Abstract
Cell-to-cell communication is integral to the evolution of multicellularity. In plant development, peptide signals relay information coordinating cell proliferation and differentiation. These peptides are often encoded by gene families and bind to corresponding families of receptors. The precise spatiotemporal expression of signals and their cognate receptors underlies developmental patterning, and expressional and biochemical changes over evolutionary time have likely contributed to the refinement and complexity of developmental programs. Here, we discuss two major plant peptide families which have central roles in plant development: the CLAVATA3/ENDOSPERM SURROUNDING REGION (CLE) peptide family and the EPIDERMAL PATTERNING FACTOR (EPF) family. We discuss how specialization has enabled the CLE peptides to modulate stem cell differentiation in various tissue types, and how differing activities of EPF peptides precisely regulate the stomatal developmental program, and we examine the contributions of these peptide families to plant development from an evolutionary perspective.
Introduction
Coordinated behavior within cell communities depends on cell-to-cell communication. From the yeast mating pheromones that guide cell–cell fusion [1] to the myriad of secreted signals that maintain homeostasis and modulate development in animals, all eukaryotic kingdoms employ peptides as intercellular signals. In plants, several families of peptides with diverse biological roles have been discovered. For example, systemins in the Solanacea mediate the response to abiotic and biotic stresses [2], and self-incompatibility in Brassica is regulated by the S-locus cysteine rich protein (SRC) [3]. Multiple peptide families coordinate cell behaviors during development [4,5] and developing cells are exposed to a mixture of signaling molecules. The orchestration of signals within specific developmental contexts demands regulated expression and regulated activity of both the peptides and their cognate receptors. At the same time, many cells are both recipient and producer of signals; thus, cell–cell cross-talk involving feedback loops becomes central for ensuring tissue integrity. Numerous mechanisms, including post-translational modifications of signals [6] and complex downstream signal transduction, contribute to the ultimate readout of this communication.
In this review we focus on the roles of two specific gene families in organizing and maintaining stem-cell-like populations in the meristems and in the stomatal lineage and consider recent results that illustrate how signaling strategies are used during plant development.
From clavata Mutants to the CLE Family of Signal Peptides
The Arabidopsis clavata (clv) mutants were originally identified by their shared club-shaped fruit phenotype (clava = club) [7,8]. This phenotype is the result of an expansion of the stem-cell population in the floral meristem, and analyses of the shoot meristem revealed a similar stem-cell defect there [9-11]. Genetic evidence indicates that three CLV loci act together [12] and the corresponding genes encode components of a signaling pathway: CLV1, a leucine rich repeat-receptor like kinase (LRR-RLK); CLV2, an LRR-containing accessory receptor protein; and CLV3, a novel signal peptide [10,13,14].
CLV3 belongs to a family of 32 CLE genes in Arabidopsis encoding small (<15 kDa) proteins with a characteristic amino-terminal stretch of hydrophobic amino acids that acts as a signal peptide for secretion, and a 14 amino acid signature CLE motif near the carboxyl terminus [15,16]. Biochemical evidence based on CLV3 and CLE2 suggests that CLE proteins are processed into active 12 or 13 amino acid peptides (CLEp) from the CLE motif (Figure 1A) [17-20]. Treatment of full-length preCLV3 protein with cauliflower protein extracts revealed an endopeptidase cleavage site preceding Arg70. Progressive processing by a carboxy-peptidase results in CLV3 peptides with gradually shorter carboxy-terminal extensions and may explain differences in the lengths of CLV3 peptides identified [21,22]. Interestingly, expression of a mutated form of CLV3 with Arg70Ala could still partially suppress the clv3 phenotype, indicating that either cleavage can occur independently of Arg70 in vivo or that the uncleaved protein has retained some activity [22]. The conserved prolines found at positions 4, 7, and 9 (Figure 1A,B) are common sequence elements shared among almost all CLE peptides, and these residues can be hydroxylated (Pro4 and Pro7) or arabinosylated (Pro7). While the role of the hydroxyproline Pro4 and Pro7 residues is not yet clear, Pro7 arabinosylation can enhance CLV3p binding to CLV1, highlighting the potential to regulate ligand–receptor interaction by post-translational modifications of CLE peptides [19,20].
Figure 1. Scheme of CLE and EPF structure and maturation.
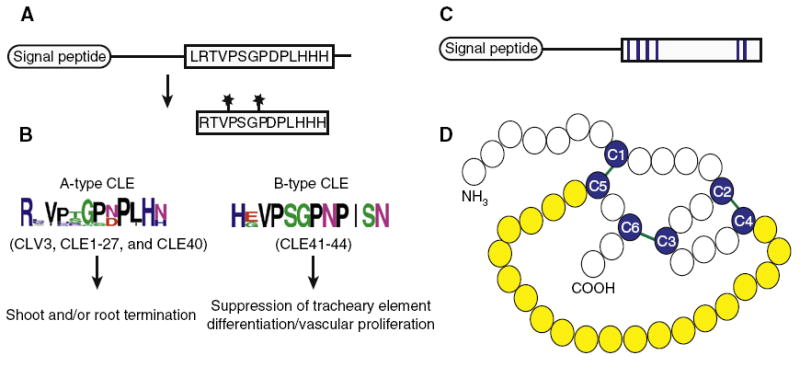
(A) Full length CLE is processed to release the active CLEp consisting of the CLE motif (here represented by CLV3). Stars represent sites of post-translational modifications (B) Consensus sequences of A-type and B-type CLE peptide sequences and activities of each class are shown. Sequence alignments were generated by ClustalW2 and logos were made with WebLogo. (C,D) EPF family protein structure using STOMAGEN as a representative example. (C) Full length STO MAGEN is processed to produce a cysteinerich active peptide (conserved cysteines shown in blue). (D) Experimentally determined structure of STOMAGEN with conserved cysteines (blue) and intramolecular disulfide bonds (green). The variable loop residues exposed at the surface are highlighted in yellow. Image modeled after [62].
While mutations disrupting CLV3, CLE40, or CLE41 produce discernable phenotypes, putative null alleles of several other CLE genes (CLE1, CLE7, CLE16, and CLE18) do not [23], suggesting either functional redundancy or no role for many CLEs under the conditions tested. To overcome this limitation, overexpression and exogenous feeding assays have been used to study CLE activity [21,24]. These studies have led to the proposal that there are two major classes of CLEs (Figure 1B) [24]. A-type CLEs (CLV3, CLE1-27, and CLE40) can induce termination of the root and/or shoot meristem activity through terminal differentiation of stem cells [18,24]. Specificities, however, vary: different CLEs can induce termination of only the shoot meristem, of only the root meristem, or of both [25-27]. For example, both CLV3 and CLE17 are co-expressed in stem cells of the shoot meristem, but only overexpression of CLV3 and not of CLE17 induces shoot meristem termination [23]. One potential explanation for this difference is that the predicted CLE17p lacks the amino-terminal histidine residue which, in CLV3p, is critical for binding to the CLV1 receptor [28]. In contrast to A-type CLEs, B-type CLEs (CLE41–44) do not induce termination of the root/shoot meristems. In Arabidopsis, B-type CLEs can suppress xylem differentiation [18,24,26] and in Zinnia, TDIF (tracheary element differentiation inhibitory factor, the homolog of CLE41/CLE44) suppresses tracheary element differentiation from cultured mesophyll cells [18]. Notably, some A-type CLEs enhance the vascular cell proliferation-stimulating activity of B-type CLEs [24], suggesting that overlapping functions also exist between the two classes of CLE peptides.
In summary, there are clear differences between activities of different CLEps in the aforementioned gain-of-function experiments, consistent with binding of individual CLEps to specific sets of receptors; however, there is also evidence for functional overlap among CLEps, indicating the potential of different CLEps to interact with common sets of receptors.
CLE Gene Members Are Differentially Expressed during Plant Development
The potential of different CLEps to interact with overlapping sets of receptors suggests that a key determinant of CLEp action is the specific tissue distribution of the peptides and their cognate receptors. The comprehensive analysis of A-type CLE promoter-driven GUS reporter expression patterns conducted by Jun et al. [23] highlights this aspect. In this study, individual CLEs display distinct and highly specialized expression patterns, which indicate localized function and the potential to relay precise positional information. However, a single tissue might express a complex mixture of different CLEps. This is seen, for example, in the vasculature, which expresses 14 A-type CLE genes in an overlapping domain with the B-type CLE genes, thereby providing an opportunity for both antagonistic and synergistic interactions among CLEs [24,29]. To fully understand CLE activity, it will be necessary to determine the range of CLEp movement, the expression patterns of cognate receptors, and the transcriptional responses downstream of cell-type-specific ligand–receptor interactions.
CLE Functions in Stem Cell Regulation
Three well-studied cases indicate that regulatory modules utilizing CLE peptides have evolved in stem cell regulation of shoot, root, and vascular meristems and highlight the interactions between CLE signaling and downstream transcription factors of the WUSCHEL related homeobox (WOX) protein family.
CLV3–WUS Modulates Stem Cell Proliferation in the Shoot Meristem
The most extensively studied CLE, CLV3, is exclusively expressed in a small cell group of the apical layers of the shoot and floral meristem central zone, coincident with the location of predicted long-term stem cells (Figure 2A) [13]. CLV3 acts with the homeodomain transcription factor WUSCHEL (WUS) in a regulatory feedback loop, with WUS required for the expression of CLV3 in the apical stem cells and CLV3 delimiting the expression of WUS to the organizing center (OC) of the shoot meristem [9,11]. WUS appears to have numerous transcriptional targets in the OC cells, including the cytokinin signaling inhibitors it represses [30]. Ultimately, maintenance of stem cell pluripotency is the developmental read-out of this feedback loop. Elegant live-cell imaging studies allowed separation of the local CLV3/WUS effects on cell fate in the center of the shoot meristem from longer range effects on cell proliferation at the periphery [31,32].
Figure 2. Models for CLE-dependent stem cell maintenance in the shoot, root, and vascular meristems.
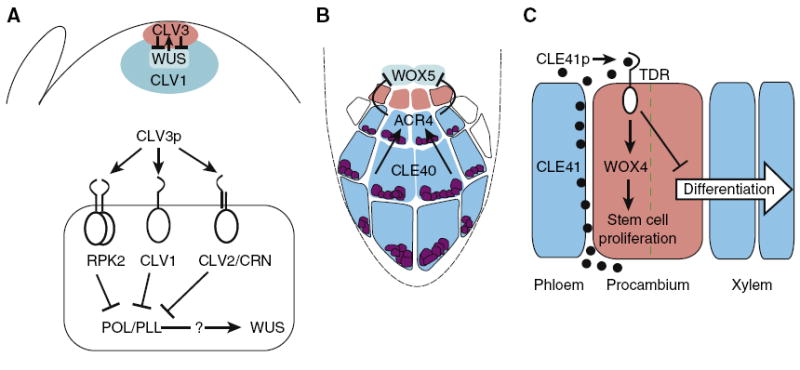
(A) Upper panel: WUS expression in the organizing center (turqoise) is required for expression of CLV3 in the stem cells (red), and CLV3 in turn restricts the expression of WUS. The CLV1 receptor (blue) is largely expressed underneath the two outermost cell layers of the shoot meristem. Lower panel: CLV3p interaction with several receptor complexes which repress the expression of WUS via repressing POL/PLL. (B) CLE40 expression in the columella root cap (blue) acts through ACR4 to restrict WOX5 expression to the quiescent center (turquoise). Columella stem cells are indicated in red. (C) CLE41 is secreted by phloem cells and perceived by TDR, which represses differentiation and upregulates the expression of WOX4 to support stem cell proliferation (red). The dashed green line indicates the plane of division set up by the adjacent CLE41 expression. Where CLE41 concentration is low, the procambial stem cell daughter undergoes differentiation.
The mechanism that determines the range of CLV3 spread such that WUS expression is limited to the OC, but not inactivated there, is central to understanding CLE function in general and meristem homeostasis in particular. Overexpression of CLV3 throughout the meristem causes differentiation of the shoot meristem, suggesting that OC cells are competent to abolish WUS expression if the CLV3 concentration is high enough [9,33,34]. Overexpression of CLV3 in the epidermal layer of the meristem also terminates the meristem, suggesting that CLV3 can act across several cell layers from the epidermis into the OC [33]. In the same study, both the long range effects of unmodified CLV3 and the spread of a CLV3–GFP fusion protein were efficiently blocked by coexpression of CLV1. In light of the finding that normal CLV3 propeptide is proteolytically processed, one plausible explanation for the latter finding is that fusion of GFP to the carboxyl terminus of CLV3 blocked processing from this side. As CLV1 is normally expressed in deeper cell layers of the shoot meristem but not in the epidermis, the observed preferential lateral spread of CLV3–GFP in the outermost cell layers led to the hypothesis that the CLV1 receptor delimits the movement of CLV3 into cells beyond the third outermost cell layer by ligand sequestering and that this contributes to allowing WUS expression in the OC. [33]. However, a recent paper monitoring CLV1–GFP localization in the shoot meristem suggests an alternative model [35]. In this work, using live imaging of the shoot meristem, internalization of CLV1–2XGFP (see Intracellular Transduction of CLV3 section) has been taken as a proxy for CLV3 presence. This study presents evidence that CLV3 can broadly diffuse through the shoot meristem, including into the fourth outermost cell layer. In this model, WUS might be expressed in the OC because the effects of CLV3/CLV1 signaling are antagonized there and only experimentally produced CLV3p levels are high enough to terminate WUS expression. However, since most CLV1 receptor appears already internalized in wild type and is only rarely detected at the plasma membrane (see below), it remains to be shown how overexpression of CLV3 can have such dramatic effects, causing WUS repression and stem cell termination in a CLV1-dependent manner [9,33]. Direct localization of radioactively labeled or tagged CLV3p in the shoot meristem could assist in distinguishing between these models. While CLV3 has been the focus of most attention, the shoot meristem also expresses other CLE genes (CLE 16, 17, 27, 40) [13,23,36], the function of which has yet to be revealed.
Signal Perception in the Shoot Meristem
In addition to CLV1, CLAVATA2 (CLV2), CORYNE (CRN), BAM1, BAM2, and RPK2/Toadstool2 (RPK2) encode membrane-associated proteins that play roles in regulating the response to CLV3 in the shoot meristem. CLV2 encodes a LRR-receptor like transmembrane protein that lacks an intracellular kinase domain and that may interact with CRN, a membrane-associated putative serine/threonine kinase that lacks an extracellular LRR [37-40]. CLV2 and CRN are expressed more broadly than CLV1 and could play roles in responding to CLE peptides other than CLV3p [14,37]. BAM1 and BAM2, which encode LRR-RLKs, are broadly expressed in the apex and may confer different activities inside and outside the central zone of the shoot meristem. On one hand, the genes seem to act antagonistically to CLV1 and CLV2 as the double bam1;bam2 mutant has a smaller meristem in comparison to the enlarged meristems of clv mutants [41]. On the other hand, broad expression of CLV1 can replace BAM function and BAM-overexpression can partially rescue clv1, consistent with CLV1 and BAM activating overlapping downstream processes [41]. In line with this view, higher-order complexes of CLV1–BAM and CLV2–CRN, as well as larger multimers of CLV2–CRN– CLV1, have been demonstrated in vitro [38-40] and binding assays have demonstrated that CLV1, CLV2, BAM1, and BAM2 can all bind CLV3p [38]. Finally, analysis of the rpk2 single mutant and the clv1;clv2;rpk2 triple mutant, which phenocopies clv3, indicates that RPK2, which is expressed in the shoot and root meristem, may also transmit the CLV3 signal [42]. Thus, of the multiple receptors expressed in the shoot meristem, some may be dedicated to one pathway, while others are broader in their expression and function.
Intracellular Transduction of CLV3 Signaling
The signaling cascade that connects CLV3p perception by cell surface receptors with WUS transcriptional suppression is under intensive investigation. Biochemical evidence showing that, in response to CLV3, CLV1 is phosphorylated and a MAPK pathway is activated indicates that phospho-signaling is involved in the CLV signaling pathway [43]. In a recent paper, Nimchuk and colleagues [35] show that a functional CLV1–2xGFP fusion protein is found at the plasma membrane of inflorescence meristem cells only in clv3 mutants, whereas in wild type the tagged receptor is detected in lytic vacuoles. The authors suggest an attractive model in which binding of CLV3p to the CLV1 receptor at the plasma membrane results in endocytosis of CLV1 and subsequent trafficking to lytic vacuoles. One interesting question in the future will be to see how common ligand-mediated receptor internalization is in CLE and other peptide signaling pathways. Genetic and biochemical evidence also link the protein phosphatases POLTERGEIST (POL) and POLTERGEIST-LIKE1 (PLL1) as signaling intermediates between CLV3 perception and WUS regulation (Figure 2A) [44-48]. In the root meristem, POL/PLL1 are also required to maintain expression of the WUS homologue WOX5. The broad developmental defects of the pol;pll1 double mutant indicate that other developmental steps may also be influenced by this mode of regulation [44,45,49]. Interestingly, in response to stimulation by CLV3, CLV1 was found in a complex with the kinase-associated protein phosphatase (KAPP) and a Rho-GTPase related protein. KAPP has been demonstrated to interact biochemically with and negatively regulate a number of other LRR-RLKs [46,47]. Increases in the abundance of KAPP enhance internalization of the somatic embryogenesis receptor kinase 1 (SERK1) [50]. The connection between internalization and phosphorylation of CLV1 and the identification of downstream targets of phospho-regulation will be a key step in elucidating the molecular mechanisms involved in stem cell maintenance.
The CLE40–WOX5 Interaction Regulates Homeostasis of Columella Stem Cells in the Root
The regulation of stem cell development in the root meristem by CLE40 and WOX5 is reminiscent of regulation in the shoot meristem. CLE40 is expressed in the differentiating columella cells (CCs) of the distally located root cap and restricts WOX5 expression to the central quiescent center (QC) [51]. WOX5 in turn is required to maintain the columella stem cells (CSCs), located between the QC and CCs (Figure 2B) [52]. Loss of CLE40 function results in a subtle root wave phenotype, a lateral expansion of the WOX5 expression domain, and an additional layer of CSC-like cells [36,51]. Plants mutant for the receptor kinase ARABIDOPSIS HOMOLOG OF CRINKLY4 (ACR4) show a similar lateral expansion of the WOX5 expression domain, an additional layer of CSCs, and are partially insensitive to CLE40p, indicating that CLE40 activity is mediated in part through ACR4 [53]. Similar to the shoot meristem, ligand sequestering by the receptor has been hypothesized to protect the QC from CLE40 suppression, but low expression levels of ACR4 in the QC might also allow WOX5 expression there [51]. Direct binding of CLE40 to ACR4 has not yet been demonstrated, but such physical interaction would be intriguing since the ectodomain of ACR4 is different from the LRR-containing ectodomains of known CLE receptors like CLV1, BAM, and RPK2 [54,55]. As in the shoot meristem, several other CLEs and RLKs are expressed in the root together with CLE40 and ACR4 [23,27,36,51], suggesting a complex CLE signaling network. Despite the similarities between the roles of CLV3/WUS and CLE40/WOX5 in stem cell regulation in shoot and root meristems, there are also important differences. For example, in the shoot meristem, CLV3 is expressed within the stem cell region and partially overlaps with WUS expression [32], leading to models postulating both paracrine and autocrine functions [33]. By contrast, in the root meristem, CLE40 is expressed in differentiating CCs separated by one layer of stem cells from the WOX5-expressing QC. One unifying concept could be that, in both cases, distal cells act as sources of CLE-mediated positional information that confines WOX expression to proximal stem cell organizers. In this view, the expression of CLV3 in stem cells of the shoot meristem would be a coincidental rather than an inherent feature of stem cell identity.
The CLE41–PXY/TDR–WOX4 Signaling Pathway Regulates the Proliferation of Stem Cells in the Vasculature
The B-type CLEs CLE41 and CLE44 are normally expressed in the vasculature and when exogenously applied to Arabidopsis they can repress the differentiation of procambial cells into xylem cells [29]. A receptor for CLE41 is PHLOEM INTERCALATED WITH XYLEM/TDIF RECEPTOR (PXY/TDR), an RLK expressed in the Arabidopsis procambium [29,56,57]. In the pxy/tdr mutant the organization of the vasculature is disrupted: procambial cells proliferate more slowly, and xylem and phloem cells are no longer separated spatially by the procambium but are instead interspersed [29,56]. One proposal of how CLEs can relay positional information is seen in this context. When CLE41 was provided from spatially distinct sources, oriented cell divisions of procambial cells were dependent not only on the amount of CLE41p but also on the direction from which it first approached the target cells [57]. Like in the shoot and root meristems, a CLEp–WOX pathway appears to be involved in the regulation of vascular stem cells. CLE41 is secreted from the phloem and acts through PXY/TDR in the adjacent procambial cells to stimulate WOX4, which in turn functions in maintaining the procambial cells [57,58]. Outside the range of CLE41 activity procambial daughter cells can differentiate to xylem (Figure 2C). Notably, in the vasculature CLE41 appears to positively affect WOX4, in contrast to the repressive relationship between CLV3 and CLE40 and their associated WOX genes in shoot and root meristems, respectively. A clearer understanding of the regulation of vascular development will rely on a model incorporating the activity of other CLE peptides expressed from these tissues.
Stomatal Development and the EPF family
Peptide signals are also key regulators of the stem-cell-like divisions used in stomatal development, but here, members of a different class of peptides, the epidermal patterning factor (EPF) family, play a predominant role. The stomatal lineage is initiated when a multipotent epidermal (protodermal) cell divides asymmetrically and gives rise to a meristemoid and a larger daughter cell. Subsequent division and differentiation events generate specialized stomatal guard cells, whose ultimate function is to regulate plant/atmosphere gas exchange (Figure 3). Although not organized into a niche like the apical meristematic populations, stomata require positional information to orient the asymmetric divisions that not only create guard cells, but enforce patterning rules that ensure that two stomata are not in direct physical contact. Four EPF family members have been characterized with respect to stomatal development: EPF1, EPF2, STOMAGEN, and CHALLAH (CHAL) [59-64]. Collectively, these ligands influence both the frequency and orientation of asymmetric divisions. Genetic and biochemical studies have revealed important structural elements of EPF peptides, functional diversification among family members, and potential receptors through which these peptides signal.
Figure 3. Model of stomatal development and proposed receptor–ligand interactions.
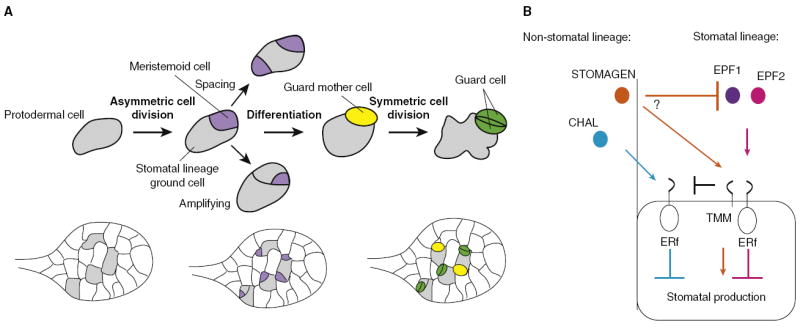
(A) Scheme of Arabidopsis stomatal development illustrated in isolated cells, and in the context of the developing leaf below. A protodermal cell (grey) enters the stomatal lineage via an asymmetric division generating a meristemoid (purple) and larger daughter cell known as a stomatal lineage ground cell (SLGC). The meristemoid may undergo additional asymmetric ‘amplifying’ divisions (producing additional SLGCs) and neighboring cells that undergo asymmetric divisions will orient their division such that two meristemoids are not physically adjacent to each other. The meristemoid differentiates into a guard mother cell (yellow) and the subsequent symmetric division gives rise to two guard cells (green). (B) Summary of EPF ligand–receptor interactions. EPF1 and EPF2 inhibit stomatal development through TMM and ERf receptors. STOMAGEN promotes stomatal development and may compete with EPF1 and EPF2 to do so. CHAL inhibits stomatal development through ERf receptors, and TMM dampens CHAL signaling.
EPF Family Processing and Structure
The cysteine-rich EPF family is much smaller than the CLE family, comprising 11 members in Arabidopsis [60]. Biochemical analysis of STOMAGEN has provided insight into EPF peptide processing and structure. STOMAGEN encodes a 102 amino acid protein, with a 31 amino acid signal peptide at the amino terminus. The mature processed STOMAGEN peptide is larger than the CLE peptides, consisting of the carboxy-terminal 45 amino acids of the propeptide (Figure 1C,D) [62,64]. The region flanking the cleavage site is highly conserved among EPF family members, indicating that other EPFs are likely subject to similar post-translational processing. Although a putative protease (SDD1) is a negative regulator of stomatal development [65], genetic evidence suggests that it acts independently of characterized EPF ligands and no protease has yet been shown to process the EPFs [59-63,66]. All EPF family members possess a carboxy-terminal region with six (and some eight) cysteines with conserved spacing. In STOMAGEN, disulfide bridges form between cysteine residues 13 and 20, between 8 and 41, and between 16 and 43, yielding a predicted ‘knot’ structure whose surface-exposed loop (illustrated in yellow in Figure 1D) shows the largest degree of sequence variation among family members [62]. This variable loop would be in a position to engage other proteins such as cell-surface receptors and changes in this region could explain the diversification of biochemical activity among EPF family members [62].
EPF Family Receptors
Genetic interaction studies in Arabidopsis suggest that LRR-containing receptors mediate EPF signaling. The ERECTA family receptors (ERf) belong to class XIIIb of the LRR receptor-like kinase family and mediate the activity of EPF1, EPF2 and CHAL [59-61,66]. ERf might also mediate STOMAGEN activity but this has not yet been tested directly. The TOO MANY MOUTHS (TMM) receptor-like protein is expressed early in the developing epidermis [67] and mediates the activities of EPF1, EPF2, and STOMAGEN but dampens the activity of CHAL [59-64,66]. Unlike TMM, ERf has a broad expression pattern and has been implicated in additional developmental programs, including a general role in regulating cell proliferation [68,69]. This situation is reminiscent of the shoot meristem but reversed in that the shoot meristem LRR-kinases are more spatially restricted than the LRR accessory proteins.
Stomatal Lineage Intrinsic EPFs Have Distinct Temporal–Spatial Roles
The discrete steps of stomatal development have enabled the careful functional characterization of several EPF family members. Although EPF1 and EPF2 inhibit stomatal development through common receptors, they act at slightly different developmental stages and thus their consequences on epidermal patterning are distinct [59-61]. EPF2 is expressed in protodermal cells that have not yet divided and, consistent with this, regulates early decisions that impact both stomatal and ground cell proliferation. Overexpression of EPF2 inhibits asymmetric divisions into the stomatal lineage (divisions of grey protodermal cells in Figure 3A) whereas loss of EPF2 increases asymmetric divisions, resulting in increased production of both stomatal guard cells and neighboring stomatal lineage ground cells (hereafter referred to as ‘ground cells’) [60,61]. EPF1 is expressed at a later stage and is first visible in meristemoids (purple cells in Figure 3A). In EPF1 overexpression lines, protodermal cells divide asymmetrically but the resulting meristemoids do not differentiate further. The most characteristic defect associated with epf1 is the incorrect orientation of asymmetric divisions (such as the spacing divisions depicted in Figure 3A), resulting in pairs of physically adjacent stomata [59], which is consistent with EPF1 providing a local orienting signal similar to CLE41 in the vasculature. The epf1; epf2 double mutants display an additive phenotype [60,61], and promoter swap experiments are consistent with EPF1 and EPF2 also having distinct biochemical activities rather than solely expressional differences [60].
STOMAGEN Opposes EPF1/2 and Promotes Stomatal Development
Another EPF ligand, STOMAGEN, antagonizes EPF1 and EPF2 function and is a positive regulator of stomatal development. STOMAGEN is expressed in the leaf mesophyll layers [62,64] and overexpression (or the application of chemically synthesized peptide) increases both stomatal density and the presence of physically adjacent stomata [62-64]. RNA interference (RNAi) knockdown of STOMAGEN, conversely, results in the production of fewer stomata and ground cells [63,64].
How might this ‘positive’ ligand interact with the negative regulators? Do these ligands compete for binding to a common receptor, do they interact with each other, or are different receptor combinations of ERf and TMM engaged by different ligands? The current data are somewhat conflicting. Like EPF1 and EPF2, STOMAGEN requires TMM to exert its cell fate and cell proliferation-promoting effects [62,64], suggesting a shared receptor. RNAi knockdown of STOMAGEN in epf1 and epf2 single mutants or epf1; epf2 double mutants reduces stomatal density, suggesting independent ligand activities [64], and consistent with this, STOMAGEN overexpression can enhance the increased stomata phenotypes of epf1 or epf2 single mutants [63]. In other studies, however, application of synthetic STOMAGEN could not enhance the stomatal density in epf1; epf2 double mutants, a result consistent with the ligands competing for a common receptor [62]. RNAi knockdown of STOMAGEN reduces ground cell density in both WT and epf1 mutants; however, this same treatment fails to reduce (in fact, further increases) the epf2 mutant’s increased ground cell density. These results position STOMAGEN independent of EPF1 but indicate that STOMAGEN requires EPF2 to inhibit asymmetric cell divisions [64]. The opposing effects of STOMAGEN and EPF1/2 on the stomatal development parallel the in vitro effects of CLEs on vascular differentiation in Zinnia extracts where some CLEs suppress vascular differentiation while at least one is capable of promoting vascular differentiation [18,29]. Thus far, however, antagonism among CLEs has yet to be demonstrated for the regulation of a developmental process in vivo.
CHAL Reveals That EPF Ligands Can Have Different Interactions with LRR-Receptors
Another EPF, CHALLAH (CHAL), is similar to EPF1 and EPF2 in that it can inhibit stomatal development; however, it does so in an organ-specific way and only in the absence of TMM [66]. CHALLAH was identified as a stem-specific suppressor of tmm. Although tmm mutants have excess stomata on leaves, they do not produce stomata on stems. Loss of CHAL restores stomata to tmm stems without affecting leaf stomatal development, thereby providing an explanation for the opposite phenotypes of tmm in the stem and leaf [66]. CHAL is expressed in inner tissue layers of the stem and hypocotyl, but not in the epidermis or in leaves. Additionally, overexpression studies reveal that CHAL requires ERf to inhibit stomatal development but that, in contrast to the other EPFs, whose functions are dependent on both ERf and TMM, CHAL’s overexpression effects are enhanced in the absence of TMM. This unusual relationship leads to the model that TMM may act as a buffer for the ERf system, absorbing excess CHAL and preventing it from interfering with epidermal pattern [66].
Given the varying effects on stomatal development and different genetic interactions with receptors, it is of interest to determine the basis of the diverse biochemical activities among characterized EPF members and to know whether variation in the surface exposed loop between the fourth and fifth conserved cysteines is sufficient to account for this diversity. Domain swap experiments using EPF peptides may elucidate the specificity conferred through the loop region: for example, would replacing CHAL’s variable region with the corresponding region from STOMAGEN generate a positive regulator of stomatal development that requires TMM to exert its effects?
While four of the EPF family members have been characterized, the functions of an additional seven members are not known. Overexpression screens indicate that EPFL4 and EPFL5 are also capable of inhibiting stomatal development, but their expression patterns are not consistent with a role in stomatal development and neither epfl4 nor epfl5 single mutants, nor the epfl4;epfl5 double mutants, have a detectable epidermal phenotype [60]. Given the pathogen resistance, inflorescence architecture and reproductive development (among other) phenotypes associated with ERf mutants, a reasonable hypothesis is that the remaining EPF ligands mediate other, non-stomatal ERf functions.
Potential Peptide Signals to Transcriptional Feedback Loops in Stomatal Development
There are other parallels between the CLEs and EPFs in their relationships with downstream targets. The basic helix-loop-helix (bHLH) transcription factor SPEECHLESS (SPCH) is a positive regulator of stomatal lineage initiation; it is expressed in many protodermal cells and is necessary (though not sufficient) for these cells to undergo stomatal precursor generating divisions [70,71]. EPF2 and SPCH expression overlaps, but EPF2 is not expressed in spch mutants [60]. This expression pattern, combined with the behavior of mutants, hints at a potential negative feedback loop to regulate epidermal cell proliferation. In this scenario, protodermal cells capable of dividing through SPCH activity turn on EPF2, which in turn inhibits divisions (perhaps non-cell autonomously) and potentially in a threshold-dependent manner [60]. The negative feedback loop in the stomatal lineage is not yet understood mechanistically, but it would allow for the proper balance of proliferation and differentiation, analogous to the effects of CLV3 and WUS in the shoot meristem.
Evolutionary Perspective of CLE and EPF Signaling
While most of the functional characterization of the CLE and EPF family peptide ligands has been carried out in Arabidopsis, representatives of both classes are found in diverse plant groups. The evolutionary history of CLE and EPF family members garnered from phylogenetic analysis has the potential to reveal the contributions of these corresponding gene families to functional innovations in higher plants.
Based on their broad presence in basal and derived plant species, CLE genes and their cognate receptors appear to have originated early in plant evolution. One CLE-like gene has been identified in the flagellate algae Chlamydomonas and one in the moss Physcomitrella patens. In Selaginella molendorffi, 15 CLEs have been identified that cluster with most Arabidopsis CLE clades including the branch containing CLV3 [16]. Homologues of the receptor kinase CLV1 are also present in Physcomitrella. By contrast, homologues of CRN and CLV2, though found in several angiosperms, have not been detected in Physcomitrella or Selaginella, consistent with a later origin of these functions. However, confirmation of this idea requires additional data from intermediate clades [72]. In rice, 47 putative CLE genes have been identified and the CLV3 ortholog FLORAL ORGAN NUMBER4 (FON4) regulates stem cells in shoot and floral meristems [73], indicating both structural and functional conservation between monocot and dicot plants. Interestingly, rice, but not Arabidopsis, encodes proteins possessing multiple CLE domains [74]. An issue to be addressed in the future is whether CLE domains of these proteins are processed into separate, active peptides. Such a situation might simply double the signal dose but also has the potential to couple different signal specificities.
Can the strikingly similar CLE functions in apical and vascular meristems be taken as indications of an ancient association between CLE and stem cells? Indeed, it is tempting to speculate that expansion of the CLE family correlates in part with the evolution of multicellular stem cell pools (in contrast to the single apical stem cells found in basal plants). However, while the discernable phenotypes of clv mutants have paved the way to recognize the role of CLE peptides in stem cell regulation and the function of most peptides have been tested in this context, the variety of expression patterns, including in stomatal precursors [23], indicates that the endogenous functions of CLE peptides may have more facets. One striking observation is that CLE function in stem cell regulation is mediated by different WOX genes. Arabidopsis contains 14 functional WOX genes, most of which are expressed in highly regulated and dynamic patterns during development; in many cases these coincide with CLE expression patterns [23,75] and WOX genes are present throughout the plant kingdom. An intriguing question for future studies is whether the link between positional information provided by localized CLE function and WOX regulation holds true beyond Arabidopsis meristems.
The EPF family also has an ancient origin. Physcomitrella patens, Selaginella moellindorffii, and rice all encode EPF homologues that bear resemblance to the stomatal-specific EPF1/2 and other EPFs that more closely resemble the non-stomatal-expressed CHAL [76]. Selaginella and rice also contain EPF sequences that most closely resemble STOMAGEN (Figure 4). No obvious EPF homologues were found in Chlamydomonas; however the sophisticated approaches used to identify CLE homologues in this alga [16] have not yet been applied to the EPF family.
Figure 4. Alignment of EPFs.

CLUSTAL_W2 alignment of characterized Arabidopsis EPFs and homologues in Physcomitrella patens (Pp), Selaginella moellindorffii (Sm), and Oryza sativa (Os). Alignment uses the putative active peptides based on the mature STOMAGEN sequence.
TMM and ERf homologues are also found in a variety of plant species, including Physcomitrella [77,78]. The presence of ligand and receptor homologues in diverse and basal plant species suggests that cell-to-cell signaling may be an ancient and widely used mechanism in stomatal development. Once the entire suite of Arabidopsis EPFs is functionally characterized, a comprehensive sequence analysis may be useful in predicting which specific EPF functions are conserved across species. As with the CLEs, it will be especially interesting to determine the original activities of the EPF family: did the EPF family evolve before or coincident with stomatal development? When did oppositely acting EPFs emerge? What is the relationship between the expansion of the EPF genes and antagonistic interactions between TMM and the ER family?
Perspectives
We have discussed how two distinct peptide classes participate in plant development, focusing on unique roles in cell fate specification and tissue organization. Future inquiry will no doubt reveal additional classes of peptide hormones as predicted by in silico gene screening [79] and also by the vast numbers of uncharacterized receptor kinases which could be potential receptors for both unidentified and known orphan peptide hormones [4,55]. These studies will reveal whether CLE and EPF peptides provide useful paradigms for peptide signaling strategies in plant development.
One puzzling lesson from studying signaling in plants is the remarkable complexity of these signal systems. Why do higher plants express multiple classes of signal peptides and myriads of cognate receptors, together with many other signals such as hormones or steroids? The numbers of signal components appear associated with the increasing complexity of the plant body in evolution, with recent basal plants containing small gene families in comparison with a vastly expanded signaling network in higher plants. But the number of cell types and of tissues in higher plants is relatively small in comparison to higher animals, which seem to employ a far more modest signaling inventory. The necessity to adapt to environmental changes might account for some of the signal complexity in higher plants, but definite answers to these questions are currently hampered by the fact that, for the majority of signal components, such as peptides and predicted receptors, a biological function has yet to be assigned.
Acknowledgments
We gratefully acknowledge financial support from the Deutsche Forschungsgemeinschaft (German Excellence Initiative BIOSS, ERA-PG, SFB592) and the BMFT (FRISYS) to the T.L. laboratory and from the US National Science Foundation (IOS-0845521) and National Institutes of Health (1RO1GM086632-01) to the D.B. laboratory. K.A.D. was supported by a Graduate Research Fellowship from the National Science Foundation. We apologize to all colleagues whose work could not be mentioned due to space constraints.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dohlman HG, Thorner JW. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu Rev Biochem. 2001;70:703–754. doi: 10.1146/annurev.biochem.70.1.703. [DOI] [PubMed] [Google Scholar]
- 2.Ryan CA, Pearce G, Scheer J, Moura DS. Polypeptide hormones. Plant Cell Suppl. 2002;14:S251–S264. doi: 10.1105/tpc.010484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takayama S, Isogai A. Self-incompatibility in plants. Annu Rev Plant Biol. 2005;56:467–489. doi: 10.1146/annurev.arplant.56.032604.144249. [DOI] [PubMed] [Google Scholar]
- 4.Butenko MA, Vie AK, Brembu T, Aalen RB, Bones AM. Plant peptides in signalling: looking for new partners. Trends Plant Sci. 2009;14:255–263. doi: 10.1016/j.tplants.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Rowe MH, Bergmann DC. Complex signals for simple cells: the expanding ranks of signals and receptors guiding stomatal development. Curr Opin Plant Biol. 2010;13:548–555. doi: 10.1016/j.pbi.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shinohara H, Matsubayashi Y. Arabinosylated glycopeptide hormones: new insights into CLAVATA3 structure. Curr Opin Plant Biol. 2010;13:515–519. doi: 10.1016/j.pbi.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 7.Koornneef M, Van Eden J, Hanhart CJ, Stam P, Braaksma FJ, Feenstra WJ. Linkage map of Arabidopsis thaliana. J Hered. 1983;74:265–272. [Google Scholar]
- 8.Leyser HMO, Furner IJ. Characterization of three shoot apical meristem mutants of Arabidopsis thaliana. Development. 1992;116:397–403. [Google Scholar]
- 9.Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289:617–619. doi: 10.1126/science.289.5479.617. [DOI] [PubMed] [Google Scholar]
- 10.Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89:575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- 11.Schoof H, Lenhard M, Haecker A, Mayer KF, Jurgens G, Laux T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- 12.Kayes JM, Clark SE. CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development. 1998;125:3843–3851. doi: 10.1242/dev.125.19.3843. [DOI] [PubMed] [Google Scholar]
- 13.Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- 14.Jeong S, Trotochaud AE, Clark SE. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell. 1999;11:1925–1934. doi: 10.1105/tpc.11.10.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cock JM, McCormick S. A large family of genes that share homology with CLAVATA3. Plant Physiol. 2001;126:939–942. doi: 10.1104/pp.126.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oelkers K, Goffard N, Weiller GF, Gresshoff PM, Mathesius U, Frickey T. Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biol. 2008;8:1. doi: 10.1186/1471-2229-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fiers M, Golemiec E, van der Schors R, van der Geest L, Li KW, Stiekema WJ, Liu CM. The CLAVATA3/ESR motif of CLAVATA3 is functionally independent from the nonconserved flanking sequences. Plant Physiol. 2006;141:1284–1292. doi: 10.1104/pp.106.080671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito Y, Nakanomyo I, Motose H, Iwamoto K, Sawa S, Dohmae N, Fukuda H. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science. 2006;313:842–845. doi: 10.1126/science.1128436. [DOI] [PubMed] [Google Scholar]
- 19.Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science. 2006;313:845–848. doi: 10.1126/science.1128439. [DOI] [PubMed] [Google Scholar]
- 20.Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y. A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat Chem Biol. 2009;5:578–580. doi: 10.1038/nchembio.182. [DOI] [PubMed] [Google Scholar]
- 21.Ni J, Clark SE. Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiol. 2006;140:726–733. doi: 10.1104/pp.105.072678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ni J, Guo Y, Jin H, Hartsell J, Clark SE. Characterization of a CLE processing activity. Plant Mol Biol. 2010;75:67–75. doi: 10.1007/s11103-010-9708-2. [DOI] [PubMed] [Google Scholar]
- 23.Jun J, Fiume E, Roeder AH, Meng L, Sharma VK, Osmont KS, Baker C, Ha CM, Meyerowitz EM, Feldman LJ, et al. Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant Physiol. 2010;154:1721–1736. doi: 10.1104/pp.110.163683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitford R, Fernandez A, De Groodt R, Ortega E, Hilson P. Plant CLE peptides from two distinct functional classes synergistically induce division of vascular cells. Proc Natl Acad Sci USA. 2008;105:18625–18630. doi: 10.1073/pnas.0809395105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strabala TJ, O’Donnell PJ, Smit AM, Ampomah-Dwamena C, Martin EJ, Netzler N, Nieuwenhuizen NJ, Quinn BD, Foote HC, Hudson KR. Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiol. 2006;140:1331–1344. doi: 10.1104/pp.105.075515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinoshita A, Nakamura Y, Sasaki E, Kyozuka J, Fukuda H, Sawa S. Gain-of-function phenotypes of chemically synthetic CLAVATA3/ESR-related (CLE) peptides in Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 2007;48:1821–1825. doi: 10.1093/pcp/pcm154. [DOI] [PubMed] [Google Scholar]
- 27.Meng L, Feldman LJ. CLE14/CLE20 peptides may interact with CLAVATA2/CORYNE receptor-like kinases to irreversibly inhibit cell division in the root meristem of Arabidopsis. Planta. 2010;232:1061–1074. doi: 10.1007/s00425-010-1236-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kondo T, Nakamura T, Yokomine K, Sakagami Y. Dual assay for MCLV3 activity reveals structure-activity relationship of CLE peptides. Biochem Biophys Res Commun. 2008;377:312–316. doi: 10.1016/j.bbrc.2008.09.139. [DOI] [PubMed] [Google Scholar]
- 29.Hirakawa Y, Shinohara H, Kondo Y, Inoue A, Nakanomyo I, Ogawa M, Sawa S, Ohashi-Ito K, Matsubayashi Y, Fukuda H. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc Natl Acad Sci USA. 2008;105:15208–15213. doi: 10.1073/pnas.0808444105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leibfried A, To JP, Busch W, Stehling S, Kehle A, Demar M, Kieber JJ, Lohmann JU. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature. 2005;438:1172–1175. doi: 10.1038/nature04270. [DOI] [PubMed] [Google Scholar]
- 31.Yadav RK, Tavakkoli M, Reddy GV. WUSCHEL mediates stem cell homeostasis by regulating stem cell number and patterns of cell division and differentiation of stem cell progenitors. Development. 2010;137:3581–3589. doi: 10.1242/dev.054973. [DOI] [PubMed] [Google Scholar]
- 32.Reddy GV, Meyerowitz EM. Stem-cell homeostasis and growth dynamics can be uncoupled in the Arabidopsis shoot apex. Science. 2005;310:663–667. doi: 10.1126/science.1116261. [DOI] [PubMed] [Google Scholar]
- 33.Lenhard M, Laux T. Stem cell homeostasis in the Arabidopsis shoot meristem is regulated by intercellular movement of CLAVATA3 and its sequestration by CLAVATA1. Development. 2003;130:3163–3173. doi: 10.1242/dev.00525. [DOI] [PubMed] [Google Scholar]
- 34.Muller R, Borghi L, Kwiatkowska D, Laufs P, Simon R. Dynamic and compensatory responses of Arabidopsis shoot and floral meristems to CLV3 signaling. Plant Cell. 2006;18:1188–1198. doi: 10.1105/tpc.105.040444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nimchuk ZL, Tarr PT, Ohno C, Qu X, Meyerowitz EM. Plant stem cell signaling involves ligand-dependent trafficking of the CLAVATA1 receptor kinase. Curr Biol. 2011;21:1–8. doi: 10.1016/j.cub.2011.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hobe M, Muller R, Grunewald M, Brand U, Simon R. Loss of CLE40, a protein functionally equivalent to the stem cell restricting signal CLV3, enhances root waving in Arabidopsis. Dev Genes Evol. 2003;213:371–381. doi: 10.1007/s00427-003-0329-5. [DOI] [PubMed] [Google Scholar]
- 37.Muller R, Bleckmann A, Simon R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell. 2008;20:934–946. doi: 10.1105/tpc.107.057547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo Y, Han L, Hymes M, Denver R, Clark SE. CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification. Plant J. 2010;63:889–900. doi: 10.1111/j.1365-313X.2010.04295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bleckmann A, Weidtkamp-Peters S, Seidel CA, Simon R. Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol. 2010;152:166–176. doi: 10.1104/pp.109.149930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu Y, Wang Y, Li R, Song X, Wang Q, Huang S, Jin JB, Liu CM, Lin J. Analysis of interactions among the CLAVATA3 receptors reveals a direct interaction between CLAVATA2 and CORYNE in Arabidopsis. Plant J. 2010;61:223–233. doi: 10.1111/j.1365-313X.2009.04049.x. [DOI] [PubMed] [Google Scholar]
- 41.DeYoung BJ, Bickle KL, Schrage KJ, Muskett P, Patel K, Clark SE. The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 2006;45:1–16. doi: 10.1111/j.1365-313X.2005.02592.x. [DOI] [PubMed] [Google Scholar]
- 42.Kinoshita A, Betsuyaku S, Osakabe Y, Mizuno S, Nagawa S, Stahl Y, Simon R, Yamaguchi-Shinozaki K, Fukuda H, Sawa S. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development. 2010;137:3911–3920. doi: 10.1242/dev.048199. [DOI] [PubMed] [Google Scholar]
- 43.Betsuyaku S, Takahashi F, Kinoshita A, Miwa H, Shinozaki K, Fukuda H, Sawa S. Mitogen-activated protein kinase regulated by the CLAVATA receptors contributes to shoot apical meristem homeostasis. Plant Cell Physiol. 2011;52:14–29. doi: 10.1093/pcp/pcq157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu LP, Simon EJ, Trotochaud AE, Clark SE. POLTERGEIST functions to regulate meristem development downstream of the CLAVATA loci. Development. 2000;127:1661–1670. doi: 10.1242/dev.127.8.1661. [DOI] [PubMed] [Google Scholar]
- 45.Yu LP, Miller AK, Clark SE. POLTERGEIST encodes a protein phosphatase 2C that regulates CLAVATA pathways controlling stem cell identity at Arabidopsis shoot and flower meristems. Curr Biol. 2003;13:179–188. doi: 10.1016/s0960-9822(03)00042-3. [DOI] [PubMed] [Google Scholar]
- 46.Trotochaud AE, Hao T, Wu G, Yang Z, Clark SE. The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein. Plant Cell. 1999;11:393–406. doi: 10.1105/tpc.11.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams RW, Wilson JM, Meyerowitz EM. A possible role for kinase-associated protein phosphatase in the Arabidopsis CLAVATA1 signaling pathway. Proc Natl Acad Sci USA. 1997;94:10467–10472. doi: 10.1073/pnas.94.19.10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song SK, Lee MM, Clark SE. POL and PLL1 phosphatases are CLAVATA1 signaling intermediates required for Arabidopsis shoot and floral stem cells. Development. 2006;133:4691–4698. doi: 10.1242/dev.02652. [DOI] [PubMed] [Google Scholar]
- 49.Song SK, Hofhuis H, Lee MM, Clark SE. Key divisions in the early Arabidopsis embryo require POL and PLL1 phosphatases to establish the root stem cell organizer and vascular axis. Dev Cell. 2008;15:98–109. doi: 10.1016/j.devcel.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shah K, Russinova E, Gadella TW, Jr, Willemse J, De Vries SC. The Arabidopsis kinase-associated protein phosphatase controls internalization of the somatic embryogenesis receptor kinase 1. Genes Dev. 2002;16:1707–1720. doi: 10.1101/gad.220402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stahl Y, Wink RH, Ingram GC, Simon R. A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr Biol. 2009;19:909–914. doi: 10.1016/j.cub.2009.03.060. [DOI] [PubMed] [Google Scholar]
- 52.Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007;446:811–814. doi: 10.1038/nature05703. [DOI] [PubMed] [Google Scholar]
- 53.De Smet I, Vassileva V, De Rybel B, Levesque MP, Grunewald W, Van Damme D, Van Noorden G, Naudts M, Van Isterdael G, De Clercq R, et al. Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science. 2008;322:594–597. doi: 10.1126/science.1160158. [DOI] [PubMed] [Google Scholar]
- 54.Hirakawa Y, Kondo Y, Fukuda H. Regulation of vascular development by CLE peptide-receptor systems. J Integr Plant Biol. 2010;52:8–16. doi: 10.1111/j.1744-7909.2010.00904.x. [DOI] [PubMed] [Google Scholar]
- 55.Shiu SH, Bleecker AB. Plant receptor-like kinase gene family: diversity, function, and signaling. Sci STKE. 2001;2001:re22. doi: 10.1126/stke.2001.113.re22. [DOI] [PubMed] [Google Scholar]
- 56.Fisher K, Turner S. PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Curr Biol. 2007;17:1061–1066. doi: 10.1016/j.cub.2007.05.049. [DOI] [PubMed] [Google Scholar]
- 57.Etchells JP, Turner SR. The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development. 2010;137:767–774. doi: 10.1242/dev.044941. [DOI] [PubMed] [Google Scholar]
- 58.Hirakawa Y, Kondo Y, Fukuda H. TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell. 2010;22:2618–2629. doi: 10.1105/tpc.110.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hara K, Kajita R, Torii KU, Bergmann DC, Kakimoto T. The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 2007;21:1720–1725. doi: 10.1101/gad.1550707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hara K, Yokoo T, Kajita R, Onishi T, Yahata S, Peterson KM, Torii KU, Kakimoto T. Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis Leaves. Plant Cell Physiol. 2009;50:1019–1031. doi: 10.1093/pcp/pcp068. [DOI] [PubMed] [Google Scholar]
- 61.Hunt L, Gray JE. The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr Biol. 2009;19:864–869. doi: 10.1016/j.cub.2009.03.069. [DOI] [PubMed] [Google Scholar]
- 62.Kondo T, Kajita R, Miyazaki A, Hokoyama M, Nakamura-Miura T, Mizuno S, Masuda Y, Irie K, Tanaka Y, Takada S, et al. Stomatal density is controlled by a mesophyll-derived signaling molecule. Plant Cell Physiol. 2010;51:1–8. doi: 10.1093/pcp/pcp180. [DOI] [PubMed] [Google Scholar]
- 63.Hunt L, Bailey KJ, Gray JE. The signalling peptide EPFL9 is a positive regulator of stomatal development. New Phytol. 2010;186:609–614. doi: 10.1111/j.1469-8137.2010.03200.x. [DOI] [PubMed] [Google Scholar]
- 64.Sugano SS, Shimada T, Imai Y, Okawa K, Tamai A, Mori M, Hara-Nishimura I. Stomagen positively regulates stomatal density in Arabidopsis. Nature. 2010;463:241–244. doi: 10.1038/nature08682. [DOI] [PubMed] [Google Scholar]
- 65.Berger D, Altmann T. A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 2000;14:1119–1131. [PMC free article] [PubMed] [Google Scholar]
- 66.Abrash EB, Bergmann DC. Regional specification of stomatal production by the putative ligand CHALLAH. Development. 2010;137:447–455. doi: 10.1242/dev.040931. [DOI] [PubMed] [Google Scholar]
- 67.Nadeau JA, Sack FD. Control of stomatal distribution on the Arabidopsis leaf surface. Science. 2002;296:1697–1700. doi: 10.1126/science.1069596. [DOI] [PubMed] [Google Scholar]
- 68.Shpak ED, McAbee JM, Pillitteri LJ, Torii KU. Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science. 2005;309:290–293. doi: 10.1126/science.1109710. [DOI] [PubMed] [Google Scholar]
- 69.Shpak ED, Berthiaume CT, Hill EJ, Torii KU. Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development. 2004;131:1491–1501. doi: 10.1242/dev.01028. [DOI] [PubMed] [Google Scholar]
- 70.MacAlister CA, Ohashi-Ito K, Bergmann DC. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature. 2007;445:537–540. doi: 10.1038/nature05491. [DOI] [PubMed] [Google Scholar]
- 71.Pillitteri LJ, Sloan DB, Bogenschutz NL, Torii KU. Termination of asymmetric cell division and differentiation of stomata. Nature. 2007;445:501–505. doi: 10.1038/nature05467. [DOI] [PubMed] [Google Scholar]
- 72.Miwa H, Tamaki T, Fukuda H, Sawa S. Evolution of CLE signaling: origins of the CLV1 and SOL2/CRN receptor diversity. Plant Signal Behav. 2009;4:477–481. doi: 10.4161/psb.4.6.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chu H, Qian Q, Liang W, Yin C, Tan H, Yao X, Yuan Z, Yang J, Huang H, Luo D, et al. The floral organ number4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiol. 2006;142:1039–1052. doi: 10.1104/pp.106.086736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sawa S, Kinoshita A, Betsuyaku S, Fukuda H. A large family of genes that share homology with CLE domain in Arabidopsis and rice. Plant Signal Behav. 2008;3:337–339. doi: 10.4161/psb.3.5.5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van der Graaff E, Laux T, Rensing SA. The WUS homeobox-containing (WOX) protein family. Genome Biol. 2009;10:248. doi: 10.1186/gb-2009-10-12-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rychel AL, Peterson KM, Torii KU. Plant twitter: ligands under 140 amino acids enforcing stomatal patterning. J Plant Res. 2010;123:275–280. doi: 10.1007/s10265-010-0330-9. [DOI] [PubMed] [Google Scholar]
- 77.Peterson KM, Rychel AL, Torii KU. Out of the mouths of plants: the molecular basis of the evolution and diversity of stomatal development. Plant Cell. 2010;22:296–306. doi: 10.1105/tpc.109.072777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lehti-Shiu MD, Zou C, Hanada K, Shiu SH. Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol. 2009;150:12–26. doi: 10.1104/pp.108.134353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ohyama K, Ogawa M, Matsubayashi Y. Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. Plant J. 2008;55:152–160. doi: 10.1111/j.1365-313X.2008.03464.x. [DOI] [PubMed] [Google Scholar]


