Abstract
Recombination events between non-identical sequences most often involve heteroduplex DNA intermediates that are subjected to mismatch repair. The well-characterized long-patch mismatch repair process, controlled in eukaryotes by bacterial MutS and MutL orthologs, is the major system involved in repair of mispaired bases. Here we present evidence for an alternative short-patch mismatch repair pathway that operates on a broad spectrum of mismatches. In msh2 mutants lacking the long-patch repair system, sequence analysis of recombination tracts resulting from exchanges between similar but non-identical (homeologous) parental DNAs showed the occurrence of short-patch repair events that can involve <12 nucleotides. Such events were detected both in mitotic and in meiotic recombinants. Confirming the existence of a distinct short-patch repair activity, we found in a recombination assay involving homologous alleles that closely spaced mismatches are repaired independently with high efficiency in cells lacking MSH2 or PMS1. We show that this activity does not depend on genes required for nucleotide excision repair and thus differs from the short-patch mismatch repair described in Schizosaccharomyces pombe.
Keywords: gene conversion/mismatch repair/Saccharomyces cerevisiae/short-patch mismatch repair
Introduction
The long-patch mismatch repair (MMR) process is involved in elimination of replicative errors, in heteroduplex DNA repair during recombination and in prevention of recombination between divergent sequences. In Escherichia coli, it is controlled by MutS, MutL and MutH, whereas in eukaryotes several MutS and MutL homologs have been described (for reviews see Kolodner, 1996; Modrich and Lahue, 1996). The biological importance of this system is evidenced by the severe mutator phenotype of the mutants, and in mammals defects in MMR result in cancer predisposition (for reviews see Modrich and Lahue, 1996; Peltomaki and Vasen, 1997).
However, the long-patch system is not the only mechanism that is able to correct mispaired bases. Besides systems specialized in the repair of specific mismatches, the existence of secondary generalized MMR processes has been suggested by a number of genetic, molecular and enzymic studies (reviewed in Kolodner, 1996). The existence of an MMR process that can at least partially substitute for, or complement the long-patch mechanism should also be a factor of genetic stability. Indeed, Fleck et al. (1999) have recently demonstrated in the yeast Schizosaccharomyces pombe the presence of a short-patch MMR activity involved in recombination and mutagenesis. This system is independent of the long-patch MMR pathway and is controlled by nucleotide excision repair (NER) genes. It corrects not only C/C mismatches, which are poorly recognized by the long-patch MMR process, but also other types of mismatch.
In this study, we present molecular and genetic evidence for the existence of short-patch MMR activity in Saccharomyces cerevisiae. It was revealed by DNA sequence analysis of tracts generated by recombination between parental alleles differing by ∼10% of silent mutations and by two non-silent mutations allowing prototroph selection. The sequence differences allow the localization of repair events that occurred on the DNA heteroduplex intermediates. In many cases, and especially in mutants deficient in long-patch MMR, we observed ‘patched’ sequences best explained by short-patch repair events. If this interpretation were right, we reasoned that this activity should repair independently two closely spaced mismatches. Homologous recombination assays show that this is indeed the case and that this short-patch MMR activity is not dependent on NER genes, in contrast to that described in S.pombe.
Results
Genetic system
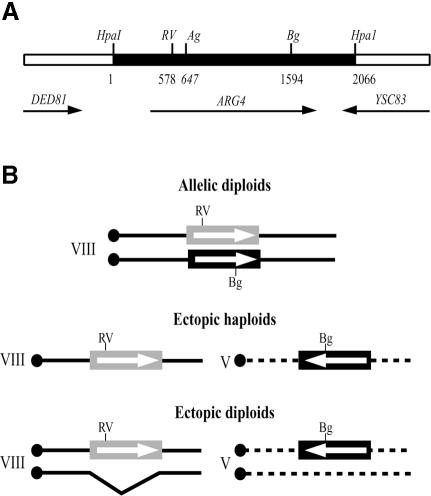
To construct a genetic system allowing the study of heteroallelic recombination between homeologous alleles, the ARG4 gene from Saccharomyces douglasii (ARG4D) was integrated into the genome of S.cerevisiae. The two ARG4C and ARG4D genes, shown in Figure 1A, differ by ∼8% of substitutions in the open reading frame (ORF) and 20% of base substitutions and small insertions/deletions in the intergenic regions (Adjiri et al., 1994). A 2.1 kb HpaI fragment containing arg4D mutated at the BglII restriction site (arg4D-Bg, a 4 bp insertion) was integrated into the S.cerevisiae genome. Two types of strains were used in this study (Figure 1B): allelic diploids and ‘ectopic cells’ containing the alleles in ectopic locations. In allelic diploids, both alleles are located at the natural position on chromosome VIII. One of the chromosomes carries the HpaI segment of S.douglasii bearing arg4D-Bg while the other allele, from S.cerevisiae, is mutated at the EcoRV site (arg4C-RV, a 2 bp deletion). The cells containing the ectopic genes have the arg4C-RV allele at its natural position on chromosome VIII and the arg4D-Bg allele integrated into chromosome V. The two genes are in opposite orientation with respect to the centromeres. In this configuration, the formation of a wild-type gene by a reciprocal exchange between the mutated sites would be lethal, due to the formation of acentric and dicentric chromosomes. Ectopic diploids were obtained by crossing ectopic haploids with cells bearing a deletion of the HpaI–ARG4–HpaI segment, so that only ectopic alleles can interact to form an ARG+ gene.
Fig. 1. Structure of the ARG4 chromosomal region and schematic representation of the different genetic systems used. (A) The organization of this region is similar in S.cerevisiae and S.douglasii. The ARG4 ORFs differ by 8% of base substitutions, and the intergenic regions by ∼20% of substitutions and small insertions/deletions. The HpaI segment, shown in black, is the region substituted in S.cerevisiae by the corresponding fragment of S.douglasii. Frameshift mutations were introduced at the EcoRV (RV, –2 bp), the AgeI (Ag, +4 bp) or the BglII site (Bg, +4 bp). Coordinates are given with respect to the S.cerevisiae sequence, number 1 being the first nucleotide of the HpaI site. (B) Diagram of the genetic constructs involving RV and Bg.
Rationale of the molecular analyses
Owing to the presence of a large number of silent polymorphic sites that differentiate both alleles, our genetic system allows the structure of recombinant tracts to be characterized precisely. It permits it to be asked whether the absence of the long-patch MMR uncovers another repair mechanism, which may confer tract profiles different from those found in wild-type cells.
We chose to study ectopic recombinants in order to be able to amplify one of the alleles specifically. Our analyses were focused on the ARG+ alleles located on chromosome VIII and not at the ectopic position (see Materials and methods). Among the mitotic ARG+ recombinants, the wild-type allele was found to be equally distributed between the two loci. After meiosis, >90% of the ARG+ alleles were on chromosome VIII, indicating a preferential conversion of the RV mutated site. This difference between mitotic and meiotic recombinants probably reflects, as for allelic diploids, a polarity of meiotic gene conversion due to initiation by a double-strand break in the promotor region of ARG4 (Nicolas et al., 1989). The polarity results in a much higher frequency of conversion at the EcoRV than at the BglII site. Because the system excludes the recovery of ARG+ clones resulting from cross-overs between the mutated sites, we know that we analyzed the recipient allele converted by the ectopic donor carrying the EcoRV+ site. Assuming that the event occurred through formation of a heteroduplex DNA, the heteroduplex region must have covered the mutated RV site, forming an RV/EcoRV+ mismatch, and is expected to most often not reach the 1 kb distant BglII+ site. If the RV/EcoRV+ mismatch is repaired to EcoRV+/EcoRV+, both strands of the heteroduplex encode a wild-type ARG4+ gene or its complementary information and are recovered in daughter cells on selective medium. If this mismatch remains unrepaired before replication, only one strand encodes ARG4+ and is recovered in the colony.
In wild-type cells, the RV/EcoRV+ mismatch is expected to be repaired, leading to the rescue of both strands of the heteroduplex. On the contrary, in msh2 mutants, this mismatch is expected often to remain unrepaired, and the progeny of only one strand will be recovered. However, data in the literature indicate that in msh2 cells, both strands of a heteroduplex appeared to be recovered in a significant proportion of cases. Notably, an analysis of meiotic segregation of the arg4-RV heterozygous mutation in msh2 cells showed that non-Mendelian events include 30–50% of EcoRV+/EcoRV+ or RV/RV convertants (Alani et al., 1994). These could be generated either through double-strand gap repair (Orr-Weaver et al., 1981; Szostak et al., 1983), or by the action of an alternative MMR process. We reasoned that in the latter case, repair of the RV/EcoRV+ mismatch would not necessarily be concerted with repair of the many mismatches present on the heteroduplex DNA. Mismatches left unrepaired would segregate upon replication and the ARG+ colony should in fact be sectored for silent mutations. The sequence profile of ARG4 amplified from the whole colony should then indicate the presence of two different DNAs, uncovered by superimposed bases corresponding to the sites of unrepaired mismatches. The finding of such a sequence profile would demonstrate the involvement of an intermediary heteroduplex and would allow detection of an alternative MMR process.
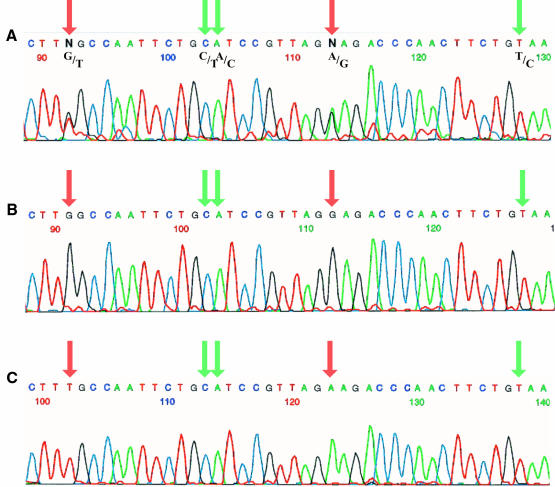
We found such cases and an example is shown in Figure 2. This recombinant was selected as an ARG+ mitotic colony from msh2 ectopic cells. On the first line is shown the sequence of a 40 bp tract of ARG4 amplified from the whole original clone. The presence of overlapping bases (red arrows) at two sites reveals an intermediary heteroduplex containing two unrepaired mismatches that were resolved by replication, as evidenced by the two different sequences found in subclones (middle and lower sequences). In between these two unrepaired mismatches, the two adjacent C/T and A/C mismatches (or G/A and T/G, green arrow) were co-repaired and the excision tract was at most 20 nucleotides long, indicating a short-patch repair event. Another mismatch, T/C (or A/G) at the right, was also repaired. Examination of the whole sequence allows the organization of the patches on each strand to be determined and the original recombinant molecule to be visualized.
Fig. 2. Sequence profile uncovering short-patch repair. (A) A partial sequence of ARG4 amplified from a single recombinant. The arrows point to sequence differences between the parental alleles, which are indicated underneath. Red arrows correspond to sites where the two parental bases are superimposed indicating an absence of MMR and uncovering a mixed clone. Green arrows indicate sites where only one of the parental bases is found. (B and C) The sequence of two subclones derived from the initial mixed clone.
Molecular evidence for the existence of a short-patch MMR process
Evidence for short-patch repair tracts was first obtained from mitotic cells. To investigate whether similar results are found after meiosis, we sequenced a few meiotic recombinants as well, which also revealed a short-patch repair activity. However, due to the small number of meiotic tracts analyzed in msh2 cells, this sample should not be considered as representative of the recombinant cell population.
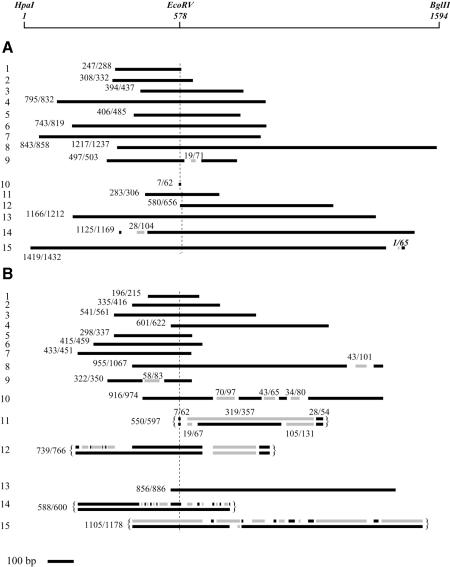
In Figure 3 are shown the length and position of the mitotic and meiotic recombinant tracts, as indicated in the legend. Their exact size is unknown since their borders may be located anywhere between two polymorphic sites. Therefore, we have indicated the minimal and maximal size of the entire rearranged region and that of the intervening tracts.
Fig. 3. Length, position and structure of recombinant tracts. Donor DNA is in black and recipient DNA in gray. The maximal and minimal size (in base pairs) of the whole rearranged region is indicated on the left, and that of the internal rearrangements above the sequence. Gaps are regions of identity between the alternate S.cerevisiae and S.douglasii tracts. (A) Wild-type cells. Tracts nos 1–9 were obtained from mitotic events in ectopic haploids (FF18248). Tracts nos 10–15 were obtained from meiotic events in ectopic diploids (FF181387). (B) msh2 cells. The 12 first cases correspond to mitotic events in ectopic haploids (FF181378); the three others are meiotic events in ectopic cells (nos 13 and 14, FF181396) and in allelic cells (no. 15, Ec182). For recombinants 11, 12, 14 and 15, both strands of the original recombinant were recovered and sequenced. No. 15 was selected as an ARG+/arg– sectored colony. Cases nos 12, 14 and 15 are detailed in Figure 4.
For wild-type cells (Figure 3A), out of nine recombination tracts from spontaneous mitotic events (cases 1–9), one is discontinuous, with a short intervening tract of S.cerevisiae covering four polymorphic sites. Among the five meiotic tracts (nos 10–15), two are discontinuous (nos 14 and 15) and include five and one polymorphic sites, respectively.
In the msh2 context (Figure 3B), among the 12 mitotic tracts (cases 1–12), five are discontinuous (nos 8–12), and among the three meiotic ones, two are discontinuous (nos 14 and 15). Case 15 was screened as a monosporic sectored ARG+/arg– colony derived from allelic diploids. Allelic diploids, rather than ectopic ones, were used to screen ARG+/arg– sectors because the rates of recombination between the homeologous genes are much higher in allelic cells (our unpublished results), allowing easier screening of sectors by replica-plating of unselected monosporic clones.
The sequence of both strands of the intermediary recombination structure could be determined in two mitotic (nos 11 and 12) and two meiotic (nos 14 and 15) cases. As expected, recombinants 11, 12 and 14, selected as ARG+, contained the EcoRV+ site on both strands of the intermediary heteroduplex DNA, while in case 15 a mismatch at this site persisted. These four sectored clones are not due to an artifact where two independent colonies would have been analyzed. Indeed, for three of them, the recombinant tract on each strand starts and ends at the same position. In the fourth case (no. 15), both terminal regions of the intermediary heteroduplex DNA appear as unrepaired. However, five unlinked heterozygous markers segregated in this cross and an identical genotype was found for all ARG+ and arg– subclones tested. The probability for independent events to yield the same reassortment of markers is very low (3%), which strongly argues that the initial colony derived from a single spore.
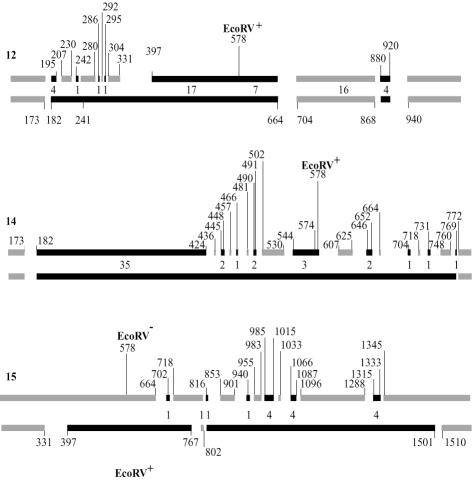
For these four recombinants, the recovery and sequencing of both strands of the original duplex allow localization of PMS tracts (unprocessed heteroduplex DNA detected as a post-meiotic or post-mitotic segregant), conversion tracts (donor sequence on both strands) and restoration tracts (recipient sequence on both strands). In several cases, the patched structure of the tract would be very difficult to interpret differently than by short-patch MMR, at least for regions consisting of short alternate patches of PMS, conversion or restoration. Recombinants 12, 14 and 15 are detailed in Figure 4. They display regions of alternate short-patch PMS and conversion tracts, presumably formed by independent MMR events with preferential repair of the recipient DNA strand. Indeed, in these three recombinants a single restoration event was detected (no. 15), and mismatches were most often repaired by excision on the recipient strand.
Fig. 4. Detailed structure of recombinant tracts. These three recombinants were recovered in msh2 cells. They correspond to one mitotic and two meiotic events (case nos 12, 14 and 15 in Figure 3B). The numbers are the coordinates, as defined in the legend of Figure 1, and indicate the position of the first and last polymorphic site that delimits rearranged segments. Numbers between the two strands of each duplex are the numbers of mismatches that were repaired in the conversion and restoration tracts.
In 14 repair events, a single mismatch was involved (11 cases shown in Figure 4 and three additional cases from Figure 3). Three transversions and 11 transitions were generated: two T→A and one C→A; three G→A, five A→G, two C→T and one T→C. Only G→C and C→G transversions generated by repair of G/G or C/C mismatches are not found in our sample, which does not mean that these mismatches are not repairable. Since each individual substitution can be generated by repair of two alternative mismatches, we cannot determine which one was involved. Besides G/G and C/C, all other types of mismatch could have been involved in the observed repair events. Cases where adjacent mismatches were co-repaired do not allow the determination of which of them was the initial target since the tract could have resulted from a single or from several independent repair events. The maximal size of an excision tract corresponds to the distance between unrepaired flanking mismatches. For events that involved a single mismatch, this size ranges from 11 to 53 nucleotides. Thus, the excision tracts are very short.
The absence of any significant difference concerning the short-patch repair events among the cases recovered from mitotic and meiotic cells suggests that the processes involved in their formation are similar during mitosis and meiosis.
Differential effect of msh2 and pms1 mutations on recombination between homologous alleles carrying close or distant mutations
Recombination in heteroallelic cells can result in the formation of a wild-type gene either by a reciprocal exchange between the mutations or by non-reciprocal transfer covering one mutation. In both mitotic and meiotic yeast cells, the majority of the events are due to conversion resulting from heteroduplex DNA formation followed by MMR (Roman, 1956; Fogel et al., 1981; Petes et al., 1991). If two sites are close together, the probability of them being included in the same heteroduplex and being repaired by a single long-patch excision tract is high. Such events will not form wild-type genes. However, if a short-patch repair process is able to act independently on these sites, a wild-type gene can be generated. Therefore, mutations like msh2 or pms1, which eliminate the long-patch repair, may uncover a short-patch repair mechanism by increasing prototroph frequency. Such a hyper-recombination effect is not expected if the mutated sites are located far apart, since they would rarely be included together in a heteroduplex DNA.
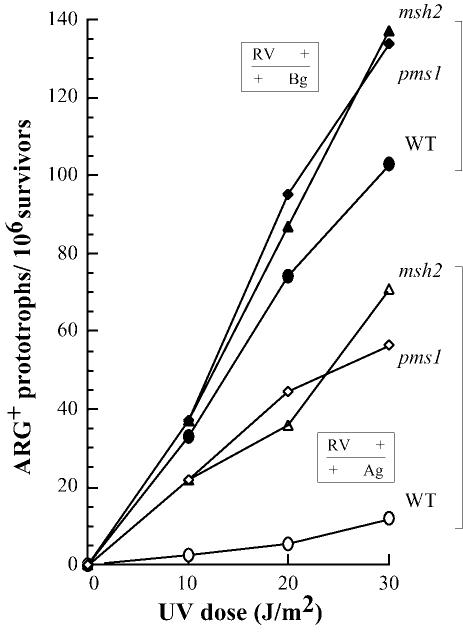
We tested the effect of msh2 and pms1 mutations on recombination in homologous heteroallelic cells containing mutations that are either closely spaced (RV and Ag, 69 bp apart) or distant (RV and Bg, 1016 bp apart). The RV mutation is a 2 bp deletion and the Bg and Ag mutations are each a 4 bp insertion. The alleles are located in an ectopic position, the Ag or the Bg allele being integrated on chromosome V. Mitotic recombination was measured in haploids, and meiotic recombination in diploids obtained by crossing the same haploids with cells deleted for the ARG4 locus. UV-induced and spontaneous rates of mitotic recombination were measured. As seen in Figure 5, both the msh2 and the pms1 mutations increased the UV induction of intragenic ARG+ recombinants as compared with the wild-type response. This increase is modest when the arg4 mutations are far apart (1.1–1.3 according to the dose) and was much more pronounced when the sites are close together (6–9.5). Comparable results were obtained for spontaneous mitotic rates (Table I): increases over the wild-type levels in long-patch MMR defective mutants were found to be 1.1 (pms1) and 1.6 (msh2) for cells with distant mutated sites, while for cells with close sites the increase was 6.3 (pms1) and 7.3 (msh2). In meiotic cells (Table I), the msh2 mutation exhibits a hyper-recombination effect (four times) only when the alleles are located close together
Fig. 5. UV induction of ARG+ recombinants in wild-type and msh2 cells, heteroallelic for distant or close mutations in ARG4. Open symbols: heteroallelic cells with closely spaced mutations (wild type, Ec153; msh2, Ec144000; pms1, FF181617); closed symbols: heteroallelic cells with distant mutations (wild type, Ec160; msh2, FF181379; pms1, FF181614).
Table I. Mitotic and meiotic rates of recombination between close or distant mutations.
| RV-Bg | RV-Age | ||||
|---|---|---|---|---|---|
| (A) | |||||
| WT | msh2 | pms1 | WT | msh2 | pms1 |
| (FF181246) | (FF181379) | (FF181614) | (Ec153) | (Ec144000) | (FF181617) |
| 3 (1) | 4.9 (1.6) | 3.2 (1.1) | 0.3 (1) | 2.2 (7.3) | 1.9 (6.3) |
| msh2 | msh2-rad1 | msh2-rad14 | msh2 | msh2-rad1 | msh2-rad14 |
| (FF181379) | (Ec168) | (Ec167) | (Ec144000) | (Ec144001) | (Ec144003) |
| 4.9 (1) | 3.5 (0.7) | 5.8 (1.2) | 2.2 (1) | 2.2 (1) | 2.8 (1.3) |
| pms1 | pms1-rad1 | pms1- rad14 | pms1 | pms1-rad1 | pms1- rad14 |
| (181614) | (FF181615) | (FF181616) | (FF181617) | (FF181619) | (FF181620) |
| 3.2 (1) | 4.6 (1.4) | 4.4 (1.6) | 1.9 (1) | 1.6 (0.8) | 1.7 (0.9) |
| WT | rad1 | rad14 | WT | rad1 | rad14 |
| (FF181246) | (Ec160) | (Ec166) | (Ec153) | (Ec153000) | (Ec153002) |
| 3 (1) | 2.9 (1) | 3.2 (1) | 0.3 (1) | 0.36 (1.2) | 0.27 (0.9) |
| (B) | |||||
| WT | msh2 | WT | msh2 | ||
| (Ec183) | (Ec184) | (Ec185) | (Ec186) | ||
| 1.2 (1) | 1 (0.8) | 0.2 (1) | 0.75 (3.8) |
(A) Mitotic rates for 107 divisions, between arg4-RV and arg4-Bg (columns 1–3) or between arg4-RV and arg4-Ag (columns 4–6). In parentheses is the increase over the wild-type, msh2 or pms1 rates. (B) Meiotic rates for 103 asci.
These results would be expected if a short-patch repair activity, uncovered in the msh2 or pms1 mutants, acts independently on mismatches separated by 69 bp.
The short-patch repair is not controlled by genes required for NER
In S.pombe, a short-patch MMR activity independent of MSH2 and PMS1 was shown to be controlled by NER genes (Fleck et al., 1999). msh2 and pms1 mutations display a stimulation effect on recombination between closely spaced alleles, largely abolished by mutations in SWI10, RHP14 and RAD16 genes involved in an NER pathway. It led us to ask whether the hyper-recombination effect of msh2 and pms1 that we observed is also dependent on the NER process. We therefore studied the effects of mutations in RAD14 or RAD1, two genes essential for the removal of UV-induced dimers (Unrau et al., 1971; Reynolds and Friedberg, 1981). RAD14 and RAD1 are the respective S.cerevisiae orthologs of the S.pombe RHP14 and RAD16 genes (Carr et al., 1994; Fleck et al., 1999).
As seen in Table I, neither rad1 nor rad14 mutation decreases significantly the elevated spontaneous recombination rates between the close RV and Ag sites observed in msh2 or pms1 mutants. Therefore, these NER genes do not control the short-patch MMR responsible for the differential hyper-recombination effect of pms1 or msh2 described above.
Discussion
In this study, we provide evidence for a short-patch MMR activity in S.cerevisiae, independent of the long-patch MMR system and of the NER pathway. It was first uncovered by sequence analysis of intragenic recombinant tracts that involved parental homeologous arg4 alleles, and further supported by a recombination assay involving heteroalleles with either distant or close mutations.
Molecular evidence for an alternative short-patch mismatch repair process
In wild-type cells, 12 of 15 tracts analyzed were found to be continuous, while three others contained a short intervening recipient sequence (Figure 3A). A majority of continuous tracts have also been found in previous studies on meiotic and mitotic conversion. This was observed not only when the parental DNAs differ by a few mutations (Petes et al., 1991), but also when they are homeologous (Harris et al., 1993; Chen and Jinks-Robertson, 1998, 1999). In both meiotic and mitotic cells, the tract continuity is believed to result from a long-patch excision, governed by MSH2, that can cover most if not all of the heteroduplex DNA (Bishop and Kolodner, 1986; Detloff and Petes, 1992). If the ‘Msh2’ repair process is the only one that repairs mismatches, continuous recombinant tracts will also be found in an msh2 mutant, with only one of the DNA strands being recombined. We found that this is not always the case.
In msh2 cells, ∼50% of the tracts analyzed contain one or more interruptions. The four cases where both strands were recovered indicate that the alternate stretches of parental DNAs correspond essentially to alternate regions of conversion and unrepaired heteroduplex DNA tracts (nos 11, 12, 14 and 15 in Figure 3B). Events nos 12, 14 and 15 (Figure 4) uncover regions with very short alternate tracts close together. We do not see any alternative for their formation other than by short-patch MMR. This repair favors the donor DNA, as does repair by the long-patch system. For example, event no. 14 has the recipient strand patched while the donor strand is continuous. In the four cases where both strands were recovered, there are 23 apparent stretches of conversion and four of restoration.
The preferential repair by excision of the recipient strand may well account for the relatively high frequency of continuous tracts (and of tracts with few patches) found in msh2 mutants. Indeed, by selecting ARG+ cells, the donor strand, most often unaffected by repair, should be preferentially recovered. This may not be the only explanation for the recovery of continuous tracts. It could also be that not all cells underwent short-patch repair, that replication segregated the strands before repair has occurred, or, finally, that some events involved double-strand gaps and no intermediary heteroduplex DNA (Orr-Weaver et al., 1981; Szostak et al., 1983).
Our data further show that this activity is not or not always processive along the DNA. If some repaired tracts involve several mismatches in a row, others involve a single mismatch. Rather long regions of unrepaired heteroduplex DNA are also observed, e.g. in case no. 15 (Figure 4) where three regions of 156, 192 and 267 bp, which contain 10, 13 and 24 mismatches, respectively, are left unrepaired. The nature of the mismatch does not explain why some of them escape repair, since identical mismatches are sometimes but not always repaired. It seems that mismatches are repaired individually with a limited probability. The size of the excision tract can be very short, with an upper limit of 11 nucleotides (conversions at positions 241, 286 and 295 in event no. 12, Figure 4).
The spectrum of action of the system is large. The different base substitutions found in tracts that involve a single mismatch indicate that all mismatches except C/C or G/G are potential substrates for short-patch repair. We cannot conclude that C/C and C/G are not repaired. First, the C→G or G→C mutations that differentiate the parental sequences and that could form these mismatches are rare, and, secondly, G→C or C→G transversions are observed in tracts uncovering repair of several mismatches. Specifically designed experiments will be needed to obtain the answer. The pertinence of this question lies in the fact that C/C mismatches are the only ones that are not a good substrate for the long-patch MMR (Kramer et al., 1989; Detloff et al., 1991).
Recombinant tracts are not longer in msh2 than in wild-type cells
Alani et al. (1994) proposed that MMR proteins control heteroduplex DNA length by binding to mismatches, which would explain the abolition by msh2 of meiotic conversion gradients seen in S.cerevisiae. If so, one can predict that recombinant tracts in homeologous systems will be longer in msh2 than in wild-type cells. We compared the recombinant tract length observed in wild-type and msh2 cells (Figure 3A). In wild-type haploid cells, the minimal length of mitotic tracts ranges from 247 to 1217 bp, with a mean of 605 bp, while in msh2 cells, it ranges from 196 to 955 bp, with a mean of 525 bp. These mean values are not significantly different, indicating that the mitotic tract length is not regulated by MSH2. For meiotic events, the small number of cases analyzed makes the conclusion less definitive. In wild-type cells, the tract lengths range from 7 to 1419 bp, with a mean size of 763 bp, while in msh2 cells the three tracts are 580, 856 and 1105 bp long, respectively (Figure 3B). These values fall into the range observed for tracts analyzed in spores from wild-type diploids, which argues that meiotic tract length is not under the control of MSH2.
The possible control by MSH2 of heteroduplex length was also recently questioned by Chen and Jinks-Robertson (1998, 1999) who used an artificial system of inverted 350 bp homeologous repeats. These authors reached different conclusions than ours. They found that meiotic conversion tracts were ∼50% longer in msh2 than in wild-type cells. For mitotic events, they obtained a similar result in haploid cells, but not for diploid cells in which msh2 had no effect. It might be that the differences in the results obtained in the two laboratories relate to the different genetic systems used.
Genetic evidence for short-patch repair activity
Providing that an independent short-patch MMR mechanism exists, the elimination of the long-patch MMR by mutations should confer a hyper-recombination phenotype in homologous heteroallelic cells containing closely spaced mutations. Our results (Figure 5; Table I) show that the absence of Msh2 increases recombination by a factor of 4–9 when sites are close together, as compared with a factor of at most 1.6 for distant sites. The effect of pms1 was not tested in meiosis, but is similar to that of msh2 in mitotic cells. These results confirm the existence of a short-patch repair mechanism independent of MSH2 and PMS1. They also indicate that this process efficiently repairs mismatches due to short insertions or deletions.
The efficiency of this system is quite high. For instance, after UV treatment, the induction of recombinants in wild-type cells is ∼10 times less when the mutations are close to each other than if they are distant. In msh2 or pms1 cells, this induction is only 50% smaller than in cells with the distant alleles. In other words, the distance effect on recombination is largely suppressed. Furthermore, the short-patch MMR events are probably more frequent than detected genetically since not all the possible repair events lead to the formation of a wild-type ARG4 gene. In this respect, it may be relevant to recall the effects of MMR mutations on meiotic segregation of heterozygous markers. The pms1 and msh2 mutations increase the class of post-meiotic segregation (unrepaired heteroduplex DNA) at the expense of the conversion class. However, the level of residual conversions remains high and the proportion of conversions among the non-Mendelian segregants can reach 50% (Williamson et al., 1985; Alani et al., 1994). This was notably reported by Alani et al. (1994) for the same EcoRV mutation that is present in our strains. A likely explanation is that these convertants are formed by an alternative efficient MMR activity that could well be the same as the short-patch system that we report here.
In wild-type cells, mismatches are predominantly repaired by the long-patch system, as indicated earlier in this discussion. This predominance of long- versus short-patch repair may be due to a close link between the long-patch MMR system and recombination as well as replication. Besides binding to mismatches (Alani et al., 1995), Msh2 was also reported to bind Holliday junctions (Alani et al., 1997; Marsischky et al., 1999) and proliferating cell nuclear antigen (Umar et al., 1996). Msh2 may bind mismatches and initiate their repair before any other repair protein can act. In support of this view is the very rapid PMS1-dependent repair observed after the formation of a mismatch during mating-type switching (Haber et al., 1993). However, in the literature, there are data that could indicate the activity of short-patch repair in wild-type cells. In most analyses of conversion tracts, a minority of them were reported to be ‘complex’ and in some cases could have involved short-patch repair. Weng and Nickoloff (1998) postulated a short-patch activity to explain some of the tracts that they observed. In our study, the three tracts from wild-type cells that had a short stretch of recipient DNA could reflect restoration events by a short excision tract (Figure 3A).
Fleck et al. (1999) also found a hyper-recombination effect of msh2 and pms1 in heteroallelic cells bearing mutations at sites located close together. They further demonstrated that this effect depends on a short-patch MMR system governed by NER genes. In our test, we found no effect of rad1 or rad14 mutations coupled to either msh2 or pms1 mutations. We only studied mitotic cells, while in S.pombe the test involved meiotic recombination. However, from our data, we see no reason to believe that the process might differ during meiotic and mitotic recombination and we therefore conclude that the short-patch MMR activities in the two yeasts are under different genetic control. We noticed in the work of Fleck et al. that heteroallelic cells bearing closely spaced mutations (which would generate non-C/C mismatches) produce prototrophs at higher rates when defective for both the NER and the long-patch systems than when wild type. This suggests the activity of another short-patch MMR system. It might be that this activity involves the UVE1 gene. Uve1p is a nuclease involved in an alternative NER pathway (Bowman et al., 1994; Freyer et al., 1995). The mutant exhibits a strong mutator phenotype and the protein possesses an in vitro nicking activity on the 5′ side of mismatches (Kaur et al., 1999). Similar activities were reported in S.cerevisiae cell extracts (Chang and Lu, 1991) and are possibly involved in the short-patch MMR described here. When genes controlling the S.cerevisiae system are identified, it will be possible to ask whether they also play a role in mutation avoidance.
Besides this potential role, a short-patch MMR activity is also expected to have important biological consequences related to recombination, at least in mutants affected in the long-patch repair pathway. First, as shown in S.pombe (Fleck et al., 1999) and in this study, it can separate efficiently two closely spaced mutations. Secondly, it can generate highly patched products during a single round of recombination between diverged DNAs. Not only the rate at which sequence diversification occurs, but also the structure of the outcome products will be different from those observed in cells where the MSH2-controlled long-patch MMR process predominates. This is likely to play a role in adaptation and evolution.
Materials and methods
Yeast strains, plasmids and media
Table II gives the list of strains used and their relevant genotype. All strains are MGD derivatives. The parental strains, kindly provided by A.Nicolas, used to derive our strains are ORT118-2 (MATa, arg4-EcoRV, leu2, trp1, ura3, cyh2) and ORT126 (MATα, arg4-BglII, trp1, ura3, ade2, his3). The corresponding strains containing an arg4-HpaI deletion (2.1 kb) are MGD131-2C and MGD131-102-a. These strains and their pms1::TRP1 or msh2::Tn10LUK7-7 derivatives have been described previously (Rocco et al., 1992; Alani et al., 1994). The replacement of the arg4C allele by the corresponding arg4D allele was performed by Adjiri (Adjiri, 1993).
Table II. Strains list.
| Strain | Genotype |
|---|---|
| FF181248 | MATα arg4C-RV ura3-52-arg4D-Bg-URA3 |
| FF181378 | MATα arg4C-RV ura3-52-arg4D-Bg-URA3 msh2Δ |
| FF181246 | MATα arg4D-RV ura3-52-arg4D-Bg-URA3 |
| Ec160 | MATα arg4C-RV ura3-52-arg4C-Bg-URA3 rad1::LEU2 |
| Ec166 | MATα arg4D-RV ura3-52-arg4D-Bg-URA3 rad14::LEU2 |
| FF181379 | MATα arg4D-RV ura3-52-arg4D-Bg-URA3 msh2Δ |
| Ec168 | MATα arg4D-RV ura3-52-arg4D-Bg-URA3 msh2Δ rad1::LEU2 |
| Ec167 | MATα arg4D-RV ura3-52-arg4D-Bg-URA3 msh2Δ rad14::LEU2 |
| FF181614 | MATα arg4C-RV ura3-52-arg4C-Bg-URA3 pms1::TRP1 |
| FF181615 | MATα arg4C-RV ura3-52-arg4C-Bg-URA3 pms1::TRP1 rad1::LEU2 |
| FF181616 | MATα arg4C-RV ura3-52-arg4C-Bg-URA3 pms1::TRP1 rad14::LEU2 |
| Ec153 | MATα arg4C-RV ura3-52-arg4C-Ag-URA3 |
| Ec153000 | MATα arg4C-RV ura3-52-arg4C-Ag-URA3 rad1::LEU2 |
| Ec153002 | MATα arg4C-RV ura3-52-arg4C-Ag-URA3rad14::LEU2 |
| Ec144000 | MATα arg4C-RV ura3-52-arg4C-Ag-URA3 msh2::Tn10LUK7-7 |
| Ec144001 | MATα arg4C-RV ura3-52-arg4C-Ag-URA3 msh2::Tn10LUK7-7 rad1::LEU2 |
| Ec144003 | MATα arg4C-RV ura3-52-arg4C-Ag-URA3 msh2::Tn10LUK7-7 rad14::LEU2 |
| FF181617 | MATα arg4C-RV ura3-52-arg4C-Ag-URA3 pms1::TRP1 |
| FF181619 | MATα arg4C-RV ura3-52-arg4C-Ag-URA3 pms1::TRP1 rad1::LEU2 |
| FF181620 | MATα arg4C-RV ura3-52-arg4C-Ag-URA3 pms1::TRP1 rad14::LEU2 |
| Ec182 | MATa/MATα, arg4D-RV/arg4C-Bg, msh2::URA3/msh2::Tn10LUK7-7 |
| FF181384 | MATa/MATα, arg4C-RV/arg4Δ, ura3-52-arg4D-Bg-URA3/ura3-52 |
| FF181396 | MATa/MATα, arg4C-RV/arg4Δ, ura3-52-arg4D-Bg-URA3/ura3-52 msh2Δ/msh2::URA3 |
| Ec183 | MATa/MATα, arg4D-RV/arg4Δ, ura3-52-arg4D-Bg-URA3/ura3-52 |
| Ec184 | MATa/MATα, arg4D-RV/arg4Δ, ura3-52-arg4D-Bg-URA3/ura3-52, msh2Δ/msh2::URA3, PMS1/pms1::TRP1 |
| Ec185 | MATa/MATα, arg4C-RV/arg4Δ, ura3-52-arg4C-Ag-URA3/ura3-52 |
| Ec186 | MATa/MATα, arg4C-RV/arg4Δ, ura3-52-arg4C-Ag-URA3/ura3-52, msh2::URA3/msh2::Tn10LUK7-7, PMS1/pms1::TRP1 |
To construct ectopic strains, the 2.1 kb HpaI–HpaI fragments containing the desired allele of ARG4 were first cloned into the NruI site of YIp5. This integrative vector contains the yeast URA3 reporter gene (Johnston and Davis, 1984). The plasmid was digested by NcoI, a unique site in URA3, in order to direct its integration into the ura3-52 chromosomal allele by transformation. The plasmid bearing the arg4-AgeI mutation (‘Ag’) has a 3.5 kb HindIII–HindIII fragment of S.cerevisiae integrated into YIp5. The mutation was created by filling in the ends of the AgeI-cut plasmid and re-ligation, resulting in a 4 bp insertion. On these different plasmids, the orientation of arg4 with respect to URA3 is such that, after integration, the two arg4 alleles are in reverse orientation with respect to the centromeres. Ectopic diploids were obtained by crossing the ectopic haploids with cells bearing a genomic deletion of the HpaI region. These diploids are heterozygous for leu2, ade2, his3, cyh2. This is not indicated in Table II for the sake of clarity. Diploids containing the arg4 alleles in allelic position (Ec182) are also heterozygous for the same markers.
The different plasmids used are pFA4 (arg4D-Bg), pNM22 (arg4C-Bg) and pNM23 (arg4C-Ag). They have the HpaI–arg4–HpaI fragment (2.1 kb) integrated into YIp5. pNM3, pNM4 and pNM14 have a larger HindIII fragment (3.5 kb from the ARG4C region) integrated into YIp5, with the internal HpaI fragment containing ARG4C, argD-RV or argD-Bg, respectively. To disrupt RAD14 and RAD1 we used the plasmids pBM190 (rad14::LEU2) (Bankmann et al., 1992) and pWJ153 (rad1::LEU2), given by R.Rothstein. pEN63 (msh2::URA3), from E.Alani, was used in some cases to delete MSH2 and to derive cells that had lost URA3, indicated in Table II as msh2Δ.
Standard yeast genetic and molecular techniques were used (Guthrie and Fink, 1991). YPD medium and SD medium, supplemented with the desired nutrients, were used for mitotic growth. Pre-sporulation and sporulation procedures were according to Resnick et al. (1983).
Determination of meiotic and mitotic recombination rates
The frequency of meiotic ARG+ recombinants was determined by plating sporulated cultures on synthetic medium lacking arginine and diluted aliquots on YPD supplemented with 10 mg/l cycloheximide. Because the recessive cyh mutation is heterozygous in our diploid cells and segregates during sporulation, the cycloheximide in the medium selects colonies derived from asci.
Spontaneous rates of recombination were determined by the method of the median (Lea and Coulson, 1949). The median was from seven independent cultures.
For UV induction of recombinants, UV light (260 nm) was applied to cell suspensions (2 × 107/ml) in saline (0.9% NaCl). The UV fluence was 1 J/m2, as determined with a Latarjet dosimeter. Aliquots were plated on medium lacking arginine for recombinant selection and on rich medium for survival determination. The survival of the different strains was similar and was ∼50% at 30 J/m2.
All experiments were repeated at least three times.
Selection and sequencing of the ARG+ alleles
To select spontaneous mitotic recombinants from ectopic haploids, cells were plated on rich medium and the colonies were replica-plated on medium lacking arginine. ARG+ papillae derived from individual colonies were picked, ensuring that they originated from independent recombination events. Meiotic recombinants were selected as described above. They derived from ectopic diploids, except for one case (no. 15) where allelic diploids were used to select ARG+/arg– sectored monosporic clones. Individual cells from each recombinant colony were plated on rich medium and their genotype analyzed by replica-plating. Since five independent markers segregated, finding the same genotype for all cells of a colony greatly minimized the possibility that it derived from two spores.
The genomic DNA from ARG+ clones was extracted and the allele present on chromosome VIII was specifically PCR amplified using primers that anneal on sequences located outside of the HpaI region. The upper primer (arg4EU) was GTTGGCGCAGGCAATTAATT and the lower one (arg4EL) was AGAATGGCCGGTTCAGACAT. The amplified product was submitted to restriction digestion by EcoRV and BglII, allowing it to be determined whether this allele codes for ARG4+. The whole amplified HpaI–ARG4–HpaI fragments were sequenced by the dideoxy method using a Perkin Elmer ABI310 sequencer. Some of the sequences were determined by the E.S.G.S. Company, group Cybergen.
Acknowledgments
Acknowledgements
We thank Serge Gangloff, Christine Soustelle, Julie Smith and Xavier Veaute for helpful discussions and critical reading of the manuscript. This work was supported by the Commissariat à l’Energie Atomique and the Centre National de la Recherche Scientifique, and by grants from Electricité de France and from the Ministère de l’Education Nationale, de la Recherche et de la Technologie. E.C. was supported by the Ligue Nationale contre le Cancer, and by the Association pour la Recherche sur le Cancer.
References
- Adjiri A. (1993) Etude des effets des hétérologies sur la recombinaison intragénique chez Saccharomyces cerevisiae. PhD thesis, Université Paris-Sud, Orsay, France.
- Adjiri A., Chanet,R., Mezard,C. and Fabre,F. (1994) Sequence comparison of the ARG4 chromosomal regions from the two related yeasts, Saccharomyces cerevisiae and Saccharomyces douglasii. Yeast, 10, 309–317. [DOI] [PubMed] [Google Scholar]
- Alani E., Reenan,R.A. and Kolodner,R.D. (1994) Interaction between mismatch repair and genetic recombination in Saccharomyces cerevisiae. Genetics, 137, 19–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani E., Chi,N.W. and Kolodner,R. (1995) The Saccharomyces cerevisiae Msh2 protein specifically binds to duplex oligonucleotides containing mismatched DNA base pairs and insertions. Genes Dev., 9, 234–247. [DOI] [PubMed] [Google Scholar]
- Alani E., Lee,S., Kane,M.F., Griffith,J. and Kolodner,R.D. (1997) Saccharomyces cerevisiae MSH2, a mispaired base recognition protein, also recognizes Holliday junctions in DNA. J. Mol. Biol., 265, 289–301. [DOI] [PubMed] [Google Scholar]
- Bankmann M., Prakash,L. and Prakash,S. (1992) Yeast RAD14 and human xeroderma pigmentosum group A DNA-repair genes encode homologous proteins. Nature, 355, 555–558. [DOI] [PubMed] [Google Scholar]
- Bishop D.K. and Kolodner,R.D. (1986) Repair of heteroduplex plasmid DNA after transformation into Saccharomyces cerevisiae. Mol. Cell. Biol., 6, 3401–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman K.K., Sidik,K., Smith,C.A., Taylor,J.S., Doetsch,P.W. and Freyer,G.A. (1994) A new ATP-independent DNA endonuclease from Schizosaccharomyces pombe that recognizes cyclobutane pyrimidine dimers and 6-4 photoproducts. Nucleic Acids Res., 22, 3026–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr A.M. et al. (1994) The RAD16 gene of Schizosaccharomyces pombe: a homolog of the RAD1 gene of Saccharomyces cerevisiae. Mol. Cell. Biol., 14, 2029–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D.Y. and Lu,A.L. (1991) Base mismatch-specific endonuclease activity in extracts from Saccharomyces cerevisiae. Nucleic Acids Res., 19, 4761–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. and Jinks-Robertson,S. (1998) Mismatch repair proteins regulate heteroduplex formation during mitotic recombination in yeast. Mol. Cell. Biol., 18, 6525–6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. and Jinks-Robertson,S. (1999) The role of the mismatch repair machinery in regulating mitotic and meiotic recombination between diverged sequences in yeast. Genetics, 151, 1299–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detloff P. and Petes,T.D. (1992) Measurements of excision repair tracts formed during meiotic recombination in Saccharomyces cerevisiae. Mol. Cell. Biol., 12, 1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detloff P., Sieber,J. and Petes,T.D. (1991) Repair of specific base pair mismatches formed during meiotic recombination in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol., 11, 737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleck O., Lehmann,E., Schar,P. and Kohli,J. (1999) Involvement of nucleotide-excision repair in MSH2 PMS1-independent mismatch repair. Nature Genet., 21, 314–317. [DOI] [PubMed] [Google Scholar]
- Fogel S., Mortimer,R.K. and Lusnak,K. (1981) Mechanisms of Meiotic Gene Conversion, or ‘Wanderings on a Foreign Strand’. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Freyer G.A., Davey,S., Ferrer,J.V., Martin,A.M., Beach,D. and Doetsch,P.W. (1995) An alternative eukaryotic DNA excision repair pathway. Mol. Cell. Biol., 15, 4572–4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C. and Fink,G.R. (1991) Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego, CA. [Google Scholar]
- Haber J.E., Ray,B.L., Kolb,J.M. and White,C.I. (1993) Rapid kinetics of mismatch repair of heteroduplex DNA that is formed during recombination in yeast. Proc. Natl Acad. Sci. USA, 90, 3363–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris S., Rudnicki,K.S. and Haber,J.E. (1993) Gene conversions and crossing over during homologous and homeologous ectopic recombination in Saccharomyces cerevisiae. Genetics, 135, 5–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. and Davis,R.W. (1984) Sequences that regulate the divergent GAL1–GAL10 promoter in Saccharomyces cerevisiae. Mol. Cell. Biol., 4, 1440–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur B., Fraser,J.L., Freyer,G.A., Davey,S. and Doetsch,P.W. (1999) A Uve1p-mediated mismatch repair pathway in Schizosaccharomyces pombe. Mol. Cell. Biol., 19, 4703–4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner R. (1996) Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev., 10, 1433–1442. [DOI] [PubMed] [Google Scholar]
- Kramer B., Kramer,W., Williamson,M.S. and Fogel,S. (1989) Heteroduplex DNA correction in Saccharomyces cerevisiae is mismatch specific and requires functional PMS1 genes. Mol. Cell. Biol., 9, 4432–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea D.E. and Coulson,C.A. (1949) The distribution in the numbers of mutants in bacterial populations. J. Genet., 49, 264–285. [DOI] [PubMed] [Google Scholar]
- Marsischky G.T., Lee,S., Griffith,J. and Kolodner,R.D. (1999) Saccharomyces cerevisiae MSH2/6 complex interacts with Holliday junctions and facilitates their cleavage by phage resolution enzymes. J. Biol. Chem., 274, 7200–7206. [DOI] [PubMed] [Google Scholar]
- Modrich P. and Lahue,R. (1996) Mismatch repair in replication fidelity, genetic recombination and cancer biology. Annu. Rev. Biochem., 65, 101–133. [DOI] [PubMed] [Google Scholar]
- Nicolas A., Treco,D., Schultes,N.P. and Szostak,J.W. (1989) Identification of an initiation site for meiotic gene conversion in the yeast Saccharomyces cerevisiae. Nature, 338, 35–39. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T.L., Szostak,J.W. and Rothstein,R.J. (1981) Yeast transformation: a model system for the study of recombination. Proc. Natl Acad. Sci. USA, 78, 6354–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltomaki P. and Vasen,H.F. (1997) Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology, 113, 1146–1158. [DOI] [PubMed] [Google Scholar]
- Petes T.D., Malone,R.E. and Symington,L.S. (1991) Recombination in yeast. In Broach,J.R., Pringle,J.R. and Jones,E.W. (eds), The Molecular and Cellular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis and Energetics. Vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 407–521. [Google Scholar]
- Resnick M.A., Stasiewicz,S. and Game,J.C. (1983) Meiotic DNA metabolism in wild-type and excision-deficient yeast following UV exposure. Genetics, 104, 583–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds R.J. and Friedberg,E.C. (1981) Molecular mechanisms of pyrimidine dimer excision in Saccharomyces cerevisiae: incision of ultraviolet-irradiated deoxyribonucleic acid in vivo. J. Bacteriol., 146, 692–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocco V., de Massy,B. and Nicolas,A. (1992) The Saccharomyces cerevisiae ARG4 initiator of meiotic gene conversion and its associated double-strand DNA breaks can be inhibited by transcriptional interference. Proc. Natl Acad. Sci. USA, 89, 12068–12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman H. (1956) Studies of recombination in yeast. Cold Spring Harbor Symp. Quant. Biol., 21, 175–185. [DOI] [PubMed] [Google Scholar]
- Szostak J.W., Orr-Weaver,T.L., Rothstein,R.J. and Stahl,F.W. (1983) The double-strand-break repair model for recombination. Cell, 33, 25–35. [DOI] [PubMed] [Google Scholar]
- Umar A., Buermeyer,A.B., Simon,J.A., Thomas,D.C., Clark,A.B., Liskay,R.M. and Kunkel,T.A. (1996) Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell, 87, 65–73. [DOI] [PubMed] [Google Scholar]
- Unrau P., Wheatcroft,R. and Cox,B.S. (1971) The excision of pyrimidine dimers from DNA of ultraviolet irradiated yeast. Mol. Gen. Genet., 113, 359–362. [DOI] [PubMed] [Google Scholar]
- Weng Y.S. and Nickoloff,J.A. (1998) Evidence for independent mismatch repair processing on opposite sides of a double-strand break in Saccharomyces cerevisiae. Genetics, 148, 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson M.S., Game,J.C. and Fogel,S. (1985) Meiotic gene conversion mutants in Saccharomyces cerevisiae. I. Isolation and characterization of pms1-1 and pms1-2. Genetics, 110, 609–646. [DOI] [PMC free article] [PubMed] [Google Scholar]