Abstract
RNA phage GA coat and lysis protein expression are translationally coupled through an overlapping termination and initiation codon UAAUG. Essential for this coupling are the proximity of the termination codon of the upstream coat gene to the initiation codon of the lysis gene (either a <3 nucleotide separation or physical closeness through a possible hairpin structure) but not the Shine–Dalgarno sequence. This suggests that the ribosomes completing the coat gene translation are exclusively responsible for translation of the lysis gene. Inactivation of ribosome recycling factor (RRF), which normally releases ribosomes at the termination codon, did not influence the expression of the reporter gene fused to the lysis gene. This suggests the possibility that RRF may not release ribosomes from the junction UAAUG. However, RRF is essential for correct ribosomal recognition of the AUG codon as the initiation site for the lysis gene.
Keywords: initiation/recycling/ribosome/ribosome recycling factor (RRF)/translational coupling
Introduction
Translational coupling is the mechanism by which an appropriate quantitative ratio of a certain protein to another protein is obtained. It also functions to attain the sequential expression of these genes. Depending on the origin of the ribosomes that translate the coupled downstream gene, translational coupling can be classified into three categories. In the first category, the downstream cistron becomes available for translation by free ribosomes through the action of the ribosomes translating the upstream cistron (hereafter called upstream ribosomes); however, the downstream gene is translated exclusively by free ribosomes (Berkhout and van Duin, 1985). In the second category, the downstream gene becomes available in the same way but both the free and the upstream ribosomes translate the downstream cistron (Baughman and Nomura, 1983; Berkhout et al., 1987; Schmidt et al., 1987; Das and Yanofsky, 1989; de Smit and van Duin, 1990). The third category is the case where the downstream cistron is translated exclusively by the upstream ribosomes, as suggested previously (Ivey-Hoyle and Steege, 1989). Coupling of the expression of the coat and lysis gene in the GA RNA phage, as shown here, belongs to this latter group. These two genes share one nucleotide between the termination and the initiation codon, UAAUG (Inokuchi et al., 1986).
Ribosome recycling factor (RRF, originally called ribosome releasing factor) is an essential factor (Janosi et al., 1994) that disassembles the post-termination complex (Janosi et al., 1996a,b; Kaji et al., 1998; Kaji and Hirokawa, 2000). Structurally, RRF is a near perfect mimic of tRNA (Selmer et al., 1999). The mechanism of RRF action has been proposed on the basis of its structure and that of various mutations (Janosi et al., 2000). The disassembly depends on EF-G (Hirashima and Kaji, 1973) or RF3 (Grentzmann et al., 1998), GTP (Hirashima and Kaji, 1972a; Karimi et al., 1999) and RRF (Hirashima and Kaji, 1972b, 1973). RRF releases ribosomes from mRNA as 70S ribosomes (Hirashima and Kaji, 1972a; Ogawa and Kaji, 1975) or as 50S subunits leaving the 30S subunits on the mRNA (Pavlov et al., 1997; Karimi et al., 1999) depending on such factors as mRNA configuration, Mg2+ and IF3 concentration. When 30S subunits remain on the ribosome, tRNA also stays on it and IF3 is necessary to release this tRNA from the 30S subunit (Gualerzi et al., 1971; Karimi et al., 1999). RRF stimulates in vitro protein synthesis 4- (Kung et al., 1977) to 7- (Ryoji et al., 1981b; Pavlov et al., 1997) fold. In the absence of RRF, the ribosome of the post-termination complex not only remains on the mRNA, but translates the 3′ portion of the mRNA downstream from the termination codon (Ryoji et al., 1981a; Janosi et al., 1998). In addition to disassembling the post-termination complex, RRF maintains translational accuracy during the chain elongation (Janosi et al., 1996b). The gene coding for RRF (frr) in Escherichia coli is located near 4 minutes on the chromosome (Ichikawa et al., 1989). Every organism so far examined has a homologue of frr, except for archeons. RRFs from Pseudomonas aeruginosa (Ohnishi et al., 1999), yeast (Kanai et al., 1998) and spinach (Rolland et al., 1999) have been characterized recently.
Here we present evidence that the lysis gene of the GA phage is translated exclusively by the ribosomes that completed the upstream coat gene translation. Due to the overlap of the termination and the initiation codons, UAAUG, inactivation of RRF does not influence the translation of the reporter gene fused to the lysis gene, suggesting that RRF may not release ribosomes from mRNA at the post-termination complex of the coat cistron. However, RRF plays a key role in this translational coupling because it makes the ribosomes correctly initiate the translation of the lysis gene from the initiation codon AUG. This is a new RRF function, which hitherto has not been described.
Results
UAA and AUG must be in close proximity for translational coupling between the coat and the lysis protein genes
The termination codon and the initiation codon of the coat and lysis protein genes of the GA phage are positioned in a uniquely close proximity sharing one nucleotide. To study the role of this junction sequence in the translational coupling of these two genes, we used various plasmids derived from plasmid pLWT (plasmid for lysis protein wild-type). This plasmid carries the lac promoter, the 3′ portion of the coat gene and the lysis gene in the wild-type setting (Table I). We wanted to estimate how far the termination codon could be separated from the initiation codon without losing expression of the lysis gene. For this purpose, the termination codon was placed either eight [pL-8AA (UAA at 8 nucleotides from AUG)], 20 (pL-20AA), 33 [pL-33GA (UGA at 33 nucleotides from AUG)] or 80 (pL-80AA) nucleotides upstream from AUG as shown in Table I. Lysis protein production was measured by lysis of the cells harbouring these plasmids upon induction of transcription by isopropyl-β-d-thiogalactopyranoside (IPTG). As can be seen from line 4 (pL-20AA) of this table, the termination codon has to be within at least 20 nucleotides from the initiation codon. This does not mean that a distance >20 nucleotides abolishes the translational coupling completely. As discussed below and in Figure 1, as long as the physical distance between AUG and UAA is made close by loop formation, the translational coupling should take place.
Table I. Effects of the distance between the UAA and AUG codons on lysis gene expression.
Bold triplets indicate the termination codon of the coat gene. Double underlining shows the initiation codon AUG of the lysis gene. Italicized letters represent the base substitutions. Lower case letters show the nucleotides inserted. Dashes denote the same nucleotides as pLWT. Numbers indicate the nucleotide position from the 5′ terminus of GA RNA (Inokuchi et al., 1986).
aThe number of plus signs reflects the ratio of the cell density, OD660(IPTG+)/OD660(IPTG–), which is shown in parentheses, after 3 h incubation as described in Materials and methods.
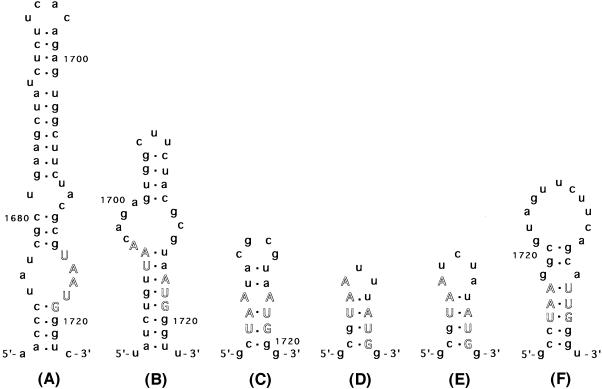
Fig. 1. Possible hairpin structures which bring the termination and initiation codons close to each other in the initiation region of the lysis gene. (A) pLWT, (B) pL-20AA, (C) pL-8AA, (D) pL+3U, (E) pL+5U and (F) KU-1. The numbers in (A), (B), (C) and (F) indicate the nucleotide positions from the 5′ terminus of GA or KU1 RNA (Inokuchi et al., 1986; Groeneveld et al., 1996). Termination and initiation codons of the coat and lysis genes are represented in white upper case letters. Dots indicate base pairs including G⋅U pairing.
In the next series of experiments shown in Table I, the relative distance between UAA and AUG was gradually increased as shown in the series of plasmids indicated by pL+0 (+ 0 nucleotide between them) to pL+5U (+ 5 nucleotides with 3′U). It is clear from Table I that the insertion of >2 nucleotides between them abolished the expression of the lysis gene, with the exception of plasmids pL+3U and pL+5U. We conclude that the translational coupling requires the termination codon to be within a distance of two nucleotides from the initiation codon. This is in contrast to the results where UAA can be as far away as 20 nucleotides as indicated in lines 1–5 of Table I.
To understand the seemingly contradictory results of these two series of experiments, we examined the possible hairpin structure of mRNA of these constructs near the initiation codon. As can be seen from Figure 1, in every positive case where the distance is >2 nucleotides, it is possible to construct a hairpin structure that brings the termination codon close to the AUG codon. We conclude that AUG and UAA must be very close for the coupling to occur. Although a portion of these loops could be broken down by the translating ribosomes, by the time the ribosome starts breaking the loop, it is already near the AUG codon. In some cases, the portion of the loop is in the part that is not translated (see, for example, Figure 1B).
It can be seen in Table I that the plasmid pL+0 (UAaAUG) produced less cell lysis than the plasmids pL+1C, +1U or +2C. This is probably because the UAAA sequence was leaky in that ribosomes may not terminate at this sequence as efficiently as with UAA (Martin et al., 1988). Thus, in pL+0, the ribosomes coming to the UAaAUG often must read through the UAA codon and terminate its coat synthesis at the UGA codon that is six nucleotides downstream. This would result in less lysis protein synthesis as shown in line 5 of Table III.
Table III. The nature of the termination and initiation codon does not influence the translational coupling, but the initiation codon in appropriate positions relative to the termination codon is important.
Bold triplets indicate the termination codon of the coat gene. Double underlining shows the initiation codon of the lysis gene. Italicized letters represent the base substitutions. Dashes denote the same nucleotides as pLWT.
aThe number of plus signs reflects the ratio of the cell density, OD660(IPTG+)/OD660(IPTG–), which is shown in parentheses, after 3 h incubation as described in Materials and methods.
bE.coli 594(Su–) cells were used because JM109 cells have an amber (UAG) suppressor tRNA.
cIn E.coli JM109, termination takes place at UGA downstream from AUG because UAG is read as a sense codon in Su+ cells.
Evidence that the lysis gene is translated exclusively by the ribosomes that translated the upstream coat gene—lack of Shine–Dalgarno (SD) sequence and the effect of deletion of the upstream gene
From the sequence analysis of the GA phage, it has been suggested that the ribosomes reading the upstream sequence are responsible for translation of the lysis gene (Inokuchi et al., 1986). To investigate this point further, plasmids without the upstream coat gene with [pL-7SD (added SD sequence at 7 nucleotides upstream of AUG)] and without [pL-21d or pL-9d (without 9 or 21 nucleotides upstream of UAA due to deletion of the entire upstream gene)] an SD sequence were constructed. As shown in Table II, elimination of the upstream gene abolished the expression of the lysis gene, suggesting that the ribosomes reading the upstream gene are responsible for the translation of the lysis gene. Furthermore, the lysis gene was translated from the construct lacking the upstream gene provided an artificial strong SD sequence was added upstream of the initiation codon of the lysis gene (pL-7SD). It appears that the SD sequence allows the free ribosomes to translate the lysis gene in the absence of the upstream coat gene because the initiation codon of the lysis gene is no longer hidden by the secondary structure of the RNA.
Table II. The upstream coat gene is essential for lysis gene expression.
Bold triplets indicate the termination codon of the coat gene. Double underlining shows the initiation codon AUG of the lysis gene. Italicized letters represent the base substitutions. Dashes denote the same nucleotides as pLWT. Asterisks show the deletions.
aThe number of plus signs reflects the ratio of the cell density, OD660(IPTG+)/OD660(IPTG–), which is shown in parentheses, after 3 h incubation as described in Materials and methods.
In addition to the possible hairpin structure in the vicinity of the initiation codon as shown in Figure 1A, the lack of a strong SD sequence appeared to be responsible for preventing free ribosomes from translating the lysis gene. As discussed in the preceding paragraph, upon addition of a strong SD sequence, lysis protein was produced even in the absence of the upstream gene (pL-7SD). To ensure that the weak SD-like sequence GAGUGG at nucleotide 1699 (12 nucleotides upstream from AUG) is not functioning, this was changed to CCAUGG in the plasmid pLSD (–) (wild-type minus SD-like sequence). As can be seen in Table II (line 2), this change did not abolish expression of the lysis gene. Together with this result and the effect of removal of the upstream sequence, we conclude that the upstream ribosomes, but not free ribosomes, must be responsible for production of the lysis protein.
The position but not the nature of the initiation and termination codon influences lysis protein production
In the experiment described in Table III, the initiation codon of the lysis gene was changed from AUG to GUG or to no initiation codon, keeping all other elements the same as those of the wild-type. As shown in Table III, substitution of AUG with GUG, a rarer initiation codon, did not influence production of the lysis protein. In a similar manner, substitution of the termination codon, UAA, with UAG or UGA did not influence translation of the lysis protein gene.
However, an inhibition of lysis protein production was observed when the initiation codon was eliminated [pLWTAUG (–)]. Furthermore, as pointed out in Table I, the position of the initiation codon relative to the termination codon is critical. Thus, in the presence of amber suppressor tRNA (in the strain JM109), substitution of UAA with UAG caused the termination not at UAG but at UGA, six nucleotides downstream of UAG. When the termination codon was thus moved six nucleotides downstream, the lysis protein was not produced (pLWTAG in JM109, last line in Table III). This suggests that ribosomes finishing translation at UGA could not go back four nucleotides to initiate lysis protein synthesis.
Only 25–30% of the upstream ribosomes participate in translating the lysis gene
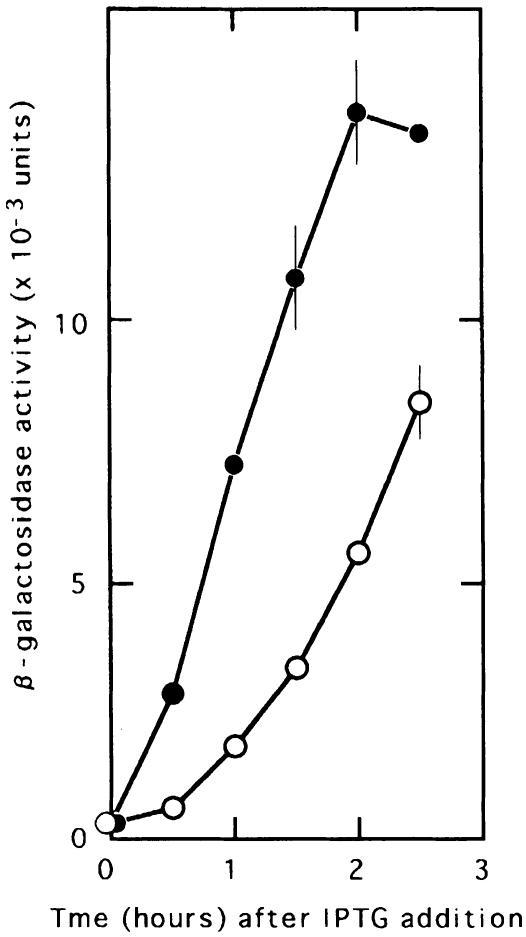
In the experiment described in Figure 2, a reporter gene fused to the lysis gene measured the expression of the lysis protein. Plasmid pL84Z has the β-galactosidase gene (lacZ) without the N-terminal nine amino acids and the initiation signals 84 nucleotides downstream from the AUG codon of the lysis gene (Table IV). The rest of the construct is identical to the pLWT that was used in the preceding experiments. To measure the amount of ribosomes translating the coat gene, a similar plasmid (pCLZ) (which does not have UAAUG but with UAA replaced with UAC and UG removed, resulting in the coat-lysis fusion gene) was constructed. Since β-galactosidase is fused directly to the upstream gene, the amount of translation of the upstream gene can be measured by the amount of β-galactosidase expressed from this construct. On the other hand, the amount of β-galactosidase expressed by pL84Z would represent the lysis gene translated by the portion of the ribosomes that completed the upstream gene translation through the coupling mechanism.
Fig. 2. A large proportion of the upstream ribosomes drop off at UAAUG, the border between the coat and lysis genes. Escherichia coli JM109 cells harbouring the plasmid pL84Z (open circle, represents coupled lysis gene translation) or pCLZ (closed circle, represents the upstream coat gene translation) were grown at 37°C to an OD660 of 0.15. β-galactosidase synthesis was induced by addition of IPTG (2 mM) at 37°C.
Table IV. Constructs of the plasmids carrying the reporter lacZ gene.
Bold triplets indicate the termination codon of the coat gene. Italicized letters represnt the base substitutions. Lower case letters represent the lacZ gene. Underlining indicates the nucleotide insertions. Double underlining indicates the initiation codons of the lysis gene. Dashes denote the same nucleotide as pLWT. Asterisks indicate the deletions.
We reasoned that the ratio of β-galactosidase expressed by pL84Z to that by pCLZ should indicate the percentage of the upstream ribosomes translating the lysis gene. As can be seen from Figure 2, the average ratio was ∼25–30%, indicating that the major portion of the upstream ribosomes are released at the junction UAAUG.
Inactivation of RRF does not influence translation of the reporter gene fused to the lysis gene
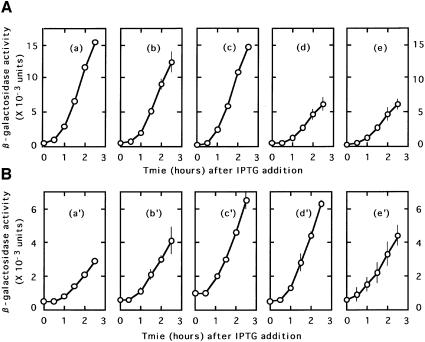
Under normal circumstances where the termination codon is not so close to the initiation codon, RRF is supposed to release ribosomes from mRNA during the process of disassembly of the post-termination complex (Hirashima and Kaji, 1972a; Ogawa and Kaji, 1975). It is therefore of interest to determine whether RRF is responsible for the release of 70–75% of the upstream ribosomes as described in the preceding section. The amount of β-galactosidase activity produced from pL84Z as shown in Figure 2 represents the translation by the ribosomes that survived the loss of ribosomes from the post-termination complex of the coat gene. If ribosomes were released at UAAUG by RRF, one would expect that in vivo inactivation of RRF would make all the upstream ribosome stay on the mRNA.
In testing this possibility, we have to be aware of the fact that the in vivo inactivation of RRF reduces protein synthesis in general because RRF is essential for protein synthesis. Therefore, we have to measure the amount of β-galactosidase produced from pL84Z relative to that from a plasmid expressing the lacZ gene without the translational coupling. To accomplish this, the β-galactosidase activity expressed from pL-7SD84Z was measured in parallel. Plasmid pL-7SD84Z has no upstream gene but has the SD sequence seven nucleotides upstream of the lysis gene initiation codon. The lacZ gene is fused at the 85th nucleotide downstream from the initiation codon.
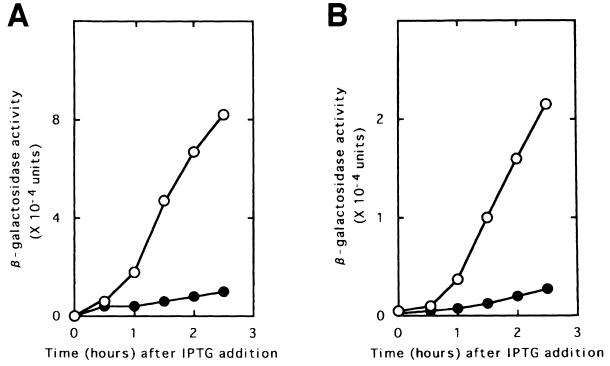
As shown in Figure 3A, the amount of β-galactosidase produced from pL-7SD84Z in LJ14 (which carries a temperature-sensitive RRF) at the non-permissive temperature (39°C) was ∼10% of that produced in the parental wild-type MC1061. Since the lysis gene in this plasmid is not under the control of the upstream gene, the percentage value represents reduction of β-galactosidase synthesis due to the general effect of RRF inactivation on protein synthesis. In Figure 3B, a similar experiment was carried out with pL84Z. It is clear that the amount of β-galactosidase produced in LJ14 by this plasmid was also ∼10% of that produced in MC1061. The results shown in Figure 3A and B indicate that the reporter gene expression is not influenced by inactivation of RRF. It should be mentioned that, in the wild type, ∼25–30% of the upstream ribosome gives the reporter gene expression. Inactivation of RRF causes, theoretically, 33% of all the ribosome remaining on the mRNA to read the reporter gene, because only a third of the ribosome remaining on the mRNA will be in-frame with the reporter gene. Therefore, the results can mean either that the ribosome is released without RRF or that the ribosome is released by RRF but is quickly re-bound to the initiation codon. We prefer the former interpretation as mentioned in the Discussion. Here, we call this release ‘mechanical release’ because it is not enzymatic.
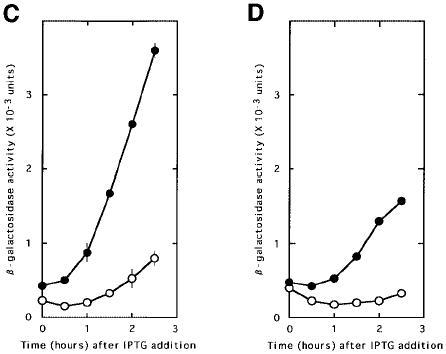
Fig. 3. RRF does not release ribosomes from the post-termination complex formed at UAAUG. Escherichia coli MC1061 (with the wild-type RRF, open circle) or LJ14 (with a temperature-sensitive RRF, closed circle) harbouring various plasmids as indicated below were grown at 27°C to an OD660 of 0.15. They were transferred to 39°C (non-permissive temperature for temperature-sensitive RRF) in the presence of IPTG (2 mM) for induction of reporter gene transcription (β-galactosidase, expressed as means ± SE shown by bars). (A) pL-7SD84Z, representing β-galactosidase synthesis with an added SD sequence without control by the coupling mechanism. (B) pL84Z, representing lysis gene synthesis through coupling. (C) pLAUG(–)86Z, representing lysis gene translation without the AUG of UAAUG expressed by the Z gene at 86 nucleotides downstream from UAA. (D) pLAUG(–)5Z, representing lysis gene translation without the AUG of UAAUG expressed by the Z gene at five nucleotides downstream from UAA. Note that the ratio of β-galactosidase in MC1061 to that in LJ14 of (A) was identical to that of (B). The reporter gene expression in LJ14 is more than that in MC1061 in (C) and (D), indicating the release of ribosomes from UAA by RRF with these constructs.
On the other hand, as shown in Figure 3C and D, if the reporter gene is placed downstream of UAA without AUG and RRF is inactivated, the amount of β-galactosidase from pLAUG(–)86Z or from pLAUG(–)5Z produced in LJ14 was much higher than in MC1061. This indicates that, as expected, RRF does release ribosomes from mRNA at UAA when AUG is not close, and inactivation of RRF resulted in the increased amount of the downstream reporter gene expression. Plasmid pLAUG(–)86Z gave a higher value for the reporter gene expression than pLAUG(–)5Z. This is because the reinitiation of the unscheduled translation downstream of the termination codon may occur as far downstream as 45 nucleotides away from the termination codon (Janosi et al., 1998). If the termination codon is too close to the reporter gene (five nucleotides versus 86 nucleotides), the unscheduled reinitiation of translation due to the inactivation of RRF may take place at a position deep in the Z gene. This would result in a product that may not be active because of too much truncation of the N-terminal portion of β-galactosidase.
RRF makes ribosomes recognize the initiation codon at UAAUG situated between the coat and lysis gene
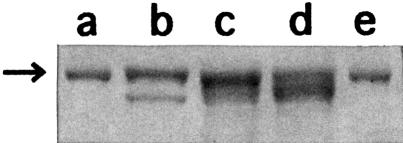
In the experiment described in Figure 4, the lysis–β-galactosidase fusion protein synthesized in LJ14 cells (lane c) and in MC1061 cells (lane b) from plasmid pL84Z at the non-permissive temperature was purified and an equal amount of each preparation was subjected to SDS–gel electrophoresis. It appears from Figure 4 that the major band corresponding to the fusion protein (expected mol. wt 120 kDa, shown by the arrow) in lane c is more diffuse and moved slightly faster than that in lane b. This indicates that the fusion protein in lane c is a mixture of proteins with various molecular weights (∼116 kDa) smaller than that produced in wild-type MC1061 (120 kDa, lane b). This suggests that, in the absence of RRF, the ribosomes reaching the sequence UAAUG terminate the translation of the coat gene but fail to recognize AUG properly and start translation randomly downstream from the termination codon without any initiation signal such as the SD or AUG (Janosi et al., 1998).
Fig. 4. Heterogeneous smaller fusion proteins (lysis protein–β-galactosidase) were produced in the absence of RRF. Escherichia coli MC1061 cells carrying pL84Z (lane b) and LJ14 cells carrying pL84Z (lane c) or pLAUG(–)86Z (lane d) were induced by IPTG to transcribe the coat–lysis–lacZ gene at 39°C for 3 h. The lysis–lacZ fusion protein was purified as described in Materials and methods. The protein (1.2 µg) was electrophoresced on a 7.5% SDS–polyacrylamide gel and stained with Coomassie Brilliant Blue. The arrow indicates the position of the full-size fusion protein (120 kDa). Lanes a and e show the position of the marker protein [β-galactosidase from E.coli (116 kDa)].
Lane d represents the β-galactosidase fusion made from the construct pLAUG(–)86Z in LJ14 at 39°C. This plasmid lacks the initiation codon AUG of the lysis gene. The mixture of fusion proteins made from this construct in the absence of RRF appears similar to that made from the plasmid with AUG shown in lane c. They are smaller and more diffuse than in lane b. This means that, in the absence of RRF, the ribosomes behave identically on the mRNA regardless of the presence of AUG. In other words, the ribosomes disregard the AUG of UAAUG in the absence of RRF. It should be noted that the density of this band does not reflect the amount of the fusion protein produced in the cells because an approximately equal amount of each sample was applied to the gel. The actual amount of the fusion protein synthesized in the absence of RRF is much smaller than that in the presence of RRF, as can be seen from Figure 3.
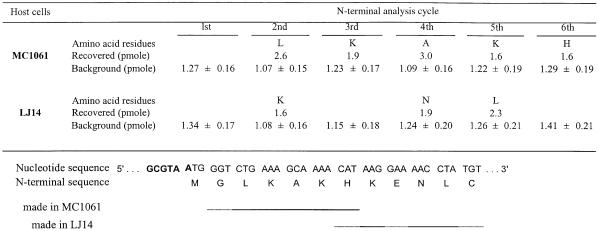
The finding that, in the absence of RRF, ribosomes do not recognize the AUG of UAAUG as the initiation site of the lysis gene was supported further by a separate experiment where N-terminal analysis of the β-galactosidase fusion protein was performed. As shown in Table V, except for the first round, we were able to detect amino acids corresponding to the β-galactosidase fusion at levels significantly higher than background. Thus, we confirmed the N-terminal amino acid sequence (G)-L-K-A-K-H for the control fusion protein (Inokuchi et al., 1986). In this experiment, six cycles of N-terminal amino acid analysis were performed for the fusion protein formed in MC1061 (in the presence of RRF) and in LJ14 at the non-permissive temperature (in the absence of RRF). It can be seen in Table V that, with the fusion protein synthesized in the absence of RRF, the first, third and sixth cycles of the analysis failed to provide any corresponding amino acid residues at levels higher than background. In the second, fourth and fifth cycles, K, N and L were detected as slightly higher levels than background. These amino acids correspond to the 8th, 10th and 11th amino acids of the fusion protein. This is consistent with our previous results showing that ribosomes may slide downstream on the mRNA upon inactivation of RRF. Although these results are statistically significant, the amount recovered after subtraction of background is much less than that from the fusion protein synthesized in the presence of RRF. These data strongly suggest that the N-terminal sequence of the protein produced in the absence of RRF is heterogeneous while fusion protein with a discrete N-terminus is formed with the wild-type.
Table V. N-terminal amino acid analysis of fusion β-galactosidase.
After SDS–PAGE, the β-galactosidase fusion protein was transfered to a PVDF membrane (PE Biosystems). The membrane was stained, cut out and then used for the amino acid sequence analysis. Six amino acid residues at the N-terminus were analysed. The Gln, Thr and Glu residues were not identified because their peaks and those of the chemical reagents used for analysis overlapped.
The minor band corresponding to a mol. wt of 110 kDa present in lane b of Figure 4 was found to have the N-terminal sequence G-L-K-(A)-K-H. This corresponds to the N-terminal sequence of the β-galactosidase fusion as described above. This indicates that the unexpected band present in the wild-type extract was derived from the C-terminal truncation of the 120 kDa fusion protein. It is noteworthy that this unexpected band also appears diffuse in LJ14 (lane c), again suggesting the random initiation of the fusion protein in the absence of RRF.
The effect of the distance between UAA and AUG on proper recognition of AUG by ribosomes with the help of RRF
In the experiment described in Figure 5A, plasmids having various distances between UAA and AUG and the downstream β-galactosidase gene (see Table IV) were placed in MC1061 that has wild-type RRF at 39°C. The amount of β-galactosidase produced by these plasmids with zero and one nucleotide insertions was identical to that produced by pL84Z with UAAUG (the amount of β-galactosidase in a equals that in b, which is identical to that in c). Therefore, release of ribosomes from mRNA at the junction point remained the same as long as UAA was not further than one nucleotide away from AUG. Comparison of these data with those presented in Table I revealed that, in a similar fashion, proper recognition of AUG by ribosomes with the help of RRF could take place even if AUG is one nucleotide away from UAA. In this condition, active lysozyme synthesis that requires correct initiation was observed (lines 6–10 of Table I). When the distance was three or five nucleotides, production of the fusion protein decreased significantly (Figure 5A, c>d, c>e), suggesting that the ribosome releasing action of RRF takes over, ribosomes are released and hence the downstream gene expression decreases. Under these conditions, no lysis protein activity was detected (lines 11–18 of Table I, except for the cases where hairpin formation is possible), suggesting that RRF can no longer place ribosomes in the correct position to read the initiation codon AUG.
Fig. 5. The distance between UAA and AUG determines proper recognition of AUG by ribosomes with the help of RRF. Various plasmids with varying distances between UAA and AUG were used. Plasmids pL84Z (UAAUG, in a and a′), pL+084Z (UAaAUG, in b and b′), pL+1C84Z (UAacAUG in c and c′), pL+3C84Z (UAauucAUG in d and d′) and pL+5A84Z (UAaucuaaAUG in e and e′) were placed in E.coli MC1061 (A) or in LJ14 (B). Cells were grown at 27°C and switched to 39°C (non-permissive temperature for LJ14) at the beginning of the experiment. The transcription of mRNA from the plasmid was induced by the addition of IPTG and the β-galactosidase activity (means ± SE shown by bars) was measured.
Since the major portion of the upstream ribosomes may be mechanically dropped off from UAAUG (Figures 2 and 3), we investigated the junction sequences that influence this mechanical release of ribosomes from mRNA. In the experiment described in Figure 5B, the same plasmids used in Figure 5A were placed in LJ14 at 39°C and the β-galactosidase activity was measured. It should be pointed out that very little if any RRF activity is present under these conditions. Therefore, the expression of the reporter gene represents the translation by those upstream ribosomes that survived the release from UAAUG without involving RRF. It is clear from this figure that the reporter gene is expressed progressively more as the distance between UAA and AUG increased from –1 to +1 (a′<b′<c′). Increasing the distance from two to three nucleotides did not increase the induced β-galactosidase further (Figure 5B, d′). When the distance reached five nucleotides, the amount of β-galactosidase was slightly reduced (e′<d′). This is perhaps due to accidental drop off of the initiating ribosomes before they reach the initiation site. These data indicate that the UAAUG configuration gives the maximum mechanical release of ribosomes. In confirmation of our preceding report that ribosomes can change the reading frame in the absence of RRF (Janosi et al., 1998), the β-galactosidase fusion was observed regardless of the reading frame from the termination codon UAA.
An important finding in Figure 5 is that the proper recognition of AUG by the ribosomes with the help of RRF can take place even when a portion of the ribosomes are released by RRF. In other words, these two activities of RRF are not mutually exclusive. Thus, RRF functions to keep the amount of released ribosomes to the same level as that with UAAUG even when the overlap was eliminated or one nucleotide was inserted between UAA and AUG (a = b = c) while the mechanical release decreased (a′<b′<c′). This means that RRF started functioning as a releasing factor as soon as the overlap of UAA and AUG was eliminated, while the proper recognition of AUG still takes place under these conditions as shown in Table I.
Discussion
We propose that the lysis gene of RNA phage GA is translated exclusively by ribosomes coming from the upstream coat gene on the basis of the following observations. (i) The initiation codon AUG of the lysis gene is well hidden by the possible hairpin structure. (ii) There is no functional SD or SD-like sequence near the initiation codon AUG. (iii) Elimination of the upstream sequence completely abolished the expression of the downstream lysis gene. Furthermore, introduction of the SD sequence into this construct restored lysis gene expression even without the upstream coat gene. (iv) RRF may not release ribosomes from the unusual gene border sequence UAAUG. (v) The ribosomes translating the upstream coat gene must come into close proximity with the initiation codon of the downstream lysis gene either by a nucleotide distance of not more than two nucleotides or by RNA secondary structure.
Having established that the lysis gene is translated exclusively by the upstream ribosomes, this system is an ideal system to study the role of RRF in the behaviour of ribosomes in translational coupling with the unusual sequence UAAUG at the gene border. Two new findings of general significance were made in this system regarding the mode of action of RRF. First, inactivation of RRF did not increase expression of the downstream reporter gene fused with the lysis gene, suggesting the possibility that ribosomes may not be released from the junction sequence UAAUG by the action of RRF. This is in contrast to the events at normal termination codons where ribosomes are released by RRF. Inactivation of RRF makes the ribosomes remain on the mRNA and induces the downstream unscheduled translation reinitiation giving rise to the expression of the downstream reporter gene (Janosi et al., 1998). Elimination of AUG from the junction sequence UAAUG restored the release of ribosomes by RRF, indicating that RRF functions within this system as a ribosome releasing factor if a proper mRNA configuration is present. Clearly, RRF may be inhibited from performing its normal function, i.e. releasing ribosomes from mRNA, because of the unusual overlapping codons UAAUG. As soon as the overlap was eliminated, RRF displayed releasing activity.
Secondly, inactivation of RRF produced abnormal fusion proteins between the lysis protein and β-galactosidase. They had smaller molecular weights and were heterogeneous without discrete N-terminals. The products are almost identical to those made from the construct without the initiation triplet AUG. This means that ribosomes could not recognize AUG of UAAUG as the initiation site in the absence of RRF. This is because ribosomes (probably as 70S ribosomes; Petersen et al., 1978), in the absence of RRF, slide down the mRNA and reinitiate randomly downstream of the termination codon (Janosi et al., 1998). Since the intact N-terminus is required for the full activity of the lysis protein (Y.Inokuchi, unpublished observation), loss of RRF activity would lead to complete loss of active lysis protein. This finding establishes the important new role of RRF in inducing ribosomal recognition of the initiation signal near the termination codon without releasing ribosomes from the mRNA. These two functions of RRF, forcing ribosomes to recognize AUG correctly and releasing other ribosomes from the junction point, are not mutually exclusive. Three roles for RRF discovered thus far are: disassembly of the post-termination complex (Hirashima and Kaji, 1972a; Ogawa and Kaji, 1975); error prevention (Janosi et al., 1996b); and the recognition of the initiation codon as described here. These roles may be three different manifestations of one activity of RRF.
How does RRF force the ribosomes to recognize the initiation codon at the UAAUG sequence? We propose a possible mechanism on the basis of the near perfect structural mimicry of tRNA by RRF (Selmer et al., 1999). It has been proposed that RRF binds to the A-site of the ribosome and is translocated to the P-site by EF-G just as tRNA is translocated from the A-site to the P-site (Selmer et al., 1999). At the time at which the coat protein is released from the ribosome, UAA is at the A-site. After the release of the complete coat protein, UAA will be translocated to the P-site and the A-site will be occupied by UGG. RRF will then occupy the A-site before the tryptophanyl-tRNA coded for by UGG occupies the A-site. This is possible because of the abundance of RRF in the cell (Ryoji, 1981). Once the A-site is occupied by RRF, we postulate that RRF could translocate three or two nucleotides depending on the mRNA sequence near the termination codon. For example, when the termination codon is close to the SD sequence or poly(A) sequence, it may not translocate three nucleotides to the P-site and release tRNA (Karimi et al., 1999). This must be because RRF lacks the portion corresponding to the CCA end of tRNA and does not have the anticodon.
If RRF is translocated only two nucleotides, this will place AUG precisely at the empty P-site. The empty P-site is now available for binding of formylmethionyl-tRNA. This enables the proper initiation of the lysis protein. This can happen only with the AUG codon when the P-site is empty because only N-blocked aminoacyl-tRNA available in the cell is formylmethionyl-tRNA. It is known that N-blocked aminoacyl-tRNA has a special affinity for the P-site (RajBhandary and Chow, 1995). This role of RRF in recognition of the initiation codon is perhaps of general significance in all translational coupling in prokaryotes including ribosomal protein synthesis (Sor et al., 1987) and synthesis of metabolic enzymes (Das and Yanofsky, 1989). It is possible that the downstream box (Resch et al., 1996; Sprengart et al., 1996; O’Connor et al., 1999) of the lysis gene, 5′-AUGggUCugAAAG-3′ (the nucleotides complementary to nucleotides 1469–1483 of 16S rRNA are in upper case) may play a role in this abnormal initiation.
Our data suggest the possibility that without the participation of RRF, 70–80% of the upstream ribosomes leave mRNA from the junction between the coat and the lysis gene. How does this happen? It is possible that in the above hypothesis, there may be a finite possibility that ribosomes may slip 2 nucleotides to place AUG in the A-site without involving RRF. The P-site remains empty or occupied by RF1 or RF2. The P-site becomes empty because RFs will be removed by RF3 and GTP (Freistroffer et al., 1997; Grentzmann et al., 1998). This configuration, an empty P-site and aminoacyl-tRNA at the A-site, never occurs under normal physiological conditions and may lead to disassembly of the complex resulting in the mechanical release of the ribosome from mRNA. The unusual overlapping codon configuration must be responsible for this release because the mechanical release of ribosomes was maximum with UAAUG, and removal of the overlap reduced the mechanical release. Alternatively, one may argue that there is no such thing as the mechanical release and all the ribosomes are released from mRNA by RRF at UAAUG but 25–30% of them are recaptured at AUG due to the special configuration of the junction. However, this interpretation does not explain the data presented in Figure 5B, which show various amounts of the reporter gene translation in the absence of RRF depending on the sequence of the junction.
The release of ribosomes at this junction may be of physiological importance. Each phage consists of 180 coat proteins and the burst size of the GA phage is ∼500 at 37°C (Furuse, 1982). Therefore, each cell would produce ∼90 000 coat proteins. If all the ribosomes from the upstream coat gene engage in the synthesis of the lysis protein, the amount of the lysis protein may be too great and cell lysis may occur prematurely. An RNA phage such as MS2 (Furuse, 1982) has a larger burst size than the GA phage but, as if to compensate for it, has less efficient initiation of lysis protein synthesis (Adhin and van Duin, 1990).
The data presented here support the general concept that the behaviour of ribosomes at the post-termination complex is influenced greatly by many factors including the configuration of mRNA at the post-termination complex. In fact, the distance between the termination and the initiation codon determines the amount of downstream gene reading (Das and Yanofsky, 1989; Ivey-Hoyle and Steege, 1989). When ribosomes have to go 34 nucleotides upstream in order to find the proper AUG (de Smit and van Duin, 1990), 95% of the upstream ribosomes drop off (Adhin and van Duin, 1990). The presence of a strong SD sequence, the local concentration of IF3 and the distance between the termination and initiation codons are important factors in determining the behaviour of ribosomes at the termination codon (Ivey-Hoyle and Steege, 1989; Pavlov et al., 1997; Karimi et al., 1999).
Materials and methods
Bacteria
Escherichia coli JM109 (recA1, endA1, gyrA96, thi, hsdR17, supE44, relA1, Δ(lac-proAB)/F′ traD36, proAB+, lacIq, lacZΔM15) and 594/F′lacIq (Inokuchi and Hirashima, 1987) were used as recipients of the plasmids for transformation. The E.coli strains MC1061/F′lacIq and LJ14/F′lacIq were constructed by introducing plasmid F′ (proAB, lacIq, lacZΔM15, Tn10) into MC1061 and LJ14 (Janosi et al., 1998). These strains are designated simply as MC1061 or LJ14 in the text.
Materials
Restriction endonucleases, DNA ligase and TaKaRa Ex Taq polymerase were purchased from TaKaRa Shuzo.
Plasmids
The plasmids used in this study are listed in Tables I–IV. Plasmid pUC8 is a vector carrying a lac promoter and an α-peptide sequence of lacZ into which a 33 bp multiple cloning site polylinker is inserted. Plasmid pUC18 is similar to pUC8 except that it carries a 54 bp polylinker. Plasmid pKK223-3 is a vector carrying a tac promoter and a 33 bp multiple cloning site polylinker. Plasmid pMC1871 is a vector carrying the lacZ gene lacking its promoter, ribosome-binding site and the 5′-terminal non-essential part of β-galactosidase. These vectors were purchased from Pharmacia.
Plasmid pGA416 (pUC8 carrying the cDNA region corresponding to the coat and replicase genes of phage GA RNA; Inokuchi et al., 1986) was digested with NaeI (which cuts at nucleotide position 1649 of the GA phage cDNA) and SpeI (position 3439). The resulting cDNA fragment carrying the distal part of the coat gene and the replicase β-subunit gene was inserted into plasmid pUC18 DNA cleaved with SmaI and XbaI. The plasmid thus constructed was designated pLWT. For construction of plasmid pL-33GA, pGA416 DNA was digested with NaeI and BglII (position 2453) and the cDNA fragment (corresponding to the region 1469–2453) was inserted into pUC8 DNA cleaved with SmaI and BamHI. We constructed plasmid pL-80AA by cleaving pLWT DNA with EcoRI, filling the recessed 3′ termini by DNA polymerase I (Klenow fragment), and then ligating the DNA end. For construction of pL-8AA and pL-20AA, the PCR-amplified NaeI–MluI fragment (positions 1649 and 1710) was replaced with the corresponding region of pLWT DNA. For construction of pLWTAG and pLWTGA, the MluI–BglII-cut DNA of pL+2C DNA was replaced with the PCR-amplified MluI–BglII DNA fragment having UAGUG (pLWTAG) or UGAUG (pLWTGA) in place of UAAUG.
For construction of plasmids pL-21d and pL-9d, the PCR-amplified DNA fragment containing the GA cDNA region 1694–2459 (pL-21d) or 1706–2459 (pL-9d) was inserted into pKK223-3 at the PstI and HindIII site, respectively. For this purpose, the PstI site at one end of the DNA and the HindIII site at the other end were added to the PCR-amplified fragment.
Plasmid pL-7SD is identical to pL-80AA except that it lacks the region upstream from position 1709 of the GA cDNA sequence. This plasmid was constructed by substituting the KpnI (on pUC18)–BglII-cut DNA fragment of pL-80AA DNA with amplified DNA having an artificial SD sequence (5′-GGAG-3′). Plasmids with nucleotide insertions between the UAA and the AUG codon listed in Table I were constructed by substituting the MluI–BglII-digested DNA fragment of pLWT with DNA fragments that were PCR amplified using appropriate primers containing the UAAUG region with various insertions. For construction of plasmid pL-7SD84Z, pL-7SD DNA was cleaved with NruI (position 1796) and PstI (on pUC18) and the lacZ DNA was inserted. The lacZ DNA fragment was prepared by PCR using pMC1871 DNA as a template and the primers 5′-tttcgcgatcgtcgttttacaacgtcgtg-3′ and 5′-ttctgcagcctgcccggttattattatttttgac-3′.
Plasmid pL-7SD84Z encodes for a chimeric protein in which Val9 of β-galactosidase is joined to Ala27 of the lysis protein through an extra isoleucine residue. Plasmid pL84Z or pLWTAUG(–)86Z was made by replacing the NruI–PstI fragment of pLWT or pLWTAUG(–) DNA with the NruI–PstI fragment of pL-7SD84Z DNA carrying the lacZ gene. In a similar way, plasmids pL+084Z, pl+1C84Z, pL+3C84Z and pL+5A84Z carrying lacZ were constructed from pL+0, pL+1C, pL+3C and pL+5A, respectively. In constructing plasmid pCLZ carrying the coat–lysis–lacZ fusion gene, the MluI–BglII DNA fragment of pLWT was replaced with a PCR-amplified DNA fragment having the mutation at UAAUG, and then the NruI–PstI fragment of the resultant plasmid was replaced with that of pL-7SD84Z DNA. For construction of pLAUG(–)5Z, the nucleotide sequence 5′-UAGGUCUG-3′ next to the UAA of pLWTAUG(–) DNA was changed to 5′-UAUACGUA-3′ to create an SnaBI site (TACGTA). The lacZ DNA fragment with the codon GUA instead of GUC for Val9 of β-galactosidase was joined to the SnaBI site.
Assay of β-galactosidase
The β-galactosidase activity of the fusion protein was measured according to Miller (Miller, 1992) except that an OD660 of cell density was used instead of OD600. Miller units were calculated using the conversion factor, OD660/OD600 = 0.57.
Cell lysis
Escherichia coli JM109 cells harbouring the plasmid carrying the lysis gene were grown at 37°C in LB broth [1.0% tryptone (Difco), 0.5% yeast extract, 1.0% sodium chloride] containing 50 µg/ml ampicillin to a cell density (OD660) of ∼0.15. At that time, the culture was divided into two parts and 2 mM (final concentration) IPTG was added to one part to induce transcription of the lysis gene. After 3 h incubation, the cell density (OD660) was measured. The number of plus signs in the tables reflects the ratio of the cell density, OD660 (IPTG+)/OD660 (IPTG–) after 3 h incubation. When cell lysis was observed and the ratio was <0.3, this was scored as ++, while + indicates that the ratio was between 0.3 and 0.7.
MC1061 cells harbouring pL84Z and LJ14 cells harbouring pL84Z or pLAUG(–)86Z were grown at 27°C. At a cell density of OD660 = 0.13, IPTG (final 2 mM) was added to the culture and the incubation was continued at 39°C for another 3 h. The cells were pelleted and sonicated in a buffer (100 mM Tris–HCl pH 7.4, 10 mM EDTA, 1 mM phenylmethylsulfonyl fluoride). The fusion protein was purified using ProtoSorb lacZ Immunoaffinity Adsorbent (Promega). For N-terminal amino acid sequence analysis of the β-galactosidase fusion protein, the protein was transferred from an SDS–polyacrylamide gel to a PVDF membrane, which was then cut out and analysed using an Applied Biosystems Procise Sequencer.
Acknowledgments
Acknowledgements
The authors thank colleagues of Y.I. for their help in isolating mutants of the lysis gene, and H.Nagasawa (University of Tokyo) for his technical advice on amino acid sequence analysis, G.Hirokawa for critical reading of this manuscript and James Kocsis of Jefferson Medical College for scientific as well as linguistic advice.
References
- Adhin M.R. and van Duin,J. (1990) Scanning model for translational reinitiation in eubacteria. J. Mol. Biol., 213, 811–818. [DOI] [PubMed] [Google Scholar]
- Baughman G. and Nomura,M. (1983) Localization of the target site for translational regulation of the L11 operon and direct evidence for translational coupling in Escherichia coli. Cell, 34, 979–988. [DOI] [PubMed] [Google Scholar]
- Berkhout B. and van Duin,J. (1985) Mechanism of translational coupling between coat protein and replicase genes of RNA bacteriophage MS2. Nucleic Acids Res., 13, 6955–6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B., Schmidt,B.F., van Strien,A., van Boom,J., van Westrenen,J. and van Duin,J. (1987) Lysis gene of bacteriophage MS2 is activated by translation termination at the overlapping coat gene. J. Mol. Biol., 195, 517–524. [DOI] [PubMed] [Google Scholar]
- Das A. and Yanofsky,C. (1989) Restoration of a translational stop–start overlap reinstates translational coupling in a mutant trpB′–rpA gene pair of the Escherichia coli tryptophan operon. Nucleic Acids Res., 17, 9333–9340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Smit M.H. and van Duin,J. (1990) Control of prokaryotic translational initiation by mRNA secondary structure. Prog. Nucleic Acid Res. Mol. Biol., 38, 1–35. [DOI] [PubMed] [Google Scholar]
- Freistroffer D.V., Pavlov,M.Y., MacDougall,J., Buckingham,R.H. and Ehrenberg,M. (1997) Release factor RF3 in E.coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP-dependent manner. EMBO J., 16, 4126–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse K. (1982) Phylogenetic studies on RNA coliphages. PhD thesis, Keio University School of Medicine. [Google Scholar]
- Grentzmann G., Kelly,P.J., Laalami,S., Shuda,M., Firpo,M.A., Cenatiempo,Y. and Kaji,A. (1998) Release factor RF-3 GTPase activity acts in disassembly of the ribosome termination complex. RNA, 4, 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groeneveld H., Oudot,F. and van Duin,J.V. (1996) RNA phage KU1 has an insertion of 18 nucleotides in the start codon of its lysis gene. Virology, 218, 141–147. [DOI] [PubMed] [Google Scholar]
- Gualerzi C., Pon,C.L. and Kaji,A. (1971) Initiation factor dependent release of aminoacyl-tRNAs from complexes of 30S ribosomal subunits, synthetic polynucleotide and aminoacyl tRNA. Biochem. Biophys. Res. Commun., 45, 1312–1319. [DOI] [PubMed] [Google Scholar]
- Hirashima A. and Kaji,A. (1972a) Factor-dependent release of ribosomes from messenger RNA—requirement for two heat-stable factors. J. Mol. Biol., 65, 43–58. [DOI] [PubMed] [Google Scholar]
- Hirashima A. and Kaji,A. (1972b) Purification and properties of ribosome-releasing factor. Biochemistry, 11, 4037–4044. [DOI] [PubMed] [Google Scholar]
- Hirashima A. and Kaji,A. (1973) Role of elongation factor G and a protein factor on the release of ribosomes from messenger ribonucleic acid. J. Biol. Chem., 248, 7580–7587. [PubMed] [Google Scholar]
- Ichikawa S., Ryoji,M., Siegfried,Z. and Kaji,A. (1989) Localization of the ribosome-releasing factor gene in the Escherichia coli chromosome. J. Bacteriol., 171, 3689–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inokuchi Y. and Hirashima,A. (1987) Interference with viral infection by defective RNA replicase. J. Virol., 61, 3946–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inokuchi Y., Takahashi,R., Hirose,T., Inayama,S., Jacobson,A.B. and Hirashima,A. (1986) The complete nucleotide sequence of the group II RNA coliphage GA. J. Biochem., 99, 1169–1180. [DOI] [PubMed] [Google Scholar]
- Ivey-Hoyle M. and Steege,D.A. (1989) Translation of phage-f1 gene-VII occurs from an inherently defective initiation site made functional by coupling. J. Mol. Biol., 208, 233–244. [DOI] [PubMed] [Google Scholar]
- Janosi L., Shimizu,I. and Kaji,A. (1994) Ribosome recycling factor (ribosome releasing factor) is essential for bacterial growth. Proc. Natl Acad. Sci. USA, 91, 4249–4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janosi L., Hara,H., Zhang,S. and Kaji,A. (1996a) Ribosome recycling by ribosome recycling factor (RRF)—an important but overlooked step of protein biosynthesis. Adv. Biophys., 32, 121–201. [DOI] [PubMed] [Google Scholar]
- Janosi L., Ricker,R. and Kaji,A. (1996b) Dual functions of ribosome recycling factor in protein biosynthesis: disassembling the termination complex and preventing translational errors. Biochimie, 78, 959–969. [DOI] [PubMed] [Google Scholar]
- Janosi L. et al. (1998) Evidence for in vivo ribosome recycling, the fourth step in protein biosynthesis. EMBO J., 17, 1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janosi L., Mori,H., Sekine,Y., Abragan,J., Janosi,R., Hirokawa,G. and Kaji,A. (2000) Mutations influencing the frr gene coding for ribosome recycling factor (RRF). J. Mol. Biol., 295, 815–829. [DOI] [PubMed] [Google Scholar]
- Kaji A. and Hirokawa,G. (2000) Disassembly of post termination complex by RRF (ribosome recycling factor), a possible new target for antimicrobial agents. In Garret,R.A., Douthwaite,S.R., Liljas,A., Matheson,A.T., Moore,P.B. and Noller,H.F. (eds), The Ribosome: Structure, Function, Antibiotics and Cellular Interactions. American Society of Microbiology Press, Washington, DC, pp. 527–539. [Google Scholar]
- Kaji A., Teyssier,E. and Hirokawa,G. (1998) Disassembly of the post-termination complex and reduction of translational error by ribosome recycling factor (RRF)—A possible new target for antibacterial agents. Biochem. Biophys. Res. Commun., 250, 1–4. [DOI] [PubMed] [Google Scholar]
- Kanai T., Takeshita,S., Atomi,H., Umemura,K., Ueda,M. and Tanaka,A. (1998) A regulatory factor, Fil1p, involved in derepression of the isocitrate lyase gene in Saccharomyces cerevisiae. A possible mitochondrial protein necessary for protein synthesis in mitochondria. Eur. J. Biochem., 256, 212–220. [DOI] [PubMed] [Google Scholar]
- Karimi R., Pavlov,M.Y., Buckingham,R.H. and Ehrenberg,M. (1999) Novel roles for classical factors at the interface between translation termination and initiation. Mol. Cell, 3, 601–609. [DOI] [PubMed] [Google Scholar]
- Kung H.-F., Treadwell,B.V., Spears,C., Tai,P.-C. and Weissbach,H. (1977) DNA-directed synthesis in vitro of β-galactosidase: requirement for a ribosome release factor. Proc. Natl Acad. Sci. USA, 74, 3217–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R., Weiner,M. and Gallant,J. (1988) Effects of release factor context at UAA codons in Escherichia coli. J. Bacteriol., 170, 4714–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.H. (1992) Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- O’Connor M., Asai,T., Squires,C.L. and Dahlberg,A.E. (1999) Enhancement of translation by the downstream box does not involve base pairing of mRNA with the penultimate stem sequence of 16S rRNA. Proc. Natl Acad. Sci. USA, 96, 8973–8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa K. and Kaji,A. (1975) Requirement for ribosome releasing factor for the release of ribosomes at the termination codon. Eur. J. Biochem., 58, 411–419. [DOI] [PubMed] [Google Scholar]
- Ohnishi M., Janosi,L., Shuda,M., Matsumoto,H., Hayashi,T., Terawaki,Y. and Kaji,A. (1999) Molecular cloning, sequencing, purification and characterization of Pseudomonas aeruginosa ribosome recycling factor, RRF. J. Bacteriol., 181, 1281–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov M.Yu., Freistroffer,D., MacDougall,J., Buckingham,R.H. and Ehrenberg,M. (1997) Fast recycling of Escherichia coli ribosomes requires both ribosome recycling factor (RRF) and release factor RF3. EMBO J., 16, 4134–4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen H.U., Joseph,E., Ullman,A. and Danchin,A. (1978) Formylation of initiator tRNA methionine in prokaryotic protein synthesis: in vivo polarity in lactose operon expression. J. Bacteriol., 135, 453–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RajBhandary U.L. and Chow,M. (1995) Initiator tRNAs and initiation of protein synthesis. In Söll,D. and RajBhandary,U.L. (eds), tRNA: Structure, Biosynthesis, and Function. ASM Press, Washington, DC, pp. 511–528. [Google Scholar]
- Resch A., Tedin,K., Grundling,A. and Blasi,U. (1996) Downstream box–anti-downstream box interactions are dispensable for translation initiation of leaderless mRNAs. EMBO J., 15, 4740–4748. [PMC free article] [PubMed] [Google Scholar]
- Rolland N. et al. (1999) Plant ribosome recycling factor homologue is a chloroplastic protein and is bactericidal in Escherichia coli carrying temperature-sensitive ribosome recycling factor. Proc. Natl Acad. Sci. USA, 96, 5464–5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoji M. (1981) Studies on the roles of ribosome releasing factor of Escherichia coli. PhD thesis, University of Pennsylvania School of Medicine. [Google Scholar]
- Ryoji M., Berland,R. and Kaji,A. (1981a) Reinitiation of translation from the triplet next to the amber termination codon in the absence of ribosome-releasing factor. Proc. Natl Acad. Sci. USA, 78, 5973–5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoji M., Karpen,J.W. and Kaji,A. (1981b) Further characterization of ribosome releasing factor and evidence that it prevents ribosomes from reading through a termination codon. J. Biol. Chem., 256, 5798–5801. [PubMed] [Google Scholar]
- Schmidt B.F., Berkhout,B., Overbeek,G.P., van Strien,A. and van Duin,J. (1987) Determination of the RNA secondary structure that regulates lysis gene expression in bacteriophage MS2. J. Mol. Biol., 195, 505–516. [DOI] [PubMed] [Google Scholar]
- Selmer M., Al-Karadaghi,S., Hirokawa,G., Kaji,A. and Liljas,A. (1999) Crystal structure of Thermotoga maritima ribosome recycling factor: a tRNA mimic. Science, 286, 2349–2352. [DOI] [PubMed] [Google Scholar]
- Sor F., Bolotin-Fukuhara,M. and Nomura,M. (1987) Mutational alterations of translational coupling in the L11 ribosomal protein operon of Escherichia coli. J. Bacteriol., 169, 3495–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprengart M.L., Fuchs,E. and Porter,A.G. (1996) The downstream box: an efficient and independent translation initiation signal in Escherichia coli. EMBO J., 15, 665–674. [PMC free article] [PubMed] [Google Scholar]