Abstract
Through mutational analysis in Drosophila, we have identified the gene multiple asters (mast), which encodes a new 165 kDa protein. mast mutant neuroblasts are highly polyploid and show severe mitotic abnormalities including the formation of mono- and multi-polar spindles organized by an irregular number of microtubule-organizing centres of abnormal size and shape. The mast gene product is evolutionarily conserved since homologues were identified from yeast to man, revealing a novel protein family. Antibodies against Mast and analysis of tissue culture cells expressing an enhanced green fluorescent protein–Mast fusion protein show that during mitosis, this protein localizes to centrosomes, the mitotic spindle, centromeres and spindle midzone. Microtubule-binding assays indicate that Mast is a microtubule-associated protein displaying strong affinity for polymerized microtubules. The defects observed in the mutant alleles and the intracellular localization of the protein suggest that Mast plays an essential role in centrosome separation and organization of the bipolar mitotic spindle.
Keywords: centrosome/Drosophila/MAPs/microtubules/mitosis
Introduction
The mitotic spindle is a specialized structure required during mitosis for chromosome segregation, made up of microtubules (MTs) organized from microtubule-organizing centres (MTOCs) that in most animal cells correspond to centrosomes (reviewed in Zimmerman et al., 1999).
Assembly of the mitotic spindle starts during late G2, after centrosome replication. This transition is marked by a dramatic change in MT behaviour thought to be triggered by activation of maturation-promoting factor (MPF) (Verde et al., 1992). At this stage, centrosomes begin to nucleate more and highly dynamic MTs from well defined asters (Saxton et al., 1984) that are involved in centrosome separation and migration to opposite poles of the cell (Saunders and Hoyt, 1992). It is now thought that the formation and maintenance of a bipolar spindle involves at least three families of molecular motors. These include the bipolar kinesins, C-terminal kinesins and cytoplasmic dynein (Robinson et al., 1999; Sharp et al., 1999, 2000).
Recent studies on a new family of non-motor microtubule-associated proteins (MAPs), the dis1-TOG family, have suggested that these proteins may also play important roles in the organization of the bipolar spindle. This family includes the human ch-TOG (Charrasse et al., 1998), Xenopus XMAP215 (Vasquez et al., 1994; Tournebize et al., 2000), Drosophila melanogaster Msps (Cullen et al., 1999), Caenorhabditis elegans ZYG-9 (Kemphues et al., 1986; Matthews et al., 1998), Schizosaccharomyces pombe p93dis1 (Nabeshima et al., 1995), Saccharomyces cerevisiae Stu2p (Wang and Huffaker, 1997) and Dictyostelium discoideum DdCP224 (Gräf et al., 2000) proteins. These MAPs localize to either the centrosome/spindle pole body, spindle MTs or both during mitosis or meiosis and have been implicated in the control of microtubule dynamics, stability of the mitotic apparatus, duplication of the centrosome and cytokinesis. Although all members of the dis1-TOG family have been shown to bind microtubules, the MT-binding domain has only been determined for Stu2p, p93dis1 and ch-TOG, and this region falls outside the conserved domains (Nakaseko et al., 1996; Wang and Huffaker, 1997; Charrasse et al., 1998). The MT-binding domain of ch-TOG is also found in other MAPs including tau2, MAP4 and MAP2b (Charrasse et al., 1998).
Here we report the identification and characterization of a new Drosophila gene that we have named multiple asters (mast). Mutations in mast cause abnormal chromosome segregation associated with irregular centrosome separation and severely disrupted spindles. We show that the gene encodes a conserved 165 kDa MAP, defining a new conserved family of proteins that is related to the dis1-TOG family. We also show that the mast gene product localizes to centrosomes, interphase and spindle MTs, centromeres and the spindle midzone. Our data suggest that Mast is required for centrosome segregation and organization of the bipolar spindle.
Results
Identification and characterization of the multiple asters (mast) mutations
The first multiple asters (mastP1) mutant allele was identified by Omel’yanchuk et al. (1997). Subsequently, we identified two other P-element-induced alleles, mastP2 and mastP3, from the Berkeley Drosophila Genome Project (BDGP) database. A fourth allele, mastP4, an imprecise excision allele, was obtained after remobilization of the P-element in mastP1.
mastP1 and mastP4 cause late larval/pupal lethality when homozygous or hemizygous over Df(3L)31A. mastP3 causes early embryonic lethality of homozygous individuals, suggesting the presence of a second mutation, since mastP3/Df(3L)31A die during late larval/pupal stages. The mastP2 allele is semi-lethal, and viable adults homozygous, hemizygous or heterozygous over the other alleles are obtained. These adults are sterile; moreover, testes and ovaries of mastP2 and mastP2/Df(3L)31A adults are rudimentary.
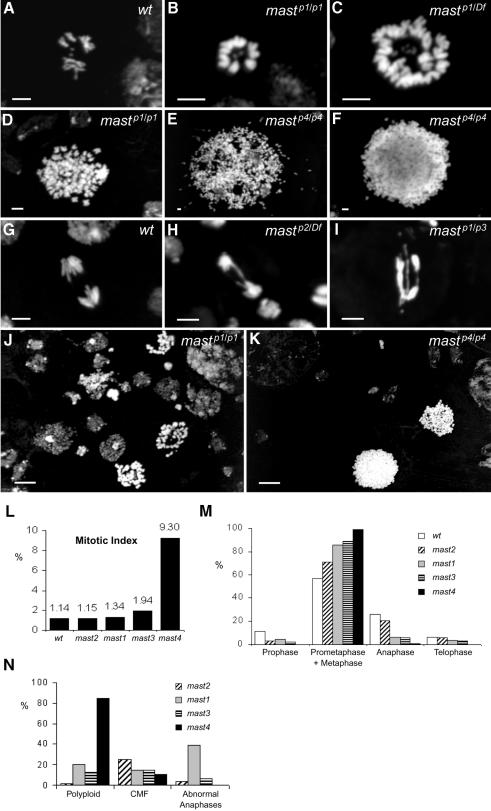
Neuroblasts of homozygous or hemizygous larvae carrying mast mutant alleles show severe mitotic abnormalities (Figure 1), including highly condensed chromosomes (Figure 1B–F, J and K) that frequently are organized in circular arrangements (Figure 1B and C), very few and irregular anaphases (Figure 1H and I) and highly polyploid cells (Figure 1C–F, J and K). Quantification of mitotic parameters shows that mastP1 and mastP2 do not cause a significant increase in the mitotic index. However, mastP3/Df(3L)31A shows an elevated mitotic index and mastP4 a severe mitotic arrest (Figure 1L). Quantification of mitotic figures with respect to mitotic progression indicates that all mutant alleles cause a decrease in the number of cells in prophase, a significant increase of cells in prometaphase/metaphase and a decrease in the proportion of cells in anaphase or telophase (Figure 1M). Quantification of the different types of mitotic abnormalities suggests that most alleles cause either a severe increase in the proportion of polyploid cells, metaphases with a circular chromosome configuration or abnormal anaphases (Figure 1N). The effects on viability, mitotic phenotype and mitotic progression allowed us to order the alleles from least affected to very severe according to the following series: mastP2 < mastP1 < mastP3 < mastP4. Taken together, these results indicate that mutations in mast cause severe abnormalities in chromosome segregation, leading cells to arrest at prometaphase/metaphase. However, the arrest can be overcome and cells undergo multiple rounds of proliferation since most of them are polyploid.
Fig. 1. Cytological analysis and quantification of mitotic phenotypes in mast mutant neuroblasts. Third instar larval brains were dissected from wild-type (A and G) or mast mutant (B–F and H–K) individuals. Wild-type cells in metaphase (A) or anaphase (G) are shown for comparison. mast mutant cells show either diploid (B) or polyploid (C) circular mitotic figures (CMF) with chromosomes organized with their centromeres facing a central region where the small fourth chromosomes are located. Most cells show highly condensed chromosomes (B–F, J and K). mast mutant cells at anaphase (H and I) can also be found and occasionally show chromatin bridges and abnormal segregation. In the most severe mastP4 allele, cells show extensive polyploidy with most chromosomes organized in a sphere-like conformation (F). (L) Quantification of mitotic index. (M) Quantification of mitotic cells with respect to different stages of mitosis. (N) Quantification of the abnormal mitotic parameters in all alleles. Bar = 5 µm except in (J) and (K), 50 µm.
Molecular cloning of the mast gene
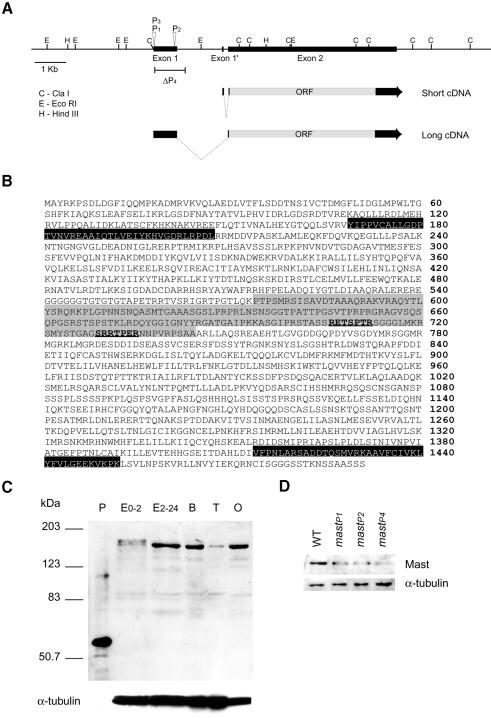
In order to identify the mutated gene, we characterized the locus at the molecular level. We mapped by in situ hybridization a single P-element insertion in the mastP1 allele to the 78C1–C2 cytological region (data not shown) and cloned both sides of the insertion by plasmid rescue and inverse PCR. DNA sequence analysis with the BDGP databases indicated that it was a new gene and identified a number of expressed sequence tags (ESTs) that had already been partially sequenced. We fully sequenced the largest cDNA (LD11488) of 5938 bp (these sequence data have been submitted to the DDBJ/EMBL/GenBank databases under accession No. AF250842). DNA sequence comparisons between the LD11488 cDNA and the corresponding genomic sequence, as well as sequences from cDNAs isolated from adult heads and larva/early pupa, allowed us to determine the intron–exon organization of the gene (Figure 2A). The results indicate that there are at least two types of transcripts: a 5.2 kb transcript, present in cDNA libraries from adult heads and larva/early pupa, and a second transcript of 5.9 kb present in cDNA libraries prepared from embryos. Since in situ hybridization to polytene chromosomes with the larger cDNA hybridizes to a single site (data not shown), the simplest interpretation is that the two transcripts are produced by tissue-specific alternative splicing. The cDNA sequence contains a single open reading frame of 1491 amino acids, coding for a protein with a predicted molecular mass of 165.5 kDa and a pI of 9.17 (Figure 2B).
Fig. 2. Molecular characterization and expression analysis of mast. (A) Molecular map of the mast locus. Wedges represent the insertion of a P{1ArB}-element in mastP1 (P1) and a P{EP}-element in mastP2 (P2) or mastP3 (P3), and black boxes represent the exons. The bar under the map corresponds to the deleted region in mastP4 (ΔP4). Arrows represent the two transcripts corresponding to the short cDNA from adult heads or larva/pupa, and the long cDNA from embryos. The open reading frame is represented by a grey box. Note that neither exon 1 or exon 1′ are coding exons. (B) Predicted amino acid sequence of Mast. Black boxes represent HEAT repeats, and predicted sites of phosphorylation by p34cdc2 are bold underlined. The grey region defines the conserved MAP-4 microtubule-binding domain. (C) Developmental expression of Mast. Protein samples prepared from successive developmental stages of the wild-type strain were loaded in equal amounts (see α-tubulin as control). P, rMast1; E0-2, 0–2 h embryos; E2-24, 2–24 h embryos; B, third instar larval brains; T, adult testes; O, adult ovaries. The anti-Mast antibody specifically recognizes the recombinant protein and a band of 165 kDa, corresponding to Mast, in all extracts. Note that in early embryo extracts the antibody recognizes an additional band of higher molecular weight. (D) Expression of Mast in different mutant alleles. Protein samples from brains of wild-type or homozygous mutant third instar larvae were loaded in equal amounts (see α-tubulin as control).
We also characterized the molecular lesions in other mast alleles. The mastP1 and mastP3 alleles were obtained in different screens; however, they carry a P insertion at the same genomic position, 2317 bp upstream of the predicted ATG. The P-element in mastP2 is inserted 1679 bp from the ATG. The fourth allele, mastP4, was obtained after remobilization of the P-element in mastP1. Southern analysis of mastP4 shows that it has lost part of the P-element (including the ry+ gene). However, all genomic restriction fragments upstream from the insertion are unchanged, and it has a deletion of up to 1 kb of genomic sequence downstream from the insertion site (Figure 2A). Furthermore, western analysis shows that a residual amount of full-size protein is expressed in mastP4 homozygotes, confirming that the coding region of mast is not affected (see Figure 2D). As a result of the remobilization of the insertion in mastP1, we also obtained 39 independent lines that had lost the insertion, were viable and show normal mitotic parameters. These results, together with the genetic complementation data, suggest that the late larval lethality and mitotic phenotypes are the result of mutations in the locus mast.
Analysis of the protein sequence shows that Mast contains a 170 amino acid domain that shares limited homology with the proline-rich domain of MAP4 (Figure 2B), which is thought to be involved in the high efficiency binding to MTs (Aizawa et al., 1991). It also contains two regions with significant homology to the HEAT repeat (Andrade and Bork, 1995), at positions 169–207 and 1414–1452 (Figure 2B). This motif was identified in Huntingtin, a protein associated with Huntington’s disease, and is also present in the 65 kDa regulatory subunit of protein phosphatase 2A (Hemmings et al., 1990) and in proteins of the dis1-TOG family (Tournebize et al., 2000). The Mast protein also contains two cyclin-dependent kinase p34cdc2 consensus phosphorylation sites (Kennelly and Krebs, 1991) (Figure 2B).
Expression of the mast gene in wild-type and mutant tissues
In order to analyse the expression of the mast gene, we raised antibodies (Rb726) against a fragment of the Mast protein. The immunopurified antibody (IP726α) recognizes the bacterially expressed protein and, in extracts from embryos, larvae and adult tissues, a large protein of 165 kDa (Figure 2C). The protein is present during early embryogenesis, but it is highly expressed during late embryogenesis, in larval brains and ovaries, and significantly reduced in testes. In early embryos, we also find a slightly larger protein that is recognized specifically by IP726α. We have also analysed the level of the Mast protein in homozygous mutant individuals and found that it is reduced in both mastP1 and mastP2 and barely detectable in mastP4 when compared with the wild-type control (Figure 2D).
Evolutionary conservation of Mast
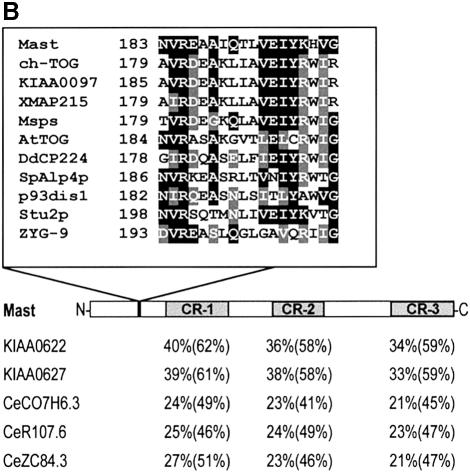
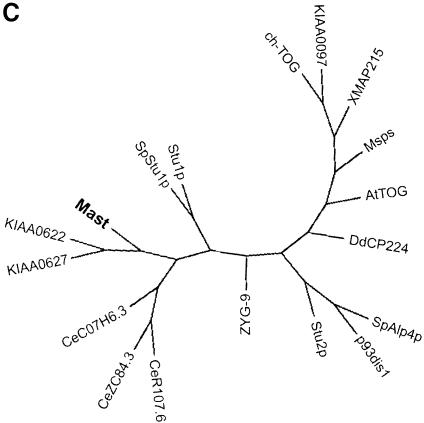
In order to determine whether the Mast protein is evolutionarily conserved, we performed BLAST searches against current databases (see Materials and methods). Mast shares significant identity with proteins encoded by two human cDNAs (KIAA0622 and KIAA0627; Ishikawa et al., 1998), three putative proteins in C.elegans (C07H6.3, R107.6 and ZC84.3; Wilson et al., 1994) and also limited identity with Stu1p from S.cerevisiae (Pasqualone et al., 1994) and its putative homologue in S.pombe, which we have called SpStu1p. Multiple alignment of the Mast sequence with those most closely related from other species (Figure 3A) shows that all the proteins share identity throughout their sequence; however, three regions (CR-1, CR-2 and CR-3) are more highly conserved (Figure 3A and B). These results suggest that Mast and its homologues define a new evolutionarily conserved protein family that we have named Stu1-Mast.
Fig. 3. Protein sequence alignment and phylogenetic analysis. (A) Multiple sequence alignment of the predicted protein sequences closely related to Mast, including two from human (KIAA0622 and KIAA0627), three from C.elegans (CeC07H6.3, CeR107.6 and CeZC84.3), one from S.pombe (SpStu1p) and one from S.cerevisiae (Stu1p), revealed three regions of more significant identity. (B) Conserved regions are represented in grey boxes and the percentage identity and similarity (in parentheses) of the most conserved proteins are indicated below. Additionally, a small domain of 18 amino acids that is highly conserved between Mast and members of the dis1-TOG family is represented. (C) Phylogenetic unrooted tree with all proteins that share significant sequence identity with Mast.
Database searches also indicate that Mast shares identity with proteins from the dis1-TOG family, especially at the N-terminal half of the protein (amino acids 1–494), where they are 20–25% identical and 40–45% similar. Inside this region, there is a small domain of 18 amino acid residues that is highly conserved among these proteins and falls inside the first HEAT repeat of Mast (Figure 3B). Phylogenetic analysis including all sequences from the two groups suggests that they are evolutionarily close, but distinct, since they are positioned in different branches of the dendrogram (Figure 3C).
Localization of Mast during the cell cycle
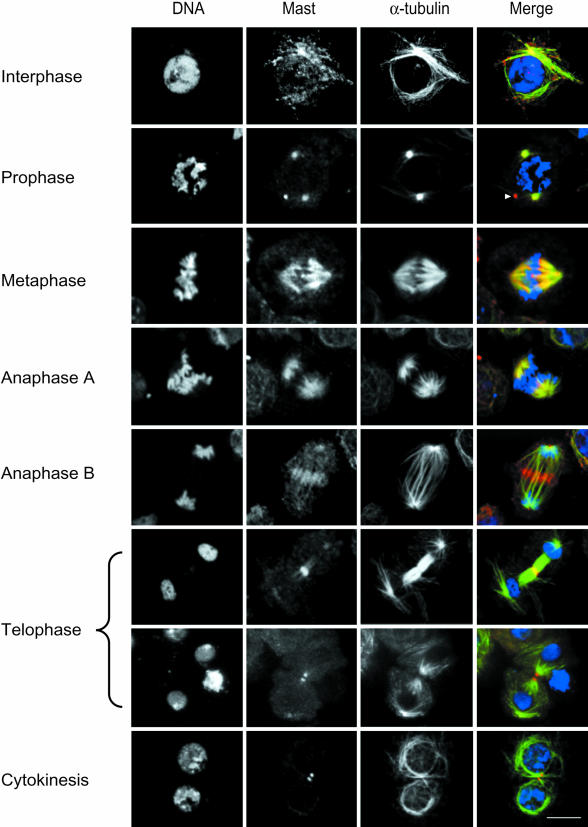
To determine the intracellular localization of Mast during different stages of the cell cycle, we used IP726α for indirect immunofluorescence in S2 Drosophila tissue culture cells (Figure 4). At interphase, Mast is focused on MTOCs and shows a punctuate pattern co-localizing with α-tubulin. At prophase, Mast accumulates at the MTOCs, as shown by α-tubulin co-staining (Figure 4). Double immunostaining with either anti-centrosomin (CNN) or γ-tubulin antibodies also shows co-localization with Mast (data not shown). In some cells, Mast also associates with a rod-like structure present in interphase or mitotic cells (Figure 4). This structure is stained by IP726α when cells are prepared by different fixation methods and in different cell types. At metaphase, Mast localizes to centrosomes, the mitotic spindle and centromeres, maintaining this localization during anaphase A. Later, at anaphase B, Mast appears concentrated at the spindle midzone, associated with polar MTs, and shows faint centrosomal localization until late telophase. During very late telophase, it localizes at either side of the midbody, and centrosomal localization is barely detectable. After cytokinesis, Mast can be seen associated again with the rod-like structure (data not shown). This structure might correspond to the remains of the midbody.
Fig. 4. Immunolocalization of Mast in S2 Drosophila culture cells. Individual images for DNA, Mast and α-tubulin are shown. In the merged images, DNA is in blue, Mast in red and α-tubulin in green. Cells in interphase show Mast localized in a punctuate cytoplasmic pattern. At prophase, Mast is found at the centrosomes and most of the time is also associated with an unidentified rod-like structure (arrowhead). During metaphase, Mast associates with spindle microtubules and it is also concentrated at the centromeres. During anaphase, Mast is found at the spindle poles, microtubules and at the spindle midzone. At early telophase, the whole spindle midzone is labelled and some Mast is still present at the spindle poles, and at later stages Mast localizes at either side of the midbody even after cytokinesis is almost complete. Bar = 10 µm.
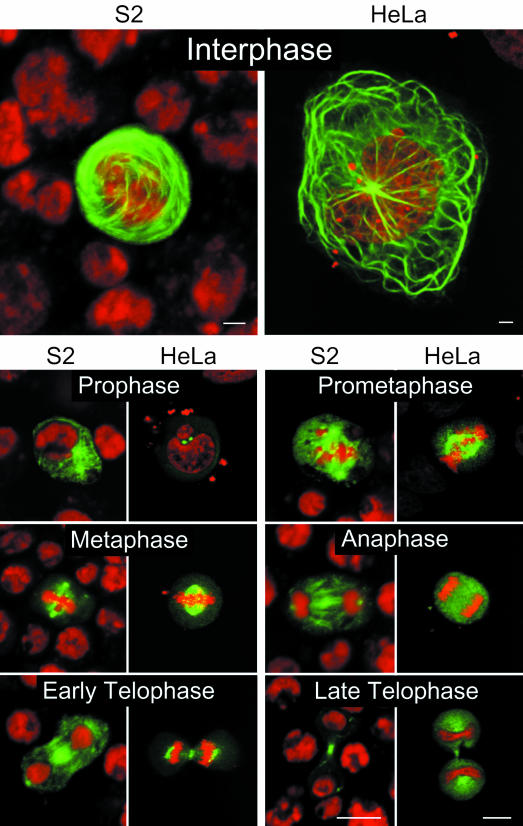
To confirm the specificity of the immunofluorescence labelling, the coding region of mast was cloned in both Drosophila and mammalian transfection vectors to express an enhanced green fluorescent protein (EGFP)–Mast fusion protein in S2 and HeLa cells. As a control, we expressed EGFP alone and verified that it has a homogeneous distribution in S2 or HeLa cells (data not shown). However, the EGFP–Mast fusion protein follows a pattern of localization in both cell types similar to that described in the previous section (Figure 5). In interphase, EGFP–Mast signal is strongly associated with MTOCs and with a fibrillar network that resembles MT bundles. This extensive fibrillar network is not observed in transfected cells treated with colchicine (data not shown). At prophase, the protein localizes to the cytoplasm and shows accumulation at the centrosome. Later, during prometaphase/metaphase, spindle association is clearly evident, as well as localization to the centrosomes and centromeres. During early anaphase, EGFP–Mast appears more diffuse, although, at later stages, centrosomes, spindle MTs and the spindle midzone show accumulation of the protein. In telophase, both S2 and HeLa cells show EGFP signal at the midbody and centrosomes. These results indicate that Mast associates with MTs and centrosomes during most of the cell cycle but undergoes accumulation in additional structures during mitosis.
Fig. 5. Transfection of EGFP–Mast in Drosophila (S2) and human (HeLa) culture cells. DNA is shown in red and EGFP–Mast in green. During interphase, both S2 and HeLa cells show strong EGFP–Mast signal associated with a fibrillar network that resembles microtubule bundles. In prophase, EGFP–Mast is restricted to the centrosomes. At prometaphase and metaphase, EGFP–Mast signal accumulates at the spindle poles, spindle microtubules and the centromeres. During anaphase, spindle poles, microtubules and a more diffuse cytoplasmic signal are observed. Finally, at telophase, EGFP–Mast localizes to the centrosomes and spindle midzone. Bar = 10 µm.
Microtubule-binding assays
Indirect and direct localization show that Mast associates with MTs throughout the cell cycle. To determine whether Mast is a MAP, we performed subcellular fractionation and followed the protein using IP726α. The results show that during sequential purification of MTs from embryo extracts, Mast remains tightly bound to the insoluble fraction of polymerized MTs and can be partially released from the polymer after incubation in high salt (Figure 6). These in vitro results support the intracellular localization data, suggesting that Mast associates with MTs.
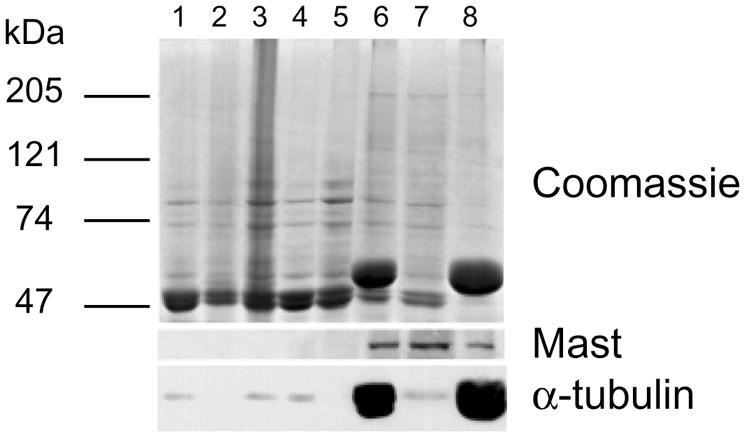
Fig. 6. Mast binds to microtubules in vitro. Microtubules were purified from 0- to 3-h-old embryos by sequential rounds of polymerization and depolymerization. Lane 1, crude extract; lane 2, low speed pellet; lane 3, supernatant. The supernatant was centrifuged at high speed and the resulting supernatant (lane 4) was incubated on ice to depolymerize microtubules, followed by incubation with taxol and GTP at 20°C to repolymerize microtubules. After saccharose gradient centrifugation, the soluble material (lane 5) was separated from microtubules and associated proteins (lane 6). MAPs (lane 7) were extracted from microtubules (lane 8) with 0.5 M NaCl. Samples (30 µg) from each purification stage were separated by SDS–PAGE and the gel stained with Coomassie Blue (top panel) or immunoblotted with IP726α (middle panel) and anti-α-tubulin antibodies (bottom panel).
Localization of Mast in cells arrested with colchicine
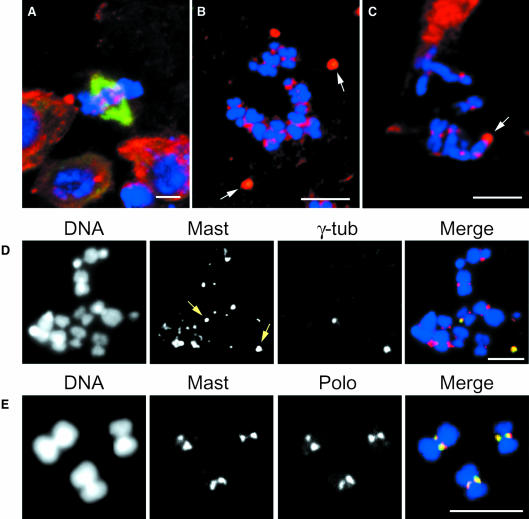
Since Mast is a MAP, we wanted to determine whether its localization to the various compartments of the mitotic apparatus depends upon active MT polymerization. Accordingly, S2 tissue culture cells were incubated in the presence of colchicine for various periods of time, fixed and immunostained with IP726α and with an α-tubulin (Figure 7A and B) or γ-tubulin (Figure 7D) antibody. The results show that after 8 or 16 h incubation, no significant spindle MTs are present in these cells (Figure 7B–D). In all cells analysed, Mast shows co-localization with γ-tubulin (Figure 7D), suggesting that microtubules are not required to maintain its localization at the centrosome. Furthermore, in the absence of MTs, Mast shows strong accumulation at the primary constriction of highly condensed chromosomes (Figure 7B–D). In order to determine whether this localization corresponds to the centromere, isolated mitotic chromosomes from S2 cells were stained for both Mast and the mitotic kinase Polo that was previously shown to accumulate at this site (Logarinho and Sunkel, 1998). The results obtained show that Mast and Polo co-localize at the centromere of isolated chromosomes (Figure 7E).
Fig. 7. Immunolocalization of Mast after microtubule depolymerization. S2 cells were grown for 0, 8 or 16 h in the presence of colchicine. Cells were stained to reveal Mast (red), and α-tubulin (A–C) or γ-tubulin (D) (green). Isolated chromosomes (E) were stained to reveal Mast (red) and Polo (green). DNA is shown in blue. Control cells (A) show a well-organized bipolar spindle and Mast localization to the spindle poles and the centromeres. In cells incubated in colchicine for short (8 h, B) or longer periods (16 h, C and D), microtubules depolymerize and Mast remains associated with both centrosomes (arrows) and centromeres. Mast staining co-localizes with γ-tubulin at the centrosomes (D) and with Polo at the centromeres (E). Bar = 5 µm.
Organization of the mitotic apparatus in mast mutant neuroblasts
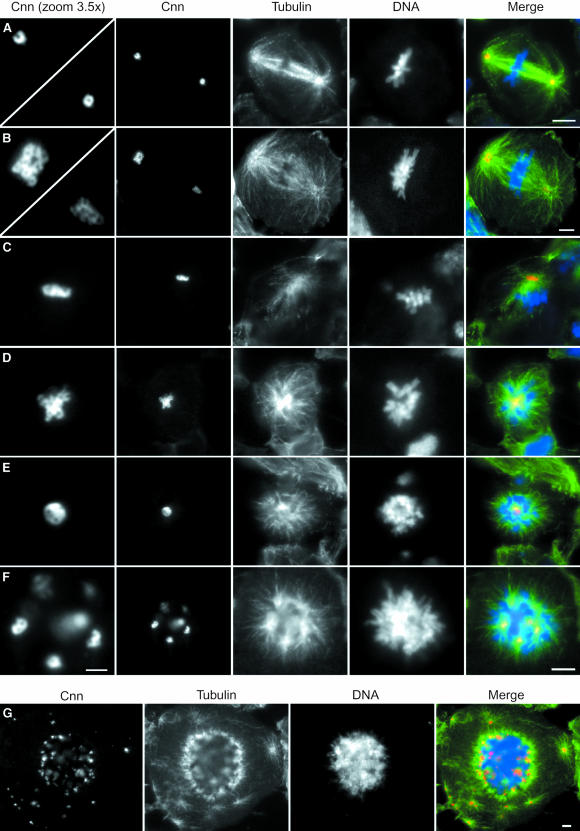
To characterize the organization of the mitotic apparatus in mast mutant cells, neuroblasts from mutant larvae were immunostained to visualize the spindle (α-tubulin) and centrosomes (CNN, γ-tubulin or CP190). Since the results with all three centrosomal markers are very similar, only CNN staining is shown (Figure 8). The wild-type control cell at metaphase shows a typical neuroblast bipolar spindle (Figure 8A). CNN stains the centrosomes in a ring-like pattern that is seen clearly in the amplified image shown in the left panel. All other images are from mast mutant cells that display various degrees of disorganization of the mitotic apparatus. Extensive analysis of mutant neuroblasts stained with α-tubulin to reveal mitotic MTs suggests that the overall morphology of MTs is not the same as that of the wild-type controls. A comparison between control (Figure 8A) and mutant (Figure 8B) cells at metaphase highlights the differences. While in the wild-type cell MTs are generally straight and form tight bundles, in mutant cells MTs are generally irregular in shape and do not always appear straight. Occasionally, mutant cells are able to organize bipolar spindles; however, both poles appear associated with large asters (Figure 8B). CNN staining shows that spindle poles contain an irregular number of ring-like structures. A higher magnification of the MTOCs indicates that in this cell, one of the poles contains at least nine centrosomes while the other pole contains several poorly defined centrosomes. Many cells were found to contain only a single MTOC with a large associated aster and multiple ring-like structures that are stained by CNN (Figure 8C and D). Some cells contain a single aster and show no organized ring-like CNN-positive structures but a rather diffuse staining in the form of a ball that localizes to the middle of the condensed chromosome mass (Figure 8E). We also observe many cells in which chromosomes are organized into a sphere with MTs emanating from a number of irregular sized foci that are all CNN positive and occasionally display pairs of ring-like structures reminiscent of two adjacent normal centrosomes (Figure 8F).
Fig. 8. Centrosomes and mitotic spindles in mast mutant cells. Squashed preparations of brains isolated from either wild-type (A) or various mast mutant allelic combinations (B–G) were incubated with an anti-CNN antibody to reveal the centrosome, anti-α-tubulin antibody to visualize the microtubules and DAPI to stain DNA. The first column shows a magnified view of the CNN staining of centrosomes shown in the second column (A–F). In the last column, individual images of CNN (red), α-tubulin (green) and DNA (blue) have been merged. (A) During metaphase, wild-type neuroblasts show a normal bipolar spindle organized from ring-like CNN-positive structures. (B) Most mast mutant cells show abnormal spindle organization even if a polyploid cell is able to organize a bipolar structure. The spindle poles are highly unequal in size and show many ring-like structures tightly associated. (C) Monopolar spindle in a mast mutant cell organized from two associated ring-like structures. (D) Monopolar spindle in a mast mutant cell organized by an aggregate of CNN-positive centrosomal material that shows no clear internal organization. (E) Mutant cell with highly condensed chromosomes organized into a ball-like structure around a mass of centrosomal material. Note the extensive microtubule network emanating from the single large centrosome. (F) Mutant cell with highly condensed chromosomes organized around several CNN-positive aggregates from which asters are irradiated. Note, in the magnified view of the centrosome, two pairs of ring-like structures resembling normal centrosomes. (G) Highly polyploid cell exhibiting a large number of centrosomal aggregates within the chromosome mass, as well as multiple cytoplasmatic CNN-positive aggregates. Note that all CNN-positive aggregates are associated with microtubule asters. Bar = 2 µm in the ‘CNN zoom 3.5×’ column and 5 µm in all other figures.
Finally, in various combinations of mutant alleles, we observed highly polyploid cells that contain CNN-positive centrosome aggregates inside the chromosome mass as well as other CNN-positive aggregates in the surrounding cytoplasm (Figure 8G). These cytoplasmic aggregates are also capable of nucleating MT asters. These results suggest that mast mutant cells are capable of replicating their centrosomes but either cannot segregate them properly or they segregate but later collapse, forming aggregates. Since it has not been possible to determine the exact centrosome number and ploidy in the same cells, we cannot exclude the possibility that mast mutant cells over-replicate their centrosomes. Nevertheless, these abnormal centrosome aggregates do nucleate MT asters but rarely are able to organize bipolar spindles. However, the circular organization of condensed chromosomes located around a large centrosome from which MTs are nucleated suggests that chromosomes do interact with these MT asters.
Spindle checkpoint in mast mutant neuroblasts
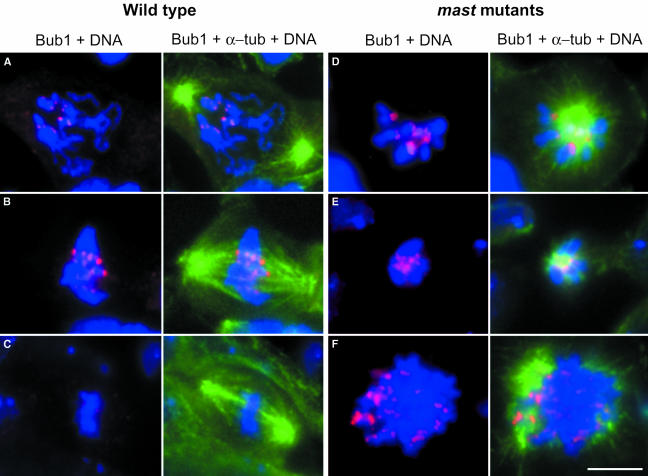
Mutations in mast cause mitotic arrest; however, many cells progress into subsequent cell cycles and become polyploid. To determine whether mast mutant cells are able to activate the spindle checkpoint, we have stained mast mutant cells with antibodies against the checkpoint protein Bub1 (Basu et al., 1999) and α-tubulin to visualize the spindle (Figure 9). In wild-type controls (Figure 9A–C), Bub1 localizes to centromeres during prophase and prometaphase and is virtually absent during metaphase. However, condensed mitotic chromosomes in mutant cells that show monopolar spindles (Figure 9D and E) or that are highly polyploid (Figure 9F) consistently show strong Bub1 accumulation at the centromere. Furthermore, incubation of mastP1 mutant cells in colchicine results in a significant prometaphase arrest (>98%), and no premature sister chromatid separation is observed (data not shown). These results suggest that mast mutant cells have an active spindle checkpoint response.
Fig. 9. Immunolocalization of a spindle checkpoint protein in mast mutant cells. Wild-type (A–C) or mast mutant (D–F) neuroblasts were stained with antibodies against Bub1 (red) and α-tubulin (green). DNA is shown in blue. In wild-type cells, Bub1 accumulates at the centromeres during prophase (A) and prometaphase (B) and is severely reduced as the chromosomes align in the metaphase plate (C). In mast mutant cells, Bub1 shows strong accumulation at the centromeres of chromosomes in monopolar (D and E) and polyploid (F) cells. Bar = 5 µm.
Discussion
We have identified a new Drosophila gene, multiple asters (mast), which encodes an essential protein required for the organization and function of the mitotic spindle. Mast is a MAP that appears to define a new family of proteins conserved from yeast to man. Mutations in mast cause severe alterations in chromosome segregation. The Mast protein shows co-localization with interphase MTs and, during mitosis, associates with centrosomes, the mitotic spindle, centromeres and the spindle midzone. We propose that the Mast protein is required for accurate centrosome separation and organization of the bipolar spindle during mitosis.
Mast is part of a new family of proteins
Database searches and phylogenetic analysis have shown that Mast is part of a new conserved family of proteins that so far contains two human, three C.elegans and two yeast members. Mast is more closely related to the human and the three C.elegans proteins than to either the S.pombe or S.cerevisiae proteins. Indeed, extensive identity is observed between the Drosophila, human and C.elegans members throughout the protein sequence. Interestingly, when the EGFP–Mast fusion protein is expressed in HeLa cells, it shows a pattern of cell cycle-dependent localization like that in Drosophila cells. Furthermore, antibodies raised against one of the human proteins also show a very similar immunolocalization pattern to that described for Mast (H.Maiato and C.E.Sunkel, unpublished observations). The strong sequence conservation together with the localization data suggest that the human and Drosophila proteins might be functionally related.
Mast binds microtubules and localizes to multiple compartments of the mitotic apparatus
Sequence analysis of the Mast protein revealed that it shares conservation with both bovine and mouse MAP4. The homology is restricted to a domain rich in proline and basic residues thought to be involved in MT binding (Aizawa et al., 1991). This domain falls outside the conserved regions CR-1, CR-2 and CR-3. Although we have not determined experimentally whether this region of the protein is responsible for MT binding, our results indicate that Mast co-sediments with polymerized MTs in a salt-stable complex. The immunolocalization and transfection studies provided further evidence that Mast is a MAP since the protein shows clear co-localization with both interphase and spindle MTs. In agreement with this, Stu1p was also demonstrated to interact with MTs (Pasqualone and Huffaker, 1994). However, Mast also localizes to other compartments of the mitotic apparatus even in the presence of MT poisons. The presence of two putative p34cdc2 phosphorylation sites suggests that Mast might undergo specific post-translational modifications during the G2/M transition, allowing it to reach specific mitotic structures. A number of MAPs have been shown to be phosphorylated upon entry into mitosis, allowing for modifications in MT dynamics to take place (Verde et al., 1992).
Mast shares limited homology with members of the dis1-TOG family; however, like Mast, all these proteins localize to centrosomes and/or to spindle MTs during mitosis. In some aspects, Mast shows patterns of localization closer to those of ZYG-9, p93dis1 and XMAP215. These three proteins co-localize with interphase MTs and, during mitosis, ZYG-9 and p93dis1 localize to the centrosome in the absence of MTs. Nevertheless, during late stages of mitosis, Mast shows strong localization to the spindle midzone, like Msps and ch-TOG, two other members of the dis1-TOG family. Mast and ZYG-9, however, do show some unique features since both proteins remain localized to the centromeres of mitotic chromosomes when cells are incubated in the presence of MT-depolymerizing agents. The localization of Mast to MTs, centrosomes, centromeres and the spindle midzone suggests very strongly that this protein might play a role in the regulation of MT dynamics, as has been shown in vitro and in cell-free extracts for some members of the dis1-TOG family (Tournebize et al., 2000).
Mast is required for centrosome segregation and bipolar spindle organization
Genetic analysis of the mast locus shows that this protein is required for the organization of the mitotic spindle. Mutant neuroblasts show highly abnormal mitotic figures, rarely organize bipolar spindles and most of them contain one or more MTOCs of irregular size and shape. Detailed analysis of the abnormal MTOCs indicates that these are composed mostly of multiple CNN-positive ring-like structures that are present in all normal centrosomes. Therefore, it is unlikely that mutations in mast cause an abnormal organization of the centrosome itself. Furthermore, the abnormal MTOCs observed in mast mutant cells contain many CNN-positive ring-like structures, suggesting that centrosome replication is not affected. One possible interpretation of our data could be that in mutant cells centrosomes do segregate but at a later stage collapse, forming a monopolar configuration as was shown for mutations in KLP61F (Sharp et al., 1999). However, in strong mutant alleles, multiple MTOCs of irregular size are associated with MT asters that are found dispersed throughout the cell. In this context, mutations in mast display a phenotype similar to that of zyg-9, which shows numerous cytoplasmic clusters of short MTs during meiosis (Kemphues et al., 1986). However, ZYG-9 is thought to be required for the organization of long MTs during meiosis, and mutations in mast do not appear to cause a significant reduction in MT length (see Figure 8). Nevertheless, the morphology of MTs in mast mutant cells is not normal, appearing to be much more irregular in shape. Furthermore, disruption of STU1 in S.cerevisiae causes severe defects in spindle assembly (Pasqualone et al., 1994). Accordingly, the highly abnormal pattern of centrosome segregation observed as a result of mutations in mast could be due to abnormal MT organization.
Mast and the control of mitotic progression
If Mast is essential for normal centrosome segregation and bipolar spindles are rarely observed, it might be expected that the spindle checkpoint (Nicklas, 1997) would prevent these cells from advancing into a new cycle of proliferation. However, despite the increase in mitotic index, mutations in mast cause the formation of highly polyploid cells. Mutant cells respond to the spindle checkpoint when arrested with colchicine in prometaphase, since premature sister chromatid separation is never observed. Furthermore, mutant cells with abnormal spindle morphology and highly condensed chromosomes show strong accumulation of the spindle checkpoint protein Bub1. This staining pattern is similar to that observed in chromosomes of cells arrested in prometaphase after MT depolymerization (Basu et al., 1999), suggesting that in mast mutant cells the interactions that occur between MTs and chromosomes are unable to inactivate the spindle checkpoint, leading to a prolonged prometaphase arrest. However, since highly polyploid cells are formed, these cells must have undergone multiple cell cycles in the absence of chromosome segregation and cytokinesis. Therefore, it is most likely that after some time, these cells adapt; they either become insensitive or override the spindle checkpoint and progress into a new cycle of proliferation. Indeed, the length of time that different cell types remain in M phase in the presence of microtubule inhibitors varies widely (Kung et al., 1990).
Materials and methods
Genetic variants
mastP1 was described initially as the v401 allele of the gene v40 (Fedorova et al., 1997). It has a P{1ArB} insertion in position 78C1–C2. Two other mutant alleles, mastP2 and mastP3, EP(3)3515 and EP(3)3403, respectively, were obtained from BDGP. mastP4 was obtained by mobilization of the P-element of mastP1 as described below. Df(3L)31A, a deficiency for the region 78A–78E, was obtained from Bloomington Drosophila Stock Centre. All lines were balanced over TM6B. Oregon-R strain was used as wild-type control. All stocks were grown at 25°C under standard conditions and media.
Mobilization of the P-element of mastP1 and generation of mastP4
mastP1/TM6, Tb, Hu, e females were crossed with Δ(2–3), Sb, e/TM6, Ubx, e males. Resulting Δ(2–3), Sb, e/mastP1 males were crossed individually with red1, mbcC1, e1/TM3, Sb1, ry1, e1 females (Umea). Excision of the P insertion was recognized by the loss of ry+. From 35 000 chromosomes scored, we obtained 39 lines that had lost the insertion and were viable, fertile and had no mitotic phenotype. Among the non-viable ry– lines, we identified one line, mastP4, in which homozygotes died as late larvae/pupae and which cytological analysis revealed to have very severe mitotic defects.
Cytological analysis
The analysis of the mitotic phenotype was carried out according to González and Glover (1993). For quantification of the mitotic parameters, the numbers of cells present in 50 randomly chosen optic fields (100×) per brain were counted. Five brains were scored for each allele. All the calculations were based on the sum of the total number of cells present in the five brains scored.
Molecular analysis
Genomic fragments adjacent to a P-element insertion were cloned by plasmid rescue and inverse PCR and sequenced. Three cDNA clones were identified in the BDGP database: LD11488 (0–24 h mixed stage embryonic library), LP08134 (larval–early pupal library) and GH26741 (adult head library). The genomic sequence (AC014071), from Celera Genomics, includes the complete mast gene. The LD11488 cDNA clone was sequenced completely with the T7Sequencing Kit™ (Pharmacia). This cDNA is 5938 bp long, with a 769 bp 5′-untranslated region and a 3′-untranslated region of 679 bp.
Database searches and sequence analysis
BLAST (Altschul et al., 1997) searches were performed in non-redundant GenBank CDS translations, PDB, SwissProt, SPupdate and PIR databases. All the unpublished protein sequences that were used in this study can be found under the following accession numbers: CeC07H6.3 (CE01153), SpStu1p (CAA15921), At-TOG (AAD15450) and SpAlp4p (CAA22843). Phylogenetic analysis and unrooted dendrogram elaboration were performed using PHYLIP (Retief, 2000).
Antibodies and western analysis
In order to express a segment of the Mast protein in Escherichia coli, the LD11488 cDNA was digested with BglII and SacI. The resulting 1281 bp fragment was subcloned in the pQE-32 vector (Qiagen), resulting in plasmid pQE-Mast1. The recombinant Mast1 protein was purified from inclusion bodies isolated from E.coli transformed with pQE-Mast1 after isopropyl-β-d-thiogalactopyranoside (IPTG) induction and used for immunization in rabbits (Diagnostics Scotland, Edinburgh, UK). The polyclonal Rb726 serum was subsequently immunopurified (IP726α) against the recombinant protein immobilized on nitrocellulose (according to Sambrook et al., 1989). For western blotting, total protein extracts from embryos, brains, testes and ovaries were prepared in SDS–PAGE sample buffer as previously described (Sunkel et al., 1995), separated by SDS–PAGE and blotted to nitrocellulose. Membranes were incubated with IP726α (1:100) or with an anti-α-tubulin monoclonal antibody (Amersham) (1:500) and detection was with horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG (Vector, UK) (1:1000). The signal was developed with the ECL Chemiluminescent Detection System (Amersham, UK).
Immunofluorescence in S2 culture cells
Drosophila S2 cells were grown in Schneider’s medium (Gibco-BRL) with 5% fetal bovine serum (FBS), at 25°C. For microtubule depolymerization, cells were incubated with 30 µM colchicine (Sigma, USA) for 8 or 16 h. Cells were spun against a slide at 1000 r.p.m. for 5 min, fixed in 3.7% formaldehyde in PHEM (60 mM PIPES, 25 mM HEPES pH 7.0, 10 mM EGTA, 4 mM MgSO4) for 12 min and washed three times for 5 min with phosphate-buffered saline (PBS). Blocking was performed in PBS-TF (1% Triton X-100, 10% FBS in PBS) with 0.5 mg/ml RNase, for 1 h at room temperature. Slides were then incubated with IP726α and a mouse anti-α-tubulin antibody (Amersham) in PBS-TF, at a 1:10 and 1:200 dilution, respectively, for 45 min at room temperature and washed in PBS. Fluorescein isothiocyanate (FITC) –α-rabbit conjugated IgG (Vector, UK) and Cy5–α-mouse conjugated IgG (Jackson Laboratories, USA) were used as secondary antibodies, diluted 1:1000 and 1:200, respectively. Cells were washed as before, DNA stained with propidium iodide and the preparation mounted in Vectashield (Vector, UK). Preparations were observed with a Bio-Rad MRC600 confocal microscope and images processed with Photoshop 5.5 (Adobe Systems).
Cell transfection and GFP analysis
EGFP–Mast constructs prepared done by linker-mediated subcloning. A linker corresponding to the 5′ end of the mast coding sequence including a 5′-protruding extremity compatible with BglII and a 3′-protruding end compatible with BstXI was produced by the annealing of complementary oligonucleotides. A 5128 bp BstXI–XhoI mast cDNA fragment was ligated in the presence of the linker to the plasmids pMTEGFP-C1 (T.Megraw, unpublished) or pEGFP-C1 (Clontech), both digested with BglII and SalI, resulting in plasmids pMTEGFP-Mast and pEGFP-Mast. These plasmids were used to transfect, respectively, Drosophila S2 and human HeLa cells. Cells were transfected using 1 µg of plasmid, prepared by QIAGEN midiprep (Qiagen), and the FuGENE™ 6 Transfection Reagent (Boehringer Mannheim). After 24 h of growth, expression of EGFP–Mast in S2 cells was induced from the metallothionein promoter by 1.0 mM CuSO4. Significant expression was detected 9 h after induction. Cells were spun, fixed and washed as described above. The expression of EGFP–Mast in transfected HeLa cells is constitutive and detected soon after transfection. Cells were grown for 24 h, washed twice for 5 min with PBS, fixed in 3% formaldehyde in PBS for 10 min, and washed as above. S2 and HeLa preparations were mounted in Vectashield (Vector, UK) after DNA staining with propidium iodide, observed with a Bio-Rad MRC600 confocal microscope and images processed as above.
Chromosome isolation
S2 cells were arrested in mitosis with 30 µM colchicine (Sigma, USA) for 16 h. Chromosomes were then prepared as previously described (Bousbaa et al., 1997). Immunofluorescence was performed as above, using IP726α and a monoclonal anti-Polo antibody (Logarinho and Sunkel, 1998).
Microtubule-binding assays
Microtubules were purified from 0- to 3-h-old Drosophila embryos as described by Saunders et al. (1997). Samples of the various steps were collected and 30 µg of total protein of each extract were separated by SDS–PAGE and stained with Coomassie Blue or subjected to western blotting and probed with IP726α or an anti-α-tubulin monoclonal antibody (Amersham) as described above.
Immunofluorescence in brains
The whole brain of third instar larva was prepared for immunostaining as previously described (Bonaccorsi et al., 2000). Immunostaining of microtubules and centrosomes was performed with antibodies against α-tubulin (Amersham) and CNN (Heuer et al., 1995) diluted 1:200 and 1:500, respectively. The antibodies were visualized with FITC–α-mouse conjugated IgG (Vector, UK) or Cy3–α-rabbit conjugated IgG (Jackson Laboratories, USA). To visualize the Bub1 protein, we used antibodies as described previously (Basu et al., 1999). The preparations were analysed in a Zeiss Axioskop microscope, and images acquired with a SPOT 2 camera (Diagnostic Instruments, USA) and processed as above.
Acknowledgments
Acknowledgements
We would like to thank T.Megraw for the EGFP plasmids and advice on cell transfection, and T.Kaufman for the anti-CNN antibody. We are grateful to E.Bronze-da-Rocha for help with HeLa cell cultures, all the staff of C.E.S.’s laboratory for helpful comments and stimulating discussions, M.J.Falcão and T.Lopes for laboratory assistance, and A.Santos for general technical support. C.L. and H.M. hold PhD studentships and P.S. holds a postdoctoral research fellowship from the PRAXIS XXI programme of the FCT of Portugal. H.M. is a student of the Gulbenkian PhD Programme in Biology and Medicine. C.E.S. is financed by grants from FCT of Portugal and the TMR programme of the EU.
References
- Aizawa H., Emori,Y., Mori,A., Murofushi,H., Sakai,H. and Suzuli,K. (1991) Functional analysis of the domain structure of microtubule-associated protein-4 (MAP-U). J. Biol. Chem., 266, 9841–9846. [PubMed] [Google Scholar]
- Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade M.A. and Bork,P. (1995) HEAT repeats in the Huntington’s disease protein. Nature Genet., 11, 115–116. [DOI] [PubMed] [Google Scholar]
- Basu J., Bousbaa,H., Logarinho,E., Li,Z., Williams,B.C., Lopes,C., Sunkel,C.E. and Goldberg,M.L. (1999) Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila. J. Cell Biol., 146, 13–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaccorsi S., Giansanti,M.G. and Gatti,M. (2000) Spindle assembly in Drosophila neuroblasts and ganglion mother cells. Nature Cell Biol., 2, 54–56. [DOI] [PubMed] [Google Scholar]
- Bousbaa H., Correia,L., Gorbsky,G.J. and Sunkel,C.E. (1997) Mitotic phosphoepitopes are expressed in Kc cells, neuroblasts and isolated chromosomes of Drosophila melanogaster. J. Cell Sci., 110, 1979–1988. [DOI] [PubMed] [Google Scholar]
- Charrasse S.M., Schroeder,M., Gauthier-Rouviere,C., Ango,F., Cassimeris,L., Gard,D.L. and Larroque,C. (1998) The TOGp protein is a new human microtubule-associated protein homologous to the Xenopus XMAP215. J. Cell Sci., 111, 1371–1383. [DOI] [PubMed] [Google Scholar]
- Cullen C.F., Deák,P., Glover,D.M. and Ohkura,H. (1999) mini spindles: a gene encoding a conserved microtubule-associated protein required for the integrity of the mitotic spindle in Drosophila. J. Cell Biol., 146, 1005–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova S.A., Chubykin,V.L., Gusachenko,A.M. and Omel’yanchuk, L.V. (1997) Mutation chromosome bows (chbv40) associated with the abnormal chromosome spindle in Drosophila melanogaster. Genetika, 33, 1502–1509. [PubMed] [Google Scholar]
- González C. and Glover,D.M. (1993) Techniques for studying mitosis in Drosophila. In Fantes,P. and. Brookes,R. (eds), The Cell Cycle: A Practical Approach. Oxford University Press, Oxford, pp. 163–168. [Google Scholar]
- Gräf R., Daunderer,C. and Schliwa,M. (2000) Dictyostelium DdCP224 is a microtubule-associated protein and a permanent centrosomal resident involved in centrosome duplication. J. Cell Sci., 113, 1747–1758. [DOI] [PubMed] [Google Scholar]
- Hemmings B.A., Adams-Pearson,C., Maurer,F., Mueller,P., Goris,J., Merlevede,W., Hofsteenge,J. and Stone,S.R. (1990) α-Forms and β-forms of the 65-kDa subunit of protein phosphatase 2A have a similar 39 amino acid repeating structure. Biochemistry, 29, 3166–3173. [DOI] [PubMed] [Google Scholar]
- Heuer J., Li,K. and Kaufman,T. (1995) The Drosophila homeotic target gene centrosomin (cnn) encodes a novel centrosomal protein with leucine zippers and maps to a genomic region required for midgut morphogenesis. Development, 121, 3861–3876. [DOI] [PubMed] [Google Scholar]
- Ishikawa K., Nagase,T., Suyama,M., Miyajima,N., Tanaka,A., Kotani,H., Nomura,N. and Ohara,O. (1998) Prediction of the coding sequences of unidentified human genes. X. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res., 5, 169–176. [DOI] [PubMed] [Google Scholar]
- Kemphues K.J., Wolf,N., Wood,W.B. and Hirsh,D. (1986) Two loci required for cytoplasmic organization in early embryos of Caenorhabditis elegans.Dev. Biol., 113, 449–460. [DOI] [PubMed] [Google Scholar]
- Kennelly P.J. and Krebs,E.G. (1991) Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J. Biol. Chem., 266, 15555–15558. [PubMed] [Google Scholar]
- Kung A.L., Sherwood,S. and Schimke,R.T. (1990) Cell line-specific differences in the control of cell cycle progression in the absence of mitosis. Proc. Natl Acad. Sci. USA, 87, 9553–9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logarinho E. and Sunkel,C.E. (1998) The Drosophila POLO kinase localises to multiple compartments of the mitotic apparatus and is required for the phosphorylation of MPM2 reactive epitopes. J. Cell Sci., 111, 2897–2909. [DOI] [PubMed] [Google Scholar]
- Matthews L.R., Carter,P., Thierry-Mieg,D. and Kemphues,K. (1998) ZYG-9, a Caenorhabditis elegans protein required for microtubule organisation and function, is a component of meiotic and mitotic spindle poles. J. Cell Biol., 141, 1159–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K., Kurooka,H., Takeuchi,M., Kinoshita,K., Nakaseko,Y. and Yanagida,M. (1995) p93dis1, which is required for sister chromatid separation, is a novel microtubule and spindle pole body-associating protein phosphorylated at the Cdc2 target sites. Genes Dev., 9, 1572–1585. [DOI] [PubMed] [Google Scholar]
- Nakaseko Y., Nabeshima,K., Kinoshita,K. and Yanagida,M. (1996) Dissection of fission yeast microtubule associating protein p93dis1: regions implicated in regulated localisation and microtubule interaction. Genes Cells, 1, 633–644. [DOI] [PubMed] [Google Scholar]
- Nicklas R.B. (1997) How cells get the right number of chromosomes. Science, 275, 632–637. [DOI] [PubMed] [Google Scholar]
- Omel’yanchuk L.V., Volkova,E.I. and Fedorova,S.A. (1997) Characterisation of insertion mutations leading to mitosis abnormalities in Drosophila melanogaster by means of the reporter gene-containing transposon. Genetika, 33, 1494–1501. [PubMed] [Google Scholar]
- Pasqualone D. and Huffaker,T.C. (1994) STU1, a suppressor of a β-tubulin mutation, encodes a novel and essential component of the yeast mitotic spindle. J. Cell Biol., 127, 1973–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retief J.D. (2000) Phylogenetic analysis using PHYLIP. Methods Mol. Biol., 132, 243–258. [DOI] [PubMed] [Google Scholar]
- Robinson J.T., Wojcik,E.J., Sanders,M.A., McGrail,M. and Hays,T.S. (1999) Cytoplasmic dynein is required for the nuclear attachment and migration of centrosomes during mitosis in Drosophila. J. Cell Biol., 146, 597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Saunders R.D., Avides,M.C., Howard,T., González,C. and Glover,D.M. (1997) The Drosophila gene abnormal spindle encodes a novel microtubule-associated protein that associates with the polar regions of the mitotic spindle. J. Cell Biol., 137, 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W.S. and Hoyt,M.A. (1992) Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell, 70, 451–458. [DOI] [PubMed] [Google Scholar]
- Saxton W.M., Stemple,D.L., Leslie,R.J., Salmon,E.D., Zavortink,M. and McIntosh,J.R. (1984) Tubulin dynamics in cultured mammalian cells. J. Cell Biol., 99, 2175–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp D.J., Yu,K.R., Sisson,J.C., Sullivan,W. and Scholey,J.M. (1999) Antagonistic microtubule-sliding motors position mitotic centrosomes in Drosophila early embryos. Nature Cell Biol., 1, 51–54. [DOI] [PubMed] [Google Scholar]
- Sharp D.J., Brown,H.M., Kwon,M., Rogers,G.C., Holland,G. and Scholey,J.M. (2000) Functional coordination of three mitotic motors in Drosophila embryos. Mol. Biol. Cell, 11, 241–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkel C.E., Gomes,R., Sampaio,P., Perdigão,J. and González,C. (1995) γ-Tubulin is required for the structure and function of the microtubule organising centre in Drosophila neuroblasts. EMBO J., 14, 28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournebize R. et al. (2000) Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nature Cell Biol., 2, 13–19. [DOI] [PubMed] [Google Scholar]
- Vasquez R.J., Gard,D.L. and Cassimeris,L. (1994) XMAP from Xenopus eggs promotes rapid plus end assembly of microtubules and rapid microtubule polymer turnover. J. Cell Biol., 127, 985–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F., Dogterom,M., Stelzer,E., Karsenti,E. and Leibler,S. (1992) Control of microtubule dynamics and length by cyclin A- and cyclin B-dependent kinases in Xenopus egg extracts. J. Cell Biol., 118, 1097–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.J. and Huffaker,T.C. (1997) Stu2p: a microtubule-binding protein that is an essential component of the yeast spindle pole body. J. Cell Biol., 139, 1271–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R. et al. (1994) 2.2 Mb of contiguous nucleotide sequence from chromosome III of C.elegans. Nature, 368, 32–38. [DOI] [PubMed] [Google Scholar]
- Zimmerman W., Sparks,C.A. and Doxsey,S.J. (1999) Amorphous no longer: the centrosome comes into focus. Curr. Opin. Cell Biol., 11, 122–128. [DOI] [PubMed] [Google Scholar]