Abstract
In the yeast Saccharomyces cerevisiae, the MADS-box protein Mcm1, which is highly related to mammalian SRF (serum response factor), forms a ternary complex with SFF (Swi five factor) to regulate the cell cycle expression of genes such as SWI5, CLB2 and ACE2. Here we show that the forkhead protein Fkh2 is a component of SFF and is essential for ternary complex formation on the SWI5 and ACE2 promoters. Fkh2 is essential for the correct cell cycle periodicity of SWI5 and CLB2 gene expression and is phosphorylated with a timing that is consistent with a role in this expression. Furthermore, investigation of the relationship between Fkh2 and a related forkhead protein Fkh1 demonstrates that these proteins act in overlapping pathways to regulate cell morphology and cell separation. This is the first example of a eukaryotic transcription factor complex containing both a MADS-box and a forkhead protein, and it has important implications for the regulation of mammalian gene expression.
Keywords: cell cycle regulation/forkhead/SFF/transcription/yeast
Introduction
The cell division cycle consists of an ordered series of events. An important element of regulation is to limit the expression and stability of key cell cycle controllers, such as the cyclins, to times when they are required in the cell cycle (reviewed in Morgan, 1995). Model organisms, such as Saccharomyces cerevisiae, have been used to understand the temporal control of cell cycle events. In S.cerevisiae, as in other eukaryotes, the regulation of gene expression is important in ensuring ordered cell cycle progression. For example, different cyclin genes are transcribed in G1, S and G2 phases (Koch and Nasmyth, 1994). Several genes that are expressed in G2 have identical cell cycle expression patterns, including the mitotic cyclin genes CLB1 and CLB2 (Ghiara et al., 1991; Surana et al., 1991), and the SWI5 (Nasmyth et al., 1987) and ACE2 genes (Dohrmann et al., 1992), which encode transcription factors.
Analysis of the regulation of the SWI5 and CLB2 genes identified a promoter element that is bound by the transcription factor Mcm1 and an unidentified protein termed SFF (Swi five factor) to form a ternary complex (Lydall et al., 1991; Maher et al., 1995). Mcm1 is a member of the eukaryotic MADS-box transcription factor family, which includes the yeast proteins, ArgRI, Rlm1 and Smp1, and the human proteins SRF and MEF2 (reviewed in Shore and Sharrocks, 1995). Mcm1 is an essential protein that is involved in transcriptional activation and repression in yeast to control a wide range of cellular processes, including, for example, cell type, the osmotic stress response and the regulation of the cell division cycle (Shore and Sharrocks, 1995). Apart from G2-specific transcription, Mcm1 has also been linked with the regulation of M/G1-specific transcription (McInerny et al., 1997). In the regulation of G2-specific transcription by Mcm1–SFF, although the identity of SFF was unknown, the binding site for SFF was defined by mutational analyses to a region close to the Mcm1-binding site (Lydall et al., 1991; Maher et al., 1995). This led to the proposal that the Mcm1–SFF ternary complex regulates a group of genes in G2 that includes SWI5, ACE2, CLB1 and CLB2. As Mcm1 has other non-cell cycle-related functions in yeast, it was suggested that SFF plays a role in the cell cycle regulation of the activity of the complex on the SWI5 promoter. Interestingly, the Mcm1–SFF complex has been proposed to be functionally analogous to the mammalian SRF–TCF complex, in that the MADS-box protein (Mcm1 or SRF) acts to recruit a key co-regulatory protein (SFF or TCF) (Lydall et al., 1991). However, Mcm1–SFF acts in G2 in the cell cycle, whilst the SRF–TCF complex is involved in regulating cell cycle entry (Treisman, 1994).
Forkhead proteins, also known as winged helix proteins, are a conserved family of eukaryotic transcription factors that are characterized by a region of homology of 110 amino acids, which spans a monomeric DNA-binding motif (for reviews see Lai et al., 1993; Kaufmann and Knöchel, 1996). In metazoans, they are involved in embryogenesis and development. Furthermore, human forkhead proteins have been linked directly with the cell cycle (Pati et al., 1997). Several forkhead transcription factors, Fhl1, Hcm1, Fkh1 and Fkh2, have been identified in S.cerevisiae (Zhu et al., 1993; Hermann-Le Denmat et al., 1994; Zhu and Davis, 1998). Fhl1 is implicated in the function of RNA polymerase III activity (Hermann-Le Denmat et al., 1994), while Hcm1 has been linked with the mitotic function of calmodulin (Zhu et al., 1993; Zhu and Davis, 1998). The Fkh1 and Fkh2 proteins have been suggested to regulate pseudohyphal growth (Zhu and Davis, 1998). Analysis of the predicted sequences of the proteins suggests that Fkh1 and Fkh2 are highly related and form a separate subclass of yeast forkhead proteins (Kaufmann and Knöchel, 1996). In addition to the forkhead domain, Fkh1 and Fkh2 share homology at their N-termini, but Fkh2 possesses a longer C-terminal domain. Fkh1 and Fkh2 also possess an FHA (forkhead-associated) domain close to their N-termini. FHA domains have been identified in at least 20 otherwise unrelated proteins and are implicated in recognizing phosphothreonine residues (Bucher and Bairoch, 1994; Durocher et al., 1999; Yaffe and Cantley, 1999). Interestingly, analysis of the expression patterns of these four genes during the cell cycle revealed that HCM1 was regulated at G1 while FKH1 was regulated in G2 (Spellman et al., 1998). Hcm1 was previously linked with the cell cycle (Zhu et al., 1993; Zhu and Davis, 1998), but these data suggested that Fkh1, and possibly its homologue Fkh2, may also function in cell cycle processes.
We have investigated the relationship between the Fkh1, Fkh2 and Hcm1 proteins by constructing all possible combinations of gene deletions. Analysis of the mutants revealed that Fkh2 affects nuclear migration and the formation of the mitotic spindle. In addition, the fkh1Δfkh2Δ double mutant has more severe phenotypes than either of the single mutants, suggesting that they act in overlapping pathways to regulate cell morphology and cell separation. Furthermore, we demonstrate that Fkh2 is important for the cell cycle expression of the SWI5 and CLB2 genes and that Fkh2 is a component of the previously identified cell cycle-dependent regulatory activity SFF. Moreover, our data suggest that Fkh2 is regulated by cell cycle-dependent phosphorylation. Thus we have identified Fkh2 as a critical component of the Mcm1–SFF complex in the regulation of cell cycle-dependent gene expression.
Results
Deletion of the FKH1, FKH2 and HCM1 genes
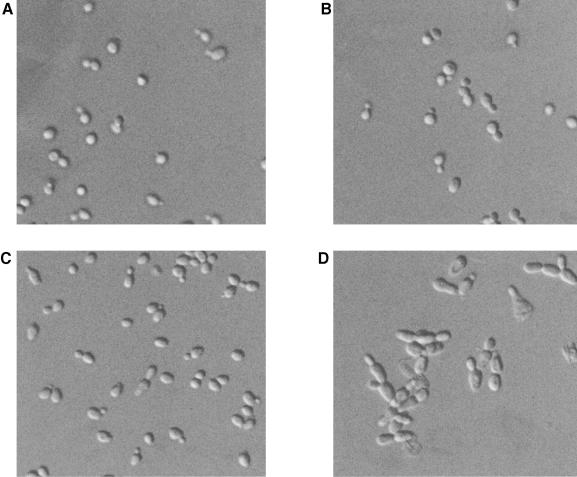
To investigate the role of the Fkh1, Fkh2 and Hcm1 proteins in S.cerevisiae, a diploid strain heterozygous for all three gene loci was constructed. This strain was sporulated, and viable spores containing all possible gene deletion combinations were obtained. However, unexpectedly, the fkh1Δfkh2Δ and fkh1Δfkh2Δhcm1Δ spores were found to have a low viability of 50 and 35%, respectively, suggesting that the function of Fkh1 and Fkh2 was linked. The cell morphology of the fkh1Δ and fkh2Δ single mutants looked very similar to that of the wild-type (Figure 1), although cell volume analysis indicated they were ∼10% larger than wild-type (data not shown). Furthermore, growth rate analysis suggested that fkh2Δ cells, unlike fkh1Δ cells, have an ∼10% longer generation time than wild-type cells. In agreement with the loss of spore viability, the fkh1Δfkh2Δ double mutant showed more severe defects than either of the single mutants. The fkh1Δfkh2Δ double mutant displayed very misshapen and elongated cells, which formed chains and clumps of cells (Figure 1) that were difficult to separate. These data strongly suggested that Fkh1 and Fkh2 share an overlapping function(s) in pathways that affect cell morphology and cell separation. Deletion of the HCM1 gene had no effect on any of these phenotypes (data not shown).
Fig. 1. Fkh1 and Fkh2 act in overlapping pathways affecting cell morphology and cell separation. Differential interference contrast photographs of the wild-type W303-1a (A), fkh1Δ (B), fkh2Δ (C) and fkh1Δfkh2Δ (D) strains.
Fkh1 and Fkh2 affect nuclear migration and spindle structure
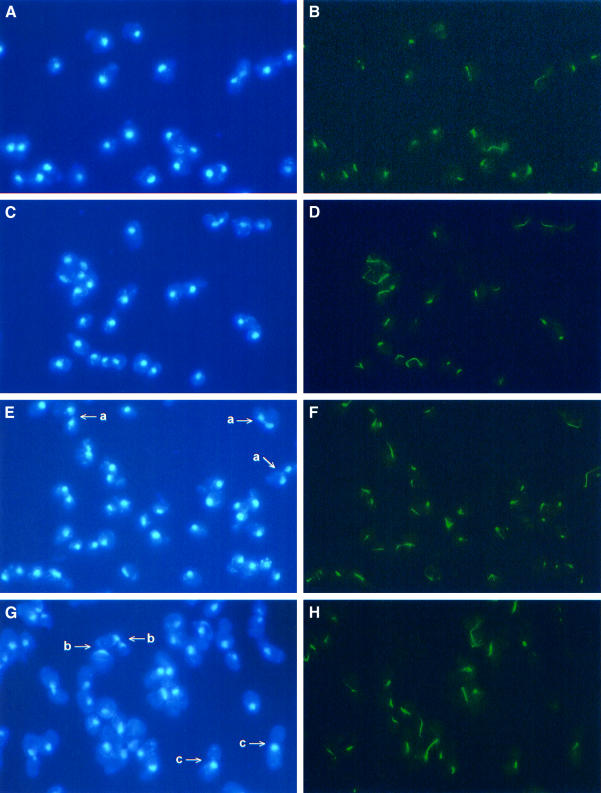
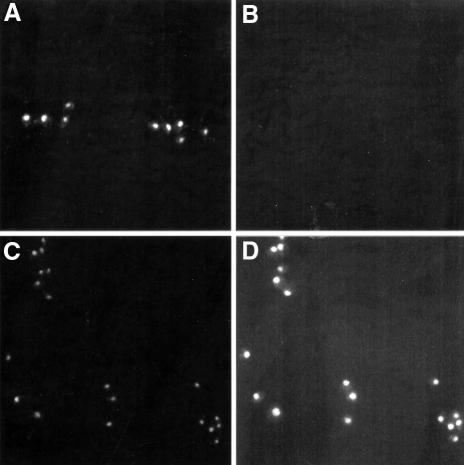
The genetic analysis suggested that Fkh1 and Fkh2 are important for late cell cycle events. To examine this further, nuclear migration and mitotic spindle formation were investigated in fkh1Δ, fkh2Δ and fkh1Δfkh2Δ mutant cells. Cells of the fkh1Δ strain had nuclear morphologies and spindle characteristics similar to the wild-type (Figure 2). In contrast, the fkh2Δ mutant showed clear defects, which included, for example, large budded cells with a small spindle, cells with a fragmented spindle with the nuclei still not completely separated, an increased frequency of cells with separated nuclei with a very long spindle, and multibudded cells undergoing nuclear division with a long spindle. Cells of the fkh1Δfkh2Δ double mutant had a higher percentage of some of these phenotypes. However, in addition, a significant number of cells were large budded cells with the nucleus at the bud neck with no apparent spindle (Figure 2G and H, and data not shown). There were also some cells containing two or more nuclei and with an abnormal spindle (data not shown). Taken together, these data suggested that Fkh1 and Fkh2 regulate the expression of genes linked with nuclear migration and spindle formation, with Fkh2 perhaps having a more predominant role.
Fig. 2. Fkh2 affects spindle elongation and nuclear migration. Fluorescent detection of the nucleus (DAPI staining: A, C, E and G) and the mitotic spindle (indirect immunofluorescence: B, D, F and H) in the wild-type (A and B), fkh1Δ (C and D), fkh2Δ (E and F) and fkh1Δfkh2Δ (G and H) strains. Examples of defects observed: (a) multibudded cells undergoing nuclear division with a long spindle; (b) large budded cells with the nucleus at the bud neck with no apparent spindle; and (c) large budded cells with the nucleus at the bud neck with a very short spindle.
Fkh2 affects G2-specific gene expression
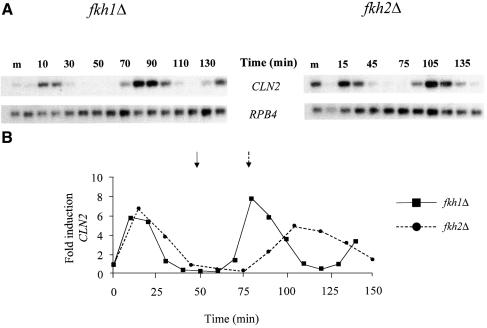
The studies above suggest that Fkh1 and Fkh2 function in the expression of genes whose products influence several late cell cycle events. Previous work had identified a group of genes, CLB1, CLB2, SWI5 and ACE2, which are expressed periodically in G2 (Nasmyth et al., 1987; Ghiara et al., 1991; Surana et al., 1991; Dohrmann et al., 1992), and two of these, SWI5 and CLB2, have been shown to be regulated by the Mcm1–SFF ternary complex (Lydall et al., 1991; Maher et al., 1995). To investigate whether Fkh1 or Fkh2 had any role in cell cycle-regulated gene expression, the fkh1Δ and fkh2Δ strains were synchronized with α-factor and the expression of the G1-regulated gene CLN2 examined during the cell cycle. The fkh1Δ mutant had similar cell cycle-regulated expression of CLN2 to that of the wild-type (Figure 3, and data not shown). The fkh2Δ mutant also showed normal cell cycle induction and approximate timing of expression of CLN2 in the first cycle, although the timing of expression in the next cycle was later (Figure 3). This effect on CLN2 gene expression in the fkh2Δ strain suggested that the cell cycle was longer than that of the fkh1Δ strain, in agreement with the growth studies (see above).
Fig. 3. Cell cycle expression of the CLN2 gene. (A) Northern blot analysis of RNA isolated from α-factor-synchronized cultures of the fkh1Δ and fkh2Δ strains using probes specific for the CLN2 and RPB4 transcripts. Following release from the α-factor block, RNA was isolated from cells of the fkh1Δ and the fkh2Δ strain at 10 and 15 min intervals, respectively. The first lane on each blot contains RNA isolated from midlog (m) cultures of each strain. (B) The panel below the northern blot shows the fold induction of CLN2 expression. Samples were first quantitated relative to the RPB4 loading control, then fold induction during the cell cycle was calculated relative to the time 0 value for each strain. The arrows indicate when maximum budding was achieved in the first cycle of the fkh1Δ strain (solid arrow) and the fkh2Δ strain (dotted arrow) following α-factor release.
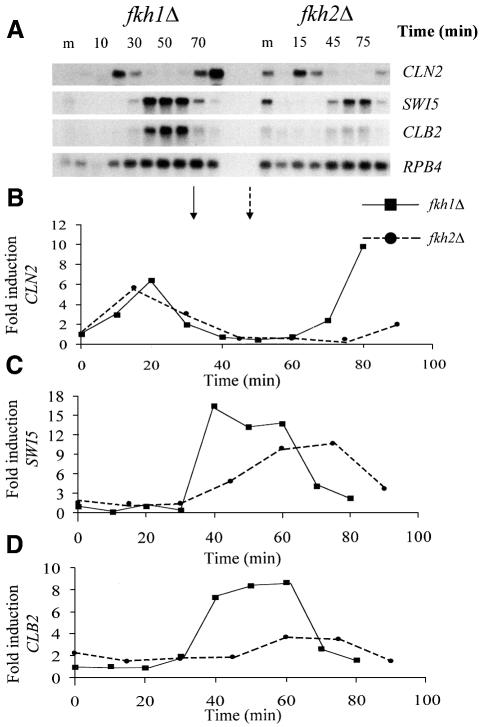
To compare the effects of Fkh1 and Fkh2 on SWI5 and CLB2 expression, the RNA from the first cycle of the fkh1Δ and fkh2Δ synchronized cultures was analyzed on the same membrane (Figure 4). As can be seen, the cell cycle induction of SWI5 is inhibited in the fkh2Δ strain although there is also a small increase in the basal level of expression (Figure 4). Furthermore, deletion of the FKH2 gene has a more severe influence on the induction and timing of CLB2 expression (Figure 4). These data strongly suggest that Fkh2 functions in the regulation of several Mcm1–SFF-dependent genes.
Fig. 4. Fkh2 affects the cell cycle expression of the SWI5 and CLB2 genes. (A) Northern blot analysis of RNA isolated from α-factor-synchronized cultures of the fkh1Δ and fkh2Δ strains using probes specific for the CLN2, SWI5, CLB2 and RPB4 transcripts. RNA from the 0–80 min time points of the fkh1Δ strain and the 0–90 min time points of the fkh2Δ strain, spanning approximately the first cycle following α-factor release, were analysed on the same membrane. The fkh1Δ and fkh2Δ RNA samples were collected at 10 and 15 min intervals, respectively. The midlog sample for each strain is indicated (m). The panels below the northern blots show the fold induction of the CLN2 (B), SWI5 (C) and CLB2 (D) transcripts. Samples were first quantitated relative to the RPB4 loading control, then fold induction during the cell cycle was calculated using the time 0 value for the fkh1Δ strain only. This allowed direct comparisons between the strains. The arrows indicate when maximum budding was achieved for the fkh1Δ strain (solid arrow) and the fkh2Δ strain (dotted arrow) following α-factor release.
Cell cycle localization of Fkh2
These data suggested that Fkh2 affects the expression of the SWI5 and CLB2 genes in the cell division cycle. Hence we next examined whether Fkh2 is indeed present in the nucleus and whether this changes in the cell division cycle. A strain was constructed that contained the Fkh2 protein tagged with 13 Myc epitopes [Fkh2(Myc)] that is expressed from its own chromosomal locus. The localization of the tagged protein was observed subsequently by immunofluorescence using the anti-Myc antibody and 4′,6-diamidino-2-phenylindole (DAPI) staining (Figure 5). The Fkh2(Myc) protein was found to be localized in the nucleus and this was unaffected by cell cycle position.
Fig. 5. Fkh2 is localized to the nucleus during the entire cell division cycle. Nuclear staining by DAPI treatment (A and C) and immunofluorescence using the anti-Myc antibody (B and D) of the W303-1a wild-type strain, containing the wild-type version of Fkh2 (A and B), or the strain AP16, containing the Myc-tagged version of Fkh2 (C and D).
The SFF consensus sequence is identical to a forkhead consensus binding site
The data above show that Fkh2 is a nuclear protein and that it may regulate the expression of SWI5 and CLB2. Analysis of SFF binding to the SWI5 and CLB2 promoters had identified bases that when mutated abolished Mcm1–SFF-dependent ternary complex formation (Lydall et al., 1991; Maher et al., 1995). Scrutiny of promoters of several genes that are expressed periodically during G2 (Spellman et al., 1998) revealed potential Mcm1-binding sites. We have identified additional conserved sequences to the 3′ side of the potential Mcm1 sites with homology to the region important for SFF binding in the SWI5 and CLB2 promoters (Figure 6). Interestingly, this potential SFF consensus sequence is identical to the core consensus binding site for many forkhead proteins (Kaufmann et al., 1995). Mutations in the SWI5 and CLB2 promoters that prevent SFF binding, a C→A substitution at position 296 in the SWI5 promoter and a C→A substitution at position 306 in the CLB2 promoter, lie in the forkhead-like protein-binding site we have identified (Figure 6). Furthermore, equivalent C→A substitutions of the forkhead consensus binding site prevent forkhead proteins from binding (Kaufmann et al., 1995). Taken together with the expression studies, this analysis suggested that Fkh2 may be a component of SFF.
Fig. 6. Identification of a forkhead consensus sequence in the promoters of genes which are co-expressed in the cell division cycle. Promoter analysis of several genes that have a similar expression pattern to SWI5 (Spellman et al., 1998) shows two conserved regions, which correspond to the Mcm1 consensus sequence (bold) and the SFF/forkhead consensus sequence (bold/italics). A296* and 306A* correspond to point mutations C→A in the SWI5 and CLB2 promoters, respectively, which abolish SFF binding. The sequences of wild-type and mutant SWI5 and CLB2 promoters were from Lydall et al. (1991) and Maher et al. (1995), respectively. All other promoter sequences were obtained from the Saccharomyces Genomic Database. The Mcm1 and forkhead consensus sequences were from Wynne and Treisman (1992) and Kaufmann et al. (1995), respectively.
Fkh2 is required for the formation of complexes on G2-specific gene promoters
Gel mobility shift assays were used next to investigate whether the formation of the Mcm1–SFF complex on the SWI5 promoter was dependent on the presence of any of the forkhead genes. Extracts from wild-type yeast strains were tested for their ability to form complexes with a SWI5 promoter fragment containing the binding site for the Mcm1–SFF ternary complex (Figure 7A). Several major complexes were obtained with extracts from the wild-type strain (Figure 7B, lane 1). The upper two complexes were dependent on SFF, as a point mutation within the SFF-binding site abolishes the formation of these complexes (Figure 7B, lane 9; Lydall et al., 1991). Competition assays were carried out with a site that is specific for Mcm1 and lacks potential SFF-binding sites (PPAL). This site abolishes the formation of the majority of the complexes, demonstrating that the SFF cannot bind in the absence of Mcm1 (Figure 7C, lanes 1–3; Lydall et al., 1991). In contrast, a control competitor only affects the formation of the non-specific faster migrating complexes (Figure 7C, lanes 4 and 5).
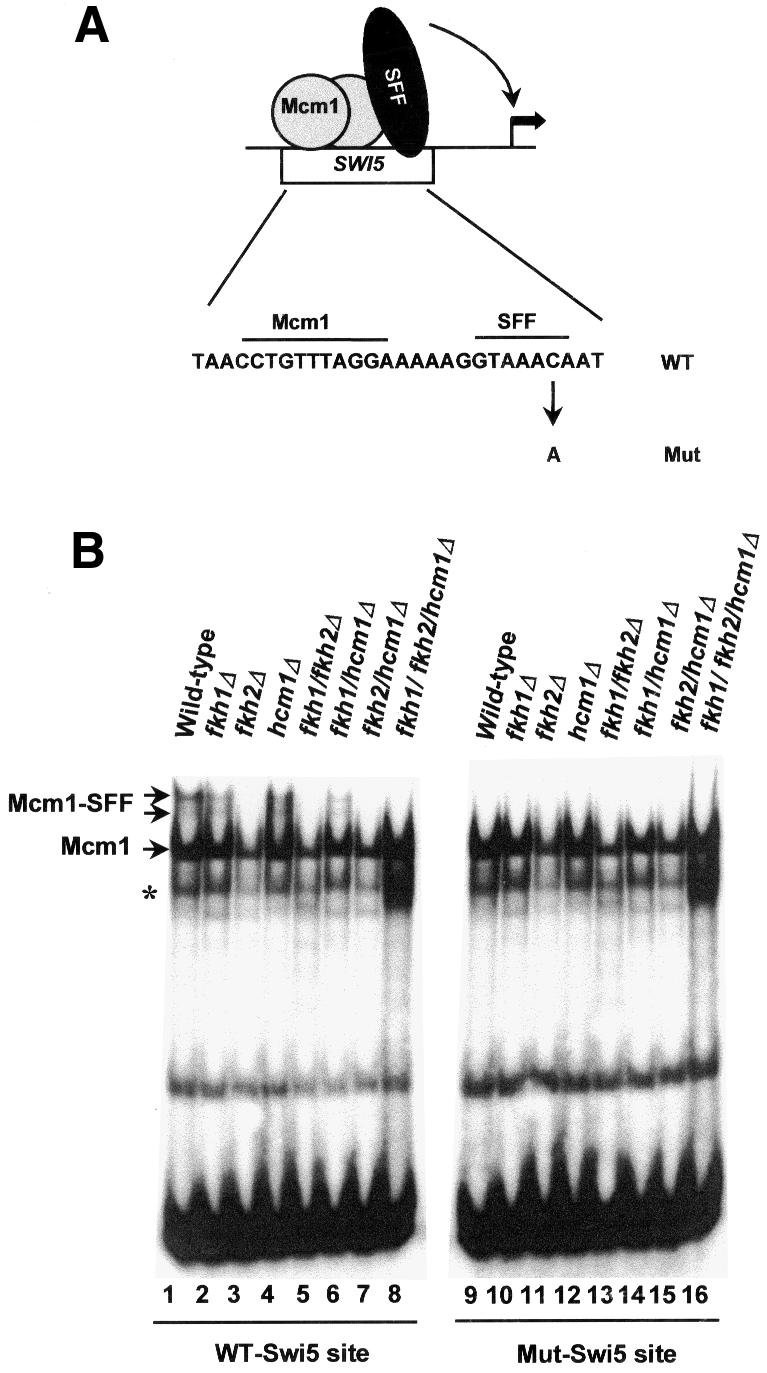
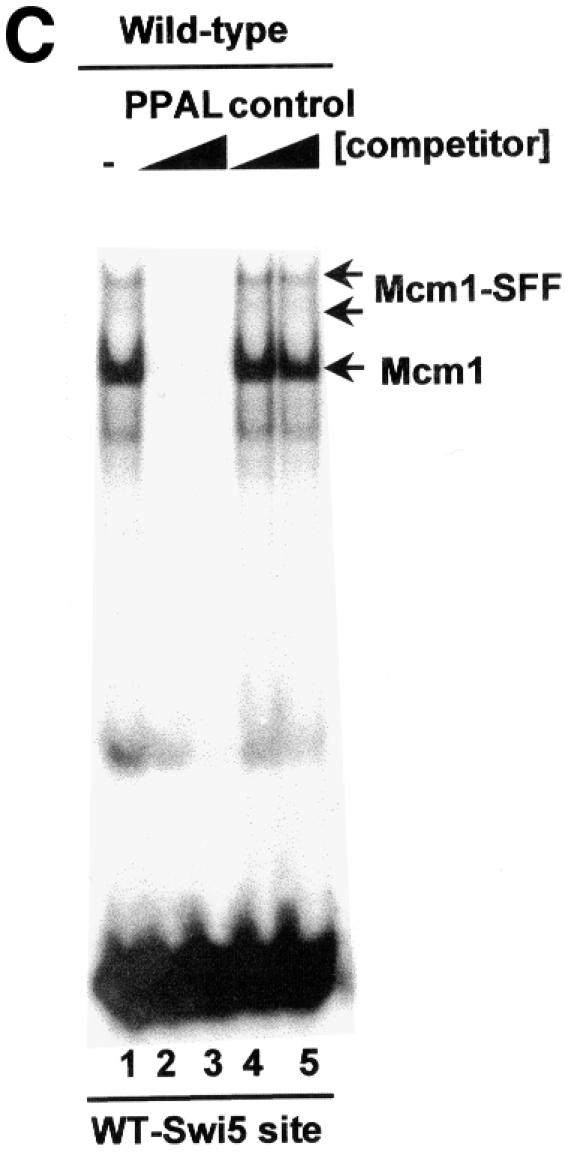
Fig. 7. Fkh2 is required for the formation of the Mcm1–SFF complex at the SWI5 promoter. (A) Schematic illustration of the Mcm1–SFF complex on the SWI5 promoter. The sequence of the wild-type site is shown below (Mcm1- and SFF-binding sites are indicated) and the mutation that blocks SFF binding is indicated. (B) Gel mobility shift assay of extracts from wild-type yeast (W303-1a) or strains that contain deletions of the indicated combinations of forkhead genes using the wild-type or mutant SWI5 promoter fragments. (C) Competition analysis of complex formation on the wild-type SWI5 site with extracts from wild-type yeast (W303-1a) and no competitor (lane 1), a 10- and 100-fold excess of PPAL competitor DNA (lanes 2 and 3) or a 10- and 100-fold excess of control E74 competitor DNA (lanes 4 and 5). The bands corresponding to the Mcm1 and Mcm1–SFF complexes are indicated. The asterisk represents a band that probably arises from partially degraded Mcm1.
Having established the assay conditions, extracts from mutant yeast strains that harbour deletions in one or more genes encoding forkhead proteins were analyzed on the wild-type and mutant SWI5-binding sites. The Mcm1–SFF ternary complex was absent in extracts from any mutant deleted for the FKH2 gene, demonstrating that Fkh2 was essential for this complex (Figure 7B, lanes 2–8). In contrast, Mcm1 binding was detected in extracts from all the mutant strains. Similarly, Mcm1 binding to the mutant SWI5 promoter fragment was detectable in all cases, whereas no Mcm1–SFF complexes could be detected on this site (Figure 7B, lanes 10–16).
These data demonstrate that Fkh2 is essential for SFF binding at the SWI5 promoter. To investigate whether Fkh2 represents part of the Mcm1–SFF complexes at other promoters, we next examined the ACE2 promoter. An ACE2 promoter fragment, which contains potential Mcm1- and SFF-binding sites (Figure 6), was examined by gel mobility shift assays using extracts from wild-type strains (Figure 8). A pattern of complexes formed with this probe that closely resembled those found on the SWI5 promoter (compare Figure 8, lane 1 and Figure 7B, lane 1). The dependence of these complexes on Mcm1 was confirmed by competition analysis (data not shown). To investigate the role of the forkhead proteins in these complexes, extracts from all of the deletion mutants were examined (Figure 8, lanes 2–8). As was observed with the SWI5 promoter, the low mobility bands corresponding to the Mcm1–SFF complex were lost with extracts derived from any strain containing an FKH2 gene deletion. Another low mobility complex is apparent that is only lost when extracts derived from the triple mutant strain are used (Figure 8, compare lanes 7 and 8). It is not clear whether this extra band is related to the SFF site or another site in the fragment.
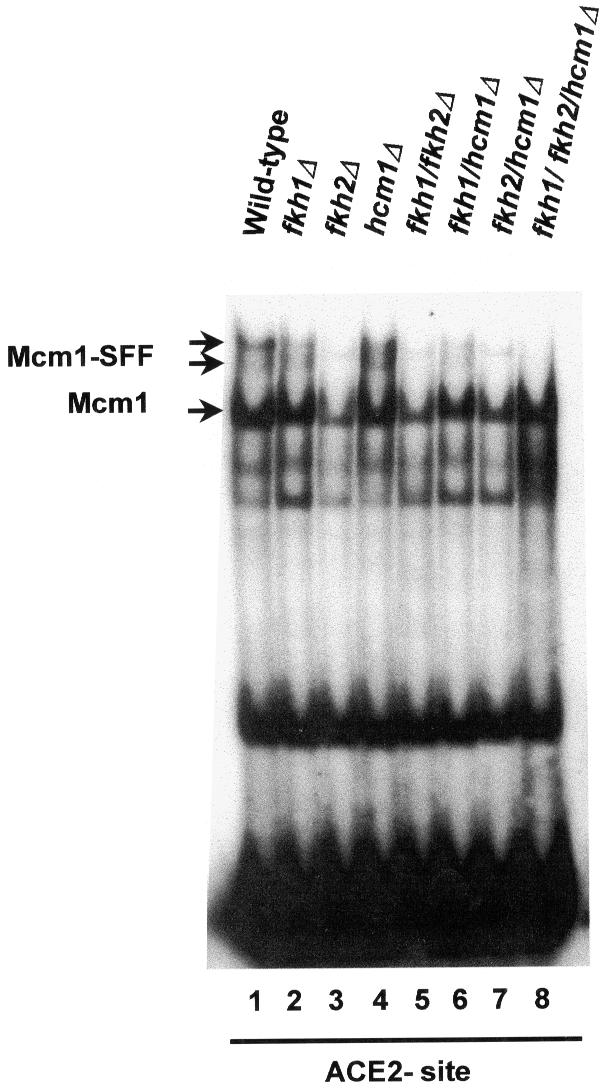
Fig. 8. Fkh2 is required for the formation of the Mcm1–SFF complex at the ACE2 promoter. Gel mobility shift assay of extracts from wild-type yeast (W303-1a) or strains that contain deletions of the indicated combinations of forkhead genes using the wild-type ACE2 promoter fragments. The bands corresponding to the Mcm1 and Mcm1–SFF complexes are indicated.
Collectively, these data demonstrate that Fkh2 is essential for the formation of Mcm1–SFF-dependent ternary complexes on the SWI5 and ACE2 promoters.
Fkh2 corresponds to the SFF component of the ternary complex on the SWI5 and ACE2 promoters
The data presented above indicate that Fkh2 is required for the formation of the Mcm1–SFF complex on the SWI5 and ACE2 promoters. However, it is possible that this effect was indirect and that Fkh2, for example, regulates the expression and/or activity of SFF.
The similarity of the SFF-binding sites to consensus motifs for forkhead proteins (Figure 6) strongly suggests that Fkh2 binds directly to these promoters. To demonstrate direct binding of Fkh2 to the SWI5 and ACE2 promoters, AP16, a strain containing the Myc epitope-tagged version of Fkh2 expressed from its normal chromosomal locus, was utilized. Gel mobility shift assays were performed with extracts from the wild-type [Fkh2(WT)]- and the Fkh2(Myc)-expressing strains and the wild-type and mutant SWI5 promoter fragments. The Mcm1–SFF ternary complex was observed with extracts from both strains on the wild-type SWI5 promoter fragment. However, extracts containing Fkh2(Myc) gave rise to a Mcm1–SFF complex that migrated more slowly than with the wild-type Fkh2 protein (Figure 9, compare lanes 1 and 3), consistent with the increase in size caused by the addition of 130 amino acids when the Fkh2 protein was tagged with 13 Myc epitopes, thereby suggesting that Fkh2 is a component of SFF. To demonstrate this conclusively, the formation of the Mcm1–SFF complex was examined by adding anti-Myc antibody to the extracts derived from wild-type- and Fkh2(Myc)-expressing strains, prior to binding to the SWI5 site. The addition of antibody did not affect Mcm1 binding in either strain. However, in contrast, the formation of the Mcm1–SFF complex was affected from cell extracts derived from the Myc-tagged Fkh2 but not the wild-type control (Figure 9, lanes 2 and 4). As expected, no Mcm1–SFF complexes were detected with either extract using the mutant SWI5 promoter fragment, and neither did anti-Myc antibody addition affect Mcm1 binding (Figure 9, lanes 5–8). Similar results were obtained by analyzing the formation of ternary Mcm1–SFF complexes on the ACE2 promoter (data not shown). Thus, the Fkh2 protein is a key component of SFF on the SWI5 and ACE2 promoters.
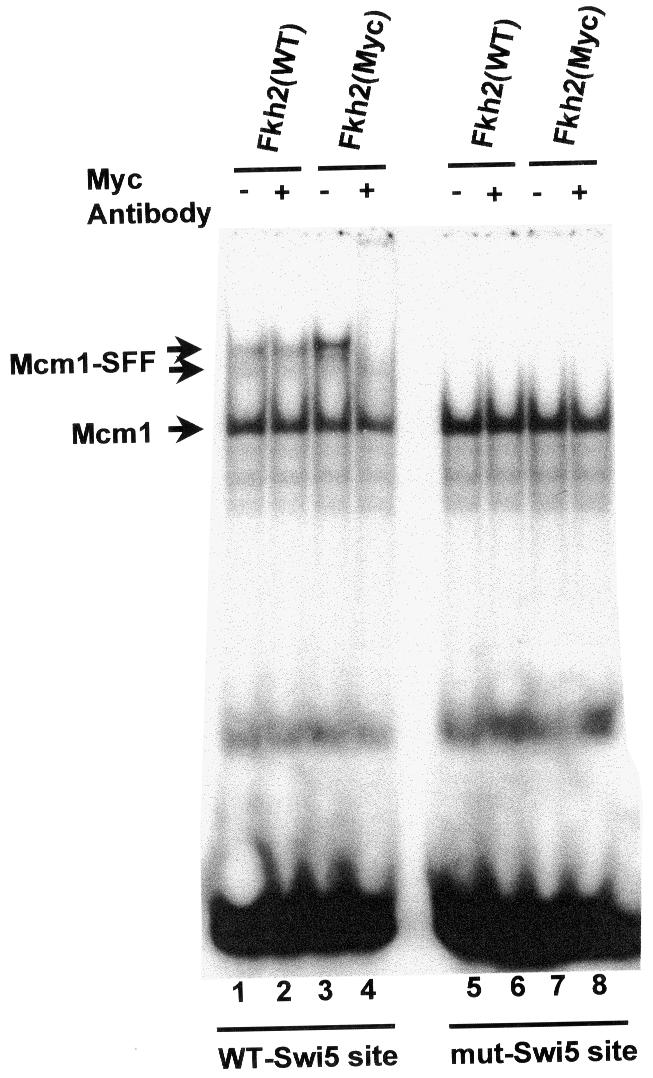
Fig. 9. The SFF component of the Mcm1–SFF complex is Fkh2. Gel mobility shift assay of extracts from yeast strains that contain the wild-type version of Fkh2 [Fkh2(WT); W303-1a] or a Myc-tagged version of Fkh2 [Fkh2(Myc); AP16] using the wild-type or mutant SWI5 promoter fragments. Anti-Myc antibody is added to the binding reactions prior to incubation with DNA where indicated. The bands corresponding to the Mcm1 and Mcm1–SFF complexes are indicated.
Fkh2 is phosphorylated in a cell cycle-dependent manner
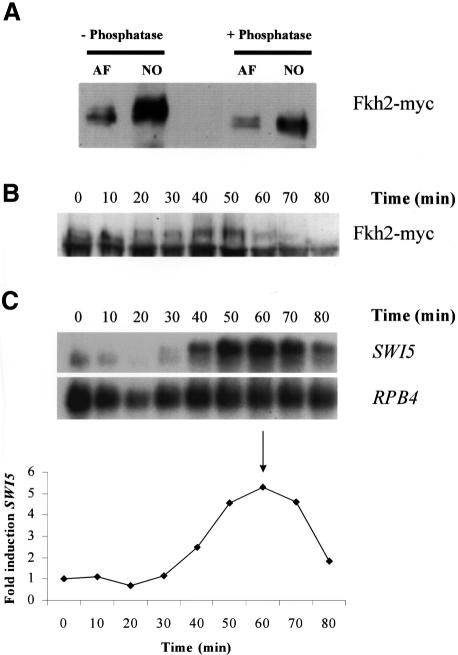
These studies have demonstrated that Fkh2 is a component of SFF. Our data also suggest that a component of Mcm1–SFF is likely to be a target of the cell cycle machinery. Moreover, since Mcm1 regulates the expression of several genes in a non-cell cycle-dependent manner, a component specifically of SFF may be the target for this regulation. To investigate any possible connection between the cell cycle machinery and SFF, we next examined whether Fkh2 was phosphorylated during the cell cycle. Western blot analysis was carried out on crude extracts isolated from cultures of the strain expressing Fkh2(Myc) following a cell cycle block with α-factor or nocodazole. This revealed that Fkh2 is indeed phosphorylated at different time points in the cell division cycle (Figure 10A). Specific bands of the expected size for the Fkh2(Myc) protein were detected in extracts from the α-factor- and nocodazole-blocked cells. Significantly, the band from the nocodazole-treated culture was broad and generally of lower mobility than that from the α-factor-arrested cells. When treated with phosphatase, the band from the nocodazole-treated cells collapsed to a sharper band(s) of higher mobility. Fkh2 is therefore phosphorylated at the nocodazole arrest point. There is also a suggestion of a minor phosphorylated species in the α-factor-treated cells.
Fig. 10. Fkh2 is phosphorylated in a cell cycle-dependent manner. (A) Western blot of extracts from cultures of AP16 blocked with either α-factor or nocodazole, using the anti-Myc antibody. Samples were either untreated or treated with phosphatase. Western (B) and northern (C) blot analysis of protein and RNA isolated from an α-factor-synchronized culture of the AP16 strain using the anti-Myc antibody (western) and probes specific for the SWI5 and RPB4 transcripts (northern). Cells were collected at 10 min intervals. The panel below the northern blot shows the fold induction of the SWI5 transcript. Samples were first quantitated relative to the RPB4 loading control then fold induction during the cell cycle was calculated using the time 0 value. The arrow in the graph indicates when maximum budding was achieved (solid arrow) following α-factor release.
Nocodazole blocks cells in metaphase consistent with the notion that phosphorylation of Fkh2 might be linked to Mcm1–SFF-regulated gene expression. To explore further the relationship between the cell cycle-dependent phosphorylation and gene expression, northern and western blot analyses were performed with RNA and protein isolated from an α-factor-synchronized culture of the Fkh2(Myc)-expressing strain (Figure 10B and C). Between 10 and 20 min after release from α-factor, the mobility of Fkh2 decreases; a higher mobility band present at 10 min disappears and a lower mobility band appears. The appearance of this band precedes the increase in SWI5 mRNA and increases in intensity, reaching a peak at 40–50 min, at about the time of maximal increase in the SWI5 transcript levels. Thereafter, the upper phosphorylated species decreases in amount and the faster mobility band reappears (Figure 10B, and data not shown).
Taken together, these data suggest that Fkh2 is phosphorylated in a cell cycle-dependent manner. The timing of at least one of the phosphorylation events correlates with SWI5 expression suggesting a causal relationship. The basis of this phosphorylation of Fkh2, and whether this is controlled by the cell cycle machinery, is currently under investigation but is consistent with a model where the activity of Mcm1–SFF is regulated by a protein kinase(s) during the cell division cycle.
Discussion
Previous studies have identified a cell cycle transcription factor complex, Mcm1–SFF, which regulates the expression of the SWI5 and CLB2 genes in G2 of the cell division cycle (Lydall et al., 1991; Maher et al., 1995). SFF was an unidentified protein(s) that could bind to these promoters only in a ternary complex with Mcm1. Mutations that prevented the binding of SFF, but not Mcm1, were correlated with loss of cell cycle expression, which led to the suggestion that SFF was essential for cell cycle regulation of these genes. Here we have demonstrated that the forkhead protein, Fkh2, and the MADS-box protein, Mcm1, form the Mcm1–SFF complex and regulate the cell cycle expression of SWI5 and CLB2. MADS-box proteins and forkhead proteins are found from yeast to mammals and regulate gene expression in a variety of cellular and developmental processes (reviewed in Lai et al., 1993; Shore and Sharrocks, 1995; Kaufmann and Knöchel, 1996). However, this is the first example of these two different types of transcription factor forming a complex and this also has important implications for the regulation of cellular processes in higher eukaryotes.
MADS-box proteins interact with other transcription factors to form regulatory complexes, and it is the interplay between these proteins that determines transcriptional activity when bound to promoters (Shore and Sharrocks, 1995). The multiple roles of Mcm1 in several cellular processes suggest that it is the accessory binding proteins that provide the specific activating and repressing functions. Indeed, our studies of SWI5 and CLB2 expression suggest that Fkh2 is important for the activating activity of the Mcm1–SFF complex.
How is the Mcm1–SFF transcription factor complex regulated during the cell cycle? Previously it was reported that the Mcm1–SFF complex may be detectable in extracts prepared from G1 cells (Lydall et al., 1991). This is consistent with a model where Mcm1–SFF binds to the SWI5 promoter throughout the cell cycle and that the Mcm1–SFF complex undergoes cell cycle regulation to activate transcription. In agreement with this, our studies have shown that the Fkh2 protein can be detected in the nucleus of cells throughout the cell cycle. What other mechanism(s) could be responsible for the regulation of Mcm1–SFF? Previous studies have shown that the activity of Cdc28–Clb2 is required for the periodic activation of expression of the SWI5 and CLB2 genes, leading to the suggestion that a positive feedback loop acts to regulate G2 periodic gene expression (Amon et al., 1993). It is possible that the Mcm1–SFF transcription factor complex is phosphorylated directly by Cdc28–Clb2. However, although Mcm1 is known to be a phosphoprotein (Kuo et al., 1997), it also regulates the expression of several genes in a non-cell cycle-dependent manner. Hence Fkh2 is a more likely target for specific cell cycle phosphorylation. Indeed, examination of the predicted sequence of the Fkh2 protein revealed several S/TP motifs, which may be potential targets for Cdc28–Clb2. Furthermore, we have found that Fkh2 becomes phosphorylated with a cell cycle timing that is consistent with the role of this protein in the activation of gene expression at G2/M (Figure 10). The molecular basis of this phosphorylation of Fkh2 is under investigation, but our data suggest that the activity of the Mcm1–SFF transcription factor complex may be regulated by a cell cycle-regulated protein kinase(s). Moreover, these data are also consistent with the positive feedback loop model described above (Figure 11).
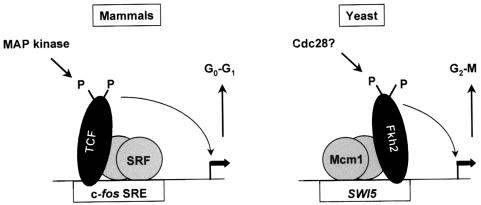
Fig. 11. Comparison of the mammalian SRF–TCF and yeast Mcm1–Fkh2 complexes. The yeast Mcm1–SFF complex is very similar to the mammalian SRF–TCF complex. Both complexes contain winged helix–turn–helix proteins, the forkhead protein (Fkh2) and the ETS-domain protein (TCF). In addition, Fkh2 and TCF are recruited to promoters by the highly related MADS-box proteins Mcm1 and SRF. Furthermore, Fkh2 (see previously) and TCF (Treisman, 1994) are modified by phosphorylation. Indeed, there are numerous S/TP motifs in the C-terminal region of Fkh2, which is highly reminiscent of TCF’s structure. Although the SRF–TCF complex is thought to function to promote cell cycle entry, while the Mcm1–Fkh2 complex regulates cell cycle expression in G2/M, these similarities suggest that the activity of these complexes may be regulated in a similar manner.
The fkh1Δfkh2Δ double mutant has severe effects on cellular morphology and cell separation compared with either of the single mutants. Sequence comparisons between Fkh1 and Fkh2 suggest that these two proteins are members of the same forkhead protein family subclass (Kaufmann and Knöchel, 1996). However, the cell morphology defects of the fkh1Δfkh2Δ mutant suggest that these proteins have distinct functions that only partially overlap in the regulation of the expression of genes required for late cell cycle processes. Moreover, we also show that the cell cycle periodicity and timing of SWI5 and CLB2 gene expression are affected following deletion of the FKH2 gene. These results are in good agreement with a recent study that suggested that CLB2 expression was affected in a fkh2Δ strain although the basis for this effect was not characterized (Hollenhorst et al., 2000). In previous studies, it was found that a small fragment containing the SWI5 and CLB2 SFF site can confer cell cycle-dependent transcription upon a reporter construct, although the influence of deletion of the SFF site on periodic transcription in the context of the chromosomal promoter has not been determined. Hence, other as yet unidentified proteins may contribute to the residual periodic expression of the SWI5 and CLB2 promoters in fkh2Δ cells. It is also possible that the residual periodicity observed in the fkh2Δ strain is linked to the function of Fkh1. Indeed, expression studies with synchronized cultures of the fkh1Δfkh2Δ double mutant suggest that this is the case (Hollenhorst et al., 2000; data not shown). Although we can find no binding of Fkh1 to the SFF site on the SWI5 and ACE2 promoters, in vivo Fkh1 may bind to this site or another site on these promoters. Alternatively, Fkh1 may have an indirect effect on the expression of SWI5 and CLB2 by affecting the expression of other G2 phase-regulated genes. It is worth noting, for example, that there are a large number of genes expressed coincidentally with SWI5, CLB2 and ACE2, and analysis of their promoters has revealed potential binding sites for Mcm1 and a forkhead protein, which may prove to be Fkh1 (Figure 6, and data not shown). Moreover, there is a correlation between effects on the expression of these G2-regulated genes and the cell separation/cell morphology pathways. For example, the absence of the ACE2 gene causes cell separation defects resulting in clumps of cells (King and Butler, 1998). It is also significant that loss of ACE2 results in invasion of solid media by haploid cells, a phenotype we have also observed only in the fkh1Δfkh2Δ double mutant (data not shown). Furthermore, the effects of deleting the FKH1 and FKH2 genes on pseudohyphal growth are suppressed by overexpressing the CLB2 gene (Hollenhorst et al., 2000).
Significantly, the yeast Mcm1–SFF complex has been proposed to represent a functionally analogous complex to the mammalian SRF–TCF complex that plays a pivotal role in regulating the induction of immediate-early genes such as the proto-oncogene c-fos in response to mitogenic stimuli (Lydall et al., 1991). Thus the SRF–TCF complex is thought to function to promote cell cycle entry. Similarly, the Mcm1–SFF complex plays a role in regulating cell cycle progression, albeit at a later time point. Molecularly, their mechanisms of complex assembly and function appear to be similar, with both TCF (in mammals) and SFF (in yeast) being recruited to promoters by the highly related MADS-box proteins SRF and Mcm1 (Figure 11). Both SFF and TCF are critical for the transcriptional activity of these complexes. The identification of Fkh2 as a component of SFF has uncovered a number of other intriguing parallels between these mammalian and yeast complexes. Firstly, whilst the TCFs and Fkh2 are members of different transcription factor families, ETS-domain (Sharrocks et al., 1997; Graves and Petersen, 1998) and forkhead (Lai et al., 1993; Kaufmann and Knöchel, 1996), respectively, their DNA-binding domains are related and form part of the larger winged helix–turn–helix family. Indeed, ETS-domain proteins appear unique to the metazoan lineage. Secondly, both Fkh2 (see previously) and TCF (Treisman, 1994) are modified by phosphorylation. In the case of Fkh2, there are numerous S/TP motifs in its C-terminal region, which is highly reminiscent of TCF’s structure. Hence these similarities in structure and regulation suggest that studies of the mechanisms of regulation of activity and interaction between these highly conserved proteins will shed new light on the roles and activities of these proteins in eukaryotes.
Materials and methods
Yeast strains and growth conditions
Saccharomyces cerevisiae strains (Table I) were grown at 30°C in YPD medium for non-selective growth, or in SD medium for selective growth (Sherman et al., 1986). The deletion strains used in this study, AP1, AP3, AP5, AP7, AP9, AP11 and AP13, were from the cross between AP-H1F1 and AP-F2. Sporulation medium was 1% potassium acetate, 0.1% yeast extract, 0.05% dextrose and 2% agar (Sherman et al., 1986). Kanamycin selection of yeast transformants used YPD plates containing 200 µg/ml G418 sulfate (Calbiochem).
Table I. Saccharomyces cerevisiae strains used in this study.
| Strain | Genotype |
|---|---|
| W303-1a | MATa ade2-1 trp1-1 can1-100 leu2-3,112 his3-11 ura3 |
| W303-1b | MATα ade2-1 trp1-1 can1-100 leu2-3,112 his3-11 ura3 |
| AP-H1 | MATa ade2-1 trp1-1 can1-100 leu2-3,112 his3-11 ura3 hcm1::LEU2 |
| AP-H1F1 | MATa ade2-1 trp1-1 can1-100 leu2-3,112 his3-11 ura3 hcm1::LEU2 fkh1::HIS3 |
| AP-F2 | MATα ade2-1 trp1-1 can1-100 leu2-3,112 his3-11 ura3 fkh2::URA3 |
| AP1 | MATa ade2-1 trp1-1 can1-100 leu2-3,112 his3-11 ura3 hcm1::LEU2 |
| AP3 | MATa ade2-1 trp1-1 can1-100 leu2-3,112 his3-11 ura3 fkh1::HIS3 |
| AP5 | MATa ade2-1 trp1-1 can1-100 leu2-3,112 his3-11 ura3 fkh2::URA3 |
| AP7 | MATa ade2-1 trp1-1 can1-100 leu2-3,112 his3-11 ura3 hcm1::LEU2 fkh1::HIS3 |
| AP9 | MATa ade2-1 trp1-1 can1-100 leu2-3,112 his3-11 ura3 hcm1::LEU2 fkh2::URA3 |
| AP11 | MATa ade2-1 trp1-1 can1-100 leu2-3,112 his3-11 ura3 fkh1::HIS3 fkh2::URA3 |
| AP13 | MATa ade2-1 trp1-1 can1-100 leu2-3,112 his3-11 ura3 hcm1::LEU2 fkh1::HIS3 fkh2::URA3 |
| AP16 | MATa ade2-1 trp1-1 can1-100 leu2-3,112 his3-11 ura3 FKH2-13×Myc KanR |
Cell synchrony
α-factor peptide (Novagen) was added (5 µg/ml) to midlog cultures in YPD and cells were incubated for 3–4 h. α-factor release was achieved by filtration and saline wash. Samples were collected every 10–15 min and stored for RNA or protein extraction. Nocodazole (Sigma) was added (15 µg/ml) to midlog cultures in YPD and cells were incubated for 3 h to achieve >90% large buds.
Yeast techniques
Yeast cells were transformed using the lithium acetate method described by Schiestl and Gietz (1989). Genomic DNA was isolated as described by Hoffman and Winston (1987).
Deletion of the HCM1, FKH1 and FKH2 genes
A SpeI–EcoRV-digested PCR fragment, isolated using oligonucleotides Hcm1-1 5′-TAGTTACATCTACTAGTTGAGCTTCTTTTATTGACCG-3′ and Hcm1-2 5′-GAAAATGCTATGTGATATCATAACTCCAGCCCAGT-3′, carrying the HCM1 gene and 85 nucleotides upstream and 118 nucleotides downstream of the gene, was ligated into Bluescript (Stratagene). A 1.5 kb MscI–EcoRI fragment, internal to the HCM1 gene, was then replaced with the LEU2 gene from YDp-L (Berben et al., 1991). A SpeI–HindIII disruption cassette was used to disrupt the HCM1 gene in W303-1a, creating AP-H1. Disruption cassettes for the FKH1 and FKH2 genes were obtained by PCR using the YDp-H (HIS3) and YDp-U (URA3) vectors (Berben et al., 1991), and then introduced into the strains AP-H1 and W303-1b, respectively, creating AP-H1F1 and AP-F2. Gene replacements were confirmed by PCR.
Myc tagging of FKH2
Fkh2 was tagged at the C-terminus with 13 Myc epitopes following the method of Longtine et al. (1998) using the pFA6a-13Myc-KanMX6 vector. A PCR product carrying a 13 Myc epitopes-Kan cassette was introduced into W303-1a and kanamycin-resistant transformants isolated. Tagging of FKH2 was confirmed by PCR in the new strain AP16.
RNA analysis
RNA extraction was performed following the protocol described by Aves et al. (1985), from cells of midlog and α-factor-synchronized cultures. Northern blots were performed as described previously (Morgan et al., 1995). Probes for RNA hybridization were either restriction fragments or PCR-generated fragments internal to the gene concerned labelled with [α-32P]dCTP (3000 Ci/mmol) with the Promega ‘Prime-a-Gene Labelling System’. The invariant RPB4 transcript was used as a loading control. Probed membranes were autoradiographed with Fuji Medical X-ray film (Super RX). Membranes were quantitated using a phosphorimager (Bio-imaging analyser Fujifilm Bas-1500) and Tina 2.0 software (raytest).
Light and fluorescence microscopy
For light microscopy, cells were photographed on Ektachrome 400T using an Axiophot (Zeiss) microscope set up for differential interference contrast. For fluorescence microscopy, nuclear [DAPI and fluorescein isothiocyanate (FITC)] and mitotic spindle (FITC) fluorescence were detected by exciting cells with a UV light at ∼365 nm (DAPI) and 450–490 nm (FITC) at 1000× magnification. Pictures were taken using colour Fuji Provia 1600 ASA film. Cells to be analyzed were treated essentially as described by Kilmartin and Adams (1984) except that lyticase (5 mg/ml in 50% glycerol, for 30 min at 30°C) was used. Monoclonal rat anti-tubulin antibody YOL1/34 and sheep anti-rat–FITC conjugate (both from Harlan Sera-Lab Ltd, UK), monoclonal mouse anti-Myc (9E10) IgG (BAbCO) and goat anti-mouse–FITC conjugate (Sigma) were used following the manufacturers’ instructions. DAPI (0.5 µg/ml; Sigma) was used for nuclear visualization. Slides were mounted using VECTASHIELD®.
Gel mobility shift assays
Yeast total cell extracts were prepared as described previously (Lydall et al., 1991). Double-stranded DNA probes were prepared by annealing pairs of synthetic oligonucleotides and radiolabelling with [α-32P]dCTP (3000 Ci/mmol) by fill-in of 5′ overhangs using Klenow enzyme, followed by gel purification: wild-type SWI5 (ADS490/ADS809; top strand: 5′-CTAGTTAACCTGTTTAGGAAAAAGGTAAACAATAAC-3′), mutant SWI5 [mut-SWI5; containing the A296 mutation (Lydall et al., 1991)] (ADS493/ADS810; top strand: 5′-CTAGTTAACCTGTTTAGGAAAAAGGTAAAAAATAAC-3′) and wild-type ACE2a (ADS799/ADS800 5′-CTAGATATCTCAAAACGGCAAAATGTAAACATTGGCA-3′). Gel mobility shift assays were carried out as described previously (Lydall et al., 1991). Supershift assays were carried out as for gel mobility shift assays except that the protein extract was pre-incubated with buffer or 200 ng of anti-Myc (9E10) mouse monoclonal IgG (Santa Cruz Biotech) prior to the addition of the DNA. Competition assays were carried out as for gel mobility shift assays except for the presence of 0-, 10- or 100-fold excess of cold competitor DNA prepared by annealing pairs of synthetic oligonucleotides. Specific competitor DNA contains a palindromic consensus Mcm1-binding site PPAL (ADS442/ADS443; top strand: 5′-CTAGGTAAATTTCCTAATTAGGAAAGTAC-3′) (Bruhn and Sprague, 1994) whereas the non-specific control competitor contains the consensus ETS protein-binding site E74 (ADS107/ADS108; 5′-CTAGAGCTGAATAACCGGAAGTAACTCAT-3′). The Mcm1-binding sites are underlined whereas the SFF-binding sites are shown in italics. The base altered in the A296 mutant in the SFF-binding motif in the SWI5 promoter is shown in bold.
Cell cycle phosphorylation assays
Protein extracts were obtained from cell cultures, synchronized with α-factor or treated with nocodazole, using 150 µl of lysis buffer [20 mM HEPES, 350 mM NaCl, 10% (v/v) glycerol, 0.1% Tween-20 containing protease inhibitors]. Sodium vanadate (2 mM) and sodium fluoride (50 mM) were also included when we wished to preserve the phosphorylation state of the extracts. Approximately 20 µg of protein were separated by SDS–PAGE, transferred to nitrocellulose membrane, blocked with 10% (w/v) bovine serum albumin (BSA) in 0.1% Tween-20 in Tris-buffered saline (TTBS) for 30 min at room temperature, then incubated with 1/1000 9E10 anti-Myc mouse monoclonal antibody (BAbCO) in 5% BSA in TTBS for 2–16 h. The membrane was washed with TTBS and incubated with 1/2000 horseradish peroxidase-conjugated anti-mouse IgG (Sigma) in 5% milk in TTBS for 45 min. Membranes were washed with TTBS then developed with ECL reagent (Amersham) and exposed to Fuji X-ray RX film. For phosphatase treatment, protein samples were treated with alkaline phosphatase (Roche) for 30 min at 37°C prior to western blot analysis as described above.
Acknowledgments
Acknowledgements
We thank David Lydall, Janet Quinn and Simon Whitehall for valuable discussions and advice on the manuscript. We also thank Trevor Jowett for his help with microscopy, and the Molecular Biology Centre, University of Newcastle, for DNA sequencing. A.P. was funded by a CRC PhD studentship, A.G.W. by an MRC studentship, S.J.R., E.A.V. and B.A.M. by the BBSRC and MRC, and F.L.L. and A.D.S. by the BBSRC and The Wellcome Trust, and a Lister Institute of Preventative Medicine Research Fellowship to A.D.S.
References
- Amon A., Tyers,M., Futcher,B. and Nasmyth,K. (1993) Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell, 74, 993–1007. [DOI] [PubMed] [Google Scholar]
- Aves S.J., Durkacz,B.W., Carr,A. and Nurse,P. (1985) Cloning, sequencing and transcriptional control of the Schizosaccharomyces pombe cdc10 ‘start’ gene. EMBO J., 4, 457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berben G., Dumont,J., Gilliquet,V., Bolle,P.A. and Hilger,F. (1991) The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast, 7, 475–477. [DOI] [PubMed] [Google Scholar]
- Bruhn L. and Sprague,G.F.,Jr (1994) MCM1 point mutants deficient in expression of α-specific genes: residues important for interaction with α1. Mol. Cell. Biol., 14, 2534–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher P. and Bairoch,A. (1994) A generalized profile syntax for biomolecular sequence motifs and its function in automatic sequence interpretation. Intell. Syst. Mol. Biol., 2, 53–61. [PubMed] [Google Scholar]
- Dohrmann P.R., Butler,G., Tamai,K., Dorland,S., Greene,J.R., Thiele,D.J. and Stillman,D.J. (1992) Parallel pathways of gene regulation: homologous regulators SWI5 and ACE2 differentially control transcription of HO and chitinase. Genes Dev., 6, 93–104. [DOI] [PubMed] [Google Scholar]
- Durocher D., Henckel,J., Fersht,A.R. and Jackson,S.P. (1999) The FHA domain is a modular phosphopeptide recognition motif. Mol. Cell, 4, 387–394. [DOI] [PubMed] [Google Scholar]
- Ghiara J.B., Richardson,H.E., Sugimoto,K., Henze,M., Lew,D.J., Wittenberg,C. and Reed,S.I. (1991) A cyclin B homolog in S.cerevisiae: chronic activation of the cdc28 protein kinase by cyclin prevents exit from mitosis. Cell, 65, 163–174. [DOI] [PubMed] [Google Scholar]
- Graves B.J. and Petersen,J.M. (1998) Specificity within the ets family of transcription factors. In van de Woude,G. and Klein,G. (eds), Advances in Cancer Research. Academic Press, San Diego, CA. [DOI] [PubMed] [Google Scholar]
- Hermann-Le Denmat S., Werner,M., Sentenac,A. and Thuriaux,P. (1994) Suppression of yeast RNA polymerase III mutations by FHL1, a gene coding for a fork head protein involved in rRNA processing. Mol. Cell. Biol., 14, 2905–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman C.S. and Winston,F. (1987) A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene, 57, 267–272. [DOI] [PubMed] [Google Scholar]
- Hollenhorst P.C., Bose,M.E., Mielke,M.R., Muller,U. and Fox,C.A. (2000) Forkhead genes in transcriptional silencing, cell morphology and the cell cycle: overlapping and distinct functions for FKH1 and FKH2 in Saccharomyces cerevisiae. Genetics, 154, 1533–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann E. and Knöchel,W. (1996) Five years on the wings of fork head. Mech. Dev., 57, 3–20. [DOI] [PubMed] [Google Scholar]
- Kaufmann E., Müller,D. and Knöchel,W. (1995) DNA recognition site analysis of Xenopus winged helix proteins. J. Mol. Biol., 248, 239–254. [DOI] [PubMed] [Google Scholar]
- Kilmartin J.V. and Adams,A.E.M. (1984) Structural rearrangements of tubulin and actin during cell cycle of the yeast Saccharomyces.J. Cell Biol., 98, 922–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King L. and Butler,G. (1998) Ace2p, a regulator of CTS1 (chitinase) expression, affects pseudohyphal production in Saccharomyces cerevisiae. Curr. Genet., 34, 183–191. [DOI] [PubMed] [Google Scholar]
- Koch C. and Nasmyth,K. (1994) Cell cycle regulated transcription in yeast. Curr. Opin. Cell Biol., 6, 451–459. [DOI] [PubMed] [Google Scholar]
- Kuo M.H., Nadeau,E.T. and Grayhack,E.J. (1997) Multiple phosphorylated forms of the Saccharomyces cerevisiae Mcm1 protein include an isoform induced in response to high salt concentrations. Mol. Cell. Biol., 17, 819–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E., Clark,K.L., Burley,S.K. and Darnell,J.E.,Jr (1993) Hepatocyte nuclear factor 3/fork head or ‘winged helix’ proteins: a family of transcription factors of diverse biologic function. Proc. Natl Acad. Sci. USA, 90, 10421–10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie,A., Demarini,D.J., Shah,N.G., Wach,A., Brachat,A., Philippsen,P. and Pringle,J.R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast, 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Lydall D., Ammerer,G. and Nasmyth,K. (1991) A new role for MCM1 in yeast: cell cycle regulation of SWI5 transcription. Genes Dev., 5, 2405–2419. [DOI] [PubMed] [Google Scholar]
- Maher M., Cong,F., Kindelberger,D., Nasmyth,K. and Dalton,S. (1995) Cell cycle-regulated transcription of the CLB2 gene is dependent on Mcm1 and a ternary complex factor. Mol. Cell. Biol., 15, 3129–3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerny C.J., Partridge,J.F., Mikesell,G.E., Creemer,D.P. and Breeden,L.L. (1997) A novel Mcm1-dependent element in the SWI4, CLN3, CDC6 and CDC47 promoters activates M/G1-specific transcription. Genes Dev., 11, 1277–1288. [DOI] [PubMed] [Google Scholar]
- Morgan B.A., Bouquin,N., Merrill,G.F. and Johnston,L.H. (1995) A yeast transcription factor bypassing the requirement for SBF and DSC1/MBF in budding yeast has homology to bacterial signal transduction proteins. EMBO J., 14, 5679–5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan D.O. (1995) Principles of CDK regulation. Nature, 374, 131–133. [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Seddon,A. and Ammerer,G. (1987) Cell cycle regulation of SWI5 is required for mother-cell-specific HO transcription in yeast. Cell, 49, 549–558. [DOI] [PubMed] [Google Scholar]
- Pati D., Keller,C., Groudine,M. and Plon,S.E. (1997) Reconstitution of a MEC1-independent checkpoint in yeast by expression of a novel human fork head cDNA. Mol. Cell. Biol., 17, 3037–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl R.H. and Gietz,R.D. (1989) High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet., 16, 339–346. [DOI] [PubMed] [Google Scholar]
- Sharrocks A.D., Brown,A.L., Ling,Y. and Yates,P.R. (1997) The ETS-domain transcription factor family. Int. J. Biochem. Cell Biol., 29, 1371–1387. [DOI] [PubMed] [Google Scholar]
- Sherman F., Fink,G.R. and Hicks,J.B. (1986) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Shore P. and Sharrocks,A.D. (1995) The MADS-box family of transcription factors. Eur. J. Biochem., 229, 1–13. [DOI] [PubMed] [Google Scholar]
- Spellman P.T., Sherlock,G., Zhang,M.Q., Iyer,V.R., Anders,K., Eisen,M.B., Brown,P.O., Botstein,D. and Futcher,B. (1998) Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell, 9, 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surana U., Robitsch,H., Price,C., Schuster,T., Fitch,I., Futcher,A.B. and Nasmyth,K. (1991) The role of CDC28 and cyclins during mitosis in the budding yeast S.cerevisiae. Cell, 65, 145–161. [DOI] [PubMed] [Google Scholar]
- Treisman R. (1994) Ternary complex factors: growth regulated transcriptional activators. Curr. Opin. Genet. Dev., 4, 96–101. [DOI] [PubMed] [Google Scholar]
- Wynne J. and Treisman,R. (1992) SRF and Mcm1 have related but distinct DNA binding specificities. Nucleic Acids Res., 20, 3297–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe M.B. and Cantley,L.C. (1999) Grabbing phosphoproteins. Nature, 402, 30–31. [DOI] [PubMed] [Google Scholar]
- Zhu G. and Davis,T. (1998) The fork head transcription factor Hcm1p participates in the regulation of SPC110, which encodes the calmodulin-binding protein in the yeast spindle pole body. Biochim. Biophys. Acta, 1448, 236–244. [DOI] [PubMed] [Google Scholar]
- Zhu G., Muller,E.G.D., Amacher,S.L., Northrop,J.L. and Davis,T.N. (1993) A dosage-dependent suppressor of a temperature-sensitive calmodulin mutant encodes a protein related to the fork head family of DNA-binding proteins. Mol. Cell. Biol., 13, 1779–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]