Abstract
Vulval induction in Caenorhabditis elegans has helped define an evolutionarily conserved signal transduction pathway from receptor tyrosine kinases (RTKs) through the adaptor protein SEM-5 to RAS. One component present in other organisms, a guanine nucleotide exchange factor for Ras, has been missing in C.elegans. To understand the regulation of this pathway it is crucial to have all positive-acting components in hand. Here we describe the identification, cloning and genetic characterization of C.elegans SOS-1, a putative guanine nucleotide exchanger for LET-60 RAS. RNA interference experiments suggest that SOS-1 participates in RAS-dependent signaling events downstream of LET-23 EGFR, EGL-15 FGFR and an unknown RTK. We demonstrate that the previously identified let-341 gene encodes SOS-1. Analyzing vulval development in a let-341 null mutant, we find an SOS-1-independent pathway involved in the activation of RAS signaling. This SOS-1-independent signaling is not inhibited by SLI-1/Cbl and is not mediated by PTP-2/SHP, raising the possibility that there could be another RasGEF.
Keywords: Caenorhabditis elegans/proto-oncogene/ras pathway/tyrosine kinase/vulval development
Introduction
Polypeptide hormones bind cell surface receptors containing tyrosine kinase activity to regulate cell proliferation, differentiation, migration and metabolism. All receptor tyrosine kinases exhibit a similar molecular architecture and apparently are activated by a common mechanism. Ligand binding to the extracellular domain induces receptor dimerization, which in turn leads to activation of the catalytic kinase domain (Lemmon and Schlessinger, 1994). Autophosphorylation of tyrosine residues that are located in the C-terminal region leads to the physical recruitment of cytoplasmic signaling molecules with phosphotyrosine (pTyr)-recognition modules, such as SH2 (Src homology 2) and PTB (phosphotyrosine binding) domains (Pawson, 1995). These receptor-binding proteins include the adapter Grb2, which leads to Ras activation, Ras-GTPase activating protein, phospholipase C-(PLC)-γ, the tyrosine phosphatase SHP-2, the regulatory subunit of phosphatidylinositol-3-OH kinase (Ullrich and Schlessinger, 1990; Heldin, 1995; Pawson, 1995), the scaffolding protein Shc (Pelicci et al., 1992) and the proto-oncoprotein c-Cbl (Meisner et al., 1997). The association between proteins with SH2/PTB domains and receptor pTyrs serves as the initial step in the recruitment of an activated RTK signaling complex. Different SH2/PTB domains have distinct specificities for the pTyr residues dependent on the surrounding amino acid residues (Panayotou and Waterfield, 1993; Songyang et al., 1993, 1994; Kavanaugh and Williams, 1994; Kavanaugh et al., 1995). Association of RTKs with substrates is therefore dependent upon the amino acid context of the pTyr, and that, in part, determines which proteins join the signaling complex and what cellular responses are evoked (Schlessinger and Ullrich, 1992; Pawson and Saxton, 1999). The achievement of a precise amplitude and duration of the signaling required for the reproducible developmental consequences observed can be obtained by regulating the production of signaling molecules and their presentation to their substrates. Excessive or prolonged RTK signaling has been implicated in many cases of malignant transformation (Alroy and Yarden, 1997; Cheng et al., 1998). Therefore, proper mechanisms must be employed to strongly control the signaling from RTKs to prevent spontaneous, ligand-independent activation and downregulate signaling after it has been activated.
EGF RTK signaling is used multiple times during the normal development of C.elegans, mediating vulval induction, viability, male spicule development, hermaphrodite ovulation, and differentiation of the ventral uterus and posterior ectoderm (Ferguson and Horvitz, 1985; Aroian and Sternberg, 1991; Chamberlin and Sternberg, 1994; Clandinin et al., 1998; Jiang and Sternberg, 1998; Chang et al., 1999). The high resolution of the vulval induction assay makes it very sensitive, allowing one to identify mutants and to reflect the relative strength of RAS-dependent signaling (Han and Sternberg, 1991). All the other developmental events are also RAS-dependent, with the exception of hermaphrodite ovulation, which is instead mediated through an inositol trisphosphate signaling pathway (Clandinin et al., 1998).
The wild-type vulva is derived from three of six multipotential vulval precursor cells (VPCs), in response to an inductive signal from the anchor cell (AC) in the somatic gonad (Horvitz and Sternberg, 1991). The induced VPCs undergo three rounds of division and subsequent morphogenesis to form the vulva. VPCs that do not receive adequate signal from the anchor cell divide only once and become part of the worm’s large syncytial epidermis. The inductive signal, LIN-3, is an EGF-like growth factor produced by the anchor cell. LIN-3 activates LET-23, a C.elegans homolog of EGF RTK, in the VPCs (Aroian et al., 1990; Hill and Sternberg, 1992). LET-23 activation initiates a signaling cascade that involves downstream effectors such as SEM-5, LET-60, LIN-45, MEK-2 and SUR-1/MPK-1, which are the nematode counterparts of the mammalian Grb2 adaptor, Ras GTP-binding protein, Raf serine/threonine kinase, MAPK kinase and MAPK, respectively (Beitel et al., 1990; Han and Sternberg, 1990; Clark et al., 1992a; Han et al., 1993; Lackner et al., 1994; Wu and Han, 1994; Kornfeld et al., 1995; Wu et al., 1995). Loss-of-function (lf) mutations in these signaling proteins result in less than three VPCs undergoing vulval differentiation, whereas gain-of-function (gf) mutations result in more than three VPCs undergoing vulval differentiation.
In addition to the main RAS-dependent signaling pathway, several accessory proteins have been identified. ptp-2, which encodes an SH2 domain-containing protein similar to the mammalian SHPs and Drosophila Corkscrew, appears to affect vulval induction only in a sensitized genetic background and behaves as a positive effector of the LET-23 pathway (Gutch et al., 1998). Loss of function in ptp-2 suppresses the multiple vulva (Muv) phenotype induced by a loss-of-function lin-15 mutation, and an activated let-23 or let-60 mutation. lin-15 encodes two novel proteins that somehow inhibit RAS-dependent vulval signaling, and lin-15 mutation formally acts as if LET-23 was activated in a ligand-independent manner (Clark et al., 1994; Huang et al., 1994).
FGFR is another RTK whose developmental roles are dependent upon RAS. Genes involved in the FGFR pathway were defined by mutations affecting sex myoblast migration, including EGL-17 FGF, EGL-15, SEM-5 and several RAS–MAP kinase cascade components (Stern and Horvitz, 1991; Clark et al., 1992a; Sundaram et al., 1996; Chen et al., 1997). Others, including soc-1 and soc-2, were defined by their ability to suppress a clear (Clr) phenotype, which appears to be caused by excess EGL-15 signaling (Selfors et al., 1998).
Mutations in let-60/mek-2/mpk-1 cause a delay in pachytene exit during meiosis resulting in sterility (Church et al., 1995). The inductive signal, cognate receptor, and their signal transducers used for let-60/mek-2/mpk-1-mediated germline meiotic cell cycle progression are still unknown (Sternberg and Han, 1998).
Ras-dependent RTK signaling pathways are well conserved at the level of their mechanistic aspects throughout metazoan evolution. Ras is activated following the recruitment of Sos to the plasma membrane, which stimulates guanine nucleotide exchange. Recruitment of Sos follows receptor activation and is dependent upon the adaptor protein Grb2. Sos contains two structural domains, CDC25 and DH, which act as exchange factors specifically upon the closely related Ras and Rho family GTPases, respectively (Chardin et al., 1993; Nimnual et al., 1998). An EGF RTK substrate, Eps8, and its associated protein, E3b1, have been shown recently to regulate the transduction of signals from Ras to Rac through Sos (Scita et al., 1999). Despite our understanding of Sos function, obtained primarily from work on flies and mammals, the identification of the exchange factor for RAS in C.elegans has remained elusive. For our goal of understanding how receptor tyrosine kinase signaling is regulated, it is necessary to identify all the positive- and negative-acting components. To this end we cloned the previously unidentified C.elegans Sos homolog, SOS-1, and show that it is encoded by the let-341 gene. We demonstrate SOS-1 activity is required for normal RAS activation. In addition to a requirement for SOS-1, we show that there is a SOS-1 independent pathway operating in vulval induction. This SOS-1 independent pathway does not utilize PTP-2. This raises the possibility that there remain other activators of RAS signaling that are yet to be identified.
Results
Isolation of a Sos homolog in C.elegans
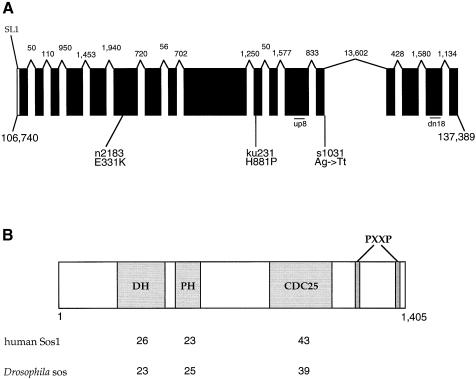
The C.elegans Sequencing Consortium (1998) identified an incomplete genomic sequence, which GeneFinder predicts to encode a C.elegans homolog of Sos. By using reverse transcription–PCR (RT–PCR) and 5′ and 3′ rapid amplification of cDNA ends (RACE), we have been able to obtain a nearly complete cDNA of this gene and confirm that it encodes a Sos homolog (Figure 1). We will refer to this gene as sos-1 (for son of sevenless homolog). A SL1 trans-spliced leader sequence, which is immediately followed by the putative initiation methionine, has been identified in the 5′ end of the cDNA. However, the cDNA is lacking the extreme 3′ sequence as the termination codon is still missing. This is due to a highly adenylated sequence in the 3′ terminus, which presumably prevents the recovery of the 3′ end by use of a poly(dT) primer (Figure 1A). The longest cDNA fragment was sequenced and this 4.2 kb cDNA contains a single long open reading frame (ORF) (DDBJ/EMBL/GenBank accession No. AF251308). We obtained two shorter splice variants predicted to encode proteins that end prematurely immediately after exon 12 and 13 (see Materials and methods). Sequence analysis of the corresponding genomic region encompassing this 4.2 kb cDNA revealed that the coding region of this gene spans a region >30.6 kb and includes a long 13.6 kb intron. Conceptual translation of the 4.2 kb ORF predicted a protein of 1413 amino acids with signature motifs that match the guanine nucleotide exchange factor for Ras. The predicted structure of SOS-1 contains one Dbl homology (DH) domain followed by one pleckstrin homology (PH) domain at the N-terminus and a C-terminal CDC25 catalytic domain followed by two PxxP motifs (Figure 1B). CDC25 domain is the most conserved region in RAS-binding exchange factors among species (Figure 1C).
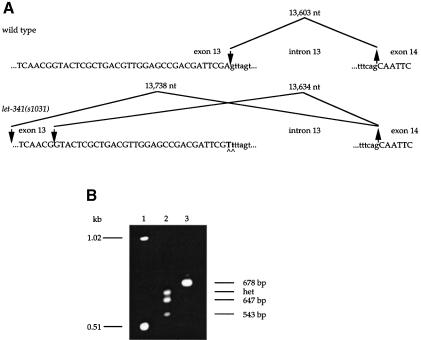
Fig. 1. (A) Genomic structure of the sos-1 locus. The solid bars represent exons, which were inferred by comparing the nucleotide sequence of a sos-1 cDNA with that of the genomic DNA; the 5′ untranslated region is depicted as an open box. Sizes of the 16 introns are noted by the numbers above each intron. The start and tentative end of the let-341 sos-1 coding sequences are indicated by the corresponding nucleotide numbers from YAC Y61A9LA. Diagonal line labeled SL1 is the trans-spliced leader attachment site. The positions of let-341 mutations and corresponding amino acid or splice site changes are shown. Exonic sequence is shown in uppercase lettering, intronic sequence in lowercase. The approximate locations of primers up8 and dn18 used to analyze s1031 transcripts are indicated. (B) Comparative analyses of signature motifs in SOS-1 homologs. The protein predicted to be encoded by the sos-1 gene is depicted schematically. The numbers listed below represent the percent identity between DH domain, PH domain and CDC25 catalytic domain of SOS-1 and those domains of human Sos1 and Drosophila sos. Motifs were identified using the ProfileScan program. (C) Sequence alignment of CDC25 domains of RAS-binding exchange factors from C.elegans (Ce), Drosophila melanogaster (Dm) and Homo sapiens (Hs), with residue numbers of C.elegans (Ce) SOS-1 indicated. Residue identity between species is highlighted in black, and similarity is highlighted in gray. The arrows indicate the start of deletions in the s1031 transcripts.
Phenotypic effects displayed by the progeny of SOS-1 dsRNA-injected hermaphrodites
To understand the biological functions of SOS-1, we used double-stranded RNA interference (RNAi) to transiently knock out gene expression during animal development. RNAi has been shown to robustly and efficiently interfere with, if not remove, gene expression in many model systems including C.elegans, Drosophila, Trypanosoma brucei and plants (Fire et al., 1998; Sharp, 1999). A 1.6 kb SOS-1 cDNA, which represents the sequences encoding the C-terminal portion of the DH and the entire PH domains, was subcloned into the pCRII-TOPO vector. The nucleotide sequence of this 1.6 kb cDNA is not similar to any other gene identified by the C.elegans Sequencing Consortium (as of April 2000). Sense and antisense RNA were synthesized from T7 and Sp6 promoters, respectively, and sense/antisense annealing was carried out in vitro prior to microinjection.
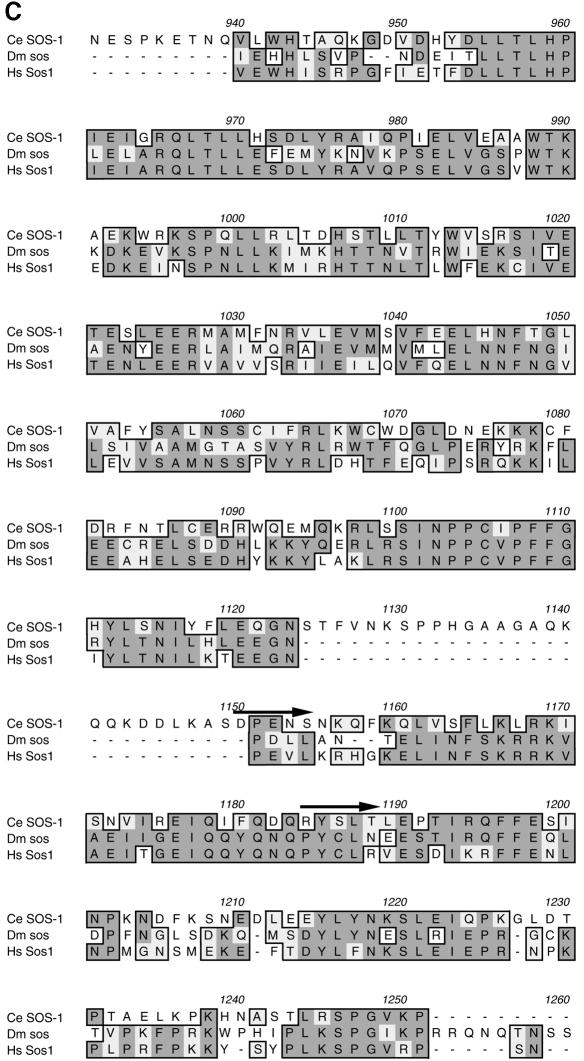
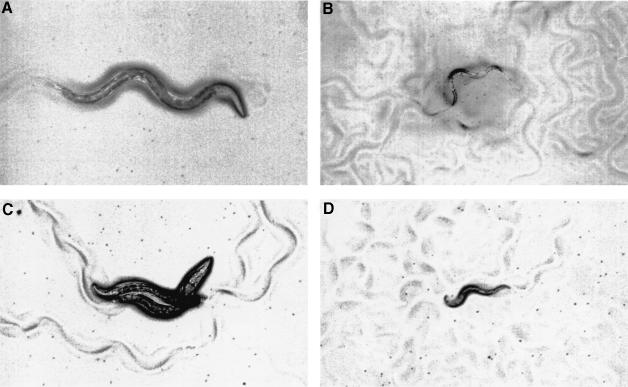
RNAi reveals that SOS-1 participates in those functions mediated by LET-60 RAS. Progeny of hermaphrodites injected with sos-1 dsRNA showed a low penetrance of L1-L2 lethality (8%; n = 66). Animals escaping this early larval lethality generally displayed slow growth and a scrawny body morphology, resembling the phenotype of hypomorphic egl-15 mutants, an FGFR homolog (61/61 animals examined) (Figure 4B). When examined under Nomarski optics, uterine cells are generally mis-positioned or mis-specified (data not shown). During oogenesis, a significant retardation of pachytene exit is observed (11/11 animals examined) (Figure 3D). By young adult stage, the proximal germline nuclei, which normally undergo spermatogenesis in wild-type animals, remain arrested in the pachytene stage of meiosis I. The more distal nuclei, which presumably undergo oogenesis, also remain arrested in the pachytene phase. Clumped pachytene nuclei can be easily found in the distal arm of the gonad, which might be the result of the diminished size of the gonad.
Fig. 4. sos-1 RNAi-affected animals display the phenotypes associated with reduction-of-function FGFR pathway mutants. All the animals were photographed at the same magnification. (A) Wild type (N2). (B) sos-1 RNAi-affected N2 animal, which confers a scrawny body morphology. (C) clr-1(e1745ts) mutants, which confer a temperature-sensitive Clr phenotype. The biological basis of the Clr phenotype is not understood but the abnormality appears to result from clear fluid accumulating in the pseudocoelom (Kokel et al., 1998). (D) sos-1 RNAi-affected clr-1(e1745ts) mutant, which confers a non-Clr phenotype. Animals shown in (A) and (B) are young adults and were raised at 20°C. L4 larvae shown in (C) and (D) were raised at 20°C and then switched to the nonpermissive temperature of 25°C for 10 h before being photographed.
Fig. 3. sos-1 RNAi affects male spicule development and hermaphrodite oocyte maturation. (A) Nomarski photomicrograph of a him-5(e1490) male shows a long and straight spicule, denoting by an arrow head. (B) A male progeny of him-5(e1490) hermaphrodite injected with sos-1 dsRNA shows a short and crumpled spicule, denoting by an arrow head. (C and D) Fluorescence micrograph of wild-type (bottom left) and sos-1 RNAi-affected (bottom right) hermaphrodites stained with DAPI. Regions of pachytene nuclei are indicated by a white bar and nuclei in diakinesis by a semi-gray bar. Arrow indicates the first diakinesis nucleus seen in the sos-1 RNAi-affected hermaphrodite. All animals shown are in their young adult stage. Dorsal is up and anterior to the right in all panels. The scale bar is 20 µm.
In addition, adult sos-1 [RNAi] males generally exhibit short and crumpled spicules (20/20 animals examined) (Figure 3B). let-60(n1046gf), but neither a let-23(sa62gf) nor a lin-15(n765ts) mutation, suppresses the lethality and scrawny phenotype conferred by the sos-1 RNAi, implying that SOS-1 functions downstream of LET-23 and upstream of LET-60. The hyperinduced vulval phenotypes observed in let-60(n1046gf), let-23(sa62gf) and lin-15(n765ts) mutants are suppressed by the sos-1 RNAi. However, the suppression of let-60(n1046gf) is weaker than that of let-23(sa62gf) and lin-15(n765ts) based on the distributions of vulval induction (Figure 2; Table I). Since all these phenotypes are also exhibited by mutants in RAS signaling pathway, we conclude that SOS-1 acts on RAS during larval development.
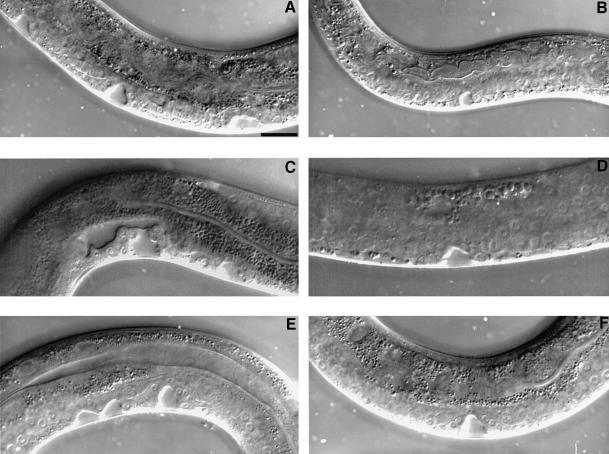
Fig. 2. Suppression of excessive vulval signaling by sos-1 RNAi. Nomarski photomicrographs of the vulval induction in the L4 stage hermaphrodites. (A) lin-15(n765) mutant, which displays a total of six VPCs adopting vulval fates and forms three vulval invaginations. (B) sos-1 RNAi-affected lin-15(n765) mutant, which displays a one cell induction that forms one vulval invagination. (C) let-23(sa62) mutant, which displays a total of four VPCs undergoing vulval differentiation and forms two vulval invaginations. (D) sos-1 RNAi-affected let-23(sa62) mutant, which displays only two induced vulval cells that form one vulval invagination. (E) let-60(n1046) mutant, which displays a total of five VPCs adopting vulval fates and forms three vulval invaginations. (F) sos-1 RNAi-affected let-60(n1046) mutant, which displays a wild-type vulval induction and invagination. Dorsal is up and anterior to the right in all panels. The scale bar is 20 µm.
Table I. SOS-1 mediates the ectopic vulval induction during excessive let-23 signaling.
| Genotype | SOS-1 RNAi | % <3 | % = 3 | % >3 | Numbers of animals | Average vulval induction |
|---|---|---|---|---|---|---|
| let-60(n1046) | – | 0 | 0 | 100 | 20 | 5.0 |
| let-60(n1046) | RNAi | 0 | 62 | 38 | 21 | 3.3 |
| let-23(sa62) | – | 0 | 7 | 93 | 15 | 4.2 |
| let-23(sa62) | RNAi | 14 | 72 | 14 | 14 | 2.9 |
| lin-15(n765) | – | 0 | 0 | 100 | 20 | 5.3 |
| lin-15(n765) | RNAi | 33 | 67 | 0 | 12 | 2.5 |
| let-60(n1046); let-341(s1031) | – | 41 | 52 | 7 | 54 | 2.7 |
| let-60(n1046); let-341(s1031) | RNAi | 39 | 52 | 9 | 23 | 2.6 |
Vulval induction was scored at 20°C using Nomarski optics during the early-mid L4 stage. The complete genotypes for strains listed are: let-60(n1046)unc-31(e169) [for let-60(n1046)]; let-23(sa62); him-5(e1490) [for let-23(sa62)]; lin-15(n765) [for lin-15(n765)]; let-60(n1046); let-341(s1031)unc46(e177) [for let-60(n1046); let-341(s1031)]. Average vulval induction represents average number of VPCs undergoing vulval differentiation per animal; 3.0 is wild-type level.
SOS-1 is encoded by let-341
sos-1 maps to the left arm of chromosome V in a region that is close to the estimated position of let-341 on the genetic map. let-341 alleles were originally isolated in a screen for essential genes on the left arm of V (Johnsen and Baillie, 1991). In addition, let-341 alleles were also isolated as extragenic suppressors of the Multivulval phenotype of lin-15(lf) mutants (Clark et al., 1992b) and in a screen for suppressors of the Clr (soc) phenotype of clr-1(lf) mutants (M.Stern, personal communication). Genetic studies suggest that let-341 has genetic properties consistent with it encoding a RAS exchange factor, namely that it acts downstream of lin-15 and upstream of let-60 (Clark et al., 1992b). We therefore tested whether let-341 mutants have lesions in sos-1. Molecular lesions associated with three let-341 alleles were found to affect the sos-1 ORF, indicating that SOS-1 transcript corresponds to let-341 gene (Figure 1A). let-341(s1031) mutants display a 100% embryonic or early larval lethality, which can be suppressed by a let-60(gf) mutation. Sequence analysis reveals that s1031 contains an AG→TT mutation at the splice donor of intron 13 in sos-1. The n2183 mutation, provided by M.Stern, changes a glutamic acid to lysine at codon 331 within the DH domain (Figure 1). The ku231 allele, provided by Z.Chen and M.Han, results in the substitution of proline for a conserved histidine at codon 881 right before the start of the CDC25 catalytic domain.
s1031 represents a null allele for sos-1
As the molecular lesion associated with s1031 is predicted to alter splicing, we looked at the sos-1 mRNA produced in this mutant. We could not detect wild-type sos-1 mRNA in s1031 mutants by RT–PCR (Figure 5). The subsequent sequence analysis revealed that in s1031 animals, upstream cryptic splice donors are used and consequently various deletions in the regions encoding the C-terminus of the CDC25 catalytic domain occur (Figures 1C and 5). The truncated transcripts are predicted to encode either a dominant negative or a loss-of-function SOS-1 protein, suggesting that s1031 completely eliminates SOS-1 activity. Consistent with s1031 being a null allele, a sem-5 null or severe reduction-of-function mutation does not fur ther reduce the vulval induction in let-60(n1046); let-341(s1031) mutant animals (Table II). To strengthen this conclusion further, we use sos-1 RNAi to remove any residual sos-1 activity in let-60(n1046); let-341(s1031) mutants. The average vulval induction displayed by the progeny of sos-1 dsRNA-injected hermaphrodites is 2.6 (n = 23), which is not statistically different from that displayed by animals without sos-1 dsRNA injection [2.7 (n = 54)] (Table I). Together, these three lines of observation provide compelling evidence that s1031 is a null allele.
Fig. 5. let-341(s1031) is a putative null allele of sos-1. (A) The sequence of the let-341(s1031) mutation. The upper panel shows the genomic sequence around the boundaries of exon 13–intron 13 and intron 13–exon 14. Exons are shown in uppercase lettering, introns in lowercase. The large downward-pointing arrow indicates the 5′ location of the wild-type splice. The upward-pointing arrow indicates the 3′ splice junction used. The lower panel shows the same information in the presence of the s1031 mutation. The mutation is indicated in bold and by carets. The 5′ splice donors have shifted upstream. (B) RT–PCR analysis of wild-type and s1031 RNA. PCR amplification for 35 rounds of wild-type and s1031 reverse-transcribed RNA was performed with primers up8 and dn18 (see Figure 1A). Lanes: 1, 1 kb ladder; 2, amplification of s1031 cDNA; 3, amplification of wild-type cDNA. Correctly spliced mRNA yields a product of 678 bp (correct joining of exons 12–16). In s1031 animals, a 647 bp product results from usage of a upstream cryptic splice donor at nucleotide 119 792 of YAC Y61A9LA and removal of 31 bp from exon 13. Exon 13 skipping in s1031 animals results in a 543 bp product. A putative heteroduplex (het), expected to form from two fast migrating species, is indicated.
Table II. A SOS-1-independent activation of ras signaling in the vulva.
| Genotype |
% <3 | % = 3 | % >3 | Numbers of animals | Average vulval induction | ||||
|---|---|---|---|---|---|---|---|---|---|
| let-60 | let-341 | gap-1 | sem-5 | gonad | |||||
| 1. n1046 | + | + | + | + | 0 | 15 | 85 | 20 | 4.4 |
| 2. n1046 | + | + | + | – | 89 | 0 | 11 | 18 | 1.3 |
| 3. n1046 | s1031 | + | + | + | 41 | 52 | 7 | 54 | 2.7 |
| 4. n1046 | + | + | ay73 | + | 0 | 87 | 13 | 30 | 3.2 |
| 5. n1046 | s1031 | + | ay73 | + | 50 | 25 | 25 | 8 | 2.8 |
| 6. n1046 | s1031 | + | + | – | 95 | 5 | 0 | 22 | 0.8 |
| 7. n1046 | s1031 | n1691 | + | + | 0 | 35 | 65 | 20 | 3.9 |
| 8. n1046 | s1031 | n1691 | + | – | 54 | 15 | 31 | 13 | 2.4 |
Vulval induction was scored at 20°C using Nomarski optics during the early-mid L4 stage. let-341(s1031) was marked with unc-46(e177). sem-5(ay73) is a putative null allele resulting in a Q10Stop (Clark et al., 1992a; M.Stern, personal communication). gap-1(n1691) corresponds to a Q149Stop, which defines a putative null allele. The distributions generated from each genotype were entered into the InStat program (GraphPad Software, version 2.0) and compared in pairs using the Mann–Whitney test to generate two-tailed P values. The differences resulting from the following comparisons are considered very significant: 2 versus 3, P <0.0001; 2 versus 4, P <0.0001; 2 versus 5, P = 0.0021; 3 versus 6, P <0.0001; 3 versus 7, P <0.0001; 7 versus 8, P = 0.0048. With a P value of 0.7448, 3 versus 5 is considered not significant.
Since s1031 is a null allele, we tested whether this mutation alters vulval induction. We used a let-60(gf) mutation to rescue the lethality of s1031 and found that loss of let-341 sos-1 function reduces vulval induction. Specifically, 41% of let-60(n1046); let-341(s1031) mutant animals exhibit less than wild-type vulval induction, further supporting a role of SOS-1 during vulval induction.
Ligand-dependent and SOS-1-independent activation of RAS signaling in the vulva
To test whether lin-3-induced vulval signaling is solely mediated by LET-341 SOS-1, we compared the vulval induction in a mutant lacking lin-3 to a mutant lacking let-341 SOS-1. Since LIN-3 produced by the gonadal AC is the only inductive source for vulval differentiation, ablation of gonad eliminates the ligand for LET-23-mediated vulval signaling. The extent of vulval differentiation in let-60(gf); let-341(null) mutants is significantly higher than that in gonad-ablated let-60(gf) mutants (Table II). This observation suggests that not all of the inductive signaling is mediated through SOS-1. This SOS-1-independent pathway could also be SEM-5 independent, since let-60(gf); sem-5(null) and let-60(gf); let-341(null); sem-5(null) mutants also produce significantly more vulval differentiation than do gonad-ablated let-60(gf) mutants. A transgenic line that bears multiple copies of let-60(n1046gf) DNA displays completely ligand-independent signaling (Sundaram and Han, 1995). Importantly, this transgenic array does not confer 100% induction in all six VPCs. It has been shown that a weak lin-45 mutation suppresses the vulval induction conferred by this transgenic line while a weak sem-5 mutation or the gonad ablation does not. This indicates that gonad ablation does not affect signaling parallel to LET-60 RAS (Sundaram and Han, 1995). Thus, the ligand-dependent and SOS-1-independent pathway are most likely to act at the level of LET-60 RAS.
To address the question of whether LET-23 is required for the SOS-1-independent activation of LET-60 RAS, we assayed the extent of vulval induction in let-23(sy17 null); let-60(n1046) double mutants. The average vulval induction of sy17; n1046 strain shows no difference from that of n1046 single mutants (Table III). Our interpretation of this observation is that there is a LET-23-dependent negative regulation for vulval signaling, which can be lifted by the presence of the gonadal signal. This model is supported by our other experimental results. Because LET-23 has both positive and negative effects on vulval signaling, we cannot rule out a LET-23-independent response to the gonadal signal.
Table III. sli-1 regulates let-23 signaling upstream of ras, in a SOS-1-dependent manner.
| Genotype |
Average vulval induction | Proportion of animals with>3 cells induced | ||||
|---|---|---|---|---|---|---|
| ptp-2 | let-23 | let-60 | let-341 | sli-1 | ||
| 1. + | + | n1046 | + | + | 4.4a | 17/20 |
| 2. + | sy17 | n1046 | + | + | 4.6 | 19/20 |
| 3. + | + | n1046 | + | sy143 | 5.5a | 20/20 |
| 4. + | + | n1046 | s1031 | + | 2.7 | 4/54 |
| 5. + | + | n1046 | s1031 | sy143 | 2.9 | 1/20 |
| 6. op194 | + | n1046 | s1031 | + | 3.0 | 2/20 |
Vulval induction was scored at 20°C using Nomarski optics during the early-mid L4 stage. ptp-2(op194) and let-341(s1031) were marked with unc-4(e120) and unc-46(e177), respectively. ptp-2(op194) is a putative null allele resulting in a genomic deletion of 1458 nucleotides (Gutch et al., 1998). sli-1(sy143) is also a putative null allele resulting in a Q152Stop (Yoon et al., 1995).
aThe difference of average vulval induction between let-60(n1046) and let-60(n1046); sli-1(sy143) mutants is highly significant (P = 0.0002) based on the Mann–Whitney test. The most significant difference occurs in the P8.p induction with 33% of P8.p induced for let-60(n1046) and 78% for let-60(n1046); sli-1(sy143). The differences result from the following comparisons are considered not significant: 4 versus 5, P = 0.1629; 4 versus 6, P = 0.1313.
To test whether this putative SOS-1-independent pathway is under regulation by SLI-1, we crossed a sli-1 mutation to a genetic background where the SOS-1-dependent pathway has been removed. SLI-1, similar to mammalian proto-oncoprotein c-Cbl, inhibits LET-23 mediated signaling during vulval development (Jongeward et al., 1995; Yoon et al., 1995). We found that a sli-1(lf) mutation fails to enhance the vulval induction in let-60(gf); let-341(null) mutants, arguing that SLI-1 does not negatively regulate this other pathway parallel to SOS-1 (Table III). This result also supports our interpretation that let-341(s1031) represents a loss-of-function rather than a severe reduction-of-function mutation, because we expect that fluctuation in the ras-dependent signaling caused by a sli-1 mutation would result in a dramatic change in vulval induction under the condition where a low level activity of LET-23/SEM-5/SOS-1 is present (C.Yoon, C.Chang and P.Sternberg, unpublished).
Gonad-ablated let-60(gf); let-341(null) animals have less vulval differentiation than do let-60(gf); let-341(null) or gonad-ablated let-60(gf) animals. Therefore, gonad ablation not only removes a ligand-dependent and SOS-1-independent positive signaling pathway, but also reveals a ligand-independent activation of RAS by SOS-1. Although this ligand-independent activation of RAS by SOS-1 does not contribute significantly the vulval induction in gonad-ablated let-60(gf) animals, it could explain the difference of vulval induction between let-60(gf); let-341(null) and let-60(gf); sem-5(null) mutants (Table II).
PTP-2 is not involved in SOS-1-independent vulval signaling
Based on the observation that the lf mutation in ptp-2 reduces the vulval induction conferred by a let-60(gf) mutation, ptp-2 was previously interpreted to act in the vulval signaling pathway as a positive effector downstream of, or in parallel to, let-60 ras (Gutch et al., 1998). To determine whether the response of PTP-2 to receptor activation is parallel to that of SOS-1, we generated triple mutants with activated RAS but lacking PTP-2 and SOS-1 [ptp-2(null); let-60(gf); let-341(null)], and assessed their vulval differentiation. The ptp-2 mutation did not further reduce the vulval induction observed in let-60(gf); let-341(null) mutants, suggesting that PTP-2 does not mediate SOS-1-independent vulval signaling (Table III).
Lifting inhibition by GAP-1 does not explain the SOS-1-independent signaling activity
Besides a SOS-1-independent activation of RAS, an alternative explanation for the elevated signaling activity in the let-60(gf); let-341(null) mutant animals is that activation of LET-23 by the ligand may relieve the inhibition of RAS by GAP-1, GTPase activating protein (Hajnal et al., 1997). In this model, LET-23 would be able to relieve GAP-1-mediated inhibition of RAS upon activation by the ligand in let-60(gf); let-341(null) double mutants, while let-60(gf) single mutants with an ablated gonad display constitutively active GAP-1 and cause a further decrease in the vulval induction. To test this model, we constructed the triple mutant let-60(gf); let-341(null); gap-1(null) and compared the average vulval induction in the conditions with and without the gonad. We find that gonad ablation significantly reduces the vulval induction of let-60(gf); let-341(null); gap-1(null) animals [from 3.9 (n = 20) to 2.4 (n = 13); P = 0.0048] (Table II), indicating that the anchor cell signal is not working solely through let-341 and gap-1, which are absent already in let-60(gf); let-341(null); gap-1(null) animals. This result rules out the possibility that the gonad-dependent SOS-1-independent signaling is mediated solely through gap-1.
A role for SOS-1 in FGFR-mediated signaling during development
Hyperactive EGL-15 FGF receptor signaling, conferred by either an activated egl-15 or a lf clr-1 mutation, results in a Clr phenotype. Decreased EGL-15 signaling, conferred by a partial lf egl-15 mutation, results in a scrawny body morphology phenotype (Kokel et al., 1998). clr-1, which encodes a receptor tyrosine phosphatase, is believed to attenuate signaling from FGFR by dephosphorylation. A scrawny phenotype was observed in the SOS-1 RNAi-affected animals (Figure 4B), suggesting that SOS-1 normally mediates FGFR signaling. To test this possibility, we removed SOS-1 function in the temperature-sensitive clr-1(e1745ts) mutants by SOS-1 RNAi and assessed their Clr phenotype. We found that the Clr phenotype of clr-1(e1745ts) mutants was suppressed by SOS-1 RNAi (77/77 animals examined) (Figure 4C and D), indicating a positive role for SOS-1 in FGFR-mediated signaling. Indeed, the let-341(ku231) mutation also suppresses the Clr phenotype of clr-1(e1745ts) mutants (data not shown) and let-341(n2183) was isolated as a soc mutant, as mentioned before. Since many sem-5 alleles were also isolated as suppressors of the Clr phenotype of clr-1(lf) mutant (Clark et al., 1992a), it is likely that SEM-5-dependent recruitment of SOS-1 to the active FGFR (EGL-15) plays an analogous role in activating RAS as with the EGFR (LET-23).
Discussion
SOS-1 participation during vulval development
We have found the C.elegans homolog of Sos, and demonstrated by RNAi and by phenotypic analysis of a null allele that it is required for vulval induction. sos-1 RNAi does not affect vulval induction in the wild-type animals at an appreciable frequency (1/50 animals examined vulvaless; data not shown). It might be that vulval induction occurs late in development or a certain property of vulval lineages renders this developmental process more resistant to sos-1 RNAi. However, the effect of sos-1 RNAi on vulval signaling becomes obvious in sensitized genetic backgrounds. Although SOS-1 acts downstream of let-23 and upstream of let-60, sos-1 RNAi affects excessive vulval signaling in both let-23(gf) and let-60(gf) mutants. The fact that vulval signaling was affected in a greater degree in let-23(gf) than let-60(gf) mutants is consistent with our expectation that sos-1 RNAi abrogates both ligand-dependent and -independent activation by let-23(gf), while only abrogating ligand dependent activation by let-60(gf). The involvement of SOS-1 during vulval development is further confirmed by our observation that 41 percent of the let-60(gf); let-341(null) animals examined displayed less than a wild-type extent of vulval differentiation (Table II).
SOS-1 involvement in oocyte maturation and uterine differentiation
Consistent with the observation that sos-1 RNAi prevents meiotic prophase progression, sos-1 RNAi-affected hermaphrodites also display a late onset sterile phenotype in the adult stage (10/10 animals examined; data not shown). Furthermore, this sterile phenotype can be suppressed by a let-60(gf) mutation (data not shown). Taken together, these results identify SOS-1 as an activator for LET-60 RAS in regulating oogenesis. It remains unknown what is required upstream of SOS-1 to permit exit from pachytene (Church et al., 1995).
As judged by anatomy, we can not tell whether abnormalities in the dorsal and ventral uterine lineages are caused by a mis-determination of cell fates, defects in cell migration, or both. Most of the adult hermaphrodites affected by sos-1 RNAi have an abnormal everted vulva that consists of vulval cells and somatic gonadal tissues that have been extruded out of the body cavity (data not shown). Uterine cells are known to be required for the proper attachment between the uterus and vulva (Newman and Sternberg, 1996), a process that has been suggested to prevent the vulva and uterus from slipping out of the body cavity during vulval eversion (Seydoux et al., 1993).
SOS-1-dependent and -independent signaling
The finding that the sli-1(null) mutation fails to enhance the vulval induction in let-60(gf); let-341(null) mutants suggests that the synergy between sli-1(null) and let-60(gf) in the vulva is dependent upon SOS-1. This result is consistent with our previous data suggesting that sli-1 acts upstream of let-60 ras to inhibit signaling from let-23 (C.Yoon, C.Chang and P.Sternberg, unpublished). The finding that the extent of vulval induction in a let-60(gf); let-341(null) background is higher than in gonad ablated let-60(gf) animals reveals that there is a ligand-dependent and SOS-1-independent signaling by EGF RTK. The simplest explanation is that there is a SOS-1-independent positive signaling pathway in let-60(gf); let-341(null) animals. Alternatively, gonad-ablated let-60(gf) animals may be subject to a form of negative regulation in the vulva that is independent of SOS-1 and is lifted in a let-60(gf); let-341(null) background, perhaps by the presence of the ligand. A SOS-1-independent positive signaling pathway elicited by LIN-3 might involve another guanine nucleotide exchange factor for RAS or the DOS/PTP-2-cascade. It has been previously shown that there may be more than one activator of LET-60 RAS. Genetic analysis of sur-5 revealed that a sur-5(lf) mutation selectively suppresses one group of let-60(dn) alleles but not the other, supporting the hypothesis that there could be more than one activator for LET-60 RAS (T.Gu et al., 1998). let-60(dn) alleles have been shown to affect signaling by titrating upstream activators rather than downstream effectors (Han and Sternberg, 1991). At least seven additional putative guanine nucleotide exchange factors have been predicted from the C.elegans genome sequence (as of April 2000), and one or more of these could be the hypothetical activator.
Regulation of RTK pathways by PTP-2
Csw and SHP-2 are PTP-2 homologs in Drosophila and vertebrates, respectively. Csw is normally required for Sevenless, Breathless, Torso and DER pathways, while SHP-2 has been shown to function in EGFR, PDGFR, FGFR and HGFR signaling pathways. The best characterized substrates for Csw/SHP-2 are Daughter of sevenless (Dos)-type proteins (Herbst et al., 1996; Raabe et al., 1996; H.H.Gu et al., 1998). It is believed that dephosphorylation of Dos by Csw generates a positive downstream signal. However, it remains unclear how Dos dephosphorylation creates such a positive signal. It is conceivable that PTP-2 might dephosphorylate specific phospho-tyrosyl residue(s) on a Dos-like protein, thus controlling the composition of an assembled signaling complex. Based on our results that PTP-2 function could be dependent upon SOS-1, several models for PTP-2 action can be proposed (Figure 6). PTP-2 could modulate the SEM-5-directed recruitment of SOS-1 or the catalytic activity of SOS-1. Alternatively, PTP-2 could release a negative regulator, such as SLI-1, from the signaling complex.
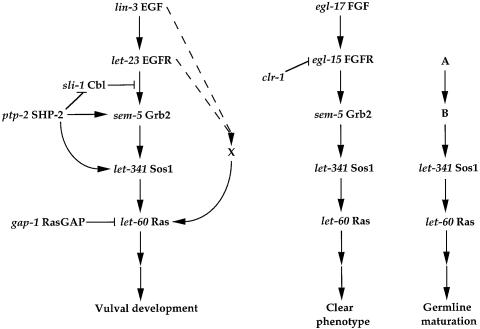
Fig. 6. Models for EGFR, FGFR and an unknown RTK pathways. The order of linear pathways was either determined by double mutants analyses or inferred from biochemical assays in other model systems. For example, the relationship between sem-5 Grb2 and let-341 Sos1 was adapted from the previous molecular characterization of mammalian counterparts. Arrows indicate a positive effect and bars indicate a negative effect. For simplicity, only events upstream of RAS are presented. The dashed pathway X is insufficient to allow signaling in the absence of let-60 activity. The putative relationships between ptp-2 and target genes are yet to be defined. A, B and X are hypothetical receptor tyrosine kinase, adaptor and RasGEF, respectively.
SOS-1 acts in multiple, RAS-dependent signaling pathways
SEM-5 recruitment of SOS-1 to the cell membrane seems to be a common mechanism involved in transducing signals from the activated EGFR and FGFR to RAS (Figure 6). Although SOS-1 also participates in signaling during the maturation of germline cells, how it is directed and to what signal it responds are unclear. Regulation of signaling upstream of RAS has been intensively studied from EGFR by using vulval development as a major paradigm. Several negative regulators have been previously implicated as affecting RAS activation by LET-23, including SLI-1/Cbl, UNC-101/AP47 and GAP-1/RasGAP. Biochemical studies suggest that RasGAP inhibits the signaling by directly acting on Ras (Boguski and McCormick, 1993), while Cbl desensitizes the signaling through binding to active receptors (Meisner et al., 1997). It is not clear how the UNC-101 adaptor antagonizes the RAS signaling pathway. In this report, we have provided evidence for the existence of a SOS-1-independent signaling pathway, which is dependent upon ligand, independent of PTP-2, and is not negatively regulated by SLI-1. Recently, Cbl was shown to function as a ubiquitin-protein ligase for active RTKs (Joazeiro et al., 1999). The failure to see an effect of SLI-1 on the SOS-1-independent pathway argues that promoting the degradation of active EGFR might not be the major activity of SLI-1. SLI-1 and another negative regulator of LET-23 EGFR signaling, ARK-1, both have proline-rich domains that can interact with the SH3 domain of SEM-5 (C.Yoon, C.Chang, P.Sternberg, unpublished; Hopper et al., 2000). Competition between SOS-1 and negative regulators for binding to the SEM-5 adaptor is a potential mechanism of direct negative regulation of LET-60 RAS activation. Alternatively, the SH3 domains of SEM-5 may, in addition to the recruitment of SOS-1, also recruit negative regulators simultaneously with SOS-1. It is less likely that ptp-2 activates signaling by inhibiting gap-1 function, since a ptp-2 mutation did not further reduce the vulval induction in let-60(gf); let-341(null) mutants, and a let-60(gf); let-341(null) mutant is sensitive to gap-1 regulation (Tables II and III). Regulation of Sos1 by phosphorylation has been proposed previously (Buday et al., 1995; Hu and Bowtell, 1996). Enhanced phosphorylation of Sos1, while not altering its catalytic activity for Ras, weakens its binding affinity for Grb2 or uncouples the Sos1–Grb2 complex from tyrosine kinase substrates (Buday and Downward, 1993). Although Sos1 is phosphorylated mostly on serine and threonine, regulated tyrosine phosphorylation has never been ruled out for Sos1. Therefore, it is possible that protein tyrosine kinases/phosphatases might modulate RAS signaling strength by directly regulating the phosphorylation state of SOS-1.
Materials and methods
Strains
Strains were handled according to the standard protocol and maintained at 20°C (Brenner, 1974). The following alleles were used: for LGII, let-23(sa62), ptp-2(op194) (Gutch et al., 1998), unc-4(e120), clr-1(e1745) (Hedgecock et al., 1990); for LGIV, let-60(n1046) (Ferguson and Horvitz, 1985), unc-31(e169); for LGV, him-5(e1490), let-341(s1031) (Johnsen and Baillie, 1991), let-341(n2183) (M.Stern, unpublished), let-341(ku231) (Z.Chen and M.Han, unpublished); for LGX, lin-15(n765), sem-5(ay73) (M.Stern, unpublished), gap-1(n1691) (Hajnal et al., 1997), sli-1(sy143).
Construction of double and triple mutants
Double and triple mutants were constructed using standard genetic methods, and markers used are indicated in the tables. For let-60(n1046); let-341(s1031) and let-60(n1046); sli-1(sy143) double mutants and for let-60(n1046); let-341(s1031); sem-5(ay73), let-60(n1046); let-341 (s1031); gap-1(n1691), let-60(n1046); let-341(s1031); sli-1(sy143) and ptp-2(op194); let-60(n1046); let-341(s1031) triple mutants, the presence of let-341, sli-1, sem-5, and gap-1 mutations was confirmed by sequencing the appropriate region of genomic DNA from each strain.
sos-1 cDNA isolation and allele sequencing
Partial sos-1 cDNA was derived from an incomplete genomic sequence, which was provided by C.elegans Sequencing Consortium (1998) and predicted to encode a Sos homolog using the program GeneFinder. RT–PCRs were performed to rectify the GeneFinder prediction and confirmed our further derivation based on the comparative analyses. Two splice variants were recovered in addition to the major transcript. One variant results from failure to use the normal splice donor site of intron 12. The other results from usage of an alternative splice acceptor site in intron 13 (AG/GAATTGCCGAACATATACTAATCACTCTAA). 5′ and 3′ RACE reactions were carried out according to the manufacturer’s instructions to obtain sequences corresponding to the 5′ and 3′ ends of the sos-1 transcript (5′/3′ RACE kit, Boehringer Mannheim).
To identify molecular lesions, PCR was performed on both reverse-transcribed RNA and genomic DNA from mutants, the products were purified and sequenced. The consequence of splice site mutation was determined by PCR amplification of reverse-transcribed RNA from mutants and sequencing purified PCR fragments directly.
RNAi
dsRNA corresponding to sos-1 was generated by in vitro transcription using 1.6 kb sos-1 cDNA corresponding to the nucleotides 1045–2613 relative to the initiation codon inserted into pCRII-TOPO (Invitrogen). Transcripts were prepared using T7 and Sp6 RNA polymerase and annealed prior to injection (Fire et al., 1998). Progeny of injected animals were assayed at 20°C unless indicated otherwise.
Vulval induction assay
The extent of vulval induction was determined by examining vulval anatomy in the early to mid-L4 stage of development under Nomarski optics using a Plan 100 objective (Sternberg and Horvitz, 1986). Wild-type animals have three VPCs generating vulval progeny; vulvaless animals have fewer than three VPCs generating vulval progeny; multivulva or hyperinduced animals have more than three VPCs generating vulval progeny. In some cases, a VPC generates one daughter that makes vulval progeny and another daughter that becomes nonvulval epidermis; such VPCs are scored as one-half VPC differentiating into vulval tissue.
Cell ablation
Ablations of the AC were performed with a laser microbeam as described by Sulston and White (1980).
DAPI staining
Animals were washed in M9 buffer before picked into a small drop of Vectashield with 4′,6-diamidino-2-phenylindole (Vector Laboratories) in a depression slide. After 30–60 min at room temperature, animals were dried in a vacuum for 1 h before mounted on agarose pads for observation.
Acknowledgments
Acknowledgements
We thank M.Stern for providing his unpublished let-341(n2183) and sem-5(ay73) alleles and for discussion of FGF receptor signaling; Z.Chen and M.Han for their generosity in providing their unpublished let-341(ku231) allele; and N.Moghal and J.Alberola-Ila for critical reading of this manuscript. Some strains were provided by the Caenorhabditis Genetics Center, supported by the National Center for Research Resources of the National Institutes of Health. This work has been supported by US Public Health Service grant (HD23690) to P.W.S., an investigator with the Howard Hughes Medical Institute. C.C. is funded by a NIH predoctoral training grant (GM07616).
References
- Alroy I. and Yarden,Y. (1997) The ErbB signaling network in embryogenesis and oncogenesis: signal diversification through combinatorial ligand-receptor interactions. FEBS Lett., 410, 83–86. [DOI] [PubMed] [Google Scholar]
- Aroian R.V. and Sternberg,P.W. (1991) Multiple functions of let-23, a C. elegans receptor tyrosine kinase gene required for vulval induction. Genetics, 128, 251–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroian R.V., Koga,M., Mendel,J.E., Ohshima,Y. and Sternberg,P.W. (1990) The let-23 gene necessary for Caenorhabditis elegans vulval induction encodes a tyrosine kinase of the EGF receptor subfamily. Nature, 348, 693–699. [DOI] [PubMed] [Google Scholar]
- Beitel G., Clark,S. and Horvitz,H.R. (1990) The Caenorhabditis elegans ras gene let-60 acts as a switch in the pathway of vulval induction. Nature, 348, 503–509. [DOI] [PubMed] [Google Scholar]
- Boguski M.S. and McCormick,F. (1993) Proteins regulating Ras and its relatives. Nature, 366, 643–654. [DOI] [PubMed] [Google Scholar]
- Brenner S. (1974) The genetics of Caenorhabditis elegans.Genetics, 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buday L. and Downward,J. (1993) Epidermal growth factor regulates P21ras through the formation of a complex of receptor, GRB2 adaptor protein and Sos nucleotide exchange factor. Cell, 73, 611–620. [DOI] [PubMed] [Google Scholar]
- Buday L., Warne,P.H. and Downward,J. (1995) Downregulation of the Ras activation pathway by MAP kinase phosphorylation of Sos. Oncogene, 11, 1327–1331. [PubMed] [Google Scholar]
- Chamberlin H.M. and Sternberg,P.W. (1994) The lin-3/let-23 pathway mediates inductive signaling during male spicule development in Caenorhabditis elegans.Development, 120, 2713–2721. [DOI] [PubMed] [Google Scholar]
- Chang C., Newman,A.P. and Sternberg,P.W. (1999) Reciprocal EGF signaling back to the uterus from the induced C. elegans vulva coordinates morphogenesis of epithelia. Curr. Biol., 9, 237–246. [DOI] [PubMed] [Google Scholar]
- Chardin P., Camonis,J.H., Gale,N.W., van Aelst,L., Schlessinger,J., Wigler,M.H. and Bar-Sagi,D. (1993) Human Sos1: a guanine nucleotide exchange factor for Ras that binds to GRB2. Science, 260, 1338–1343. [DOI] [PubMed] [Google Scholar]
- Chen E.B., Branda,C.S. and Stern,M.J. (1997) Genetic enhancers of sem-5 define components of the gonad-independent guidance mechanism controlling sex myoblast migration in Caenorhabditis elegans hermaphrodites. Dev. Biol., 182, 88–100. [DOI] [PubMed] [Google Scholar]
- Cheng A.M. et al. (1998) Mammalian Grb2 regulates multiple steps in embryonic development and malignant transformation. Cell, 95, 793–803. [DOI] [PubMed] [Google Scholar]
- Church D., Guan,K.-L. and Lambie,E.J. (1995) Three genes of the MAP kinase cascade, mek-2, mpk-1/sur-1 and let-60 ras, are required for meiotic cell cycle progression in Caenorhabditis elegans.Development, 121, 2525–2535. [DOI] [PubMed] [Google Scholar]
- Clandinin T.R., DeModena,J.A. and Sternberg,P.W. (1998) Inositol trisphosphate mediates a RAS-independent response to LET-23 receptor kinase activation in C. elegans.Cell, 92, 523–533. [DOI] [PubMed] [Google Scholar]
- Clark S.G., Stern,M.J. and Horvitz,H.R. (1992a) C. elegans cell-signalling gene sem-5 encodes a protein with SH2 and SH3 domains. Nature, 356, 340–344. [DOI] [PubMed] [Google Scholar]
- Clark S.G., Stern,M.J. and Horvitz,H.R. (1992b) Genes involved in two Caenorhabditis elegans cell-signaling pathways. Cold Spring Harbor Symp. Quant. Biol., 57, 363–373. [DOI] [PubMed] [Google Scholar]
- Clark S.G., Lu,X. and Horvitz,H.R. (1994) The C. elegans locus lin-15, a negative regulator of a tyrosine kinase signaling pathway, encodes two different proteins. Genetics, 137, 987–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson E. and Horvitz,H.R. (1985) Identification and characterization of 22 genes that affect the vulval cell lineages of Caenorhabditis elegans.Genetics, 110, 17–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans.Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Gu H.H., Pratt,J.C., Burakoff,S.J. and Neel,B.G. (1998) Cloning of p97/Gab2, the major SHP2-binding protein in hematopoietic cells, reveals a novel pathway for cytokine-induced gene activation. Mol. Cell, 2, 729–740. [DOI] [PubMed] [Google Scholar]
- Gu T., Satoshi,O. and Han,M. (1998) Caenorhabditis elegans SUR-5, a novel but conserved protein, negatively regulates LET-60 Ras activity during vulval induction. Mol. Cell. Biol., 18, 4556–4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutch M.J., Flint,A.J., Keller,J., Tonks,N.K. and Hengartner,M.O. (1998) The Caenorhabditis elegans SH2 domain-containing protein tyrosine phosphatase PTP-2 participates in signal transduction during oogenesis and vulval development. Genes Dev., 12, 571–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnal A., Whitfield,C.W. and Kim,S.K. (1997) Inhibition of Caenorhabditis elegans vulval induction by gap-1 and by let-23 receptor tyrosine kinase. Genes Dev., 11, 2715–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M. and Sternberg,P.W. (1990) let-60, a gene that specifies cell fates during C. elegans vulval induction, encodes a ras protein. Cell, 63, 921–931. [DOI] [PubMed] [Google Scholar]
- Han M. and Sternberg,P.W. (1991) Analysis of dominant negative mutations of the Caenorhabditis elegans let-60 ras gene. Genes Dev., 5, 2188–2198. [DOI] [PubMed] [Google Scholar]
- Han M., Golden,A., Han,Y. and Sternberg,P.W. (1993) C. elegans lin-45 raf gene participates in let-60 ras stimulated vulval differentiation. Nature, 363, 133–140. [DOI] [PubMed] [Google Scholar]
- Hedgecock E.M., Culotti,J.G. and Hall,D.H. (1990) The unc-5, unc-6 and unc-40 genes guide circumferential migrations of pioneer axons and mesodermal cells on the epidermis in C. elegans. Neuron, 4, 61–85. [DOI] [PubMed] [Google Scholar]
- Heldin C.-H. (1995) Dimerization of cell surface receptors in signal transduction. Cell, 80, 213–223. [DOI] [PubMed] [Google Scholar]
- Herbst R., Carroll,P.M., Allard,J.D., Schilling,J., Raabe,T. and Simon,M.A. (1996) Daughter of Sevenless is a substrate of the phosphotyrosine phosphatase corkscrew and functions during Sevenless signaling. Cell, 85, 899–909. [DOI] [PubMed] [Google Scholar]
- Hill R.J. and Sternberg,P.W. (1992) The lin-3 gene encodes an inductive signal for vulval development in C. elegans. Nature, 358, 470–476. [DOI] [PubMed] [Google Scholar]
- Hopper N.A., Lee,J. and Sternberg,P.W. (2000) ARK-1 inhibits EGFR signaling in C. elegans. Mol. Cell, in press. [PubMed] [Google Scholar]
- Horvitz H.R. and Sternberg,P.W. (1991) Multiple intercellular signalling systems control the development of the C. elegans vulva. Nature, 351, 535–541. [DOI] [PubMed] [Google Scholar]
- Hu Y. and Bowtell,D.D. (1996) Sos1 rapidly associates with Grb2 and is hypophosphorylated when complexed with the EGF receptor after EGF stimulation. Oncogene, 12, 1865–1872. [PubMed] [Google Scholar]
- Huang L.S., Tzou,P. and Sternberg,P.W. (1994) The lin-15 locus encodes two negative regulators of Caenorhabditis elegans vulval development. Mol. Biol. Cell, 5, 395–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L. and Sternberg,P.W. (1998) Interactions of EGF, Wnt and HOM-C genes specify P12 neuroectoblast fate in C. elegans.Development, 125, 2337–2347. [DOI] [PubMed] [Google Scholar]
- Joazeiro C.A., Wing,S.S., Huang,H., Leverson,J.D., Hunter,T. and Liu,Y.C. (1999) The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science, 286, 309–312. [DOI] [PubMed] [Google Scholar]
- Johnsen R.C. and Baillie,D.L. (1991) Genetic analysis of a major segment [LGV(left)] of the genome of Caenorhabditis elegans.Genetics, 129, 735–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongeward G.D., Clandinin,T.R. and Sternberg,P.W. (1995) sli-1, a negative regulator of let-23-mediated signaling in C. elegans.Genetics, 139, 1553–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanaugh W.M. and Williams,L.T. (1994) An alternative to SH2 domains for binding tyrosine-phosphorylated proteins. Science, 266, 1862–1865. [DOI] [PubMed] [Google Scholar]
- Kavanaugh W.M., Turck,C.W. and Williams,L.T. (1995) PTB domain binding to signaling proteins through a sequence motif containing phosphotyrosine. Science, 268, 1177–1179. [DOI] [PubMed] [Google Scholar]
- Kokel M., Borland,C.Z., DeLong,L., Horvitz,H.R. and Stern,M.J. (1998) clr-1 encodes a receptor tyrosine phosphatase that negatively regulates an FGF receptor signaling pathway in Caenorhabditis elegans. Genes Dev., 12, 1425–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld K., Guan,K.-L. and Horvitz,H.R. (1995) The Caenorhabditis elegans gene mek-2 is required for vulval induction and encodes a protein similar to the protein kinase MEK. Genes Dev., 9, 756–768. [DOI] [PubMed] [Google Scholar]
- Lackner M.R., Kornfeld,K., Miller,L.M., Horvitz,H.R. and Kim,S.K. (1994) A MAP kinase homolog, mpk-1, is involved in ras-mediated induction of vulval cell fates in Caenorhabditis elegans. Genes Dev., 8, 160–173. [DOI] [PubMed] [Google Scholar]
- Lemmon M.A. and Schlessinger,J. (1994) Regulation of signal transduction and signal diversity by receptor oligomerization. Trends Biochem. Sci., 19, 459–463. [DOI] [PubMed] [Google Scholar]
- Meisner H., Daga,A., Buxton,J., Fernandez,B., Chawla,A., Banerjee,U. and Czech,M.P. (1997) Interactions of Drosophila Cbl with epidermal growth factor receptors and role of Cbl in R7 photoreceptor. Mol. Cell. Biol., 17, 2217–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A.P. and Sternberg,P.W. (1996) Coordinated morphogenesis of epithelia during development of the Caenorhabditis elegans uterine-vulval connection. Proc. Natl Acad. Sci. USA, 93, 9329–9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimnual A.S., Yatsula,B.A. and Bar-Sagi,D. (1998) Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science, 279, 560–563. [DOI] [PubMed] [Google Scholar]
- Panayotou G. and Waterfield,M.D. (1993) The assembly of signalling complexes by receptor tyrosine kinases. Bioessays, 15, 171–177. [DOI] [PubMed] [Google Scholar]
- Pawson T. (1995) Protein modules and signalling networks. Nature, 373, 573–580. [DOI] [PubMed] [Google Scholar]
- Pawson T. and Saxton,T.M. (1999) Signaling networks–do all roads lead to the same genes? Cell, 97, 675–678. [DOI] [PubMed] [Google Scholar]
- Pelicci G. et al. (1992) A novel transforming protein (SHC) with an SH2 domain is implicated in mitogenic signal transduction. Cell, 70, 93–104. [DOI] [PubMed] [Google Scholar]
- Raabe T., Riesgo-Escovar,J., Liu,X., Bausenwein,B.S., Deak,P., Maröy,P. and Hafen,E. (1996) DOS, a novel pleckstrin homology domain-containing protein required for signal transduction between Sevenless and Ras1 in Drosophila.Cell, 85, 911–920. [DOI] [PubMed] [Google Scholar]
- Schlessinger J. and Ullrich,A. (1992) Growth factor signaling by receptor tyrosine kinases. Neuron, 9, 383–391. [DOI] [PubMed] [Google Scholar]
- Scita G., Nordstrom,J., Carbone,R., Tenca,P., Giardina,G., Gutkind,S., Bjarnegard,M., Betsholtz,C. and Di Fiore,P.P. (1999) EPS8 and E3B1 transduce signals from Ras to Rac. Nature, 401, 290–293. [DOI] [PubMed] [Google Scholar]
- Selfors L.M., Schultzman,J.L., Borland,C.Z. and Stern,M.J. (1998) soc-2 encodes a leucine-rich repeat protein implicated in fibroblast growth factor receptor signaling. Proc. Natl Acad. Sci. USA, 95, 6903–6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G., Savage,C. and Greenwald,I. (1993) Isolation and characterization of mutations causing abnormal eversion of the vulva in Caenorhabditis elegans. Dev. Biol., 157, 423–436. [DOI] [PubMed] [Google Scholar]
- Sharp P.A. (1999) RNAi and double-strand RNA. Genes Dev., 13, 139–141. [PubMed] [Google Scholar]
- Songyang Z. et al. (1993) SH2 domains recognize specific phosphopeptide sequences. Cell, 72, 767–778. [DOI] [PubMed] [Google Scholar]
- Songyang Z. et al. (1994) Specific motifs recognized by the SH2 domains of Csk, 3BP2, fps/fes, GRB-2, HCP, SHC, Syk and Vav. Mol. Cell. Biol., 14, 2777–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern M.J. and Horvitz,H.R. (1991) A normally attractive cell interaction is repulsive in two C. elegans mesodermal cell migration mutants. Development, 113, 797–803. [DOI] [PubMed] [Google Scholar]
- Sternberg P.W. and Horvitz,H.R. (1986) Pattern formation during vulval development in Caenorhabditis elegans.Cell, 44, 761–772. [DOI] [PubMed] [Google Scholar]
- Sternberg P.W. and Han,M. (1998) Genetics of RAS signaling in C. elegans. Trends Genet., 14, 466–472. [DOI] [PubMed] [Google Scholar]
- Sulston J.E. and White,J.G. (1980) Regulation and cell autonomy during postembryonic development of Caenorhabditis elegans. Dev. Biol., 78, 577–597. [DOI] [PubMed] [Google Scholar]
- Sundaram M. and Han,M. (1995) The C. elegans ksr-1 gene encodes a novel Raf-related kinase involved in ras-mediated signal transduction. Cell, 83, 889–901. [DOI] [PubMed] [Google Scholar]
- Sundaram M., Yochem,J. and Han,M. (1996) A Ras-mediated signal transduction pathway is involved in the control of sex myoblast migration in Caenorhabditis elegans.Development, 122, 2823–2833. [DOI] [PubMed] [Google Scholar]
- Ullrich A. and Schlessinger,J. (1990) Signal transduction by receptors with tyrosine kinase activity. Cell, 61, 203–212. [DOI] [PubMed] [Google Scholar]
- Wu Y. and Han,M. (1994) Suppression of activated Let-60 Ras protein defines a role of C. elegans Sur-1 MAP kinase in vulval differentiation. Genes Dev., 8, 147–159. [DOI] [PubMed] [Google Scholar]
- Wu Y., Han,M. and Guan,K.-L. (1995) MEK-2, a Caenorhabditis elegans MAP kinase kinase, functions in Ras-mediated vulval induction and other developmental events. Genes Dev., 9, 724–755. [DOI] [PubMed] [Google Scholar]
- Yoon C.H., Lee,J., Jongeward,G.D. and Sternberg,P.W. (1995) Similarity of sli-1, a regulator of vulval development in Caenorhabditis elegans, to the mammalian proto-oncogene, c-cbl.Science, 269, 1102–1105. [DOI] [PubMed] [Google Scholar]