Abstract
Yth1p is the yeast homologue of the 30 kDa subunit of mammalian cleavage and polyadenylation specificity factor (CPSF). The protein is part of the cleavage and polyadenylation factor CPF, which includes cleavage factor II (CF II) and polyadenylation factor I (PF I), and is required for both steps in pre-mRNA 3′-end processing. Yth1p is an RNA-binding protein that was previously shown to be essential for polyadenylation. Here, we demonstrate that Yth1p is also required for the cleavage reaction and that two protein domains have distinct roles in 3′-end processing. The C-terminal part is required in polyadenylation to tether Fip1p and poly(A) polymerase to the rest of CPF. A single point mutation in the highly conserved second zinc finger impairs both cleavage and polyadenylation, and affects the ability of Yth1p to interact with the pre-mRNA and other CPF subunits. Finally, we find that Yth1p binds to CYC1 pre-mRNA in the vicinity of the cleavage site. Our results indicate that Yth1p is important for the integrity of CPF and participates in the recognition of the cleavage site.
Keywords: mRNA/pre-mRNA 3′-end processing/RNA–protein interaction/zinc finger
Introduction
In eukaryotes, the 3′-ends of mRNAs are generated by endonucleolytic cleavage of a primary transcript and subsequent polyadenylation of the 5′ cleavage product by poly(A) polymerase. This RNA processing reaction depends on cis-acting signals on the RNA and on trans-acting protein factors (for review see Wahle and Rüegsegger, 1999; Zhao et al., 1999a).
Fractionation of yeast extracts led to the separation of protein factors that are required for 3′-end formation in vitro (Chen and Moore, 1992). Cleavage was obtained by combining fractions containing cleavage factor I (CF I) and cleavage factor II (CF II) activity. Specific polyadenylation was reconstituted with combinations of fractions containing CF I, poly(A) polymerase (Pap1p) and polyadenylation factor I (PF I). Further purification showed that CF I could be separated into two activities, which were named CF IA and CF IB (Kessler et al., 1996). CF IA is required for both processing steps and is composed of the proteins Rna14p, Pcf11p, Rna15p and Clp1p (Minvielle-Sebastia et al., 1994, 1997; Kessler et al., 1996; Amrani et al., 1997b). In addition to these four polypeptides, poly(A)-binding protein I (Pab1p) was detected in purified CF IA (Minvielle-Sebastia et al., 1997). Biochemical and genetic evidence indicates an involvement of Pab1p in poly(A) length control (Amrani et al., 1997a; Minvielle-Sebastia et al., 1997). A single polypeptide, Nab4p/Hrp1p, constitutes CF IB. This factor is needed for cleavage site selection and for polyadenylation (Kessler et al., 1997; Minvielle-Sebastia et al., 1998). A multiprotein complex that harbours CF II–PF I activity (CPF) has been isolated by affinity purification (Preker et al., 1997; Ohnacker et al., 2000). This complex includes Pap1p, Fip1p, Pta1p, Pfs1p, Pfs2p and the yeast counterparts of the four subunits of the mammalian cleavage and polyadenylation specificity factor CPSF. Yhh1p/Cft1p, Ydh1p/Cft2p, Ysh1p/Brr5p and Yth1p are the homologues of the 160, 100, 73 and 30 kDa subunits of CPSF (Preker et al., 1995, 1997; Chanfreau et al., 1996; Jenny et al., 1996; Stumpf and Domdey, 1996; Barabino et al., 1997; Zhao et al., 1997). A subcomplex that only carried CF II activity was shown to contain the yeast homologues of the three large CPSF subunits and Pta1p (Zhao et al., 1997, 1999b).
The yeast pre-mRNA 3′-end processing signals are not well conserved (Guo and Sherman, 1996). In addition to the cleavage and polyadenylation site, two cis-acting elements, the efficiency element and the positioning element, are found upstream of the cleavage site. Computational sequence analyses of Saccharomyces cerevisiae 3′ UTRs highlighted the existence of putative signals, which are located immediately upstream and downstream of the cleavage site (Graber et al., 1999; van Helden et al., 2000).
The 3′-end processing machinery must contain proteins that recognize signals on pre-mRNA and confer specificity to the processing reaction. Among the RNA-binding components of the 3′-end processing complex are two proteins that contain an RNA-binding domain of the RNP type: Rna15p and Nab4p/Hrp1p (Minvielle-Sebastia et al., 1991; Kessler et al., 1997). The binding site of Rna15p on the pre-mRNA has not yet been identified. In contrast, UV cross-linking and SELEX experiments with Nab4p/Hrp1p showed that this protein binds the efficiency element in GAL7 pre-mRNA and has a preference for UAUAUA-containing sequences (Kessler et al., 1997; Chen and Hyman, 1998; Valentini et al., 1999). Cross-linking experiments with the CF II subunit Ydh1p/Cft2p suggested that the interaction of this protein with GAL7 pre-mRNA depends on the presence of the efficiency element as well as on sequences downstream of the cleavage site (Zhao et al., 1997). Thus, although most of the essential components of the yeast 3′-end processing machinery and some cis-acting signals have been identified, the assignment of processing factors to the individual recognition signals is far from complete. Consequently, the fashion in which specificity of cleavage site selection is ensured is poorly understood. In addition, since Pap1p itself is neither a specific nor a processive enzyme in the absence of other polyadenylation factors (Lingner et al., 1991), the mechanism that enables the polymerase to accomplish specific and processive poly(A) synthesis remains to be elucidated.
Yth1p is the yeast homologue of the 30 kDa subunit of mammalian CPSF, a factor required for both cleavage and polyadenylation. Yth1 protein has been shown to interact with Fip1p, which in turn binds to Pap1p (Preker et al., 1995; Barabino et al., 1997). The first YTH1 mutant allele, yth1-1, caused a polyadenylation defect (Barabino et al., 1997). Thus, Yth1p seemed to be required solely for the second step of pre-mRNA 3′-end processing. In addition, although Yth1p was found to interact with RNA (Barabino et al., 1997), its binding site remained unidentified. In this paper, we analyse the nature of two different mutations, which cause distinct pre-mRNA 3′-end processing phenotypes in vitro. We show that the deletion of 50 amino acids at the C-terminus of Yth1-1 protein impairs the interaction with Fip1p and the association of Fip1p and poly(A) polymerase with CPF. In addition, we demonstrate that the highly conserved second zinc finger of Yth1p is an essential domain involved in protein–RNA and protein–protein interactions. Mutations in this zinc finger caused cleavage and polyadenylation defects in vitro. Restoration of cleavage activity with recombinant wild-type protein demonstrated the dependence of the cleavage reaction on Yth1p. Finally, we show that recombinant Yth1p binds near the cleavage and polyadenylation site on CYC1 pre-mRNA. We therefore conclude that Yth1p is required for both steps of pre-mRNA 3′-end formation and suggest that it is important for the specificity of this processing reaction.
Results
In vivo depletion of Yth1p impairs both steps of pre-mRNA 3′-end processing
In vivo depletion of Yth1p was initially chosen to investigate the requirement of the protein in both pre-mRNA 3′-end processing steps. Park and Szostak (Park et al., 1992) described the generation of a conditional allele based on the use of a protein destabilizing element in combination with an inducible and repressible promoter. A ubiquitin coding sequence is fused to the N-terminus of the open reading frame (ORF) of interest with an arginine at the fusion breakpoint. In vivo, deubiquitylation exposes an arginine instead of a methionine at the N-terminus, which targets the protein for rapid degradation (Bachmair et al., 1986).
We therefore constructed a plasmid expressing a ubiquitin–R-Yth1 fusion protein under the control of a UAS-GAL-CYC1 hybrid promoter (pGUR-YTH1), which supports transcription in the presence of galactose, but not in the presence of glucose. This plasmid was introduced into a strain deleted for the chromosomal copy of the YTH1 gene (SB3; Barabino et al., 1997). To analyse the in vitro phenotype of Yth1p depletion, extracts were prepared from cells either grown in the presence of galactose or shifted to glucose-containing medium. Extracts from cells grown in galactose and therefore expressing R-Yth1p were able efficiently to cleave and polyadenylate the pre-mRNA (Figure 1A and B, lanes 2 and 3). However, extracts prepared from cells grown on glucose-containing medium showed a strong reduction of both cleavage and polyadenylation activity (Figure 1A and B, lanes 4 and 5). Western blot analysis of both extracts confirmed that the shift to glucose-containing medium effectively resulted in the depletion of the Yth1 protein (Figure 1C, compare lanes 1 and 2) without affecting the levels of other 3′-end processing factors such as Fip1p or Rna15p (Figure 1C, compare lanes 3 and 4). These results indicate a direct involvement of Yth1p in cleavage as well as in polyadenylation.
Fig. 1. In vivo depletion of Yth1 protein affects both pre-mRNA 3′-end processing steps. (A) Cleavage assay with CYC1 pre-mRNA. (B) Polyadenylation assay with pre-cleaved CYC1 RNA, which ends at the natural cleavage site. Cells expressing a ubiquitin–R-Yth1 fusion protein under the control of a UAS-GAL-CYC1 hybrid promoter (pGUR-YTH1) were grown in either galactose- or glucose–containing medium. Increasing amounts of extract (10 and 20 µg, respectively) prepared from these cells (galactose, lanes 2 and 3; glucose, lanes 4 and 5) were incubated with the RNA under standard assay conditions for 1 h. The position of the precursors (lane 1), the 5′ cleavage product and the polyadenylated RNA are indicated. HpaII-digested pBR322 fragments served as marker (M). They range from 309 to160 nucleotides in (A) and from 309 to 180 nucleotides in (B). (C) Western blot of extracts prepared from cells expressing a ubiquitin–R-Yth1 fusion protein under the control of a UAS-GAL-CYC1 hybrid promoter (pGUR-YTH1), which were grown in either galactose- or glucose-containing medium (lanes 1 and 3, and 2 and 4, respectively). Ten micrograms of extract were fractionated on a 12% SDS–polyacrylamide gel, blotted onto a nitrocellulose membrane and probed first with α-Yth1p, and subsequently with α-Fip1p and α-Rna15p antibodies.
The highly conserved second zinc finger in Yth1p is an essential domain
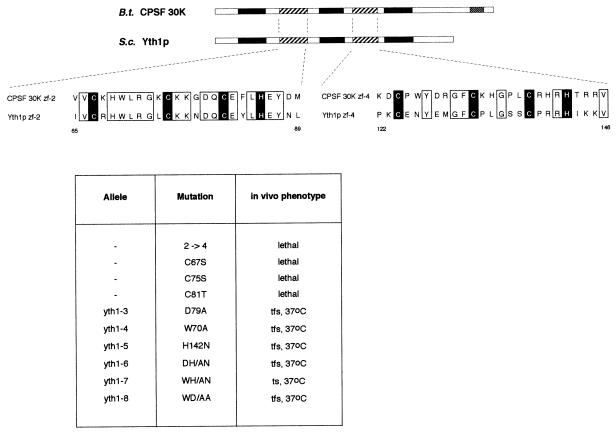
Yth1p shares a five zinc finger structure with all its homologues. However, the overall sequence identity with bovine CPSF 30K is only 26%. The second zinc finger (ZF2) represents a notable exception since this region is 76% identical and 96% similar to the corresponding part of CPSF 30K and its homologues in higher eukaryotes. In comparison, ZF4 is only 50–57% identical and 63% similar in the same organisms. This remarkable evolutionary conservation suggests that this domain may mediate an important biological function.
To address the function of ZF2 we carried out site-directed mutagenesis and generated the mutants listed in Figure 2. The plasmids carrying the mutations were introduced into SB3 by plasmid shuffling. The resulting mutant strains were plated on 5-fluoro-orotic acid (5-FOA) medium to select for the loss of the plasmid carrying the wild-type gene and thus to test for the ability of the mutant alleles to sustain viability. The initial mutant made was a substitution of ZF2 with a copy of ZF4 (mutant yth1-2→4, Figure 2), which, in contrast to a deletion of ZF2, should preserve the overall structure of the protein. We chose ZF4 because the spacings between the three conserved cysteine and the histidine residues are identical in ZF2 and ZF4. This substitution is lethal, supporting the idea that ZF2 has an essential function in vivo. Mutations in the conserved residues C67, C75 and C81 to serine or to threonine are also lethal. In contrast, mutations of three other conserved residues [W70A, D79A and H142N (ZF4)] either alone or in combination gave rise to five temperature–formamide-sensitive (tfs) alleles (yth1-3 to yth1-6 and yth1-8) and one temperature-sensitive (yth1-7) allele (see Figure 2).
Fig. 2. Mutagenesis in the highly conserved second zinc finger of Yth1p. In the schematic alignment of Yth1p and CPSF 30K, zinc fingers 1, 3 and 5 are represented by black boxes and zinc fingers 2 and 4 by striped boxes. The C-terminal zinc knuckle motif of CPSF 30K is represented by a stippled box. In the amino acid sequence alignment of the second and fourth zinc fingers of CPSF 30K and Yth1p, the cysteine and histidine residues are indicated white on black; all other identical residues are boxed. Bos taurus CPSF 30K (DDBJ/EMBL/GenBank accession No. U96448; Barabino et al., 1997), S.cerevisiae YTH1 (systematic name: YPR107c; Barabino et al., 1997). The results from the site-directed mutagenesis of zinc finger 2 are summarized in the table below the alignments.
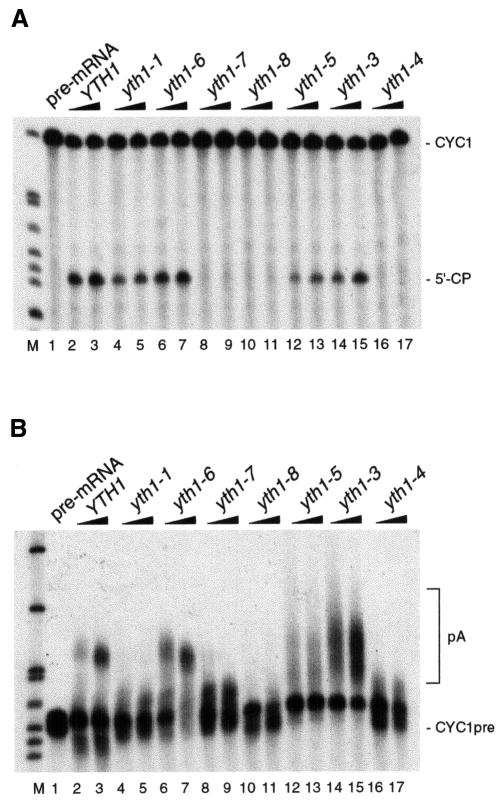
In order to define the role of the second zinc finger in 3′-end processing, extracts prepared from the viable mutant strains were tested for either specific cleavage or polyadenylation in vitro. While the double mutant allele yth1-6 (Figure 3A and B, lanes 6 and 7) did not affect processing activity, extracts prepared from yth1-3 and yth1-5 cells carrying the corresponding single mutations showed abnormal polyadenylation (Figure 3B, lanes 12–15). In these extracts, poly(A) tails ranged from very short to abnormally long. Although we have not characterized the phenotype of these strains in more detail, this observation suggests a role for Yth1p in the control of the poly(A) tail length. Cleavage and polyadenylation activity was completely abolished in extracts from yth1-4, yth1-7 and yth1-8 mutant cells (Figure 3A and B, lanes 16 and 17, and 8–11, respectively). The processing defect caused by the yth1-4, yth1-7 and yth1-8 alleles corroborated the evidence for a requirement of Yth1p in both steps of 3′-end processing. Since the single point mutation W70A was sufficient to prevent both processing steps completely, we chose the yth1-4 allele for further experiments. Cleavage and polyadenylation activity could be complemented by mixing yth1-4 extract with extract from either rna14-1 or rna15-1 mutant cells (Minvielle-Sebastia et al., 1994), which by themselves are unable to cleave and polyadenylate the pre-mRNA substrate (results not shown). Taken together, these results uncover a specific requirement for the second zinc finger of Yth1p in both steps of pre-mRNA 3′-end processing.
Fig. 3. YTH1 mutant extracts are impaired in cleavage and polyadenylation. (A) Cleavage assay. (B) Polyadenylation assay. Extracts prepared from wild-type cells (lanes 2 and 3) and from yth1 mutant strains were added to the reactions as indicated on top of each lane. The position of the precursors (lane 1) and either the 5′ cleavage product or of the polyadenylated RNA are indicated. End-labelled HpaII-digested pBR322 fragments served as marker (M).
Yth1p is essential for cleavage activity in yeast extracts
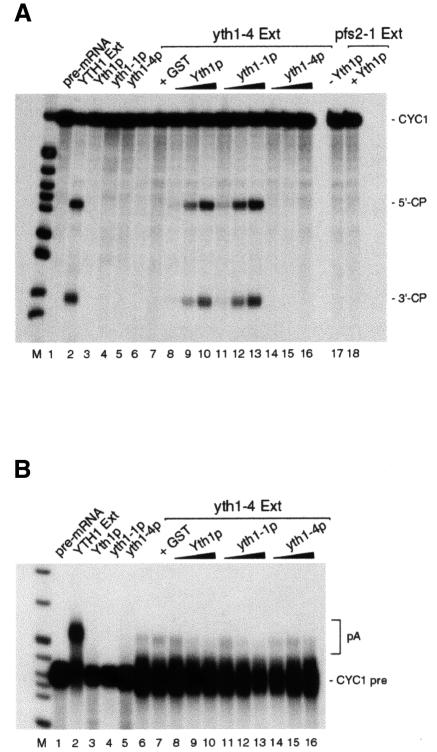
To confirm that Yth1p is required for both cleavage and polyadenylation, we tried to reconstitute processing activity in the yth1-4 extract by complementation with glutathione S-transferase (GST)-tagged wild-type and mutant proteins. While the proteins alone did not show any activity (Figure 4A and B, lanes 3–5), both wild-type GST–Yth1 and mutant GST–Yth1-1 proteins could efficiently reconstitute cleavage when combined with the mutant extract (Figure 4A, lanes 8–13). As expected, GST–Yth1-4p could not reconstitute processing, confirming that the cleavage defect in this extract is caused by the mutation in the Yth1 protein (Figure 4, lanes 14–16). Reconstitution of cleavage is specific for the yth1 mutant extract, since addition of recombinant Yth1p to a pfs2-1 mutant extract (Ohnacker et al., 2000) did not restore activity (Figure 4A, lane 18).
Fig. 4. Reconstitution of cleavage activity in yth1-4 extract by complementation with recombinant Yth1 protein. (A) Cleavage assay. (B) Polyadenylation assay. Wild-type YTH1 yeast extract (lane 2) served as positive control. Two hundred nanograms of recombinant GST–Yth1p, GST–Yth1-1p or GST–Yth1-4p were incubated with the pre-mRNA (lanes 3–5). yth1-4 mutant extract was incubated with CYC1 RNA either alone (lane 6) or in combination with GST (lane 7), 5–200 ng of GST–Yth1p (lanes 8–10), GST–Yth1-1p (lanes 11–13) or GST–Yth1-4p (lanes 14–16). In (A), an aliquot of a pfs2-1 mutant extract was tested either alone or in combination with 200 ng of Yth1p (lanes 17 and 18, respectively). The position of the precursors, the 5′- and the 3′-cleavage products and the polyadenylated reaction products is indicated on the right. End-labelled HpaII-digested pBR322 fragments served as marker (M).
Surprisingly, polyadenylation could not be reconstituted by addition of the wild-type Yth1 protein (Figure 4B, lanes 8–10). Possibly, the endogenous mutant Yth1 protein can prevent the recombinant wild-type protein from establishing interactions with other CF II–PF I subunits, such as Fip1p, which are required for polyadenylation. Irrespective of the reason for the failure of Yth1p to restore polyadenylation activity, the results depicted in Figure 4 show that Yth1p has an essential function in the cleavage reaction.
Protein–protein and RNA–protein interactions are disrupted in YTH1 mutants
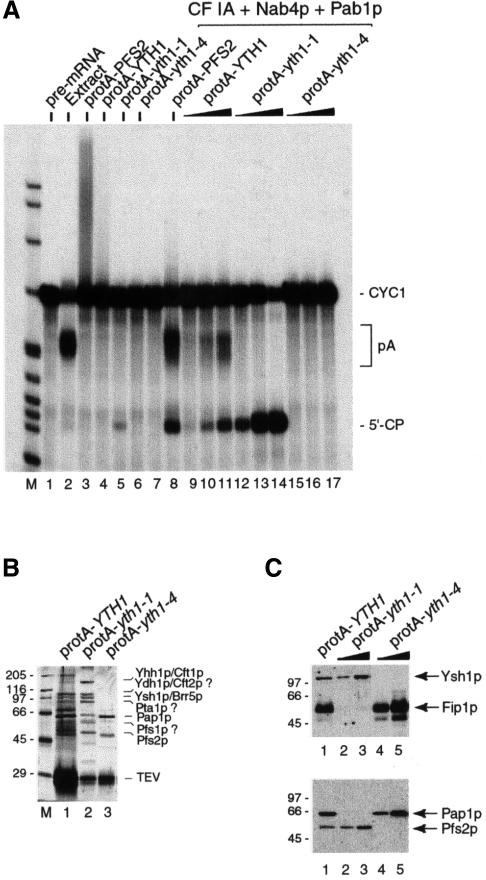
Previously, we reported the isolation of the yth1-1 allele, a deletion of the last 50 amino acids of Yth1p that caused an in vitro polyadenylation defect (Figure 2, lanes 4 and 5; Barabino et al., 1997). In order to understand the molecular mechanism underlying the different in vitro 3′-end processing defects of the yth1-1 and yth1-4 mutants, we isolated the Yth1p-containing complexes formed in these cells. For this purpose, we generated strains in which a tag consisting of protein A and the TEV protease cleavage site was fused to the wild-type and mutant YTH1 ORFs. Extracts from these strains were incubated with IgG–agarose. After extensive washing, the bound fraction was eluted by cleavage with TEV protease. The eluates were then tested for 3′-end processing activity in a coupled cleavage and polyadenylation assay (Figure 5A). As a control, the CF II–PF I complex (CPF, for cleavage and polyadenylation factor) was isolated from a strain expressing a protein A fusion of the Pfs2 protein (Ohnacker et al., 2000). While the Pfs2 and the Yth1 eluates showed the unspecific elongation of the pre-mRNA previously described for PF I (Preker et al., 1997), the Yth1-1 eluate retained a slight cleavage activity, probably due to contaminating CF I. The Yth1-4 eluate instead was completely inactive. Cleavage and polyadenylation activity was restored by combining a mixture of purified CF IA, recombinant Nab4p/Hrp1p and Pab1p (Figure 5A, lane 7) with either the Pfs2 eluate (Figure 5A, lane 8) or increasing amounts of the wild-type Yth1 eluate (Figure 5A, lanes 9–11). As expected, addition of the Yth1-1 eluate could only restore cleavage but not polyadenylation (Figure 5A, lanes 12–14), whereas the Yth1-4 eluate in combination with the other factors did not cleave the pre-mRNA (Figure 5A, lanes 15–17). These results confirmed that the proteins eluted from the IgG–agarose reflected the activity of the original unfractionated extracts.
Fig. 5. Mutations in Yth1p affect the integrity of the CPF complex. (A) Reconstitution of in vitro pre-mRNA 3′-end processing with the protein complexes associated with wild-type and mutant Yth1p. The multiprotein complexes associated with either protein A–TEV-tagged Pfs2p, Yth1p, Yth1-1p or Yth1-4p were isolated by batch adsorption to IgG–agarose and cleavage with TEV protease as described in Materials and methods. Comparable amounts of the different IgG eluates were added to the coupled cleavage and polyadenylation assay as indicated on top of each lane. In lanes 8–17, the eluates were combined with a mixture of purified CF IA, 10 ng of Nab4p/Hrp1p and 100 ng of Pab1p in order to reconstitute 3′-end processing in vitro. A wild-type yeast extract served as positive control (lane 2). The position of the precursor (lane 1), the 5′ cleavage product and the polyadenylated RNA is indicated. End-labelled HpaII-digested pBR322 fragments served as marker (M). (B) The CF II–PF I complex co-purifies with protein A-tagged Yth1 protein. The multiprotein complexes associated with protein A–TEV-tagged wild-type or mutant Yth1p were isolated from extracts prepared from strains SB16 (wild-type, lane1), SB17 (yth1-1, lane 2) and SB18 (yth1-4, lane 3). The isolated material was analysed on an SDS–polyacrylamide gel, which was subsequently stained with silver. About six times more of the wild-type Yth1 eluate was fractionated on the gel than of the mutant eluates. (C) Immunoblot analysis of the IgG eluates prepared from SB16 (lane 1), SB17 (yth1-1, lanes 2 and 3) and SB18 (lanes 4 and 5) extracts. The eluates were fractionated on a 10% SDS–polyacrylamide gel, blotted on a nitrocellulose membrane and probed with α-Ysh1p and α-Fip1p antibodies (top), or α-Pap1p and α-Pfs2p antibodies. The sizes of protein standards (in kilodaltons) are indicated on the left.
Analysis of the proteins that co-purify with the tagged wild-type and mutant Yth1 proteins indicated that the wild-type eluate contained all the proteins previously described to be present in the CPF complex (Figure 5B, lane 1). However, we observed that the CPF complex associated with the wild-type Yth1 protein was pulled out inefficiently in comparison to the two mutants. In the yth1 mutants, only partial complexes were formed. In particular, Pap1p could not be detected in the IgG eluate isolated from the yth1-1 extract (Figure 5B, lane 2) whereas all the CF II subunits appeared to be still present, consistent with the finding that cleavage is unaffected in the yth1-1 extract. The opposite situation was observed for the Yth1-4 eluate (Figure 5B, lane 3). In this eluate, Pap1p was present but the CF II subunits could no longer be detected. Western blot analysis with specific antibodies (Figure 5C) confirmed the strong under-representation of both Pap1p and Fip1p in the Yth1-1 eluate and of the other CF II–PF I components in the Yth1-4 eluate. These results suggest that in the Yth1-1 extract the specific interaction between Yth1p and Fip1p is disrupted because of the deletion of the last C-terminal 50 amino acids of Yth1p. This in turn would lead to the observed defect in polyadenylation. In the yth1-4 extract instead, the single amino acid substitution in the second zinc finger of Yth1p results in the destabilization of as yet unidentified interactions with other CF II subunit(s), which most likely causes the loss of cleavage activity.
To identify additional interaction partners among the CPF subunits and to confirm that the C-terminal portion of Yth1p indeed represents the Fip1p interaction domain, we carried out GST pull-down experiments. Purified GST-tagged Yth1 wild-type and mutant proteins were incubated with in vitro translated, 35S-labelled Ysh1p, Fip1p, Pfs2p and Pfs1p. Of these proteins, Ysh1p and, as expected, Fip1p were found to bind Yth1p under the conditions tested (Figure 6A, lanes 3 and 8). The interaction with Ysh1p shows that Yth1p contacts at least one CF II subunit. This contact is maintained in the Yth1-4 mutant (Figure 6A, lane 5). Therefore, the under-representation of the CF II subunits in the Yth1-4p-associated complex (Figure 5B, lane 3) must result from the destabilization of another interaction. The Yth1-1 protein also interacted with Ysh1p, but its ability to bind Fip1p was strongly reduced (Figure 6A, lanes 4 and 9). This confirms that the C-terminal region of Yth1p is responsible for the interaction with Fip1p and its association with CPF.
Fig. 6. Mutations in Yth1p impair protein–protein and RNA–protein interactions. (A) Pull-down experiment with glutathione–Sepharose. GST–tagged Yth1, Yth1-1 and Yth1-4 proteins were tested for interaction with either in vitro translated, 35S-labelled Ysh1p (lanes 3–5) or Fip1p (lanes 8–10). Lanes 1 and 6, 5% of the inputs; lanes 2 and 7, GST bound to glutathione–Sepharose. (B) UV cross-linking to CYC1 RNA. Increasing amounts of recombinant Yth1, Yth1-1 and Yth1-4 proteins were incubated with 100 fmol of pre-mRNA as described. To verify that comparable amounts of each protein had been loaded, the gel was first stained with Coomassie Blue. The amount of cross-linked protein was quantified as described in Materials and methods. Lanes 1, 3 and 5, 100 ng; lanes 2, 4 and 6, 200 ng; lane 7, pre-mRNA incubated without protein. (C) UV cross-linking with Yth1-2→4 protein to CYC1 RNA. Increasing amounts of recombinant Yth1 and Yth1-2→4 proteins were incubated with 150 fmol of pre-mRNA as described. An asterisk indicates a band that does not correspond to Yth1-2→4p as assessed by western blot analysis. The band corresponding to Yth1p is indicated by an arrowhead. Lanes 2 and 4, 50 ng; lanes 3 and 5, 100 ng; lane 1, pre-mRNA incubated without protein.
We had previously shown that Yth1p is an RNA-binding protein (Barabino et al., 1997). To determine whether either the yth1-1 or the yth1-4 mutation affects RNA binding, we carried out UV cross-linking assays on CYC1 pre-mRNA. As shown in Figure 6B, efficient cross-linking was found with both the Yth1 and the Yth1-1 proteins. Binding of Yth1-4p to the substrate was reduced 2- to 3-fold compared with the wild-type protein (Figure 6B, compare lanes 1 and 2 with 5 and 6). This observation suggests that the W/A mutation in the second zinc finger affects not only protein–protein interaction(s), but also reduces the ability of the protein to bind RNA. The importance of ZF2 for the interaction with RNA is supported by UV cross-link experiments with the mutant Yth1-2→4 protein. Only a contaminating Escherichia coli polypeptide, but not the recombinant protein, cross-linked to CYC1 RNA (Figure 6C, lanes 4 and 5). Taken together, these results demonstrate that the highly conserved second zinc finger of Yth1p is an essential domain involved in protein–RNA and protein–protein interactions.
Yth1p binds near the cleavage site
In order to determine whether Yth1p is able to recognize specific RNA sequences within the substrate, we performed RNase H protection experiments. In this assay, the uniformly labelled RNA substrate is first incubated with the recombinant protein under standard in vitro cleavage conditions. Then, the pre-mRNA is challenged with the addition of a complementary single-stranded DNA oligonucleotide and RNase H. Pre-mRNA, which associates with the oligonucleotide and is accessible to RNase H, is cleaved at the site of hybridization. The degree of protection can be determined by comparing the amount of intact pre-mRNA remaining after oligonucleotide and RNase H treatment with that present in a mock reaction to which no protein had been added. We therefore designed a series of overlapping antisense DNA oligonucleotides that cover a region of the CYC1 pre-mRNA from the efficiency element to 10 nucleotides downstream of the cleavage site (listed in Figure 7A). Most oligonucleotides, with the exception of #7 and #8, hybridized efficiently to the RNA. The result of a typical experiment is presented in Figure 7B. After 15 min of pre-incubation with Yth1p, significant levels of pre-mRNA persist only in the presence of oligonucleotides that hybridize to a region surrounding the cleavage site (Figure 7B, lanes 12, 14 and 16). Although protection in this region could be observed also with the full-length CYC1 pre-mRNA, the effect was stronger with a ‘short’ CYC1 substrate. The protection in the cleavage site region was confirmed by similar experiments performed with the GAL-7 pre-mRNA where Yth1p prevents binding of an oligonucleotide that covers the 14 nucleotides immediately upstream of the cleavage site (results not shown). Weak protection could also be observed with oligonucleotide #8 in the region partly overlapping with the proposed efficiency element (Figure 7, lane 4). However, the efficiency element has been previously defined to be the binding site of Nab4/Hrp1p (Kessler et al., 1997; Chen and Hyman, 1998). Therefore, in the presence of Nab4/Hrp1p, Yth1p may preferentially bind near the cleavage site.
Fig. 7. Yth1 protein binds in the vicinity of the cleavage site. (A) Sequence of the ‘short’ CYC1 substrate (see Materials and methods) and position of the antisense DNA oligonucleotides #7–14. The nucleotides that define the efficiency and positioning elements (Russo et al., 1991) are indicated in bold. The region encompassing the deletion in the cyc1-512 substrate is boxed. Vector sequences are indicated in lower case. (B) RNase H protection assay with recombinant Yth1p and ‘short’ CYC1 pre-mRNA. The pre-mRNA substrate was pre-incubated either in the absence (–) or in the presence (+) of ∼200 ng of recombinant Yth1 protein before addition of the corresponding DNA oligonucleotide (indicated at the top) and RNase H. End-labelled HpaII-digested pBR322 fragments served as marker (M).
Discussion
The C-terminal domain of Yth1p tethers Fip1p and poly(A) polymerase to the CPF complex
Previous work showed that Yth1p, the yeast homologue of the 30 kDa subunit of CPSF, directly interacts with Fip1p, which in turn binds to poly(A) polymerase (Preker et al., 1995; Barabino et al., 1997). Furthermore, the yth1-1 mutant allele, which has a C-terminal deletion of 50 amino acids, causes a polyadenylation defect in vitro. Here, we show that the yth1-1 mutation drastically weakens the interaction with Fip1p. Moreover, in the complex associated with protein A-tagged Yth1-1p, Fip1p and Pap1p are heavily under-represented in comparison with the other CF II–PF I subunits. We conclude that the interaction between Yth1p and Fip1p is required for the stable association of Fip1p and poly(A) polymerase to CPF and that the C-terminal portion of Yth1p plays an essential role in this context.
The significance of this interaction is underscored by the observation that isolated Pap1p polyadenylates any given RNA in a non-specific and distributive manner (Lingner et al., 1991). In combination with Fip1p, the polymerase was found to function even less efficiently (Zhelkovsky et al., 1998). In contrast, as part of the CPF complex, poly(A) polymerase works more efficiently than it does alone and its processivity is increased (Preker et al., 1997). Thus, we propose that the interaction between Fip1p and Yth1p helps to ensure processive and specific polyadenylation by tethering Pap1p to CPF.
The mechanism by which poly(A) tail length control is achieved is not completely understood. Pab1p (Amrani et al., 1997a; Minvielle-Sebastia et al., 1997) and poly(A) nuclease (PAN; Brown and Sachs, 1998) have been suggested to be required for this process. The inhibition of Pap1p by Fip1p in non-specific polyadenylation assays (Zhelkovsky et al., 1998) has led to the idea that Fip1p might also be involved in poly(A) tail length control. Interestingly, extracts prepared from certain yth1 mutant strains produced heterogeneous poly(A) tails. Thus, it is conceivable that a Yth1p–Fip1p–Pap1p subcomplex within CPF may participate in determining poly(A) tail length. The mechanism by which this is accomplished remains to be investigated.
Yth1p is essential for the cleavage reaction
In the past, several observations have supported the idea that Yth1p might be involved only in poly(A) tail synthesis but not in the preceding endonucleolytic cleavage reaction. Yth1p was found to be a component of a multiprotein complex, CPF, which has CF II–PF I activity (Preker et al., 1997; Ohnacker et al., 2000) but it was not detected by silver staining in a subcomplex of this factor, which only retained CF II activity (Zhao et al., 1997). Purified CF II fractions were found to contain Pta1p and the homologues of CPSF subunits, Yhh1/Cft1p, Ydh1/Cft2p and Ysh1/Brr5p. Yth1p was not detected in co-immunoprecipitates that were obtained from partially purified CF II with anti-Yhh1/Cft1p antibodies (Zhao et al., 1999b). Finally, the yth1-1 mutation caused an in vitro polyadenylation defect, whereas cleavage activity remained unaffected (Barabino et al., 1997). However, none of these observations provides compelling evidence against an involvement of Yth1p in the cleavage reaction. First, Yth1p stains poorly with silver (Preker et al., 1997) and thus may have escaped detection in purified CF II fractions. Secondly, since the co-immunoprecipitates obtained from partially purified CF II could not be tested for activity, it was not possible to assess the requirement of Yth1p for CF II activity. Finally, the yth1-1 mutation might be detrimental to only one of several functions of the protein.
In this report, we present the results of a mutational analysis of the second zinc finger of Yth1p. The high degree of evolutionary conservation of this domain suggests that ZF2 mediates an essential function. Consistent with this idea, the replacement of ZF2 with ZF4 (yth1-2→4) as well as mutations in the conserved C residues are lethal. Other point mutations in this domain cause conditional growth defects. The in vitro 3′-end processing defect of these mutants (yth1-4, yth1-7 and yth1-8) indicates a role for Yth1p not only in polyadenylation, but also in cleavage. Most importantly, cleavage activity in yth1-4 mutant extracts could be restored by addition of recombinant Yth1p, demonstrating that Yth1p is directly required for the cleavage reaction in yeast extracts.
The highly conserved second zinc finger is required for the interactions with CF II subunits and with the pre-mRNA
The analysis of the polypeptide composition of the complex associated with the Yth1-4 protein allows speculation as to why processing is defective in this strain. The observation that the CF II components are under-represented in this complex implies that the physical contact between Yth1p and the other CF II subunits may be important for cleavage activity. Since in GST pull-down experiments Yth1-4p still retains the ability to interact with Ysh1p, another interaction must be weakened, possibly with Yhh1/Cft1p and/or Ydh1/Cft2p, the other two homologues of CPSF subunits.
In addition, Yth1-4p binds RNA less efficiently than the wild-type protein. The inability of recombinant Yth1-2→4p to bind RNA confirms the importance of the second zinc finger for the interaction with the substrate. Therefore, the cleavage defect of the yth1-4 mutant extract may be caused at least in part by a reduction of RNA-binding capacity. The finding that recombinant Yth1p binds near the cleavage site on the pre-mRNA suggests that it helps to stabilize the binding of the cleavage complex at the poly(A) site. The region protected by Yth1p in RNase H protection assays is quite U-rich, in agreement with its previously described binding preference for poly(U) (Barabino et al., 1997). Interestingly, two computational studies revealed the frequent occurrence of U-rich stretches immediately upstream and downstream of the cleavage site (Graber et al., 1999; van Helden et al., 2000). Possibly, this U-rich element could put the cleavage site in an easily accessible environment and guide the processing machinery to this location. Thus, Yth1p might be directly involved in the recognition of the cleavage site. However, because of the variability of the different sequence elements, Yth1p is certainly not the only determinant of site-specific cleavage and polyadenylation. Ydh1/Cft2p and possibly other homologues of the CPSF subunits may participate in binding of CPF to the pre-mRNA. It will be interesting to see whether the binding preference we observed for Yth1p holds true for the entire CPF complex.
It is also possible that Yth1p is directly involved in the endonucleolytic cleavage of the pre-mRNA. The Drosophila homologue of Yth1p, the clipper protein, was reported to have an endoribonucleolytic activity specific for RNA hairpins (Bai and Tolias, 1996). We could not detect a similar enzymatic activity for recombinant Yth1p (results not shown). However, it is conceivable that the pre-mRNA must adopt a particular folding, possibly induced by other subunits of the 3′-end processing complex, before endonucleolytic cleavage at the poly(A) site can occur.
Parallels between yeast CPF and mammalian CPSF
The existence of homologues of components of the mammalian pre-mRNA 3′-end processing apparatus, and in particular of the four CPSF subunits in yeast CPF (Ohnacker et al., 2000), suggests that important aspects of the protein–protein and RNA–protein interactions are conserved in eukaryotes. CPSF is required for both cleavage and polyadenylation. Its 160 and 30 kDa subunits were found to be involved in the interaction with the pre-mRNA (Keller et al., 1991). Furthermore, CPSF binds to poly(A) polymerase (Murthy and Manley, 1995), increases its processivity (Bienroth et al., 1993) and is involved in poly(A) length control (Wahle, 1995). Owing to the conservation of the protein components mentioned above, yeast CPF would be expected to show similar properties. Our results are consistent with this view. The requirement for Yth1p in both 3′-end processing steps, its role in tethering Pap1p via Fip1p to CPF, and its possible involvement in poly(A) tail length control show that Yth1p is a true functional homologue of a CPSF subunit.
Materials and methods
Yeast strains, methods and media
Most media were prepared as described previously (Guthrie and Fink, 1991). When strains were tested for formamide sensitivity, 3% formamide (v/v) was added (Aguilera, 1994). Yeast transformations were carried out with the lithium acetate method described previously (Gietz et al., 1992). The strains SB10 (yth1-3), SB12 (yth1-4), SB11 (yth1-5), SB14 (yth1-6), SB13 (yth1-7), SB15 (yth1-8), SB16 (pNOPPATA-YTH1), SB17 (pNOPPATA-yth1-1) and SB18 (pNOPPATA-yth1-4) were derived from strain SB3 (MATa, ade2, leu2, ura3, trp1, his3, yth1::TRP1, pHH1-YTH1) (Barabino et al., 1997) by plasmid shuffling. In addition, strains SB5 [same as SB3 but pGUR-YTH1 (CEN4 ADE2 YTH1)] and SB7 [same as SB3 but pFA-yth1-1 (CEN4 ADE2 yth1-1)] were employed in this study (Barabino et al., 1997).
Generation of conditional alleles of the YTH1 gene
Replacement of ZF4 with ZF2 and site-directed mutagenesis of ZF2 were performed according to Mikaelian and Sergeant (1992). For details, see Supplementary Materials and methods available at The EMBO Journal Online. After sequencing, the NotI (filled in)–EcoRI fragment containing the entire ORF was subcloned into the centromeric vector pHA linearized with Bsu36I (filled in) and NotI. pHA, which contains a constitutive promoter, was derived from pHH1 (centromeric vector containing an HA tag; L.Minvielle-Sebastia, unpublished data) by replacement of the URA3 cassette with the ADE2 cassette. The constructs were transformed into SB3 and three colonies for each transformation were replicated on synthetic complete (SC) medium containing 5-FOA to select for the loss of the URA3-marked YTH1 plasmid. After growth for 36 h at 24°C, only the following plasmids were able to complement the YTH1 null allele: pHH1-yth1D/A (yth1-3), pHH1-yth1W/A (yth1-4), pHH1-yth1H/N (yth1-5), pHH1-yth1DH/AN (yth1-6), pHH1-yth1WH/AN (yth1-7) and pHH1-yth1WD/AA (yth1-8). These cells were then streaked on YPD plates and grown at 16, 24, 30 and 37°C, and on YPD containing 3% formamide (YPDF) at 35°C to test for conditional growth defects.
In vitro 3′-end processing assays
Yeast cultures were grown in 1 l of YPAD to an OD600 of 2–6. The harvested cells were frozen in liquid nitrogen and homogenized in a mortar as described elsewhere (Ansari and Schwer, 1995). The subsequent centrifugation steps, the ammonium sulfate precipitation and the dialysis were essentially carried out as described in Butler et al. (1990). Between 10 and 30 µg of protein were used in the assays.
Extracts from strain SB5, grown on either galactose medium or glucose medium, were prepared as described elsewhere (Jenny et al., 1996).
32P-labelled CYC1 RNA and CYC1 pre-cleaved RNA were prepared as run-off transcripts and gel purified (Minvielle-Sebastia et al., 1994; Preker et al., 1995).
In vitro polyadenylation assays were carried out as described previously (Minvielle-Sebastia et al., 1994). When only cleavage was assayed, magnesium acetate was replaced by EDTA, which results in inhibition of poly(A) polymerase activity.
Affinity purification of proteins associated with wild-type and mutant Yth1p
A tag consisting of protein A followed by the TEV protease cleavage site has been described as a tool for the one-step affinity purification of protein complexes from yeast extracts (Senger et al., 1998; Ohnacker et al., 2000). To generate an expression construct for the protein A–TEV–Yth1p fusion protein, a NotI (filled in)–SalI fragment containing the YTH1 ORF was cloned into the NcoI (filled in)–SalI site of pRS315-LEU2-ProtA-TEV (Senger et al., 1998). Constructs for the expression of protein A–TEV–yth1-1p (strain SB17) and protein A–TEV–yth1-4p (strain SB18) were generated in a similar way. Extracts from strains expressing these fusion proteins were subjected to batch adsorption to IgG–agarose. All incubation steps were performed at 4°C with gentle mixing. Four hundred microlitres of IgG–agarose (Sigma), 2.2 ml of extract, 1.1 ml of 80% glycerol and 6.6 ml of buffer IPPD-150 (10 mM Tris–HCl pH 7.9, 150 mM NaCl, 0.1% NP-40, 0.5 mM dithiothreitol) including 1 mM phenylmethylsulfonyl fluoride, 0.4 µg/ml leupeptin and 0.7 µg/ml pepstatin were mixed in a tube and incubated for 3 h. The washes and the elution of the bound proteins by cleavage with TEV protease were essentially carried out under the conditions described in Ohnacker et al. (2000).
Recombinant proteins and purified processing factors
The complete coding sequences of YTH1, yth1-1, yth1-4 and yth1-2→4 were cloned into a GST expression vector derived from pGEX-4T3 (a gift from B.Dichtl). The recombinant proteins were expressed in E.coli BL21 (DE3) pLysS (Studier, 1991). Growth of the bacteria, induction of expression, homogenization of the cells, binding of proteins to GST–Sepharose (Pharmacia), washing steps, and elution of the bound material were carried out according to the manufacturer’s instructions. The proteins were further purified on a Smart Mini Q column (Pharmacia).
CF IA was co-purified with CF II–PF I by IgG–agarose batch adsorption of proteins from a strain expressing protein A–TEV-tagged Pfs2p (Ohnacker et al., 2000). Subsequently, CF IA was separated from CF II–PF I by chromatography on a Mini Q column. Purified CF II as well as recombinant Nab4p/Hrp1p were gifts from L.Minvielle-Sebastia (Minvielle-Sebastia et al., 1997, 1998). Recombinant Pab1p was kindly provided by A.B.Sachs (University of California, Berkeley, CA).
In vitro protein-binding assays
For the in vitro expression of Ysh1/Brr5 and Fip1 proteins, plasmid DNAs were transcribed and translated in rabbit reticulocyte lysate with the TNT coupled transcription–translation system (Promega) in a total volume of 50 µl according to the manufacturer’s instructions. Purified GST or GST–Yth1 fusion proteins (0.2–0.5 µg) were incubated with the in vitro translated, 35S-labelled proteins (2–5 µl of the in vitro translation reaction corresponding to ∼1–2 fmol) in a total volume of 25 µl for 1 h at 25°C. Five hundred microlitres of binding buffer (1× phosphate-buffered saline, 0.05% NP-40, 0.5 mg/ml bovine serum albumin) were added together with 15 µl of GST–Sepharose. After incubation for 1.5 h at 4°C, the resin was pelleted and washed three times with 1 ml of IPP250 (20 mM Tris–HCl pH 7.9, 250 mM NaCl, 0.05% NP-40). Proteins were eluted in 20 µl of SDS–PAGE sample buffer and resolved by 10% SDS–PAGE. The signal was enhanced by treatment with Amplify solution (Amersham).
In vitro RNA-binding assays
UV cross-linking assays were performed essentially according to Minvielle-Sebastia et al. (1998) on full-length CYC1 pre-mRNA. The amount of cross-linked protein was determined with the IPLabgel software (version 1.5, Signal Analytics Corp.).
The ‘short’ CYC1 and ‘short’ cyc1-512 RNAs (a gift from B.Dichtl, manuscript in preparation) were prepared from linearized pGem3 constructs as run-off transcripts with SP6 polymerase (Boehringer). The ‘short’ cyc1-512 pre-mRNA contains 38 additional nucleotides at the 5′ end, so that its size is comparable to that of the wild-type RNA. RNase H protection experiments were carried out as follows. A master mix containing ∼100–400 µg of recombinant protein and 30 fmol of 32P-labelled RNA per sample was incubated under standard cleavage conditions for 15 min at 30°C. Subsequently, 10 µl aliquots were transferred to Eppendorf tubes containing 10 pmol of DNA oligonucleotide (Microsynth) and 0.7 U of RNase H (Promega). Incubation for 1 h at 30°C was followed by proteinase K digestion at 65°C for 45 min. The RNAs were separated on a 12% polyacrylamide–8 M urea gel and visualized by autoradiography.
Supplementary data
Supplementary data to this paper (Materials and methods) are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are grateful to Bernhard Dichtl for providing the RNA substrates used in the RNA-binding experiments and for critically reading the manuscript. We thank U.Rüegsegger and E.Wahle for critical comments. This work was supported by the University of Basel, the Swiss National Science Foundation, the European Community (via the Bundesamt für Bildung und Wissenschaft, Bern) and the Louis-Jeantet-Foundation for Medicine.
References
- Aguilera A. (1994) Formamide sensitivity: a novel conditional phenotype in yeast. Genetics, 136, 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani N., Minet,M., Le Gouar,M., Lacroute,F. and Wyers,F. (1997a) Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol. Cell. Biol., 17, 3694–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani N., Minet,M., Wyers,F., Dufour,M.-E., Aggenbeck,L.P. and Lacroute,F. (1997b) PCF11 encodes a third protein component of the yeast cleavage and polyadenylation factor I. Mol. Cell. Biol., 17, 1102–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari A. and Schwer,B. (1995) SLU7 and a novel activity, SSF1, act during the PRP16-dependent step of yeast pre-mRNA splicing. EMBO J., 14, 4001–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmair A., Finley,D. and Varshavsky,A. (1986) In vivo half-life of a protein is a function of its amino-terminal residue. Science, 234, 179–186. [DOI] [PubMed] [Google Scholar]
- Bai C. and Tolias,P.P. (1996) Cleavage of RNA hairpins by a developmentally regulated CCCH zinc finger protein. Mol. Cell. Biol., 16, 6661–6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabino S.M., Hübner,W., Jenny,A., Minvielle-Sebastia,L. and Keller,W. (1997) The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev., 11, 1703–1716. [DOI] [PubMed] [Google Scholar]
- Bienroth S., Keller,W. and Wahle,E. (1993) Assembly of a processive messenger RNA polyadenylation complex. EMBO J., 12, 585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.E. and Sachs,A.B. (1998) Poly (A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol. Cell. Biol., 18, 6548–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J.S., Sadhale,P.P. and Platt,T. (1990) RNA processing in vitro produces mature 3′ ends of a variety of Saccharomyces cerevisiae mRNAs. Mol. Cell. Biol., 10, 2599–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G., Noble,S.M. and Guthrie,C. (1996) Essential yeast protein with unexpected similarity to subunits of mammalian cleavage and polyadenylation specificity factor (CPSF). Science, 274, 1511–1514. [DOI] [PubMed] [Google Scholar]
- Chen J. and Moore,C. (1992) Separation of factors required for cleavage and polyadenylation of yeast pre-mRNA. Mol. Cell. Biol., 12, 3470–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. and Hyman,L.E. (1998) A specific RNA–protein interaction at yeast polyadenylation efficiency elements. Nucleic Acids Res., 26, 4965–4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D., St Jean,A., Woods,R.A. and Schiestl,R.H. (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res., 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber J.H., Cantor,C.R., Mohr,S.C. and Smith,T.F. (1999) Genomic detection of new yeast pre-mRNA 3′-end processing signals. Nucleic Acids Res., 27, 888–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z. and Sherman,F. (1995) 3′-end-forming signals of yeast mRNA. Mol. Cell. Biol., 15, 5983–5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z. and Sherman,F. (1996) 3′-end-forming signals of yeast mRNA. Trends Biochem. Sci., 21, 477–480. [DOI] [PubMed] [Google Scholar]
- Guthrie C. and Fink,G.R. (1991) Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego, CA. [Google Scholar]
- Jenny A., Minvielle-Sebastia,L., Preker,P.J. and Keller,W. (1996) Sequence similarity between the 73 kDa protein of mammalian CPSF and a subunit of yeast polyadenylation factor I. Science, 274, 1514–1517. [DOI] [PubMed] [Google Scholar]
- Keller W., Bienroth,S., Lang,K.M. and Christofori,G. (1991) Cleavage and polyadenylation factor (CPF) specifically interacts with the pre-mRNA 3′ processing signal AAUAAA. EMBO J., 10, 4241–4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler M.M., Zhao,J. and Moore,C.L. (1996) Purification of the Saccharomyces cerevisiae cleavage/polyadenylation factor I. Separation into two components that are required for both cleavage and polyadenylation of mRNA 3′ ends. J. Biol. Chem., 271, 27167–27175. [DOI] [PubMed] [Google Scholar]
- Kessler M.M., Henry,M.F., Shen,E., Zhao,J., Gross,S., Silver,P.A. and Moore,C.L. (1997) Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev., 11, 2545–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J., Kellermann,J. and Keller,W. (1991) Cloning and expression of the essential gene for poly(A) polymerase from S.cerevisiae. Nature, 354, 496–498. [DOI] [PubMed] [Google Scholar]
- Mikaelian I. and Sergeant,A. (1992) A general and fast method to generate multiple site directed mutations. Nucleic Acids Res., 20, 376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle-Sebastia L., Winsor,B., Bonneaud,N. and Lacroute,F. (1991) Mutations in the yeast RNA14 and RNA15 genes result in an abnormal mRNA decay rate; sequence analysis reveals an RNA-binding domain in the RNA15 protein. Mol. Cell. Biol., 11, 3075–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle-Sebastia L., Preker,P. and Keller,W. (1994) RNA14 and RNA15 proteins as components of a yeast pre-mRNA 3′-end processing factor. Science, 266, 1702–1705. [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia L., Preker,P.J., Wiederkehr,T., Strahm,Y. and Keller,W. (1997) The major yeast poly(A)-binding protein is associated with cleavage factor IA and functions in premessenger RNA 3′-end formation. Proc. Natl Acad. Sci. USA, 94, 7897–7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle-Sebastia L., Beyer,K., Krecic,A.M., Hector,R.E., Swanson,M.S. and Keller,W. (1998) Control of cleavage site selection during mRNA 3′ end formation by a yeast hnRNP. EMBO J., 17, 7454–7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy K.G.K. and Manley,J.L. (1995) The 160-kD subunit of human cleavage–polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev., 9, 2672–2683. [DOI] [PubMed] [Google Scholar]
- Ohnacker M., Barabino,S.M., Preker,P.J. and Keller,W. (2000) The WD-repeat protein Pfs2p bridges two essential factors within the yeast pre-mRNA 3′-end-processing complex. EMBO J., 19, 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E.C., Finley,D. and Szostak,J.W. (1992) A strategy for the generation of conditional mutations by protein destabilization. Proc. Natl Acad. Sci. USA, 89, 1249–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker P.J., Lingner,J., Minvielle-Sebastia,L. and Keller,W. (1995) The FIP1 gene encodes a component of a yeast pre-mRNA poly adenylation factor that directly interacts with poly(A) polymerase. Cell, 81, 379–389. [DOI] [PubMed] [Google Scholar]
- Preker P.J., Ohnacker,M., Minvielle-Sebastia,L. and Keller,W. (1997) A multisubunit 3′ end processing factor from yeast containing poly(A) polymerase and homologues of the subunits of mammalian cleavage and polyadenylation specificity factor. EMBO J., 16, 4727–4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo P., Li,W.Z., Hampsey,D.M., Zaret,K.S. and Sherman,F. (1991) Distinct cis-acting signals enhance 3′ endpoint formation of CYC1 mRNA in the yeast Saccharomyces cerevisiae. EMBO J., 10, 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger B., Simos,G., Bischoff,F.R., Podtelejnikov,A., Mann,M. and Hurt,E. (1998) Mtr10p functions as a nuclear import receptor for the mRNA-binding protein Npl3p. EMBO J., 17, 2196–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F.W. (1991) Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J. Mol. Biol., 219, 37–44. [DOI] [PubMed] [Google Scholar]
- Stumpf G. and Domdey,H. (1996) Dependence of yeast pre-mRNA processing on CFT1: a sequence homolog of the mammalian AAUAAA binding factor. Science, 274, 1517–1520. [DOI] [PubMed] [Google Scholar]
- Valentini S.R., Weiss,V.H. and Silver,P.A. (1999) Arginine methylation and binding of Hrp1p to the efficiency element for mRNA 3′-end formation. RNA, 5, 272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helden J., del Olmo,M. and Pérez-Ortín,J.E. (2000) Statistical analysis of yeast genomic downstream sequences reveals putative polyadenylation signals. Nucleic Acids Res., 28, 1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E. (1995) Poly(A) tail length control is caused by termination of processive synthesis. J. Biol. Chem., 270, 2800–2808. [DOI] [PubMed] [Google Scholar]
- Wahle E. and Rüegsegger,U. (1999) 3′-end processing of pre-mRNA in eukaryotes. FEMS Microbiol. Rev., 23, 277–295. [DOI] [PubMed] [Google Scholar]
- Zhao J., Kessler,M.M. and Moore,C.L. (1997) Cleavage factor II of Saccharomyces cerevisiae contains homologues to subunits of the mammalian cleavage/polyadenylation specificity factor and exhibits sequence-specific, ATP-dependent interaction with precursor RNA. J. Biol. Chem., 272, 10831–10838. [DOI] [PubMed] [Google Scholar]
- Zhao J., Hyman,L. and Moore,C. (1999a) Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev., 63, 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Kessler,M., Helmling,S., O’Connor,J.P. and Moore,C. (1999b) Pta1, a component of yeast CF II, is required for both cleavage and poly(A) addition of mRNA precursor. Mol. Cell. Biol., 19, 7733–7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelkovsky A., Helmling,S. and Moore,C. (1998) Processivity of the Saccharomyces cerevisiae poly(A) polymerase requires interactions at the carboxyl-terminal RNA binding domain. Mol. Cell. Biol., 18, 5942–5951. [DOI] [PMC free article] [PubMed] [Google Scholar]