW60(pPPY511) was associated with the recovery of very low numbers of bacteria 96 h after inoculation, whereas RW60(pPPY511, pDAHR15) achieved bacterial numbers similar to RW60 alone, confirming the phenotypes observed (Figure 6).
Fig. 6. Bacterial multiplication in leaves of bean cv. Canadian Wonder inoculated with bacterial suspensions of 2 × 108 cells/ml of race 7 (open), RW60 (stippled), RW60 (pPPY511) (grey) and RW60(pPPY511, pDAHR15) (black); bars, ± SEM.
Analysis of segregating plant populations
The phenotypes caused by RW60, RW60(avrPphF) and RW60(avrPphF, avrPphC) were very distinct, as shown in Figure 4A and B. We were, therefore, able to analyse the segregation of reactions in crosses between cvs Canadian Wonder and Tendergreen. Analyses of F2 populations summarized in Tables III and IV are based on the following proposals: (i) Canadian Wonder has two R genes, RF matching avrPphF and Rβ2 matching a β avr gene unmasked in RW60; (ii) Tendergreen has Rβ1 matching a second β avr gene in RW60, but does not harbour RF; (iii) the plasmid-cured strain RW60 causes a rapid HR on cv. Tendergreen and hr on cv. Canadian Wonder due to the avrβ1–Rβ1 and avrβ2–Rβ2 interactions, respectively; (iv) avrPphF acts as a virulence determinant on cv. Tendergreen. The phenotypes observed corresponded to those expected from these proposals, the parental genotypes predicted being Rβ1, Rβ1; rβ2, rβ2; rF, rF for Tendergreen, and rβ1, rβ1; Rβ2, Rβ2; RF, RF for Canadian Wonder.
Table III. Segregation of resistance to RW60(avrβ1, avrβ2) amongst the F2 progeny of a cross between Tendergreen (Rβ1, Rβ1; rβ2, rβ2)a and Canadian Wonder (rβ1, rβ1; Rβ2, Rβ2)a.
aRβ1 confers a rapid HR and Rβ2 a slow hr.
bThe predicted phenotype would be a susceptible reaction, unless there are other β avr genes in RW60.
Table IV. Segregation of resistance to RW60 (avrβ1, avrβ2, avrPphFa) amongst the F2 progeny of a cross between Tendergreen (Rβ1, Rβ1; rβ2, rβ2; rF, rF) and Canadian Wonder (rβ1, rβ1; Rβ2, Rβ2; RF, RF).
aavrPphF present on pPPY511. The parental phenotypes were rapid HR and susceptibility on cvs Canadian Wonder and Tendergreen, respectively.
bDominant means homozygous (RR) or heterozygous (Rr).
cPredicted to develop a susceptible reaction due to suppression of Avrβ1 by AvrPphF or with all R genes recessive, if no other avr–R gene interactions occurring.
The reaction of F2 progeny to RW60 (Table III) indicated that the avrβ2 gene, although conferring a slower hr, was epistatic to avrβ1. Phenotypes segregated hr:HR 31:7, closely approximating the predicted 13:3 ratio (χ2 = 0.0025). Reaction to RW60(avrPphF) clearly indicated the presence of the matching dominant RF gene in Canadian Wonder with phenotypes observed, HR:hr or S, 27:11 (Table IV). Amongst the 38 F2 progeny examined, two plants developed a rapid HR when challenged by RW60 (indicating avrβ1–Rβ1 interaction), but were susceptible to RW60(avrPphF), indicating the absence of RF. The virulence function of avrPphF therefore appeared to be directly related to blocking the proposed avrβ1–Rβ1 interaction.
Nineteen of the 38 progeny were also tested with the RW60(avrPphF, avrPphC) transconjugant. Amongst the 64 possible genotypes predicted from the Canadian Wonder × Tendergreen cross only three, rβ1, rβ1; rβ2, rβ2; RF, RF and rβ1, rβ1; rβ2, rβ2; RF, rF (or rF, RF) would be expected to produce a rapid HR when challenged by RW60(avrPphF) but be susceptible to RW60(avrPphF, avrPphC). One plant was indeed detected with this phenotype, clearly supporting the gene-specific virulence function of avrPphC masking the avrPphF–RF interaction, and also the probable absence of other R genes, which would have prevented the establishment of a susceptible response.
avrPphF acts as a virulence gene in soybean
Race 7 of Pph typically causes a rapid HR in soybean leaves, but in certain cvs such as Choska it was found to cause a weak susceptible response recognized by yellowing and water soaking at inoculation sites in leaves. The plasmid-cured strain RW60 caused a null response on all cvs of soybean tested. avrPphF was able to restore virulence to RW60 in cvs Choska and Osumi (Figure 4C; Table II). Restoration of virulence by avrPphF was also demonstrated by increases in bacterial populations in leaves of cv. Osumi (Figure 7).
Fig. 7. Bacterial growth in leaves of soybean cv. Osumi inoculated with bacterial suspensions of 108 cells/ml of P.s. pv. glycinea (open), RW60 (stippled) and RW60 (pPPY511) (grey); bars, ± SEM.
Discussion
The pattern of avirulence conferred by avrPphF in wild-type races of Pph fits that predicted for an α avr gene matching the R1 gene for resistance in Phaseolus (Taylor et al., 1996). The cloning strategy adopted failed to isolate the fourth avr gene predicted to match R4 (Table I), possibly because the gene may have lethal effects in E.coli. The first avr gene cloned from Pph (Shintaku et al., 1989), and designated avrPphA according to the nomenclature proposed by Vivian and Mansfield (1993), did not appear to match any of the R genes recognized in Phaseolus. The proposed gene-for-gene pattern of race structure (Taylor et al., 1996) has, therefore, not been entirely confirmed by molecular genetics.
The avrPphF locus is organized into an operon with a characteristic ‘hrp box’ promoter indicating regulation by HrpL. Both ORFs within the operon were required for either avirulence or virulence functions. The requirement for more than one transcriptional unit for the function of an avr gene was described in detail for avrE from P.s. pv. tomato by Lorang and Keen (1995). The avrE locus is linked to the right end of the hrp region and comprises two convergently transcribed units, which are both required for the avirulence phenotype. avrBs1 from Xanthomonas campestris pv. vesicatoria comprises an operon of two ORFs, but only the second is required for avirulence activity (Ronald and Staskawicz, 1988). Here, we show that avrPphF is organized into an operon with two ORFs, both of which are needed for function.
The mechanism by which avrPphF causes effects in plant cells remains unknown. According to published reports, all of the bacterial avr genes that have been tested elicit the HR if they are expressed transiently in plant cells; examples are avrPphB and avrPphE from Pph (Stevens et al., 1998). The implication from in planta expression is that the Avr proteins act as elicitors following their delivery into plant cells by the hrp-dependent type III secretion apparatus (Alfano and Collmer, 1997; Bonas and Van den Ackerveken, 1997; Galan and Collmer, 1999). In planta expression of the ORFs from avrPphF should reveal whether one functions as the elicitor per se and one perhaps acts as a chaperone for protein delivery. The two ORFs were always found together, arranged into the avrPphF operon, but only in strains expressing the A1 phenotype (Table I; Figure 2). The adjacent IS100 homologue was distributed throughout strains irrespective of the race designation. IS100 is present in species of Yersinia and several copies have been located in plasmids encoding type III secretion systems or other virulence factors (Buchrieser et al., 1998; Hu et al., 1998). Sequences hybridizing to the insertion sequence were, however, not widely distributed amongst other pathovars of P.syringae.
Given the pathogenicity functions attributed to other avr genes from P.syringae, e.g. avrE and avrRpm1 (Lorang et al., 1994; Ritter and Dangl, 1995), the finding that avrPphF was able to restore virulence to compromised plasmid-cured strains such as RW60 was not unexpected. However, avrPphF is, to our knowledge, the first gene to be shown to have cultivar- and gene-specific virulence activity. By contrast, the discovery that avrPphF also acts as a β avr gene, i.e. has functions revealed only in the absence of the PAI, operating in a defined gene-for-gene manner with a matching R gene in Canadian Wonder, was most unexpected. The interaction between avrPphC and avrPphF blocking the HR in cv. Canadian Wonder would appear to occur in the plant, possibly with a receptor for the AvrPphF protein, because avrPphC has no effect on the expression of the HR caused by avrPphF in cv. Red Mexican. It is important to emphasize that the virulence and β avirulence functions of avrPphF are only observed in the absence of other genes from the PAI. The AvrPphF proteins appear to block the HR caused by β avr genes such as avrβ1, which is predicted to be present in the chromosome of Pph. The β avr function of avrPphF is in turn blocked by avrPphC. In contrast to avrPphF, the other genes recognized to act as virulence determinants, virPphA and to a lesser extent ORF4 from the PAI, do not have differential effects in the cultivars tested (Jackson et al., 1999).
The ability of certain Avr proteins to express virulence functions, blocking the phenotype conferred by other avr genes, may involve three different types of interaction: (i) between the Avr proteins themselves; (ii) between Avr and masked R proteins; (iii) downstream of hypothetical Avr–R protein interactions to interfere with gene-specific signalling cascades leading to the HR. A possible target for (iii) would be one of the MAP kinases which have been implicated in plant defence (Grant and Mansfield, 1999; Romeis et al., 1999). The virulence determinant YopJ from Yersinia has recently been found to bind and inactivate MAP kinase kinases in mammalian cells, leading to suppression of defence responses (Orth et al., 1999). There are no reports of direct interactions between Avr proteins, but Avr–R protein binding has been demonstrated between AvrPto and Pto (Tang et al., 1996). However, the action of intermediates linking Avr and R proteins has also been proposed (Grant and Mansfield, 1999).
The multiple function of avr genes found in Phaseolus was also extended to the Pph–soybean interaction. Whether or not soybean should continue to be considered as a non-host to Pph (Yucel et al., 1994) is questionable as certain strains clearly show weak pathogenicity. Interestingly, however, genes recognized for virulence or avirulence function in Phaseolus have so far all displayed the opposite activity in soybean leaves, as summarized in Table V. Pathogenicity towards the legume hosts has probably involved interaction with common virulence targets. As targets mutated to activate resistance (i.e. acting as R gene products), perhaps by hyperactivation of signal transduction cascades blocked by Vir factors, the pathogen would need to acquire further vir genes whose products would override the activation of resistance. Restoration of virulence could be achieved by suppression of signal transduction, or, as outlined above, simply by blocking any direct avr–R gene interactions.
Table V. Genes with dual avirulence and virulence functions, either eliciting or blocking the HR, respectively, identified using strains of P.s. pv. phaseolicola lacking the 154 kb plasmid which contains the putative pathogenicity island.
| Gene designation | Plant in which phenotype observeda | |
|---|---|---|
| Avirulence | Virulence | |
| avrPphF | Red Mexicanb, Canadian Wonderb | Tendergreen, Soybean |
| avrPphC | Soybeanc | Canadian Wonder |
| virPphAd | Soybean | Canadian Wonder, Red Mexican, Tendergreen |
aBean cultivars are named.
bAvirulence due to interaction with different R genes.
An intriguing question is whether or not the avr or vir genes now identified from a highly evolved pathogen such as Pph represent virulence determinants that have had a fundamental role in the evolution to parasitism from a saprophytic ancestry. The layers of interactions and possibly interconnected targets that exist amongst the numerous avr and vir genes in the present populations of plant pathogens may make it difficult to distinguish the evolutionary significance of individual effector proteins. It is possible that gene-specific virulence, as shown by avrPphF, represents a second wave of activity, whereas virPphA, apparently with less cultivar specificity, may have more direct effects on signal transduction pathways and represent the first class of virulence factor. The co-evolution of hosts and pathogens would mean that clear distinction between primary and secondary waves of activity may have become very blurred, and it is certainly impossible to predict the function of avirulence and virulence factors we have identified. In order to unravel the complex web of interacting effectors, it is now a priority to identify binding partners in the plant for the Avr proteins recognized to have multiple functions.
Materials and methods
Bacterial strains and plasmids
Principal bacterial strains, cosmids and plasmids used are listed in Table VI. Isolates and transconjugants of Pph were grown routinely on King’s medium B (KB) agar at 25°C and E.coli strains on Luria–Bertani (LB) agar or in LB broth at 37°C (King et al., 1954; Miller, 1972). Antibiotics, obtained from Sigma, were usually used at the following concentrations (µg/ml): rifampicin 60; tetracycline 20; kanamycin 40; ampicillin 100; spectinomycin 40; nalidixic acid 50; chloramphenicol 25.
Table VI. Bacterial strains and plasmids used in this studya.
| Strain/plasmid | Relevant properties | Source or reference |
|---|---|---|
| Bacteria. P.syringae pv. phaseolicolaPrincipal isolates used | ||
| 1375A | race 5, wild-type isolate | D.Teversonb |
| 1375AN | race 5, NalR from 1375A | D.Teverson |
| 1375AR | race 5, RifR from 1375A | D.Teverson |
| 1281A | race 1, wild-type isolate | D.Teverson |
| 1281AR | race 1, RifR from 1281A | D.Teverson |
| 1448A | race 6, wild-type isolate | Fillingham et al. (1992) |
| 1448AR | race 6, RifRfrom 1448A | Fillingham et al. (1992) |
| 1449B | race 7, wild-type isolate | D.Teverson |
| 1449BR | race 7, RifR from 1449B | D.Teverson |
| RW60 | Vir–, pAV511–, ApS, RifR | Jackson et al. (1999) |
| Additional isolates | ||
| 882 | race 2 | D.Teverson |
| 1301A | race 3 | Hitchin et al. (1989) |
| 1302A | race 4 | Jenner et al. (1991) |
| 52A | race 5 | Hitchin et al. (1989) |
| 2656A | race 8 | D.Teverson |
| 2709A | race 9 | D.Teverson |
| P.cichorii 2379 | lettuce pathogen | NCPPBc |
| P.s. pv. cornafaciens 1354 | oat pathogen | Harper et al. (1987) |
| P.s. pv. pisi 299A | pea pathogen | J.Taylorb |
| P.s. pv. glycinea 1139 | soybean pathogen | this study |
| P.s. pv. maculicola 1820 | brassica pathogen | NCPPB |
| P.s. pv. tabaci 11528 | tobacco pathogen | J.Turnerd |
| P.s. pv. tomato DC3000 | tomato pathogen | Whalen et al. (1991) |
| P.s. pv. syringae 281 | lilac pathogen | NCPPB |
| E.coli | ||
| C2110 | NalR, polA1 | Leong et al. (1982) |
| DH5α | NalR, recA, lacZΔM15 | Bethesda Research Laboratories |
| HB101 | SmR, recA | Boyer and Roulland-Dussoix (1969) |
| Plasmids | ||
| pBluescript SK+ | ApR, multiple cloning sites and priming sites | Stratagene |
| pHoKmGus | ApR, KmR, tnpA, promoterless β-glucuronidase gene in Tn3, pWB15A replicon | Bonas et al. (1989) |
| pLAFR3 | TcR, IncP1 replicon, Tra–, Mob+, cosmid | Staskawicz et al. (1987) |
| pDSK600 | SpR, IncQ replicon, 3× lac UV5promoter | Murillo et al. (1994) |
| pRK2013 | KmR, ColE1 replicon, Tra+, Mob+, helper plasmid | Figurski and Helinski (1979) |
| pSShe | CmR, tnpA+, pACYC184 replicon | Stachel et al. (1985) |
| Clones containing avrPphF | ||
| pPPY501 | pLAFR3-based genomic clone from race 5 strain 1375 | this study |
| pPPY502 | pLAFR3-based genomic clone from race 5 strain 1375 | this study |
| pPPY503 | 5.3 kb BamHI–HindIII subclone of pPPY502 harbouring avrPphF | this study |
| pPPY505 | 1.8 kb BamHI fragment from pPPY503 in pBluescript II SK+ | this study |
| pPPY507 | 1.1 kb EcoRI fragment in pLAFR3 | this study |
| pPPY511 | insert as in pPPY505 but in pDSK600 | this study |
| pPPY512 | avrPphF ORF2 in pDSK600 | this study |
| pPPY514 | avrPphF ORF1 in pDSK600 | this study |
| Additional plasmids | ||
| pMS2330 | pLAFR1 based genomic clone harbouring avrPphA from HB33 | Shintaku et al. (1989) |
| pAV511 | native plasmid from 1449B, ∼154 kb | Jackson et al. (1999) |
| pAV520 | genomic clone harbouring pAV511 sequences in pLAFR3 | Jackson et al. (1999) |
| pAV521 | genomic clone harbouring pAV511 sequences in pLAFR3. | Jackson et al. (1999) |
| pDAHR15 | 1.4 kb ClaI–BamHI fragment containing avrPphC in pDSK600 | Yucel et al. (1994) |
aNalR, RifR, SmR, ApR, KmR, TcR and CmR indicate resistance to nalidixic acid, rifampicin, streptomycin, ampicillin, kanamycin, tetracycline and chloramphenicol, respectively.
bHorticulture Research International, Wellesbourne, UK.
cNational Collection of Plant Pathogenic Bacteria, York, UK.
dUniversity of East of Anglia, Norwich, UK.
Pathogenicity tests and in planta bacterial population counts
Pods and leaves of French bean were inoculated as described previously (Harper et al., 1987; Hitchin et al., 1989). The inoculum concentration routinely used in bean leaves was 2 × 108 cells/ml. Soybean plants were inoculated using a 1 ml syringe (without needle) to infiltrate bacterial suspensions of 108 cells/ml into the underside of fully expanded primary leaves. Bacterial multiplication in leaves was examined by cutting tissue from the inoculation sites with a 0.6-cm-diameter borer, homogenization in 10 mM MgCl2, and serial dilution of the homogenate, which was then spread onto LB agar with appropriate antibiotics to allow colony development at 25°C.
General molecular techniques
Basic procedures were carried out as described in Sambrook et al. (1989). The methods of Jenner et al. (1991) and Mansfield et al. (1994) were followed for the construction of the genomic library from race 5 strain 1375A, preparation of minipreps, Southern blotting and hybridization. The library was screened for determinants of avirulence by conjugation of individual clones into race 6 strain 1448AR with the helper plasmid pRK2013. Transconjugants were tested for pathogenicity on pods of cvs Red Mexican and A53 as described by Harper et al. (1987). Transposon mutagenesis of pAV521 and pPPY503, and marker exchange of avrPphF::Tn3gusA33 into race 7 were carried out as described by Mansfield et al. (1994); the position of the transposon A33 in the target DNA was confirmed by sequencing.
PCR and DNA sequencing
Standard PCRs were performed with Taq polymerase and buffers from Bioline using a Perkin Elmer Gene Amp 2400. Sequencing was performed using the dideoxy chain termination method with the Sequenase enzyme (Pharmacia) as described in the manufacturer’s instructions. Primers used for the amplification of avrPphF ORF1 were: ORF1F, 5′-ATGAAGAATTCGTTCGACCG-3′; ORF1R, 5′-TCAGACCGAACTCTCAGACA-3′. For the amplification of avrPphF ORF2, the primers used were: ORF2F, 5′-ATGGGTAATATCTGCAATTCG-3′; ORF2R, 5′-GGCCAGTTATAGGAGCTAAT-3′.
Construction of non-polar mutations
For the generation of the non-polar mutants, race 7ΔORF1 and race 7ΔORF2, the following steps were followed. ORF1 was deleted from position 332–439 in pPPY505 by a double digest with AatII–AgeI. Digested pPPY505 was filled in using Klenow polymerase and re-ligated. Deletions were selected by PCR and the deleted form cloned into pOK (Huguet et al., 1998). ORF2 was deleted from position 767–1157 in pPPY505 by a double digest with Csp45I–Bst98I. The plasmid with the digested insert was filled in and re-ligated. Cloned deletions were again selected by PCR and the deleted form was then introduced to pOK. Mutated genes were introduced to race 7 strain 1449BR by homologous recombination in two steps as described by Kaniga et al. (1991) and Huguet et al. (1998). In both mutants, the presence of the correct deletion in race 7 was verified by PCR.
Acknowledgments
Acknowledgements
Many thanks are due to Elisabeth Huguet and Ulla Bonas for advice on the creation of non-polar mutants, to Noel Keen for clones containing avrPphC and to Suresh Patil for avrPphA. We also wish to acknowledge support from the BBSRC, EC grants BIO-CT97-2244 and TMR, CICYT grant BIO97-0598 and the British Council-MEC Acciones Integradas programme. Experiments with genetically modified strains were carried out under MAFF licences PHL30/2609 and PHL63/2851.
References
- Alfano J.R. and Collmer,A. (1997) The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins and death. J. Bacteriol., 179, 5655–5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonas U. and Van den Ackerveken,G.F.J.M. (1997) Recognition of bacterial avirulence proteins occurs inside the plant cell: a general phenomenon in resistance to bacterial diseases? Plant J., 12, 1–7. [DOI] [PubMed] [Google Scholar]
- Bonas U., Stall,R.E. and Staskawicz,B. (1989) Genetic and structural characterisation of the avirulence gene avrBs3 from Xanthomonas campestris pv. vesicatoria. Mol. Gen. Genet., 218, 127–136. [DOI] [PubMed] [Google Scholar]
- Boyer H.W. and Roulland-Dussoix,D. (1969) A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol., 41, 459–472. [DOI] [PubMed] [Google Scholar]
- Buchrieser C., Prentice,M. and Carniel,E. (1998) The 102-kilobase unstable region of Yersinia pestis comprises a high-pathogenicity island linked to a pigmentation segment which undergoes internal rearrangement. J. Bacteriol., 180, 2321–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collmer A. (1998) Determinants of pathogenicity and avirulence in plant pathogenic bacteria. Curr. Opin. Plant Biol., 1, 329–335. [DOI] [PubMed] [Google Scholar]
- Dangl J.L. (1994) The enigmatic avirulence genes of phytopathogenic bacteria. In Dangl,J.L. (ed.), Bacterial Pathogenesis of Plants and Animals: Molecular and Cellular Mechanisms. Springer-Verlag, Berlin, Germany, pp. 99–118. [DOI] [PubMed] [Google Scholar]
- De Ley J. (1968) DNA base composition and hybridization in the taxonomy of phytopathogenic bacteria. Annu. Rev. Phytopathol., 6, 63–90. [Google Scholar]
- De Wit P.J.G.M. (1995) Fungal avirulence genes and plant resistance genes: unraveling the molecular basis of gene-for-gene interactions. Adv. Bot. Res., 21, 148–185. [Google Scholar]
- Figurski D.H. and Helinski,D.R. (1979) Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl Acad. Sci. USA, 76, 1648–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingham A.J., Wood,J., Bevan,J.R., Crute,I.R., Mansfield,J.W., Taylor,J.D. and Vivian,A. (1992) Avirulence genes from Pseudomonas syringae pathovars phaseolicola and pisi confer specificity toward both host and non-host species. Physiol. Mol. Plant Pathol., 40, 1–15. [Google Scholar]
- Flor H.H. (1942) Inheritance of pathogenicity in Melampsora lini.Phytopathology, 32, 653–669. [Google Scholar]
- Flor H.H. (1971) Current status of the gene-for-gene concept. Annu. Rev. Phytopathol., 9, 275–296. [Google Scholar]
- Galan J.E. and Collmer,A. (1999) Type III secretion machines: bacterial devices for protein delivery into host cells. Science, 284, 1322–1328. [DOI] [PubMed] [Google Scholar]
- Grant M.R. and Mansfield,J.W. (1999) Early events in host–pathogen interactions. Curr. Opin. Plant Biol., 2, 312–319. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack K.E. and Jones,J.D.G. (1996) Resistance gene-dependent plant defense responses. Plant Cell, 8, 1773–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper S., Zewdie,N., Brown,I.R. and Mansfield,J.W. (1987) Histological, physiological and genetical studies of the responses of leaves and pods of Phaseolus vulgaris to three races of Pseudomonas syringae pv. phaseolicola and Pseudomonas syringae pv. coronafaciens. Physiol. Mol. Plant Pathol., 31, 153–172. [Google Scholar]
- Hitchin F., Jenner,C., Harper,S., Mansfield,J., Barber,C. and Daniels,M. (1989) Determinant of cultivar specific avirulence cloned from Pseudomonas syringae pv. phaseolicola race 3. Physiol. Mol. Plant Pathol., 34, 309–322. [Google Scholar]
- Hobohm U. and Sander,C. (1995) A sequence property approach to searching protein databases. J. Mol. Biol., 251, 390–399. [DOI] [PubMed] [Google Scholar]
- Hu P., Elliott,J., McCready,P., Skowronski,E., Garnes,J., Kobayashi,A., Brubaker,R.R. and Garcia,E. (1998) Structural organization of virulence-associated plasmids of Yersinia pestis. J. Bacteriol., 180, 5192–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huguet E., Hahn,K., Wengelnik,K. and Bonas,U. (1998) hpaA mutants of Xanthomonas campestris pv. vesicatoria are affected in pathogenicity but retain the ability to induce host-specific hypersensitive reaction. Mol. Microbiol., 29, 1379–1390. [DOI] [PubMed] [Google Scholar]
- Innes R.W., Bent,A.F., Kunkel,B.N., Bisgrove,S.R. and Staskawicz,B.J. (1993) Molecular analysis of avirulence gene avrRpt2 and identification of a putative regulatory sequence common to all known Pseudomonas syringae avirulence genes. J. Bacteriol., 175, 4859–4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J.W. et al. (1999) Identification of a pathogenicity island, which contains genes for virulence and avirulence, on a large native plasmid in the bean pathogen Pseudomonas syringae pathovar phaseolicola. Proc. Natl Acad. Sci. USA, 96, 10875–10880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner C., Hitchin,E., Mansfield,J., Walters,K., Betteridge,P. and Taylor,J. (1991) Gene-for-gene interactions between Pseudomonas syringae pv. phaseolicola and Phaseolus. Mol. Plant-Microbe Interact., 4, 553–562. [PubMed] [Google Scholar]
- Jones D.A. and Jones,J.D.G. (1996) The role of leucine-rich repeat proteins in plant defences. Adv. Bot. Res., 24, 91–167. [Google Scholar]
- Kaniga K., Delor,I. and Cornelis,G.R. (1991) A wide-host-range suicide vector for improving reverse genetics in Gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene, 109, 137–141. [DOI] [PubMed] [Google Scholar]
- King E., Ward,M. and Raney,D. (1954) Two simple media for the demonstration of phycocyanin and fluorescein. J. Lab. Clin. Med., 44, 301–307. [PubMed] [Google Scholar]
- Kobayashi D., Tamaki,S.J. and Keen,N.T. (1989) Cloned avirulence genes from the tomato pathogen Pseudomonas syringae pv. tomato confer specificity on soybean. Proc. Natl Acad. Sci. USA, 86, 157–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi D., Tamaki,S., Trollinger,D.J., Gold,S. and Keen,N.T. (1990) A gene from Pseudomonas syringae pv. glycinea with homology to avirulence gene D from P.s. pv. tomato but devoid of the avirulence phenotype. Mol. Plant Microbe Interact., 3, 103–111. [DOI] [PubMed] [Google Scholar]
- Leong S., Ditta,G. and Helinski,D. (1982) Heme biosynthesis in Rhizobium. J. Biol. Chem., 257, 8724–8730. [PubMed] [Google Scholar]
- Lorang J and Keen,N.T. (1995) Characterization of avrE from Pseudomonas syringae pv. tomato: a hrp-linked avirulence locus consisting of at least two transcriptional units. Mol. Plant Microbe Interact., 8, 49–57. [DOI] [PubMed] [Google Scholar]
- Lorang J., Shen,H., Kobayashi,D., Cooksey,D. and Keen,N.T. (1994) avrA and avrE in Pseudomonas syringae pv. tomato PT23 play a role in virulence on tomato plants. Mol. Plant Microbe Interact., 7, 508–515. [Google Scholar]
- Mansfield J., Jenner,C., Hockenhull,R., Bennett,M.A. and Stewart,R. (1994) Characterization of avrPphE, a gene for cultivar-specific avirulence from Pseudomonas syringae pv. phaseolicola which is physically linked to hrpY, a new hrp gene identified in the halo-blight bacterium. Mol. Plant Microbe Interact., 7, 726–739. [DOI] [PubMed] [Google Scholar]
- Mansfield J., Bennett,M., Bestwick,C. and Woods-Tör,A. (1997a) Phenotypic expression of gene-for-gene interaction involving fungal and bacterial pathogens: variation from recognition to response. In Crute,I.R., Holub,E.B. and Burdon,J.J. (eds), The Gene-for-Gene Relationship in Plant–Parasite Interactions. CAB International, Wallingford, UK, pp. 265–291. [Google Scholar]
- Mansfield J., Tsiamis,G., Puri,N., Bennett,M., Jenner,C., Stevens,C., Teverson,D., Lyons,N. and Taylor,J. (1997b) Analysis of gene-for-gene interactions between Pseudomonas syringae pv. phaseolicola and Phaseolus. In Rudolph,K., Burr,T.J., Mansfield,J.W., Stead,D., Vivian,A. and von Kietzell,J. (eds), Pseudomonas syringae Pathovars and Related Pathogens. Kluwer Academic, Dordrecht, The Netherlands, pp. 385–391. [Google Scholar]
- Miller J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Murillo J., Shen,H., Gerhold,D., Sharma,A., Cooksey,D.A. and Keen,N.T. (1994) Characterization of pPT23B, the plasmid involved in syringolide production by Pseudomonas syringae pv. tomato PT23. Plasmid, 31, 275–287. [DOI] [PubMed] [Google Scholar]
- Orth K., Palmer,L.E., Bao,Z.Q., Stewart,S., Rudolph,A.E., Bliska,J.B. and Dixon,J.E. (1999) Inhibition of the mitogen-activated protein kinase kinase superfamily by a Yersinia effector. Science, 285, 1920–1923. [DOI] [PubMed] [Google Scholar]
- Person C., Samborski,D.J. and Rohringer,R. (1962) The gene-for-gene concept. Nature, 194, 561–562. [DOI] [PubMed] [Google Scholar]
- Pirhonen M.U., Lidell,M.C., Rowley,D.L., Lee,S.W., Jin,S., Liang,Y., Silverstone,S., Keen,N.T. and Hutcheson,S.W. (1996) Phenotypic expression of Pseudomonas syringae avr genes in E.coli is linked to the activities of the hrp-encoded secretion system. Mol. Plant Microbe Interact., 9, 252–260. [DOI] [PubMed] [Google Scholar]
- Reuber T.L. and Ausubel,F.M. (1996) Isolation of Arabidopsis genes that differentiate between resistance responses mediated by the RPS2 and RPM1 disease resistance genes. Plant Cell, 8, 241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter C. and Dangl,J.L. (1995) The avrRpm1 gene from Pseudomonas syringae pv. maculicola is required for avirulence on Arabidopsis. Mol. Plant Microbe Interact., 8, 444–453. [DOI] [PubMed] [Google Scholar]
- Ritter C. and Dangl,J.L. (1996) Interference between two specific pathogen recognition events mediated by distinct plant disease resistance genes. Plant Cell, 8, 251–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis T., Piedras,P., Zhang S., Klessig,D.F., Hirt,H. and Jones,J.D. (1999) Rapid Avr9- and Cf9-dependent activation of MAP kinases in tobacco cell cultures and leaves. Convergence of resistance gene, elicitor, wound and salicylate responses. Plant Cell, 11, 273–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald P.C. and Staskawicz,B.J. (1988) The avirulence gene avrBs1 from Xanthomonas campestris pv. vesicatoria encodes a 50kD protein. Mol. Plant Microbe Interact., 1, 191–198. [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Shintaku M.H., Kluepfel,D.A., Yacoub,A. and Patil,S.S. (1989) Cloning and partial characterization of an avirulence determinant from race 1 of Pseudomonas syringae pv. phaseolicola. Physiol. Mol. Plant Pathol., 35, 313–322. [Google Scholar]
- Singer M. and Berg,P. (1991) Genes and Genomes: A Changing Perspective. University Science Books, Mill Valley, CA. [Google Scholar]
- Stachel S.E., An,G., Flores,C.W. and Nester,E.W. (1985) A Tn3lacZ transposon for the random generation of β-galactosidase gene fusions: Application to the analysis of gene expression in Agrobacterium. EMBO J., 4, 891–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskawicz B., Dahlbeck,D., Keen,N. and Napoli,C. (1987) Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol., 169, 5789–5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens C., Bennett,M.A., Athanassopoulos,E., Tsiamis,G., Taylor,J.D. and Mansfield,J.W. (1998) Sequence variations in alleles of the avirulence gene avrPphE.R2 from Pseudomonas syringae pv. phaseolicola lead to loss of recognition of the AvrPphE protein within bean cells and a gain in cultivar-specific virulence. Mol. Microbiol., 29, 165–177. [DOI] [PubMed] [Google Scholar]
- Tang X.Y., Frederick,R.D., Zhou,J., Halterman,D.A., Jia,Y. and Martin,G.B. (1996) Initiation of plant-disease resistance by physical interaction of AvrPto and Pto kinase. Science, 274, 2060–2063. [DOI] [PubMed] [Google Scholar]
- Taylor J.D., Teverson,D.M., Allen,D.J. and Pastor-Corrales,M.A. (1996) Identification and origin of races of Pseudomonas syringae pv. phaseolicola from Africa and other bean growing areas. Plant Pathol., 45, 469–478. [Google Scholar]
- Van den Ackerveken G.F.J.M., Marois,E. and Bonas,U. (1996) Recognition of the bacterial avirulence protein AvrBs3 occurs inside the host plant cell. Cell, 87, 1307–1316. [DOI] [PubMed] [Google Scholar]
- Vivian A. and Mansfield,J. (1993) A proposal for a uniform genetic nomenclature for avirulence genes in phytopathogenic pseudomonads. Mol. Plant Microbe Interact., 6, 9–10. [Google Scholar]
- Vivian A., Gibbon,M.J. and Murillo,J. (1997) The molecular genetics of specificity determinants in plant pathogenic bacteria. In Crute,I.R., Holub,E.B. and Burdon,J.J. (eds), The Gene-for-Gene Relationship in Host–Parasite Interactions. CAB International, Wallingford, UK, pp. 293–328. [Google Scholar]
- Whalen M.C., Innes,R.W., Bent,A.F. and Staskawicz,B.J. (1991) Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell, 3, 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J.R., Vivian,A., Jenner,C., Mansfield,J.W. and Taylor,J.D. (1994) Detection of a gene in pea controlling nonhost resistance to Pseudomonas syringae pv. phaseolicola. Mol. Plant Microbe Interact., 7, 534–537. [Google Scholar]
- Yucel I., Slaymaker,D., Boyd,C., Murillo,J., Buzzell,R.I. and Keen,N.T. (1994) Avirulence gene avrPphC from Pseudomonas syringae pv. phaseolicola 3121: a plasmid-borne homologue of avrC closely linked to an avrD allele. Mol. Plant Microbe Interact., 7, 677–679. [DOI] [PubMed] [Google Scholar]