Abstract
AIDS chemotherapy is limited by inadequate intracellular concentrations of the active triphosphate form of nucleoside analogues, leading to incomplete inhibition of viral replication and the appearance of drug-resistant virus. Drug activation by nucleoside diphosphate kinase and inhibition of HIV-1 reverse transcriptase were studied comparatively. We synthesized analogues with a borano (BH3–) group on the α-phosphate, and found that they are substrates for both enzymes. X-ray structures of complexes with nucleotide diphosphate kinase provided a structural basis for their activation. The complex with d4T triphosphate displayed an intramolecular CH…O bond contributing to catalysis, and the Rp diastereoisomer of thymidine α-boranotriphosphate bound like a normal substrate. Using α-(Rp)-boranophosphate derivatives of the clinically relevant compounds AZT and d4T, the presence of the α-borano group improved both phosphorylation by nucleotide diphosphate kinase and inhibition of reverse transcription. Moreover, repair of blocked DNA chains by pyrophosphorolysis was reduced significantly in variant reverse transcriptases bearing substitutions found in drug-resistant viruses. Thus, the α-borano modification of analogues targeting reverse transcriptase may be of generic value in fighting viral drug resistance.
Keywords: CH…O bond/crystallography/HIV/nucleoside diphosphate kinase/pyrophosphorolysis
Introduction
The fight against human immunodeficiency virus (HIV) relies on the association of drugs directed towards two viral enzymes, the reverse transcriptase (Lightfoote et al., 1986) and the protease. Most drugs targeting reverse transcriptase are nucleoside analogues, such as 3′-azido 3′-deoxythymidine (AZT, Zidovudine) and 2′,3′-didehydro 2′,3′-dideoxythymidine (d4T, Stavudine). Antiviral nucleoside analogues per se are neither substrates nor inhibitors of reverse transcriptase. They must first be converted into triphosphates and compete with natural 2′-deoxynucleotides (dNTPs) for incorporation into viral DNA by reverse transcriptase. Because they lack a 3′-OH group, DNA chain elongation terminates, which accounts for the observed antiviral effect (Mitsuya et al., 1985; Larder, 1992; Balzarini, 1999). Their potency as a drug depends on their uptake by the infected cell, their phosphorylation by cellular kinases and their ability to block viral DNA synthesis. A single enzyme, nucleoside diphosphate kinase (NDPK), produces the triphosphate derivatives. In vitro, NDPK is 104 and 103 times less efficient on AZT and d4T diphosphate, respectively, than on thymidine diphosphate (Bourdais et al., 1996; Schneider et al., 1998, 2000). The efficacy of incorporation is limited by the low concentration of the triphos phate analogues relative to dNTPs in the infected cell. The poor activation of the drugs has a dramatic consequence: incomplete suppression of viral DNA synthesis allows the selection of resistance mutations (Larder, 1992). The emergence of variant viruses is the most serious threat against the efficacy of currently used combinations of three anti-retroviral drugs (tritherapies). The advent of second-generation drugs is urgent: ∼20 and 3% of HIV-1 isolates in western urban areas have nucleoside- and multidrug-resistant phenotypes, respectively (Wainberg and Friedland, 1998; Boden et al., 1999).
We conducted a structural and biochemical study of nucleoside analogue activation, which gives a basis to the rational design of novel drugs overcoming these limitations. Here we consider analogues where an oxygen of the α-phosphate is replaced with a borano (BH3–) group yielding α-(Rp)-boranophosphate. X-ray structures at 1.9 Å resolution show that the analogues bind NDPK like normal substrates. Biochemical data demonstrate that their activation by NDPK is enhanced, and that they provide reverse transcription inhibitors of increased efficiency. Moreover, repair of the analogue-terminated DNA chain by pyrophosphorolysis in drug-resistant reverse transcriptase mutants is greatly impeded.
Results
Synthesis of nucleotide analogues
We synthesized nucleotide analogues of thymidine, AZT and d4T containing a borano group (BH3–) on the α-phosphate. The borano group is isoionic to oxygen, and it generates a chiral centre when present on the α-phosphate of a dNTP (Figure 1). The two diastereo isomers were separated by reverse-phase high-performance liquid chromatography (HPLC). The same single diastereoisomer of unknown absolute configuration acted as an efficient substrate for both NDPK and HIV-1 reverse transcriptase.
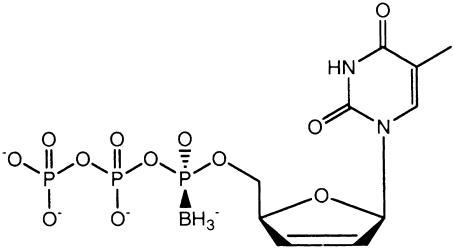
Fig. 1. Chemical formula of the α-(Rp)-borano-d4T triphosphate diastereoisomer.
Stereochemistry of α-borano TDP in complex with NDPK
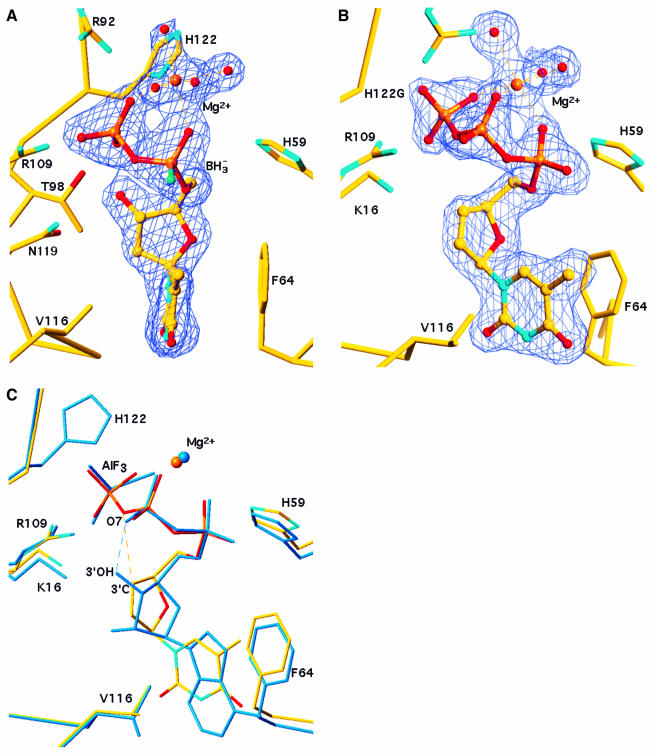
To determine the absolute configuration of the active diastereoisomer, we crystallized a complex of α-borano-thymidine diphosphate (TDP) with NDPK. NDPK transfers the γ-phosphate of a nucleoside triphosphate onto a nucleoside diphosphate via a phospho-histidine intermediate. Eukaryotic NDPKs are closely related. Human NDPK has a very similar structure to the Dictyostelium enzyme, and essentially the same active site (Dumas et al., 1992; Moréra et al., 1995; Webb et al., 1995). The crystals of Dictyostelium NDPK in complex with α-borano-TDP were isomorphous to a previously determined complex with ADP and aluminium fluoride, where AlF3 mimicks the γ-phosphate of ATP undergoing transfer onto the catalytic histidine (Xu et al., 1997b). The 1.92 Å resolution X-ray structure (Table I) demonstrates that the α-borano analogue binds like ADP and makes the same interactions (Figure 2A). The presence of boron with five electrons instead of oxygen with eight electrons is apparent in the electron density, which is consistently weaker at the Rp than the Sp position of the α-phosphate. Moreover, a Mg2+ ion ligates the α- and β-phosphates of the analogue in the same way as for natural nucleotides. As only the Rp diastereoisomer could be crystallized and the α-borano group does not interact with the protein, metal ligation by oxygen atoms of the phosphates is likely to determine the stereochemistry of recognition of the α-borano group by NDPK.
Table I. Summary of crystallographic data collection and refinement statistics.
| Complex | d4T triphosphate | α-borano-TDP |
|---|---|---|
| Resolution (Å) | 1.85 | 1.92 |
| Unique reflections | 37 405 | 33 469 |
| Average redundancy | 5.0 | 5.6 |
| Rsym (%)a | 5.6 (32.8) | 10.5 (32.7) |
| Completeness | 99.3 | 97.1 |
| Range for refinement (Å) | 30–1.85 | 30–1.92 |
| Total reflections used | 35 535 | 32 052 |
| Protein atoms | 3446 | 3446 |
| Solvent molecules | 346 | 400 |
| Ligand atoms | 84 | 75 |
| Average B-factor (Å2) | 24.3 | 28 |
| R.m.s.d. bond lengths (Å) | 0.012 | 0.010 |
| R.m.s.d. bond angles (°) | 1.7 | 1.5 |
| Rcryst (%)b | 20.9 | 22.8 |
| Rfree (%) | 24.2 | 27.4 |
aRsym = Σhi |I(h)i – <I(h)>|/Σhi I(h)i. Outer shell of resolution in parentheses.
bRcrys = Σh ||Fo| – |Fc||/Σh |Fo| calculated with no I/σ cut-off; Rfree is derived from 4% of the data set excluded from refinement.
Fig. 2. Nucleotide analogues bound to NDPK. (A) α-(Rp)-borano thymidine diphosphate bound to the wild-type Dictyostelium enzyme. The 2Fo – Fc electron density map at 1.92 Å resolution is contoured at 1σ. The borano group (BH3– in green) points towards the reader. An Mg2+ ion ligates the α- and the β-phosphate, and four water molecules (red spheres) complete the octahedral geometry. The thymine base is sandwiched between F64 and V116. The geometry of the boranophosphate group was taken from the crystal structure of a dimethyl ester (Summers et al., 1998) where the P–B and P–O bond lengths are 1.90 and 1.51 Å, respectively. (B) d4T triphosphate bound to the H122G variant. The 2Fo – Fc electron density map at 1.85 Å resolution is contoured at 1σ. The Mg2+ ion ligates all three phosphates. (C) Comparison with the natural substrate in the ADP–AlF3 complex (PDB file 1KDN; Xu et al., 1997b). Bonds in d4T triphosphate are atom-coloured, the ADP–AlF3 complex is in blue. AlF3 mimics the γ-phosphate undergoing transfer to His122, on top of the figure. In ADP, a hydrogen bond links the 3′-OH of the ribose to oxygen O7 of the β-phosphate. In d4T, the 3′-CH group makes a short (3.2 Å) contact with the equivalent oxygen, which can be interpreted as a CH…O bond (dashes). The superposition is based on all Cα positions. The figure was made with TURBO (Roussel and Cambillau, 1991).
A CH…O bond in d4T triphosphate bound to NDPK
To check whether this structural model is valid for antiviral nucleoside analogues used in AIDS therapy, we prepared a complex of d4T triphosphate with NDPK and determined its crystal structure at 1.85 Å resolution. A similar analysis has been performed on AZT derivatives bound to thymidylate kinase (Lavie et al., 1997) and to NDPK (Xu et al., 1997a). The present study makes use of the H122G mutant of the Dictyostelium enzyme. This mutant lacks the catalytic histidine and cannot autophos phorylate, but it binds ATP and transfers its γ-phosphate efficiently onto externally supplied imidazole (Admiraal et al., 1999). Again, the complex with d4T triphosphate was isomorphous with the ADP–AlF3 complex. The analogue binds like ADP, the additional γ-phosphate occupying the AlF3 position, and the Mg2+ ion interacts with all three phosphate groups (Figure 2B and C). Interactions with the protein are the same as for ADP–AlF3, with an important exception. In a natural substrate, the 3′-OH of ribose or deoxyribose receives two hydrogen bonds from protein groups, and donates one to the oxygen atom that is labelled O7 in Figure 2C and bridges the β- and γ-phosphates. The latter bond is particularly important in catalysis (Xu et al., 1997b; Gonin et al., 1999).
Instead of a 3′-OH, d4T carries a double bond in the sugar ring. We find that polar protein groups pack around the double bond making short contacts (3.2–3.4 Å) with the C2′ and C3′ atoms. The shortest contact is between C3′ and O7, the oxygen that bridges the β- and γ-phosphates. The comparison of our structural analysis with NDPK bound to AZT triphosphate suggests a reason why d4T derivatives are better substrates of NDPK than AZT and other analogues lacking a 3′-OH group. In AZT, the bulky 3′-azido substituent interferes with the proper positioning of a lysine side chain (K16 in Figure 2C) that is involved in catalysis (Xu et al., 1997a). With d4T triphosphate, this side chain and all other protein catalytic groups have the same position as in complexes with natural substrates such as ADP, GDP or TDP (Cherfils et al., 1994; Moréra et al., 1995).
Also, we noted the short distance between the modified sugar ring and the bridging oxygen O7. This oxygen is the leaving group when the γ-phosphate is transferred. In a natural substrate, the hydrogen bond to the sugar 3′-OH activates it (Figure 2C). In d4T triphosphate, the 3′-CH group points towards O7 in the plane of the double bond. The distance and geometry of the contact are consistent with a CH…O bond. CH…O bonds are observed in small molecule crystals, in proteins and in DNA (Neidle and Taylor, 1977; Derewenda et al., 1995; Mandel-Gutfreund et al., 1998). Although much weaker than the bond with the 3′-OH of a natural substrate, the CH…O7 bond may explain why d4T diphosphate is phosphorylated 10-fold faster than dideoxyTDP (Schneider et al., 1998, 2000). The dideoxy sugar has methylene groups instead of a double bond and their hydrogens are not oriented properly for an interaction with the bridging oxygen.
Activation of α-borano nucleotide analogues by NDPK
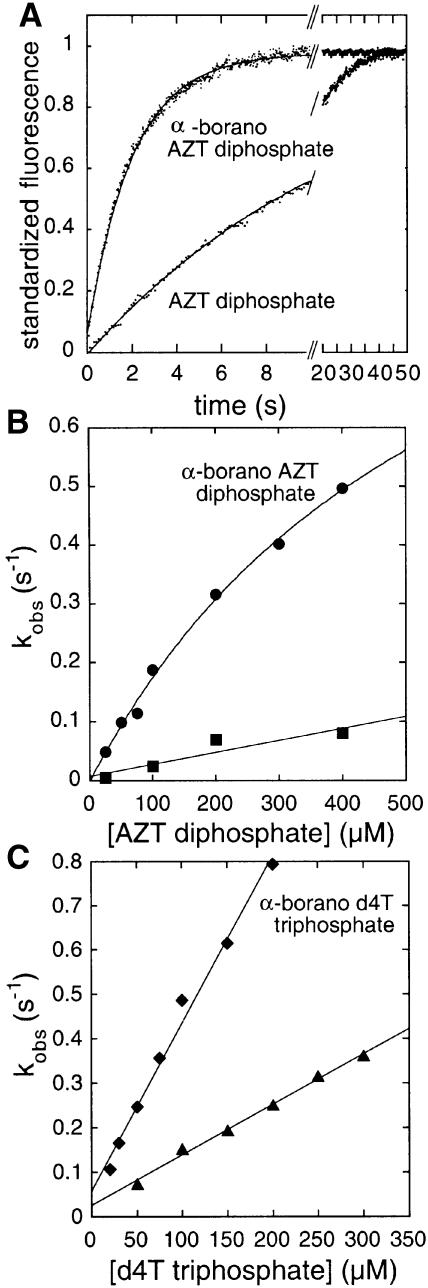
We examined pre-steady-state kinetic parameters of the phosphorylation of the α-borano diphosphate derivatives of AZT and d4T by Dictyostelium NDPK. Protein fluorescence is quenched upon phosphorylation of the protein by an NTP substrate and enhanced upon its dephosphorylation by NDP (Deville-Bonne et al., 1996). With AZT diphosphate as the phosphate acceptor, the reaction is 104-fold slower than with TDP (Schneider et al., 1998). When α-borano-AZT diphosphate is tested in this way, the Sp isomer is inactive but the Rp isomer is a better phosphate acceptor than AZT diphosphate (Figure 3A). Its phosphorylation follows an exponential time course with a rate constant kobs that increases with nucleotide concentration. The initial slope yields the catalytic efficiency of the reaction (Schneider et al., 1998), which is 10-fold higher for the α-borano compound (Figure 3B). A 10-fold enhancement of catalytic efficiency is also observed for the Rp isomer of α-borano TDP relative to TDP (Table II). As α-borano-d4T diphosphate was not available, we used α-borano-d4T triphosphate to phosphorylate NDPK and study phosphate transfer in the reverse direction. Again, the presence of the α-borano group in the Rp position results in an enhancement of the catalytic efficiency of the reaction relative to d4T triphosphate (Figure 3C; Table II).
Fig. 3. Enhancement of AZT and d4T activation by NDPK in the presence of the Rp α-borano group. (A) Kinetics of phosphorylation of AZT diphosphate and α-borano-AZT diphosphate by phosphorylated human NDPK A. The phosphorylated enzyme (1 µM), prepared as described (Deville-Bonne et al., 1996), is mixed with 400 µM AZT diphosphate or α-borano-AZT diphosphate in 50 mM Tris–HCl pH 7.5, 5 mM MgCl2, 75 mM KCl, 1 mM DTT and 5% glycerol (final concentrations) at 20°C. The decrease in fluorescence is monitored with a Hi-Tech SF-61DX2 stopped flow device (λexc = 304 nm, excitation slit = 2 nm, emission filter <320 nm). The solid lines represent the best fit of each curve to a monoexponential. (B) Concentration dependence of the pseudo-first order phosphorylation rate constant kobs: α-borano-AZT diphosphate (circles) and AZT diphosphate (squares). Data were analysed according to the classical model of a fast binding reaction followed by a slow, rate-limiting phosphotransfer (Schneider et al., 1998). The catalytic efficiency is kmax/KD = 2000 M–1s–1 for α-borano-AZT diphosphate and 200 M–1s–1 for AZT diphosphate. As no saturation is reached, the maximum rate of phosphotransfer, kmax, is not measurable in this experiment. (C) Concentration dependence of the pseudo-first order rate constant kobs for NDPK A phosphorylation by α-borano-d4T triphosphate (diamonds) and d4T triphosphate (triangles). The catalytic efficiency is kmax/KD = 6000 M–1s–1 for α-borano-d4T triphosphate and 800 M–1s–1 for d4T triphosphate.
Table II. NDPK binding and activation of α-(Rp)-borano thymidine nucleotide analogues.
| Nucleotide | Catalytic efficiency (M–1s–1)a | KD (µM)b | |
|---|---|---|---|
| TTP | – |
1 × 106 |
1.2 |
| AZT triphosphate | – | 80 | 30.0 |
| α-borano |
375 |
2.0 |
|
| d4T triphosphate | – | 800 | 2.0 |
| α-borano |
6000 |
0.25 |
|
| TDP | – | 2 × 106 | – |
| α-borano |
27 × 106 |
– |
|
| AZT diphosphate | – | 200 | – |
| α-borano |
2000 |
– |
|
| d4T diphosphate |
– |
2600 |
– |
| α-borano | ND | – |
aThe phosphotransfer reaction between human NDPK A and thymidine derivatives was studied in both directions as described in Figure 3. Note that the α-borano group increases the catalytic efficiency 4- to 10-fold.
bThe F64W-H122G variant of Dictyostelium NDPK is designed to measure NTP binding in the absence of phosphotransfer. Protein fluorescence (λexc = 310 nm, λem = 330 nm) increases by 50% upon binding saturating amounts of NTP at 20°C in 50 mM Tris–HCl, 5 mM MgCl2 and 75 mM KCl pH 7.5. KD values are estimated by fitting the binding curve to a quadratic equation.
The binding affinity for the analogues was measured using a variant of Dictyostelium NDPK that lacks the catalytic histidine and where a tryptophan replaces the phenylalanine stacking on the base in the active site (F64 in Figure 2B). In this variant, protein fluorescence changes upon nucleotide binding whilst no phosphorylation takes place (Deville-Bonne et al., 1996; Schneider et al., 2000). Binding isotherms show a 10-fold decrease in KD for the α-borano analogue relative to d4T triphosphate (not shown). Similar experiments with TTP, AZT triphosphate and their α-borano analogues also indicate a 10-fold decrease in KD when the borano group is present in the Rp position (Table II). We conclude that the increase in catalytic efficiency of α-boranophosphate derivatives results from an improvement in the affinity for the enzyme.
Inhibition of reverse transcription by α-borano nucleotide analogues
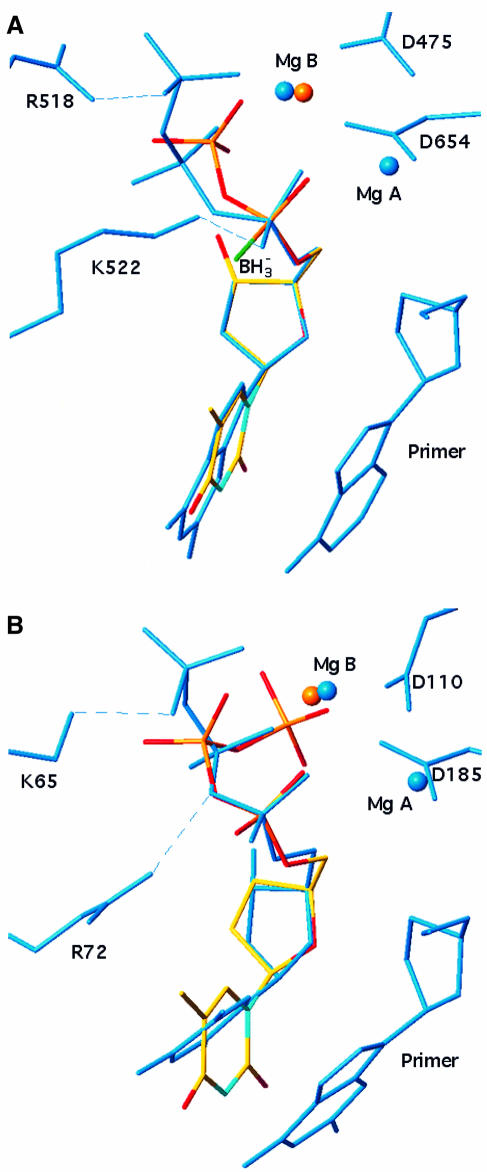
Examination of crystal structures of nucleotide substrates bound either to NDPK (this work) or to DNA polymerases such as reverse transcriptase (Huang et al., 1998) and bacteriophage T7 DNA polymerase (Doublié et al., 1998) shows that their NDP moieties have similar conformations. This is illustrated in Figure 4 by comparing d4T triphosphate from Figure 2B with dideoxyGTP bound to T7 DNA polymerase (Figure 4A; Doublié et al., 1998) and with deoxyTTP bound to reverse transcriptase (Figure 4B; Huang et al., 1998). Common features are the conformation of the α- and β-phosphates and the position of one of the bound Mg2+ ions. The similarity is especially striking in the case of T7 polymerase, which is a higher resolution (2 Å) structure than for reverse transcriptase, making details of the nucleotide conformation and the Mg2+ binding mode more reliable. In all three enzymes, the same oxygen of the α-phosphate ligates the metal, explaining why the other α-phosphate oxygen can be modified without interfering with activation by NDPK or incorporation in DNA by reverse transcriptase.
Fig. 4. Conformation of the nucleotide substrate in NDPK, T7 DNA polymerase and HIV reverse transcriptase. d4T triphosphate from the NDPK complex in Figure 2B is shown in atom-type coloured bonds superimposed onto (A) dideoxyGTP in the ternary complex with bacteriophage T7 DNA polymerase–DNA (PDB file 1T7P) and (B) deoxyTTP in the ternary complex with HIV reverse transcriptase–DNA (PDB file 1RTD). Least-square fitting was performed on atom N1 of the base and common atoms in the sugar and the α-phosphate. In T7 polymerase and reverse transcriptase, relevant active site residues, DNA and the ligand are in blue bonds, and blue spheres represent two Mg2+ ions bound. In NDPK, the red sphere is the single Mg2+ ion bound to d4T triphosphate. It is located <1 Å away from one of the two Mg2+ of the polymerases.
When the α-borano analogues of AZT and d4T triphosphate were tested for their ability to inhibit reverse transcription (Ueno et al., 1995), potent inhibition was observed only with the Rp analogue. The fact that only one diastereoisomer is a DNA polymerase substrate is consistent with results obtained by others using DNA-dependent DNA polymerases (Eckstein and Thomson, 1995). Chain terminator analogues produce a pattern of DNA polymerization inhibition similar to that of true competitive inhibitors (Reardon and Miller, 1990). The apparent inhibition constant Ki was determined for each analogue in triplicate. We used Lineweaver–Burk and IC50 analysis (Copeland, 1996). For AZT triphosphate, the two methods gave Ki values of 28 and 22 nM; they gave 2.8 and 8.2 nM for the α-borano analogue. For d4T triphos phate, we obtained 35 and 49 nM, and 15 and 20 nM for the α-borano analogue. Thus, Ki decreased by a factor of 2.2–10 in the presence of the α-borano group.
Michaelis–Menten parameters were measured using a primer extension assay and synthetic DNA-primed DNA template (Reardon and Miller, 1990) (Table III). The catalytic efficiency of incorporation (kcat/Km) of the α-borano analogues increases 9-fold for α-borano-AZT triphosphate and 3-fold for α-borano-d4T triphosphate. This results from a decrease in the Km values while kcat is unchanged (Table III). Using this primer–template system, we performed pre-steady-state kinetics that allow the measurement of KD for the analogues and their incorporation rate constant kpol (Kati et al., 1992; Hsieh et al., 1993) (Figure 5). The catalytic efficiency kpol/KD is ∼50% higher when the borano group is present (Table III). Unlike NDPK, this is due to an increase in kpol whereas the affinity is not significantly changed.
Table III. Kinetic parameters of wild-type reverse transcriptase for thymidine nucleotide analogues.
| TTP | AZT triphosphate |
d4T triphosphate |
|||
|---|---|---|---|---|---|
| – | α-borano | – | α-borano | ||
| Km (µM)a | 1.7 | 2.4 | 0.2 | 1.2 | 0.44 |
| kcat (s–1)a | 0.05 | 0.08 | 0.06 | 0.055 | 0.056 |
| kcat/Km (µM–1s–1) | 0.029 | 0.033 | 0.30 | 0.046 | 0.127 |
| KD (µM)b | 11c | 7.1 | 7.6 | 21.3 | 18.7 |
| kpol (s–1)b | 5.4c | 12.8 | 18.4 | 10.8 | 16 |
| kpol/KD (µM–1s–1) | 0.49c | 1.8 | 2.4 | 0.51 | 0.85 |
aA single-nucleotide incorporation and gel assay was used as described (Reardon and Miller, 1990) except that incubations were at 37°C, and the heteropolymeric DNA primer–template system as described (Canard et al., 1998). Km and kcat were obtained using Lineweaver–Burk or Eadie–Hofstee plots in which values of correlation coefficients were >0.98.
bKD and kpol were obtained as described in Figure 5 (Kati et al., 1992). Standard deviations were <16%.
cDeterminations carried out at 25°C and taken from Reardon and Miller (1990).
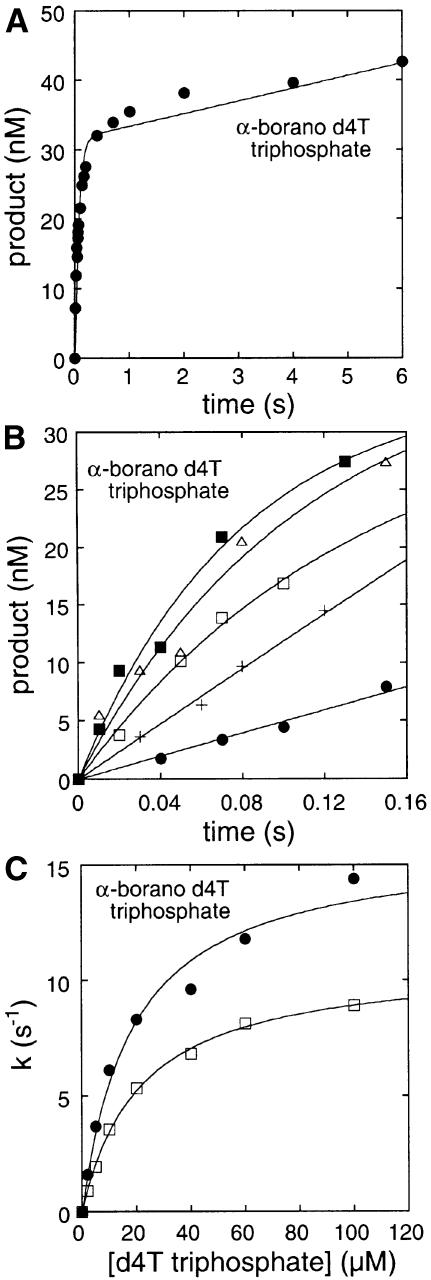
Fig. 5. Pre-steady-state kinetics of α-borano-d4T monophosphate and d4T monophosphate incorporation into DNA as described (Kati et al., 1992). (A) Pre-steady-state kinetics of a single α-borano-d4T monophosphate addition. A 5′-32P-labelled 21mer was annealed to a 31mer DNA template (100 nM) specifying a single thymidine insertion site immediately adjacent to the 3′ end of the primer, and reverse transcriptase (30 nM active sites) was allowed to bind (Canard et al., 1998). The reaction was initiated by the addition of 100 µM α-borano-d4T triphosphate, 6 mM MgCl2 in a rapid quench apparatus (Kin Tek, State college, PA) at 37°C, and quenched at various times with 0.3 M EDTA. Products were analysed by denaturing gel electrophoresis and quantitated using photostimulatable plates and a Fuji Imager. Data were fitted to the burst equation: product = A(1 – e–kt) + ksst, where A is the amplitude, k is the apparent pre-steady-state rate and kss is the steady-state rate. (B) Partial data set of pre-steady-state rates determined as described above using 2.5 (filled circles), 5 (crosses), 10 (open circles), 20 (open triangles) and 60 µM (filled squares) α-borano-d4T triphosphate, 6 mM MgCl2. (C) First-order rates determined as described above of α-borano-d4T triphosphate and d4T triphosphate plotted against nucleotide analogue concentration to determine KD (µM) and kpol (per second) using a hyperbolic fit of the data (Kati et al., 1992).
Targeting drug-resistant reverse transcriptase using α-borano analogues
In the crystal structure of reverse transcriptase in complex with DNA and deoxyTTP, the positive charges of R72 and K65 side chains interact with α- and γ-phosphate groups of the nucleotide, respectively (Figure 4B). As these side chains would be closest to the borano group in an Rp α-borano substrate analogue, the R72A and K65A substitutions were made in order to characterize putative interactions with the α-borano group. Unexpectedly and unlike wild-type reverse transcriptase, R72A reverse transcriptase discriminates against AZT triphosphate and d4T triphosphate 350- and 13-fold, respectively (Table IV). The activity of R72A reverse transcriptase is at least 25-fold lower than that of wild type (Table IV; Sarafianos et al., 1995), and mutations at position 72 are never observed in viral isolates. K65A reverse transcriptase also discriminates 6- to 7-fold against analogues. Therefore, both R72A and K65A confer resistance to AZT triphosphate and d4T triphosphate in vitro. In contrast, neither variant reverse transcriptase discriminates against α-borano analogues (Table IV). We conclude that an intricate pattern of interactions between residues 72 and 65, the phosphate groups and the sugar 3′ position of the substrate nucleotide mediates the AZT triphosphate and d4T triphosphate resistance observed here in vitro.
Table IV. Inhibition of wild-type and drug-resistant HIV-1 reverse transcriptase by nucleotide analogues.
| Reverse transcriptase | Wild type | R72A | K65A | K65R |
|---|---|---|---|---|
| Relative catalytic efficiencya | 1 | 0.04 | 0.46 | 0.96 |
| AZT triphosphate | ||||
| Ki (nM)b | 21.8 | 6500 | 125 | 20.5 |
| α-borano Ki (nM)b | 8.25 | 32.5 | 9 | 9 |
| Ratio | 2.6 | 200 | 13.9 | 2.2 |
| d4T triphosphate | ||||
| Ki (nM)b | 49.2 | 500 | 275 | 60 |
| α-borano Ki (nM)b | 20.3 | 5 | 27.5 | 19.5 |
| Ratio | 2.4 | 100 | 10 | 3 |
aOligo(dT)12–18–poly(rA) primer–template, TTP as the nucleotide substrate (Ueno et al., 1995). The catalytic efficiency is measured relative to wild-type reverse transcriptase (0.63 µM–1s–1).
bThese Ki values were determined as described (Ueno et al., 1995), from three and two independent experiments for wild-type and variant reverse transcriptases, respectively.
This prompted us to examine the K65R variant, a substitution found in HIV-1 strains cross-resistant to deoxynucleosides (Gu et al., 1994; Foli et al., 1996). Introduction of an α-borano group in AZT triphosphate and d4T triphosphate enhances the affinity of the analogue for K65R reverse transcriptase by a factor of 2 and 3, respectively (Table IV), indicating that the α-borano substitution might also be of interest on other non-thymine nucleoside analogues.
Reverse transcriptase-catalysed pyrophosphorolytic repair of blocked DNA chains
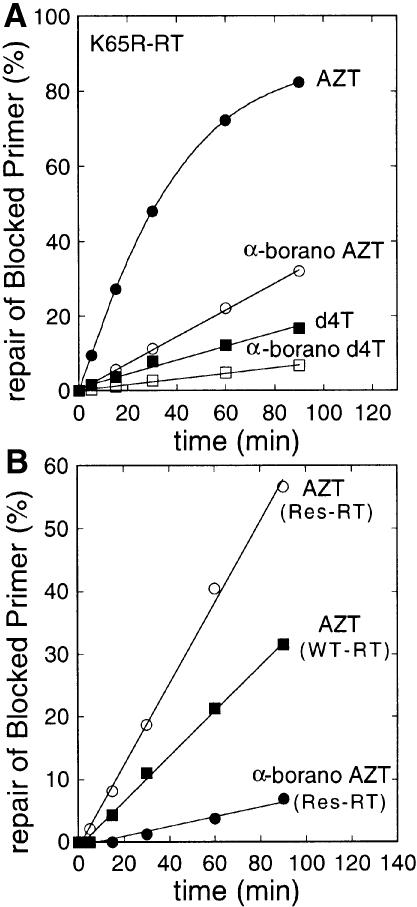
Drug resistance to nucleoside analogues by HIV sometimes correlates poorly with the loss of affinity of the corresponding nucleotide for reverse transcriptase in vitro (Lacey et al., 1992; Larder, 1992; Gu et al., 1994; Arion et al., 1996). For instance, AZT resistance has been shown to involve a reverse transcriptase-catalysed repair mechanism by excision of the analogue from the viral DNA chain (Arion et al., 1998; Meyer et al., 1999). The DNA 3′ end can be unblocked by the nucleophilic attack of either pyrophosphate or ATP. Repair of AZT-terminated DNA by the two drug-resistant reverse transcriptases used above was examined using a gel assay (Figure 6). During DNA polymerization in the presence of added pyrophos phate, the appearance of full-length extension products past the analogue insertion site indicates nucleotide incorporation made possible through pyrophosphorolysis of the terminal AZT nucleotide. Strikingly, the rate of the excision reaction by K65R reverse transcriptase decreases 5-fold in the presence of an α-borano group (Figure 7A). A similar effect is observed when comparing d4T triphos phate and its α-borano counterpart. When the variant reverse transcriptase carrying the AZT resistance mutations D67N/K70R/T215F/K219Q is used, increased pyro phosphorolysis of the unsubstituted analogue (∼2-fold) is observed (Figure 7B). Again, incorporation of the α-borano analogue reduces pyrophosphorolysis 9-fold.
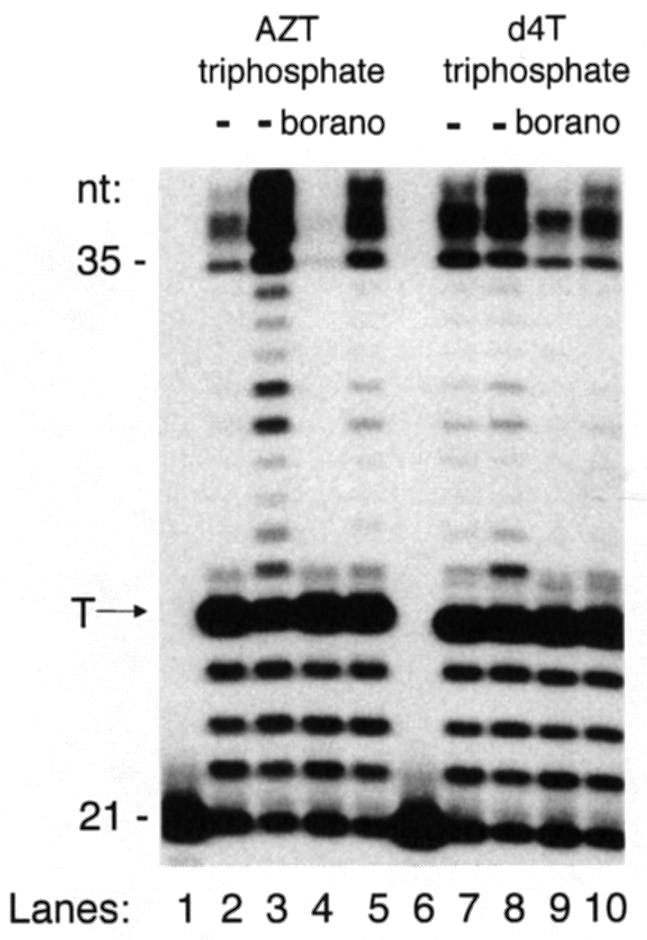
Fig. 6. Pyrophosphorolytic repair of incorporated α-borano thymidine 5′-monophosphate analogues by drug-resistant reverse transcriptase. Incorporation of α-borano thymidine 5′-triphosphosphate analogues in the presence of pyrophosphate. A 5′-32P-labelled 21mer was annealed to a 35mer DNA template (50 nM) specifying a single thymidine insertion site (‘T’ arrow) four bases away from the 3′ end of the primer. Polymerization was initiated by the addition of K65R reverse transcriptase (100 nM) and nucleotides (50 µM each of dATP, dCTP, dGTP and analogue-TP, and 5 µM TTP) for 15 min at 37°C. The pyrophosphorolytic repair reaction was initiated by the addition of 1 mM pyrophosphate (PPi). Aliquots were withdrawn during the time course of the reaction, and products analysed by denaturing gel electrophoresis. This autoradiograph of the gel shows the unextended primer (control, lane 1), and products at zero (even lanes) and 60 min (odd lanes). Over time, the amount of products >25 nucleotides does not change when PPi is omitted (not shown), but increases in the presence of PPi due to pyrophosphorolytic repair of the analogue-terminated DNA chain followed by DNA polymerization. Lanes 2, 3, 6 and 7, unsubstituted analogues; lanes 4, 5, 8 and 9, α-borano analogues.
Fig. 7. Time course of the pyrophosphorolytic repair reaction under reaction conditions shown above. (A) Variant K65R reverse transcriptase. Polymerization products were quantitated from an experiment similar to that in Figure 6 using photostimulatable plates and a Fuji Imager. The percentage repair of blocked primer is the ratio of extension products >25 nucleotides over those >24 nucleotides. (B) Same as in (A) but using wild-type and AZT-resistant (D67N/K70R/T215F/Y/K219Q) reverse transcriptase in conjunction with either AZT triphosphate or α-borano-AZT triphosphate as indicated.
We conclude that the presence of the α-borano group confers increased stability towards pyrophosphorolytic repair to the blocked DNA chain.
Discussion
The present study shows that the mode of binding of the nucleotide substrate has common features in NDPK and in DNA polymerases such as that of bacteriophage T7 and HIV reverse transcriptase. The two classes of enzymes, which are otherwise very different, catalyse a nucleophilic attack on either the γ-phosphate (NDPK) or the α-phosphate (the polymerases) of the substrate nucleotide. The common features of the bound substrate observed in X-ray structures make a modification of the α-phosphate compatible with both the activation into a triphosphate and its incorporation into the growing DNA chain. The substitution of a BH3– group for an oxygen yieding an α-(Rp)-phosphate actually enhances the rate of phosphorylation by NDPK and makes the product a more efficient inhibitor of reverse transcriptase. In contrast, the Sp derivative is inactive.
Our study shows that stereospecificity originates from the requirement that the nucleotide analogue interacts with a Mg2+ ion, which has the same ligation geometry in NDPK as in reverse transcriptase. This requirement applies to natural substrates such as thymidine nucleotides, and also to the α-boranophosphate derivatives of AZT and d4T, two thymidine analogues that are major drugs against AIDS. In addition, we find that the sugar modification in d4T makes the diphosphate derivative a better substrate for NDPK than AZT or dideoxy analogues. Whilst all these analogues lack a 3′-OH group involved in catalysis by NDPK, the geometry of the C2′–C3′ double bond in d4T allows the formation of an intramolecular CH…O hydrogen bond that partly substitutes for the missing 3′-OH...O. The 10-fold rate enhancement is highly significant, and may suffice to change the rate-limiting step for activation, i.e. the formation of the monophosphate form for d4T and the di- and triphosphate form for AZT (Lavie et al., 1997; Schneider et al., 2000). Moreover, the implication of a CH…O bond in enzymatic catalysis is a rare observation deserving notice in this context.
In the NDPK reaction, the α-phosphate is involved only through its contribution to Mg2+ ligation. We find that its modification into a boranophosphate enhances catalysis if it does not prevent metal binding, i.e. for the Rp diastereoisomer. The affinity of the enzyme for α-(Rp)-boranophosphate derivatives is improved by an order of magnitude, increasing the catalytic efficiency for the natural substrate TDP, and also for AZT and d4T analogues. Taking place in vivo, this 10-fold increase in efficiency would be of great value in the case of AZT, which is converted very poorly into a triphosphate by cellular enzymes.
The better activation by NDPK is only one of the elements making α-boranophosphate nucleotide ana logues attractive. The major limitation of current therapeutic protocols against AIDS is the emergence of resistant reverse transcriptase variants in viral isolates. Novel nucleoside analogues should either elicit less resistance or retain activity on reverse transcriptase variants. To design such second-generation drugs, nucleoside activation and reverse transcription inhibition should be taken as a whole, as the low concentration of the active form of the drugs in HIV-infected cells contributes to the emergence of drug resistance. Due to incomplete inhibition of viral replication, escape mutants are selected quickly, taking over wild-type viral populations, and conferring a drug resistance phenotype to the virus. It is shown here that the presence of a borano group on the α-phosphate of both AZT and d4T derivatives results in an enhanced activation by NDPK and an enhanced incorporation into DNA by reverse transcriptase.
Moreover, α-borano derivatives exhibit increased stability towards repair mechanisms that contribute to drug resistance. Resistance observed in vivo cannot be attributed simply to a loss of affinity of reverse transcriptase for nucleotide analogues. Mutant reverse transcriptase isolates from AZT-resistant viruses often display little in vitro resistance to inhibition of DNA synthesis. Rather, the hallmark of AZT resistance is an increased pyrophos phorolysis allowing repair of the blocked DNA chain. The reason may be a longer lifetime of the reverse transcriptase–DNA complex allowing pyrophosphorolysis, and also polyphosphate synthesis, to proceed at the active site (Arion et al., 1998; Canard et al., 1998; Meyer et al., 1999). Consistent with these findings, AZT-resistant reverse transcriptase (D67N/K70R/T215F/K219Q) exhibits an increased processivity of DNA synthesis (Caliendo et al., 1996). Our assay demonstrates that pyrophosphorolysis increases 2-fold in AZT-resistant reverse transcriptase, and that it is greatly reduced after incorporation of the α-borano analogue. Therefore, compounds of this class are good candidates to target selectively reverse transcriptases made drug resistant by mutations enhancing repair of analogue-terminated DNA chains.
In contrast, mutations such as M184V or L74V leading to resistance to dideoxynucleotides, d4T or 3TC tend to reduce processivity (Back et al., 1996; Sharma and Crumpacker, 1999). Nevertheless, α-borano analogues may still prove useful in that case. Resistance may be due to the analogue-terminated DNA chain being released more readily from the enzyme and made available for repair by cellular exonucleases. As the 3′–5′ borano–phosphodiester bond is resistant to cleavage by nucleases (Eckstein and Thomson, 1995; He et al., 1999), the α-borano group should have a protective effect against repair.
These experiments suggest that α-borano analogues may be of use in vivo against variant viruses that have become resistant by several repair mechanisms. The design of an α-borano d4T diphosphate pro-drug using nanoparticle-based biovectors and/or phosphotriester nucleotide analogues is being explored. It would bypass two steps of intracellular phosphorylation, including the step catalysed by thymidine kinase that appears to be rate limiting in forming d4T triphosphate. More generally, boranophosphate nucleotide analogues should be of interest in the study of all enzymes carrying out phosphotransfer reactions, of which NDPK and reverse transcriptase are two examples.
Materials and methods
Synthesis of nucleotide analogues
AZT di- and triphosphate were prepared from AZT monophosphate as described (Hoard and Ott, 1965). α-borano-AZT 5′-diphosphate was synthesized according to the procedure described (Tomasz et al., 1992), but the phosphitylation step of the nucleotide was performed as described (Meier et al., 1992). α-borano-AZT 5′-triphosphate and α-borano-d4T 5′-triphosphate were synthesized as described (He et al., 1998). NMR assignments are available upon request. Diastereoisomers were purified (>99%) by reverse-phase HPLC on a C18 column using a 0–15% acetonitrile gradient in 10 mM triethyl ammonium acetate buffer pH 6.8.
Enzyme purification
Human NDPK A was purified as a recombinant protein overexpressed in Escherichia coli (BL21-DE3). Plasmid pJC20 was a kind gift from M.Konrad (Göttingen, Germany). After cell breakage in a French press, and centrifugation (10 000 g, 20 min), the cell extract was treated with 0.15% polymin P (BASF) in order to precipitate most of the nucleic acids, then NDPK A was purified by two successive steps of ion-exchange chromatography on a POROS HQ column (PerSeptive Biosystem; 2.6 × 12 cm) with a linear KCl gradient (0.02–1 M) in 50 mM Tris–HCl pH 8.1, 1 mM EDTA and 1 mM dithiothreitol (DTT), followed by a ceramic hydroxyapatite column (American International Chemical; 1.6 × 10 cm) with a linear potassium phosphate gradient (0.01–1 M) in 10 mM potassium phosphate pH 6.5 and 1 mM DTT. After concentration and desalting, the enzyme was dialysed extensively against 50 mM Tris–HCl pH 7.5, 1 mM DTT, 20 mM KCl and 50% glycerol, then stored at –20°C. NDPK A was >97% pure as judged by SDS–PAGE.
The double mutant F64W-H122G NDPK from Dictyostelium was overexpressed in E.coli (XL1-Blue) and purified as previously described for the single mutant F64W NDPK. Enzyme concentration expressed as 17 kDa subunits was determined either colorimetrically or using an absorbance coefficient of ΔA280 = 0.85 for a 1 mg/ml solution. F64W-H122G mutant NDPK had no measurable NDPK activity and a very weak ATPase activity (kcat/Km = 450 M–1s–1). The affinity of NDP and NTP derivatives for NDPK was determined by following the variation of the intrinsic fluorescence upon nucleotide binding as described (Schneider et al., 1998). The correction for the contribution of ATPase activity was <2%.
HIV-1 reverse transcriptases were expressed in E.coli, and p66–p66 homodimers were purified as described (Canard et al., 1999). Wild-type reverse transcriptase used in pre-steady-state kinetic experiments was purifed as p66–p51 heterodimers as described (Canard et al., 1998).
Crystallographic methods
Crystals of the H122G variant of Dictyostelium NDPK (Admiraal et al., 1999) grew in drops containing 19% PEG 400, 50 mM Tris–HCl pH 7.5, 20 mM MgCl2, 6 mg/ml protein and 7.5 mM d4T triphosphate over wells containing 38% PEG 400. They belong to space group P3121 with a = b = 70.02 Å and c = 152.10 Å and a trimer in the asymmetric unit. Diffraction data were collected from a cryocooled crystal at λ = 1.364 Å on station D41 of the LURE-DCI synchrotron radiation source (Orsay, France) using an MAResearch Image plate detector, and processed using the HKL program package (Otwinowski and Minor, 1997). The initial model was the protein moiety of the ADP–AlF3 complex (PDB file 1KDN). It was refined with CNS using anisotropic overall B-factors and a bulk solvent mask (Brünger et al., 1998). Statistics on data collection and the refined model are given in Table I.
Co-crystals of α-borano-TDP with wild-type Dictyostelium NDPK were obtained by seeding with crystals of the ADP–AlF3 complex (Xu et al., 1997b). The drops contained 20% PEG 550 monomethylester, 50 mM Tris–HCl pH 7.5, 25 mM MgCl2, 7.5 mg/ml protein and 11 mM α-borano-TDP over wells containing 28% PEG 550 in the same buffer. The first crystals were then used for macro-seeding under the same conditions. They belong to space group P3121 with a = b = 70.36 Å and c = 153.79 Å and a trimer in the asymmetric unit. Before flash-freezing in liquid propane, PEG 550 was replaced with 39% PEG 400. A flash-frozen crystal was maintained at 100 K in an MSC cryostream while diffraction data were collected on a rotating anode source (Rigaku) using the R-Axis IIc image plate system. Diffracted amplitudes were indexed and integrated using MOSFLM (Leslie, 1992) and scaled with SCALA of the CCP4 package. All subsequent steps were the same as for the α-borano-TDP complex. For model refinement with CNS, the geometry of the boranophosphate group was taken from the crystal structure of a dimethyl ester (Summers et al., 1998) where the P–B and P–O bond lengths are 1.90 and 1.51 Å, respectively. Statistics on data collection and the refined model are given in Table I.
Nucleotide analogue incorporation by reverse transcriptase and pyrophosphorolysis assays
Standard reverse transcriptase and nucleotide selectivity assays were as described (Canard et al., 1999). Analysis of data was performed according to published procedures (Henderson, 1972; Cheng and Prusoff, 1973; Copeland, 1996) as described in the text. The primer–template system used for primer extension assays has been described (Canard et al., 1998). It was used for both steady-state and pre-steady-state kinetic assays. Experimental conditions are given in the figure legends. The primer–template system used for pyrophosphorolysis assays was a 21mer (5′-ATACTTTAACCATATGTACC-3′) annealed to a 35mer (5′-GGT CCGTTGCATGCGGATACATATGGTTAAAGTAT-3′) DNA template. The single template ‘A’ specifying a single thymidine insertion site four bases away from the 3′ end of the primer is shown in bold. Experimental conditions are given in the figure legends.
Acknowledgments
Acknowledgements
We thank Tom Ellenberger, Jeff Stock, Herman van Tilbeurgh, Christian Cambillau and Yves Bourne for critical reading of the manuscript, and Fabrice Agou for help in the purification of NDPK. We thank the Agence Nationale de la Recherche contre le SIDA, the Ensemble contre le SIDA and the Association pour la Recherche sur le Cancer for financial support. We acknowledge help from staff of the LURE-DCI synchrotron radiation centre (Orsay, France) in data collection.
Note added in proof
Co-ordinates have been deposited at the Protein Data Bank (accession Nos 1F6T and 1F3F).
References
- Admiraal S.J., Schneider,B., Meyer,P., Janin,J., Veron,M., Deville-Bonne,D. and Herschlag,D. (1999) Nucleophilic activation by positioning in phosphoryl transfer catalyzed by nucleoside diphosphate kinase. Biochemistry, 38, 4701–4711. [DOI] [PubMed] [Google Scholar]
- Arion D., Borkow,G., Gu,Z., Wainberg,M.A. and Parniak,M.A. (1996) The K65R mutation confers increased DNA polymerase processivity to HIV-1 reverse transcriptase. J. Biol. Chem., 271, 19860–19864. [DOI] [PubMed] [Google Scholar]
- Arion D., Kaushik,N., McCormick,S., Borkow,G. and Parniak,M.A. (1998) Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry, 37, 15908–15917. [DOI] [PubMed] [Google Scholar]
- Back N.K., Nijhuis,M., Keulen,W., Boucher,C.A., Oude Essink,B.O., van Kuilenburg,A.B., van Gennip,A.H. and Berkhout,B. (1996) Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J., 15, 4040–4049. [PMC free article] [PubMed] [Google Scholar]
- Balzarini J. (1999) Suppression of resistance to drugs targeted to human immunodeficiency virus reverse transcriptase by combination therapy. Biochem. Pharmacol., 58, 1–27. [DOI] [PubMed] [Google Scholar]
- Boden D. et al. (1999) HIV-1 drug resistance in newly infected individuals. J. Am. Med. Assoc., 282, 1135–1141. [DOI] [PubMed] [Google Scholar]
- Bourdais J., Biondi,R., Sarfati,S., Guerreiro,C., Lascu,I., Janin,J. and Veron,M. (1996) Cellular phosphorylation of anti-HIV nucleosides. Role of nucleoside diphosphate kinase. J. Biol. Chem., 271, 7887–7890. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Caliendo A.M., Savara,A., An,D., DeVore,K., Kaplan,J.C. and D’Aquila,R.T. (1996) Effects of zidovudine-selected human immunodeficiency virus type 1 reverse transcriptase amino acid substitutions on processive DNA synthesis and viral replication. J. Virol., 70, 2146–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canard B., Sarfati,S.R. and Richardson,C.C. (1998) Enhanced binding of azidothymidine-resistant human immunodeficiency virus 1 reverse transcriptase to the 3′-azido-3′-deoxythymidine 5′-monophosphate-terminated primer. J. Biol. Chem., 273, 14596–14604. [DOI] [PubMed] [Google Scholar]
- Canard B., Chowdhury,K., Sarfati,R., Doublié,S. and Richardson,C.C. (1999) The motif D loop of human immunodeficiency virus type 1 reverse transcriptase is critical for nucleoside 5′-triphosphate selectivity. J. Biol. Chem., 274, 35768–35776. [DOI] [PubMed] [Google Scholar]
- Cheng Y. and Prusoff,W.H. (1973) Relationship between the inhibition constant (Ki) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol., 22, 3099–3108. [DOI] [PubMed] [Google Scholar]
- Cherfils J., Moréra,S., Lascu,I., Véron,M. and Janin,J. (1994) X-ray structure of nucleoside diphosphate kinase complexed with thymidine diphosphate and Mg++ at 2 Å resolution. Biochemistry, 33, 9062–9069. [DOI] [PubMed] [Google Scholar]
- Copeland R.A. (1996) Enzymes. A Practical Introduction to Structure, Mechanism and Data Analysis. Wiley-VCH, New York, NY. [Google Scholar]
- Derewenda Z.S., Lee,L. and Derewenda,U. (1995) The occurrence of C-H...O hydrogen bonds in proteins. J. Mol. Biol., 252, 248–262. [DOI] [PubMed] [Google Scholar]
- Deville-Bonne D., Sellam,O., Merola,F., Lascu,I., Desmadril,M. and Veron,M. (1996) Phosphorylation of nucleoside diphosphate kinase at the active site studied by steady-state and time-resolved fluorescence. Biochemistry, 35, 14643–14650. [DOI] [PubMed] [Google Scholar]
- Doublié S., Tabor,S., Long,A.M., Richardson,C.C. and Ellenberger,T. (1998) Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature, 391, 251–258. [DOI] [PubMed] [Google Scholar]
- Dumas C., Lascu,I., Morera,S., Glaser,P., Fourme,R., Wallet,V., Lacombe,M.L., Veron,M. and Janin,J. (1992) X-ray structure of nucleoside diphosphate kinase. EMBO J., 11, 3203–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckstein F. and Thomson,J.B. (1995) Phosphate analogs for study of DNA polymerases. Methods Enzymol., 262, 189–202. [DOI] [PubMed] [Google Scholar]
- Foli A. et al. (1996) In vitro selection and molecular characterization of human immunodeficiency virus type 1 with reduced sensitivity to 9-[2-(phosphonomethoxy)ethyl]adenine (PMEA). Antiviral Res., 32, 91–98. [DOI] [PubMed] [Google Scholar]
- Gonin P., Xu,Y., Milon,L., Dabernat,S., Morr,M., Kumar,R., Lacombe,M.L., Janin,J. and Lascu,I. (1999) Catalytic mechanism of nucleoside diphosphate kinase investigated using nucleotide analogues, viscosity effects and X-ray crystallography. Biochemistry, 38, 7265–7272. [DOI] [PubMed] [Google Scholar]
- Gu Z., Fletcher,R.S., Arts,E.J., Wainberg,M.A. and Parniak,M.A. (1994) The K65R mutant reverse transcriptase of HIV-1 cross-resistant to 2′,3′-dideoxycytidine, 2′,3′-dideoxy-3′-thiacytidine and 2′,3′-dideoxy inosine shows reduced sensitivity to specific dideoxynucleoside triphosphate inhibitors in vitro. J. Biol. Chem., 269, 28118–28122. [PubMed] [Google Scholar]
- He K., Hasan,A., Krzyzanowska,B. and Shaw,B.R. (1998) Synthesis and separation of diastereomers of ribonucleoside 5′-(α-P-borano) triphosphates. J. Org. Chem., 63, 5769–5773. [DOI] [PubMed] [Google Scholar]
- He K., Porter,K.W., Hasan,A., Briley,J. and Shaw,B.R. (1999) Synthesis of 5-substituted 2′-deoxycytidine 5′-(α-P-borano)triphosphates, their incorporation into DNA and effects on exonuclease. Nucleic Acids Res., 27, 1788–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson P.J. (1972) A linear equation that describes the steady-state kinetics of enzymes and subcellular particles interacting with tightly bound inhibitors. Biochem. J., 127, 321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoard D.E. and Ott,D.G. (1965) Conversion of mono- and oligodeoxyribonucleotides to 5′-triphosphates. J. Am. Chem. Soc., 87, 1785–1788. [DOI] [PubMed] [Google Scholar]
- Hsieh J.C., Zinnen,S. and Modrich,P. (1993) Kinetic mechanism of the DNA-dependent DNA polymerase activity of human immunodeficiency virus reverse transcriptase. J. Biol. Chem., 268, 24607–24613. [PubMed] [Google Scholar]
- Huang H., Chopra,R., Verdine,G.L. and Harrison,S.C. (1998) Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science, 282, 1669–1675. [DOI] [PubMed] [Google Scholar]
- Kati W.M., Johnson,K.A., Jerva,L.F. and Anderson,K.S. (1992) Mechanism and fidelity of HIV reverse transcriptase. J. Biol. Chem., 267, 25988–25997. [PubMed] [Google Scholar]
- Lacey S.F., Reardon,J.E., Furfine,E.S., Kunkel,T.A., Bebenek,K., Eckert,K.A., Kemp,S.D. and Larder,B.A. (1992) Biochemical studies on the reverse transcriptase and RNase H activities from human immunodeficiency virus strains resistant to 3′-azido-3′-deoxythymidine. J. Biol. Chem., 267, 15789–15794. [PubMed] [Google Scholar]
- Larder B. (1992) Reverse transcriptase inhibitors and drug resistance. In Skalka,A.M. and Goff,S.P. (eds), Reverse Transcriptase. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 205–222. [Google Scholar]
- Lavie A., Vetter,I.R., Konrad,M., Goody,R.S., Reinstein,J. and Schlichting,I. (1997) Structure of thymidylate kinase reveals the cause behind the limiting step in AZT activation. Nature Struct. Biol., 4, 601–604. [DOI] [PubMed] [Google Scholar]
- Leslie A.G.W. (1992) Recent changes to the MOSFLM package for film and image plate data. Joint CCP4 and ESF-EACMB Newsletter on Protein Crystallography, Warrington, UK, Vol. 26. [Google Scholar]
- Lightfoote M.M., Coligan,J.E., Folks,T.M., Fauci,A.S., Martin,M.A. and Venkatesan,S. (1986) Structural characterization of reverse transcriptase and endonuclease polypeptides of the acquired immunodeficiency syndrome retrovirus. J. Virol., 60, 771–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel-Gutfreund Y., Margalit,H., Jernigan,R.L. and Zhurkin,V.B. (1998) A role for CH…O interactions in protein–DNA recognition. J. Mol. Biol., 277, 1129–1140. [DOI] [PubMed] [Google Scholar]
- Meier C., Neumann,J.M., André,F., Henin,Y. and Huynh-Dinh,T. (1992) O-alkyl-5′,5′-dinucleoside phosphates as prodrugs of 3′-azidothymidine and cordycepin. J. Org. Chem., 57, 7300–7308. [Google Scholar]
- Meyer P.R., Matsuura,S.E., Mian,A.M., So,A.G. and Scott,W.A. (1999) A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol. Cell, 4, 35–43. [DOI] [PubMed] [Google Scholar]
- Mitsuya H., Weinhold,K.J., Furman,P.A., St Clair,M.H., Lehrman,S.N., Gallo,R.C., Bolognesi,D., Barry,D.W. and Broder,S. (1985) 3′-Azido-3′-deoxythymidine (BW A509U): an antiviral agent that inhibits the infectivity and cytopathic effect of human T-lymphotropic virus type III/lymphadenopathy-associated virus in vitro. Proc. Natl Acad. Sci. USA, 82, 7096–7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moréra S., Lacombe,M.L., Xu,Y., LeBras,G. and Janin,J. (1995) X-ray structure of human nucleoside diphosphate kinase B complexed with GDP at 2 Å resolution. Structure, 3, 1307–1314. [DOI] [PubMed] [Google Scholar]
- Neidle S. and Taylor,G. (1977) Nucleic acid binding drugs. Part IV. The crystal structure of the anti-cancer agent daunomycin. Biochim. Biophys. Acta, 479, 450–459. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Reardon J.E. and Miller,W.H. (1990) Human immunodeficiency virus reverse transcriptase. Substrate and inhibitor kinetics with thymidine 5′-triphosphate and 3′-azido-3′-deoxythymidine 5′-triphosphate. J. Biol. Chem., 265, 20302–20307. [PubMed] [Google Scholar]
- Roussel A. and Cambillau,C. (1991) Silicon Graphics Directory. Silicon Graphics, Mountain View, CA. [Google Scholar]
- Sarafianos S.G., Pandey,V.N., Kaushik,N. and Modak,M.J. (1995) Site-directed mutagenesis of arginine 72 of HIV-1 reverse transcriptase. Catalytic role and inhibitor sensitivity. J. Biol. Chem., 270, 19729–19735. [DOI] [PubMed] [Google Scholar]
- Schneider B., Xu,Y.W., Sellam,O., Sarfati,R., Janin,J., Veron,M. and Deville-Bonne,D. (1998) Pre-steady state of reaction of nucleoside diphosphate kinase with anti-HIV nucleotides. J. Biol. Chem., 273, 11491–11497. [DOI] [PubMed] [Google Scholar]
- Schneider B., Biondi,R., Sarfati,R., Agou,F., Guerreiro,C., Deville-Bonne,D. and Veron,M. (2000) The mechanism of phosphorylation of anti-HIV d4T by nucleoside diphosphate kinase. Mol. Pharmacol., 57, 948–953. [PubMed] [Google Scholar]
- Sharma P.L. and Crumpacker,C.S. (1999) Decreased processivity of human immunodeficiency virus type 1 reverse transcriptase (RT) containing didanosine-selected mutation Leu74Val: a comparative analysis of RT variants Leu74Val and lamivudine-selected Met184Val. J. Virol., 73, 8448–8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers J.S., Roe,D., Boyle,P.D., Colvin,M. and Shaw,B.R. (1998) Structural studies of a borane-modified phiosphate diester linkage: ab initio calculations on the dimethylboranophosphate anion and the single-crystal X-ray structure of its diisopropylammonium salt. Inorg. Chem., 37, 4158–4159. [DOI] [PubMed] [Google Scholar]
- Tomasz J., Shaw,B.R., Porter,K. and Sood,A. (1992) 5′-p-borane-substituted thymidine monophosphate and triphosphate. Angew. Chem. Engl. Ed., 31, 1373–1375. [Google Scholar]
- Ueno T., Shirasaka,T. and Mitsuya,H. (1995) Enzymatic characterization of human immunodeficiency virus type 1 reverse transcriptase resistant to multiple 2′,3′-dideoxynucleoside 5′-triphosphates. J. Biol. Chem., 270, 23605–23611. [DOI] [PubMed] [Google Scholar]
- Wainberg M.A. and Friedland,G. (1998) Public health implications of antiretroviral therapy and HIV drug resistance. J. Am. Med. Assoc., 279, 1977–1983. [DOI] [PubMed] [Google Scholar]
- Webb P.A., Perisic,O., Mendola,C.E., Backer,J.M. and Williams,R.L. (1995) The crystal structure of a human nucleoside diphosphate kinase, Nm23-H2. J. Mol. Biol., 251, 574–587. [DOI] [PubMed] [Google Scholar]
- Xu Y.W., Sellam,O., Morera,S., Sarfati,S., Biondi,R., Veron,M. and Janin,J. (1997a) X-ray analysis of azido-thymidine diphosphate binding to nucleoside diphosphate kinase. Proc. Natl Acad. Sci. USA, 94, 7162–7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.W., Morera,S., Janin,J. and Cherfils,J. (1997b) AlF3 mimics the transition state of protein phosphorylation in the crystal structure of nucleoside diphosphate kinase and MgADP. Proc. Natl Acad. Sci. USA, 94, 3579–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]