SUMMARY
Both microRNAs and alternative pre-mRNA splicing have been implicated in the development of the nervous system (NS), but functional interactions between these two pathways are poorly understood. We demonstrate that the neuron-specific microRNA miR-124 directly targets PTBP1 (PTB/hnRNP I) mRNA, which encodes a global repressor of alternative pre-mRNA splicing in nonneuronal cells. Among the targets of PTBP1 is a critical cassette exon in the pre-mRNA of PTBP2 (nPTB/brPTB/PTBLP), an NS-enriched PTBP1 homolog. When this exon is skipped, PTBP2 mRNA is subject to nonsense-mediated decay (NMD). During neuronal differentiation, miR-124 reduces PTBP1 levels, leading to the accumulation of correctly spliced PTBP2 mRNA and a dramatic increase in PTBP2 protein. These events culminate in the transition from non-NS to NS-specific alternative splicing patterns. We also present evidence that miR-124 plays a key role in the differentiation of progenitor cells to mature neurons. Thus, miR-124 promotes NS development, at least in part by regulating an intricate network of NS-specific alternative splicing.
INTRODUCTION
Cellular differentiation requires global changes in regulatory networks at all levels of gene expression. Posttranscriptional gene regulation affords a mechanism for rapid changes in the cellular proteome and is widely used during the development of the mammalian nervous system (NS). Among the well-studied examples of posttranscriptional control are the regulation of alternative pre-mRNA splicing (Black, 2003; Licatalosi and Darnell, 2006) and repression of protein synthesis by microRNAs (miRNAs) (Kosik, 2006).
Approximately half of all mammalian genes encode alternatively spliced pre-mRNAs, and a large fraction of the alternative splicing events occur exclusively in the NS (Black, 2003; Licatalosi and Darnell, 2006; Lipscombe, 2005; Maniatis and Tasic, 2002; Yeo et al., 2004). This process generates a vast combinatorial diversity of NS-specific proteins that function in axon guidance, synaptic connectivity, and synaptic transmission (Licatalosi and Darnell, 2006; Lipscombe, 2005; Ule and Darnell, 2006).
The RNA-binding protein PTBP1 (also known as PTB or hnRNP I) has been described as a repressor of NS-specific splicing. PTBP1 binds to pyrimidine-rich sequences in pre-mRNAs and inhibits the splicing of nearby neuron-specific alternative exons (Black, 2003; Sharma et al., 2005; Spellman and Smith, 2006; Wagner and Garcia-Blanco, 2001). PTBP1 is expressed at high levels in nonneuronal cells but is downregulated in the NS, thus allowing the inclusion of neuron-specific exons in mature mRNAs (Ashiya and Grabowski, 1997; Lillevali et al., 2001; Markovtsov et al., 2000). The mechanism by which PTBP1 is downregulated in neurons is not known.
The reduced expression of PTBP1 coincides with an increased expression of its NS-enriched homolog, PTBP2 (a.k.a. brPTB, nPTB, or PTBLP) (Kikuchi et al., 2000; Lillevali et al., 2001; Markovtsov et al., 2000; Polydorides et al., 2000). PTBP2 has been implicated in the regulation of alternative splicing of the Src N1 exon and the glycine receptor α2 exon E3A (Markovtsov et al., 2000; Polydorides et al., 2000). Recent studies have shown that PTBP2 mRNA translation is inhibited by a muscle-specific miRNA, miR-133, in differentiating C2C12 myoblasts, which results in low levels of PTBP2 protein in muscle (Boutz et al., 2007). However, the mechanisms by which PTBP2 protein is downregulated in other tissues, and up-regulated in the NS, are not understood.
miRNAs are ~22 nt noncoding RNAs that play important roles in the posttranscriptional regulation of gene expression (Bartel, 2004; Kim, 2005; Kloosterman and Plasterk, 2006; Valencia-Sanchez et al., 2006). miRNAs act by repressing translation and/or by destabilizing target mRNAs. In metazoans, individual miRNAs can downregulate hundreds of mRNA targets by interacting with partially complementary sequences within their 3′ untranslated regions (3′UTR) (Lewis et al., 2005; Lim et al., 2005). The miRNA/mRNA interaction normally requires 6–8 base pairs (bp) of perfect complementarity between the miRNA 5′ terminus (seed sequence) and a cognate miRNA target site (MTS) in the mRNA 3′UTR (Brennecke et al., 2005; Doench and Sharp, 2004; Farh et al., 2005; Lewis et al., 2005; Rajewsky, 2006).
Although a number of miRNAs are expressed in the NS (Kim et al., 2004; Krichevsky et al., 2003; Miska et al., 2004; Sempere et al., 2004), relatively little is known about their functions (Ashraf et al., 2006; Chang et al., 2004; Kosik, 2006; Li et al., 2006; Schratt et al., 2006). One of the most conserved and abundantly expressed NS-specific miRNAs is miR-124 (Lagos-Quintana et al., 2002). In human and mouse, this miRNA is encoded by three distinct genes (Conaco et al., 2006; Deo et al., 2006). miR-124 is expressed in neurons, but not astrocytes, and the levels of miR-124 increase over time in the developing NS (Krichevsky et al., 2003; Miska et al., 2004; Smirnova et al., 2005). In combination with the NS-specific miRNA miR-9, miR-124 stimulates neuronal and represses glial differentiation of ES cells in vitro (Krichevsky et al., 2006). When introduced into nonneural cells, miR-124 downregulates a number of nonneuronal mRNAs (Lim et al., 2005). Conversely, inhibition of miR-124 with an antisense 2′-OMe-RNA increases levels of nonneuronal transcripts in primary cortical neurons (Conaco et al., 2006).
Two recent studies show that miR-124 downregulates a subset of predicted mRNA targets in vivo (Cao et al., 2007; Visvanathan et al., 2007). In addition, one of these studies describes a stimulatory effect of miR-124 on neuronal differentiation in the developing spinal cord and in the pluripotent embryonal carcinoma cell line P19 (Visvanathan et al., 2007). This stimulation is, at least in part, due to miR-124-induced repression of SCP1, a small phosphatase specific for phosphoserines in the C-terminal domain of RNA polymerase II (Visvanathan et al., 2007). SCP1 has been implicated in the function of the NRSF/REST complex, a global repressor of NS-specific transcription in nonneuronal cells (Yeo et al., 2005). Thus, miR-124 may control the global de-repression of NS-specific gene expression at the level of transcription.
Here we show that miR-124 induces a switch from general to neuron-specific alternative splicing, by directly targeting the mRNA of PTBP1. Among the pre-mRNAs controlled by PTBP1 at the level of splicing are PTBP2 and Gabbr1 (GABA-B receptor 1). In these two cases, PTBP1-mediated exon skipping leads to nonsense-mediated decay (NMD) of the corresponding mRNA species. We show that PTBP2 also functions as a splicing repressor of PTBP1-controlled exons. However, the repressive effect of PTBP2 is significantly weaker than that of PTBP1. We also demonstrate that miR-124 enhances neuronal differentiation of P19 cells, and that PTBP1 downregulation contributes to this stimulatory effect. We conclude that miR-124 promotes NS development at least in part by repressing PTBP1 and triggering a downstream switch to global NS-specific alternative splicing.
RESULTS
miR-124 Induces Neuronal Differentiation and NS-Specific Alternative Splicing in Neuroblastoma Cell Lines
To investigate the function of miR-124 in neuronal cells, we transfected mouse neuroblastoma cell lines, CAD and Neuro2a (N2a), with the RIPmiR-124 plasmid (dsRed, intron, pre-miR-124), in which the miRNA sequence is inserted in an intron of a recombinant dsRed2 gene expressed under the control of a CMV promoter. Using this expression system, miRNA producing cells could be easily visualized and FACS sorted to obtain populations expressing physiological levels of miRNA (Figure 1A and Figure S1 in the Supplemental Data available with this article online; also see Supplemental Results). Normally, the neuronal differentiation of CAD and N2a cells requires serum starvation or retinoic acid (RA) treatment, respectively. However, when either cell line was transfected with RIPmiR-124 striking neuron-like differentiation was observed in the absence of any additional treatment (Figures S2–S4, and the top two panels in Figure 1G; also see Supplemental Results). By contrast, the empty RI vector (dsRed, intron) and plasmids expressing other miRNAs did not induce visible differentiation.
Figure 1. miR-124 Targets the Global Splicing Repressor PTBP1 and Promotes NS-Specific Alternative Splicing.
(A) CAD cells were transfected with the RI or RIPmiR-124 plasmids and FACS sorted to enrich dsRed-positive cells and postcultured for 12 hr or 60 hr. The expression of miR-124 was analyzed by northern blotting (2.5 mg/lane total RNA) along with RNA from newborn (P0) mouse brain (2.5 mg/lane) and a synthetic miR-124 22-mer RNA (Dharmacon; 5 fmol/lane). The mature miR-124 species (21–24 nt) and the hairpin-like miR-124 precursor (pre) (Kim, 2005) are indicated on the left. Bottom panel, U6 snRNA loading control.
(B) Table based on microarray data listing genes upregulated by miR-124 >2-fold. NS enrichment of exon-included and exon-skipped forms was calculated as a fraction of corresponding cDNA/EST clones originating from the NS.
(C) Luciferase reporter constructs containing PTBP1 and PTBP2 3′UTRs. Positions of the miR-124 MTSs are indicated.
(D) Interactions of miR-124 with the conserved 8- and 9-mers within the PTBP1 and PTBP2 3′UTRs.
(E) N2a cells were cotransfected with RIPmiR-124 luciferase reporter constructs containing the shown 3′UTRs, and the luciferase activity was measured 24 hr posttransfection. For each experiment, luciferase activity was normalized to the corresponding RI vector control. Data are the means of three independent transfections, each done in triplicate, ± SD.
(F) Immunoblot showing that miR-124 induces a decrease in PTBP1 protein and an increase in PTBP2 protein. CAD cells were transfected with RIPmiR or RI in the presence of pcDNA3 or pCMV-fPTBP, sorted, and postcultured for 60 hr. Tubulin (Tub) is a loading control. Graphs on the right show quantitation of the corresponding band intensities performed by detecting chemiluminescence with a CCD camera.
(G) PTBP1 downregulation is required for the miR-124-induced neuronal differentiation of CAD cells. Cells were prepared as in (F) and imaged using dsRed fluorescence.
To investigate the mechanism of the miR-124-induced differentiation, RNA from RIPmiR-124- and RI-transfected CAD cells was analyzed using gene expression microarrays. The expression of miR-124 at physiological levels led to a >1.5-fold (q < 1%) downregulation of 342 annotated genes, at least some of which could be bona fide miR-124 targets (Figure S5B and Table S2; Supplemental Results).
Surprisingly, miR-124 expression also led to a >1.5-fold (q < 1%) upregulation of 61 mRNAs. Twelve genes that were upregulated >2-fold were further investigated (Figure 1B). Examination of cDNA and expressed sequence tag (EST) sequences revealed that at least ten of these genes contain alternatively spliced exons, and that the exon-included transcripts of these genes are enriched in the NS compared to their exon-skipped counterparts (Figure 1B). In eight genes, the upregulated probe sets correspond to an alternatively spliced exon, while probe sets specific to other regions of the same genes showed little or no change (Figure S6). A systematic analysis of the microarray data identified six additional genes, in which miR-124 expression altered the patterns of pre-mRNA splicing (Figure S6 and Table S3). A similar effect on alternative splicing was observed when cells were transfected with a synthetic siRNA-like miR-124 precursor (data not shown).
miR-124 Targets mRNAs of the Splicing Regulators PTBP1 and PTBP2
We considered the possibility that changes in alternative splicing induced by miR-124 were due to the downregulation of a splicing repressor. In fact, the splicing repressor PTBP1 was among the top-scoring downregulated genes in miR-124-treated cells (Figure S5A). The PTBP1 3′UTR contains one 8-mer miR-124 MTS conserved in mammals, one rodent-specific 7-mer, and four 6-mers, which may potentially function as miR-124 MTSs (Figures 1C and 1D). Interestingly, the 3′UTR of PTBP2 mRNA also contains conserved miR-124-specific MTSs (Figures 1C and 1D).
To determine whether miR-124 can directly target PTBP1 and PTBP2 mRNAs, we cotransfected N2a cells with luciferase reporters containing PTBP1 or PTBP2 3′UTRs and either RIPmiR-124 or control RI plasmids (Figure 1E). Cotransfection with RIPmiR-124 decreased the expression of the PTBP1 3′UTR-containing reporter to ~42% of the level of the RI-transfected control. The reporter containing the PTBP2 3′UTRs was downregulated to ~74% of the RI level. Downregulation was not observed when the luciferase reporter was fused with 3′UTRs from an SV40 virus mRNA or the mouse NRSF/REST mRNA, both of which lack discernible miR-124 binding sites. On the other hand, the 3′UTR of the SCP1 mRNA, a known miR-124 target (Visvanathan et al., 2007), decreased luciferase activity to ~36% of the RI control (Figure 1E). Site-directed mutagenesis experiments confirmed that the putative MTSs were specifically targeted by miR-124 (Figure S7). miR-124 also inhibited the PTBP1 and PTBP2 3′UTR reporters in HEK293T cells, whereas two other miRNAs, miR-9 and miR-31, did not have a significant effect (Figure S8). Thus, both PTBP1 and PTBP2 mRNAs are specifically targeted by miR-124; however, PTBP1 is a much better target than PTBP2.
miR-124 Expression Leads to a Decrease in Endogenous PTBP1 Protein and an Increase in PTBP2
We also examined the effect of miR-124 on endogenous PTBP1 and PTBP2 protein levels. The PTBP1 levels decreased ~4.6-fold in CAD cells transfected with RIPmiR-124 (Figure 1F, lanes 1 and 2). Surprisingly, the levels of endogenous PTBP2 protein increased ~6-fold, despite the presence of miR-124 MTSs in the 3′UTR of PTBP2 mRNA (Figure 1F, lanes 1 and 2). To determine whether the downregulation of PTBP1 is required for miR-124-induced neuronal differentiation, we cotransfected CAD cells with RIPmiR-124 and a Flag-PTBP1 expression construct lacking the PTBP1 3′UTR (pCMV-fPTBP1; Figure 1F, lanes 3 and 4). A partial rescue of the endogenous PTBP1 protein level dramatically inhibited miR-124-induced neurite outgrowth (Figure 1G). Attempts to completely restore the PTBP1 levels in RIPmiR-124a-transfected cells adversely affected cell viability (data not shown). Thus, endogenous PTBP1 mRNA is targeted by miR-124, and a decrease in PTBP1 protein levels is required for miR-124-dependent neuronal differentiation of CAD cells.
PTBP1 Downregulation Is Required for miR-124-Induced Alternative Splicing
PTBP1 is a splicing repressor that binds to pyrimidine-rich sequences in pre-mRNA and prevents the inclusion of adjacent exons in mature mRNA (Black, 2003; Spellman and Smith, 2006; Wagner and Garcia-Blanco, 2001). Examination of the nucleotide sequences surrounding the 5′ ends of alternatively spliced exons upregulated by miR-124 revealed that potential PTBP1 binding sites (YUCUUY and YUCUCY) are present, on average, ~3 times more frequently (p = 8.05E-4; Wilcoxon test) than at similar positions in the control set of exons from the same genes (Figure S9).
To determine whether downregulation of PTBP1 was required for the inclusion of these exons, we analyzed the effect of miR-124 on the abundance of NS-specific (N) and general (G) forms of Mtap4, Rufy3, and Cdc42 mRNAs using reverse transcriptase-quantitative PCR (RT-qPCR) (Figure 2A). Mtap4, Rufy3, and Cdc42 genes have 3′ alternative exons and are top hits on the microarray list (Figure 1B). miR-124 stimulated N splicing for all three pre-mRNAs, while having little effect on the G splicing (Figure 2A). However, when the levels of PTBP1 were partially supplemented by expression of pCMV-fPTBP1, both the basal level of the N form and the degree of its induction by miR-124 significantly decreased (Figure 2A). A similar observation was made with Rtn4/Nogo pre-mRNA that contains an alternatively spliced cassette exon (Figures S10A and S10B). We also showed that putative PTBP1 binding sites upstream of the NS-specific exon 4a of Mtap4 pre-mRNA were required for PTBP1-dependent repression of exon inclusion using minigene transfection experiments (Supplemental Results and Figures S11A–S11C).
Figure 2. Downregulation of PTBP1 Is Necessary and Sufficient for the miR-124-Mediated Splicing Switch.
(A) PTBP1 downregulation is necessary for the miR-124 mediated splicing of NS-specific 3′ alternative exons. Transfected CAD cells were prepared as in Figure 1F and analyzed by RT-qPCR. (Top) Structure of the alternatively spliced regions of Mtap4, Rufy3, and Cdc42 genes. Blue, NS-specific alternative 3′ exons induced by miR-124; peach, other exons. Black arrowheads, polyadenylation signals. (Bottom) RT-qPCR analysis of the N splicing (primers F1/R1) and G splicing (primers F1/R2). The color codes are as shown to the left of the first histogram.
(B) RNAi depletion of PTBP1 (blue bars) stimulates N splicing and represses G splicing in Mtap4, Rufy3, and Cdc42 pre-mRNAs. Yellow bars, samples transfected with a control siRNA. In all RT-qPCR analyses, data are means of three amplifications ± SD, normalized to RI vector + pcDNA3 in (A) or control siRNA transfection in (B).
A Decrease in PTBP1 Levels Is Sufficient for miR-124-Induced Alternative Splicing
To determine whether the miR-124-induced decrease in PTBP1 levels is sufficient for the splicing switch, we knocked down PTBP1 using RNAi and repeated the RT-qPCR analyses (Figure 2B). A nearly complete depletion of PTBP1 protein (see Figure 3C) stimulated N splicing of Mtap4, Rufy3, and Cdc42 pre-mRNAs 4.4- to 15.1-fold, similar to the effects observed with miR-124 (Figure 2B). G splicing was diminished 1.4- to 2.5-fold in all three cases. We also confirmed that PTBP1 is sufficient for the alternative splicing switch in Rtn4/Nogo (Figures S10C and S10D) and Pdlim7 pre-mRNAs (data not shown). The changes in alternative splicing induced by PTBP1 siRNA were not due to off-target effects (Figure S12). Additional studies revealed that miR-124 also induced the inclusion of exons known to be regulated by PTBP1 (Figure S13). We conclude that the decrease in PTBP1 levels is both necessary and sufficient for the miR-124-induced inclusion of several alternative NS-specific exons.
Figure 3. PTBP1 Controls the Abundance of PTBP2 mRNA by Repressing Alternative Exon Splicing and Triggering NMD.
(A) Ptbp2 gene diagram showing the alternatively spliced exon 10 (blue). The correct stop codon and the premature termination codon (PTC) generated as a result of exon 10 skipping are indicated by red and purple arrowheads, respectively.
(B) Both miR-124 and PTBP1-specific siRNA reduce PTBP1 mRNA levels and induce a reciprocal increase in PTBP2 mRNA. The RT-qPCR analysis was carried out using primers specific to constitutively spliced regions of the two genes (Ptbp1 F1/R1 and Ptbp2 F2/R2, respectively; Table S1). Data are means of three amplifications ± SD. The mean of the RI transfection experiment was set to 1.
(C and D) (C) Time course of PTBP1 protein downregulation by RNAi. Only PTBP1 knockdown leads to the accumulation of PTBP2 protein. (D) RT-PCR showing the effects of PTBP1 and Rent1 knockdowns on the distribution of exon 10 included (i) and skipped (s) forms of PTBP2 mRNA.
(E) Rent1 knockdown consistently increased the fraction of the (s) form. Quantifications are averages of four independent experiments ± SD.
miR-124 Regulates the Abundance of PTBP2 and Gabbr1 mRNAs by Preventing PTBP1-Dependent Exon Skipping and NMD
Ptbp2 and GABA-B receptor 1 (Gabbr1) genes contain alternatively spliced cassette exons (Rahman et al., 2004; Schwarz et al., 2000; Figure 3A and Figure S14A), and the levels of PTBP2 and Gabbr1 mRNAs are significantly upregulated by miR-124 (Figure 1B). PTBP2 cassette exon 10 has been shown to control the abundance of PTBP2 mRNA (Ni et al., 2007). When this exon is skipped, PTBP2 mRNA acquires a premature termination codon (PTC) and is subject to NMD, whereas the exon-included mRNA has a complete open reading frame and is therefore stabilized (Ni et al., 2007). Similarly, skipping alternative exon 15 in Gabbr1 mRNA should generate an in-frame nonsense codon (Figure S14A), and low level expression of the corresponding exon-skipped form has been detected mainly outside of the NS (Schwarz et al., 2000).
PTBP1 protein is known to control the abundance of its own mRNA through an alternative exon skipping and subsequent NMD (Wollerton et al., 2004). Because PTBP2 exon 10 and Gabbr1 exon 15 contain adjacent PTBP1 consensus binding sites (Figure S9), it seems likely that PTBP1 can also repress these exons, thus subjecting the exon-skipped mRNAs to NMD. Consistent with this possibility, PTBP1 downregulation by RNAi induced a robust accumulation of both PTBP2 and Gabbr1 mRNAs (Figure 3B and Figure S14B). Comparable changes in mRNA abundance were detected when cells were transfected with RIPmiR-124 (Figure 3B and data not shown). Moreover, the time course of PTBP1 downregulation by siRNA closely followed the time course of PTBP2 protein accumulation (Figure 3C).
To directly test whether PTBP1-controlled exon skipping and NMD contributed to the PTBP2 and Gabbr1 regulation, we knocked down expression of either PTBP1 or an essential NMD component, Rent1/Upf1 (Figure 3C and Figure S15), and examined the changes in the ratios between the corresponding exon-included (i) and skipped (s) mRNA forms by RT-PCR (Figures 3D and 3E and Figure S14C). As expected from the PTBP1/NMD model, PTBP1 knockdown dramatically upregulated the (i) forms of both PTBP2 and Gabbr1 mRNAs, while Rent1 knockdown led to a detectable accumulation of the (s) form. We also carried out minigene experiments confirming that putative PTBP1 binding sites upstream of the PTBP2 exon 10 were required for the repression of this exon (Supplemental Results and Figures S11D–S11F). Thus, PTBP1 controls the abundance of at least two NS-enriched mRNAs through a mechanism involving alternative splicing and NMD.
PTBP2 Is a Weak Repressor of Several PTBP1-Controlled Exons
We also studied the potential involvement of PTBP2 in the regulation of the miR-124-induced exons. For this purpose, we knocked down both PTBP1 and PTBP2 by RNAi in CAD cells and assayed the effect of this treatment on alternative splicing (Figure 4). Interestingly, the depletion of both PTBP1 and PTBP2 resulted in ~3 times more efficient derepression of the NS-specific exons in Cltb, Mtap4, and Cdc42 pre-mRNAs compared to depletion of PTBP1 alone. The knockdown of PTBP2 also stimulated the inclusion of the N1 exon in Src pre-mRNA but had no detectable effect on Rufy3 splicing. However, in the latter case PTBP1 knockdown alone converted most of the mRNA into the N splicing form (Figure 4C, bottom). Thus, it may not be possible to detect an effect of PTBP2 on Rufy3 splicing in the absence of PTBP1. We conclude that, at least in the pre-mRNAs examined, PTBP2 functions as a weak repressor of PTBP1-regulated exons.
Figure 4. PTBP2 Is a Weak Repressor of PTBP1-Regulated Exons.
(A) CAD cells were transfected with siRNAs against PTBP1 and/or PTBP2 as indicated, and the protein levels were analyzed by immunoblotting 72 hr posttransfection. Knockdown of both PTBP1 and PTBP2 induced a more complete derepression of (B) cassette NS exons and (C) 3′-terminal alternative exons than knockdown of PTBP1 alone. RT-qPCR in (C) was carried out as in Figure 2. Data are means of three amplifications ± SD, normalized to control siRNA transfection. For multiplex RT-PCR, we used mixtures of the corresponding F1/R1/R2 primers (see Figure 2A and Table S1).
miR-124 and PTBP1 Display Reciprocal Patterns of Expression in the Mouse Embryo
To determine whether miR-124 can repress PTBP1 in vivo, we visualized the expression of mature miR-124 in the developing mouse embryo using in situ hybridization (ISH) with an miR-124-specific locked nucleic acid (LNA) probe. Consistent with an earlier report (Deo et al., 2006), miR-124 is expressed in both the central and peripheral NS (CNS and PNS; Figures 5A, 5B, and 5D and Figure S16). Throughout the CNS, miR-124 is expressed at low levels in the neural precursors residing in the neuroepithelial layer (NL) in the spinal cord and the ventricular zone (VZ) in the forebrain. On the other hand, miR-124 expression is substantially higher in differentiating and mature neurons residing in the spinal cord mantle layer (ML) and in the differentiated forebrain regions from subventricular zone (SVZ) to cortical plate (CP).
Figure 5. Reciprocal Patterns of miR-124 and PTBP1 Expression during Mouse Development Correlate with Overlapping Patterns of miR-124 and NS-Specific Alternative Splicing.
(A) Mature miR-124 expression in an E12.0 embryo visualized by ISH with an miR-124-specific LNA probe.
(B) ISH of E10.5 transverse sections at the cervical level showing that PTBP1 mRNA and miR-124 are expressed in a reciprocal pattern, while the expression of miR-124 and PTBP2 mRNA overlap. The exception to this is in the NL, where both PTBP1 and PTBP2 mRNA are present. However, at the protein level PTBP1 and PTBP2 are not coexpressed in the NL, suggesting that PTBP1 is translationally repressed.
(C) Immunofluorescence analysis shows that PTBP1 protein (green) is largely excluded from the spinal cord, whereas PTBP2 protein (red) is expressed throughout the spinal cord but excluded from cells expressing high levels of PTBP1 protein.
(D) ISH of horizontal sections of diencephalon at E13.5 showing miR-124 expression in differentiating neurons, opposite to PTBP1 expression in the VZ containing neural precursor cells, both dorsally (Neurog2-positive region) and ventrally (Mash1). PTBP2 expression in the VZ is relatively low, but it is higher in the subventricular zone (SVZ), which contains cells committed to differentiation. The pattern is characterized by the SVZ marker Dlx5.
(E) ISH of horizontal sections of forebrain at E13.5 showing PTBP1 is expressed in the VZ of the telencephalon, where little or no miR-124 is detected. As expected, the N splicing patterns of Cdc42 and Rufy3 are opposite to that of PTBP1 expression, and identical to that of miR-124 expression. By contrast, the G forms of Cdc42 and Rufy3 are enriched or at least not excluded in the regions expressing PTBP1. sc, spinal cord; rh, rhombencephalon; me, mesencephalon; di, diencephalon; te, telencephalon; NL, neuroepithelial layer; ML, mantle layer; DRG, dorsal root ganglia; VZ, ventricular zone; SVZ, subventricular zone; CP, cortical plate; and LV, lateral ventricle.
We also hybridized adjacent sections with probes specific to PTBP1 and PTBP2 mRNAs (Figures 5B, 5D, and 5E and Figure S16). Consistent with the experiments described above, the expression patterns of PTBP1 mRNA and miR-124 RNA are mutually exclusive. Conversely, PTBP2 mRNA was enriched in CNS and PNS. Surprisingly, detectable levels of PTBP2 mRNA were coexpressed with PTBP1 mRNA in the neural progenitor zones (NL and VZ; Figures 5B and 5D). However, immunostaining data showed that the levels of PTBP1 protein in this region were extremely low, which suggests that PTBP1 mRNA may be subject to translational repression in this region (Figure 5C).
We also examined the expression of the N and G forms of Mtap4, Rufy3, and Cdc42 mRNAs in embryonic fore-brain using splice form-specific probes (Figure 5E and the top row in Figure 6A). As expected of bona fide PTBP1 targets, all three N splice forms were excluded from the PTBP1-expressing VZ, but expressed in the regions containing differentiating and mature neurons, which lack detectable PTBP1 expression. Conversely, the G forms of these mRNAs were not excluded from the regions expressing PTBP1. Cdc42-G showed a dramatic enrichment in the PTBP1-expressing cells (Figure 5E). Additional analyses of the spatiotemporal correlation between miR-124 expression and patterns of PTBP1-cotrolled alternative splicing are presented in the Supplemental Results and Figure S17. Taken together, these data provide further evidence for the conclusion that miR-124 downregulates PTBP1 and thus derepresses NS-specific splicing.
Figure 6. NS-Specific miRNAs Control PTBP1 Expression In Vivo.
(A) At E14.5, the mutant forebrain (bottom row) is substantially reduced in size and enveloped by a thin lamina of Foxg1-positive cells expressing high levels of PTBP1 mRNA and the Mtap4 G splice form but low levels of the Mtap4 N form. Wild-type control sections are shown on the top. In each panel, dashed lines mark the lateral and diencephalic boundaries of the telencephalon. LV, lateral ventricle; VZ, ventricular zone; di, diencephalon; and me, mesenchyme. Rostral, left; lateral, top.
(B) Quantitation of PTBP1 expression from (A). To quantify, PTBP1 mRNA levels in the lateral telencephalic wall of Dicer mutant and the lateral layer of wild-type VZ were normalized to corresponding levels in mesenchymal cells and the normalized values were averaged from six mutant and six wild-type forebrain sections (±SD).
(C–E) miR-124 reduces PTBP1 expression in cortical neurons. (C) Primary cortical cultures from E15.5 embryos contain ~95% Tuj1-positive cells. (D) Transfection of these cells with 2′-OMe-RNA against miR-124, but not miR-1, inactivates the endogenous miR-124. (E) RT-qPCR data showing that miR-124 knockdown induces an increase in the PTBP1 and Vamp3 mRNA levels, but not the levels of Rps17 mRNA. Hprt mRNA was used as a normalization control, and the corresponding expression levels in neurons transfected with anti-miR-1 were set to 1. Results are averaged from two independent transfection experiments each done in triplicate ± SD.
The Dicer/miRNA Pathway Is Involved in the Regulation of PTBP1 Expression In Vivo
To determine whether the miRNA processing machinery is required to establish PTBP1 expression patterns in the embryonic NS, we knocked out the essential miRNA biogenesis component, Dicer, in the telencephalic region of the brain. This was accomplished by crossing a mouse line containing a conditional Dicer allele (DicerloxP) with a Foxg1-cre line that expresses Cre recombinase throughout the telencephalon (Harfe et al., 2005; Harris et al., 2006; Hebert and McConnell, 2000).
At E13.5–E14.5, Foxg1-cre/DicerloxP/loxP embryos exhibited reduced forebrains compared to their wild-type littermates. The affected tissue appeared to have the normal number of cells undergoing mitosis in the VZ (Figure S18A). However, the expression of neuronal differentiation markers (Tuj1, MAP2, and Dlx5) along the lateral telencephalic wall was uneven and visibly weaker than in the wild-type mouse (Figures S18B, S18D, and S18E). These uneven patterns are most likely due to an increase in apoptosis, because we detect strong staining of cleaved caspase-3 in this region (Figure S18C). As expected, miR-124 was absent from the E13.5–E14.5 Dicer null telencephala (Figure 6A and Figure S18F).
Strikingly, in Foxg1-cre/DicerloxP/loxP embryos, high levels of PTBP1 protein were observed throughout the entire span of the lateral telencephalic wall (Figures S18D and S18E). This pattern differed dramatically from that of the wild-type embryos, where telencephalic expression of PTBP1 protein was restricted to the VZ. Cells ectopically expressing PTBP1 in Dicer mutant forebrain were MAP2 negative, but a fraction of them expressed Tuj1, an earlier marker of neuronal development than MAP2 (Figures S18D and S18E). Coexpression of PTBP1 and Tuj1 was not observed in the wild-type littermates.
We also found that the levels of PTBP1 mRNA were elevated ~2.5-fold in Foxg1-positive regions of E14.5 Dicer mutants compared to the PTBP1 levels in the VZ of the wild-type littermates (Figures 6A and 6B). The elevated PTBP1 expression correlated with a decrease in the levels of PTBP2, a low abundance of the N splice forms of Mtap4, Rufy3, and Cdc42, and normal or high abundance of the corresponding G forms (Figure 6A, Figure S18F, and data not shown). These data are consistent with the conclusion that the Dicer/miRNA pathway controls PTBP1 expression in the mouse brain in vivo.
miR-124 Promotes Downregulation of PTBP1 mRNA in Cortical Neurons
To further investigate the role of miR-124 in vivo, we isolated cortical neurons from E15.5 mouse embryos (Figure 6C) and transfected them with antisense 2′-OMe-RNAs complementary to miR-124, or with a control 2′-OMe-RNA complementary to muscle-specific miRNA miR-1, as described in Conaco et al. (2006). The amounts of anti-miR-124 antisense RNA were in excess to the endogenous mature miR-124 and pre-miR-124 (Figure 6D). RT-qPCR analyses revealed an ~1.6-fold (p < 0.005) increase in PTBP1 mRNA from the anti-miR-124-transfected cells as compared to the anti-miR-1-transfected control (Figure 6E). A similar effect of anti-miR-124 was observed with Vamp3 mRNA (Figure 6E), the top-scoring hit on the list of mRNAs downregulated by miR-124 in CAD cells (Figure S5A). By contrast, the expression level of Rps17 mRNA, which does not have miR-124 MTSs, was not affected. We conclude that miR-124 is required for the downregulation of PTBP1 in developing neurons.
PTBP1 Repression Is Essential for Efficient Neuronal Differentiation of P19 Cells
To determine whether miR-124-dependent PTBP1 down-regulation is required during neuronal differentiation, we used P19 cells, an embryonal carcinoma cell line that can be induced to differentiate into neurons by RA. RA also promotes an upregulation of miR-124 expression in this system (Sempere et al., 2004). We found that the RA-induced accumulation of miR-124 correlates with a reciprocal decrease in PTBP1 protein levels, an increase in PTBP2 levels, and a switch to NS-specific alternative splicing (Figures S19A–S19C). Differentiated P19 cells containing high levels of PTBP2 were positive for the neuronal marker MAP2, whereas MAP2-negative cells contained large amounts of PTBP1 and little PTBP2 (Figure S19D). Notably, miR-124 was sufficient to induce the switch from PTBP1 to PTBP2 (Figure S19E).
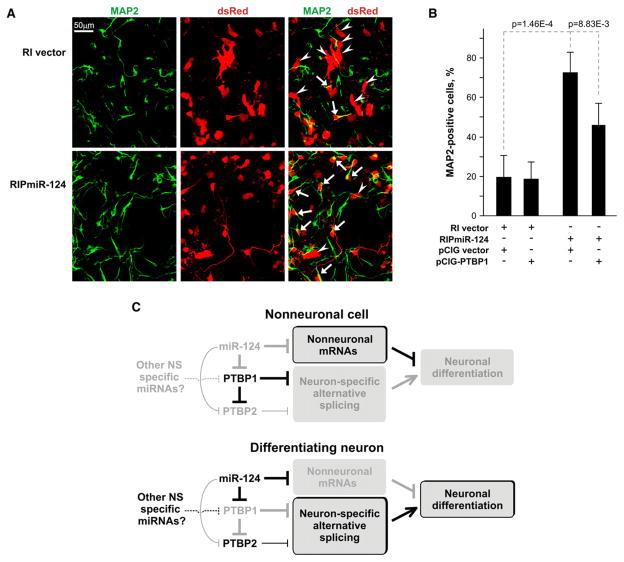
We did not observe neuronal differentiation induced by miR-124 expression alone. However, when RIPmiR-124-transfected cells were posttreated with RA, miR-124 dramatically enhanced RA-induced neuronal differentiation: 72.7% of transfected dsRed-expressing cells were MAP2-positive at 2 days after RA treatment versus only 19.5% in the corresponding RI vector control (p = 1.46E-4) (Figures 7A and 7B). A comparable enhancement of neuronal differentiation by miR-124 was detected when we stained cells with NeuN, another marker of differentiated neurons (data not shown). When P19 cells were co-transfected with RIPmiR-124 and a PTBP1-expressing plasmid lacking the natural PTBP1 3′UTR only 45.8% of the cells were MAP2 positive. These results, together with the neuroblastoma differentiation data (Figure 1G, Figures S2–S4), strongly support the conclusion that the downregulation of PTBP1 is required for the stimulatory effect of miR-124 on neuronal development.
Figure 7. miR-124 Stimulates Neuronal Differentiation at Least in Part by Reducing PTBP1 Levels.
(A) miR-124 stimulates neuronal differentiation of P19 cells. Cells were transfected with RIPmiR-124 plasmid or RI vector, induced with RA for 4 days, and plated on polylysine/laminin-coated coverslips. Two days after plating, the cells were fixed and stained with antibodies against MAP2. Arrows, dsRed-positive cells, which are also MAP2 positive (neurons); arrowheads, dsRed-positive cells, which do not express MAP2 (immature neurons and glia).
(B) PTBP1 downregulation is required for an optimal stimulatory effect of miR-124. The experiment was carried out as in (A), but cells were cotransfected with RIPmiR-124 plasmid or RI vector and pCIG vector or pCIG-PTBP1 expression plasmid prior to RA treatment. Both pCIG and pCIG-PTBP1 plasmids also encode EGFP protein that localizes to the nucleus. The fraction of MAP2-positive cells among cells expressing both dsRed and EGFP was averaged over seven randomly selected fields ± SD.
(C) Regulatory networks controlled by miR-124. In nonneuronal cells, high levels of PTBP1 repress NS-specific alternative splicing, which helps maintain a nonneuronal state. During neuronal differentiation, high levels of miR-124 downregulate PTBP1, leading to a switch to NS-specific alternative splicing and neuronal differentiation. A decrease in the PTBP1 levels also promotes exon inclusion in PTBP2 mRNA and accumulation of PTBP2 protein. PTBP2 fine-tunes the splicing pattern by weakly repressing the inclusion of a subset of NS-specific exons. In addition to PTBP1, miR-124 also downregulates several other nonneuronal mRNAs, including those of SCP1, laminin γ1, and integrin β1 (Cao et al., 2007; Visvanathan et al., 2007), as well as proteins involved in cell proliferation (Figure S5; Wang, 2006).
DISCUSSION
NS-specific alternative pre-mRNA splicing plays an important role in the establishment of neuronal identity (Black, 2003; Licatalosi and Darnell, 2006; Lipscombe, 2005; Ule and Darnell, 2006). However, the regulatory network that underlies the transition from nonneuronal to NS-specific alternative splicing is poorly understood. Similarly, several brain-specific miRNAs have been identified, but relatively little is known about their functions (Kosik, 2006). Here we show that miR-124 is a critical regulator of NS-specific splicing, and that it functions by controlling a switch in the ratio of the splicing repressors PTBP1 and PTBP2. This, in turn, leads to the initiation of a global NS-specific splicing program and promotes neuronal differentiation (Figure 7C).
We show that PTBP1 protein is present at high levels in neural progenitor cells and outside of the NS where little or no miR-124 is expressed. In nonneuronal cells PTBP1 represses the inclusion of alternative exon 10 in PTBP2 mRNA and destabilizes PTBP2 mRNA via the NMD pathway. In differentiating and mature neurons, expression of miR-124 reduces PTBP1 levels, triggering NS-specific splicing events and increasing the abundance of PTBP2 protein by allowing the inclusion of exon 10 in PTBP2 mRNA.
Similar to previously studied examples of miRNA/mRNA interactions, the 3′UTR of PTBP1 mRNA contains several miR-124 target sites that render the expression of a luciferase reporter sensitive to miR-124. Rescue of the PTBP1 3′UTR reporter activity required the inactivation of the strongest 8-mer MTS and four additional 6- and 7-mer MTSs. We therefore conclude that the regulation of PTBP1 by miR-124 requires several closely positioned MTSs in the 3′ UTR of PTBP1, which may function synergistically to achieve optimal sensitivity to miR-124 levels (Doench and Sharp, 2004). Considering that one consequence of the upregulation of miR-124 is a dramatic increase in the level of PTBP2 protein, it was surprising to find that the 3′UTR of PTBP2 is also targeted by miR-124, albeit weakly. This miR-124-mediated repression of PTBP2 expression is due primarily to a single 9-mer MTS in the PTBP2 3′UTR. At present, the functional significance of this miR-124 targeting of PTBP2 is not understood.
Data presented in this study and in a number of earlier reports (Black, 2003; Spellman and Smith, 2006; Wagner and Garcia-Blanco, 2001) implicate PTBP1 as a global repressor of NS-specific alternative splicing. Our microarray data indicate that at least 16 exons are upregulated by miR-124 in CAD cells. It is likely that even more exons are affected, because the microarray we used was not designed to specifically detect alternative splicing events. In all the cases we examined closely (Cdc42, Gabbr1, Mtap4, Rufy3, Pdlim7, Ptbp2, Rtn4/Nogo, and Tpm3), siRNA knockdown of PTBP1 was sufficient to induce NS-specific alternative splicing confirming that they are regulated by PTBP1.
PTBP2 was identified as an NS-enriched homolog of PTBP1 that interacts with the global splicing regulator Nova, and this interaction was shown to interfere with the inclusion of a Nova-induced exon in the glycine receptor α2 mRNA (Polydorides et al., 2000). In addition, PTBP2 weakly represses Src N1 cassette exon splicing in vitro, whereas PTBP1 acts as a stronger repressor (Markovtsov et al., 2000). Here we provide in vivo evidence that PTBP2 acts as a weak repressor of NS-specific exons in several pre-mRNAs (Figure 4). Understanding the role of PTBP2 in NS-specific alternative splicing is complicated by the recent finding that Nova can function as either an activator or repressor of alternative splicing depending on the position of its binding sites relative to the regulated exons (Ule et al., 2006). Thus, the increase in PTBP2 protein in response to miR-124 could, in principle, lead to either the repression or activation of exon inclusion via the inhibition of Nova by PTBP2.
Recently, the muscle-specific miRNA miR-133 was shown to repress both PTBP1 and PTBP2 protein synthesis in differentiating myoblasts (Boutz et al., 2007). In this system, PTBP2 appears to be the primary target, opposite to the miRNA regulation in the NS, where miR-124 efficiently downregulates PTBP1 expression but only weakly represses PTBP2 mRNA. Thus, individual 3′UTRs may contain arrays of MTSs for distinct tissue-specific miR-NAs, which could fine-tune gene expression levels in corresponding tissues.
To determine whether the miRNA processing machinery, and thus microRNAs in general, are required for the downregulation of PTBP1 in the mouse brain, we made use of a conditional knockout mouse in which the Dicer gene is inactivated in the embryonic telencephalon. Examination of these mice revealed that the forebrain was substantially smaller than in the wild-type mice, likely the consequence of a high-frequency apoptosis, consistent with previously described conditional Dicer mutant phenotypes (Cobb et al., 2005; Harfe et al., 2005; Harris et al., 2006; Muljo et al., 2005). However, many of the surviving cells expressed high levels of PTBP1 mRNA and protein, which correlated with a decrease in the levels of PTBP2; a low abundance of the NS-specific splice forms of Mtap4, Rufy3, and Cdc42; and normal or high abundance of the corresponding general splice forms (Figures 6A and 6B, Figure S18, and data not shown). These data indicate that the Dicer/miRNA pathway controls the level of PTBP1 expression and is essential for normal brain development. However, further experiments will be required for a more complete understanding of the role of miRNAs in the developing NS.
Evidence that miR-124 plays an important role in brain development was provided by the observation that expression of miR-124 at physiological levels in either CAD or N2a cells induces neurite outgrowth under conditions in which spontaneous differentiation is not normally observed (Figure 1G and Figures S2–S4). Notably, miR-124 reduces the levels of PTBP1 mRNA >2-fold and complementation of PTBP1 is sufficient to inhibit miR-124-induced neurite outgrowth in CAD cells (Figure 1G and Figure S5A). In addition, miR-124 may directly target mRNAs encoding proteins required for cell proliferation (Figures S5E and S5F; Wang, 2006). Consistent with the latter observation, miR-124 induces cell-cycle arrest in neuroblastoma cells (Figures S4B and S4C). Thus, miR-124 likely contributes to neuronal differentiation by repressing a range of mRNAs.
We nóte, however, that the role of miR-124 in neuronal differentiation is controversial. An earlier study concluded that miR-124 does not play an essential role in neuronal differentiation in the chick spinal cord (Cao et al., 2007). However, a more recent study in the same system showed that miR-124 stimulates neurogenesis and that anti-miR-124 2′-OMe-RNA oligonucleotides decreased the expression of neuronal markers and increased the number of proliferating cells (Visvanathan et al., 2007). We have shown that miR-124 enhances the neurodifferentiation of P19 cells stimulated by RA but does not promote detectable differentiation in the absence of RA. We therefore conclude that miR-124 is necessary, but not sufficient, for neuronal differentiation. Consistent with this view, neurons undergoing differentiation express dramatically higher levels of miR-124 compared to neuronal progenitor cells (Figure 5 and Figure S16) (Cao et al., 2007; Deo et al., 2006; Visvanathan et al., 2007).
We have shown that downregulation of PTBP1 protein levels is required for optimal miR-124-induced differentiation of both CAD and P19 cells. Two observations are consistent with the possibility that corresponding changes in alternative splicing are required for neuronal differentiation. First, most of the alternatively spliced forms induced by miR-124 are enriched in the NS (Figure 1B). Second, several of the corresponding genes encode proteins essential for neuronal morphogenesis and function (Table S4). However, PTBP1 is known to function in processes other than alternative splicing, including regulation of mRNA translation and stability (Bushell et al., 2006; Cho et al., 2005; Kim et al., 2000; Knoch et al., 2004; Pautz et al., 2006; Pilipenko et al., 2001). Therefore, further studies will be required to determine whether any of these additional activities contribute to the PTBP1-mediated inhibition of neuronal differentiation.
Our conclusion that miR-124 regulates the expression of a global repressor of NS-specific alternative splicing in nonneuronal cells has an interesting parallel at the level of transcription. As previously mentioned, miR-124 was recently shown to target SCP1 mRNA during neuronal differentiation (Visvanathan et al., 2007). SCP1 has been implicated in the function of the NRSF/REST complex, a global repressor of NS-specific transcription in nonneuronal cells (Yeo et al., 2005). Thus, miR-124, and possibly other tissue-specific miR-NAs, can contribute to establishing correct cell identity by controlling the levels of global repressors of gene expression.
EXPERIMENTAL PROCEDURES
RIPmiR Transfections and Cell Sorting
Cells were transfected with plasmid DNA and Lipofectamine 2000 (Invitrogen) as recommended. Following a 12–16 hr incubation, cells were replated at ~5 × 104/cm2 and incubated for another 24 hr. Cells were resuspended in PBS, 5% FBS, and 1 mM sodium pyruvate and sorted using a MoFlo instrument (Dako). For microarray analyses, total RNA was isolated immediately after sorting. Alternatively, cells were postcultured after sorting on poly-D-lysine-coated plates.
RT-PCR and RT-qPCR
Total RNA was purified using Trizol (Invitrogen) or RNeasy kits (QIAGEN) as recommended. RNA samples were treated with 50 units/ml of RQ1 DNase (Promega) at 37ºC for 20 min to remove genomic DNA. Reverse transcription (RT) was done using SuperScript III (Invitrogen) and random decamers at 50ºC for 1 hr. cDNA samples were analyzed by regular PCR (Taq DNA polymerase; NEB) or qPCR (brilliant SYBR green QPCR kit; Stratagene) with appropriate primers (Table S1). qPCR reactions were carried out in triplicate, and the data were normalized against the levels of Rps17 or Hprt mRNA. When appropriate, PCR products were analyzed by electrophoresis in agarose gels.
siRNA, Pre-miRNA, and 2′-OMe-RNA Transfections
CAD cells were seeded in 24-well plates at 1 to 2 × 105/well in 0.5 ml antibiotic-free medium and immediately transfected with 100 nM of an appropriate siRNA, pre-miRNA, or 2′-OMe-RNA using Lipofectamine 2000. Primary cortical neurons were transfected with 500 nM of anti-miR-124 or anti-miR-1 2′-OMe-RNAs as described (Conaco et al., 2006). Mouse PTBP1 and Rent1 mRNAs were knocked down using corresponding ON-TARGETplus siRNAs (Dharmacon). In RNAi experiments, BLOCK-iT siRNA (Invitrogen) was used as a negative control.
Microarray Experiments
CAD cells were transfected with RIPmiR-124 or empty RI vector, and a population of dsRed-positive cells was obtained by FACS sorting as described. Total RNA was isolated from the sorted cells using the RNeasy Mini kit (QIAGEN) as recommended. cDNA synthesis and cRNA synthesis and labeling, as well as microarray hybridization, washing, staining, and scanning, were performed as described in the Affymetrix GeneChip Expression Analysis Technical Manual. The microarray analysis was performed using Affymetrix GeneChip 430 2.0. Raw image files (CEL) from three RIPmiR-124 and three RI vector chips were normalized by robust multichip analysis (RMA) (Bioconductor release 1.9) using PM-only models. Probe sets with the expression level < 32 across all samples were excluded from further analyses. Normalized expression values were analyzed with SAM (significance analysis of microarray) using the permuted unpaired two-class test. The likelihood of overrepresentation in the gene ontology (GO) biological process categories was generated using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) 2.1 (Dennis et al., 2003; http://david.abcc.ncifcrf.gov/).
Supplementary Material
Acknowledgments
We thank Robert Darnell, Catherine Dulac, Philip Choi, Susan McConnell, and Michael McManus for mouse lines and reagents. We are grateful to Snezhana “Nyusha” Oliferenko, Ivo Lieberam, Jan Stenman, Weisheng Chen, Emiko Morimoto, Stefanie Schalm, Bill McCallum, Polina Kehayova, and Qiao Zhou for valuable comments. Brian Tilton provided technical assistance with FACS. This study was supported by an NIH grant 2R01NS043915-27 (T.M.). E.V.M. is a Leukemia and Lymphoma Society Fellow, and M.A.C. is a Milton Safenowitz Post-Doctoral Fellow of the ALS Association.
Footnotes
Accession Numbers
Microarray data have been deposited into the GEO database under number GSE8498.
Supplemental Data include 19 figures, four tables, Supplemental Results, Supplemental Experimental Procedures, and Supplemental References and can be found with this article online at http://www.molecule.org/cgi/content/full/27/3/435/DC1/.
References
- Ashiya M, Grabowski PJ. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA. 1997;3:996–1015. [PMC free article] [PubMed] [Google Scholar]
- Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Boutz PL, Chawla G, Stoilov P, Black DL. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev. 2007;21:71–84. doi: 10.1101/gad.1500707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushell M, Stoneley M, Kong YW, Hamilton TL, Spriggs KA, Dobbyn HC, Qin X, Sarnow P, Willis AE. Polypyrimidine tract binding protein regulates IRES-mediated gene expression during apoptosis. Mol Cell. 2006;23:401–412. doi: 10.1016/j.molcel.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Cao X, Pfaff SL, Gage FH. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007;21:531–536. doi: 10.1101/gad.1519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Johnston RJ, Jr, Frokjaer-Jensen C, Lockery S, Hobert O. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature. 2004;430:785–789. doi: 10.1038/nature02752. [DOI] [PubMed] [Google Scholar]
- Cho S, Kim JH, Back SH, Jang SK. Polypyrimidine tract-binding protein enhances the internal ribosomal entry site-dependent translation of p27Kip1 mRNA and modulates transition from G1 to S phase. Mol Cell Biol. 2005;25:1283–1297. doi: 10.1128/MCB.25.4.1283-1297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb BS, Nesterova TB, Thompson E, Hertweck A, O’Connor E, Godwin J, Wilson CB, Brockdorff N, Fisher AG, Smale ST, Merkenschlager M. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201:1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci USA. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Deo M, Yu JY, Chung KH, Tippens M, Turner DL. Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev Dyn. 2006;235:2538–2548. doi: 10.1002/dvdy.20847. [DOI] [PubMed] [Google Scholar]
- Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci USA. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KS, Zhang Z, McManus MT, Harfe BD, Sun X. Dicer function is essential for lung epithelium morphogenesis. Proc Natl Acad Sci USA. 2006;103:2208–2213. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert JM, McConnell SK. Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Dev Biol. 2000;222:296–306. doi: 10.1006/dbio.2000.9732. [DOI] [PubMed] [Google Scholar]
- Kikuchi T, Ichikawa M, Arai J, Tateiwa H, Fu L, Higuchi K, Yoshimura N. Molecular cloning and characterization of a new neuron-specific homologue of rat polypyrimidine tract binding protein. J Biochem (Tokyo) 2000;128:811–821. doi: 10.1093/oxfordjournals.jbchem.a022819. [DOI] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kim YK, Hahm B, Jang SK. Polypyrimidine tract-binding protein inhibits translation of bip mRNA. J Mol Biol. 2000;304:119–133. doi: 10.1006/jmbi.2000.4179. [DOI] [PubMed] [Google Scholar]
- Kim J, Krichevsky A, Grad Y, Hayes GD, Kosik KS, Church GM, Ruvkun G. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci USA. 2004;101:360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Knoch KP, Bergert H, Borgonovo B, Saeger HD, Altkruger A, Verkade P, Solimena M. Polypyrimidine tract-binding protein promotes insulin secretory granule biogenesis. Nat Cell Biol. 2004;6:207–214. doi: 10.1038/ncb1099. [DOI] [PubMed] [Google Scholar]
- Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of micro-RNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. Splicing regulation in neurologic disease. Neuron. 2006;52:93–101. doi: 10.1016/j.neuron.2006.09.017. [DOI] [PubMed] [Google Scholar]
- Lillevali K, Kulla A, Ord T. Comparative expression analysis of the genes encoding polypyrimidine tract binding protein (PTB) and its neural homologue (brPTB) in prenatal and postnatal mouse brain. Mech Dev. 2001;101:217–220. doi: 10.1016/s0925-4773(00)00566-9. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lipscombe D. Neuronal proteins custom designed by alternative splicing. Curr Opin Neurobiol. 2005;15:358–363. doi: 10.1016/j.conb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- Markovtsov V, Nikolic JM, Goldman JA, Turck CW, Chou MY, Black DL. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol Cell Biol. 2000;20:7463–7479. doi: 10.1128/mcb.20.20.7463-7479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O’Brien G, Shiue L, Clark TA, Blume JE, Ares M., Jr Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pautz A, Linker K, Hubrich T, Korhonen R, Altenhofer S, Kleinert H. The polypyrimidine tract-binding protein (PTB) is involved in the post-transcriptional regulation of human inducible nitric oxide synthase expression. J Biol Chem. 2006;281:32294–32302. doi: 10.1074/jbc.M603915200. [DOI] [PubMed] [Google Scholar]
- Pilipenko EV, Viktorova EG, Guest ST, Agol VI, Roos RP. Cell-specific proteins regulate viral RNA translation and virus-induced disease. EMBO J. 2001;20:6899–6908. doi: 10.1093/emboj/20.23.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polydorides AD, Okano HJ, Yang YY, Stefani G, Darnell RB. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc Natl Acad Sci USA. 2000;97:6350–6355. doi: 10.1073/pnas.110128397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman L, Bliskovski V, Kaye FJ, Zajac-Kaye M. Evolutionary conservation of a 2-kb intronic sequence flanking a tissue-specific alternative exon in the PTBP2 gene. Genomics. 2004;83:76–84. doi: 10.1016/s0888-7543(03)00207-6. [DOI] [PubMed] [Google Scholar]
- Rajewsky N. microRNA target predictions in animals. Nat Genet Suppl. 2006;38:S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Schwarz DA, Barry G, Eliasof SD, Petroski RE, Conlon PJ, Maki RA. Characterization of gamma-aminobutyric acid receptor GABAB(1e), a GABAB(1) splice variant encoding a truncated receptor. J Biol Chem. 2000;275:32174–32181. doi: 10.1074/jbc.M005333200. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian micro-RNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Falick AM, Black DL. Polypyrimidine tract binding protein blocks the 5′ splice site-dependent assembly of U2AF and the prespliceosomal E complex. Mol Cell. 2005;19:485–496. doi: 10.1016/j.molcel.2005.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova L, Grafe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Regulation of miRNA expression during neural cell specification. Eur J Neurosci. 2005;21:1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- Spellman R, Smith CW. Novel modes of splicing repression by PTB. Trends Biochem Sci. 2006;31:73–76. doi: 10.1016/j.tibs.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Ule J, Darnell RB. RNA binding proteins and the regulation of neuronal synaptic plasticity. Curr Opin Neurobiol. 2006;16:102–110. doi: 10.1016/j.conb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Garcia-Blanco MA. Polypyrimidine tract binding protein antagonizes exon definition. Mol Cell Biol. 2001;21:3281–3288. doi: 10.1128/MCB.21.10.3281-3288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Systematic identification of microRNA functions by combining target prediction and expression profiling. Nucleic Acids Res. 2006;34:1646–1652. doi: 10.1093/nar/gkl068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollerton MC, Gooding C, Wagner EJ, Garcia-Blanco MA, Smith CW. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol Cell. 2004;13:91–100. doi: 10.1016/s1097-2765(03)00502-1. [DOI] [PubMed] [Google Scholar]
- Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo M, Lee SK, Lee B, Ruiz EC, Pfaff SL, Gill GN. Small CTD phosphatases function in silencing neuronal gene expression. Science. 2005;307:596–600. doi: 10.1126/science.1100801. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.