Abstract
Isothiocyanates (ITCs) derived from cruciferous vegetables induce apoptosis in cancer cells. We demonstrate that certain naturally-occurring ITCs selectively deplete mutant p53, but not the wild type, and do so via a transcription-independent mechanism. Direct p53 binding followed by conformational changes appears to be a mechanism by which mutant p53 is depleted. Structure-activity relationship studies (SARs) using naturally-occurring and synthetic ITCs show that depletion is influenced by the ITC side chain moiety. Furthermore, we show that cells with p53 mutations are more sensitive to cytotoxicity induced by phenethyl isothiocyanate (PEITC) than those with the wild type. 2,2-diphenylethyl ITC, a synthetic ITC and one of the most potent depletors of mutant p53 studied, induces apoptosis to the greatest extent in mutant p53 breast cancer cells. Collectively, this study shows that mutant p53 depletion may be an important novel target for cancer chemoprevention and therapy by natural and synthetic ITCs.
Keywords: Isothiocyanate(s), p53, protein depletion, structure-activity relationships
Introduction
Isothiocyanates (ITCs) have been shown to induce apoptosis in cultured tumor cells and in tissues of treated animals. Induction of apoptosis is believed to be an important mechanism for the chemopreventive activities of ITCs (1); however, the early events in the induction of apoptosis by ITCs have not yet been fully investigated. Our studies have shown that, once inside the cell, ITCs bind to proteins, and that protein binding affinities are closely associated with the ability to induce apoptosis (2). Recently, we identified tubulin as one of the binding targets for ITCs and found that covalent binding to the cysteine residues in tubulin causes conformation changes, leading to its degradation (3).
The tumor suppressor p53 plays a pivotal role in many cellular processes, including DNA repair, cell cycle arrest and apoptosis (4). About half of all human cancers carry p53 mutations (4). Most p53 mutations are within the central core DNA-binding domain (DBD); this disrupts DNA binding to p53, and severely abrogates the tumor suppressor function. Mutant p53 impairs the DNA-damage response, and renders tumor cells more resistant to drug-induced apoptosis (5). Evidence also shows that mutant p53 promotes tumorigenesis through gain of function by transactivating growth-related genes or by suppressing specific target genes (6). Mutant p53 overexpression in deficient cells causes enhanced plating efficiency in soft agar and increased tumorigenicity in nude mice (6). Also, mice harboring a p53 point mutation have a high frequency of tumor formation and metastasis compared to p53-deficient mice (7, 8). Humans with germ-line p53 mutation (Li-Fraumeni Syndrome) are at significantly increased risk of cancer development (9). Therefore, abolishing mutant p53 may offer a promising approach for cancer prevention and therapy. A previous study using p53 −/− mouse embryonic fibroblasts demonstrated that apoptosis induced by phenethyl ITC (PEITC) is p53-dependent (10). However, other studies have shown that PEITC induces apoptosis in prostate and colon cancer cells with different p53 statuses (11, 12).
In this study, we report an intriguing observation that PEITC, which is abundant in watercress, can selectively deplete mutant p53 protein, but not the wild type. This is a potentially important finding because of the role of mutant p53 protein in human cancers. We found that PEITC-induced mutant p53 depletion occurs post-transcriptionally. To discover which ITCs most efficiently deplete mutant p53, we conducted structure-activity relationship studies (SARs) of both synthetic and naturally-occurring ITCs by focusing on the following structural characteristics of the ITC side chain moiety: 1) lipophilicity, 2) electronic factor, 3) steric factor, and 4) alkyl chain length. We also studied aliphatic ITCs, including sulforaphane (SFN) and allyl ITC (AITC), which are abundant in broccoli and mustard, respectively. To explore the possible mechanisms by which ITCs deplete mutant p53, we studied covalent modification of cysteine residues in purified mutant p53 DNA binding domain (DBD) protein by ITCs and its sequential conformational changes. Induction of apoptosis by ITCs was then compared in cells with either mutant or wild type p53, and the SARs for apoptosis induction in a mutant p53 breast cancer cell line were also examined.
Results
PEITC causes rapid depletion of mutant p53 by a post-transcriptional mechanism
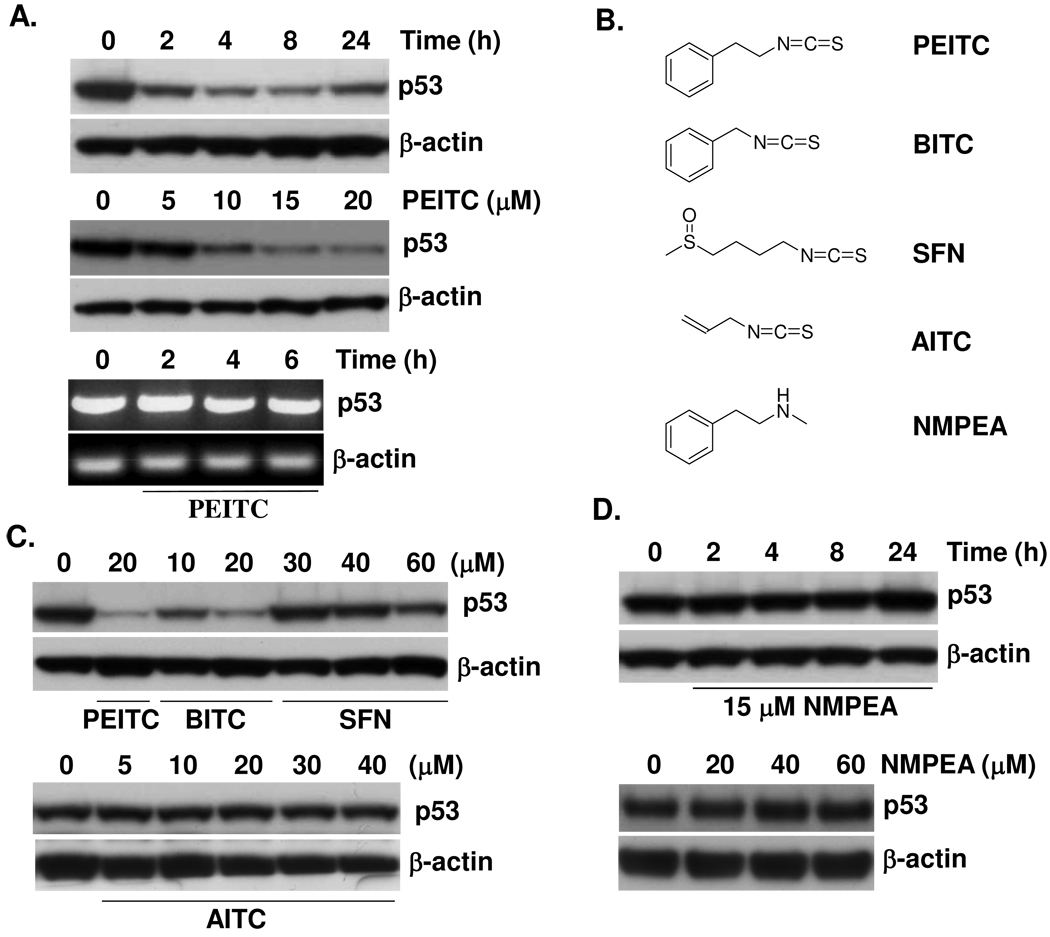
Treatment of human H596 (G245C) non-small cell lung cancer cells with 15 µM PEITC caused a rapid depletion of mutant p53 protein. Depletion occurred as early as 2 h and peaked at 4 h; it was sustained during 24 h treatment and was concentration-dependent (Fig. 1A). The expression of other proteins, including Bax, DR5, Hsp90, and JNK, was not altered (not shown). To determine whether depletion occurs at the post-transcriptional or transcriptional level, reverse transcription-PCR was performed on RNA derived from DMSO and PEITC-treated H596 cells. As shown in Fig. 1A, 2, 4 and 6 h after PEITC treatment, the mutant p53 mRNA levels remained unchanged. Therefore, PEITC depleted mutant p53 without causing changes in the p53 mRNA expression levels. These results strongly suggest that PEITC depletes mutant p53 at the post-transcriptional level. We next examined the effects of other widely studied naturally-occurring ITCs, benzyl ITC (BITC), SFN and AITC, on mutant p53 protein levels. H596 cells were incubated with PEITC, BITC, SFN or AITC for 2 h. Fig. 1C shows that BITC at 10 µM significantly reduced the level of mutant p53, while at 20 µM it showed a similar potency as PEITC. However, neither SFN nor AITC significantly affected mutant p53 protein levels, even at 40 µM; although at 60 µM SFN slightly reduced mutant p53 levels. To further investigate the structural requirement of ITCs to deplete mutant p53, N-methylphenethylamine (NMPEA), an analog of PEITC lacking an ITC functional group was used. Fig. 1D shows that NMPEA at 15 µM failed to alter the mutant p53 protein level even up to 24 h or at 60 µM for 2 h. Because the ITC functional group is highly electrophilic, these results indicate that the ITC group is essential for the effect, possibly through binding to target proteins. Moreover, the structure of the side-chain moiety appears to dictate the potency of mutant p53 depletion by ITCs.
Figure 1. PEITC causes rapid depletion of mutant p53 in H596 cells.
(A) PEITC decreased mutant p53 protein in a time and dose-dependent manner. H596 cells were treated with 15 µM PEITC for various times, or incubated with various concentrations of PEITC for 2 h. p53 and β-actin levels were determined by immunoblotting. Lower panel: H596 cells were treated with 15 µM PEITC for 2, 4 and 6 h, mRNA was then isolated and RT-PCR for p53 and β-actin mRNA was performed. (B) Structures of ITCs and NMPEA. (C) Effects of PEITC, BITC, SFN and AITC on mutant p53 protein in H596 cells. (D) NMPEA has no effect on p53 mutant protein in H596 cells.
PEITC depletes mutant p53 in a variety of cancer cells
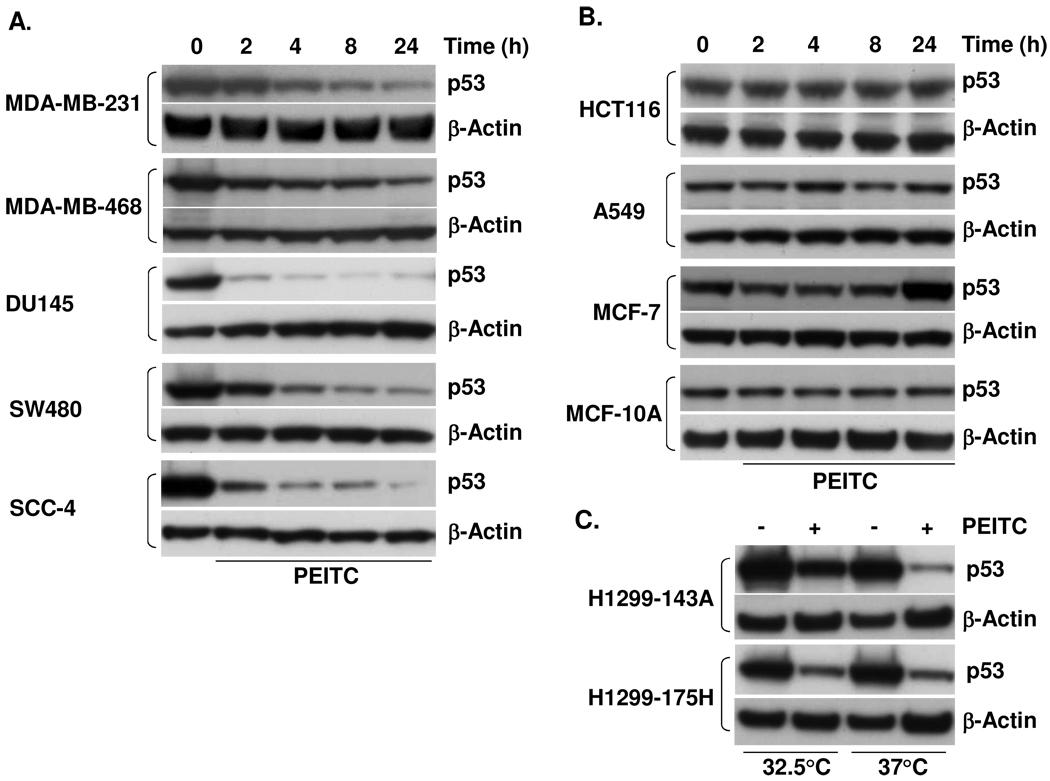
We next examined whether the depletion of mutant p53 by PEITC is cell type-specific. Various tumor cells with mutant p53 were used, including MDA-MB-231 (breast cancer, R280K), MDA-MB-468 (breast cancer, R273H), DU145 (prostate cancer, P274L/V223F), SW480 (colon cancer, R273H) and SCC-4 (oral cancer, P151S). Notably, while the types of mutations among the cell lines vary, all are located in the core DNA-binding domain. The five cell lines were treated with 10 µM or 15 µM PEITC, depending on the observed cytotoxicity. Fig. 2A shows that PEITC depleted mutant p53 in all five cell lines in a time-dependent manner, and certain cells appeared more responsive than others. These results clearly show that the effects of PEITC on mutant p53 are not specific to a certain p53 mutation.
Figure 2. PEITC depletes p53 protein in human cancer cells with different p53 mutation, but not in cells with wild type p53.
(A) Cell lines harboring mutant p53 were treated with 15 µM PEITC (MDA-MB-231, MDA-MB-468, SCC-4) or 10 µM PEITC (DU145, and SW480) for the indicated times. (B) Cells with wild type p53 were treated with 20 µM PEITC (HCT116, A549, and MCF-7) or 15 µM PEITC (MCF-10A) for the indicated times. (C) H1299-143A cells with p53 mutant conformation (at 37°C) are more sensitive to PEITC-induced depletion than that with wild type conformation (at 32.5°C). H1299-175H cells are used as control. Both cells were treated with 20 µM PEITC for 24 h.
Wild type p53 is not a target for depletion by PEITC in cancer cells
To determine whether wild type p53 is affected by PEITC treatment, we used A549 lung cancer cells, HCT116 colon cancer cells, MCF-7 breast cancer cells and MCF-10A human normal mammary epithelial cells, all expressing wild type p53. Cell lines were treated with 15 or 20 µM PEITC for 2, 4, 8 and 24 h. Contrary to mutant p53, wild type p53 expression did not decrease over the treatment period. In fact, it appeared marginally elevated at 24 h in MCF-7 cells (Fig. 2B), suggesting that PEITC may moderately activate p53 in cells with wild type p53. To investigate whether the conformation of p53 contributes to the differential response to PEITC, we used H1299 lung cancer cells (null p53) stably transfected with temperature-sensitive p53 mutant 143Ala (H1299-143A) (13). p53 mutant 143Ala can have two conformations: at 32.5°C it possesses a strong DNA-binding ability and generates transcriptional activity, thus, functioning like wild type p53, and at 37°C its ability to bind DNA and activate transcription is severely diminished or lost, acting as mutant p53. After treatment of H1299-143A cells with PEITC for 24 h, p53 mutant 143A protein was depleted more robustly at 37°C than at 32.5°C (Fig. 2C). To rule out the possibility that lower temperature caused the effect, H1299 cells stably expressing non-temperature-sensitive p53 mutant R175H (H1299-175H) were treated with PEITC for 24 h; the same extent of depletion was observed at both temperature (Fig. 2C). Therefore, our data suggest that the differential response between mutant and wild type p53 to PEITC is likely due to the difference in their conformation.
SARs for the depletion of mutant p53 by ITCs
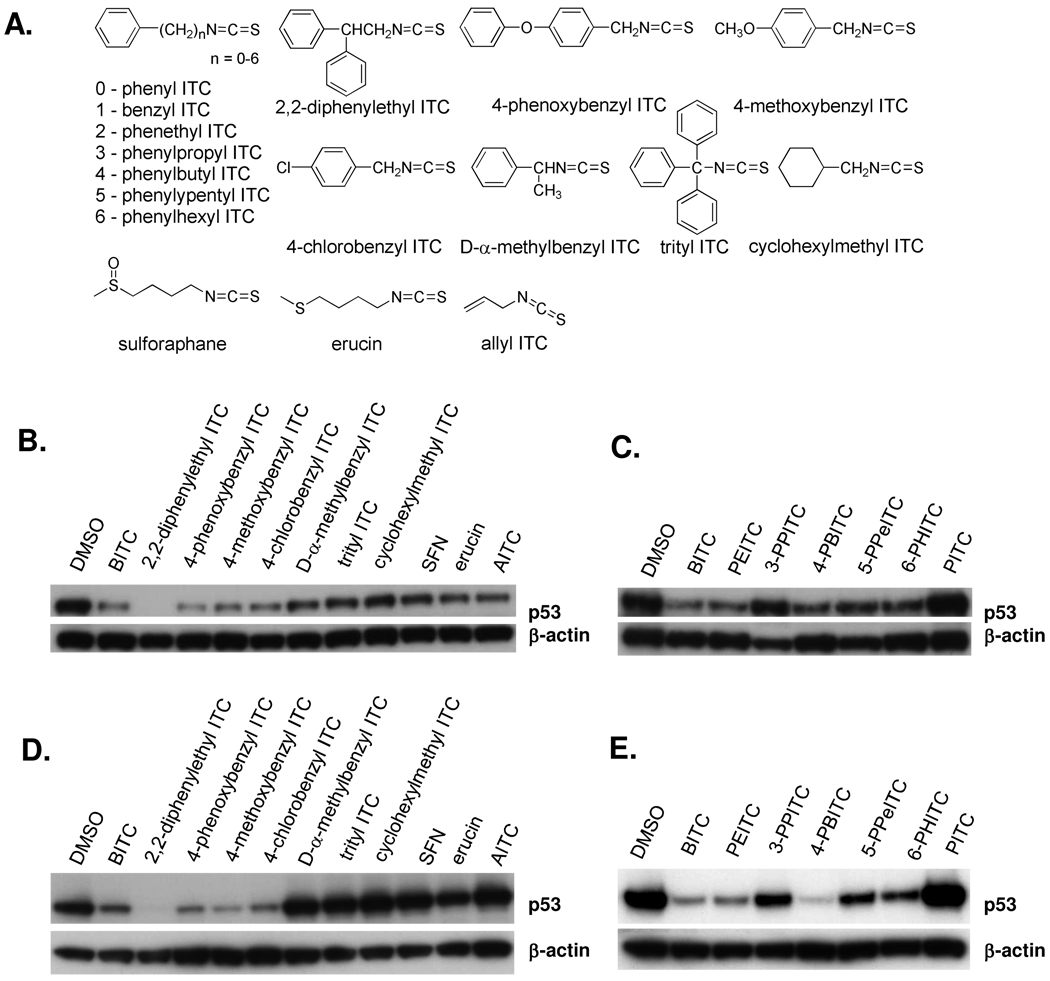
BITC and PEITC are arylalkyl ITCs of different alkyl chain length with relatively high lipophilicities, whereas SFN, an alkyl ITC, is considerably more water-soluble. Previously, we showed BITC, PEITC and SFN exhibit differential activity toward total protein binding and apoptosis induction, following an order of BITC>PEITC>SFN (2). Moreover, BITC and PEITC can modify proteins and tubulin to a greater extent than SFN, and the degree of binding correlates with the potency of the ITC to induce mitosis arrest and apoptosis (2, 3). Importantly, the same order of potency was seen toward mutant p53 depletion. NMPEA, an analog of PEITC without the ITC functional group, is completely devoid of all of these activities. Here, we studied the structural features of the ITC side chain moiety with relation to the extent of depletion of mutant p53. Compounds of more than one aromatic ring with greater lipophilicity than BITC and PEITC were used. These compounds are 2,2-diphenylethyl ITC and 4-phenoxybenzyl ITC. We also examined the effects of an electron-withdrawing and an electron-releasing substituent at the para position of the aromatic ring by using 4-chlorobenzyl ITC and 4-methoxybenzyl ITC. To investigate whether covalent binding by ITCs is a trigger of mutant p53 depletion, we selected ITCs with bulky substituent(s) adjacent to the –N=C=S to create steric hindrance, including D-α-methylbenzyl ITC and trityl ITC. Aromatic ITCs with varying alkyl chain length from phenyl ITC to 6-phenylhexyl ITC (6-PHITC) were used. Structures of the ITCs are shown in Fig. 3A. The aliphatic compounds used include cyclohexylmethyl ITC, SFN, erucin and AITC.
Figure 3. Structure-activity relationships for the depletion of mutant p53 by ITCs.
(A) Structures of the naturally-occurring and synthetic ITCs used. (B and C) Depletion of mutant p53 protein by ITCs in H596 non-small cell lung cancer cells treated with 20 µM ITCs, as determined by immunoblotting. (D and E) MDA-MB-231 breast cancer cells were treated with 10 µM ITCs.
As shown in Figure 3 (B–E), mutant p53 is depleted similarly in H596 (Fig. 3B and C) and MDA-MB-231 (Fig. 3D and E) cancer cells treated with 10 or 20 µM ITCs, respectively, for 24 h. Also, at equimolar concentrations BITC and PEITC deplete mutant p53 to approximately the same extent, while SFN depletes p53 significantly less in H596 or MD-MBA-231 cells, compared to DMSO control. The presence of a phenyl ring appears important as its electron-withdrawing ability reduces the electron density around the carbon atom of the –N=C=S functional group rendering it more electrophilic (14). The aliphatic ITCs, cyclohexylmethyl ITC, SFN, erucin and AITC, all have little to no mutant p53 depleting activity, whereas 2,2-diphenylethyl ITC and 4-phenoxybenzyl ITC deplete mutant p53 to an even greater extent than BITC (Fig. 3B and D). Thus, increasing the lipophilicity of the side chain moiety by the addition of aromatic groups increases the activity of the ITC to deplete mutant p53. Introducing an electron-withdrawing or releasing substituent at the para position dose not exert much effect, as no significant difference was seen with 4-chlorobenzyl ITC and 4-methoxybenzyl ITC when compared with BITC. In MDA-MB-231 cells, 4-chlorobenzyl ITC may deplete to a greater extent than BITC; however, 4-methoxybenzyl ITC is just as efficient as BITC, if not more. D-α-methylbenzyl ITC and trityl ITC deplete little to no mutant p53; these ITCs have bulky substituents at the α-carbon adjacent to the ITC functional group. These differ from 2,2-diphenylethyl ITC, which does not have its bulky substituents on the α-carbon. Bulky substituents adjacent to the –N=C=S group could hinder binding of the ITC to proteins. The alkyl chain length also seems to influence the activity (Fig. 3C and E). PITC does not deplete mutant p53, whereas BITC, PEITC, 3-PPITC, 4-PBITC, 5-PPeITC and 6-PHITC all deplete mutant p53 to different degrees. Interestingly, depletion is greatest when one, two or four methylene groups are inserted between the phenyl ring and the –N=C=S functional group (BITC, PEITC and 4-PBITC, respectively), and it is lowest when three methylene groups are present (3-PPITC). The large contrast of PPITC compared with PEITC and PBITC is notable.
Covalent modification of mutant (G245C) p53 DBD by ITCs and subsequent conformational changes
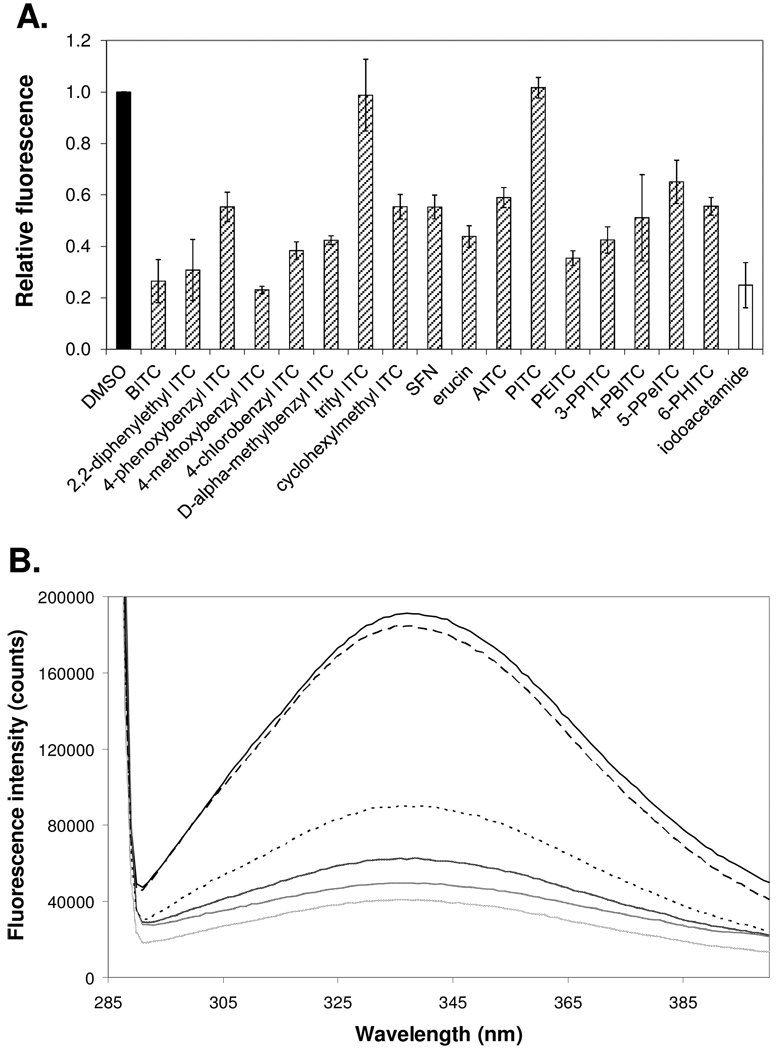
As in the case of tubulin degradation, it is possible that mutant p53 protein is depleted by ITCs as a result of direct binding to its cysteine residues and subsequent conformational changes. We used a monochlorobimane fluorometric assay (15) to study cysteine modification of mutant (G245C) p53 DBD by ITCs, and compared intrinsic tryptophan fluorescence intensities as an indicator of conformational changes (16). The G245C point mutation is especially relevant because it is the p53 mutation present in the H596 non-small cell lung cancer cell line studied here. Fig. 4A shows the relative amount of free thiols present, as indicated by relative fluorescence intensities, in the mutant (G245C) p53 DBD after 1 h incubation with DMSO (negative control), ITCs or iodoacetamide (positive control). BITC, 2,2-diphenylethyl ITC, 4-methoxybenzyl ITC, 4-chlorobenzyl ITC, PEITC, and 3-PPITC show greater binding affinities for mutant (G245C) p53 DBD than do trityl ITC, cyclohexylmethyl ITC, SFN, AITC and PITC (p<0.05). These affinities correlate well with their respective activity to deplete mutant p53. However, the binding affinities are not always in agreement with the depleting potencies of ITCs. For example, 4-phenoxybenzyl ITC was one of the most effective depletors, but it did not display stronger binding to mutant p53 than the weaker depletors, such as D-α-methylbenzyl ITC. These results suggest that binding affinity to mutant p53 can only partially explain depleting activities and other factor(s) may be involved.
Figure 4. Cysteine binding in mutant (G245C) p53 DNA-binding domain by ITCs and subsequent conformational changes.
(A) Cysteine binding in the mutant (G245C) p53 DNA-binding domain by DMSO (control), ITCs or iodoacetamide, measured using a monochlorobimane fluorometric assay (λex 380 nm; λem 485 nm). (B) Intrinsic fluorescence emission spectra of mutant (G245C) p53 DNA-binding domain at 32°C (black solid line) incubated with, from highest intensity to lowest, DMSO (black large-dashed line), SFN (black small-dashed line), D-α-methylbenzyl ITC (dark grey line), BITC (medium grey line) or 4-phenoxybenzyl ITC (light grey line).
Using intrinsic tryptophan fluorescence emission spectroscopy, we demonstrated that the conformation of mutant p53 changes to different degrees after its reaction with BITC, 4-phenoxybenzyl ITC, D-α-methylbenzyl ITC and SFN (Fig. 4B). These ITCs were chosen because their binding affinities correlate either well or poorly with depletion. 4-Phenoxybenzyl ITC, a potent depletor, induces the greatest conformational change, despite its lower binding affinity for mutant p53 protein. BITC causes the second largest conformational change, followed by D-α-methylbenzyl ITC and finally SFN. D-α-methylbenzyl ITC shows a lower binding affinity than BITC (p<0.05), but a higher binding affinity than 4-phenoxybenzyl ITC (p<0.05); however, D-α-methylbenzyl ITC does not deplete mutant p53 to the same degree as 4-phenoxybenzyl ITC. The fact that D-α-methylbenzyl ITC induces less mutant p53 protein conformational change than 4-phenoxybenzyl ITC may explain why it does not deplete as efficiently. Similarly, despite its binding to mutant p53, SFN causes the least conformational change among ITCs tested, which correlates with its lack of depleting activity.
Mutant p53 cells are more sensitive to apoptosis induced by PEITC than wild type cells
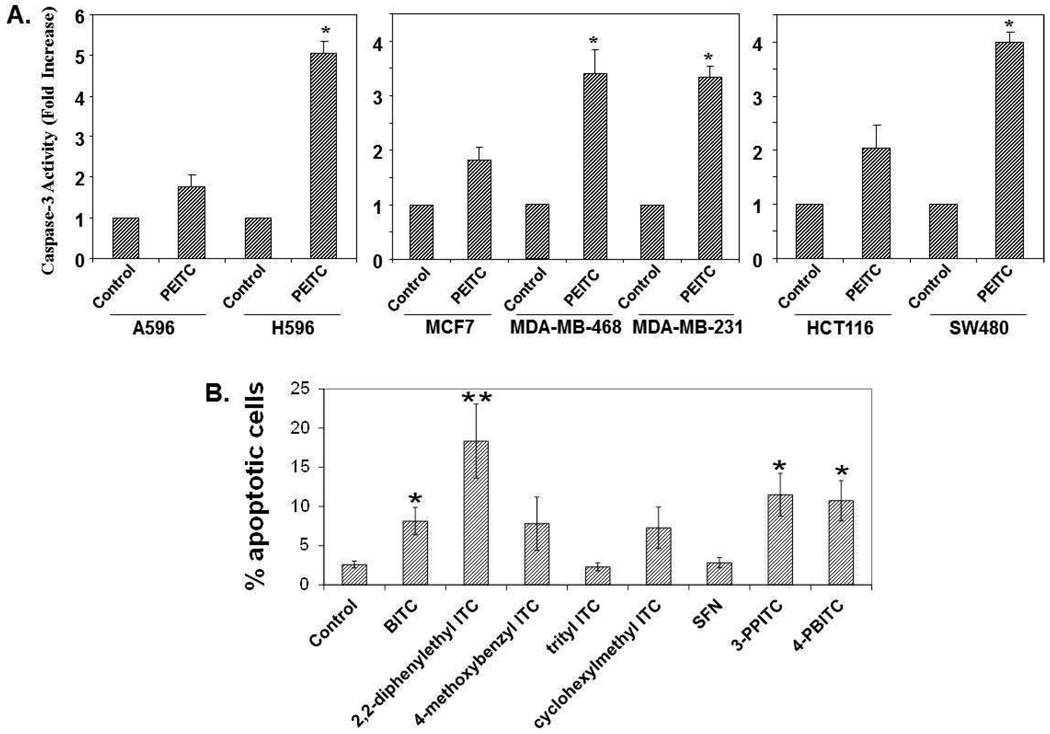
The role of p53 in the induction of apoptosis by ITCs is not well established and likely to be cell-specific. Mutant p53 renders cancer cells resistance to chemotherapeutic drugs; if ITCs can deplete mutant p53, then ITCs should be more cytotoxic in cells with mutant p53 protein. To examine this possibility, we assessed the cytotoxicity induced by PEITC in cancer cells containing either wild type or mutant p53. Three sets of cell lines from lung, breast, and colon were used: A549 (wild type p53) and H596 (mutant p53) lung cancer cells, MCF-7 (wild type p53) and MDA-MB-231 and MDA-MB-468 (mutant p53) breast cancer cells, HCT116 (wild type p53) and SW480 (mutant p53) colon cancer cells. After treating with PEITC for 24 h, apoptosis was determined by a caspase-3 activity assay. As shown in Fig. 5A, all the mutant p53 cancer cells appeared significantly more sensitive than the wild type cells to PEITC-induced apoptosis.
Figure 5. Cells with mutant p53 are more sensitive to PEITC-induced apoptosis than wild type cells, and induction of apoptosis by various ITCs in a cell line with mutant p53.
(A) A549 (wt p53) and H596 (mutant p53) non-small cell lung cancer cells, MCF-7 (wt p53), MDA-MB-231 and MDA-MB-468 (mutant p53) breast cancer cells, HCT116 (wt p53) and SW480 (mutant p53) colon cancer cells were treated with DMSO and PEITC for 24 h. Caspase-3 activity was analyzed by a caspase-3 colorimetric assay kit. The data represents the mean ± S.D. of three independent experiments (*P<0.01). (B) Apoptosis in MDA-MB-231 breast cancer cells treated with 10 µM ITCs measured using FITC-Annexin V and propidium iodide. Shown are cells in early apoptosis (FITC-Annexin V+ / PI−). *p<0.05 compared to DMSO (control), ** p<0.05 compared to BITC.
Induction of apoptosis by various ITCs in a mutant p53 cell line
To examine the correlation between depletion of mutant p53 and apoptosis by ITCs, FITC-annexin V staining assay was used to quantify apoptotic cells, we treated MDA-MB-231 cancer cells with DMSO or 10 µM of various ITCs for 24 h, Fig. 5B shows that treatment with BITC, 2,2-diphenylethyl ITC, 3-PPITC or 4-PBITC, which all deplete mutant p53 (Figure 3B–E), results in a greater induction of apoptosis than treatment with DMSO (p<0.05). 2,2-diphenylethyl ITC, the most potent depletors of mutant p53 studied, is the strongest inducer of apoptosis (p<0.05 compared to BITC). The percentage of apoptotic cells induced by trityl ITC, cyclohexylmethyl ITC or SFN, all of which deplete little to no mutant p53, does not differ from control. However, the correlation is not consistently evident. For example, 3-PPITC, which does not deplete mutant p53 to the same extent as BITC or 4-PBITC, induces the same level of apoptosis as these ITCs.
Discussion and Conclusions
In this study we demonstrate that, depending on the structure, ITCs can robustly deplete mutant p53 protein in a variety of cancer cells in a time- and dose-dependent manner. The depletion appears to be specific to mutant p53, because wild type p53 is not affected by the same treatments. This is the first report of mutant p53 as a potential novel target for ITCs. Depletion of mutant p53 may be an important event in cancer prevention as ITCs may suppress gain-of-function properties through mutant p53 depletion. Furthermore, depletion of mutant p53 may reduce drug-resistance, and lead to new strategies for treating cancer in the clinic.
Our data show that PEITC causes mutant p53 depletion at the post-transcriptional level, since mutant p53 mRNA remains unchanged by PEITC treatment. Together with the fact that MNPEA, an analog without ITC functional group, is devoid of depleting activity, these results suggest a possible mechanism involving protein modification via covalent binding by the ITC functional group. Previously, we showed that the potency of inhibition of tumorigenesis by aromatic ITCs is related to the lipophilicity and alkyl chain length of the side chain moiety (14, 17). The present study also showed that depletion is influenced by the structure of the side chain moiety of ITC compounds. The lipophilicity and the steric hindrance appear to be the most important factors in determining the activity of ITCs toward mutant p53 depletion. We demonstrated that increasing lipophilicity (by adding aromatic rings) and creating spatial hindrance (by introducing bulky substituents) at the α-carbon significantly enhances or decreases the p53 depleting activity, respectively. These results again support a model for mutant p53 depletion in which ITCs bind to the protein via -N=C=S through hydrophobic interactions. The role of the aromatic ring is further suggested by the observations that aliphatic ITCs have little or no depleting activity. The alkyl chain length in arylalkyl ITCs also affects their activity. The sharp decline in depleting activity of PITC compared to BITC and PPITC compared to PEITC and PBITC suggest that the subtle changes of alkyl chain length in arylalkyl ITCs can markedly influence activity, possibly due to specific spatial requirements for interactions with the protein targets.
Studies have shown that inhibition of endogenous mutant p53 with RNA interference(RNAi) sensitizes cancer cells to chemotherapeutic agent-induced cell death in vitro and in vivo (18). We found in human lung, breast and colon cancer cells that mutant p53 confers cells with increased sensitivity to PEITC-induced cytotoxicity. Xiaoet al also showed that MDA-MB-231 breast cancer cells with mutant p53 were more sensitive to BITC-induced apoptosis than the wild type MCF-7 cells and the normal human mammary epithelial MCF-10A cells (19). The discovery of 2,2-diphenylethyl ITC, a synthetic compound, as one of the most potent depletors of mutant p53, which induces apoptosis to a greater extent than BITC, is potentially important. We are currently evaluating the efficacy of this compound as a chemopreventive and therapeutic agent in rodents with mutant p53.
We previously showed that BITC and PEITC have greater binding affinities than SFN toward intracellular proteins and that protein binding affinities of ITCs correlate well with their potencies of apoptosis induction (2). We also reported that ITCs covalently bind in vitro and in vivo to the cysteine residues in tubulin, causing its conformation change followed by aggregate formation and degradation (3). The levels of modification of tubulin by BITC, PEITC and SFN correlate with their potencies of apoptosis induction. Pretreatment of cells with tubulin binding agents, for example, taxol, diminishes the binding by ITCs and consequently the downstream effects. The exact protein targets of ITCs for mutant p53 depletion are not yet identified; it is possible certain p53 chaperone or other proteins, such as Hsp90, could serve as targets. It is well-known that cysteines in p53 are potential sites of covalent modification (20), and that the modification could lead to a change in p53 function. In this study, we explored the relationships of mutant p53 depletion and direct covalent modification of cysteine residues by ITCs in the mutant p53 DNA-binding domain and the subsequent conformational changes. We found the binding affinities to mutant p53 DBD of some, but not all, ITCs appear to correlate with their ability to deplete. Certain ITCs, such as 4-phenoxybenzyl ITC, exhibit potent depleting, yet low binding activity to mutant p53. However, this disparity may be partially explained by protein conformational changes as a result of the binding event. Our studies show that binding to mutant p53 may constitute an important step for its depletion by ITCs. Wild type p53 is tightly regulated through MDM2-mediated ubiquitination and 26S proteasome degradation process. We had preliminary data showing that PEITC-induced depletion of mutant p53 cannot be abrogated by MDM2 inhibitor Nutlin-3 or 26S proteasome inhibitor MG132 and bortezomib in H596 cells (see supplement data), suggesting that PEITC induces the depletion likely through a novel mechanism not by a MDM2-ubiquitin-mediated 26S proteasome-independent pathway. Additional in vivo studies seem to be warranted to fully understand the underlying mechanisms of mutant p53 depletion by ITCs and its functional consequences.
Experimental Section
Cell culture and Materials
Human H596, A549, HCT116, MDA-MB-231, MDA-MB-468, MCF-7, DU145, MCF-10A, SW480 and SCC-4 cells were obtained from ATCC (Manassas, VA). H1299-175H cells were kindly provided by Dr. Maria Laura Avantaggiati (Georgetown University). Cells were maintained in DMEM or RPMI-1640 medium supplemented with 10% FBS (Hyclone). DMSO, PEITC, BITC, SFN, AITC, NMPEA and monochlorobimane were purchased from Sigma-Aldrich (St. Louis, MO). 2,2-diphenylethyl ITC, 4-phenoxybenzyl ITC, 4-methoxybenzyl ITC, 4-chlorobenzyl ITC, D-α-methylbenzyl ITC, trityl ITC and 3-phenylpropyl ITC were purchased from Trans World Chemicals (Rockville, MD). Erucin and 4-phenylbutyl ITC were from LKT Laboratories, Inc. (St. Paul, MN). SFN was a generous gift from Dr. Stephen Hecht (University of Minnesota, MN) and 5-phenylpentyl ITC and 6-phenlhexyl ITC were generous gifts from Dr. Arun K. Sharma (Penn State Hershey College of Medicine, PA). All the compounds are with purities ≥95% as determined by gas liquid chromatography or HPLC. The p53 DO-1 and β-actin antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), horseradish peroxidase-labeled goat ant-mouse secondary antibodies were purchased from GE-Healthcare (Pittsburgh, PA) and Bio-Rad Protein Assay was from Bio-Rad (Hercules, CA).
Immunoblot analysis
Whole cell lysates were prepared in lysis buffer (20 mM HEPES, pH 7.4, 2 mM EGTA, 50 mM b-glycerol phosphate, 1% Triton X-100, 10% glycerol, 1 mM DTT, 1 mM PMSF, 10 mg/ml leupeptin, 10 mg/ml aprotinin, 1 mM Na3VO4 and 5 mM NaF). For immunoblot analysis, equal amounts of protein were resolved on 4–12% NuPAGE BisTris gels (Invitrogen) and transferred to PVDF membranes (Millipore). The membranes were blocked with 5% milk in TBST, and were then probed with p53 (DO-1) or β-actin antibody. Proteins were detected using enhanced chemiluminescence (ECL) reagents (GE Healthcare).
RT-PCR
Total RNA was extracted from the cells using the RNeasy kit (Qiagen). Reverse transcription-PCR was performed using SuperScript One-Step RT-PCR kit (Invitrogen) according to the manufacturer's instructions. Primers for p53 were: 5'-TTCTTGCATTCTGGGACAGCC-3' (sense) and 3'-GGCCTCATTCAGCTCTCGGAAC-5' (antisense). Primers for β-Actin were: 5'-TGGGCATGGGTCAGAAGGAT-3' (sense) and 3'-GAGGCGTACAGGGATAGCAC-5' (antisense). β-Actin was used as an internal standard for RNA normalization.
Expression and purification of wild type and mutant p53 DNA-binding domain
BL21 (DE2) Escherichia coli were transformed with pET29b wild type or mutant (G245C) p53 DNA-binding domain (DBD) plasmids. The E. coli were then cultured in LB media supplemented with 100 µM ZnCl2 and 25 µg/mL kanamycin at 37 °C until OD600 = 0.6–0.8. Protein expression was induced by 0.1 mM isopropyl β-D-1-thiogalactopyranoside for 16 hrs at 37°C for wild type and 16°C for mutant p53. Bacterial pellets were then harvested by centrifugation at 5000×g, 10 min, 4°C and lysed via sonication in 50 mM Tris, 50 mM KCl, 5 mM dithiothreitol, 1 mM PMSF, pH 7 buffer. The soluble fraction was passed through a Capto S cation exchange column (GE Healthcare) and eluted with 50 mM Tris, 200 mM KCl, 5mM DTT, pH 7. Eluted protein was dialyzed overnight against a non-salt dialysis buffer (50 mM Tris, 5mM DTT, pH 7) and loaded onto a HiTrap Heparin HP column (GE Healthcare). p53 DBD was purified via a ÄKTApurifier-10™ protein liquid chromatography system (GE Healthcare) with an increasing KCl salt gradient and purity was tested by SDS-PAGE and coomassie blue staining. Purified protein was dialyzed in PBS and protein concentrations were determined prior to performing fluorescence experiments.
Monochlorobimane fluorometric assay
Using a white 96-well plate, 100 µL of purified mutant (G245C) p53 DNA-binding domain protein in PBS was treated with DMSO, 100 µM ITCs in DMSO or 500 µM iodoacetamide (positive binding control). An additional well containing 100 µL PBS was used to determine background. Samples were incubated with compounds in the dark for 1 h at 32°C, after which 5 µL of 10 mM monochlorobimane was added (in dark) and samples were further incubated in the dark at 32°C for 30 min. Fluorescence readings were then obtained using a BIO-TEK Synergy HT plate reader (λex 380 nm; λem 485 nm). Data shown are the average of three experiments with standard deviation. A two-sample t-test was performed, assuming unequal variances, in Microsoft Excel and * indicates p<0.05.
Fluorescence emission spectroscopy
A SPEX FluoroMax-2 spectrofluorometer was used to collect emission spectra. λex 280 nm; λem 285–400 nm; integration time 0.5 sec nm−1; 3 scans averaged; slit width 5 nm. Data was collected after 6 µM of mutant (G245C) p53 DBD (250 µL) was incubated for 1h at 32°C alone or with DMSO or 60 µM ITCs.
Caspase-3 activity assay
Caspase-3 activity was measured by detection of the cleavage of a colorimetric caspase-3 substrate, N-acetyl-Asp-Glu-Val-Asp (DEVD)-p-nitroaniline, using an assay kit (R&D Systems, Inc., Minneapolis, MN). The assay was performed according to the instructions of the manufacturer. In brief, cells were treated with drugs for 24 h. The cells were collected, and lysed in ice-cold lysis buffer provided by the manufacturer. The same amount of protein extracts (100–200 µg) were incubated in a reaction buffer containing N-acetyl-DEVD-p-nitroaniline at 37 °C for 2 to 4 h. The levels of the resulting proteolytic fragment p-nitroanilide were measured as optical density at 405 nm with a plate reader. The data represent the mean ± SD of three independent experiments.
Annexin V apoptosis assay
MDA-MB-231 breast cancer cells were treated at 40–50% confluency with DMSO, 10 µM BITC, 2,2-diphenylethyl ITC, 4-methoxybenzyl ITC, trityl ITC, cyclohexylmethyl ITC, SFN, 3-PPITC or 4-PBITC in DMSO for 24 h, after which time cells were collected, washed twice with cold PBS and resuspended in 1X binding buffer from FITC Annexin V apoptosis kit (BD Pharmingen). Then, 100 µL of each sample was added to a 5 mm tube and the Annexin V apoptosis kit protocol was followed. Samples were immediately analyzed with flow cytometry. Data shown are the average of three experiments with standard deviation. A two-sample t-test was performed, assuming unequal variances, in Microsoft Excel and * indicates p<0.05.
Supplementary Material
Acknowledgements
We would like to thank Dr. Karen Creswell and Ms. Annie Park of the Georgetown University Lombardi Comprehensive Cancer Center Flow Cytometry Core Facilities for their help with the apoptosis studies. We also would like to thank Dr. Kepher H. Makambi for helpful statistical analysis and thank Dr. Emily J. Greenspan for critical reading of the manuscript. This research was funded by Ruth L. Kirschstein National Research Service Award T32 CA9686 (A. J. D.) and NIH grant number CA100853.
Abbreviations
- DMSO
Dimethyl sulfoxide
- ITC(s)
isothiocyanate(s)
- PEITC
phenethyl isothiocyanate
- BITC
benzyl isothiocyanate
- SFN
sulforaphane
- AITC
allyl ITC
- PITC
phenyl ITC
- 3-PPITC
3-phenylpropyl ITC
- 4-PBITC
4-phenylbutyl ITC
- 5-PPeITC
5-phenylpentyl ITC
- 6-PHITC
6-phenylhexyl ITC
- NMPEA
N-methylphenethylamine
- DBD
DNA-binding domain
Footnotes
Supporting Information Available: We have preliminary data showing that PEITC induces mutant p53 depletion likely through a MDM2-ubiquitin-mediated 26S proteasome-independent pathway. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.WHO. Lyon, France: IARC Press; IARC Handbook of Cancer Prevention, Vol. 9: Cruciferous vegetables, isothiocyanates and indoles. 2004
- 2.Mi L, Wang X, Govind S, Hood BL, Veenstra TD, Conrads TP, Saha DT, Goldman R, Chung FL. The role of protein binding in induction of apoptosis by phenethyl isothiocyanate and sulforaphane in human non-small lung cancer cells. Cancer Res. 2007;67:6409–6416. doi: 10.1158/0008-5472.CAN-07-0340. [DOI] [PubMed] [Google Scholar]
- 3.Mi L, Xiao Z, Hood BL, Dakshanamurthy S, Wang X, Govind S, Conrads TP, Veenstra TD, Chung FL. Covalent binding to tubulin by isothiocyanates: a mechanism of cell growth arrest and apoptosis. J. Biol. Chem. 2008;283:22136–22146. doi: 10.1074/jbc.M802330200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hussain SP, Harris CC. Molecular epidemiology of human cancer: contribution of mutation spectra studies of tumor suppressor genes. Cancer Res. 1998;58:4023–4037. [PubMed] [Google Scholar]
- 5.Hofseth LJ, Hussain SP, Harris CC. p53: 25 years after its discovery. Trends Pharmacol. Sci. 2004;25:77–81. doi: 10.1016/j.tips.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Strano S, Dell’Orso S, Mongiovi AM, Monti O, Lapi E, Di Agostino S, Fontemaggi G, Blandino G. Mutant p53 proteins: between loss and gain of function. Head Neck. 2007;29:488–496. doi: 10.1002/hed.20531. [DOI] [PubMed] [Google Scholar]
- 7.Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, Valentin-Vega YA, Terzian T, Caldwell LC, Strong LC, El-Naggar AK, Lozano G. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 8.De Flora S, Balansky RM, D'Agostini F, Izzotti A, Camoirano A, Bennicelli C, Zhang Z, Wang Y, Lubet RA, You M. Molecular alterations and lung tumors in p53 mutant mice exposed to cigarette smoke. Cancer Res. 2003;63:793–800. [PubMed] [Google Scholar]
- 9.Hisada M, Garber JE, Fung CY, Fraumeni JF, Jr, Li FP. Multiple primary cancers in families with Li-Fraumeni syndrome. J. Natl. Cancer Inst. 1998;90:606–611. doi: 10.1093/jnci/90.8.606. [DOI] [PubMed] [Google Scholar]
- 10.Huang C, Ma WY, Li J, Hecht SS, Dong Z. Essential role of p53 in phenethyl isothiocyanate-induced apoptosis. Cancer Res. 1998;58:4102–4106. [PubMed] [Google Scholar]
- 11.Xiao D, Singh SV. Phenethyl isothiocyanate-induced apoptosis in p53-deficient PC-3 human prostate cancer cell line is mediated by extracellular signal-regulated kinases. Cancer Res. 2002;62:3615–3619. [PubMed] [Google Scholar]
- 12.Pappa G, Lichtenberg M, Iori R, Barillari J, Bartsch H, Gerhäuser C. Comparison of growth inhibition profiles and mechanisms of apoptosis induction in human colon cancer cell lines by isothiocyanates and indoles from Brassicaceae. Mutat. Res. 2006;599:76–87. doi: 10.1016/j.mrfmmm.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Raffo AJ, Drew L, Mao Y, Tran A, Petrylak DP, Fine RL. Fas-mediated apoptosis is dependent on wild type p53 status in human cancer cells expressing a temperature-sensitive p53 mutant alanine-143. Cancer Res. 2003;63:1527–1533. [PubMed] [Google Scholar]
- 14.Jiao D, Eklind KI, Choi C-I, Desai DH, Amin SG, Chung F-L. Structure-activity relationships of isothiocyanates as mechanism-based inhibitors of 4-(methylnitrosamino)-1-(3-pyridy)-1-butanone-induced lung tumorigenesis in A/J Mice. Cancer Res. 1994;54:4327–4333. [PubMed] [Google Scholar]
- 15.Kamencic H, Lyon A, Paterson PG, Juurlink BH. Monochlorobimane fluorometric method to measure tissue glutathione. Anal. Biochem. 2000;286:35–37. doi: 10.1006/abio.2000.4765. [DOI] [PubMed] [Google Scholar]
- 16.Winterfeld S, Imhof N, Roos T, Bar G, Kuhn A, Gerken U. Substrate-induced conformational change of the Escherichia coli membrane insertase YidC. Biochemistry. 2009;48:6684–6691. doi: 10.1021/bi9003809. [DOI] [PubMed] [Google Scholar]
- 17.Morse MA, Eklind KI, Hecht SS, Jordan. KG, Choi C-I, Desai DH, Amin SG, Chung F-L. Structure-activity relationships for inhibition of 4-(methylnitrosamino)-l-(3-pyridyl)-l-butanone lung tumorigenesis by arylalkyl isothiocyanates in A/J mice. Cancer Res. 1991;57:1846–1850. [PubMed] [Google Scholar]
- 18.Bossi G, Lapi E, Strano S, Rinaldo C, Blandino G, Sacchi A. Mutant p53 gain of function: reduction of tumor malignancy of human cancer cell lines through abrogation of mutant p53 expression. Oncogene. 2006;25:304–309. doi: 10.1038/sj.onc.1209026. [DOI] [PubMed] [Google Scholar]
- 19.Xiao D, Vogel V, Singh SV. Benzyl Isothiocyanate-induced apoptosis in human breast cancer cells is initiated by reactive oxygen species and regulated by Bax and Bak. Mol. Cancer Ther. 2006;5:2931–2945. doi: 10.1158/1535-7163.MCT-06-0396. [DOI] [PubMed] [Google Scholar]
- 20.Na HK, Surh YJ. Transcriptional regulation via cysteine thiol modification: a novel molecular strategy for chemoprevention and cytoprotection. Mol. Carcinog. 2006;45:368–380. doi: 10.1002/mc.20225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.