Abstract
Using a coupled, in vitro transcription and polyadenylation system we have investigated the molecular mechanism of transcriptional termination by RNA polymerase II (PolII). We showed previously that specific G-rich sequences pause transcription and then activate polyadenylation. We show that physiological pause sites activate polyadenylation in our in vitro system. We also investigate the mechanism of PolII transcriptional termination, and show that these transcripts are either directly released from the transcription complex or are 3′ end processed while still attached to the complex. We also show that 3′ product (generated by cleavage/polyadenylation) remains associated with the transcription complex, but is rapidly degraded on it.
Keywords: cleavage/3′ end processing/polyadenylation/transcriptional termination
Introduction
Transcriptional termination by RNA polymerase II is known to be a coupled process in which RNA processing at the poly(A) site of the nascent transcript dictates downstream termination (Proudfoot and Whitelaw, 1986; Connelly and Manley, 1988; Edwalds-Gilbert et al., 1993; Birse et al., 1997, 1998). It is not known how these two spatially separate molecular events interconnect, although recent studies have shown that cleavage/polyadenylation factors associate with the elongating PolII through the C-terminal domain of the largest subunit (Dantonel et al., 1997; McCracken et al., 1997). Also, the C-terminal domain directly activates 3′ processing in vitro, suggesting that PolII may play a direct role in coordinating polyadenylation (Hirose and Manley, 1998). To understand these basic molecular events fully, it is necessary to develop an in vitro system that couples polyadenylation and transcription. We have recently developed such a system, and showed that specific G-rich sequences both pause PolII and activate cleavage/polyadenylation at an upstream poly(A) signal in a transcription-dependent manner (Yonaha and Proudfoot, 1999). Although these G-rich sequences were originally identified as binding sites for the transcription factor MAZ (Ashfield et al., 1994), we have shown that they are also capable of acting as intrinsic pausing sequences to pure PolII (Yonaha and Proudfoot, 1999). This suggests that such sequences may have a general role in modulating co-transcriptional RNA processing events. Indeed, these G-rich sequences have recently been shown to modulate the efficiency of alternative splicing when inserted into an α-tropomysin gene construct (Roberts et al., 1998). The results presented in this paper apply our coupled in vitro transcription–polyadenylation system to investigate the mechanism of transcriptional termination. To do this we have used immobilized DNA templates that allow separation of RNAs attached to the ternary complex of DNA and PolII from released RNA. Our results demonstrate that transcription termination occurs at all stages in intimate association with the transcription complex. We also demonstrate that physiological pause sites function in our in vitro system.
Results
Physiological pause sequences activate 3′ end processing in the coupled system
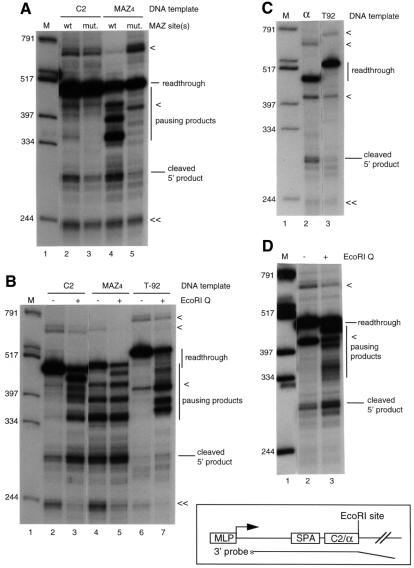
We have previously shown that specific G-rich sequences (four MAZ sites), strongly activate cleavage/polyadenylation at an upstream synthetic poly(A) site (SPA; Levitt et al., 1989) by intrinsic PolII pausing in vitro (Yonaha and Proudfoot, 1999). However, they are artificially constructed sequences used to allow maximum PolII pausing. It is therefore important to test more physiological pause sequences using our coupled in vitro system. It was previously reported that the 3′ flanking sequence of the human C2 gene contains a single MAZ site just downstream of C2 poly(A) signal and functions as a PolII pause site (Ashfield et al., 1991, 1994). We therefore constructed linearized templates containing MLP and SPA followed by the C2 pause site with either a wild-type or mutant MAZ site (Figure 1, diagram). Using these DNA templates, the in vitro coupled reaction was carried out for 2 h and transcripts were analyzed by S1 protection with homologous DNA probes (Figure 1A). With the wild-type C2 template, most transcripts read through the C2 sequences although a weak pausing product at the MAZ site (lower band) and two other pausing products were also visible (upper bands). Weak activation of 3′ end processing was also observed (lane 2). On the other hand, the mutant C2 template did not result in pausing at the MAZ site and gave little cleaved 5′ product (lane 3). As before, the original MAZ4 template produced strong pausing products at the MAZ sites (predominantly the first MAZ site) and strong activation of cleavage (lane 4), which was not observed when the MAZ4 sequence was mutated (lane 5; Yonaha and Proudfoot, 1999). These results indicate that the C2 pause site activates 3′ end processing, although to a lesser extent than the artificial MAZ sites. We also carried out the coupled reaction with a template containing a single artificial MAZ site just downstream of SPA, but observed only weak pausing product at the MAZ site and no significant activation of cleavage/polyadenylation (data not shown). This suggests that the C2 pause site contains additional sequence elements required for activation of 3′ end processing.
Fig. 1. Physiological sequences activate 3′ end processing. (A) The C2 pause site activates 3′ end processing. The wild-type C2 template and the DNA probe are indicated in the diagram. The MAZ site in the C2 pause site and the MAZ4 sequences, GGGGGAGGGGG, was mutated to GGTGAAAGGTG. Lane 1, marker; lane 2, wild-type C2 template; lane 3, mutant C2 template; lane 4, wild-type SPA and wild-type MAZ4 template; lane 5, wild-type SPA and mutant MAZ4 template. (B) A ‘roadblock’ to PolII elongation in the C2 pause site by Gln111 activates 3′ end processing. The EcoRI site is just downstream of the C2 pause site and the MAZ4 sites on the C2 and MAZ4 templates, respectively. The EcoRI site is 92 bp downstream of the poly(A) site on the T-92 template (see Figure 2, diagram). Lane 1, marker; lanes 2 and 3, wild-type C2 template; lanes 4 and 5, wild-type SPA and wild-type MAZ4 template; lanes 6 and 7, T-92 template. The reaction mixtures were preincubated with 0.05 mg of Gln111 EcoRI (lanes 3, 5 and 7) or not (lanes 2, 4 and 6). (C) The α pause site activates 3′ end processing. The α template and DNA probe are indicated in the diagram. Lane 1, marker; lane 2, α template; lane 3, T-92 template. (D) A ‘roadblock’ to PolII elongation in the α pause site by Gln111 activates 3′ end processing. The reaction mixture with the α template was preincubated with 0.05 mg of Gln111 EcoRI (lane 3) or not (lane 2). The coupled reactions were carried out for 2 h and S1 protected DNA products were analyzed on 6% polyacrylamide–8 M urea gel. < indicates a partial digestion product; << indicates a specific hybridization product from the 3′ flanking region near the end of the DNA templates (see Figure 2, diagram).
We next investigated the effect of a ‘roadblock’ to PolII elongation, by a high-affinity DNA-binding protein, on 3′ end processing with templates containing the pause sites. To do this, a mutant EcoRI enzyme (Gln111; Wright et al., 1989), which binds to but does not cleave at EcoRI sites, was added to the coupled reactions with the wild-type C2 template, the wild-type MAZ4 template and T-92 template. The C2 and MAZ4 templates have an EcoRI site immediately following the pause sequences while T-92 has an EcoRI site 92 bp downstream of the SPA (Figure 1B). As before, addition of Gln111 to T-92 template promoted a strong build-up of transcripts ∼30 nucleotides upstream from the EcoRI site, but did not activate cleavage/polyadenylation (lanes 6 and 7; Yonaha and Proudfoot, 1999). Note that the strong partial S1 digestion product (denoted by <) varied in intensity in different experiments. The possibility that spacing of the Gln111-induced ‘roadblock’ from the poly(A) signal might be significant was tested previously by moving the EcoRI site closer to the poly(A) signal (T-36 template in Yonaha and Proudfoot, 1999). No activation of 3′ cleavage was observed with this closer spaced construct. As shown in Figure 1B, lane 3, addition of Gln111 to the reaction mixture with the wild-type C2 template, which has the EcoRI site just downstream of the C2 pause site, generates pausing product ∼30 nucleotides upstream of the EcoRI site and greatly enhances pausing at the single MAZ site. Significantly, this enhanced pausing activates 3′ end processing 3-fold (compare lane 2 with lane 3). In contrast, addition of Gln111 to the reaction mixture with wild-type MAZ4 template, which also has an EcoRI site just downstream of the MAZ4 sequences, produced weak pausing upstream of the EcoRI site and did not affect cleavage/polyadenylation. This result was expected since the MAZ4 sites have strong intrinsic pausing activity, which activates 3′ processing (lanes 4 and 5).
It has been reported previously that a 92 bp sequence from the 3′ flanking region of the human α2 globin gene (α pause site) slows down PolII elongation and consequently activates 3′ end processing in vivo (Enriquez-Harris et al., 1991). A template containing the α pause site downstream of the SPA was therefore constructed and the coupled reaction carried out. As shown in Figure 1C, the α template weakly activated 3′ end processing, but did not give an apparent pausing product. Next, Gln111 was added to the reaction mixture with the α template (Figure 1D). As with C2, addition of Gln111 to the reaction mixture generated not only a pausing product ∼30 nucleotides upstream of the EcoRI sites but also a heterogeneous pausing product over the α pause site coupled with a 2.6-fold enhancement of cleavage/polyadenylation. The α pause site has an A-rich region containing a nearly perfect (CA4)6 repeat sequence, suggesting that the repeat sequence is a potential pause site. These results demonstrate that two different physiological pause sites derived from the 3′ flanking regions of the human C2 and α2-globin genes weakly activate 3′ end processing. In both cases this activation is significantly enhanced by inducing a ‘roadblock’ to PolII elongation using Gln111 binding to downstream EcoRI sites.
Transcripts paused at MAZ sites are resistant to sarkosyl treatment
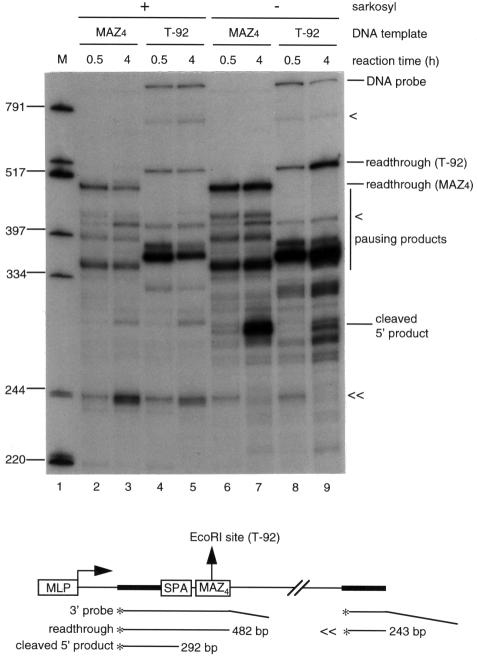
To characterize further the nature of PolII pausing in our coupled in vitro system, we analyzed the stability of transcripts paused at the MAZ4 sites to the detergent sarkosyl, which is known to disrupt initiation of transcription, but has little effect on transcriptional elongation (Hawley and Roeder, 1985). To do this, we carried out the in vitro coupled reaction with the wild-type MAZ4 template and T-92 template in the presence of 0.1% sarkosyl (Figure 2). Gln111 was added to the T-92 reactions to block PolII elongation. Reactions were initiated for 5 min and then sarkosyl was added to the reactions shown in lanes 2–5. We confirmed that addition of 0.1% sarkosyl before starting the reaction inhibited transcriptional initiation (data not shown). Transcription was continued for a total of 0.5 or 4 h. Sarkosyl addition blocks accumulation of transcripts (compare lanes 2–5 with 6–9), presumably through inhibition of re-initiation, and also inhibits 3′ end processing. Sarkosyl has little effect on transcriptional elongation; this can be seen from the accumulation of an S1 product after 4 h (<<, lanes 3 and 5) corresponding to transcripts that have read through to near the end of the linear DNA templates. This product is detectable due to duplication of the polylinker sequence that is detected by the S1 probe (see diagram in Figure 2). It is particularly notable that the MAZ4 products are not significantly reduced after 4 h in the presence of sarkosyl. Although the more dominant Gln111 product is somewhat reduced after 4 h, no significant increase in readthrough transcription is detectable. This may be, in part, due to RNA degradation occurring during the 4 h incubation period. These results demonstrate that the MAZ products are not truly transcriptional pause sites, but instead appear to represent irreversibly halted transcripts that may then be subjected to 3′ end processing.
Fig. 2. Transcripts paused at MAZ sites are resistant to sarkosyl treatment. Lane 1, marker; lanes 2, 3, 6 and 7, wild-type SPA and wild-type MAZ4 site template; lane 4, 5, 8 and 9, T-92 template. The templates are indicated in the diagram with 3′ probe. The reaction mixture with the T-92 template was preincubated with 0.05 mg of Gln111 EcoRI. The coupled in vitro transcription–polyadenylation reaction was initiated for 5 min at 30°C without RNase inhibitor, then 0.1% sarkosyl was added to the reactions shown in lanes 2–5 and reactions were continued. The total reaction times are indicated. S1-protected DNA products were analyzed on 6% polyacrylamide–8 M urea gel. < and << are as in Figure 1 (see the diagram).
To confirm that the S1 protected bands at the MAZ sites correspond to authentic 3′ termini, transcripts obtained with the MAZ4 template were hybridized to DNA probes and digested with exonuclease VII, a single-strand-specific exonuclease. Exonuclease VII digestion showed the same products obtained with S1 nuclease at each MAZ site, confirming that they are authentic 3′ ends (data not shown).
3′ end processing occurs on the ternary complex
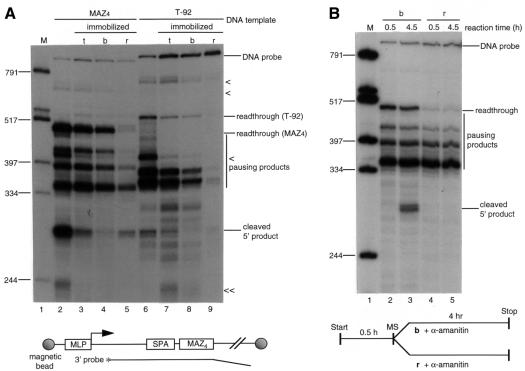
To analyze the mechanism of the PolII transcriptional termination in greater detail, we have used immobilized templates with the coupled system. The MAZ4 and T-92 templates were end-labelled with biotin and attached at both ends to streptavidin-coated magnetic beads (Figure 3A, diagram) to allow separation of transcripts still attached to the template and PolII (lanes b) from those released into the supernatant (lanes r) (see Materials and methods). As shown in Figure 3A, coupled reactions with normal and immobilized templates were carried out and transcripts were fractionated by magnetic selection. Transcripts obtained were analyzed as before by S1 nuclease. Lanes 2–5 show the experiment with the MAZ4 template. The unfractionated reaction (t) with immobilized template gives lower cleavage/polyadenylation activity (lane 3) than normal template (lane 2), indicating that magnetic beads partially inhibit polyadenylation activity. However, after magnetic selection (lanes 4 and 5), almost all cleaved 5′ product was present in the supernatant fraction. About 30% of paused transcript at the MAZ sites was also released from template, while readthrough transcript was wholly template-bound. With the T-92 template (lanes 6–9), almost all of the Gln111 pausing products remained in the template-bound fraction. These results indicate that the transcripts halted at MAZ sites (which activate polyadenylation) appear to be associated with the ternary complex less stably than readthrough transcripts or those paused by Gln111, even though they are resistant to sarkosyl treatment. In effect, the transcripts halted and then released from the MAZ sites are terminated independently of polyadenylation.
Fig. 3. Transcript analysis using immobilized templates. (A) Transcripts paused at MAZ sites are partially released from the ternary complex. Lane 1, marker; lane 2, wild-type SPA and wild-type MAZ4 site template; lanes 3–5, immobilized wild-type SPA and wild-type MAZ4 template (indicated in the diagram with 3′ probe); lane 6, T-92 template; lanes 7–9, immobilized T-92 template. The reaction mixture with T-92 and immobilized T-92 templates was preincubated with 0.05 mg of Gln111 EcoRI. The coupled in vitro transcription–polyadenylation reaction was carried out for 3 h. Transcripts with immobilized templates were then fractionated into template-bound (b) and supernatant fractions (r) by magnetic selection. The unfractionated total reaction (t) is shown. These transcripts were analyzed by S1 nuclease protection using 3′ probe on 6% polyacrylamide–8 M urea gel. (< and << are as in Figure 1). (B) Activation of polyadenylation occurs on the ternary complex. Transcripts produced during 30 min incubation with the immobilized wild-type SPA and wild-type MAZ4 template were fractionated. The bound fraction was mixed with supernatant obtained after 30 min incubation with magnetic beads. α-amanitin (2 µg/ml) was added to both fractions to halt transcription and incubated at 30°C for a further 4 h. The released fraction was supplemented with additional nuclear extract in a separate experiment (data not shown). RNAs were analyzed by S1 nuclease protection using 3′ probe on a 6% polyacrylamide–8 M urea gel. b and r are as in (A). The diagram below outlines the experimental approach. MS denotes magnetic selection.
We next wished to determine whether transcripts released at MAZ sites are capable of further cleavage and polyadenylation. We therefore modified the coupled reaction as follows. Transcription of the immobilized MAZ4 templates was carried out for 30 min, enough time to produce paused transcripts but not enough for cleavage products to accumulate. These transcripts were fractionated and then incubated for 4 h with reaction mixtures containing 2 µg/ml of α-amanitin, which blocks all transcription (both initiation and elongation) but allows the cleavage/polyadenylation reaction to occur (Figure 3B). Surprisingly, template-bound transcripts were processed (lane 3), while released transcripts were not (lane 5). It should be noted that the coupled system was carried out using 4 mM MgCl2, and that 3′ processing of the released transcript would not be expected to occur under such conditions (see Yonaha and Proudfoot, 1999). It is also possible that the released fraction lacked essential factors retained in the bound fraction so that additional nuclear extract was added. However, still no cleavage/polyadenylation was detected (data not shown). These results demonstrate that activation of cleavage/polyadenylation in the coupled system occurs on the assembled ternary complex (nascent RNA, DNA template and PolII). The released transcripts are not capable of 3′ end processing under these conditions. Finally, it should be noted that although the cleavage products observed in these studies were not directly tested for the presence of poly(A) tails, they have previously been shown to be polyadenylated RNAs (Yonaha and Proudfoot, 1999).
3′ product remains associated with the ternary complex and is rapidly degraded
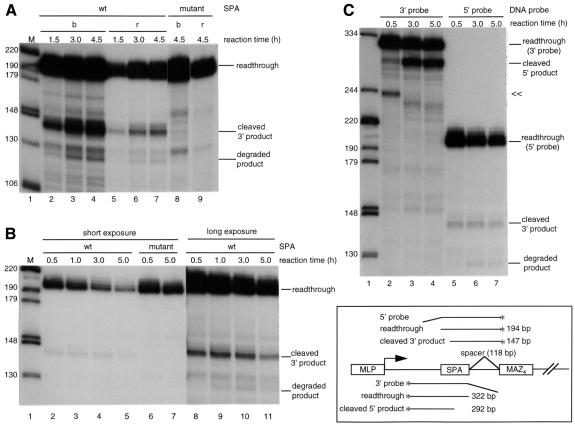
To characterize the mechanism of PolII termination fully, it is necessary to follow the fate of the 3′ product separated from the 5′ polyadenylated RNA. Such analysis has proved technically impossible in vivo as this 3′ product is highly unstable and consequently undetectable. However, as described in these studies, we can track the fate of the 3′ product in our in vitro system. To allow easier identification of the 3′ product, we constructed immobilized templates containing a 118 bp spacer between the wild-type or mutant SPA and MAZ4 sites (Figure 4, diagram). The insertion of the spacer does not affect the enhancement of cleavage/polyadenylation activity by MAZ sites (data not shown). The coupled reactions with spacer-containing templates were performed; transcripts were fractionated and then analyzed by S1 nuclease protection using a 5′ end-labelled probe that hybridizes to the spacer and SPA sequences. As shown in Figure 4A, the wild-type SPA template gave a cleaved 3′ product, while the mutant SPA template did not (compare lanes 4 and 8). Interestingly, >80% of the cleaved 3′ product was present in the template-bound fraction. Moreover, the wild-type SPA template gives a major degradation product that is ∼15 bp shorter than the cleaved 3′ product, while the mutant SPA does not. Enhanced degradation products are also apparent below the major degradation product, consistent with 5′→3′ exonuclease activity. More than 80% of these degraded products were also retained in the template-bound fraction. These results indicate that 3′ product generated by 3′ end processing remains associated with the ternary complex during its subsequent destruction.
Fig. 4. 3′ product remains associated with the ternary complex and is rapidly degraded. (A) Coupled reactions with immobilized wild-type (lanes 2–7) or mutant (lanes 8 and 9) SPA, 118 bp spacer and wild-type MAZ4 site template were carried out for the times indicated. The AATAAA sequence of the SPA was mutated to GCGGCG. The transcripts were fractionated and analyzed by S1 nuclease protection using the 5′ probe on a 9% polyacrylamide–8 M urea gel. b and r are as in Figure 3A. (B) Analysis of the stability of the cleaved 3′ product. The wild-type (lanes 2–5 and 8–11) or mutant (lanes 6 and 7) SPA, the 118 bp spacer and wild-type MAZ4 templates were transcribed in the coupled system without RNase inhibitor for 0.5 h. Transcription was halted by addition of 2 µg/ml α-amanitin and the reactions were continued. The total reaction times are indicated. The transcripts were analyzed by S1 nuclease protection using the 5′ probe on a 9% polyacrylamide–8 M urea gel. Lanes 8–11 are longer exposures of lanes 2–5, respectively. (C) Comparison of stability of the 5′ and 3′ products. The same reaction as in (B) with wild-type SPA, 118 bp spacer and wild-type MAZ4 template. Transcripts were analyzed by S1 nuclease protection using the 3′ (lanes 2–4) or 5′ probe (lanes 5–7) on a 7.5% polyacrylamide–8 M urea gel. The DNA templates and the DNA probes used are indicated in the diagram. << is as in Figure 1. Note that the read-through product detected with the 5′ and 3′ probes used in Figure 4 will include all RNAs that have not cleaved at SPA.
Finally, we wished to investigate the stability of the 3′ product and therefore modified the coupled reaction as follows. Templates containing either the wild-type or mutant SPA, separated from the MAZ4 sites by the 118 bp spacer (not immobilized), were transcribed in the coupled system for 30 min, the time taken for the cleaved product to begin to accumulate. Transcription was then halted by addition of 2 µg/ml α-amanitin and the reactions were continued (Figure 4B). Following S1 mapping using the 5′ probe (as in Figure 4A) 88% of readthrough product remained at 5 h reaction time using the mutant SPA template, while only 20% did so with the wild-type SPA template (compare lanes 6 and 2 with 7 and 5, respectively). These data indicate that >60% of RNA containing wild-type SPA sequence was cleaved and polyadenylated during a 4.5 h reaction. The 3′ product did not accumulate; instead, degraded products were only visible after long exposure (lanes 8–11). These results suggest that the 3′ product is unstable and rapidly degraded. We also compared the relative stability of 3′ and polyadenylated 5′ products. The same reaction as in Figure 4B was carried out using template containing the wild-type SPA, 118 bp spacer and MAZ4 sites, and the transcripts were analyzed by S1 nuclease protection using a 5′ or 3′ probe (Figure 4C). In contrast to the lack of accumulation of 3′ product (lanes 5–7), the 3′ probe revealed significant accumulation of 5′ product (lanes 2–4; 14-fold increase after 5 h reaction), since it is polyadenylated and stabilized. Moreover the << product (described in Figure 2) disappears after 3 h reaction time, indicating that the 3′ end region of run-off RNA is also rapidly degraded. Overall, these results clearly demonstrate that the 3′ product is considerably less stable than polyadenylated 5′ product.
Discussion
Two PolII termination models were originally proposed to explain the requirement of a functional poly(A) site in the termination. The ‘antiterminator’ model postulated that an antitermination factor leaves the polymerase as it passes a poly(A) signal, so destabilizing it (Logan et al., 1987) while the ‘torpedo’ model suggested that cleavage at the poly(A) signal results in degradation of the nascent, uncapped transcripts, leading to termination (Connelly and Manley, 1989; Proudfoot, 1989). Recent studies in different in vivo systems now backed up by the in vitro analysis presented in this paper are most consistent with a mechanism of PolII termination that incorporates aspects of both original models into one unified process.
All models for PolII transcriptional termination predict that closely spaced genes require a pause site to slow down elongating polymerases and so give time for the termination mechanism to occur. Indeed, studies in Schizosccaromyces pombe have recently illustrated the importance of transcriptional pause sites in promoting efficient PolII transcriptional termination (Birse et al., 1997; Aranda and Proudfoot, 1999). We previously obtained indirect evidence for the presence of pause sites in two different human genes. The C2 complement and α2 globin genes both contain such pause sites, which promote PolII termination by activating 3′ end processing in vivo. These observations were based on experiments carried out using a combination of nuclear run-on and poly(A) site competition assays (Ashfield et al., 1991, 1994; Enriquez-Harris et al., 1991). In this report, we analyzed these physiological pause sites using the coupled in vitro transcription–polyadenylation system, and demonstrate that they slightly activate 3′ end processing but do not show strong pausing activity. However, by using a Gln111-induced ‘roadblock’ to PolII elongation following these physiological pause sites, both pausing and cleavage/polyadenylation are strongly activated. Perhaps the closely positioned promoters and associated DNA-binding factors of the downstream gene, which for C2 is factor B and α2 is α1 globin, might act like Gln111 to provide a transcriptional roadblock that could then enhance pausing and upstream polyadenylation.
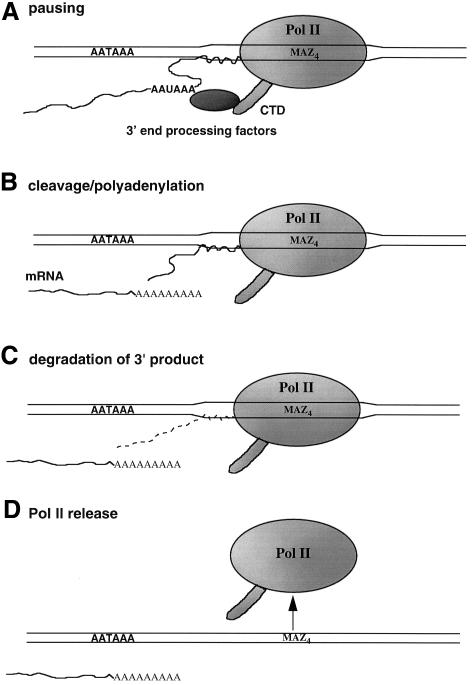
We have investigated the mechanism of PolII transcriptional termination using our coupled in vitro system with immobilized templates, and show that 3′ end processing occurs on the ternary complex (nascent RNA, DNA and PolII). We also show that 3′ product generated by processing remains associated with the complex and is rapidly degraded on it. It is interesting to relate these in vitro results to recent experiments that have investigated PolII termination using in vivo systems. RT–PCR analysis of nascent transcripts isolated from the BR1 giant chromosome of Chironomus tentans (Baurén et al., 1998) and electron microscopic analysis of mini-chromosomes injected into Xenopus laevis oocytes (Osheim et al., 1999) both indicate that transcript termination occurs in the 3′ flanking region of the gene prior to cleavage at the poly(A) signal. However, later experiments showed that efficient termination depended on the presence of a poly(A) signal. This laboratory has also shown that, for the human ε and β-globin genes, transcription proceeds up to the site of termination prior to cleavage at the upstream poly(A) signal. These studies also predicted that cleavage at the poly(A) site may occur concomitantly with the PolII termination process (Dye and Proudfoot, 1999). The data presented here are entirely consistent with these in vivo studies. As illustrated in Figure 5, we demonstrate that our in vitro transcripts first arrest at the MAZ4 sequence and are then subject to 3′ end processing at the upstream poly(A) site, but only when part of the ternary complex. The attached 3′ product is then degraded. We predict that this final RNA degradation process removes all attached transcripts from the arrested PolII and so promotes PolII release.
Fig. 5. Diagram illustrating the different steps associated with PolII termination. Steps A–C were directly investigated in these studies.
Future experiments using our in vitro termination system should allow us to analyze directly the release of PolII from the termination site, and to determine which other factors may be required to mediate this process.
Materials and methods
Coupled in vitro transcription–polyadenylation and RNA mapping by S1 nuclease protection analysis
Coupled in vitro transcription–polyadenylation and S1 nuclease protection assays were carried out as described by Yonaha and Proudfoot (1999). After 5 min of transcription, 0.1% sarkosyl was added to the reactions where indicated, and transcription was allowed to continue for the times indicated. To construct the C2 and α templates, the C2 gene 3′ flanking regions between the StyI and BamHI sites and the α2 gene 3′ flanking region 319–411 bp downstream of α2 poly(A) site were inserted between the KpnI and EcoRI sites of pMLPIII (Yonaha and Proudfoot, 1999) containing the SPA. To construct the wild-type or mutant SPA, 118 bp spacer and MAZ4 template, an oligonucleotide (118 bp) containing four copies of the sequence ATTCGATCGGTTCGGGGCGAGC and four wild-type MAZ site oligonucleotides were inserted in tandem between the KpnI and EcoRI sites of pMLPIII containing SPA or SPA mutant sequences. These plasmids were linearized with HindIII and treated with Klenow fragment. 3′ DNA probes were constructed as follows: BamHI–EcoRI DNA fragments of the various pMLPIII plasmids were inserted into pGEM3SapI. They were then digested with SapI and AlwNI, except for the C2 plasmids, which were digested with SapI and FspI. The resulting fragments were filled in with [α-32P]dGTP using Klenow DNA polymerase. 5′ DNA probes were constructed as follows: wild-type or mutant SPA and the spacer oligonucleotide were inserted in tandem between the BamHI and EcoRI sites of pUC19. These clones were digested with HindIII and XhoI, which is positioned just upstream of the EcoRI site. The fragments were gel purified, treated with calf intestinal alkaline phosphatase and labeled with [γ-32P]ATP using T4 polynucleotide kinase.
Immobilization of templates
Various pMLPIII plasmids were digested with HindIII. For immobilization, biotin-14-dATP (Gibco-BRL) was incorporated into the HindIII restriction site using Klenow enzyme. One hundred-and-fifty milligrams of streptavidin-coated magnetic beads (Promega) were prepared by washing four times in 150 µl wash buffer (50 mM NaCl, 10 mM Tris–HCl pH 7.9, 5 mM EDTA). Beads were drained and then blocked by incubation for 10 min at room temperature in 100 µl of wash buffer plus 200 µg tRNA (Sigma). Beads were drained and resuspended in 300 µl of RNase protection buffer (300 mM NaCl, 10 mM Tris–HCl pH 7.9, 5 mM EDTA). The biotinylated templates (10 µg) were added to the mixture and incubated with agitation at room temperature for 30 min. After magnetic selection, the immobilized templates were suspended in 40 µl of TE.
Acknowledgments
Acknowledgements
We thank Paul Modrich for Gln111 EcoRI. We are grateful to members of the Proudfoot laboratory for many helpful discussions. This work was supported by the Wellcome Trust (grant no. 032773).
Note added in proof
Recent analysis has revealed that readthrough transcripts of DNA templates containing the MAZ4 sequence are subject to partial S1 digestion at the MAZ4 sequences. Consequently a small fraction of the MAZ4 bands observed in these studies will be due to internal RNA:DNA duplex breathing rather than authentic 3′ termini. These results also emphasize the unusual structure of this nucleic acid sequence, which may well account for its interesting behaviour in our in vitro system.
References
- Aranda A. and Proudfoot,N.J. (1999) Definition of transcriptional pause elements in fission yeast. Mol. Cell. Biol., 19, 1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashfield R., Enriquez-Harris,P. and Proudfoot,N.J. (1991) Transcriptional termination between the closely linked human complement genes C2 and Factor B: common termination factor for C2 and c-myc? EMBO J., 10, 4197–4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashfield R., Patel,A., Bossone,S.A., Brown,H., Campbell,R.D., Marcu,K.B. and Proudfoot,N.J. (1994) MAZ-dependent termination between closely spaced human complement genes. EMBO J., 13, 5656–5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baurén G., Belikov,S. and Wieslander,L. (1998) Transcriptional termination in the Balbiani ring 1 gene is closely coupled to 3′-end formation and excision of the 3′ terminal intron. Genes Dev., 12, 2759–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse C.E., Lee,B.A., Hansen,K. and Proudfoot,N.J. (1997) Transcriptional termination signals for RNA polymerase II in fission yeast. EMBO J., 16, 3633–3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse C.E., Minvielle-Sebastia,L., Lee,B.A., Keller,W. and Proudfoot,N.J. (1998) Coupling termination of transcription to messenger RNA maturation in yeast. Science, 280, 298–301. [DOI] [PubMed] [Google Scholar]
- Connelly S. and Manley,J.L. (1988) A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev., 2, 440–452. [DOI] [PubMed] [Google Scholar]
- Connelly S. and Manley,J.L. (1989) RNA polymerase II transcription termination is mediated specifically by protein binding to a CCAAT box sequence. Mol. Cell. Biol., 9, 5245–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantonel J., Murthy,K.G.K., Manley,J.L. and Tora,L. (1997) Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature, 389, 399–402. [DOI] [PubMed] [Google Scholar]
- Dye M.J. and Proudfoot,N.J. (1999) Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol. Cell, 3, 371–378. [DOI] [PubMed] [Google Scholar]
- Edwalds-Gilbert G., Prescott,J. and Falck-Pedersen,E. (1993) 3′ RNA processing efficiency plays a primary role in generating termination competent RNA polymerase II elongation complexes. Mol. Cell. Biol., 13, 3472–3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggermont J. and Proudfoot,N.J. (1993) Poly(A) signals and transcriptional pause sites combine to prevent interference between RNA polymerase II promoters. EMBO J., 12, 2539–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enriquez-Harris P., Levitt,N., Briggs,D. and Proudfoot,N.J. (1991) A pause site for RNA polymerase II is associated with termination of transcription. EMBO J., 10, 1833–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D.K. and Roeder,R.G. (1985) Separation and partial characterization of three functional steps in transcription initiation by human RNA polymerase II. J. Biol. Chem., 260, 8163–8172. [PubMed] [Google Scholar]
- Hirose Y. and Manley,J.L. (1998) RNA polymerase II is an essential mRNA polyadenylation factor. Nature, 395, 93–96. [DOI] [PubMed] [Google Scholar]
- Levitt N., Briggs,D., Gil,A. and Proudfoot,N.J. (1989) Definition of an efficient synthetic poly(A) site. Genes Dev., 3, 1019–1025. [DOI] [PubMed] [Google Scholar]
- Logan J., Falck-Pedersen,E., Darnell,J.E. and Shenk,T. (1987) A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse βmaj-globin gene. Proc. Natl Acad. Sci. USA, 84, 8306–8310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S., Fong,N., Yankulov,F., Ballantyne,S., Pan,G., Greenblatt,J., Patterson,D.S., Wickens,M. and Bentley,D.L. (1997) The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature, 385, 357–361. [DOI] [PubMed] [Google Scholar]
- Osheim Y.N., Proudfoot,N.J. and Beyer,A.L. (1999) EM visualization of transcription by RNA polymerase II: downstream termination requires a poly(A) signal but not transcript cleavage. Mol. Cell, 3, 379–387. [DOI] [PubMed] [Google Scholar]
- Proudfoot N.J. (1986) Transcriptional interference and termination between duplicated α-globin gene constructs suggest a novel mechanism for gene regulation. Nature, 322, 562–565. [DOI] [PubMed] [Google Scholar]
- Proudfoot N.J. (1989) How RNA polymerase II terminates transcription in higher eukaryotes. Trends Biochem. Sci., 14, 105–110. [DOI] [PubMed] [Google Scholar]
- Roberts G.C., Gooding,C., Mak,H.Y., Proudfoot,N.J. and Smith,C.W.J. (1998) Co-transcriptional commitment to alternative splice site selection. Nucleic Acids Res., 26, 5568–5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw E. and Proudfoot,N. (1986) α-Thalassaemia caused by a poly(A) site mutation reveals that transcriptional termination is linked to 3′ end processing in the human α2 globin gene. EMBO J., 5, 2915–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright D.J., King,K. and Modrich,P. (1989) The negative charge of Glu-111 is required to activate the cleavage center of EcoRI endonuclease. J. Biol. Chem., 264, 11816–11821. [PubMed] [Google Scholar]
- Yonaha M. and Proudfoot,N.J. (1999) Specific transcriptional pausing activates polyadenylation in a coupled in vitro system. Mol. Cell, 3, 593–600. [DOI] [PubMed] [Google Scholar]