Abstract
GTP hydrolysis by elongation factor G (EF-G) is essential for the translocation step in protein elongation. The low intrinsic GTPase activity of EF-G is strongly stimulated by the ribosome. Here we show that a conserved arginine, R29, of Escherichia coli EF-G is crucial for GTP hydrolysis on the ribosome, but not for GTP binding or ribosome interaction, suggesting that it may be directly involved in catalysis. Another conserved arginine, R59, which is homologous to the catalytic arginine of Gα proteins, is not essential for GTP hydrolysis, but influences ribosome binding and translocation. These results indicate that EF-G is similar to other GTPases in that an arginine residue is required for GTP hydrolysis, although the structural changes leading to GTPase activation are different.
Keywords: fast kinetics/gtpase/mechanism of GTP hydrolysis/protein synthesis
Introduction
Elongation factor G (EF-G) is a large, multi-domain GTPase that catalyzes the movement of tRNA and mRNA during translocation on the ribosome. The GTPase activity of EF-G, which intrinsically is unmeasurably low (Parmeggiani and Sander, 1981), is enhanced strongly upon binding to the ribosome. The molecular mechanism of GTPase activation and GTP hydrolysis is not known. So far, GTPase mechanisms were elucidated for members of two subfamilies of the GTPase superfamily: small Ras-like GTPases (Mittal et al., 1996; Ahmadian et al., 1997; Rittinger et al., 1997a,b; Scheffzek et al., 1997) and heterotrimeric G proteins (Coleman et al., 1994; Sondek et al., 1994). The GTPase activity of the small G proteins Ras and Rho is enhanced by GTPase activating proteins (GAPs), which introduce a catalytic arginine into the active site of the GTPase (‘arginine finger’), thereby stabilizing the transition state of the GTPase reaction and greatly accelerating the rate of GTP cleavage. The GTPase activity of the Gα subunits of some heterotrimeric G proteins is enhanced by proteins called regulators of G protein signaling (RGS) (De Vries and Farquhar, 1999), which stabilize the GTPase transition state of G proteins but do not donate catalytic residues for the GTPase reaction (Tesmer et al., 1997). Rather, the catalytic arginine of the heterotrimeric G proteins is a constituent of the Gα subunit itself (Coleman et al., 1994; Sondek et al., 1994).
The GTPase activity of EF-G seems to be stimulated by a mechanism resembling the RGS type. The major stimulating interaction is that with ribosomal protein L7/12, which in experiments with isolated protein stimulates the GTPase activity of EF-G several hundred-fold (Savelsbergh et al., 2000). On the ribosome, the effect of L7/12 may be augmented by other interactions (see Discussion). The single, highly conserved arginine of L7/12 is not essential for the stimulation, indicating that the function of L7/12 may be to induce or stabilize the GTPase transition state of EF-G by promoting the re-orientation of catalytic residues within the EF-G molecule. According to cryoelectron microscopy, EF-G interacts with the stalk of the 50S ribosomal subunit (Agrawal et al., 1998; Stark et al., 2000), which comprises protein L7/12, based on immunoelectron microscopic and biochemical data (for review see Traut et al., 1995).
The presumed catalytic arginine of EF-G could be located either in the G domain itself or in domain 5, which, according to the crystal structure of EF-G⋅GDP from Thermus thermophilus (Ævarsson et al., 1994; Czworkowski et al., 1994), interacts with the G domain near the nucleotide binding site. However, the deletion of domain 5 had no effect on GTP hydrolysis (A.Savelsbergh, N.B.Matassova, M.V.Rodnina and W.Wintermeyer, submitted), leaving arginine residues within the G domain as candidates for catalysis. There are eight conserved arginines in the G domain of EF-G, of which two, R29 and R59, are located in the vicinity of the β-phosphate of GDP in the crystal structure and are, therefore, potential candidates for a catalytic arginine. R29 of Escherichia coli EF-G (R31 in T.thermophilus) is located in helix A and is in close proximity to the GDP binding site. R59 is located in the effector loop (R61 in T.thermophilus) and, according to sequence comparisons, is homologous to the catalytic arginine of Gα proteins. In the present work, we replaced R29 and R59 of E.coli EF-G with alanine, lysine or methionine, and studied the effect of the mutations on the activity of the factor in GTP hydrolysis and translocation on the ribosome.
Results
GTPase activity of EF-G mutants on the ribosome
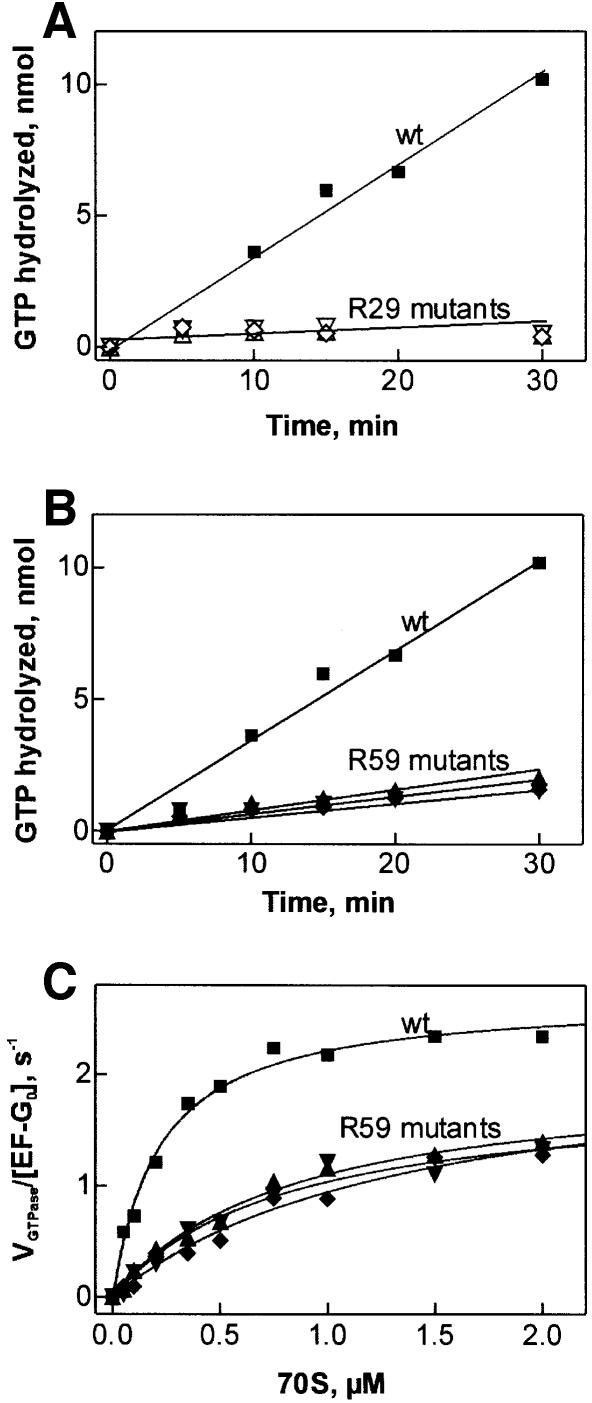
Mutagenesis was performed and EF-G mutants were expressed and purified as described in Materials and methods. The ribosome-stimulated GTP hydrolysis by the EF-G mutants was first measured under multiple-turnover conditions, using catalytic amounts of EF-G and excess of both ribosomes and GTP, where efficient GTP cleavage was observed with wild-type EF-G. As shown in Figure 1A, the replacements of arginine 29 with methionine, lysine or alanine almost completely abolished the GTPase activity of EF-G. Also, the R59 mutants showed a reduced GTPase activity (Figure 1B), although the extent of inhibition was much less than with the R29 mutants. To quantify the effects of the R59 mutations on the affinity to the ribosomes and efficiency of GTP cleavage, the steady-state kinetic parameters, KM and kcat, were measured in the presence of constant saturating concentration of GTP and varying concentrations of the ribosomes, both in excess over EF-G (Figure 1C). The R59 mutations decreased KM 4- to 7-fold, whereas the kcat of GTP hydrolysis was reduced at most 2-fold (Table I). Overall, the catalytic efficiency of GTP cleavage, kcat/KM, was reduced from 12 µM–1s–1 for wild-type EF-G to ∼2 µM–1s–1 for the R59 mutants.
Fig. 1. Effect of R29 and R59 mutations on turnover GTP hydrolysis by EF-G on the ribosome. (A) R29 mutations; (B) R59 mutations; (C) Michaelis–Menten titrations with R59 mutants. Wild-type EF-G (squares), R29K (open upward triangles), R29M (open downward triangles), R29A (open diamonds), R59K (filled upward triangles), R59M (filled downward triangles), R59A (filled diamonds).
Table I. Effect of R59 mutations in EF-G on the steady-state kinetic parameters of ribosome-stimulated GTP hydrolysis.
| EF-G | KM, µM | kcat, s–1 | kcat/KM, µM–1s–1 |
|---|---|---|---|
| wild type | 0.22 ± 0.02 | 2.7 ± 0.1 | 12 ± 1 |
| R59K | 0.8 ± 0.1 | 2.0 ± 0.1 | 2.5 ± 0.5 |
| R59M | 0.8 ± 0.2 | 1.9 ± 0.2 | 2.4 ± 0.8 |
| R59A | 1.4 ± 0.4 | 2.2 ± 0.3 | 1.6 ± 0.7 |
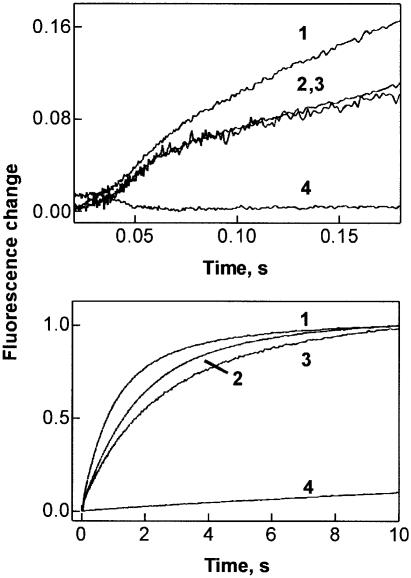
Analogous results were obtained when GTP hydrolysis was followed by monitoring the release of inorganic phosphate (Pi) in a stopped-flow apparatus. This technique allowed us to measure both rapid single-round and subsequent slower multiple-round GTP hydrolysis and Pi release in one experiment. EF-G⋅GTP was rapidly mixed with ribosomes, and the liberation of Pi was measured by the fluorescence increase of labeled phosphate binding protein, MDCC-PBP. After a short lag phase, a rapid increase of MDCC fluorescence, due to Pi liberation after the first round of GTP hydrolysis, was observed for both wild-type EF-G and the R59 mutants (Figure 2, upper panel). This result shows that the R59 mutants were about as active as wild-type EF-G in one round of GTP hydrolysis and subsequent release of Pi. The rapid increase of the signal was followed by a further slower increase due to multiple rounds of GTP hydrolysis (lower panel). With the R29 mutants, virtually no Pi release was observed, in keeping with the inhibition of GTP hydrolysis shown above. The conclusion from these results is that R29 is required for GTP hydrolysis, whereas R59 is not involved in GTP hydrolysis and rather influences the binding of EF-G to the ribosome.
Fig. 2. Kinetics of Pi release from EF-G following GTP hydrolysis on the ribosome. Upper panel, short time window; lower panel, long time window. EF-G or EF-G mutants (2 mM, final concentration) were rapidly mixed with ribosomes (0.2 mM) and liberated Pi was monitored by the fluorescence of MDCC-labeled PBP (Materials and methods). 1, wild-type EF-G; 2, R59K; 3, R59M; 4, R29A.
GTP binding to R29 mutants of EF-G
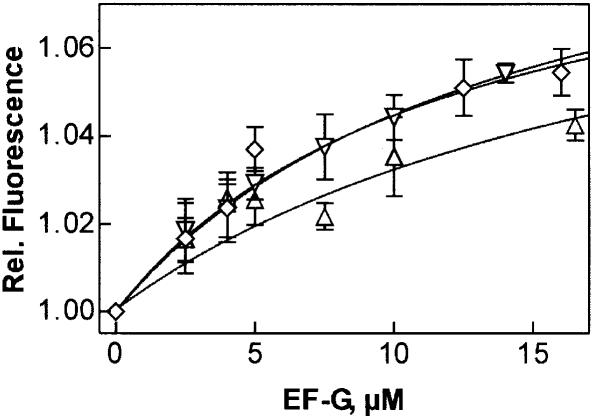
The lack of GTPase activity of EF-G R29 mutants may result from a deficiency in GTP binding. Therefore, the nucleotide binding properties of the mutant proteins were tested using a fluorescent GTP derivative, mant-GTP, which shows a fluorescence increase upon binding to EF-G (Savelsbergh et al., 2000). A similar fluorescence increase due to mant-GTP binding was observed with the R29 mutants of EF-G (not shown), indicating that a complex was formed. To estimate the Kd of the interaction of mant-GTP with EF-G, titration experiments were performed. For wild-type EF-G, a Kd value of 25 ± 4 µM for mant-GTP was obtained (not shown). The Kd for unmodified GTP was somewhat lower, 12 µM, as determined by a chase titration of the complex, EF-G⋅mant-GTP, with unmodified GTP. Titration experiments with R29 mutants showed that affinities for mant-GTP were lowered such that measuring fluorescence changes in steady-state proved difficult. Reliable fluorescence amplitudes were obtained in stopped-flow experiments monitoring time courses of mant-GTP binding to EF-G. From the concentration dependence of the fluorescence amplitudes (Figure 3), Kd values of ∼50 ± 30 µM were estimated for the R29 mutants. Thus, the mutations of R29 lowered the affinity for mant-GTP binding ∼2-fold. Assuming the same affinity decrease for unmodified GTP, the R29 mutants should bind GTP with a Kd of 20–30 µM, i.e. are expected to be saturated with GTP at the conditions of the GTP hydrolysis experiments (1 mM GTP). Thus, the lack of GTPase activity of the R29 mutants cannot be attributed to impaired GTP binding; rather, it suggests that the GTP cleavage step is affected.
Fig. 3. Interaction of mant-GTP with R29 mutants of EF-G. The fluorescence change of mant-GTP upon binding to the EF-G mutants was measured by stopped flow. R29K (upward triangles), R29M (downward triangles), R29A (diamonds).
Activity of EF-G mutants in translocation
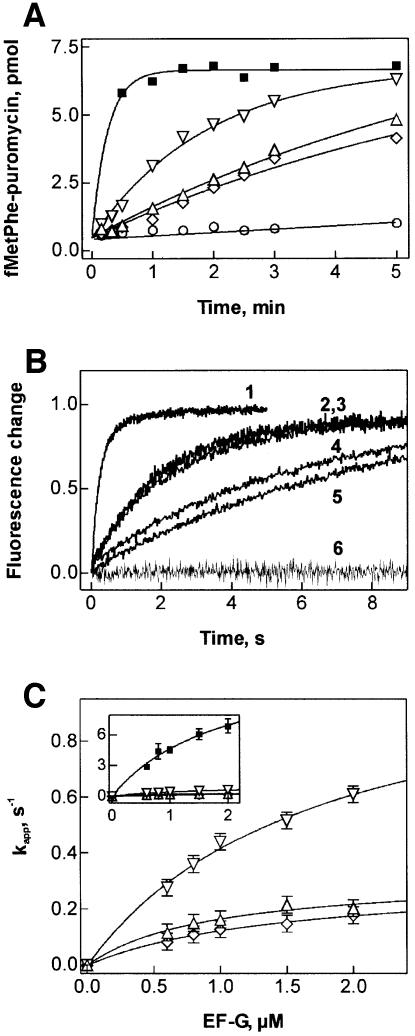
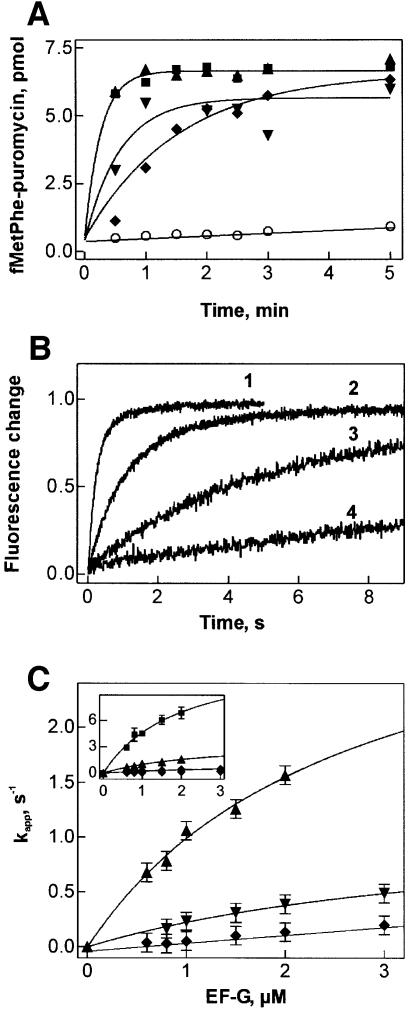
To test the ability of mutant EF-G to promote translocation, pretranslocation complexes were used that contained fMetPhe-tRNAPhe in the A site and deacylated tRNAfMet in the P site; the extent of translocation was determined by the puromycin assay. Multiple-turnover translocation was assayed using catalytic amounts of EF-G or EF-G mutants. The mutations of R29 significantly decreased the rate of turnover translocation (Figure 4A). The rates observed with EF-G(R29A) and EF-G(R29K) were about the same, while the reaction with EF-G(R29M) was somewhat faster. The turnover reaction with wild-type EF-G could not be resolved, as translocation was complete within 30 s.
Fig. 4. Translocation activity of R29 mutants of EF-G. (A) Multiple-turnover translocation. Pretranslocation complex (0.2 µM) was incubated with wild-type EF-G (squares), R29K (upward triangles), R29M (downward triangles), R29A (diamonds) (0.04 µM) or without factor (circles) in the presence of GTP (1 mM). Translocation was measured by the formation of fMetPhe-puromycin. (B) Single-round translocation measured by stopped flow. Pretranslocation complex containing fMetPhe-tRNAPhe(Prf16/17) in the A site (0.1 µM, final concentration) was rapidly mixed with EF-G (0.8 µM) in the presence of GTP (1 mM), caged GTP (0.1 mM) or without nucleotide. Time courses were evaluated by single-exponential fitting with the kapp values given. 1, wild-type EF-G (4.4 s–1); 2, wild-type EF-G with caged GTP (0.5 s–1); 3, R29M (0.4 s–1); 4, R29K (0.15 s–1); 5, R29A (0.11 s–1); 6, R29K without nucleotide; EF-G(R29M) and EF-G(R29A) without nucleotide were the same (not shown). (C) Dependence of kapp of translocation on the concentration of mutant and wild-type (inset) EF-G. For kapp values measured at 2 µM factor see Table II. Symbols are as in (A).
Single-round translocation was measured by stopped flow, monitoring the fluorescence change of proflavin-labeled fMetPhe-tRNAPhe(Prf 16/17) accompanying translocation from the A to the P site with excess of EF-G over the ribosomes. The rate of a single round of translocation with EF-G(R29M), 0.4 s–1, was reduced ∼10-fold compared with wild-type EF-G (4.4 s–1); both were measured in the presence of GTP at a factor concentration of 0.8 µM (Figure 4B). With EF-G(R29A) and EF-G(R29K), translocation was even slower, 0.11 and 0.15 s–1, respectively. Interestingly, a 10-fold reduced translocation rate was also observed with wild-type EF-G in the presence of a non-hydrolyzable GTP analog, caged GTP, whereas with another non-hydrolyzable GTP analog, GDPNP, the rate was somewhat lower, 0.2 s–1 (not shown). In the absence of nucleotide, no single-round translocation was observed with the R29 mutants (Figure 4B), supporting the notion that the mutant proteins were able to bind GTP.
To distinguish between the effects on the rate of catalysis and on affinity to the ribosome, the rate of translocation was measured at increasing concentrations of EF-G R29 mutants (Figure 4C). At the maximum concentration of factor that could be attained in these experiments, 2 µM, the rate of translocation was 10–40 times lower than with wild-type EF-G (Table II). Half-saturation levels were reached at 0.5–1 µM concentration of factor in all cases, indicating that the affinity to the ribosome was not significantly affected by these mutations.
Table II. Effect of R29 and R59 mutations on the translocation rate.
| EF-G | kapp, s–1 |
|---|---|
| wild type | 7 ± 1 |
| R29K | 0.20 ± 0.05 |
| R29M | 0.6 ± 0.1 |
| R29A | 0.18 ± 0.01 |
| R59K | 1.6 ± 0.1 |
| R59M | 0.4 ± 0.1 |
| R59A | 0.14 ± 0.01 |
Under multiple-turnover conditions, EF-G(R59K) was as fast as the wild-type factor within the time resolution of the experiment (Figure 5A), whereas the replacements R59A and R59M led to a considerable decrease in the rate of translocation. The same tendency was found for single-round translocation, except that now a difference in the rate of translocation with wild-type EF-G and EF-G(R59K) became apparent (Figure 5B). The rates of single-round translocation increased with the concentration of R59 mutants (Figure 5C). As saturation was not reached in these experiments, kapp values for translocation at 2 µM factor have to be compared (Table II). The rate of translocation with EF-G(R59K) was decreased five times relative to the wild type, and this factor is likely to remain at saturating concentrations of factor. The mutations to methionine and alanine had much stronger effects, reducing the translocation rate 20- and 50-fold, respectively.
Fig. 5. Translocation activity of EF-G R59 mutants. (A) Multiple-turnover translocation. EF-G wt (squares), R59K (upward triangles), R59M (downward triangles), R59A (diamonds), no EF-G (circles). (B) Single-round translocation. Time courses were evaluated by single-exponential fitting with the kapp values given. 1, wild-type EF-G(4.4 s–1); 2, R59K (0.8 s–1); 3, R59M (0.17 s–1); 4, R59A (0.04 s–1). (C) Dependence of kapp of translocation on the concentration of mutant and wild-type (inset) EF-G. For kapp values measured at 2 µM factor see Table II. Symbols are as in (A). Experiments were performed as in Figure 4.
Discussion
GTPase activation in EF-G
Arginine residues play a crucial role in the catalysis of GTP hydrolysis by several GTPases such as Ras, Rho or the Gα subunits of heterotrimeric G proteins. In the case of Ras and Rho, the catalytic arginine is provided in trans by the respective GAP (Ahmadian et al., 1997; Rittinger et al., 1997a,b; Scheffzek et al., 1997). Upon interaction with their respective target GTPase, both RasGAPs and RhoGAPs stabilize the switch II regions and introduce an arginine residue (‘arginine finger’) into the nucleotide binding site that neutralizes the developing negative charge on the pentavalent transition state of the leaving phosphate group. Such arginine residues have also been identified in the catalytic domains of two yeast GAPs activating the Ypt GTPases, again suggesting an arginine-finger mechanism of GTPase activation (Albert et al., 1999).
In contrast, the strong stimulation by L7/12 of the GTPase activity of EF-G does not involve an extrinsic arginine residue donated by L7/12 (Savelsbergh et al., 2000). This does not exclude the possibility that the stimulation by L7/12 on the ribosome is augmented by other ribosomal components, and that other ribosomal proteins may donate a catalytic arginine in trans. In addition to L7/12, the factor binding site contains proteins L10 and L11 bound to the 1060 region of 23S rRNA as well as the α-sarcin–ricin stem–loop region (SRL) around nucleotide 2660 of 23S rRNA and two proteins, L6 and L14, adjacent to it (Ban et al., 1999). Each one of the four proteins, L10, L11, L6 and L14, contains conserved arginine residues. However, L10 does not contribute to the GTPase activation of EF-G, as the rate of GTP hydrolysis by EF-G is the same in the presence of L7/12 alone and in the presence of the complex of L10 and four copies of L7/12 (L8 complex) (Savelsbergh et al., 2000). Also, L11 is unlikely to be important for GTPase stimulation, because there are viable strains of bacteria that completely lack L11 (Stöffler et al., 1980). Finally, the contact of EF-G with the SRL region was not yet formed in the state stalled immediately after GTP cleavage, as shown by both electron cryomicroscopy (Stark et al., 2000) and footprinting analysis (Rodnina et al., 1999; A.Savelsbergh, N.B.Matassova, M.V.Rodnina and W.Wintermeyer, submitted). Taken together, these results indicate that the ribosomal proteins at or close to the factor binding site are unlikely to take part in GTPase activation by providing catalytic arginines in trans. Thus, if a catalytic arginine is required for GTP hydrolysis, then it would probably have to be intrinsic to EF-G.
Examples where catalytic arginine residues are constituents of G proteins are provided by the Gα subunits of heterotrimeric G proteins. The GTPase activity of these proteins is stimulated by RGSs (De Vries and Farquhar, 1999), and the crystal structure of RGS4 bound to Gαi1 activated by AlF4– revealed that RGS4 stabilizes the switch regions and re-orients the catalytic arginine (R178 of Gαi1) (Tesmer et al., 1997). The homolog of the catalytic arginine in Gα is R59 in the effector region of E.coli EF-G, a residue that is strictly conserved in EF-G as well as in other translation factors, EF-Tu and IF2. However, mutations of the homologous arginine (R59) in EF-Tu from T.thermophilus did not significantly affect the GTPase activity of the factor (Zeidler et al., 1995). In line with this result, our data show that the R59 mutants of EF-G retain GTPase activity. Thus, R59 does not seem to be involved in the stabilization of the GTPase transition state of EF-G.
The replacement of another conserved arginine residue in E.coli EF-G, R29, with lysine, methionine or alanine, completely abolished the GTPase activity of EF-G, whereas nucleotide binding and ribosome interaction of the factor were not affected. Thus, R29 has an essential role in GTP hydrolysis. This may be direct, as discussed below, or indirect, in that R29 may be involved in the interaction of EF-G with the ribosome that activates the GTPase or in the stabilization of the conformation of EF-G in the transition state of GTP hydrolysis. However, in such a case one would expect that lysine could at least partially replace arginine in these functions, which is not observed. Therefore, this possibility seems to be less likely, although it cannot be excluded at present.
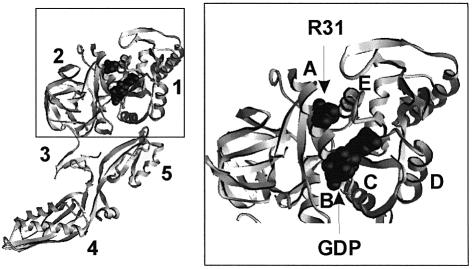
The other possibility is that R29 participates directly in the stabilization of the GTPase transition state, i.e. plays a role similar to that of the catalytic arginine residue of Gα. The homologous residue (R31) in T.thermophilus EF-G is located in helix A, and the distance of the guanidino group of R31 to the β-phosphate of GDP is ∼11 Å (Figure 6). Therefore, the direct participation of R29 of E.coli EF-G in the catalysis of GTP cleavage would require a conformational change of the G domain to bring R29 closer to the phosphates of GTP. An internal rotation of helix A would probably be insufficient to bring about such a movement of R29, and a more extensive re-orientation of helix A would be required. Examples of an activation by induced conformational changes of the catalytic site are provided by the heterotrimeric G proteins, where the activating proteins (RGS) bind at the effector regions of the Gα subunit, or, alternatively, by the ARF GTPases in which the activating protein (ARFGAP) binds at a distance from the nucleotide binding site (Goldberg, 1999). In the case of EF-G, however, GTPase activation, presumably by the binding of ribosomal protein L7/12, would have to induce a different structural change, involving a re-orientation of helix A, in order to position R29 into the proper position for GTP hydrolysis.
Fig. 6. Location of R31 (R29 in E.coli EF-G) in the crystal structure of EF-G from T.thermophilus. 1–5, domains of EF-G. A–E, helices in domain 1 (G domain). The positions of R31 and GDP are shown in space-filling representation. The location of R61 (R59 in E.coli EF-G) is not known, because this part of EF-G (effector region) is not resolved in the available crystal structures.
Although the model of R29 acting as the catalytic arginine in EF-G consistently explains the available data, there are some caveats. One is that the model cannot be extended to the related elongation factor, EF-Tu, because EF-Tu has alanine at the homologous position. Thus, the model implies that the two factors differ with respect to the molecular details of how GTP hydrolysis is catalyzed. In the case of EF-Tu, it is still possible that a catalytic arginine might be provided in trans by the ribosome. Alternatively, GTP hydrolysis by EF-G and EF-Tu might not involve a catalytic arginine at all, but follow a different mechanism, perhaps similar to that of ATP hydrolysis in myosin (Smith and Rayment, 1996). In such a case, R29 of EF-G would have an essential function in the stabilization of the transition state of GTP hydrolysis.
GTP hydrolysis and translocation
Previously it was shown by rapid kinetic methods that the rate of translocation is decreased ∼50 times when GTP is replaced with a number of non-hydrolyzable GTP analogs or GDP (Rodnina et al., 1997), indicating an important role of GTP hydrolysis for EF-G function in translocation. The present work with a GTPase-inactive mutant of EF-G avoids the use of GTP analogs, thereby providing complementary information. The data demonstrate that abolishing GTP hydrolysis by mutating R29 of EF-G inhibits translocation to about the same extent as replacing GTP with non-hydrolyzable analogs or GDP (Rodnina et al., 1997), in keeping with an important role of GTP hydrolysis for attaining rapid translocation. The same parallel was seen in turnover, because the turnover of EF-G, i.e. the dissociation from the ribosome after GTP hydrolysis and/or translocation, was inhibited either by replacing GTP with non-hydrolyzable analogs or by blocking GTP hydrolysis by mutating R29.
Mutations of R59 of EF-G have only small effects on GTP hydrolysis, whereas they inhibit translocation up to 50-fold. Interestingly, the rate of translocation is decreased only 5-fold when R59 is replaced with lysine, indicating that for proper function a positively charged amino acid is required at position 59. This suggests that R59 may be involved in a salt bridge that forms at some point during translocation and helps to stabilize the transition state. The interaction could be either intermolecular, involving the ribosome, or intramolecular, involving another domain of EF-G (possibly domain 5) and stabilizing an intermediate conformation of EF-G. Alternatively, the presence of a charged residue in the solvent-exposed effector loop may be important for the conformational dynamics of the factor.
The translocation activity of the other two mutants, R59M and R59A, is as low as that of the R29 mutants, although the inhibition has different reasons. In one case (R29 mutants), it is due to the lack of GTP hydrolysis, whereas in the other (R59 mutants) it may be caused by an inhibition of a conformational change that in wild-type EF-G is induced by GTP hydrolysis and/or Pi release. Thus, the present results support the model of EF-G function (Rodnina et al., 1997; Stark et al., 2000), in which conformational changes of the G domain caused by GTP hydrolysis and Pi release lead to changes of the overall conformation of EF-G, which, in turn, promote translocation by inducing a structural change of the ribosome.
Materials and methods
Buffers and reagents
Buffer A is 50 mM Tris–HCl pH 7.5, 70 mM NH4Cl, 30 mM KCl, 7 mM MgCl2 and 1 mM dithiothreitol (DTT). Buffer B is buffer A at pH 8.0 containing 8 M urea, 5 mM β-mercaptoethanol and 0.1 mM phenylmethylsulfonyl fluoride (PMSF). Buffer C is buffer A containing 5 mM β-mercaptoethanol and 0.1 mM PMSF. Buffer D is buffer C containing 500 mM imidazole. Ni-NTA was obtained from Qiagen.
2′(3′)-O-(N-methylanthraniloyl)guanosine 5′-triphosphate (mant-GTP) was purchased from Molecular Probes. GTP was purchased from Roche Diagnostics.
Ribosomes, mRNAs, tRNAs and factors
70S ribosomes from E.coli MRE 600 were prepared as described (Rodnina and Wintermeyer, 1995). f[3H]Met-tRNAfMet, [14C]Phe-tRNAPhe, MFTI-mRNA, EF-Tu and initiation factors were prepared and purified as described (Rodnina et al., 1994a,b, 1999).
Preparation of EF-G mutants
The plasmid used as a template for mutagenesis, pET24 EF-G, contained the E.coli EF-G gene and an additional eight amino acids containing six histidines at the C-terminus. The plasmid was provided by Kevin S.Wilson and Harry Noller, University of California, Santa Cruz, CA. EF-G(R29) mutants were generated using the Chameleon™ Double-stranded site-directed mutagenesis kit (Stratagene). EF-G(R59) mutants were made by PCR mutagenesis. All mutations were verified by DNA sequencing.
EF-G mutants were expressed in E.coli BL21(DE3)pLysS (Novagen) and purified from cell extracts by affinity chromatography on Ni-NTA–agarose in buffer B. For renaturation, the urea in buffer B was gradually diluted out with buffer C, and the proteins were finally eluted in buffer D. Proteins were transferred into buffer A and concentrated by ultrafiltration on Centricon 10 filters (Amicon). Protein concentrations were determined by the Bradford method using bovine serum albumin as a standard. The purity of the proteins was >90% according to SDS–PAGE. Wild-type EF-G, containing a histidine tag at the C-terminus, was expressed and purified in the same way as the EF-G mutants.
GTPase activity
To measure the time courses of GTP hydrolysis, ribosomes (0.2 µM) were mixed with EF-G (0.04 µM) and the reaction was started by addition of [γ-32P]GTP (1 mM). Samples (50 µl) were quenched at the time points indicated by adding 25 µl of 2 M HClO4/6 mM potassium phosphate, and [32P]phosphate was determined after extraction into 500 µl of isopropyl acetate in the presence of sodium molybdate (20 mM). Scintillation liquid (Quickszint 501/30% Triton X-100) was added to the organic phase and counted in the scintillation counter (2500 TR; Packard-Bell). Steady-state kinetic parameters were determined under conditions of initial velocity. EF-G (0.04 µM) was incubated with [γ-32P]GTP (50 µM) and increasing amounts of the ribosomes for 6 min at 37°C. Samples were quenched as described above and analyzed by thin-layer chromatography on PEI-cellulose in 0.5 M potassium phosphate pH 3.5. The extent of hydrolysis was quantified using a Bio-Rad phosphoimager. Blanks for [γ-32P]GTP hydrolysis of EF-G and ribosomes were measured separately and subtracted. The steady-state kinetic parameters were evaluated by fitting a Michaelis–Menten equation using TableCurve software (Jandel Scientific).
Dissociation of Pi
Pi release from EF-G after GTP hydrolysis was measured in a stopped-flow apparatus (Applied Photophysics), monitoring the fluorescence change of PBP labeled with a coumarin derivative, MDCC (Brune et al., 1994). Ribosomes (0.2 µM, concentration after mixing) were mixed with EF-G (2 µM), GTP (100 µM) and MDCC-PBP (2.5 µM). The fluorescence of MDCC was excited at 425 nm and monitored after passing a KV450 filter (Schott). The rate constant of Pi binding to MDCC-PBP, k1 = 96 µM–1s–1, was determined in separate experiments (A.Savelsbergh, N.B.Matassova, M.V.Rodnina and W.Wintermeyer, submitted). The concentration of MDCC-PBP (2.5 µM) was chosen such that the rate of Pi binding to MDCC-PBP (k1′ = k1⋅[MDCC-PBP] = 240 s–1) was faster than the rate of GTP hydrolysis in EF-G (120 s–1 at 2 µM EF-G; Rodnina et al., 1997) and thus the uptake of Pi liberated from the factor by MDCC-PBP did not significantly affect the measurements. To minimize phosphate contaminations, all solutions and the stopped-flow apparatus were pre-incubated with 0.2 mM 7-methylguanosine and 0.1 U/ml purine nucleoside phosphorylase (Brune et al., 1994).
Mant-GTP binding to EF-G mutants
Mant-GTP binding to R29 mutants of EF-G was monitored by the fluorescence change of mant-GTP by stopped flow. The fluorescence of mant-GTP was excited at 349 nm and measured after passing a KV408 filter (Schott). Experiments were performed in buffer A at 20°C by rapidly mixing equal volumes (60 µl each) of mant-GTP (0.5 µM, concentration after mixing) with varying concentrations of EF-G and monitoring the time courses of the fluorescence change. The data were evaluated by fitting an exponential function with the characteristic time constant, kapp, the amplitude, A, and another variable for the final signal, F∞, according to the equation F = F∞ + A⋅exp(–kapp⋅t), where F is the fluorescence at time t. The results of several (typically five) experiments were used to calculate the average A and the standard deviation. Calculations were performed using TableCurve software (Jandel Scientific).
Ribosome complex formation and translocation assays
To prepare 70S initiation complexes, ribosomes (0.5 µM) were programmed with a 3-fold excess of MFTI-mRNA by incubating with a 1.5-fold excess of E.coli IF1, IF2, IF3, f[3H]Met-tRNAfMet and 1 mM GTP in buffer A for 45 min at 37°C. Ternary complex, EF-Tu⋅GTP⋅[14C]Phe-tRNAPhe, was prepared by incubating 0.55 µM EF-Tu with 1 mM GTP, 3 mM phosphoenol pyruvate, 0.5 mg/l pyruvate kinase and 1.2 µM [14C]Phe-tRNAPhe for 15 min at 37°C. Ternary complex was added to the initiation complex and incubated for 1 min at 37°C to form pretranslocation complexes. To 40 µl of pretranslocation complex (0.25 µM), 10 µl of EF-G (0.2 µM) was added to induce translocation. The extent of translocation was measured by the formation of fMetPhe-puromycin (1 mM puromycin, 10 s at 37°C; Rodnina et al., 1997).
Single-round translocation was measured by stopped flow (Rodnina et al., 1997). Pretranslocation complex (0.1 µM, final concentration after mixing) with f[3H]Met[14C]Phe-tRNAPhe(Prf16/17) in the A site and tRNAfMet in the P site was mixed with EF-G (0.8 µM) in the presence of GTP (1 mM) or caged GTP (0.1 mM) in buffer A at 37°C. For translocation experiments with caged GTP or without nucleotide, initiation complexes were purified from GTP by ultracentrifugation through 0.2 ml sucrose cushions (1.1 M sucrose in buffer A) at 55 000 r.p.m. in the Sorvall ultracentrifuge (RC M120 GX, rotor S55S). To form the pretranslocation complex, ribosome pellets were resuspended in buffer A and added to ternary complex, EF-Tu⋅GTP⋅[14C]Phe-tRNAPhe(Prf16/17), purified by gel filtration (Rodnina et al., 1994a). Proflavin fluorescence was excited at 460 nm and measured after passing a KV500 filter (Schott). The results of several (4–8) experiments were averaged. The data were evaluated by fitting an exponential function with the characteristic time constant, kapp, and the fluorescence amplitude, A, as described above for nucleotide binding experiments.
Acknowledgments
Acknowledgements
We thank Andreas Savelsbergh for MDCC-labeled PBP and for the help with experiments of Figure 2; Kevin Wilson and Harry Noller for the plasmid construct coding for EF-G with a C-terminal histidine tag; Yuri Semenkov and Vladimir Katunin for gifts of purified tRNAs; Martin Webb for the E.coli strain overproducing mutant PBP; Roger Goody for caged GTP; and Simone Möbitz and Petra Striebeck for expert technical assistance. The work was supported by the Alfried Krupp von Bohlen und Halbach-Stiftung, the European Union, the Fonds der Chemischen Industrie, and the Deutsche Forschungsgemeinschaft. D.M. acknowledges a fellowship of the Werner Richard–Dr Carl Dörken-Stiftung.
References
- Ævarsson A., Brazhnikov,E., Garber,M., Zheltonosova,J., Chirgadze,Y., al-Karadaghi,S., Svensson,L.A. and Liljas,A. (1994) Three-dimensional structure of the ribosomal translocase: elongation factor G from Thermus thermophilus. EMBO J., 13, 3669–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal R.K., Penczek,P., Grassucci,R.A. and Frank,J. (1998) Visualization of elongation factor G on the Escherichia coli 70S ribosome: the mechanism of translocation. Proc. Natl Acad. Sci. USA, 95, 6134–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadian M.R., Stege,P., Scheffzek,K. and Wittinghofer,A. (1997) Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nature Struct. Biol., 4, 686–689. [DOI] [PubMed] [Google Scholar]
- Albert S., Will,E. and Gallwitz,D. (1999) Identification of the catalytic domains and their functionally critical arginine residues of two yeast GTPase-activating proteins specific for Ypt/Rab transport GTPases. EMBO J., 18, 5216–5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N., Nissen,P., Hansen,J., Capel,M., Moore,P.B. and Steitz,T.A. (1999) Placement of protein and RNA structures into a 5 Å-resolution map of the 50S ribosomal subunit. Nature, 400, 841–847. [DOI] [PubMed] [Google Scholar]
- Brune M., Hunter,J.L., Corrie,J.E. and Webb,M.R. (1994) Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry, 33, 8262–8271. [DOI] [PubMed] [Google Scholar]
- Coleman D.E., Berghuis,A.M., Lee,E., Linder,M.E., Gilman,A.G. and Sprang,S.R. (1994) Structures of active conformations of Gia1 and the mechanism of GTP hydrolysis. Science, 265, 1405–1412. [DOI] [PubMed] [Google Scholar]
- Czworkowski J., Wang,J., Steitz,T.A. and Moore,P.B. (1994) The crystal structure of elongation factor G complexed with GDP, at 2.7 Å resolution. EMBO J., 13, 3661–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries L. and Farquhar,M.G. (1999) RGS proteins: more than just GAPs for heterotrimeric G proteins. Trends Cell Biol., 9, 138–144. [DOI] [PubMed] [Google Scholar]
- Goldberg J. (1999) Structural and functional analysis of the ARF1–ARFGAP complex reveals a role for coatomer in GTP hydrolysis. Cell, 96, 893–902. [DOI] [PubMed] [Google Scholar]
- Mittal R., Ahmadian,M.R., Goody,R.S. and Wittinghofer,A. (1996) Formation of a transition-state analog of the Ras GTPase reaction by Ras-GDP, tetrafluoroaluminate and GTPase-activating proteins. Science, 273, 115–117. [DOI] [PubMed] [Google Scholar]
- Parmeggiani A. and Sander,G. (1981) Properties and regulation of the GTPase activities of elongation factors Tu and G and of initiation factor 2. Mol. Cell. Biochem., 35, 129–158. [DOI] [PubMed] [Google Scholar]
- Rittinger K., Walker,P.A., Eccleston,J.F., Nurmahomed,K., Owen,D., Laue,E., Gamblin,S.J. and Smerdon,S.J. (1997a) Crystal structure of a small G protein in complex with the GTPase-activating protein rhoGAP. Nature, 388, 693–697. [DOI] [PubMed] [Google Scholar]
- Rittinger K., Walker,P.A., Eccleston,J.F., Smerdon,S.J. and Gamblin,S.J. (1997b) Structure at 1.65 Å of RhoA and its GTPase-activating protein in complex with a transition-state analogue. Nature, 389, 758–762. [DOI] [PubMed] [Google Scholar]
- Rodnina M.V. and Wintermeyer,W. (1995) GTP consumption of elongation factor Tu during translation of heteropolymeric mRNAs. Proc. Natl Acad. Sci. USA, 92, 1945–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina M.V., Fricke,R. and Wintermeyer,W. (1994a) Transient conformational states of aminoacyl-tRNA during ribosome binding catalyzed by elongation factor Tu. Biochemistry, 33, 12267–12275. [DOI] [PubMed] [Google Scholar]
- Rodnina M.V., Semenkov,Y.P. and Wintermeyer,W. (1994b) Purification of fMet-tRNA(fMet) by fast protein liquid chromatography. Anal. Biochem., 219, 380–381. [DOI] [PubMed] [Google Scholar]
- Rodnina M.V., Savelsbergh,A., Katunin,V.I. and Wintermeyer,W. (1997) Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature, 385, 37–41. [DOI] [PubMed] [Google Scholar]
- Rodnina M.V., Savelsbergh,A., Matassova,N.B., Katunin,V.I., Semenkov,Y.P. and Wintermeyer,W. (1999) Thiostrepton inhibits turnover but not GTP hydrolysis by elongation factor G on the ribosome. Proc. Natl Acad. Sci. USA, 96, 9586–9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelsbergh A., Mohr,D., Wilden,B., Wintermeyer,W. and Rodnina,M.V. (2000) Stimulation of the GTPase activity of translation elongation factor G by ribosomal protein L7/12. J. Biol. Chem., 275, 890–894. [DOI] [PubMed] [Google Scholar]
- Scheffzek K., Ahmadian,M.R., Kabsch,W., Wiesmuller,L., Lautwein,A., Schmitz,F. and Wittinghofer,A. (1997) The Ras–RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science, 277, 333–338. [DOI] [PubMed] [Google Scholar]
- Smith C.A. and Rayment,I. (1996) X-ray structure of the magnesium(II)⋅ADP⋅vanadate complex of the Dictyostelium discoideum myosin motor domain to 1.9 Å resolution. Biochemistry, 35, 5404–5417. [DOI] [PubMed] [Google Scholar]
- Sondek J., Lambright,D.G., Noel,J.P., Hamm,H.E. and Sigler,P.B. (1994) GTPase mechanism of G proteins from the 1.7-Å crystal structure of transducin α–GDP–AlF4. Nature, 372, 276–279. [DOI] [PubMed] [Google Scholar]
- Stark H., Rodnina,M.V., Wieden,H.-J., van Heel,M. and Wintermeyer,W. (2000) Large-scale movement of elongation factor G and extensive conformational changes of the ribosome during translocation. Cell, 100, 301–309. [DOI] [PubMed] [Google Scholar]
- Stöffler G., Cundliffe,E., Stöffler-Meilicke,M. and Dabbs,E.R. (1980) Mutants of Escherichia coli lacking ribosomal protein L11. J. Biol. Chem., 255, 10517–10522. [PubMed] [Google Scholar]
- Tesmer J.J., Berman,D.M., Gilman,A.G. and Sprang,S.R. (1997) Structure of RGS4 bound to AlF4-activated Gia1: stabilization of the transition state for GTP hydrolysis. Cell, 89, 251–261. [DOI] [PubMed] [Google Scholar]
- Traut R.R., Dey,D., Bochkariov,D.E., Oleinikov,A.V., Jokhadze,G.G., Hamman,B. and Jameson,D. (1995) Location and domain structure of Escherichia coli ribosomal protein L7/L12: site specific cysteine cross-linking and attachment of fluorescent probes. Biochem. Cell Biol., 73, 949–958. [DOI] [PubMed] [Google Scholar]
- Zeidler W., Egle,C., Ribeiro,S., Wagner,A., Katunin,V., Kreutzer,R., Rodnina,M., Wintermeyer,W. and Sprinzl,M. (1995) Site-directed mutagenesis of Thermus thermophilus elongation factor Tu. Replacement of His85, Asp81 and Arg300. Eur. J. Biochem., 229, 596–604. [DOI] [PubMed] [Google Scholar]