Abstract
Stem cell factor (SCF) is an early-acting hematopoietic cytokine that elicits multiple biological effects. SCF is dimeric and occurs in soluble and membrane-bound forms. It transduces signals by ligand- mediated dimerization of its receptor, Kit, which is a receptor tyrosine kinase related to the receptors for platelet-derived growth factor (PDGF), macrophage colony-stimulating factor, Flt-3 ligand and vascular endothelial growth factor (VEGF). All of these have extracellular ligand-binding portions composed of immunoglobulin-like repeats. We have determined the crystal structure of selenomethionyl soluble human SCF at 2.2 Å resolution by multiwavelength anomalous diffraction phasing. SCF has the characteristic helical cytokine topology, but the structure is unique apart from core portions. The SCF dimer has a symmetric ‘head-to-head’ association. Using various prior observations, we have located potential Kit-binding sites on the SCF dimer. A superimposition of this dimer onto VEGF in its complex with the receptor Flt-1 places the binding sites on SCF in positions of topographical and electrostatic complementarity with the Kit counterparts of Flt-1, and a similar model can be made for the complex of PDGF with its receptor.
Keywords: crystal structure/helical cytokine/hematopoiesis/multiwavelength anomalous diffraction (MAD)/stem cell factor
Introduction
Stem cell factor (SCF) is an early-acting hematopoietic cytokine that binds at the cell surface to its receptor, Kit, whereby it produces other biological effects in addition to those on hematopoiesis (see reviews by Galli et al., 1994; Lev et al., 1994; Besmer, 1997; Broudy, 1997). SCF, which is produced by various fibroblast-type cells including bone marrow stromal cells, has also been called Kit ligand (KL), mast cell growth factor (MGF) and steel factor. The biochemistry and molecular biology that identified SCF and Kit as a ligand–receptor pair were preceded by an array of elegant animal biology studies that anticipated the underlying molecular mechanisms responsible for the genetics (Russell, 1979). Mice with mutations in the steel (Sl) locus (gene for SCF) or in the dominant-spotting (W) locus (c-kit, the gene for Kit) show complex phenotypes that include varying degrees of macrocytic anemia, sterility, lack of coat pigmentation and mast cell deficiency. Kit mutations in man are responsible for the autosomal dominant congenital pigmentation disorder, piebaldism. Consistent with these phenotypes, in the last 10 years a host of in vitro and in vivo experiments have clearly established Kit-mediated roles for SCF in early stages of hematopoiesis, in gametogenesis, in melanocyte proliferation and function and in mast cell proliferation, maturation and activation (Galli et al., 1994; Lev et al., 1994; Besmer, 1997; Broudy, 1997). Potential therapeutic applications of SCF include the treatment of anemias, boosting the mobilization of hematopoietic stem/progenitor cells to the peripheral blood for harvest and transplantation, and increasing the efficiency of gene transduction for gene therapy (Galli et al., 1994; McNiece and Briddell, 1995; Glaspy, 1996; Broudy, 1997).
SCF is expressed as membrane-associated forms of either 248 or 220 amino acids (Galli et al., 1994; Lev et al., 1994; Besmer, 1997; Broudy, 1997). The two forms are a consequence of alternative mRNA splicing that includes or excludes exon 6. Exon 6 encodes a proteolytic cleavage site such that soluble SCF1–165 is released from the 248 amino acid precursor. Residues 166–189 represent a tether to the membrane, residues 190–221 represent a hydrophobic transmembrane segment and residues 222–248 represent a cytoplasmic domain. The 220 residue form lacks the cleavage site and tends to remain membrane bound. Soluble SCF exists as a non-covalently associated homodimer (Arakawa et al., 1991). Each SCF monomer contains two intra-chain disulfide bridges, Cys4–Cys89 and Cys43–Cys138 (Langley et al., 1992). The N-terminal 141 residues of SCF have been identified as a functional core, SCF1–141, which includes the dimer interface and portions that bind and activate the receptor Kit (Langley et al., 1994).
It has been proposed that SCF is a member of the helical cytokine structural superfamily characterized by a double-crossover four-helix bundle topology (Bazan, 1991). Three-dimensional structures are known for many of the family members and, from a comparison of the structures and sequences, the members have been classified into three subgroups (Sprang and Bazan, 1993): short-chain, long-chain and interferon-like.
Most helical cytokines signal through members of the hematopoietic cytokine receptor superfamily, which are without intrinsic kinase activity (Heldin, 1995). SCF, in contrast, signals through a class III receptor tyrosine kinase (i.e. Kit). This class of kinases also includes the receptors for platelet-derived growth factor (PDGF), macrophage colony-stimulating factor (M-CSF) and Flt-3 ligand, and it is related to the class V receptor tyrosine kinases (Flt-1, Flk-1/KDR and Flt-4) for vascular endothelial growth factors (VEGFs) and less closely to the class IV receptors (FGFRs) for fibroblast growth factors (FGFs) (Fantl et al., 1993; Heldin, 1995; Rousset et al., 1995). The receptors in these classes have ‘split’ kinase domains intracellularly and multiple immunoglobulin (Ig)-like domains extracellularly.
The structures of PDGF (Oefner et al., 1992), M-CSF (Pandit et al., 1992) and VEGF (Muller et al., 1997) have all been determined by X-ray crystallography, as has the complex of VEGF with domain 2 of its receptor, Flt-1 (Wiesmann et al., 1997). Recently, instructive structures have also been reported for FGF2 bound to a two-domain fragment of FGFR1 (Plotnikov et al., 1999) and for the comparable complex between FGF1 and FGFR2 (Stauber et al., 2000). The ligands for the class III and class V receptors are all dimeric, whereas the class IV receptors use both FGF monomers and heparin as ligands. SCF, like the other dimeric ligands, initiates signal transduction by direct dimerization of its receptor, Kit, and the two juxtaposed receptors undergo tyrosine autophosphorylation (Heldin, 1995; Broudy, 1997), which initiates downstream intracellular signaling.
Here we report the crystal structure of the core fragment of recombinant human stem cell factor, SCF1–141, as determined at 2.2 Å resolution from multiwavelength anomalous diffraction (MAD) measurements. Incorporating data from mutagenesis and other structure–function studies, we locate putative receptor-binding sites on the surface of the symmetric SCF dimer. From a comparison of these results with structural and functional data for the related ligand–receptor systems, we model the complex of SCF with the receptor Kit and suggest a similar mode of association between other class III and class V receptors and their ligands.
Results and discussion
Structure determination
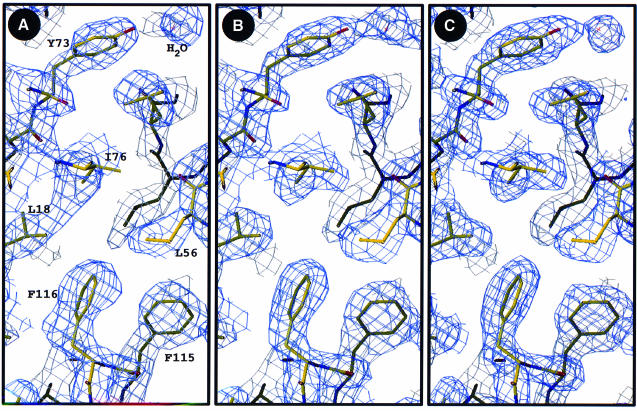
Both native and selenomethionyl (SeMet) human SCF1–141 were expressed as recombinant proteins in Escherichia coli (Langley et al., 1994). Crystals grew in space group P212121 with four SCF subunits and 39% solvent in the asymmetric unit. Our attempts to solve the structure of SCF1–141 by molecular replacement from other cytokine structures gave good rotation solutions, but no significant translation function peaks. We then evaluated experimental phases in a MAD experiment. Four-wavelength data were measured from a single, frozen SeMetSCF1–141 crystal and analyzed with MADSYS (Hendrickson, 1985). Twelve selenium sites were found in four congruent sets associated with the respective SCF subunits in the crystal. A MAD-phased electron density map was calculated at 2.3 Å resolution (Figure 1A) and improved by molecular averaging (Figure 1B) and refinement (Figure 1C).
Fig. 1. Representative electron density distributions. (A) MAD-phased experimental map calculated at 2.3 Å resolution. (B) The experimental map after 4-fold averaging. (C) The current 2Fo – Fc map superimposed with the model refined at 2.2 Å resolution. Each map is contoured at 1.0σ. Figures were drawn by O (Jones et al., 1991).
An atomic model was fitted to the experimental maps and refined at 2.2 Å resolution to an R-value of 0.199 (|F| >2σ) with stereochemical ideality typified by the r.m.s. deviation from bond ideality of 0.016 Å. There are no residues in energetically disallowed regions of the Ramachandran plot. This model for SeMetSCF1–141 has 3804 non-hydrogen atoms from 448 amino acid residues, 264 water molecules, three Ca2+ ions and one polyethylene glycol (PEG) moiety. All four polypeptide chains (designated A, B, C and D) are sufficiently disordered before residue 11 to preclude modeling of this portion, and none of them is fully ordered through to the end. Specifically, A92–103, B130–136, B139–141, C92–103, C127–141, D91–103 and D128–141 are all disordered. This disorder is such that, of the eight disulfide bridges, only two are seen. To test whether the reducing agent used to crystallize SeMetSCF1–141 (see Materials and methods) might have broken these bonds and caused the disorder, we also refined the structure of native SCF1–141 crystallized without reducing agent. The two crystals were nearly isomorphous, and the two structures showed the same pattern of order–disorder.
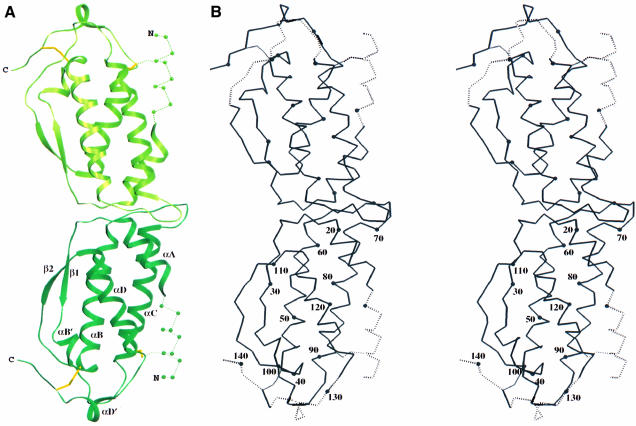
Structure of SCF
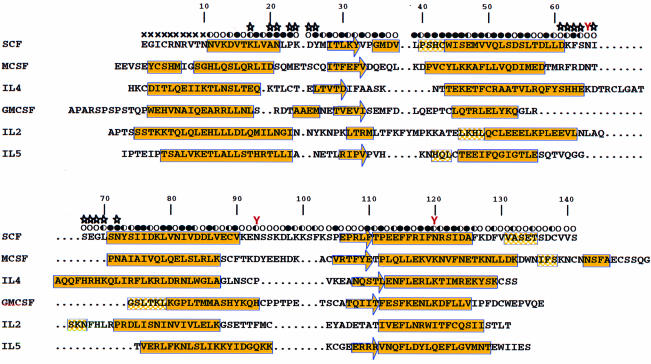
The four independent SCF subunits in the crystal are similar but distinctive, and identification of the AB and CD pairs as molecular dimers is unmistakable. None of the SCF subunit copies is complete, but each flexible portion except for the N-terminus is stabilized by lattice contacts to another subunit. Thus, through the combination of chains A and B, there are images for all residues except 1–10, and the position of Cys89, to which Cys4 must bridge, determines the approximate course of this disordered segment. The overall structure of this composite SCF dimer is shown in Figure 2A, and the Cα backbone for the actual AB dimer is drawn in stereo in Figure 2B. Topologically, the SCF structure is similar to that of other short-chain helical cytokines (Rozwarski et al., 1994), with a core of four helices (αA, αB, αC and αD) and two β-strands, β1 between αA and αB and β2 between αC and αD. Apart from the tight β2–αD connection, however, the segments outside these core elements are unique in conformation if not in length. In particular, there is an additional one-turn helix, αB′, between β1 and αB, there is an exceptional hairpin loop between αB and αC at the dimer interface, and there is another extra one-turn helix, αD′, in the C-terminal extension. The bounds of secondary structure elements are given in Figure 3.
Fig. 2. Overall structure of an SCF dimer. (A) Ribbon diagram. (B) Cα stereodiagram of the AB dimer. Figures were drawn using SETOR (Evans, 1993).
Fig. 3. Structure-based sequence alignment of SCF with other short-chain helical cytokines of human species. Dots denote gaps. The secondary structure elements were assigned according to the output of the PROCHECK program (Laskowski et al., 1993) except the helix assignment for residues 35–38, which was identified by inspection of the hydrogen bond pattern. Secondary structures are shown in yellow with filled boxes referring to α-helices, half-filled boxes to 310-helices and arrows to β-strands. The solvent accessibility of the SCF dimer is indicated for each residue by an open circle if the fractional solvent accessibility is >0.4, a half-filled circle if it is 0.1–0.4, and a filled circle if it is <0.1. Residues at the SCF dimer interface are identified by stars, and the N-linked glycosylation sites by red Ys above the Asn residues.
The core SCF dimer has its subunits arranged in a head-to-head manner with the opposed four-helix bundle axes nearly coincident (Figure 2). This gives the molecule an elongated shape, ∼85 × 30 × 20 Å. Approximately 855 Å2 of surface area from each protomer is buried into the dimer interface. The interface is dominated by contacts from the C-terminal end of αA and the αA–β1 connection of one monomer to the αB–αC loop of the other monomer (Figure 2), and the reciprocal pair is related by an approximate dyad axis of symmetry. The actual symmetry operators have rotational and translational components of 176.3° and 0.33 Å, respectively, for the AB dimer and 177.4° and 0.04 Å, respectively, for the CD dimer. The two dimers thereby deviate significantly and similarly (with A matched to C and B to D) from true 2-fold symmetry. Nevertheless, since interatomic contacts at the interface are symmetric, we presume that these deviations reflect flexibility rather than inherent asymmetry.
The crystal structure is compatible with solution biochemistry. Consistent with the relative rates of in vitro oxidation of methionyl residues (Hsu et al., 1996), Met36 and Met48 are buried in the hydrophobic core whereas Met27 is solvent accessible. Furthermore, as predicted on the basis of fluorescence spectroscopy studies (Arakawa et al., 1991), Trp41 is buried within the hydrophobic core.
Natural SCF and Chinese hamster ovary (CHO) cell-expressed recombinant SCF are heavily glycosylated, with both N- and O-linked carbohydrates. All four potential N-linked sites of human SCF1–165 are in the SCF1–141 portion that we have crystallized (Langley et al., 1992; Lu et al., 1992). Although the recombinant proteins expressed in bacteria are non-glycosylated, both human and rat SCF expressed in E.coli and then refolded in vitro have native structures, as judged by biophysical methods and in vitro biopotency assays (Arakawa et al., 1991; Langley et al., 1992). The crystal structure of our recombinant SCF is compatible with the glycosylation pattern found for SCF from mammalian cells. Thus, the potential site at Asn72, which is unglycosylated in both human and rat SCF expressed from mammalian cells, is buried in the dimer interface, whereas the site at Asn120, which is fully glycosylated in both species, is solvent accessible. Other sites (Asn65 in both human and rat, human Asn93 and rat Asn109) are glycosylated in some molecules but not others. These sites are also accessible in the atomic model. Asn93 is located in the highly flexible region between αC and β2, and its side chain is disordered.
Although natural SCF is a non-covalently associated dimer, recombinant human SCF produced in E.coli can fold alternatively in vitro into a covalently linked dimer. These dimers have Cys4–Cys89′ and Cys43–Cys138′ intermolecular disulfide bonds (Lu et al., 1996). The disulfide-linked SCF dimer and native non-covalently associated SCF dimer are similar with regard to biochemical and biophysical properties, biopotency and receptor-binding affinity. It was proposed that the disulfide-linked dimer arises from a double swap of αA and αD helices between the monomers (Lu et al., 1996). The crystal structure of SCF, however, suggests that a single swap at the αB–αC loop near residue 68 is more likely.
Comparison with other short-chain helical cytokines
Although SCF has the characteristic features of the short-chain helical cytokines, as among other members, both sequence and structure are highly divergent. If anything, SCF resembles the others less than they resemble one another (Table I). Our comparison of SCF with other short-chain helical cytokine structures [granulocyte–macrophage colony-stimulating factor (GM-CSF) (Diederichs et al., 1991), M-CSF (Pandit et al., 1992), interleukin (IL)-2 (McKay, 1992), IL-4 (Wlodaver et al., 1992) and IL-5 (Milburn et al., 1993)] shows greatest structural similarity with M-CSF or IL-4, but even here fewer than half of the residues can be superimposed (Table I). Sequence similarities are essentially random. Our structure-based sequence alignment (Figure 3) has pairwise identities ranging from 6.7 to 18.8% (Table I) and not even a single residue in SCF is conserved in all of the others. Nevertheless, the core elements are remarkably similar in structure.
Table I. Structural and sequence comparisons of short-chain helical cytokines.
| SCF | M-CSF | IL-4 | GM-CSF | IL-2 | IL-5 | |
|---|---|---|---|---|---|---|
| SCF | 14.1 (13.0) | 12.7 (12.3) | 12.5 (23.5) | 18.8 (16.4) | 6.7 (21.1) | |
| M-CSF | 64 (1.76) | 14.8 (18.9) | 13.8 (18.3) | 17.5 (17.1) | 10.5 (18.6) | |
| IL-4 | 63 (1.58) | 54 (1.82) | 26.6 (25.0) | 14.5 (22.2) | 18.9 (18.9) | |
| GM-CSF | 48 (1.63) | 58 (1.81) | 64 (1.56) | 9.8 (26.0) | 20.4 (14.7) | |
| IL-2 | 48 (1.70) | 57 (1.58) | 69 (1.33) | 61 (1.48) | 14.5 (22.2) | |
| IL-5 | 45 (1.70) | 38 (1.72) | 53 (1.32) | 49 (1.33) | 62 (1.37) |
Structural comparisons and sequence comparisons between the short-chain helical cytokines are given in the lower left and upper right triangles, respectively. Structural comparisons are given as the maximum number of equivalent α-carbon atoms between two short-chain helical cytokines, and the r.m.s. deviation (Å) (in parentheses). Sequence comparisons are given as the percentage of sequence identity from sequence alignment based on structural superimposition, and that based on the sequence alignment from the BESTFIT program of the GCG package (in parentheses). The latter alignment is based only on maximizing the percentage of identity, similarities and length of the matching sequences, and the sequences submitted to the BESTFIT program were restricted within the region as defined in the PDB files, including the disordered residues. With the advantage of the relatively large number of independent data points (15 pairs), we analyzed the correlation between sequence identity and structural deviation. Without any restriction of structural alignment, the correlation coefficient (C) between structural deviation and sequence identity is –0.21 and the Student’s t probability (P) is 0.44, suggesting little correlation between a specific sequence and the tertiary fold. With the restriction of structural alignment, however, C is –0.30 and P is 0.28, indicating that the structure-based sequence identity and structural deviation are weakly connected (as also observed in another highly diverged protein family, hemoglobin; Aronson et al., 1994).
Core portions aside, SCF differs markedly from other short-chain helical cytokines, as indeed they differ from one another (Figure 3; Rozwarski et al., 1994). First, helix αA of SCF is unusually shortened at its N-terminus. Its disordered extension must deviate toward αC, as in M-CSF but not in the others, by virtue of the Cys4–Cys89 disulfide bridge in common with M-CSF. Secondly, the conformation of the αA–β1 connection is distinctive as required for the dimer interface, and the β1–αB connection uniquely has αB′. Again at the dimer interface, the αB–αC hairpin loop extends out distinctively along the dyad axis. Thirdly, the unusually long αC–β2 loop of SCF is both highly flexible (only one ordered copy) and with a unique path when ordered. Finally, the C-terminal extension after αD compares only to that of M-CSF, and then only in its general direction of exit out past αB and the β-strands.
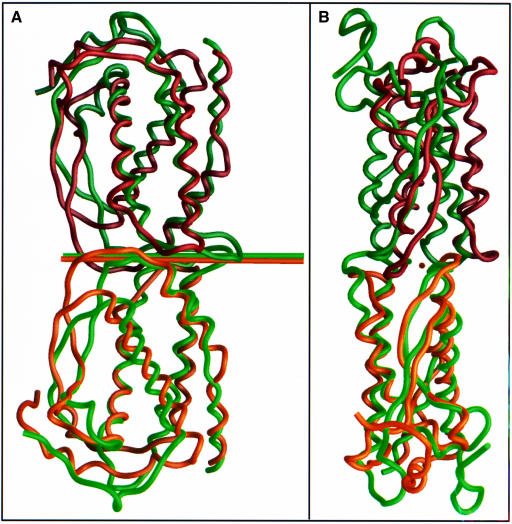
SCF and M-CSF are similar in gene structure, alternative splicing, proteolytic maturation, disulfide bridging, dimer assembly and receptor type (these similarities also extend to the Flt-3 ligand; Lyman and Jacobsen, 1998). Despite negligible sequence identity, an alignment and secondary structure prediction prompted by these relationships (Bazan, 1991) fits the actual structure amazingly well, except for shifts in αB and in the αC–β2 loop. Here reality confounds logic; unexpectedly, comparable glycosylation sites (Asn120 in SCF and Asn122 in M-CSF) are displaced by one helical turn, and comparable disulfide bridges (Cys43–Cys138 in SCF and Cys48–Cys139 in M-CSF) are not superimposable structurally (Figure 4).
Fig. 4. Comparison of SCF dimer (shades of green) and M-CSF dimer (shades of brown). (A) View as in Figure 2. (B) View perpendicular to that in (A), along the dyad axis of M-CSF. Symmetry axes are shown as lines in (A) and dots in (B). Figures were drawn by GRASP (Nicholls et al., 1991).
Comparison with other cytokine dimers
Helical cytokines dimerize in various ways (Sprang and Bazan, 1993). Among the five dimeric helical cytokines for which crystal structures have been described previously [M-CSF, IL-5, ciliary neurotrophic factor (CNTF), interferon-γ (IFN-γ) and IL-10], only IFN-γ and IL-10 are similar dimers. These latter two have a ‘tip-to-tip’ packing with helix axes approximately perpendicular. Otherwise, the only salient feature in common is having the subunits oriented with bundle axes aligned in parallel and helix dipoles positioned to compensate. There is ‘head-to-head’ packing of the four-helix bundles in M-CSF, ‘tail-to-tail’ packing in IL-5 and ‘side-to-side’ packing in CNTF. Moreover, IFN-γ, IL-10 and IL-5 are all interdigitated dimers with helices swapped between subunits.
SCF, in keeping with its relationship to M-CSF, is a non-interdigitated ‘head-to-head’ dimer (Figure 4). The interfaces between protomers are completely different, however. One αA–β1 loop of M-CSF is situated between the αA–β1 and αB–αC loops of the other protomer, whereas in SCF each αA–β1 loop interacts only with the αB–αC loop of the partner. This staggered mode of M-CSF dimerization (Figure 4B) is dictated by the position of the Cys31–Cys31′ intermolecular disulfide bond in M-CSF. The dyad axes are oriented similarly in the two cases (perpendicular to the bundle axis and parallel to the αAαD and αBαC helix planes), but whereas the dyad axis in SCF nearly intersects the bundle axis, that in M-CSF is offset toward the αAαD helix pair (Figure 4). Thus, when one protomer of an SCF dimer is superimposed onto a protomer from M-CSF, the superimposition of the two mates requires a translation of 3.8 Å but a rotation of only 4.7°.
Location of the binding site for the receptor Kit
SCF binds with high affinity (nM range) to its receptor (Philo et al., 1996; Broudy, 1997). Various structure–function studies and analyses help to define residues of SCF that may be involved in this binding. These studies include mutagenesis, immunochemical mapping, comparative analyses of inter-species ligand–receptor interactions and analyses of glycosylation.
From studies of truncation and point mutants, Langley et al. (1994) demonstrated that the N-terminal residues 1–4 and 1–10 and the Cys4–Cys89 disulfide bond are required for receptor binding and bioactivity, and that the Cys43–Cys138 disulfide bond and C-terminal residues past 127 are not required for receptor binding but may have some roles in bioactivity. Moreover, alterations at Asn10 and Asn11 brought about by chemical isomerization or by mutagenesis have positive or negative effects on bioactivity, depending on the substitution (Hsu et al., 1998). A quadruple mutant of SCF (Arg121Asn, Asp124Asn, Lys127Asp and Asp128Lys) was found to be defective in bioactivity (Matous et al., 1996). The molecular cause of this deficiency may be specific to Lys127 or due to indirect electrostatic effects. Arg121 and Asp124 are adjacent to the main N-linked glycosylation site, which is not involved in binding (see below), and Asp128 is absent in the 1–127 truncation mutant that retains full receptor-binding activity (Langley et al., 1994). Moreover, a study of human–murine SCF chimeras narrowed the important receptor recognition epitopes to within residues 1–35 and 79–97 (Matous et al., 1996), and the epitope of a neutralizing monoclonal antibody was mapped to the region of residues 60–95 (Mendiaz et al., 1996) and 79–97 (Matous et al., 1996).
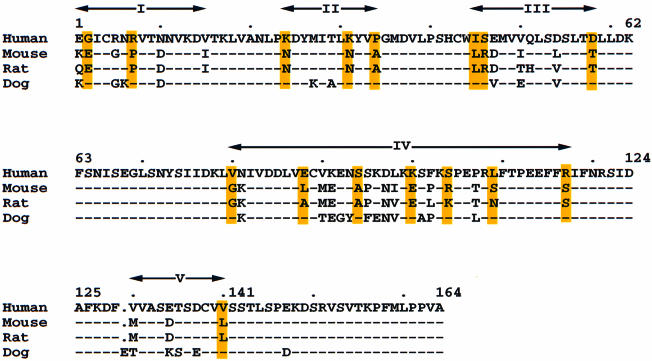
Although SCF molecules from different mammalian species are very similar (>75% identity), there are substantial differences in inter-species receptor activation. Human SCF activates mouse Kit very poorly, rodent SCF has only slightly lower potency than human SCF in binding/activating human Kit (Martin et al., 1990; Lev et al., 1992), and dog SCF activates human Kit slightly better than does human SCF itself (K.E.Langley, unpublished data). It is likely that the receptor-binding regions involve residues that are different between man and mouse but conserved between man and dog. These residues can be classified into five groups in the sequence (Figure 5). Most residues in group III (45–58) are buried and those in group II (24–34) are close to the dimer interface. The residues in groups I (1–15), IV (80–117) and V (130–140) are more likely to be involved in direct receptor binding.
Fig. 5. Sequence alignments of SCF from human, mouse, rat and dog (Anderson et al., 1990; Huang et al., 1990; Martin et al., 1990; Shull et al., 1992). The residues that are conserved in human and dog but different from rat and mouse are shadowed in yellow. Five regions of divergent sequence are identified (Roman numerals). Dots denote gaps, and dashes indicate residues identical to the human residues.
Human SCF expressed in CHO cells is ∼30% carbohydrate by weight (Arakawa et al., 1991). The main glycosylation site is at Asn120 (Langley et al., 1992; Lu et al., 1992). Glycosylation at this site, which is near the center of the αD helix, does not appear to influence biological activity; therefore, the area around this residue cannot be involved in receptor binding. Glycosylation of human SCF at either Asn65 or Asn93 lowers the biological activity ∼10-fold; therefore, these residues may be near but not directly at the binding site.
Taken together, these observations indicate that the receptor-binding site may include residues from the first few N-terminal residues, the 79–95 region (mainly located on the αC helix) and the C-terminal end of αD (around 127). These regions are contiguous on the SCF surface in our atomic model. The putative receptor-binding site of M-CSF was mapped to a similar region (Taylor et al., 1994).
Structural characteristics of SCF–Kit and related ligand–receptor complexes
Class III and class V receptor tyrosine kinases are distinguished from one another by the number of Ig repeats in their extracellular ligand-binding portions (five for class III and seven for class V) and by the length of kinase insert. These Ig-like receptors share similar signal transduction pathways, chromosomal localization and gene organization (Rousset et al., 1995), but their ligands come with completely unrelated topologies as typified by PDGF and VEGF (cystine knot) on the one hand, versus M-CSF, SCF and Flt-3 ligand (helical cytokine) on the other. Even receptors of the same class have unrelated ligands; thus, both SCF and PDGF use class III receptors and VEGF and Flt-3 ligand use class V receptors. The amino acid sequences of the ligands are extremely dissimilar even when the fold is the same, as for PDGF versus VEGF (25% identity) and M-CSF versus SCF (14% identity).
Although Ig-like receptors have very similar kinase portions (∼70% amino acid sequence identity between III and V), their Ig-like domains are dissimilar in sequence both between repeats within a molecule and also at comparable positions between different receptors (Rousset et al., 1995). Nevertheless, there are features of the receptor–ligand interaction that the class III and class V receptors have in common. First, for every studied example, the ligand-binding function has been localized to the first three Ig-like domains and, where defined further, to domains D2 and D3 specifically (Heidaran et al., 1990; Blechman et al., 1993; Lev et al., 1993; Wang et al., 1993; Davis-Symyth et al., 1996; Barleon et al., 1997). Secondly, the ligands for all of these receptors are functional as dimers; M-CSF, VEGF and PDGF are covalently linked dimers, while SCF and Flt-3 ligand are non-covalently associated dimers. In each case, signaling occurs through ligand-mediated receptor oligomerization (Heldin, 1995). For SCF–Kit, it has been shown directly by biophysical methods that complexes containing two SCF subunits and two Kit extracellular domain molecules can form in solution (Philo et al., 1996).
The structure of domain D2 of receptor Flt-1 in complex with VEGF (Wiesmann et al., 1997) provides a template for ligand interactions with PDGF-related receptors. Wiesmann et al. (1997) modeled the interaction of VEGF with D1D2D3D4(Flt-1) and discussed the likelihood that other ligand complexes with class III and class V receptors may be similar. In light of the structure of SCF and the identified location of receptor-binding sites, we have modeled the SCF–Kit complex by analogy.
The D2(Flt-1) domain is similar in structure to telokin, as predicted (Harpaz and Chothia, 1994), and thereby also to both domains in the structure of vascular cell adhesion molecule (VCAM)-1 (Jones et al., 1995). To test the validity of VCAM-1 as a model for D2D3(Flt-1) and D2D3(Kit), we used a prediction-based threading program (Fisher and Eisenberg, 1996) to thread the sequences of the Ig-like domains of Flt-1 and Kit into the telokin and VCAM-1 structures. Fits were achieved with moderate to very high confidence of similarity. The resulting structure-based sequence alignment of D2D3(Kit) with the VCAM-1 template (five gaps) has a continuous domain boundary, and residues Cys151 and Cys183 in D2(Kit) are positioned properly to make an additional disulfide bridge between strands C and F.
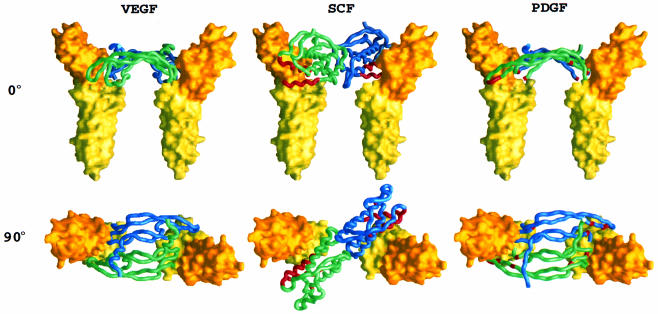
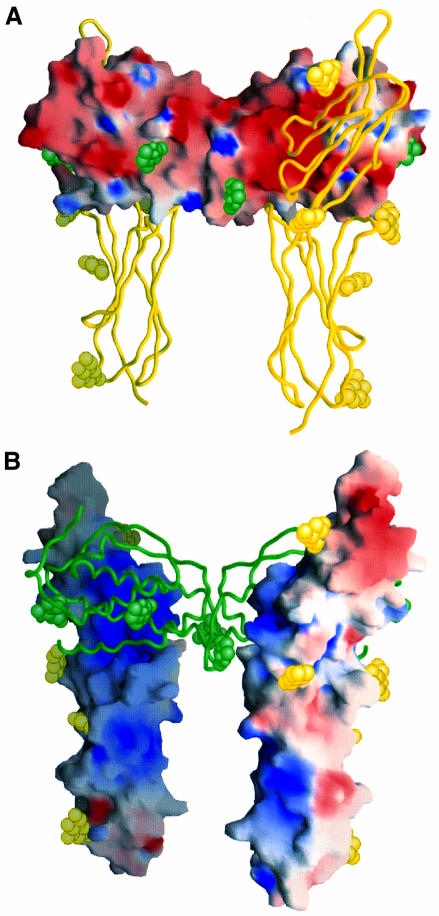
We next constructed a model of the VEGF–D2D3(Flt-1) receptor complex from a rigid-body superimposition of VEGF (Muller et al., 1997) and VCAM-1 so as to mimic the reported VEGF–D2(Flt-1) structure (Wiesmann et al., 1997). Then, keeping the dyad symmetric receptor pair fixed, we successively replaced VEGF with other Ig-like receptor ligands of known three-dimensional structure: PDGF (Oefner et al., 1992), M-CSF (Pandit et al., 1992) and SCF (this work). Each was placed on the dyad axis and positioned to optimize contacts between the VEGF-binding site on the receptor and the putative receptor-binding regions of the ligands. Remarkably, these disparate dimeric ligands have similar spacings between binding sites and a satisfactory fit is possible for each (Figure 6). We also constructed simple homology models of the various receptors with changes in the backbone only to accommodate insertions and deletions. The model for SCF complexed with D2D3(Kit) shows a striking electrostatic complementarity between a highly negative binding surface on SCF and a positive surface on Kit (Figures 7A and B). The glycosylation sites on both molecules are also compatible with unimpeded interaction.
Fig. 6. Ligand (worm structures)–receptor (yellow structures) models of: VEGF–Flt-1, SCF–Kit and PDGF–PDGF receptor. The VCAM-1 structure (PDB code: 1vca) was used, without any modification, to approximate the receptor models. Receptor models are presented as yellow surfaces. Background portions of ligands are colored light blue for one protomer and green for the other; receptor-interacting residues identified from site-directed mutagenesis experiments [VEGF (Muller et al., 1997), PDGF (Fenstermaker et al., 1993)] and other experimental data (SCF; see text) are colored magenta. Figures were drawn by GRASP (Nicholls et al., 1991).
Fig. 7. Electrostatic and carbohydrate surfaces of SCF and homology-modeled receptor Kit. (A) Electrostatic surface of SCF and worm of D2D3(Kit). (B) Electrostatic surface of the Kit fragment and worm of SCF. Negative potential is colored red and positive potential blue, with greatest saturations at –10 and +10 kT, respectively. Carbohydrate moieties are represented by CPK models of β-d-N-acetylglucose (green for SCF moieties and yellow for potential Kit moieties). Figures were drawn by GRASP (Nicholls et al., 1991).
The receptor Kit is activated by both soluble and membrane-bound forms of SCF, and signaling from the membrane-bound form appears to have in vivo roles (see review by Lyman and Jacobsen, 1998). Moreover, as in the case of D4(Flt-1) (Barleon et al., 1997), D4(Kit) may be involved in inter-receptor contacts in the signaling dimer (Blechman et al., 1995) [although this proposal for Kit has been questioned (Philo et al., 1996; Lemmon et al., 1997)]. The model that we have constructed for the SCF–Kit complex is compatible with these properties (Figure 7). The C-termini of the SCF dimer are directed oppositely from those of Kit, as would be appropriate for a cell–cell contact, and the receptor units would cross naturally at D4. It is noteworthy that the ligands of two other Ig-like class III receptors also have membrane-bound forms (M-CSF and Flt-3 ligand) (Kawasaki and Ladner, 1990; Lyman and Jacobsen, 1998).
Materials and methods
SCF expression, purification and analyses
Human SCF1–141 was expressed recombinantly in E.coli as described previously (Langley et al., 1994). For SeMetSCF1–141, the expression vector was transfected into the methionine auxotrophic E.coli strain FM5. Fermentation was carried out at 30°C in 8 l of minimal medium consisting of ammonium sulfate (10 g/l), glucose (5 g/l), methionine (0.125 g/l), phosphate salts, magnesium, citric acid, trace metals and vitamins. When an OD600 of 3–5 was reached, a feed medium was added that consisted of the following components in a total volume of 1 l: 100 g of ammonium sulfate, 450 g of glucose, 2 g of methionine, magnesium, trace metals and vitamins. At an OD600 of 12.4, induction medium (1 l containing 100 g of ammonium sulfate, 300 g of glucose and 1 g of SeMet) was added and fermentation proceeded at 30°C. Five hours later (at an OD600 of ∼16), the temperature was raised to 42°C to induce SCF expression and additional SeMet (1 g) was added. Cells were harvested 4 h after the temperature shift (OD600 of ∼16). SeMetSCF1–141 expression was estimated as 0.5 g/l. Both SCF1–141 and SeMetSCF1–141 were purified with minor modifications to previously described procedures (Langley et al., 1992, 1994). Both retain the initiating methionine (or SeMet) residue [position (–1)] (Langley et al., 1994). N-terminal amino acid sequencing was performed as described (Lu et al., 1991). About 90% SeMet was present in SeMetSCF1–141 at each of the Met positions, based on amino acid analysis and N-terminal sequencing results (i.e. lack of recovery of Met residues for SeMetSCF1–141 in comparison with SCF1–141; data not shown).
Crystallization
Crystals were obtained by use of the hanging drop vapor diffusion method under aerobic conditions. The initial crystals were grown by mixing 1 µl of protein solution (44 mg/ml SCF1–141 or 38 mg/ml SeMetSCF1–141 in 10 mM sodium phosphate pH 6.5, 80 mM NaCl) with 1 µl of crystallization reservoir solution. The crystallization reservoir solution included 25% (w/w) PEG400, 240 mM CaCl2 and 100 mM HEPES pH 7.4 for SCF1–141, and 22% (w/w) PEG400, 220 mM CaCl2, 100 mM HEPES pH 7.2 and 5–10 mM dithiothreitol (DTT) for SeMetSCF1–141. Crystallization trays were incubated at 20°C and crystals reached full size in ∼3 days with typical dimensions of 0.5 × 0.2 × 0.2 mm. Microseeding and lower concentrations of DTT solution (∼2 mM) were needed to reproduce SeMetSCF1–141 crystals subsequently. An extant SeMetSCF1–141 crystal was washed with its reservoir solution and then crushed to produce microseeds, which were stored in 50 µl of a stabilizing solution of 32% (w/w) PEG400, 260 mM CaCl2, 100 mM HEPES pH 7.4 at room temperature. For microseeding experiments, the seed stock was diluted by 10- to 10 000-fold with crystallization reservoir solution. A 1 µl aliquot of this prepared precipitant was mixed with 1 µl of the protein solution to make the droplet. The crystal for MAD phasing was grown from a crystallization reservoir solution containing 2 mM DTT.
Diffraction measurements
X-ray diffraction data from SCF1–141 crystals were recorded on two Hamlin–Xuong area detectors at 293K at a home source. The data were integrated using the UCSD software package and scaled using AGROVATA and ROTAVATA as implemented in the CCP4 suite (CCP4, 1994). The MAD experiments for SeMetSCF1–141 were conducted at the X4A synchrotron beam line of Brookhaven National Laboratory using Fuji image plates. A single crystal was frozen at 110K using paratone-N (Exxon) as a cryoprotectant. The MAD data were collected at four wavelengths (before the edge, at the SeK edge, at the peak and after the peak) in oscillations of 1.3–1.5° without overlap. The SeMetSCF1–141 crystal was oriented such that the b-axis was parallel to the oscillation axis, and a mirror geometry was used during data collection. The MAD data were processed using DENZO and Scalepack (Otwinowski, 1993; Gewirth, 1995) (Table II).
Table II. MAD data collection and phasing statistics.
| Data collection (25–2.0 Å)a | ||||
| Wavelength (Å) |
Unique reflections |
Completeness (%) |
Signal (I/σ) |
Rsym (%) |
| λ1 = 0.9919 (pre-edge) | 65 810 | 95.1 | 18.4 | 6.7 |
| λ2 = 0.9793 (inflection) | 65 759 | 95.0 | 16.7 | 5.8 |
| λ3 = 0.9791 (peak) | 65 665 | 94.9 | 15.2 | 6.7 |
| λ4 = 0.9686 (remote) |
65 689 |
94.9 |
16.0 |
5.6 |
| Anomalous diffraction ratios (20–2.6 Å)b | ||||
| |
λ1 |
λ2 |
λ3 |
λ4 |
f′ (e) |
f″(e) |
| λ1 | 0.035 | 0.051 | 0.042 | 0.035 | –4.0 | –0.5 |
| (0.030) | ||||||
| λ2 | 0.052 | 0.033 | 0.051 | –10.3 | 3.8 | |
| (0.029) | ||||||
| λ3 | 0.070 | 0.041 | –8.1 | 5.6 | ||
| (0.031) | ||||||
| λ4 | 0.055 | –3.9 | 3.8 | |||
| (0.030) | ||||||
| MAD phasing (25–2.6 Å)c | ||||||
| R(°|FT|) = 0.044 | R(°|FA|) = 0.39 | <Δ(ΔΦ)> = 41.6° | <σ(ΔΦ)> = 18.7° | |||
aUnique reflections are determined by point group 222 (not mmm) to distinguish Bijvoet-related reflections. Rsym = 100 × Σhkl Σi|Ii – <I>|/Σhkl Σi Ii, where Ii is the ith measurement of reflection hkl and <I> is the weighted mean of all measurements of I.
bAnomalous diffraction ratios = <Δ|F|2>1/2/<|F|2>1/2, where Δ|F| is the absolute value of the Bijvoet (diagonal elements) or dispersive difference (off-diagonal elements), respectively. Values in parentheses are for centric data.
cR = Σhkl Σ ||Fi| – <F>|/Σ|F|. °FT is the structure factor due to normal scattering from all the atoms. °FA is the structure factor due to normal scattering from the anomalous scatterers only, and ΔΦ is the phase difference between °FT and °FA. Δ(ΔΦ) is the difference between two independent determinations of ΔΦ.
Molecular replacement attempts
Structure determination by the molecular replacement method was attempted for the home source data set. The MERLOT (Fitzgerald, 1988) and AMoRe (CCP4, 1994) programs were used with various four-helix bundle structures as search models, and a good rotation solution was obtained. The rotation solution agreed well with the orientation of helical bundles (approximately along the b-axis of the unit cell) that was deduced from native Patterson maps. Dissimilarities among the helical cytokines and the multiplicity of subunits (four) hampered detection of any significant translation function peaks.
Phase evaluation
The processed MAD data were passed through the MADSYS programs (Hendrickson, 1985). Algebraic and probabilistic MAD phasing procedures (Hendrickson, 1985; Pähler et al., 1990) were applied for phase determination (Table II). Selenium sites were located using the HASSP program (CCP4, 1994) in FA Patterson and difference Fourier maps and refined by MADSYS programs. The choice of enantiomer was determined by comparison of the electron density maps computed from the two enantiomorphic selenium structures to maximum Bragg spacings of 2.6 Å. The phases were improved by 4-fold non-crystallographic symmetry (NCS) averaging. The rotation–translation matrices of the NCS axes were determined by TOSS (Hendrickson, 1979) from the selenium sites and subsequently refined by LSQRHO (W.A.Hendrickson, unpublished) and RAVE (Kleywegt and Jones, 1994), and the averaging procedure by DM (CCP4, 1994).
Model building and refinement
The initial model of SeMetSCF1–141 was built into the averaged map at 2.3 Å by using program O (Jones et al., 1991). The model includes 98 core residues for each of the four molecules in an asymmetric unit. The remote wavelength after the SeK peak was used for the refinement with the Bijvoet difference applied to Se scattering factors. The R-value for this model, before any refinement, was 42.1% in the resolution range 10.0–2.3 Å. NCS restraints were applied during the initial rounds of refinement. After several iterations of least square and simulated annealing refinement with X-PLOR (Brünger et al., 1987) and manual rebuilding against SIGMAA (Read, 1986) and 2|Fo| – |Fc| maps, the crystallographic R-value is 19.9% for the current model (Table III). The sites of Ca2+ ions, a component of the crystallization medium, were located from a Bijvoet difference Patterson map at the remote wavelength before the SeK edge. The SCF1–141 model was obtained by subjecting the refined SeMetSCF1–141 model to refinement against the area-detector data set from the SCF1–141 crystal using the X-PLOR program (Brünger et al., 1987). The atomic coordinates have been deposited in the Brookhaven Protein Data Bank with accession code of 1scf.
Table III. Lattice and refinement statistics.
| SeMetSCF1–141 (λ4) | SCF1–141 | |
|---|---|---|
| Lattice | ||
| space group | P212121 | P212121 |
| cell constants (a,b,c) (Å) | 71.8, 82.6, 88.2 | 73.0, 84.7, 88.8 |
| Zaa | 4 | 4 |
| Refinementb | ||
| resolution range (Å) | 20.0–2.2 | 8.0–3.3 |
| completeness (%) | 96.6 | 98.6 |
| unique reflectionsc | 49 851 | 7990 |
| R-valued (|F|>2σ) (%) | 19.9 | 20.8 |
| Rfreee (%) | 24.2 | 27.3 |
| Rsymf (%) | 5.6 | 15.2 |
| Model parameter | ||
| total non-H atoms | 3804 | 3502 |
| total residues | 448 | 447 |
| total water molecules | 264 | 0 |
| total metal ions | 3 | 0 |
| r.m.s. bond length/angle | 0.016/2.5° | 0.017/3.0° |
| average B-factor (Å2) | 32.1 | 18.7 |
| main-chain r.m.s. B (bond/angle) (Å2) | 1.2/1.6 | 1.9/2.2 |
| side-chain r.m.s. B (bond/angle) (Å2) | 2.1/2.4 | 3.0/3.3 |
aZa, number of molecules in the asymmetric unit.
bThe reflection data higher than the resolution range were not included in the refinement due to poor Rsym in these resolution shells.
cUnique reflections are determined by point group 222 for the SeMetSCF1–141 data set to distinguish Bijvoet-related reflections and by point group mmm for the SCF1–141 data set.
dR-value = Σhkl||Fo| – |Fc||/Σhkl|Fo|
eA subset of the data (6%) was excluded from the refinement and used for the free R-value calculation.
fRsym for the SeMetSCF1–141 data set was calculated in the resolution range of 25–2.2 Å and the for SCF1–141 data set in the resolution range 13–3.3 Å.
Structure analysis
Solvent accessibilities were defined compared with the corresponding Gly-X-Gly peptide (Shrake and Rupley, 1973), as calculated by X-PLOR (Brünger et al., 1987). Structural superimpositions were based on α-carbon atoms alone, with r.m.s. deviations calculated only from atom pairs identified as equivalent. The coordinates were taken from the Brookhaven Protein Data Bank with entry codes: M-CSF, 1hmc (Pandit et al., 1992); IL-4, 1rcb (Wlodaver et al., 1992); GM-CSF, 1gmf (Diederichs et al., 1991); IL-2, 3ink (McKay, 1992); and IL-5, 1hul (Milburn et al., 1993). Initial segments of equivalence between two structures were defined according to equivalent secondary structure elements. These structures were then superimposed using the program TOSS (Hendrickson, 1979), and the number of equivalent atoms was extended using the Lsq_imp command in program O (Jones et al., 1991) with the criteria of a cut-off distance of 3.0 Å and a minimum fragment length of three consecutive residues. Different initial equivalent segments did give different results in the structural alignment, as Rozwarski et al. (1994) observed in their study. We tried several initial sets of equivalent segments for each alignment and retained the one that generated the greatest number of equivalent atoms after the Lsq_imp extension.
Acknowledgments
Acknowledgements
We thank C.Ogata, Y.Liu and H.Wu for help in synchrotron data collection. This work was supported in part by NIH grant GM-34102. Beamline X4A at the National Synchrotron Light Source, a Department of Energy facility, is supported by the Howard Hughes Medical Institute.
References
- Anderson D.M. et al. (1990) Molecular cloning of mast cell growth factor, a hematopoietin that is active in both membrane bound and soluble forms. Cell, 63, 235–243. [DOI] [PubMed] [Google Scholar]
- Arakawa T. et al. (1991) Glycosylated and unglycosylated recombinant-derived human stem cell factor are dimeric and have extensive regular secondary structure. J. Biol. Chem., 266, 18942–18948. [PubMed] [Google Scholar]
- Aronson H.-E.G., Royer,W.E.J. and Hendrickson,W.A. (1994) Quantification of tertiary structural conservation despite primary sequence drift in the globin fold. Protein Sci., 3, 1706–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barleon B., Totzke,F., Herzog,C., Blanke,S., Kremmer,E., Siemeister,G., Marme,D. and Matiny-Baron,G. (1997) Mapping of the sites for ligand binding and receptor dimerization at the extracellular domain of the vascular endothelial growth factor receptor flt-1. J. Biol. Chem., 272, 10382–10388. [DOI] [PubMed] [Google Scholar]
- Bazan J.F. (1991) Genetic and structural homology of stem cell factor and macrophage colony-stimulating factor. Cell, 65, 9–10. [DOI] [PubMed] [Google Scholar]
- Besmer P. (1997) Kit-ligand–stem cell factor. In Garland,J.M., Quesenberry,P.J. and Hilton,D.J. (eds), Colony-Stimulating Factors: Molecular and Cellular Biology. 2nd edn. Marcel Dekker, Inc., New York, NY, pp. 369–404. [Google Scholar]
- Blechman J.M., Lev,S., Brizzi,M.F., Leitner,O., Pegoraro,L., Givol,D. and Yarden,Y. (1993) Soluble c-kit proteins and antireceptor monoclonal antibodies confine the binding site of the stem cell factor. J. Biol. Chem., 268, 4399–4406. [PubMed] [Google Scholar]
- Blechman J.M., Lev,S., Barg,J., Eisenstein,M., Vaks,B., Vogel,Z., Givol,D. and Yarden,Y. (1995) The fourth immunoglobulin domain of the stem cell factor receptor couples ligand binding to signal transduction. Cell, 80, 103–113. [DOI] [PubMed] [Google Scholar]
- Broudy V.C. (1997) Stem cell factor and hematopoiesis. Blood, 90, 1345–1364. [PubMed] [Google Scholar]
- Brünger A.T., Kuriyan,J. and Karplus,M. (1987) Crystallographic R-factor refinement by molecular dynamics. Science, 235, 458–460. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 252–270. [DOI] [PubMed] [Google Scholar]
- Davis-Symyth T., Chen,H., Park,J., Presta,L.G. and Ferrara,N. (1996) The second immunoglubulin-like domain of the VEGF tyrosine kinase receptor Flt-1 determines ligand binding and may initiate a signal transduction cascade. EMBO J., 15, 4919–4927. [PMC free article] [PubMed] [Google Scholar]
- Diederichs K., Boone,T. and Karplus,P.A. (1991) Novel fold and putative receptor binding site of granulocyte–macrophage colony-stimulating factor. Science, 254, 1779–1782. [DOI] [PubMed] [Google Scholar]
- Evans S.V. (1993) SETOR: hardware lighted three dimensional solid model representations of macromolecules. J. Mol. Graph., 11, 134–138. [DOI] [PubMed] [Google Scholar]
- Fantl W.J., Johnson,D.E. and Williams,L.T. (1993) Signalling by receptor tyrosine kinases. Annu. Rev. Biochem., 62, 453–481. [DOI] [PubMed] [Google Scholar]
- Fenstermaker R.A. et al. (1993) A cationic region of the platelet-derived growth factor (PDGF) A-chain (Arg159–Lys160–Lys161) is required for receptor binding and mitogenic activity of the PDGF-AA homodimer. J. Biol. Chem., 268, 10482–10489. [PubMed] [Google Scholar]
- Fisher D. and Eisenberg,D. (1996) Fold recognition using sequence-derived predictions. Protein Sci., 5, 947–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald P.M.D. (1988) MERLOT, an integrated package of computer programs for the determination of crystal structures by molecular replacement. J. Appl. Crystallogr., 21, 273–278. [Google Scholar]
- Galli S.J., Zsebo,K.M. and Geissler,E. (1994) The kit ligand, stem cell factor. Adv. Immunol., 55, 1–96. [DOI] [PubMed] [Google Scholar]
- Gewirth D. (1995) The HKL Manual. Yale University, New Haven, CT. [Google Scholar]
- Glaspy J. (1996) Clinical applications of stem cell factor. Curr. Opin. Hematol., 3, 223–229. [DOI] [PubMed] [Google Scholar]
- Harpaz Y. and Chothia,C. (1994) Many of the immunoglobulin superfamily domains in cell adhesion molecules and surface receptors belong to a new structural set which is close to that containing variable domains. J. Mol. Biol., 238, 528–539. [DOI] [PubMed] [Google Scholar]
- Heidaran M.A., Pierce,J.H., Jensen,R.A., Matsui,T. and Aaronson,S.A. (1990) Chimeric α- and β-platelet-derived growth factor (PDGF) receptors define three immunoglobulin-like domains of the α-PDGF receptor that determine PDGF-AA binding specificity. J. Biol. Chem., 265, 18741–18744. [PubMed] [Google Scholar]
- Heldin C.-H. (1995) Dimerization of cell surface receptors in signal transduction. Cell, 80, 213–223. [DOI] [PubMed] [Google Scholar]
- Hendrickson W.A. (1979) Transformations to optimize the superposition of similar structures. Acta Crystallogr. A, 35, 158–163. [Google Scholar]
- Hendrickson W.A., Smith,J.L. and Sheriff,S. (1985) Direct phase determination based on anomalous scattering. Methods Enzymol., 115, 41–55. [DOI] [PubMed] [Google Scholar]
- Hsu Y.-R., Narhi,L.O., Spahr,C., Langley,K.E. and Lu,H.S. (1996) In vitro methionine oxidation of Escherichia coli-derived human stem cell factor: effects on the molecular structure, biological activity, and dimerization. Protein Sci., 5, 1165–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y.-R., Chang,W.-C., Mendiaz,E.A., Hara,S., Chow,D.T., Mann,M.B., Langley,K.E. and Lu,H.S. (1998) Selective deamidation of recombinant human stem cell factor during in vitro aging: isolation and characterization of the aspartyl and isoaspartyl homodimers and heterodimers. Biochemistry, 37, 2251–2262. [DOI] [PubMed] [Google Scholar]
- Huang E., Nocka,K., Beier,D.R., Chu,T.-Y., Buck,J., Lahm,H.-W., Wellner,D., Leder,P. and Besmer,P. (1990) The hematopoietic growth factor KL is encoded by the Sl locus and is the ligand of the c-kit receptor, the product of the W locus. Cell, 63, 225–233. [DOI] [PubMed] [Google Scholar]
- Jones E.Y., Harlos,K., Bottomley,M.J., Robinson,R.C., Driscoll,P.C., Edwards,R.M., Clements,J.M., Dudgeon,T.J. and Stuart,D.I. (1995) Crystal structure of integrin-binding fragment of vascular cell adhesion molecule-1 at 1.8 Å resolution. Nature, 373, 539–544. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.-Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kawasaki E.S. and Ladner,M.B. (1990) Molecular biology of macrophage colony-stimulating factor. Immunol. Ser., 49, 155–176. [PubMed] [Google Scholar]
- Kleywegt G.J. and Jones,T.A. (1994) Halloween… masks and bones. In Bailey,S., Hubbard,R. and Waller,D. (eds), From First Map to Final Model, Proceedings of the CCP4 Study Weekend. Daresbury Laboratory, Warrington, UK, pp. 59–66. [Google Scholar]
- Langley K.E. et al. (1992) Purification and characterization of soluble forms of human and rat stem cell factor recombinantly expressed by Escherichia coli and by Chinese hamster ovary cells. Arch. Biochem. Biophys., 295, 21–28. [DOI] [PubMed] [Google Scholar]
- Langley K.E. et al. (1994) Properties of variant forms of human stem cell factor recombinantly expressed in Escherichia coli. Arch. Biochem. Biophys., 311, 55–61. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Lemmon M.A., Pinchasi,D., Zhou,M., Lax,I. and Schlessinger,J. (1997) Kit receptor dimerization is driven by bivalent binding of stem cell factor. J. Biol. Chem., 272, 6311–6317. [DOI] [PubMed] [Google Scholar]
- Lev S., Yarden,Y. and Givol,D. (1992) Dimerization and activation of the Kit receptor by monovalent and bivalent binding of the stem cell factor. J. Biol. Chem., 267, 15970–15977. [PubMed] [Google Scholar]
- Lev S., Blechman,J., Nishikawa,S.-I., Givol,D. and Yarden,Y. (1993) Interspecies molecular chimeras of Kit help define the binding site of the stem cell factor. Mol. Cell. Biol., 13, 2224–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev S., Blechman,J.M., Givol,D. and Yarden,Y. (1994) Steel factor and c-kit protooncogene: genetic lessons in signal transduction. Crit. Rev. Oncog., 5, 141–168. [DOI] [PubMed] [Google Scholar]
- Lu H.S., Clogston,C.L., Wypych,J., Fausset,P., Lauren,S., Mendiaz,E.A., Zsebo,K. and Langley,K.E. (1991) Amino acid sequence and post-translational modification of stem cell factor isolated from buffalo rat liver cell-conditioned medium. J. Biol. Chem., 266, 8102–8107. [PubMed] [Google Scholar]
- Lu H.S. et al. (1992) Post-translational processing of membrane-associated recombinant human stem cell factor expressed in Chinese hamster ovary cells. Arch. Biochem. Biophys., 298, 150–158. [DOI] [PubMed] [Google Scholar]
- Lu H.S., Jones,M.D., Shieh,J.-H., Mendiaz,E.A., Feng,D., Watler,P., Narhi,L.O. and Langley,K.E. (1996) Isolation and characterization of a disulfide-linked human stem cell factor dimer. J. Biol. Chem., 271, 11309–11316. [DOI] [PubMed] [Google Scholar]
- Lyman S.D. and Jacobsen,S.E.W. (1998) c-Kit ligand and Flt3 ligand: stem/progenitor cell factors with overlapping yet distinct activities. Blood, 91, 1101–1134. [PubMed] [Google Scholar]
- Martin F.H. et al. (1990) Primary structure and functional expression of rat and human stem cell factor DNAs. Cell, 63, 203–211. [DOI] [PubMed] [Google Scholar]
- Matous J.V., Langley,K.E. and Kaushansky,K. (1996) Structure–function relationships of stem cell factor: an analysis based on a series of human–murine SCF chimera and the mapping of a neutralizing monoclonal antibody. Blood, 88, 437–444. [PubMed] [Google Scholar]
- McKay D.B. (1992) Unraveling the structure of IL-2. Science, 257, 412–413. [DOI] [PubMed] [Google Scholar]
- McNiece I.K. and Briddell,R.A. (1995) Stem cell factor. J. Leukoc. Biol., 58, 14–22. [DOI] [PubMed] [Google Scholar]
- Mendiaz E.A., Chang,D.G., Boone,T.C., Grant,J.R., Wypych,J., Aguero,B., Egrie,J.C. and Langley,K.E. (1996) Epitope mapping and immunoneutralization of recombinant human stem cell factor. Eur. J. Biochem., 239, 842–849. [DOI] [PubMed] [Google Scholar]
- Milburn M.V., Hassell,A.M., Lambert,M.H., Jordan,S.R., Proudfoot,A.E., Graber,P. and Wells,T.N. (1993) A novel dimer configuration revealed by the crystal structure at 2.4 Å resolution of human interleukin-5. Nature, 363, 172–176. [DOI] [PubMed] [Google Scholar]
- Muller Y.A., Li,B., Christinger,H.W., Wells,J.A., Cunningham,B.C. and de Vos,A.M. (1997) Vascular endothelial growth factor: crystal structure and functional mapping of the kinase domain receptor binding site. Proc. Natl Acad. Sci. USA, 94, 7192–7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Oefner C., DíArcy,A., Winker,F.K., Eggimann,B. and Hosang,M. (1992) Crystal structure of human platelet-derived growth factor BB. EMBO J., 11, 3921–3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z. (1993) Oscillation data reduction program. In Sawyer,N.I.L. and Bailey,S. (eds), Data Collection and Processing. Science and Engineering Research Council, Warrington, UK, pp. 55–62. [Google Scholar]
- Pähler A., Smith,J.L. and Hendrickson,W.A. (1990) A probability representation for phase information from multiwavelength anomalous dispersion. Acta Crystallogr. A, 46, 537–540. [DOI] [PubMed] [Google Scholar]
- Pandit J., Bohm,A., Jancarik,J., Halenbeck,R., Koths,K. and Kim,S.-H. (1992) Three-dimensional structure of dimeric human recombinant macrophage colony-stimulating factor. Science, 258, 1358–1362. [DOI] [PubMed] [Google Scholar]
- Philo J.S., Wen,J., Wypych,J., Schwartz,M.G., Mendiaz,E.A. and Langley,K.E. (1996) Human stem cell factor dimer forms a complex with two molecules of the extracellular domain of its receptor, Kit. J. Biol. Chem., 271, 6895–6902. [DOI] [PubMed] [Google Scholar]
- Plotnikov A.N., Schlessinger,J., Hubbard,S.R. and Mohammadi,M. (1999) Structural basis for FGF receptor dimerization and activation. Cell, 98, 641–650. [DOI] [PubMed] [Google Scholar]
- Read R.J. (1986) Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. A, 42, 140–149. [Google Scholar]
- Rousset D., Agnes,F., Lachaume,P., Andre,C. and Galibert,F. (1995) Molecular evolution of the genes encoding receptor tyrosine kinase with immunoglobulin-like domains. J. Mol. Evol., 41, 421–430. [DOI] [PubMed] [Google Scholar]
- Rozwarski D.A., Gronenborn,A.M., Clore,G.M., Bazan,J.F., Bohm,A., Wlodawer,A., Hatada,M. and Karplus,P.A. (1994) Structural comparisons among the short-chain helical cytokines. Structure, 2, 159–173. [DOI] [PubMed] [Google Scholar]
- Russell E.S. (1979) Hereditary anemias of the mouse: a review for geneticists. Adv. Genet., 20, 357–459. [PubMed] [Google Scholar]
- Shrake A. and Rupley,J.A. (1973) Environment and exposure to solvent of protein atoms. Lysozyme and insulin. J. Mol. Biol., 79, 351–371. [DOI] [PubMed] [Google Scholar]
- Shull R.M., Suggs,S.V., Langley,K.E., Okino,K.H., Jacobsen,F.W. and Martin,F.H. (1992) Canine stem cell factor (c-kit ligand) supports the survival of hematopoietic progenitors in long-term canine marrow culture. Exp. Hematol., 20, 1118–1124. [PubMed] [Google Scholar]
- Sprang S.R. and Bazan J.F. (1993) Cytokine structural taxonomy and mechanisms of receptor engagement. Curr. Opin. Struct. Biol., 3, 815–827. [Google Scholar]
- Stauber D.J., DiGabriele,A.D. and Hendrickson,W.A. (2000) Structural interactions of fibroblast growth factor with its ligands. Proc. Natl Acad. Sci. USA, 97, 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor E.W., Fear,A.L., Bohm,A., Kim,S.-H. and Koths,K. (1994) Structure–function studies on recombinant human macrophage colony-stimulating factor (M-CSF). J. Biol. Chem., 269, 31171–31177. [PubMed] [Google Scholar]
- Wang Z., Myles,G.M., Brandt,C.S., Lioubin,M.N. and Rohrschneider,L. (1993) Identification of the ligand-binding regions in the macrophage colony-stimulating factor receptor extracellular domain. Mol. Cell. Biol., 13, 5348–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesmann C., Fuh,G., Christinger,H.W., Eigenbrot,C., Wells,J.A. and de Vos,A.M. (1997) Crystal structure at 1.7 Å resolution of VEGF in complex with domain 2 of the Flt-1 receptor. Cell, 91, 695–704. [DOI] [PubMed] [Google Scholar]
- Wlodawer A., Pavlovsky,A. and Gustchina,A. (1992) Crystal structure of human recombinant interleukin-4 at 2.25 Å. FEBS Lett., 309, 59–64. [DOI] [PubMed] [Google Scholar]