Abstract
The non-Mendelian element [URE3] of yeast is considered to be a prion form of the Ure2 protein. The [URE3] phenotype occurs at a frequency of 10–5 in haploid yeast strains, is reversible, and its frequency is increased by overexpressing the URE2 gene. We created a new mutant of the Ure2 protein, called H2p, which results in a 1000-fold increase in the rate of [URE3] occurrence. To date, only the overexpression of various C-terminal truncated mutants of Ure2p gives rise to a comparable level. The h2 allele is, thus, the first characterized URE2 allele that induces prion formation when expressed at a low level. By shuffling mutated and wild-type domains of URE2, we also created the first mutant Ure2 protein that is functional and induces prion formation. We demonstrate that the domains of URE2 function synergistically in cis to induce [URE3] formation, which highlights the importance of intramolecular interactions in Ure2p folding. Additionally, we show using a green fluorescent protein (GFP) fusion protein that the h2 allele exhibits numerous filiform structures that are not generated by the wild-type protein.
Keywords: aggregation/GFP/prion/Saccharomyces cerevisiae/[URE3]
Introduction
The genetic properties of some non-Mendelian determinants of yeast, such as [URE3] (Lacroute, 1971; Aigle and Lacroute, 1975) and [PSI] (Cox, 1965; Ter-Avanesyan et al., 1994), support the hypothesis that they are yeast prions (Wickner, 1994). This hypothesis is based on the genetic properties of these elements: they are dominant and display a non-Mendelian segregation; their phenotype is reversible; they depend on the expression of the regular form of the protein (Ure2p and Sup35p, respectively); and their frequency is increased by the overexpression of the normal protein (reviewed by Wickner et al., 1995).
The URE2 gene codes for a 354-amino-acid protein that contains two domains. The N-terminal domain is responsible for inducing [URE3] formation de novo. This N-terminal domain was originally defined as the first 65 amino acids of Ure2p, the prion forming domain (PFD) (Masison and Wickner, 1995), but has now been more widely defined as the first 94 Ure2p amino acids that are able to induce [URE3] (Komar et al., 1999). The C-terminal domain of the Ure2 protein contains the catalytic domain (Coschigano and Magasanik, 1991), which inhibits the activity of the transcription factor Gln3p in the presence of a rich nitrogen source (Courchesne and Magasanik, 1988). Ure2p thus contributes to nitrogen catabolism repression by allowing cells to import poor nitrogen sources, like allantoin, when necessary, and by maintaining an optimal level of glutamine for growth. Recently, it was also demonstrated that Ure2p plays a role in the TOR kinases signaling pathway, which regulates a range of cellular functions including cell proliferation and response to nutrients (Cardenas et al., 1999).
The prion conversion process of the mammalian prion protein, PrP, remains poorly understood. The yeast [PSI] model, however, provides a clearer picture of prion formation: when the yeast Sup35 protein converts to the prion form, it becomes insoluble and pellets when centrifuged at 15 000 g (Paushkin et al., 1996). Autocatalytic conversion of the yeast Sup35 protein to the [PSI] prion has been demonstrated in vitro, and the [PSI] phenotype is associated with the aggregation state of the Sup35 protein (Paushkin et al., 1996, 1997).
Polymorphisms of the different prion proteins have been widely studied, especially polymorphisms of the mammal prion protein PrP. Several amino acid changes of PrP influence incubation time and/or susceptibility to transmissible spongiform encephalopathy (TSE) in animal models or human. The single 101L mutation in mouse PrP generated in transgenic mice alters the incubation time of a TSE infection (Manson et al., 1999). This mutation is equivalent to the P102L substitution in the human PrP that is associated with Gerstmann–Straussler–Scheinkel syndrome (GSS), a hereditary form of TSE in humans (Hsiao et al., 1992). Other amino acid substitutions alter susceptibility to TSE. In sheep, polymorphisms at amino acids 136, 154 and 171 in the PrP gene affect both the occurrence of the disease and the mean survival time of affected animals (Goldmann et al., 1994; Hunter et al., 1994). The presence of a valine or a methionine at position 129 of human PrP was described as influencing susceptibility to and the incubation period of two hereditary forms of TSE: fatal familial insomnia and a subtype of familial Creutzfeld–Jakob disease (Goldfarb et al., 1992). Also the incidence of iatrogenic transmission of Creutzfeld–Jakob disease by growth hormone treatment has been associated with polymorphisms of the PrP gene. Early in 1993, a homozygous genotype with a valine at position 129 of the PrP gene was described as playing a role in the disease incidence in French patients treated with extractive growth hormone (Labauge et al., 1993; Masson et al., 1994). The molecular mechanism by which these different forms of the PrP protein influence incubation times and phenotypic expression remains unknown, although thermodynamic and kinetic models have been proposed to explain the role of these single mutations (Cohen, 1999).
In the yeast Saccharomyces cerevisiae, several mutants have been identified that enhance or inhibit [URE3] or [PSI] formation. The PNM2 mutation that eliminates the [PSI] element was identified as a single transition in the SUP35 gene that generates a glycine to aspartic acid substitution in the PFD of the Sup35 protein (Doel et al., 1994). Trans acting factors that influence prion phenotypes in yeast have also been described. For example, the Hsp104 chaperone protein is required to maintain the [PSI] phenotype (Chernoff et al., 1995). Nonsense mutations in the URE2 gene that lead to the expression of the N-terminal domain of Ure2p or missense mutations that result in the expression of an out-of-frame catalytic domain of Ure2p favor the formation of [URE3] (Wickner, 1994; Masison et al., 1997). Until now, however, alleles of URE2 or SUP35 that induce high levels of [URE3] and [PSI] prion phenotype, respectively, were all proteins that lost their functional acitivity.
Here we describe the creation of a mutated allele of URE2, called h2, which induces [URE3] formation at a high frequency when expressed on a monocopy plasmid. We characterized this mutant using genetic and cellular approaches, and analyzed the effects of mutated domains on Ure2 activity and prion formation by shuffling the different mutated domains. In this process, we obtained the first prion-inducing Ure2 protein that is still functional. Finally, by expressing H2–GFP (green fluorescent protein) fusion protein in yeast, we studied the aggregation patterns of protein encoded by the h2 allele.
Results
Selection of inducing alleles
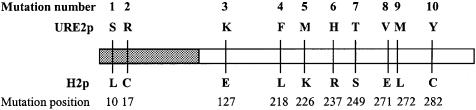
In order to select alleles of URE2 that induce [URE3] at a high frequency, we transformed the CC30 yeast strain with a pFL39-URE2 mutagenized library, and 105 cells were replicated on ureidosuccinate (USA) medium, which permits the selection of the [URE3] phenotype. Eight clones presenting three or more [USA+] papillae, thus likely to present a high frequency of [URE3] appearance, were selected. Each clone was then replated on USA-containing medium and one was selected for further analysis as it presented a high level of [USA+] clones. The transforming plasmid from this clone was re-introduced into the wild-type strain CC30. The transformed strain then exhibited a 1000-fold increase in [USA+] frequency. The sequence of the URE2 allele carried on the transforming plasmid and called h2 revealed 14 mutations, of which 10 lead to amino acid substitutions in Ure2p (Figure 1). Two substitutions were in the PFD (defined as the domain spanning amino acids 1–94), and eight mutations were located in the catalytic domain of Ure2p. Complementation assays demonstrated that the h2 allele was not able to complement the Δure2 strain AF36.
Fig. 1. Amino acid substitutions are indicated with solid lines and position; above is the wild-type URE2 sequence and below is the h2 allele. The shaded box indicates amino acids 1–94 including the PFD; the white box indicates the catalytic part of the protein.
To characterize the [USA+] clones induced by the h2 allele, we first plated these [USA+] colonies on a non-selective medium in order to cure the clones of plasmids. In parallel, in the same way we analyzed [USA+] clones induced by a multicopy plasmid carrying the PFD of URE2. Table I shows that 24 [USA+]-independent clones induced by the overexpression of a plasmid carrying the PFD reverted to [USA–] when the inducing plasmid was lost. In contrast, among 15 [USA+] clones induced by the h2 allele, five remained [USA+] after the loss of the plasmid, suggesting the formation of [URE3], as the [USA+] phenotype was maintained after the loss of the h2 allele. We analyzed this phenotype further and demonstrated that it was dominant by crossing these clones with the CC30 strain. After sporulation, tetrads showed a characteristic non-Mendelian inheritance of the [USA+] phenotype (data not shown).
Table I. Phenotype of cells after loss of plasmids inducing the [USA+] phenotype.
| Inducing plasmid | Number of [USA+] clones analyzed | Number of [USA+] clones after loss of the plasmid |
|---|---|---|
| pY2L-Nter | 24 | 0 |
| pFL39-h2 | 15 | 5 |
N- and C-terminal mutations have a cis synergistic effect
By shuffling different regions of the h2 and URE2 genes, we generated new URE2 alleles which encode proteins that combine either the mutated PFD of H2p and a wild-type Ure2p catalytic domain (hM 1,2), or a wild-type Ure2p PFD with the mutated catalytic domain of H2p (hM 3-10). These constructs were expressed on a monocopy plasmid and tested for their ability both to complement a Δure2 strain and to induce the [URE3] phenotype. Results are shown in Table II, lines 3 and 4. The two alleles increased the frequency of [URE3] clones 10 and 6 times, respectively. The hM 3-10 allele carrying the mutated catalytic domain was not able to complement a Δure2 strain. However, the hM 1,2, carrying a wild-type catalytic domain of URE2, was still functional.
Table II. Complementation test and [URE3] induction by different mutated alleles of URE2.
| Allele | USA growth | [URE3] induction |
|---|---|---|
| URE2 | – | 1 |
| h2 | + | 1000 |
| hM 1,2 | – | 10 |
| hM 3-10 | + | 6 |
| hM 1 | – | 1 |
| hM 2 | +/– | 1 |
| hM 3 | + | 1 |
| hM 4 | – | 1 |
| hM 5 | +/– | 1 |
| hM 6 | – | 1 |
| hM 7 | + | 1 |
| hM 8 | +/– | 1 |
| hM 9 | – | 1 |
| hM 10 | + | 1 |
| hM 1,3 | +/– | 1 |
| hM 1,7 | + | 1 |
| hM 1,8 | – | 10 |
| hM 1,9 | – | 1 |
| hM 2,3 | + | 12 |
| hM 2,7 | +/– | 1 |
| hM 2,8 | +/– | 1 |
| hM 2,9 | + | 1 |
| hM 1,2,3 | + | 500 |
Numbers written after hM indicate the position of mutations re-inserted, as indicated on Figure 1.
The absence of growth (–) indicates the functionality of the URE2 allele. +/– indicates very slow growth.
[URE3] induction is given as 1 for the inducing effect of the wild-type URE2 gene, corresponding to a frequency of 10–5. Standard deviation is <10%.
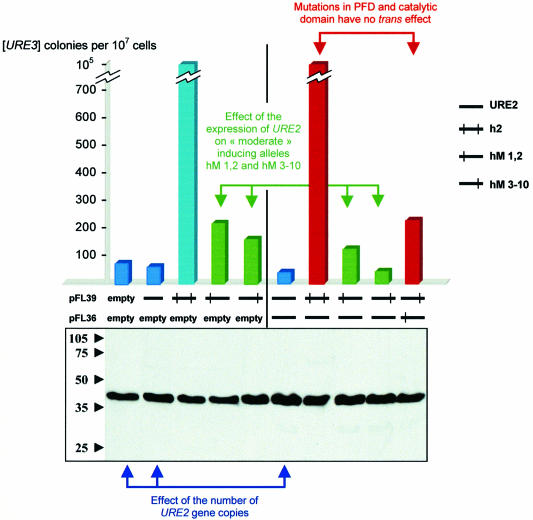
The [URE3]-inducing effect of the two hybrid alleles, hM 1,2 and hM 3-10, was lower than that observed with the entire h2 allele. To determine whether the two mutated domains could interact in trans, we expressed the hM 1,2 and hM 3-10 alleles in the same cell simultaneously. As the rate of [URE3] appearance partially depends on the concentration of Ure2p, we performed 10 separate co-transformations. The western blot performed with these strains indicated that the concentration of Ure2p was proportional to the copy number of the plasmids (Figure 2). Each transformant was then plated on a medium containing USA. The results shown in Figure 2 lead to several observations. Whereas the overexpression of Ure2p increases [URE3] appearance (Masison and Wickner, 1995), two copies of the URE2 gene cause a decrease in the rate of [URE3] (Figure 2, blue arrow). This effect is even stronger with three copies. However, the concentration of Ure2p in cells is higher in this latter case (Figure 2, compare lane 1 with lanes 2 and 6 on the western blot). In the same way, co-expressing a URE2 gene with hM 1,2 or hM 3-10 constructs partially inhibited their inducing effect (Figure 2, green arrow). However, the inducing effect of the h2 allele is not significantly affected by the presence of the expression of a wild-type URE2 gene.
Fig. 2. Cis and trans effect of the co-expression of different mutated alleles of URE2. The CC30 strain was co-transformed with different plasmids based on pFL39 and pFL36 leading to the expression of URE2; h2; hM 1,2; hM 3-10. The 10 different combinations were analyzed for their inducing properties by plating 107 cells on USA-selective medium. Standard deviation is not >10%. Bottom, the expression levels of Ure2p in each co-transformed strain were compared by western blot analysis. Numbers indicate the molecular weights (in kilodaltons).
When strain CC30 was co-transformed by vectors expressing hM 1,2 and hM 3-10 constructs, the level of [USA+] clones was not greater than that observed for the cells transformed by hM 1,2 alone. The low level of [USA+] clones compared with the control (co-transformation of the CC30 strain by h2 and URE2 genes; Figure 2, red arrow) indicates that the mutated domains have a synergistic inducing effect when located on the same protein.
Effect of the mutations of the h2 allele
In order to assess the individual input of each mutation in the [URE3]-inducing effect, each mutation was re-introduced by directed mutagenesis of the URE2 gene. We showed that no single mutation had a significant effect on the rise of [URE3]. We further investigated the effect of the mutations by combining different pairs of mutations, always selecting one mutation located in the PFD of the protein and a second mutation in the catalytic domain. We observed that several pairs of mutations had a significant effect on [URE3] induction, confirming that the two domains of the protein play a synergistic role in the inducing properties of Ure2p (Table II). Furthermore, we noticed that most of the inducing properties of the h2 allele were determined by the first three mutations, which are located in the PFD and in the catalytic domain of Ure2p (Figure 1; mutant allele hM 1,2,3).
We also tested each construct for its ability to complement the Δure2 strain AF36. One of the inducing mutation pairs (hM 1,8) retained the catalytic function of the URE2 gene, but not the second (hM 2,3) (Table II). Thus, different mutations in the URE2 gene produce either functional or non-functional prion-inducing proteins.
h2–GFP fusions present several aggregation patterns
It has been reported several times that the [URE3] phenotype may be related to an aggregation state of the Ure2p protein in the cytoplasm of the cells (Edskes et al., 1999). To study Ure2p aggregation, we constructed a gene that generates a protein fusion between GFP and the protein encoded by the h2 allele. This construct was expressed under the control of the URE2 promoter, on a monocopy or a multicopy plasmid (pYeFc1L and pYeFc2L expression vectors, respectively). As controls, we constructed the same GFP fusion with the wild-type URE2 gene.
These plasmids were then used to transform the CC30 strain in order to measure the prion-inducing effect. The h2–GFP allele was still able to induce [URE3] at a high frequency, but its effect was somewhat reduced compared with that of the h2 allele (Table III, line 2). The decrease in induction observed when expressing the h2–GFP fusion construct was also observed for the URE2–GFP fusion protein expressed on the multicopy plasmid pYeFc 2L that was used as a control. By transforming the CC34 strain with the monocopy and multicopy plasmids bearing the H2–GFP fusion protein, we ensured that the h2–GFP fusion did not cure [URE3] in a strain initially carrying the [URE3] prion (Table III, line 3). The C-terminal GFP fusion is able to decrease slightly the inducing effect of URE2, but when overexpressed on the pYeFc 2L plasmid, it has a destabilizing effect on a pre-existing [URE3] phenotype, as previously described (Edskes et al., 1999).
Table III. Number of [URE3] clones per 106 cells on USA medium.
| GFP fusion construct | Control | pYeFc1L URE2–GFP | pYeFc1L h2–GFP | pYeFc2L URE2–GFP | pYeFc2L h2–GFP |
|---|---|---|---|---|---|
| CC30 | 39 | 60 | 129 | 50 | ∼1000 |
| CC34 | [URE3] | [URE3] | [URE3] | cured | [URE3] |
[URE3] indicates that all cells were able to grow on USA medium, showing that the [URE3] phenotype of the CC34 strain was not affected by the expression of the different constructs. Only the multicopy expression of the URE2–GFP fusion cured the CC34 strain, which is consistent with what was previously reported.
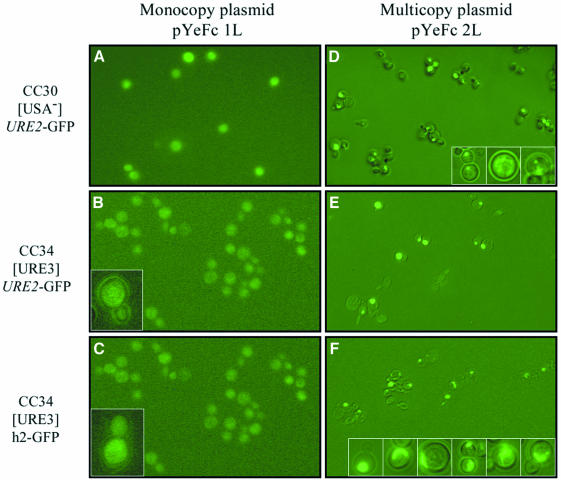
The proteins encoded by these constructs exhibited different expression patterns that did not depend on the [URE3] phenotype but on the expression levels of each construct (Figure 3). When expressed on the monocopy plasmid pYeFc 1L, the URE2–GFP and h2–GFP fusion constructs generated, in 100% of cells, a diffuse green color in both strains CC30 and CC34 (Figure 3A, B and C).
Fig. 3. Microscopy observation of CC30 and CC34 strains transformed by monocopy and multicopy plasmids expressing URE2–GFP and h2–GFP. (A and D) Cellular distribution of URE2–GFP in the CC30 [USA–] strain expressed on a monocopy and a multicopy plasmid, respectively. (B and E) Cellular distribution of URE2–GFP in the CC34 [URE3] strain expressed on a monocopy and a multicopy plasmid, respectively. (C and F) Cellular distribution of h2–GFP in the CC34 [URE3] strain expressed on a monocopy and a multicopy plasmid, respectively. Insets in (B) and (C) show the diffuse pattern exhibited by the expression on a monocopy plasmid of the wild-type and mutant URE2 gene, whatever the [URE3] or [USA–] phenotype. Insets in (D) show the spots and clumps typically observed by expressing the wild-type URE2 gene on a multicopy plasmid, and insets in (F) show the typical filiform and ramified structures observed by expressing the h2 allele on a multicopy plasmid.
However, when expressed on the multicopy plasmid pYeFc 2L, the GFP fusion proteins produced by the two constructs showed different patterns of expression (Figure 3D, E and F). When transformed by the control URE2–GFP, ∼30% of the cells presented a diffuse expression of GFP and ∼70% showed either one big spot or several small green spots, localized in the cytoplasm, either in the CC30 [USA–] strain (Figure 3D, insets) or in the CC34 [URE3] strain (Figure 3E). However, the CC34 [URE3] strain transformed with the h2–GFP construct presented a diffuse color in ∼10% of the cells and a green spot in 40% of the cells (Figure 3F). In addition, 50% of these cells presented a new coloration pattern, with some long cytoplasmic filiform structures that sometimes branched out (Figure 3F, insets). Furthermore, these aggregation patterns were observed regardless of the [URE3] phenotype in both the CC30 and CC34 strains.
Discussion
The molecular and biochemical ways in which the [URE3] phenotype is generated from the Ure2 protein remain unclear. The prion hypothesis postulates that [URE3] is the product of an autocatalytic inactivation of Ure2p. This inactivation implies a Ure2p–Ure2p interaction, leading to the inactivation of this protein. As a consequence, USA can be imported into the cell, leading to a [USA+] phenotype. However, this [USA+] phenotype can also be due to classical mutations affecting the URE2 gene. In this case, the [USA+] phenotype would segregate as expected for a nuclear mutation. Only the segregation of the [USA+] phenotype or its curability allows the prion state to be distinguished from mutations that affect Ure2p expression. A third case may also exist that could lead to the same [USA+] phenotype but with distinct genetic properties. This case may be found upon expression of a Ure2p ‘inhibitor’. Such a molecule would inhibit Ure2p, thus permitting the entry of USA. The [USA+] phenotype would only exist, however, while this inhibitor was expressed. After the loss of such an inhibitor, Ure2p should regain activity and give rise to the [USA–] phenotype. This kind of inactivation would not correspond to an autocatalytic process and such a mechanism, even if it involves the PFD as the inhibitor molecule, could not correspond to [URE3]. Both [USA+] clones obtained by overexpression of the PFD or the h2 allele in our experiment were therefore analyzed after the loss of the inducing construct. Surprisingly, the PFD can inactivate Ure2p when expressed in these strains, but this inactivation proved to be transitory, given that no [USA+] clone retained that phenotype after the loss of the plasmid. In contrast, at least in the genetic background of our strains, the h2 element can promote an inactivation phenotype, [USA+], which can be maintained even after the loss of the inducing element. Indeed, h2 efficiently promotes the conversion of Ure2p to its prion form Ure2p[URE3]. Thus, several ways to inactivate the Ure2 protein exist; some are connected to the prion mechanisms, which are persistent, and others are transitory inactivation mechanisms.
The protein encoded by the h2 allele of URE2 differs from the wild-type gene by 10 mutations located both in the PFD and the catalytic domain of the protein. By shuffling sequences between wild-type and mutated genes, we generated several new URE2 alleles including one that both encodes a functional Ure2p and induces [URE3]. This allele is particularly exciting as all the inducing proteins so far reported can not fulfill the functional criteria. These previous constructs may act by destabilizing Ure2p through an artificial mechanism that would never occur in the yeast cell. In contrast, the functional inducing allele that we generated mimics the mechanism behind [URE3] formation. The balance between functional Ure2pC and Ure2p[URE3] in this case merely favors Ure2p[URE3]. The existence of this allele emphasizes the ‘protein only’ model for [URE3] formation.
To dissect the role of the mutations in h2, the functional inducing allele (hM 1,2) and its counterpart (hM 3-10) obtained by the complementary shuffling were co-expressed. Co-expression of the hM 1,2 and hM 3-10 alleles induces the [URE3] phenotype at a level similar to that observed with the expression of each allele alone. Thus, there is no trans synergistic effect of the mutated domains of URE2, whereas this effect is maximal when the two mutated domains are carried by the same molecule. These data suggest that mutations of the H2 protein modify the intramolecular interactions necessary for its folding. Co-expression in yeast of various inducing constructs also generated additional data (Figure 2): when expressed on a monocopy plasmid, the URE2 gene did not induce [URE3], as it does when overexpressed. On the contrary, it inhibited [URE3] appearance, and the inhibition effect was proportional to the number of URE2 gene copies, at least from one to three. This result suggests that the catalytic domain of Ure2p, known for its inhibiting properties on the PFD, exceeds the effect of the PFD when the number of copies of the URE2 gene is low. This inhibitory effect, however, appears to be overcome by overexpression of the PFD on a multicopy plasmid. This inhibitory effect of the catalytic domain is also observed on the weaker inducing alleles hM 1,2 and hM 3-10.
To analyze further the role of each mutation in the h2 allele, we re-introduced these 10 mutations into URE2 as single mutations. Although some of them abolish the function of Ure2p, they do not behave as [URE3] inducers. The inducing effect of h2, therefore, does not appear to be the consequence of the loss of function of Ure2p. Genetic analysis of URE2 alleles carrying two of the h2 mutations showed that two pairs of mutations had an inducing effect on [URE3] formation. One of these pairs (hM 2,3) abolishes the catalytic function of URE2, whereas the other one (hM 1,8) preserves it. In both cases, the changes occurring in the catalytic domain are important since an acidic amino acid replaces a neutral or a basic amino acid. The more drastic change, E127K, by itself provokes the loss of URE2 function. In the PFD (which is dispensable for the catalytic function of Ure2p), the changes (S10L and R17C) involve two basic amino acids. The synergistic effect could be due to intramolecular interactions involving electrostatic forces.
Wickner has proposed that the mechanism responsible for the formation of the prion form relies on a polymerization of the normal protein into filaments that lead to a conformational change of the protein and then its loss of function (Edskes et al., 1999). In the case of the [PSI] determinant, many arguments support this hypothesis: the crystal form of Sup35 protein is indeed able to convert the normal form of Sup35p into a prion form efficiently. Moreover, the formation of Sup35p fibrils is strictly related to the appearance of the prion phenotype. However, in the case of the mammal prion protein PrP, no data show any absolute connection between fibrils and the converted resistant form PrPSc. In the genetic background of our yeast strains, we observed a similar phenomenon.
We found, while expressing the fusion URE2–GFP in our strains, that there is no strict connection between the existence of aggregates and the [URE3] phenotype (E.Guillemet, E.Fernandez-Bellot, C.Thual and C.Cullin, submitted). We also observed that some factors that increase the appearance of [URE3] also stimulate aggregate formation. The results presented here indicate that aggregate formation is not related to phenotype but to URE2 expression level; there is no correlation between the frequency of [URE3] induction and the frequency of cells presenting aggregates. When the inducing allele h2 is expressed under the control of the URE2 promoter, the number of cells presenting aggregates is not significantly greater than when the wild-type URE2 gene is expressed. Moreover, in the [USA+] clones, not all the cells present aggregates as would be expected.
The nature of the structures observed in vivo remains to be characterized. If the [URE3] phenotype corresponds to a certain aggregation level, the expression of GFP fusion proteins demonstrated that this level is not observable by optical microscopy. Moreover, the spots observed when Ure2–GFP or H2–GFP are expressed on a multicopy plasmid, if they are correlated with an increase in [URE3], are not directly dependent on [URE3] formation. It is possible that various levels of aggregation in vivo, based on what we previously observed in vitro (Thual et al., 1999), correspond to various partially or completely inactive states of protein, and that these states may or may not be autocatalytic.
Materials and methods
Construction of a mutant URE2 library and the selection of prion-inducing alleles
The URE2 gene, carried on a pFL39 plasmid, was amplified by PCR in the presence of 200 µM each of dGTP, dTTP and dCTP, and several concentrations of dATP ranging from 1 to 50 µM, using Taq DNA Polymerase (Promega). When amplification efficiency was poor, the corresponding PCR product was cloned by gap-repair into the pFL39 plasmid.
Construction of hM 1,2 and hM 3-10 alleles
The 5′ sequences corresponding to the PFD of the h2 allele were used to replace the wild-type sequences by digesting the pFL39-h2 and pFL39-URE2 plasmids with BsiWI and NotI. The eluted pFL39 vector containing the wild-type 3′ sequences of the URE2 gene was then ligated with a fragment containing the h2 5′ sequences to give the pFL39 hM 1,2 vector, and reciprocally the eluted pFL39 vector containing the 3′ region of the h2 gene was ligated with a fragment containing wild-type 5′ sequences to generate the pFL39 hM 3-10 vector.
Construction of pFL39-hM 1,2,3
The pFL39-hM 1,2,3 vector, containing the first three mutations of the h2 allele, was constructed by digesting pFL39-h2 with XhoI and ApaI. The 0.9 kb fragment containing the mutated 5′ region of the h2 gene was ligated into pFL39-URE2 digested with XhoI and ApaI.
Construction of pFL vectors with leucine as the selectable marker
The pFL39 vectors containing different constructs were cut by BglII to remove the selectable marker tryptophan. A BglII–BglII fragment containing the LEU2 gene was then inserted to give the pFL36 vectors containing the different URE2 alleles.
Expression levels of Ure2p in co-transformed strains
Yeast cultures were grown on a solid medium selective for plasmid markers. The cells were harvested, washed in water, and lysed by vortexing with glass beads in 50 mM Tris–HCl pH 7.5, 100 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (PMSF; Sigma). Cell debris was removed by centrifugation at 15 000 g for 15 min. Total protein extract (50 µg) of each lysate was deposed on an SDS–polyacrylamide gel, according to standard procedures. After transfer from the gel to nitrocellulose membrane (Schleicher and Schuell), the blot was probed with a polyclonal rabbit antibody against Ure2p. Coomassie Blue staining was also performed to confirm that an equal amount of protein was loaded in the lanes.
Directed mutagenesis of the URE2 gene
The different mutations of the h2 allele were re-inserted into the URE2 gene by PCR-directed mutagenesis, using Advantaq DNA polymerase (Clontech). The URE2 gene was amplified with the ure2Xho primer (5′-GCTTCCTTACTCGAGGTTG-3′) and the hMxR primers (x corresponding to the number of the mutation; hM1R: 5′-GGAATTCAACA CTTGGTTG-3′; hM2R: 5′-GTTTACTTGACAGAGCGCATTGG-3′; hM3R: 5′-CTATAGCAACTTCGAATCCATTAG-3′; hM4R: 5′-GAC GTTTGGAGGAACAACCATG-3′; hM5R: 5′-CTTGTCCAATCTT TGGCGCATG-3′; hM6R: 5′-CTTTTGTGAACGGAAGTATCTG-3′; hM7R: 5′-AACCTCATCCGAATATCTTTCTA-3′; hM8R: 5′-CTAA TTCCATCTCCAGCGCTTC-3′; hM9R: 5′-GTCTAATTCCAACACC AGcgct-3′; hM10R: 5′-ACCAGCTGAGCATGCAGCCGC-3′), and with the ure2Nco primer (5′-CATTATTCCATGGGACAAAGGCC-3′) and the hMxD primers (hM1D: 5′-CAACCAAGTGTTGAATCT CTC-C-3′; hM2D: 5′-CCAATGCGCTCTGTCAAGTAAAC-3′; hM3D: 5′-ctaATGGATTCGAAGTTGCTATAG-3′; hM4D: 5′-CATGGTTG TTCcTCCAAACGTC-3′; hM5D: 5′-CATGCGCCAAAGATTGGA CAAG-3′; hM6D: 5′-CAGATACTTCCGTTCACAAAAG-3′; hM7D: 5′-tagAAAGATATTCGGATGAGGTT-3′; hM8D: 5′-GAAGCGCT GGAGatggaattag-3′; hM9D: 5′-AGCGCTGGTGTTGGAATTA GAC-3′; hM10D: 5′-GCGGCTGCATGCTCAGCTGGt-3′). The amplification products carrying the same mutation were mixed and re-amplified with the ure2Xho and ure2Nco primers to reconstitute a whole URE2 gene carrying a single mutation. Alleles carrying two mutations were constructed in the same way.
Strains, media and complementation tests
CC30 strain: MATa, trp1-1, ade2-1, leu2-3,112, his3-11,15, ΔURA2::HIS3. CC34 strain: MATa, trp1-1, ade2-1, leu2-3,112, his3-11,15, ΔURA2::HIS3, [URE3]. The CC34 strain carries the [URE3] element described originally by Aigle and Lacroute (1975), which was transmitted to the CC30 strain by cytoduction. Functional complementation tests were carried out with the AF36 strain: MATa; trp1-1; ade2-1; leu2-3,112; his3-15,15; URA2::HIS3; cyh2r; URE2::CYH2. Transformations were carried out as previously described (Gietz et al., 1995). Complete medium (YPGA) was prepared with 1% bacto-yeast extract, 2% bacto-peptone, 2% glucose, 2.5% bacto-agar, 20 mg/ml adenine. Minimal medium (W0) was prepared with 0.7% nitrogen base without amino acids (Difco), 2% glucose, 2.5% agar, supplemented with amino acids. [URE3] selection is based on the USA uptake in the ura2– strain, caused by the inactivation of Ure2p after its conversion to the prion form. [URE3] colonies may then grow on a minimal medium supplemented with appropriate amino acids, except uracil, and 15 mg/l USA, as previously described (Lacroute, 1971).
Construction of h2–GFP fusions
The pYeFc 1L-URE2–GFP and pYeFc 2L-URE2–GFP plasmids were constructed by digesting pYeFc 1L-V10-URE2–GFP and pYeFc 2L-V10-URE2–GFP vectors with BamHI and PvuII to remove the V10 (PGK) promoter and the first 850 nucleotides of the URE2 gene. A fragment containing the URE2 promoter and the first 850 nucleotides of the URE2 gene was amplified by PCR with primers 208 (5′-CCGGCGCGGATCCCTACCGTCCTCTATGTCTCC-3′) and 209 (5′-GTGTTGTACCAGCTGAGTATGCAGCCGC-3′), with plasmid pFL39-URE2 as the template. After digesting this fragment with BamHI and PvuII, it was re-ligated with the digested pYeFc URE2–GFP plasmids to give the pYeFc 1L-URE2–GFP and pYeFc 2L-URE2–GFP plasmids. Plasmids pYeFc 1L-h2–GFP and pYeFc 2L-h2–GFP were constructed in the same way by amplifying the h2 allele under the control of the URE2 promoter with pFL39-h2 as the template.
Microscopy observations
Cells were grown overnight on selective medium and were suspended in a 50 µl Dabco solution [218 mM diazabicyclo-2-2-2-octane (Sigma), 25% phosphate-buffered saline, 75% glycerol]. Cells were photographed using a DMRB microscope (Leica, Germany) with a PL APO 63× objective.
Acknowledgments
Acknowledgements
We gratefully thank Professor François Lacroute for constructing the mutagenized library and his helpful suggestions, and Ranjiv Khush for critical reading of the manuscript. The work was supported by grants from Action Concertée Coordonnée Science du Vivant No. 10 (9510001) to C.C. and an EC Contract No. BI104-98-6045, Maintenance and transmission of yeast prions: a model system.
References
- Aigle M. and Lacroute,F. (1975) Genetical aspects of [URE3], a non-mitochondrial, cytoplasmically inherited mutation in yeast. Mol. Gen. Genet., 136, 327–335. [DOI] [PubMed] [Google Scholar]
- Cardenas M.E., Cutler,N.S., Lorenz,M.C., Di Como,C.J. and Heitman,J. (1999) The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev., 13, 3271–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff Y.O., Lindquist,S.L., Ono,B., Inge,V.S. and Liebman,S.W. (1995) Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science, 268, 880–884. [DOI] [PubMed] [Google Scholar]
- Cohen F.E. (1999) Protein misfolding and prion diseases. J. Mol. Biol., 293, 313–320. [DOI] [PubMed] [Google Scholar]
- Coschigano P.W. and Magasanik,B. (1991) The URE2 gene product of Saccharomyces cerevisiae plays an important role in the cellular response to the nitrogen source and has homology to glutathione S-transferases. Mol. Cell. Biol., 11, 822–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne W.E. and Magasanik,B. (1988) Regulation of nitrogen assimilation in Saccharomyces cerevisiae: roles of the URE2 and GLN3 genes. J. Bacteriol., 170, 708–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox B.S. (1965) ψ, a cytoplasmic supressor of super-supressor in yeast. Heredity (Edinb.), 20, 505–521. [Google Scholar]
- Doel S.M., McCready,S.J., Nierras,C.R. and Cox,B.S. (1994) The dominant PNM2-mutation which eliminates the ψ factor of Saccharomyces cerevisiae is the result of a missense mutation in the SUP35 gene. Genetics, 137, 659–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes H.K., Gray,V.T. and Wickner,R.B. (1999) The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc. Natl Acad. Sci. USA, 96, 1498–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D., Schiestl,R.H., Willems,A.R. and Woods,R.A. (1995) Studies on the transformation of intact yeast cells by the LiAc/SS–DNA/PEG procedure. Yeast, 11, 355–360. [DOI] [PubMed] [Google Scholar]
- Goldfarb L.G. et al. (1992) Fatal familial insomnia and familial Creutzfeldt–Jakob disease: disease phenotype determined by a DNA polymorphism. Science, 258, 806–808. [DOI] [PubMed] [Google Scholar]
- Goldmann W., Hunter,N., Smith,G., Foster,J. and Hope,J. (1994) PrP genotype and agent effects in scrapie: change in allelic interaction with different isolates of agent in sheep, a natural host of scrapie. J. Gen. Virol., 75, 989–995. [DOI] [PubMed] [Google Scholar]
- Hsiao K., Dlouhy,S.R., Farlow,M.R., Cass,C., Da Costa,M., Conneally,P.M., Hodes,M.E., Ghetti,B. and Prusiner,S.B. (1992) Mutant prion proteins in Gerstmann–Straussler–Scheinker disease with neurofibrillary tangles. Nature Genet., 1, 68–71. [DOI] [PubMed] [Google Scholar]
- Hunter N., Goldmann,W., Smith,G. and Hope,J. (1994) The association of a codon 136 PrP gene variant with the occurrence of natural scrapie. Arch. Virol., 137, 171–177. [DOI] [PubMed] [Google Scholar]
- Komar A.A., Melki,R. and Cullin,C. (1999) The [URE3] yeast prion: from genetics to biochemistry. Biochemistry (Mosc.), 64, 1401–1407. [PubMed] [Google Scholar]
- Labauge P., Pages,M., Blard,J.M., Chatelain,J. and Laplanche,J.L. (1993) Valine homozygous 129 PrP genotype in a French growth-hormone-related Creutzfeldt–Jakob disease patient. Neurology, 43, 447. [DOI] [PubMed] [Google Scholar]
- Lacroute F. (1971) Non-Mendelian mutation allowing ureidosuccinic acid uptake in yeast. J. Bacteriol., 106, 519–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manson J.C. et al. (1999) A single amino acid alteration (101L) introduced into murine PrP dramatically alters incubation time of transmissible spongiform encephalopathy. EMBO J., 18, 6855–6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masison D. and Wickner,R.B. (1995) Prion-inducing domain of yeast ure2p and protease resistance of ure2p in prion-containing cells. Science, 270, 93–95. [DOI] [PubMed] [Google Scholar]
- Masison D.C., Maddelein,M.L. and Wickner,R.B. (1997) The prion model for [URE3] of yeast: spontaneous generation and requirements for propagation. Proc. Natl Acad. Sci. USA, 94, 12503–12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson C., Delalande,I., Deslys,J.P., Henin,D., Fallet-Bianco,C., Dormont,D. and Leys,D. (1994) Creutzfeldt–Jakob disease after pituitary-derived human growth hormone therapy: two cases with valine 129 homozygous genotype. Neurology, 44, 179–180. [DOI] [PubMed] [Google Scholar]
- Paushkin S.V., Kushnirov,V.V., Smirnov,V.N. and Ter-Avanesyan,M.D. (1996) Propagation of the yeast prion-like [ψ+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J., 15, 3127–3134. [PMC free article] [PubMed] [Google Scholar]
- Paushkin S.V., Kushnirov,V.V., Smirnov,V.N. and Ter-Avanesyan,M.D. (1997) In vitro propagation of the prion-like state of yeast Sup35 protein. Science, 277, 381–383. [DOI] [PubMed] [Google Scholar]
- Ter-Avanesyan M.D., Dagkesamanskaya,A.R., Kushnirov,V.V. and Smirnov,V.N. (1994) The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [ψ+] in the yeast Saccharomyces cerevisiae. Genetics, 137, 671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thual C., Komar,A.A., Bousset,L., Fernandez-Bellot,E., Cullin,C. and Melki,R. (1999) Structural characterization of Saccharomyces cerevisiae prion-like protein Ure2. J. Biol. Chem., 274, 13666–13674. [DOI] [PubMed] [Google Scholar]
- Wickner R.B. (1994) [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science, 264, 566–569. [DOI] [PubMed] [Google Scholar]
- Wickner R.B., Masison,D.C. and Edskes,H.K. (1995) [PSI] and [URE3] as yeast prions. Yeast, 11, 1671–1685. [DOI] [PubMed] [Google Scholar]