Abstract
Geminiviruses replicate in nuclei of mature plant cells after inducing the accumulation of host DNA replication machinery. Earlier studies showed that the viral replication factor, AL1, is sufficient for host induction and interacts with the cell cycle regulator, retinoblastoma (pRb). Unlike other DNA virus proteins, AL1 does not contain the pRb binding consensus, LXCXE, and interacts with plant pRb homo logues (pRBR) through a novel amino acid sequence. We mapped the pRBR binding domain of AL1 between amino acids 101 and 180 and identified two mutants that are differentially impacted for AL1–pRBR interactions. Plants infected with the E-N140 mutant, which is wild-type for pRBR binding, developed wild-type symptoms and accumulated viral DNA and AL1 protein in epidermal, mesophyll and vascular cells of mature leaves. Plants inoculated with the KEE146 mutant, which retains 16% pRBR binding activity, only developed chlorosis along the veins, and viral DNA, AL1 protein and the host DNA synthesis factor, proliferating cell nuclear antigen, were localized to vascular tissue. These results established the importance of AL1–pRBR interactions during geminivirus infection of plants.
Keywords: AL1/cell cycle/differentiation/plant DNA virus/pRb
Introduction
Geminiviruses are a large family of plant viruses with circular, single-stranded DNA genomes that replicate through double-stranded DNA intermediates in nuclei of mature cells (reviewed by Hanley-Bowdoin et al., 1999; Gutierrez, 2000). The double-stranded form also serves as the transcription template for the production of a small number of viral proteins, none of which acts as a DNA polymerase during viral replication. Instead, geminiviruses rely on the nuclear DNA replication machinery of their hosts. In mature plants, most cells have exited the cell cycle, undergone differentiation and no longer contain detectable levels of replicative enzymes (Coello et al., 1992; Nagar et al., 1995). To overcome this barrier, geminiviruses reprogram their hosts to create a replication-competent environment (Nagar et al., 1995). Mammalian DNA tumor viruses also depend on host replicative enzymes and can induce their synthesis in quiescent cells by modifying cell cycle and transcriptional controls (Chellappan et al., 1992; Nevins, 1992; Jansen-Durr, 1996). As a consequence, DNA viruses have provided valuable insight into the mechanisms that mediate and control DNA replication, transcription and cell division in animals. Geminiviruses have the same potential for plants and can be used to compare fundamental cellular processes across kingdoms.
Geminiviruses fall into three subgroups—the begomoviruses, curtoviruses and mastreviruses—based on their genome structures and insect vectors. Tomato golden mosaic virus (TGMV) is a typical begomovirus with a bipartite genome. Only one of the seven proteins, AL1 (also designated C1 or Rep), encoded by TGMV is required for viral replication (Elmer et al., 1988). The AL1 protein specifically binds to double-stranded DNA during origin recognition (Fontes et al., 1994a) and acts as an endonuclease and a ligase to initiate and terminate rolling circle replication (Laufs et al., 1995; Orozco and Hanley-Bowdoin, 1996). AL1 also hydrolyzes ATP during an uncharacterized step of viral replication (Desbiez et al., 1995; Orozco et al., 1997) and interacts with itself and the viral replication enhancer, AL3 (Settlage et al., 1996). The functional domains for DNA binding, DNA cleavage/ligation and oligomerization have been mapped to the N-terminal half of the AL1 protein (Orozco et al., 1997; Orozco and Hanley-Bowdoin, 1998).
In mature plants, DNA replication and the corresponding enzymes are confined to meristems, developing leaves and roots, and the cambium (Martinez et al., 1992; Staiger and Doonan, 1993). Some geminiviruses are restricted to the phloem (Esau, 1977; Horns and Jeske, 1991; Sanderfoot and Lazarowitz, 1996) and may replicate in procambial cells using pre-existing plant machinery. Other geminiviruses like TGMV are not confined to vascular tissue and, instead, are found in terminally differentiated cells throughout the leaf, stem and root (Nagar et al., 1995; Lucy et al., 1996). Because mature plant cells are not competent for DNA replication, a likely early step in geminivirus infection is virus-induced synthesis of host DNA replication enzymes (Hanley-Bowdoin et al., 1999). This idea is strongly supported by the accumulation of proliferating cell nuclear antigen (PCNA), the processivity factor of DNA polymerase δ, in differentiated cells of TGMV-infected plants but not in equivalent healthy cells (Nagar et al., 1995). PCNA is also expressed in differentiated cells of transgenic plants expressing TGMV AL1, establishing that AL1 is sufficient for host induction.
Several lines of evidence suggest that TGMV infection and the AL1 protein induce the accumulation of host replication machinery using plant cell cycle controls. First, the PCNA promoter is active in mature leaves of TGMV-infected plants but only functions in young tissue of healthy plants (E.Egelkrout, D.Robertson and L.Hanley-Bowdoin, unpublished data). In uninfected cells, PCNA promoter activity is highest during late G1 of the cell cycle (Sekine et al., 1999). Secondly, infected plant cells incorporate high levels of bromodeoxyuridine into TGMV and host DNA, indicative of progression into S phase and DNA replication (S.Nagar, L.Hanley-Bowdoin and D.Robertson, in preparation). Thirdly, a high proportion of TGMV-infected cells contain condensed chromatin characteristic of early mitotic prophase (Bass et al., 2000). Lastly, TGMV AL1 (Ach et al., 1997a) and the related mastrevirus RepA proteins (Grafi et al., 1996; Xie et al., 1996; Horvath et al., 1998; Liu et al., 1999) bind to plant homologues of the cell cycle regulator, retinoblastoma (pRb). By analogy with mammalian DNA viruses (Weinberg, 1995), these interactions may bypass a pRb phosphorylation requirement for cell cycle entry and G1 progression during geminivirus infection.
In animals, pRb is part of a small gene family that also includes p107 and p130 (Herwig and Strauss, 1997). Plant pRb homologues (pRBR, retinoblastoma-related) have been cloned from maize (Grafi et al., 1996; Xie et al., 1996; Ach et al., 1997a), tobacco (Nakagami et al., 1999), Chenopodium (Fountain et al., 1999) and Arabidopsis (this paper; reviewed by Durfee et al., 2000). There is considerable evidence in animal systems that pRb family members negatively regulate cell cycle progression and facilitate differentiation (reviewed by Sidle et al., 1996). Experiments showing that a maize pRb homologue is preferentially expressed in mature leaf tissue (Huntley et al., 1998) suggest that pRBR may serve similar functions in plants. The pRb family members of plants and animals display strong sequence homology across a large central domain known as the A/B pocket (Murray, 1997). This region is involved in a variety of protein interactions (Taya, 1997), including interactions with SV40 large T-antigen, adenovirus E1A and human papillomavirus E7 (Lee et al., 1998), all of which bind pRb through a conserved LXCXE motif (Dyson et al., 1992). Plant cyclin D (Soni et al., 1995; Ach et al., 1997a; Huntley et al., 1998; Nakagami et al., 1999) and mastrevirus RepA (Xie et al., 1995; Collin et al., 1996; Grafi et al., 1996; Horvath et al., 1998; Liu et al., 1999) also interact with pRBR through LXCXE sequences. In contrast, TGMV AL1 and the Rep proteins of other members of the begomovirus and curtovirus subgroups, which include nearly all dicot-infecting geminiviruses, do not have LXCXE motifs, and it is not clear how they interact with pRBR. To address this question and the biological significance of pRBR interaction during geminivirus infection, we mapped the pRBR-binding domain of TGMV AL1 and tested the impact of site-directed mutations in this region on pRBR binding and TGMV infection.
Results
TGMV AL1 and SV40 large T-antigen interact with a plant pRb homologue pRBR differently
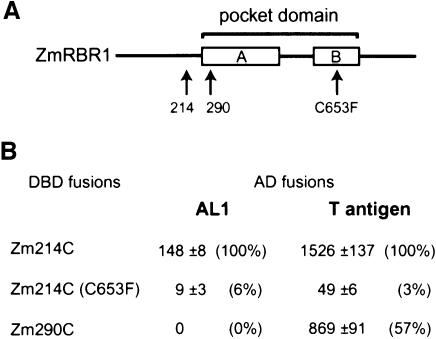
To understand how AL1 interacts with pRBR, we used yeast two-hybrid assays to compare the pRBR binding requirements of TGMV AL1 and SV40 large T-antigen. For these experiments, amino acids 214–866 (Zm214C) or 290–866 (Zm290C) of the maize pRb homologue, RRB1 (Ach et al., 1997a; by convention renamed ZmRBR1), were fused downstream of the Gal4 DNA binding domain (DBD; Figure 1). The Zm214C construct contains an intact A/B pocket (ZmRBR1 amino acids 273–722) while the Zm290C construct lacks the first 17 amino acids of the A box (Ach et al., 1997a). The RBR1 clones were cotransformed into yeast with plasmids corresponding to the Gal4 activation domain (AD) fused to either full-length AL1 or to large T-antigen amino acids 87–708, which include the LXCXE motif. When the Zm214C construct was used, β-galactosidase activity indicative of protein interaction was detected for both AL1 and large T-antigen (Figure 1). The relative activity was 10-fold less for AL1 than for large T-antigen. Both viral proteins were severely impaired in their interactions with a Zm214C variant carrying a C653F mutation (Ach et al., 1997a), which destabilizes pRb conformation to block protein interactions generally through the pocket domain (Lee et al., 1998). When the Zm290C construct was tested, interaction between large T-antigen and ZmRBR1 was detected, albeit at about half of that observed with Zm214C. In contrast, AL1 failed to interact with Zm290C. The use of a two-hybrid construct corresponding to full-length ZmRBR1 resulted in reduced activity relative to Zm214C for both large T-antigen and AL1 (data not shown), indicating that additional N-terminal pRBR sequences do not enhance interactions with the viral proteins. Together, these results showed that AL1 interacts with pRBR differently than large T-antigen and requires a longer pocket domain for binding.
Fig. 1. AL1 and large T-antigen interact with ZmRBR1 differently. (A) Diagram of the maize pRb homologue ZmRBR1 showing the pocket domain with the A and B boxes. Arrows mark the N-terminal truncations at positions 214 and 290 and the C653F mutation. (B) Mean β-galactosidase specific activities (1 unit = 1.0 mmol product/min/mg protein at pH 7.3 at 37°C) from two-hybrid assays containing the indicated GAL4 DBD–ZmRBR1 fusions and GAL4 AD fusions for full-length TGMV AL1 or SV40 large T-antigen (amino acids 87–708) are given. Two standard errors are given for each value. Relative activities are in parentheses.
The pRBR binding domain of TGMV AL1
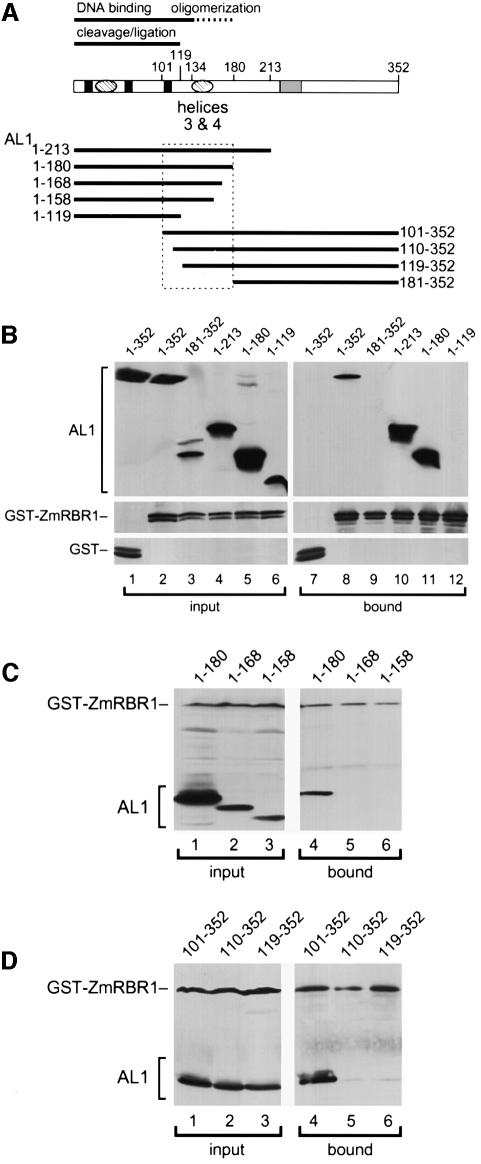
pRb, p107 and p130 interact with a variety of cellular proteins, some of which lack LXCXE motifs and, instead, bind through α-helical regions (Taya, 1997). Secondary structure prediction analysis identified two putative sets of α-helices in the N-terminal half of TGMV AL1 (Figure 2A; Orozco et al., 1997) that may be involved in pRBR binding. To test if the AL1 N-terminus mediates the interaction, we examined the pRBR binding properties of a series of truncated AL1 proteins (Figure 2A). The various AL1 proteins were co-expressed with a GST–ZmRBR1 fusion in insect cells and fractionated by glutathione affinity chromatography. As shown previously (Ach et al., 1997a), full-length AL11–352 was in the bound fraction in the presence of GST–ZmRBR1 (Figure 2B, lanes 2 and 8) but not GST alone (lanes 1 and 7). No other proteins were detected in the bound fractions by silver staining (our unpublished results), indicating that it is unlikely that AL1–pRBR interaction is bridged by a third protein in insect cells. The truncated proteins, AL11–213 (Figure 2B, lanes 4 and 10) and AL11–180 (lanes 5 and 11), also co-purified with GST–ZmRBR1 whereas AL1181–352 (lanes 3 and 9) and AL11–119 (lanes 6 and 12) did not. These results localized the pRBR binding domain to the N-terminal half of TGMV AL1. The inability of AL11–119 to bind GST–ZmRBR1 supported a potential role for helices 3 and 4 in the interaction.
Fig. 2. Mapping the pRBR binding domain of TGMV AL1. (A) Diagram of the AL1 protein showing the positions of the three DNA cleavage motifs (solid boxes), two predicted pairs of α-helices (hatched ovals) and the ATP binding site (hatched box). The DNA binding and cleavage/ligation domains are indicated by solid lines and the oligomerization domain is shown as a dashed line. Solid lines below the diagram mark the sizes of truncated AL1 proteins, which are designated by their N- and C-terminal amino acids. The boxed region indicates the limits of the pRBR binding domain. (B) Total protein extracts from insect cells co-expressing GST or GST–ZmRBR1 with different AL1 proteins (top) were incubated with glutathione–Sepharose, washed and eluted. Input (lanes 1–6) and bound (lanes 7–12) proteins were resolved by SDS–PAGE and analyzed by immunoblotting. The top panels were visualized using an anti-AL1 antibody while the bottom panels were visualized using an anti-GST antibody. The extracts in lanes 1 and 7 contained GST and full-length AL11–352. The extracts in lanes 2–6 and 8–12 contained GST–ZmRBR1 and full-length AL11–352 (lanes 2 and 8), AL1181–352 (lanes 3 and 9), AL11–213 (lanes 4 and 10), AL11–180 (lanes 5 and 11) or AL11–119 (lanes 6 and 12). (C) Input (lanes 1–3) and bound (lanes 4–6) fractions are shown for interactions between GST–ZmRBR1 and the C-terminal truncations (top) corresponding to AL11–180 (lanes 1 and 4), AL11–168 (lanes 2 and 5) and AL11–158 (lanes 3 and 6). (D) Input (lanes 1–3) and bound (lanes 4–6) fractions are shown for interactions between GST–ZmRBR1 and the N-terminal truncations (top) corresponding to AL1101–352 (lanes 1 and 4), AL1110–352 (lanes 2 and 5) and AL1119–352 (lanes 3 and 6). In (C) and (D), the blots were probed with both anti-AL1 and anti-GST antibodies.
To define better the limits of the pRBR binding domain, we analyzed a set of AL1 proteins with closely spaced truncations. In Figure 2C, the C-terminally truncated proteins, AL11–168 (lanes 2 and 5) and AL11–158 (lanes 3 and 6), did not co-purify with GST–ZmRBR1 while AL11–180 (lanes 1 and 4) was in the bound fraction. In Figure 2D, the N-terminally truncated proteins, AL1110–352 (lanes 2 and 5) and AL1119–352 (lanes 3 and 6), also failed to bind GST–ZmRBR1 whereas AL1101–352 (lanes 1 and 4) co-purified with the ZmRBR1 fusion. Together, these data mapped the N-terminal border of the pRBR binding domain between TGMV AL1 amino acids 101 and 110 and the C-terminal border between amino acids 168 and 180. Although this 80-amino-acid region includes helices 3 and 4, the inability of AL11–168, AL1110–352 and AL1120–352 to interact with GST–ZmRBR1 indicated that these helices are not sufficient for pRBR binding and that flanking sequences also contribute to the interaction. Given that the pRBR binding domain fully encompasses the AL1 oligomerization domain (Figure 2A; Orozco et al., 2000), one possibility is that formation of AL1 oligomers is a requirement for interaction with pRBR.
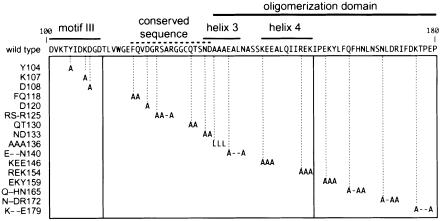
The sequence requirements for AL1–pRBR interaction were characterized using a series of site-directed mutations in the pRBR binding domain of TGMV AL1 (Figure 3). Mutations Y104, K107 and D108 targeted residues in motif III, the active site for DNA cleavage and ligation (Laufs et al., 1995). Mutations FQ118, D120, RS-R125, QT130 and ND133 modified a conserved sequence that is part of the AL1 DNA binding domain (Orozco and Hanley-Bowdoin, 1998). Mutations AAA136, E--N140, KEE146 and REK154 were in helices 3 and 4, which are part of the AL1 oligomerization domain (Orozco et al., 2000). Mutations EKY159, Q-HN165, N-DR and K--E179 were in a region that contributes the primary oligomerization contacts. All of the mutations except AAA136 were alanine substitutions and predicted to be structurally neutral. For AAA136, alanine residues were replaced by leucines that may have affected the overall conformation of the protein. The mutant AL1 proteins were fused to the Gal4 AD and expressed in yeast. Immunoblot analysis verified that the mutant proteins accumulated to levels similar to an AD–wild-type AL1 fusion (Orozco et al., 2000).
Fig. 3. Site-directed mutations in the pRBR binding domain of TGMV AL1. The AL1 sequence between amino acids 100 and 180 is shown, and the locations of motif III, a conserved sequence and the oligomerization domain are indicated by the solid and dotted lines above the sequence. The predicted α-helices 3 and 4 are also marked. The boxed region indicates the positions of the site-directed substitutions. Mutations (on the left) are designated by the wild-type sequence and the position of the last altered amino acid. Dashes indicate amino acids that were not changed.
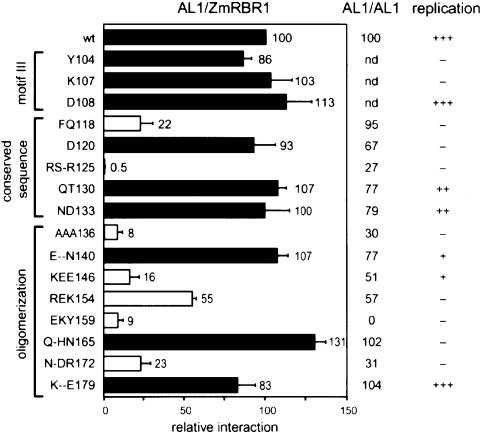
The effects of the mutations on AL1–pRBR interactions were analyzed in yeast two-hybrid assays using the DBD–Zm214C fusion (Figure 4). The Y104, K107 and D108 mutations in motif III had no significant impact on AL1–ZmRBR1 interactions. Mutations in the conserved region showed differential effects. The D120, QT130 and ND133 mutants displayed wild-type levels of ZmRBR1 interaction while FQ118 and RS-R125 were reduced. The FQ118 mutation had no effect on AL1–AL1 interactions in two-hybrid assays (Figure 4; Orozco et al., 2000), establishing the specificity of the mutation for pRBR binding. In contrast, the RS-R125 mutation reduced AL1 oligomerization and interfered with its DNA binding and cleavage activities (data not shown), indicating that it is pleiotropic in character. Mutations in the AL1 oligomerization domain also differentially impacted AL1–ZmRBR1 interactions (Figure 4). The pRBR binding activities of E--N140 and K--E179 were similar to wild-type AL1 while Q-HN165 was slightly enhanced for pRBR binding. The AAA136, KEE146, REK154, EKY159 and N-DR172 mutants were all impaired for pRBR binding. Comparison of the combined effects of the mutations on pRBR binding and AL1 oligomerization revealed that these mutants were of two classes. The first class, which includes REK154, EKY159 and N-DR172, were similarly impaired for pRBR interactions and AL1 oligomerization. The correlation between these interactions was not attributable to reduced AD–mutant AL1 protein production (Orozco et al., 2000) and, instead, is consistent with AL1 oligomerization being a prerequisite for pRBR binding. The second class, represented by AAA136 and KEE146, was more severely impacted for pRBR interactions than for AL1 oligomerization. For these mutants, the reduced pRBR binding activity was most likely to be due to a composite effect of altered AL1 oligomerization and impaired AL1–pRBR contacts. The locations of the AAA136 and KEE146 mutations in helices 3 and 4, respectively, further supported the idea that AL1 may bind pRBR through an α-helical region.
Fig. 4. Mutations in the pRBR binding domain of AL1 impair interactions with ZmRBR1. A ZmRBR1 expression cassette corresponding to Zm214C fused to the GAL4 DBD was co-transformed into yeast with cassettes for either wild-type or mutant AL1 fused to the GAL4 AD (on the left). Interactions between ZmRBR1 and the AL1 proteins were assayed by measuring β-galactosidase activity in total protein extracts and normalized to wild type (100). The open bars indicate mutants impaired for ZmRBR1 binding, whereas the filled bars mark mutants with activity similar to or greater than wild-type AL1. The error bars correspond to two standard errors. The locations of motif III, the conserved sequence and the oligomerization domain are shown on the left. The effects of the mutations on AL1 oligomerization activity and TGMV replication in transient assays are indicated on the right (Orozco et al., 2000).
TGMV AL1 binds a pRb homologue from a dicot species
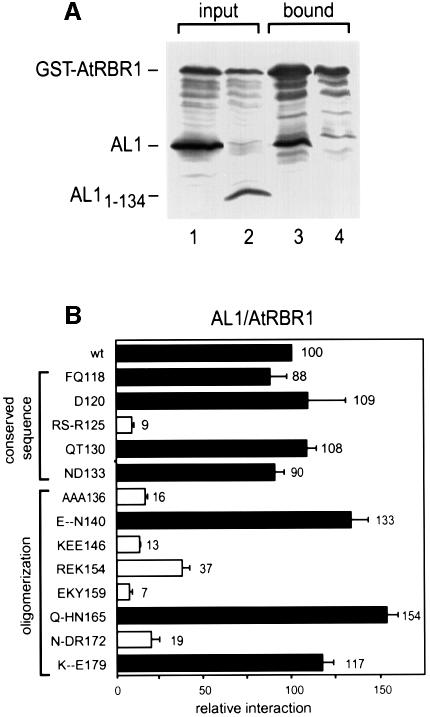
Because TGMV and other members of the begomovirus subgroup only infect dicot species and the RepA proteins of monocot-infecting geminiviruses bind to maize ZmRBR1 through LXCXE motifs (Grafi et al., 1996; Xie et al., 1996), it was important to examine the interactions between AL1 and a dicot pRBR protein. For these experiments, coding sequences for a full-length Arabidopsis pRb homologue (AtRBR1; DDBJ/EMBL/GenBank accession No. AF245395) and GST were fused and expressed in insect cells with either full-length TGMV AL11–352 (Figure 5A, lane 1) or truncated AL11–134 (lane 2). After glutathione affinity chromatography, AL11–352 (Figure 5A, lane 3) but not AL11–134 (lane 4) was detected in the bound fraction. Thus, TGMV AL1 interacts with pRBR proteins of dicot as well as monocot origins. The failure of GST–AtRBR1 to interact with AL11–134 is consistent with the deletion of helices 3 and 4 as well as the AL1 oligomerization domain, both of which are required for ZmRBR1 binding.
Fig. 5. AL1 interacts with AtRBR1. (A) Total protein extracts from insect cells co-expressing GST–AtRBR1 with either full-length AL11–352 (lanes 1 and 3) or AL11–134 (lanes 2 and 4) were incubated with glutathione–Sepharose, washed and eluted. Input (lanes 1 and 2) and bound (lanes 3 and 4) proteins were resolved by SDS–PAGE and analyzed by immunoblotting with anti-AL1 and anti-GST antibodies. (B) An AtRBR1 expression cassette corresponding to At319C fused to the GAL4 DBD was cotransformed into yeast with cassettes for either wild-type or mutant AL1 fused to the GAL4 AD (on the left) and analyzed as described in Figure 4. The locations of the conserved sequence and the oligomerization domain are indicated on the left.
We also examined the impact of various AL1 site-directed mutations (Figure 3) on AtRBR1 binding by co-expressing a DBD fusion with AtRBR1 amino acids 319–958 (At319C) with an intact pocket domain and AD–AL1 fusions in yeast. As observed with ZmRBR1, mutations in the conserved sequence and the oligomerization domain of AL1 differentially impacted AtRBR1 binding. The At319C and Zm214C DBD fusions displayed very similar binding profiles with the various AL1 mutants (cf. Figures 4 and 5B), indicating that AL1 contacts the Arabidopsis and maize pRBR proteins similarly. The only exception was the FQ118 mutant, which showed 88 and 22% of wild-type activity with the At319C and Zm214C constructs, respectively. This difference may reflect the 68% divergence between the A/B pockets and C-terminal domains of Arabidopsis and maize pRBR.
Impaired AL1–pRBR binding alters symptoms and tissue specificity of TGMV infection
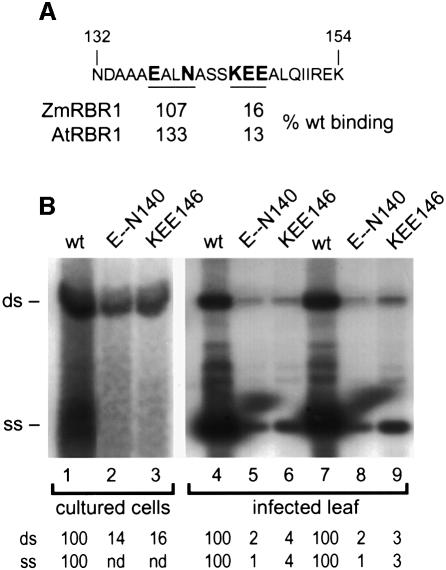
In animal systems, interaction between pRb and viral proteins induces quiescent cells to re-enter the cell cycle and support viral DNA replication (Nevins, 1992). Because TGMV infects differentiated plant cells (Nagar et al., 1995), we examined the impact of altering AL1–pRBR interactions on geminivirus infection. For these experiments, it was essential to use an AL1 mutant that was impaired for pRBR interactions but still supported viral DNA replication. Transient replication assays using plant expression cassettes encoding mutant AL1 proteins showed that D108, QT130, ND133, E--N140, KEE146 and K--E179 are able to support TGMV DNA synthesis (Figure 3; Orozco et al., 2000). Of these mutants, only KEE146 is impaired for pRBR interaction in two-hybrid assays (Figures 4 and 5B). We controlled for the reduced replication phenotype of KEE146 by using E--N140, which is also attenuated for viral DNA accumulation but wild type for pRBR binding (Figures 4 and 5B). The mutations were transferred into the AL1 open reading frame (ORF) of a TGMV A replicon, and replication of the mutant A components was assessed in tobacco protoplasts. Similar levels of double-stranded DNA synthesis were detected for both mutants with the KEE146 (Figure 6B, lane 3) and E--N140 (lane 2) A components accumulating to 14 and 16% of wild-type TGMV A (lane 1), respectively. No single-stranded viral DNA accumulation was detected for either mutant. These results showed that the KEE146 and E--N140 mutations impact TGMV A replication similarly in cycling cells.
Fig. 6. Two TGMV A mutants altered in the AL1 pRBR binding domain replicate to similar levels. (A) The sequence between AL1 amino acids 132 and 154 is shown, with the mutated amino acids in E--N140 and KEE146 indicated by bold face type. The relative binding activities with ZmRBR1 and AtRBR1 are given for the two mutants. (B) In lanes 1–3, tobacco protoplasts were transfected with TGMV A replicons with either wild-type (lane 1) or mutant AL1 ORFs corresponding to E--N140 (lane 2) or KEE146 (lane 3). Total DNA was isolated from 7 × 106 cells 72 h post-transfection and analyzed on DNA gel blots. In lanes 4–9, N.benthamiana plants were bombarded with DNAs corresponding to TGMV A and B replicons. The AL1 ORFs of the A components were either wild type (lanes 4 and 7) or carried the E--N140 (lanes 5 and 8) or KEE146 mutation (lanes 6 and 9). Total DNA (2.5 µg/lane) was isolated from systemically infected leaves from two plants for each construct at 18 days post-infection and analyzed on DNA gel blots. TGMV DNA was detected using a radiolabeled probe specific for the A component. The positions of double- (ds) and single- (ss) stranded forms of TGMV A are marked on the left. The relative accumulations of the DNA forms are given at the bottom of each lane with wild type set at 100. In lanes 2 and 3, single-stranded DNA was not detected (nd).
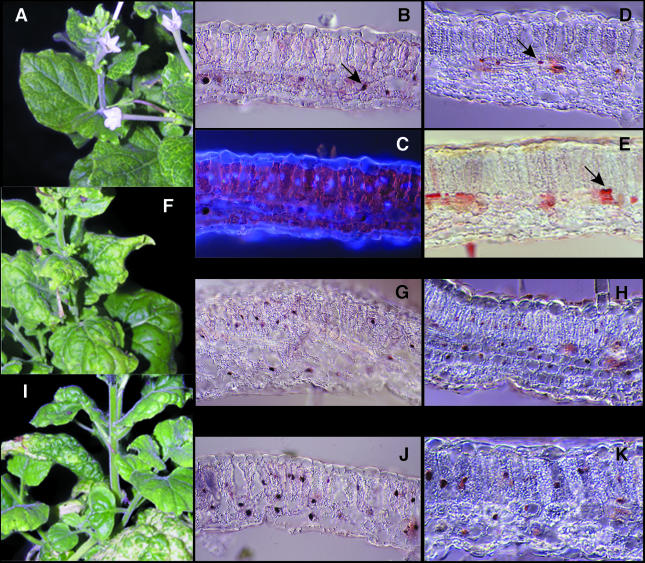
Wild-type or mutant A component DNA was co-bombarded with TGMV B DNA onto Nicotiana benthamiana plants. Plants inoculated with wild-type virus developed symptoms by 1 week post-inoculation, showing typical chlorosis along veins, leaf curling and stunting of new growth (Figure 7I). The E--N140 mutant, which was delayed ∼2 days relative to wild-type virus, developed essentially wild-type symptoms (Figure 7F). In contrast, the symptoms produced by the KEE146 mutant, which appeared ∼1 week late, were very mild (Figure 7A). KEE146-inoculated plants only developed chlorosis along the veins and displayed no leaf curling or stunting. The mild symptoms were seen in 10 of 10 plants and maintained over a 4-week infection period. To verify that the different symptoms caused by the mutant viruses were not due to differential replication in plants, total DNA was isolated from systemically infected leaves 18 days post-inoculation and analyzed on DNA gel blots. Double- and single-stranded forms of TGMV A DNA were detected for each virus (Figure 6B, lanes 4–9). However, the levels of both DNA forms were severely reduced in plants inoculated with the mutant viruses, both of which accumulated to <5% of wild-type levels. These results demonstrated that low levels of viral DNA replication are sufficient to cause wild-type symptoms in the case of the E--N140 mutant. Thus, the attenuated symptoms seen with the KEE146 mutant were not due to reduced replication.
Fig. 7. A pRBR binding mutant displays altered symptoms and accumulation patterns in plants. Nicotiana benthamiana plants infected with the pRBR binding mutant KEE146 (A–E), the control mutant E-N140 (F, G and H) or wild-type TGMV (I, J and K) were analyzed at 18 days post-infection. (A), (F) and (I) show symptomatic plants. In (B), (G) and (J), viral DNA was detected in mature leaves by in situ hybridization with a TGMV B probe labeled with digoxigenin and visualized using an anti-digoxigenin antibody conjugated to alkaline phosphatase. In (D), (H) and (K), TGMV AL1 protein was detected using an anti-AL1 antibody and peroxidase detection. In (E), the host DNA synthesis factor, PCNA, was detected using an anti-human PCNA antibody and peroxidase detection. Total DNA was visualized in (C) by DAPI staining, with the same section depicted in (B) and (C). Arrows in (B), (D) and (E) show nuclei infected with the KEE146 mutant. All sections were analyzed at 400× magnification.
To understand better the basis of the different symptoms, we compared the patterns of viral DNA and AL1 protein accumulation in plants infected with wild-type and mutant TGMV (Figure 7). As reported earlier (Nagar et al., 1995; Bass et al., 2000), wild-type TGMV DNA (Figure 7J) and AL1 protein (Figure 7K) were found in nuclei of epidermal, mesophyll and vascular cells. Even though the intensities of the signals were less than for wild-type virus, the E--N140 mutant displayed the same distribution patterns (Figure 7G and H). In contrast, viral DNA (Figure 7B) and AL1 protein (Figure 7D) corresponding to the KEE146 mutant were found almost exclusively in vascular cells and at very low frequency (<1%) in other cell types. DAPI staining of total DNA verified that nuclei could be readily detected in the mesophyll and epidermal cells of KEE146-infected leaves (cf. Figure 7B and C). The same patterns of viral DNA and AL1 accumulation were observed between 14 and 24 days post-inoculation (data not shown), indicating that the KEE146 mutant remained in vascular tissue throughout infection.
TGMV infection induces the accumulation of the host DNA synthesis factor, PCNA, in mature plant cells (Nagar et al., 1995). Immunohistochemical analysis of leaves systemically infected with the KEE146 mutant only detected PCNA protein in vascular-associated nuclei (Figure 7E). This contrasts with PCNA patterns in wild-type TGMV-infected plants, which were shown previously to accumulate the host factor in mesophyll and epidermal as well as vascular cells (Nagar et al., 1995). Together, these data established that there is a strong correlation between the ability of TGMV AL1 to bind pRBR efficiently, to induce PCNA accumulation and to support infection throughout the leaf.
Discussion
DNA viruses with small genomes do not have sufficient coding capacity to specify the DNA polymerases and accessory factors required for their replication and, instead, recruit the replication machinery of their hosts. As a consequence, these viruses can only amplify in cells that contain the requisite replication enzymes, which are typically confined to actively cycling cells. To overcome this limitation, mammalian DNA tumor viruses encode proteins that interact with components of the host transcriptional apparatus and cell cycle regulatory network (Nevins, 1992). They target basal transcription factors (Gruda et al., 1993), the histone transacetylase p300 (Eckner et al., 1994), and the tumor suppressors, pRb (Dyson et al., 1992) and p53 (Werness et al., 1990). These protein interactions cause quiescent animal cells to re-enter the cell division cycle and synthesize the enzymes necessary for viral DNA replication. The discovery that TGMV AL1 also induces accumulation of host DNA replication machinery in mature cells (Nagar et al., 1995) and binds the plant pRb homologue pRBR (Ach et al., 1997a) suggested that geminiviruses use similar mechanisms to reprogram their plant hosts. In this paper, we showed that TGMV AL1–pRBR binding strongly influences the symptoms and tissue specificity of geminivirus infection, establishing the importance of this host interaction for plant DNA viruses.
Co-purification experiments using truncated AL1 proteins and a GST–ZmRBR1 fusion (Figure 2) and yeast two-hybrid assays (data not shown) established that the pRBR binding domain of TGMV AL1 is between amino acids 101 and 180. This region overlaps the domains responsible for DNA cleavage/ligation, DNA binding and AL1 oligomerization (Figure 2A), all of which are required for AL1 replication activity. The ability of AL1101–352 but not AL1110–352 to bind ZmRBR1 in insect cells established that motif III, which corresponds to the active site for DNA cleavage (Orozco and Hanley-Bowdoin, 1998), is also involved in AL1–pRBR interaction. Even though site-directed mutations in motif III had no detectable effects on pRBR binding, the failure of a truncated AL1 protein lacking motif III to interact with ZmRBR in yeast indicated that this region contributes specific contacts that cannot be provided by fusion to the Gal4 DBD. For most DNA viruses, including the adenoviruses and papillomaviruses (Dyson et al., 1992) of animals and the mastreviruses and nanoviruses of plants (Horvath et al., 1998; Liu et al., 1999; Aronson et al., 2000), initiation of replication and pRb binding are mediated by different proteins. In polyomaviruses, large T-antigen acts in both processes but the regions responsible for these activities are distinct and separable in vivo (Gjorup et al., 1994). Thus, the arrangement of TGMV AL1 is unique and raises important questions regarding how host induction and replication are coordinated. One possible scenario is that when AL1 is bound to pRBR, it is not functional for TGMV replication. The plant pRBR protein is >100 kDa in mass and its binding is likely to impact the accessibility of motif III. In this case, AL1–pRBR binding would ensure that initiation of rolling circle replication does not occur until the required host enzymes are available. Alternatively, pRBR may be part of the TGMV replication initiation complex but this idea is counter to the well-documented negative effect of pRb on DNA replication in other systems (Xie et al., 1996; Sterner et al., 1998; Reynisdottir et al., 1999).
Experiments in both insect cells and yeast indicated that there is a link between the ability of AL1 to bind pRBR and to form oligomers. However, our results do not provide any information regarding the stoichiometry of the AL1 oligomers that bind pRBR. This is an important question because recombinant AL1 occurs as an octamer in solution (Orozco et al., 1997) and binds DNA minimally as a dimer (Orozco and Hanley-Bowdoin, 1998). In addition, because pRBR is likely to be a monomeric protein, it is not clear how AL1 oligomerization contributes to the interaction. It is possible that stable complex formation requires the concerted interaction of more than one AL1 subunit with pRBR. This model predicts that different AL1 subunits contact pRBR through different amino acids, and that the pRBR binding domain represents a composite of the various contacts. Alternatively, oligomerization may induce a conformational change in AL1 that is required for pRBR binding. Previous experiments showed that different AL1 complexes are likely to mediate TGMV replication and transcriptional repression in vivo (Orozco et al., 2000). The dependency of pRBR binding on oligomerization further supports the idea that AL1 functional status is modulated by AL1–AL1 interactions.
Low stringency searches of both plant and animal databases failed to detect any proteins except other geminivirus Rep proteins with significant homology to the pRBR binding domain of TGMV AL1. In addition, direct sequence comparison with known pRBR binding proteins, including plant E2F (Ramirez-Parra et al., 1999; Sekine et al., 1999) and Msi1 proteins (Ach et al., 1997b), which do not contain LXCXE motifs, did not uncover any related amino acid sequences. Thus, it is likely that dicot-infecting geminiviruses interact with pRBR through a novel or previously uncharacterized motif. However, like many pRb binding proteins (Taya, 1997), TGMV AL1–pRBR interactions are mediated at least in part by two predicted α-helices between amino acids 132 and 154 (Orozco et al., 1997). The importance of the helices, which are designated as 3 and 4, is consistent with their strong conservation with respect to position, length and spacing across all geminivirus AL1 homologues (Orozco et al., 2000). Our data suggested that the two helices make different contributions to pRBR binding. With the exception of AAA136, helix 3 mutants displayed wild-type pRBR binding activity. The bulkier leucine side chains in AAA136 are not predicted to disrupt helical structure but may interfere with tertiary folding of the pRBR binding domain and/or the ability of AL1 to contact pRBR. In contrast, mutations in helix 4 generally reduced AL1–pRBR interactions, suggesting that this region is directly involved in pRBR binding. The stronger sequence conservation of helix 4 versus helix 3 is also consistent with different roles for the two helices in pRBR binding (Orozco et al., 2000).
TGMV AL1 and pRBR interact in insect cells and yeast, but there is no direct proof for their interaction in plant cells. Immunoblot and immunolocalization experiments showed that both proteins are most abundant in mature tissues and localize to plant nuclei (Nagar et al., 1995; Ach et al., 1997a; Huntley et al., 1998), thereby providing opportunity for interaction. However, we have been unable to detect AL1–pRBR complexes in extracts from infected leaves or cultured cells transfected with expression cassettes for the two proteins. This was not unexpected given that both proteins are recalcitrant to extraction under native conditions (our unpublished results). In contrast, genetic experiments provided strong data for the functional significance of AL1–pRBR interactions. Two TGMV AL1 mutants that only differed with respect to their pRBR binding properties resulted in different symptoms and tissue distributions in infected plants. The E--N140 mutant, which was wild type for pRBR binding, accumulated throughout the leaf and showed wild-type symptoms, whereas the KEE146 mutant, which was impaired for pRBR interaction, localized to vascular tissue and only produced chlorosis along the veins. Our results differed from those reported by Liu et al. (1999), who did not detect an effect of a pRBR binding mutation on bean yellow dwarf virus (BYDV) infection. However, TGMV and BYDV belong to different geminivirus subgroups and interact with their hosts through distinct mechanisms. This difference is underscored by the observation that BYDV RepA but not Rep, the functional homologue of TGMV AL1, interacts with pRBR. In addition, RepA binds pRBR through an LXCXE motif and mutations in this motif retained at least 5% of wild-type activity. We showed that SV40 large T-antigen, which also has an LXCXE motif, binds pRBR with 10-fold higher activity than TGMV AL1. If BYDV RepA and SV40 large T-antigen bind pRBR with similar strengths, the RepA mutant may have retained sufficient activity to support wild-type infection. Lastly, the tissue specificity of BYDV infection is not known. If BYDV is a phloem-associated geminivirus, a reduction in pRBR binding activity may not visibly alter symptoms.
Our data do not formally exclude the possibility that the TGMV KEE146 mutant is impaired for virus movement but this is unlikely for several reasons. Begomovirus A components depend on their cognate B DNAs for movement functions (Sanderfoot and Lazarowitz, 1996). The nuclear shuttling and cell-to-cell trafficking activities of the BR1 and BL1 proteins encoded by B DNA account for all of the functions required for geminivirus movement. Microinjection experiments indicated that BR1 and BL1 are sufficient for nucleic acid movement between plant cells (Noueiry et al., 1994). There is also no evidence indicating that the AL1 homologue from any geminivirus has a role in movement. Finally, TGMV carrying the E-N140 mutation, which is within 10 amino acids of the KEE146 mutation, is able to move out of vascular cells. This is striking given the low E-N140 accumulation, establishing that movement and wild-type symptoms do not require high levels of TGMV DNA.
Viral DNA and AL1 protein corresponding to the KEE146 mutant accumulate in vascular but not in mesophyll or epidermal cells. We also detected PCNA in infected cells, indicating that the KEE146 mutant can induce the production of host DNA replication machinery in vascular tissue. This tissue specificity may reflect a different mechanism or a lower threshold for induction in vascular versus mesophyll and epidermal cells. Recent results indicating that a TGMV AL1 mutant with no pRBR binding activity cannot support infection (our unpublished data) suggested that pRBR binding is a general feature of host activation in all tissue types, including the phloem. The KEE146 mutant retained ∼16% of wild-type pRBR binding activity, which may be sufficient to induce vascular cells but not other cell types to re-enter the cell division cycle. One possibility is that pRBR is present at a lower concentration in vascular tissue and, thus, more readily inactivated. Alternatively, vascular cells may express a distinct pocket protein that has a higher affinity for the KEE146 mutant. There is also evidence that vascular cells are predisposed to return to the cell division cycle (Martinez et al., 1992; Hemerly et al., 1993; Umeda et al., 1999) and, as such, may be more readily activated than surrounding cell types.
DNA blot analysis indicated that the KEE146 and E--N140 mutants accumulate to similar levels in infected plants but two observations suggest that the amount of KEE146 DNA on a per cell basis is significantly higher. First, the E--N140 mutant was distributed across many more cells than the KEE146 mutant. Secondly, in situ hybridization signals for KEE146 and wild-type TGMV DNA were of similar intensity while the E--N140 signal was considerably weaker. The quantitative differences cannot be attributed to different replication efficiencies because the two mutants accumulated to similar levels in protoplasts. In animals, pRb–E2F complexes negatively regulate expression of proteins required for progression through as well as entry into S phase (Ohtani et al., 1995; Schulze et al., 1995). Hence, once a KEE146-infected cell is induced to re-enter the cell cycle, it may be less efficient at exiting S phase and allow higher levels of viral DNA to accumulate. Future experiments that assess the molecular consequences of AL1–pRBR interactions will address this hypothesis and provide valuable insight into pRBR function during plant cell division and differentiation.
Materials and methods
Baculoviruses
Baculoviruses corresponding to GST (pNSB314) and GST–ZmRRB1 (renamed ZmRBR1) were described earlier (Orozco and Hanley-Bowdoin, 1996; Ach et al., 1997a). Arabidopsis thaliana cDNAs encoding a pRb-like protein were isolated by low stringency hybridization using the maize RRB1 cDNA (Ach et al., 1997a). The longest clone, p4a1-1, was isolated from a 3-day-old hypocotyl cDNA library, sequenced and found to contain a single ORF with the capacity to encode a full-length protein of 1013 amino acids (DDBJ/EMBL/GenBank accession No. AF245395). The AtRBR1 locus encoding this ORF maps to chromosome 3 at 18.4 cM, ∼100 kb distal to the FAD2 locus (between FAD2 and mi207). A baculovirus construct for expression of AtRBR1 was generated by PCR amplification of the p4a1-1 ORF. The PCR product was cloned into pBluescript SK(–) using EcoRV and NotI ends to give AtRbSO1. A fragment with NotI and repaired XhoI ends was then cloned into pNSB314 (Orozco and Hanley-Bowdoin, 1996) with NotI and repaired EcoRI ends to construct a baculovirus transfer vector containing a GST–AtRBR1 fusion.
Recombinant baculoviruses corresponding to full-length TGMV AL11–352 (pNSB244) and the truncated proteins, AL11–213 (pNSB392), AL11–180 (pNSB517), AL11–168 (pNSB708), AL11–158 (pNSB646), AL11–119 (pNSB388), AL1119–352 (pNSB516) and AL1181–352 (pNSB469), were described elsewhere (Orozco et al., 1997, 2000; Orozco and Hanley-Bowdoin, 1998). The baculovirus transfer vector, pMON27025 (Luckow et al., 1993), was modified by treatment with AatII and Klenow to remove the endogenous AatII site followed by insertion of the complementary linker oligonucleotides, 5′-GATCCATGGACGTC and 5′-GATCGACGTCCATG, into the BamHI site to create an ATG codon and an AatII site. An AatII–HindIII fragment from pNSB148 (Gladfelter et al., 1997) was then cloned in-frame to create the AL1101–352 transfer vector (pNSB583). The complementary oligonucleotides, 5′-CCATATGACTCTTGTATGGGGAGAATT and 5′-TTAAGGTATACTGATAACATACCCCTC, were inserted into EcoRI-digested pNSB148 to generate an in-frame ATG upstream of AL1 amino acid 110. A repaired NdeI–BamHI fragment from the resulting clone was inserted into pMON27025 with repaired BamHI ends to give the AL1110–352 transfer vector (pNSB840). An AL11–134 transfer vector (pNSB828) was generated by NotI digestion of pNSB593, a transfer vector carrying the AL1 mutant ND133 (Orozco et al., 2000), followed by insertion of the complementary linker oligonucleotides, 5′-GGCCTTAGTGATGATGG TGATGGTGATGGTGGTGATG and 5′-GGCCCATCACCACCATCA CCATCACCATCATCACTAA.
Plasmids
ZmRBR1 coding sequences were fused to the Gal4 DBD of pAS1 (Clontech, Palo Alto, CA). Zm214C and Zm290C were described earlier (Ach et al., 1997a). pTD1-1, which contains SV40 large T-antigen amino acids 87–708, was from Clontech. Plasmid pNSB904 encoding the mutant, Zm214C(C653F), was generated by replacing the SpeI–BamHI fragment of Zm214C with the equivalent fragment from Zm290C(C653F) (Ach et al., 1997a). For AtRBR1, sequences encoding amino acids 319 to C were fused downstream of the Gal4 DBD by cloning the SmaI fragment of SOAtRb-bac33 into pAS2-1 (Clontech) to give pNSB886.
Yeast expression cassettes for AL1 were generated using pACT2 (Clontech), which contains the Gal4 AD. Cassettes for wild-type AL1 (pNSB809), FQ118 (pNSB872), D120 (pNSB871), RS-R125 (pNSB786), QT130 (pNSB788), ND133 (pNSB790), E--N140 (pNSB893), KEE146 (pNSB894), REK154 (pNSB759), EKY159 (pNSB760), Q-HN165 (pNSB761), N-DR172 (pNSB762) and K--E179 (pNSB763) were described previously (Orozco et al., 2000). Cassettes for Y104 (pNSB815), K107 (pNSB818) and D108 (pNSB816) were made by replacing the AatII–BamHI fragment of pNSB735 (Orozco et al., 2000) with the equivalent fragments from pNSB684, pNSB780 and pNSB683, respectively (Orozco and Hanley-Bowdoin, 1998). To make the AAA136 cassette (pNSB758), site-directed mutagenesis was performed using pNSB148 and the primer, 5′-GAAGCAATTTAAGGCCTCTAGTAGA AGGTCGTTAGATG. The NdeI–SalI fragment of the mutant was inserted into the same sites of pMON1549 (Fontes et al., 1994a) to give pNSB676. The AatII–BamHI fragment of pNSB735 (Orozco et al., 2000) was replaced by the equivalent fragment from pNSB676, resulting in pNSB758.
TGMV A replicons carrying mutant AL1 coding sequences were made using pMON1565, a plasmid that contains 1.5 copies of TGMV A (Orozco and Hanley-Bowdoin, 1996). The mutant replicons, E--N140 (pNSB899) and KEE146 (pNSB896), were generated by replacing the SalI–NcoI fragment of pMON1565 with those of pNSB640 and pNSB641, respectively (Orozco et al., 2000).
Protein interaction assays
Recombinant proteins were produced in Spodoptera frugiperda Sf9 cells using a baculovirus expression system according to published protocols (Orozco and Hanley-Bowdoin, 1996). Protein extracts from cells co-expressing GST–ZmRBR1 or GST–AtRBR1 with full-length or truncated versions of TGMV AL1 were assayed for interaction by co-purification on glutathione–Sepharose (Settlage et al., 1996). Co-purification was monitored by SDS–PAGE followed by immunoblotting using the ECL detection system (Amersham Life Science, Arlington Heights, IL). Primary antibodies were rabbit polyclonal anti-GST (Upstate Biotechnology Inc.) and anti-AL1 (Settlage et al., 1996) antisera.
Interactions between Gal4 fusion proteins were evaluated in Saccharomyces cerevisiae strain Y187 by measuring β-galactosidase activity as described previously (Orozco et al., 2000). The different constructs were tested in a minimum of four independent transformants in at least two experiments. The relative activities of the mutant proteins were normalized against wild-type AL1, which was set to 100%.
Replication and infectivity assays
For transient replication assays, protoplasts were isolated from Nicotiana tabacum (BY-2) suspension cells, electroporated and cultured according to published methods (Fontes et al., 1994a). Cells were transfected with 15 µg of wild-type or mutant TGMV A replicon DNA. Total DNA was extracted 3 days post-transfection, digested with XhoI and DpnI, and analyzed for viral DNA accumulation by DNA gel blot hybridization using a TGMV A specific probe (Fontes et al., 1994b). Viral DNA was quantified by phosphoimager analysis.
For infectivity assays, N.benthamiana plants at the 6-leaf stage were bombarded as described previously (Nagar et al., 1995). Replicon DNA (5 µg of each plasmid) corresponding to wild-type or mutant TGMV A was precipitated onto 1.0 µm microprojectiles in the presence of a wild-type TGMV B replicon (pTG1.4B) (Fontes et al., 1994b) and bombarded into plants. Total DNA was isolated from symptomatic leaf tissue (0.5 g) at 18 days post-bombardment (Dellaporta et al., 1983), digested with XhoI, and analyzed on DNA gel blots.
In situ analyses
Systemically infected leaves were fixed, embedded in agar and sectioned as described by Bass et al. (2000). Leaf cross-sections (50–60 µm) were cut using a vibratome, agitated gently in 100% methanol for 30–45 min at 25°C, and transferred to 1× SSC. For in situ DNA hybridization, the sections were washed three times in 1× SSC and incubated at 37°C for 30–60 min in prehybridization solution (47% deionized formamide, 4× SSC, 1 mM EDTA, 100 µg/ml salmon sperm DNA, 100 µg/ml yeast tRNA, 5× Denhardt’s). A digoxigenin-labeled probe, which was synthesized using a TGMV B template and the Dig High Prime labeling kit (Boehringer Mannheim, Indianapolis, IN), was added in fresh hybridization buffer to a final concentration of 1 ng/µl. Hybridization was performed at 37°C for 16 h, and the sections were washed at 37°C once in fresh hybridization buffer, three times in 2× SSC and twice in 1× SSC. The hybridized probe was detected at 25°C using the Detection Starter kit (Boehringer Mannheim).
Total DNA was detected by DAPI staining (Bass et al., 2000). The AL1 and PCNA proteins were immunolocalized using mouse monoclonal antiserum against TGMV AL1 or human PCNA (PC10; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and visualized (Nagar et al., 1995) using the Vectastain® Elite ABC horseradish peroxidase kit (Vector Laboratories, Burlingame, CA). The sections were mounted on glass slides in phosphate-buffered saline containing 90% glycerol and visualized using a Nikon Eclipse 800 compound microscope with differential interference contrast optics and recorded on Kodak Elite 100 slide film using a Nikon U-III camera system.
Acknowledgments
Acknowledgements
We thank the Arabidopsis Biological Resource Center (ABRC) at Ohio State University for supplying DNA blots for genomic mapping and cDNA libraries. We also thank Dr Tim Petty for critical reading of the manuscript. The research was supported by grants to L.H.-B. from the National Science Foundation (MCB-9809953) and the DuPont Company (Educational Aid Grant program) and to W.G. from the Department of Energy (85ER13375). W.G. also acknowledges support of this work from funds of a UC Berkeley Chancellor’s Professorship.
References
- Ach R.A., Durfee,T., Miller,A.B., Taranto,P., Hanley-Bowdoin,L., Zambriski,P.C. and Gruissem,W. (1997a) RRB1 and RRB2 encode maize retinoblastoma-related proteins that interact with a plant D-type cyclin and geminivirus replication protein. Mol. Cell. Biol., 17, 5077–5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ach R.A., Taranto,P. and Gruissem,W. (1997b) A conserved family of WD-40 proteins binds to the retinoblastoma protein in both plants and animals. Plant Cell, 9, 1595–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson M.N., Meyer,A.D., Gyorgyey,J., Katul,L., Vetten,H.J., Gronenborn,B. and Teimchenko,T. (2000) Clink, a nanovirus-encoded protein, binds both pRB and SKP1. J. Virol., 74, 2967–2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass H., Nagar,S., Hanley-Bowdoin,L. and Robertson,D. (2000) Chromosome condensation induced by geminivirus infection of mature plant cells. J. Cell Sci., 113, 1149–1160. [DOI] [PubMed] [Google Scholar]
- Chellappan S., Kraus,V.B., Kroger,B., Munger,K., Howley,P.M., Phelps,W.C. and Nevins,J.R. (1992) Adenovirus E1A, simian virus 40 tumor antigen and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc. Natl Acad. Sci. USA, 89, 4549–4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coello P., Rodriguez,R., Garcia,E. and Vazquez-Ramos,J.M. (1992) A DNA polymerase from maize axes—its purification and possible role. Plant Mol. Biol., 20, 1159–1168. [DOI] [PubMed] [Google Scholar]
- Collin S., Fernandez-Lobato,M., Gooding,P.S., Mullineaux,P.M. and Fenoll,C. (1996) The two nonstructural proteins from wheat dwarf virus involved in viral gene expression and replication are retinoblastoma-binding proteins. Virology, 219, 324–329. [DOI] [PubMed] [Google Scholar]
- Dellaporta S.L., Wood,J. and Hicks,J.B. (1983) Maize DNA minipreps. Maize Genet. Coop. Newsl., 57, 26–29. [Google Scholar]
- Desbiez C., David,C., Mettouchi,A., Laufs,J. and Gronenborn,B. (1995) Rep protein of tomato yellow leaf curl geminivirus has an ATPase activity required for viral DNA replication. Proc. Natl Acad. Sci. USA, 92, 5640–5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee T., Feiler,H. and Gruissem,W. (2000) Retinoblastoma-related proteins in plants: homologs or orthologs of their metazoan counterparts? Plant Mol. Biol., in press. [DOI] [PubMed] [Google Scholar]
- Dyson N., Guida,P., Munger,K. and Harlow,E. (1992) Homologous sequences in adenovirus E1A and human papillomavirus E7 proteins mediate interaction with the same set of cellular proteins. J. Virol., 66, 6893–6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckner R., Ewen,M.E., Newsome,D., Gerdes,M., Decaprio,J.A., Lawrence,J.B. and Livingston,D.M. (1994) Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev., 8, 869–884. [DOI] [PubMed] [Google Scholar]
- Elmer J.S., Brand,L., Sunter,G., Gardiner,W.E., Bisaro,D.M. and Rogers,S.G. (1988) Genetic analysis of tomato golden mosaic virus II. Requirement for the product of the highly conserved AL1 coding sequence for replication. Nucleic Acids Res., 16, 7043–7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau K. (1977) Virus-like particles in the nuclei of phloem cells in spinach leaves infected with the curly top virus. J. Ultrastruct. Res., 61, 78–88. [DOI] [PubMed] [Google Scholar]
- Fontes E.P.B., Eagle,P.A., Sipe,P.A., Luckow,V.A. and Hanley-Bowdoin,L. (1994a) Interaction between a geminivirus replication protein and origin DNA is essential for viral replication. J. Biol. Chem., 269, 8459–8465. [PubMed] [Google Scholar]
- Fontes E.P.B., Gladfelter,H.J., Schaffer,R.L., Petty,I.T.D. and Hanley-Bowdoin,L. (1994b) Geminivirus replication origins have a modular organization. Plant Cell, 6, 405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fountain M.D., Murray,J.A.H. and Beck,E. (1999) Isolation of a full-length cDNA encoding a retinoblastoma (accession No. AJ011681) protein from suspension cultured photoautotrophic Chenopodium rubrum L. cells. Plant Physiol., 119, 363.10232959 [Google Scholar]
- Gjorup O.V., Rose,P.E., Holman,P.S., Bockus,B.J. and Schaffhausen, B.S. (1994) Protein domains connect cell cycle stimulation directly to initiation of DNA replication. Proc. Natl Acad. Sci. USA, 91, 12125–12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter H.J., Eagle,P.A., Fontes,E.P.B., Batts,L.A. and Hanley-Bowdoin,L. (1997) Two domains of the AL1 protein mediate geminivirus origin recognition. Virology, 239, 186–197. [DOI] [PubMed] [Google Scholar]
- Grafi G., Burnett,R.J., Helentjaris,T., Larkins,B.A., Decaprio,J.A., Sellers,W.R. and Kaelin,W.G. (1996) A maize cDNA encoding a member of the retinoblastoma protein family: involvement in endoreduplication. Proc. Natl Acad. Sci. USA, 93, 8962–8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruda M.C., Zabolotny,J.M., Xiao,J.H., Davidson,I. and Alwine,J.C. (1993) Transcriptional activation by simian virus 40 large T-antigen: interactions with multiple components of the transcription complex. Mol. Cell. Biol., 13, 961–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez C. (2000) DNA replication and cell cycle in plants: learning from geminiviruses. EMBO J., 19, 792–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley-Bowdoin L., Settlage,S.B., Orozco,B.M., Nagar,S. and Robertson,D. (1999) Geminiviruses: models for plant DNA replication, transcription and cell cycle regulation. Crit. Rev. Plant Sci., 18, 71–106. [PubMed] [Google Scholar]
- Hemerly A.S., Ferreira,P., Engler,J.D., Van Montagu,M., Engler,G. and Inze,D. (1993) Cdc2A expression in Arabidopsis is linked with competence for cell division. Plant Cell, 5, 1711–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herwig S. and Strauss,M. (1997) The retinoblastoma protein: a master regulator of cell cycle, differentiation and apoptosis. Eur. J. Biochem., 246, 581–601. [DOI] [PubMed] [Google Scholar]
- Horns T. and Jeske,H. (1991) Localization of abutilon mosaic virus (AbMV) DNA within leaf tissue by in situ hybridization. Virology, 181, 580–588. [DOI] [PubMed] [Google Scholar]
- Horvath G.V., Pettko-Szandtner,A., Nikovics,K., Bilgin,M., Boulton,M., Davies,J.W., Gutierrez,C. and Dudits,D. (1998) Prediction of functional regions of the maize streak virus replication-associated proteins by protein–protein interaction analysis. Plant Mol. Biol., 38, 699–712. [DOI] [PubMed] [Google Scholar]
- Huntley R. et al. (1998) The maize retinoblastoma protein homologue ZmRb-1 is regulated during leaf development and displays conserved interactions with G1/S regulators and plant cyclin D (CycD) proteins. Plant Mol. Biol., 37, 155–169. [DOI] [PubMed] [Google Scholar]
- Jansen-Durr P. (1996) How viral oncogenes make the cell cycle. Trends Genet., 12, 270–275. [DOI] [PubMed] [Google Scholar]
- Laufs J., Traut,W., Heyraud,F., Matzeit,V., Rogers,S.G., Schell,J. and Gronenborn,B. (1995) In vitro cleavage and joining at the viral origin of replication by the replication initiator protein of tomato yellow leaf curl virus. Proc. Natl Acad. Sci. USA, 92, 3879–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.O., Russo,A.A. and Pavletich,N.P. (1998) Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature, 391, 859–865. [DOI] [PubMed] [Google Scholar]
- Liu L., Saunders,K., Thomas,C.L., Davies,J.W. and Stanley,J. (1999) Bean yellow dwarf virus RepA, but not Rep, binds to maize retinoblastoma protein and the virus tolerates mutations in the consensus binding motif. Virology, 256, 270–279. [DOI] [PubMed] [Google Scholar]
- Luckow V.A., Lee,S.C., Barry,G.F. and Olins,P.O. (1993) Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol., 67, 4566–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucy A.P., Boulton,M.I., Davies,J.W. and Maule,A.J. (1996) Tissue specificity of Zea mays infection by maize streak virus. Mol. Plant Microbe Interact., 9, 22–31. [Google Scholar]
- Martinez M.C., Jorgensen,J.E., Lawton,M.A., Lamb,C.J. and Doerner,P.W. (1992) Spatial pattern of cdc2 expression in relation to meristem activity and cell proliferation during plant development. Proc. Natl Acad. Sci. USA, 89, 7360–7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J.A.H. (1997) The retinoblastoma protein is in plants! Trends Plant Sci., 2, 82–84. [Google Scholar]
- Nagar S., Pedersen,T.J., Carrick,K., Hanley-Bowdoin,L. and Robertson,D. (1995) A geminivirus induces expression of a host DNA synthesis protein in terminally differentiated plant cells. Plant Cell, 7, 705–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H., Sekine,M., Murakami,H. and Shinmyo,A. (1999) Tobacco retinoblastoma-related protein phosphorylated by a distinct cyclin-dependent kinase complex with Cdc2/cyclin D in vitro. Plant J., 18, 243–252. [DOI] [PubMed] [Google Scholar]
- Nevins J.R. (1992) E2F—a link between the Rb tumor suppressor protein and viral oncoproteins. Science, 258, 424–429. [DOI] [PubMed] [Google Scholar]
- Noueiry A.O., Lucas,W.J. and Gilbertson,R.L. (1994) Two proteins of a plant DNA virus coordinate nuclear and plasmodesmal transport. Cell, 76, 925–932. [DOI] [PubMed] [Google Scholar]
- Ohtani K., Degregori,J. and Nevins,J.R. (1995) Regulation of the cyclin E gene by transcription factor E2F1. Proc. Natl Acad. Sci. USA, 92, 12146–12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco B.M. and Hanley-Bowdoin,L. (1996) A DNA structure is required for geminivirus origin function. J. Virol., 270, 148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco B.M. and Hanley-Bowdoin,L. (1998) Conserved sequence and structural motifs contribute to the DNA binding and cleavage activities of a geminivirus replication protein. J. Biol. Chem., 273, 24448–24456. [DOI] [PubMed] [Google Scholar]
- Orozco B.M., Miller,A.B., Settlage,S.B. and Hanley-Bowdoin,L. (1997) Functional domains of a geminivirus replication protein. J. Biol. Chem., 272, 9840–9846. [DOI] [PubMed] [Google Scholar]
- Orozco B.M., Kong,L.-J., Batts,L.A., Elledge,S. and Hanley-Bowdoin,L. (2000) The multifunctional character of a geminivirus replication protein is reflected by its complex oligomerization properties. J. Biol. Chem., 275, 6114–6122. [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra E., Xie,Q., Boniotti,M.B. and Gutierrez,C. (1999) The cloning of plant E2F, a retinoblastoma-binding protein, reveals unique and conserved features with animal G1/S regulators. Nucleic Acids Res., 27, 3527–3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynisdottir I., Bhattacharyya,S., Zhang,D. and Prives,C. (1999) The retinoblastoma protein alters the phosphorylation state of polyomavirus large T antigen in murine cell extracts and inhibits polyomavirus origin DNA replication. J. Virol., 73, 3004–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot A.A. and Lazarowitz,S.G. (1996) Getting it together in plant virus movement: Cooperative interactions between bipartite geminivirus movement proteins. Trends Cell Biol., 6, 353–358. [DOI] [PubMed] [Google Scholar]
- Schulze A., Zerfass,K., Spitkovsky,D., Middendorp,S., Berges,J., Helin,K., Jansen-Durr,P. and Henglein,B. (1995) Cell cycle regulation of the cyclin A gene promoter is mediated by a variant E2F site. Proc. Natl Acad. Sci. USA, 92, 11264–11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine M., Masaki,I., Uemukai,K., Maeda,Y., Nakagami,H. and Shinmyo,A. (1999) Isolation and characterization of the E2F-like gene in plants. FEBS Lett., 460, 117–122. [DOI] [PubMed] [Google Scholar]
- Settlage S.B., Miller,B. and Hanley-Bowdoin,L. (1996) Interactions between geminivirus replication proteins. J. Virol., 70, 6790–6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidle A., Palaty,C., Dirks,P., Wiggan,O., Kiess,M., Gill,R.M., Wong,A.K. and Hamel,P.A. (1996) Activity of the retinoblastoma family proteins, pRB, p107 and p130, during cellular proliferation and differentiation. Crit. Rev. Biochem. Mol. Biol., 31, 237–271. [DOI] [PubMed] [Google Scholar]
- Soni R., Carmichael,J.P., Shah,Z.H. and Murray,J.A.H. (1995) A family of cyclin D homologs from plants differentially controlled by growth regulators and containing the conserved retinoblastoma protein interaction motif. Plant Cell, 7, 85–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger C. and Doonan,J. (1993) Cell division in plants. Curr. Opin. Cell Biol., 5, 226–231. [DOI] [PubMed] [Google Scholar]
- Sterner J.M., Dew-Knight,S., Musahl,C., Kornbluth,S. and Horowitz,J.M. (1998) Negative regulation of DNA replication by the retinoblastoma protein is mediated by its association with MCM7. Mol. Cell. Biol., 18, 2748–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taya Y. (1997) RB kinases and RB-binding proteins: New points of view. Trends Biochem. Sci., 22, 14–17. [DOI] [PubMed] [Google Scholar]
- Umeda M., Umeda-Hara,C., Yamaguchi,M., Hashimoto,J. and Uchimiya,H. (1999) Differential expression of genes for cyclin-dependent protein kinases in rice plants. Plant Physiol., 119, 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg R.A. (1995) The retinoblastoma protein and cell cycle control. Cell, 81, 323–330. [DOI] [PubMed] [Google Scholar]
- Werness B.A., Levine,A.J. and Howley,P.M. (1990) Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science, 248, 76–79. [DOI] [PubMed] [Google Scholar]
- Xie Q., Suarez-Lopez,P. and Gutierrez,C. (1995) Identification and analysis of a retinoblastoma binding motif in the replication protein of a plant DNA virus: Requirement for efficient viral DNA replication. EMBO J., 14, 4073–4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q., Sanz-Burgos,P., Hannon,G.J. and Gutierrez,C. (1996) Plant cells contain a novel member of the retinoblastoma family of growth regulatory proteins. EMBO J., 15, 4900–4908. [PMC free article] [PubMed] [Google Scholar]