Abstract
The Caenorhabditis elegans POU protein UNC-86 specifies the HSN motor neurons, which are required for egg-laying, and six mechanosensory neurons. To investigate how UNC-86 controls neuronal specification, we characterized two unc-86 mutants that do not respond to touch but show wild-type egg-laying behavior. Residues P145 and L195, which are altered by these mutations, are located in the POU-specific domain and abolish the physical interaction of UNC-86 with the LIM homeodomain protein, MEC-3. This results in a failure to maintain mec-3 expression and in loss of expression of the mechanosensory neuron-specific gene, mec-2. unc-86-dependent expression of genes in other neurons is not impaired. We conclude that distinct residues in the POU domain of UNC-86 are involved in modulating UNC-86 activity during its specification of different neurons. A structural model of the UNC-86 POU domain, including base pairs and amino acid residues required for MEC-3 interaction, revealed that P145 and L195 are part of a hydrophobic pocket which is similar to the OCA-B-binding domain of the mammalian POU protein, Oct-1.
Keywords: Caenorhabditis elegans/MEC-3/neural specification/POU domain/UNC-86
Introduction
Cell type-specific activation of effector genes is a prerequisite for the development of all multicellular organisms. This specificity is accomplished by the interactions of particular transcription factors with their binding sites in the promoters of these effector genes. During neurogenesis, a multitude of distinct neurons that express cell type-specific repertoires of effector genes are generated.
The POU domain proteins are a class of transcription factors that generate some of the diverse expression patterns of genes in both neural and non-neural tissues. The nematode protein UNC-86 and the mammalian proteins Pit-1 and Oct-1 are among the best studied POU domain transcription factors. All POU proteins differentially regulate the expression of distinct effector genes in different cell types. Cell type-specific gene activation by Pit-1 and Oct-1 is, at least in part, achieved by their interactions with other proteins. While the largest number of interacting proteins has been described for Pit-1 (Schaufele et al., 1992; Bradford et al., 1997; Palomino et al., 1998; Tremblay et al., 1998; Cohen et al., 1999), the best studied of the POU protein interactions is that of Oct-1 with the B-cell-specific Bob1/OBF-1/OCA-B protein (henceforth referred to as OCA-B) (Luo et al., 1992). The recently solved co-crystal structure of a ternary complex of the Oct-1 POU domain, DNA and the OCA-B peptide indicates that OCA-B establishes contacts with both the Oct-1 POU domain and the DNA (Chasman et al., 1999). OCA-B thus recognizes a combined protein–DNA interface.
The Caenorhabditis elegans POU homeobox gene, unc-86, is exclusively expressed in the nervous system. The anatomical details of all C.elegans neurons are known, as is, in many cases, their function. This allows the in vivo analysis of gene expression and function. After a neuroblast division, the unc-86 promoter responds to asymmetry cues in 27 different lineages to help specify 57 of the 302 neurons in the animal (Baumeister et al., 1996). Since UNC-86 protein is required for both the generation and differentiation of all six mechanosensory neurons, loss-of-function mutations in unc-86 result in a loss of the animal’s ability to respond to mechanical stimulation (Chalfie and Sulston, 1981). In contrast to mechanosensory neurons, the HSN cells are generated in unc-86 loss-of-function mutants, but fail to differentiate fully and do not produce the neurotransmitter serotonin (Desai et al., 1988). Since serotonin is required for stimulating the contractions of the vulval muscles, unc-86 mutant animals fail to lay eggs properly (Sulston and Horvitz, 1981). The consequences of loss of unc-86 therefore vary in different neural cell types, suggesting that UNC-86 activity is modulated differently in these cells.
The LIM homeodomain protein MEC-3 is the only known protein that interacts with UNC-86. UNC-86 binds to at least three regulatory sequences, CS1–CS3, in the mec-3 promoter to establish mec-3 expression in 10 terminally differentiating neurons, possibly by interacting with an ‘establishment’ cofactor (Way et al., 1991; Xue et al., 1992; Wang and Way, 1996). MEC-3 then interacts with UNC-86 on its own promoter to maintain the mec-3 expression. An UNC-86–MEC-3 heterodimer also regulates the expression of downstream genes required for the function of mechanosensory neurons (Duggan et al., 1998). It is not known whether similar interactions occur in the HSN motor neurons.
Here owe report the characterization of two unc-86 alleles that result in defective mechanosensation but do not affect egg-laying. The mechanosensory neurons in these animals did not display wild-type UNC-86 activity, as shown by behavioral assays and their failure to express marker genes. All other neurons we examined revealed wild-type expression of unc-86-dependent genes. We identified single missense mutations in the coding region of these two unc-86 alleles. By in vitro and in vivo studies we found that both UNC-86 variants fail to interact physically with MEC-3. We demonstrate that the loss of interaction with MEC-3 is caused by amino acid substitutions in the POU domain at positions previously implicated in facilitating protein interactions of both Oct-1 and Pit-1. The side chains at these positions, together with adjacent base pairs in the UNC-86–DNA complex, appear to form an extended protein interaction surface. Our data support the notion that distinct POU proteins can interact with various proteins and DNA target sites using a common mechanism. The data also suggest that the unc-86 activity in the 47 other neurons of C.elegans that do not express mec-3 is mediated via a distinct interaction surface.
Results
Touch sensation and egg-laying behavior of the unc-86(u5) and unc-86(u168) animals
The unc-86(u5) and unc-86(u168) alleles were isolated in a genetic screen for animals defective in mechanosensation (Chalfie and Au, 1989). Although unc-86(u5) and unc-86(u168) animals have a fully penetrant mechanosensation defect indistinguishable from that of an unc-86(n846) null mutant (Table I), they display normal egg-laying. The null allele unc-86(n846) results in an egg-laying defect characterized by the retention of substantially more eggs (34 ± 6) in the uterus of the second-day adult animals than in wild type (13 ± 3). In contrast, unc-86(u5) and unc-86(u168) mutant animals retain 15 ± 4 and 13 ± 3 eggs, respectively, and are, in this respect, indistinguishable from wild type. This implies that the mutations in the unc-86(u5) and unc-86(u168) alleles affect unc-86 function in mechanosensory neurons but not in the HSN motor neurons. We therefore studied the unc-86(u5) and unc-86(u168) alleles in more detail to understand the differential control of unc-86 activity in these distinct neurons.
Table I. Characterization of different unc-86 alleles.
| Wild type | n846 | u5 | u168 | |
|---|---|---|---|---|
| Mec phenotype | + | – | – | – |
| Egl phenotype | + | – | + | + |
| UNC-86 antibody staining | + | – | + | + |
| Q-lineage gcy-32 expression | + | reiterated | + | + |
| mec-2 expression | + | – | – | – |
| mec-7 expression | + | – | + | + |
| mec-3 expression | ||||
| establishment | + | – | + | + |
| maintenance | + | – | – | – |
| tph-1 expression | + | ADF/HSN | + | + |
| (NSM, ADF, HSN) |
unc-86(u5) and unc-86(u168) affect gene expression in mechanosensory neurons
We were interested in the cause of the mechanosensation defect in unc-86(u5) and unc-86(u168) animals. unc-86 is required both for the correct execution of the cell lineage that generates mechanosensory neurons and for their correct differentiation, so unc-86(u5) and unc-86(u168) could affect mechanosensation at either level. To distinguish between these two possibilities, we examined the mechanosensory neurons AVM and PVM, which arise and differentiate after the animal hatches.
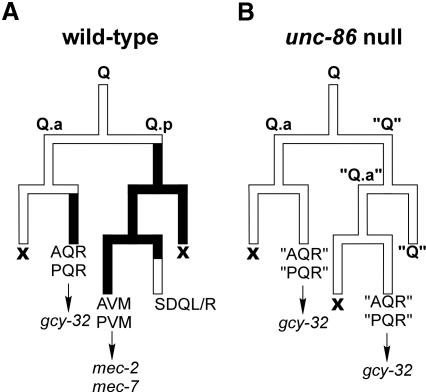
The two Q neuroblasts generate a total of six neurons, including the mechanosensory neurons AVM and PVM (Figure 1A). AVM, PVM and their sister cells SDQL/R are descendants of the posterior daughters (Q.p) of the Q neuroblasts. Their anterior daughters (Q.a) generate the two interneurons AQR and PQR.
Fig. 1. The lineage of the Q-neuroblast can be followed by monitoring the expression of cell type-specific markers [adapted from Finney and Ruvkun (1990)]. (A) Solid black bars represent unc-86 expression. The expression of marker genes is indicated. (B) In unc-86(n846) animals, Q.a is unaffected, but Q.p reiterates the fate of its mother. 'AQR', 'PQR', 'Q' and 'Q.a' designate the supernumerary copies of the respective neurons in unc-86 null mutant animals, which do not express unc-86. X denotes programmed cell death.
We visualized particular cells of the Q-lineage using reporter genes (Figure 1A). To examine the posterior branch of the Q-lineage and the generation of the mechanosensory neurons AVM and PVM, we used mec-2::GFP (green fluorescent protein) and mec-7::GFP reporter gene constructs. mec-2 and mec-7 encode a stomatin-like protein and a β-tubulin, respectively (Savage et al., 1989; Huang et al., 1995). MEC-2 and MEC-7 are essential structural components of mechanosensory neurons. To test the integrity of the anterior branch of the Q-lineage, we used gcy-32::GFP. gcy-32 encodes a guanyl cyclase that is expressed in the AQR/PQR interneurons of the Q-lineage and in the URX interneurons of different origin (Yu et al., 1997).
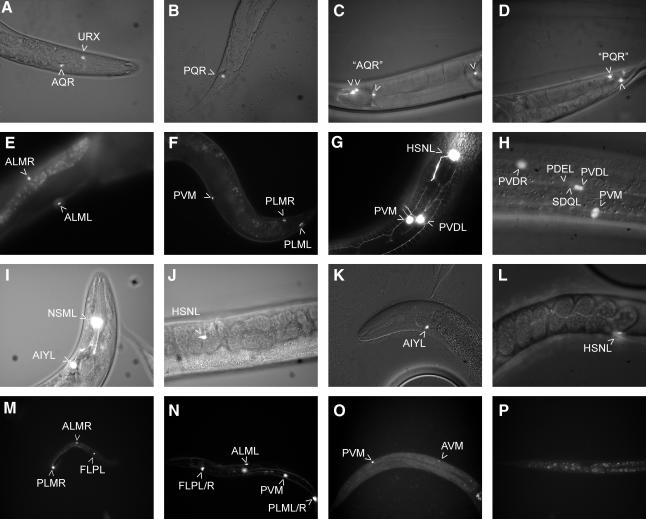
In an unc-86(n846) null mutant animal, the fate of Q.a is not affected (Figure 1B). However, its posterior sister, Q.p, is transformed and reiterates the fate of its mother cell, Q. As a consequence, the Q.p daughters, AVM/PVM and SDQL/R, are not generated and extra copies of AQR and PQR are produced (Chalfie et al., 1981; Sulston and Horvitz, 1981). Accordingly, unc-86(n846) animals do not express the mec-2::GFP and mec-7::GFP reporter genes (Table I) but express gcy-32::GFP in the additional AQR and PQR neurons (Figure 2C and D). In contrast, in both unc-86(u5) and unc-86(u168) mutant animals, the numbers and positions of the AQR/PQR and the AVM/PVM cells were indistinguishable from those in the wild type (Figure 2A, B, G and H). These data were corroborated by Nomarski optics, DAPI staining (data not shown) and indirect immunofluorescence staining with a polyclonal UNC-86 antibody. This antibody stains all 57 neurons previously shown to express unc-86 (Finney and Ruvkun, 1990), in both wild-type and unc-86(u5) and unc-86(u168) mutant animals (Table I). This suggests that, unlike the null allele unc-86(n846), the unc-86(u5) and unc-86(u168) alleles do not cause any lineage defects. Strikingly, however, mec-2::GFP, which in wild-type animals is exclusively expressed in the six mechanosensory neurons, is not expressed in the unc-86(u5) and unc-86(u168) allelic animals (Figure 2E and F). This suggests that the unc-86(u5) and unc-86(u168) alleles affect not the generation but the differentiation of these neurons.
Fig. 2. Morphological characterization of unc-86(u5), unc-86(u168) and unc-86(n846) animals. (A and B) Wild-type animals expressing a gcy-32::GFP construct from the extrachromosomal array adEx1295 (Yu et al., 1997). Expression can be detected in the bilaterally symmetrical URX neurons and the asymmetrical AQR and PQR neurons. (C and D) unc-86(n846) animals have multiple AQR/PQR-like neurons expressing gcy-32::GFP. The cells are located on the right anterior and on the left posterior side of the animal, respectively. Some AQR/PQR-like neurons failed to migrate to their expected positions. (E and F) Wild-type animals carrying an integrated array with mec-2::GFP (uIs9). GFP expression can only be detected in the mechanosensory neurons ALML/R, PLML/R and AVM/PVM. Expression is absent in unc-86(u5) and unc-86(u168) animals (data not shown). (G and H) unc-86(u5) animal carrying the extrachromosomal array byEx104 containing mec-7::GFP. Expression is indistinguishable from that of wild-type animals (data not shown). GFP expression was also detected in the HSN motor neurons, in some head neurons and in the ventral and dorsal cord neurons of L1 larvae (data not shown). Expression of the mec-7::GFP reporter gene is dependent on unc-86 in all neurons except the ventral and dorsal cord neurons (data not shown). Ectopic expression of a mec-7 reporter gene construct has been reported before and is probably due to the titration of a transcriptional repressor (Mitani et al., 1993). (I and J) Wild-type animal carrying the extrachromosomal array byEx105 containing a ttx-3::GFP and a tph-1::GFP reporter construct. ttx-3::GFP is expressed in the bilaterally symmetrical AIY interneurons and was used as an injection marker (Hobert et al., 1997). tph-1::GFP is strongly expressed in the serotonergic neurons NSM and HSN, and weakly in AIM, ADF and RIH (Sze et al., 2000). Expression in wild-type animals was indistinguishable from that in unc-86(u5) and unc-86(u168) animals (data not shown). (K and L) unc-86(n846) animal carrying byEx105. Expression in NSM, AIM and RIH was not detected, even though these neurons are generated in unc-86(n846) animals (Baumeister et al., 1996). The expression of tph-1::GFP in HSN was weakened; HSN neurons often failed to migrate to their proper position and had axonal outgrowth defects, as reported previously (Baum et al., 1999). (M and N) Wild-type animals carrying the extrachromosomal array byEx131 containing mec-3::GFP. mec-3::GFP is expressed in the six mechanosensory neurons ALML/R, PLML/R, AVM/PVM and in the PVDL/R (data not shown) and FLPL/R neurons. (O and P) unc-86(u168) animal carrying byEx131. Expression was detected in the AVM/PVM neurons shortly after they were generated. The expression was only transient. In L2 animals and later stages, there was no mec-3::GFP expression. Expression in unc-86(u5) animals was identical (data not shown).
Maintenance of mec-3 expression is abolished in unc-86(u5) and unc-86(u168) animals
One of the key steps in the differentiation of mechanosensory neurons is expression of the LIM homeobox gene mec-3, which is initiated by UNC-86 and then maintained by a heterodimer of UNC-86 and MEC-3 (see Intro duction). In order to test whether mec-3 is expressed in unc-86(u5) and unc-86(u168) animals, we used a mec-3::GFP reporter gene construct which, in wild-type animals, is expressed in the same 10 neurons that express mec-3 (Figure 2M and N).
In unc-86(u5) and unc-86(u168) animals, expression of mec-3::GFP was detected in AVM and PVM mechanosensory neurons at the L1 larval stage (Figure 2O), suggesting that unc-86-dependent establishment of mec-3 expression was not affected by these mutations. However, GFP expression quickly faded and was no longer detectable in L2 or later stages (Figure 2P). In contrast to AVM and PVM, which are generated in L1, the ALM and PLM mechanosensory neurons are generated during embryogenesis. We occasionally observed embryonic mec-3:: GFP staining that we attributed to the expression in these neurons. However, none of these additional cells showed GFP fluorescence in L1 or later stages. mec-3::GFP expression in the two PVD neurons was very weak in wild-type animals and was not detected in unc-86(u5) or unc-86(u168) animals. Taken together, these findings suggest that the maintenance, but not the establishment, of mec-3 expression is affected in unc-86(u5) and unc-86(u168) animals. This closely resembles the phenotype of a mec-3 null mutant, in which the expression of mec-3 is initially turned on by UNC-86 but cannot be maintained due to the lack of a functional MEC-3 protein (Way and Chalfie, 1989). Since the UNC-86–MEC-3 heterodimer also regulates the expression of additional downstream genes in mechanosensory neurons (Duggan et al., 1998), the lack of mec-2 expression may be a direct consequence of the failure to maintain mec-3 expression.
unc-86(u5) and unc-86(u168) do not affect the expression of other unc-86-dependent genes
Although no target genes of unc-86 in cells other than mechanosensory neurons have so far been identified, we wanted to determine whether the expression of additional marker genes in unc-86-expressing neurons was affected by the unc-86(u5) and unc-86(u168) alleles.
The gcy-32::GFP reporter gene is expressed in the AQR/PQR neurons and in the two URX interneurons (Figure 2A and B). All four neurons express unc-86. Even though the URX neurons are generated in unc-86(n846) animals (Baumeister et al., 1996), they fail to express gcy-32::GFP (Figure 2C and D), suggesting that gcy-32 expression is regulated by unc-86, either directly or indirectly. In contrast, expression of gcy-32::GFP in unc-86(u5) and unc-86(u168) animals was indistinguishable from that in the wild type (Table I). This suggests that unc-86 function in the URX neurons is not affected in unc-86(u5) and unc-86(u168) mutant animals.
unc-86 expression in the HSN motor neurons is a prerequisite for serotonin production in these cells (Desai et al., 1988), so the candidate target genes of unc-86 in HSN may encode key enzymes in serotonin synthesis. Therefore, we tested a reporter gene construct that consisted of a translational fusion of GFP to the gene tph-1, which encodes a tryptophan hydroxylase (Sze et al., 2000). tph-1::GFP was expressed strongly in the serotonergic neurons NSM and HSN (Figure 2I and J) and weakly in AIM, RIH and ADF (data not shown). Expression in NSM, AIM and RIH was completely absent in unc-86(n846) animals (Figure 2K) and only weak in HSN (Figure 2L). We also observed migration and axonal outgrowth defects of HSN similar to those reported previously (Baum et al., 1999). However, tph-1::GFP expression in all serotonergic neurons and the migration and wiring of HSN in unc-86(u5) and unc-86(u168) animals were indistinguishable from those in wild-type animals. This is in accordance with the wild-type egg-laying behavior of unc-86(u5) and unc-86(u168) animals.
We propose that mutations in the unc-86(u5) and unc-86(u168) alleles selectively affect the interaction of UNC-86 with the LIM homeodomain protein MEC-3 in mechanosensory neurons.
unc-86(u5) and unc-86(u168) code for amino acid substitutions in the POU domain that abolish interaction with MEC-3
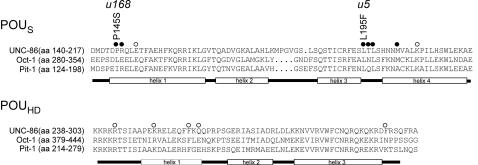
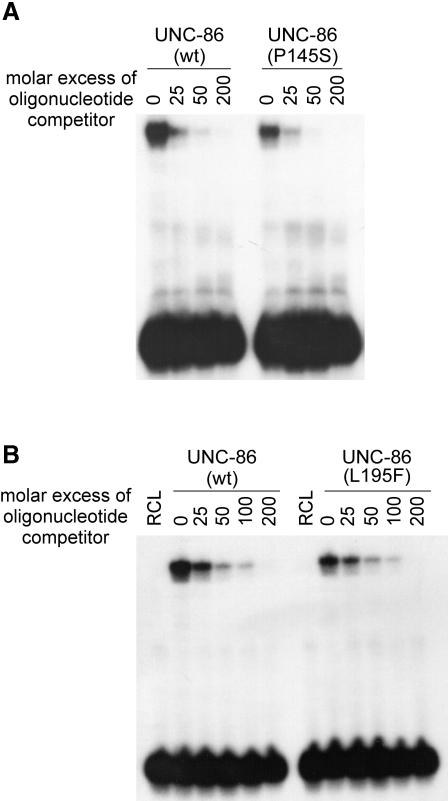
We isolated and sequenced the unc-86 cDNAs of unc-86(u5) and unc-86(u168) animals. Both cDNAs contain missense mutations that result in amino acid substitutions in the POU-specific domain. unc-86(u168) codes for a P145S substitution and unc-86(u5) for an L195F substitution (Figure 3). Since the POU domain is crucial for DNA binding, we were interested in whether these exchanges altered the DNA-binding properties of the mutant protein. Therefore, we tested the two UNC-86 variants in electrophoresis mobility shift assays (EMSAs) with the CS1 DNA element from the mec-3 promoter (see Introduction). Both mutant proteins were able to bind to this DNA in vitro (Figure 4).
Fig. 3. Positions of amino acid exchanges in UNC-86 and sequence comparison with the POU-specific domain (POUS) and POU homeodomain (POUHD) of the murine Oct-1 (SwissProt accession No. P25425) and Pit-1 (SwissProt accession No. S11663). Amino acids are numbered from the N-terminus of the proteins. The helices, as determined by the crystal structures of Oct-1 and Pit-1, are depicted below the amino acid sequence. Filled circles indicate amino acids that impaired ternary complex formation. Open circles indicate amino acids that had no effect on ternary complex formation. T196 was identified as a mediator of ternary complex formation in a suppressor screen with MEC-3 mutants (data not shown). Positions 146, 149, 197, 202, 206, 242, 295, 296 and 297 were described previously (Röckelein et al., 2000).
Fig. 4. P145S and L195F substitutions do not affect the DNA binding of UNC-86. (A and B) Equal amounts of in vitro translated mutant and wild-type UNC-86 protein were incubated at room temperature with radiolabeled double-stranded CS1 DNA and increasing amounts of unlabeled CS1 DNA. The resulting protein–DNA complexes were separated from the unbound probe on a non-denaturing polyacrylamide gel and detected with a phosphoimager. RCL, reticulocyte lysate.
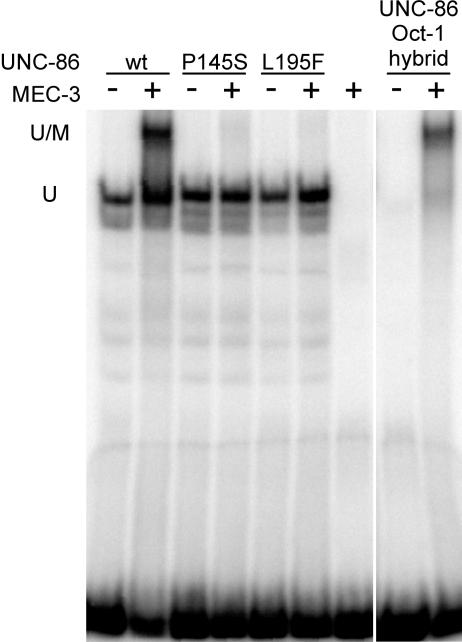
It has been shown previously that the POU domain, in addition to binding DNA, also mediates the interaction with MEC-3 (Xue et al., 1993). Therefore, we tested ternary complex formation between both UNC-86 variants, MEC-3 and the CS1 DNA in vitro and in our previously established in vivo system in Saccharomyces cerevisiae (Röckelein et al., 2000). In the absence of MEC-3, both UNC-86 variants activated the lacZ reporter gene to a level that was indistinguishable from that obtained with wild-type UNC-86 (Table II). The co-expression of wild-type unc-86 and mec-3 resulted in a synergistic 47-fold increase in β-galactosidase activity compared with unc-86 alone. In contrast, expression of the P145S or L195F mutant with mec-3 resulted in an only 8- and 4-fold increase, respectively. In EMSAs, neither mutant could form a complex with MEC-3 on DNA in vitro (Figure 5). Therefore, the amino acid substitutions P145S and L195F severely impair the synergistic interaction of UNC-86 with MEC-3, whereas they affect neither the DNA binding of UNC-86 nor its intrinsic ability to activate transcription.
Table II. Effects of UNC-86 variants P145S (u168) and L195F (u5) and the UNC-86–Oct-1 hybrid on lacZ reporter gene activation by UNC-86 in the yeast strain RB101.
| UNC-86 variant | Relative activity (%)a | Relative activitywith MEC-3 (%)a | Fold increaseb |
|---|---|---|---|
| Wild type | 2.1 ± 0.2 | 100 ± 6 | 47 |
| P145S (u168) | 1.53 ± 0.04 | 11.5 ± 0.6 | 8 |
| L195F (u5) | 1.8 ± 0.1 | 7.4 ± 0.3 | 4 |
| UNC-86–Oct-1 hybridc | 0.01 ± 0.002 | 0.25 ± 0.02 | 25 |
aActivity shown as the mean ± 2 SE relative to the activity of wild-type UNC-86 with MEC-3, which is set at 100%.
bRatio of the mean relative activity with MEC-3 and the mean relative activity of the UNC-86 variant alone.
cThe POU domain of UNC-86 is substituted with the Oct-1 POU domain (see Materials and methods).
Fig. 5. UNC-86 P145S and L195F and an UNC-86–Oct-1 hybrid were assessed for ternary complex formation with MEC-3 on CS1 DNA. Equal amounts of in vitro translated UNC-86 wild-type, mutant and hybrid proteins were incubated at room temperature with or without ∼50 ng of recombinant MEC-3 protein. Retardation of radiolabeled CS1 DNA was monitored after non-denaturing polyacrylamide gel electrophoresis using a phosphoimager. U denotes a binary complex of UNC-86 bound to the CS1 DNA, while U/M denotes a ternary complex of UNC-86, MEC-3 and CS1 DNA.
The POU domain of Oct-1 confers interaction with MEC-3
P145 and L195 in UNC-86 correspond to L285 and L332 in the Oct-1 POU domain. Both positions form part of a hydrophobic pocket involved in interactions of Oct-1 with its B-cell-specific cofactor, OCA-B (Chasman et al., 1999). We wanted to test whether the Oct-1 POU domain could functionally substitute for the UNC-86 POU domain, so we analyzed a hybrid UNC-86 protein containing the Oct-1 POU domain for its ability to form a ternary complex with MEC-3 on CS1 DNA in vitro. The UNC-86–Oct-1 hybrid protein showed a strongly impaired DNA binding, but was able to form a ternary complex with MEC-3 on DNA (Figure 5). This suggests that the POU domains of Oct-1 and UNC-86, in spite of only 50% sequence identity, have sufficient structural similarity to allow association of the Oct-1 POU domain with MEC-3. In addition, protein interaction with MEC-3 is able to stabilize the binding of the hybrid POU protein on DNA.
Next, we wanted to test whether the ternary complex of the UNC-86–Oct-1 hybrid protein and MEC-3 on the CS1 DNA was transcriptionally active, so we analyzed the hybrid protein in the presence and absence of MEC-3 for reporter gene activation in our yeast system. The complex of the hybrid protein and MEC-3 displayed <1% of the activity obtained with UNC-86 and MEC-3 on the CS1 DNA. Furthermore, the hybrid protein alone was not able to activate transcription (Table II). This is consistent with the strongly impaired DNA binding seen in the EMSA. We have shown previously that the POU domain of UNC-86 does not contain an activation domain (Röckelein et al., 2000), so differences in the level of reporter gene activation cannot be explained by a lack of activating properties of the Oct-1 POU domain. These data indicate that the POU domain may have additional roles in forming a transcriptionally active complex besides mediating the DNA and protein interactions. Whether this reflects a lack of recruitment of yeast endogenous co-activators by the Oct-1 POU domain, or whether the POU domain of Oct-1 recruits transcriptional repressor in yeast, will be the subject of further investigations.
Binding site requirements for UNC-86 and MEC-3 interaction
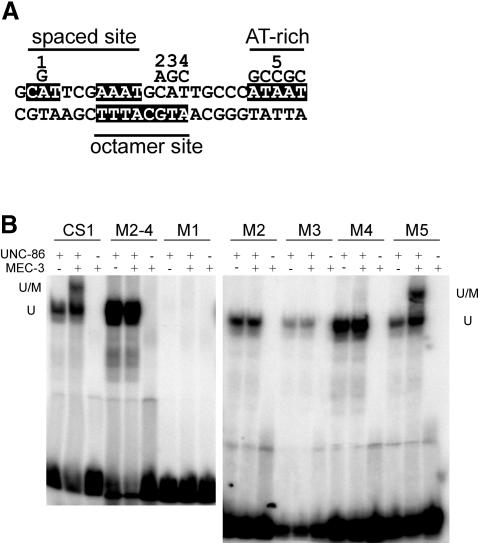
The CS1 mec-3 promoter element contains both a spaced POU consensus binding site (CATnnnA/TAAT) (Wang and Way, 1996) and an overlapping motif on the complementary strand (ATGCATTT) that is similar to a POU octamer site (Verrijzer et al., 1992) (Figure 6A). Mutating the ATG of the octamer consensus sequence severely impaired the interaction of UNC-86 with MEC-3 both in EMSAs in vitro (Figure 6B) and in yeast in vivo (Röckelein et al., 2000), whereas neither the DNA binding nor the transcriptional activation mediated by UNC-86 was affected. Single base substitutions showed that each nucleotide of the ATG is essential for the recruitment of MEC-3 into the ternary complex (Figure 6B). This may suggest that either UNC-86 binds to the spaced site and the ATG is directly contacted by MEC-3, or that UNC-86 can bind both the octamer and the spaced site, but the formation of a ternary complex is only possible on the octamer site. In the latter case, it is conceivable that MEC-3 may bind to an AT-rich sequence adjacent to the octamer site (Figure 6A). To distinguish between these possibilities, we introduced nucleotide exchanges at various positions and tested UNC-86 binding and ternary complex formation with MEC-3 (Figure 6B). Neither UNC-86 nor UNC-86–MEC-3 bound to a CS1 element in which the CAT of the spaced consensus site was mutated (Figure 6B). In contrast, mutating the AT-rich motif adjacent to the octamer site affected neither UNC-86 binding nor ternary complex formation with MEC-3 (Figure 6B), suggesting that the AT-rich motif is not contacted by either protein. Therefore, we conclude that the UNC-86 POU domain probably interacts only with the CATtcgAAAT sequence in the CS1 element, whereas the UNC-86–MEC-3 heterodimer also contacts the immediately adjacent ATG.
Fig. 6. Binding site requirements of UNC-86 on the CS1 mec-3 promoter element. (A) Substitutions as indicated. Each of the depicted base substitutions was assessed for UNC-86 binding and ternary complex formation with MEC-3 in EMSA. (B) Experimental set-up and labeling as in Figure 5.
The POU-specific domain and the CS1 DNA comprise a protein–DNA interface for the interaction with MEC-3
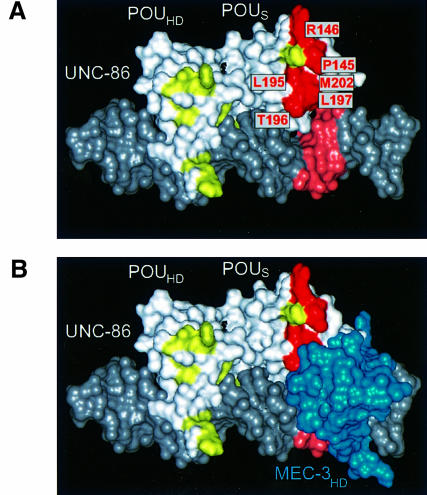
Computer models based on genetic data have been used successfully to predict protein–DNA interactions in the absence of other structural data (Baumeister et al., 1992). Therefore, we examined the location of P145 and L195F, and additional residues in UNC-86 that have been implicated in MEC-3 interactions (Röckelein et al., 2000), in a structural model of the UNC-86 POU domain.
We modeled the POU domain as binding to the CATtcgAAAT sequence in the CS1 DNA based on the backbone coordinates of the Pit-1 POU domain (Jacobson et al., 1997). We were intrigued by the fact that, in this model, all residues that had been implicated in MEC-3 contacts are clustered near the ATG that is also essential for MEC-3 binding (Figure 7A). We suggest that these residues, together with the ATG nucleotides, form part of an extended binding surface for the MEC-3 protein. Next, we added the MEC-3 homeodomain to the model (Figure 7B). In this extended model, the MEC-3 homeodomain can bind to the ATG and is located near to the residues that affect MEC-3 binding.
Fig. 7. Model of the ternary complex formation of UNC-86 and MEC-3 on DNA (InsightII; Molecular Simulations). (A) van der Waals representation of the UNC-86 POU-specific domain (POUS) and POU homeodomain (POUHD) binding to the CS1 DNA of the mec-3 promoter, generated based on the backbone coordinates of the Pit-1 POU domain crystal structure (PDB accession No. 1AU7). Amino acids that are essential for ternary complex formation are shown in red and labeled with the residue and its position in UNC-86. Amino acids that did not impair the interaction after mutation are depicted in yellow. The ATG nucleotides of the CS1 DNA required for binding MEC-3, but not UNC-86, are shown in light red. (B) van der Waals representation of the MEC-3 homeodomain (MEC-3HD), shown in blue, modeled based on the backbone coordinates of the POU homeodomain of Pit-1 (PDB accession No. 1AU7).
Discussion
In this paper, we report the molecular analysis of the first mutations in UNC-86 that selectively impair one of the supposedly many functions of this POU protein in the C.elegans nervous system. The P145 and L195 residues that are affected in the unc-86(u168) and unc-86(u5) alleles, respectively, correspond to L285 and L332 in the Oct-1 POU domain, which form part of a hydrophobic pocket involved in interactions of Oct-1 with the B-cell-specific protein OCA-B (Chasman et al., 1999). In previous experiments (Röckelein et al., 2000), we had already identified other residues in UNC-86 that selectively affected the interaction with the LIM homeodomain protein MEC-3, and several of these also correspond to residues which, in the Oct-1 POU-specific domain, comprise part of the OCA-B-binding surface (Gstaiger et al., 1996; Babb et al., 1997; Sauter and Matthias, 1998).
These two functionally unrelated POU proteins obviously employ similar mechanisms to ensure the specificity of their protein interactions even though they recognize structurally dissimilar proteins. The Oct-1- and UNC-86-interacting proteins, OCA-B and MEC-3, are not homologous and there is no other homologue to OCA-B in the worm genome. OCA-B and MEC-3 both recognize a DNA–protein interface consisting of site-specific DNA nucleotides and selected amino acids in the POU-specific domain of Oct-1 and UNC-86, respectively. Our data suggest that the contact surface on the POU-specific domain is sufficiently similar between Oct-1 and UNC-86 to allow the physical interaction of MEC-3 with the POU domain of Oct-1. However, there are considerable differences in the way in which the POU proteins, and the proteins with which they interact, recognize DNA, suggesting that the base pair composition of the DNA-binding site may play an important role in discriminating between potential co-activators. Oct-1 binds to the octamer motif ATGCAAAT (Verrijzer et al., 1992), which differs from the UNC-86 spaced binding site CATtcgAAAT in the mec-3 promoter. Consequently, swapping the POU domains prevented the binding of the UNC-86–Oct-1 hybrid protein to the UNC-86 binding site. Based on our experimental and modeling data, we suggest that MEC-3 interacts with three bases adjacent to, but separate from, the UNC-86 binding site, whereas the OCA-B-binding site is embedded in the octamer-like motif (Cepek et al., 1996). This difference correlates with the size of the DNA-binding motifs of MEC-3 and OCA-B. Owing to its small size, the uncommon OCA-B DNA-binding peptide fold can be accommodated between the bulky POU-specific domain and homeodomain of Oct-1 (Chasman et al., 1999). The helix–turn–helix motif in the homeodomain of MEC-3 is a significantly larger DNA-binding module, which might require a binding site that does not overlap with that of the POU domain.
The hydrophobic pocket in the POU-specific domain of Oct-1 and UNC-86 that is contacted by both OCA-B and MEC-3, respectively, also participates in the homodimerization of the Pit-1 POU protein (Jacobson et al., 1997). Therefore, it may represent a conserved domain in functionally unrelated POU proteins for homodimerization and for interaction with such diverse proteins as OCA-B and MEC-3 in such distantly related organisms as nematodes and mammals.
Mutations in the hydrophobic pocket of UNC-86 prevent heterodimerization with MEC-3, and thus result in the loss of expression of MEC-3-dependent downstream genes in mechanosensory neurons. Most strikingly, however, the mutations do not affect all aspects of UNC-86 function. The 27 cell lineages that depend on unc-86 activity are not altered in the unc-86(u5) and unc-86(u168) alleles and generate all 57 neurons as in wild type. unc-86(u5) and unc-86(u168) did not impair the function of unc-86 in the HSN motor neurons or the expression of the unc-86 downstream genes gcy-32 and tph-1 in URX and serotonergic neurons, respectively. There are several possible explanations for this cell type-specific effect. First, proteins that potentially interact with UNC-86 in other neurons may respond less sensitively to changes in the hydrophobic pocket. We cannot formally exclude this, since no other proteins have yet been identified that interact with UNC-86. Secondly, UNC-86 may not require additional protein interactions for its activity in neurons other than mechanosensory neurons. The regulation of UNC-86 activity could, instead, be accomplished by post-translational modifications, such as phosphorylation. Indeed, UNC-86 can be phosphorylated in vitro (our unpublished data) and phosphorylation through a variety of serine/threonine kinases has been shown to be important for regulating the activity of Pit-1 and Oct-1 (Kapiloff et al., 1991; Segil et al., 1991; Caelles et al., 1995; Inamoto et al., 1997). However, the phosphorylation of UNC-86 alone is unlikely to generate a level of combinatorial complexity sufficient for discriminating the various UNC-86 functions in 47 additional neurons. Thirdly, UNC-86 could use other protein interaction interfaces for its activity in other neurons. There is evidence for a protein interaction in Oct-1 that is accomplished via an alternative interface. The herpes simplex virus VP16 protein (Pomerantz et al., 1992) contacts amino acids in the first helix of the homeodomain. The interfaces used by VP16 and OCA-B are mutually exclusive, since Oct-1 can associate with both proteins at the same time (Babb et al., 1997). Since we have not yet identified mutations in the homeodomain of UNC-86 that selectively impair the physical interaction with MEC-3 (Röckelein et al., 2000), the homeodomain could be used for contacting additional proteins in other neurons. Furthermore, alternative interfaces could arise through the phosphorylation of particular residues in an existing interface. Interestingly, we identified T196 as a key residue for ternary complex formation, which can both influence protein and DNA interactions of UNC-86 (our unpublished data). The putative phosphorylation of T196 could have a pronounced influence on the DNA-binding and interaction properties of UNC-86. Future experiments will address this aspect.
It has been suggested that the bases adjacent to the UNC-86 binding site in the CS1 DNA are bound by an unknown cofactor, which, together with UNC-86, is responsible for the establishment of mec-3 expression in mechanosensory neurons (Wang and Way, 1996). While this factor has remained elusive, our data show that this site can be occupied by MEC-3 itself to generate an activating ternary complex. The establishment of mec-3 expression was not affected in unc-86(u5) and unc-86(u168) animals. This suggests that the putative ‘establishment cofactor’ of UNC-86 does not depend on the integrity of the MEC-3 contact interface. However, it may also be possible that the establishment of mec-3 expression is achieved by UNC-86 alone and is prevented in other neurons by the action of repressors, such as egl-44, egl-46 or sem-4 (Mitani et al., 1993).
In summary, we have identified two mutations in the POU-specific domain of UNC-86 that abolish the physical interaction of UNC-86 with MEC-3. The P145S and L195F mutants we described here represent the first UNC-86 mutants that allow the correlation of the defective interaction with MEC-3 in vitro and the resulting loss of co-activation by MEC-3 in yeast with a selective defect in the mechanosensory neurons of C.elegans. These mutations affect a hydrophobic pocket in the POU-specific domain that is conserved relative to the mammalian Oct-1 protein and that is apparently selectively required for the interaction with MEC-3. It will be interesting to identify other proteins that interact with UNC-86 and use distinct interaction surfaces.
Materials and methods
Plasmid constructions
The construction of pBY175 and pBY868 containing wild-type unc-86 and mec-3, respectively, in a yeast expression vector is described elsewhere (Röckelein et al., 2000). All unc-86 and mec-3 mutants were constructed by site-directed PCR mutagenesis (Ho et al., 1989). Mutations were confirmed by single-strand sequencing. Primers RB699 (5′-ACTGAAGCTTCTCGCGAATTGCGGCCG-3′) and RB700 (5′-GATCGGATCCGGAGCTGAAAGTACAG-3′) were used to amplify a 1.7 kb genomic fragment from the 5′ region up to the third exon of zk1290.2 (tph-1). The fragment was subcloned into the pEGFP-N3 vector (Clontech), yielding plasmid pBY668. pBY848 (mec-7::GFP) was cloned by inserting a PstI–KpnI fragment from plasmid pPD52.102 into pPD95.85 (both from A.Fire). The UNC-86–Oct-1 hybrid was made by amplifying the POU domain of the murine Oct-1 cDNA by PCR and subcloning into the vector pBY531 via BamHI and MluI sites, which were introduced into the unc-86 cDNA by silent mutagenesis. This results in an exchange of amino acids 143–280 of UNC-86 (Baumeister et al., 1996) with amino acids 283–421 of Oct-1 (Sturm et al., 1988). Thus, the POU homeodomain of the hybrid contains only the first 43 amino acids Oct-1. Plasmid pBY1107, encoding a LIM-less mec-3 without the acidic domain fused to His6 tags and thioredoxin, is described elsewhere (Röckelein et al., 2000).
Growth of worms, construction of transgenic animals and indirect immunofluorescence staining
All C.elegans strains were cultured as described (Wood, 1988). Transgenic animals were constructed as described (Mello et al., 1991). All plasmids were injected at 20 ng/µl, except pRF4, which was injected at 120 ng/µl. For tph-1 expression analysis, pBY668 and pBY218, which carries a ttx-3::GFP reporter gene specifically expressed in AIY neurons (Hobert et al., 1997), were injected into the Bristol N2 strain, generating strain BR1163 byEx105 (pBY668; pBY218). For mec-7 expression analysis, pBY848 was injected into the Bristol N2 strain, generating strain BR1315 byEx104 (pBY848). For mec-3 expression analysis, pPD118.17 containing a mec-3::GFP reporter gene and pRF4 were injected into Bristol N2, generating strain BR1841 byEx131 (pPD118.17; pRF4). For gcy-32::GFP expression analysis, the extrachromosomal array adEx1295 [lin-15(+); gcy-32::GFP] from strain DA1295 (Yu et al., 1997) was used. Extrachromosomal arrays were transferred into an unc-86 genetic background by crossing and picking animals, which were either Mec and Rol or Mec and showing GFP fluorescence. For mec-2 expression analysis, unc-86(u5), unc-86(u168) and unc-86(n846) males were crossed with BR754 uIs9V (mec-2::GFP; pRF4) (M.Chalfie, personal communication). Homozygous unc-86III; uIs9V animals were obtained by picking Mec and Rol animals that had 100% Rol progeny. If an unc-86 mutant strain failed to show expression of a reporter gene, the integrity of the array was verified by backcrossing with Bristol N2 to reconstitute GFP expression. Indirect immunofluorescence staining using anti-UNC-86 polyclonal serum was carried out as described (Finney and Ruvkun, 1990).
Behavioral assays of unc-86 mutant animals
Animals were tested for mechanosensation as described (Chalfie and Sulston, 1981). Egg-laying behavior was determined on the second day of adulthood in synchronized animals as follows. Animals were washed off a non-starved plate with M9 buffer (Wood, 1988), treated with an alkaline hypochlorite solution (0.5% NaOCl, 0.25 M KOH) for 5 min and washed five times with M9 buffer. Worms were allowed to hatch in M9 buffer overnight without food and then grown on NGM plates with food for 78 h at 20°C. The number of eggs in utero was then determined for 50 animals per strain.
Identification of mutations in the unc-86(u5) and unc-86(u168) alleles
Non-starved mixed-stage animals were washed off four 9 cm NGM plates (Wood, 1988) with M9 buffer and washed several times with M9 to remove bacteria. Six hundred microliters of RLT buffer (Qiagen) were added and animals were vortexed in the presence of glass beads (250–500 µm). Total RNA was isolated from the supernatant using an RNeasy Kit (Qiagen) following the manufacturer’s instructions. unc-86 cDNAs were isolated by reverse transcription of unc-86 mRNAs using MMLV reverse transcriptase (MBI-Fermentas) and an unc-86-specific primer, followed by PCR using Pfu DNA polymerase (Stratagene). cDNAs were subcloned into the pBluescript II SK– vector and sequenced on both strands by TopLab.
Purification of recombinant proteins
Escherichia coli carrying pBY1107, which encodes MEC-3 lacking the LIM and the acidic domains fused at the N-terminus to a His6 tag and thioredoxin, were induced with 1 mM isopropyl-β-d-thiogalactopyrano side (IPTG) at 37°C for 4 h. Recombinant MEC-3 was purified from the soluble fraction using Ni2+-NTA–agarose (Qiagen) following the manufacturer’s instructions. The resulting protein was >99% pure as judged by Coomassie staining after SDS–PAGE. The protein was dialyzed against EMSA buffer (Röckelein et al., 2000) before use. Escherichia coli producing His6::UNC-86 expressed from a pET-28 plasmid were harvested after 4 h of induction with 1 mM IPTG at 37°C. Recombinant His6::UNC-86 protein was purified from inclusion bodies using Ni2+-NTA–agarose according to the manufacturer’s instructions.
Production of antiserum
One rabbit was injected with >200 µg of recombinant UNC-86 mixed with phosphate-buffered saline (PBS) and RAS adjuvant (Ribi Immunochem Research) and injected again after 3, 6 and 9 weeks. Sera were tested in western blots of whole cell lysates of GST::UNC-86-producing E.coli. The antiserum used for indirect immunofluorescence studies was pre-adsorbed on fixed unc-86(n846) mutant animals. The antiserum stained all 57 unc-86-expressing neurons in wild-type animals and no additional cells (Finney and Ruvkun, 1990).
Yeast media and methods
Construction of the S.cerevisiae strain RB101 is described elsewhere (Röckelein et al., 2000). RB101 was grown at 30°C in liquid or solid SC medium (Sherman et al., 1986). Plasmids were transformed into RB101 as described (Gietz and Woods, 1994) and transformants were allowed to grow on SC medium lacking the amino acids required for plasmid selection. Liquid β-galactosidase assays using chlorophenol red–β-d- galactopyranoside (CPRG; Roche) were performed as described (Clontech, 1997) at 20°C.
EMSAs
EMSAs were performed as described (Röckelein et al., 2000) for 30 min at room temperature with both wild-type CS1 (5′-GCATTCGAAATGCATTGCCCATAATG-3′), M1 (5′-GCGTTCGAAATGAGCTGCCCATAATG-3′), M2 (5′-GCATTCGAAATGAATTGCCCATAATG-3′), M3 (5′-GCATTCGAAATGCGTTGCCCATAATG-3′), M4 (5′-GCATTCGAAATGCACTGCCCATAATG-3′) and M5 (5′-GCATTCGAAATGCATTGCCCGCCGCG-3′) double-stranded oligonucleotides (mutated bases underlined). UNC-86 and the UNC-86–Oct-1 hybrid used in the EMSA were in vitro translated using pCITE (Novagen), the TNT T7 expression system (Promega) and [35S]methionine (Amersham) according to the manufacturers' instructions. Approximately 50 ng of recombinant, dialyzed MEC-3 fused to thioredoxin were used in the EMSA.
Acknowledgments
Acknowledgements
We thank Marty Chalfie for the gift of the uIS9, unc-86(u5) and unc-86(u168) strains and Andy Fire for plasmids. We thank the members of the Baumeister laboratory for their help and support and R.Grosschedl for comments on the manuscript. Some strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH Center for Research Resources. This work was financed through a grant from the Deutsche Forschungsgemeinschaft to R.B.
Note added in proof
We thank J.Sze and G.Ruvkun for communicating that a tph-1::GFP fusion is only expressed in the ADF neurons of an unc-86 null mutant.
References
- Babb R., Cleary,M.A. and Herr,W. (1997) OCA-B is a functional analog of VP16 but targets a separate surface of the Oct-1 POU domain. Mol. Cell. Biol., 17, 7295–7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum P.D., Guenther,C., Frank,C.A., Pham,B.V. and Garriga,G. (1999) The Caenorhabditis elegans gene ham-2 links Hox patterning to migration of the HSN motor neuron. Genes Dev., 13, 472–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister R., Muller,G., Hecht,B. and Hillen,W. (1992) Functional roles of amino acid residues involved in forming the α-helix–turn–α-helix operator DNA binding motif of Tet repressor from Tn10. Proteins, 14, 168–177. [DOI] [PubMed] [Google Scholar]
- Baumeister R., Liu,Y. and Ruvkun,G. (1996) Lineage-specific regulators couple cell lineage asymmetry to the transcription of the Caenorhabditis elegans POU gene unc-86 during neurogenesis. Genes Dev., 10, 1395–1410. [DOI] [PubMed] [Google Scholar]
- Bradford A.P., Wasylyk,C., Wasylyk,B. and Gutierrez-Hartmann,A. (1997) Interaction of Ets-1 and the POU-homeodomain protein GHF-1/Pit-1 reconstitutes pituitary-specific gene expression. Mol. Cell. Biol., 17, 1065–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caelles C., Hennemann,H. and Karin,M. (1995) M-phase-specific phosphorylation of the POU transcription factor GHF-1 by a cell cycle-regulated protein kinase inhibits DNA binding. Mol. Cell. Biol., 15, 6694–6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepek K.L., Chasman,D.I. and Sharp,P.A. (1996) Sequence-specific DNA binding of the B-cell-specific coactivator OCA-B. Genes Dev., 10, 2079–2088. [DOI] [PubMed] [Google Scholar]
- Chalfie M. and Au,M. (1989) Genetic control of differentiation of the Caenorhabditis elegans touch receptor neurons. Science, 243, 1027–1033. [DOI] [PubMed] [Google Scholar]
- Chalfie M. and Sulston,J. (1981) Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev. Biol., 82, 358–370. [DOI] [PubMed] [Google Scholar]
- Chalfie M., Horvitz,H.R. and Sulston,J.E. (1981) Mutations that lead to reiterations in the cell lineages of C. elegans. Cell, 24, 59–69. [DOI] [PubMed] [Google Scholar]
- Chasman D., Cepek,K., Sharp,P.A. and Pabo,C.O. (1999) Crystal structure of an OCA-B peptide bound to an Oct-1 POU domain/octamer DNA complex: specific recognition of a protein–DNA interface. Genes Dev., 13, 2650–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clontech (1997) Clontech Manual: MATCHMAKER One-Hybrid System Protocol. Clontech Laboratories, Palo Alto, CA. [Google Scholar]
- Cohen L.E., Hashimoto,Y., Zanger,K., Wondisford,F. and Radovick,S. (1999) CREB-independent regulation by CBP is a novel mechanism of human growth hormone gene expression. J. Clin. Invest., 104, 1123–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai C., Garriga,G., McIntire,S.L. and Horvitz,H.R. (1988) A genetic pathway for the development of the Caenorhabditis elegans HSN motor neurons. Nature, 336, 638–646. [DOI] [PubMed] [Google Scholar]
- Duggan A., Ma,C. and Chalfie,M. (1998) Regulation of touch receptor differentiation by the Caenorhabditis elegans mec-3 and unc-86 genes. Development, 125, 4107–4119. [DOI] [PubMed] [Google Scholar]
- Finney M. and Ruvkun,G. (1990) The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell, 63, 895–905. [DOI] [PubMed] [Google Scholar]
- Gietz R.D. and Woods,R.A. (1994) High efficiency transformation with lithium acetate. In Guthrie,C. and Fink,G.R. (eds), Guide to Yeast Genetics and Molecular Biology. Oxford University Press, Oxford, UK, pp. 121–134. [Google Scholar]
- Gstaiger M., Georgiev,O. and Schaffner,W. (1996) Fine mapping of protein interaction surfaces with a PCR-based mutagenesis screen in yeast. Trends Genet., 12, 393–394. [DOI] [PubMed] [Google Scholar]
- Ho S.N., Hunt,H.D., Horton,R.M., Pullen,J.K. and Pease,L.R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene, 77, 51–59. [DOI] [PubMed] [Google Scholar]
- Hobert O., Mori,I., Yamashita,Y., Honda,H., Ohshima,Y., Liu,Y. and Ruvkun,G. (1997) Regulation of interneuron function in the C. elegans thermoregulatory pathway by the ttx-3 LIM homeobox gene. Neuron, 19, 345–357. [DOI] [PubMed] [Google Scholar]
- Huang M., Gu,G., Ferguson,E.L. and Chalfie,M. (1995) A stomatin-like protein necessary for mechanosensation in C. elegans. Nature, 378, 292–295. [DOI] [PubMed] [Google Scholar]
- Inamoto S., Segril,N., Pan,Z.-Q., Kimura,M. and Roeder,R.G. (1997) The cyclin-dependent kinase-activating kinase (CAK) assembly factor, MAT1, targets and enhances CAK activity on the POU domains of octamer transcription factors. J. Biol. Chem., 272, 29852–29858. [DOI] [PubMed] [Google Scholar]
- Jacobson E.M., Li,P., Leon-del-Rio,A., Rosenfeld,M.G. and Aggarwal,A.K. (1997) Structure of Pit-1 POU domain bound to DNA as a dimer: unexpected arrangement and flexibility. Genes Dev., 11, 198–212. [DOI] [PubMed] [Google Scholar]
- Kapiloff M.S., Farkash,Y., Wegner,M. and Rosenfeld,M.G. (1991) Variable effects of phosphorylation of Pit-1 dictated by the DNA response elements. Science, 253, 786–789. [DOI] [PubMed] [Google Scholar]
- Luo Y., Fujii,H., Gerster,T. and Roeder,R.G. (1992) A novel B cell-derived coactivator potentiates the activation of immunoglobulin promoters by octamer-binding transcription factors. Cell, 71, 231–241. [DOI] [PubMed] [Google Scholar]
- Mello C.C., Kramer,J.M., Stinchcomb,D. and Ambros,V. (1991) Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J., 10, 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitani S., Du,H., Hall,D.H., Driscoll,M. and Chalfie,M. (1993) Combinatorial control of touch receptor neuron expression in Caenorhabditis elegans. Development, 119, 773–783. [DOI] [PubMed] [Google Scholar]
- Palomino T., Sanchez-Pacheco,A., Pena,P. and Aranda,A. (1998) A direct protein–protein interaction is involved in the cooperation between thyroid hormone and retinoic acid receptors and the transcription factor GHF-1. FASEB J., 12, 1201–1209. [DOI] [PubMed] [Google Scholar]
- Pomerantz J.L., Kristie,T.M. and Sharp,P.A. (1992) Recognition of the surface of a homeodomain protein. Genes Dev., 6, 2047–2057. [DOI] [PubMed] [Google Scholar]
- Röckelein I., Röhrig,S., Donhauser,R., Eimer,S. and Baumeister,R. (2000) Identification of amino acid residues in the Caenorhabditis elegans POU protein UNC-86 that mediate UNC-86/MEC-3/DNA ternary complex formation. Mol. Cell. Biol., 20, 4806–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter P. and Matthias,P. (1998) Coactivator OBF-1 makes selective contacts with both the POU-specific domain and the POU homeodomain and acts as a molecular clamp on DNA. Mol. Cell. Biol., 18, 7397–7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage C., Hamelin,M., Culotti,J.G., Coulson,A., Albertson,D.G. and Chalfie,M. (1989) mec-7 is a β-tubulin gene required for the production of 15-protofilament microtubules in Caenorhabditis elegans. Genes Dev., 3, 870–881. [DOI] [PubMed] [Google Scholar]
- Schaufele F., West,B.L. and Baxter,J.D. (1992) Synergistic activation of the rat growth hormone promoter by Pit-1 and the thyroid hormone receptor. Mol. Endocrinol., 6, 656–665. [DOI] [PubMed] [Google Scholar]
- Segil N., Roberts,S.B. and Heintz,N. (1991) Mitotic phosphorylation of the Oct-1 homeodomain and regulation of Oct-1 DNA binding activity. Science, 254, 1814–1816. [DOI] [PubMed] [Google Scholar]
- Sherman S., Fink,G.K. and Hicks,J.B. (1986) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sturm R.A., Das,G. and Herr,W. (1988) The ubiquitous octamer binding protein Oct-1 contains a POU domain with a homeobox subdomain. Genes Dev., 2, 1582–1599. [DOI] [PubMed] [Google Scholar]
- Sulston J.E. and Horvitz,H.R. (1981) Abnormal cell lineages in mutants of the nematode Caenorhabditis elegans. Dev. Biol., 82, 41–55. [DOI] [PubMed] [Google Scholar]
- Sze J.Y., Victor,M., Loer,C., Shi,Y. and Ruvkun,G. (2000) Food and metabolic signalling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature, 403, 560–564. [DOI] [PubMed] [Google Scholar]
- Tremblay J.J., Lanctot,C. and Drouin,J. (1998) The pan-pituitary activator of transcription, Ptx1 (pituitary homeobox 1), acts in synergy with SF-1 and Pit1 and is an upstream regulator of the LIM-homeodomain gene Lim3/Lhx3. Mol. Endocrinol., 12, 428–441. [DOI] [PubMed] [Google Scholar]
- Verrijzer C.P., Alkema,M.A., van Weperen,W.W., Van Leeuwen,H.C., Strating,M.J.J. and der Vliet,P. (1992) The DNA binding specifity of the bipartite POU domain and its subdomains. EMBO J., 11, 4993–5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. and Way,J.C. (1996) Promoter sequences for the establishment of mec-3 expression in the nematode Caenorhabditis elegans. Mech. Dev., 56, 183–196. [DOI] [PubMed] [Google Scholar]
- Way J.C. and Chalfie,M. (1989) The mec-3 gene of Caenorhabditis elegans requires its own product for maintained expression and is expressed in three neuronal cell types. Genes Dev., 3, 1823–1833. [DOI] [PubMed] [Google Scholar]
- Way J.C., Wang,L., Run,J.Q. and Wang,A. (1991) The mec-3 gene contains cis-acting elements mediating positive and negative regulation in cells produced by asymmetric cell division in Caenorhabditis elegans. Genes Dev., 5, 2199–2211. [DOI] [PubMed] [Google Scholar]
- Wood W.B. (1988) The Nematode C. elegans. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Xue D., Finney,M., Ruvkun,G. and Chalfie,M. (1992) Regulation of the mec-3 gene by the C. elegans homeoproteins UNC-86 and MEC-3. EMBO J., 11, 4969–4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue D., Tu,Y. and Chalfie,M. (1993) Cooperative interactions between the Caenorhabditis elegans homeoproteins UNC-86 and MEC-3. Science, 261, 1324–1328. [DOI] [PubMed] [Google Scholar]
- Yu S., Avery,L., Baude,E. and Garbers,D.L. (1997) Guanylyl cyclase expression in specific sensory neurons: a new family of chemosensory receptors. Proc. Natl Acad. Sci. USA, 94, 3384–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]