Abstract
In Escherichia coli, FtsZ, a homologue of eukaryotic tubulins, and ZipA, a membrane-anchored protein that binds to FtsZ, are two essential components of the septal ring structure that mediates cell division. Recent data indicate that ZipA is involved in the assembly of the ring by linking FtsZ to the cytoplasmic membrane and that the ZipA–FtsZ interaction is mediated by their C-terminal domains. We present the X-ray crystal structures of the C-terminal FtsZ-binding domain of ZipA and a complex between this domain and a C-terminal fragment of FtsZ. The ZipA domain is a six-stranded β-sheet packed against three α-helices and contains the split β–α–β motif found in many RNA-binding proteins. The uncovered side of the sheet incorporates a shallow hydrophobic cavity exposed to solvent. In the complex, the 17-residue FtsZ fragment occupies this entire cavity of ZipA and binds as an extended β-strand followed by α-helix. An alanine-scanning mutagenesis analysis of the FtsZ fragment was also performed, which shows that only a small cluster of the buried FtsZ side chains is critical in binding to ZipA.
Keywords: bacterial cell division/crystal structure/FtsZ/single isomorphous replacement/ZipA
Introduction
Cell division in bacteria is mediated by the septal ring, a membrane-associated organelle that drives the formation of the septum (Lutkenhaus and Addinall, 1997). The septal ring, of which FtsZ is a key component, assembles at the division site well before membrane constriction and remains associated with the ingrowing cell wall until septal closure (Bi and Lutkenhaus, 1991). This assembly, often designated as the FtsZ ring, is believed to be functionally analogous to the contractile ring in eukaryotic cells. In vitro studies showed that, in the presence of GTP/GDP, FtsZ polymerizes into large protofilaments similar to those found in the walls of microtubules (de Boer et al., 1992; RayChaudhuri and Park, 1992; Mukherjee and Lutkenhaus, 1994, 1998; Erickson et al., 1996). Recently published crystal structures of FtsZ and tubulin revealed that FtsZ is indeed a tubulin-like GTPase with a structure closely related to those of α- and β-tubulin (Löwe and Amos, 1998; Nogales et al., 1998).
To date, the precise mechanism of the ring assembly and how it affects cell wall invagination remains unknown. One of the functions of the FtsZ ring is to recruit other cell division proteins to the septum. In Escherichia coli, apart from FtsZ, eight additional division proteins are now known: FtsA, FtsI, FtsK, FtsL, FtsN, FtsQ, FtsW and ZipA (for review see Rothfield and Justice, 1997; Rothfield et al., 1999;). Among them, ZipA (Z interacting protein A) is a newly identified division factor that is recruited to the septal ring at a very early stage of the division cycle (Hale and de Boer, 1997; Liu et al., 1999). Unlike FtsZ itself, which has a widespread phylogenetic distribution, ZipA is not that highly conserved and is apparently present in a subset of Gram-negative genomes (RayChaudhuri, 1999). No convincing homology is seen in Gram-positive and archaeal genomes.
Escherichia coli ZipA, a 36.4 kDa membrane-anchored protein, has been discovered from a search for proteins that interact directly with FtsZ (Hale and de Boer, 1997). It contains three domains: a short N-terminal membrane-anchored domain, a central P/Q domain that is rich in proline and glutamine and a C-terminal domain, which comprises almost half the protein (residues 185–328). As discussed below, this large domain appears to be involved in interaction with FtsZ. Requirement of ZipA for cell division in E.coli was demonstrated by the formation of non-septate filaments after overexpression or depletion of ZipA (Hale and de Boer, 1997), indicating a ZipA-induced block in septum formation and suggesting that disruption of the ZipA–FtsZ interaction disrupts cell division.
Based on sequence similarity, the majority of FtsZs contain two main regions. An N-terminal region (∼320 residues) is highly conserved and consists of two domains as revealed by the X-ray structure (Löwe and Amos, 1998). This region is required to bind and hydrolyse GTP and is sufficient for polymerization (Wang et al., 1997). It is followed by a variable spacer region and a conserved segment of ∼15 amino acids at the extreme C-terminus. The structure of this part of FtsZ has not been determined. Although this region is not required for the GTPase activity of FtsZ (Wang and Lutkenhaus, 1996), a mutant of Caulobacter crescentus FtsZ missing its last 24 amino acids has a dominant lethal cell phenotype (Din et al., 1998). Similar results on C-terminal deletions were obtained with Bactillus subtilis FtsZ (Wang et al., 1997), suggesting that the conserved C-terminal segment found in at least 11 FtsZ sequences identified to date has an important function.
A series of experiments has been performed recently to develop a clearer knowledge of the cell division cycle in bacteria. Depletion studies of both FtsZ and ZipA (Liu et al., 1999; Hale and de Boer, 1999) showed that in the absence of FtsZ, ZipA fails to assemble in the ring structure and in contrast, in the majority of ZipA-depleted filaments FtsZ rings are still present. These results suggest that ZipA, rather than being a nucleating factor for the FtsZ ring, functions concurrently with or soon after initial ring formation. However, although depletion of ZipA does not prevent formation of FtsZ rings, it does lead to a significant reduction in the number of rings per unit of cell mass (Hale and de Boer, 1999), suggesting a stabilizing role of ZipA in growth of the nucleated FtsZ oligomers. It is also speculated that ZipA, as it is anchored to the cytoplasmic membrane while binding FtsZ, may function as an FtsZ receptor that tethers FtsZ protofilaments to the membrane during invagination of the septum (Rothfield and Justice, 1997). Furthermore, two-hybrid experiments and a co-sedimentation assay (Liu et al., 1999) indicated that in E.coli the interaction between ZipA and FtsZ is mediated by the C-terminal regions of the proteins: the C-terminal domain of ZipA (residues 176–328) is sufficient for binding to FtsZ, and the binding site on FtsZ for ZipA is identified as part of a 60-residue region at the C-terminus of the protein.
In this study, we present the high resolution crystal structures of the C-terminal domain of E.coli ZipA (ZipA/M185, residues 185–328), determined at 1.5 Å, and a 1:1 complex between this domain and a peptide representing the E.coli FtsZ fragment (residues 367–383), determined at 1.95 Å. ZipA/M185 is the domain that binds FtsZ with the same affinity as a longer version of ZipA (residues 23–328) (E.Glasfeld, unpublished data). The 17 amino acid peptide is a conserved segment at the C-terminus of FtsZ that competes with the full length FtsZ for binding to ZipA (E.Glasfeld, unpublished data). The structures reported here provide the first three-dimensional information available for ZipA and the first insight into the nature of the interaction of ZipA with FtsZ. This structural information combined with the mutational analysis of the FtsZ fragment, which is also described here, may be useful in designing new drugs against infectious diseases caused by bacterial organisms.
Results
Structure determination
The C-terminal domain of ZipA (ZipA/M185, residues 185–328) was expressed in E.coli and purified to homogeneity as described (see Materials and methods). Crystals were grown that belonged to space group P21 (unit cell dimensions a = 49.89 Å, b = 41.74 Å, c = 71.16 Å, β = 98.26°), with two molecules per crystallographic asymmetric unit. Diffraction data were collected from a crystal of the native protein and from a crystal of a selenomethionine (Se-Met) substituted form of the protein. Both the native and the heavy-atom derivative (Se-Met) data were measured in house at λ = 1.5418 Å. The structure was determined to a resolution of 2 Å by Se-Met single isomorphous replacement with anomalous scattering (SIRAS), and refined to 1.5 Å resolution (Rwork = 19.8%, Rfree = 21.7%), using a native data set collected at the Advanced Light Source. The final model of ZipA/M185 contains two copies of the protein: residues A190–A328 and B189–B328 and 422 water molecules.
A mixture of ZipA/M185 with a synthetic peptide corresponding to the E.coli FtsZ residues 367–383 (KEPDYLDIPAFLRKQAD) was prepared for cocrystallization trials as described (see Materials and methods). The 1:1 complex crystallized in the space group P21 (a = 36.53 Å, b = 38.9 Å, c = 54.54 Å, β = 75.89°), with one complex per asymmetric unit. The structure was determined by molecular replacement, with ZipA/M185 as a search model, and refined to 1.95 Å (Rwork = 20.5%, Rfree = 25.1%) using the data collected in house. The final model contains ZipA/M185 residues 185–328, FtsZ-peptide residues 367–383 and 204 water molecules.
Details of the crystallizations, data collection and structural determination are given in Materials and methods and summarized in Table I.
Table I. Statistics for data collection, phasing and refinement.
| Structure | ZipA/M185 | ZipA/M185:FtsZ peptide | ||
|---|---|---|---|---|
| Data collection | ||||
| Native (R-axis) | Se-Met (R-axis) | Native (ALS) | Native (R-axis) | |
| Resolution range (Å) | 25–1.9 (1.97–1.9) | 25–1.85 (1.92–1.85) | 20–1.5 (1.55–1.5) | 25–1.95 (2.02–1.95) |
| Completeness (%) | 99.1 (98.8) | 99.4 (99.9) | 99.5 (97.8) | 95.6 (88.6) |
| Total observations 93 040 | 193 437 | 178 183 | 41 423 | |
| Unique reflections | 22 875 | 24 555 | 46 702 | 10 556 |
| Average I/σ(I) | 36.4 (14.4) | 36.4 (21.2) | 19.8 (4.2) | 26.9 (8.6) |
| Rsyma (%) | 3.2 (10.1) | 5.0 (9.6) | 6.3 (19.9) | 5.5 (14.6) |
| Phasing (20–2 Å)b | ||||
| Number of sites | 16 | |||
| Phasing powerc (centric/acentric) | 1.48/1.91 | |||
| Cullis R-factord (centric/acentric) | 0.57/0.64 | |||
| Isomoprhous difference (centric/acentric) | 38.1/23.2 | |||
| Anomalous difference (observed/calculated) | 3.4/1.4 | |||
| FOMe (centric/acentric) | 0.66/0.47 | |||
| Model refinement | ||||
| Maximum resolution (Å) | 1.5 | 1.95 | ||
| Rwork/Rfreef (%) | 19.8/21.7 | 20.5/25.1 | ||
| R.m.s.d. | ||||
| bonds (Å) | 0.009 | 0.008 | ||
| angles (°) | 1.34 | 1.42 | ||
aRsym= Σ |Ih – <Ih>| /ΣIh, where <Ih> is the average intensity over symmetry equivalents. Numbers in parentheses reflect statistics for the last shell.
bPhasing statistics as reported by MLPHARE.
cPhasing power = Σ|FH| /Σ|||FPHobs| – |FPHcalc||, where FH is the calculated heavy-atom structure factor amplitude.
dCullis R-factor = Σ||FPHobs| – |FPHcalc||/Σ||FPHobs| – |FP||, where |FPHobs| – |FPHcalc| is lack of closure and |FPHobs| – |FP| is isomorphous difference.
eFigure of merit = <ΣP(α)eiα/Σ|P(α)|>, where α is the phase and P(α) is the phase probability distribution.
fRwork = Σ||Fobs| – |Fcalc|/Σ|Fobs|, Rfree is equivalent to Rwork, but calculated for a randomly chosen 5% (or 10%) of reflections omitted from the refinement process.
Structure of ZipA/M185
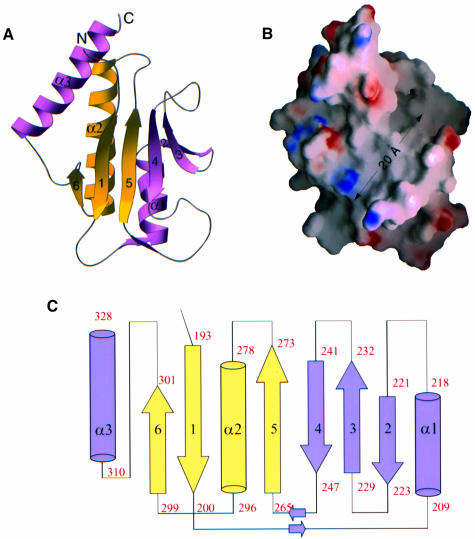
The polypeptide chain of the Zip/M185 monomer folds into one domain of α/β topology, which forms a six-stranded antiparallel β-sheet packed against three α-helices (Figure 1A). The core of the domain represents a well known structural motif, the split β–α–β fold (Orengo and Thorton, 1993), which consists of a three-stranded antiparallel β-sheet (β1, β5, β6) and one α-helix (α2), with topology β1, β5, α2, β6. This fold, found in many ribosomal proteins, is the ‘common’ motif for RNA-binding domains (Liljas and Garber, 1995; Yonath and Franceschi, 1997). In these domains, the connection between the first (β1) and the second (β5) strand is variable and sometimes constitutes a separate domain (Nikonov et al., 1996). In the structure of ZipA/M185, the insert between β1 and β5 (residues 200–264) is composed of one α-helix (α1) and three antiparallel strands (β2, β3, β4) directly adjacent to strand β5, thus extending the β-sheet. The third α-helix (α3) is found C-terminal to the split motif. The connectivity scheme for the whole domain is β1–α1–β2–β3–β4–β5–α2–β6–α3 (Figure 1C). The connections between the secondary structural elements are mostly short type 3-β-turns except for the linkages between the split motif and the insert. These linkages are long irregular loops (residues 200–209 and 248–264) at the bottom of the domain (Figure 1A), which pack together through two antiparallel mini-strands along their courses.
Fig. 1. Structure and topology of ZipA/M185. (A) Ribbon diagram of the ZipA/M185 monomer. For clarity, β-strands have been labeled by number only. The β–α–β split motif is shown in yellow (1–5–α2–6). The insert in the split motif (α1–2–3–4) and helix α3, which immediately follows the motif, are colored purple. Prepared using the program RIBBONS (Carson, 1991). (B) Surface potential representation of ZipA/M185 (GRASP, Nicholls et al., 1991). Regions with electrostatic potential less than –11.5 kBT are red, while those greater than +10.5 kBT are blue (kB, Boltzmann constant, T, absolute temperature). The view is from the uncovered side of the β-sheet showing the cavity of neutral charge, which extends to ∼20 Å across the sheet and has space to accommodate a ligand. The orientation of the ZipA/M185 molecule is similar to that in (A). (C) Topological diagram showing the β–α–β fold of ZipA/M185. The diagram is arranged to coincide with the orientation in (A) and (B). β-strands are represented as arrows, while α-helices are cylinders. The color coding and secondary structural element numbering are the same as in (A). The assignment of the secondary structure of ZipA/M185 was done using the algorithm of Kabsch and Sander, as implemented in PROCHECK (Laskowski et al., 1993).
As in many proteins sharing the canonical split motif, one side of the β-sheet of ZipA/M185 is covered by the α-helices and the opposite side is open to solvent. The interior where the β-strands make extensive contacts with the three helices (α1, α2 and α3), as well as the interfaces where the helices contact each other, are mainly hydrophobic. The exposed sides of the α-helices are of polar and hydrophilic residues, with electrostatic potential on their surface dominated by an acidic patch. The uncovered side of the β-sheet incorporates a large but shallow solvent-exposed cavity, 20 Å long and 12 Å wide, extending diagonally across the sheet (Figure 1B). The surface of this β-sheet is walled in on either side by the loops connecting strands and is lined with amino acids from four strands (β3, β4, β5, β1) and from the β2–β3, β4–β5 and β6–α3 connections. Most of them are non-polar side chains, which, together with the backbone, determine the shape and surface properties of the cavity. Lys250 and Arg305 are the only charged residues on both walls that interrupt the hydrophobic nature of the cavity (Figure 1B). While Lys250 is projecting away, the side chain of Arg305 is oriented across the cavity, thereby closing off part of the left entrance to the hydrophobic volume (see below and Figure 2B). In both ZipA/M185 monomers, the volume within the cavity contains a moderate number of water molecules, with which the guanidinium group of this same Arg305 forms an extensive network of hydrogen bonds.
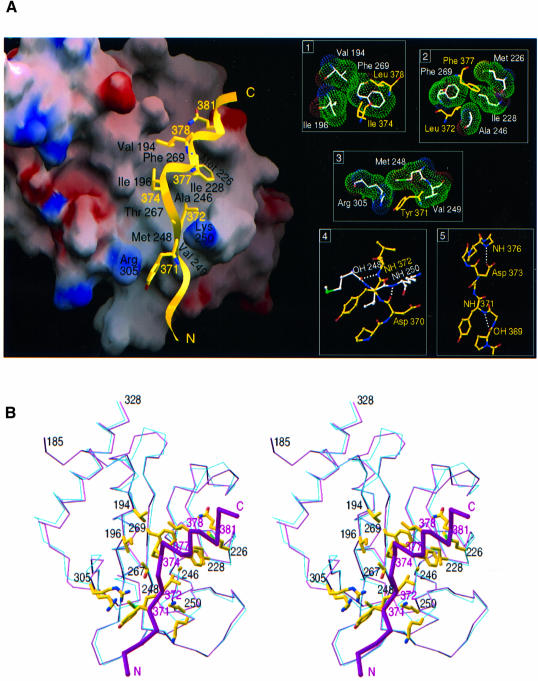
Fig. 2. Overview of the complex between ZipA/M185 and the FtsZ fragment. (A) Electrostatic potential surface of the FtsZ-binding cavity of ZipA/M185 (GRASP, Nicholls et al., 1991) as it is seen in complex with the FtsZ peptide (yellow ribbons). Contouring scheme for potentials is the same as in Figure 1B. FtsZ residues interacting with the cavity are shown. ZipA/M185 residues that contribute most of the total contacts are designated by name and number (black); those in the peptide, by number only (yellow). The peptide binds as an extended β-strand (residues 367–373) followed by α-helix (residues 374–383). Inserts 1–3, on the right, show peptide side chains (yellow) in hydrophobic pockets of ZipA/M185 (with side chains in white and van der Waals surface in green). Interaction between the peptide (yellow) and ZipA/M185 (white) backbones and internal hydrogen bonding within the peptide (see text) are shown in inserts 4 and 5, respectively. Hydrogen bonds are indicated by dashed white lines. Lys250 of ZipA/M185 does not make contacts with acidic residues Asp370 and Asp373 of the peptide; instead, both Asp370 and Lys250 are involved in hydrogen bonding with water molecules (not shown), and the side chain of Asp373 is hydrogen bonded to the NH group of Ala376. (B) Stereo diagram showing superposition of Cα traces of the ZipA domains as they are seen in the uncomplexed structure (blue lines) and in complex with the FtsZ peptide (purple lines). The most significant changes are found in the segment from the β4–β5 loop (residues 247–251) and in the β6–α3 loop (residues 303–306). From this orientation, the side-chain rearrangement of both Lys250 and Arg 305, upon peptide binding, is evident. Representative contact residues from ZipA/M185 (designated by number in black) and from the peptide (designated by number in purple) are shown, with the Cα trace of the peptide shown as magenta sticks. The orientation is similar to that in (A), and allows for direct comparison of the ZipA/M185–FtsZ interactions. (B) was prepared using the program RIBBONS (Carson, 1991).
In the crystals, ZipA/M185 molecules pack tightly together. When superimposed, the two monomers are very close in structure; a root-mean-square (r.m.s.) deviation is 0.79 Å for 139 Cα pairs. Each monomer reveals different crystal contacts, with some interactions between ZipA/M185 copies in the vicinity of surface areas of the cavity. The non-crystallographic symmetry does not reveal any possible model for a ring-shaped formation.
Structure of the FtsZ fragment bound to ZipA/M185
A 17-residue FtsZ peptide (367KEPDYLDIPAFLRKQAD383) is bound by the hydrophobic surface of the ZipA/M185 cavity, on the solvent-exposed side of the β-sheet. Upon binding, two segments of the peptide adopt different conformations: residues 367–373 have φ and ψ angles that are typical for an extended β-strand, while residues 374–383 are in a regular helical conformation (Figure 2A). This conformation directs six side chains of the 30 Å long peptide towards interactions with the hydrophobic surface of the ZipA/M185 cavity. The solvent accessible area buried upon peptide binding is 536.4 Å2 for ZipA/M185 and 660 Å2 for the peptide, using a probe radius of 1.4 Å in SURFACE (CCP4, 1994).
Direct interatomic contacts are made between 11 ZipA/M185 amino acid residues and seven peptide residues. Most of these are hydrophobic contacts (inserts 1–3 in Figure 2A) but also include two hydrogen bonds between the backbones of the interacting molecules (insert 4). Residues in contact are concentrated in the span from 370 to 381 of the peptide (Asp370, Tyr371, Leu372, Ile374, Phe377, Leu378 and Gln381) and are distributed over six segments of ZipA/M185: β1 (Val194, Ile196), β4 (Ala246), β5 (Thr267, Phe269), β2–β3 (Met226, Ile228), β4–β5 (Met248, Val249, Lys250) and β6–α3 (Arg305). In ZipA/M185, these 11 residues have the most extensively buried side chains and contribute 78% of the total contacts. In the FtsZ peptide, among the side chains buried upon interaction with ZipA/M185 (Figure 2A), there are four (Tyr371, Ile374, Leu378, Gln381) that project across and two (Leu372, Phe377) that are oriented down into the cavity. Ile374, Phe377 and Leu378 are deeply buried and account for 38.2% of the total contacts. With the exception of Gln381, which contacts the cavity through the hydrophobic methylene groups of its side chain, the peptide residues close to its N- and C-termini (residues 367–369 and 379–383) extend on either end of the binding site and make no contacts with the ZipA/M185 domain. As a result, ∼55% of the peptide surface (818 Å2) remains solvent accessible in the complex.
Although the overall structure of ZipA/M185 is unchanged in this complex, small but significant local changes do occur; in pairwise superposition, r.m.s.d. is 0.93 Å for the Cα atoms of the uncomplexed and complexed molecules (Figure 2B). Such changes are restricted to the binding site. In particular, the intercalation of peptide residue Tyr371 into the hydrophobic volume of the cavity is accompanied by a slight displacement (∼0.8 Å) of the β6–α3 loop toward the peptide. Upon this rearrangement, the side chain of Arg305 is swung out of the cavity and into solvent, such that the guanidinium group of Arg305 is optimally positioned to be stacked on the Tyr371 ring (3.2 Å). A much larger structural change occurs in the segment of the β4–β5 loop. Although the hydrogen-bonding pattern between the strands is maintained, residues 248–250 rotate as a rigid group by ∼2.5 Å towards the floor of the cavity. Here Lys250 is still exposed, but its side chain flips to avoid a close contact introduced by the peptide. This conformational adjustment in the ZipA/M185 structure wedges the position of the peptide backbone at this point, by forming two hydrogen bonds to the peptide. These two hydrogen bonds are made between main-chain carbonyl or nitrogen atoms of peptide residues Asp370 and Leu372, and those of ZipA/M185 residues Lys250 and Met248 (insert 4 in Figure 2A). The formation of hydrogen bonds, which involve only main-chain atoms of the interacting molecules, provides a component to the ZipA–FtsZ interaction that depends less on the sequence and more on the overall folding and represents a common finding in receptor–ligand interactions (Schreuder et al., 1997).
Within the bound peptide, we observe, apart from the classical intrahelical hydrogen bonding, two additional internal hydrogen bonds (insert 5 in Figure 2A). One is a main chain–main chain hydrogen bond formed between Pro369 and Tyr371 at the N-terminal region of the peptide. Another such bond is a side-chain-to-backbone hydrogen bonding between Asp373 and Ala376, which is associated with the capping interaction at the helix N-terminus (Richardson and Richardson, 1988). Helix capping is often observed near the ends of helices, where the first NH groups necessarily lacking intrahelical hydrogen bonds are capped by alternative hydrogen bond partners (Aurora and Rose, 1998). In the FtsZ peptide, this alternative bond is provided by the NH group of Ala376 from the helix and by the side chain of Asp373 from the adjacent strand. Numerous studies demonstrate that capping hydrogen bonds stabilize α-helices in both proteins and peptides (Aurora and Rose, 1998). This suggests that the capping motif observed in the structure of the peptide, as well as the extra pattern of internal hydrogen bonding at its N-terminus, can be involved in stabilization of the conformation of the ZipA-bound peptide.
The aligned sequences of C-terminal regions of ZipA (Figure 3A) and those of FtsZ (Figure 3B) and the structure of this complex show that most of the residues in the ZipA–FtsZ interface are well conserved within each subset. A few differences in residues that occupy less conservative positions can be accommodated without any change in the principal structural features of the interaction. Likewise, FtsZ side chains that project away from the binding site are variable, excluding two consensus residues Asp373 (or Glu373) and Pro375 (Figure 3B). A preference for the acidic residue and proline at these positions has an important effect on the conformation of the bound peptide. As noted above, the side chain of Asp373 compensates for the lack of the unsatisfied hydrogen bond within the initial helical turn. To facilitate the helical capping without altering the structure of the peptide backbone, an acidic residue at position 373 should be favored over other side chains. Proline, often found at helix ends, by adopting restricted conformations can account for the hinge point, where the course of the peptide is altered away from the extended conformation. At the same time, the pyrrolidine ring lacks an NH group, thereby obviating the need for a hydrogen-bond acceptor. Thus, steric and chemical characteristics of proline are likely to decrease the flexibility of the peptide helix at this point.
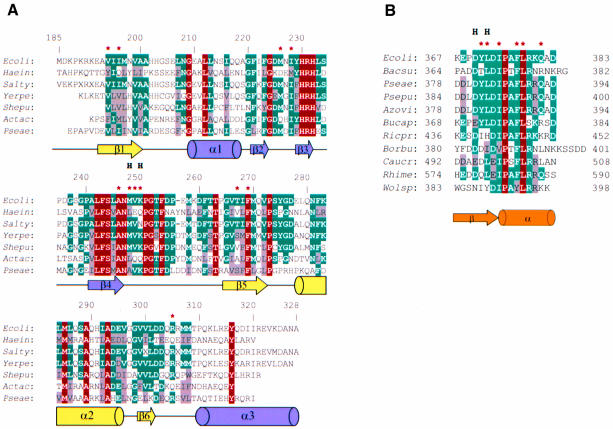
Fig. 3. Sequence alignments. (A) Alignment of amino acid sequences of ZipA C-terminal regions from seven species: Escherichia coli (Ecoli); Haemophila influenzae (Haein); Salmonella typhimurium (Salty); Yersinia pestis (Yerpe); Shewanella putrefaciens (Shepu); Actinobacillus actinomycetemcomitans (Actac) and Pseudomonas aeruginosa (Pseae). The numbering is sequential for E.coli ZipA. The sequences share between 34 and 89% identity (X substitutes low-complexity sequences) and were identified by BLAST search (Altschul et al., 1997) using completely and partially sequenced genomes. Residues are highlighted as highly conserved (red) through to moderately well-conserved (blue and gray). Arrows and cylinders below the sequences indicate secondary structure elements observed in the crystal structure of E.coli ZipA, with coloring as in Figure 2. Lines represent areas of turns or loops. The asterisks mark residues involved in interaction with the FtsZ fragment. Residues with main-chain atoms hydrogen bonded to FtsZ are marked by H. (B) Sequence alignment of FtsZ C-terminal fragments from 11 species (BLAST search using complete genomes): E.coli (Ecoli); Bacillus subtilis (Bacsu); Pseudomonas aeruginosa (Pseae); Pseudomonas putida (Psepu); Azotobacter vinelandi (Azovi); Buchnera aphidicola (Bucap); Rickettsia prowazekii (Ricpr); Borrelia burgdorferi (Borbu); Caulobacter crescentus (Caucr); Rhizobium meliloti (Rhime) and Wolbachia sp. (Wolsp). Residues are highlighted using the same definition as in (A). The conformation of the bound FtsZ fragment is represented by an arrow and a cylinder below the sequences. The asterisks mark residues interacting with ZipA/M185 and marked by H are residues with the main-chain atoms hydrogen bonded to ZipA/M185.
Besides the interactions described above, there are some other indirect contacts between the bound peptide and ZipA/M185. Most of them involve hydrophilic and polar residues interacting through well-ordered water molecules. In the cavity itself six trapped water molecules are seen in the complex.
Alanine-scanning analysis of the FtsZ peptide
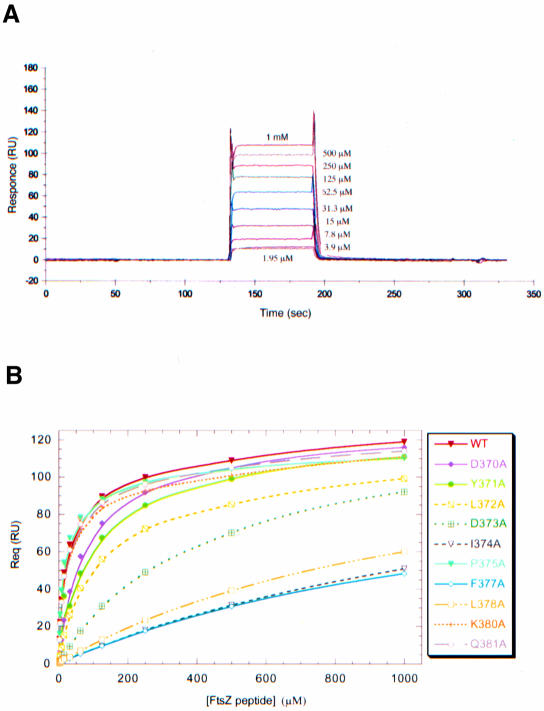
To characterize further the binding of the FtsZ fragment to ZipA/M185 we used a surface plasmon resonance (SPR) based assay (Szabo et al., 1995), in which ZipA/M185 was covalently immobilized to a biosensor chip (see Materials and methods). As detected by a 100-fold difference in the dissociation constants, the FtsZ peptide shows less binding to immobilized ZipA/M185 (KD ∼20 µM; Figure 4A and Table II) than the full length FtsZ for soluble ZipA (residues 23–328) (KD ∼0.2 µM; ELISA assay, E.Glasfeld, unpublished data). The observation that the binding affinities derived from interaction between FtsZ and ZipA (23–328) and those derived using FtsZ and biotin-tagged ZipA/M185 are essentially the same implies that ZipA/M185 retains the binding properties of ZipA (23–328). To check whether the weaker binding was due to the ZipA/M185 immobilization, we applied isothermal titration calorimetry (ITC; Fisher and Singh, 1995), in which attachment to a surface is not required. The dissociation equilibrium constants derived from the calorimetric approach were slightly higher (KD ∼35 µM) but consistent with those derived using biosensor assays. Thus, one conceivable reason for this decreased affinity is that the peptide can be assumed to form a substantially smaller contact interface with ZipA/M185 compared with the full length FtsZ. Another possibility is that the rest of FtsZ could be required for a proper folding of its C-terminal fragment upon or prior to ZipA binding.
Fig. 4. Biosensor assays of FtsZ peptides binding to ZipA/M185. (A) BIAcore sensorgrams of wild-type FtsZ peptide (residues 367–383) binding to immobilized ZipA/M185. Binding curves are expressed in resonance units (RU) as a function of time (s). One representative set of a triplicate experiment is shown, with concentrations of injected peptide indicated on the sensograms. (B) Plots of equilibrium binding responses (Req) versus the concentrations of FtsZ peptides. Each binding curve (colored lines) was derived from biosensor experiments performed in triplicate for each FtsZ variant (experimental sensorgrams obtained for mutants are not shown). In those experiments in which the binding responses closely approach or do not reach equilibrium, the maximal binding response (Rmax) was estimated to be 110–115 RU at higher concentrations of injected peptides.
Table II. Alanine-scanning mutation analysis of FtsZ peptide.
In order to determine which contact side chains of the FtsZ peptide contribute to the binding affinity for ZipA/M185, we designed and analyzed 10 single-site alanine substitutions in the FtsZ peptide using the structure of the complex as a guideline. By measuring binding affinities of these mutants relative to the wild type, we calculated the relative reduction in binding to ZipA/M185 as a consequence of introduced mutations (Figure 4B and Table II). This analysis identified seven side chains that when converted to alanine disrupt binding affinity by a factor ranging from 3- to 70-fold (Table II). Five of these buried side chains (Tyr371, Leu372, Ile374, Phe377 and Leu378) are in direct contact with ZipA/M185, but only three of them (Ile374, Phe377 and Leu378) were found to account for virtually all the binding affinity, as each of these mutants individually caused a 48- to 70-fold reduction in binding. Consistent with this, these highly conserved residues (Figure 3B) are deeply or almost completely buried in hydrophobic pockets (with 80–94% of the surface area buried for each residue) near the center of the contact interface (Figure 2A). Mutants at less conservative positions (Tyr371 and Leu372) cause 4- to 5-fold reductions in binding or, as in the case of Gln381, do not affect the binding at all. These contact side chains are located toward the periphery of the interface and contribute less to the total contacts. An alanine replacement of Gln381 appears to be isofunctional, as this residue interacts with ZipA/M185 through the innermost aliphatic portion of its side chain.
Aspartic acids Asp370 and Asp373 are the only two side chains that do modulate binding, but are not involved in direct interaction with the ZipA/M185 molecule. Yet their functional importance is evident and can be accounted for by the structure. The subtle effect of the D370A mutant (3.2-fold decrease in binding) is correlated with a decrease in the number of van der Waals contacts that are seen near but outside of the contact interface. Those involve hydrogen bonds between the charged Asp370 and ZipA/M185 backbone atoms through the bridging water molecules (not shown). Another charged-to-alanine substitution (D373A) results in the conformer, which reduces binding affinity by more than 18-fold. Its dramatic effect on binding supports the conclusion that loss of the capping hydrogen bond, the formation of which is ensured by acidic residue at position 373, diminishes the stability of the bound peptide.
Overall this analysis identified four most disruptive alanine mutants: three at hydrophobic residues (Ile372, Phe377 and Leu378), which form extensive well-packed hydrophobic contacts with ZipA/M185, and one at an acidic residue (Asp373), which is part of the helical capping motif within the structure of the bound FtsZ peptide. Unexpectedly, the alanine mutation of absolutely conserved Pro375 resulted in a non-disruptive mutant (P375A), which, in fact, has a slight enhancing effect on binding. It is possible that the functional role of Pro375, as well as of other solvent-exposed side chains (Arg379, Lys380, Asp383), can be revealed only in the context of the entire complex formation wherein other regions of FtsZ could participate in interaction with either its own C-terminal fragment or ZipA, or both of them. Moreover, the functional importance of the main chain–main chain contacts that may contribute significantly to the overall binding, as well as trapped water molecules, which are also present at the contact interface, can not be tested directly by this mutational approach and therefore remains unknown.
Structural similarities with other proteins
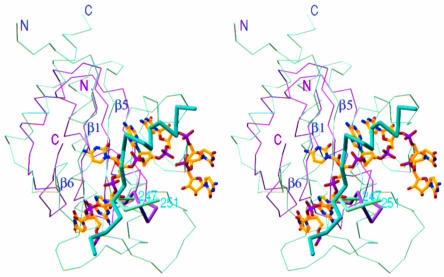
In a large number of proteins sharing the β–α–β split fold, ZipA represents the first example of this structural class observed in proteins that are part of the cell division machinery. Although this structural motif is the most abundant element in RNA-binding proteins and is associated with their common function as RNA-interacting proteins, in ZipA/M185, this motif is involved in a protein–protein interaction. Comparison of the FtsZ-binding domain of ZipA with the RNA-binding domain of the U1A spliceosomal protein (Burd and Dreyfuss, 1994) reveals that they are quite close topologically: the insert in the split motif of ZipA/M185 and that of U1A are in similar locations. Moreover, the RNA fragment, as seen in the U1A–RNA complex (Outbridge et al., 1994; Allain et al., 1996), is bound by residues on the surface of the β-sheet involving the connecting loops of the split β–α–β motif. When the ZipA/M185 and U1A domains are superimposed, the FtsZ fragment and the RNA fragment occupy similar positions on the uncovered sides of their β-sheets (Figure 5). In addition, the RNA-binding loop, which is an α-helical turn connecting two β-strands in U1A, has a structural homologue in ZipA/M185, the backbone of which is hydrogen bonded to the backbone of the peptide.
Fig. 5. A comparison of the FtsZ-binding domain of ZipA with the RNA-binding domain of the U1A spliceosomal protein. Cα stereo superposition of ZipA/M185 (green) and U1A (purple) is based on Cα atoms in the β strands of the β–α–β split motif (β1, β5, β6) and includes a ball-and-stick representation of bound RNA fragment and a Cα-model of FtsZ-peptide (sticks in cyan). The RNA-binding loop in U1A and the peptide-binding loop in ZipA/M185 (residues 247–251, designated by number in cyan) are highlighted. The N- and C-termini for the superimposed domains are indicated. Figure prepared using the program RIBBONS (Carson, 1991).
As expected, the specific features involved in nucleotide binding are not observed in ZipA/M185. The interactions between ZipA/M185 and the FtsZ peptide are mainly hydrophobic in nature, without any charged residues, which are normally essential for nucleotide ligands. Nonetheless, the observation that the FtsZ-binding region on ZipA/M185 has a structure reminiscent of RNA-binding motifs is rather striking, raising the intriguing questions of why and how evolution has selected the β–α–β split fold for such diverse ligands and associated with them such different biological functions. These issues are discussed in more detail below.
Discussion
In vivo studies demonstrated that the recently discovered membrane-anchored protein ZipA (Hale and de Boer, 1997) localizes at the site of cell division at a very early stage of the division cycle. They also suggested that the interaction between ZipA and the key cell division protein FtsZ occurs within a complex ring-like structure consisting of multiple components (Hale and de Boer, 1999; Liu et al., 1999). It is believed that in the cell at least 10 molecules of FtsZ are available for each ZipA (Hale and de Boer, 1997), but it is not known whether the FtsZ ring is enclosed by one large or by multiple smaller polymeric molecules. The precise molecular architecture of this complex assembly, as well as the stoichiometry of FtsZ’s interaction with ZipA within this assembly remain to be established. In vitro studies further showed that, in the absence of assembly promoting agents such as DEAE–dextran or cationic lipids (Mukherjee and Lutkenhaus, 1994; Erickson et al., 1996), ZipA can induce association of FtsZ protofilaments into side-by-side arrays of long thick bundles or sheets (RayChaudhuri, 1999). Moreover, the very recent results of de Boer and his colleagues showed that the C-terminal FtsZ-binding domain of ZipA (186–325) alone can promote extensive bundling formation of the FtsZ polymers in vitro and that the conserved C-terminal fragment of FtsZ is required for this formation (P.de Boer, personal communication). It is not known whether the ZipA-mediated bundling of FtsZ filaments represents the physiological organization of the FtsZ ring in bacterial cells and, if it does, how the interaction of the C-terminal domain of ZipA with the FtsZ polymers stimulates association between FtsZ protofilaments.
The answers to all these questions will ultimately require the detailed structural characterization of ZipA, other FtsZ-binding partners and their complexes with FtsZs, which we have initiated by X-ray analysis of ZipA/M185 and its complex with the FtsZ fragment. This analysis reveals many molecular aspects of ZipA–FtsZ association, including the atomic structure of ZipA/M185 and the explicit interaction between the C-terminal regions of the two division proteins, and thus can be regarded as a starting point in dissecting the molecular mechanism of bacterial cell division.
First, our results demonstrate that the identifying feature of the complex formation between the C-terminal regions of ZipA and FtsZ is the hydrophobic burial of highly exposed, predominantly non-polar surfaces. The low hydrogen-bonding ability of these binding surfaces results in the formation of only two hydrogen bonds, which involve main-chain atoms of the interacting molecules and thus represent the contacts that do not depend on amino acid side-chain specificity. Although detailed predictions are premature, this type of interaction has an advantage in implying less geometric constraints on binding partners and could provide a rationale for how other FtsZ-interacting proteins, such as FtsA, may bind specifically this same conserved C-terminal region of FtsZ (Wang et al., 1997; Din et al., 1998; Ma and Margolin, 1999).
Secondly, our studies reveal unexpected structural relationships between bacterial cell division protein ZipA and RNA-binding proteins belonging to the structural class with the β–α–β split topology. The fact that the FtsZ-binding region on ZipA bears some structural resemblance to the RNA-binding motifs and that such diverse ligands as protein fragments and RNA hairpins occupy similar positions on the exposed β-sheet surfaces of the folded domains raises the question of whether the structural similarities observed between these evolutionarily unrelated proteins can have some functional implica tions. Speculative rather than practical considerations can be made based on structure–function relationships that have been proposed for proteins containing RNA-binding domains (RBDs) of the β–α–β topology (Outbridge et al., 1994; Allain et al., 1996). For these proteins, the surface of the β-sheet is seen as a RNA-binding and RNA-folding platform, as formation of the complex orders the flexible RNA loop and changes the conformation of bound RNA. Thus, a single RBD can directly modify overall RNA structures, which is akin to chaperone activity. Furthermore, many of these proteins have a modular structural organization; they can contain several RBDs and auxiliary P/Q domains that are rich in proline, glutamine or glycine. RNA-binding experiments demonstrated that, when bound to RNA, these multi-RBD proteins can facilitate or hinder the formation of specialized high-order complexes, like spliceosomes, or form stable ribonucleoprotein particles by protein–protein interactions (Burd and Dreyfuss, 1994; Mattaj and Nagai, 1995). By analogy with this situation we can speculate that, if the in vitro FtsZ bundling activity of ZipA is physiologically relevant and the FtsZ-binding domain of ZipA is indeed sufficient for promoting an FtsZ protofilament association, then, similar to the RBDs, the β–α–β domain of ZipA should retain some structural properties that would be responsible for the unusual stability of this high-order ring-like structure. It is possible that engagement of the FtsZ C-terminal tail by the β-sheet surface of ZipA orders or stabilizes an otherwise flexible FtsZ tail, reorganizing the rest of the FtsZ structure for functional interaction. Such chaperone-like activity of ZipA might allow the next step in FtsZ–ZipA assembly, including lateral association between nucleated FtsZ protofilaments, to occur. Alternatively, ZipA may trigger FtsZ bundling formation simply by shielding in its binding crevice of the β-sheet the otherwise exposed hydrophobic surface of the 17-residue long region of FtsZ, thereby maximizing surface complementary between FtsZ protofilaments. Understanding how the FtsZ filaments and structural modules of ZipA (the membrane anchor domain, the central P/Q rich domain and the C-terminal FtsZ-binding domain) combine to support cell division in bacteria can provide a rationale behind the modular organization of ZipA and hence help to determine the precise arrangement of FtsZ and ZipA when assembled with other cell division proteins as a unit to form a septal ring structure.
Finally, the solution of these structures resulted in mutational analysis of the FtsZ fragment, from which it became apparent that most critical interactions on the FtsZ side are provided by a small cluster of the deeply buried hydrophobic side chains (Ile374, Phe377 and Leu378). These residues contribute most of the binding energy and therefore represent the major ZipA-binding determinants. Given mutation data obtained on numerous protein–protein interfaces (reviewed by Wells, 1996), it is very likely that functionally important residues on ZipA are those that are in direct contact with the key binding determinants of FtsZ. On this basis it seems possible to derive a limited site of interaction of FtsZ with ZipA that can be used as a framework for rational drug design. Small molecules designed to bind to this simplified site could form sufficient key interactions to maintain the complex, thereby hindering the binding of FtsZ. These data could be combined with further mutational and structural studies to assist in designing new drugs against infectious diseases. However, since ZipA has been found in only some Gram-negative eubacteria these new potential antibacterial agents will not represent broad-spectrum antibiotics.
Materials and methods
Expression and purification of ZipA/M185 and Se-Met ZipA/M185
ZipA/M185 was cloned into a pET derived vector and expressed in BL21DE2pLysS E.coli. Cells were grown in a Biostat C-10 (10 l) vessel (B. Braun Biotech) using rich media at 37°C and induced for 4 h with 1 mM IPTG. Se-Met labelled expression of Zipa/M185 was carried out in LeMaster media in BL21DE3pLysS E.coli at 37°C. Cultures were induced for 4 h with 1 mM IPTG.
Cells expressing ZipA/M185 were resuspended in buffer containing 25 mM HEPES pH 7.5, 2 mM dithiothreitol, and 0.1 mM phenylmethylsulfonyl fluoride, and lysed by passage through a Microfluidizer (Microfluidics Corporation, Newton, MA). Cleared lysate was loaded onto a QAE Toyopearl column and eluted protein was passed through a hydroxyapatite column (Bio-Rad). ZipA/M185-containing fractions were then subjected to FPLC anion exchange chromatography using a Mono Q column (Pharmacia) followed by a TSK-G3000SW size exclusion column. The final product was exchanged into buffer containing 20 mM Tris pH 8.0, concentrated to 25 mg/ml, and used for crystallization. Se-Met ZipA/M185 was purified following the same procedure as for native ZipA/M185.
Crystallization of ZipA/M185 and Se-Met ZipA/M185
Crystallization conditions for ZipA/M185 were determined from the sparse matrix screens (Hampton Research). Screening was done using hanging drop vapor diffusion by combining 1 µl of protein solution (25 mg/ml) with 1 µl of well solution at both 18 and 4°C. Initially, ill-formed crystals of ZipA/M185 grew spontaneously at 18°C in a mother liquor consisting of 25% PEG 6000 and 100 mM 2-morpholinoethanesulfonic acid (MES) pH 6.0. To produce diffraction quality crystals, streak seeding was used to seed pre-equilibrated (∼3 h) 2 µl drops containing 25 mg/ml ZipA/M185, 20% PEG 6000, and 100 mM MES pH 6.0. Monoclinic plate-like crystals (space group P21; a = 49.89 Å, b = 41.74 Å, c = 71.16 Å, β = 98.26°; two molecules per asymmetric unit; 37% solvent content) developed overnight and reached their maximum size (0.5 × 0.8 × 0.3 mm3) in 3–4 days. Se-Met ZipA/M185 crystallized under the same conditions using a similar seeding technique, with the native crystals as seeds. Differences in cell dimensions were <0.6%.
Crystallization of ZipA/M185 with the FtsZ peptide
A molar excess of a synthetic 17 amino acid peptide that encompasses the conserved C-terminal region of E.coli FtsZ (367KEPDYLDIPAFLRKQAD383) was added to the ZipA/M185 protein (25 mg/ml) such that the final mixture contained 1.3:1 FtsZ peptide versus ZipA/M185. Crystallization conditions were again found using PEG 6000 as precipitant (PEG 6K Grid Screen, Hampton Research), except that co-crystals appeared under basic pH (30% PEG 6000 and 100 mM Bicine, pH 9.0). Poor quality crystals grew spontaneously in 4–5 days as clusters of thin elongated plates. Since these crystals were not consistently reproducible, we applied a streak seeding technique using crystals of the ZipA/M185 alone as seeds. As in the case of ZipA/M185, 2 µl drops were pre-equilibrated (∼3 h) prior to cross-seeding. The best monocrystals grew over a period of 2–4 days with a maximum size of 0.2 × 0.2 × 1.0 mm3. They belonged to space group P21 (a = 36.53 Å, b = 38.9 Å, c = 54.54 Å, β = 75.89°) with 1:1 complex per asymmetric unit and 32% solvent content.
Data collection and processing
Prior to data collection, all crystals were flash cooled under a gaseous nitrogen stream at 100 K. Both native and Se-Met crystals of ZipA/M185 were soaked (∼1 min) in a solution containing mother liquor at pH 6.0, 15% ethylene glycol and 35% PEG 4000. Using our in-house RAXIS IV mounted on a Rigaku RUH2R rotating anode, two data sets were collected for phase determination: the 1.9 Å data for the native ZipA/M185 crystals (180 frames with 1° oscillation) and the 1.85 Å data for the Se-Met form of the protein (360 frames with 1° oscillation). For each data set, a single crystal was used. For refinement purposes the high resolution native data set (1.5 Å) was collected at beamline 5.0.2 at the Advanced Light Source (Berkley) using a Quantum 4 CCD detector (Area Detector Systems). These data were obtained from the same crystal that we used for in-house data collection.
As in the case of ZipA/M185, a co-crystal of ZipA/M185:FtsZ-peptide was soaked (∼1 min) in a solution containing 15% ethylene glycol, 35% PEG 4000 plus the mother liquor at pH 9.0. The 1.95 Å data set was collected from a single crystal (180 frames with 1° oscillation) using our in-house RAXIS IV imaging plate system,.
All the data were integrated with DENZO and then scaled and merged with SCALEPACK (Otwinowski, 1993). Most of the subsequent processing used the CCP4 programs (CCP4, 1994). The statistics from refinement are given in Table I.
SIRAS and refinement
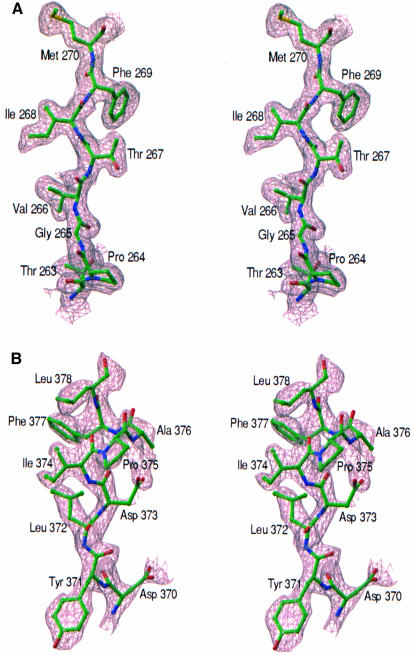
The data from Se-Met derivative were scaled to the native data to a resolution of 1.9 Å (SCALEIT in CCP4) and isomorphous difference Patterson synthesis along with the anomalous Patterson were calculated at 2 Å. Sixteen selenium sites were located using these Pattersons and from a double difference Fourier analysis (FFT in CCP4). The N-terminal Se-Met in both ZipA/M185 molecules was disordered. Refinement of occupancies, coordinates, as well as anomalous scatterer parameters, and phase calculation were performed with MLPHARE (Otwinowski, 1991). Phasing statistics generated by MLPHARE are given in Table I. The initial SIRAS map calculated at 2 Å was solvent-flattened using DM (Cowtan and Main, 1996), assuming 35% solvent content. Experimental maps were also calculated using SHARP (de la Fortelle and Bricogne, 1997) and subsequent density modification by SOLOMON (CCP4, 1994). These maps were produced using all 16 sites that were identified with MLPHARE phases. The final map (Figure 6A) was significantly better in terms of connectivity and resolution than that obtained by MLPHARE and DM. Because both algorithms produced clearly interpretable maps, all density-modified and unmodified SIRAS maps were used to build 100% complete models using X-AUTOFIT within QUANTA (Molecular Simulations, Inc., San Diego, CA).
Fig. 6. Electron density maps. (A) Experimental map of ZipA/M185 at 2 Å resolution, calculated with SIRAS phases improved by density modification. The map, contoured at 1.2 σ, is superimposed on the refined coordinates of residues B263–B270. (B) Difference electron density in the region of the FtsZ peptide bound to ZipA/M185. Electron density is from an Fobs – Fcalc map (25–1.95 Å, contoured at 1.7σ) calculated using model phases, with the peptide atoms omitted from all calculations. Figure prepared using the program RIBBONS (Carson, 1991).
This model was then used as the initial model for refinement against the 1.5 Å resolution native data set. Refinement and map calculations were done in CNS (Brünger et al., 1998). At all stages, data from 20.0 to 1.5 Å, with |Fobs|> 0, were included, with 5% of omitted reflections for Rfree calculation. The minimization included a bulk-solvent correction coupled with simulated annealing, positional and individual B factor refinement. Water molecules were located from electron density > 3σ in Fo – Fc maps. The final model (Rwork = 19.8%, and Rfree = 21.7%) contains residues A190–A328, B189–B328 and 422 water molecules. All non-glycine φ and ψ angles lie in the allowed regions of the Ramachandran plot, with 93.7% in the most favored regions and 6.3% in additional allowed regions. Residues A185–A189, B185–B188 were not detected in the electron density maps because of disordering.
Molecular replacement and refinement
ZipA/M185 was located using the final model of the ZipA/M185 monomer (residues B190–B328) in rotation and translation searches with AMoRe (Navaza, 1994). This model provided unambiguous rotation and translation function solutions. The rigid body refined model gave R factor of 44.2% and correlation coefficient of 55.6% for all data between 12 and 3 Å. The search model was immediately subjected to simulated annealing refinement coupled with a bulk solvent correction as implemented in CNS (Brünger et al., 1998). This resulted in Rwork = 32% and Rfree = 38.7% for 25–1.95 Å data, with 10% randomly selected reflections for Rfree calculation. This refined model was used to calculate the 1.95 Å Fo – Fc map, which showed clear electron density for the bound FtsZ peptide (Figure 6B). All 17 amino acid residues of the FtsZ peptide were fitted into this map and the refined model of ZipA/M185 was rebuilt using the 1.95 Å 3Fo – 2Fc map. After three cycles of rebuilding, minimization (positional plus individual B factor refinement) converged to Rwork of 20.5% and Rfree = 25.1%. The final model contains ZipA/M185 residues 185–328, FtsZ peptide residues 367–383 and 204 water molecules. All non-glycine φ and ψ angles lie in the allowed regions of the Ramachandran plot, with 94.1% in the most favored regions and 5.9% in additionally allowed regions. Side chains for peptide residues 367–368 have weak electron densities, therefore the polyalanines represent this region in the model. The N-terminus of ZipA/M185 (residues 185–189) stabilized and was clearly visible in all electron density maps, probably because of the tighter crystal packing.
Biosensor-based analysis
A BIAcore 2000 biosensor system (Pharmacia Biosensor, Uppsala) was used to assay interactions between ZipA/M185 and variants of the FtsZ peptide. Soluble ZipA/M185 molecules were immobilized to the biosensor CM5 chip by standard amine coupling chemistry. The peptide was injected over the chip in 10 mM HEPES pH 7.5, 150 mM NaCl, 3 mM EDTA and 0.005% polysorbate 20 v/v, at a flow rate of 10 µl/min. Binding between ZipA/M185 and the peptide resulted in changes in the SPR signal that are read out in real time as resonance units (RU). The equilibrium dissociation constant (KD column in Table II) was derived from sensorgram data (shown only for the wild-type peptide binding to immobilized ZipA/M185 in Figure 4A) using a steady affinity model by fitting the plots of Req (the equilibrium binding response) versus the concentration of the injected peptide (Figure 4B).
Protein Data Bank coordinates
The coordinates have been deposited in the Protein Data Bank. The accession code for ZipA/M185 is 1F46 and the code for the ZipA/M185–FtsZ peptide complex is 1F47.
Acknowledgments
Acknowledgements
The authors wish to thank K.Malakian for growth of E.coli, as well as T.McDonagh, A.Tam and S.Menon for assistance with BIAcore and ITC assays. We are grateful to R.Chopra, K.Svenson and the staff at Advanced Light Source for help with data collection. We also thank K.Svenson and R.Mathew for help with crystallization screening, and F.X.Sullivan and J.Tobin for critical reading of the manuscript.
References
- Allain F.H.-T., Gubser,C.C, Howe,P.W.A., Nagai,K., Neuhaus,D. and Varani,G. (1996) Specificity of ribonucleoprotein interaction determined by RNA folding during complex formation. Nature, 380, 646–650. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurora R. and Rose,G.D. (1998) Helix capping. Protein Sci., 7, 21–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E. and Lutkenhaus,J. (1991) FtsZ ring structure associated with division in Escherichia coli. Nature, 354, 161–164. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Burd C.G. and Dreyfuss,G. (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science, 265, 615–621. [DOI] [PubMed] [Google Scholar]
- Carson M. (1991) Ribbons 2.0. J. Appl. Crystallogr., 24, 958–961. [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for X-ray crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cowtan K.D. and Main,P. (1996) Phase combination and cross validation in iterated density modification calculations. Acta Crystallogr. D, 42, 43–48. [DOI] [PubMed] [Google Scholar]
- de Boer P., Crossley,R. and Rothfield,L. (1992) The essential bacterial cell-division protein FtsZ is a GTPase. Nature, 359, 254–256. [DOI] [PubMed] [Google Scholar]
- de la Fortelle E. and Bricogne,G. (1997) Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol., 276, 494–523. [DOI] [PubMed] [Google Scholar]
- Din N., Quardokus,E.M., Sackett,M.J. and Brun,Y.V. (1998) Dominant C-terminal deletions of FtsZ that affect its ability to localize in Caulobacter and its interaction with FtsA. Mol. Microbiol., 27, 1051–1063. [DOI] [PubMed] [Google Scholar]
- Erickson H.P., Taylor,D.W., Taylor,K.A. and Bramhill,D. (1996) Bacterial cell division protein FtsZ assembles into protofilament sheet and minirings, structural homologs of tubulin polymers. Proc. Natl Acad. Sci. USA, 93, 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher H.F and Singh,N. (1995) Calorimetric methods for interpreting protein–ligand interactions. Methods Enzymol., 259, 194–221. [DOI] [PubMed] [Google Scholar]
- Hale C.A. and de Boer,P.A.J. (1997) Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E.coli. Cell, 88, 175–185. [DOI] [PubMed] [Google Scholar]
- Hale C.A. and de Boer,P.A.J. (1999) Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J. Bacteriol., 181, 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A., Moss,D.S. and Thornton,J.M. (1993) Mainchain bond lengths and bond angles in protein structures. J. Mol. Biol., 231, 1049–1067. [DOI] [PubMed] [Google Scholar]
- Liljas A. and Garber,M. (1995) Ribosomal proteins and elongation factors. Curr. Opin. Struct. Biol., 5, 721–727. [DOI] [PubMed] [Google Scholar]
- Liu Z., Mukherjee,A. and Lutkenhaus,J. (1999) Recruitment of ZipA to the division site by interaction with FtsZ. Mol. Microbiol., 31, 1853–1861. [DOI] [PubMed] [Google Scholar]
- Löwe J. and Amos,L. (1998) Crystal structure of the bacterial cell-division protein FtsZ. Nature, 391, 203–206. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J. and Addinall,S.G. (1997) Bacterial cell division and the Z ring. Annu. Rev. Biochem., 66, 93–116. [DOI] [PubMed] [Google Scholar]
- Ma X. and Margolin,W. (1999) Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol., 181, 7531–7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I.W. and Nagai,K. (1995) Recruiting proteins to the RNA world. Nature Struct. Biol., 2, 518–522. [DOI] [PubMed] [Google Scholar]
- Mukherjee A. and Lutkenhaus,J. (1994) Guanine nucleotide-dependent assembly of FtsZ into filaments. J. Bacteriol., 176, 2754–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A. and Lutkenhaus,J. (1998) Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J., 17, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaza Z. (1994) AMoRe—an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Nikonov S. et al. (1996) Crystal structure of the RNA binding ribosomal protein L1 from Thermus thermophilus. EMBO J., 15, 1350–1359. [PMC free article] [PubMed] [Google Scholar]
- Nogales E., Wolf,S.G. and Downing,K.H. (1998) Structure of the alphabeta tubulin dimer by electron crystallography. Nature, 391, 199–203. [DOI] [PubMed] [Google Scholar]
- Orengo C.A. and Thorton,J.M. (1993) Alpha plus beta folds revisited: some favored motifs. Structure, 1, 105–120. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. (1991) Isomorphous Replacement and Anomalous Scattering (Daresbury Laboratory, UK).
- Otwinowski Z. (1993) In Sawyer, L., Isaacs,M. and Bailey,S.W. (eds), Data Collection and Processing. Science and Engineering Council, Daresbury,UK, pp. 56–62. [Google Scholar]
- Outbridge C., Ito,N., Evans,P.R., Teo,C.-H. and Nagai,K. (1994) Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature, 372, 432–438. [DOI] [PubMed] [Google Scholar]
- RayChaudhuri D. (1999) ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J., 18, 2372–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RayChaudhuri D. and Park,J.T. (1992) Escherichia coli cell-division gene FtsZ encodes a novel GTP-binding protein. Nature, 359, 251–254. [DOI] [PubMed] [Google Scholar]
- Richardson J.S. and Richardson,D.C. (1988) Amino acid preferences for specific locations at the ends of α-helices. Science, 240, 1648–1652. [DOI] [PubMed] [Google Scholar]
- Rothfield L.I. and Justice,S.S. (1997) Bacterial cell division: the cycle of the ring. Cell, 88, 581–584. [DOI] [PubMed] [Google Scholar]
- Rothfield L.I., Justice,S. and Garcia-Lara,J. (1999) Bacterial cell division. Annu. Rev. Genet., 33, 423–448. [DOI] [PubMed] [Google Scholar]
- Schreuder H., Tardif,C., Trump-Kallmeyer,S., Soffientini,A., Sarubbi,E., Akeson,A., Bowin,T., Yanofsky,S. and Barrett,R.W. (1997) A new cytokine-receptor binding mode revealed by the crystal structure of the IL-1 receptor with an antagonist. Nature, 386, 194–199. [DOI] [PubMed] [Google Scholar]
- Szabo A., Stolz,L. and Granzow,R. (1995) Surface plasmon resonance and its use in biomolecular interaction analysis (BIA). Curr. Opin. Struct. Biol., 5, 699–705. [DOI] [PubMed] [Google Scholar]
- Wang X. and Lutkenhaus,J. (1996) Characterization of the ftsZ gene from Mycoplasma pulmonis, an organism lacking a cell wall. J. Bacteriol., 178, 2314–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Huang,J., Mukherjee,A., Cao,C. and Lutkenhaus,J. (1997) Analysis of the interaction of FtsZ with itself, GTP and FtsA. J. Bacteriol., 179, 5551–5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J.A. (1996) Binding in the growth hormone receptor complex. Proc. Natl Acad. Sci. USA, 93, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonath A. and Franceschi,F. (1997) New RNA recognition features revealed in ancient ribosomal proteins. Nature Struct. Biol., 4, 3–5. [DOI] [PubMed] [Google Scholar]