Abstract
The immunologic and virologic outcome of therapeutic DNA-vaccines administered during antiretroviral therapy (ART) using electroporation with or without (interleukin) IL-2 treatment was evaluated in the SIV-mac239/macaque model. Rhesus macaques inoculated with pathogenic SIVmac239 were treated with ART [(R(-9-(2-phosphonomethoxypropyl) adenine) (PMPA), FTC, Zerit®] from weeks 13 to 41 postinfection (wpi). Group 1 (n = 7) received ART only, groups 2 and 3 (each n = 6) additionally received SIVmac239-derived gp140Env, GagPol, and TatRevNef plasmids by in vivo electroporation at 22, 26, 30, and 34 wpi, and group 3 also IL-2 for 14 days after each vaccination. Endpoints evaluated were viral load, Gag181–189-specific CD8+ T-cell responses in MamuA01+ animals, lymphoproliferative responses, and CD4 T-cell counts. Viremia in all animals dropped below 200 RNA copies/ml during ART. Frequencies of Gag181–189-specific CD8+ T cells prior to ART were detectable in all three groups (1.27–3.01%) and increased significantly (p < 0.01) postvaccination with maximum responses after the fourth immunization (0.2% versus 3.49–7.15%). Gag181–189-specific CD8+ T-cell frequencies increased post-ART cessation in all groups and remained at significantly higher levels (p < 0.001) until the end of the study (75 wpi) in both groups of vaccinated animals. Lymphoproliferative responses were detected against Gag in a limited number of animals after vaccination and post-ART. However, plasma RNA viral loads rebounded after ART termination to similar levels in all three groups, but remained below 105 copies/ml until the end of the study, which could be a late effect of the triple drug therapy.
Introduction
Currently, the only licensed treatment for HIV-1-infected patients is highly active antiretroviral therapy (HAART). However, these drugs are costly and need to be taken according to strict dosing schedules. Furthermore, HAART can cause serious side effects, which are difficult to manage in the light of the lifelong dependency on these drugs.1 Virus reservoirs inaccessible to HAART remain in certain host tissues and thus are a source of viral rebound if therapy is discontinued or inadequate.2,3 Consequently, in the event of HAART cessation, most patients experience a rebound of plasma viremia to high levels with the concomitant emergence of drug-resistant strains that evolved in the viral reservoir during the treatment period.4 In addition, HAART treatment alone does not reconstitute anti-HIV-1 immune responses, particularly in chronically infected patients.5,6
Therapeutic vaccination in nonhuman primate models has been shown to boost antiviral immune responses during HAART, resulting in reduced viral levels for up to 3–6 months.7,8 Patients are most likely to be diagnosed with HIV infection in the chronic phase, when a substantial loss of memory CD4+ T cells, including HIV-1-specific memory CD4+ T cells, has already occurred. Consequently, these patients are unable to mount a potent anti-HIV-1 response9 and, in general, do not recover this response during HAART.10 Therefore, immune restoration by therapeutic vaccination could be a very important tool for the improved control of HIV replication in combination with HAART or as standalone therapy for those patients who cannot tolerate these drugs.
In the present study, the effect of DNA vaccination with or without interleukin (IL)-2 protein treatment on virologic and immunologic surrogate markers was evaluated in chronically infected rhesus macaques under ART. We have previously shown that the potency of HIV-1 DNA vaccines can be remarkably enhanced by the use of electroporation.11 The objective of this study was to determine whether DNA vaccination enhanced by electroporation could induce strong immune responses in rhesus macaques under ART and whether these immune responses have the potential to reduce the rebound in viremia after ART cessation. The second goal was to evaluate the immune-enhancing effect of IL-2 administration concomitant to therapeutic DNA vaccination, as IL-2 was previously shown to increase the frequency of CD25+ (IL-2R) T cells, thus boosting antigen-specific cellular immune responses when given in combination with a prophylactic HIV-1 Gag DNA vaccine.12
Materials and Methods
Plasmid DNA cassettes
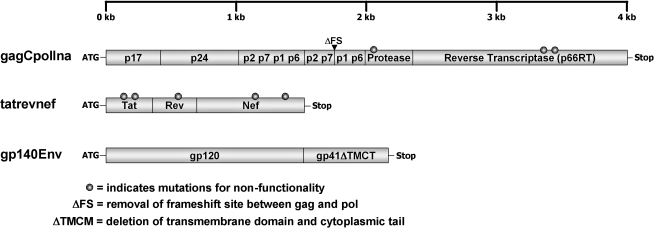
A panel of codon-optimized expression cassettes based on the amino acid sequences of SIVmac239 (Genbank # M33262) Gag, Pol, Tat, Rev, Nef, and Env antigens was designed (Fig. 1). All gene cassettes were cloned into the eukaryotic expression vector pCMVKm2 that contained the cytomegalovirus immediate early enhancer/promoter and bGH terminator (Chiron Corporation, Emeryville, CA)13 To further enhance the translation efficiency of all expression cassettes, an optimal “Kozak” consensus sequence (GCCACCAUGG) prior to the start codon sequence (AUG) was inserted for optimal recognition of the translational start site by the ribosome.14
FIG. 1.
Diagram of expression cassettes used in animal studies. All genes were based on the amino acid sequences of SIV-mac239 Gag, Pol, Tat, Rev, Nef, and Env and codon optimized for increased expression in primate cells. The cassettes were cloned into the eukaryotic expression vector pCMVKm2 that contained the cytomegalovirus immediate early enhancer/promoter. The gagCpolIna and tatrevnef genes were rendered nonfunctional by selected mutations as detailed in the Materials and Methods section.
The simian immunodeficiency virus (SIV) gagCpolIna polyprotein expression cassette was designed using a similar strategy as previously described for HIV-1.15 Briefly, the entire integrase coding sequence in pol was deleted to prevent integration of viral genome into cellular DNA, and the catalytic triad and primer grip regions of the reverse transcriptase (RT) coding sequences were deleted to inactivate RT enzymatic activity.16,17 In addition, the frameshift region was removed by insertion of an extra (T) nucleotide at the p1 “slippery sequence” (TTTTTTA) in order to express gag and pol genes in frame. The pol region included p1p6Pol-coding sequences up to RNAse H for gagpol. To include p1p6Gag, the p2p7p1p6 fragment was added to get gag-complete plus pol. Protease was inactivated (Ina) by the introduction of a specific point mutation,18 resulting in the gagCpolIna cassette (Fig. 1).
The nucleotide sequences coding for tat, rev, and nef were assembled in frame to express a single unprocessed polyprotein. All genes were rendered nonfunctional for safety reasons. The mutations introduced in Tat (C50G and C66S) were based on mutations in the cysteine-rich region of the shorter HIV-1 Tat sequence (C22G and C37S).19 For the inactivation of the Rev function, the previously described HIV-1 revM10 mutant was prepared for the SIV rev analogue.20 The SIV-mac239 premature stop codon within nef was removed (Stop93E; TAA → GAG) to express the full-length protein. To abolish CD4 and MHC downregulation, two other mutations were introduced in nef (D155L and Y223F).21
The SIVmac239 Envelope protein was expressed as secreted gp140 without the transmembrane region and cytoplasmic tail using tissue plasminogen activator as the signal leader sequence.
Mouse studies and cytokine flow cytometry
Three groups of 6- to 8-week-old female BALB/c mice (n = 5) were vaccinated at weeks 0 and 4 bilaterally in the tibialis anterior muscles with 45 μg of gagCpolIna, tatrevnef, or gp140env plasmid DNA in isotonic saline. Spleens were harvested 2 weeks after the last vaccination, and isolated splenocytes were subsequently stimulated in duplicate samples with SIVmac239 Gag protein (2 μg/well) or SIVmac239-based Gag, Pol, Env, or TatRevNef overlapping peptide pools (0.25 μg each peptide/well; obtained from the NIH AIDS Research and Reference Reagent Program). Unstimulated cells and spleen cells from naïve mice were used as background or negative controls, respectively. After 5 h of stimulation, cells were washed and incubated with anti-CD16/32 (Pharmingen, San Diego, CA), fixed in 1% (w/v) paraformaldehyde, and stored overnight at 4ºC. On the following day, cells were stained with fluorescein isothiocyanate (FITC) conjugated anti-CD8 monoclonal antibody (mAb) or with Cy5.5-conjugated anti-CD4 mAb (Pharmingen), washed with phosphate-buffered saline (PBS), treated with 0.5% (w/v) saponin (Sigma, St. Louis, MO), and then incubated with phycoerythrin-conjugated mouse interferon-γ (IFN-γ) mAb (Pharmingen) in the presence of 0.1% (w/v) saponin. Analysis of IFN-γ-secreting CD4+ and CD8+ T cells was performed with a FACSCalibur flow cytometer (Becton Dickinson Immunocytometry Systems, San Jose, CA). The background values (unstimulated sample) were then subtracted from each stimulated sample to generate the final values for % IFN-γ-secreting CD4+ or CD8+ T cells.
Rhesus macaques and treatment/vaccination schedule
Thirty Indian rhesus macaques (Macaca mulatta) were included at the beginning of the study. Housing and handling was performed in accordance with the standards of the Association for Assessment and Accreditation of Laboratory Animal Care, the Animal Welfare Act as amended, and the Public Health Service Policy. All animals were tested seronegative for SIV, simian retrovirus, simian T-cell lymphotropic virus type 1, and herpesvirus B before the study. The MamuA01 MHC allele status was determined by discriminatory polymerase chain reaction (PCR).22 All animals were inoculated intravenously with 1,000 MID50 (50% monkey infectious doses) of pathogenic SIVmac239.
Antiretroviral treatment consisted of 30 mg/kg once daily in the first month, then 20 mg/kg once daily R(-9-(2-phosphonomethoxypropyl) adenine) (PMPA) subcutaneously; 50 mg/kg once daily FTC (emtricitabine) subcutaneously, and 1.2 mg/kg twice daily Zerit® (2′-3′-didehydro-2′-3′-dideoxythymidine) orally. IL-2 (Proleukin®, Chiron) was administered at 50,000 IU/kg twice a day subcutaneously, which was within the nontoxic dose range estimated elsewhere.23
Vaccinations were performed by injection of 0.5 ml DNA in saline bilaterally into the quadriceps and deltoid muscles (2 mg each of gagCpolIna, tatrevnef, and gp140Env plasmids combined to 6 mg total DNA/2 ml saline). Immediately after each DNA injection, a four-needle array electrode (electrode gap 0.86 cm) was inserted at the exact site of injection and a total of two pulses of 206 V/60 ms each (4 Hz) were applied with a MedPulser DNA Delivery system (Genetronics, Inc., a subsidiary of Inovio Biomedical Corp., San Diego, CA).24,25
Plasma viral load determination
Plasma viral load was determined by real time RT-PCR on an ABI 7700 sequence detector (Applied Biosystems, Foster City, CA). Briefly, viral RNA was isolated from plasma using RNA-STAT 60 reagent (Tel-test, Friendswood, TX) following the manufacturer's instructions. The isolated RNA samples were reverse transcribed and amplified by RT-PCR as previously described26 with the following modifications: The RNA samples were added to a mix of One-Step RT-PCR Master Mix (Applied Biosystems) with primers and probe designed to amplify a conserved region of the SIVmac239 gag gene: (1) forward primer, SIV-F 5′ AGTATGGGCAGC-AAATGAAT 3′; (2) reverse primer, SIV-R 5′ TTCTCTT-CTGCGTGAATGC 3′; and (3) the probe, SIV-P 6FAM-AGATTTGGATTAGCAGAAAGCCTGTTGGA-TAMRA. The reaction samples were reverse-transcribed at 48ºC for 30 min, held at 95ºC for 10 min, and then a cDNA-PCR reaction was run for 40 cycles at 95ºC for 15 s and 60ºC for 1 min. The signal was compared with a standard curve of known concentrations to determine the viral copies present in each sample; all samples were performed in triplicate along with the standard RNA templates.
MHC class I tetramer staining
Peripheral blood mononuclear cells (PBMCs) were isolated from whole ethylenediamine-tetraacetic acid (EDTA) blood using Histopaque cell separation solution (Sigma-Aldrich, St. Louis, MO) and analyzed by four-color flow cytometry for SIV-specific CD8+ T cells. Two million PBMCs were stained for 30 min at room temperature with anti-CD3-FITC and anti-CD8-PerCP monoclonal antibody conjugates (BD BioSciences, San Jose, CA) and phycoerythrin-conjugated MamuA01 Gag181-189 (p11C peptide) tetramer (iTAg MHC Tetramer; Beckman Coulter, Miami, FL). Samples were washed twice with PBS and fixed in 2% paraformaldehyde in PBS. At least 100,000 cells in the lymphocyte gate were collected on a FACSCalibur flow cytometer and analyzed using the Cell Quest software package (BD Biosciences). The corresponding antibody isotype control conjugates for the CD3 and CD8 antibodies were used to determine any background staining.
Lymphoproliferation assay
The lymphoproliferation assay was performed using SIV Gag and Env peptide pools provided by the NIH AIDS Research and Reference Reagent Program (Bethesda, MD). The peptides were 15-mers with 11 amino acid overlap, spanning the full Gag and Env sequences of SIVmac239. The Gag peptides pool consisted of 125 peptides and the Env peptides pool consisted of 218 peptides. Unstimulated cells were used as negative controls and Concanavalin A (Con A)- or phytohemaglutinin (PHA)-stimulated cells were used as positive controls. Briefly, PBMCs were isolated from EDTA blood by Histopaque separation (Sigma-Aldrich). Two hundred thousand (2 × 105) PBMCs/well in 100 μl were seeded in triplicate in a 96-well round-bottom plate and cultured at 37ºC with 5% CO2 for 4 days with either 100 μl of phytohemagglutinin (PHA) (20 mg/ml), Con A (20 mg/ml), SIV Gag or Env pool (2 μg/ml per peptide), or complete RPMI alone. [3H]-Thymidine (1 μCi; PerkinElmer, Wellesley, MA) was added to each well and incubated overnight at 37ºC. Cells were harvested with an Inotech Cell Harvester System. Radioactivity incorporated by the cells and plotted onto the glass fiber filters was measured using a 1450 Microbeta Jet counter (Gaithersburg, MD). Results were expressed as stimulation indices (SI) calculated by dividing the mean counts per minute (c.p.m.) of cells exposed to the Gag or Env peptides, PHA, or ConA by the mean c.p.m. of cells incubated with medium alone. An SI greater than 5 was considered positive.
Lymphocyte immunophenotyping
The CD4 lymphocyte subset was determined by flow cytometry. Whole blood collected in EDTA (110 μl) was incubated for 15 min at room temperature with a mixture of appropriate antibody conjugates. The monoclonal antibody conjugates used were anti-CD2-FITC, anti-CD20-PE, anti-CD3-FITC, and anti-CD4-PE (BD Biosciences). The corresponding mouse isotype control conjugates were also used to determine any background staining and to set up the gating. Cells were then incubated for 10 min at 37ºC with 2 ml of erythrocytes lysing buffer (BD Biosciences). Cells were washed twice with PBS resuspended in 2% paraformaldehyde in PBS. Ten thousand (10,000) events were collected on a FACSCalibur flow cytometer in the lymphocyte gate, and data were analyzed using Cell Quest software (BD Biosciences). The absolute number of cells per milliliter of blood was calculated for each lymphocyte subset, CD3+ CD4+ T lymphocytes, and CD2− CD20+ B lymphocytes, by multiplying the percentage of each lymphocyte population obtained from the FACS analysis to the absolute number of total lymphocytes obtained from cell blood counts using the VetScan HMT Hematology Analyzer (Abaxis Inc., Union City, CA).
Statistical analysis
Prior to vaccination, 19 animals were selected based on optimal viral control under ART, randomized using a random number generator algorithm,27 and assigned to groups 1, 2, and 3 (Table 1). Due to the high magnitude of the response variable “Viral Load,” the log transformation has been used. An analysis of variance (ANOVA) model was applied to the response variables “Viral Load,” and percentage of “Tetramer Responses” with three factors (“Time,” “Group,” and “Animal”) and factor “Animal” was nested in factor “Group.” A one-way ANOVA model was applied to tetramer responses at 13 weeks postinfection (wpi). Due to the high magnitude of the response variable “CD4 counts,” the log transformation has been used. An ANOVA model was applied to log “CD4 counts” with three factors (“Time,” “Group,” and “Animal”), and factor “Animal” was nested in factor “Group.” An ANOVA model was applied to log “CD4 counts” with four factors: “Time Group” (15–22 weeks and 24–36 weeks), “Time,” “Group,” and “Animal.” Factor “Animal” is nested in factor “Group,” and factor “Time” is nested in factor “Time Group.” The ANOVA results are summarized in Table 2. For the evaluation of the variables “Viral Load” and “CD4 counts” between two groups, t-test was performed using log-transformed values. These p values are indicated in the results section. All reported p values are two-sided. A probability value of p < 0.05 was considered statistically significant.
Table 1.
Overview of Treatment Groups, MamuA01 Status, Peak Viremia, and RNa Viral Load at Randomization (22 wpi)
| Group | Treatment | Animal number | MamuA0l status | Peak viremia prior ART (2–4 wpi) | Viral load at randomization (22 wpi) |
|---|---|---|---|---|---|
| 1 | ART only | #3447 | + | 4.6 × 106 | 543 |
| #3451 | − | 6.2 × 106 | <200 | ||
| #3452 | − | 4.1 × 106 | <200 | ||
| #3453 | − | 6.7 × 106 | 260 | ||
| #3511 | + | 1.2 × 107 | <200 | ||
| #3515 | + | 1.7 × 107 | <200 | ||
| #3684 | − | 1.5 × 107 | 620 | ||
| 2 | ART + DNA | #3444 | + | 3.0 × 106 | 333 |
| #3522 | + | 1.7 × 107 | <200 | ||
| #3524 | + | 1.3 × 107 | <200 | ||
| #3526 | − | 1.7 × 107 | 393 | ||
| #3527 | − | 2.1 × 107 | <200 | ||
| #3530 | − | 3.1 × 107 | 340 | ||
| 3 | ART + DNA + IL-2 | #3449 | − | 2.7 × 107 | 383 |
| #3512 | − | 2.5 × 106 | <200 | ||
| #3514 | + | 6.1 × 106 | <200 | ||
| #3516 | + | 4.1 × 107 | <200 | ||
| #3518 | − | 6.5 × 106 | 277 | ||
| #3525 | + | 3.8 × 107 | <200 |
Table 2.
Significancy of Differences in Viral Load, Tetramer Responses, and CD4 T Cell Counts Between Groups 1, 2, and 3
| |
|
Significancyaof difference in parameters between |
||
|---|---|---|---|---|
| Parameter evaluated | Time period (wpi) | Grp. 1 and 2b | Grp. 1 and 3 | Grp. 2 and 3 |
| Viral loadc | 43–50 | P = 0.2686 | P = 0.2320 | P = 0.0277 |
| 60–75 | P = 0.0550 | P < 0.0001 | P = 0.0019 | |
| Tetramerd | 13 | P = 0.5785 | P = 0.8371 | P = 0.7223 |
| 24–41 | P = 0.0038 | P < 0.0001 | P = 0.0433f | |
| 43–71 | P < 0.0001 | P < 0.0001 | P = 0.7740 | |
| CD4 T cellse | 15–22 | P < 0.0001 | P = 0.0359 | P < 0.0001 |
| 24–36 | P < 0.0001 | P < 0.0001 | P = 0.2198 | |
| 47–71 | P < 0.0001 | P < 0.0001 | P < 0.0001 | |
Significancy of variables between groups were determined by ANOVA as described in the Methods section. A probability value of P < 0.05 was considered statistically significant.
Grp. 1: ART only; Grp.2: ART + DNA; Grp.3: ART + DNA + IL-2.
Viral load as viral RNA copies/ml plasma.
SIVmac239Gag181–189-specific CD8+ T cell responses in MamuA01+ vaccinated and unvaccinated control animals measured by tetramer staining.
Number of CD4 T cells per μl whole blood.
Borderline significance between Grp.2 and 3 24–41 wpi based on a single high responding animal.
Results
Immunogenicity of DNA vaccines in mice
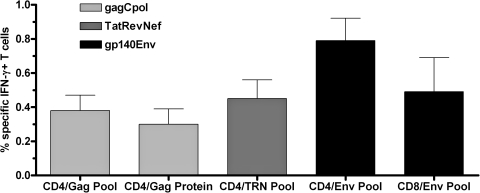
Prior to evaluation of the DNA vaccines in nonhuman primates, all constructs were tested in BALB/c mice. Splenocytes of vaccinated mice were harvested 2 weeks after the last vaccination and stimulated with SIVmac239 Gag protein or SIVmac239 Gag, Tat, Rev, Nef, Env, and Pol peptide pools. As shown in Figure 2, CD4+ T-cell responses were detected against the Gag, TatRevNef polyprotein, and Env antigens. The frequency of specific IFN-γ-secreting CD4+ T cells was between 0.3% (Gag protein) and 0.8% (Env pool). CD4 T-cell responses to Gag showed a small increase if stimulation was performed with the Gag peptide pool (0.4%) instead of Gag protein. H-2Dd-restricted CD8+ T-cell responses were only detectable against Env (0.8% specific IFN-γ-secreting T cells). Responses to Pol were not detectable, likely due to MHC-restriction in the inbred BALB/c strain. However, expression of the GagPol polyprotein (beside TatRevNef, and Env) was confirmed by Western blot (data not shown).
FIG. 2.
Quantitative analysis of % interferon-γ (IFN-γ)-secreting CD4 or CD8 T cells of BALB/c mice vaccinated with plasmid DNAs as indicated in the figure legend (gagCpolIna, tatrevnef, or gp140Env). Three groups of mice (n = 5) received two immunizations with DNA at weeks 0 and 4 and spleens were harvested 2 weeks after the second immunization. Pooled splenocytes were stimulated for 5 h with either SIVmac239 Gag protein or SIVmac239 peptide pools (Gag, TatRevNef (TRN), and Env) and cells were then stained for either CD4 or CD8 and intracellular IFN-γ on the next day as described in the Materials and Methods and analyzed by flow cytometry. Only detectable responses are shown in the graph (see text for details). Error bars represent the standard deviation from the mean of duplicate samples.
Infection and selection of rhesus macaques
Initially, 30 naïve rhesus macaques were inoculated with SIVmac239 (1,000 MID50 intravenously). All animals became infected and viral loads within the acute phase (4 weeks) of infection ranged from 2.53 × 106 to 4.07 × 107 RNA copies/ml (Table 1). Two animals were rapid progressors and had to be euthanized due to the development of AIDS-like symptoms 8 and 13 wpi, respectively. Another two animals were euthanized 4 and 5 weeks after start of ART treatment, respectively, due to opportunistic infections, and one animal died from cardiac arrest during anesthesia. At week 22, another six animals were excluded from the study due to incomplete virus control under ART. The remaining 19 animals, including nine MamuA01+ animals, were randomly distributed into three groups according to their MamuA01 status and viral load (Table 1).
Viral control and immune responses under ART before and after vaccination
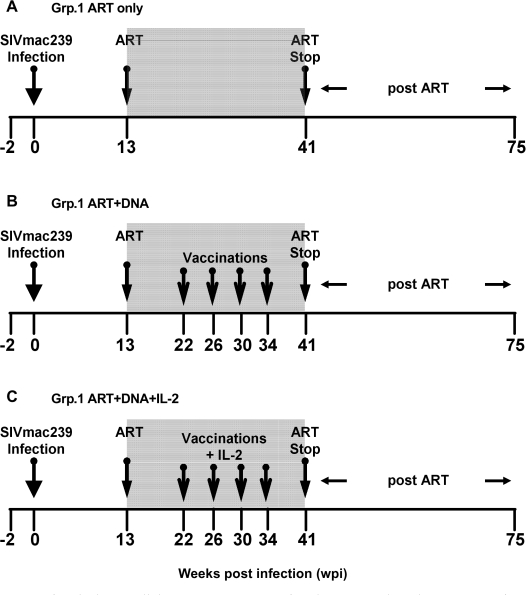
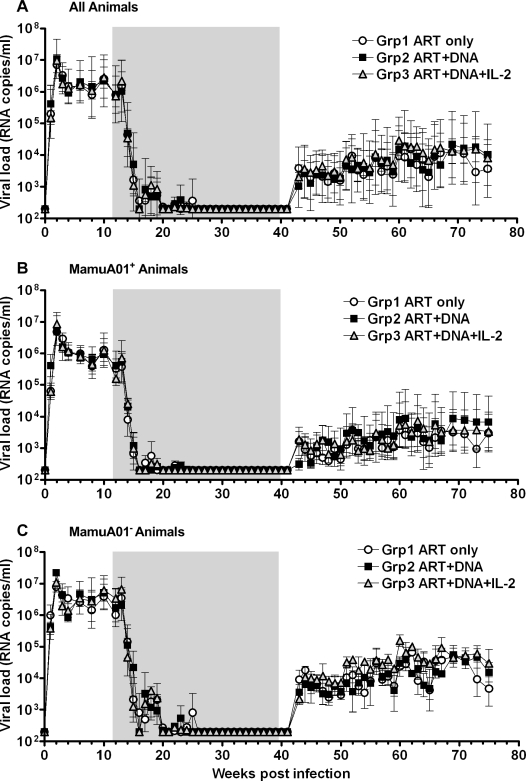
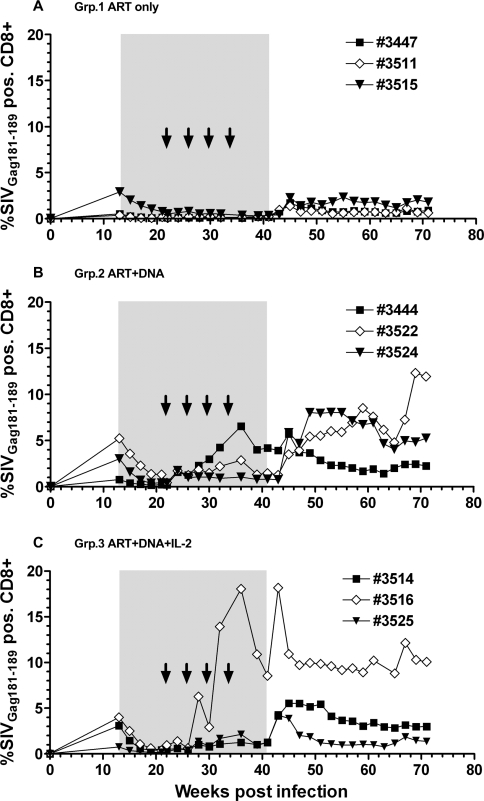
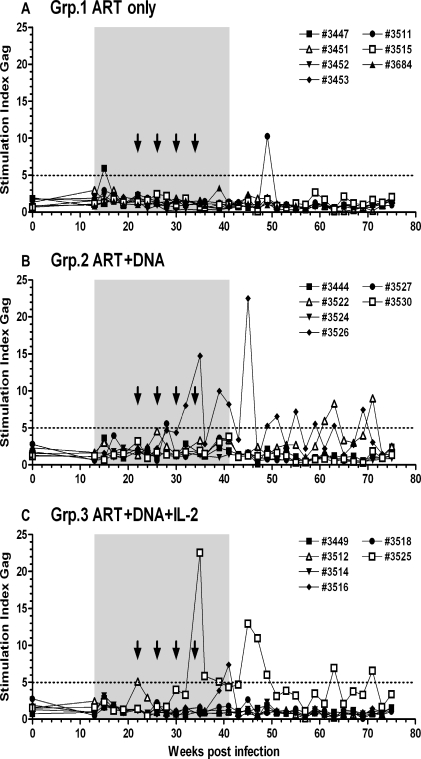
ART (PMPA, FTC, and Zerit®) was administered in the chronic phase of infection (13 wpi) and was continued until 41 wpi (Fig. 3). Viral load was very efficiently controlled by ART, dropping below the assay detection limit (<200 copies/ml) in most of the animals by 20 wpi (Fig. 4). The peak viral load at 2 wpi was comparable between MamuA01+ and MamuA01− animals (mean ± SD: 6.77 ± 0.52 versus 7.09 ± 0.30 log10; p > 0.05; t-test); however, the viral set-point at 12 wpi was about sixfold lower in MamuA01+ macaques (mean ± SD: 5.44 ± 0.54 versus 6.25 ± 0.66 log10; p < 0.01; t-test). In addition, virus control under ART was achieved earlier in MamuA01+ animals (compare Figs. 4B and 4C, 16–20 wpi). Comparable frequencies of SIVGag181–189-specific CD8+ T cells were detectable in all groups at the time of ART initiation at 13 wpi as a result of exposure to viral antigens during the infection phase (means 1.27–3.01%; Table 2 and Fig. 5). During ART, the levels of tetramer-positive CD8+ T cells in the control animals declined and remained low (maximum 0.79%) from week 21 until ART was terminated (41 wpi). Animals in groups 2 and 3 were vaccinated with DNA by in vivo electroporation starting at 22 wpi every 4 weeks with the last vaccination at 34 wpi. Animals in group 3 received low-dose IL-2 protein therapy in addition to the immunization twice daily for 14 days following each vaccination. All vaccinated MamuA01+ animals developed significantly higher levels of SIVGag181–189-specific CD8+ T cells upon vaccination than the unvaccinated control animals during the vaccination period (24–41 wpi; mean ± SD: group 1: 0.28 ± 0.05%; group 2: 2.08 ± 0.68%; group 3: 3.32 ± 2.34%; Table 2 and Fig. 5). A weak but significant difference in frequency of SIVGag181–189-specific CD8+ T cells was detected in vaccinated animals with IL-2 treatment (group 3) compared with the vaccinated animals without IL-2 treatment (group 2) between 24 and 41 wpi (Table 2 and Fig. 5). However, the significant difference was caused by a single animal with a high response in group 3 (#3516), which had a substantial influence on the mean response due to the small group size (n = 3). Vaccinations had only limited impact on lymphoproliferative responses, as these were detectable (SI > 5) only against Gag peptide pools in two vaccinated animals (#3526, group 2 and #3525, group 3) during and after the vaccinations (32 to 39 wpi). Another three animals (#3447, group 1; #3527, group 2; and #3516, group 3) had Gag responses detectable only at a single time point during the entire study (15, 28, and 41 wpi; Fig. 6). Measurements of CD4 lymphoproliferative responses against Env peptide pools remained undetectable in all animals at any time point (SI < 5; data not shown).
FIG. 3.
Overview of study design. All rhesus macaques were infected intravenously with 1,000 MID50 of SIVmac239. Antiretroviral therapy (ART) was initiated at week 13 using a triple drug combination (PMPA R (-9-(2-phosphonomethoxypropyl) adenine); FTC (emtricitabine), and Zerit® (2′-3′-didehydro-2′-3′-dideoxythymidine). Animals in group 1 (n = 7) were treated only with ART during the time frame shown (A). Group 2 (n = 6) received simian immunodeficiency virus (SIV) DNA vaccination by in vivo electroporation four times in 4-week intervals in addition to ART (B), and animals of group 3 (n = 6) received SIV DNA vaccination by in vivo electroporation four times in 4-week intervals followed by interleukin (IL)-2 administration from days 2–16 after vaccination in addition to ART (B). The gray shaded area represents the time under ART; numbers represent weeks pre-/postinfection.
FIG. 4.
Mean of viral loads expressed as RNA copies/ml of plasma for treatment groups 1, 2, and 3 for all animals (A), for MamuA01+ (B), and MamuA01− (C) animals. The gray shaded box represents time under antiretroviral therapy.
FIG. 5.
SIVmac239Gag181–189-specific CD8+ T-cell responses in MamuA01+ vaccinated and unvaccinated control animals measured by tetramer staining. Peripheral blood mononuclear cells (PBMCs) were isolated and analyzed by four-color flow cytometry for simian immunodeficiency virus (SIV)-specific CD8+ T cells. PBMCs were stained with anti-CD3-fluorescein isothiocyanate and anti-CD8-PerCP monoclonal antibody conjugates and phycoerythrin-conjugated MamuA01 Gag181–189 (p11C peptide) tetramer. T-cell responses were measured at time of infection and then from antiretroviral therapy (ART) initiation (13 weeks postinfection) until week 71 postinfection. Time under ART is marked as gray shaded area; vaccinations are indicated as arrows. Longitudinal values for individual animals are shown with their corresponding animal number depicted in each figure legend.
FIG. 6.
Lymphoproliferative responses in untreated (A: antiretroviral therapy [ART] only), DNA vaccinated (B: ART+DNA), and DNA vaccinated and interleukin (IL)-2 treated (C: ART+DNA+IL-2) animals. Time under ART is marked as gray shaded area; vaccinations are indicated as arrows. Longitudinal values for individual animals are shown with their corresponding animal number depicted in each figure legend.
Immune responses and viral rebound after ART cessation
Seven weeks after the final vaccination, ART was discontinued (41 wpi). Virus rebounded in all groups from 43 to 50 wpi but to levels more than 2 logs lower than during the pre-ART set-point at 12 wpi, which was a statistically significant difference (mean ± SD: 3.37 ± 0.66 versus 5.89 ± 0.73 log10 viral RNA copies/ml plasma; p < 0.0001; t-test; Fig. 4). The virus load continued to increase until week 60, when it reached a plateau until the termination of the study at week 75 but still remained significantly lower than at set-point prior to ART (60–75 wpi; mean ± SD: all groups: 3.99 ± 0.94 log10; p < 0.0001; t-test). The viral rebound upon ART cessation resulted in significant lower viral loads in MamuA01+ animals from all groups versus MamuA01− animals in the evaluated time periods from 43–50 wpi (mean ± SD: 2.93 ± 0.57 versus 3.75 ± 0.47 log10; p < 0.001; t-test) and 60–75 wpi (mean ± SD: 3.54 ± 1.01 versus 4.45 ± 0.59 log10; p < 0.0001; t-test; Figs. 4B and 4C). However, viral load was comparable between unvaccinated controls (group 1) and DNA vaccinated animals (group 2) at 43–50 wpi (mean ± SD: 3.36 ± 0.15 versus 3.27 ± 0.15 log10) and 60–75 wpi (3.85 ± 0.27 versus 3.97 ± 0.30 log10), and between group 1 and group 3 (DNA + IL-2) at 43–50 wpi (3.36 ± 0.15 versus 3.46 ± 0.13 log10). Significant, but small, differences in viral RNA copies were detected between group 2 and group 3 at 43–50 wpi (3.27 ± 0.15 versus 3.46 ± 0.13 log10) and at 60–75 wpi (3.97 ± 0.30 versus 4.16 ± 0.17 log10), as well as between group 1 and group 3 at 60–75 wpi (3.85 ± 0.27 versus 4.16 ± 0.17 log10; Table 2 and Fig. 4). Nevertheless, the differences were very small between these groups with a maximum twofold difference in viral load between two groups (0.31 log10 group 1 versus group 3 at 60–75 wpi). Two weeks post-ART cessation (43 wpi) the level of SIVGag181–189-specific CD+ T-cell responses in MamuA01+ animals peaked in group 3 (DNA + IL-2; mean ± SD: 8.86 ± 8.06%) whereas the animals in group 2 (DNA only) reached that peak 2 weeks later (mean ± SD: 5.03 ± 1.32%; Fig. 5). The control animals (group 1) also demonstrated elevated SIVGag181–189-specific CD8 T-cell frequencies as result to increased viral antigen exposure post ART (peak 45 wpi; mean ± SD: 1.79 ± 0.45%). However, these frequencies remained significantly lower in comparison to the vaccinated animals throughout the end of the study (43–71 wpi; mean ± SD: group 1: 1.12 ± 0.24%; group 2: 5.02 ± 1.17%; group 3: 5.13 ± 1.58%; Table 2 and Fig. 5). Lymphoproliferative responses against Gag post-ART cessation were detectable in all groups (Fig. 6). In the ART-only control group (group 1), two animals had a response at one time point (#3447; 15 wpi and #3511; 49 wpi; Fig. 6A). On the contrary, one animal in each of the vaccinated groups (#3526 and #3525) that responded during the vaccination period had also repeatedly detectable responses after ART cessation (Figs. 6B and 6C). One animal in group 2 (Fig. 6B; #3522) had repeated lymphoproliferative responses only after ART was discontinued (63, 65, and 71 wpi).
Impact of treatments on CD4 T-cell counts
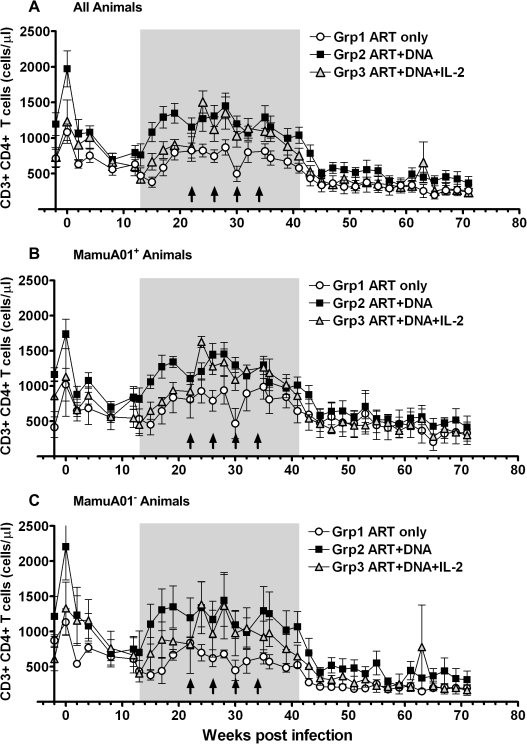
The number of CD4+ T cells in the peripheral blood decreased in all animals after the acute infection from their pre-infection levels (weeks −2 and 0; mean ± SD: group 1: 897 ± 261/μl; group 2: 1,583 ± 550/μl; group 3: 981 ± 353/μl), reaching their lowest level between 8 and 15 wpi (mean ± SD: group 1, 15 wpi: 380 ± 188/μl; group 2, 8 wpi: 697 ± 228/μl; group 3, 13 wpi: 427 ± 149/μl; Fig. 7). The difference between the CD4 T-cell numbers for each group before challenge and at the lowest levels between 8 and 15 wpi reached statistical significance (groups 1 and 2: p < 0.01; group 3: p < 0.05; t-test). The CD4 T-cell numbers recovered significantly during ART and reached a plateau at 22 wpi (mean ± SD: group 1: 824 ± 277/μl; p < 0.01; t-test) or 19 wpi (group 2: 1,449 ± 442/μl; group 3: 1,353 ± 553/μl; p < 0.01; t-test) either close to the baseline CD4 levels or above (group 3). During the prevaccination period of 15 to 22 wpi, differences in the CD4 levels under ART were detectable. The CD4 counts of the DNA vaccinated animals (group 2) were significantly greater than the CD4 levels of unvaccinated (group 1) or animals treated with DNA vaccine plus IL-2 (group 3). In contrast, the difference in CD4 counts between group 1 and group 3 animals was only weakly significant (mean ± SD: group 1: 645 ± 207/μl; group 2: 1,218 ± 122/μl; group 3: 817 ± 104/μl; Table 2 and Fig. 7). The CD4 T-cell numbers increased to the highest levels since preinfection from 24 wpi on and peaked at either 24 wpi (group 3; mean ± SD: 1,506 ± 382/μl) or at 28 wpi (mean ± SD: group 1: 868 ± 268/μl; group 2: 1,449 ± 442/μl). In addition, the time period under ART and before vaccination (15–22 wpi) was compared to the period under ART with vaccination (24–36 wpi) within each group. A significant difference in CD4 T-cell numbers between these periods was observed for group 3, which received IL-2 for 14 days after each vaccination. Although the CD4 counts for group 2 animals were comparable to group 3 animals during the vaccination period from 24–36 wpi, this group had already higher CD4 counts before immunization compared with groups 1 and 3 (Table 2 and Fig. 7).
FIG. 7.
Measurement of absolute numbers of CD4 T cells in peripheral blood monocytes (PBMCs). Mean values are shown for all monkeys (A) and separately for MamuA01+ (B) and MamuA01− animals (C). The time under antiretroviral therapy is marked as gray shaded area, vaccinations are indicated as arrows.
The frequency of CD4 T cells started to drop slightly from 36 wpi until ART cessation at 41 wpi when the CD4 counts decreased considerably until 45 wpi as a result of increased virus replication (mean ± SD: group 1: 336 ± 245/μl; group 2: 501 ± 152/μl; group 3: 400 ± 177/μl). The CD4 T-cell numbers subsequently continued to decline slowly until the final measurement at 71 wpi (mean ± SD: group 1: 263 ± 197/μl; group 2: 366 ± 227/μl; group 3: 234 ± 144/μl). The CD4 counts during that period from 47 to 71 wpi remained comparable between all groups (Table 2 and Fig. 7).
Until study termination at week 75 postchallenge, all animals remained healthy without signs of opportunistic infections. The viral load and CD4 counts remained comparable between all groups despite significantly increased SIVGag181–189-specific CD8 T-cell responses in both vaccinated groups as compared with the unvaccinated controls.
Discussion
The goal of this study was to obtain a significant reduction in viral load as a result of DNA vaccination after ART discontinuation in rhesus macaques chronically infected with SIVmac239. The triple-drug combination (PMPA, FTC, Zerit®) resulted in a drastic decline of viral load (>10,000-fold) to below 200 viral RNA copies/ml in most animals (Table 1 and Fig. 4). However, vaccination appeared to have no differential impact on viral loads after ART cessation, despite significantly increased frequencies of Gag181-189-specific CD8+ T cells in vaccinated MamuA01+ animals compared with the controls (Table 2 and Figs. 4 and 5). Interestingly, all animals including the unvaccinated controls demonstrated a comparable viral rebound post-ART (Fig. 4) that was significantly lower (p < 0.0001) during the 34-week period after ART cessation (mean: 3.72 log10) than the viral load at set point before ART (12 wpi; mean: 5.89 log10). Viral loads for all animals remained on average below 104 RNA copies/ml (mean ± SD: 5.80 × 103 ± 7.29 × 103) after ART cessation (43 wpi) until week 50 and remained below 105 RNA copies/ml for most animals for another 6 months until the end of the study at 75 wpi (mean ± SD: 4.02 × 104 ± 6.13 × 104). MamuA01+ animals had a more efficient viral control during 16–19 wpi under ART (Figs. 4B versus 4C) and lower viral rebound than MamuA01− animals (43–50 wpi; 2.93 versus 3.75 log10; p < 0.001), which is in agreement with previous findings for that MHC haplotype.28 In addition, none of the animals were MamuB17 positive, which is also associated with improved virus control in SIVmac239-infected rhesus macaques.29 Furthermore, neutralizing antibody responses were evaluated in all animals post-ART using SIVmac239CS23 pseudovirus in TZM-bl cells. The lower limit of detection in the assay was a titer of <20. One animal in each group had high titers (>1,000) and two animals each in group 2 (ART + DNA) and 3 (ART + DNA + IL-2) had low titers (>40) at several time points. Therefore, potent virus neutralization was not evident, except in three animals, which were equally distributed among the three groups.
SIV GAG181–189-specific CD8+ T-cell responses in vaccinated MamuA01+ animals were significantly boosted during ART as a result of in vivo DNA electroporation and remained significantly higher than those of unvaccinated animals throughout the remainder of the study (Table 2 and Fig. 5). SIV Gag-specific lymphoproliferative responses were detectable at multiple time points during and after vaccinations in one animal of each vaccinated group (Fig. 6, #3526 and #3525), and one-time responses were detected by one or two animals in each group (Fig. 6). A comparable pattern of potent CD8 T-cell responses and less efficient lymphoproliferative responses has recently been reported in a prophylactic vaccine study using in vivo electroporation. The reason for low lymphoproliferative responses was not the delivery method per se, but rather a result of the gagpol fusion cassette versus separate gag and pol cassettes as immunogens.11 If the separate administration of gag and pol DNAs in our study could have altered the outcome in favor of better virus control after ART was stopped remains to be evaluated. Nevertheless, animals that received IL-2 in addition to vaccination had significantly increased CD4 counts but the viral rebound post-ART cessation was comparable to unvaccinated controls and animals that received only the vaccine without IL-2 (Table 2 and Fig. 7). Although the frequency of GAG181–189 tetramer-positive CD8+ T cells was only measured in MamuA01+ animals, the potency of in vivo electroporation of DNA vaccines has been previously demonstrated in genetically diverse macaques in numerous studies, resulting in strong CD4+ and CD8+ T-cell responses30,31 to HIV-1 Gag, Pol or Env antigens.11,25,32
Because all animals in our study demonstrated a comparable slow viral load rebound independent of treatment (DNA, IL-2, unvaccinated), the possibility exists that the particular drug regimen (PMPA, FTC, Zerit®) could have contributed to that effect. The beneficial impact of PMPA on viral control has been reported in several in vivo studies, in particular related to immune augmentation and improved viral suppression.33–38 The highly efficient CD8+ T cell-mediated suppression of SIV in combination with PMPA (Tenofovir) therapy despite a resistance mutation to PMPA (K65R) has been reported,39 and SIV-infected rhesus macaques with previously undetectable viremia under PMPA demonstrated a very moderate viral rebound (maximum 103 RNA copies/ml plasma) upon drug interruption. Based on these studies, a preexisting antiviral response prior to PMPA treatment is therefore a possible factor for the subsequent control of virus post-ART cessation. All MamuA01+ animals in our study had detectable CD8 T-cell responses throughout the study until study termination, although these responses were significantly lower for the unvaccinated control animals (Table 2 and Fig. 5). Nevertheless, a related outcome has recently been described in a therapeutic vaccination study using recombinant modified vaccinia virus ankara with a beneficial effect of PMPA on viral load post-ART cessation in acutely SIV-infected rhesus macaques.40 It remains to be seen whether the solely T cell-based vaccine approach applied in our study was inefficient in itself or if the benefit of the triple-drug regimen on the twofold log virus reduction masked the potential contribution of CD8 T cells in the vaccinated animals to control the virus post-ART cessation. Both possibilities need to be taken into consideration especially since a reduction of viral load post-ART cessation of about 1 log was seen in chronically infected, vaccinated animals in a comparable study.41 The excellent viral control in all animals in combination with detectable CD8 T-cell responses for all animals (MamuA01+) and potential synergistic immune-augmentation effects of PMPA could have contributed to the slow rebound post-ART. The CD4 T cells continued to decline slowly during the off-ART period until the latest evaluated time point at 71 wpi (Fig. 7), and all animals remained healthy without detectable opportunistic infections until study termination (75 wpi).
An up to 1,000-fold viral load reduction as a result of a different therapeutic vaccination strategy has recently been shown in rhesus macaques and humans using ex vivo pulsed autologous dendritic cells.42–44 These promising results demonstrated the potential and possibility to develop therapeutic vaccination treatments for HIV-1-infected patients in the future. However, pulsing ex vivo dendritic cells remains at present more an experimental procedure and is not feasible for large-scale administration of therapeutic vaccines. Delivery of vaccines using recombinant viruses, such as adenovirus, is currently more practical. However, recent preliminary results from a phase II clinical trial (STEP) using a solely T cell-based prophylactic vaccine approach using recombinant adenovirus (Ad5) resulted in a potentially increased susceptibility of HIV-1 infections in vaccinated volunteers and the viral load was not reduced in these infected individuals (http://www.merck.com/newsroom/press_releases/re-search_and_development/2007_1107.html).
In summary, the vaccination strategy of DNA in combination with electroporation and IL-2 elicited T-cell responses and improved CD4 counts but did not have a beneficial effect on viral loads. Therefore, more research is needed to define the ideal components, induced immune responses (T cell and/or B cell), and delivery technologies for effective future vaccine candidates.
Acknowledgments
This work was supported by NIH/NIAID contracts N01-AI-15451 and N01-AI-15443. PMPA and FTC were kindly provided by Gilead Corporation, and Zerit® was a gift from the AIDS Research and Reference Reagent Program, NIAID, NIH. We would also like to thank Ping Shi for statistical analysis, Ron Desrosiers for donating the SIVmac239 challenge stock, and the laboratory of David Montefiori for performing the neutralizing antibody assays.
References
- 1.Boyd M. Reiss P. The long-term consequences of antiretroviral therapy: a review. J HIV Ther. 2006;11:26–35. [PubMed] [Google Scholar]
- 2.Blankson JN. Persaud D. Siliciano RF. The challenge of viral reservoirs in HIV-1 infection. Annu Rev Med. 2002;53:557–593. doi: 10.1146/annurev.med.53.082901.104024. [DOI] [PubMed] [Google Scholar]
- 3.Wong JK. Hezareh M. Gunthard HF, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 4.Noe A. Plum J. Verhofstede C. The latent HIV-1 reservoir in patients undergoing HAART: an archive of pre-HAART drug resistance. J Antimicrob Chemother. 2005;55:410–412. doi: 10.1093/jac/dki038. [DOI] [PubMed] [Google Scholar]
- 5.Imami N. Gotch F. Prospects for immune reconstitution in HIV-1 infection. Clin Exp Immunol. 2002;127:402–411. doi: 10.1046/j.1365-2249.2002.01822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burton CT. Mela CM. Rosignoli G. Westrop SJ. Gotch FM. Imami N. Immune modulation and reconstitution of HIV-1-specific responses: novel approaches and strategies. Curr Med Chem. 2006;13:3203–3211. doi: 10.2174/092986706778742972. [DOI] [PubMed] [Google Scholar]
- 7.Fuller DH. Rajakumar PA. Wu MS, et al. DNA immunization in combination with effective antiretroviral drug therapy controls viral rebound and prevents simian AIDS after treatment is discontinued. Virology. 2006;348:200–215. doi: 10.1016/j.virol.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 8.Hel Z. Venzon D. Poudyal M, et al. Viremia control following antiretroviral treatment and therapeutic immunization during primary SIV251 infection of macaques. Nat Med. 2000;6:1140–1146. doi: 10.1038/80481. [DOI] [PubMed] [Google Scholar]
- 9.Brenchley JM. Ruff LE. Casazza JP. Koup RA. Price DA. Douek DC. Preferential infection shortens the life span of human immunodeficiency virus-specific CD4+ T cells in vivo. J Virol. 2006;80:6801–6809. doi: 10.1128/JVI.00070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grabar S. Le Moing V. Goujard C, et al. Clinical outcome of patients with HIV-1 infection according to immunologic and virologic response after 6 months of highly active antiretroviral therapy. Ann Intern Med. 2000;133:401–410. doi: 10.7326/0003-4819-133-6-200009190-00007. [DOI] [PubMed] [Google Scholar]
- 11.Otten GR. Schaefer M. Doe B, et al. Potent immunogenicity of an HIV-1 gag-pol fusion DNA vaccine delivered by in vivo electroporation. Vaccine. 2006;24:4503–4509. doi: 10.1016/j.vaccine.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 12.Barouch DH. Craiu A. Kuroda MJ, et al. Augmentation of immune responses to HIV-1 and simian immunodeficiency virus DNA vaccines by IL-2/Ig plasmid administration in rhesus monkeys. Proc Natl Acad Sci U S A. 2000;97:4192–4197. doi: 10.1073/pnas.050417697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chapman BS. Thayer RM. Vincent KA. Haigwood NL. Effect of intron A from human cytomegalovirus (Towne) immediate-early gene on heterologous expression in mammalian cells. Nucleic Acids Research. 1991;19:3979–3986. doi: 10.1093/nar/19.14.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.zur Megede J. Otten GR. Doe B, et al. Expression and immunogenicity of sequence-modified human immunodeficiency virus type 1 subtype B pol and gagpol DNA vaccines. J Virol. 2003;77:6197–6207. doi: 10.1128/JVI.77.11.6197-6207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel PH. Jacobomolina A. Ding JP, et al. Insights into DNA polymerization mechanisms from structure and function analysis of HIV-1 reverse transcriptase. Biochemistry. 1995;34:5351–5363. doi: 10.1021/bi00016a006. [DOI] [PubMed] [Google Scholar]
- 17.Palaniappan C. Wisniewski M. Jacques PS. Le Grice SF. Fay PJ. Bambara RA. Mutations within the primer grip region of HIV-1 reverse transcriptase result in loss of RNase H function. J Biol Chem. 1997;272:11157–11164. doi: 10.1074/jbc.272.17.11157. [DOI] [PubMed] [Google Scholar]
- 18.Konvalinka J. Litterst MA. Welker R, et al. An active-site mutation in the human immunodeficiency virus type 1 proteinase (PR) causes reduced PR activity and loss of PR-mediated cytotoxicity without apparent effect on virus maturation and infectivity. J Virol. 1995;69:7180–7186. doi: 10.1128/jvi.69.11.7180-7186.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scriba TJ. zur Megede J. Glashoff RH. Treurnicht FK. Barnett SW. van Rensburg EJ. Functionally-inactive and immunogenic Tat, Rev and Nef DNA vaccines derived from sub-Saharan subtype C human immunodeficiency virus type 1 consensus sequences. Vaccine. 2005;23:1158–1169. doi: 10.1016/j.vaccine.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 20.Endres CL. Bergquam EP. Axthelm MK. Wong SW. Suppression of simian immunodeficiency virus replication by human immunodeficiency virus type 1 trans-dominant negative rev mutants. J Virol. 1995;69:5164–5166. doi: 10.1128/jvi.69.8.5164-5166.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swigut T. Iafrate AJ. Muench J. Kirchhoff F. Skowronski J. Simian and human immunodeficiency virus Nef proteins use different surfaces to downregulate class I major histocompatibility complex antigen expression. J Virol. 2000;74:5691–5701. doi: 10.1128/jvi.74.12.5691-5701.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knapp LA. Lehmann E. Piekarczyk MS. Urvater JA. Watkins DI. A high frequency of Mamu-A*01 in the rhesus macaque detected by polymerase chain reaction with sequence-specific primers and direct sequencing. Tissue Antigens. 1997;50:657–661. doi: 10.1111/j.1399-0039.1997.tb02927.x. [DOI] [PubMed] [Google Scholar]
- 23.Nacsa J. Edghill-Smith Y. Tsai WP, et al. Contrasting effects of low-dose IL-2 on vaccine-boosted simian immunodeficiency virus (SIV)-specific CD4+ and CD8+ T cells in macaques chronically infected with SIVmac251. J Immunol. 2005;174:1913–1921. doi: 10.4049/jimmunol.174.4.1913. [DOI] [PubMed] [Google Scholar]
- 24.Rabussay D. Dev N. Fewell J. Smith LC. Widera G. Zhang L. Enhancement of therapeutic drug and DNA delivery into cells by electroporation. J Phys D Appl Phys. 2003;36:348. [Google Scholar]
- 25.Widera G. Austin M. Rabussay D, et al. Increased DNA vaccine delivery and immunogenicity by electroporation in vivo. J Immunol. 2000;164:4635–4640. doi: 10.4049/jimmunol.164.9.4635. [DOI] [PubMed] [Google Scholar]
- 26.Suryanarayana K. Wiltrout TA. Vasquez GM. Hirsch VM. Lifson JD. Plasma SIV RNA viral load determination by real-time quantification of product generation in reverse transcriptase-polymerase chain reaction. AIDS Res Hum Retroviruses. 1998;14:183–189. doi: 10.1089/aid.1998.14.183. [DOI] [PubMed] [Google Scholar]
- 27.Wichman BA. Hill IC. Algorithm AS 183: an efficient and portable pseudo-random number generator. Appl Stat. 1982;31:188–190. [Google Scholar]
- 28.Zhang ZQ. Fu TM. Casimiro DR, et al. Mamu-A*01 allele-mediated attenuation of disease progression in simian-human immunodeficiency virus infection. J Virol. 2002;76:12845–12854. doi: 10.1128/JVI.76.24.12845-12854.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yant LJ. Friedrich TC. Johnson RC, et al. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol. 2006;80:5074–5077. doi: 10.1128/JVI.80.10.5074-5077.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Selby M. Goldbeck C. Pertile T. Walsh R. Ulmer J. Enhancement of DNA vaccine potency by electroporation in vivo. J Biotechnol. 2000;83:147–152. doi: 10.1016/s0168-1656(00)00308-4. [DOI] [PubMed] [Google Scholar]
- 31.Mir LM. Therapeutic perspectives of in vivo cell electropermeabilization. Bioelectrochemistry. 2001;53:1–10. doi: 10.1016/s0302-4598(00)00112-4. [DOI] [PubMed] [Google Scholar]
- 32.Otten G. Schaefer M. Doe B, et al. Enhancement of DNA vaccine potency in rhesus macaques by electroporation. Vaccine. 2004;22:2489–2493. doi: 10.1016/j.vaccine.2003.11.073. [DOI] [PubMed] [Google Scholar]
- 33.Metzner KJ. Binley JM. Gettie A. Marx P. Nixon DF. Connor RI. Tenofovir treatment augments anti-viral immunity against drug-resistant SIV challenge in chronically infected rhesus macaques. Retrovirology. 2006;3:97. doi: 10.1186/1742-4690-3-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Rompay KK. Antiretroviral drug studies in nonhuman primates: a valid animal model for innovative drug efficacy and pathogenesis experiments. AIDS Rev. 2005;7:67–83. [PubMed] [Google Scholar]
- 35.Van Rompay KK. Johnson JA. Blackwood EJ, et al. Sequential emergence and clinical implications of viral mutants with K70E and K65R mutation in reverse transcriptase during prolonged tenofovir monotherapy in rhesus macaques with chronic RT-SHIV infection. Retrovirology. 2007;4:25. doi: 10.1186/1742-4690-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Rompay KK. Singh RP. Brignolo LL, et al. The clinical benefits of tenofovir for simian immunodeficiency virus-infected macaques are larger than predicted by its effects on standard viral and immunologic parameters. J Acquir Immune Defic Syndr. 2004;36:900–914. doi: 10.1097/00126334-200408010-00003. [DOI] [PubMed] [Google Scholar]
- 37.Van Rompay KK. McChesney MB. Aguirre NL. Schmidt KA. Bischofberger N. Marthas ML. Two low doses of tenofovir protect newborn macaques against oral simian immunodeficiency virus infection. J Infect Dis. 2001;184:429–438. doi: 10.1086/322781. [DOI] [PubMed] [Google Scholar]
- 38.Van Rompay KK. Miller MD. Marthas ML, et al. Prophylactic and therapeutic benefits of short-term 9-[2-(R)-(phosphonomethoxy)propyl]adenine (PMPA) administration to newborn macaques following oral inoculation with simian immunodeficiency virus with reduced susceptibility to PMPA. J Virol. 2000;74:1767–1774. doi: 10.1128/jvi.74.4.1767-1774.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Rompay KK. Singh RP. Pahar B, et al. CD8+-cell-mediated suppression of virulent simian immunodeficiency virus during tenofovir treatment. J Virol. 2004;78:5324–5337. doi: 10.1128/JVI.78.10.5324-5337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uberla K. Rosenwirth B. Ten Haaft P. Heeney J. Sutter G. Erfle V. Therapeutic immunization with modified vaccinia virus ankara (MVA) vaccines in SIV-infected rhesus monkeys undergoing antiretroviral therapy. J Med Primatol. 2007;36:2–9. doi: 10.1111/j.1600-0684.2006.00190.x. [DOI] [PubMed] [Google Scholar]
- 41.von Gegerfelt AS. Rosati M. Alicea C, et al. Long-lasting decrease in viremia in macaques chronically infected with simian immunodeficiency virus SIVmac251 after therapeutic DNA immunization. J Virol. 2007;81:1972–1979. doi: 10.1128/JVI.01990-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andrieu JM. Lu W. A dendritic cell-based vaccine for treating HIV infection: background and preliminary results. J Intern Med. 2007;261:123–131. doi: 10.1111/j.1365-2796.2006.01738.x. [DOI] [PubMed] [Google Scholar]
- 43.Lu W. Arraes LC. Ferreira WT. Andrieu JM. Therapeutic dendritic-cell vaccine for chronic HIV-1 infection. Nat Med. 2004;10:1359–1365. doi: 10.1038/nm1147. [DOI] [PubMed] [Google Scholar]
- 44.Lu W. Wu X. Lu Y. Guo W. Andrieu JM. Therapeutic dendritic-cell vaccine for simian AIDS. Nat Med. 2003;9:27–32. doi: 10.1038/nm806. [DOI] [PubMed] [Google Scholar]